Abstract
Summary
High-throughput screening (HTS) enables systematic testing of thousands of chemical compounds for potential use as investigational and therapeutic agents. HTS experiments are often conducted in multi-well plates that inherently bear technical and experimental sources of error. Thus, HTS data processing requires the use of robust quality control procedures before analysis and interpretation. Here, we have implemented an open-source analysis application, Breeze, an integrated quality control and data analysis application for HTS data. Furthermore, Breeze enables a reliable way to identify individual drug sensitivity and resistance patterns in cell lines or patient-derived samples for functional precision medicine applications. The Breeze application provides a complete solution for data quality assessment, dose–response curve fitting and quantification of the drug responses along with interactive visualization of the results.
Availability and implementation
The Breeze application with video tutorial and technical documentation is accessible at https://breeze.fimm.fi; the R source code is publicly available at https://github.com/potdarswapnil/Breeze under GNU General Public License v3.0.
Contact
swapnil.potdar@helsinki.fi
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Advances in automated liquid dispensing, assay miniaturization, systems biology as well as ex vivo disease models have boosted the development of high-throughput cell-based functional testing platforms. Thousands of conventional cell lines have been screened with hundreds of compounds in several large-scale projects, such as the Cancer Cell Line Encyclopedia (Barretina et al., 2012), Genomics of Drug Sensitivity in Cancer (Yang et al., 2013), Cancer Therapeutics Response Portal (Seashore-Ludlow et al., 2015) and Genentech Cell Line Screening Initiative (Haverty et al., 2016). Similar drug testing efforts have been applied on primary cell models to generate individualized drug profiles for drug repurposing, patient stratification and for the identification of potential drug combinations (Kodack et al., 2017; Lee et al., 2018; Pemovska et al., 2013, 2015; Saeed et al., 2017). One common end-point in cell-based drug testing is cell viability and/or toxicity readouts generated over multiple concentrations in microwell plates (96-, 384- and 1536-well formats), where plate layout and placement of controls play an important role to minimize the risk of experimental errors influencing data quality (Mpindi et al., 2015). Therefore, quality control (QC) process is required to ensure compliant and reproducible drug testing readouts from the assay. The dose–response curve-fitting process enables direct translation of the raw cell viability measurements based on several intensity scoring technologies to clinically interpretable dose values. The fitted dose–response curve is then used to summarize and quantify the observed response into a single metric, such as IC50 and EC50 dose or as an absolute area under the curve (AUC) or drug sensitivity score (DSS; Yadav et al., 2015). To date, there are several, freely available analysis tools available for handling the drug screening data, such as cellHTS (https://www.dkfz.de/signaling/cellHTS/) (Boutros et al., 2006), HTS Navigator (Fourches et al., 2014), Knime (https://knime.com), PharmacoGx (Smirnov et al., 2016) as well as commercial solutions such as Dotmatics (https://dotmatics.com) and Genedata Screener (https://genedata.com). Although these are useful resources for drug testing data analysis, we believe there is a room for improvement in the alignment of the data flow process through QC, dose–response curve fitting, multiparametric scoring and interactive visualizations. Breeze is an easy to use publicly available tool, which includes comprehensive plate QC statistics with a diverse collection of drug quantification metrics and interactive visualization options, which offers the users the ability to perform comparison of drug response profiles across multiple samples (Fig. 1). Systematic use of standardized data quality processing and drug quantification methods enables direct comparison of responses from a large number of studies (Mpindi et al., 2016).
Fig. 1.
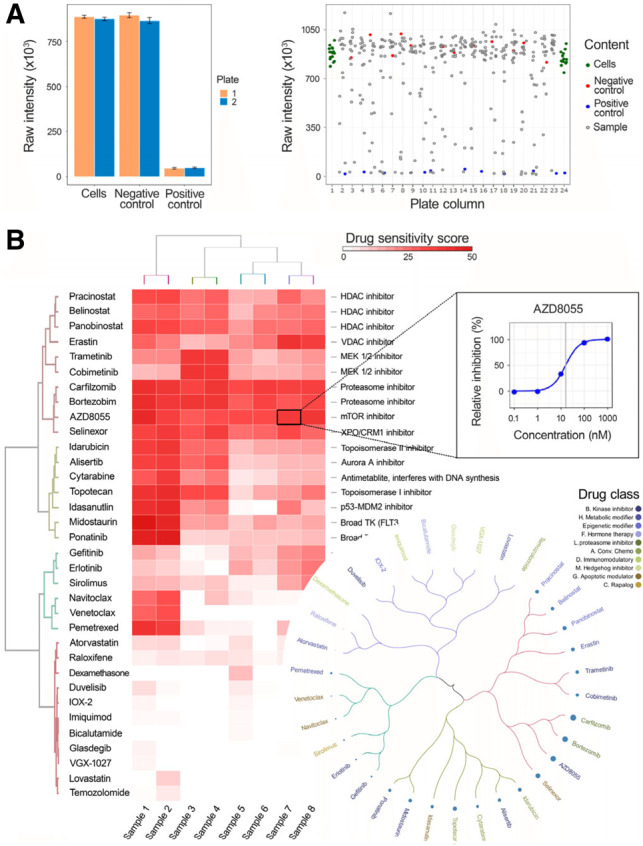
Example outcome plots of Breeze application. (A) The technical quality is visualized by listing QC metrics such as Z’ and SSMD, along with bar plots, plate heat maps and scatterplot of signal intensities of plate wells help in identifying distribution of controls and pinpoint outliers. (B) Drug quantification visualization in form of interactive heatmap and circular plots. The heatmap shows efficacies of the drugs (higher in red and lower in white) across different samples along with drug names and potential drug targets. Hovering over the heatmap shows individual dose–response curves of specific drugs
2 Materials and methods
2.1 Data submission and processing
The Breeze application allows multiple data input formats, which need to comprise of drug names, concentration ranges and phenotypic measurements. These measurements can both be provided as raw data or pre-calculated percent inhibition (PI) values. In case, when the raw data are provided, PI for each data point is calculated based on the values of positive and negative controls on the corresponding plate. Detailed description of input data format is given in technical documentation. A template of the data input structure is available for download to facilitate data processing and analysis.
2.2 Quality control
Common technical errors in HTS assays include spatial plate variability and/or striping due to dispensing errors as well as edge effects due to uneven evaporation of the plate edges. Hence, assessing and quantifying the potential errors of the raw data is a crucial first step of the analysis. The standard QC metrics Z’ (Zhang, 1999) and SSMD (Zhang, 2007) explore the distribution of the positive and negative control wells (Chen et al., 2016). However, those metrics may not capture all spatial plate effects and hence Breeze generates a comprehensive table of different metrics along with several visualizations. The QC table includes parameters such as Z’, SSMD, signal/background ratio, SD, coefficient of variation and central tendency of controls (Supplementary Fig. S1). The QC visualizations in Breeze include interactive plate heatmaps (Supplementary Fig. S3), scatterplots and barplots (Fig. 1A and Supplementary Figs S1, S2 and S4–S6).Visualizations are helpful in interpretation and spotting technical problems such as issues in dispensing cells, drugs, reagents on culture conditions, edge effects, striping, patterning as well as observing signal window, performance, and distribution of compounds and controls, variations across plates and outliers.
2.3 Curve fitting
Curve fitting is an important part of the dose–response data analysis and involves arranging the PI values at each point of the concentration range and fitting these points using four-parameter logistic curve (Findlay and Dillard, 2007; Vølund, 1978). In order to quantify the quality of curve fitting, a standard error of the estimate is calculated for each dose–response curve. The resulting curve fit images (Fig. 1B, top-right) and the fitting parameters are exported in the Excel files.
2.4 Quantification of drug responses
Breeze offers several possibilities to summarize dose–response relationship into a single metric including IC50, EC50, AUC and DSS. The DSS scoring metric is adding normalization to standard AUC (Yadav et al., 2015). This standardization facilitates the correlation of drug sensitivity and resistance testing results across several studies. The results are depicted by interactive visualizations such as heatmaps and bar plots (Fig. 1B and Supplementary Fig. S7). The heatmap provides a comprehensive overview of the data based on distance matrix methods such as Pearson, Euclidean, Manhattan, Spearman and so on. The bar plots list the top-responding drugs in a sample (Supplementary Fig. S8), while the circular tree correlates drug response patterns among group of samples (Supplementary Fig. S9). The user can also upload the DSS of the reference/control screen, to calculate differential response of drugs, between samples and control.
2.5 Tutorial and feedback
Breeze is implemented using R and PHP and hosted with Apache HTTP Server. To facilitate its usage, a step-by-step video tutorial and example input data are available on the website. The user may leave their comments or suggestions using a feedback form. Breeze source code is provided at https://github.com/potdarswapnil/Breeze, to run analyses independently and potentially extend functionality.
3 Conclusion
The Breeze application facilitates a quick and robust analysis of drug testing data by integrating systematic QC procedures and drug response quantification to a standard metric that enables method comparison across several studies. The interactive visualizations, intuitive graphics and easily exportable results provide a framework for reproducible and quality processing of drug testing data. Breeze is a unique computational environment, which provides extensive functionality in terms of QC, summary metrics and visualization of different aspects of HTS experiments.
Supplementary Material
Acknowledgements
We would like to thank all FIMM Breeze application users for valuable input and beta testing.
Funding
The FIMM High-Throughput Biomedicine Unit is financially supported by the University of Helsinki and Biocenter Finland. The work has received funding from the Sigrid Juselius Foundation, Cancer Society of Finland, Academy of Finland [292611, 326238 and 310507 to T.A.], the Magnus Ehrnrooth Foundation, Biocentrum Helsinki and St. Baldrick’s Foundation.
Conflict of Interest: none declared.
References
- Barretina J. et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature, 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M. et al. (2006) Analysis of cell-based RNAi screens. Genome Biol., 7, R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay J.W.A., Dillard R.F. (2007) Appropriate calibration curve fitting in ligand binding assays. AAPS J., 9, E260–E267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. et al. (2016) mQC: a heuristic quality-control metric for high-throughput drug combination screening. Sci. Rep., 6, 37741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourches D. et al. (2014) HTS navigator: freely accessible cheminformatics software for analyzing high-throughput screening data. Bioinformatics, 30, 588–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverty P.M. et al. (2016) Reproducible pharmacogenomic profiling of cancer cell line panels. Nature, 533, 333–337. [DOI] [PubMed] [Google Scholar]
- Kodack D.P. et al. (2017) Primary patient-derived cancer cells and their potential for personalized cancer patient care. Cell Rep., 21, 3298–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H. et al. (2018) Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell, 173, 515–528.e517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpindi J.P. et al. (2015) Impact of normalization methods on high-throughput screening data with high hit rates and drug testing with dose-response data. Bioinformatics, 31, 3815–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpindi J.P. et al. (2016) Consistency in drug response profiling. Nature, 540, E5–E6. [DOI] [PubMed] [Google Scholar]
- Pemovska T. et al. (2015) Axitinib effectively inhibits BCR-ABL1(T315I) with a distinct binding conformation. Nature, 519, 102–105. [DOI] [PubMed] [Google Scholar]
- Pemovska T. et al. (2013) Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov., 3, 1416–1429. [DOI] [PubMed] [Google Scholar]
- Saeed K. et al. (2017) Comprehensive drug testing of patient-derived conditionally reprogrammed cells from castration-resistant prostate cancer. Eur. Urol., 71, 319–327. [DOI] [PubMed] [Google Scholar]
- Seashore-Ludlow B. et al. (2015) Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov., 5, 1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov P. et al. (2016) PharmacoGx: an R package for analysis of large pharmacogenomic datasets. Bioinformatics, 32, 1244–1246. [DOI] [PubMed] [Google Scholar]
- Vølund A. (1978) Application of the four-parameter logistic model to bioassay: comparison with slope ratio and parallel line models. Biometrics, 34, 357–365. [PubMed] [Google Scholar]
- Yadav B. et al. (2015) Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci. Rep., 4, 5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. et al. (2013) Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res., 41 (Database issue), D955–D961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.H. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen., 4, 67–73. [DOI] [PubMed] [Google Scholar]
- Zhang X.D. (2007) A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics, 89, 552–561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


