Summary
Effective laboratory markers for the estimation of disease severity and predicting the clinical progression of coronavirus disease‐2019 (COVID‐19) is urgently needed. Laboratory tests, including blood routine, cytokine profiles and infection markers, were collected from 389 confirmed COVID‐19 patients. The included patients were classified into mild (n = 168), severe (n = 169) and critical groups (n = 52). The leukocytes, neutrophils, infection biomarkers [such as C‐reactive protein (CRP), procalcitonin (PCT) and ferritin] and the concentrations of cytokines [interleukin (IL)‐2R, IL‐6, IL‐8, IL‐10 and tumor necrosis factor (TNF)‐α] were significantly increased, while lymphocytes were significantly decreased with increased severity of illness. The amount of IL‐2R was positively correlated with the other cytokines and negatively correlated with lymphocyte number. The ratio of IL‐2R to lymphocytes was found to be remarkably increased in severe and critical patients. IL‐2R/lymphocytes were superior compared with other markers for the identification of COVID‐19 with critical illness, not only from mild but also from severe illness. Moreover, the cytokine profiles and IL‐2R/lymphocytes were significantly decreased in recovered patients, but further increased in disease‐deteriorated patients, which might be correlated with the outcome of COVID‐19. Lymphopenia and increased levels of cytokines were closely associated with disease severity. The IL‐2R/lymphocyte was a prominent biomarker for early identification of severe COVID‐19 and predicting the clinical progression of the disease.
Keywords: clinical progression, COVID‐19, cytokine, IL‐2R/lymphocyte ratio, SARS‐CoV‐2
IL‐2R/lymphocyte was a superior marker for the identification of severe COVID‐19 patients.
![]()
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), a new family of coronavirus, can cause severe respiratory infections [1]. The disease caused by SARS‐CoV‐2 has been recently named as the coronavirus disease‐2019 (COVID‐19) by the World Health Organization (WHO). Since the first report in December 2019, a severe outbreak caused by SARS‐CoV‐2 in China has rapidly spread to multiple regions of the world [2, 3, 4]. WHO has defined COVID‐19 as a pandemic due to the speed and scale of transmission.
Among a total of 81 385 case records reported by the Chinese Center for Disease Control and Prevention, most COVID‐19 patients were classified as mild (81%), 14% were severe and 5% were critical. The overall case‐fatality rate (CFR) was 2·3%, and no deaths were reported in mild and severe cases. However, the CFR among critical cases was 49% [5]. Some patients, particularly those of older age and with pre‐existing co‐morbid conditions, could develop to critical illness, and died of multiple organ failure in a short period of time [5, 6]. Thus, efficient indicators for evaluation of disease severity and clinical progression still need further investigation, which is urgently needed for reducing COVID‐19 mortality.
Previous studies have shown that many laboratory biomarkers, such as lymphocyte numbers, lactate dehydrogenase and D‐dimer, are out of normal ranges in COVID‐19 patients [6, 7]. These laboratory biomarkers are routine and are the most available in clinics. Another study compared plasma cytokine levels between patients admitted to the intensive care unit (ICU) and those not requiring ICU admission, and found that many cytokines, such as interleukin (IL)‐2, IL‐7, IL‐10, granulocyte–colony‐stimulating factor (G‐CSF), interferon gamma‐inducible protein‐10 (IP‐10P, chemotactic protein‐1 (MCP1), MIP1A and tumor necrosis factor (TNF)‐α, had higher levels in ICU patients compared with non‐ICU patients [8]. However, available markers for predicting the progression of patients with COVID‐19 are limited.
In this study, we analyzed the differences of different biomarkers, including blood routine, cytokine profiles, procalcitonin (PCT), C‐reactive protein (CRP) and ferritin, among COVID‐19 patients with different severity illness. Our data showed that IL‐2R/lymphocytes could be used for early identification of severe COVID‐19.
Methods
Patients
This study recruited COVID‐19 patients from 24 January to 15 February 2020, admitted at Tongji Hospital, Wuhan, China. Patients who had clinical symptoms (fever, cough or shortness of breath) and radiological characteristics (unilateral pneumonia, bilateral pneumonia or ground‐glass opacity), and with positive SARS‐CoV‐2 real‐time reverse transcription–polymerase chain reaction (RT–PCR) results, were continuously recruited. Patients younger than 18 years were excluded. Symptoms, signs and laboratory tests during the hospital stay were collected. This study was approved by the ethical committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China (grant number: TJ‐C20200128, date: 2020‐2‐12).
Grouping criteria
COVID‐19 patients were classified into three groups with different illness severity, according to the guidelines of diagnosis and treatment for COVID‐19 made by the Chinese National Health Commission, as follows – (1) mild: patients had typical symptoms and radiological findings; (2) severe: patients met one of the following criteria – respiratory distress (respiration rate ≥ 30 times/min), blood oxygen saturation (SpO2) ≤ 93% in the resting state and arterial partial pressure of O2 to fraction of inspired oxygen (PaO2/FiO2) ratio ≤ 300 mmHg; and (3) critical: the patients met one of the following criteria – respiratory failure requiring mechanical ventilation, septic shock and/or multiple organ dysfunction or failure needing ICU treatment. Clinical information related to patient classification was collected from the medical records of the patients.
Real‐time RT–PCR
Throat or nasal‐swab specimens from the upper respiratory tract of all patients on admission were collected and maintained in a viral transport medium. Sputum specimens were also collected in some patients. SARS‐CoV‐2 infection was confirmed using TaqMan one‐step RT–PCR kits from Shanghai Huirui Biotechnology Company Ltd and Shanghai BioGerm Medical Biotechnology Company Ltd, both of which were approved by the China Food and Drug Administration. In brief, RNA was extracted from clinical samples with commercial RNA extraction kits. Then, 5 μl of RNA template was used for real‐time RT–PCR, which targeted the open reading frame (ORF)1ab and nucleoprotein gene. Conditions for applications of real‐time RT–PCR were as follows: 50°C for 15 min and 95°C for 5 min, 45 cycles of amplification at 95°C for 10 s and 55°C for 45 s. The positive COVID‐19 real‐time RT–PCR result was defined if both ORF1ab and nucleoprotein cycle thresholds were < 35.
Cytokine profile analysis
Serum samples were collected from the included participants on admission or at the time of disease aggravation. In those who recovered from illness, the serum samples prior to hospital discharge were also collected. The levels of IL‐1β, IL‐2R, IL‐8, IL‐10, and TNF‐α in serum were measured according to an automatic procedure of a solid‐phase two‐site chemiluminescent immunometric assay via IMMULITE 1000 Analyzer (Siemens, Munich, Germany). The level of IL‐6 was measured using the electrochemiluminescence method (Roche Diagnostics, Basel, Switzerland).
Statistical analysis
The results are presented as mean ± standard deviation (s.d.). Differences of markers among the three groups were compared by analysis of variance (ANOVA). The differences of markers between different time‐points from the same patients were analyzed using the paired Student’s t‐test. Spearman’s rank correlation test for non‐parametric data was employed to analyze the relationship between two factors. All statistically significant variables were taken as candidates for multivariable logistic regression analyses, and the regression equation (predictive model) was obtained. The regression coefficients of the predictive model were regarded as the weights for the respective variables and a score for each patient was calculated. Receiver operating characteristic (ROC) analysis was performed to determine the best threshold value of the laboratory markers for distinguishing critical illness patients from mild or severe illness patients. Area under the curve (AUC), sensitivity and specificity were reported. Statistical analyses were performed using GraphPad Prism version 6, MedCalc version 11.6 and SPSS version 22.0. Statistical significance was determined as P < 0·05.
Results
This study included 389 hospitalized patients with confirmed COVID‐19. The patients were classified into three groups with different severity of illness. The basic characteristics of mild (n = 168), severe (n = 169) and critical (n = 52) patients are shown in Table 1. The ages of severe (64·7 ± 12·2) and critical (65·7 ± 14·2) patients were significantly higher than mild patients (56·6 ± 13·9). The percentage of males was higher than females in severe and critical groups. The most common symptoms at onset of illness were fever, cough, fatigue, expectoration and shortness of breath. Other symptoms were chest distress, diarrhea, muscle ache, headache, nausea and vomiting. More than half the patients had chronic diseases, including hypertension (32·9%), diabetes (10%), cardiovascular (8·2%) diseases and chronic kidney disease (3·3%) (Table 1).
Table 1.
Baseline characteristics of 389 patients with COVID‐19 pneumonia
| Total (n = 389) | Mild (n = 168) | Severe (n = 169) | Critical (n = 52) | |
|---|---|---|---|---|
| Age (years) | ||||
| Mean (s.d.) | 61·3 (13·8) | 56·7 (13·9) | 64·6 (12·2) | 65·4 (14·2) |
| Sex | ||||
| Male | 200 (51·4%) | 74 (44%) | 94 (55·6%) | 32 (61·5%) |
| Female | 189 (48·6%) | 94 (56%) | 75 (44·4%) | 20 (38·5%) |
| Signs and symptoms at admission | ||||
| Fever | 310 (79·7%) | 136 (81%) | 131 (77·5%) | 43 (82·7%) |
| Cough | 236 (60·7%) | 107 (64%) | 102 (60·4%) | 27 (51·9%) |
| Fatigue | 146 (37·5%) | 58 (34·5%) | 72 (42·6%) | 16 (30·8%) |
| Expectoration | 95 (24·4%) | 33 (19·6%) | 46 (27·2%) | 16 (30·8%) |
| Shortness of breath | 76 (19·5%) | 21 (12·5%) | 35 (20·7%) | 20 (38·5%) |
| Chest distress | 72 (18·5%) | 24 (14·3%) | 32 (18·9%) | 16 (30·8%) |
| Diarrhea | 44 (11·3%) | 22 (13·1%) | 16 (9·5%) | 6 (11·5%) |
| Muscle ache | 23 (5·9%) | 11 (6·5%) | 9 (5·3%) | 5 (9·6%) |
| Headache | 14 (3·7%) | 5 (3%) | 6 (3·6%) | 3 (5·8%) |
| Dizziness | 14 (3·7%) | 10·6% | 3 (1·8%) | 1 (1·9%) |
| Nausea and vomiting | 12 (3·0%) | 4 (2·4%) | 6 (3·6%) | 2 (3·8%) |
| Pharyngalgia | 10 (2·6%) | 7 (4·2%) | 3 (1·8%) | 0 |
| Chest pain | 10 (2·6%) | 5 (3%) | 2 (1·2%) | 3 (5·8%) |
| Comorbidities | 230 (59·1%) | 85 (50·6%) | 109 (64·5%) | 36 (69·2%) |
| Hypertension | 128 (32·9%) | 44 (26·2%) | 63 (37·3%) | 21 (40·4%) |
| Diabetes | 39 (10%) | 16 (9·5%) | 19 (11·2%) | 4 (7·7%) |
| Cardiovascular disease | 32 (8·2%) | 15 (8·9%) | 12 (7·1%) | 5 (9·6%) |
| Chronic kidney disease | 13 (3·3%) | 4 (2·4%) | 7 (4·1%) | 2 (3·8%) |
| Cerebrovascular disease | 8 (2·1%) | 3 (1·8%) | 3 (1·8%) | 2 (3·8%) |
| Malignancy | 4 (1%) | 2 (1·2%) | 2 (1·2%) | 0 |
| Chronic liver disease | 3 (0·8%) | 1 (0·6%) | 1 (0·6%) | 1 (1·9%) |
| COPD | 3 (0·8%) | 0 | 2 (1·2%) | 1 (1·9%) |
Data are presented as numbers (%) unless otherwise indicated. COVID‐19 = coronavirus disease‐2019; COPD = chronic obstructive pulmonary disease; s.d. = standard deviation.
The results of blood routine, infectious biomarkers and cytokine profiles in different groups are shown in Fig. 1. We observed that leukocytes, neutrophils, CRP, PCT and ferritin were significantly increased, while lymphocytes were significantly decreased in severe and critical patients compared with mild patients (Fig. 1a,b). These biomarkers also showed statistically significant differences between severe and critical patients. The cytokine profiles, including IL‐1β, IL‐2R, IL‐6, IL‐8, IL‐10 and TNF‐α on admission, were detected. We found that the cytokine profiles, except IL‐1β, were all significantly increased with increased severity of illness (Fig. 1c). Moreover, the concentration of IL‐2R was positively correlated with that of IL‐6, IL‐8, IL‐10 and TNF‐α (Fig. 2a–d). We also observed that the amount of IL‐2R was negatively correlated with lymphocyte numbers (Fig. 2e). We then further calculated the ratio of IL‐2R to lymphocytes and found that the value of IL‐2R/lymphocytes was remarkably increased in severe and critical patients compared with mild patients (Fig. 2f).
Fig. 1.
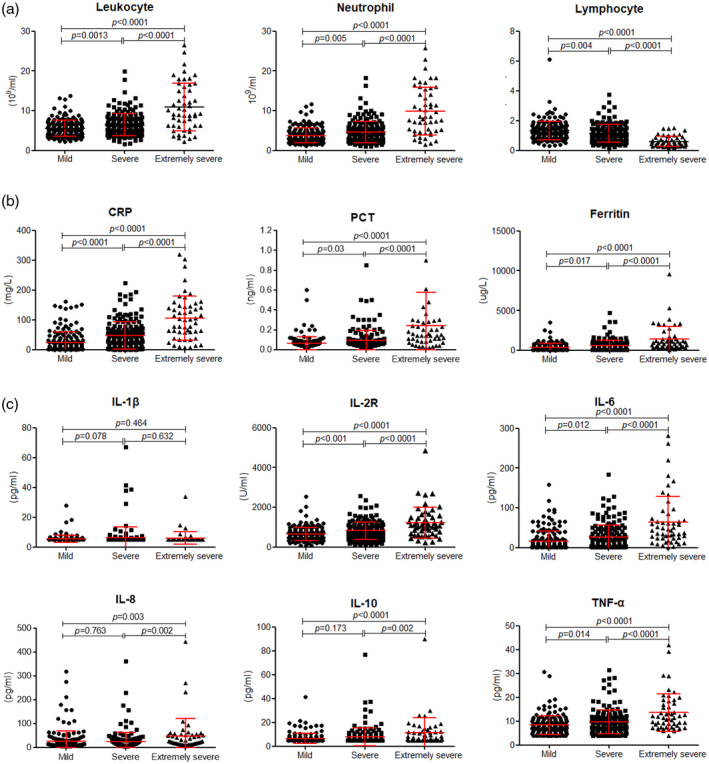
Blood cell counts, infectious biomarkers and cytokine profiles in coronavirus disease‐2019 (COVID‐19) patients. (a) The absolute counts of leukocytes, neutrophils and lymphocytes in patients with mild, severe and critical illness. (b) The levels of C‐reactive protein (CRP), procalcitonin (PCT) and ferritin in different groups. (c) The levels of interleukin (IL)‐1β, IL‐2R, IL‐6, IL‐8, IL‐10 and tumor necrosis factor (TNF)‐α in patients with mild, severe and critical illness. The normal reference ranges of IL‐1β, IL‐6, IL‐8, IL‐10 and TNF‐α were 5, 1·5, 5, 5 and 4 pg/ml, respectively. If the concentrations of the corresponding cytokines were lower than the normal range, we recorded the minimum values. Differences of markers among the three groups were compared by analysis of variance (ANOVA). Results of the test were significant with P < 0·05.
Fig. 2.
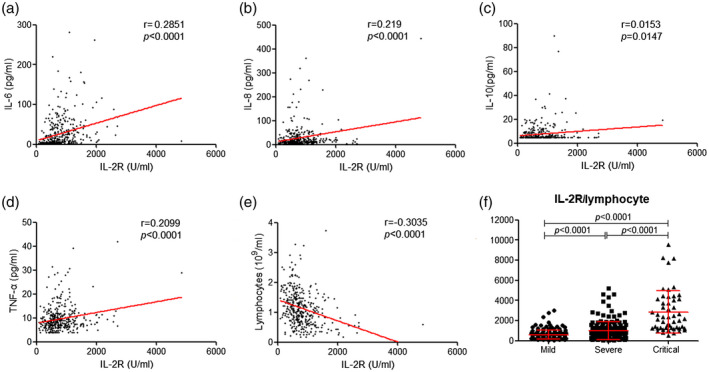
The relationship between the amounts of interleukin (IL)‐2R and other cytokines or lymphocytes. Correlation between the levels of IL‐2R and IL‐6 (a), IL‐8 (b), IL‐10 (c) and tumor necrosis factor (TNF)‐α (d). (e) Correlation between lymphocytes and IL‐2R level. (f) The value of IL‐2R/lymphocytes in mild, severe and critical patients. Spearman’s rank correlation test for non‐parametric data was employed to analyze the relationship between two factors. The differences of IL‐2R/lymphocytes among the three groups were compared by analysis of variance (ANOVA). Results of the test were significant with P < 0·05.
For the prediction of severe COVID‐19 patients, all statistically significant variables were taken as candidates for further multivariable logistic regression analyses. We found that the increase of IL‐2R/lymphocytes, CRP and ferritin were associated with increased odds, and chosen as the prediction model indicators of severe COVID‐19. Odds ratios (OR) and their corresponding confidence intervals are shown in Table 2. ROC analysis showed that IL‐2R/lymphocytes had a higher AUC (0·948) than CRP (0·886) and ferritin (0·812) to differentiate critical from mild patients. The performance of IL‐2R/lymphocytes to distinguish severe from mild patients was poor (Fig. 3a). However, IL‐2R/lymphocytes showed a sensitivity of 90·9% and a specificity of 66·5% to differentiate between critical and severe patients (Fig. 3b) and a sensitivity of 94·2% and a specificity of 82·1% in distinguishing critical from mild patients (Fig. 3c).
Table 2.
Multivariable logistic regression analysis for prediction of severe COVID‐19
| Parameter | OR | OR (97·5% CI = 2·5%) | P‐value | |
|---|---|---|---|---|
| IL‐1β | 1·0379 | 0·9739 | 1·1062 | 0·2515 |
| IL‐2R | 0·9993 | 0·9983 | 1·0004 | 0·2078 |
| IL‐6 | 1·0000 | 0·9903 | 1·0097 | 0·9936 |
| IL‐8 | 0·9985 | 0·9929 | 1·0041 | 0·5934 |
| IL‐10 | 1·0163 | 0·9769 | 1·0573 | 0·4234 |
| TNF‐α | 1·0115 | 0·9443 | 1·0834 | 0·7450 |
| Lymphocyte | 1·2623 | 0·7412 | 2·1499 | 0·3911 |
| Neutrophil | 1·0638 | 0·9467 | 1·1953 | 0·2986 |
| PCT | 2·5333 | 0·7287 | 8·8067 | 0·1437 |
| IL‐2R/lymphocytes | 1·0013 | 1·0005 | 1·0021 | 0·0020 |
| CRP | 1·0085 | 1·0008 | 1·0163 | 0·0300 |
| Ferritin | 1·0006 | 1·0001 | 1·0010 | 0·0206 |
COVID‐19 = coronavirus disease‐2019; OR = odds ratio; CI = confidence interval; PCT = procalcitonin; CRP = C‐reactive protein; IL = interleukin; TNF = tumor necrosis factor.
Fig. 3.
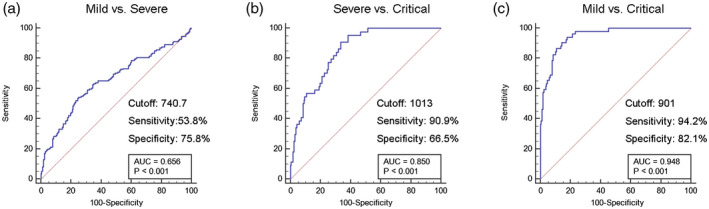
Receiver operating characteristic (ROC) analysis showing the performance of the interleukin (IL)‐2R/lymphocyte among coronavirus disease‐2019 (COVID‐19) patients with different severity of illness.
Furthermore, we monitored the change of lymphocytes and cytokine levels in the disease progression of SARS‐CoV‐2‐infected patients. In the 28 recovered patients lymphocyte numbers gradually increased to normal levels, while the levels of IL‐2R, IL‐6, IL‐8, IL‐10, TNF‐α and IL‐2R/lymphocytes were significantly decreased between the time of admission and hospital discharge (Fig. 4) In contrast, lymphocyte numbers were further decreased, while cytokines and IL‐2R/lymphocytes were significantly increased in the deteriorated patients (Fig. 5).
Fig. 4.
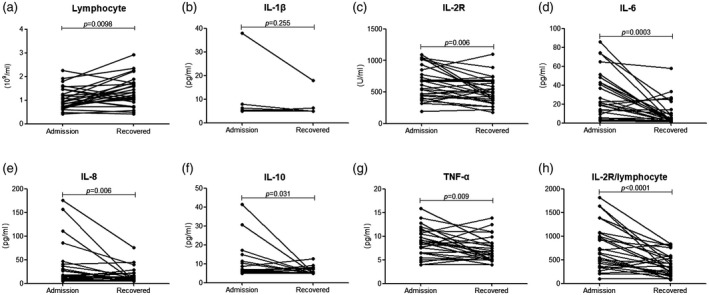
The change of various markers in recovered coronavirus disease‐2019 (COVID‐19) patients. The lymphocytes (a), [interleukin (IL)‐1β (b), IL‐2R (c), IL‐6 (d), IL‐8 (e), IL‐10 (f), tumor necrosis factor (TNF)‐α (g) and IL‐2R/lymphocytes (h)] were compared between the time of admission and hospital discharge. Differences of markers between different time‐points of the same patients were analyzed using the paired Student’s t‐test. Results of the test were significant with P < 0·05.
Fig. 5.
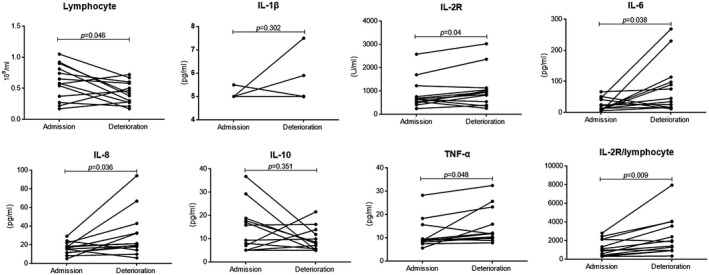
The change of various markers in deteriorated coronavirus disease‐2019 (COVID‐19) patients. The lymphocytes (a), interleukin (IL)‐1β (b), IL‐2R (c), IL‐6 (d), IL‐8 (e), IL‐10 (f), tumor necrosis factor (TNF)‐α and IL‐2R/lymphocytes (h) were compared between the time of admission and deterioration. Differences of markers between different time‐points of the same patients were analyzed using the paired Student’s t‐test. Results of the test were significant with P < 0·05.
Discussion
The prevalence of COVID‐19 began from December 2019, then quickly spread worldwide, and has now become an urgent public health challenge. In this study, we report a cohort of 389 patients with laboratory‐confirmed SARS‐CoV‐2 infection. Most patients presented with fever, cough and fatigue, which share similar clinical features with previous beta‐coronavirus infections, such as SARS and Middle East respiratory syndrome (MERS) [9, 10]. Highly pathogenic human coronaviruses (hCoV) infections frequently lead to lower respiratory tract disease, and disease severity was influenced by initial viral titers in airways, age and co‐morbid conditions [6, 11, 12]. We observed that in severe and critical patients the percentage of males was higher than females, and the mean ages of these patients were much older than mild patients. Moreover, critical patients were accompanied by high percentages of manifestations and co‐morbidities, indicating the poor prognosis and high CFR of these patients. Therefore, it is of crucial significance to find potential markers to identify critical patients early and give timely effective treatment.
Lymphopenia was observed during SARS and MERS infections [13]. Similarly, the developed lymphopenia was also very common in COVID‐19 and this phenomenon was more obvious in ICU patients [6, 7]. The mechanisms of lymphopenia in SARS‐CoV‐2 infection might be due to inhibition of the cellular immune effective function, and more cells underwent apoptosis following overactive inflammatory responses, as reported in SARS infection [14]. The results of lymphocytes might be associated with disease severity and mortality.
Massive inflammatory cell infiltration and cytokine storms could result in acute lung injury and acute respiratory distress syndrome during SARS and MERS infections, which have been specially emphasized for the pathogenesis of hCoV infections [15]. In COVID‐19, both T helper type 1 (Th1) and Th2 cell responses were initiated and the increased amounts of cytokines (e.g. IL‐1β, IFN‐γ, IP‐10, MCP1, IL‐10 and IL‐4) were associated with disease severity [6]. A recent pathological report of COVID‐19 has also shown that the counts of CD4+ T and CD8+ T cells were decreased, but with high expression of human leukocyte antigen D‐related (HLA‐DR) and CD38. The over‐activation of T cells, including the increase of proinflammatory Th17 cells and high cytotoxicity of CD8+ T cells, might lead to the severe immune injury [16]. During viral infection, adaptive immune cells not only played a crucial role in virus clearance but also in dampening innate immunity and limiting further damage to the host [17, 18]. However, exuberant inflammatory responses promoted T cell apoptosis, thus resulting in uncontrolled inflammatory responses due to diminished T cell responses. Our data demonstrate that infection biomarkers such as CRP, PCT and ferritin and the concentrations of IL‐2R, IL‐6, IL‐8, IL‐10 and TNF‐α were all significantly increased with the increased illness severity, suggesting that the magnitude of the cytokine storm results in high morbidity and mortality attributed by immunopathology.
Reliable and feasible early identification of severe patients is crucial in clinical practice, as the mortality of these patients is extremely high [5]. However, the available laboratory markers have limited value at present. During immune activation, CD25 is expressed by T cells and its soluble form (sCD25), also known as IL‐2R, is released into the bloodstream [19]. Our data show that increased IL‐2R and lymphopenia were both correlated with the severity of COVIN‐19. Moreover, the concentration of IL‐2R was positively correlated with IL‐6, IL‐8, IL‐10 and TNF‐α, and negatively correlated with lymphocytes. Therefore, the value of IL‐2R/lymphocytes was superior to reflect the imbalance of inflammation and immune response.
IL‐2R/lymphocytes, CRP and ferritin were selected as prediction markers of severe illness by multivariable logistic regression analyses. These markers show that infection, injury of immunity and an exaggerated inflammatory response might exert synthetic efforts on the progression of severe illness. The performance of IL‐2R/lymphocytes exhibited higher sensitivity and specificity in differentiating critical patients from mild or severe patients. The application of IL‐2R/lymphocytes would allow the early diagnosis and treatment of critical patients and decrease mortality. Furthermore, concentrations of cytokines, including IL‐2R, IL‐6, IL‐8 and TNF‐α, were decreased, but lymphocytes were increased in recovered patients. However, lymphocytes were further decreased with high levels of cytokines in disease‐deteriorated patients. Therefore, the monitoring these laboratory markers and cytokines could also be used to predict the progression of COVID‐19.
Several limitations of this study should be mentioned. First, not all laboratory tests have been included, as these markers are also valuable in COVID‐19, such as coagulation and chemistry markers. Secondly, the percentage of severe patients was high in the included population, as Tongji hospital was a designated hospital for admitting severe patients, so the prediction model might have some bias. Thirdly, continuous monitoring data in one patient at different disease courses is limited, and needs to be further validated in larger populations.
In summary, this study demonstrates that lymphopenia and high concentrations of cytokines were associated with the pathogenesis of COVID‐19, and the findings of the study could help in more clearly understanding the immune disorder of the disease. IL‐2R/lymphocytes are a prominent biomarker for early identification of severe patients and predicting the clinical progression of COVID‐19.
Disclosures
We declare no competing interests.
Acknowledgements
H. H. Y., Z. Y. S. and F. W. designed the experiment; B. Z., H. H., Y. L., G. X. T. and L. Y. M. collected the data; S. J. W., W. Y. L. and L. E. M. analyzed the data; H. H. Y., Z. Y. S. and F. W. wrote the manuscript. This study was funded by the National Mega Project on Major Infectious Disease Prevention (2017ZX10103005‐007).
Contributor Information
F. Wang, Email: fengwang@tjh.tjmu.edu.cn.
Z. Sun, Email: zysun@tjh.tjmu.edu.cn.
References
- 1. Zhu N, Zhang D, Wang W et al, China Novel Coronavirus I, Research Team . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bassetti M, Vena A, Giacobbe DR. The novel Chinese coronavirus (2019‐nCoV) infections: challenges for fighting the storm. Eur J Clin Invest 2020; 50:e13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rothe C, Schunk M, Sothmann P et al Transmission of 2019‐nCoV infection from an asymptomatic contact in Germany. N Engl J Med 2020; 382:970–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. The L. Emerging understandings of 2019‐nCoV. Lancet 2020; 395:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu Z, McGoogan JM. Characteristics of and Important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020; 323:1239–42. [DOI] [PubMed] [Google Scholar]
- 6. Chen N, Zhou M, Dong X et al Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020; 395:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang D, Hu B, Hu C et al Clinical Characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA 2020. 10.1001/jama.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang C, Wang Y, Li X et al Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee N, Hui D, Wu A et al A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 2003; 348:1986–94. [DOI] [PubMed] [Google Scholar]
- 10. Ranin J, Salemovic D, Brmbolic B et al Comparison of demographic, epidemiological, immunological, and clinical characteristics of patients with HIV mono‐infection versus patients co‐infected with HCV or/and HBV: a Serbian cohort study. Curr HIV Res 2018; 16:222–30. [DOI] [PubMed] [Google Scholar]
- 11. Assiri A, Al‐Tawfiq JA, Al‐Rabeeah AA et al Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis 2013; 13:752–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saad M, Omrani AS, Baig K et al Clinical aspects and outcomes of 70 patients with Middle East respiratory syndrome coronavirus infection: a single‐center experience in Saudi Arabia. Int J Infect Dis 2014; 29:301–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 2016; 14:523–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chan PK, Chen GG. Mechanisms of lymphocyte loss in SARS coronavirus infection. Hong Kong Med J 2008; 14(Suppl 4):21–6. [PubMed] [Google Scholar]
- 15. Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol 2017; 39:529–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu Z, Shi L, Wang Y et al Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020; 8:420–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao J, Zhao J, Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus‐infected mice. J Virol 2010; 84:9318–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han AR, Lee HR, Park BB et al Lymphoma‐associated hemophagocytic syndrome: clinical features and treatment outcome. Ann Hematol 2007; 86:493–8. [DOI] [PubMed] [Google Scholar]
- 19. Buono A, Lidbury JA, Wood C et al Development, analytical validation, and initial clinical evaluation of a radioimmunoassay for the measurement of soluble CD25 concentrations in canine serum. Vet Immunol Immunopathol 2019; 215:109904. [DOI] [PubMed] [Google Scholar]


