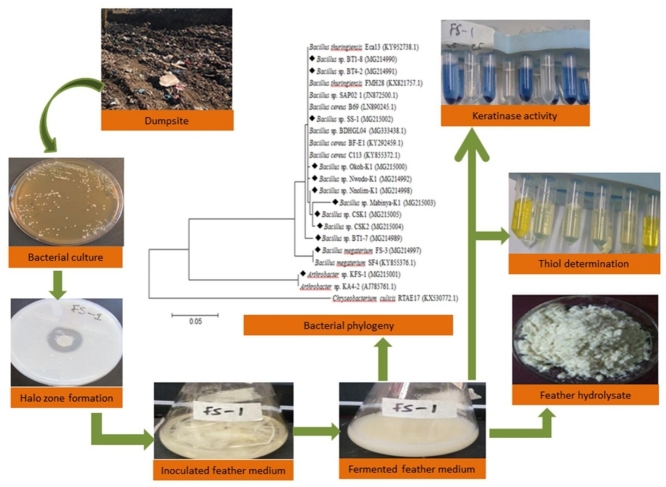
Graphical abstract
Keywords: Chicken feather, Keratinase, Thiol group, Valorisation
Highlights
-
•
Proteolytic bacteria were isolated from agro-waste dumpsites.
-
•
The isolates degraded intact feathers and produced keratinases in basal media.
-
•
Feather degradation generated high concentration of free thiol containing groups.
-
•
The remarkable thiol concentrations suggest keratinous waste valorisation potential of these bacteria.
-
•
The isolates were identified through 16S rDNA sequence as Bacillus spp. and Arthrobacter sp.
Abstract
Microbial bioconversion of carbonoclastic materials is an efficient tool for the exploitation and valorization of underutilized agro-industrial wastes. The agro-industrial sector accumulates tones of keratinous wastes biomass which may be valorized into high value products. Consequently, the keratinolytic potentials of some bacteria isolated from terrestrial milieu was evaluated. Soil samples were collected from dumpsites, keratinase producing bacteria were isolated. Bacterial species were identified through 16S rRNA gene sequences. The keratinase activity was assessed in relation to thiol formation, percentage feather degradation and quantitation of keratinase produced. Keratinolytic bacteria were identified as Bacillus spp. (accession numbers: MG214989 – MG214992, MG214997, MG214998, MG215000, MG215002–MG215005) and Arthrobacter sp. (accession numbers; MG215001). The degree of chicken feather degradation ranged from 61.5 ± 0.71 % to 85.0 ± 1.41 %. Similarly, the activity of keratinase, total protein and thiol group ranged from 198.18 ± 15.43–731.83 ± 14.14 U/mL; 0.09 ± 0.01–0.87 ± 0.05 mg/mL; and 0.69 ± 0.12–2.89 ± 0.11 mM respectively. Notably, Bacillus sp. Nnolim-K1 displayed the best keratinolytic potential with extracellular keratinase activity and feather degradation of 731.83 ± 14.14 U/mL and 85.0 ± 1.41 % respectively, and that is an indication of a potential relevance biotechnologically.
1. Introduction
Agro-industrial processes are on the upward trajectory and, expectedly so, would continue to increase as the world’s burgeoning population needs sustenance. Diverse agro-industrial activities have led to the accumulation of byproducts of the various kinds either as waste or secondary products with recyclable value. Besides, most waste products from these agro-industries are fed into the environment; thus, constituting an environmental nuisance. Sustainable biotechnology, on the other hand, drives zero waste and, strategically propels the valorisation of waste to high-value products. Microbes and microbial products with unique properties are employed in the bio-recycling or valorisation processes for high-value products. Enzymes, a product of microbial metabolism, are of high relevance in the green economy for apparent reasons of high specificity to substrates, enhanced catalytic efficiency, cost-effectiveness, and eco-friendliness [1,2]. Microbial enzymes including polymerases, proteases, lipases and amylases are high up in contribution to the bio-economy world over. Microbial proteases belong to the hydrolases group and, are predominantly multifunctional enzymes that catalyse the hydrolysis of an array of proteinaceous polymers into constituent peptides and amino acids [3,4]. The global contribution of proteases to industrial enzymes is approximately 60 %, of which the primary sources are bacteria from the genus Bacillus [5]. Several of the proteases produced by Bacillus spp. serves a different purpose and, are commercially available [6].
Microbial keratinases (EC 3.4.21/24/99.11), which is a member of proteolytic enzymes, is a group predominantly associated with keratin hydrolysis [7,8]. Keratins are fibrous, structural and insoluble proteins that constitute the epidermis and epidermal appendages, such as skin, hair, nails, hooves, horns, scales, claws, and feathers [9,10]. Keratins are classified as α-keratin and β-keratin, based on the major secondary structural elements of polypeptides, α-helices or pleated β-sheets, respectively [9]. The conformational orientations of the cysteine residues and interactions of hydrophobic groups confer mechanical stability to the polymer against biotic and abiotic factors; thus, a major contributor for the recalcitrance of keratin to decomposition [11]. The agro-industrial wastes, especially those emanating from leather and poultry processing industries are considered to have little or no economic relevance due to their structural stability, which makes valorisation difficult. Nonetheless, several strategies have been employed to harness the locked up potentials from keratinous wastes, and these strategies have included thermo-energetic processing, acid or alkaline hydrolysis [12]. Keratin waste valorisation with the above-indicated procedures has yielded products which have not been suitable for industrial applications [13]. Keratinases which may bioconvert keratin to peptides and amino acids, on the other hand, has not been a front runner in the valorisation of keratinous waste biomass; although agro wastes are thought to increase as the world population increases. So, it would be prudent to state that keratinases shall enjoy extreme importance soon [2].
The bioconversion of keratinous wastes into amino acids or peptides with functional values would be an attractive endeavour for several applications including animal feed formulations. The valorisation approach would represent a potentially sustainable strategy for the proper management of keratin-rich agro wastes [10] and food security in animal farming. Therefore, sourcing efficient and effective keratinolytic microorganisms and enzymes would be beneficial from the biotechnological and industrial viewpoint. Consequently, this study was set out to evaluate the keratinolytic potentials of some bacterial isolates autochthonous to the terrestrial milieu of Raymond Mhlaba Municipality, South Africa. The bacterial isolates remarkably degraded intact chicken feathers with formation of high concentrations of thiol groups in the fermentation medium through extracellular production of keratinolytic enzymes.
2. Materials and methods
2.1. Keratin – substrate preparation
The keratin substrate was prepared from chicken feathers obtained from a local poultry processing farm. The feathers were thoroughly washed and rinsed with distilled water, and subsequently, dried at 60 °C for 48 h. The dried feathers were milled into a fine powder with a pulveriser fitted with a 2 mm mesh and stored in an airtight container at room temperature.
2.2. Samples collection and bacteria isolation
The soil samples were collected in April 2018 from dumpsites with deposits of keratinous wastes in Alice (geographical coordinates 32° 47’ 0” S, 26° 50’ 0” E), in the Ramond Mhlaba Local Municipality, Eastern Cape Province, South Africa. The sampling sites (dumpsites) receive a collection of municipal wastes that come from both households and agro-farms. The samples were aseptically transported to the laboratory and processed within 6 h of collection. Approximately 1 g of the soil samples was inoculated in 500 mL Erlenmeyer flask containing 99 mL sterile basal medium with the following constituents (g/L): K2HPO4, 0.3; KH2PO4, 0.4; MgCl2, 0.2; CaCl2, 0.22; NH4Cl, 0.5; chicken feather powder (CFP), 10 [14] and initial pH of 6.0. The flask was incubated in a rotary shaker for 5 days at 30 °C. The culture broth (100 μL) was spread plated on a CFP agar plate supplemented with nystatin (50 mg/L) to inhibit fungal growth. The CFP agar consisted of the following constituents (g/L): K2HPO4, 0.3; KH2PO4, 0.4; MgCl2, 0.2; CaCl2, 0.22; NH4Cl, 0.5; CFP, 10; and bacteriological agar, 15. After 48 h of incubation at 30 °C, distinct colonies were picked and purified.
2.3. Isolates screening for proteolytic activity
Pure bacterial culture was inoculated onto freshly prepared CFP agar plates and incubated at 30 °C for 18 h. A loopful of bacterial colonies was transferred into sterile microtubes and was washed twice with sterile saline (8.5 g/L NaCl). The pellet was re-suspended in sterile saline and the optical density adjusted to 0.1 (corresponding to 0.5 McFarland’s standard) at 600 nm, and isolates were screened for the ability to hydrolyse casein in skimmed milk (SM) as described by Riffel and Brandelli [15] with modification. The proteolytic potentials of the isolates were examined on SM agar with the following composition (g/L): K2HPO4, 0.3; KH2PO4, 0.4; MgCl2, 0.2; CaCl2, 0.22; NH4Cl, 0.5; skimmed milk, 10; and bacteriological agar, 15. The medium pH was adjusted to 6.0. The plates were inoculated at the center with 10 μL of the standardised bacterial suspension (comparable to 1 × 108 CFU/mL) and incubated at 30 °C for 24 h. The halo zone formation of was measured to the nearest millimeter after the incubation period.
2.4. Keratinolytic activity screening
Bacterial strains with proteolytic activity on skimmed milk agar plates were grown in a basal medium containing (g/L): K2HPO4, 0.3; KH2PO4, 0.4; MgCl2, 0.2; CaCl2, 0.22; and whole chicken feathers, 10; as the only source of carbon and nitrogen [16]. The initial medium pH was adjusted to 6.0 before sterilisation by autoclaving at 121 °C for 15 min. The submerged fermentation was carried out in triplicate using 250 mL Erlenmeyer flasks containing 100 mL working medium for 96 h at 30 °C in an orbital shaker (130 rpm). About 2% (v/v) of the bacterial suspension served as fresh inoculum for the submerged cultivation. After incubation, flasks with a complete or considerable decomposition of intact chicken feathers were selected for further studies, and respective culture broths further analysed. The cultures were filtered, and the residual chicken feathers were used to determine percentage degradation, while some aliquot of the filtrates were spun at 15,000×g for 10 min using centrifuge (HERMLE Labortechnik GmbH, Germany). Subsequently, culture supernatants were used to quantify keratinase production, protein concentration, and thiol concentration. The change in the initial pH of the culture broth was determined with JENWAY pH meter (Bibby Scientific Ltd, UK). Bacterial strains that displayed high keratin degrading capacity were maintained on CFP agar slants at 4 °C for the preparation of fresh inoculum, and in 20 % glycerol at −86 °C for long term storage.
2.5. Determination of percentage feather degradation
Chicken feather degradation by isolated bacterial strains was estimated by using feather weight loss approach [17]. Unutilized feathers were recovered by filtering the culture broth through Whatman no. 1 filter paper. Afterwards, the residues were washed with distilled water to remove bacterial cells; then dried in an oven at 60 °C for 24 h to achieve a constant weight. The dry weight of the residual feathers was determined, and the percentage of degradation was calculated with the equation as shown below.
Where RF = dry weight of residual feathers after fermentation; WF = dry weight of intact feathers before fermentation.
2.6. Assay for keratinase activity
Keratinase activity was determined following the method of Jaouadi et al. [18] with slight modification. Briefly, the reaction mixture contained 0.5 mL of diluted crude enzyme preparation and 0.5 mL of 10 g/L keratin azure (Sigma Aldrich, USA) in 100 mM Tris-HCl buffer (pH 7.5). The mixture was shaking incubated for 1 h at 37 °C, with shaking (220 rpm). After that, the reaction mixture was placed on ice for 10 min to stop the reaction. The unutilized substrate was removed by centrifugation at 15,000×g for 10 min and filtration through Millipore cellulose filters (0.45 μm). The free azo dye was determined by measuring the absorbance of the filtrate at 595 nm using SYNERGYMx 96 wells microplate reader (BioTek, USA). The control followed the same protocol except that the mixture contained the enzyme and buffer without the substrate. One unit of keratinase was defined as the amount of enzyme causing an increase of 0.01 in absorbance at 595 nm per hour under the assay protocol described.
2.7. Determination of total protein concentration
The protein concentration of the crude extract was estimated using the Bradford method [19], and bovine serum albumin (BSA) served as a protein standard.
2.8. Thiol concentration determination
The presence of free sulfur-containing groups in the fermentation broth was determined using a previously described method [20]. Briefly, 4 mg/mL of 5,5’-dithiobis (2-nitrobenzoic acid) (DTNB) (Sigma-Aldrich, USA) was dissolved in phosphate buffer (0.1 M; pH 8.0). Then, 50 μL of this solution were mixed with 500 μL of distilled water, and 250 μL of crude extract was subsequently added to this mixture and allowed to stand at room temperature for 5 min for stable colour development. The yellow-coloured sulfide that developed upon reduction of DTNB was measured spectrophotometrically at 412 nm. Broth from the un-inoculated medium was also assayed as described above and served as a control.
2.9. Molecular identification of keratin-degrading bacteria
The genomic DNA of the keratinolytic bacterial isolates was extracted during the exponential growth phase using the ZR Fungal/Bacterial DNA Kit™ (Zymo Research). The 16S target region was amplified using the polymerase chain reaction (PCR) under standard conditions. The set of universal primers used for the 16S rRNA gene sequence amplification were 27f (5’-AGAGTTTGATCMTGGCTCAG-3’) and 1492 r (5’-CGGTTACCTTGTTACGACTT-3’) as forward and reverse primers respectively [21]. PCR was performed using a master mix with constituents including DreamTaq™ DNA polymerase (Thermo Scientific™). The amplicons were gel extracted (Zymo Research, Zymoclean™ Gel DNA Recovery Kit), and sequenced in forward and reverse directions on the ABI PRISM™ 3500xl Genetic Analyzer. The basic local alignment search tool (BLASTn) program on the National Center for Biotechnology Information (NCBI) was used to compare the nucleotide sequences with reference sequences in the database [22]. The sequences were aligned; trimmed and the phylogenetic tree constructed with muscle, by exploring the construct/text neighbour-joining tool in molecular evolutionary genetics analysis (MEGA) software, version 7.0.26 [23]. The partial 16S rDNA nucleotide sequences (ranged between 822 bp and 858 bp) of these keratinolytic strains were submitted at GenBank through the NCBI under the accession numbers MG214989 – G214992, MG214997 – MG214998, MG215000 – MG215005.
2.10. Statistical analysis
All experiments were performed in triplicate and the results were presented as mean values with standard deviations. Data were subject to analysis of variance, and compared at P < 0.05.
3. Results
3.1. Keratinolytic activity screening – qualitative evaluation
Eighteen bacterial isolates showed protease activity with varied halo zone formation on the skimmed milk agar plates (Table 1). The halo zones were measured to the nearest millimetre and ranged from 21.5 ± 2.1 (mm) for isolate coded as FS-3 to 37.0 ± 1.41 (mm) for isolate coded as FS-2. The proteolytic activity of these 18 strains served as an index for selection in the subsequent experimentation. Out of the 18 isolates positive for protease activity; twelve showed keratinolytic potentials as observed from the fermentation flasks (supplementary file). On the contrary, the remaining six isolates, HS-1, SS-5, SS-2, SS-4, SS-7, and FS-2 demonstrated good proteolytic activity, with diameter of halo zone that ranged from 28.0 ± 0.71 mm for HS-1 to 37.0 ± 1.41 mm for FS-2 on the skimmed milk agar but were unable to degrade chicken feathers as sole source of carbon and nitrogen (data not shown).
Table 1.
Halo zone formation on skimmed milk agar through hydrolysis by proteolytic bacteria.
| S/N | Isolate code | Diameter (mm) | Feather degradation |
|---|---|---|---|
| 1. | FS-3 | 21.5 ± 2.12a | + |
| 2. | FS-1 | 25.5 ± 0.71b | + |
| 3. | FS-4 | 26.5 ± 0.71bc | + |
| 4. | SS-3 | 28.0 ± 0.00bcd | + |
| 5. | HS-1 | 28.0 ± 1.41bcd | – |
| 6. | BT1-7 | 28.5 ± 0.71bcde | + |
| 7. | BT4-2 | 29.0 ± 1.41bcde | + |
| 8. | SS-5 | 29.0 ± 0.00bcde | – |
| 9. | SS-2 | 30.0 ± 0.00cde | – |
| 10. | SS-4 | 30.0 ± 2.83cde | – |
| 11. | BT5-7 | 30.5 ± 3.54cde | + |
| 12. | SS-1 | 30.5 ± 0.71cde | + |
| 13. | SS-6 | 30.5 ± 0.71cde | + |
| 14. | SS-7 | 30.5 ± 0.71cde | – |
| 15. | BT1-8 | 32.0 ± 1.41def | + |
| 16. | HS-2 | 32.5 ± 3.54ef | + |
| 17. | SS-8 | 35.5 ± 3.54fg | + |
| 18. | FS-2 | 37.0 ± 1.41g | – |
Values are presented as mean and standard deviation, number of replicate, n = 3. The values without the same superscript letters down the column are significantly different (P < 0.05). (+) indicates isolates that could degrade intact chicken feathers. (-) indicates isolates that could not degrade intact chicken feathers.
3.2. Keratinolytic activity screening - quantitative evaluation
The extracellular keratinase activity determined from the cell-free extracts ranged from 198.2 ± 15.4 U/mL for SS-1 to 731.8 ± 14.1 U/mL for FS-4 (Table 2). The total protein concentrations estimation also ranged from 0.09 ± 0.02 mg/mL to 0.87 ± 0.05 mg/mL for FS-3 and BT1-8 respectively. The thiols produced during the degradation of chicken feather ranged from 0.69 ± 0.1 mM for BT4-2 to 2.89 ± 0.1 mM for FS-4. The slightly acidic medium (pH 6.0) which favored the production of keratinases changed to alkaline during fermentation; varied pH increases were observed and the values ranging from 7.4 for BT5-7 and BT4-2 to 8.6 for SS-8 as shown in Table 2. In addition, the feather degradation potential varied with test isolate, and ranged from 61.5 ± 0.7 % for SS-1 to 85.0 ± 1.4 % for FS-4.
Table 2.
Evaluation of keratinolytic potentials of the study bacterial isolates.
| S/N | Isolate code | Keratinase activity (U/mL) | Protein concentration (mg/mL) | Thiol concentration (mM) | Final pH | Feather degradation (%) |
|---|---|---|---|---|---|---|
| 1. | FS-4 | 731.83 ± 14.14h | 0.47 ± 0.01d | 2.89 ± 0.11g | 8.3 ± 0.01h | 85.0 ± 1.41f |
| 2. | SS-8 | 510.91 ± 5.15g | 0.67 ± 0.00e | 2.09 ± 0.23ef | 8.6 ± 0.01i | 81.0 ± 1.41de |
| 3. | SS-6 | 452.73 ± 28.28f | 0.49 ± 0.03d | 1.89 ± 0.02de | 8.2 ± 0.02g | 82.0 ± 2.83def |
| 4. | HS-2 | 400.01 ± 7.71e | 0.45 ± 0.02cd | 2.27 ± 0.08f | 8.2 ± 0.01h | 83.0 ± 1.41ef |
| 5. | BT5-7 | 372.12 ± 2.10de | 0.32 ± 0.03b | 0.81 ± 0.13a | 7.4 ± 0.02a | 67.0 ± 1.41b |
| 6. | FS-1 | 349.09 ± 2.57cd | 0.47 ± 0.01c | 1.29 ± 0.07b | 7.6 ± 0.01c | 74.5 ± 2.12c |
| 7. | SS-3 | 334.55 ± 12.86c | 0.27 ± 0.01b | 1.68 ± 0.19cd | 8.1 ± 0.01f | 80.0 ± 1.41de |
| 8. | BT4-2 | 331.82 ± 14.14c | 0.14 ± 0.02a | 0.69 ± 0.12a | 7.4 ± 0.01b | 64.5 ± 0.71ab |
| 9. | BT1-7 | 292.73 ± 20.57b | 0.72 ± 0.04f | 1.53 ± 0.11bc | 7.9 ± 0.04e | 68.5 ± 2.12b |
| 10. | FS-3 | 287.28 ± 5.14b | 0.09 ± 0.02a | 1.64 ± 0.04cd | 8.2 ± 0.01h | 78.5 ± 0.71d |
| 11. | BT1-8 | 281.81 ± 25.71b | 0.87 ± 0.05g | 2.71 ± 0.09g | 8.3 ± 0.02h | 81.5 ± 3.54def |
| 12. | SS-1 | 198.18 ± 15.43a | 0.12 ± 0.01a | 0.80 ± 0.04a | 7.7 ± 0.02d | 61.5 ± 0.71a |
The values are presented as mean and standard deviation of triplicate experiments; the values without the same superscript letters down the column are significantly different (P < 0.05).
3.3. Keratinolytic bacterial isolates identification
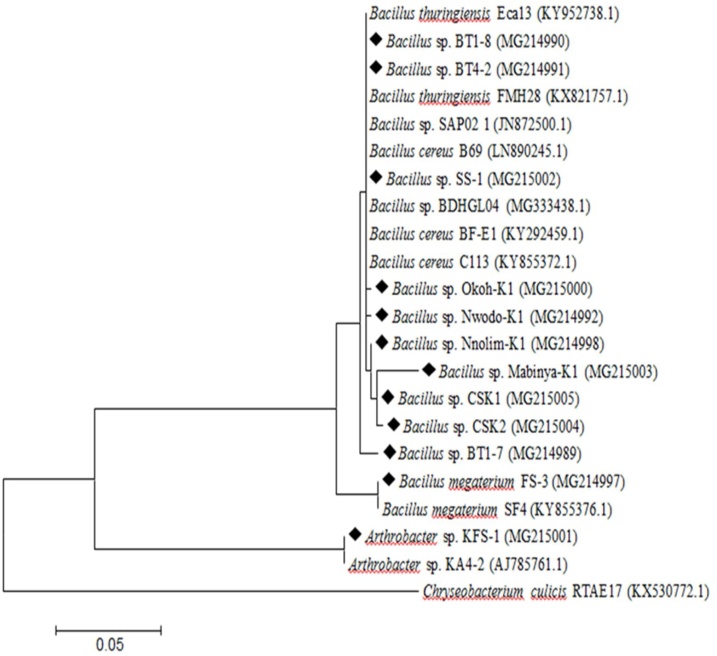
Bacterial isolates with keratinolytic potentials were identified through 16S rRNA gene sequence. The sequence BLAST analysis showed a high percentage of sequence homology with related sequences in NCBI GenBank. Eleven isolates coded as FS-4, SS-8, SS-6, HS-2, BT1-7, SS-3, BT4-2, BT5-7, FS-3, BT1-8 and SS-1 were identified as Bacillus spp.; FS-3 showed 100 % sequence homology with Bacillus megaterium and B. megaterium SF4 (KY855376.1). Conversely, the isolate with the code FS-1 showed 99 % sequence homology with Arthrobacter sp. KA4-2 (AJ_785761.1). The summary of the keratinolytic bacteria identification is presented in Table 3. A significant proportion of the isolates belonged to the Bacillus cereus sensu lato group, and the dendrogram clustering of the phylogeny is shown in Fig. 1.
Table 3.
Summary of keratinolytic bacteria identification through 16S rRNA gene sequence.
| S/N | Isolate code |
Reference sequence | Sequence similarity (%) | Sequence identity | Sequence length (bp) | NCBI accession number |
|---|---|---|---|---|---|---|
| 1. | FS-4 | Bacillus thuringiensis Ta3 (MK517627.1) | 100 | Bacillus sp. Nnolim-K1 | 851 | MG214998 |
| 2. | SS-8 |
Bacillus cereus KBB4 (MN032406.1) |
99 | Bacillus sp. CSK1 | 853 | MG215004 |
| 3. | SS-6 | Bacillus thuringiensis C4 (MN173385.1) | 100 | Bacillus sp. CSK2 | 853 | MG215005 |
| 4. | HS-2 |
Bacillus cereus BF-E1 (KY292459.1) |
100 | Bacillus sp. Okoh-K1 | 858 | MG215000 |
| 5. | BT1-7 | Bacillus thuringiensis Md1-10 (MF581416.1) | 100 | Bacillus sp. BT1-7 | 826 | MG214989 |
| 6 | SS-3 | Bacillus cereus 4 F (MK104469.1) | 100 | Bacillus sp. Mabinya-K1 | 832 | MG215003 |
| 7 | BT4-2 | Bacillus thuringiensis FMH28(KX821757.1) | 100 | Bacillus sp. BT4-2 | 858 | MG214991 |
| 8 | BT5-7 |
Bacillus sp. BDHGL04 (MG333438.1) |
100 | Bacillus sp. Nwodo-K1 | 823 | MG214992 |
| 9 | FS-3 |
Bacillus megaterium SF4 (KY855376.1) |
100 | Bacillus megaterium FS-3 | 836 | MG214997 |
| 10 | BT1-8 | Bacillus thuringiensis Eca13(KY952738.1) | 100 | Bacillus sp. BT1-8 | 856 | MG214990 |
| 11 | SS-1 |
Bacillus cereus B69 (LN890245.1) |
100 | Bacillus sp. SS-1 | 858 | MG215002 |
| 12 | FS-1 | Arthrobacter sp. KA4-2 (AJ_785761.1) | 99 | Arthrobacter sp. KFS-1 | 822 | MG215001 |
Fig. 1.
A phylogenetic tree of keratinolytic bacterial isolates and the closely related genera based on 16S rRNA gene sequence. NCBI accession numbers are shown in parentheses. The tree was generated using neighbor-joining technique, and the evolutionary distances calculated based on the maximum composite likelihood and the bootstrap value was set as 1000 replicates. The tagged bacterial strains represent keratinolytic isolates, and those without tags are reference strains from GenBank. The tree was rooted on Chryseobacterium culicis as an out-group.
4. Discussion
Green technology promotes environmental sustainability and has significantly spurred a continuum in the search for microbes with industrial and biotechnological prospects from diverse ecological niche. Consequently, a collection of bacterial strains was isolated from dumpsites using basal medium with chicken feather as the only carbon and nitrogen source. There are reports on bacteria with remarkable keratinolytic activity from soil samples with deposition of keratinous wastes [[24], [25], [26]]. Similarly, accounts of keratinolytic bacteria isolated from aquatic milieu also abound [27,28]. The bacteria isolated from dump sites were qualitatively screened, and eighteen (18) of the isolates showed proteolytic activity through the hydrolysis of protein with subsequent formation of halo zones on the skimmed milk agar plates. This preliminary screening on the skimmed milk for protein hydrolysis served as a vital step in the identification of isolates with proteolytic potentials [15,29].
Subsequently, only twelve out of the eighteen bacterial isolates were able to simultaneously hydrolyse skimmed milk and utilize chicken feather as sole source of carbon and nitrogen. Consequently, these bacterial species possess keratinolytic properties as they were able to hydrolyse keratin and utilize the hydrolysate as nutrient, and this characteristic may only be attributed to the diversity of their genetic composition. A similar observation has been made with keratin degrading bacterial species isolated from a different ecological niche [16,30].
The Basic Local Alignment Search Tool (BLAST) analysis of the 16S rRNA gene sequences of the keratinolytic bacteria showed ten out of the twelve test bacteria to possess a high sequence homology with Bacillus cereus sensu lato group and the bacteria were respectively identified as Bacillus spp. with suffix unique to the respective strain. Although, these bacteria were identified as Bacillus sp.; the varied degree for which feather was hydrolysed is an indication of uniqueness for the respective bacteria and strain difference. Furthermore, two bacterial isolates coded as FS-3 and FS-1 respectively showed 100 and 99 % sequence similarity with Bacillus megaterium SF4 (KY855376.1) and Arthrobacter sp. KA4-2 (AJ_785761.1), respectively, hence, they were identified as Bacillus megaterium FS-3 and Arthrobacter sp. KFS-1.
Bacillus represents about 92 % of the isolates that showed significant keratinolytic potentials and this genus was previously reported for similar properties [[31], [32], [33]], including the most extensively studied Bacillus licheniformis PWD-1 [34]. Bacillus species are the leading microbes noted for the production of keratinases and, this pattern may be as a result of the excellent plasticity the bacterial strains have shown towards the degradation of vast arrays of recalcitrant biomass for nutrients [8]. Arthrobacter sp. has similarly been documented to possess the potentials to serve as an essential source of biomolecules of interest including enzymes [35], and antimicrobial compounds [36]. The Arthrobacter sp. KFS-1 reported in this study effectively degraded chicken feathers with high extracellular keratinase production.
The varied degree in the keratinolytic activity observed among the isolated bacterial species would be a reflection on their different optimal fermentation process conditions. Consequently, the extracellular keratinase production achieved by the various isolates was akin to the feather degradation capacity; except for a few isolates including Bacillus sp. Mabinya-K1, B. megaterium FS-3 and Bacillus sp. BT1-8 that demonstrated significant feather degradation with relatively decreased extracellular keratinase activity. The disparity observed with keratinase activity in relation to the degree of feather degradation may be attributed to, either, the negative influence of the fermentation medium and environmental conditions on the active extracellular keratinase [13], or the utilization of the degradation products faster by some microbe as a source of nutrient [37]. In addition, the enzyme specificity toward keratin degradation may have been higher with the strains showing higher degradation potentials.
Another indicator of the effective degradation of the chicken feathers was the formation of the thiol group. The hydrolysis of disulfide bridges, which substantively contribute to the mechanical stability of keratin leads to the formation of the thiol group [38]. The concentrations of thiol group formed by respective keratinolytic bacteria were comparable to the degree of feather biodegradation with slight variation, and this may be attributed to the specificity and profound activity of microbial keratinolytic enzymes in the bioconversion of the recalcitrant keratinous biomass [33,39]. Similarly, the high thiol concentration yielded by some isolates might be an indication of predilection of the microbial sulfitolytic systems for the cleavage of the polypeptides at positions rich in cystine disulfide bridges, hence, releasing free sulfur-containing groups into the medium [40]. The thiol group formation observed with the bacterial strains under investigation is significantly higher than most reported keratinolytic bacteria including Chryseobacterium sp. kr6 [41], Stenotrophomonas maltophilia [38], and Bacillus sp. MBRL 575 [30] that yielded respective maximum thiol concentrations of 75 μM, 84 μM and 44.5 μM during feather biodegradation. This observation therefore underpins the dexterity of the study bacterial isolates in keratin-rich wastes bioconversion to functional proteins and their biotechnological prospects in agro sector of the economy.
The optimization of the fermentation process variables for the keratinolytic bacteria are critical for the harnessing of optimal potentials, and the bacterial species showed significant keratinolytic activities within the ambits of moderate conditions which is an indication that a cost effective process could be in play at a pilot scale production. Factors that may be in play with regard to the pH change may be medium ammonification resulting from the chicken feather dismemberment into various soluble proteinaceous molecules or associated byproducts of metabolism [42]. The acidic fermentation medium has been reported to be ambient for microbial keratinase production [43,44]. However, the pH drift to neutral or alkaline may be necessary or inimical for the effective keratinolysis. An alkaline pH has been likewise reported among active feather degrading bacteria [33,45].
5. Conclusion
In conclusion, the Bacillus spp. and Arthrobacter sp. identified via 16S rRNA gene sequence showed excellent keratinase production potential, and the keratinases showed varied activity which may have been as a result of enzyme specificity and yield accordingly. Thiol group formation likewise showed efficient hydrolysis of cysteine disulfide linkages of the feather keratin by the microbial sulfolytic systems. The considerable degradation of chicken feather and extracellular keratinase produced in basal medium was an indication that the bacterial species showed potentials as industrially important organisms. The valorization of agro-waste biomass (chicken feather) into digestible proteins using microbial-based technology represents sustainable development from both economic and environmental perspectives, and the bacterial species reported in this study have shown the potentials. The keratinases may have novel properties, and the enzyme hydrolysis products may also be novel; this is subject to further studies as it was beyond the scope of the reported work. In addition, the gene(s) coding for the enzyme(s) would be of high importance particularly, if cloning and expression in an industrially suitable vector for large-scale keratinase production is put into perspective.
Author statement
Nonso Nnolim and Uchechukwu Nwodo: Conceptualization, Uchechukwu Nwodo: Resources and Funding acquisition, Nonso Nnolim: Methodology, Data curation, Manuscript draft. Uchechukwu Nwodo and Anthony Okoh: Supervision, Review and Editing.
Declaration of Competing Interest
The authors declare no financial or commercial conflict of interest.
Acknowledgements
The Department of Science and Technology (DST) and the Technology Innovation Agency (TIA) supported this work under SIIP enzyme and microbial technologies (grant number: DST/CON/0177/2018). We also acknowledge the support of the South African Medical Research Council (SAMRC).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2020.e00483.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Ahuja S.K., Ferreira G.M., Moreira A.R. Utilization of enzymes for environmental applications. Crit. Rev. Biotechnol. 2004;24:125–154. doi: 10.1080/07388550490493726. [DOI] [PubMed] [Google Scholar]
- 2.Baweja M., Tiwari R., Singh P.K., Nain L., Shukla P. An alkaline protease from Bacillus pumilus MP 27: functional analysis of its binding model toward its applications as detergent additive. Front. Microbiol. 2016;7:1195. doi: 10.3389/fmicb.2016.01195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guleria S., Walia A., Chauhan A., Shirkot C.K. Purification and characterization of detergent stable alkaline protease from Bacillus amyloliquefaciens SP1 isolated from apple rhizosphere. J. Basic Microbiol. 2016;56:138–152. doi: 10.1002/jobm.201500341. [DOI] [PubMed] [Google Scholar]
- 4.Johannes T.W., Zhao H. Directed evolution of enzymes and biosynthetic pathways. Curr. Opin. Microbiol. 2006;9:261–267. doi: 10.1016/j.mib.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Bouacem K., Bouanane-Darenfed A., Laribi-Habchi H., Elhoul M.B., Hmida-Sayari A., Hacene H., Ollivier B., Fardeau M.L., Jaouadi B., Bejar S. Biochemical characterization of a detergent-stable serine alkaline protease from Caldicoprobacter guelmensis. Int. J. Biol. Macromol. 2015;81:299–307. doi: 10.1016/j.ijbiomac.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Sundararajan S., Kannan C.N., Chittibabu S. Alkaline protease from Bacillus cereus VITSN04: potential application as a dehairing agent. J. Biosci. Bioeng. 2011;111:128–133. doi: 10.1016/j.jbiosc.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Brandelli A. Bacterial keratinases: useful enzymes for bioprocessing agroindustrial wastes and beyond. Food Bioproc. Technol. 2008;1:105–116. [Google Scholar]
- 8.Daroit D.J., Brandelli A. A current assessment on the production of bacterial keratinases. Crit. Rev. Biotechnol. 2014;34:372–384. doi: 10.3109/07388551.2013.794768. [DOI] [PubMed] [Google Scholar]
- 9.Fontoura R., Daroit D.J., Correa A.P., Meira S.M., Mosquera M., Brandelli A. Production of feather hydrolysates with antioxidant, angiotensin-I converting enzyme-and dipeptidyl peptidase-IV-inhibitory activities. New Biotechnol. 2014;31:506–513. doi: 10.1016/j.nbt.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Jin H.S., Park S.Y., Kim K., Lee Y.J., Nam G.W., Kang N.J., Lee D.W. Development of a keratinase activity assay using recombinant chicken feather keratin substrates. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma B., Qiao X., Hou X., Yang Y. Pure keratin membrane and fibers from chicken feather. Int. J. Biol. Macromol. 2016;89:614–621. doi: 10.1016/j.ijbiomac.2016.04.039. [DOI] [PubMed] [Google Scholar]
- 12.Yusuf I., Ahmad S.A., Phang L.Y., Yasid N.A., Shukor M.Y. Effective production of keratinase by gellan gum-immobilised Alcaligenes sp. AQ05-001 using heavy metal-free and polluted feather wastes as substrates. 3 Biotech. 2019;9(2019):32. doi: 10.1007/s13205-018-1555-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yusuf I., Ahmad S.A., Phang L.Y., Syed M.A., Shamaan N.A., Khalil K.A., Dahalan F.A., Shukor M.Y. Keratinase production and biodegradation of polluted secondary chicken feather wastes by a newly isolated multi heavy metal tolerant bacterium-Alcaligenes sp. AQ05-001. J. Environ. Manage. 2016;183:182–195. doi: 10.1016/j.jenvman.2016.08.059. [DOI] [PubMed] [Google Scholar]
- 14.Letourneau F., Soussotte V., Bressollier P., Branland P., Verneuil B. Keratinolytic activity of Streptomyces sp. S. K1-02: a new isolated strain. Lett. Appl. Microbiol. 1998;26:77–80. doi: 10.1046/j.1472-765x.1998.00281.x. [DOI] [PubMed] [Google Scholar]
- 15.Riffel A., Brandelli A. Keratinolytic bacteria isolated from feather waste. Braz. J. Microbiol. 2006;37:395–399. [Google Scholar]
- 16.Tatineni R., Doddapaneni K.K., Potumarthi R.C., Vellanki R.N., Kandathil M.T., Kolli N., Mangamoori L.N. Purification and characterization of an alkaline keratinase from Streptomyces sp. Bioresour. Technol. 2008;99:1596–1602. doi: 10.1016/j.biortech.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 17.Reddy M.R., Reddy K.S., Chouhan Y.R., Bee H., Reddy G. Effective feather degradation and keratinase production by Bacillus pumilus GRK for its application as bio-detergent additive. Bioresour. Technol. 2017;243:254–263. doi: 10.1016/j.biortech.2017.06.067. [DOI] [PubMed] [Google Scholar]
- 18.Jaouadi B., Abdelmalek B., Fodil D., Ferradji F.Z., Rekik H., Zaraî N., Bejar S. Purification and characterization of a thermostable keratinolytic serine alkaline proteinase from Streptomyces sp. strain AB1 with high stability in organic solvents. Bioresour. Technol. 2010;101:8361–8369. doi: 10.1016/j.biortech.2010.05.066. [DOI] [PubMed] [Google Scholar]
- 19.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Ellman G.L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 21.Turner S., Pryer K.M., Miao V.P., Palmer J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis 1. J. Eukaryot. Microbiol. 1999;46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaouadi N.Z., Rekik H., Badis A., Trabelsi S., Belhoul M., Yahiaoui A.B., Aicha H.B., Toumi A., Bejar S., Jaouadi B. Biochemical and molecular characterization of a serine keratinase from Brevibacillus brevis US575 with promising keratin-biodegradation and hide-dehairing activities. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhange K., Chaturvedi V., Bhatt R. Potential biofilm dispersal by a partially purified keratinase produced by Stenotrophomonas maltophilia strain Kb2. Biocatal. Agric. Biotechnol. 2015;4:801–805. [Google Scholar]
- 26.Barman N.C., Zohora F.T., Das K.C., Mowla M.G., Banu N.A., Salimullah M., Hashem A. Production, partial optimization and characterization of keratinase enzyme by Arthrobacter sp. NFH5 isolated from soil samples. AMB Express. 2017;7:181. doi: 10.1186/s13568-017-0462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaturvedi V., Bhange K., Bhatt R., Verma P. Production of kertinases using chicken feathers as substrate by a novel multifunctional strain of Pseudomonas stutzeri and its dehairing application. Biocatal. Agric. Biotechnol. 2014;3:167–174. [Google Scholar]
- 28.Bouacem K., Bouanane-Darenfed A., Jaouadi N.Z., Joseph M., Hacene H., Ollivier B., Fardeau M.L., Bejar S., Jaouadi B. Novel serine keratinase from Caldicoprobacter algeriensis exhibiting outstanding hide dehairing abilities. Int. J. Biol. Macromol. 2016;86:321–328. doi: 10.1016/j.ijbiomac.2016.01.074. [DOI] [PubMed] [Google Scholar]
- 29.Saha S., Dhanasekaran D. Isolation and screening of keratinolytic actinobacteria form keratin waste dumped soil in Tiruchirappalli and Nammakkal, Tamil Nadu, India. Curr. Res. J. Biol. Sci. 2010;2:124–131. [Google Scholar]
- 30.Kshetri P., Ningthoujam D.S. Keratinolytic activities of alkaliphilic Bacillus sp. MBRL 575 from a novel habitat, limestone deposit site in Manipur, India. SpringerPlus. 2016;5:595. doi: 10.1186/s40064-016-2239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lateef A., Oloke J.K., Kana E.G., Sobowale B.O., Ajao S.O., Bello B.Y. Keratinolytic activities of a new feather-degrading isolate of Bacillus cereus LAU 08 isolated from Nigerian soil. Int. Biodeterior. Biodegrad. 2010;64:162–165. [Google Scholar]
- 32.Gegeckas A., Gudiukaitė R., Citavicius D. Keratinolytic proteinase from Bacillus thuringiensis AD-12. Int. J. Biol. Macromol. 2014;69:46–51. doi: 10.1016/j.ijbiomac.2014.05.024. [DOI] [PubMed] [Google Scholar]
- 33.Nnolim N.E., Okoh A.I., Nwodo U.U. Bacillus sp. FPF-1 produced keratinase with high potential for chicken feather degradation. Molecules. 2020;25:1505. doi: 10.3390/molecules25071505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams C.M., Richter C.S., Mackenzie J.M., Shih J.C. Isolation, identification, and characterization of a feather-degrading bacterium. Appl. Environ. Microbiol. 1990;56:1509–1515. doi: 10.1128/aem.56.6.1509-1515.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shanati T., Lockie C., Beloti L., Grogan G., Ansorge-Schumacher M.B. Two enantiocomplementary Ephedrine Dehydrogenases from Arthrobacter sp. TS-15 with broad substrate specificity. ACS Catal. 2019;9:6202–6211. [Google Scholar]
- 36.Undabarrena A., Beltrametti F., Claverías F.P., González M., Moore E.R., Seeger M., Cámara B. Exploring the diversity and antimicrobial potential of marine actinobacteria from the comau fjord in Northern Patagonia, Chile. Front. Microbiol. 2016;7:1135. doi: 10.3389/fmicb.2016.01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taskin M., Kurbanoglu E.B. Evaluation of waste chicken feathers as peptone source for bacterial growth. J. Appl. Microbiol. 2011;111:826–834. doi: 10.1111/j.1365-2672.2011.05103.x. [DOI] [PubMed] [Google Scholar]
- 38.Jeong J.H., Lee O.M., Jeon Y.D., Kim J.D., Lee N.R., Lee C.Y., Son H.J. Production of keratinolytic enzyme by a newly isolated feather-degrading Stenotrophomonas maltophilia that produces plant growth-promoting activity. Process Biochem. 2010;45:1738–1745. [Google Scholar]
- 39.Łaba W., Żarowska B., Chorążyk D., Pudło A., Piegza M., Kancelista A., Kopeć W. New keratinolytic bacteria in valorization of chicken feather waste. AMB Express. 2018;8:9. doi: 10.1186/s13568-018-0538-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He Z., Sun R., Tang Z., Bu T., Wu Q., Li C., Chen H. Biodegradation of feather waste keratin by the keratin-degrading strain Bacillus subtilis 8. J. Microbiol. Biotechnol. 2018;28:314–322. doi: 10.4014/jmb.1708.08077. [DOI] [PubMed] [Google Scholar]
- 41.Riffel A., Lucas F., Heeb P., Brandelli A. Characterization of a new keratinolytic bacterium that completely degrades native feather keratin. Arch. Microbiol. 2003;179:258–265. doi: 10.1007/s00203-003-0525-8. [DOI] [PubMed] [Google Scholar]
- 42.Tiwary E., Gupta R. Medium optimization for a novel 58 kDa dimeric keratinase from Bacillus licheniformis ER-15: biochemical characterization and application in feather degradation and dehairing of hides. Bioresour. Technol. 2010;101:6103–6110. doi: 10.1016/j.biortech.2010.02.090. [DOI] [PubMed] [Google Scholar]
- 43.Abdel-Fattah A.M., El-Gamal M.S., Ismail S.A., Emran M.A., Hashem A.M. Biodegradation of feather waste by keratinase produced from newly isolated Bacillus licheniformis ALW1. J. Genetic Eng. Biotechnol. 2018;16:311–318. doi: 10.1016/j.jgeb.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J.M., Lim W.J., Suh H.J. Feather-degrading Bacillus species from poultry waste. Process Biochem. 2001;37:287–291. [Google Scholar]
- 45.He Z., Sun R., Tang Z., Bu T. Biodegradation of feather waste keratin by the keratin-degrading strain Bacillus subtilis 8. J. Microbiol. Biotechnol. 2018;28:314–322. doi: 10.4014/jmb.1708.08077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.