Abstract
This study was designed to assess the significance of interleukin-2 receptor (CD25) and inteleukin-3 receptor (CD123) expression in cytogenetically normal acute myeloid leukemia (CN-AML) patients. The current study includes 80 CN-AML (≤ 60 years) before the start of therapy. Blast cells expression for CD25 and CD123 were identified by flowcytometry in fresh bone marrow samples. CD25+/CD123-; CD25-/CD123+. CD25+/CD123+, CD25-/CD123- expression were as follow: 10/80 (12.5%); 18/80 (22.5%); 17/80; (21.25%), 35/80 (43.5%) respectively. The total CD25 expression was detected in 27/80 (33.75%), and CD123 expression was detected in 35/80 (43.75%%). CN-AML patients showed CD25+/CD123+ co-expression had the lowest induction remission rate and the shortest overall survival as compared to those lack co-expressions (P <0.01; P = 0.023 respectively). Also, there is strong positive association between CD25+/CD123+ co-expression and FLT3 mutations (P<0.001) and negative one with NPM1 mutation (P<0.001). In conclusion: CD25+/CD123+ co-expression in CN-AML patients define a subgroup of patients with adverse outcome. Identification of CD25/CD123 expression in CN-AML patents at diagnosis could be included in risk stratification. There is strong association between CD25+/CD123+ positive expression and FLT3 mutations.
Keywords: CD25, CD123, AML, Prognosis
1. Introduction
Acute myeloid leukemia (AML) is the commonest acute leukemia in the adults age. AML is a heterogeneous disease which caused by various molecular and cytogenetic abnormalities driven leukemogenesis, including chromosome abnormalities and gene mutations. It was ranked as the six highest cancer-related death in male population [1].
The risk stratification of AML was primarily based on Cytogenetic findings and molecular genetic alterations which provide significant prognostic information for determining the response to chemotherapy and survival outcome. However; about 40% of AML patients lack cytogenetic abnormalities [2]. In this group of patient's current efforts have to be done for better stratifications. The most important recurrent genetic aberrations govern the prognostic stratification of that subgroup of patients are the FMS-like tyrosine kinase 3 (FLT3) and Nucleophosmin 1 (NPM1) mutations. In the context of this evolving molecular risk assessment, antigen-expression profiles have been identified as surrogates for certain leukemic genotypes. Furthermore, a few single antigens per se have been found to be predictive of clinical response. In most cases, however, the prognostic power of antigens has not been disassociated from underlying genetic determinants [3], [4], [5], [6], [7].
Previous reports suggest that CD25 and CD123 expression have been associated poor outcome of AML patients [3,4]. CD25 is known as interleukin 2 (IL2) receptor alpha (IL2RA). IL-2 cytokine regulates cell proliferation, differentiation, survival and apoptosis [5]. CD25 was previously reported to be frequently expressed by AML blast cells; and associated with poor outcome of AML independent of cytogenetic findings [6], [7], [8].
CD123 is known as interleukin 3 receptor, alpha (IL-3Rα). IL3R is a heterodimeric cytokine receptor composed of the alpha unit and beta unit, which well be activated by ligand binding [6]. IL-3 is one of the prominent cytokines that controls proliferation, growth and differentiation of hematopoietic cells [6,7]. Recently many authors [7,8] have shown that CD123 is expressed in several hematologic neoplasms, including B-cell acute lymphoblastic leukemia, but expressed at a low level or to be absent on normal hematopoietic stem cells [7]. Importantly, CD123 is expressed at both the level of leukemic stem cells (LSCs) and more differentiated leukemic blasts [8], [9], [10], [11], [12]. No previous study was assessed the prognostic value of CD25/CD123 expression in AML with normal cytogenetic. The aim of this study was to determine the prognostic impact of CD25/CD123 co-expression on disease characteristics as well as CN-AML patient's outcome.
2. Methods
2.1. Patient samples
This study comprised 80 CN-AML patients (age ≤ 60 years) before start of therapy. All investigated patients gave informed consent. Diagnostic bone marrow and/or peripheral blood samples were taken from the all investigated patients. The patients were subjected to both morphologic examination of the peripheral blood smear and bone marrow smear, immunophenotypic evaluation using antibodies panel included CD34PE(Clone8G12), MPOPE(clone 5B8) CD33PE (Clone D3HL60.251), CD13FITIC (Clone SJIDI), CD117PE(Clone 10uD2), HLADR Percep-Cy(Clone G4606); CD14APC(Clone rmC5-3), CD61FITC (clone HI-PL2);CD64PE (Clone22), Glycophorin FITC (Clone 11E4B-7–6) by flow cytometry. The included AML patients were followed up to 12 months or until death. The study has been approved by Mansoura faculty of medicine local ethics committee.
2.2. Exclusion criteria
Patients with acute promyelocytic leukemia or favorable [t(8;21); inv 16; t(16;16)], and unfavorable risk cytogenetics [numerical or structural deletion of chromosome 7 or 5, trisomy 8, t(9;22), 11q23 rearrangements] according to the WHO 2016 criteria.
3. Methods
3.1. Cytogenetic study
Conventional karyotyping was performed on bone marrow (BM) diagnostic aspirates after short-term culture and analyzed after G-banding [13]. The description of the karyotypes was done according to the International System for Human Cytogenetic Nomenclature.
3.2. Molecular studies of NPM1 exon12 and FLT3-ITD and FLT3-TKD
RNA was extracted from fresh EDTA-anticoagulated peripheral blood or bone marrow leukemic cells using the QIAamp blood RNA extraction kit (QIAGEN Inc., Valencia, CA). Then cDNA synthesis were carried out (Invitrogen Life Technologies, Breda, The Netherlands) followed by amplification of selected DNA sequence ( NPM1 exonn12 or FLT3 status (both ITD and TKD mutations) using previously described primers (14); PCR Cycling program for mutation detection were as follows: one cycle, five minutes at 94 °C; Thirty cycles, one minute at 94 °C, one minute at 58 °C, and one minute at 72 °C; and one cycle, seven minutes at 72 °C. PCR products were subsequently purified followed by direct sequencing using an ABI-PRISM3100 genetic analyzer (Applied Biosystems, Foster City, CA). Raw data were analyzed with GeneMapper v4.0 software (Applied Biosystems, Inc., USA).
3.3. Flow cytometric determination of CD25/CD123 cell antigen expression
For CD25 and CD123 detection, the stain/lyse/wash technique was used. Briefly, in one tube 10 μl of the CD25-PE McAb (CLONE MA-251 BD PHARMAGEN ); 10 ul of CD123-FITC MoAb ( CLONE 7G3 BD PHARMAGEN); and 10 ul of CD45 APC ( CLONE H130 BD PHARMAGEN) were added to 100 μl of ethylene diamine tetra acetic acid (EDTA) fresh bone marrow samples, mixed well, and incubated for 15 min at room temperature. The cells were then washed twice with phosphate-buffered saline (PBS); 2 ml lysing solution was added, mixed, and left for 15 min in the dark, and then the cells were washed twice with PBS. After the last wash, the cells were suspended in 500 ul of PPS and then analyzed using a flow cytometer (a FACS Canto flow cytometer with Cell Quest software; Becton Dickinson). At least 10,000 events/ tube were measured.
The blast gate was defined on the basis of CD45dim expression and side-scatter characteristics and calculated as a percentage of total gated events. For analysis of CD25 and CD123 expression, measurements included mean fluorescence intensity (MFI) on leukemic blasts (adjusted for background fluorescence using negative internal controls) and relative mean fluorescence intensity (RFI) ratio (leukemic blasts versus non-leukemic events). In patients’ samples, for CD25 (CD25+) and CD123 (CD123+) was defined as expression in ≥20% of leukemic blasts using MFI more than that detected from the background fluorescence in the negative controls (nonleukemic gated events).
3.4. Statistical analysis
Quantitative data were presented as median (minimum, maximum) or mean ± standard deviation (SD) values. Qualitative data were presented as frequencies and percentages. Fisher`s exact test was used to determine the differences between groups when the data is normally distributed. Kruskal-Wallis tests was used to compare non parametric continuous variables between more than 2 groups and Wilcoxon-Mann-Whitney rank-sum test for comparison between 2 groups. Log rank compared Kaplan-Meier survival curve. The significance level was set at P ≤ 0.05. Statistical analysis was performed with IBM® SPSS® Statistics Version 24 for Microsoft Windows, SPSS Inc. with two sided statistically significance level at P < 0.05.
4. Results
The pattern of CD25 and CD123 expression in CN-AML patients are shown in Table 1. CD25+/CD123- (single positive expression); CD25-/CD123+ (single positive expression). CD25+/CD123+ (double positive expression), CD25-/CD123- (double negative expression) were as follow: 10/80 (12.5%); 18/80 (22.5%); 17/80; (21.25%), 35/80 (43.5%) respectively. The total CD25 expression was 27/80 (33.75%), and CD123 expression was 35/80 (43.75%%).
Table 1.
CN-AML patient's characteristics.
| Item | No (%) |
|---|---|
| Sex | |
| Male | 45 |
| Female | 35 |
| FAB subtypes | |
| M0 | 3 (3.7%) |
| M1 | 15(18.7%) |
| M2 | 17(21.3%) |
| M4 | 35(43.8%) |
| M5 | 10(12.5%) |
| Induction of remission | |
| Responder | 45 |
| Non-responder | 35 |
|
CN AML Positive CD25+/CD123+ co-expression |
17 |
|
CN-AML Lack CD25+/CD123+ Co-expression |
63 |
| Deaths | |
| Alive | 47 |
| Dead | 33 |
Comparing blood cell counts and blast cell percentage in the blood smear and bone marrow in different pattern of expression reveal that; the CD25+/CD123+ group have lower hemoglobin level; lower platelets count; higher blood white cell count; higher bone marrow blasts; blood smear blast cells as compared to subgroups of CN-AML patients with other pattern of expressions and the differences were statistically significant (P = 0.014; 0.43;<0.05; 0.04 respectively) (Table 2).
Table 2.
Association between CD25 and CD123 expressions and hematological parameters.
| Pattern of expression | Hb (g/dl)M±SD | Platelets x 103/ul)Median (Range) | WBCs x 103/ul)Median (Range) | Blood blast cells (%)Median (Range) | BM blast cells (%)Median (Range) |
|---|---|---|---|---|---|
| CD25+/CD123- (n = 10) |
9.62±2.35 | 34 (4.55–249) | 17.7 (1.2–155) | 50 (10–90) | 50 (4–90) |
| C25-/CD123+ (n = 18) |
9.97±2.66 | 25.6 (4.55–249) | 20.8 (1.11–315) | 40 (12–90) | 60 (6–90) |
| CD25+/CD123+ Co-expression (n = 17) |
7.76±2.11 | 17.85 (8.01–73) | 18.6 (1.2–226) | 55 (14–90) | 87.5 (70–92) |
| CD25-/CD123- (n = 35) |
9.13±2.37 | 57.9 (8.01–160) | 12.8 (1.11–140) | 22 (5–92) | 60 (4–50) |
| P = 0.014 | P = 0.043 | P<0.05 | P<0.05 | P = 0.004 |
CN-AML subgroup patients showed CD25+/CD123+ expression had significantly lower induction of remission response rate as compared to other subgroups (Table 3). The difference was statistically significant (P<0.01). Out of 80 tested patients 20 (25%) harbored in their leukemic cells the FLT3-ITD mutations. The association between FLT3 positive mutations and the pattern of CD25 and CD123 expressions were as follow: 17 cases of them were present in CD25/CD123 positive co-expression subgroup; 11 in the CD25+/CD123- negative subgroup and 16 in CD25-/CD123+ cases and no one case in the CD25-/CD123- subgroup. The differences in the distributions of FLT3 mutations among AML patients’ subgroups was statistically significant (P<0.001) (Table 4). Moreover, NPM1 was detected in 52.5% of the studied CN-AML patients. The association studies between NPM1 and CD25 and CD123 positive co-expression reveal that there is strong association between lack of co-expression and NPM1 negative (Table 4).
Table 3.
Impact of CD25, CD123 expression pattern on the induction of remission response.
| CN-AML | CD25+/CD123- expression | CD25-/CD123+ expression | CD25+/CD123+Co-expression | CD25-/CD123-expression |
|---|---|---|---|---|
| Positive(n = 12) | Positive(n = 18) | positive(n = 17) | (n = 33) | |
| Induction of remission response | ||||
| Responder (n = 45) (56.3%) |
5 | 8 | 2 | 30 |
| Non-responder (n = 35) (43.7%) |
7 | 10 | 15 | 3 |
| P value | <0.01 | |||
Table 4.
Association of CD25 and CD123 expression and FLT3 and NPM1 mutations.
| CN-AML | CD25+/CD123- expression | CD25-/CD123+ expression | CD25+/CD123+Co-expression | CD25-/CD123-expression |
|---|---|---|---|---|
| Positive (n = 12) |
Positive (n = 18) |
positive (n = 17) |
(n = 33) | |
| FLT3 mutations | ||||
| Yes (n = 20) (25%) | 1 | 2 | 17 | 0 |
| No (n = 60) (75%) | 11 | 16 | 0 | 33 |
| P value | <0.001 | |||
| NPM1mutation | ||||
| Yes (n = 42) | 5 | 5 | 1 | 31 |
| No (n = 38) | 7 | 13 | 16 | 2 |
| P value | <0.001 | |||
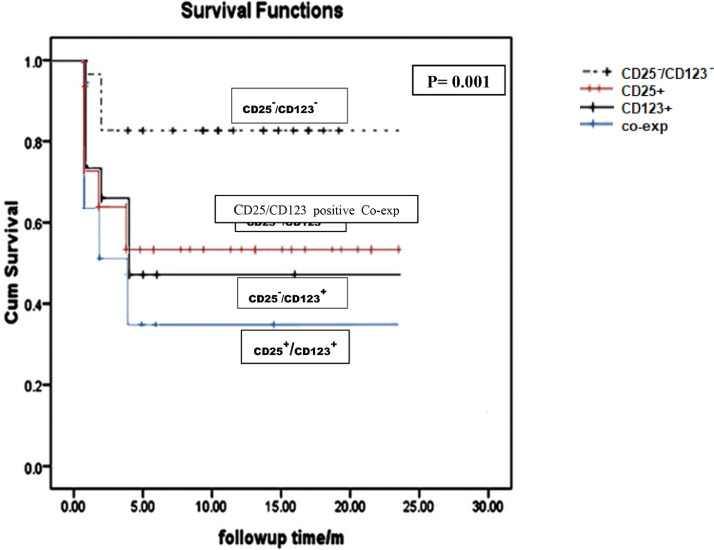
The impact of CD25 and CD123 expressions pattern on the CN-AML patients’ overall survival were studied using Kaplan Meier curve. From this curve, it is evident that CD25+/CD123+ co-expression had the lowest overall survival as compared to those lack CD25/CD123 double expression (P<0.01) (Table 5; Fig. 1).
Table 5.
Survival Analysis of CN-AML subgroups.
| CN-AML subgroups | Mean | |||
|---|---|---|---|---|
| Estimate | Std. Error | 95% Confidence Interval | ||
| Lower Bound | Upper Bound | |||
| Double negative expression | 10.686 | 0.545 | 9.618 | 11.753 |
| Double positive expression | 4.882 | 0.973 | 2.975 | 6.790 |
| Single (CD25+) expression | 7.200 | 1.522 | 4.217 | 10.183 |
| Single (CD123+) expression | 6.611 | 1.139 | 4.379 | 8.843 |
| P value | = 0.001 | |||
Fig. 1.
Impact of CD25/CD123 expressions pattern on the overall survival of CN-AML patients. CN-AML patients showed CD25+/CD123+ co-expression had the shortest OS; followed by C25-/CD123+, then CD25+/CD123- and longer one is present in CD25-/CD123- and the differences is statistically significant (P = 0.001).
5. Discussion
CD25+/CD123+ positive expression was identified in 17/80 (21.2%) of CN-AML patients. No previous study was evaluating the CD25+/CD123+ co-expression in this cohort of CN-AML patients. However; Miltiades et al. [15] studied CD25 expression in myelodysplastic syndrome AML blasts and reported that CD25 positive expression was detected in 14 patients (17%) at diagnosis. Moreover, Cerny et al. [16] found that the positive expression of CD25 at diagnosis was a strong predictor of treatment failure. Also, Khasawneh and Abdel-Wahab [14] concluded that the determination of CD25 expression status improves prognostic risk classification in AML independent of established biomarkers and could be predict patient responses and minimal residual disease.
The data concerning the impact of CD25 positive expression and the role of IL2 on the proliferation of CD25 positive blasts cells on AML patients is unclear [17]. CD25 may impart environmental signals, whereas, soluble form of CD25 may suppress the immune antitumor response through competition with the cellular surface CD25 for IL-2 which lead to increased tumor burden as well as disease severity [17,18]. The expression of CD25 on AML blast may point for specific myeloid progenitor or stage which have chemotherapy resistant characteristic [19].
The induction of remission response rate was significantly lower in CD25+/CD123+ double positive expression as compared to subgroup of CN-AML patients lack double antigens expression. This finding is similar to that reported by Thol et al. [20] and Hou et al. [21]; they observed that CD25 expression was associated with a reduced response to induction chemotherapy, irrespective of the dose of daunorubicin.
FLT3-ITD mutation was detected in 20 out of 80 CN-AML patients (25%). This finding is nearly similar to that previously reported by Niparuck et al. [1] which is 18%, but near to that described by Medinger and Passweg [22] which ranged between 28–34%. Moreover, there is strong association between FLT3-ITD mutation and CD25+/CD123+ co-expression in our study. All cases harbored FLT3-ITD mutation were positive for CD25/CD123. Similar finding was reported by Angelini et al. [23].
NPM1 mutations was directed in 52.5% of CN-AML. Similar finding was reported by Angenendt etal [24] and Kunchala et al. [25] who stated that Approximately 50–60 percent of patients with NK-AML carry NPM1 mutations. Nucleophosmin 1 (NPM1) performs diverse biological functions including molecular chaperoning, ribosome biogenesis, DNA repair, and genome stability. Moreover; mutant NPM1 (NPM1c+) acts in a dominant negative fashion and also blocks the differentiation of myeloid cells through gain-of-function for the AML phenotype
CN-AML patient's subgroup exhibited CD25+/CD123+ co-expression had significantly shorter overall survival as compared to those patients who had other pattern of expressions. The poor outcome of CD25+ AML patients has been attributed to the chemo-resistant properties of myeloblasts expressing CD25+ which suggested by previous study [25].
Previous report [8] stated that myeloblasts expressing CD123 exhibit higher proliferative activity and more resistant to apoptosis trigged by growth factor deprivation; Moreover, Myeloblasts showed CD123+ expression frequently display constitutive Stat5 phosphorylation and that its incubation with IL-3 induced Stat5 activation at a significant higher levels than that leukemic cells with normal CD123 levels [8]. Furthermore, Positive CD123 expression at diagnosis is associated with increased blast cells and total white blood cells and associated with a negative prognosis [8]. In addition; it was reported that CD123 positive myeloblasts displayed a constitutive activation of NF-kB [26].
The poor outcome of CD25+ AML patients has been attributed to the chemo-resistant properties of the CD25+ positive myeloblasts, which was reflected on both induction failure [27] and relapse [9,15]. This finding was explained by Gönen, et al. [6] who stated there is positive correlation between CD25 positive expression and presence of internal tandem duplications in FLT3 (FLT3-ITD), DNMT3A, and NPM1 mutations. Moreover; the studies done on gene expression analysis revealed that CD25+ positive expression is correlated with leukemia stem cell signatures [6]. The high frequency of relapse in AML patients was attributed to LSCs which are resistant to chemotherapy. This was attributed to that LSCs express drug efflux proteins which is the suggested cause of multidrug resistance. In addition, many stem cell characteristics including quiescence are determined by interactions with the niche [27,28]. The limitation of this study is the small AML patients’ sample. Repeating this study on large cohort cytogenetically normal AML patients is recommended to validate this study.
6. Conclusions
CD25+/CD123+ co-expression in CN-AML patients define a subgroup of patients with adverse clinical outcome. detection of CD25/CD123 expression in CN-AML patents at diagnosis could be included in risk stratification. There is strong association between CD25/CD123 co-expression and FLT3 mutations.
Declaration of Competing Interest
The authors declare that there is no conflict of interest
References
- 1.Niparuck P., Limsuwanachot N., Pukiat S., Chantrathammachart P., Rerkamnuaychoke B., Magmuang S., Phusanti S., Boonyawat K., Puavilai T., Angchaisuksiri P., Artit Ungkanont A., Chuncharunee S. Cytogenetics and FLT3-ITD mutation predict clinical outcomes in non-transplant patients with acute myeloid leukemia. Exp. Hematol. Oncol. 2019;8:3. doi: 10.1186/s40164-019-0127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gregory T., Wald D., Chen Y., Vermaat J., Xiong Y., Tse W. Molecular prognostic markers for adult acute myeloid leukemia with normal cytogenetics. J. Hematol. Oncol. 2009;23(2):23. doi: 10.1186/1756-8722-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehninger A., Kramer M., Röllig C., Thiede C., Bornhäuser M., von Bonin M., Wermke M., Feldmann A., Bachmann M., Ehninger G., Oelschlägel U. On behalf of the study alliance leukemia. distribution and levels of cell surface expression of CD33 and CD123 in acute myeloid leukemia. Blood Cancer J. 2014;4(6):e218. doi: 10.1038/bcj.2014.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morel P., Ernst L., Metes D. Functional CD32 molecules on human NK cells. Leuk Lymphoma. 1999;35:47–56. doi: 10.3109/10428199909145704. [DOI] [PubMed] [Google Scholar]
- 5.Driesen J., Popov A., Schultze J. CD25 as an immune regulatory molecule expressed on myeloid dendritic cells. Immunobiology. 2008;213:849–858. doi: 10.1016/j.imbio.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 6.Gönen M., Sun Z., Figueroa M., Patel J., Abdel-Waheb Racevskis J, Ketterling R., H Fernandez H., Rowe J., Tallman M., Melnick A., Levine R., Paietta E. CD25 expression status improves prognostic risk classification in AML independent of established biomarkers: ECOG phase 3 trial, E1900. Blood. 2012;2:2297–2306. doi: 10.1182/blood-2012-02-414425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angelova E., Audette C., Kovtun Y., Daver N., Wang S., Pierce S., Konoplev S., Khogeer H., Jorgensen J., Konopleva M., Zweidler-McKay P., Medeiros L., Kantarjian H., Jabbour E., Khoury J. CD123 expression patterns and selective targeting with a CD123-targeted antibody drug conjugate (IMGN632) in acute lymphoblastic leukemia. Haematologica. 2019;104(4):749–755. doi: 10.3324/haematol.2018.205252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Testa U., Pelosi E., Frankel A. CD123 is a membrane biomarker and a therapeutic target in hematologic malignancies. Biomarker Res. 2014;2:4. doi: 10.1186/2050-7771-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakase K., Kita K., Miwa H., Nishii K., Shikami M., Tanaka I., Tsutani H., Ueda T., Nasu K., Kyo T., Dohy H., Shiku H. Katayama N clinical and prognostic significance of cytokine receptor expression in adult acute lymphoblastic leukemia: interleukin-2 receptor alpha-chain predicts a poor prognosis. Leukemia. 2007;21:326–332. doi: 10.1038/sj.leu.2404497. [DOI] [PubMed] [Google Scholar]
- 10.Ruella M., David M, Barrett D., Kenderian S., Shestova O., Hofmann T., Perazzelli J., Klichinsky M., Aikawa V., Nazimuddin F., Kozlowski M., Scholler J., Lacey S., Melenhorst J., Morrissette J., Christian D., Hunter C., Kalos M., Porter D., June C., Grupp S., Gill S. Dual CD19 and CD123 targeting prevents antigen-loss relapses after CD19-directed immunotherapies. J. Clin. Invest. 2016;126(10):3814–3826. doi: 10.1172/JCI87366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman M., Neering S., Upchurch D., Grimes B., Howard D., Rizzieri D., Luger S., Jordan C. Nuclear factor-kappa B is constitutively activated in primitive acute human acute myelogenous leukemia cells. Blood. 2011;2(8):2301–2307. doi: 10.1182/blood.v98.8.2301. [DOI] [PubMed] [Google Scholar]
- 12.Vergez F., AS Green A., J Tamburini T., JE Sarry J., B Gaillard B., P Cornillet-Lefebvre P., M Pannetier M., Neyret A., Chapuis N., Ifrah N., Dreyfus F., Manenti S., Demur C., Delabesse E., Lacombe C., Mayeux P., Bouscary D., Recher C., Bardet V. High levels of CD34+CD38low/-CD123+ blasts are predictive of an adverse outcome in acute myeloid leukemia: a Groupe Ouest-Est des Leucemies Aigues et Maladies du Sang (GOELAMS) study. Haematologica. 2011;96:1792–1798. doi: 10.3324/haematol.2011.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stasi R., Del Poeta G., Masi M., Tribalto M., Venditti A., Papa G., Nicoletti B., Vernole P., Tedeschi B., Delaroche I., Mingarelli R., Dallapiccola B. Incidence of chromosome abnormalities and clinical significance of karyotype in de novo acute myeloid leukemia. Cancer Genet Cytogenetic. 1993;67:28–34. doi: 10.1016/0165-4608(93)90040-s. 67. [DOI] [PubMed] [Google Scholar]
- 14.Chauhan P., Ihsan R., Singh L., Gupta D., Mittal V., Kapur S. *Mutation of NPM1 and FLT3 genes in acute myeloid leukemia and their association with clinical and immunophenotypic features. Disese Markers. 2013;35:581–588. doi: 10.1155/2013/582569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miltiades P., Lamprianidou L., Vassilakopoulos T., Papageorgiou S., Galanopoulos A,Vakalopoulou S., Garypidou V., Papaioannou M., Hadjiharissi E., Pappa V., Papadaki H., panoudakis E., Tsatalas K., Kotsianidis I. Expression of CD25 antigen on CD34+ cells is an independent predictor of outcome in late-stage MDS patients treated with azacytidine. Blood Cancer J. 2014;4:e187. doi: 10.1038/bcj.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerny J., Yu M., Ramanathan H., Raffel G., Walsh W., Fortier N., Shanahan O'Rourke E, Bednarik J., Barton B., Kroll-Desrosiers A., Hao S., Woda B., Hutchinson L., Evens Rosmarin A, Nath R. Expression of CD25 independently predicts early treatment failure of acute myeloid leukaemia (AML) Br. J. Hematol. 2013;160:262–266. doi: 10.1111/bjh.12109. [DOI] [PubMed] [Google Scholar]
- 17.Terwijn M., Feller N., van Rhenen A., Kelder A., Westra G., Zweegman S., Ossenkoppele G., Schuurhuis G. Interleukin-2 receptor alpha-chain (CD25) expression on leukemic blasts is predictive for outcome and level of residual disease in AML. Eur. J. Cancer. 2009;45:1692–1699. doi: 10.1016/j.ejca.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 18.Cimino G., Amadori S., Cava M., De Sanctis V., Petti M., Di Gregorio A., Sgadari C., Vegna L., Cimino G., Mandelli F. Serum interleukin-2 (IL-2), soluble IL-2 receptors and tumor necrosis factor-alfa levels are significantly increased in acute myeloid leukemia patients. Leukemia. 1991;5:32–35. [PubMed] [Google Scholar]
- 19.Decker T., Bogner C., Oelsner M., Peschel C., Ringshausen I. Antiapoptotic effect of interleukin-2 (IL-2) in B-CLL cells with low and high affinity IL-2 receptors. Ann. Hemat. 2010;89:1125–1132. doi: 10.1007/s00277-010-0994-1. [DOI] [PubMed] [Google Scholar]
- 20.Thol F., Damm F., Ludeking A., Winschel C., Wagner K., Morgan M., Yun H., Göhring G., B Schlegelberger B., Hoelzer D., Lübbert M., Kanz L., Fiedler W., Kirchner Heil G, Krauter J., Ganser A., Heuser M. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J. Clin. Oncol. 2011;29:2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 21.Hou H., Y Kuo Y., Chou W., Chou W., Lee M., Chen C., Lin L., Tseng M., Huang C., Chiang Y., Lee F., Liu M., Liu C., Tang J., M Yao M., Huang S., Ko S., Hsu B., Wu S., Tsay W., Chen Y., Tien H. DNMT3A mutations in acute myeloid leukemia: stability during disease evolution and clinical implications. Blood. 2012;119:559–568. doi: 10.1182/blood-2011-07-369934. [DOI] [PubMed] [Google Scholar]
- 22.Medinger M., Passweg J.R. Acute myeloid leukaemia genomics. Br. J. Haemat. 2017;179:530–542. doi: 10.1111/bjh.14823. [DOI] [PubMed] [Google Scholar]
- 23.-Angelini D., Ottone T., Guerrera G. Leukemia-associated CD34/CD123/CD25/ CD99+ immunophenotype identifies FLT3-mutated clones in acute myeloid leukemia. Clin. Cancer Res. 2015;21 doi: 10.1158/1078-0432.CCR-14-3186. See comment in PubMed Commons below: 3977-85. [DOI] [PubMed] [Google Scholar]
- 24.Angenendt L., Röllig C., Montesinos P., Martínez-Cuadrón D., Barragan E., García R., Botella C., Martínez P., Ravandi F., Kadia T., Kantarjian H.M., Cortes J., Juliusson G., Lazarevic V., Höglund M., Lehmann S., Recher C., Pigneux A., Bertoli S., Dumas P.Y., Dombret H., Preudhomme C., Micol J.B., Terré C., Ráčil Z., Novák J., Žák P., Wei A.H., Tiong I.S., Wall M., Estey E., Shaw C., Exeler R., Wagenführ L., Stölzel F., Thiede C., Stelljes M., Lenz G., Mikesch J.H., Serve H., Ehninger G., Berdel W.E., Kramer M., Krug U., Schliemann C. Chromosomal abnormalities and prognosis in NPM1-mutated acute myeloid leukemia: a pooled analysis of individual patient data from nine international cohorts. J. Clin. Oncol. 2019;37(29):2632–2642. doi: 10.1200/JCO.19.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunchala P., Kuravi S., Jensen R., McGuirk J., Balusu R. When the good go bad: mutant NPM1 in acute myeloid leukemia. Blood Rev. 2018;32(3):167–183. doi: 10.1016/j.blre.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Paietta E., Neuberg D., Bennett J., Dewald G., Rowe J., Cassileth P., Cripe L., Tallman M., Wiernik P., the Eastern Cooperative Oncology Group Low expression of the myeloid differentiation antigen CD65s, a feature of poorly differentiated AML in older adults: study of 711 patients enrolled in ECOG trials. Leukemia. 2003;17:1544–1550. doi: 10.1038/sj.leu.2402999. [DOI] [PubMed] [Google Scholar]
- 27.Hwang K., Park C., Jang S., Chi H., Kim D., Lee J., Im H., Seo J. Flow cytometric quantification and immunophenotyping of leukemic stem cells in acute myeloid leukemia. Ann. Hematol. 2012;91:1541–1546. doi: 10.1007/s00277-012-1501-7. [DOI] [PubMed] [Google Scholar]
- 28.Riether C., Schürch C., Ochsenbein A. Regulation of hematopoietic and leukemic stem cells by the immune system. Cell Death Differ. 2015;22:187–198. doi: 10.1038/cdd.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]