Abstract
Motivation
Cancer heterogeneity is observed at multiple biological levels. To improve our understanding of these differences and their relevance in medicine, approaches to link organ- and tissue-level information from diagnostic images and cellular-level information from genomics are needed. However, these ‘radiogenomic’ studies often use linear or shallow models, depend on feature selection, or consider one gene at a time to map images to genes. Moreover, no study has systematically attempted to understand the molecular basis of imaging traits based on the interpretation of what the neural network has learned. These studies are thus limited in their ability to understand the transcriptomic drivers of imaging traits, which could provide additional context for determining clinical outcomes.
Results
We present a neural network-based approach that takes high-dimensional gene expression data as input and performs non-linear mapping to an imaging trait. To interpret the models, we propose gene masking and gene saliency to extract learned relationships from radiogenomic neural networks. In glioblastoma patients, our models outperformed comparable classifiers (>0.10 AUC) and our interpretation methods were validated using a similar model to identify known relationships between genes and molecular subtypes. We found that tumor imaging traits had specific transcription patterns, e.g. edema and genes related to cellular invasion, and 10 radiogenomic traits were significantly predictive of survival. We demonstrate that neural networks can model transcriptomic heterogeneity to reflect differences in imaging and can be used to derive radiogenomic traits with clinical value.
Availability and implementation
Contact
whsu@mednet.ucla.edu
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Radiogenomic mapping is the integration of traits observed on medical images and traits found at the molecular level, such as gene expression profiling (Diehn et al., 2008; Segal et al., 2007). As such, the study of radiogenomics plays a role in precision medicine, where associations can describe prognosis or therapy response (Chang et al., 2018; Naeini et al., 2013; Pope et al., 2008). Common approaches to radiogenomic mapping include dimensionality reduction and pairwise comparisons (Aerts et al., 2014; Diehn et al., 2008; Gevaert et al., 2014; Grossmann et al., 2017; Gutman et al., 2013; Jamshidi et al., 2014; Zinn et al., 2011) or predictive models, such as decision trees (Gevaert et al., 2017; Hu et al., 2017; Kickingereder et al., 2016 Segal et al., 2007; Zhang et al., 2017) or linear regression (Gevaert et al., 2012; Grossmann et al., 2017; Guo et al., 2015; Yamashita et al., 2016; Zhu et al., 2015). Markedly, these approaches often require feature selection; assume linearity; and/or depend on pairwise associations, limiting their capacity to represent complex biological relationships.
Neural networks, with their ability to automatically learn non-linear, hierarchical representations of large input spaces (Goodfellow et al., 2016; Lecun et al., 2015), are alternate approaches for radiogenomics (Chang et al., 2018; Chen et al., 2016; Ha et al., 2019; Korfiatis et al., 2017; Li et al., 2018). The models can combine low-level features into a structure of complex features to create a new, abstracted and transformed representation better suited for learning than the original input (Bengio, 2009). Current applications of neural networks have focused on diagnostic images as inputs to predict a single gene status and have excluded gene interactions (Chang et al., 2018; Ha et al., 2019; Korfiatis et al., 2017). Instead, the non-linear transformations could be applied to molecular data and identify new and/or confirm prior radiogenomic associations. Toward understanding the biological basis of imaging traits, we thus present an approach using the representational power of neural networks to model tumor transcriptomes and non-linearly map genes to tumor imaging traits. No a priori selection is used on the transcriptome.
A limitation of neural networks is that they are considered ‘black boxes’, which makes it difficult to interpret the learned relationships. The interpretability of neural networks should be examined in order to take advantage of their learning capabilities. To the best of our knowledge, no prior studies have interpreted neural networks to ascertain what radiogenomic associations are learned. Therefore, we provide approaches, called gene masking and gene saliency, for understanding radiogenomic neural networks. The approaches were based on visualization techniques, input masking and saliency maps (Simonyan et al., 2014; Zeiler and Fergus, 2014), that probe the trained neural networks to extract predictive relationships between gene expressions and imaging traits. Here, our model interpretation methods can identify cohort-level relationships, which we refer to as radiogenomic associations, and patient-level relationships, which we refer to as radiogenomic traits. We validated the extracted associations and traits generated by a network in a classification task with known relationships, such as gene expressions (model input) and molecular subtypes (model output; Verhaak et al., 2010).
As a use case, we study radiogenomic associations related to human-understandable imaging traits in magnetic resonance imaging (MRI) scans from patients with glioblastoma (GBM). GBM is a grade IV malignant brain tumor with poor prognosis, and for which imaging is heavily used in diagnosis, prognosis and treatment assessment. MRI traits, such as tumor enhancement, non-contrast enhancement, edema and necrosis (Fig. 1), describe some of the visual, phenotypic variations between patients as they are treated (Pope et al., 2005). We present here the extracted relationships found by the radiogenomic neural networks using our approach, compare against previous work to show both new and consistent findings, and establish the radiogenomic relationships’ clinical value in estimating patient survival over clinical or imaging traits alone.
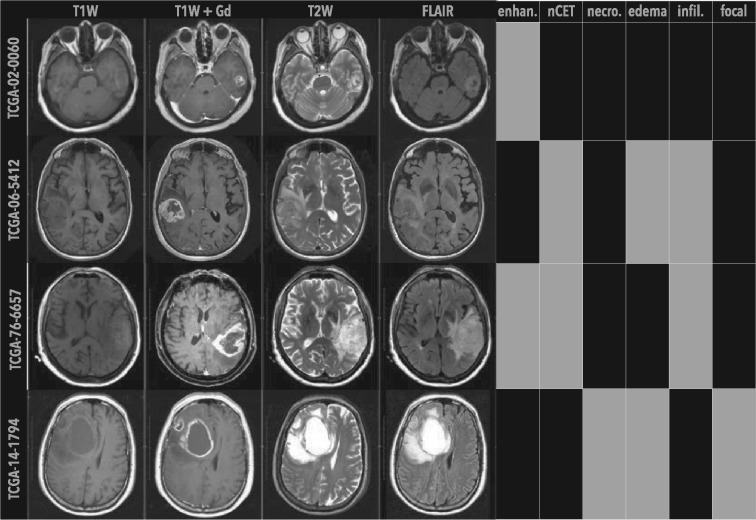
Fig. 1.
Examples of phenotypic differences observed in GBM patients. Shown are single, axial images of pre-op MRI scans from the TCGA–GBM cohort. Four MRI sequences were used to annotate tumor (white arrows) imaging traits: T1W, T1W+Gd and T2W and FLAIR images. MRI traits included enhancing (enhan.), nCET, necrosis (necro.), edema, infiltrative (infil.) and focal, where class labels were indicated by black (proportions , expansive, or focal) or gray (proportions , infiltrative, or non-focal) blocks
2 Materials and methods
2.1 Gene expression
Transcriptomes were available for 528 GBM patients as part of The Cancer Genome Atlas (TCGA) (Supplementary Table S1). Samples were untreated primary tumors and had 80% cancer and 50% necrotic cells (McLendon et al., 2008). Samples were previously analyzed by the Broad Institute on Affymetrix arrays, quantile normalized and background corrected. Level 3 data were downloaded from the Genomic Data Commons at https://gdc.cancer.gov/. Each profile had 12 042 genes.
2.2 Magnetic resonance imaging
Medical images for 262 GBM patients were downloaded from The Cancer Imaging Archive (TCIA) (Scarpace et al., 2016). Patients were matched with shared barcodes used by TCGA and TCIA. A board-certified neuroradiologist (S.E.-S., 26 years of experience) evaluated images using the Osirix medical image viewer. An electronic form was used to record MRI traits according to the Visually Accessible Rembrandt Images (VASARI) feature guide at https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project. A total of 175 patients had pre-operative (pre-op) MRIs and transcriptomes (Supplementary Table S1). Six MRI traits were annotated from the pre-op studies. The number of traits estimated for each patient was dependent on the availability of MR sequences. For example, enhancement could be estimated from patients who only had T1-weighted images with and without contrast and no FLAIR or T2-weighted images. Traits were binarized given the small sample sizes (Supplementary Table S2).
2.3 Radiogenomic modeling
To map relationships between gene expression profiles and MRI traits, feed-forward neural networks (Goodfellow et al., 2016) were used (Fig. 2a and b). Each MRI trait was a binary classification task, where the positive class was the least frequent label (Supplementary Table S2). Models were provided all 12 042 gene expressions as input vectors to classify each imaging trait, resulting in one model per trait. During training, early stopping with a patience of 200 epochs was used while monitoring the area under the curve (AUC) of the receiver operating characteristic curve. To help learning in the radiogenomic model, an autoencoder was used as many more patients had transcriptomic data. The radiogenomic neural networks were pre-trained using weights transferred from a deep transcriptomic autoencoder trained on a separate subset of 353 patients. The transcriptome dataset was the subset of TCGA–GBM patients who only had transcriptomes and no no pre-op MRI data.
Fig. 2.
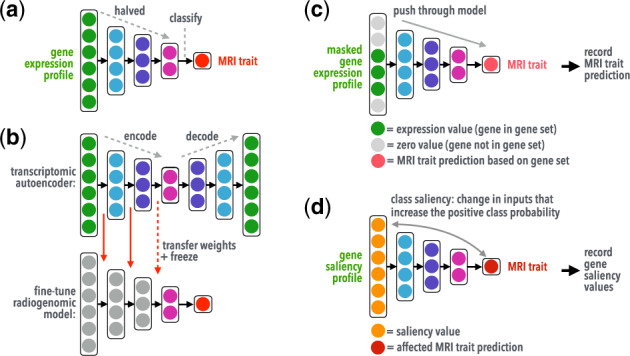
Illustration showing(a) the radiogenomic neural network's architecture, (b) transfer learning using a deep transcriptomic autoencoder, and interpretation methods using (c) gene masking and (d) gene saliency. Pretrained weights learned in the autoencoder were transferred to a radiogenomic model, where weights were frozen (non-trainable, long red arrows) and/or fine-tuned (trainable, dashed red arrow) during radiogenomic training. (Color version of this figure is available at Bioinformatics online.)
The transcriptomic autoencoder takes a gene expression profile as input, compresses the information through three encoding layers, and then decodes the information to reconstruct the transcriptome. Early stopping was used in training and monitored the mean coefficient of determination (R2) of each gene. The trained autoencoder weights, along with the gene pre-processing parameters, were used as non-random weight initialization (weights can be fine-tuned during training) and/or frozen weights (weights cannot be fine-tuned during training) in the radiogenomic models.
Performance was estimated with 10-fold cross-validation. Each gene expression was mean subtracted and divided by its SD, a process performed at each fold split. Hyperparameters were optimized via grid search. In supervised models, sample weighting and stratified fold splitting were included. The overall methods for radiogenomic model training are illustrated in Figure 3a. The autoencoder achieved a mean validation of 0.45 R2 in cross-validation and 0.61 R2 in retraining (Supplementary Fig. S2). Radiogenomic models were then constructed to have the same encoding architecture, activation function, and optimizer as the best performing autoencoder. All neural networks were trained on NVIDIA Tesla K80 and V100 GPUs through Amazon Web Services using Python 3.6, Keras 2.2.4 (Chollet et al., 2015), and TensorFlow 1.12.0 on a Ubuntu 16.04 machine. Other classifiers (Supplementary Table S3) were implemented via XgBoost 0.80 (Chen and Guestrin, 2016) and sklearn 0.20.0 (Pedregosa et al., 2011).
Fig. 3.
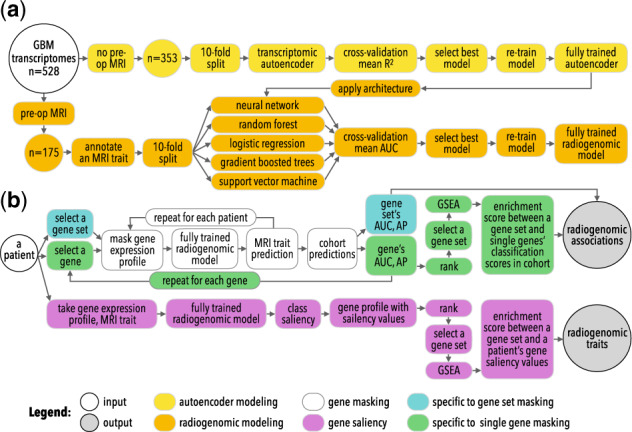
An overview of the study’s approaches to radiogenomic neural network (a) training and (b) interpretation, gene masking and gene saliency, to extract radiogenomic associations and radiogenomic traits
2.4 Bootstrapping
The best performing models of each model type (Supplementary Tables S3) were evaluated on bootstrapped datasets to measure classification performance variability within and between model types. For each bootstrap, a radiogenomic dataset was split for 10-fold cross-validation using a different seed. In each split, training, and validation samples were separately resampled with replacement to obtain bootstrapped sets. Sets were resampled if not all classes were observed. Each model was trained on the same bootstrapped dataset using the aforementioned model training methods. This process was repeated 100 times for each MRI trait.
2.5 Molecular subtype modeling
To validate the use of neural networks to model transcriptomes, a model was trained on a classification task where the relationship between model input and output were known. Previously, an 840 gene expression signature was used to define four molecular subtypes in GBM: classical, mesenchymal, neural, and proneural (Verhaak et al., 2010). A neural network was trained to predict the four subtypes from entire gene expression profiles consisting of 12 042 genes. Given there was no ground truth for radiogenomic relationships, the aforementioned model was used to test if (i) neural networks can effectively prioritize among a high-dimensional input to uncover known associations with the predicted label and (ii) if so, how can the trained model be interpreted via the proposed methods to validate if the model learned the correct associations. Of the 528 patients, 171 had subtype labels (Supplementary Tables S1 and S2). The four subtypes were modeled as a multi-class classification task via one-hot-encoding. Gene expressions pre-processing and model training were performed in the same manner as the radiogenomic models, mainly, 10-fold cross-validation and hyperparameter grid search (Supplementary Table S4). Gene masking and gene saliency were applied on the fully trained subtype neural network to demonstrate the usability of our model interpretation methods to identify which genes in the transcriptome were useful for classification.
2.6 Gene masking
Masking is a type of sensitivity analysis where the value(s) of one or more components of the input is retained while all others are replaced with zeros. The goal is to determine the impact that the kept input components have on the end classification; this procedure was previously described in Zeiler and Fergus (2014). Here, we define ‘gene masking’ to extract radiogenomic associations from a trained neural network (Fig. 2c). For each individual, the gene expression values of a particular gene set were kept while all other expressions were replaced with zeros. The masked profiles were pushed through a fully trained neural network and the output, a class probability based on using genes from the gene set, was recorded. After repeating this process for each patient, classification performance was calculated. Each gene set was evaluated by AUC and average precision (AP) to measure the strength of a radiogenomic association. As such, gene masking reported a set of genes’ abilities to predict an MRI trait in the entire cohort.
In single gene masking, each gene was masked one at a time and additionally used in gene set enrichment analysis (GSEA; Subramanian et al., 2005; ranked by AP or AUC). In gene set masking, predefined gene sets (<500 genes, see Section 2.8) from the Molecular Signatures Database (MSigDB, v6.2; Liberzon et al., 2011), molecular subtypes (Verhaak et al., 2010) and brain cell types and phenotypes (Darmanis et al., 2015; Patel et al., 2014; Zhang et al., 2016) taken from (Puchalski et al., 2018) were masked. MSigDB was also queried for gene sets that include the 22 genes characterized as potential contributors of GBM tumorigenesis (McLendon et al., 2008; Parsons et al., 2008; Supplementary Material). The top performing genes or gene sets for each MRI trait were visualized by clustering classification scores via pheatmap in R.
2.7 Gene saliency
Class saliency is a visualization technique used to compute the gradient of an output class prediction with respect to an input via backpropagation (Simonyan et al., 2014). Thus, class saliency identifies the relevant input components whose values would affect the positive class probability in a trained neural network. Here, we define ‘gene saliency’ as the genes whose change in expression would increase the model’s belief of the positive class label (Fig. 2d). In each model, salient genes are derived for each patient, ranked and used in GSEA to determine if a gene set is relevant to predicting his/her MRI trait. Subsequently, positive enrichment between a single patient’s salient genes and a gene set is defined as a ‘radiogenomic trait’. For example, the edema model was probed to identify a single patient’s salient genes. The most salient genes were the genes that increased the probability of edema being . If GSEA found the salient genes were enriched by a gene set, then the prediction of the patient’s edema was related to the gene set (i.e. the patient has the radiogenomic trait between the gene set and edema). If a patient is not enriched, then the patient does not have the radiogenomic trait. Saliency was implemented using keras-vis 0.4.1 (Kotikalapudi et al., 2017). The input range was determined by the gene expression range in the dataset and the parameter backprop_modifier was set to 'guided.' All other parameters were set to default. In the subtype neural network, gene saliency was repeated for each class as one-versus-others. Figure 3b depicts gene masking and gene saliency.
2.8 Gene set enrichment analysis
Pre-ranked GSEA was performed using fgsea (Sergushichev, 2016) using the recommended maximum size of 500 genes in a gene set, a minimum size of 15, and 10 000 permutations. Genes were ranked via single gene masking classification scores or gene saliency values for a patient. Enrichments were significant at an adjusted P-value of <0.05. Correlation between a gene expression and an imaging trait was used for comparison. Clustering of enrichment scores was performed as aforementioned.
2.9 Survival
Clinical data from TCGA were used to define patient outcomes. Overall survival (OS) and time-to-death events were in the patient file. Progression-free survival (PFS) outcomes were defined by the follow-up file, where all event types with days-to-event data were considered, but only the earliest event was used (Supplementary Material). Variables considered for survival analyses were all binary and consisted of each patient’s MRI traits (Supplementary Table S2); radiogenomic traits (result of gene saliency analysis); and covariates including gender, race (binned as white and non-white) and age at initial diagnosis (binned via the median value in all 528 patients).
Survival analyses were done in R using survival and survminer. Kaplan–Meier estimates were used to compare imaging traits versus radiogenomic traits and obtained via survfit, ggsurvplot and survdiff, where the log-rank test was used to test for significant differences between groups. Cox proportional hazards models were used to estimate univariate and adjusted hazard ratios (HRs) via coxph. Radiogenomic traits enriched in at least five patients were kept. Cox models used backward feature selection via step and were forced to keep all patient covariates, while free to choose any imaging or radiogenomic trait. For any comparison, patients with missing information were removed.
3 Results
3.1 Neural networks achieve best performance in classifying MRI traits
Using the transcriptome as input, neural networks were better at estimating MRI traits than all other classifiers (Fig. 4a), including gradient-boosted trees, random forest, support vector machines, and logistic regression. In predicting proportions of non-contrast enhancing tumor (nCET), necrosis and edema, the first hidden layer used frozen pre-trained weights and only the last two hidden layers were used for fine-tuning (Table 1). Bootstrapping showed neural networks had higher performances, where 95% CIs in Figure 4b indicate neural networks outperformed other models by more than 0.10 AUC (Supplementary Table S5 and Figs S3–S5).
Fig. 4.
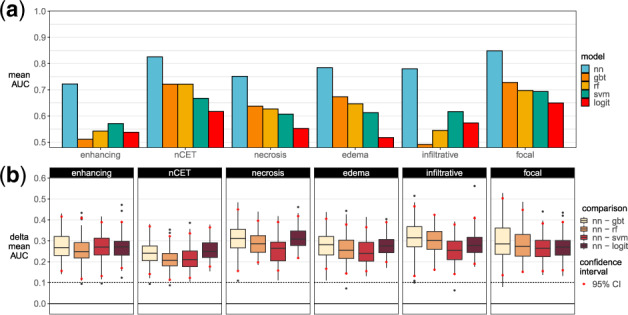
Radiogenomic models performances. (a) Observed 10-fold cross-validation performances. (b) Performance differences between a neural network and another model in 100 bootstrapped datasets. nn, neural network; gbt, gradient-boosted trees; rf, random forest; svm, support vector machines; logit, logistic regression
Table 1.
Neural network hyperparameters
| CV means |
Retrain |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Label | R 2 | epoch | R 2 | epoch | Hidden layers | optimizer | activation | dropout | ||
|
| ||||||||||
| Transcriptome | 0.45 | 467 | 0.61 | 486 | 4000/2000/1000 | Adadelta | tanh | 0.0 | ||
|
| ||||||||||
| AE |
||||||||||
| Label | AUC | epoch | AUC | epoch | Hidden layers | optimizer | activation | dropout | Layer | Frozen |
| Enhancing | 0.72 | 38 | 1.00 | 14 | 4000/2000/1000 | Adadelta | tanh | 0.6 | 3 | 0 |
| nCET | 0.83 | 38 | 1.00 | 11 | 4000/2000/1000 | Adadelta | tanh | 0.0 | 1 | 1 |
| Necrosis | 0.75 | 44 | 1.00 | 11 | 4000/2000/1000 | Adadelta | tanh | 0.0 | 1 | 1 |
| Edema | 0.78 | 109 | 1.00 | 16 | 4000/2000/1000 | Adadelta | tanh | 0.0 | 1 | 1 |
| Infiltrative | 0.78 | 70 | 1.00 | 12 | 4000/2000/1000 | Adadelta | tanh | 0.0 | 2 | 1 |
| Focal | 0.85 | 44 | 1.00 | 12 | 4000/2000/1000 | Adadelta | tanh | 0.6 | 3 | 0 |
| Subtype | 0.99 | 14 | 0.998 | 66 | 3000/1500/750 | Nadam | sigmoid | 0.4 | — | — |
Note: Layer refers to the depth of hidden layers in the radiogenomic model that used pretrained weights from the autoencoder (AE; e.g. two AE layers indicate the first two hidden layers used pretrained weights). Retrain refers to models trained on the full dataset.
3.2 Neural networks correctly learned known associations between gene expressions and molecular subtypes
To verify the relationships learned in the neural network’s layers, a molecular subtype model was trained. The model achieved a micro-averaged AUC of 0.994 in 171 patients (Supplementary Table S6). Gene masking with subtypes gene sets showed the neural network was able to predict subtype classes with high certainty (Fig. 5). For example, when the model was given the expressions of the 216 mesenchymal genes, subtype probabilities approached 1 or 0 and often corresponded to the correct subtype (0.90 AUC and 0.89 AP). Performance improved when the model was given all 840 subtype genes (0.99 AUC and 0.98 AP). Conversely, given the expressions of 200 random, non-subtype genes, the model was less certain (i.e., probabilities away from 1 or 0), and the fully trained performance of 1.0 AP dropped to 0.68 AP (Supplementary Table S7).
Fig. 5.
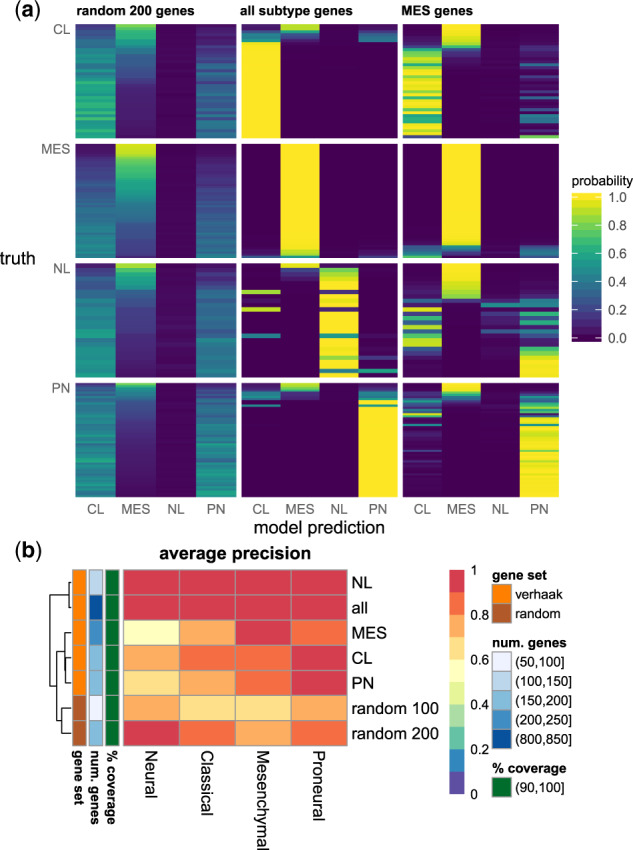
Gene masking of the subtype neural network: (a) estimated subtype probabilities, where each row was a patient and grouped by their true subtype and (b) classification performance measured by AP in gene set masking, where each row was a gene set and each column was the subtype prediction (see also Supplementary Figs S6–S8). The random gene set excluded ones in a subtype set. For visualization purposes, rows were sorted by the mesenchymal probabilities. CL, classical; MES, mesenchymal; NL, neural; PN, proneural; all, all 840 subtype genes; coverage, percent of gene set that exist in gene expression profiles
The majority of the top 20 predictive genes in each subtype class belonged to the original subtype class definition (Fig. 6a). For example, 18 of the top 20 genes for predicting the proneural subtype were a part of the proneural gene set and each had least 0.80 AP and a 0.80 AUC. Of the top 500 genes ranked by AP, 270 genes (54%) were subtype genes; this represented 32% of all subtype genes (Fig. 6b). Unsurprisingly, subtype genes were predictive of more than one subtype. GSEA also showed the most predictive single genes for each subtype prediction were significantly (adjusted P-value < 0.05) and positively enriched by the corresponding subtype gene set (Fig. 6c). This observation was corroborated in GSEA using ranked genes based on correlation (Fig. 6d) instead of gene masking.
Fig. 6.
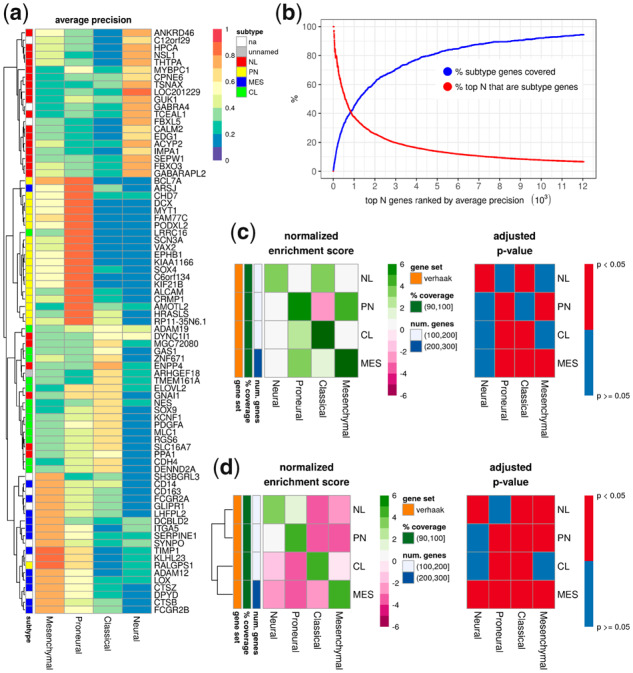
Single gene masking in the subtype model: (a) the top 20 genes used to predict each subtype; (b) the percent of subtype genes covered in the top N genes; and (c) GSEA with genes ranked by AP, where positive enrichment indicated the subtype gene set was correlated with high AP and vice versa. (d) An alternative GSEA was performed by ranking genes based on their correlation with a subtype, where positive enrichment indicated the subtype gene set was correlated with a subtype and vice versa. na, not a part of the subtype genes; unnamed, a part of the subtype genes, but not tied to a single subtype
3.3 Genes driving the prediction of MRI traits
3.3.1. Enhancing tumor
Tumor enhancement was measured on T1W with gadolinium contrast (T1W+Gd) images. Low-grade or well-differentiated brain tumors tend to generate blood vessels with intact blood–brain barriers (BBB) and do not enhance. Poorly differentiated or more aggressive tumors, like GBM, generate leaky blood vessels without an intact BBB and enhance on T1W+Gd images.
Enhancement was found to be associated with growth, immune responses, hormones, the extracellular matrix (ECM), vasculature and kinase activity in gene masking (Table 2). The ECM association included gene expressions of ECM-related proteins (Naba et al., 2012; Supplementary Fig. S18). Of the MSigDB hallmark gene sets (Liberzon et al., 2015), apical junction (cellular components, such as adherens and tight junctions), IL2/STAT5 signaling (immune response activation), complement system, early and late responses to estrogen (associated with ESR1 expression), and heme metabolism (erythroid differentiation, STAT5 activation) were most predictive of enhancement (Fig. 7a). Gene Ontology (GO) gene sets related to GBM abnormalities support the association of growth, immune system and hormones with enhancement.
Table 2.
Summary of transcriptomic drivers of MRI traits in GBM patients
| Transcriptomic drivers | ||||
|---|---|---|---|---|
|
| ||||
| Theme | Gene set (collection,a queryb) | AUC | AP | See also |
| Enhancing | ||||
| Growth/death | Growth (G, PTEN) | 0.86 | 0.84 | |
| Sensory organ development (G, EGFR, KCNK3) | 0.85 | 0.84 | Gutman et al. (2013); Jamshidi et al. (2014) | |
| Immune system | IL2/STAT5 signaling (H) | 0.77 | 0.76 | |
| Complement system (H) | 0.79 | 0.75 | ||
| Activation of immune response (G, PTEN) | 0.90 | 0.89 | ||
| Leukocyte and lymphocyte activation (G, PIK3R1) | 0.85c | 0.84c | ||
| Immune effector process (G, PIK3CA) | 0.87 | 0.84 | ||
| Hormones | Early and late responses to estrogen (H) | 0.79c | 0.73c | |
| Response to steroid hormone (G, RBI) | 0.88 | 0.88 | ||
| Reg. of hormone levels (G, PARK2) | 0.87 | 0.84 | ||
| ECM | Related to ECM proteins (C, ECM) | 0.81c | 0.75c | Diehn et al. (2008) |
| Apical junction (H) | 0.80 | 0.75 | ||
| Vasculature | Heme metabolism (H) | 0.77 | 0.65 | |
| Vasculature and heart development (G, LTBP1) | 0.80c | 0.79c | Gutman et al. (2013) | |
| Kinase activity | Multiple (G, EGFR, LTBP1, KCNK3) | 0.87c | 0.85c | Gutman et al. (2013); Jamshidi et al. (2014) |
| Edema | ||||
| EMT | EMT (H) | 0.80 | 0.80 | |
| Positive reg. of locomotion (G, EGFR, POSTN) | 0.80 | 0.81 | Zinn et al. (2011) | |
| Taxis (G, CXCL12, KIF5C) | 0.80 | 0.80 | Zinn et al. (2011) | |
| Apical junction (H) | 0.77 | 0.74 | ||
| Related to cell adhesion (G, CDKN2A, CTNNA2) | 0.79c | 0.80c | Zinn et al. (2011) | |
| Growth/death | p53 pathway (H) | 0.77 | 0.77 | |
| Autophagy (G, CDKN2A) | 0.75 | 0.76 | ||
| Myogenesis (H) | 0.75 | 0.76 | ||
| Urogenital system development (G, PTEN) | 0.81 | 0.81 | ||
| Muscle structure development (G, COL6A3) | 0.80 | 0.80 | Zinn et al. (2011) | |
| Response to growth factor (G, EGFR, POSTN) | 0.76 | 0.81 | Zinn et al. (2011) | |
| Vasculature | Heme metabolism (H) | 0.77 | 0.73 | |
| Differentiation | Central nervous sys. neuron differentiation (G, PTEN) | 0.79 | 0.81 | |
| Cell differentiation (G, MET) | 0.79 | 0.81 | ||
| Stem cell differentiation (G, CDK6) | 0.80 | 0.80 | ||
| Immune system | Reg. of cell activation (G, CDKN2A) | 0.83 | 0.82 | |
| Neg. reg. of immune system (G, CDKN2A, CXCL12) | 0.82 | 0.81 | Zinn et al. (2011) | |
| Immune effector process (G, PIK3CA) | 0.80 | 0.81 | ||
| Other | Glycolysis (H) | 0.76 | 0.70 | |
| nCET | ||||
| Cell cycle | Mitotic spindle (H) | 0.78 | 0.70 | |
| DNA repair (H) | 0.77 | 0.66 | ||
| G2M checkpoint (H) | 0.72 | 0.63 | ||
| Reg. of mitotic cell cycle (G, TP53) | 0.78 | 0.71 | ||
| Growth/death | p53 pathway (H) | 0.76 | 0.65 | |
| Urogenital and vasculature development (G, PTEN) | 0.81c | 0.74c | ||
| Neg. reg. of cell cycle (G, TP53) | 0.80 | 0.74 | ||
| Reproductive system development (G, EGFR) | 0.80 | 0.72 | ||
| UV | UV response down (H) | 0.78 | 0.59 | |
| Response to radiation (G, TP53) | 0.83 | 0.73 | ||
| Other | Glycerphospholip metabolism process (G, EGFR) | 0.75 | 0.74 | |
| Small molecule catabolic process (G, PTEN) | 0.78 | 0.73 | ||
| Necrosis | ||||
| Vasculature | Heme metabolism (H) | 0.72 | 0.61 | |
| Growth/death | Apoptosis (H) | 0.71 | 0.58 | |
| Apoptotic signaling pathway (G, IL4, TP53) | 0.72 | 0.63 | Gevaert et al. (2014) | |
| Related to TP53 | 0.76c | 0.66c | ||
| Gland development (G, EGFR) | 0.76 | 0.65 | ||
| Immune system | IL6/JAK/STAT3 signaling (H) | 0.67 | 0.56 | |
| Leukocyte cell adhesion (G, IL4, ITGA5) | 0.76 | 0.59 | Gevaert et al. (2014); Jamshidi et al. (2014) | |
| Reg. of leukocyte proliferation (G, CDKN2A) | 0.77 | 0.61 | Gutman et al. (2013) | |
| Others | Xenobiotic metabolism (H) | 0.76 | 0.65 | |
| Related to PTEN | 0.76c | 0.65c | ||
| Reg. of homeostatic process (G, NF1) | 0.78 | 0.65 | ||
| Glycolysis (H) | 0.76 | 0.56 | ||
| Focal | ||||
| Growth/death | Reg. of anatomical structure size (G, PTEN) | 0.96 | 0.88 | |
| Response to growth factor (G, EGFR) | 0.92 | 0.83 | ||
| Transport | Secretion by cell (G, NF1) | 0.95 | 0.88 | |
| Neg. reg. of transport (G, PTEN) | 0.96 | 0.87 | ||
| Reg. of cytoplasmic transport (G, TP53) | 0.95 | 0.85 | ||
| Monovalent inorganic cation transport (G, PARK2) | 0.95 | 0.81 | ||
| Response to | Steroid hormone, lipid and organic cyclic compound (G, RB1) | 0.95c | 0.84c | |
| Vasculature | Vasculature development (G, PTEN) | 0.93 | 0.84 | |
| Muscle and circulatory system process (G, PIK3CA) | 0.94c | 0.83c | ||
| Oxygen | Hypoxia (H) | 0.85 | 0.61 | |
| Others | Genes down-regulated by KRAS (H) | 0.88 | 0.64 | |
| Protein heterodimerization activity (G, TP53) | 0.97 | 0.85 | ||
| Neg. reg. of intracellular signaling transduction (G, PTEN) | 0.96 | 0.84 | ||
| Synaptic signaling (G, PTEN) | 0.92 | 0.82 | ||
| Infiltrative | ||||
| Oxygen | Reactive oxygen species pathway (H) | 0.71 | 0.50 | |
| Response to oxygen levels (G, TP53) | 0.67 | 0.60 | ||
| Transport | Neg. reg. of transport (G, PTEN, NFKBIA) | 0.74 | 0.64 | Colen et al. (2014) |
| Healing | Wound healing (G, NF1) | 0.80 | 0.64 | |
| Hemostasis (G, PIK3CA) | 0.78 | 0.59 | ||
| Growth/death | Developmental growth (G, PTEN) | 0.74 | 0.62 | |
| Spinal cord development (G, NF1) | 0.66 | 0.60 | ||
| Response to | Response to drug (G, MDM2, MYC) | 0.75 | 0.60 | |
| Response to inorganic substance (G, PTEN) | 0.73 | 0.61 | Colen et al. (2014) | |
| Others | DNA repair (H) | 0.70 | 0.58 | |
| Ligase activity (G, MDM2) | 0.75 | 0.64 | ||
| Ubiquitin-like protein transferase activity (G, MDM2) | 0.70 | 0.59 | ||
| Ubiquitin-like protein ligase binding (G, NFKBIA) | 0.67 | 0.57 | Colen et al. (2014) | |
| Related to protein and transcription factor complex (G, TP53) | 0.75c | 0.62c | ||
| WNT signaling pathway (G, PTEN, MYC) | 0.73 | 0.62 | Colen et al. (2014) | |
Note: The top five gene sets ranked by AUC or AP and compared against gene sets related to prior GBM work in gene abnormalities (McLendon et al., 2008; Parsons et al., 2008) or radiogenomics.
MSigDB: H, hallmark; G, Gene Ontology; C, canonical.
Gene sets queried in MSigDB using keyword(s) reported in Colen et al. (2014), Diehn et al. (2008), Gevaert et al. (2014), Gutman et al. (2013), Jamshidi et al. (2014) and Zinn et al. (2011).
Averaged.
neg., negative; reg., regulation; sys., system.
Fig. 7.
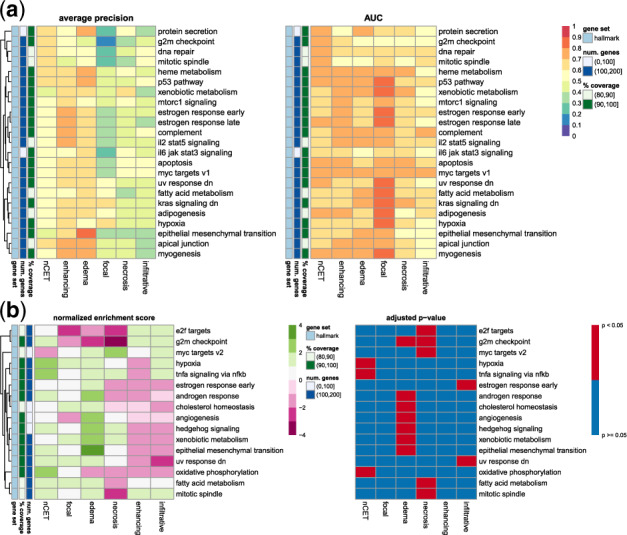
Gene masking of the radiogenomic models with the MSigDB hallmark gene sets. (a) Model performance in gene set masking. Shown are the top five gene sets ranked by AP in each MRI trait (see also Supplementary Fig. S9). (b) Enrichment among genes ranked by AP in single gene masking. Positive enrichment indicated gene sets were predictive of an MRI trait and negative enrichment indicated the opposite. Shown are hallmarks with at least one significant enrichment
In single gene masking, enhancement was best predicted by SNTB1 (0.67 AUC and 0.58 AP) and B4GALT6 (0.64 AUC and 0.60 AP; Supplementary Fig. S10). SNTB1, a cytoskeletal protein, was down-regulated in a GBM cells study (Mongiardi et al., 2016) and a potential binder to PTPRZ, a protein contributing to glioma cell growth (Bourgonje et al., 2014).
In previous GBM radiogenomic studies, enhancement was associated with hypoxia, ECM, angiogenesis in 22 patients (Diehn et al., 2008); Biocarta pathways and genes, C1orf172, CAMSAP2, KCNK3 and LTBP1 in 23 patients (Jamshidi et al., 2014); and EGFR copy number amplification in 75 patients (Gutman et al., 2013). Gene sets involving the ECM, EGFR, C1orf172, KCNK3 and LTBP1 were confirmed to have performances greater than 0.70 in both AUC and AP (Supplementary Fig. S12a) in gene masking.
3.3.2. Edema
Tumor edema was identified as abnormal hyperintensity on fluid-attenuated inversion recovery (FLAIR) or T2-weighted (T2W) images. Edema often co-occurs with enhancement, implying a more aggressive tumor and does not typically occur in low-grade brain tumors. Edema also results from leaky capillaries and usually surrounds the tumor, spreading within the white matter. Edema suggests an inflammatory and/or immune response to a malignant tumor, which is essentially a foreign body when highly dedifferentiated.
Edema was associated with epithelial mesenchymal transition (EMT, metastasis and invasion), cell differentiation and growth (Table 2). p53 pathway (cell cycle, death), myogenesis, apical junction, heme metabolism and glycolysis (cell metabolism) were the top hallmark gene sets (Fig. 7a). GO terms relating to cell differentiation, death and adhesion with 0.80 AP (Supplementary Fig. S11b). Similar to the enhancement model, vasculature, immune system and EGFR-related processes (albeit through different GO terms) were a part of the most predictive gene sets.
Growth and metastasis were also found to be predictive of edema in single gene masking. RAI2, ANXA2 and POSTN, all related to cell growth, were the top three most predictive genes with 0.68–0.70 AUC and 0.60–0.64 AP. The top three ranked by AP were MTSS1, LAMA5 and KLHDC3, with 0.65–0.66 AP and 0.63–0.67 AUC (Supplementary Fig. S10). Both MTSSI and LAMA5 were associated with metastasis (Agarwala et al., 2018).
GSEA showed significant enrichment in EMT, angiogenesis, androgen response (hormone), hedgehog signaling (including MTSSI) and xenobiotic metabolism (drug metabolism; Fig. 7b). The appearance of drug metabolism could be due to the use of symptomatic relief drugs prior to surgery, such as corticosteroids for patients with neurologic symptoms caused by edema (Omuro and DeAngelis, 2013; Pitter et al., 2016).
Previously, POSTN was associated with edema in 78 GBM patients; the authors suggested POSTN was regulated by miR-219 and contributed to cell migration or invasion (Zinn et al., 2011). GO gene sets related to the study’s top five upregulated genes and microRNAs were found to be predictive of edema (Supplementary Fig. S13). Table 2 shows an overlap of gene set patterns between the study’s findings and gene masking of the edema model. In particular, gene sets associated POSTN, cell taxis and cell adhesion added to the association between edema and EMT.
3.3.3. Non-contrast enhancing tumor
nCET was best identified with T1W with/without contrast, FLAIR and T2W images. nCET is typically lower-grade tumor (better cellular differentiation, more closely resembling normal brain tissue), generating vessels with an intact BBB and absent of contrast enhancement. Although the abnormality on images is a mass-like neoplastic tissue, it is not rapidly dividing or aggressively dedifferentiating.
Cell cycle, growth and radiation response were themes among the most predictive gene sets for nCET. Mitotic spindle, ultraviolet (UV) response down (genes down-regulated in response to UV radiation), DNA repair and p53 pathway were predictive of nCET (Fig. 7a). Of the gene sets related to GBM genomic alterations, the nCET model had a mix of ones found in the enhancing and edema model (Supplementary Fig. S11c), and supported transcription patterns found in hallmark gene sets in Table 2.
SGPL1 (0.73 AUC and 0.56 AP) and DDR1 (0.66 AUC and 0.61 AP) were the top performing genes in single gene masking of the nCET model (Supplementary Fig. S10), where hypoxia, TNFA signaling via NFKB and oxidative phosphorylation were significantly enriched (Fig. 7b). The latter two gene sets were also identified in gene set masking (Supplementary Fig. S9).
3.3.4. Necrosis
Tumor necrosis was evaluated as the area of fluid signal intensity on T1W+Gd images. As tumors proliferate, they create new blood supply (angiogenesis) and/or expand to recruit blood from adjacent tissue. Subsequently, necrosis occurs, typically within the central portions of an aggressive tumor as the outer rim of enhancing surviving cells can be observed on MR images.
Vasculature, apoptosis, immune system and homeostasis were associated with necrosis (Table 2). Predictive GO terms for necrosis included several TP53- and PTEN-related processes. Gene masking found some gene sets related to IL4, WWTR1, RUNX3, ITGA5 and CDKN2A [found in previous radiogenomic studies (Gevaert et al., 2014; Gutman et al., 2013; Jamshidi et al., 2014) Supplementary Fig. S14] support the association between necrosis and apoptosis and the immune system. Earlier, drug metabolism was predictive of edema, but was also predictive of necrosis. Besides corticosteroids, antiepileptics can be prescribed patients who experience seizures from tumors (Omuro and DeAngelis, 2013).
In single gene masking, CACNB2 (0.67 AUC and 0.51 AP) and PACSIN3 (0.64 AUC and 0.51 AP) were the most predictive for necrosis (Supplementary Fig. S10) and the MYC targets hallmark was significantly enriched (Fig. 7b).
3.3.5. Focal versus non-focal
Focal versus non-focal traits were determined via T1W+Gd and FLAIR or T2W images. Focal tumors appear in one region. Non-focal tumors included those described as multifocal, multicentric or with gliomatosis cerebri. A multifocal tumor is one with separate enhancing regions that appear connected on FLAIR/T2W images with contiguous hyperintensity spreading via white matter tracts. A multicentric GBM has multiple enhancing or non-enhancing tumors growing synchronously without contiguity on FLAIR/T2W. Gliomatosis cerebri is a rare, diffusely infiltrating subtype and involve at least three cerebral lobes.
Focal traits were associated with growth, transport, vasculature and hypoxia (Table 2). Several of these involved PTEN and interestingly, RB1 may be related to the use of corticosteroids. Secretion by cell includes genes potentially overlapped by others, such as NF1, EGF and VEGF.
In general, focal traits were better predicted by GO gene sets related to GBM genes (Supplementary Fig. S11e) than hallmark gene sets (Fig. 7) or single genes (Supplementary Fig. S10). These highly predictive GO gene sets (0.90 AUC and 0.80 AP) indicated broad tumor characteristics, e.g. proliferation (growth, response to growth factors, secretion of growth factors) resulting in the need for angiogenesis (vasculature development), were used by the focal model to determine focal versus non-focal tumors.
3.3.6. Expansive versus infiltrative
Expansive versus infiltrative was measured as the ratio of T1/FLAIR abnormality. Expansive tumors have similar distribution on T1W as on FLAIR or T2W; the closer the two, the better defined the tumor margins and the better for surgical resection. Infiltrative tumors have FLAIR/T2W abnormality that is large compared with T1W abnormality, where the tumor is spreading through white matter tracts to cause large edema relative to the core tumor mass. Infiltrative traits indicate ill-defined tumor margins, less successful surgical debulking and worse prognosis.
Infiltrative traits were best predicted by gene sets related to oxygen, transport, healing and growth (Table 2). Gene masking showed that GO gene sets were more predictive than hallmark gene sets (Fig. 7). Of the top GO gene sets, would healing and hemostasis were the most predictive and several included TP53, MDM2 and PTEN. Notably, MDM2 transcription is regulated by TP53. Previous radiogenomic studies related to expansiveness or infiltrative traits found associations with MYC, NFKBIA and immune cell gene modules (Colen et al., 2014). The infiltrative model was masked with related gene sets and was able to predict infiltrative tumors with 0.50–0.70 AP (Supplementary Fig. S15). In single gene masking, ZBTB48 (0.68 AUC and 0.53 AP) and PRTN3 (0.67 AUC and 0.57 AP) as the best single gene predictors (Supplementary Figs S16–S24).
For more gene masking (see Supplementary Figs S16–S24).
3.4 Radiogenomic traits: patient-specific associations
Gene masking was used to identify cohort-level radiogenomic associations as genes were ranked by their overall classification performance among all tumors. In contrast, gene saliency was measured for each patient and identified patient-level radiogenomic associations, termed ‘radiogenomic traits’. For salient genes in the subtype model (see Supplementary Fig. S25).
Classical subtype genes were salient in predicting larger proportions of necrosis, where 77 patients were enriched with classical genes in the necrosis model (Fig. 8). Similarly, neural and proneural gene sets were associated with greater edema and nCET proportions, respectively.
Fig. 8.
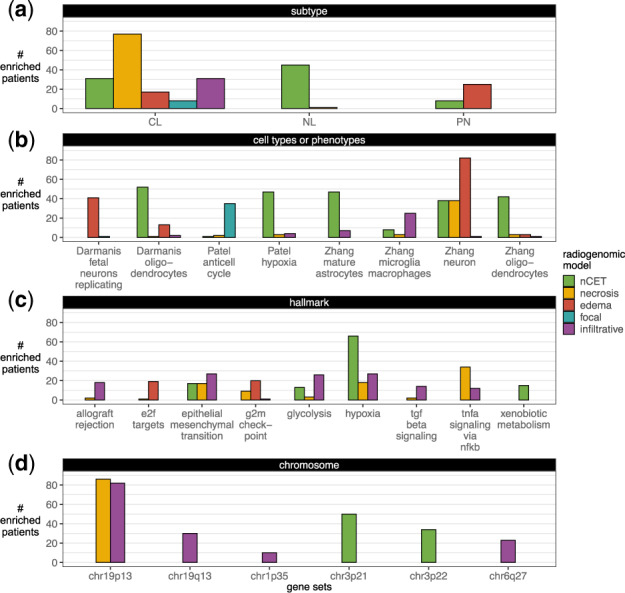
Radiogenomic traits. In gene saliency, each patient’s genes were considered enriched for a gene set at an adjusted P-value of <0.05. (a) Subtype (Verhaak et al., 2010), (b) cell types or phenotypes (Darmanis et al., 2015; Patel et al., 2014; Zhang et al., 2016) and MSigDB’s (c) hallmark and (d) chromosome gene sets with at least ten enriched patients are shown. For more gene saliency results (see Supplementary Fig. S26)
The nCET model showed more than 40 patients had salient genes enriched by oligodendrocytes, mature astrocytes, and hypoxia gene sets. Larger edema proportions were associated with neurons and replicating fetal neurons genes. The anti-cell cycle genes [negatively correlated with the cell cycles genes, some of which were a part of the hypoxia gene set in Patel et al. (2014)] were associated with prediction of the non-focal class (35 patients enriched). Patients were not significantly enriched with cell type or phenotype in the enhancing model, possibly reflecting more tumor heterogeneity in patients with more enhancing and aggressive tumor.
The hypoxia and nCET association (66 enriched patients) were consistent with the aforementioned anti-cell and hypoxia findings and with gene masking analysis, where vasculature development was predictive of nCET at the cohort-level. The association between hypoxia and greater proportions of nCET may be linked to lower-grade cells in the beginning stages of aggressive tumor growth and therefore responding to the beginning stages of hypoxic conditions and driving angiogenesis. Xenobiotic metabolism was enriched in 15 patients for predicting larger nCET, suggesting larger tumor sizes may be related to an increased dosage of drugs administered prior to surgery (Pitter et al., 2016).
Interestingly, the edema model showed only two hallmarks enriched by more than 10 patients. Although gene masking showed the edema model had high overall performance with the EMT gene set, other genes that are not associated with a predefined gene set may have been more influential in predicting each individual patient’s edema proportions. In fact, the EMT hallmark was more associated with the radiogenomic model’s belief of an infiltrative tumor in 27 patients. This subset of patients supports the hypothesis that tumor cells with alterations in EMT-related genes are driving the observation of higher edema proportions than tumor cell proportions. Glycolysis and hypoxia hallmarks were also moderately (<25 patients) associated with infiltrative tumors. TNFA signaling via NFKB was associated with larger proportions of necrosis in 34 patients.
Chromosomal aberrations have been reported in GBM (Ohgaki and Kleihues, 2007; Verhaak et al., 2010). There were 86 and 82 patients who were enriched by the chr19p13 gene set in predicting their necrosis and infiltrative traits, respectively. Genes in chr19q13, chr1p35 and chr6q27 were also salient to infiltrative tumors. chr3p21 and chr3p22 gene sets were also salient for greater nCET proportions.
3.4.1. Radiogenomic traits with survival implications
Of the 175 patients with radiogenomic data, 127 had all six MRI traits labeled and outcomes data. Given that only a subset of these individuals had clinical, imaging, and transcriptomic data to perform a survival analysis, we tested whether this subset of patients had any differences in outcomes compared with the transcriptome cohort (n = 528). There were no OS or PFS differences between the transcriptome cohort, the subset of patients with MRI traits and the subset of patients with all six MRI traits (Supplementary Fig. S27). In building the multivariable Cox models, 3 clinical traits, 6 MRI traits, and 54 radiogenomic traits were considered (Supplementary Fig. S29).
Figure 9 shows patients had significant differences when dichotomized by their radiogenomic traits. Patients that had neural genes as among the most salient genes for predicting larger nCET had better PFS compared with those who did not. Likewise, patients had significantly worse OS when chr3p21 genes were important in predicting nCET proportion was , and when chr19p13 or chr1p35 genes were important in predicting infiltration. In contrast, dichotomizing patients based solely on individual MRI traits had no OS or PFS differences, except in the counterintuitive case of expansive versus infiltrative (Supplementary Fig. S28). Infiltrative tumors had a univariate HR of 0.92 when estimating PFS (Table 3). However, after adjusting for patient covariates and radiogenomic traits, infiltrative tumors had an adjusted HR of 1.61 and correctly follow the intuition that infiltrative tumors would have a higher probability of progression than expansive tumors.
Fig. 9.
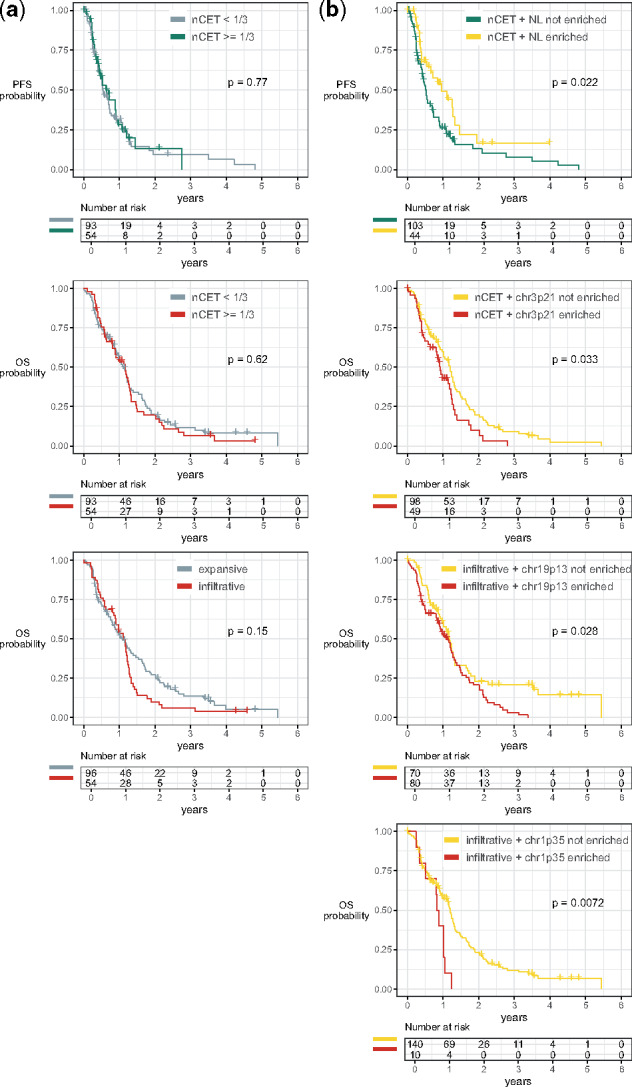
OS and PFS dichotomized by (left) imaging traits and (right) radiogenomic traits. Patients split in (b) had a median PFS of 0.96 versus 0.52 years (161-day difference). Similarly, the median OS was (d) 1.19 versus 0.91 years (101-day difference), (f) 1.18 versus 1.14 years (15-day difference) and (g) 1.19 versus 0.85 years (125-day difference). No differences were found in (a, c, e)
Table 3.
Cox regression analysis of traits associated with OS and PFS
| Univariate HR (95% CI) | Adjusted HR (95% CI) | P-value | |
|---|---|---|---|
| OS (n = 127, deaths = 107) | |||
| Clinical | |||
| Gender is male | 0.95 (0.64–1.41) | 0.80 (0.53–1.20) | 0.280 |
| Race is white | 1.69 (0.85–3.37) | 2.30 (1.09–4.85) | 0.029* |
| Diagnosis age is below median | 0.82 (0.56–1.21) | 0.79 (0.53–1.20) | 0.273 |
| Radiogenomic | |||
| Infiltrative + chr1p35 | 2.06 (0.98–4.31) | 2.06 (0.95–4.43) | 0.066 |
| Edema + endothelial | 2.07 (0.76–5.67) | 4.36 (1.47–12.9) | 0.008* |
| Necrosis + GBM core astrocytes | 0.46 (0.14–1.49) | 0.11 (0.03–0.45) | 0.002* |
| Necrosis + EMT | 1.40 (0.80–2.43) | 3.45 (1.75–6.82) | <0.001* |
| nCET + myogenesis | 2.85 (0.89–9.07) | 10.7 (2.48–46.5) | 0.002* |
| Necrosis + MYC targets (v2) | 0.76 (0.31–1.87) | 0.31 (0.10–0.98) | 0.045* |
| Infiltrative + mTORC1 signaling | 1.87 (0.76–4.61) | 2.38 (0.93–6.10) | 0.071 |
| PFS (n = 127, progressions = 88) | |||
| Clinical | |||
| Gender is male | 1.03 (0.65–1.62) | 0.80 (0.48–1.34) | 0.394 |
| Race is white | 1.28 (0.62–2.65) | 1.96 (0.89–4.30) | 0.094 |
| Diagnosis age is below median | 0.98 (0.64–1.50) | 1.25 (0.77–2.04) | 0.373 |
| Imaging | |||
| Tumor was infiltrative | 0.92 (0.58–1.45) | 1.61 (0.96–2.71) | 0.072 |
| Radiogenomic | |||
| Infiltrative + chr1p35 | 2.06 (0.98–4.31) | 4.20 (1.89–9.33) | <0.001* |
| Infiltrative + EMT | 2.57 (1.23–5.39) | 2.24 (1.27–3.96) | 0.006* |
| Necrosis + MYC targets (v2) | 0.21 (0.03–1.49) | 0.06 (0.01–0.47) | 0.008* |
| Edema + fetal neurons replicating | 1.48 (0.93–2.35) | 2.50 (1.44–4.33) | 0.001* |
| Infiltrative + TGF-β signaling | 1.51 (0.80–2.86) | 1.91 (0.94–3.92) | 0.076 |
| Edema + chr18p11 | 2.22 (0.80–6.15) | 5.79 (1.87–18.0) | 0.002* |
| Edema + G2M checkpoint | 0.75 (0.36–1.56) | 0.46 (0.20–1.05) | 0.066 |
| nCET + chr22q13 | 0.35 (0.09–1.42) | 0.36 (0.08–1.60) | 0.180 |
| Infiltrative + chr6q27 | 1.33 (0.76–2.33) | 1.79 (0.98–3.30) | 0.060 |
| Necrosis + p53 pathway | 1.06 (0.39–2.91) | 2.44 (0.81–7.39) | 0.114 |
Note: Both OS and PFS adjusted models had P < 0.001 in the likelihood ratio test.
P < 0.05.
Males and non-white races had better OS and PFS, while patients diagnosed below the median age had better OS, but worse PFS. The final Cox model consisted of six significant traits, five radiogenomic traits and race when estimating OS (Table 3). The final PFS model had five significant radiogenomic traits. In comparison, a Cox model with clinical and imaging traits had no significant factors in estimating OS or PFS. The survival analyses suggest that radiogenomic traits extracted from neural network models have prognostic value.
4 Discussion
We demonstrate how deep neural networks can be used to discover radiogenomic associations. First, we predict imaging traits using gene expression profiles, showing that our neural network-based approaches outperform other classifiers. Neural networks had better performances due to a combination of factors: the use of non-linear activation functions (versus linear relationships in logistic regression), multiple layers of transformation (versus a single, shallow transformations of support vector machines), and ability to use all gene expressions simultaneously (versus using a subset of genes at a time in random forests and gradient-boosted trees). Although neural networks have a large number of parameters to learn, we leverage transfer learning to train radiogenomic neural networks using a transcriptomic autoencoder modeled on a much larger cohort to address the impedance of relatively small radiogenomic datasets. Second, we present methods based on input masking and class saliency to facilitate interpretation of radiogenomic relationships, providing a way to understand the results of an otherwise ‘black box’ method, which is the main criticism against neural networks. Third, using our model interpretation methods, we identify pertinent gene expressions that may act as transcriptomic drivers for each imaging trait. We put forth a set of potential imaging surrogates to provide a clearer biological basis of commonly assessed imaging phenotypes in GBM and relate them to trends in patients’ OS and PFS.
Gene masking identifies cohort-level radiogenomic associations, where association strength was measured by the model’s classification when only a gene subset was used. Radiogenomic associations have common themes related to major GBM candidate driver genes and terms, such as cell growth and vasculature. Each MRI trait also show specificity toward components of these general themes, such as different functionalities of EGFR or cell death by autophagy in the edema model compared with apoptosis in the necrosis model. We identify unique associations between imaging traits and different themes: edema with cell invasion and differentiation; enhancement with immune system processes and hormones; nCET with cell cycle and UV response; necrosis with apoptosis; and focal with cell transport and response to some compounds.
Prior radiogenomic studies have mainly reported cohort-level associations. In reality, multiple gene expression profiles, when influenced by different environmental factors, may lead to the same observed imaging trait. Toward this end, gene saliency is used to identify patient-level radiogenomic traits. In gene saliency, each patient has his/her own list of relevant genes for each imaging trait; it is then determined if the patient’s salient genes are significantly associated with a gene set. We describe subsets of patients with common radiogenomic relationships that are not apparent in gene masking, such as the association between infiltrative traits and EMT genes or larger nCET proportions and drug metabolism. Some of these radiogenomic traits are significant factors in predicting patient survival.
Furthermore, we validate our modeling approach by training a neural network to predict molecular subtypes. We report an experiment that evaluates the model’s ability to learn meaningful relationships. Not only does the subtype model achieve near-perfect classification, the model is able to select genes relevant to each subtype among the input of 12 042 genes. In the radiogenomic models, we validate our radiogenomic associations with prior GBM studies in radiogenomics and genomics and found corresponding relationships. We also identify new findings that have not been widely reported in radiogenomics due to the ability of gene saliency to provide patient-specific radiogenomic traits and the inclusion of the entire gene profile in our models. These results support the neural network’s abilities to identify associations with existing domain knowledge and to suggest potential starting points for further investigation.
We recognize the limits of radiogenomic analysis, particularly in terms of small sample size, limited tissue sampling of a heterogeneous tumor and limited follow-up information. Sample size is an inherent challenge in radiogenomics. TCGA tends to have the most radiogenomic data but lacks detailed clinical data. Although larger cohorts do exist, tumors are across multiple grades (Aerts et al., 2014) and do not use molecular profiling (Chang et al., 2018). These limitations may be addressed as the cost of high-throughput platforms decrease and multiple tumor regions are sampled (Puchalski et al., 2018). With 528 gene expression profiles and a radiogenomic subset of 175, we show that neural networks can model transcriptomic heterogeneity to reflect phenotypic differences in imaging. The VASARI feature set is also limited in that it provides a gross categorization of imaging features and only one experienced reader’s annotations of the imaging data is used. A more comprehensive analysis that includes quantitative (radiomic) traits may be warranted. Finally, the radiogenomic associations and traits are only hypothesized and not experimentally proven, though we attempt to compare our discovered associations with those that have been previously reported in literature. To validate the identified associations, cell and animal studies would allow controlled experiments between genes and imaging phenotypes (Zinn et al., 2018).
5 Conclusion
Using a neural network-based approach to radiogenomic mapping, we highlight the representational and discriminative capacity of neural networks to model the high-dimensional, non-linear and correlative nature of gene expressions to predict typical GBM imaging traits. We demonstrate the use of neural network interpretation techniques based on input masking and class saliency to understand what the model has learned and to extract relevant radiogenomic relationships. The learned radiogenomic associations and traits may point to potential transcriptomic drivers of imaging traits and could further clarify the understanding of the relationship between two complementary and often unintegrated datasets. We show that integrated patient traits between imaging and gene expression are better indicators of survival than imaging or clinical traits alone. As such, prognostication and treatments may be further individualized, where targeted pathways could be considered in the selection of an appropriately tailored chemotherapeutic agent.
Supplementary Material
Acknowledgements
We are thankful for the feedback and discussions from the faculty and students of the Medical & Imaging Informatics group; the biostatisticians, Drs James Sayre and Audrey Winter; and The Cancer Imaging Archive members.
Funding
This work was supported in part by the National Institutes of Health [F31CA221061 and T32EB016640 to N.F.S. and R01CA157553 to N.F.S., W.H. and S.E.-S.]; the National Science Foundation [No. 1722516 to W.H.]; Amazon Web Services and UCLA Department of Computational Medicine partnership to W.H.; and UCLA’s Integrated Diagnostics Program to N.F.S. and W.H.
Conflict of Interest: none declared.
References
- Aerts H.J. et al. (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun., 5, 4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwala R. et al. (2018) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 46, D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengio Y. (2009) Learning Deep Architectures for AI, Vol. 2. Now Publishers, Inc, Boston, MA, USA. [Google Scholar]
- Bourgonje A.M. et al. (2014) Intracellular and extracellular domains of protein tyrosine phosphatase PTPRZ-B differentially regulate glioma cell growth and motility. Oncotarget, 5, 8690–8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K. et al. (2018) Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Clin. Cancer Res., 24, 1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Guestrin C. (2016) XGBoost: a scalable tree boosting system. In: Proceedings 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 785–794. ACM, San Francisco, CA, USA.
- Chen Y. et al. (2016) Gene expression inference with deep learning. Bioinformatics, 32, 1832–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet F. et al. (2015). Keras. Available at: https://github.com/fchollet/keras.
- Colen R.R. et al. (2014) Imaging genomic mapping of an invasive MRI phenotype predicts patient outcome and metabolic dysfunction: a TCGA Glioma Phenotype Research Group project. BMC Med. Genomics, 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S. et al. (2015) A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA, 112, 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M. et al. (2008) Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc. Natl. Acad. Sci. USA, 105, 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert O. et al. (2012) Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data-methods and preliminary results. Radiology, 264, 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert O. et al. (2014) Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology, 273, 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert O. et al. (2017) Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci. Rep., 7, 41674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I. et al. (2016) Deep Learning. MIT Press, Cambridge, MA, USA. [Google Scholar]
- Grossmann P. et al. (2017) Defining the biological basis of radiomic phenotypes in lung cancer. Elife, 6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W. et al. (2015) Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J. Med. Imaging, 2, 041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman D.A. et al. (2013) MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology, 267, 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha R. et al. (2019) Predicting breast cancer molecular subtype with MRI dataset utilizing convolutional neural network algorithm. J. Digit. Imaging, 32, 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L.S. et al. (2017) Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro. Oncol., 19, 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshidi N. et al. (2014) Illuminating radiogenomic characteristics of glioblastoma multiforme through Integration of MR imaging, messenger RNA expression, and DNA copy number variation. Radiology, 270, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickingereder P. et al. (2016) Radiogenomics of glioblastoma: machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology, 281, 907–918. [DOI] [PubMed] [Google Scholar]
- Korfiatis P. et al. (2017) Residual deep convolutional neural network predicts MGMT methylation status. J. Digit. Imaging, 30, 622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotikalapudi R. et al. (2017) keras-vis. https://github.com/raghakot/keras-vis.
- Lecun Y. et al. (2015) Deep learning. Nature, 521, 436–444. [DOI] [PubMed] [Google Scholar]
- Li S. et al. (2018). A novel radiogenomics framework for genomic and image feature correlation using deep learning. In: 2018 IEEE International Conference on Bioinformatics and Biomedicine, Vol. 1, pp. 1–8. IEEE. [Google Scholar]
- Liberzon A. et al. (2011) Molecular signatures database (MSigDB) 3.0. Bioinformatics, 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A. et al. (2015) The molecular signatures database hallmark gene set collection. Cell Syst., 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon R. et al. (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature, 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongiardi M.P. et al. (2016) c-MYC inhibition impairs hypoxia response in glioblastoma multiforme. Oncotarget, 7, 33257–33271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naba A. et al. (2012) The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell. Proteomics, 11, M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeini K.M. et al. (2013) Identifying the mesenchymal molecular subtype of glioblastoma using quantitative volumetric analysis of anatomic magnetic resonance images. Neuro. Oncol., 15, 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki H., Kleihues P. (2007) Genetic pathways to primary and secondary glioblastoma. Am. J. Pathol., 170, 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omuro A., DeAngelis L.M. (2013) Glioblastoma and other malignant gliomas. JAMA, 310, 1842. [DOI] [PubMed] [Google Scholar]
- Parsons D.W. et al. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science, 321, 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.P. et al. (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science, 344, 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F. et al. (2011) Scikit-learn: machine learning in Python. J. Mach. Learn. Res., 12, 2825–2830. [Google Scholar]
- Pitter K.L. et al. (2016) Corticosteroids compromise survival in glioblastoma. Brain, 139, 1458–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope W.B. et al. (2005) MR imaging correlates of survival in patients with high-grade gliomas. Am. J. Neuroradiol., 26, 2466–2474. [PMC free article] [PubMed] [Google Scholar]
- Pope W.B. et al. (2008) Relationship between gene expression and enhancement in glioblastoma multiforme: exploratory DNA microarray analysis. Radiology, 249, 268–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchalski R.B. et al. (2018) An anatomic transcriptional atlas of human glioblastoma. Science, 360, 660–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpace L. et al. (2016) Radiology data from The Cancer Genome Atlas Glioblastoma Multiforme [TCGA-GBM] collection. The Cancer Imaging Archive. doi: 10.7937/K9/TCIA.2016.RNYFUYE9.
- Segal E. et al. (2007) Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat. Biotechnol., 25, 675–680. [DOI] [PubMed] [Google Scholar]
- Sergushichev A.A. (2016) An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv, 060012.
- Simonyan K. et al. (2014) Deep inside convolutional networks: visualising image classification models and saliency maps. arXiv Prepr. arXiv1312.6034.
- Subramanian A. et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak R.G. et al. (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell, 17, 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K. et al. (2016) MR imaging–based analysis of glioblastoma multiforme: estimation of IDH1 mutation status. Am. J. Neuroradiol., 37, 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiler M.D., Fergus R. (2014). Visualizing and understanding convolutional networks. In: European Conference on Computer Vision, pp. 818–833. Springer.
- Zhang B. et al. (2017) Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro. Oncol., 19, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. (2016) Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron, 89, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. et al. (2015) Deciphering genomic underpinnings of quantitative MRI-based radiomic phenotypes of invasive breast carcinoma. Sci. Rep., 5, 17787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn P.O. et al. (2011) Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS One, 6, e25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn P.O. et al. (2018) A coclinical radiogenomic validation study: conserved magnetic resonance radiomic appearance of periostin-expressing glioblastoma in patients and xenograft models. Clin. Cancer Res., 24, 6288–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.