Abstract
As of April 9, 2020, a novel coronavirus (SARS‐CoV‐2) had caused 89,931 deaths and 1,503,900 confirmed cases worldwide, which indicates an increasingly severe and uncontrollable situation. Initially, little was known about the virus. As research continues, we now know the genome structure, epidemiological and clinical characteristics, and pathogenic mechanisms of SARS‐CoV‐2. Based on this knowledge, potential targets involved in the processes of virus pathogenesis need to be identified, and the discovery or development of drugs based on these potential targets is the most pressing need. Here, we have summarized the potential therapeutic targets involved in virus pathogenesis and discuss the advances, possibilities, and significance of drugs based on these targets for treating SARS‐CoV‐2. This review will facilitate the identification of potential targets and provide clues for drug development that can be translated into clinical applications for combating SARS‐CoV‐2.
Abbreviations
- 3CLpro
3C‐like main protease
- 6‐HB
six‐helical bundle.
- ACE2
angiotensin converting enzyme 2
- ASOs
antisense oligonucleotides
- ATA
aurintricarboxylic acid
- Basigin
basic immunoglobulin
- CatB/L
cysteine proteases cathepsin B and L
- CC50
half‐cytotoxic concentration
- COVID‐19
SARS‐CoV‐2 caused coronavirus disease 2019
- CoVs
coronaviruses
- DPP‐4
dipeptidyl peptidase‐4
- ECMO
extracorporeal membrane oxygenation
- EMMPRIN
extracellular MMP inducer
- G‐CSF
granulocyte‐colony stimulating factor
- GSK3
glycogen synthase kinase 3
- HR
heptad repeat
- IP10
IFN‐γ‐inducible protein 10
- IRF3
IFN regulatory factor 3
- MERS‐CoV
Middle East respiratory syndrome coronavirus
- Mpro
main protease
- nsps
nonstructural proteins
- PARP1
poly‐ADP‐ribose polymerase 1
- PARS
pneumonia‐associated respiratory syndrome
- PLpro
papain‐like protease
- RBD
receptor binding domain
- RBM
receptor binding motif
- RBV
ribavirin
- RdRP
RNA‐dependent RNA polymerase
- SARS‐CoV
severe acute respiratory syndrome coronavirus
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- scFv
single chain variable fragment
- siRNAs
small interfering RNAs
- SPR
surface plasmon resonance
- TCM
traditional Chinese medicine
1. INTRODUCTION
In December 2019, a new coronavirus called severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) caused an epidemic in Wuhan, a major city in central China. This novel coronavirus had not been reported before and it took about 1 month to demonstrate that this virus had the ability to infect humans. Subsequently, it spread rapidly throughout China and caused outbreaks in many other countries, including Japan, South Korea, Italy, and the United States (Zhu et al., 2020). On January 31, 2020, the World Health Organization (WHO) declared that SARS‐CoV‐2 caused coronavirus disease 2019 (COVID‐19), a public health emergency of international concern, and then designated it a pandemic on March 11, 2020. According to the latest data as of April 9, 2020, from the WHO, there are 89,931 deaths and 1,503,900 confirmed cases worldwide, with the mortality reaching approximately 5.980%, and the epidemic has followed a rapid growth model. A series of initiatives and statistical data hinted at the unusually severe effects of SARS‐CoV‐2, which induced serious attention and active prevention, worldwide. Although we are confident that a treatment will eventually be found, no specific vaccines or ideal drugs for SARS‐CoV‐2 have been identified for clinical use at present, so it is critical to understand the exact pathogenic mechanism and develop effective drugs and vaccines to respond to outbreaks of SARS‐CoV‐2 (Cabrini, Landoni, & Zangrillo, 2020). Several studies demonstrated ACE2 as an important therapeutic target of SARS‐CoV‐2 entry and infection, and many potential targets were subsequently proposed, such as the spike (S) protein and transmembrane serine protease 2 (TMPRSS2). Additionally, many potential drugs have been suggested, and their assessment as treatments for the virus is underway. Also, several vaccines that have proven safety, efficacy, and quality are also in early clinical trials.
However, data from these assessments and trials are from many sources and in diverse forms, which makes it difficult to generate a comprehensive understanding for reference. Therefore, in this review, we comprehensively summarize the potential therapeutic targets involved in the processes of virus transmission, infection, and pathogenesis, based on recent studies. Additionally, we discuss in depth the advancements, possibilities, and significance of promising drugs based on these targets in the treatment of SARS‐CoV‐2. We hope this review will facilitate the identification of potential targets and provide clues for drug discovery and development that can be translated into clinical applications for combating SARS‐CoV‐2.
2. EPIDEMIOLOGICAL AND CLINICAL CHARACTERISTICS OF SARS‐COV‐2
Coronaviruses (CoVs) are non‐segmented, positive‐sense, single‐stranded RNA viruses that belong to the subspecies Coronaviridae. The length of CoV genomes ranges from 26 to 32 kilobases, which makes it the largest viral RNA known (Livingston, Bucher, & Rekito, 2020). Currently, six kinds of CoVs are known to cause human diseases, which can be classified into two groups: slightly pathogenic and highly pathogenic CoVs. Among the highly pathogenic CoVs, both severe acute respiratory syndrome coronavirus (SARS‐CoV, 2002, southern China) and Middle East respiratory syndrome coronavirus (MERS‐CoV, 2012, Saudi Arabia), which mainly infect the lower airways and cause serious fatal pneumonia, are known and can affect humans (Cui, Li, & Shi, 2019). The third known highly pathogenic CoV has been identified as the pathogen causing outbreaks of SARS‐like clinical symptoms in Wuhan city of China and was officially designated SARS‐CoV‐2 by the WHO.
SARS‐CoV‐2 belongs to lineage B of the β‐CoVs, a subgroup of Sarbecovirus, which contains a positive‐sense single‐stranded RNA as the hereditary substance and is wrapped in a nucleocapsid protein in the core region and a peripheral envelope consisting of the spike protein, envelope protein, and membrane protein (Figure 1a) (Chan et al., 2020). The genomic organization of SARS‐CoV‐2 was consistent with a single‐stranded positive‐sense RNA, which contains 5′‐methylated caps and 3′‐polyadenylated tails and is arranged in the following order: 5′ end; replicase ORF1a/b; spike (S); envelope (E); membrane (M); nucleoprotein (N); accessory proteins such as ORF 3, 6, 7a, 7b, 8 and 9b; and the 3′ end (Figure 1b). Phylogenetic analysis studies showed that SARS‐CoV‐2 has the closest relationship with bat‐derived SARS‐CoV but is not completely identical to SARS‐CoV (approximately 79% identity) or MERS‐CoV (approaching 50% identity) (Paraskevis et al., 2020).
FIGURE 1.
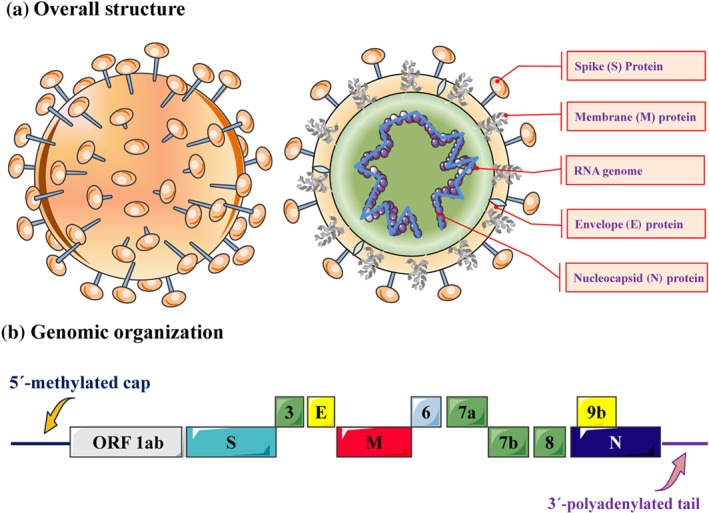
The genomic organization and virus structure of SARS‐CoV‐2. (a) The structure of SARS‐CoV‐2 consists of a single‐stranded positive‐sense RNA as the genetic material, which is surrounded by the nucleocapsid protein in the core area and a peripheral envelope consisting of the spike (S) protein, envelope ( E) protein, and membrane (M) protein. (b) The genomic organization of SARS‐CoV‐2 is based on a single‐stranded positive‐sense RNA, which contains a 5′‐methylated cap and 3′‐polyadenylated tail and is arranged in the following order: the 5′ end; open reading frame (ORF) 1a/b; spike (S); envelope ( E ) ; membrane (M); nucleocapsid protein (N); accessory proteins such as orf 3, 6, 7a, 7b, 8 and 9b; and the 3′ end
To date, studies have shown that SARS‐CoV‐2 is spread between individuals mainly by means of the respiratory system and droplets (Phan et al., 2020). At present, several studies have put forward that SARS‐CoV‐2 can be transmitted not only by droplets but also potentially via the oral‐fecal route (Huang et al., 2020), but the latter requires formal proof. Long‐term observation and statistical analysis demonstrated that the incubation time is approximately 2 to 14 days after infection, with a median period of 4 days (interquartile range, 2 to 7 days) (Livingston, Bucher, & Rekito, 2020). The lungs and immune organs are the two main targets attacked by SARS‐CoV‐2. In addition, the virus also attacks other organs, such as the heart, kidney, oesophagus, and many other specific cell types (including alveolar cells, myocardial cells, renal proximal tubule cells, and oesophageal epithelial cells), and the most likely reason is related to infections and ACE2 distribution (Yang et al., 2020; Zou et al., 2020).
Epidemiological surveys have found that the clinical features of SARS‐CoV‐2 infection are similar to those of SARS‐CoV and are characterized by fever (>37.3°C), dry cough, dyspnoea, or shortness of breath in most patients, whereas non‐respiratory clinical symptoms such as diarrhoea, sore throat, muscle ache, headache, and vomiting have also been reported in a minority of patients (Meo et al., 2020). Confirmed patients have also developed acute respiratory distress syndrome, while critical illness may present with respiratory and lung function failure, even multiple organ dysfunction and septic shock, which require extracorporeal membrane oxygenation (ECMO) and intensive care support (Wang, Hu, et al., 2020). The major pathological characteristics of the lung include pulmonary alveolitis and bronchiolitis with epithelial cell proliferation, desquamation, squamous metaplasia, and the production of mucus and oedema fluid. Meanwhile, massive diffuse pulmonary interstitial fibrosis accompanied by excessive inflammation, a certain amount of hyaline degeneration, and variable levels of pulmonary haemorrhagic infarction have been observed at the lesion site (Luo et al., 2020). Moreover, several kinds of immune cells, including focal monocytes, lymphocytes, plasma cells, and several multinucleate giant cells along with intracytoplasmic viral inclusion bodies, infiltrate into the pulmonary interstitium (Cheng et al., 2020). In addition, immunohistochemical studies indicated positive results for immune cells, and pathological staining also showed extensive pulmonary interstitial fibrosis. Eventually, these abnormal pathological changes result in lung function failure or multiple organ failure. Although there has been some progress in the investigation and research of SARS‐CoV‐2, knowledge about this virus is still insufficient (Chen et al., 2020). At present, the number of new confirmed cases and mortality are rapidly increasing daily, with only symptomatic treatment available. This situation means that research to confirm potential therapeutic targets and discover promising drugs, as soon as possible, is the current priority in the response to the SARS‐CoV‐2 outbreak.
3. POTENTIAL THERAPEUTIC TARGETS AND PROMISING DRUGS FOR THE TREATMENT OF SARS‐COV‐2
We know that the outbreaks of SARS‐CoV‐2 will be controlled with ideal drugs or effective vaccines and these therapies will eventually end the pandemic. However, what worries us most is that many existing, traditional, models of drug and vaccine development are inadequate for this outbreak. A long drawn out process, over several months or years, cannot rescue, in a timely manner, those patients who are dying or the failing economy. Vaccine and drug research and development should be given most attention because there is currently no ideal solution to clear SARS‐CoV‐2 infection, despite the immediate pressing need to identify symptomatic strategies to ease suffering and prevent potential death (Morse, Lalonde, Xu, & Liu, 2020). Therefore, accelerating research for potential therapeutic target confirmation, promising drug discovery, and clinical verification development will speed up efforts to combat SARS‐CoV‐2. Based on the current studies, we summarize the potential therapeutic targets involved in virus transmission, infection, and pathogenesis processes. Additionally, we discuss the advances, possibilities, and significance of promising drugs based on these targets for combating SARS‐CoV‐2 (Figure 2).
FIGURE 2.
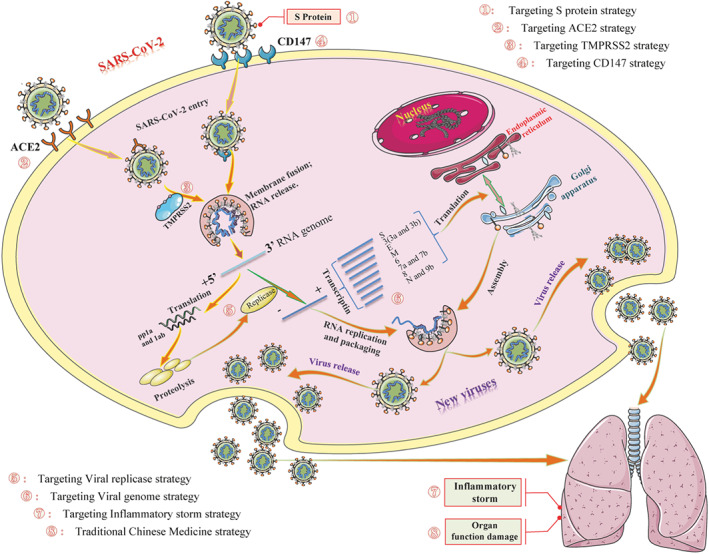
Possible life cycle of SARS‐CoV‐2 and potential intervention strategy. Diagram showing the possible transmission, infection, and pathogenesis cycle of SARS‐CoV‐2. In the infection phase, SARS‐CoV‐2 attaches to the cellular receptor via the spike (S) protein or the transmembrane glycoprotein CD147 to enter the host cell by the endosomal pathway. At this point, the S protein is activated and cleaved by transmembrane protease serine 2 (TMPRSS2) to trigger membrane fusion. Then, SARS‐CoV‐2 releases its nucleocapsid into the cytoplasm to induce translation of ORF1a/b into the large replicase polyproteins 1a (pp1a) and pp1ab and replicates its genomic RNA. Subsequently, pp1a and pp1ab produce various non‐structural proteins (nsps), in which the RNA‐dependent RNA polymerase (RdRP), papain‐like protease (PLpro), and 3C‐like protease (3CLpro) are encoded. These replicases synthesize the full‐length negative‐sense antigenome template to produce new genomic RNA and further form the assembled virion, which is then released into the extracellular space by exocytosis. Uncontrolled replication promotes SARS‐CoV‐2 infection, leading to immune disturbances and inflammatory cytokine storms and eventually resulting in multiorgan functional damage, particularly in the lungs
3.1. Potential for targeting the spike protein and promising drugs
In the initial stage of infection, the spike (S) protein of SARS‐CoV‐2 first combines with a cell surface receptor (Wrapp et al., 2020). The S protein consists of two subunits: the S1 (bulb) and S2 (stalk) subunits. S1 is responsible for receptor binding, while S2 is responsible for membrane fusion (Liu et al., 2020). More specifically, S1 combines with the cognate receptor to induce a strong conformational change in S2, thereby resulting in the fusion of the virus envelope and host cellular membrane, followed by release of the nucleocapsid into the cytoplasm (Sekimukai et al., 2020). In view of this mechanism, targeting the S protein is likely to block SARS‐CoV‐2 infection, which can be regarded as the primary factor that needs to be studied in depth. Based on past experience, SARS‐CoV‐2 S protein‐neutralizing antibodies (nAbs) are most likely to become the preferred research and development strategy that should be modelled and considered by researchers and drug R&D institutions to provide passive immunization against the infection (Ahmed, Quadeer, & McKay, 2020). According to the functional analysis of the S2 subunit, the interaction between heptad repeat 1 (HR1) and HR2 of S2 can form a six‐helical bundle (6‐HB), which facilitates the fusion process of the viral and host cell membranes. Drawing on experience with SARS‐CoV, Xia et al. designed HR1‐ and HR2‐derived peptides, designated SARS‐CoV‐2‐HR1P (aa 924‐965) and SARS‐CoV‐2‐HR2P (aa 1168‐1203), respectively (Xia et al., 2020). As SARS‐CoV‐2 and SARS‐CoV S‐HR2 sequences are 100% identical, SARS‐CoV‐2‐HR2P most likely acts as a membrane fusion inhibitor in a way similar to SARS‐HR2P, the reported SARS‐CoV fusion inhibitor (Liu et al., 2004). A fusion experiment of SARS‐CoV‐2‐HR2P showed a potent fusion inhibitory effect with a half maximal inhibitory concentration (IC50) of 0.18 μM, while SARS‐CoV‐2‐HR1P exhibited no obvious fusion inhibitory activity up to 40 μM, indicating that SARS‐CoV‐2‐HR2P may be a promising therapeutic agent for SARS‐CoV‐2. However, the safety and actual effect await verification (Xia et al., 2020). Research did not end there, so in a subsequent experiment, a pan‐coronavirus fusion inhibitor denoted as EK1 was designed based on the properties of the HR1 region (Xia et al., 2019). The results showed that EK1 also exhibited a potent fusion inhibitory effect with an IC50 of 0.19 μM, suggesting that EK1 is also a promising treatment as a SARS‐CoV‐2 drug pending verification and development. According to other research results, we know that SARS‐CoV‐2 enters the host cell by binding to a membrane protein, the angiotensin‐converting enzyme 2 (ACE2) via the S protein receptor binding domain (RBD) (Yan et al., 2020). Based on this observation, Zhang et al. synthesized a first‐in‐class peptide binder, named 23‐mer peptide binder (SBP1), by using automated fast‐flow peptide synthesis technology. After careful analysis, they found that SBP1 can potentially restrict the entry process of SARS‐CoV‐2 into human cells through binding to the SARS‐CoV‐2‐RBD with extremely low affinity (K D value = 47 nM) (Zhang, Pomplun, Loftis, Loas, & Pentelute, 2020). Moreover, the human protein‐derived sequence of SBP1 prevented the binder from being immunogenic, which further promoted the peptide binder SBP1 as a promising preclinical drug lead for anti‐SARS‐CoV‐2 drug discovery and development.
3.2. Potential ACE2 targets and promising drugs
Receptor binding is one of the major determinants of tissue tropism and host range for CoVs. A study showed that CoVs utilize existing cell surface enzymes as their binding receptors, such as the ACE2 receptor for SARS‐CoV and the dipeptidyl peptidase 4 (DPP4) receptor for MERS‐CoV (Fung & Liu, 2019). Recently, a study found that SARS‐CoV‐2 can enter into the same set of cell lines as SARS‐CoV, suggesting that they have a similar receptor, ACE2. The results of sequence analysis showed that some but not all SARS‐CoV‐2 clusters can use ACE2 for host cell entry (Cao, Li et al., 2020). According to receptor binding motif (RBM) analysis, the majority of amino acid residues essential for ACE2 binding were retained in the S protein of SARS‐CoV‐2, which was consistent with a previous conclusion that the virus used ACE2 for host cell entry (Yan et al., 2020). In agreement with these findings, SARS‐CoV‐2 uses ACE2 for cellular entry, similar to SARS‐CoV. Therefore, targeting ACE2 can prevent the replication of SARS‐CoV‐2, which can be regarded as a potential therapeutic strategy that needs to be studied in depth. A recent hypothesis proposed that ACE inhibitors (ACEIs), such as captopril and enalapril, or angiotensin AT1 receptor antagonists, including losartan and valsartan, might be beneficial for those patients who experienced pneumonia induced by SARS‐CoV‐2 (Sun, et al., 2020). However, unfortunately, this is just a plausible speculation without basic or clinical research verification. At present, researchers envision administering a type of antibody that could bind to the host cell membrane ACE2 protein, thus preventing the entry and subsequent infection by SARS‐CoV‐2. This promising intervention strategy was shown to significantly block viral entry and replication in experiments, but additional tests are needed to identify any anti‐SARS‐CoV‐2 infection effects (Li et al., 2003). Alternatively, people could simply develop a single chain variable fragment (scFv) that binds to the ACE2 protein to inhibit its binding with SARS‐CoV‐2. In addition, VHH domains or nanobodies from camelids are two possible choices for consideration as well (Desmyter et al., 2013). A potentially more promising strategy was proposed by a researcher, who claimed that an antibody‐like molecule could be created that would bind to the virus itself rather than protecting host cells against infection. Based on this strategy, researchers proposed using clinical‐grade soluble human ACE2 (hrsACE2) to bind the S protein of SARS‐CoV‐2, thereby neutralizing the virus. Experiments using SARS‐CoV indicated that this strategy is quite promising and it would be feasible to inhibit SARS‐CoV‐2 infection of cells, including Vero‐E6 cells, human capillary organoids, and human kidney organoids, using the hrsACE2, in a dose‐dependent manner (Vanessa, 2020). As ACE2 is already a valuable therapeutic target and hrsACE2 shows good therapeutic properties in vitro, it may be valuable to fuse hrsACE2 to an immunoadhesin to form an immunoglobulin‐Fc domain to prolong the availability of the circulating molecule and boost immune system functions to fight SARS‐CoV‐2 infection. This research offers solid evidence that hrsACE2‐immunoglobulin‐Fc may similarly suppress SARS‐CoV‐2 both in vitro and potentially in patients (Kruse, 2020). Overall, the therapeutic strategies outlined above indicated that ACE2 must be a potential therapeutic target in the treatment of SARS‐CoV‐2. Meanwhile, an ACE2 antibody, ACE2‐scFv, ACE2 nanobody, and ACE2‐Fc may be promising anti‐SARS‐CoV‐2 drugs after animal testing and clinical trials.
3.3. Potential TMPRSS2 targets and promising drugs
In addition to the S protein and ACE2, another study suggested that proteases may help activate the S protein by priming it to promote SARS‐CoV‐2 cellular entry. The endosomal cysteine proteases cathepsin B and L (CatB/L) and the serine protease TMPRSS2 are two critical proteases contributing to the pathogenicity of coronavirus infections (Iwata‐Yoshikawa et al., 2019). Research has found that SARS‐CoV‐2 can use TMPRSS2 rather than CatB/L for S protein priming, and the spread of SARS‐CoV‐2 might also be intimately associated with TMPRSS2 activity. In addition, an in vitro study found that the serine protease inhibitor camostat mesylate could significantly block the activity of TMPRSS2, which can efficiently prevent the virus from entering Caco‐2 (TMPRSS2+) cells rather than 293T (TMPRSS2−) cells (Hoffmann et al., 2020). This result indicates that blocking TMPRSS2 should be considered a potential therapeutic target for the treatment of SARS‐CoV‐2‐infected patients. More remarkably, camostat mesylate has already been approved for safety for the treatment of pancreatic inflammatory disease in Japan, which reminds us that camostat mesylate should be given sufficient consideration as a promising therapeutic drug for combating SARS‐CoV‐2 without safety concerns (Gibo et al., 2005). According to a new finding from the University of Tokyo, a comparable drug named nafamostat mesylate can prevent TMPRSS2‐triggered SARS‐CoV‐2 membrane fusion at a concentration less than one‐tenth that of camostat mesylate, suggesting that nafamostat mesylate may also be a promising inhibitor of SARS‐CoV‐2 infection by targeting TMPRSS2 (Inoue, 2020). As these two drugs have been prescribed in Japan for many years and have adequate clinical data with regard to safety, we expect that they can enter clinical trials for treating SARS‐CoV‐2 as soon as possible. More importantly, this process of diverting the two TMPRSS2 inhibitors into treating SARS‐CoV‐2 reminds us that searching for new therapeutic activities in marketed drugs that have been confirmed to be safe (drug repurposing) appears to be a good strategy and extremely worthwhile.
3.4. Potential CD147 targets and promising drugs
Previous studies have focused on ACE2 in host cells and its partner molecule TMPRSS2. Although progress has been made in determining the structures and binding domains and in drug screening, it is unclear whether there are other receptors on the host cell membrane for S protein binding, which reminds us of the challenges of identifying drug targets for SARS‐CoV‐2. Therefore, as a result of in‐depth study, another receptor was discovered recently to be involved in SARS‐CoV‐2 infection. CD147, usually referred to as extracellular MMP inducer (EMMPRIN) or basic immunoglobulin (Basigin), is a transmembrane glycoprotein belonging to the immunoglobulin family, which is closely associated with many disease processes, such as tumour progression, plasmodium invasion, and virus infection (Lu et al., 2018; Nguyen & Kamil, 2018; Zhang et al., 2018). Previous studies showed that CD147 can promote the entry of SARS‐CoV into host cells, while the CD147‐antagonistic peptide (AP)‐9 has high binding activity to effectively prevent SARS‐CoV infection of HEK293 cells (Chen et al., 2005). Due to the similar genetic sequences, virus structures, and clinical characteristics of SARS‐CoV‐2 and SARS‐CoV, CD147 may be a potential target for the treatment of SARS‐CoV‐2. To test this hypothesis, Wang et al. used SARS‐CoV‐2‐infected Vero E6 cells to detect the localization and binding status of CD147 and S protein during infection and observed that the S protein and CD147 were present mainly in the viral inclusion bodies of Vero E6 cells by immuno‐electron microscopy (Wang, Chen et al., 2020). During the experiment, a surface plasmon resonance (SPR) assay confirmed the interaction between CD147 and the S protein via the receptor binding domain, and the affinity constant was 1.85 × 10−7 M. Moreover, a coimmunoprecipitation (Co‐IP) assay again confirmed the interaction between CD147 and the S protein. These data indicated that SARS‐CoV‐2 may also enter the host cell through binding to the CD147 receptor. However, further research is needed to confirm that CD147 acts as a co‐receptor, a secondary receptor or an equally important new receptor. Additionally, whether the combined treatment is better than alternative treatment needs to be confirmed by additional in vivo and in vitro experiments. Thus, targeting CD147 provides a new therapeutic strategy worth exploring and extending further. Meplazumab, a commonly studied humanized anti‐CD147 antibody for treating malaria, is reported to effectively prevent viruses from entering human host cells through binding to the CD147 receptor by abolishing CD147 activity. An in vitro study showed a good therapeutic effect against the virus in a dose‐dependent manner, with a 50% maximal effect concentration (EC50) of 24.86 μg·ml−1 and IC50 of 15.16 μg·ml−1. Meanwhile, a clinical trial has achieved satisfactory effects, indicating that CD147 is indeed a novel potential therapeutic target for treating SARS‐CoV‐2 and that meplazumab should be considered a promising drug after larger experimental research investigation and clinical trial verification (Bian et al., 2020). Moreover, previous research and the development of original antibody drugs targeting the CD147 receptor, such as metuximab, metuzumab, and meplazumab, all indicate that these drugs show good safety in preclinical research and clinical administration (Chen et al., 2005; Li, Xi et al., 2020; Wang et al., 2019). Additionally, the tissue distribution specificity of CD147 in tumour tissues, inflamed tissues and pathogen‐infected cells, indicates relatively low cross‐reactivity with normal cells (Kosugi, Maeda, Sato, Maruyama, & Kadomatsu, 2015; Su & Yang, 2018). Therefore, the route of viral entry involving CD147 suggests a novel potential target with promising druggability for specific anti‐SARS‐CoV‐2 drug (such as metuximab, metuzumab, and meplazumab) verification and exploration.
3.5. Potential viral replicase targets and promising drugs
In a review of past research, we found that coronavirus genomic RNA acts as the transcript to induce ORF1a translation in a cap‐dependent manner to form the polyprotein ppan after entering and uncoiling in the host cell. Near the end of ORF1a, there are two important structures, that is, a slippery sequence and an RNA pseudoknot, which were reported to be important for inducing 25–30% of ribosomes to undergo frame shifting, thereby promoting ORF1b translation to provide the longer polyproteins pp1a and pp1ab (Masters, 2006). After autoproteolytic separation, the polyproteins pp1a and pp1ab produce several non‐structural proteins, that is, the well‐known nsps. Through the functional study of nsps, researchers found that nsp12 encodes RNA‐dependent RNA polymerase (RdRP) (Xu et al., 2003), while nsp5 and nsp3 encode the main protease (Mpro) and papain‐like protease (PLpro) (Ziebuhr, Snijder, & Gorbalenya, 2000), respectively.
During the replication process of RNA viruses, RdRP determines the fidelity and the rates of replication and mutation of the virus to condition its adaptation to the environment and even to a new host, thus influencing the evolution of the virus (Hukowska‐Szematowicz, 2020). Targeting RdRP has become recognized as one of the most important means for the treatment of SARS‐CoV. Although SARS‐CoV shares approximately 82% sequence identity with SARS‐CoV‐2 at the level of genomic RNA, their RdRP enzymes share approximately 96% sequence identity at the protein level. A high level of protein sequence conservation of RdRP indicates that almost all of the potent agents developed targeting SARS‐CoV RdRP are likely to inhibit the RdRP of SARS‐CoV‐2 (Kirchdoerfer & Ward, 2019). Although not yet approved for clinical trials, several agents targeting RdRP‐mediated generation and RdRP‐catalysed polymerization of SARS‐CoV have potential research value for SARS‐CoV‐2. Aurintricarboxylic acid (ATA), a known anionic polymer, is known to combine with several protein targets, such as gp120 and E‐selectin, and to prevent the replication of SARS‐CoV through binding to RdRP with an IC50 of 0.2 mg·ml−1 (Cushman et al., 1991; He et al., 2004; Yap, Zhang, Andonov, & He, 2005), which indicates its potential for anti‐SARS‐CoV‐2 drug development. Ribavirin (RBV), a broad‐spectrum antiviral drug, has been studied in MERS‐ and SARS‐infected patients to combat MERS/SARS‐CoV, but its efficacy against SARS‐CoV‐2 is unclear. However, whether to choose RBV as a candidate drug is unclear, as some studies have shown a worsening of patient outcomes after RBV administration, indicating that RBV (at least itself) is not a good choice for anti‐SARS‐CoV‐2 drug development (Stockman, Bellamy, & Garner, 2006; Zhang, Huang, Zheng, & Dai, 2020). Favipiravir is a new type of RdRP inhibitor. Research shows that favipiravir is activated through phosphoribosylation in cells to form favipiravir‐RTP, which is considered a substrate for viral RNA polymerase, thereby binding with RdRP to inhibit its activity (Furuta, Komeno, & Nakamura, 2017). A favipiravir clinical trial (ChiCTR2000029600) for treating SARS‐CoV‐2 infection achieved the expected results. In the clinical trial, a total of 80 patients were divided into two groups, that is, a control group and a favipiravir intervention group. The results showed that favipiravir has a better anti‐SARS‐CoV‐2 effect than similar drugs (lopinavir/ritonavir). In addition, another prospective, multicentre, open‐label, randomized superiority trial (ChiCTR200030254) has come to the similar conclusion that favipiravir (1,600 mg bid on the first day; 600 mg bid from the second day to the end of treatment) should be regarded as one of the most promising candidate drugs for combating SARS‐CoV‐2 due to its higher rate of 7‐day clinical recovery and more effectively decreasing the incidence of clinical symptoms, such as fever and cough, than the currently recommended drug, arbidol (Chen, Huang et al., 2020).
Moreover, no significant adverse reactions were observed, and relatively high patient compliance was noted in the favipiravir treatment group, indicating that favipiravir is a potential anti‐SARS‐CoV‐2 drug that requires particular attention (Dong, Hu, & Gao, 2020). Remdesivir is also a nucleotide analogue inhibitor of RdRP (Brown et al., 2019). Recently, Wang et al. found that remdesivir can effectively prevent the infection of SARS‐CoV‐2 at a very low concentration with a relatively high efficiency of selection (EC50 = 0.77 μM and a half‐cytotoxic concentration [CC50] > 100 μM [SI > 129.87]), suggesting that remdesivir is very likely to be a potential drug against SARS‐CoV‐2 infection (Wang, Cao et al., 2020). Furthermore, a report that remdesivir achieved the expected treatment effect in a patient infected with SARS‐CoV‐2 in the United States attracted much attention (Holshue et al., 2020). Currently, remdesivir has become the most promising drug that has entered the Phase III clinical trial stage, shedding new light on the treatment of SARS‐CoV‐2‐induced disease. Based on the above research, RdRP is one of the most promising therapeutic targets, and RdRP inhibitors (such as aurintricarboxylic acid, favipiravir, and remdesivir) will be the most promising drugs for SARS‐CoV‐2.
PLpro is a protease responsible for processing the polypeptide translated from the virus genetic material to form structural and non‐structural proteins, which play important roles in the processes of virus replication, assembly, and generation (Ma‐Lauer et al., 2016). When infected with MERS‐CoV or SARS‐CoV, PLpro also acts as a de‐ubiquitinase to promote the de‐ubiquitination of proteins, including IFN regulatory factor 3 (IRF3) and NF‐κB, to reduce the immune function and immunoreaction of host cells (Bekes et al., 2016; Clasman, Everett, Srinivasan, & Mesecar, 2020). These capacities determine the important roles of PLpro in the virus replication and infection processes. A recent study demonstrated that PLpro in SARS‐CoV‐2 shares approximately 83% sequence identity with PLpro in SARS‐CoV at the protein level. The residue variations between the two original sequences are present throughout the entire surface of PLpro (Morse, Lalonde, Xu, & Liu, 2020). These amino acid composition differences are supposed to influence the interaction between the two PLpro enzymes and their ligands (Baez‐Santos, St, & Mesecar, 2015; Ratia, Kilianski, Baez‐Santos, Baker, & Mesecar, 2014). However, there were no distinct differences in the three secondary structure components that formed the active sites of the two PLpro proteins, which indicated that PLpro should be considered a potential enzyme target for the treatment of SARS‐CoV‐2. As research continues, several X‐ray crystallography studies have helped to characterize PLpro enzymes and identify PLpro‐related inhibitors (Hilgenfeld, 2014). To date, different classes of SARS‐CoV PLpro inhibitors have been identified, including thiopurine compounds, small molecule inhibitors, natural products (tanshinones, geranylated flavonoids, and diarylheptanoid inhibitors), zinc ions, zinc conjugates, and one or more naphthalene inhibitors (Baez‐Santos, St, & Mesecar, 2015). The identified 6‐mercaptopurine, 6‐thioguanine, N‐ethylmaleimide, and mycophenolic acid could act as MERS‐CoV PLpro inhibitors to suppress its proteolytic activity and de‐ubiquitination independently (Cheng et al., 2015). To date, no inhibitor targeting PLpro has been confirmed to be effective against SARS‐CoV‐2. However, the successful experience of designing inhibitors targeting SARS‐CoV and MERS‐CoV PLpro enzymes with antiviral activity and selectivity shows that there is a good possibility that specific inhibitors targeting SARS‐CoV‐2 PLpro could be as effective.
The main viral protease is also known as 3C‐like main protease (3CLpro) due to its similar cleavage site specificity to that of picornavirus 3C protease (3Cpro) (Zhao, Weber, & Yang, 2013). Previous studies demonstrated that 3CLpro is one of the most crucial proteases for RdRP generation, virus replication, and infection (Zhou et al., 2019). Similar to the RdRP protein, SARS‐CoV‐2 and SARS‐CoV share approximately 96% 3CLpro sequence identity at the protein level (Morse et al., 2020). Under natural conditions, 3CLpro easily forms a dimer through hydrogen bonds. In‐depth structure analysis found that each monomer contains two main regions, the N‐terminal catalytic region and the C‐terminal region, and that the difference is dependent mainly on the catalytic region located on the protein surface (Lee et al., 2005). Because SARS‐CoV‐2 may share with SARS‐CoV similar possible interactions of substrates or inhibitors towards its active sites, imperceptible structural alterations from A to S residues are not expected to significantly affect the binding characteristics of substrates and inhibitors to those active sites. Therefore, 3CLpro is expected to become another potential enzyme target for the treatment of SARS‐CoV‐2. Lopinavir and ritonavir, two inhibitors of 3CLpro, were originally designed to treat HIV (Pillaiyar, Meenakshisundaram, & Manickam, 2020) and are now used mainly for HIV in combination with other drugs, that is, the commonly discussed antiviral cocktails. At present, several studies have shown that the two 3CLpro inhibitors have anti‐CoV properties both in vitro and in MERS‐CoV‐infected primates (not humans), and they have also shown activity in non‐randomized trials of SARS‐CoV patients (Wu et al., 2004). Based on these results, lopinavir and ritonavir may also be promising drugs for SARS‐CoV‐2 treatment. Interestingly, treatment with a formulation of lopinavir and ritonavir in tablet form (800/200 mg·day−1) called Kaletra can quickly improve clinical symptoms by inhibiting SARS‐CoV‐2 replication (Han et al., 2020). Moreover, a joint research team reported that there are approximately 30 candidate agents with potential antiviral activity against SARS‐CoV‐2 after virtual drug screening (in silico and enzyme activity test) on January 25, 2020, including lopinavir and ritonavir. Even though this combination has produced a high screening score, its effectiveness remains controversial due to the accidental consequences caused by the small sample size or combination with other promising drugs. To date, several clinical trials (such as ChiCTR2000029539, ChiCTR2000029603, and ChiCTR2000029759) have been approved in China to test the effectiveness of HIV protease inhibitors such as lopinavir and ritonavir in patients infected with SARS‐CoV‐2. However, an increasing number of clinical trials (such as NCT04252885 and ChiCTR2000029308) have not achieved the desirable results (no benefit was observed), indicating that this combination has most likely failed (Cao et al., 2020; Zhu, Lu, et al., 2020). Although hopes are fading for treatment with lopinavir and ritonavir, targeting 3CLpro is still a potential therapeutic strategy for combating SARS‐CoV‐2. As research has continued, a growing number of low MW inhibitors have been found to have high potential therapeutic effects against SARS‐CoV‐2 by targeting 3CLpro. Sun et al. found nine promising low MW inhibitors for the treatment of SARS‐CoV‐2 after optimization based on targeting 3CLpro by 32,297 potential antiviral medicinal plant compounds (Sun, Wong, & Gou, 2020). Moreover, Alessandro et al. screened multiple promising therapeutic drugs for SARS‐CoV‐2, such as indinavir, atazanavir, and cobicistat, among FDA‐approved drugs, by virtual screening. In addition, Zhang, Lin, et al. (2020) reported that α‐ketoamides can inhibit coronavirus and enterovirus replication as low MW inhibitors; among them, compound 11b is likely to prevent SARS‐CoV‐2 replication with an IC50 of 0.67 μΜ (Zhang, Wu, et al., 2020). Based on the above research, we believe that low MW inhibitors that potently inhibit 3CLpro are promising drugs for SARS‐CoV‐2.
In addition to RdRP, PLpro, and 3CLpro, helicases and glycogen synthase kinase 3 (GSK3) should also be of interest because they are essential components during the coronavirus replication process in host cells and could act as viable targets for anti‐SARS/MERS chemical therapies, according to studies that have already been confirmed by researchers (Adedeji et al., 2012; Adedeji et al., 2014; Mizutani et al., 2006; Wu et al., 2009). Although there have been no related approved inhibitor‐based anti‐SARS‐CoV‐2 therapies so far, those candidate compounds, especially remdesivir, could serve as potential leads and clinical medicines for developing effective anti‐SARS‐CoV‐2 drugs with interpretation of the crystal structure of replicases.
3.6. Targeting the viral genome and promising drugs
With the publication of the RNA genome sequence of SARS‐CoV‐2 (GenBank: MN908947), one strategy could aim to target the viral RNA genome itself for degradation, in addition to targeting the surface proteins and viral replicases. Therefore, using small interfering RNAs (siRNAs), RNA aptamers or antisense oligonucleotides (ASOs) against the SARS‐CoV RNA genome may provide important insights and promising therapeutic targets for SARS‐CoV‐2 treatment (Ahn et al., 2009; Asha et al., 2018; Qureshi, Tantray, Kirmani, & Ahangar, 2018; Shum & Tanner, 2008).
Recent studies have shown that two double‐stranded RNAs (dsRNAs) specifically bind to two separate regions of the mRNA of SARS‐CoV protein M, which was described in the patent application CN101173275. siRNA‐M1 binds to the 220‐241 position in the nucleic acid sequence of protein M mRNA, corresponding to two chemical substance registration numbers (CAS RNs), 1023405‐01‐7 and 1023405‐02‐8, while siRNA‐M2 binds to the 460‐480 position, corresponding to 1023405‐03‐9 and 1023405‐04‐0. Expression data analysis showed that the interference efficiencies of both siRNA‐M1 and siRNA‐M2 for the SARS‐CoV M protein gene were greater than 70% (Liu, Xiao, et al., 2020). In addition, the patent application CN1569233 describes promising siRNAs targeting SARS‐CoV genes that encode main components, including RdRP, the nucleoprotein N, and proteolytic enzymes. These siRNAs are reported to have a significant inhibitory or killing effect of approximately 50–90% of the SARS‐CoV virus BJ01 strain, in which the most effective are proteolytic enzyme‐targeted siRNAs. Based on these findings, siRNA may be a potential biological agent for consideration as a treatment strategy to kill SARS‐CoV‐2.
Two patents in Korea present the use of RNA aptamers for inhibiting SARS‐CoV. One (KR2009128837) showed that RNA aptamers can combine with the helicase of SARS‐CoV to inhibit the unwinding of double‐stranded DNA. The other one (KR2012139512) indicated the potential therapeutic value of RNA aptamers with unique affinity for the SARS‐CoV nucleocapsid. Based on the above results, RNA aptamers may be another potential biological agent for combating SARS‐CoV‐2, similar to the treatment of SARS‐CoV.
ASOs have also been designed to detect SARS‐CoV infections and to prevent or cure SARS‐CoV‐related disease (Lim et al., 2006). Before the occurrence of SARS‐CoV‐2, a patent application submitted by Ionis Pharmaceuticals (WO2005023083) showed hybrid DNA/RNA ASOs designed for disrupting the pseudoknot in the site of the SARS‐CoV RNA frame shift. In addition to inhibiting the virus directly, ASOs are also expected to target the disease‐related proteins involved in the inflammatory cytokine storm process, which could be considered a promising therapeutic strategy for combating SARS‐CoV‐2 (Liu et al., 2020). Therefore, ASOs may be a class of biological agents useful for the treatment of SARS‐CoV‐2, similar to the situation for SARS‐CoV.
In addition, nucleoside analogues (such as EIDD‐1931 and EIDD‐2801) should be considered as a class of biological agents with potential antiviral effects. Although the viral genome may be a potential target of siRNAs, RNA aptamers, ASOs, and nucleoside analogues for SARS‐CoV‐2, as was observed for SARS‐CoV, this approach presents several challenges. One of the most important challenges is the delivery of oligonucleotides into the lungs (Youngren‐Ortiz, Gandhi, Espana‐Serrano, & Chougule, 2016). In addition, even if siRNAs, RNA aptamers, ASOs, and other biological resources are effective clinically, a further significant problem will be how to scale up the production of these biological agents to treat large numbers of infected patients (Liu & Gou 2020). Therefore, we have a long way to go to reach the goals for the production of these potential biological agents (siRNAs, RNA aptamers, and ASOs).
3.7. Targeting the inflammatory cytokine storm and promising drugs
Although many strategies have been used to block the attachment, entry, replication, and release processes to inhibit SARS‐CoV‐2 infection, how to prevent viral evasion from host immune responses and virus‐induced cytopathic effects is considered one of the most urgent problems that need to be solved in SARS‐CoV‐2‐induced pneumonia‐associated respiratory syndrome (PARS) patients. Studies from SARS‐CoV‐induced deaths and animal models have shown that an aberrant and excessive host inflammatory cytokine storm results in a strong immunopathological response and lethal disease (Hui & Zumla, 2019; Rockx et al., 2009; Smits et al., 2010), which was also confirmed in SARS‐CoV‐2‐induced PARS patients based on pathological features and autopsies. The phrase “inflammatory cytokine storm” refers to the dysfunction of the immune system and an excessive inflammatory response that becomes uncontrollable (Chen, Zhang, Ju, & He, 2020), and it is closely associated with multiple infectious and non‐infectious conditions and diseases including graft‐versus‐host disease, autoimmune disease, severe virus infection, multiple organ dysfunction syndrome, and chimeric antigen receptor (CAR)‐T cell therapy (Channappanavar & Perlman, 2017; Giavridis et al., 2018; RIddell, 2018). During infection, CD4‐positive T cells (CD4+ T cells) are rapidly activated to form pathogenic T helper (Th) 1 cells and to produce GM‐CSF. The cytokine condition contributes to CD14+ CD16+ monocyte recruitment and IL‐6 secretion, which further aggravate the inflammatory response after SARS‐CoV‐2 infection (Li et al., 2020; Zhou et al., 2020). Recently, several clinical reports revealed that most patients infected with SARS‐CoV‐2 have increased plasma concentrations of inflammatory cytokines, such as interleukins (IL‐2, IL‐7, IL‐10), GM‐CSF, the chemokine CCL2 and TNF‐α, especially the critically ill patients (Chen, Zhou, et al., 2020; Huang et al., 2020; Peeri et al., 2020). These clinical indicators reveal the presence of severe pulmonary inflammation and the production of inflammatory cytokine storms during SARS‐CoV‐2 infection. Therefore, symptomatic treatments, especially strategies to eliminate inflammation and inflammatory cytokine storms, combined with organ support, in these critically ill patients are the most critical part of clinical management (Zumla, Hui, Azhar, Memish, & Maeurer, 2020). Therefore, the inflammatory cytokine storm is a promising research and therapeutic target that may not only identify the immunopathological mechanism but also benefit the discovery of potential drugs.
As it takes a long time to evaluate and develop specific new drugs targeting SARS‐CoV‐2, several currently marketed drugs that target inflammatory cytokine storms and reduce immunopathology could be considered at this critical moment. Tocilizumab, which can specifically bind to both the soluble IL‐6 receptor and membrane‐bound IL‐6 receptor to inhibit related signal transduction, is the first IL‐6‐ blocking antibody approved for clinical use and shows proven safety and effectiveness in therapy for rheumatoid arthritis (Safy‐Khan et al., 2020; Verhoeven et al., 2019). To date, a treatment programme utilizing tocilizumab based on conventional therapy has been administered to 20 patients (including 18 severe cases and two critical cases). The elevated body temperature was reduced to normal within 24 h, which was accompanied by varying degrees of improvement in the oxygenation index of respiratory function. After 2 weeks of careful treatment by the scheme, 19 patients recovered, and only one patient became severely ill due to critical illness (Zhou et al., 2020). This treatment programme utilizing tocilizumab based on conventional therapy has been carried out in many hospitals in Wuhan, China, and has achieved good results, which indicates that tocilizumab is a drug with great potential for targeting the inflammatory cytokine storm on the basis of conventional treatment (Xinhua Daily, 2020). Ge et al. reported a poly‐ADP‐ribose polymerase 1 (PARP1) inhibitor, CVL218, identified by their data‐driven drug repurposing framework, which could effectively inhibit SARS‐CoV‐2 replication with an EC50 of 5.12 μM. Additionally, it significantly suppressed CpG‐induced IL‐6 production by 50% and 73% in peripheral blood mononuclear cells at 1 and 3 μM concentrations after 12 h, respectively. These findings suggests that CVL218 has a significant anti‐inflammatory cytokine storm effect, which is closely associated with SARS‐CoV‐2‐induced immunopathology prevention, especially for intensive care unit (ICU) patients (Ge et al., 2020). Further in vivo pharmacokinetic and toxicokinetic studies showed that CVL218 is distributed mainly in lung tissue without apparent toxicity, which makes it a valuable candidate for the treatment of inflammatory cytokine storms induced by SARS‐CoV‐2.
Above all, the basic research and results of clinical treatment with tocilizumab and CVL218 based on conventional therapy remind researchers that exploring novel uses of already approved drugs (repurposing) may also be an effective strategy for treating SARS‐CoV‐2‐induced PARS.
3.8. Promising vaccines and passive antibodies from convalescent patient sera
Vaccination is the best strategy to prevent infections and diseases by exposure to specific pathogens, especially in vulnerable populations (Ralph et al., 2020). Therefore, it is absolutely critical to develop safe and efficient vaccines to control the spread of the pandemic and prevent the recurrence of outbreaks of SARS‐CoV‐2. In the early stages of the outbreak, the Chinese government said that at least five vaccine technologies would be explored in China, including an inactivated vaccine, a subunit protein vaccine, a nucleic acid vaccine, an adenoviral vector vaccine, and a recombinant vaccine (Zhang, 2020a). On January 23, 2020, the Coalition for Epidemic Preparedness Innovations (CEPI) declared that they would fund three vaccine technology platforms to develop effective vaccines against SARS‐CoV‐2 in the shortest period including DNA, mRNA, and “molecular clamp” platforms (Lu, 2020). It was encouraging that on March 16, 2020, the NIH announced that the first vaccine against SARS‐CoV‐2, named mRNA‐1273, entered Phase I human study (NCT04283461), which represented a total of 63 days from sequence selection to first human dosing by using the mRNA platform (Moderna, 2020). Subsequently, on the same day, another exciting development of a subunit vaccine (adenovirus Type 5 vector) against SARS‐CoV‐2 was created by experts of the Academy of Military Medical Sciences of China and was approved for clinical trials (Zhang, 2020b). The subunit vaccine, which has been approved in terms of safety, efficacy and quality by a third party, is a type of vaccine containing only a fragment of the SARS‐CoV‐2 pathogen to induce a protective immune response, according to the WHO. Since the start of the SARS‐CoV‐2 outbreak, the world's major academic institutions and biopharmaceutical companies have joined the race to develop a safe and effective prophylactic vaccine through multiple platforms such as mRNA, DNA, adenoviral vector, and recombinant protein platforms (Pang et al., 2020). On April 6, 2020, Inovio Pharmaceuticals pronounced that the FDA had accepted their investigational new drug application of a new DNA vaccine (INO‐4800). INO‐4800 is the third approved vaccine candidate worldwide and the first DNA vaccine against SARS‐CoV‐2. Meanwhile, SARS‐CoV‐ and MERS‐CoV‐related vaccine experience may accelerate the design and development of anti‐SARS‐CoV‐2 vaccines, due to the remarkable sequence homology among SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV (Lu et al., 2020). We believe that there will soon be good news regarding SARS‐CoV‐2 vaccines, as a result of the joint efforts in scientific research.
In addition to vaccines, a simple but potentially very effective strategy that could be used for combating SARS‐CoV‐2 is using convalescent patient sera, which can be obtained from patients who have recovered from virus infection. This passive strategy has been shown to be effective in the treatment of other viral diseases (Mire et al., 2016). Based on experience, patients with resolved infection will develop viral antibodies at a high titre in response to different antigens of SARS‐CoV‐2 (Li, Wang, et al., 2020). One or more passive antibodies derived from convalescent patient sera will be likely to neutralize SARS‐CoV‐2 and prevent new rounds of infection. During the outbreak of Ebola in 2014–2015, this rationale was used to treat Ebola patients with convalescent serum and achieved very good results (Kraft et al., 2015). At present, in China, many patients with resolved cases of SARS‐CoV‐2 said they are willing to donate plasma if necessary. If possible, this plasma can be transfused into other infected patients to help them overcome viral infections. As plasma donation is a mature technology, and plasma transfusion is also a part of routine medical care, this proposal is the simplest and most feasible treatment under consideration (Thorpe, Masser, Nguyen, & Davison, 2020). At the same time, we also need to consider that this proposal is not a long‐term solution because the growing number of cases is far outpacing the speed of the recovery.
3.9. The potential of molecules derived from traditional Chinese medicine to treat infections caused by SARS‐CoV‐2
As SARS‐CoV‐2 infection has become more rampant worldwide, there is an equally widespread search for potential therapeutic targets and promising drugs. In China, many researchers have been trying to find clues from traditional Chinese medicine (TCM), and now, significant progress has been achieved in the treatment of SARS‐CoV‐2. Since January 25, 2020, the early intervention of TCM has played an important role in this epidemic situation. According to statistics, a total of 60,107 patients infected with SARS‐CoV‐2 in China were treated with TCM by February 17, 2020.
A recent study screened 83 chemical structures from TCM compounds and found that theaflavin (ZINC3978446) has a low idock score (−9.11 kcal·mol−1) and binding energy (−8.8 kcal·mol−1) in the catalytic pocket of SARS‐CoV‐2 RdRP. The results from the protein‐ligand interaction profiler (PLIP) showed that theaflavin can form extra π‐cation interactions and hydrogen bonds with the catalytic pocket (at the site of Asp452, Lys545, Arg555, Thr556, Tyr619, Lys621, Cys622, Asp623, Arg624, and Asp760) of SARS‐CoV‐2 RdRp (Lung et al., 2020), which indicated that theaflavin should be recognized as a lead compound for developing promising anti‐SARS‐CoV‐2 drugs. Moreover, an in silico integrative model of absorption, distribution, metabolism, and excretion (ADME) was used to screen promising ingredients or compounds from TCM for directly inhibiting SARS‐CoV‐2 (Zhang, Pomplun, et al., 2020). Of the screened compounds, 13 (including kaempferol, moupinamide, and dihydrotanshinone I) that are components of TCMs were found to have potential anti‐SARS‐CoV‐2 activity related to similar possible targets, including PLpro, 3CLpro, and S protein. Furthermore, 125 TCMs were found to contain two or more of these 13 compounds, and 26 (such as Forsythiae fructus, Coptidis rhizome, and Mori cortex) of them are classically catalogued as treating viral respiratory infections. In addition, network pharmacology analysis predicted that these 26 TCMs have general roles, including regulating viral infection, immune/inflammation reactions, and the hypoxia response in vivo (Zhang, Wu, et al., 2020). The results of this approach showed that many ingredients or compounds of TCMs could be considered lead compounds for developing promising anti‐SARS‐CoV‐2 drugs. Therefore, researchers should also concentrate a certain effort on screening, discovery, and development of promising TCM compounds and extracts for the treatment of SARS‐CoV‐2.
4. CONCLUDING REMARKS
Although the outbreak of SARS‐CoV‐2 in China was contained by the joint efforts of the government, society, and medical staff, outbreaks in other countries and regions outside of China have become increasingly severe and uncontrollable. By April 9, 2020, the number of cumulative confirmed cases is estimated to be close to 1,503,900 with approximately 89,931 deaths, worldwide. As the epidemic spreads, scientists around the world are studying the virus to understand its pathogenesis and explore potential targets and promising drugs that would be effective in combating SARS‐CoV‐2. Although there have been some clues regarding viral pathogenesis and potential targets, there are no verified antiviral drugs with specific effects against SARS‐CoV‐2. The efficacy and safety of these promising candidate drugs in the treatment of SARS‐CoV‐2 need to be confirmed in further preclinical and clinical trials. With unremitting efforts to block the outbreak of SARS‐CoV‐2 worldwide, we hope that the infection and transmission of this virus will recede in a few months, as was observed for SARS‐CoV and MERS‐CoV. Although it will be a long and difficult road, the outbreak of SARS‐CoV‐2 worldwide has underlined the urgent need for renewed efforts to develop potential broad‐spectrum and targeted antiviral drugs to overcome this virus.
4.1. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries In http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander, Christopoulos et al., 2019; Alexander, Fabbro et al., 2019a, b).
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (No. 81803602 and 81970584), the Anhui Provincial Natural Science Foundation (No. 1708085QH207), and the Fundamental Research Funds for the Central Universities (No. WK9110000018).
Zhou H, Fang Y, Xu T, Ni W‐J, Shen A‐Z, Meng X‐M. Potential therapeutic targets and promising drugs for combating SARS‐CoV‐2. Br J Pharmacol. 2020;177:3147–3161. 10.1111/bph.15092
Contributor Information
Wei‐Jian Ni, Email: niweijian@ustc.edu.cn.
Ai‐Zong Shen, Email: sazljl@126.com.
Xiao‐Ming Meng, Email: mengxiaoming@ahmu.edu.cn.
REFERENCES
- Adedeji, A. O. , Singh, K. , Calcaterra, N. E. , DeDiego, M. L. , Enjuanes, L. , Weiss, S. , & Sarafianos, S. G. (2012). Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrobial Agents and Chemotherapy, 56(9), 4718–4728. 10.1128/AAC.00957-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adedeji, A. O. , Singh, K. , Kassim, A. , Coleman, C. M. , Elliott, R. , Weiss, S. R. , … Sarafianos, S. G. (2014). Evaluation of SSYA10‐001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and Middle East respiratory syndrome coronaviruses. Antimicrobial Agents and Chemotherapy, 58(8), 4894–4898. 10.1128/AAC.02994-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, S. F. , Quadeer A. A., & McKay M. R. (2020). Preliminary Identification of Potential Vaccine Targets for the COVID‐19 Coronavirus (SARS‐CoV‐2) Based on SARS‐CoV Immunological Studies. Viruses, 12(3), 1–15. 10.3390/v12030254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, D. G. , Jeon, I. J. , Kim, J. D. , Song, M. S. , Han, S. R. , Lee, S. W. , … Oh, J. W. (2009). RNA aptamer‐based sensitive detection of SARS coronavirus nucleocapsid protein. Analyst, 134(9), 1896–1901. 10.1039/b906788d [DOI] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Christopoulos, A. , Davenport, A. P. , Kelly, E. , Mathie, A. , Peters, J. A. , … CGTP Collaborators . (2019). THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: G protein‐coupled receptors. British Journal of Pharmacology, 176, S21–S141. 10.1111/bph.14748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , … CGTP Collaborators . (2019a). THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Catalytic receptors. British Journal of Pharmacology, 176, S247–S296. 10.1111/bph.14751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , … CGTP Collaborators . (2019b). THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Enzymes. British Journal of Pharmacology, 176, S297–S396. 10.1111/bph.14752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asha, K. , Kumar, P. , Sanicas, M. , Meseko, C. , Khanna, M. , & Kumar, B. (2018). Advancements in Nucleic Acid Based Therapeutics against Respiratory Viral Infections. Journal of Clinical Medicine, 8(1), 1–24. 10.3390/jcm8010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baez‐Santos, Y. M. , St, J. S. , & Mesecar, A. D. (2015). The SARS‐coronavirus papain‐like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Research, 115, 21–38. 10.1016/j.antiviral.2014.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekes, M. , van der Heden, V. N. G. , Ekkebus, R. , Ovaa, H. , Huang, T. T. , & Lima, C. D. (2016). Recognition of Lys48‐linked di‐ubiquitin and deubiquitinating activities of the SARS coronavirus papain‐like protease. Molecular Cell, 62(4), 572–585. 10.1016/j.molcel.2016.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, H. , Zheng, Z. , Wei, D. , Zhang, Z. , Kang, W. , Hao, C. , … Zhu, P. (2020). Meplazumab treats COVID‐19 pneumonia: An open‐labelled, concurrent controlled add‐on clinical trial. medRxiv, 2020–2023. 10.1101/2020.03.21.20040691 [DOI] [Google Scholar]
- Brown, A. J. , Won, J. J. , Graham, R. L. , Dinnon, K. R. , Sims, A. C. , Feng, J. Y. , … Sheahan, T. P. (2019). Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Research, 169, 104541–104550. 10.1016/j.antiviral.2019.104541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrini, L. , Landoni, G. , & Zangrillo, A. (2020). Minimise nosocomial spread of 2019‐nCoV when treating acute respiratory failure. Lancet, 395(10225), 685 10.1016/S0140-6736(20)30359-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, B. , Wang, Y. , Wen, D. , Liu, W. , Wang, J. , Fan, G. , … Wang, C. (2020). A trial of lopinavir‐ritonavir in adults hospitalized with severe Covid‐19. The New England Journal of Medicine, 382(19), 1787–1799. 10.1056/NEJMoa2001282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y. , Li, L. , Feng, Z. , Wan, S. , Huang, P. , Sun, X. , … Wang, W. (2020). Comparative genetic analysis of the novel coronavirus (2019‐nCoV/SARS‐CoV‐2) receptor ACE2 in different populations. Cell DISCOV, 6(11), 1–4. 10.1038/s41421-020-0147-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F. , Kok, K. H. , Zhu, Z. , Chu, H. , Too, K. , Yuan, S. , & Yuen, K. Y. (2020). Genomic characterization of the 2019 novel human‐pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect, 9(1), 221–236. 10.1080/22221751.2020.1719902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar, R. , & Perlman, S. (2017). Pathogenic human coronavirus infections: Causes and consequences of cytokine storm and immunopathology. Seminars in Immunopathology, 39(5), 529–539. 10.1007/s00281-017-0629-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Huang, J. , Cheng, Z. , Wu, J. , Chen, S. , Zhang, Y. , … Wang, X. (2020). Favipiravir versus Arbidol for COVID‐19: A randomized clinical trial. medRxiv, 2020–2023. 10.1101/2020.03.17.20037432 [DOI] [Google Scholar]
- Chen, C. , Zhang, X. R. , Ju, Z. Y. , & He, W. F. (2020). Advances in the research of cytokine storm mechanism induced by Corona Virus Disease 2019 and the corresponding immunotherapies. Zhonghua Shao Shang Za Zhi, 36(0), 1–10. E5 10.3760/cma.j.cn501120-20200224-00088 [DOI] [PubMed] [Google Scholar]
- Chen, N. , Zhou, M. , Dong, X. , Qu, J. , Gong, F. , Han, Y. , … Zhang, L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet, 395(10223), 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. , Mi, L. , Xu, J. , Yu, J. , Wang, X. , Jiang, J. , … Zhu, P. (2005). Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. The Journal of Infectious Diseases, 191(5), 755–760. 10.1086/427811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J. L. , Huang, C. , Zhang, G. J. , Liu, D. W. , Li, P. , Lu, C. Y. , & Li, J. (2020). Epidemiological characteristics of novel coronavirus pneumonia in Henan. Zhonghua Jie He He Hu Xi Za Zhi, 43(4), 327–331. E27 10.3760/cma.j.cn112147-20200222-00148 [DOI] [PubMed] [Google Scholar]
- Cheng, K. W. , Cheng, S. C. , Chen, W. Y. , Lin, M. H. , Chuang, S. J. , Cheng, I. H. , … Chou, C. Y. (2015). Thiopurine analogs and mycophenolic acid synergistically inhibit the papain‐like protease of Middle East respiratory syndrome coronavirus. Antiviral Research, 115, 9–16. 10.1016/j.antiviral.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasman, J. R. , Everett, R. K. , Srinivasan, K. , & Mesecar, A. D. (2020). Decoupling deISGylating and deubiquitinating activities of the MERS virus papain‐like protease. Antiviral Research, 174, 1–12. 10.1016/j.antiviral.2019.104661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, J. , Li, F. , & Shi, Z. L. (2019). Origin and evolution of pathogenic coronaviruses. Nature Reviews. Microbiology, 17(3), 181–192. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman, M. , Wang, P. L. , Chang, S. H. , Wild, C. , De Clercq, E. , Schols, D. , … Bowen, J. A. (1991). Preparation and anti‐HIV activities of aurintricarboxylic acid fractions and analogues: Direct correlation of antiviral potency with molecular weight. Journal of Medicinal Chemistry, 34(1), 329–337. 10.1021/jm00105a052 [DOI] [PubMed] [Google Scholar]
- Desmyter, A. , Farenc, C. , Mahony, J. , Spinelli, S. , Bebeacua, C. , Blangy, S. , … Cambillau, C. (2013). Viral infection modulation and neutralization by camelid nanobodies. Proceedings of the National Academy of Sciences of the United States of America, 110(15), E1371–E1379. 10.1073/pnas.1301336110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, L. , Hu, S. , & Gao, J. (2020). Discovering drugs to treat coronavirus disease 2019 (COVID‐19). Drug Discov Ther, 14(1), 58–60. 10.5582/ddt.2020.01012 [DOI] [PubMed] [Google Scholar]
- Fung, T. S. , & Liu, D. X. (2019). Human coronavirus: Host‐pathogen interaction. Annual Review of Microbiology, 73, 529–557. 10.1146/annurev-micro-020518-115759 [DOI] [PubMed] [Google Scholar]
- Furuta, Y. , Komeno, T. , & Nakamura, T. (2017). Favipiravir (T‐705), a broad spectrum inhibitor of viral RNA polymerase. Proceedings of the Japan Academy. Series B, Physical and Biological Sciences, 93(7), 449–463. 10.2183/pjab.93.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, Y. , Tian, T. , Huang, S. , Wan, F. , Li, J. , Li, S. , … Zeng, J. (2020). A data‐driven drug repositioning framework discovered a potential therapeutic agent targeting COVID‐19. bioRxiv, 2020–2023. 10.1101/2020.03.11.986836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavridis, T. , van der Stegen, S. , Eyquem, J. , Hamieh, M. , Piersigilli, A. , & Sadelain, M. (2018). CAR T cell‐induced cytokine release syndrome is mediated by macrophages and abated by IL‐1 blockade. Nature Medicine, 24(6), 731–738. 10.1038/s41591-018-0041-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibo, J. , Ito, T. , Kawabe, K. , Hisano, T. , Inoue, M. , Fujimori, N. , … Nawata, H. (2005). Camostat mesilate attenuates pancreatic fibrosis via inhibition of monocytes and pancreatic stellate cells activity. Laboratory Investigation, 85(1), 75–89. 10.1038/labinvest.3700203 [DOI] [PubMed] [Google Scholar]
- Han, W. , Quan, B. , Guo, Y. , Zhang, J. , Lu, Y. , Feng, G. , … Chen, Q. (2020). The course of clinical diagnosis and treatment of a case infected with coronavirus disease 2019. Journal of Medical Virology, 92(5), 461–463. 10.1002/jmv.25711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, S. D. , Sharman, J. L. , Faccenda, E. , Southan, C. , Pawson, A. J. , Ireland, S. , … Davies, J. A. (2018). The IUPHAR/BPS guide to pharmacology in 2018: Updates and expansion to encompass the new guide to immunopharmacology. Nucleic Acids Research, 46(D1), D1091–D1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, R. , Adonov, A. , Traykova‐Adonova, M. , Cao, J. , Cutts, T. , Grudesky, E. , … Li, X. (2004). Potent and selective inhibition of SARS coronavirus replication by aurintricarboxylic acid. Biochemical and Biophysical Research Communications, 320(4), 1199–1203. 10.1016/j.bbrc.2004.06.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld, R. (2014). From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. The FEBS Journal, 281(18), 4085–4096. 10.1111/febs.12936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , Schroeder, S. , Kruger, N. , Herrler, T. , Erichsen, S. , … Pohlmann, S. (2020). SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue, M. L. , DeBolt, C. , Lindquist, S. , Lofy, K. H. , Wiesman, J. , Bruce, H. , … Pillai, S. K. (2020). First case of 2019 novel coronavirus in the United States. The New England Journal of Medicine, 382(10), 929–936. 10.1056/NEJMoa2001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. , Wang, Y. , Li, X. , Ren, L. , Zhao, J. , Hu, Y. , … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, D. , & Zumla, A. (2019). Severe acute respiratory syndrome: Historical, epidemiologic, and clinical features. Infectious Disease Clinics of North America, 33(4), 869–889. 10.1016/j.idc.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukowska‐Szematowicz, B. (2020). Genetic variability and phylogenetic analysis of Lagovirus europaeus strains GI.1 (RHDV) and GI.2 (RHDV2) based on the RNA‐dependent RNA polymerase (RdRp) coding gene. Acta Biochimica Polonica, 67(1), 111–122. 10.18388/abp.2020_5161 [DOI] [PubMed] [Google Scholar]
- Inoue, J (2020). Identification of an existing Japanese pancreatitis drug, Nafamostat, which is expected to prevent the transmission of new coronavirus Infection (COVID‐19). Retrieved from https://www.u‐tokyo.ac.jp/focus/en/articles/z0508_00083.html.
- Iwata‐Yoshikawa, N. , Okamura, T. , Shimizu, Y. , Hasegawa, H. , Takeda, M. , & Nagata, N. (2019). TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. Journal of Virology, 93(6), e01815–e01818. 10.1128/JVI.01815-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer, R. N. , & Ward, A. B. (2019). Structure of the SARS‐CoV nsp12 polymerase bound to nsp7 and nsp8 co‐factors. Nature Communications, 10(1), 1–9. 10.1038/s41467-019-10280-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, T. , Maeda, K. , Sato, W. , Maruyama, S. , & Kadomatsu, K. (2015). CD147 (EMMPRIN/Basigin) in kidney diseases: From an inflammation and immune system viewpoint. Nephrology, Dialysis, Transplantation, 30(7), 1097–1103. 10.1093/ndt/gfu302 [DOI] [PubMed] [Google Scholar]
- Kraft, C. S. , Hewlett, A. L. , Koepsell, S. , Winkler, A. M. , Kratochvil, C. J. , Larson, L. , … Ribner, B. S. (2015). The use of TKM‐100802 and convalescent plasma in 2 patients with Ebola virus disease in the United States. Clinical Infectious Diseases, 61(4), 496–502. 10.1093/cid/civ334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse, R. L. (2020). Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res, 9(72), 1–11. 10.12688/f1000research.22211.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T. W. , Cherney, M. M. , Huitema, C. , Liu, J. , James, K. E. , Powers, J. C. , … James, M. N. (2005). Crystal structures of the main peptidase from the SARS coronavirus inhibited by a substrate‐like aza‐peptide epoxide. Journal of Molecular Biology, 353(5), 1137–1151. 10.1016/j.jmb.2005.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. , Fan, Y. , Lai, Y. , Han, T. , Li, Z. , Zhou, P. , … Wu, J. (2020). Coronavirus infections and immune responses. Journal of Medical Virology, 92(4), 424–432. 10.1002/jmv.25685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Wang, Y. M. , Xu, J. Y. , & Cao, B. (2020). Potential antiviral therapeutics for 2019 novel coronavirus. Zhonghua Jie He He Hu Xi Za Zhi, 43(3), 170–172. 10.3760/cma.j.issn.1001-0939.2020.03.004 [DOI] [PubMed] [Google Scholar]
- Li, J. , Xing, J. , Yang, Y. , Liu, J. , Wang, W. , Xia, Y. , … Shen, F. (2020). Adjuvant (131)I‐metuximab for hepatocellular carcinoma after liver resection: A randomised, controlled, multicentre, open‐label, phase 2 trial. The Lancet Gastroenterology & Hepatology, 5(6), 548–560. 10.1016/S2468-1253(19)30422-4 [DOI] [PubMed] [Google Scholar]
- Li, W. , Moore, M. J. , Vasilieva, N. , Sui, J. , Wong, S. K. , Berne, M. A. , … Farzan, M. (2003). Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature, 426(6965), 450–454. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, T. W. , Yuan, J. , Liu, Z. , Qiu, D. , Sall, A. , & Yang, D. (2006). Nucleic‐acid‐based antiviral agents against positive single‐stranded RNA viruses. Current Opinion in Molecular Therapeutics, 8(2), 104–107. 10.1016/j.copbio.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Liu, C. , Zhou, Q. , Li, Y. , Garner, L. V. , Watkins, S. P. , Carter, L. J. , … Albaiu, D. (2020). Research and development on therapeutic agents and vaccines for COVID‐19 and related human coronavirus diseases. ACS Central SCI, 6(3), 315–331. 10.1021/acscentsci.0c00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , & Guo, B. (2020). RNA‐based therapeutics for colorectal cancer: Updates and future directions. Pharmacological Research, 152, 1–28. 10.1016/j.phrs.2019.104550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Xiao, G. , Chen, Y. , He, Y. , Niu, J. , Escalante, C. R. , … Jiang, S. (2004). Interaction between heptad repeat 1 and 2 regions in spike protein of SARS‐associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet, 363(9413), 938–947. 10.1016/S0140-6736(04)15788-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Xiao, X. , Wei, X. , Li, J. , Yang, J. , Tan, H. , … Liu, L. (2020). Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS‐CoV‐2. Journal of Medical Virology, 92(6), 595–601. 10.1002/jmv.25726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston, E. , Bucher, K. , & Rekito, A. (2020). Coronavirus Disease 2019 and Influenza 2019‐2020. JAMA, 323(12), 1122–1122. 10.1001/jama.2020.2633 [DOI] [PubMed] [Google Scholar]
- Lu, M. , Wu, J. , Hao, Z. W. , Shang, Y. K. , Xu, J. , Nan, G. , … Bian, H. (2018). Basolateral CD147 induces hepatocyte polarity loss by E‐cadherin ubiquitination and degradation in hepatocellular carcinoma progress. Hepatology, 68(1), 317–332. 10.1002/hep.29798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Zhao, X. , Li, J. , Niu, P. , Yang, B. , Wu, H. , … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S. (2020). Timely development of vaccines against SARS‐CoV‐2. Emerg Microbes Infect, 9(1), 542–544. 10.1080/22221751.2020.1737580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung, J. , Lin, Y. S. , Yang, Y. H. , Chou, Y. L. , Shu, L. H. , Cheng, Y. C. , … Wu, C. Y. (2020). The potential chemical structure of anti‐SARS‐CoV‐2 RNA‐dependent RNA polymerase. Journal of Medical Virology, 92, 693–697. 10.1002/jmv.25761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, W , Yu, H , Gou, J , Li, X , Sun, Y , Li, J , & Liu, L (2020). Clinical pathology of critical patient with novel coronavirus pneumonia (COVID‐19). Preprints. 10.13140/RG.2.2.22934.29762 [DOI] [Google Scholar]
- Ma‐Lauer, Y. , Carbajo‐Lozoya, J. , Hein, M. Y. , Muller, M. A. , Deng, W. , Lei, J. , … von Brunn, A. (2016). p53 down‐regulates SARS coronavirus replication and is targeted by the SARS‐unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proceedings of the National Academy of Sciences of the United States of America, 113(35), E5192–E5201. 10.1073/pnas.1603435113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters, P. S. (2006). The molecular biology of coronaviruses. Advances in Virus Research, 66, 193–292. 10.1016/S0065-3527(06)66005-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meo, S. A. , Alhowikan, A. M. , Al‐Khlaiwi, T. , Meo, I. M. , Halepoto, D. M. , Iqbal, M. , … Ahmed, N. (2020). Novel coronavirus 2019‐nCoV: Prevalence, biological and clinical characteristics comparison with SARS‐CoV and MERS‐CoV. European Review for Medical and Pharmacological Sciences, 24(4), 2012–2019. 10.26355/eurrev_202002_20379 [DOI] [PubMed] [Google Scholar]
- Mire, C. E. , Geisbert, J. B. , Agans, K. N. , Thi, E. P. , Lee, A. C. , Fenton, K. A. , & Geisbert, T. W. (2016). Passive immunotherapy: Assessment of convalescent serum against Ebola virus Makona infection in nonhuman primates. The Journal of Infectious Diseases, 214(suppl 3), S367–S374. 10.1093/infdis/jiw333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani, T. , Fukushi, S. , Iizuka, D. , Inanami, O. , Kuwabara, M. , Takashima, H. , … Morikawa, S. (2006). Inhibition of cell proliferation by SARS‐CoV infection in Vero E6 cells. FEMS Immunology and Medical Microbiology, 46(2), 236–243. 10.1111/j.1574-695X.2005.00028.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moderna, I (2020). Moderna announces first participant dosed in NIH‐led phase 1 study of mRNA vaccine (mRNA‐1273) against novel coronavirus. Retrieved from https://investors.modernatx.com/news‐releases/news‐release‐details/moderna‐announces‐positive‐interim‐phase‐1‐data‐its‐mrna‐vaccine
- Morse, J. S. , Lalonde, T. , Xu, S. , & Liu, W. R. (2020). Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019‐nCoV. Chembiochem, 21(5), 730–738. 10.1002/cbic.202000047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, C. C. , & Kamil, J. P. (2018). Pathogen at the gates: Human cytomegalovirus entry and cell tropism. Viruses, 10(12), 1–17. E704 10.3390/v10120704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, J. , Wang, M. X. , Ang, I. , Tan, S. , Lewis, R. F. , Chen, J. I. , … Hsu, L. Y. (2020). Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019‐nCoV): A systematic review. Journal of Clinical Medicine, 9(3), E623–653. 10.3390/jcm9030623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis, D. , Kostaki, E. G. , Magiorkinis, G. , Panayiotakopoulos, G. , Sourvinos, G. , & Tsiodras, S. (2020). Full‐genome evolutionary analysis of the novel corona virus (2019‐nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infection, Genetics and Evolution, 79, 1–4. 10.1016/j.meegid.2020.104212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeri, N. C. , Shrestha, N. , Rahman, M. S. , Zaki, R. , Tan, Z. , Bibi, S. , … Haque, U. (2020). The SARS, MERS and novel coronavirus (COVID‐19) epidemics, the newest and biggest global health threats: What lessons have we learned? International Journal of Epidemiology, 0(0), 1–10. 10.1093/ije/dyaa033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, L. T. , Nguyen, T. V. , Luong, Q. C. , Nguyen, T. V. , Nguyen, H. T. , Le, H. Q. , … Pham, Q. D. (2020). Importation and human‐to‐human transmission of a novel coronavirus in Vietnam. The New England Journal of Medicine, 382(9), 872–874. 10.1056/NEJMc2001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar, T. , Meenakshisundaram, S. , & Manickam, M. (2020). Recent discovery and development of inhibitors targeting coronaviruses. Drug Discovery Today, 25, 668–688. 10.1016/j.drudis.2020.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi, A. , Tantray, V. G. , Kirmani, A. R. , & Ahangar, A. G. (2018). A review on current status of antiviral siRNA. Reviews in Medical Virology, 28(4), 1–11. e1976 10.1002/rmv.1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph, R. , Lew, J. , Zeng, T. , Francis, M. , Xue, B. , Roux, M. , … Kelvin, A. A. (2020). 2019‐nCoV (Wuhan virus), a novel coronavirus: Human‐to‐human transmission, travel‐related cases, and vaccine readiness. Journal of Infection in Developing Countries, 14(1), 3–17. 10.3855/jidc.12425 [DOI] [PubMed] [Google Scholar]
- Ratia, K. , Kilianski, A. , Baez‐Santos, Y. M. , Baker, S. C. , & Mesecar, A. (2014). Structural basis for the ubiquitin‐linkage specificity and deISGylating activity of SARS‐CoV papain‐like protease. PLoS Pathogens, 10(5), 1–15. e1004113 10.1371/journal.ppat.1004113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIddell, S. R. (2018). Adrenaline fuels a cytokine storm during immunotherapy. Nature, 564(7735), 194–196. 10.1038/d41586-018-07581-w [DOI] [PubMed] [Google Scholar]
- Rockx, B. , Baas, T. , Zornetzer, G. A. , Haagmans, B. , Sheahan, T. , Frieman, M. , … Katze, M. G. (2009). Early upregulation of acute respiratory distress syndrome‐associated cytokines promotes lethal disease in an aged‐mouse model of severe acute respiratory syndrome coronavirus infection. Journal of Virology, 83(14), 7062–7074. 10.1128/JVI.00127-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safy‐Khan, M. , Jacobs, J. , de Hair, M. , Welsing, P. , Edwardes, M. D. , Teitsma, X. M. , … Bijlsma, J. (2020). Effect on efficacy and safety trial outcomes of also enrolling patients on ongoing glucocorticoid therapy in rheumatoid arthritis clinical trials of tocilizumab or adalimumab or methotrexate monotherapy. Annals of the Rheumatic Diseases, 79(4), 460–463. 10.1136/annrheumdis-2019-216537 [DOI] [PubMed] [Google Scholar]
- Sekimukai, H. , Iwata‐Yoshikawa, N. , Fukushi, S. , Tani, H. , Kataoka, M. , Suzuki, T. , … Nagata, N. (2020). Gold nanoparticle‐adjuvanted S protein induces a strong antigen‐specific IgG response against severe acute respiratory syndrome‐related coronavirus infection, but fails to induce protective antibodies and limit eosinophilic infiltration in lungs. Microbiology and Immunology, 64(1), 33–51. 10.1111/1348-0421.12754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum, K. T. , & Tanner, J. A. (2008). Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. Chembiochem, 9(18), 3037–3045. 10.1002/cbic.200800491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits, S. L. , de Lang, A. , van den Brand, J. M. , Leijten, L. M. , van Icken, W. F. , Eijkemans, M. J. , … Haagmans, B. L. (2010). Exacerbated innate host response to SARS‐CoV in aged non‐human primates. PLoS Pathogens, 6(2), 1–15. e1000756 10.1371/journal.ppat.1000756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockman, L. J. , Bellamy, R. , & Garner, P. (2006). SARS: Systematic review of treatment effects. PLoS Medicine, 3(9), 1525–1531. e343 10.1371/journal.pmed.0030343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H. , & Yang, Y. (2018). The roles of CyPA and CD147 in cardiac remodelling. Experimental and Molecular Pathology, 104(3), 222–226. 10.1016/j.yexmp.2018.05.001 [DOI] [PubMed] [Google Scholar]
- Sun, M. L. , Yang, J. M. , Sun, Y. P. , & Su, G. H. (2020). Inhibitors of RAS might be a good choice for the therapy of COVID‐19 pneumonia. Zhonghua Jie He He Hu Xi Za Zhi, 43(3), 219–222. E14 10.3760/cma.j.issn.1001-0939.2020.03.016 [DOI] [PubMed] [Google Scholar]
- Sun, N , Wong, W , & Guo, J (2020). Prediction of potential 3CLpro‐targeting anti‐SARS‐CoV‐2 compounds from Chinese medicine. Preprints. 10.20944/preprints202003.0247.v1 [DOI] [Google Scholar]
- Thorpe, R. , Masser, B. M. , Nguyen, L. , & Davison, T. E. (2020). Understanding donation frequency: Insights from current plasma donors. Vox Sanguinis, 115(2), 174–181. 10.1111/vox.12861 [DOI] [PubMed] [Google Scholar]
- Vanessa, M. (2020). Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell, 181(4), 905–913. 10.1016/j.cell.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven, M. M. , de Hair, M. J. , Tekstra, J. , Bijlsma, J. W. , van Laar, J. M. , Pethoe‐Schramm, A. , … Welsing, P. M. (2019). Initiating tocilizumab, with or without methotrexate, compared with starting methotrexate with prednisone within step‐up treatment strategies in early rheumatoid arthritis: An indirect comparison of effectiveness and safety of the U‐Act‐Early and CAMERA‐II treat‐to‐target trials. Annals of the Rheumatic Diseases, 78(10), 1333–1338. 10.1136/annrheumdis-2019-215304 [DOI] [PubMed] [Google Scholar]
- Wang, D. , Hu, B. , Hu, C. , Zhu, F. , Liu, X. , Zhang, J. , … Peng, Z. (2020). Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA, 323(11), 1061–1069. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Chen, W. , Zhou, Y. , Lian, J. , Zhang, Z. , Du, P. , … Chen, Z. (2020). SARS‐CoV‐2 invades host cells via a novel route: CD147‐spike protein. bioRxiv, 2020–2023. 10.1101/2020.03.14.988345 [DOI] [Google Scholar]
- Wang, M. , Cao, R. , Zhang, L. , Yang, X. , Liu, J. , Xu, M. , … Xiao, G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019‐nCoV) in vitro. Cell Research, 30(3), 269–271. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Zhang, S. , Sun, Q. , Yang, X. , Wang, Y. , Shang, R. , … Li, Y. (2019). Dual effects of an anti‐CD147 antibody for esophageal cancer therapy. Cancer Biology & Therapy, 20(12), 1443–1452. 10.1080/15384047.2019.1647052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp, D. , Wang, N. , Corbett, K. S. , Goldsmith, J. A. , Hsieh, C. L. , Abiona, O. , … McLellan, J. S. (2020). Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science, 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. H. , Yeh, S. H. , Tsay, Y. G. , Shieh, Y. H. , Kao, C. L. , Chen, Y. S. , … Chen, P. J. (2009). Glycogen synthase kinase‐3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. The Journal of Biological Chemistry, 284(8), 5229–5239. 10.1074/jbc.M805747200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. Y. , Jan, J. T. , Ma, S. H. , Kuo, C. J. , Juan, H. F. , Cheng, Y. S. , … Wong, C. H. (2004). Small molecules targeting severe acute respiratory syndrome human coronavirus. Proceedings of the National Academy of Sciences of the United States of America, 101(27), 10012–10017. 10.1073/pnas.0403596101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, S. , Liu, M. , Wang, C. , Xu, W. , Lan, Q. , Feng, S. , … Lu, Lu (2020). Inhibition of SARS‐CoV‐2 (previously 2019‐nCoV) infection by a highly potent pan‐coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Research, 30(4), 343–355. 10.1038/s41422-020-0305-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, S. , Yan, L. , Xu, W. , Agrawal, A. S. , Algaissi, A. , Tseng, C. K. , … Lu, L. (2019). A pan‐coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Science Advances, 5(4), 1–15. 10.1126/sciadv.aav4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, S. , Zhu, Y. , Liu, M. , Lan, Q. , Xu, W. , Wu, Y. , … Lu, L. (2020). Fusion mechanism of 2019‐nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cellular & Molecular Immunology, 11, 1–3. 10.1038/s41423-020-0374-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xinhua Daily . (2020). China optimizes treatment for COVID‐19. Retrieved from http://english.www.gov.cn/news/topnews/202003/07/content_WS5e62fff1c6d0c201c2cbdb82.html
- Xu, X. , Liu, Y. , Weiss, S. , Arnold, E. , Sarafianos, S. G. , & Ding, J. (2003). Molecular model of SARS coronavirus polymerase: Implications for biochemical functions and drug design. Nucleic Acids Research, 31(24), 7117–7130. 10.1093/nar/gkg916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, R. , Zhang, Y. , Li, Y. , Xia, L. , Guo, Y. , & Zhou, Q. (2020). Structural basis for the recognition of the SARS‐CoV‐2 by full‐length human ACE2. Science, 367(6485), 1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Yu, Y. , Xu, J. , Shu, H. , Xia, J. , Liu, H. , … Shang, Y. (2020). Clinical course and outcomes of critically ill patients with SARS‐CoV‐2 pneumonia in Wuhan, China: A single‐centered, retrospective, observational study. The Lancet Respiratory Medicine, 8, 475–481. 10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap, Y. , Zhang, X. , Andonov, A. , & He, R. (2005). Structural analysis of inhibition mechanisms of aurintricarboxylic acid on SARS‐CoV polymerase and other proteins. Computational Biology and Chemistry, 29(3), 212–219. 10.1016/j.compbiolchem.2005.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren‐Ortiz, S. R. , Gandhi, N. S. , Espana‐Serrano, L. , & Chougule, M. B. (2016). Aerosol delivery of siRNA to the lungs. Part 1: Rationale for gene delivery systems. Kona: Powder Science and Technology in Japan, 33, 63–85. 10.14356/kona.2016014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Huang, S. , Zheng, F. , & Dai, Y. (2020). Controversial treatments: An updated understanding of the coronavirus disease 2019. Journal of Medical Virology, 1–22. 10.1002/jmv.25788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. H. , Wu, K. L. , Zhang, X. , Deng, S. Q. , & Peng, B. (2020). In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. Journal of Integrative Medicine, 18(2), 152–158. 10.1016/j.joim.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G. , Pomplun, S. , Loftis, A. R. , Loas, A. , & Pentelute, B. L. (2020). The first‐in‐class peptide binder to the SARS‐CoV‐2 spike protein. bioRxiv, 2020–2023. 10.1101/2020.03.19.999318 [DOI] [Google Scholar]
- Zhang, L. , Lin, D. , Kusov, Y. , Nian, Y. , Ma, Q. , Wang, J. , … Hilgenfeld, R. (2020). Alpha‐ketoamides as broad‐spectrum inhibitors of coronavirus and enterovirus replication: Structure‐based design, synthesis, and activity assessment. Journal of Medicinal Chemistry, 63(9), 4562–4578. 10.1021/acs.jmedchem.9b01828 [DOI] [PubMed] [Google Scholar]
- Zhang, M. Y. , Zhang, Y. , Wu, X. D. , Zhang, K. , Lin, P. , Bian, H. J. , … Chen, Z. N. (2018). Disrupting CD147‐RAP2 interaction abrogates erythrocyte invasion by Plasmodium falciparum. Blood, 131(10), 1111–1121. 10.1182/blood-2017-08-802918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. H . (2020a). Teamwork underway to develop vaccine. Retrieved from http://www.chinadaily.com.cn/a/202003/12/WS5e69863ca31012821727e597.html
- Zhang, Z. H. (2020b). COVID‐19 vaccine developed by military medical academy approved for clinical trials. Retrieved from http://www.chinadaily.com.cn/a/202003/17/WS5e70b732a31012821727fd62.html
- Zhao, Q. , Weber, E. , & Yang, H. (2013). Recent developments on coronavirus main protease/3C like protease inhibitors. Recent Patents on Anti‐Infective Drug Discovery, 8(2), 150–156. 10.2174/1574891x113089990017 [DOI] [PubMed] [Google Scholar]
- Zhou, J. , Fang, L. , Yang, Z. , Xu, S. , Lv, M. , Sun, Z. , … Xiao, S. (2019). Identification of novel proteolytically inactive mutations in coronavirus 3C‐like protease using a combined approach. The FASEB Journal, 33(12), 14575–14587. 10.1096/fj.201901624RR [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Fu, B. , Zheng, X. , Wang, D. , Zhao, C. , Qi, Y. , … Wei, H. (2020). Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID‐19 patients. National Science Review, 0(0), 1–5. 10.1093/nsr/nwaa041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N. , Zhang, D. , Wang, W. , Li, X. , Yang, B. , Song, J. , … Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z. , Lu, Z. , Xu, T. , Chen, C. , Yang, G. , Zha, T. , & Xue, Y. (2020). Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID‐19. The Journal of Infection, 1–3. 10.1016/j.jinf.2020.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr, J. , Snijder, E. J. , & Gorbalenya, A. E. (2000). Virus‐encoded proteinases and proteolytic processing in the Nidovirales. The Journal of General Virology, 81(Pt 4), 853–879. 10.1099/0022-1317-81-4-853 [DOI] [PubMed] [Google Scholar]
- Zou, X. , Chen, K. , Zou, J. , Han, P. , Hao, J. , & Han, Z. (2020). Single‐cell RNA‐seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019‐nCoV infection. Frontiers of Medicine, 14(2), 185–192. 10.1007/s11684-020-0754-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla, A. , Hui, D. S. , Azhar, E. I. , Memish, Z. A. , & Maeurer, M. (2020). Reducing mortality from 2019‐nCoV: Host‐directed therapies should be an option. Lancet, 395(10224), e35–e36. 10.1016/S0140-6736(20)30305-6 [DOI] [PMC free article] [PubMed] [Google Scholar]


