Abstract
Motivation
Glycan structures are commonly represented using symbols or linear nomenclature such as that from the Consortium for Functional Glycomics (also known as modified IUPAC-condensed nomenclature). No current tool allows for writing the name in such format using a graphical user interface (GUI); thus, names are prone to errors or non-standardized representations.
Results
Here we present GlycoGlyph, a web application built using JavaScript, which is capable of drawing glycan structures using a GUI and providing the linear nomenclature as an output or using it as an input in a dynamic manner. GlycoGlyph also allows users to save the structures as an SVG vector graphic, and allows users to export the structure as condensed GlycoCT.
Availability and implementation
The application can be used at: https://glycotoolkit.com/Tools/GlycoGlyph/. The application is tested to work in modern web browsers such as Firefox or Chrome.
Contact
aymehta@bidmc.harvard.edu or rcummin1@bidmc.harvard.edu
1 Introduction
Glycans (oligosaccharides) are one of the most complicated macromolecules in organisms, comprised of >50 different monosaccharides, having multiple sites of linkages (3–5), with either α- or β-anomeric configuration. In addition, glycans can range in size from monomeric to >20-mers long and can be either linear or branched (Seeberger, 2015). Representing these glycans, due to their size, with IUPAC chemical nomenclature is arduous. As a result, glycan structures are represented as linear strings such as the IUPAC, IUPAC-condensed or the Consortium for Functional Glycomics (CFG) linear nomenclature (also known as the modified IUPAC-condensed nomenclature), which are human readable (McNaught, 1997). Among these, the CFG nomenclature has been most widely adopted by the glycobiology community for its simplicity and brevity, while offering high coverage with respect to structural information (Agravat et al., 2014). It was established by the CFG (www.functionalglycomics.org) and is used extensively in glycan analysis by mass spectrometry, microarray studies and in manuscript texts (Raman et al., 2006). For the majority of glycans, the CFG nomenclature offers sufficient flexibility to represent glycan structures. More complicated machine readable formats, such as GlycoCT and WURCS, can be used to represent more intricate structures (Herget et al., 2008; Matsubara et al., 2017).
Glycan structures can be represented using symbol notations for individual monosaccharides. Recently, the symbol nomenclature for glycans (SNFG) was established in order to standardize glycan representation (Varki et al., 2015). There are a few glycan drawing tools available online, e.g. DrawGlycan-SNFG and SugarSketcher. DrawGlycan-SNFG requires standard IUPAC-condensed nomenclature as input, and does not have a graphical user interface (GUI) for drawing the glycan, and cannot generate the name from the structure (Cheng et al., 2017). SugarSketcher on the other hand provides a GUI to draw glycans, and provides output as well as input in GlycoCT format (Alocci et al., 2018), yet it cannot handle human readable formats for input directly, and does not dynamically update structures with names. Other software available for drawing glycan structures include DrawRings (http://www.rings.t.soka.ac.jp/drawRINGS-js/index.html), Polys Glycan Builder (http://glyco3d.cermav.cnrs.fr/builder.php) and Glycan Builder (http://sugarbind.expasy.org/builder). However, none of these tools allow drawing the structures using the simple CFG nomenclature.
We therefore developed GlycoGlyph, a web-based application which allows users to draw structures in SNFG format using a simple GUI or via text strings in the CFG linear nomenclature. The glycan name can also be used as an input allowing seamless name ↔ structure conversion. The availability of this novel tool will greatly improve data entry and manuscript writing by enhancing uniformity in formats, thereby enabling text mining in the future, and help to avoid errors in presenting glycan structures.
2 Materials and methods
GlycoGlyph was built using jQuery and D3.js (v5) JavaScript libraries with Bootstrap CSS. GlycoCT dictionaries for monosaccharides and substituents were adapted from SugarSketcher (Alocci et al., 2018).
3 Results
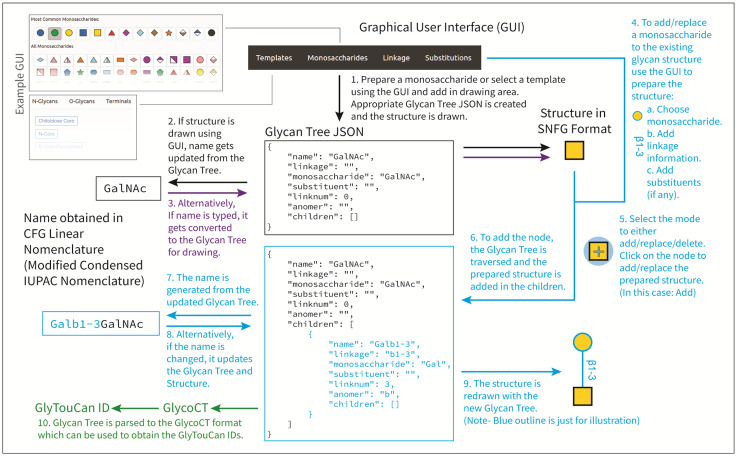
The core of GlycoGlyph involves the inter-conversion of name and structure via a glycan tree data structure (Fig. 1). The user can initiate a drawing using a template, or denote a monosaccharide by selecting one of the monosaccharides from a list (an initial monosaccharide), a linkage and add any substituents to the structure (Fig. 1). A mode to either add/replace is selected. Once the monosaccharide is selected, the user can click within the drawing area to begin or select an existing monosaccharide symbol to add/replace the prepared structure. GlycoGlyph then utilizes the D3.js data binding to the nodes in order to identify where the monosaccharide is located in the tree data to be added or replaced. It traverses the tree to add/replace the initial monosaccharide and then uses another function to generate a name from the tree data structure. Simultaneously, the symbolic structure is redrawn. The user can delete nodes using the ‘Delete’ mode. The application also offers undo/redo buttons, in the event the user makes an error. GlycoGlyph also has certain options for drawing settings, such as symbol size, fucose drawing orientation and customization for linkage text and font size. These can be accessed using the cog-wheel button. Once the structure is drawn, GlycoGlyph allows the user to save the image as an Scalable Vector Graphics (SVG) vector graphic. GlycoGlyph also provides the user with the glycan structure in GlycoCT. This tool allows the user to fetch the GlyTouCan IDs using the GlyCosMos.org API. In conclusion, GlycoGlyph makes it simple for users to draw glycan structures and generate names for the structures to be used in manuscripts, with the goal of enabling text mining.
Fig. 1.
An overview of GlycoGlyph. A step-by-step overview of how GlycoGlyph uses a glycan tree data structure to draw glycans and generate glycan names
Acknowledgements
The authors would like to thank Chao Gao, Tanya McKitrick, Jamie Heimburg-Molinaro and Alyssa McQuillan for providing useful feedback.
Funding
This work was supported by National Institutes of Health [P41GM103694 and U01GM125267].
Conflict of Interest: none declared.
References
- Agravat S.B. et al. (2014) GlycoPattern: a web platform for glycan array mining. Bioinformatics, 30, 3417–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alocci D. et al. (2018) SugarSketcher: quick and intuitive online glycan drawing. Molecules, 23, 3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. et al. (2017) DrawGlycan-SNFG: a robust tool to render glycans and glycopeptides with fragmentation information. Glycobiology, 27, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herget S. et al. (2008) GlycoCT-a unifying sequence format for carbohydrates. Carbohydr. Res., 343, 2162–2171. [DOI] [PubMed] [Google Scholar]
- Matsubara M. et al. (2017) WURCS 2.0 update to encapsulate ambiguous carbohydrate structures. J. Chem. Inf. Model., 57, 632–637. [DOI] [PubMed] [Google Scholar]
- McNaught A.D. (1997) Nomenclature of carbohydrates (recommendations 1996). Adv. Carbohydr. Chem. Biochem., 52, 43–177. [PubMed] [Google Scholar]
- Raman R. et al. (2006) Advancing glycomics: implementation strategies at the consortium for functional glycomics. Glycobiology, 16, 82R–90R. [DOI] [PubMed] [Google Scholar]
- Seeberger P.H. (2015) Monosaccharide diversity In: Varki A.et al. (eds), Essentials of Glycobiology. Cold Spring Harbor, NY, pp. 19–30. [Google Scholar]
- Varki A. et al. (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology, 25, 1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]