Summary
Centrioles are cylindrical assemblies whose peripheral microtubule array displays a 9-fold rotational symmetry that is established by the scaffolding protein SAS6. Centriole symmetry can be broken by centriole-associated structures, such as the striated fibers in Chlamydomonas that are important for ciliary function. The conserved protein CCDC61/VFL3 is involved in this process, but its exact role is unclear. Here, we show that CCDC61 is a paralog of SAS6. Crystal structures of CCDC61 demonstrate that it contains two homodimerization interfaces that are similar to those found in SAS6, but result in the formation of linear filaments rather than rings. Furthermore, we show that CCDC61 binds microtubules and that residues involved in CCDC61 microtubule binding are important for ciliary function in Chlamydomonas. Together, our findings suggest that CCDC61 and SAS6 functionally diverged from a common ancestor while retaining the ability to scaffold the assembly of basal body-associated structures or centrioles, respectively.
Keywords: centrosome, cilia, centriole, basal body, structural biology, CCDC61, SAS6, XRCC4, Chlamydomonas, microtubule, VFL3
Graphical Abstract
Highlights
-
•
CCDC61 is a paralog of SAS6, XRCC4, XLF, and PAXX
-
•
CCDC61 can form protofilaments with a 3-fold screw axis in vitro
-
•
CCDC61 binds to microtubules mainly via its coiled-coil domain
-
•
Microtubule binding of CCDC61 is important for its function in Chlamydomonas
Members of the XRCC4 superfamily of proteins have scaffolding functions in DNA repair or centriole formation. Ochi et al. describe the centrosomal protein CCDC61 as a member of this superfamily. CCDC61 oligomerizes and also binds to microtubules which plays a role in establishing accessory structures of centrioles in Chlamydomonas.
Introduction
Centrosomes are among the largest protein assemblies found in animal cells. They function primarily in the organization of the microtubule cytoskeleton and frequently constitute the dominant cellular microtubule organizing center. Due to this function, centrosomes play an important role in ensuring faithful cell division (Nigg and Raff, 2009). Centrosomes are also involved in other critical cellular processes, such as the formation of functional immunological synapses (Stinchcombe and Griffiths, 2014), the organization of actin (Farina et al., 2016), and intracellular signaling (Arquint et al., 2014).
Centrosomes consist of a pair of barrel-shaped centrioles that are surrounded by and organize the pericentriolar material (PCM), a proteinaceous matrix that anchors microtubule nucleating γ-tubulin complexes (Woodruff et al., 2014). Small electron-dense particles called centriolar satellites, which play a role in centrosomal protein delivery and cellular stress responses (Hori and Toda, 2016), are frequently found in the vicinity of centrioles. Besides their function in the recruitment and organization of the PCM, centrioles are also essential for ciliogenesis. During this process, the older (mother) centriole docks to the cell membrane and extends its peripheral microtubule array, which gives rise to a hair-like cell projection that is referred to as a cilium. In multiciliated cells, cilia formation is initiated from multiple centrioles that have been amplified around electron-dense cellular structures called deuterosomes (Spassky and Meunier, 2017). Cilia have key roles in cellular functions, such as mechanosensing, signal transduction, fluid-flow generation, and cell locomotion (Fliegauf et al., 2007).
Proteomics analyses identified over 100 different proteins associated with human centrosomes (Andersen et al., 2003). Due to a lack of structural information, the exact roles of most of these proteins for the organization and function of the centrosome, as well as their precise mechanism of action, are currently poorly understood. The highly conserved coiled-coil domain-containing protein 61 (CCDC61, also known as variable flagellar number 3, VFL3) is one of these understudied proteins. Unlike wild-type strains, the vfl3 strain of Chlamydomonas reinhardtii does not assemble two cilia per cell, but displays between none and six cilia per cell and consequently shows an altered motility (described as the Vfl− phenotype hereafter) (Wan and Goldstein, 2016, Wright et al., 1983). The vfl3 mutant has defects in the structure of the basal body complex; it is missing the associated striated fibers and contains altered rootlet microtubules (Wright et al., 1983). Basal body/centriole duplication is also compromised (Marshall et al., 2001). Recent studies on CCDC61 in the unicellular ciliate Paramecium tetraurelia showed that the protein plays a crucial role in the orientation of basal bodies and localizes at the interface between basal bodies and ciliary rootlets (Bengueddach et al., 2017). Consistent with these observations, CCDC61 was also shown to be important for the basal body orientation, and the generation of basal feet and ciliary rootlets in the multiciliated ventral epidermis of the flatworm Schmidtea mediterranea (Azimzadeh et al., 2012, Basquin et al., 2019), where its absence results in movement defects. Finally, in Xenopus laevis, the gene expression of CCDC61 was found to be upregulated by the expression of Multicilin, which promotes centriole biogenesis in multiciliated cells (Stubbs et al., 2012). These studies point toward a potential role of CCDC61 in the organization of basal bodies in cells with multiple cilia. A recent report suggests that CCDC61 might also be involved in chromatin alignment and mitotic spindle assembly, possibly by anchoring CEP170 (Bärenz et al., 2018, Pizon et al., 2020). However, how CCDC61 functions mechanistically is currently unknown.
Here, we identify CCDC61 as a highly conserved paralog of SAS6, a key organizer of the central scaffold around which centrioles are formed (Leidel et al., 2005). Our crystal structures of CCDC61 demonstrate that it adopts a SAS6-like fold and forms oligomers through two homodimerization domains in a similar way to SAS6: an N-terminal globular head and a parallel coiled-coil domain. However, instead of the spiral/ring assemblies observed with SAS6, CCDC61 assembles into linear filaments with 3-fold, left-handed screw axes in vitro. Further analysis of CCDC61 reveals that its coiled-coil domains are capable of directly interacting with microtubules. Residues important for microtubule binding are critical for correct localization of the CCDC61 ortholog VFL3 at basal bodies of Chlamydomonas as well as for ciliary function in this organism. Based on these findings, we propose that CCDC61/VFL3 plays a role in scaffolding the assembly of basal body-associated structures throughout eukaryotes.
Results
CCDC61 Is a Paralog of SAS6
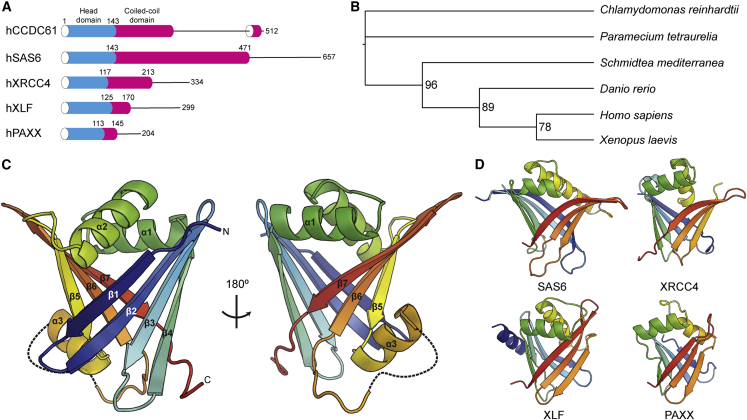
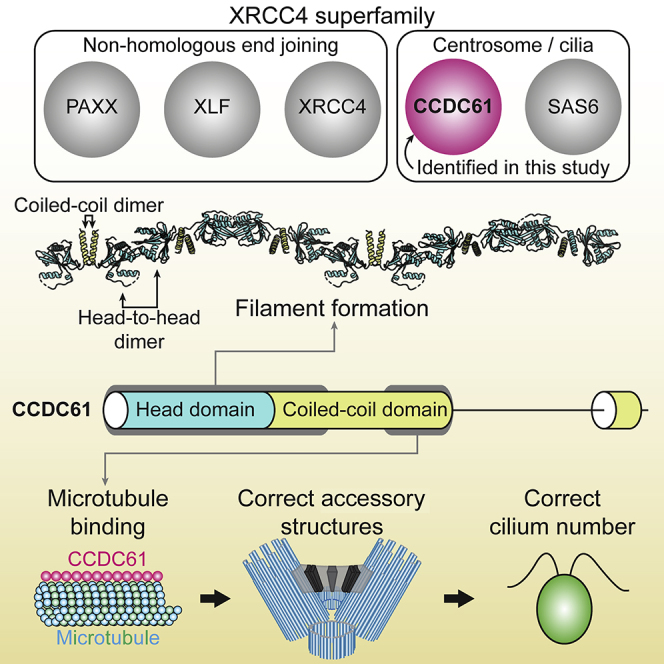
The XRCC4 protein superfamily is constituted by the centriolar protein SAS6 and the DNA repair proteins XRCC4, XLF, and PAXX. Using a similar computational approach to that used previously to identify PAXX (Ochi et al., 2015), we identified the centrosomal protein CCDC61 (Andersen et al., 2003) as an additional candidate member of this superfamily (Figures 1A and S1A). A phylogenetic analysis of CCDC61 orthologs using PSI-BLAST (Altschul et al., 1997) revealed that CCDC61 is a highly conserved protein present in most Eukaryota that possess centrioles, except for flies and nematodes (Figure 1B; Table S1). Although not present in flies, CCDC61 orthologs are readily identified in other insects that include bees, beetles, and lice (Table S1). Secondary structure analyses of CCDC61 orthologs indicate that they all have an N-terminal domain followed by a discontinuous coiled-coil domain and a low-complexity region, which includes a putative α helix (α9), predicted to be a coiled coil, at the C terminus (Figures 1A and S1B). The sequences of the N-terminal domain and α9 are particularly well conserved across species, whereas those of the coiled-coil and low-complexity region are more variable (Figure S1B).
Figure 1.
CCDC61 Is an Evolutionally Conserved Protein Paralogous to SAS6
(A) Domain architectures of the XRCC4 superfamily members. Low complexity regions are drawn by lines.
(B) A phylogenetic tree of CCDC61 orthologs. Accession numbers of the corresponding amino acid sequences are provided in Table S1. Numbers are bootstrap values.
(C) Crystal structure of hCCDC611−143. The structure is presented using a cartoon representation and a rainbow color scheme from the N terminus (N; blue) to the C terminus (C; red). Missing loops are drawn with dotted lines.
(D) Crystal structures of the XRCC4 superfamily members SAS6, XRCC4, XLF, and PAXX (PDB: 2Y3W [van Breugel et al., 2011], 1IK9 [Sibanda et al., 2001], 2QM4 [Li et al., 2008], and 3WTD [Ochi et al., 2015], respectively).
See also Figures S1, S7 and Table S1.
To gain more insight into the domain organization of CCDC61, we determined the crystal structure of the N-terminal domain of human CCDC61 (hCCDC611−143) at a resolution of 2.6 Å using X-ray crystallography. The structure was solved by the single anomalous dispersion method using seleno-methionine-substituted crystals (Figure 1C; Table 1). As indicated by our bioinformatics analyses, we found that the protein fold of CCDC61 is remarkably similar to the canonical SAS6/XRCC4-like fold, which is characterized by the presence of a seven-stranded β barrel with a helix-turn-helix motif inserted between β4 and 5 (Figures 1C and 1D). CCDC61 has an insertion of an extra α helix (α3) followed by an unstructured loop between β5 and β6 (Figures 1C and S1B), which are unique to CCDC61. We conclude that CCDC61 is a centrosomal protein that constitutes a hitherto unidentified paralog of the XRCC4 superfamily members.
Table 1.
Data Collection, Phasing and Refinement Statistics of the CCDC61 Crystal Structures
| Crystal | hCCDC611−143 |
zCCDC611−168;F129E/D130A |
zCCDC611−170 |
|---|---|---|---|
| SeMet (Peak) | Native | Native | |
| Beamline | DLS I02 | MRC LMB | DLS I03 |
| Wavelength (Å) | 0.9792 | 1.5418 | 0.9762 |
| Resolution (Å) | |||
| Overall | 29.68–2.55 | 44.81–1.97 | 68.31–2.90 |
| Outer shell | 2.66–2.55 | 2.02–1.97 | 3.08–2.90 |
| Space group | P22121 | P212121 | C2221 |
| Unit cell parameters | |||
| a, b, c (Å) | 36.877, 68.222, 180.728 | 55.36, 76.31, 83.24 | 93.09, 100.56, 135.76 |
| No. of unique reflections | 15,498 | 25,068 | 14,498 |
| Completeness (%) | 99.1 (99.7)a | 98.2 (96.6) | 100 (100) |
| Redundancy | 5.0 | 7.0 | 5.9 |
| Rmergeb (%) | 7.7 (56.8) | 10.2 (86.8) | 11.3 (84.9) |
| <I/σ> | 14.7 (2.3) | 12.6 (2.4) | 9.4 (2.0) |
| CC½ (%) | 99.8 (93.2) | 99.7 (75.6) | 99.9 (84.0) |
| Phasing method | Single anomalous diffraction | Molecular replacement | Molecular replacement |
| FOM | 0.339 | N/A | N/A |
| Overall score | 39.02 | N/A | N/A |
| Refinement | |||
| PDB: | 6HXT | 6HXV | 6HXY |
| Rcrystc (highest shell) (%) | 20.81 (35.02) | 17.32 (23.49) | 19.99 (29.73) |
| Rfreed (highest shell) (%) | 25.60 (41.38) | 23.79 (31.77) | 25.81 (34.88) |
| No. of atoms | |||
| Protein atoms | 2,900 | 2,559 | 2,335 |
| Water molecules | 66 | 299 | 27 |
| Average B factors (Å2) | 69.81 | 34.75 | 83.16 |
| Ramachandran plot (%) | |||
| Favoured | 97.2 | 97.8 | 96.6 |
| Outliers | 0.0 | 0.3 | 0.3 |
| Clashscore | 7.59 | 4.30 | 8.68 |
| MolProbity overall score | 1.72 | 1.25 | 1.75 |
| RMSD | |||
| Bond lengths (Å) | 0.004 | 0.008 | 0.008 |
| Bond angles (°) | 1.080 | 1.098 | 1.203 |
FOM, figure of merit; RMSD, root-mean-square deviation.
The statistics in parentheses are for the highest-resolution shell.
Rmerge = Σh|Ih − <I>|/ΣhIh, where Ih is the intensity of reflection h, and <I> is the mean intensity of all symmetry-related reflections.
Rcryst = Σ||Fobs| −|Fcalc||/Σ|Fobs|, Fobs and Fcalc are observed and calculated structure factor amplitudes.
Rfree as for Rcryst using a randomly selected 10% for hCCDC611−143 and zCCDC611−170, and 5% for zCCDC611−168;F129E/D130A of the data excluded from the refinement.
CCDC61 Forms Higher-Order Assemblies
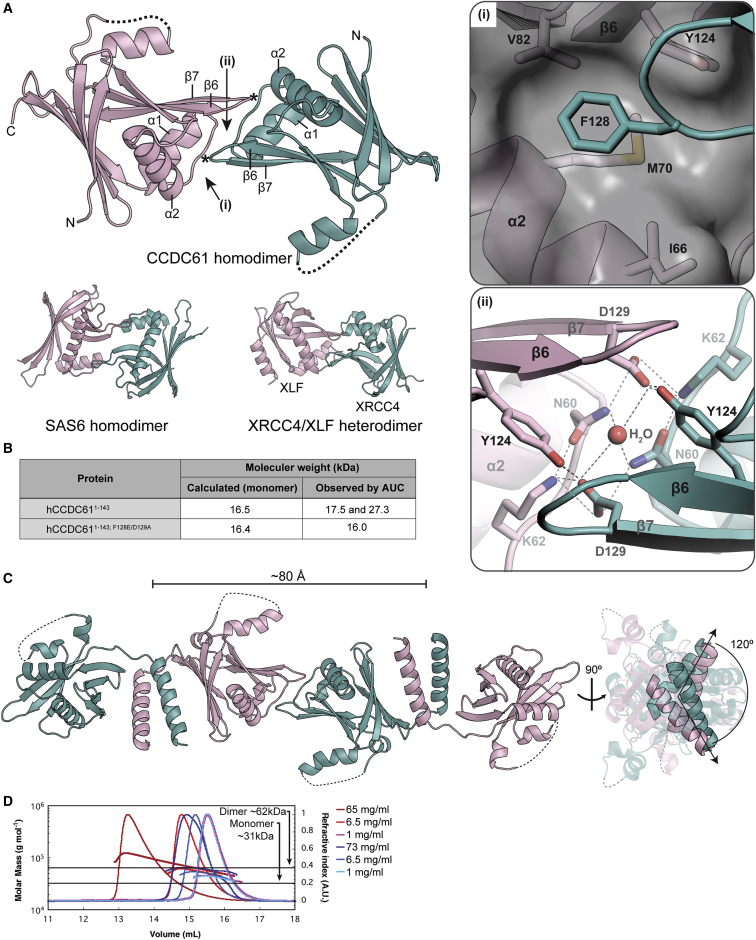
The asymmetric unit of the hCCDC611−143 crystal contained three copies of protomers that pack tightly against each other through interface regions whose residues are evolutionary conserved (Figure S2A; D1 and D2). Remarkably, one observed CCDC61 homodimer (D1) is highly similar to that formed by the SAS6 head domain (van Breugel et al., 2011, Kitagawa et al., 2011) (Figure 2A). The conserved phenylalanine F128 of hCCDC61 (asterisks in Figures 2A, close up in (i) and S1B, dark blue arrow in the alignment) makes van der Waals interactions with M70 and V82 lining a hydrophobic pocket of the homodimer partner that is constituted by α2, the β hairpin of β5 and 6, and the turns before and after α2 (Figure 2A, in (i)). The dimer interface is further stabilized by an extensive network of salt bridges and hydrogen bonds, including a β zipper formed by residues found between α1 and α2 (Figures 2A and S2B). In this network, the conserved aspartate D129 is central to hydrophilic interactions between two protomers (Figures 2A, in (ii) and S1B, light blue arrow in the alignment).
Figure 2.
CCDC61 Forms Linear Filaments via Homodimerization Mediated by the Head and Coiled-Coil Domains
(A) Crystal structure of the head-to-head homodimer of hCCDC611−143. Missing loops are drawn with dotted lines. Key residues of the interaction interface are indicated by (i) and (ii), of which magnified views are shown in the square boxes on the right. Asterisk indicates the locations of the F128 residues. Dotted lines in panel (ii) indicate hydrogen bonds. Head-to-head dimers of SAS6 and XRCC4/XLF (PDB: 2Y3V [van Breugel et al., 2011] and 3W03 [Wu et al., 2011]) are shown at the bottom.
(B) AUC results showing that hCCDC611−143 forms homodimers in solution.
(C) Crystal structure of the zCCDC611−170 tetramer. On the right, straight arrows indicate the N-to-C direction of the coiled-coil domains. The angle between the arrows is 120°.
(D) CCDC61 forms higher-order oligomers in solution. Size-exclusion chromatography with multi-angle light scattering analysis of His6-lipoyl-zCCDC611−170 (red) and His6-lipoyl-zCCDC611−170; F129E/D130A (blue) using a Superdex S200 column at room temperature. Protein concentrations (before injection onto the column) were 1, 6.5, and 65 mg/ml (lightest to darkest red, respectively) and 1, 6.8, and 73 mg/ml (lightest to darkest blue, respectively). The minimum and maximum refractive index values of each chromatography profile were normalized to 0 and 1, respectively.
See also Figures S2 and S3.
To test whether homodimer formation of hCCDC611−143 observed in crystallo is also observed in solution, we studied the oligomeric state of the protein using analytical ultracentrifugation (AUC). This analysis suggests that hCCDC611−143 exists in a monomer-dimer equilibrium with a KD of 170 ± 18 μM (Figures 2B, S2C, and S2D), suggesting a relatively weak binding affinity, similar to that observed with SAS6 (van Breugel et al., 2011, Kitagawa et al., 2011). We next mutated the key residues F128 and D129 of the SAS6-like homodimerization interface of CCDC61 (D1 dimer in Figure S2A) to glutamate and alanine, respectively (hCCDC611−143; F128E/D129A) and subjected the protein to AUC to test whether these residues are important for CCDC61 dimerization. Indeed, dimer formation was abolished in this mutant (Figures 2B and S2C), suggesting that this dimerization interface is dominant in solution, while the other hCCDC611−143 homodimer observed in the asymmetric unit of the crystal (D2 in Figure S2A) appears not to be stable under the experimental conditions.
Due to their overall structural similarity, we wondered whether the head domain of CCDC61 could interact with that of SAS6 to regulate its function. To address this question, we used the recombinant head domains of hCCDC611−143 and the 15N-labeled head domain of human SAS61−143 and performed a chemical shift perturbation experiment by nuclear magnetic resonance (NMR) spectroscopy. Our results shown in Figure S2E did not reveal an interaction between the two proteins. Thus, we conclude that the head domain of CCDC61 forms a homodimer but does not heterodimerize with SAS6.
Besides homodimerization of its head domain, SAS6 contains a second dimerization domain constituted by a parallel coiled-coil domain and, through these two interfaces, is able to assemble into a 9-fold symmetric ring structure (van Breugel et al., 2011, van Breugel et al., 2014, Cottee et al., 2015, Kitagawa et al., 2011) except for C. elegans (Hilbert et al., 2013), where its SAS6 homolog was found in vitro to form spiral assemblies instead. To find out whether CCDC61 can form a ring in a similar manner to SAS6, we determined the crystal structure of zebrafish CCDC61 (residues 1–170; zCCDC611−170), which contains both its head and parts of its coiled-coil domain, by X-ray crystallography at a resolution of 2.9 Å (Figure 2C; Table 1). In the crystal, zCCDC611−170 formed a homo-tetramer mediated by the head-to-head and the coiled-coil dimer interactions in an arrangement that would not be compatible with the assembly of a ring. A filament model of zCCDC611−170 suggests that CCDC61 would be able to form protofilaments with a left-handed 3-fold screw axis along the filament, the helical rise of which is ∼80 Å (Figures 2C and S3A). We also obtained a different crystal form of zCCDC611−170 with a hexagonal instead of an orthorhombic lattice. However, due to poor diffraction quality, we could not determine the structure of this crystal form.
To further confirm higher-order oligomer formation of zCCDC611−170 in solution, we subjected this construct, as well as its F129E/D130A mutant that disrupts the head-to-head interaction in zCCDC61, to size-exclusion chromatography with multi-angle light scattering analysis. In this experiment, the His6-lipoyl domain tag of each construct was retained to stabilize the corresponding proteins at high concentrations. The results shown in Figure 2D demonstrate that the wild-type, but not the head-to-head dimerization-deficient mutant, was able to form higher-order oligomers beyond the coiled-coil-mediated dimer. Together, these data suggest that CCDC61, like SAS6 (van Breugel et al., 2011, van Breugel et al., 2014, Kitagawa et al., 2011), is able to self-associate into ordered macromolecular assemblies.
Comparison of the structures of zCCDC61, SAS6 (Leishmania major [lmSAS6]), and Caenorhabditis elegans (ceSAS6) and human XRCC4/XLF by superposition of their head domains showed that the difference between the exact higher-order assemblies formed by these proteins originates from (1) altered relative orientation angles between their head domains and (2) altered relative orientation angles between the head and coiled-coil domains (Figures S3A and S3B). When defining as z axis (z) the rotation axis required to bring the second head domains of zCCDC61 and lmSAS6 into superposition (Figure S3B, top panel), the corresponding rotation axis between the head domains of zCCDC61 and ceSAS6 also corresponds to z, whereas that between zCCDC61 and hXRCC4/XLF is about 10° off relative to z (Figure S3B, top and lower left panels). In comparison with the relative angle between the head and coiled-coil domain of lmSAS6, the corresponding angle of the other XRCC4 superfamily members is also altered: The coiled-coil domain orientation of zCCDC61 and hXRCC4 deviate in the opposite direction to those of ceSAS6 and hXLF (Figure S3B, lower right panel). Previous observations of structures of SAS6 suggest that the relative orientation angle between the head and coiled-coil domains in particular determines the symmetry of the resulting filaments (Hilbert et al., 2013). Since both head-to-head and head-to-coiled-coil orientation angles appear to be able to change independently from each other, a confirmation of this hypothesis will require further structural information on the superfamily members. Nevertheless, we conclude that two separate dimerization domains of the XRCC4 superfamily proteins allow them to form filaments with different symmetries and helical parameters through mutations of residues involved in the head-to-head and head-to-coiled-coil interactions.
The Coiled-Coil Domain of CCDC61 Binds to Microtubules
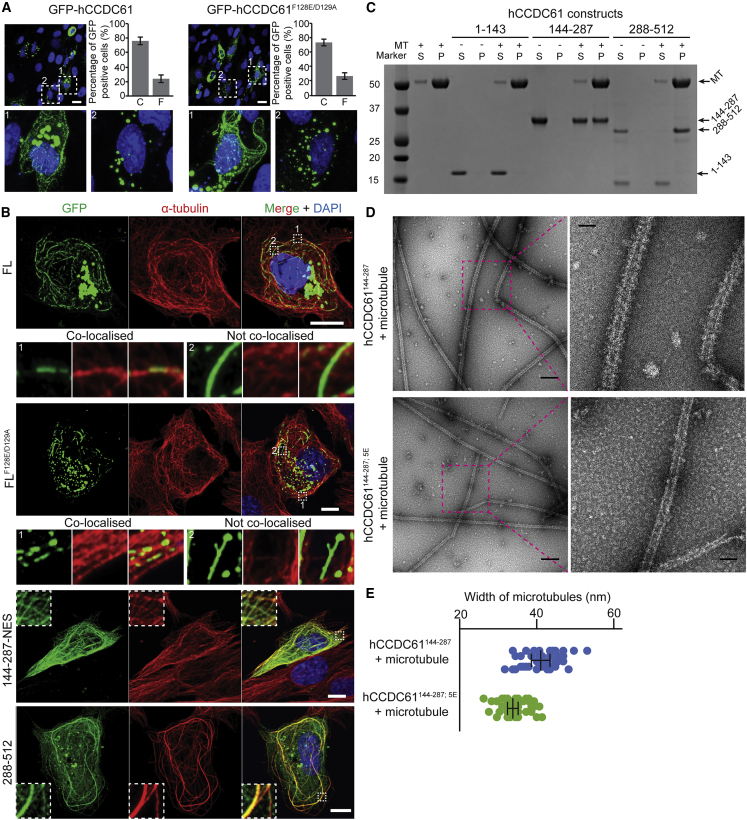
Next, we overexpressed GFP-hCCDC61 in human RPE-1 cells and performed a fluorescence-imaging experiment to better understand the behavior of full-length CCDC61. The majority of the protein-formed clusters and ∼25% of GFP-hCCDC61-expressing cells showed filament-like structures in the cytoplasmic region (Figure 3A), although the extent and type of cluster formation by GFP-hCCDC61 varied widely among cells perhaps due to differences in expression levels. We hypothesized that the observed filament formation in vivo might be mediated by head-to-head dimer formation of CCDC61 as in our crystal structure (Figure 2A). However, when we disabled head-to-head dimer formation using the GFP-hCCDC61F128E/D129A mutant, we still observed a similar variety of localization patterns compared with the wild-type experiment (Figure 3A). Some of the hCCDC61 filaments formed in vivo upon overexpression were reminiscent of cytoplasmic microtubules. Immunofluorescence experiments against GFP-hCCDC61 and microtubules in RPE-1 cells indeed showed colocalization between most, but not all, of the observed GFP-hCCDC61 filaments and microtubules (Figures 3B and S4A). Similar observations were made when we overexpressed the GFP-hCCDC61F128E/D129A mutant (Figures 3B and S4A).
Figure 3.
CCDC61 Binds Microtubules
(A) Fluorescent images of RPE-1 cells, transiently overexpressing GFP-hCCDC61 or hCCDC61F128E/D129A, showing the different CCDC61 localization patterns observed under these conditions. Bar graphs show the percentage of GFP-positive cells containing clusters-only “C” versus filament-containing cells “F” (n = 279 for GFP-hCCDC61 and n = 468 for GFP-hCCDC61F128E/D129A counted from three biological replicates). Error bars are standard deviations. Positions of blow-up images labeled with 1 (filament-containing cell) and 2 (cluster-only cell) are indicated with white-dotted squares in the top panels. Scale bars, 20 μm.
(B) Transiently overexpressed hCCDC61 colocalizes with microtubules in cells. Immunofluorescent images of RPE-1 cells transiently overexpressing GFP-hCCDC61, GFP-hCCDC61F128E/D129A, GFP-hCCDC61144−287−NES, and GFP-hCCDC61288−512. Anti-GFP staining is shown in green, microtubule staining in red. Magnified views of the regions indicated by the white-dotted squares in the merged images are shown either below (GFP-hCCDC61 and GFP-hCCDC61F128E/D129A) or as insets (GFP-hCCDC61144−287−NES and GFP-hCCDC61288−512). Displayed are representative images acquired from a total of 14, 8, 10, and 11 different RPE-1 cells for GFP-hCCDC61, GFP-hCCDC61F128E/D129A, GFP-hCCDC61144−287−NES, and GFP-hCCDC61288−512, respectively. Scale bars, 10 μm.
(C) Coiled-coil and C-terminal regions of hCCDC61 bind microtubules in vitro. Coomassie-stained SDS-PAGE gel showing a co-pelleting assay of taxol-stabilized microtubules with the head domain (1–143), PAXX-fused coiled-coil domain (144–287), or the C-terminal region (288–512) of hCCDC61. S and P indicate supernatant and pellet fraction, respectively.
(D) The coiled-coil domain of hCCDC61 directly binds microtubules. Negative-stain EM micrographs of microtubules that show their decoration with a layer of PAXX-hCCDC61144−287 that is not observed with the corresponding 5E mutant of CCDC61. Scale bars, 200 and 50 nm in the overview panels (left) and the magnified panels (right), respectively.
(E) Quantification of the widths of microtubules decorated by PAXX-hCCDC61144−287 or in the presence of PAXX-hCCDC61144−287; 5E from (D). Widths of five different positions of ten microtubules were measured for each construct. Each point (blue for PAXX-hCCDC61144−287 and green for PAXX-hCCDC61144−287; 5E) represents a measured width at each position. Error bars (standard deviations from the mean) are shown in black lines with flat arrow ends.
See also Figure S4.
The relative proportion of GFP-hCCDC61 filament- or cluster-forming cells was largely unchanged when the microtubule-destabilizing agent nocodazole or the microtubule-stabilizing agent taxol (Figure S4B) were added to cells, indicating that CCDC61 bound microtubules might be protected against the action of microtubule poisons and that the exchange rate between the different CCDC61 pools might be low. In agreement, live cell imaging of RPE-1 cells showed that GFP-hCCDC61 filaments persisted in the presence of 5 μM nocodazole over the course of 3 h (Figure S4C).
We speculated that the coiled-coil and/or the C-terminal regions of hCCDC61 are responsible for its microtubule association. To test this hypothesis, we overexpressed either the coiled-coil domain (144–287) or the C-terminal region (288–512) of hCCDC61 as GFP-tagged constructs in RPE-1 cells and carried out immunofluorescence experiments. The coiled-coil domain construct was fused to an NES to avoid its mis-localization to the nucleus. Our results indeed showed that both the coiled-coil and C-terminal regions of CCDC61 colocalize with microtubules (Figures 3B and S4A).
Since microtubule association in cells might indicate microtubule binding by CCDC61, we sought to perform a direct binding assay to address this question. To this end, we purified the hCCDC61 head domain as well as its coiled-coil and its C-terminal region as recombinant proteins and performed microtubule co-pelleting assays using taxol-stabilized microtubules in vitro. To stabilize the coiled-coil domain of hCCDC61, we fused it to the C terminus of the DNA repair protein PAXX (residues 1–137) whose head domain is structurally similar to that of CCDC61 but does not dimerize (Ochi et al., 2015). Our results suggest that both coiled-coil and C-terminal domain of CCDC61 are indeed able to directly bind to microtubules in vitro (P (pellet) in Figure 3C), whereas the head domain is unable to do so and remains in the supernatant fraction (S (supernatant) in Figure 3C).
Positively charged residues of microtubule-associated proteins frequently play a role in microtubule binding (Cooper and Wordeman, 2009). Intriguingly, the coiled-coil domain of hCCDC61 (residue 144–287; α4–7) has an overall positive charge (theoretical pI ∼10.5). To identify residues involved in microtubule binding by CCDC61, we mutated five conserved positively charged residues in α7 (K259, R263, R266, R268, and K270, Figure S1B, red arrows) and repeated the microtubule co-pelleting assay. Mutation of these residues largely abolished the microtubule binding activity of hCCDC61 (Figure S4D) without affecting the overall structure of CCDC61 or its general ability to form oligomers (Figures S4E and S4F), despite some destabilization of the mutated coiled-coil domain on its own compared with the corresponding wild-type construct in vitro (Figure S4F). The positively charged residues in the coiled-coil domain of hCCDC61 might interact with the negatively charged residues of the tubulin C termini. To test this, we removed the C-terminal tails of tubulin from taxol-stabilized microtubules using the protease subtilisin (Serrano et al., 1984) (Figure S4G) and repeated the microtubule co-pelleting assay. The result showed that the coiled-coil domain of CCDC61 indeed mainly interacts with the tubulin C termini (Figure S4H). Intriguingly, upon overexpression of the 5E mutant of GFP-hCCDC61 in RPE-1 cells, we did not observe CCDC61 filament formation (Figure S4I), while CCDC61 clusters were still observed. This indicates that in vivo and in the full-length context, the α4–7 part of CCDC61 comprises the dominant microtubule binding activity in CCDC61 and that filament formation of CCDC61 is largely mediated by its microtubule binding.
We also mixed the PAXX-stabilized α4–7 coiled-coil domain of hCCDC61 with taxol-stabilized microtubules and subjected the mixture to electron microscopic analysis using negative staining. The micrograph shown in Figure 3D demonstrates that microtubules were decorated with hCCDC61 while this decoration was not observed when we used the equivalent construct carrying the 5E mutation (average widths of microtubules: 40.98 ± 2.55 nm [mean ± standard deviation] and 33.97 ± 1.32 nm, respectively [Figure 3E]). We note that the average width of microtubules in the presence of the 5E mutant seems to be larger than the canonical diameter of microtubules (24 nm). However, this might be due to the negative staining of microtubules on carbon-coated electron microscopic grids as similar, wider microtubule widths also have been observed by others (Reid et al., 2017, Shibata et al., 2012). Together, these results suggest that CCDC61 primarily binds to microtubules by engaging the C-terminal tails of tubulin via conserved positively charged residues of α7 of CCDC61.
CCDC61 Localizes to Basal Bodies
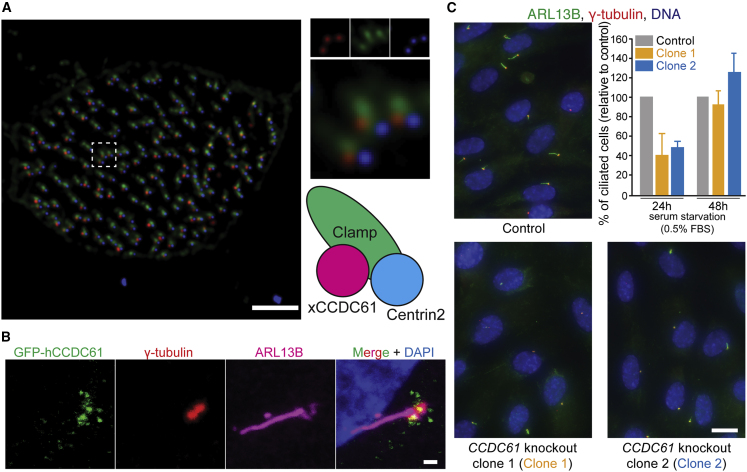
In the ciliate protist Paramecium tetraurelia, previous studies have demonstrated that CCDC61 localizes to basal body-associated substructures, such as rootlets or striated fibers (Bengueddach et al., 2017). To find out whether hCCCD61 would also be found associated with basal bodies in multicellular organisms, we checked the distribution of CCDC61 in multiciliated epithelial cells of Xenopus laevis embryos expressing Xenopus CCDC61 (xCCDC61) fused to the N terminus of RFP. Three-color imaging of xCCDC61-RFP, Centrin2-BFP (marking the distal centriole region) and Clamp-GFP (marking the rootlet) in these multiciliated frog embryos demonstrated that, like its unicellular orthologs, xCCDC61 associates with the proximal part of basal bodies in a polarized manner that is close to, but distinct from rootlets (Figure 4A). We also performed immunofluorescent imaging of GFP-hCCDC61 overexpressed in RPE-1 cells under serum-starved conditions in which these cells form a single, non-motile primary cilium. This analysis showed that hCCDC61 can also localize to the periphery of human basal bodies (Figure 4B). To investigate the function of hCCDC61 in this cell line, we generated hCCDC61-deficient RPE-1 cells using CRISPR/Cas9 (Figure S5A). Although we did not observe obvious defects in proliferation, cell-cycle progression and centrosome or centriole numbers (Figures S5B–S5D), we observed a delay in the formation of primary cilia both in these hCCDC61-deficient RPE-1 cells (Figure 4C) and also in RPE-1 cells depleted of hCCDC61 by RNA interference (Figure S5E). The delay in cilia formation that is observed in hCCDC61-deficient cells could potentially impact developmental processes. However, normal-looking cilia assemble eventually in RPE-1 cells, which suggests that hCCDC61 plays a role in ciliogenesis but is not essential for the generation of primary cilia. Together, these results suggest that the localization of CCDC61 to basal bodies is evolutionarily conserved.
Figure 4.
CCDC61 Associates with Basal Bodies and Plays a Role in Ciliogenesis
(A) xCCDC61 associates with basal bodies and rootlets in multi-ciliated epidermal cells of Xenopus embryos. A fluorescent image of a Xenopus embryo expressing xCCDC61-RFP (red), the basal body component Centrin2-BFP (blue), and the rootlet component Clamp-GFP (green). Scale bar, 3 μm.
(B) Location of hCCDC61 at the periphery of basal bodies of primary cilia. Immunofluorescent image of an RPE-1 cell transiently overexpressing GFP-hCCDC61. Co-immunofluorescent staining was performed against GFP (green), basal bodies (γ-tubulin, red), and the ciliary axoneme (ARL13B, magenta). Scale bar, 1 μm.
(C) Ciliated cells of control and CCDC61-knockout RPE-1 cells. Immunofluorescent images show representative immunofluorescent images used for quantifications of ciliogenesis of primary cilia. Scale bar, 10 μm. The bar graph shows that ciliogenesis was delayed in the CCDC61 knockout cells. Data shown correspond to three biological replicates (total cell counts n = 1,181, 1,103, and 1,008 for control, clone 1 and clone 2 cells after 24-h serum starvation respectively, and n = 1,151, 1,046 and 1,242 for control, clone 1 and clone 2 after 48-h serum starvation, respectively). Percentages are relative to control cells. Bar graphs show mean ± standard deviation.
See also Figure S5.
Characterization of Chlamydomonas Strains Carrying Mutations in the CCDC61 Ortholog VFL3
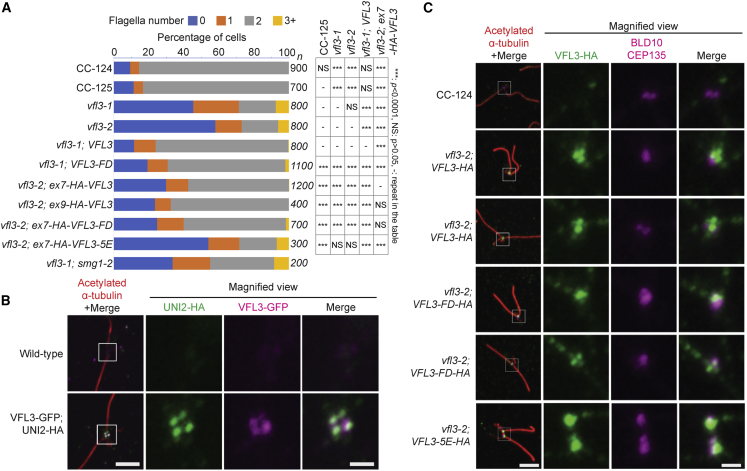
CCDC61 orthologs play an important role in the functioning of motile cilia in different model organisms (Azimzadeh et al., 2012, Bengueddach et al., 2017, Wright et al., 1983). We wondered whether the filament-forming/microtubule binding activity of CCDC61 would be functionally important in this respect. To answer this question, we used Chlamydomonas as a model organism because strains containing defective VFL3 (its CCDC61 ortholog) are available and the mutant phenotypes have been well characterized (Hoops et al., 1984, Keller et al., 2010, Marshall et al., 2001, Wright et al., 1983). Furthermore, VFL3 shares 36% sequence identity with hCCDC61 and key residues involved in head-to-head interaction and microtubule binding are conserved (Figure S1B, blue and red arrows). Through Sanger sequencing, we identified a nonsense mutation (AAG to TAG) in the VFL3 gene in the original mutant, which we named vfl3-1. The nonsense mutation (K497X; Figure S1B, green arrow) is found in exon 8. We also obtained an insertion mutant, LMJ.RY0402.091002, which has an insertion of an exogenous DNA cassette that confers paromomycin resistance, in intron 7 of VFL3, from the Chlamydomonas CLiP mutant library (Li et al., 2016). In 20 tetrads of this insertional mutant crossed to wild-type, we observed complete co-segregation of the paromomycin resistance phenotype and the Vfl− phenotype. Therefore, we considered this insertion mutant a second allele of vfl3 and renamed it vfl3-2.
In both mutant alleles, we quantitated the Vfl− phenotype. We observed that in an asynchronous culture that 7% and 6% of vfl3-1 and -2, respectively, had more than two flagella. This is a phenotype not seen in any wild-type strains. Moreover, the two mutants had an increased number of cells with no flagella (45% and 58% compared with wild-type with 10%) or one flagellum (26% and 15% versus 5% for wild-type) (Figure 5A). By immunofluorescence experiments (Figure S6A), the wild-type (CC-124) exhibited normal distal striated fibers (stained with antibodies to centrin, which is found in the distal striated fibers in Chlamydomonas [Dutcher and O'Toole, 2016]) and biciliated cells (stained by acetylated α-tubulin), whereas we noted abnormal striated fibers and abnormal cilia number in the vfl3-2 mutant strain as reported previously in vfl3-1 (Wright et al., 1983).
Figure 5.
Chlamydomonas VFL3 Protein Localizes to Basal Bodies and the Proximal Ends of Flagella
(A) Rescue of abnormal flagella numbers in vfl3 strains by wild-type VFL3. Bar chart showing flagella numbers observed in wild-type strains (CC-124 and CC125), mutant strains (vfl3-1 and vfl3-2), and the vfl3-1 and vfl3-2 strains expressing VFL3 constructs in Chlamydomonas. The numbers of cells “n” used for calculating ratio flagella numbers are shown on the right side of the chart. A χ2 test was used to determine if the number of cells with zero flagella was significantly different. NS, not significant; ∗∗∗p < 0.0001.
(B) Wild-type VFL3 protein localizes to Chlamydomonas basal bodies. In the first column, cells were stained with acetylated α-tubulin (red) for cilia and rootlet microtubules, anti-HA (green) for UNI2, and anti-GFP (magenta) for VFL3. Scale bar, 4 μm. Magnified views (4×) of the basal body regions (white boxes) are shown on the other three columns. Scale bars, 1 μm.
(C) Localization of VFL3 is affected in the 5E mutant. In the first column, cells were stained with acetylated α-tubulin (red) for cilia and rootlet microtubules, anti-HA (green) for wild-type and mutant VFL3, and anti-BLD10/CEP135 (magenta). Scale bar, 4 μm. Magnified views (4×) of the basal body regions (white boxes) are shown on the other three columns. Scale bar, 1 μm.
See also Figure S6.
Analysis of the VFL3 transcripts in vfl3-1 revealed that this mutant contains a full-length transcript (Figure S6B). In vfl3-2, the VFL3 mRNA is truncated and contains only exons 1–7, which are located upstream of the insertional cassette (Figure S6B). We found that transformation of the wild-type VFL3 gene into vfl3-1 restores normal ciliary numbers (Figures 5A; Table 2). To detect the VFL3 protein, a 3xHA epitope tag was introduced within either exon 7 (ex7-HA) or exon 9 (ex9-HA) of VFL3 and both tagged VFL3 transgenes were integrated into the vfl3-2 strain, where they gave rise to full-length VFL3 transcripts (Figure S6B). However, the tagged genes only partially rescued the mutant phenotype (Figure 5A). The number of cells with zero flagella is not restored to wild-type levels (p < 0.0001) by a χ2 test (Figure 5A). The transgene did restore the striated fiber phenotype in vfl3-2 (Figure S6A). Therefore, we confirmed that the phenotypes of the vfl3 strains are due to the VFL3 gene defects.
Table 2.
Summary of Chlamydomonas Transformation with Various Constructs
| Strain | Construct | No. of APHVIII Transformantsa | No. of vfl3 Rescued Strains |
|---|---|---|---|
| vfl3-1 | VFL3 | 875 | 3 |
| VFL3-FD | 316 | 1 (partial rescued) | |
| VFL3-5E | 449 | 0 | |
| vfl3-2 | VFL3 | 666 | 8 |
| VFL3-FD | 446 | 1 (partial rescued) | |
| VFL3-5E | 970 | 0 | |
| Ex7-HA-VFL3 | 658 | 5 | |
| Ex9-HA-VFL3 | 133 | 1 | |
| Ex7-HA-VFL3-FD | 227 | 1 (partial rescued) | |
| Ex7-HA-VFL3-5E | 221 | 0 | |
| Total | 4961 | 19 |
Transformants are identified by co-transformation with the APHVIII gene that confers resistance to the antibiotic paromomycin.
The Basic Amino Acids Involved in Microtubule Binding Are Important for VFL3 Function and Localization
Immunoblots of Chlamydomonas whole-cell extracts from various transformants that carry either ex7-HA- or ex9-HA-tagged VFL3 with an anti-HA antibody reveal a single polypeptide with the expected size of ∼85 kDa (Figures S6C and S6D), which is absent in extracts of wild-type cells (CC-124) (Figure S6C). Immunofluorescence of NFAPs (nucleoflagellar apparatus) (Wright et al., 1985) in multiple, independent transformants shows that VFL3 (Figures 5B, magenta and 5C, green) localizes to both the basal bodies (Figures 5B, green and 5C, magenta). To determine the precise location of VFL3 in the basal bodies, we co-stained VFL3 with UNI2, a protein that localizes to the distal end of the basal bodies (Figures 5B; Video S1) (Piasecki and Silflow, 2009); and BLD10/CEP135, which localizes to the cartwheel of the basal bodies at their proximal end (Matsuura et al., 2004) (Figure 5C). Our results suggest that VFL3 does not overlap completely with either UNI2 or BLD10 and is likely distributed along the full length of the basal bodies.
Cells were stained with acetylated α-tubulin (red) for cilia and rootlet microtubules, anti-HA (green) for UNI2, and anti-GFP (magenta) for VFL3. z stacked images start from the distal ends of basal bodies (labeled by positions of UNI2) and move toward the proximal ends. Scale bar, 0.8 μm.
We next asked whether filament formation and microtubule binding of VFL3 are important for its function and localization. We first generated a strain containing the F126E and D127A mutations in VFL3 (VFL3-FD), which are equivalent to the F128E/D129A mutation in hCCDC61 that disrupts its head-to-head homodimerization. We transformed the VFL3-FD transgene (untagged or HA tagged [Figure S6D]) into both vfl3-1 and vfl3-2 cells, and observed a partial rescue of the mutant phenotype in the vfl3-1 and vfl3-2 cells, respectively (Figure 5A). The untagged VFL3 transgene, and the untagged FD mutant transgene are significantly different from the mutant parent, while the untagged FD mutant is also significantly different from the strain with the wild-type transgene (p < 0.0001) based on the number of cells with zero flagella (Figure 5A) by a χ2 test. The HA-tagged FD mutant was not significantly different from the HA-tagged wild-type transgene in the vfl3-2 strain. The difference between tagged and untagged FD mutants might be due to the HA tag partially interfering with the protein function. About 2% of cells contain more than two cilia compared with less than 0.1% in wild-type cells. These data suggest that there is a partial rescue of the Vfl− phenotype by the VFL3-FD transgene. The VFL3-FD-HA protein localizes to the basal body region (Figure 5C), similar to what we observed in wild-type VFL3-HA. We also mutated the five basic amino acids residues (K266, R270, K273, R275, and R277), whose equivalents in hCCDC61 are involved in microtubule binding, to glutamates (VFL3-5E). Co-transformation of the VFL3-5E transgene into the vfl3-1 and vfl3-2 strains with the APHVIII gene, which confers resistance to the antibiotic paromomycin, failed to yield any strains with a rescued phenotype in ∼1,400 drug-resistant transformants (Table 2). To investigate whether the failure to rescue is caused by an absence of expression of the transgene or by the 5E mutation, we co-transformed an HA-tagged version of VFL3-5E into vfl3-2 and screened ∼200 drug-resistant transformants by immunoblot and immunofluorescence (Figures 5C and S6D). We obtained a single transformant that showed expression of the HA-VFL3-5E protein but the 5E mutant strain was not significantly different from the mutant vfl3-2 parent (p = 0.68) based on the number of cells with zero flagella (Figure 5A) by a χ2 test (Figure 5A). We observed accumulation of HA-VFL3-5E around the basal body region with BLD10/CEP135 (Figure 5C). Thus, our results suggest that the microtubule binding region of VFL3/CCDC61 plays a critical role in its function and accurate localization in living cells.
Discussion
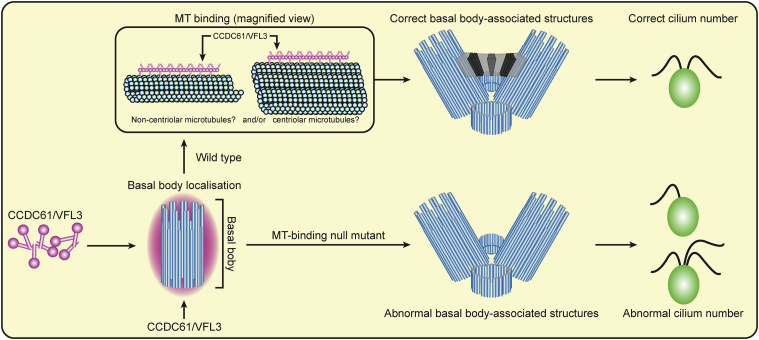
Here we provide a detailed structural and biochemical characterization as well as a functional analysis of the centrosomal protein CCDC61. CCDC61 is a paralog of the centriolar protein SAS6, forms higher-order oligomers and is capable of binding microtubules in vitro and in vivo. Furthermore, we demonstrate that the CCDC61 ortholog in Chlamydomonas, VFL3, localizes at basal bodies. Our functional studies in cells suggest that microtubule binding of VFL3/CCDC61 is important for its correct localization to basal bodies and its function in vivo, whereas its head-to-head interaction appears not to be critical but plays a role in ensuring faithful formation of basal bodies in vivo. Collectively, our data suggest that the main function of CCDC61 might lie in the organization of basal body-associated structures (Figure 6).
Figure 6.
Model of the Role of CCDC61 in Ciliary Function (in Chlamydomonas)
CCDC61 localizes to the basal body and forms filaments that bind to centriolar and/or non-centriolar microtubules. This facilitates striated fiber formation and the correct formation of basal body-associated structures, and therefore, results in the correct cilium number. A CCDC61 mutant that does not bind microtubules (MT-binding null mutant) still localizes to the basal body region. However, the mutant is incapable of facilitating striated fiber formation, leads to incorrect formation of basal body-associated structures, and therefore causes abnormal cilium numbers.
Our biochemical and structural data unambiguously demonstrate that CCDC61 belongs to the XRCC4 superfamily of proteins. Members of this superfamily have a centrosomal/centriolar function (SAS6 [Leidel et al., 2005] and CCDC61 [Andersen et al., 2003, Wright et al., 1983]) or play crucial roles in the NHEJ DNA repair pathway (XRCC4 [Li et al., 1995], XLF [Ahnesorg et al., 2006, Buck et al., 2006] and PAXX [Craxton et al., 2015, Ochi et al., 2015, Xing et al., 2015]). Their protein architecture consists of an N-terminal head domain followed by a coiled-coil and C-terminal low-complexity region. A structure-guided sequence alignment of the head domains of the human XRCC4 superfamily members showed that the sequence identities between them are below 20% (Figures S7A and S7B). However, they share a conserved sequence motif (Figure S7A, red-dotted rectangle), which has previously been named the PISA motif (Leidel et al., 2005). The motif is likely to be critical for the functions of the XRCC4 superfamily members because homozygous mutations in this motif in XRCC4, XLF, or SAS6 cause growth defects (Buck et al., 2006, Khan et al., 2014, Murray et al., 2015). Their similarity also extends to the ability of the superfamily members (except for PAXX), to form protofilaments using two dimerization interfaces provided by the head and the coiled-coil domains. Head-to-head dimerization in all these cases occurs with a low binding affinity, suggesting that these proteins need to be enriched locally and/or be stabilized by other molecules to efficiently form faithful higher-order assemblies. In agreement with this, the protein concentration of CCDC61 in HeLa cells appears to be very low (Bauer et al., 2016). SAS6 assembly, for instance, is probably aided by its interaction with CEP135 and STIL (Dzhindzhev et al., 2014, Lin et al., 2013, Ohta et al., 2014), whereas for the XRCC4/XLF complex this function is exerted by its associations with DNA ligase IV, Ku70/80 and DNA (Ochi et al., 2014). Putative CCDC61 binding proteins, such as CEP170 (Bärenz et al., 2018, Pizon et al., 2020) might play an equivalent role in CCDC61. Thus, the overarching principles of higher-order oligomerization and stabilization by other proteins appears to be conserved among the XRCC4 superfamily members. It is worth mentioning that the head domain of hCCDC61 contains a conserved surface area that is not directly involved in D1 dimer formation as described in Figures 2A and S2A. Instead, it contributes to the formation of the D2 dimer that we observed in the asymmetric unit of the hCCDC611−143 crystal, but not in solution (Figures 2B, S2A, and S2C). This surface might be involved in a protein-protein interaction between CCDC61 and another protein. Alternatively, it is possible that the D2 dimer exists in vivo under high local concentrations and that it might facilitate formation of CCDC61 filament bundles. Indeed, when we superposed two CCDC61 filaments onto the D2 structure, we found that these filaments do not clash strongly with each other (Figure S7C).
Our phylogenetic analysis demonstrates the presence of CCDC61 in most ciliated eukaryotes, except for flies and nematodes. Interestingly, the conservation pattern of CCDC61 (Table S1) is very similar to those of δ- and ε-tubulins (Hodges et al., 2010), which are important for centriolar doublet and triplet microtubule formation (Dutcher et al., 2002, Dutcher and Trabuco, 1998, Wang et al., 2017). However, our data argue against a central role of CCDC61 in centriole/centrosome duplication in human cells (Figures S5B–S5D). These results agree with findings in the flatworm S. mediterranea (Azimzadeh et al., 2012), but contrast with reports from the unicellular Chlamydomonas (Marshall et al., 2001) and Paramecium (Bengueddach et al., 2017). The difference between these studies possibly stems from the fact that centrioles are duplicated as basal bodies in these organisms, whereas they are duplicated through the centrosomal or deuterosomal pathway in humans and planaria. Thus, CCDC61 itself might not be part of the core centriole duplication machinery in these unicellular organisms, but rather be important for the maintenance of the basal body-associated architectures that are needed for the faithful recruitment of this duplication machinery to probasal bodies.
Consistent with a role of CCDC61 in basal body function, we observed localization of CCDC61 at basal bodies of primary cilia of human RPE-1 cells, motile cilia in green algae, and motile cilia of multiciliated cells of frog embryos (Figures 4A, 4B, 5B, and 5C). Our data, as well as evidence from other systems in which CCDC61 has been studied, suggest that the main function of CCDC61 is related to basal body function in cells with motile cilia. In the motile ciliate Paramecium, CCDC61 localizes at the interface between basal bodies and striated rootlets and is important for their organization (Bengueddach et al., 2017). These basal body-associated structures play a role in basal body positioning at the cell cortex and in probasal body assembly (Hoops et al., 1984). A specific role of CCDC61 in the anchoring of basal bodies in multiciliated cells is also suggested by experiments in the planarian S. mediterranea. Planaria move by gliding on a ventral array of multiciliated cells (Azimzadeh and Basquin, 2016). Knockdown of CCDC61 in S. mediterranea was found to result in an abnormal direction of locomotion (Azimzadeh et al., 2012) due to basal body mis-orientations caused by a failure to generate basal feet and ciliary rootlets correctly (Basquin et al., 2019).
This notion is in agreement with previous studies on the Chlamydomonas vfl3-1 strain that suggest that VFL3 is crucial for the faithful organization of proximal and distal striated fibers as well as rootlet microtubules (Hoops et al., 1984, Wright et al., 1983). We also confirmed this using the insertional mutant strain (vfl3-2). Interestingly, the vfl3-1 strain carries a premature stop codon after K497, which would retain the head and coiled-coil domains of VFL3 but not α9. Similarly, our transcript analysis suggests that the vfl3-2 strain may carry a truncated protein that retains the first 406 amino acids of VFL3, 22 amino acids downstream of the α8 helix. Thus, this could be indicative of a crucial role of α9 in VFL3 function, given the mutant phenotype in both strains. However, since we could not obtain reliable antibodies that detect VFL3, we cannot exclude the possibility that the truncated VFL3 gene product is destabilized in these strains explaining the observed phenotype.
Although our crystal structures of CCDC61 fragments in vitro suggest that CCDC61 forms filaments (Figure 2C), we could not visualize these in vivo. However, a VFL3/CCDC61 allele with a disabled head-to-head dimerization (VFL3-FD) was unable to completely rescue the Vfl− phenotype (Figure 5A) in Chlamydomonas, arguing for a functional role of this interface. Furthermore, since both vfl3-1 and vfl3-2 strains retain intact exons 1–7 of VFL3 (residue range 1–406), a heterodimer between the putative truncated VFL3 and the rescue construct might form and be partially functional, potentially accounting for the weak phenotype observed in the VFL3-FD strain. Intriguingly, in the filaments formed by CCDC61 in vitro, the distance between the projecting coiled-coil domains of CCDC61 that point in the same direction is about 24 nm, which corresponds to three times the 8-nm repeat of tubulin dimers in microtubules (Figure 2C). This periodicity might facilitate microtubule interaction by the CCDC61 coiled-coil domain.
Despite the weak phenotype observed in the VFL3-FD strain, we speculate that the ability of CCDC61/VFL3 to form higher-order assemblies might aid scaffold formation of the protein through which basal body-associated substructures are anchored or helped to stay in place under the mechanical stresses acting on motile cilia (Figure 6). CCDC61/VFL3 might interact with a centrosomal and basal body-specific protein that is yet to be identified, and scaffold the protein with microtubules to construct regularly aligned basal body-associated structures. Two groups recently proposed that CCDC61 interacts with CEP170 and might play a role in the subdistal appendage function of centrioles (Bärenz et al., 2018, Pizon et al., 2020). While our manuscript was under review, Pizon and colleagues also reported CCDC61 association with microtubules (Pizon et al., 2020), in agreement with our data.
Our study also raises several questions, particularly, whether, in vivo, CCDC61 forms protofilaments as observed in crystallo and, if so, what their exact role is. How does microtubule binding of CCDC61 assist in the assembly and organization of basal body-associated structures? Further research efforts are required to elucidate the exact function and the molecular mechanisms of CCDC61 that underlie the biogenesis of these structures.
STAR★Methods
Key Resource Table
| REAGENT or RESOURCE | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Rabbit anti-ARL13B | Proteintech | 17711-1-AP |
| Mouse anti-alpha-tubulin | Sigma-Aldrich | T9026 |
| Rabbit anti-acetylated-alpha-tubulin | Abcam | ab179484 |
| Mouse anti-gamma-tubulin | Sigma-Aldrich | T6557 |
| Chicken anti-GFP | Abcam | ab13970 |
| Mouse anti-GFP | Thermo Fisher Scientific | A11120 |
| Rabbit anti-HA | Gift from Dr Manu Hedge | N/A |
| Rat anti-HA | Roche | 118674230001 |
| Mouse anti-centrin | Gift from Dr Jeffrey L. Salisbury | N/A |
| Mouse anti-centrin 3 | Abnova | H00001070-M01 |
| Bacterial Strains | ||
| BL21(DE3) | New England Biolabs | C2527 |
| C41(DE3) | Miroux and Walker, 1996 | N/A |
| Rosetta (DE3) | Gift from Dr John Kilmartin | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| D-MEM Glutamax | Thermo Fisher Scientific | Catalog # 10566016 |
| D-MEM/F-12, supplied, GlutaMAX, sodium carbonate | Thermo Fisher Scientific | Catalog # 31331028 |
| D-MEM/F-12 without phenol red | Thermo Fisher Scientific | Catalog # 21041025 |
| Opti-MEM | Thermo Fisher Scientific | Catalog # 31985062 |
| CloneAmp HiFi Premix | Clontech | Catalog # 639298 |
| In-Fusion HD cloning | Clontech | Catalog # 638933 |
| RNeasy Mini Kit | Qiagen | Catalog # 74104 |
| RNase-free DNase I | Thermo Fisher Scientific | Catalog # EN0521 |
| SuperScript IV VILO Master Mix | Thermo Fisher Scientific | Catalog # 11756050 |
| QuickExtract DNA extract solution | Cambio | Catalog # QE0950 |
| 0.1% poly-L-Lysine | Sigma-Aldrich | Catalog # P8920 |
| Ni-NTA resin | Expedeon | Catalog # ANN0100 |
| Ni-NTA | Qiagen | Catalog # 30210 |
| Glutathione sepharose 4B | GE Healthcare Life Sciences | Catalog # 17075601 |
| NHS-activated sepharose 4 Fast Flow | GE Healthcare Life Sciences | Catalog # 17090601 |
| Tev protease | Homemade | N/A |
| GST-PreScission protease | Homemade | N/A |
| Tubulin | Gift from Dr Andrew Carter | N/A |
| Subtilisin A | Sigma-Aldrich | Catalog # P5380 |
| Monastrol | Sigma-Aldrich | Catalog # M8515 |
| ProLong Diamond Antifade Mountant | Thermo Fisher Scientific | Catalog # P36970 |
| Fluoromount-G | Southern Biotech | Catalog # 0100-01 |
| Hoechst 33342 | EMP Biotech | Catalog # F-0409 |
| PEI | Polysciences | Catalog # 24765 |
| Lipofectamine 3000 | Thermo Fisher Scientific | Catalog # L3000001 |
| Lipofectamine RNAiMAX | Thermo Fisher Scientific | Catalog # 13778150 |
| Deposited Data | ||
| Human XRCC4-DNA Ligase IV complex | Sibanda et al., 2001 | PDB code: 1IK9 |
| Human XLF | Li et al., 2008 | PDB code: 2QM4 |
| Human XRCC4-XLF complex | Wu et al., 2011 | PDB code: 3W03 |
| The N-terminal head domain of zebrafish SAS6 | van Breugel et al., 2011 | PDB code: 2Y3V |
| N-terminal head domain and beginning of coiled coil domain of Zebrafish SAS6 | van Breugel et al., 2011 | PDB code: 2Y3W |
| N-terminal domain of C. elegans SAS6 | Hilbert et al., 2013 | PDB code: 3PYI |
| N-terminal fragment of L. major SAS6 | van Breugel et al., 2014 | PDB code: 4CKP |
| Human PAXX | Ochi et al., 2015 | PDB code: 3WTD |
| hCCCDC611-143 structure | This paper | PDB code: 6HXT |
| zCCCDC611-168; F129E/D130A structure | This paper | PDB code: 6HXV |
| zCCCDC611-170 structure | This paper | PDB code: 6HXY |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | ATCC: CRL-3216 |
| RPE-1 | Gift from Prof. Colin A. Johnson | N/A |
| RPE-1 PuroKO | Balmus et al., 2019 | N/A |
| RPE-1 CCDC61 KO clone 1 and 2 | this paper | N/A |
| Experimental Models: Organisms/Strains | ||
| vfl3-1 | Chlamydomonas Resource Center | CC-1686 |
| vfl3-2 | this paper | N/A |
| Oligonucleotides | ||
| siRNA 1 | Thermo Fisher Scientific | siRNA ID: s59736 |
| siRNA 2 | Thermo Fisher Scientific | siRNA ID: s59737 |
| siRNA 3 | Thermo Fisher Scientific | siRNA ID: s59738 |
| Control siRNA | Thermo Fisher Scientific | siRNA ID: 4390084 |
| hCCDC61 knockout target sequence 1: GGAAGACGTAGTCCACCTGCAGG | This paper | N/A |
| hCCDC61 knockout target sequence 2: GGAGCATGCCGTGCGGGTGATGG | This paper | N/A |
| RT-PCR primer forward: TGCAGCGATTTGGAGGATTT | This paper | N/A |
| RT-PCR primer reverse: CGGAGTTGGCCAGAGATTTC | This paper | N/A |
| Primers used for site-directed mutagenesis of human and zebrafish CCDC61, and human genomic DNA PCR in Table S2 | N/A | N/A |
| Primers used to amplify Chlamydomonas VFL3 are listed in Table S3 | N/A | N/A |
| Recombinant DNA | ||
| hCCDC61 | Synthesized by GenScript | UniProt: Q9Y6R9 |
| zCCDC61 | Source BioScience | IMAGE ID: 7406569. UniProt: Q08CF3 |
| xCCDC61 | Synthesized by Thermo Fisher Scientific | NCBI accession number: XP_018084688.1 |
| PAXX | Ochi et al., 2015 | N/A |
| GFP nanobody | Synthesized by GenScript | N/A |
| pGAT3-hCCDC611-143 | this paper | N/A |
| pGAT3-hCCDC611-143; F128E/D129A | this paper | N/A |
| pSKB2LNB-zCCDC611-168; F129E/D130A | this paper | N/A |
| Lipo-zCCDC611-170 | this paper | N/A |
| Lipo-zCCDC611-170; F129E/D130A | this paper | N/A |
| pSKB2LNB-zCCDC61146-280 | this paper | N/A |
| pSKB2LNB-zCCDC61146-280; 5E | this paper | N/A |
| pSKB2LNB-PAXX1-137-hCCDC61144-287 | this paper | N/A |
| pSKB2LNB-PAXX1-137-hCCDC61144-287; 5E | this paper | N/A |
| pSKB2LNB-hSAS61-143 | this paper | N/A |
| pHAT5-GFP-nonobody | this paper | N/A |
| short-VFL3-TOPO | this paper | N/A |
| WT-VFL3-TOPO | this paper | N/A |
| pEGFP-C1-hCCDC61 | this paper | N/A |
| pEGFP-C1-hCCDC61F128E/D129A | this paper | N/A |
| pEGFP-C1-hCCDC61144-287-NES | this paper | N/A |
| pEGFP-C1-hCCDC61288-512 | this paper | N/A |
| pEGFP-C1-hCCDC611-457; F128E/D128A | this paper | N/A |
| pEGFP-C1-hCCDC611-457; F128E/D129A/5E | this paper | N/A |
| pcDNA3-3xHA-hCCDC611-457; F128E/D128A | this paper | N/A |
| pcDNA3-3xHA-hCCDC611-457; F128E/D129A/5E | this paper | N/A |
| pENTR-D-TOPO-xCCDC61 | this paper | N/A |
| pCS2+-xCCDC61-RFP | this paper | N/A |
| pCS2+-Centrin2-BFP | this paper | N/A |
| pCS2+-Clamp-GFP | Park et al., 2008 | N/A |
| AIO-GFP-hCCDC61 | this paper | N/A |
| pGAT3 | Peränen et al., 1996 | Addgene: 112589 |
| pHAT4 | Peränen et al., 1996 | Addgene: 112585 |
| pHAT5 | Peränen et al., 1996 | Addgene: 112586 |
| pSKB2LNB | Fekairi et al., 2009 | N/A |
| pcEGFP-C1 | Clontech | Catalog # 6084-1 |
| pcDNA3 | Invitrogen | Catalog # A-150228 |
| AIO-GFP | Chiang et al., 2016 | Addgene: 74119 |
| pENTR-D-TOPO | Thermo Fisher Scientific | Catalog # K240020 |
| pCR2.1-TOPO | Thermo Fisher Scientific | Catalog # K455001 |
| Software and Algorithms | ||
| Jpred | Drozdetskiy et al., 2015 | http://www.compbio.dundee.ac.uk/jpred/ |
| BackPhyre | Kelly and Sternberg, 2009 | http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index |
| HHPred | Söding et al., 2005 | https://toolkit.tuebingen.mpg.de/tools/hhpred |
| PSI-BLAST | Altschul et al., 1997 | https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE=Proteins&PROGRAM=blastp&RUN_PSIBLAST=on |
| MUSCLE | Edgar, 2004 | https://www.drive5.com/muscle/ |
| BOXSHADE | N/A | https://embnet.vital-it.ch/software/BOX_form.html |
| SIAS server | N/A | http://imed.med.ucm.es/Tools/sias.html |
| SeaView | Gouy et al., 2010 | http://doua.prabi.fr/software/seaview |
| PhyML | Guindon et al., 2010 | http://www.atgc-montpellier.fr/phyml/ |
| FigTree | N/A | http://tree.bio.ed.ac.uk/software/figtree/ |
| Modeller | Sali and Blundell, 1993 | https://salilab.org/modeller/ |
| TopMatch | Sippl and Wiederstein, 2012 | https://topmatch.services.came.sbg.ac.at/ |
| XDS | Kabsch, 2010 | http://xds.mpimf-heidelberg.mpg.de/ |
| CCP4 program suite | Winn et al., 2011 | https://www.ccp4.ac.uk/ccp4i_main.php |
| iMOSFLM | Battye et al., 2011 | Run from CCP4 program suite |
| Aimless | Evans, 2011 | Run from CCP4 program suite |
| PHENIX suite | Adams et al., 2010 | https://www.phenix-online.org/ |
| MolProbity | Run from PHENIX suite | |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PyMOL | N/A | https://pymol.org/2/ |
| Consurf | Glaser et al., 2003 | https://consurf.tau.ac.il/ |
| SEDFIT | Schuck, 2003 | http://www.analyticalultracentrifugation.com/sedfit.htm |
| Sedntrep | Dr Tomas Laue, University of New Hampshire | N/A |
| SEDPHAT | Schuck, 2003 | http://www.analyticalultracentrifugation.com/sedphat/default.htm |
| GUSSI | Brautigam, 2015 | http://biophysics.swmed.edu/MBR/software.html |
| Topspin | Bruker | N/A |
| SPARKY | T. D. Goddard and D. G. Kneller, University of California | https://www.cgl.ucsf.edu/home/sparky/ |
| CRISPR DESIGN | Hsu et al., 2013 | No longer available |
| LAS X | Leica | N/A |
| Zen | Zeiss | N/A |
| Volocity | Perkin Elmer | N/A |
| Fiji | Schindelin et al., 2012 | https://imagej.net/Fiji/Downloads |
| Photoshop | Adobe | N/A |
| Huygens Professional | Scientific Volume Imaging | N/A |
| FCS EXPRESS 6 | De Novo Software | N/A |
| Prism | GraphPad | N/A |
| Social Science Statistics | N/A | https://www.socscistatistics.com/tests/chisquare/ |
| Other | ||
| GSTrap FF 16/10 | GE Healthcare Life Sciences | Catalog # 28936550 |
| GSTrap HP | GE Healthcare Life Sciences | Catalog # 17528202 |
| HisTrap HP | GE Healthcare Life Sciences | Catalog # 17524801 |
| HisTrap FF | GE Healthcare Life Sciences | Catalog # 17525501 |
| HiTrap Q HP | GE Healthcare Life Sciences | Catalog # 17115401 |
| HiTrap Q FF | GE Healthcare Life Sciences | Catalog # 17515601 |
| HiTrap Heparin HP | GE Healthcare Life Sciences | Catalog # 17040701 |
| PD-10 desalting column | GE Healthcare Life Sciences | Catalog # 17085101 |
| Superdex 75 16/600 | GE Healthcare Life Sciences | Catalog # 28989333 |
| Superdex S200 10/300 | GE Healthcare Life Sciences | Catalog # 17517501 |
| 16 Chambered cover glass | Grace Bio-Labs | Catalog # 112358 |
| Multi-spot slide | Thermo Fisher Scientific | Catalog # 9991090 |
| 400 mesh carbon-coated copper grids | Electron Microscopy Sciences | Catalog # CF400-Cu-50 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Takashi Ochi (T.Ochi@leeds.ac.uk).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Data and Code Availability
Coordinates and structure factors of crystal structures that are presented in this paper are available in the Protein Data Bank (PDB codes: 6HXT (hCCDC611-143), 6HXV (zCCDC611-168; F129E/D130A) and 6HXY (zCCDC611-170)).
Experimental Model and Subject Details
Human Cell Culture
All cells were grown in 37°C with 5% CO2. HEK293T cells (sex: female) were grown in D-MEM, GlutaMAX (Thermo Fisher Scientific) supplied with 10% FBS. RPE-1 cells (sex: female) were grown in D-MEM/F-12 supplied, GlutaMAX, sodium carbonate (Thermo Fisher Scientific) supplied with 0, 0.5 or 10% FBS, and 100 unit of penicillin and 100 μg/ml of streptomycin. RPE-1 and RPE-1 PuroKO that used in this study have been authenticated by STR profiling. STR profiling of HEK293T revealed a 68% match between our cells and the ATCC standard; this suggest a drift in our stock (which is fairly common for HEK293T), and thus these cells were used only for protein production (Figure S4E) and not for functional assays. In addition to these cell lines, RPE-1 PuroKO/CCDC61KO cells have been confirmed as mycoplasma free.
Xenopus Embryo Culture
Xenopus embryo were prepared as described previously (Hörmanseder et al., 2017). Briefly, mature Xenopus laevis males and females were obtained from Nasco. Females were injected with 50 units pregnant mare serum gonadotropin 3 days in advance and 500 units human chorionic gonadotropin 1 day in advance in the dorsal lymph sack to induce natural ovulation. Eggs were laid in a 1x MMR buffer (5mM HEPES pH 7.8, 100mM NaCl, 2mM KCl, 1mM MgSO4, 2mM CaCl2, 0.1mM EDTA). Xenopus embryos were cultured at 14°C in the 0.1x MMR until they reached stage 27/28. Our work with Xenopus laevis is covered under the Home Office Project License PPL 70/8591 and frog husbandry and all experiments were performed according to the relevant regulatory standard.
Chlamydomonas Culture
Chlamydomonas reinhardtii strains were maintained on solid Sager and Granick (R) growth medium at 25°C. For electroporation, Chlamydomonas cells were grown in Tris-acetate phosphate (TAP) medium at 25°C under constant illumination till the cell density reached 1∼3 x 106 cells/ml. Transformants were selected on modified TAP medium (0.75 ml of Glacial acetic acid/1L TAP) supplied with 10 μg/ml hygromycin at 25°C. For immunofluorescence, Chlamydomonas cells were first resuspended in liquid M-N/5 medium for 4 hours and treated with autolysin for 30 min at 25°C before fixation of cells.
Bacterial Cell Culture
BL21(DE2) (New England Biolabs), C41(DE3) (Miroux and Walker, 1996) or Rosetta cells (a kind gift of Dr. John Kilmartin, MRC LMB, Cambridge, UK) were grown in LB or 2xTY media and used for protein expression and purification.
Method Details
Amino-acid Sequence Analysis
As summarized in Figure S1A, hCCDC61 was identified as a paralog of XRCC4 using residue 1-213 of chain A of the crystal structure of XRCC4 (PDB code: 1IK9 (Sibanda et al., 2001)) as an input for the BackPhyre webserver (Kelley and Sternberg, 2009). Candidate proteins from BackPhyre whose alignments with XRCC4 covered its head domain were further analysed using HHpred (Söding et al., 2005) and JPred (Cole et al., 2008). Criteria to be defined as XRCC4-like proteins were: a) the candidates were predicted as XRCC4-superfamily members using HHPred and b) they have secondary-structure profiles that are similar to these family members.
Orthologs of hCCDC61 (UniProt accession number: Q9Y6R9) were identified using PSI-BLAST (Altschul et al., 1997) and aligned using MUSCLE (Edgar, 2004) on SeaView (Gouy et al., 2010). A phylogenetic tree was created using the PhyML server (Guindon et al., 2010) with the default setting and 100 bootstrap replicates, and edited using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Secondary structure predictions were carried out using the JPred webserver.
The crystal structure of zebrafish SAS6 (PDB code: 2Y3W (van Breugel et al., 2011)) was used as a template to model a structure of human SAS6 using Modeller (Sali and Blundell, 1993). This model together with crystal structures of hCCDC61 (from this study), XRCC4, XLF and PAXX (PDB codes: 1IK9 (Sibanda et al., 2001), 2QM4 (Li et al., 2008) and 3WTD (Ochi et al., 2015) respectively) were superposed and aligned using TopMatch (Sippl and Wiederstein, 2012). Alignments were manually adjusted using SeaView.
Constructs and Antibodies
The human CCDC61 gene was codon optimized for E. coli and synthesized (GenScript), that of Danio rerio was purchased from Source BioScience and that of Xenopus laevis was synthesized without codon optimization (Thermo Fisher Scientific). hCCDC61 constructs were PCR amplified and cloned into a pGAT3 (Peränen et al., 1996) or pSKB2LNB vector (a pET28-derived vector resulting in a fusion protein containing a N-terminally, PreScission protease-cleavable His6 tag) (Fekairi et al., 2009) for bacterial expression and pEGFP-C1 or pcDNA3-3xHA for human expression. NES peptides (LQLPPLERLTLD (Wen et al., 1995)) were added to some of gfp-hCCDC61 constructs after short linkers (SGSS) by PCR. The zCCDC61 constructs were cloned into pSKB2LNB or a bacterial-expression plasmid encoding a N-terminal His-tagged lipoyl domain from Bacillus stearothermophilus dihydrolipoamide acetyltransferase for bacterial expression. The xCcdc61 gene was cloned into pENTR-D-TOPO vector to fuse it to RFP in pCS2+ vectors. Plasmids expressing Centrin2 and Clamp were kind gifts from Dr John Wallingford (Park et al., 2008). The Centrin2 gene followed by a bfp gene was cloned into the pCS2+ vector. The plasmid containing the bfp gene was a kind gift from Dr Keith Boyle (MRC LMB). A GFP-nanobody gene was codon optimized for E.coli and synthesized (GenScript) and cloned into pHAT5 vector (Peränen et al., 1996). Site-directed mutagenesis was carried out by PCR using forward and reverse primers encoding mutant amino acids (Table S2).
The ∼4 kb Chlamydomonas VFL3 gene, which includes ∼600 bp upstream of the start codon and ∼700 bp downstream of the stop codon, was amplified by PCR (VFL3-1F and VFL3-7R) using the CloneAmp HiFi Premix (Clontech) and cloned into the pCR2.1-TOPO vector (Thermo Fisher Scientific). No rescue of vfl3-1 was observed with this 8 Kb VFL3-TOPO plasmid (short-VFL3-TOPO), presumably due to short promoter at the 5’ end. Therefore, a ∼3.6 kb fragment upstream of VFL3, which include the 5’ UTR and part of exon 1 of VFL3, was amplified by PCR (CloneAmp HiFi, VFL3-NotI-F and VFL3-NdeI-R) and cloned into the short-VFL3-TOPO plasmid digested with NotI and NdeI by Infusion HD cloning (Clonetech). This plasmid (WT-VFL3-TOPO) contains ∼3.6 kb upstream of the VFL3 start codon and it rescues the vfl3-1 mutant phenotype.
To generate the 3x HA tagged VFL3 plasmids, a HpaI restriction site was introduced in-frame to either exon 7 or exon 9 via overlapping PCR by creating the restriction enzyme site in the primers. For the exon 7-HpaI plasmid, a 1.5 kb fragment was amplified by primers VFL3-13F-AflII and VFL3-13R-HpaI and a 0.2 kb fragment was amplified by primers VFL3-14F-HpaI and VFL3-14R-SalI from the short-VFL3-TOPO plasmid. These two fragments were gel purified and used as templates in a second round of PCR using primers VFL3-13F-AflII and VFL3-14R-SalI for a 1.7 kb fragment. This fragment was digested with AflII and SalI and replaced the corresponding fragment from the short-VFL3-TOPO plasmid. The HA epitope tag was amplified by PCR (exon 7-HpaI-HA-F and R) and cloned into the HpaI digested plasmid via Infusion HD cloning. For the exon 9-HpaI plasmid, a 0.8 kb fragment was amplified by primers VFL3-7F and VFL3-15R-HpaI and a 0.4 kb fragment was amplified by primers VFL3-15F-HpaI and VFL3-3R from the short-VFL3-TOPO plasmid. A second round PCR was used to amplify a 1.2 kb fragment with VFL3-7F and VFL3-3R. This fragment was digested with SalI and PmlI and replaced the corresponding fragment from the short-VFL3-TOPO plasmid. The HA epitope tag was amplified by PCR (exon 9-HpaI-HA-F and R) and cloned into the HpaI digested plasmid via Infusion HD cloning. The ∼3.6 kb upstream fragment described above was then introduced the exon 7-HA and exon 9-HA plasmids respectively to create exon 7-HA-VFL3 and exon 9-HA-VFL3 plasmids used in vfl3-2 rescue. A similar strategy was used to introduce a GFP-tag (Fuhrmann et al., 1999) in-frame to exon 7 of VFL3 at the HpaI site. The UNI2::HA gene was introduced into the vfl3 strains by crosses so that only one integration site was present in all of the strains.
Both FD-VFL3 and 5E-VFL3 plasmids were generated by overlapping PCR. For the FD-VFL3 plasmid, a 0.4 kb fragment was amplified by primers VFL3-8F and VFL3-FD-R and a 0.7 kb fragment was amplified by primers VFL3-FD-F and VFL3-5R from the short-VFL3-TOPO plasmid. These two fragments were gel purified and used as templates in a second round of PCR using primers VFL3-8F and VFL3-5R for a 1.1 kb fragment. This fragment was digested with AflII and BbvCI and replaced the corresponding fragment from the exon 7-HA-VFL3 plasmid. For the 5E-VFL3 plasmid, a 0.6 kb fragment was amplified by VFL3-2F and VFL3-5E-R and a 1.1 kb fragment was amplified by primers VFL3-5E-F and VFL3-6R from the short-VFL3-TOPO plasmid. These two fragments were gel purified and used as templates in a second round of PCR using primers VFL3-2F and VFL3-6R for a 1.7 kb fragment. This fragment was digested with BbvCI and SalI and replaced the corresponding fragment from the exon 7-HA-VFL3 plasmid. All primers used to amplify Chlamydomonas VFL3 are listed in Table S3.
The following primary antibodies were used: ARL13B (Proteintech, 17711-1-AP) 1/500 for immunofluorescent staining (IF), α-tubulin (Sigma-Aldrich, T9026) 1/500 for IF, acetylated α-tubulin antibody (Abcam, ab179484) 1/500 for IF, γ-tubulin (Sigma-Aldrich, T6557) 1/1000 for IF, GFP (Abcam, ab13970 or Thermo Fisher Scientific, 3E6, A11120) 1/2000 or 1/250 for IF, HA (a kind gift from Dr. Manu Hedge or Roche, 3F10, 11867423001) 1/200 for IF or 1/3000 for WB, centrin (a kind gift from Dr. Jeffrey L. Salisbury) 1/250 for IF, and Centrin 3 (Abnova, H00001070-M01) 1/500 for IF. Secondary antibodies used include Alexa-488-conjugated Donkey anti-rabbit (Thermo Fisher Scientific, A21206) 1/2000 for IF, Alexa-488-conjugated Goat anti-chicken (Thermo Fisher Scientific, A11039) 1/2000 for IF, Alexa-488-conjugated goat anti-rat antibody (Invitrogen, API83P) 1/500 for IF, Alexa-488-conjugated goat anti-mouse antibody (Molecular probes, A11001) 1/500 for IF, Alexa-555-conjugated Goat anti-mouse (Thermo Fisher Scientific, A21422) 1/2000 for IF, Alexa 594-conjugated chicken anti-mouse antibody (Invitrogen, A21201) 1/500 for IF, Alexa 594-conjugated goat anti-rabbit antibody (Molecular probes, A11037) 1/500 for IF, and Alexa 647-conjugated goat anti-mouse antibody (Thermo Fisher Scientific, A21235) 1/500 for IF, ATTO 647N-conjugated goat anti-rabbit antibody (Sigma-Aldrich , 40839) 1/2000 for IF and HRP-conjugated mouse anti-rabbit antibody (Santacruz Biotechnology, sc-2357) 1/3000 for WB.
Analysis of Chlamydomonas Transcripts
For Chlamydomonas RNA isolation, ∼5x108 cells were resuspended in 10 ml nitrogen-free medium (M-N/5) for 4 hours at 25°C. The cells were collected at 500 g for 5 minutes at room temperature and the pellet was resuspended in 1 ml of Buffer RLT (reagent from Qiagen RNeasy Mini Kit) with 10 μl of 2-mercaptoethanol. Cells were homogenized by passing through a 20-gauge needle 20 times and centrifuged at 11000 g for 5 minutes at room temperature. The supernatant was collected and RNA extraction was performed with the RNeasy Mini Kit (Qiagen) according to manufacturer’s recommendation. Five micrograms of total RNA from each strain was treated with 5 U of RNase-free DNase I (Thermo Fisher Scientific) in 50 μl reaction at 37°C for 30 minutes. The reaction was terminated by addition of 5 μL 50 mM EDTA and heat inactivation at 65°C for 10 min. One microgram of DNase-treated RNA from each strain were added to SuperScript IV VILO Master Mix (Thermo Fisher Scientific). The reverse transcription reactions were performed according to manufacturer’s recommendation with the reverse transcription temperature set to 60°C.
Protein Purification
Purification of hCCDC611-143 was carried out as follows. BL21(DE3) cells (New England Biolabs) that carried the pGAT3-hCCDC611-143 plasmid were grown in LB media at 37°C till OD600 reached between 0.6-1.0, and the gene expression was induced by 0.5 mM IPTG after cooling the cell cultures to 16°C. The cell cultures were grown at the temperature overnight. Collected cells were suspended in 8 ml of a lysis buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 1 mM EDTA, 5 mM DTT, 1x cOmplete Protease Inhibitor Cocktail EDTA-free (Roche)) per gram of cells. The lysis was carried out by sonication. Cell debris were removed by centrifuging at 32,000 g for 45 min at 4°C. The supernatant after the centrifugation was collected and loaded onto a GSTrap FF 16/10 column (GE Healthcare) equilibrated with the lysis buffer without the protease inhibitor. After washing the column with the lysis buffer, bound molecules were eluted with the same equilibrated buffer but 25 mM reduced L-glutathione. The elution was dialyzed against 5L of 30 mM Tris-HCl pH 8.0 150 mM NaCl, 0.5 mM EDTA, 5 mM 2-mercaptoethanol at 4°C overnight after tev protease was added. The cleaving of the His-GST tag leaves the extra amino-acid sequence Gly-Ser at the N-terminus of hCCDC611-143. The dialyzed sample was further dialyzed against 5L of 30 mM Tris-HCl pH 8.0 at 4°C, 150 mM NaCl, 20 mM imidazole, 2 mM 2-mercaptoethanol at 4°C for four hours. The sample was loaded onto a 5ml HisTrap HP column (GE Healthcare), and the flow through and the first 5ml wash were collected. The collected sample was diluted three-fold with 20 mM Tris-HCl pH 8.0, 2 mM DTT and loaded onto a 5ml HiTrap Q HP column (GE Healthcare) equilibrated with 20 mM Tris-HCl pH 8.0, 50 mM NaCl, 2 mM DTT. The bound molecules were eluted with a linear gradient to 600 mM NaCl. Peak fractions containing hCCDC611-143 were collected and concentrated to 2.5 ml to load onto a PD-10 column (GE Heathcare) equilibrated with 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 2%(v/v) glycerol, 2 mM DTT. Purified hCCDC611-143 was concentrated and stored at -80°C after snap-freezing in liquid nitrogen (Figure S7D). hCCDC611-143; F128E/D129A mutant and SeMet replaced hCCDC611-143 were purified in a similar way.
Purification of zCCDC611-168; F129E/D130A was carried out as follows. The supernatant of crude bacterial extracts containing zCCDC611-168; F129E/D130A was prepared in a similar way to that of hCCDC611-143. However, we used C41 cells (Miroux and Walker, 1996) instead of BL21(DE3) and 50 mM Tris-HCl pH 8.0, 300 mM NaCl, 10 mM imidazole, 2 mM 2-mercaptoethanol, 1 mM AEBSF. 5ml of Ni-NTA resin (Expedion) were added to the extracts and incubated at 4°C for 120 min. The resin was washed with 50 mM Tris-HCl pH 8.0, 300 mM NaCl, 10 mM imidazole, 2 mM 2-mercaptoethanol and the same buffer but 30 mM imidazole. Bound molecules were eluted with the same buffer but 300 mM imidazole. The GST-PreScission protease and EDTA at the final concentration of 0.5 mM were added to the elution, which was dialyzed against 5L of 30 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 2 mM 2-mercaptoethanol at 4°C overnight. The cleaving the His tag leaves the extra amino-acid sequence Gly-Pro-His at the N-terminus of zCCDC611-168; F129E/D130A. 0.5 ml of glutathione sepharose 4B were added to the dialyzed sample and incubated at 4°C for 60 min. The supernatant was collected, diluted three-fold with 20 mM Tris-HCl pH 8.0, 2 mM DTT and loaded onto a 5 ml HiTrap Q HP column equilibrated with 20 mM Tris-HCl pH 8.0, 50 mM NaCl, 2 mM DTT. Bound molecules were eluted with a linear gradient of 400 mM NaCl. Peak fractions containing zCCDC611-168; F129E/D130A were collected. The buffer of the sample was exchanged to 20 mM Tris-HCl pH8.0, 100 mM NaCl, 2 mM DTT by a PD-10 column and the protein stored at -80°C after concentration (Figure S7D).
Construct zCCDC611-170 fused to the C-terminus of a lipoyl-domain tag (Lipo-zCCDC611-170) (for SEC-MALS analysis) and its F129E/D130A mutant (for SEC-MALS analysis) were expressed in E. coli C41 in 2xTY and purified by Ni-NTA (Qiagen) beads using standard methods. Subsequently, eluates were subjected to a size exclusion chromatography step in 10 mM Tris-HCl, pH 8.0, 50 mM NaCl, 2 mM DTT and the purifications finished by ion-exchange chromatography on a HiTrap Q-FF (GE Healthcare) column using a linear salt gradient from 10 mM Tris-HCl, pH 8.0, 2 mM DTT to 10 mM Tris-HCl, pH 8.0, 2 mM DTT, 1 M NaCl. Proteins were concentrated and snap frozen in liquid nitrogen and stored at -80°C. To purify zCCDC611-170 without the lipoyl-domain tag, Lipo-zCCDC611-170 was incubated with the tev protease after the Ni-affinity purification step. Cleaving the tag leaves the extra amino-acid sequence Gly-Gly-Ser at the N-terminus of zCCDC611-170. The zCCDC611-170 solution was loaded onto a HisTrap FF column to remove the tag and tev protease and then loaded onto a HiTrap Q HP column after being diluted to 175 mM NaCl concentration by 20 mM Tris-HCl pH 8.0. The flow through fractions containing zCCDC611-170 were collected and loaded onto a PD-10 buffer exchange column equilibrated with 10 mM Tris-HCl pH8.0, 300 mM NaCl, 2 mM DTT and concentrated before being snap-frozen in liquid nitrogen. Lipo-zCCDC611-170; F129E/D130A used for Figure 2D was purified in a similar way.
Purification of zCCDC61146-280 was carried out as follows. A Ni-NTA affinity purification of zCCDC61146-280 was carried out in a similar way to that of zCCDC611-168; F129E/D130A but using 500 mM NaCl in the purification buffers. GST-PreScission protease and EDTA (at a final concentration of 0.5 mM) were added to the elution. Cleaving the tag leaves the extra amino acid sequence Gly-Pro-His-Asn at the N-terminus of the protein. 1 ml of glutathione sepharose 4B were added to the sample and incubated at 4°C for 60 min. The supernatant was collected and diluted 3-to-5 with 30 mM Tris-HCl pH 8.0, 5 mM DTT. The diluted sample was loaded on to a 5 ml HiTrap Q HP equilibrated with 30 mM Tris-HCl pH8,0, 300 mM NaCl, 5 mM DTT. The flow through was collected and concentrated to ∼3 ml before loading onto a Superdex 75 16/600 column (GE Healthcare) equilibrated with 20 m HEPES pH 7.5, 500 mM NaCl, 2 mM DTT. Fractions containing zCCDC61146-280 were collected and diluted two-fold with 20 mM HEPES pH 7.5 before loading onto a 5 ml HiTrap Heparin HP column equilibrated with 20 m HEPES pH 7.5, 200 mM NaCl, 2 mM DTT. Bound molecules were eluted with a linear gradient of 20 m HEPES pH7.5, 1 M NaCl, 2 mM DTT. Fractions containing zCCDC61146-280 were collected, and the buffer of the protein was exchanged to 20 mM HEPES pH 7.5, 200 mM NaCl, 2 mM DTT using a PD-10 column. The sample was concentrated to a desired concentration and snap frozen in liquid nitrogen before storing at -80°C (Figure S7D). zCCDC61146-280; 5E mutant was purified in a similar way to zCCDC61146-280, but a HiTrap Q HP column was used instead of the HiTrap Heparin HP column. The protein has the extra amino-acid sequence Gly-Pro-His-Asp at its N-terminus.
Purification of hCCDC61288-512 was carried out in a similar manner to zCCDC611-170. hCCDC61288-512 fused to the C-terminus of a lipoyl-domain tag was expressed in E. coli C41. All following steps were carried out at room temperature because hCCDC61288-512 tends to precipitate at 4°C. After the Ni-NTA step, the eluted proteins were loaded onto a 5ml HiTrap Heparin HP column (GE Healthcare) equilibrated with 20 mM HEPES pH 7.5, 300 mM NaCl, 2 mM DTT. Bound proteins were eluted with a linear gradient of 20 mM HEPES pH 7.5, 1 M NaCl, 2 mM DTT. Fractions containing hCCDC61288-512 were collected and the lipoyl tag was cleaved by adding tev protease, which leaves the extra amino-acid sequence Gly-Gly-Ser at the N-terminus of hCCDC61288-512. The cleaved sample was passed onto a 5 ml HisTrap HP column. The flow through was collected and dialyzed against 3L of 20 mM HEPES pH 7.5, 500 mM NaCl, 2 mM DTT. The dialysed sample was concentrated and stored at -80°C after snap frozen in liquid nitrogen (Figure 3C).
To stabilise hCCDC61144-287 and hCCDC61144-287; 5E, both constructs were fused to the C-terminus of residue 1-137 of PAXX (Ochi et al., 2015) and cloned into pSKB2LNB vector. The proteins were expressed and purified using Ni-NTA as described above. Eluted proteins were cleaved with the GST-PreScission protease and dialyzed in 2L of 30 mM Tris-HCl pH 8.0, 2 mM 2-mercaptoethanol overnight supplied with 500 mM NaCl (hCCDC61144-287) and 200 mM NaCl (hCCDC61144-287; 5E) at 4°C. The cleavage leaves the extra amino-acid sequence Gly-Pro-His at the N-terminus of these constructs. As for hCCDC61144-287, the dialyzed sample was diluted two-fold with 20 mM Tris-HCl pH8.0, 2 mM DTT and loaded on to tandemly connected 5 ml GSTrap and HiTrap Q HP equilibrated with 30 mM Tris-HCl pH 8.0, 300 mM NaCl, 2 mM DTT. The flow through was loaded onto a 5 ml HiTrap Heparin HP column equilibrated with 20 mM HEPES pH 7.5, 200 mM NaCl, 2 mM DTT. Bound proteins were eluted with a linear gradient of 20 mM HEPES pH7.5, 1 M NaCl, 2 mM DTT. The fractions containing hCCDC61144-287 were collected and passed onto a PD-10 column equilibrated with 20 mM HEPES pH 7.5, 500 mM NaCl, 2 mM DTT. The purified protein was concentrated and stored at -80°C after snap freezing in liquid nitrogen (Figure 3C). As for hCCDC61144-287; 5E, the dialyzed sample was diluted two-fold with 50 mM HEPES pH 7.5, 5 mM 2-mercaptoethanol and loaded onto a 5 ml GSTrap column. The flow through was loaded onto a 5 ml HiTrap Q HP column equilibrated with 20 mM HEPES pH 7.5, 100 mM NaCl, 2 mM DTT. Bound proteins were eluted with a linear gradient of 20 mM HEPES pH 7.5, 1 M NaCl, 2 mM DTT. The fractions containing hCCDC61144-287; 5E were collected and diluted with 20 mM HEPES pH 7.5, 200 mM NaCl, 2 mM 2-mercaptoethanol. The diluted sample supplied with 20 mM imidazole was loaded onto a 5 ml HisTrap HP column, and the flow through was collected. The protein was concentrated and diluted with 20 mM HEPES pH7.5, 200 mM NaCl, 2 mM DTT. The procedure was repeated three times. Finally, the concentrated sample was stored at -80°C after snap freezing in liquid nitrogen (Figure S4D).
For purification of 15N-labelled human SAS6, DNA encoding human SAS61-143 was cloned into pSKB2LNB vector. This construct was expressed in E. coli Rosetta in minimal medium containing 15NH4Cl and purified by standard methods using Ni-NTA (Qiagen) chromatography. The eluate was dialyzed (in the presence of GST-PreScission protease) against 10 mM Tris-HCl, pH 8.0, 2 mM DTT and further purified by ion-exchange chromatography on a HiTrap Q-FF (GE Healthcare) column using a linear salt gradient from 0 mM to 1 M NaCl in 10 mM Tris-HCl, pH 8.0, 2 mM DTT followed by size exclusion chromatography in 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM DTT (Figure S7D).
GFP nanobody was purified based on a published protocol (Kubala et al., 2010). BL21(DE3) cells that carried the pHAT5-GFP-nanobody plasmid were grown in 6L of LB media at 37°C till OD600 reached between 0.6-1.0, and the gene expression was induced by 1 mM IPTG after cooling the cell cultures to 16°C. The cell cultures were grown at the temperature overnight. Collected cells were suspended in 5 ml of a lysis buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 5%(v/v) glycerol, 0.1%(v/v) NP-40, 10 mM imidazole, 1x cOmplete Protease Inhibitor Cocktail EDTA-free) per gram of cells. The lysis was carried out by sonication. Cell debris were removed by centrifuging at 27,000 g for 30 min at 4°C. The supernatant after the centrifugation was collected and loaded onto 2x 5ml HisTrap HP columns (GE Healthcare) equilibrated with the lysis buffer without the protease inhibitor. The column was washed with 50 mM HEPES pH7.5, 1 M NaCl, 0.2%(v/v) NP-40, 50 mM imidazole and 50 mM HEPES pH7.5, 500 mM NaCl, 50 mM imidazole. The bound molecules were eluted with 50 mM HEPES pH7.5, 500 mM NaCl, 300 mM imidazole. The elution was dialyzed against 5L of 0.2 M NaHCO3 pH8.3, 500 mM NaCl at 4°C overnight. Dialyzed GFP nanobody was concentrated and stored at -80°C after snap-freezing in liquid nitrogen.
Protein Crystallization
SeMet hCCDC611-143 was crystallized at 20°C in a hanging drop containing 10 mg/ml of the protein and 8% (w/v) PEG6,000, 100 mM Tris-HCl pH 7.3 in a 1:1 ratio. zCCDC611-168; F129E/D130A was crystallized at 20°C in a sitting drop containing 10 mg/ml of the protein and 8% (w/v) PEG 6,000, 100 mM Bicine pH 9.0, 3% Trimethylamine N-Oxide in a 1:1 ratio. zCCDC611-170 was crystallized at 20°C in a sitting drop containing 4.62 mg/ml of the protein and 100 mM Citric acid pH 5.0, 1 M LiCl in a 1:1 ratio. Single crystals of the proteins were dipped into cryo-protection solutions, which were 70% reservoir and 30% ethylene glycol, and flash frozen in liquid nitrogen.
X-ray Crystallography
Diffraction images of the crystals were collected at I02 in Diamond Light Source (DLS) for SeMet hCCDC611-143, at MRC LMB using an in-house X-ray diffraction machine (RIGAKU FR-E+ SuperBright) for zCCDC611-168; F129E/D130A and at I03 in DLS for zCCDC611-170. The collected data were indexed and integrated using XDS (Kabsch, 2010) for the SeMet hCCDC611-143 data, iMOSFLM (Battye et al., 2011) for zCCDC611-168;F129E/D130A and zCCDC611-170 data, and scaled using Aimless (Evans, 2011), which were run from CCP4 program suite (Winn et al., 2011). The phenix.autosol module of PHENIX suite (Adams et al., 2010) was used to calculate phases for structure factors of the SeMet hCCDC611-143 data by the SAD method. Phases for structure factors of the zCCDC611-168; F129E/D130A or zCCDC611-170 data were determined by the phenix.phaser module using the structure of SeMet hCCDC611-143 or zCCDC611-168; F129E/D130A as a probe for molecular replacement respectively. The initial structures were build using the phenix.autobuild module. The models were refined manually using Coot (Emsley et al., 2010) and computationally using the phenix.refine module until no further improvements of the map were observed. TLS groups were selected as each chain for hCCDC611-143 and as each chain divided into two groups (from N-terminus to residue 144 and from 145 to the C-terminus) for zCCDC611-168; F129E/D130A and zCCDC611-170. Non-crystallographic symmetry restraints were not applied for the refinement of these structures. For the refinement of zCCDC611-170, E129 and A130 of zCCDC611-168; F129E/D130A were replaced with F129 and D130, and the model was refined as described above. The final structural models were validated using MolProbity (Williams et al., 2018) run from PHENIX suite. All protein-structure graphics were produced using PyMOL (Schrödinger, 2015).
Analytical Ultracentrifugation
hCCDC611-143 and hCCDC611-143; F128E/D129A at approximately 480 μM (7.9 mg/ml) in 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM DTT were subjected to velocity sedimentation at 50,000 rpm at 4°C in an An50Ti rotor using an Optima XL-I analytical ultracentrifuge (Beckmann). The data were analysed in SEDFIT 15.0 (Schuck, 2003) using a c(s) distribution model. The partial-specific volumes (v-bar), solvent density and viscosity were calculated using Sednterp (Dr Thomas Laue, University of New Hampshire). To determine the dissociation for dimerization, Kd, of hCCDC611-143 homodimer, 110 μL with total protein concentrations of 12, 4 and 1.3 mg/ml were loaded in 12 mm 6-sector cells and centrifugated at 11,600, 19,700 and 34,000 rpm at 4°C until equilibrium had been reached. Data were processed and analyzsed using SEDPHAT 13b (Schuck, 2003). Data were plotted with the program GUSSI (Brautigam, 2015).
SEC-MALS
The mass and hydrodynamic radius of CCDC61 constructs in solution was determined using SEC-MALS as described previously (van Breugel et al., 2011). SEC was in 10mM Tris-HCl pH 7.4, 150mM NaCl (1, 6.5 and 65 mg/ml of His6-lipoyl-zCCDC611-170 and 1, 6.8 and 73 mg/ml of its F129E/D130A mutant). SEC used a Superdex S200 10/300 column (GE Healthcare) running at 0.5 ml/min. The concentrations quoted are at loading and these will be at least 10 times lower during chromatography due to dilution on the column. Experiments were performed at room temperature. Since the coiled-coil regions present in His6-lipoyl-zCCDC611-170 and its mutant are very short (∼20 amino acids), coiled-coil dimerization is inefficient, explaining the presence of monomer species in the SEC-MALS runs.
Circular Dichroism (CD)
Purified zCCDC61146-280 and its 5E mutant were dialyzed against 1L of 20 mM sodium phosphate pH7.5, 500 mM NaCl, 1 mM TCEP at 4°C overnight and adjusted to a concentration of 0.375 mg/ml. The CD measurement was done at 5°C. Far-UV CD spectra at 5°C and thermal melts at 222 nm were measured using a Jasco J815 spectropolarimeter (JASCO (UK) Ltd) in 20 mM sodium phosphate pH7.5, 500 mM NaCl, 1 mM TCEP. Following dialysis into this buffer, samples of zCCDC61146-280 and its 5E mutant were diluted to 0.375 mg/ml and measured in a 1 mm pathlength cuvette. Thermal melts were performed at a heating rate of 1C°/min.
NMR
NMR data were collected at 20°C on a Bruker Avance II+ 700 MHz spectrometer, equipped with a cryogenic triple-resonance TCI probe. 2D 1H,15N BEST-Trosy data sets for 56 μM of 15N-labelled hSAS61-143 on its own and in the presence of 48 μM of hCCDC611-143 were acquired in 20 mM Tris pH8.0, 150 mM NaCl and 2 mM DTT. Data were processed using Topspin 3.0 (Bruker) and analyzed using SPARKY (T. D. Goddard and D. G. Kneller – University of California, San Francisco).
Microtubule Pelleting Assay
Taxol-stabilized microtubules were prepared as described on Anthony Hyman’s lab website (http://hymanlab.mpi-cbg.de/hyman_lab/wp-content/uploads/2012/08/Tubulin-Protocols-Mitchison.pdf). All centrifugation steps were carried out in 7 x 20 mm polycarbonate-centrifuge tubes using a TLA100 rotor and Optima TL ultracentrifuge (Beckman Culture). 20 μM Tubulin purified from pig brains (a kind gift of Dr. Andrew Carter, MRC LMB, Cambridge, UK) were polymerized at 37°C in BRB80 (80 mM PIPES pH 6.8, 1 mM MgCl2, 1 mM EGTA) supplied with 1 mM GTP and 1 mM DTT by adding 1/10 volume of 2, 20 and 200 μM of Taxol stepwise. Taxol-stabilized microtubule were pelleted by centrifugation at 70,000 rpm for 12 min at 25°C. The microtubule pellets were resuspended in the reaction buffer (20 mM Tris-HCl pH7.4, 200 mM NaCl, 1 mM DTT, 20 μM Taxol). In order to find an optimal tubulin / subtilisin ratio for removing the tubulin C-termini, we first mixed 2 mg/ml tubulins with a four-fold dilution series of subtilisin A (Sigma-Aldrich) starting from 1:1 weight ratio. This experiment was performed at 37°C for 15 min. The digestion reaction was terminated by adding 10 mM PMSF and incubated at 37°C for 5 min before being centrifuged to pellet microtubules. 20 μM of CCDC61 constructs in the reaction buffer were spun at 70,000 rpm for 12 min at 25°C. 45 μl of the supernatant of each construct were mixed with the equal volume of 20 μM of the stabilized microtubules in reaction buffer. The mixed samples were incubated at RT for 15 min. 85 μl of the mixed samples were centrifuged through a cushion of 50 μl of the reaction buffer supplied with 40%(v/v) glycerol at 70,000 rpm for 30 min at 25°C. The supernatants and pellets were analyzed using SDS-PAGE. Mixed samples before the ultracentrifugation step were also used for negative-stain EM visualization.
Knockout and Knockdown of hCCDC61
Knockout of CCDC61 in RPE-1 PuroKO cells (Balmus et al., 2019) was done using methods and reagents as described before (Chiang et al., 2016). Briefly, two target sites in exon 1 of CCDC61 (5'-GGAAGACGTAGTCCACCTGCAGG-3' and 5'-GGAGCATGCCGTGCGGGTGATGG-3') of Cas9 were designed by CRISPR DESIGN (Hsu et al., 2013). The all-in-one plasmid encoding these sites (AIO-GFP-hCCDC61) was transfected to RPE-1 cells by electroporation using the NEON transfection system (Thermo Fisher). After 48 hours, GFP-positive cells were FACS sorted in three 96-well plates at the Cambridge Stem Cell Institute, University of Cambridge. Cells were incubated at 37°C in 5% CO2 about a month till they became confluent. Genomic DNAs of the cells were extracted using QuickExtract DNA extraction solution (Cambio) and subjected to PCR using two primers (5'-TTCCAGGGTTCCATGGGTCTAGGTTTCTCTCTCATCTCCTT
-3' and 5'-CGAGGTCGACGAATTCGGCACACTCACAGCCAGCATCGAA
-3'). The PCR products were cloned into a pHAT4 (Peränen et al., 1996) vector to be sequenced. Two clones that had inserts causing premature stop codons in both alleles of the exon (Figure S5A) were selected for further studies. For counting of the centriole number, parental and CCDC61 knockout cell lines were treated with 100 μM monastrol (Sigma-Aldrich) for 4 hours. For ciliation assay, cells were serum starved in media containing 0.5% FBS for 24 and 48 hours. To determine proliferation kinetics, cells were seeded in 12 well plate at 5x103 cells/well and real-time quantitative live cell analysis was carried out for 96 hours using IncuCyte ZOOM (Essen BioScience), imaging 9 positions per well every 3 hours.
Knockdown of CCDC61 was carried out by transfecting three different Silencer Select siRNAs (siRNA IDs: s59736 as siRNA 1, s59737 as siRNA 2 and s59738 as siRNA 3) (Life Technologies) or (Ambion control siRNA) to RPE-1 cells. Briefly, RPE-1 cells were transfected with Lipofectamine RNAiMAX reagent (Thermo Fischer Scientific). The siRNAs were used at a final concentration of 60 nM and the siRNA treatments were carried out for 72 hours after transfection. To assess ciliation, 48 hours after siRNA transfection RPE-1 cells were serum starved in media containing 0.5% FBS for 24 hours. Knockdown efficiencies were assessed by reverse transcription PCR (RT-PCR) using a forward primer (5'-TGCAGCGATTTGGAGGATTT-3') and a reverse primer (5'-CGGAGTTGGCCAGAGATTTC-3').
Fluorescent and Immunofluorescent Microscopy
GFP-hCCDC61 constructs were transfected to RPE-1 cells using Lipofectamine 3000 (Thermo Fisher Scientific) by mixing 500 ng of each pEGFP-C1-hCCDC61 construct with 1 μl of P3000 reagent and 1 μl of Lipofectamine 3000 reagent in Opti-MEM (Thermo Fisher Scientific) before adding the mixture to the cells grown on a coverslip in a well of a 24-well plate. After 24 hours of transfection, cells were fixed with 4% (w/v) formaldehyde for fluorescent microscopy or cold methanol and stained with antibodies for immunofluorescent microscopy as indicated in the main text. ProLong Diamond Antifade Mountant (Thermo Fisher Scientific) was used as a mounting media. Confocal images of fixed cells were taken using a Confocal White Light Laser (WLL) Leica TCS SP8 Microscope with the HC Plan Apo 40x/1.30 63x/1.40 or 100 x/1.40 OIL (CS2) objective or a Zeiss LSM880 AxioObserver with Plan-Apochromat 40x/1.40. Image acquisition was carried out with the Leica Application Suite X (LAS X) software (Leica Microsystems) or Zen software (Zeiss). Wide-field images of fixed cells (Figure 4C) were taken using the Nikon Eclipse TE2000 Inverted Microscope with the Plan Apo VC 60 x or 100 x/1.40 OIL objective. After acquisition, images were imported into Fiji (Schindelin et al., 2012) to obtain maximum intensity projections of entire z-stacks. Fiji and Photoshop (Adobe, 2017) were used to perform level adjustment. The immunofluorescent image shown in Figure 4A was generated by deconvoluting the original image using Huygens Professional (Scientific Volume Imaging). For Chlamydomonas immunofluorescence, ∼107 Chlamydomonas cells were first resuspended in 0.5 ml M-N/5 medium for 4 hours to allow flagellar assembly. Cells were then treated with autolysin for 30 min at 25°C to remove cell walls, followed by resuspension in 1 ml MTSB (microtubule stabilization buffer, 30 mM HEPES, pH 7.4, 5 mM MgSO4, 15 mM KCl, 2 mM EGTA) at room temperature. Fifty microliters of cells were applied to a 0.1% poly-L-Lysine (Sigma-Aldrich) coated well on a multi-well slide (Thermo Fisher Scientific) for 2 minutes in the dark. Excess cell suspension was removed by pipetting. Fifty microliters of lysis buffer (MTSB + 1% NP-40) was added to each well to lyse the cells for 2 minutes at room temperature. MTSB was used to wash individual wells once and removed by pipetting. Samples were fixed with MTSB + 4% paraformaldehyde for 30 minutes at room temperature. Excess liquid was removed by pipetting before slides were submerged in cold methanol (−20°C) for 2 × 5 min and left to dry at room temperature. The remained nucleo-flagellar apparatuses attached to the wells were rehydrated with the addition of PBS (phosphate-buffered saline) for 10 minutes at room temperature. After rehydration, the samples were blocked with 100% blocking buffer (5% BSA and 1% fish gelatin in PBS) for 1 hour at room temperature, followed by inoculation with primary antibodies (diluted with 20% blocking buffer) at 4°C overnight. The samples were washed six times with 20% blocking buffer, followed by 1 hour inoculation at room temperature with secondary antibodies diluted with 20% blocking buffer. The samples were washed six times with 20% blocking buffer and mounted in Fluoromount-G (Southern Biotech). The images were captured with an UltraVIEW VoX laser spinning disk confocal microscope (PerkinElmer) and acquired by Volocity software (PerkinElmer).
Live Cell Imaging
RPE-1 cells were grown on a chambered cover glass (Grace Bio-Labs) in D-MEM/F-12 without phenol red (Thermo Fisher Scientific) supplied with 10% FBS, and 100 units of penicillin and 100 μg/ml of streptomycin and transfected with GFP-hCCDC61 using Lipofectamine 3000 (Thermo Fisher Scientific). After 24 hours, the media were replaced with the same media with 0.1% (v/v) DMSO or with 5 μg/ml nocodazole, and confocal fluorescent images of GFP positive RPE-1 cells were taken using a Zeiss LSM880 AxioObserver with Plan-Apochromat 40x/1.40, maintained at 37°C, at time points 0, 60, 120, and 180 min.
GFP Pulldown Assays
HEK293T cells were grown nearly confluent in 10 cm dishes containing D-MEM glutaMAX (Thremo Fisher Scientific) supplied with 10% FBS. A GFP construct (pEGFP-C1, pEGFP-C1-hCCDC611-457; F128E/D129A or pEGFP-C1-hCCDC611-457; F128E/D129A/5E) and HA construct (pcDNA3-3xHA-hCCDC611-457; F128E/D129A or pcDNA3-3xHA-hCCDC611-457; F128E/D129A/5E) were co-transfected into the cells using PEI (Polysciences) as indicated in Figure S4E. The cells were collected after 24 hours of the transfection. GFP-affinity resins were prepared by conjugating purified GFP nanobody to NHS-activated resins (GE Healthcare). The cells were lysed in 1 ml of 50 mM Tris pH 7.4, 200 mM NaCl, 0.2 %(v/v) NP-40, 10%(v/v) glycerol, 1x cOmplete Protease Inhibitor Cocktail EDTA-free on ice for 30 min. Debris was removed by centrifuging at 21,000 g, 4°C for 30 min. 30 μl of the GFP affinity resins were added to the supernatant and incubated on a rotating disk at 4°C for 90 min. The resins were washed five times with the lysis buffer and mixed with 30 μl of 2x SDS sample buffer. Eluted proteins were separated by SDS-PAGE and subject to western blot.
Xenopus Embryos
Xenopus Laevis embryos culture and injection were carried out as described (Hörmanseder et al., 2017). xCCDC61-RFP (0.1ng), Centrin2-BFP (0.25ng) and Clamp-GFP (0.25ng) mRNAs were injected in one cell stage embryos. Following injection embryos were cultured at 14°C to the tailbud stage (stage 27/28 (Faber and Nieuwkoop, 1994)). Embryos were then fixed for 15 min in MEMFA (100mM MOPS PH6.8, 2mM EGTA, 1mM MgSO4, 4% formaldehyde), washed 3X in 0.1X MBS (MBS (Barth-Hepes Saline) 10X stock : 88 mM NaCl, 1 mM KCl, 2,4 mM NaHCO3, 0.82 mM MgSO4, 0.33 mM Ca(NO3)2, 0.41 mM CaCl2, 10 mM HEPES pH 7.4-7.6), and equilibrated overnight at 4°C in 0.1X MBS 50% glycerol. The fixed whole embryos were mounted onto glass slides with ProLong Diamond Antifade Mountant (Thermo Fisher Scientific) and sandwiched with coverslips using a few layers of electrical tape as spacers (Werner and Mitchell, 2013) for confocal imaging.
FACS
The day before FACS experiments, 0.5x106 RPE-1 cells were seeded onto each well of a 6-well plate containing 2 ml of the RPE-1 growth media per well. On the next day, Hoechst 33342 (EMP Biotech) was added to the media at the final concentration of 2 μM and incubated at 37°C with 5% CO2 for 60 min before the cells were trypsinized and pelleted. The cells were re-suspended in 500 μl of PBS and analysed by flow cytometry using an iCyt EC800 cell analyser (Sony Biotechnology). The resulting cell-cycle distribution of cell singlets was determined using FCS EXPRESS 6 Flow software (De Novo Software).
Electron Microscopy
For negative staining, 3 μl of sample were applied onto a 400-mesh carbon-coated copper grid (EMS) that was glow discharged and incubated for 1 min at room temperature. After blotting onto filter paper, the grid was washed twice with 5 μl of water and stained with 5 μl of 2%(w/v) uranium acetate for 1 min. The grid was then blotted onto filter paper and air dried. Micrographs were collected using a Tecnai T12 (FEI) operated at 120 kV and equipped with an Orius SC200 or Ultrascan 1000 XP CCD detector (Gatan). Widths of microtubules were measured using Fiji.
Quantification and Statistical Analysis
We calculated average and standard deviation values using AVERAGE and STDEV functions in Microsoft Excel for Figures 3A, 3E, 4C, S4B, S5B, and S5D, and in GraphPad Prism for Figure S5E. For Figure 5A, χ2 analysis was performed using the website (https://wwwsocscistatistics.com/tests/chisquare/). The number of cells with no cilia and the total number of cells were used in pairwise calculations. Sample sizes n are provided in figure legends. For the statistical analysis of X-ray crystallography data, details are provided above.
Acknowledgments
T.O. is supported by a University Academic Fellow start-up fund from the University of Leeds. M.V.B. is supported by the Medical Research Council (MRC file reference MC_UP_1201/3). I.R.S. is supported by an MRC LMB César Milstein Fellowship. F.G. is supported by Cancer Research UK (C14303/A17043). F.G. acknowledges support from NIHR Cambridge Biomedical Research Centre, the University of Cambridge, and Hutchison Whampoa. S.K.D receives funds from the National Institute of General Medical Sciences (GM-032843 and GM-131909). T.L.B. is supported by Wellcome Trust program grant (09316/Z/10/Z) and Investigator Award (200814/Z/16/Z) for this research. Research in the S.P.J. laboratory is funded by Cancer Research UK (program grant C6/A18796) and a Wellcome Trust Investigator Award (206388/Z/17/Z). A.N.B. is supported by a Cancer Research UK Career Development Fellowship (C29215/A20772). J.J. is funded by a grant from the Wellcome Trust (101050/Z/13/Z) and the MRC (MR/P00479/1). Y.K. was supported by Cancer Research UK (CRUK-A12874) and is currently supported by ShanghaiTech University. R.E.G. is supported by Wellcome Trust Investigator Grant (207510/Z/17/Z). We thank Dr. Francesco Meghini for contributing to the initial characterization of CDCC61 in human cells, Dr. Andy Riddell (Wellcome - MRC Cambridge Stem Cell Institute, Cambridge, UK), Maria Daly and Dr. Fan Zhang (MRC LMB, Cambridge, UK) for FACS, Dr. Mark Skehel (MRC LMB) for mass spectrometry, Drs Jonathan Howe (MRC LMB), Mathias Pasche (MRC LMB), and Ruth Hughes (University of Leeds) for confocal microscopes, Dr. Minmin Yu (MRC LMB) for help with X-ray crystallography, Drs Shaoxia Chen, Christos Savva, and Guiseppe Cannone (MRC LMB) for help with electron microscopy, Mrs Jake Grimmett and Toby Darling (MRC LMB) for help with scientific computing, DLS I02 and I03 beamline scientists for help with collecting X-ray diffraction images, Dr. Andrew Carter (MRC LMB) for tubulin, Dr. Manu Hedge (MRC LMB) for the anti-HA antibody, Dr. Keith Boyle (MRC LMB) for a BFP-containing plasmid, Dr. Antonina Andreeva (MRC LMB) for help with the structure-guided sequence alignment, Dr. Jeffrey L. Salisbury (Mayo Clinic College of Medicine, Rochester, USA) for centrin antibodies, Dr. John Kilmartin (MRC LMB) for Rosetta (DE3) cells and Prof. Colin A. Johnson (University of Leeds) for the human RPE-1 cell line.
Author Contributions
T.O. performed computational analysis with T.L.B., X-ray crystallography with D.D.B. and M.v.B., electron microscopy, biochemistry experiments with M.v.B., gene knockout and transient expression. V.Q. performed characterizations of knockout and knockdown human cells. H.L. performed Chlamydomonas experiments. J.J. performed Xenopus experiments. I.R.S. performed FACS experiments. F.B. performed light microscopy experiments of Xenopus embryos. D.D.B. determined one of the crystal structures together with T.O. C.M.J and S.H.McL. performed biophysics experiments. S.M.V.F. performed NMR experiments. A.N.B. and Y.K. performed light microscopy experiments of human cells. S.P.J. provided resources for the knockout experiments. T.O. and M.v.B. conceptualized the project. T.O., V.Q., H.L., S.K.D., F.G., and M.v.B. conceived experiments and wrote the original draft of the manuscript. Everyone contributed for reviewing and editing the manuscript. Y.K., R.E.G., S.P.J., T.L.B., S.K.D., F.G., and M.v.B. secured funding.
Declaration of Interests
The authors declare no competing interests.
Published: May 5, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.str.2020.04.010.
Contributor Information
Takashi Ochi, Email: T.Ochi@leeds.ac.uk.
Mark van Breugel, Email: m.vanbreugel@qmul.ac.uk.
Supplemental Information
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Kunstleve R.W.G. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahnesorg P., Smith P., Jackson S.P. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Altschul S., Madden T., Schäffer A., Zhang J., Zhang Z., Miller W., Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Arquint C., Gabryjonczyk A.-M., Nigg E. Centrosomes as signalling centres. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130464. doi: 10.1098/rstb.2013.0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J., Basquin C. Basal bodies across eukaryotes series: basal bodies in the freshwater planarian Schmidtea mediterranea. Cilia. 2016;5 doi: 10.1186/s13630-016-0037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J., Wong M., Downhour D., Alvarado A., Marshall W. Centrosome loss in the evolution of planarians. Science. 2012;335:461–463. doi: 10.1126/science.1214457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmus G., Pilger D., Coates J., Demir M., Sczaniecka-Clift M., Barros A.C., Woods M., Fu B., Yang F., Chen E. ATM orchestrates the DNA-damage response to counter toxic non-homologous end-joining at broken replication forks. Nat. Commun. 2019;10:87. doi: 10.1038/s41467-018-07729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bärenz F., Kschonsak Y.T., Meyer A., Jafarpour A., Lorenz H., Hoffmann I. Ccdc61 controls centrosomal localization of Cep170 and is required for spindle assembly and symmetry. Mol. Biol. Cell. 2018;29:3105–3118. doi: 10.1091/mbc.E18-02-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basquin C., Ershov D., Gaudin N., Vu H.T.-K., Louis B., Papon J.-F., Orfila A.-M., Mansour S., Rink J.C., Azimzadeh J. Emergence of a bilaterally symmetric pattern from chiral components in the planarian epidermis. Dev. Cell. 2019;51:516–525.e5. doi: 10.1016/j.devcel.2019.10.021. [DOI] [PubMed] [Google Scholar]
- Battye G.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D. Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M., Cubizolles F., Schmidt A., Nigg E. Quantitative analysis of human centrosome architecture by targeted proteomics and fluorescence imaging. EMBO J. 2016:e201694462. doi: 10.15252/embj.201694462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengueddach H., Lemullois M., Aubusson-Fleury A., Koll F. Basal body positioning and anchoring in the multiciliated cell Paramecium tetraurelia: roles of OFD1 and VFL3. Cilia. 2017;6 doi: 10.1186/s13630-017-0050-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam C. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 2015;562:109–133. doi: 10.1016/bs.mie.2015.05.001. [DOI] [PubMed] [Google Scholar]
- van Breugel M., Hirono M., Andreeva A., Yanagisawa H., Yamaguchi S., Nakazawa Y., Morgner N., Petrovich M., Ebong I.-O., Robinson C.V. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- van Breugel M., Wilcken R., McLaughlin S., Rutherford T., Johnson C. Structure of the SAS-6 cartwheel hub from Leishmania major. eLife. 2014;3:e01812. doi: 10.7554/eLife.01812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck D., Malivert L., de Chasseval R., Barraud A., Fondaneche M.-C., Sanal O., Plebani A., Stephan J.-L., Hufnagel M., le Deist F. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Chiang T.-W., le Sage C., Larrieu D., Demir M., Jackson S. CRISPR-Cas9D10A nickase-based genotypic and phenotypic screening to enhance genome editing. Sci. Rep. 2016;6:24356. doi: 10.1038/srep24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C., Barber J.D., Barton G.J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J., Wordeman L. The diffusive interaction of microtubule binding proteins. Curr. Opin. Cell Biol. 2009;21:68–73. doi: 10.1016/j.ceb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee M., Muschalik N., Johnson S., Leveson J., Raff J., Lea S., Hyman A. The homo-oligomerisation of both Sas-6 and Ana2 is required for efficient centriole assembly in flies. eLife. 2015:e07236. doi: 10.7554/eLife.07236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craxton A., Somers J., Munnur D., Jukes-Jones R., Cain K., Malewicz M. XLS (c9orf142) is a new component of mammalian DNA double-stranded break repair. Cell Death Differ. 2015;22:890–897. doi: 10.1038/cdd.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S.K., Morrissette N.S., Preble A.M., Rackley C., Stanga J. Epsilon-tubulin is an essential component of the centriole. Mol. Biol. Cell. 2002;13:3859–3869. doi: 10.1091/mbc.E02-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S.K., O’Toole E.T. The basal bodies of Chlamydomonas reinhardtii. Cilia. 2016;5:18. doi: 10.1186/s13630-016-0039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S.K., Trabuco E.C. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell. 1998;9:1293–1308. doi: 10.1091/mbc.9.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A., Cole C., Procter J., Barton G.J. JPred4: a protein secondary structure prediction server. Nucleic. Acids Res. 2015;43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev N., Tzolovsky G., Lipinszki Z., Schneider S., Lattao R., Fu J., Debski J., Dadlez M., Glover D. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 2014;24:2526–2532. doi: 10.1016/j.cub.2014.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D. Biol. Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber J., Nieuwkoop P.D. Garland Publishing Inc.; 1994. Normal Table of Xenopus laevis (Daudin) [Google Scholar]
- Farina F., Gaillard J., Guerin C., Coute Y., Sillibourne J., Blanchoin L., Thery M. The centrosome is an actin-organizing centre. Nat. Cell Biol. 2016;18:65–75. doi: 10.1038/ncb3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S., Scaglione S., Chahwan C., Taylor E.R., Tissier A., Coulon S., Dong M.-Q., Ruse C., Yates J.R., III, Russell P. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M., Oertel W., Hegemann P. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 1999;19:353–361. doi: 10.1046/j.1365-313x.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- Glaser F., Pupko T., Paz I., Bell R., Bechor-Shental D., Martz E., Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hilbert M., Erat M.C., Hachet V., Guichard P., Blank I.D., Flückiger I., Slater L., Lowe E.D., Hatzopoulos G.N., Steinmetz M.O. Caenorhabditis elegans centriolar protein SAS-6 forms a spiral that is consistent with imparting a ninefold symmetry. Proc. Natl. Acad. Sci. U S A. 2013;110:11373–11378. doi: 10.1073/pnas.1302721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M., Scheumann N., Wickstead B., Langdale J., Gull K. Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 2010;123:1407–1413. doi: 10.1242/jcs.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops H.J., Wright R.L., Jarvik J.W., Witman G.B. Flagellar waveform and rotational orientation in a Chlamydomonas mutant lacking normal striated fibers. J. Cell Biol. 1984;98:818–824. doi: 10.1083/jcb.98.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori A., Toda T. Regulation of centriolar satellite integrity and its physiology. Cell. Mol. Life Sci. 2016:1–17. doi: 10.1007/s00018-016-2315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörmanseder E., Simeone A., Allen G., Bradshaw C., Figlmüller M., Gurdon J., Jullien J. H3K4 methylation-dependent memory of somatic cell identity inhibits reprogramming and development of nuclear transfer embryos. Cell Stem Cell. 2017;21:135–143. doi: 10.1016/j.stem.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P., Scott D., Weinstein J., Ran A., Konermann S., Agarwala V., Li Y., Fine E., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotech. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L.C., Wemmer K.A., Marshall W.F. Influence of centriole number on mitotic spindle length and symmetry. Cytoskeleton. 2010;67:504–518. doi: 10.1002/cm.20462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Khan M.A., Rupp V.M., Orpinell M., Hussain M.S., Altmüller J., Steinmetz M.O., Enzinger C., Thiele H., Höhne W., Nürnberg G. A missense mutation in the PISA domain of HsSAS-6 causes autosomal recessive primary microcephaly in a large consanguineous Pakistani family. Hum. Mol. Genet. 2014;23:5940–5949. doi: 10.1093/hmg/ddu318. [DOI] [PubMed] [Google Scholar]
- Kitagawa D., Vakonakis I., Olieric N., Hilbert M., Keller D., Olieric V., Bortfeld M., Erat M.C., Flückiger I., Gönczy P. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubala M., Kovtun O., Alexandrov K., Collins B. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 2010;19:2389–2401. doi: 10.1002/pro.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gönczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Li X., Zhang R., Patena W., Gang S., Blum S., Ivanova N., Yue R., Robertson J., Lefebvre P., Fitz-Gibbon S. An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell. 2016;28:367–387. doi: 10.1105/tpc.15.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chirgadze D.Y., Bolanos-Garcia V.M., Sibanda B.L., Davies O.R., Ahnesorg P., Jackson S.P., Blundell T.L. Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ. EMBO J. 2008;27:290–300. doi: 10.1038/sj.emboj.7601942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Otevrel T., Gao Y., Cheng H.-L., Seed B., Stamato T.D., Taccioli G.E., Alt F.W. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- Lin Y., Chang C., Hsu W., Tang C., Lin Y., Chou E., Wu C., Tang T. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J. 2013;32:1141–1154. doi: 10.1038/emboj.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W.F., Vucica Y., Rosenbaum J.L. Kinetics and regulation of de novo centriole assembly. Curr. Biol. 2001;11:308–317. doi: 10.1016/s0960-9822(01)00094-x. [DOI] [PubMed] [Google Scholar]
- Matsuura K., Lefebvre P.A., Kamiya R., Hirono M. Bld10p, a novel protein essential for basal body assembly in Chlamydomonas. J. Cell Biol. 2004;165:663–671. doi: 10.1083/jcb.200402022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux B., Walker J.E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Murray J.E., Van Der Burg M., Ijspeert H., Carroll P., Wu Q., Ochi T., Leitch A., Miller E.S., Kysela B., Jawad A. Mutations in the NHEJ component XRCC4 cause primordial dwarfism. Am. J. Hum. Genet. 2015;96:412–424. doi: 10.1016/j.ajhg.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., Raff J.W. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Ochi T., Blackford A.N., Coates J., Jhujh S., Mehmood S., Tamura N., Travers J., Wu Q., Draviam V.M., Robinson C.V. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015;347:185–188. doi: 10.1126/science.1261971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi T., Wu Q., Blundell T.L. The spatial organization of non-homologous end joining: from bridging to end joining. DNA Repair. 2014;17:98–109. doi: 10.1016/j.dnarep.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Ashikawa T., Nozaki Y., Kozuka-Hata H., Goto H., Inagaki M., Oyama M., Kitagawa D. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 2014;5:5267. doi: 10.1038/ncomms6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T., Mitchell B., Abitua P., Kintner C., Wallingford J. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat. Genet. 2008;40:871–879. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peränen J., Rikkonen M., Hyvönen M., Kääriäinen L. T7 vectors with a modified T7lacPromoter for expression of proteins in Escherichia coli. Anal. Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- Piasecki B.P., Silflow C.D. The UNI1 and UNI2 genes function in the transition of triplet to doublet microtubules between the centriole and cilium in Chlamydomonas. Mol. Biol. Cell. 2009;20:368–378. doi: 10.1091/mbc.E08-09-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizon V., Gaudin N., Poteau M., Cifuentes-Diaz C., Demdou R., Heyer V., Reina San Martin B., Azimzadeh J. hVFL3/CCDC61 is a component of mother centriole subdistal appendages required for centrosome cohesion and positioning. Biol. Cell. 2020;112:22–37. doi: 10.1111/boc.201900038. [DOI] [PubMed] [Google Scholar]
- Reid T.A., Coombes C., Gardner M.K. Manipulation and quantification of microtubule lattice integrity. Biol. Open. 2017;6:1245–1256. doi: 10.1242/bio.025320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger . Schrödinger LLC; 2015. The PyMOL Molecular Graphics System, Version 1.8. [Google Scholar]
- Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- Serrano L., de la Torre J., Maccioni R.B., Avila J. Involvement of the carboxyl-terminal domain of tubulin in the regulation of its assembly. Proc. Natl. Acad. Sci. U S A. 1984;81:5989–5993. doi: 10.1073/pnas.81.19.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata K., Miura M., Watanabe Y., Saito K., Nishimura A., Furuta K., Toyoshima Y.Y. A single protofilament is sufficient to support unidirectional walking of dynein and kinesin. PLoS One. 2012;7:e42990. doi: 10.1371/journal.pone.0042990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda B.L., Critchlow S.E., Begun J., Pei X.Y., Jackson S.P., Blundell T.L., Pellegrini L. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat. Struct. Mol. Biol. 2001;8:1015–1019. doi: 10.1038/nsb725. [DOI] [PubMed] [Google Scholar]
- Sippl M., Wiederstein M. Detection of spatial correlations in protein structures and molecular complexes. Structure. 2012;20:718–728. doi: 10.1016/j.str.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J., Biegert A., Lupas A. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N., Meunier A. The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol. 2017;18:423–436. doi: 10.1038/nrm.2017.21. [DOI] [PubMed] [Google Scholar]
- Stinchcombe J., Griffiths G. Communication, the centrosome and the immunological synapse. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130463. doi: 10.1098/rstb.2013.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs J.L., Vladar E.K., Axelrod J.D., Kintner C. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat. Cell Biol. 2012;14:140–147. doi: 10.1038/ncb2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan K.Y., Goldstein R.E. Coordinated beating of algal flagella is mediated by basal coupling. Proc. Natl. Acad. Sci. U S A. 2016;113:E2784–E2793. doi: 10.1073/pnas.1518527113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Kong D., Hoerner C., Loncarek J., Stearns T. Centriole triplet microtubules are required for stable centriole formation and inheritance in human cells. eLife. 2017;6:e29061. doi: 10.7554/eLife.29061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Meinkotht J., Tsien R., Taylor S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Werner M., Mitchell B. Using Xenopus skin to study cilia development and function. Methods Enzymol. 2013;525:191–217. doi: 10.1016/B978-0-12-397944-5.00010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff J., Wueseke O., Hyman A. Pericentriolar material structure and dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130459. doi: 10.1098/rstb.2013.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R.L., Chojnacki B., Jarvik J.W. Abnormal basal-body number, location, and orientation in a striated fiber-defective mutant of Chlamydomonas reinhardtii. J. Cell Biol. 1983;96:1697–1707. doi: 10.1083/jcb.96.6.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R.L., Salisbury J., Jarvik J.W. A nucleus-basal body connector in Chlamydomonas reinhardtii that may function in basal body localization or segregation. J. Cell Biol. 1985;101:1903–1912. doi: 10.1083/jcb.101.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Ochi T., Matak-Vinkovic D., Robinson C.V., Chirgadze D.Y., Blundell T.L. Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions. Biochem. Soc. Trans. 2011;39:1387–1392. doi: 10.1042/BST0391387. [DOI] [PubMed] [Google Scholar]
- Xing M., Yang M., Huo W., Feng F., Wei L., Jiang W., Ning S., Yan Z., Li W., Wang Q. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 2015;6:6233. doi: 10.1038/ncomms7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cells were stained with acetylated α-tubulin (red) for cilia and rootlet microtubules, anti-HA (green) for UNI2, and anti-GFP (magenta) for VFL3. z stacked images start from the distal ends of basal bodies (labeled by positions of UNI2) and move toward the proximal ends. Scale bar, 0.8 μm.
Data Availability Statement
Coordinates and structure factors of crystal structures that are presented in this paper are available in the Protein Data Bank (PDB codes: 6HXT (hCCDC611-143), 6HXV (zCCDC611-168; F129E/D130A) and 6HXY (zCCDC611-170)).