Abstract
The Middle East respiratory syndrome coronavirus (MERS‐CoV) is an emergent respiratory virus. Dromedary camels are currently the only known reservoir of MERS‐CoV and are capable of transmitting the virus within a herd. The role of semen in the transmission of MERS‐CoV has never been investigated as yet, to the best of our knowledge. Our goal was to test semen collected from dromedary camels for MERS‐CoV. A total of 67 seminal plasma samples from infertile and 13 from fertile dromedary camels were collected. The RNA was extracted from the samples and tested using commercial real‐time PCR. Nine out of sixty‐seven infertile animals (13.4%) were positive. The obtained PCR products were sequenced using the conserved MERS‐CoV‐N gene primers. MERS‐CoV‐RNA detected in seminal plasma was closely related to the lineage B. To the best of our knowledge, this is the first report about the detection of MERS‐CoV‐RNA in camel's seminal plasma. Regular testing of semen of common male camels' used for insemination should be considered to avoid a possible spread of the virus through semen.
Keywords: lineage B, MERS‐CoV, molecular, phylogenetic analysis, real‐time PCR, semen
1. INTRODUCTION
Middle East respiratory syndrome coronavirus was first identified in Saudi Arabia in late 2012 (Zaki, van Boheemen, Bestebroer, Osterhaus, & Fouchier, 2012). Since that time, there are reports of human cases, not only in Saudi Arabia but also in other countries in the Arabian Gulf area, such as UAE, Qatar and Oman. As of now, there are 2,468 MERS‐CoV human cases reported from 27 countries around the world (WHO, 2019) with a case fatality rate of 34%(WHO, 2019). Dromedary camels are the main animal reservoir for MERS‐CoV (Hemida et al., 2014). Camel to human transmission was reported in many cases (Azhar, El‐Kafrawy, et al., 2014; Azhar, Hashem, et al., 2014). Dromedary camels shed the virus, especially in their nasal secretions (Hemida et al., 2014). However, there is a discrepancy about the shedding of the virus in the body secretions of dromedary camels such as urine and milk. MERS‐CoV has not been isolated from urine, faeces nor milk of dromedaries, and recent studies showed that no viral nucleic acids were detected in the urine of positive MERS‐CoV camels (Farag et al., 2019). It was shown that MERS‐CoV can still be detected and survived in the camel milk for a prolonged time under experimental conditions (van Doremalen, Bushmaker, Karesh, & Munster, 2014). One study reported the detection of MERS‐CoV‐RNAs in the milk of some positive animals. This highlights the potential of a possible shedding of the virus in the milk of the infected animals (Reusken et al., 2014). However, this may be hampered by the milk collection technique and the possibility of faecal contamination to the camel udder (Hemida, 2019; Hemida & Alnaeem, 2019). Breeding of dromedary camels is mainly based on natural insemination (Al Eknah, 2000; Tibary, Anouassi, Sghiri, & Khatir, 2007). There are great challenges facing the implementation of artificial insemination in dromedary camels (Al Eknah, 2000; Skidmore, Morton, & Billah, 2013; Tibary et al., 2007). Male camels usually start to be used for natural insemination when they are 5–6 years old (Khanvilkar, 2009). The natural insemination season in dromedary camels usually starts in October and lasts until April (Arthur, al‐ Rahi, Hindi&, 1985). Each mature, healthy male camel can mate with 20–50 females per season (Padalino, 2015). In most of the cases, some camels’ owners seek high pedigree bull camels for natural insemination of their female camels, even if they are far away from their location (Padalino, 2015). Sometimes some camel owners transfer their female camels by vehicles over long distances to seek insemination by high pedigree camel bulls in another region. Detection of MERS‐CoV in rectal secretions of infected dromedary camels was previously reported (Mohran et al., 2016). However, testing the possibility of viral shedding in the seminal plasma has not been explored yet. The main goal of the current study was to investigate the detection of MERS‐CoV‐RNA in seminal plasma of dromedary camels.
2. MATERIALS AND METHODS
2.1. Animals
This work was conducted as a retrospective study. Sixty‐seven dromedary male camels admitted to the veterinary teaching hospital, college of veterinary medicine, King Faisal University between 2015 and 2017 were included in the study. These animals were admitted to our teaching hospital during the rutting season from November to May. These bulls were apparently healthy with no nasal discharges or any other respiratory infection‐related findings during their initial physical examination. All animals were able to mount the female camels and produce semen under artificial settings. The age of these animals ranged from 5 to 15 years. The reason for the admission was infertility (i.e. unable to achieve conception with fertile females after persistent attempts over a period of 10 months (Dictionary, 2014). Each animal in this study used as a common bull for the insemination of female animals within a herd as well as other herds around them in its area.
2.2. Seminal plasma collection's protocol
Artificial collection of semen was conducted using an electro ejaculator (miniTüb; Ideal Instruments) (Skidmore et al., 2013; Tingari, 1986). Prior to semen collection, the camels were sedated with a mixture of xylazine (Rompun®, Bayer; 0.15 mg/kg) and ketamine (Samarth®; 2.5 mg/kg) that was administered intravenously (White, Bali, & Bark, 1987). The study camels had been sexually abstinent for at least 1 month prior to semen collection. A total of sixty‐seven ejaculates were collected from mature, infertile bull. Immediately after collection, the semen samples were evaluated for motility, percentage, sperm cell concentration (106/ml), sperm abnormalities (%) and live sperm (%) using Sperm Vision® 3.5 (Minitube of America, Inc). Semen samples were centrifuged at 500 g for 15 min, and seminal plasma was stored at −80°C for further testing.
2.3. RNA extraction
RNA was independently extracted from seminal plasma by two different methods: In the first method, TRIzol® have been used, as per the manufacturer's instructions (Carossino et al., 2016). The second method, the Qiagen RNA mini‐easy RNA extraction kits, was used as per the manufacturer's instructions. The concentration of the extracted viral RNAs from both methods was measured by using a Nanodrop machine. The eluted RNAs were stored at −80°C for further testing. Testing of both batches of RNAs was done in parallel by real‐time PCR and regular PCR techniques.
2.4. Real‐Time PCR
Testing of the collected camel semen sample was done by the real‐time PCR technique using the commercial available kits (the RealStar® MERS‐CoV RT‐PCR Kit 1.0, Altona Diagnostics GmbH). Each sample was tested for two viral‐specific viral targets (the upE and the ORF1a). The amplification was done as per the manufacturer's instructions. Samples have Ct values up to 30 Cycle Threshold values (Ct) were considered as positive. Several negative controls were also tested to rule out any possibility of contamination. The RNAs free water, the master mix and the negative control of the kits have been used. The real‐time PCR techniques were conducted in two different laboratories in different buildings.
2.5. Gel‐based PCR
The RNAs from the positive real‐time PCR specimens were tested by the gel‐based PCR technique. The positive real‐time semen samples were also tested by the partial MERS‐CoV‐N, ORF1a and UpE genes oligonucleotide. The primer sequences including MERS‐CoV‐N‐F‐5′‐CCTTCG GTA CAG TGG AGC CA‐3′ and MERS‐CoV‐N‐R‐5′‐GATGGGGTTGCCAAACACAAAC‐3′. MERS‐CoV‐ORF1a‐F 5′‐TTCGATGTTGAGGGTGCTCAT −3′, and the MERS‐CoV‐ORF1a‐R −5′‐TCACACCAGTTGAAAATCCTAATTG −3′. While, the upE‐F primer sequences are 5′‐ GCAACGCGCGATTCAGTT −3′ and up‐E‐Rev 5′‐GCCTCTACACGGGACCCATA‐′ The PCR conditions and amplification parameters were conducted as previously described (Corman et al., 2012).
2.6. Sequencing of the PCR products
The amplified PCR products were purified from the gel using the QIAquick Gel Extraction Kit as per the manufacturer's instructions (Cat No/ID: 28704), as per the kit's instructions. The purified reactions were eluted in a 50‐μl elution buffer. The purified amplicons were sequenced in both directions using the Sanger approach using the Applied Biosystems® 3,500 sequencing machine. The obtained sequences were assembled into one contig by using the Sequencher 5.4.6 sequencing analysis software (© 2017 Gene Codes Corporation). All sequencing reactions were done in a sequencing facility that has not done any virology work previously. This facility is in another institute at the plant diseases department, college of Science, King Saud University.
2.7. Bioinformatics analysis
The phylogenetic trees were constructed (maximum likelihood) based on the obtained MERS‐CoV‐N gene sequences. Multiple alignments of these sequences with other sequences from GenBank were performed using the Mega‐7 package, and phylogeny was performed using the neighbour‐joining method with 1,000 bootstrap replicates, as previously described (Kumar, Stecher, & Tamura, 2016).
2.8. Statistical analysis
The non‐probability sampling strategy was applied for our specimen collection with the incidental assignment approach as previously described (Sneath, 1973). Chi‐square and Fisher's analysis were conducted to investigate the relation between MERS‐CoV positivity and some animal variables, including breed and age. The tests were conducted using SPSS 23.statistical software (IBM Corporation).
3. RESULTS
3.1. Assessment of the dromedary camel's semen quality
All bull camels in this study were able to mount the she‐camels, and semen collection was done as described above. Analysis of the collected semen and seminal plasma of the infertile dromedary camels showed cases of azoospermia, oligozoospermia, asthenozoospermia, teratozoospermia and pyospermia in 18 (28.87%), 28 (41.79%), 13 (19.40%), 7 (10.45%) and one (1.49%), respectively.
3.2. Molecular surveillance of MERS‐CoV of semen of some dromedary camels across Saudi Arabia 2015–2017
Nine out of 67 (13.2%) semen samples collected from bull dromedary camels were positive for the MERS‐CoV nucleic acids by the real‐time PCR test (Table 1). These nine animals were positive for the MERS‐CoV‐RNA by both UpE and ORF1a kits. These results were confirmed by the gel‐based PCR techniques using the truncated MERS‐CoV‐N gene. Five out of the nine positive samples (56%) were collected from Majaheem bulls, while four positive samples were collected from Waddah (13.3% and 22.2%), respectively. There was no statistically significant difference observed between the two breeds (p = .47), (Table 1). Five out of the nine positive animals were aged ≤6 years, while four positive camels were aged >6. There is no statistically significant difference observed as far as the age of the positive animals is concerned (p = .73).
TABLE 1.
Summary results of the real‐time PCR testing of dromedary camel semen collected across the kingdom 2015–2017
| N | Breed | No tested | (+Ve) | (‐Ve) | % (+Ve) | *Range of Ct values |
|---|---|---|---|---|---|---|
| 1 | Majaheem | 37 | 5 | 33 | 13.1 | 26–29 |
| 2 | Waddah | 18 | 4 | 14 | 22.2 | 28–30 |
| 3 | Sofor | 6 | 0 | 0 | 0 | 0 |
| 4 | Sudani | 1 | 0 | 0 | 0 | 0 |
| 5 | Shageh | 2 | 0 | 0 | 0 | 0 |
| 6 | Shaele | 3 | 0 | 3 | 0 | 0 |
| # | Total | 67 | 9 | 49 | 13.2 | 26–30 |
Range of the Ct values of the positive samples
3.3. Molecular characterization of MERS‐CoV detected in dromedary camel seminal plasma in Saudi Arabia
All the nine positive semen samples by real‐time PCR were tested by the PCR technique using the partial MERS‐CoV‐N gene. Phylogenetic analysis based on the generated sequences revealed a high degree of identity to the other MERS‐CoV‐RNA isolated from both human beings and animals from Saudi Arabia, UAE, and Qatar (Figure 1). The reported MERS‐CoV from semen in this study showed a high degree of identity to lineage B of MERS‐CoV (Figure 1).
FIGURE 1.
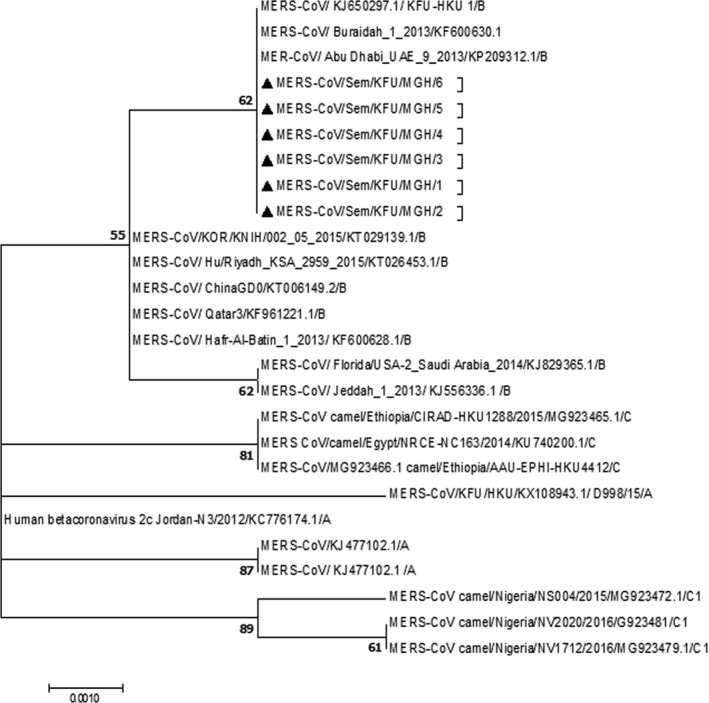
Phylogenetic analysis of the partial sequences of MERS‐CoV detected in dromedary camel semen across the Kingdome 2015–2017. The maximum likelihood phylogenetic tree of the obtained partial MERS‐CoV‐N gene. The maximum likelihood phylogenetic tree based on the partial MERS‐CoV‐N gene. The bootstrap is 1,000. The reported MERS‐CoV‐RNA from camel semen sequences were clustered together with other candidates of MERS‐CoV‐ lineage B reported in humans and camels in the Arabian Peninsula
4. DISCUSSION
The MERS‐CoV can be transmitted from camel to camel by direct contact within the same herd (Hemida et al., 2014, 2017). Theoretically, it can be transmitted from one population of camels to another through airborne infection (Hemida et al., 2017). Coronaviruses have prolonged viability in low temperature/low humidity and can survive on surfaces in the environment for a relatively long time (Chan et al., 2011). It was noted that MERS‐CoV is so far only isolated from the noses of young dromedaries, not from milk, not from urine, not from faeces, only from young dromedaries, not adults. The virus is only found in the noses for 8 days, RNA for more than 30 days (Wernery, Lau, & Woo, 2017). Viruses' transmission through the semen has been documented in many viruses affecting human beings such as HIV, Ebola and Zika virus (Council, Swanson, Spagnuolo, Wahl, & Garcia, 2015; Gornet, Bracero, & Segars, 2016; de La Vega et al., 2018). Semen also plays an important role in the transmission of many animal viruses such as Equine arteritis virus, porcine reproductive and respiratory syndrome (Johnson, Ostlund, Palmer, Fett, & Schmitt, 2012; Nathues et al., 2016). The role of dromedary camel semen in the transmission of MERS‐CoV was never tested until now. This study shows that about 13.4% of the tested camel semen samples during this time were positive for MERS‐CoV‐RNA (Table 1). About 55% of the positive samples were from the Magaheem breed of camels (Table 1), which is one of the top‐ranked native camel population in the Arabian Peninsula (Almathen, Elbir, Bahbahani, Mwacharo, & Hanotte, 2018). The other four animals were two from Waddah and two from Maghateer breed (Table 1). These findings raise the possibility of transmission of MERS‐CoV during the process of the natural insemination among bulls and she‐camels, during the rutting season of camels from November to April. Interestingly, recent studies have found a good correlation between the high prevalence of MERS‐CoV spreading among animals and humans beings where samples were collected during this time of the year (Hemida et al., 2014, 2017; Wernery et al., 2017). The RNA extraction method from seminal plasma using TRIzol resulted in high quality and more yield than the Qiagen RNA extraction kits. The obtained MERS‐CoV partial N gene sequences shared a high degree of identity with other MERS‐CoV isolated from camel and human beings in Saudi Arabia, Oman and Qatar (Al Hammadi et al., 2015; Farag et al., 2018; Hemida et al., 2014; Yusof et al., 2015). The reported sequences from semen of dromedary camels belong to lineage B of MERS‐CoV that circulated in the Arabian Peninsula (Figure 1). However, these sequences were distinct from the lineage C of MERS‐CoV and its sublineage C1, which are circulating virus strains in dromedary camels in Africa (Chu et al., 2018). Natural insemination remains the only practical method of breeding in camels. The reported MERS‐CoV sequences from semen showed a high degree of identity to the viruses isolated from both humans and camel in the area where samples collected. They share 98% identity to viruses isolated from Riyadh, Al‐Hasa, Hafr‐Elbatin, Dammam and Wadi Adasir. This suggests that the circulating strains of the virus share a common ancestor. One of the major obstacles in this study was the virus isolation, which requires biosafety level‐3 laboratory. Meanwhile, decoding the full‐length genome sequencing from these positive samples would be one of our top priority future research directions. One of the major restrictions of this study is the absence of any available swabs and sera from these animals. One of the main reasons behind this was the difficulty in convincing the camel owners to get these precious specimens from their animals. Meanwhile, this may also highlight the potential roles of males in the spreading of the virus among dromedary camels. The traditional practice of allowing bull camels from a herd to breed females from other herds in the same region and from other remote regions may contribute to the spread of the viral infection between herds in the same region. This may contribute to the overall high prevalence of MERS‐CoV among dromedary camels in the Arabian peninsula. Detection of the MERS‐CoV‐RNAs in seminal fluids of some infertile dromedary camels may draw a correlation between the infertility in male camels and the viral infection. Definitely, this will require further confirmation studies. A large‐scale surveillance longitudinal study among a large number of normal fertile bull camels, including various ages, would be another research priority in the future. This study highlights the potential roles of the dromedary camel semen in the transmission and spreading of MERS‐CoV among camels. This may have a great impact on the sustainability of the virus in the dromedary camel population not only in the Arabian Peninsula but worldwide. Testing the common male dromedary camels should be established to minimize the roles of bull camels in the transmission of the virus during the breeding seasons. To the best of our knowledge, this is the first study addressing the detection of MERS‐CoV‐RNA in the camel ejaculates. Further studies are needed to study the exact routes of MERS‐CoV transmission either among dromedary camels or between camels and humans. This will have great impacts on minimizing the viral shedding from dromedary camels to the environment and subsequently to the human; thus, the risk of human infection can be minimized.
ETHICAL APPROVAL
All animal experiments and sample collection conducted as per the King Abdul‐Aziz City of Science and Technology guidelines. This animal utilization protocol amended by King Faisal's University Animal Ethics and the National Committee of Bioethics (NCBE).
CONFLICT OF INTEREST
All authors declare there is no conflict of interest exist.
AUTHOR'S CONTRIBUTION
MGH and AAA designed experiments, conducted some laboratory experiments and analysed the data. MW conducted the field study and analysed the data; AMA conducted laboratory work and analysed the data; AAA analysed the data, conducted some statistical analysis and helped in the drafting of the Ms. All authors wrote the manuscripts and agreed to the final version of the Ms.
ACKNOWLEDGMENT
We wish to thank the King Abdul‐Aziz City for Science and Technology (KACST) for their generous funding through the MERS‐CoV research grant program (number 20‐0004), which is a part of the Targeted Research Program (TRP).
Hemida MG, Waheed M, Ali AM, Alnaeem A. Detection of the Middle East respiratory syndrome coronavirus in dromedary camel’s seminal plasma in Saudi Arabia 2015–2017. Transbound Emerg Dis. 2020;67:2609–2614. 10.1111/tbed.13610
DATA AVAILABILITY STATEMENT
Data will be available upon request.
REFERENCES
- Al Eknah, M. M. (2000). Reproduction in old world camels. Anim Reprod Sci, 60–61, 583–592. 10.1016/S0378-4320(00)00134-2 [DOI] [PubMed] [Google Scholar]
- Al Hammadi, Z. M. , Chu, D. K. , Eltahir, Y. M. , Al Hosani, F. , Al Mulla, M. , Tarnini, W. , … Poon, L. L. (2015). Asymptomatic MERS‐CoV infection in humans possibly linked to infected dromedaries imported from Oman to United Arab Emirates, may 2015. Emerging Infectious Diseases, 21, 2197–2200. 10.3201/eid2112.151132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almathen, F. , Elbir, H. , Bahbahani, H. , Mwacharo, J. , & Hanotte, O. (2018). Polymorphisms in MC1R and ASIP genes are associated with coat color variation in the Arabian camel. Journal of Heredity, 109, 700–706. 10.1093/jhered/esy024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur, G. H. , al‐Rahi, A. T. , & Hindi, A. S. A. (1985). Reproduction and genital diseases of the camel. British Veterinary Journal, 141, 650–659. [DOI] [PubMed] [Google Scholar]
- Azhar, E. I. , El‐Kafrawy, S. A. , Farraj, S. A. , Hassan, A. M. , Al‐Saeed, M. S. , Hashem, A. M. , & Madani, T. A. (2014). Evidence for camel‐to‐human transmission of MERS coronavirus. New England Journal of Medicine, 370, 2499–2505. 10.1056/NEJMoa1401505 [DOI] [PubMed] [Google Scholar]
- Azhar, E. I. , Hashem, A. M. , El‐Kafrawy, S. A. , Sohrab, S. S. , Aburizaiza, A. S. , Farraj, S. A. , … Madani, T. A. (2014). Detection of the Middle East respiratory syndrome coronavirus genome in an air sample originating from a camel barn owned by an infected patient. MBio, 5(4), e01450‐14. 10.1128/mBio.01450-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carossino, M. , Lee, P. Y. , Nam, B. , Skillman, A. , Shuck, K. M. , Timoney, P. J. , … Balasuriya, U. B. (2016). Development and evaluation of a reverse transcription‐insulated isothermal polymerase chain reaction (RT‐iiPCR) assay for detection of equine arteritis virus in equine semen and tissue samples using the POCKIT system. Journal of Virological Methods, 234, 7–15. 10.1016/j.jviromet.2016.02.015 [DOI] [PubMed] [Google Scholar]
- Chan, K. H. , Peiris, J. S. , Lam, S. Y. , Poon, L. L. , Yuen, K. Y. , & Seto, W. H. (2011). The effects of temperature and relative humidity on the viability of the SARS coronavirus. Archives of Virology, 2011, 734690. 10.1155/2011/734690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, D. K. W. , Hui, K. P. Y. , Perera, R. , Miguel, E. , Niemeyer, D. , Zhao, J. , … Peiris, M. (2018). MERS coronaviruses from camels in Africa exhibit region‐dependent genetic diversity. Proceedings of the National Academy of Sciences USA, 115, 3144–3149. 10.1073/pnas.1718769115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman, V. M. , Muller, M. A. , Costabel, U. , Timm, J. , Binger, T. , Meyer, B. , … Drosten, C. (2012). Assays for laboratory confirmation of novel human coronavirus (hCoV‐EMC) infections. Eurosurveillance, 17(49), 1–9. 10.2807/ese.17.49.20334-en [DOI] [PubMed] [Google Scholar]
- Council, O. D. , Swanson, M. D. , Spagnuolo, R. A. , Wahl, A. , & Garcia, J. V. (2015). Role of semen on vaginal HIV‐1 transmission and maraviroc protection. Antimicrobial Agents and Chemotherapy, 59, 7847–7851. 10.1128/AAC.01496-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Vega, M. A. , Soule, G. , Tran, K. N. , Tierney, K. , He, S. , Wong, G. , … Kobinger, G. P. (2018). Modeling ebola virus transmission using ferrets. mSphere, 3(5), e00309–e00318. 10.1128/mSphere.00309-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictionary, T. A. H. S. (2014). Retrieved from https://www.thefreelibrary.com/The+American+Heritage+Science+Dictionary.‐a0135119195
- Farag, E. A. , Haagmans, B. L. , Al‐Romaihi, H. , Mohran, K. , Haroun, M. , El‐Sayed, A. M. , … Reusken, C. (2019). Failure to detect MERS‐CoV RNA in urine of naturally infected dromedary camels. Zoonoses Public Health, 66, 437–438. 10.1111/zph.12583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag, E. , Sikkema, R. S. , Vinks, T. , Islam, M. M. , Nour, M. , Al‐Romaihi, H. , … Koopmans, M. (2018). Drivers of MERS‐CoV emergence in Qatar. Viruses, 11(1), 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornet, M. E. , Bracero, N. J. , & Segars, J. H. (2016). Zika virus in Semen: What we know and what we need to know. Seminars in Reproductive Medicine, 34, 285–292. 10.1055/s-0036-1592312 [DOI] [PubMed] [Google Scholar]
- Hemida, M. G. (2019). Middle East respiratory syndrome coronavirus and the one health concept. PeerJ, 7, e7556. 10.7717/peerj.7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida, M. G. , & Alnaeem, A. (2019). Some one health based control strategies for the Middle East respiratory syndrome coronavirus. One Health, 8, 100102. 10.1016/j.onehlt.2019.100102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida, M. G. , Alnaeem, A. , Chu, D. K. , Perera, R. A. , Chan, S. M. , Almathen, F. , … Peiris, M. (2017). Longitudinal study of Middle East respiratory syndrome coronavirus infection in dromedary camel herds in Saudi Arabia, 2014–2015. Emerging Microbes & Infections, 6, 1–7. 10.1038/emi.2017.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida, M. G. , Chu, D. K. , Poon, L. L. , Perera, R. A. , Alhammadi, M. A. , Ng, H. Y. , … Peiris, M. (2014). MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerging Infectious Diseases, 20, 1231–1234. 10.3201/eid2007.140571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D. J. , Ostlund, E. N. , Palmer, T. J. , Fett, K. L. , & Schmitt, B. J. (2012). Isolation of Equine rhinitis A virus from a horse semen sample. Journal of Veterinary Diagnostic Investigation, 24, 801–803. [DOI] [PubMed] [Google Scholar]
- Khanvilkar, A. V. , Samant, S. R. , & Ambore, B. N. (2009). Reproduction in Camel. Veterinary World, 2, 72–73. [Google Scholar]
- Kumar, S. , Stecher, G. , & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohran, K. A. , Farag, E. A. , Reusken, C. B. , Raj, V. S. , Lamers, M. M. , Pas, S. D. , … Koopmans, M. P. (2016). The sample of choice for detecting Middle East respiratory syndrome coronavirus in asymptomatic dromedary camels using real‐time reversetranscription polymerase chain reaction. Revue Scientifique Et Technique, 35, 905–911. [DOI] [PubMed] [Google Scholar]
- Nathues, C. , Perler, L. , Bruhn, S. , Suter, D. , Eichhorn, L. , Hofmann, M. , … Thur, B. (2016). An outbreak of porcine reproductive and respiratory syndrome virus in Switzerland following import of boar semen. Transboundary and Emerging Diseases, 63, e251–261. 10.1111/tbed.12262 [DOI] [PubMed] [Google Scholar]
- Padalino, B. , Monaco, D. , & Lacalandra, G. M. (2015). Male camel behavior and breeding management strategies: How to handle a camel bull during the breeding season? Emirates Journal of Food and Agriculture, 27, 338–349. 10.9755/ejfa.v27i4.19909 [DOI] [Google Scholar]
- Reusken, C. B. , Farag, E. A. , Jonges, M. , Godeke, G. J. , El‐Sayed, A. M. , Pas, S. D. , … Koopmans, M. P. (2014). Middle East respiratory syndrome coronavirus (MERS‐CoV) RNA and neutralising antibodies in milk collected according to local customs from dromedary camels, Qatar, April 2014. Eurosurveillance, 19(23), 1–5. 10.2807/1560-7917.ES2014.19.23.20829 [DOI] [PubMed] [Google Scholar]
- Skidmore, J. A. , Morton, K. M. , & Billah, M. (2013). Artificial insemination in dromedary camels. Animal Reproduction Science, 136, 178–186. 10.1016/j.anireprosci.2012.10.008 [DOI] [PubMed] [Google Scholar]
- Sneath, P. H. A. , Sokal, R. R. , & Freeman, W. H. (1973). Numerical Taxonomy Freeman. The Principles and Practice of Numerical Classification. New York, Stony Brook, NY: Medical Research Council Microbial Systematics Unit, Univ. Leicester, England and Dept. Ecology and Evolution, State Univ. San Francisco. [Google Scholar]
- Tibary, A. , Anouassi, A. , Sghiri, A. , & Khatir, H. (2007). Current knowledge and future challenges in camelid reproduction. Reproduction in Domestic Ruminants, 64, 297–313. 10.5661/RDR-VI-297 [DOI] [PubMed] [Google Scholar]
- Tingari, M. D. , El‐Manna, M. M. , Rahim, A. , Ahmed, A. K. , & Hamad, M. H. (1986). Studies on camel semen. I. Electroejaculation and some aspects of semen characteristics. Animal Reproduction Science, 12, 213–222. 10.1016/0378-4320(86)90042-4 [DOI] [Google Scholar]
- van Doremalen, N. , Bushmaker, T. , Karesh, W. B. , & Munster, V. J. (2014). Stability of Middle East respiratory syndrome coronavirus in milk. Emerging Infectious Diseases, 20, 1263–1264. 10.3201/eid2007.140500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernery, U. , Lau, S. K. , & Woo, P. C. (2017). Middle East respiratory syndrome (MERS) coronavirus and dromedaries. The Veterinary Journal, 220, 75–79. 10.1016/j.tvjl.2016.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, R. J. , Bali, S. , & Bark, H. (1987). Xylazine and ketamine anaesthesia in the dromedary camel under field conditions. The Veterinary Record, 120, 110–113. 10.1136/vr.120.5.110 [DOI] [PubMed] [Google Scholar]
- WHO (2019). Middle East respiratory syndrome coronavirus (MERS‐CoV) WHO. Geneva, Switzerland: WHO. http://www.who.int/emergencies/merscov/ [Google Scholar]
- Yusof, M. F. , Eltahir, Y. M. , Serhan, W. S. , Hashem, F. M. , Elsayed, E. A. , Marzoug, B. A. , … Al Muhairi, S. S. (2015). Prevalence of Middle East respiratory syndrome coronavirus (MERS‐CoV) in dromedary camels in Abu Dhabi Emirate, United Arab Emirates. Virus Genes, 50, 509–513. 10.1007/s11262-015-1174-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki, A. M. , van Boheemen, S. , Bestebroer, T. M. , Osterhaus, A. D. , & Fouchier, R. A. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine, 367, 1814–1820. 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available upon request.


