Abstract
Background
Publicly available genomic and transcriptomic data in searchable databases allow researchers to investigate specific medical issues in thousands of patients. Many studies have highlighted the role lipids play in cancer initiation and progression and reported nutritional interventions aimed at improving prognosis and survival. Therefore, there is an increasing interest in the role that fat intake may play in cancer. It is known that there is a relationship between BMI and survival in patients with cancer, and that there is an association between a high-fat diet and increased cancer risk. In some cancers, such as colorectal cancer, obesity and high fat intake are known to increase the risk of cancer initiation and progression. On the contrary, in patients undergoing treatment for melanoma, a higher BMI unexpectedly acts as a protective factor rather than a risk factor; this phenomenon is known as the obesity paradox.
Objective
We aimed to identify the molecular mechanism underlying the obesity paradox, with the expectation that this could indicate new effective strategies to reduce risk factors and improve protective approaches.
Methods
In order to determine the genes potentially involved in this process, we investigated the expression values of lipid-related genes in patients with melanoma or colorectal cancer. We used available data from 2990 patients from 3 public databases (IST [In Silico Transcriptomics] Online, GEO [Gene Expression Omnibus], and Oncomine) in an analysis that involved 3 consecutive validation steps. Of this group, data from 1410 individuals were analyzed in the IST Online database (208 patients with melanoma and 147 healthy controls, as well as 991 patients with colorectal cancer and 64 healthy controls). In addition, 45 melanoma, 18 nevi, and 7 healthy skin biopsies were analyzed in another database, GEO, to validate the IST Online data. Finally, using the Oncomine database, 318 patients with melanoma (312 controls) and 435 patients with colorectal cancer (445 controls) were analyzed.
Results
In the first and second database investigated (IST Online and GEO, respectively), patients with melanoma consistently showed significantly (P<.001) lower expression levels of 4 genes compared to healthy controls: CD36, MARCO, FABP4, and FABP7. This strong reduction was not observed in patients with colorectal cancer. An additional analysis was carried out on a DNA-TCGA data set from the Oncomine database, further validating CD36 and FABP4.
Conclusions
The observed lower expression of genes such as CD36 and FABP4 in melanoma may reduce the cellular internalization of fat and therefore make patients with melanoma less sensitive to a high dietary fat intake, explaining in part the obesity paradox observed in patients with melanoma.
Keywords: gene expression; obesity paradox; melanoma, colorectal cancer; CD36; FABPs; transcriptomic analysis; public databases
Introduction
Genomic, transcriptomic, and proteomic data from several thousand patients and corresponding healthy controls are now publicly available on the internet, for many different pathologies, including different types of cancers. This allows researchers to investigate specific questions and medical hypotheses in silico, directly in the human context, without certain ethical concerns. We previously investigated expression data from several thousand patients, and identified novel potential markers useful for improving the diagnosis of melanoma and other solid cancers [1,2], as well as novel therapeutic approaches that were then validated in vitro by classical bench science [3,4]. In this study we aimed to determine the molecular mechanisms underlying the unexpected protective role of high fat intake in melanoma, given that obesity is a known risk factor in other cancers. The role fat plays in health maintenance as well as disease initiation and progression is being extensively investigated. There is particular interest in the protective role diet may have on cancer, since different cancer types are associated with being overweight or obese; furthermore, increased cancer mortality has been linked to dyslipidemia [5]. In patients with cancer, metabolic alterations impacting carbohydrate and lipid metabolism can activate phosphoinositide 3-kinase (PI3K) pathway–dependent oncogenic signaling, leading to an inflammatory state with increased expression of specific cytokines [6]. Omega-3 polyunsaturated fatty acids (PUFAs) have a beneficial effect by counteracting inflammation in cancer cells, which PUFAs easily diffuse into via the plasma membrane, by stimulating the production of anti-inflammatory metabolites [7]. PUFAs reduce plasma lipid levels and lipoproteins by modulating hepatic lipoprotein secretion [8] and likely by also mitigating dyslipidemia effects. How obesity and diet might impact melanoma onset and therapeutic efficacy has been discussed [9]. Although obesity and abnormal lipid levels in the blood represent established risk factors in other malignancies, they do not seem to impact cutaneous melanoma [10]. In fact, they are only slightly associated with an increased risk of cutaneous melanoma in men [11], although insulin resistance and dyslipidemia seem to promote the growth of uveal melanoma. Interestingly, obesity has been associated with a better prognosis and improved survival in patients undergoing treatment for metastatic melanoma. A higher BMI appears to be a protective factor in melanoma and this phenomenon has been named the obesity paradox [12]. In several other cancers, including colorectal cancer, being overweight and having a higher BMI are known risk factors, rather than protective conditions [13]. Fat metabolism might be differently controlled in different cancer cell types, thereby explaining why dyslipidemia may play divergent roles in different cancers. Scavenger receptors including macrophage receptor with collagenous structure (MARCO) and CD36 recognize and internalize lipoproteins, making them susceptible to degradation [14,15]. Furthermore, fatty acid–binding proteins (FABPs) play an important role in cancer progression and the intracellular transportation of long-chain fatty acids [16]. These molecules exert a pivotal role in regulating lipid metabolism.
The aim of this study was to investigate the expression of genes related to lipid-handling to analyze the molecular basis of the obesity paradox observed in melanoma.
Methods
Overview
The study was carried out in 3 steps: (1) a selection step was carried out on a public database, IST (In Silico Transcriptomics) Online, to identify genes of potential interest; (2) data collected in the initial selection step were validated in a first-round validation step with another database, GEO (Gene Expression Omnibus); and (3) data were further validated in a subsequent second-round validation step with a third database (Oncomine). Table 1 shows the databases used throughout the process and the types of patients investigated in each step.
Table 1.
Schematic representation of the steps of this study.
| Study phase and data set | Cancer type and subtype | Cancer samples, n | Control samples, n | Database | |||
| Selection | IST (In Silico Transcriptomics) Online | ||||||
|
|
Melanoma and normal skin | Melanoma | 208 | 147 |
|
||
| Colorectal cancer and controls | Colorectal cancer | 991 | 64 |
|
|||
| First validation round | GEO (Gene Expression Omnibus) | ||||||
|
|
Melanoma and normal skin (GDS1375 data set) | Melanoma | 45 | 25 |
|
||
| Second validation round | Oncomine | ||||||
|
|
DNA-TCGA | Melanoma | 318 | 312 |
|
||
| DNA-TCGA | Colorectal cancer | ||||||
|
|
|
Colon adenocarcinoma | 212 | 445 |
|
||
|
|
Rectal adenocarcinoma | 90 | 445 |
|
|||
|
|
Colon mucinous adenocarcinoma | 37 | 445 |
|
|||
|
|
Cecum adenocarcinoma | 65 | 445 |
|
|||
|
|
Rectal mucinous adenocarcinoma | 7 | 445 |
|
|||
|
|
Rectosigmoid adenocarcinoma | 24 | 445 |
|
|||
First, in the selection step, the IST Online public database [17] was used to obtain gene expression data. It returns plots indicating the expression values of the given gene compared to the expression value of a second given reference gene. This can be carried out with several different cancer data sets and corresponding healthy controls. The analysis was performed with melanoma versus healthy skin and with colorectal cancer versus healthy control biopsies. In turn, we indicated our genes of interest (CD36, MARCO, FABP1, FABP2, FABP3, FABP4, FABP6, or FABP7) as the first gene and used a known housekeeping gene, beta-2 microglobulin (B2M), as the reference gene; it should be noted that the expression values of the first gene are independent from the reference gene and the values do not change if a different reference gene is chosen. We previously reported the methods used to study the expression of other molecules to identify relevant melanoma markers [2]. We analyzed data from 1410 individuals, including 208 patients with melanoma and 147 healthy controls, and 991 patients with colorectal cancer and 64 healthy controls. The first-round validation was carried out using the GEO public database [18]. The GDS1375 data set was used, which represents expression data from 45 melanoma biopsies, 18 nevi biopsies, and 7 heathy skin biopsies. The second-round validation was carried out on the DNA-TCGA data set in the Oncomine database [19]. In this case, 318 patients with melanoma were compared to 312 healthy controls, and 435 patients with colorectal cancer were compared to 445 healthy controls. The Human Protein Atlas public database was also interrogated [20].
Statistical Analysis
Within the scatterplots obtained from the IST Online database analysis, the number of patients falling above or below the chosen threshold were counted and analyzed according to the Fisher exact test using GraphPad Prism 5 (GraphPad Software, Inc).
Other statistical analyses were carried out on the expression values obtained by querying the GEO database. Data was analyzed with analysis of variance and analysis for the linear trend from healthy to nevi to melanoma samples was carried out, both with GraphPad Prism 5. The threshold for statistical significance was set at P<.001.
Results
Overview
Gene expression of CD36, MARCO, and various FABP isoforms in 355 patients (208 patients with melanoma versus 147 healthy skin controls) was analyzed, according to the transcriptome expression data reported in the IST Online database. Table 2 shows the results and indicates the statistical significance of distribution above or below the given threshold, according to the Fisher exact test, for both melanoma and colorectal cancer data for all 8 genes investigated. The threshold value was chosen as the value best discriminating the largest population within the controls. The following threshold values were used: CD36: 1000; MARCO: 150; FABP1: 100 in melanoma and 1000 in colorectal cancer; FABP2: 100; FABP3: 250; FABP4: 2000; FABP6: 200; FABP7: 500. FABP5 does not appear in the IST Online database.
Table 2.
Expression in melanoma and colorectal cancer, according to the IST (In Silico Transcriptomics) Online database. Where P values are <.001, there was a statistically significant difference between the respective cancer values versus control values, evaluated as distribution above or below the given threshold, according to the Fisher exact test.
| Gene | Regulation in melanoma versus controls | P value | Regulation in colorectal cancers versus controls | P value |
| CD36 | Downregulation | <.001 | No difference | .58 |
| MARCO | Downregulation | <.001 | No difference | .02 |
| FABP1 | No difference | .64 | Downregulation | <.001 |
| FABP2 | No difference | .15 | Downregulation | <.001 |
| FABP3 | No difference | .08 | No difference | .07 |
| FABP4 | Downregulation | <.001 | No difference | .60 |
| FABP6 | Upregulation | <.001 | Upregulation | <.001 |
| FABP7 | Downregulation | <.001 | No difference | >.99 |
Interestingly, the 4 genes that had a significant difference in melanoma were not significantly different in patients with colorectal cancer versus healthy controls, indicating that the difference observed in melanoma appears to be cancer-specific.
Figure 1 indicates the expression values of the 5 genes that had significant differences in melanoma versus healthy skin controls (CD36, MARCO, FABP4, FABP6, and FABP7). The significance of the distribution reported in Table 2 was computed by counting the number of individuals falling below or above the thresholds indicated by the dashed lines. The expression of CD36, MARCO, FABP4, FABP6, and FABP7 in melanoma samples and healthy skin biopsies is visualized, according to data retrieved from the IST Online database.
Figure 1.
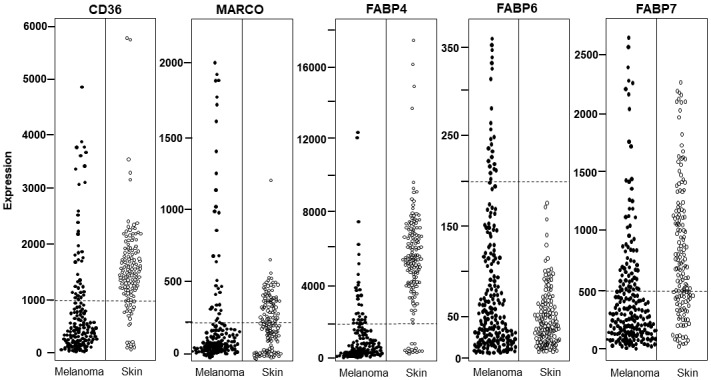
Expression of CD36, MARCO, FABP4, FABP6, and FABP7 in melanoma and healthy skin samples from the IST (In Silico Transcriptomics) Online data set. Each dot indicates one individual and dashed lines indicate the threshold used to calculate the statistical significance of the distribution difference reported in Table 2. All reported genes show a significantly different distribution in melanoma versus controls according to the Fisher exact test (P<.001).
These data indicate that melanoma samples show significantly lower expression of genes involved in fatty acid uptake (CD36 and MARCO) and intracellular fatty acid binding (FABP4 and FABP7) compared to healthy controls, and this phenomenon was not observed in a colorectal cancer data set.
First Validation Round
Data collected from the IST Online database were then validated on a different database, GEO. Expression values in melanoma were obtained from the GDS1375 data set, as detailed in the Methods section. Figure 2 shows that the expression values of CD36, MARCO, FABP4, and FABP7 are significantly decreased in melanoma samples (n=45) compared to nevi (n=18) and healthy skin (n=7) biopsies. A significant (P<.001) linear trend from healthy controls to nevi to melanoma biopsies was observed in CD36, MARCO, and FABP4. Therefore, the CD36, MARCO, FABP4, and FABP7 data obtained from the IST Online database were validated on the GEO database. FABP6, which was increased in melanoma compared to control in the IST Online database (Table 2), showed a weak, nonsignificant increase in the GEO database (Figure 2).
Figure 2.
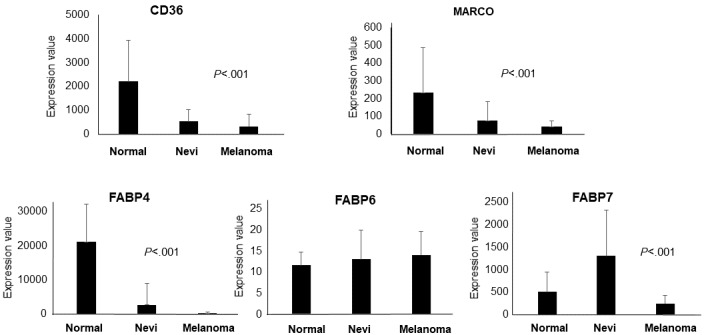
Expression of CD36, MARCO, FABP4, FABP6, and FABP7 in melanoma, nevi, and healthy skin biopsies from the GDS1375 data set on the GEO (Gene Expression Omnibus) database. Changes observed in the GEO database were in the same direction as observed in the IST (In Silico Transcriptomics) Online database for all genes except FABP6. Statistical significance was calculated by analysis of variance.
Second Validation Round
The 4 genes validated in GEO were further investigated in a third public database, Oncomine. Table 3 shows the statistical significance of the log2 copy number units change in the DNA-TCGA data set expression data from patients with melanoma and colorectal cancer compared to healthy controls, as analyzed in the Oncomine database. This analysis validated significant differences in CD36 and FABP4 expression in melanoma versus controls, and no significant difference in colorectal cancer.
Table 3.
Gene expression in the DNA-TCGA data sets analyzed in the Oncomine databasea.
| Gene | P values for the melanoma DNA-TCGA data set | P values for the colorectal cancer DNA-TCGA data set |
| CD36 | <.001 | >.99 |
| MARCO | .58 | >.99 |
| FABP4 | <.001 | >.99 |
| FABP7 | >.99 | .04 |
aThe statistical significance of the differential log2 copy number in patients with melanoma or colorectal cancer versus controls is reported. The significance threshold was set to P<.001.
A final investigation was then carried out using the Human Protein Atlas public database. Although the potential roles of the genes investigated in this study were not verified in melanoma, their roles have been confirmed in other cancers. Specifically, increased CD36 gene expression levels indicate an unfavorable prognostic value in 354 patients with stomach cancer (P<.001), and increased FABP7 gene expression levels indicate an unfavorable prognostic value in 877 patients with renal cancer (P<.001), yet indicate a favorable prognostic value in 1075 patients with breast cancer.
Discussion
In this study we investigated the expression of different genes involved in lipid metabolism and found a significant difference in melanoma versus controls. This may explain part of the mechanism behind the obesity paradox observed in patients undergoing treatment for metastatic melanoma. The mechanisms underlying the association between dyslipidemia and melanoma remain controversial; this is due to the different metabolic controls within bulk melanoma cells and cancer stem cells or metastasis-initiating cells [21,22]. Metastasis-initiating cells display high CD36 levels, which may indicate a crucial contribution of dietary lipids in the promotion of metastasis [23]. Furthermore, as we previously demonstrated, melanoma cancer stem cells show higher intracellular neutral lipids, higher lipogenesis activation, and lower autophagic flux [24]. This evidence indicates a complex molecular apparatus that allows melanoma cells to finely regulate fatty acid storage and mobilization depending on the metabolic environment and their differentiation level.
FABP4 and FABP2 have recently been reported to have a significant association with cancer progression in patients with colorectal cancer [16] and several studies demonstrate that obesity and high fat intake are risk factors in colorectal cancer [13,25-28]. On the other hand, several studies highlight the obesity paradox in melanoma, reporting significantly lower mortality in overweight patients undergoing treatment for melanoma [29], although a recent publication indicated that some caution is warranted [30]. Patients with melanoma or colorectal cancer appear to respond in opposite ways to high dietary fat intake or fat metabolism and may therefore be useful models to investigate how lipid-related gene expression may differentially regulate cancer initiation. For this reason, this study investigated lipid-related gene expression in patients with melanoma or colorectal cancer. Many genes have been identified as lipid modulators, although this field still remains poorly investigated. Genes controlling dyslipidemia in mice were recently reported [31], as well as other molecules that interfere with lipid storage [32,33], while a complete list of lipid-related genes in humans is currently lacking. In this work we investigated the expression of 8 genes (FABPs and other lipid-related genes) in melanoma and colorectal cancer biopsies, hypothesizing that differences in melanoma and colorectal cancer gene expression may partly explain the different role dietary fat plays in melanoma (ie, protective) and colorectal cancer (ie, detrimental). A significant reduction of the genes for scavenger receptors CD36 and MARCO (which are able to bind lipoproteins) and FABP4 and FABP7 translocases (which are able to bind and cell-internalize fatty acids) was found in melanoma biopsies compared to healthy controls, according to 2 independent databases, IST Online and GEO. We hypothesize that this reduced expression may lead to a reduced uptake of lipids and reduced cellular internalization. CD36 and FABP4 were also validated in a third database, Oncomine, using the DNA-TCGA data set. These genes showed no difference in control expression data compared to data from patients with colorectal cancer, for whom high fat dietary intake represents a negative prognostic factor. Therefore, we believe that the reduced gene expression observed in melanoma (571 patients and 484 controls, in 3 independent databases) might contribute to counteracting the detrimental effects of high fat intake.
More extensive analyses are ongoing in other cancers and on a larger list of relevant lipid-related genes; nevertheless, the results from this study may reveal some of the molecular mechanisms responsible for the obesity paradox observed in melanoma.
Acknowledgments
We thank Professor Eugenio Gaudio (Sapienza University) for the illuminating brainstorms rising our interest in molecular mechanisms linking cancer cell growth and lipid metabolism. This work was supported by an Italia-India Cooperation grant (Sapienza University) to CG and by RC 2018 from the Ministry of Health to AF.
Abbreviations
- B2M
beta-2 microglobulin
- FABP
fatty acid–binding protein
- GEO
Gene Expression Omnibus
- IST
In Silico Transcriptomics
- MARCO
macrophage receptor with collagenous structure
- PI3K
phosphoinositide 3-kinase
- PUFA
polyunsaturated fatty acids
Footnotes
Authors' Contributions: CG contributed to conceptualization and writing. LT and FS conducted data retrieval. FS analyzed data and LT wrote the initial draft. AF contributed to data interpretation and writing. CG and AF provided supervision.
Conflicts of Interest: None declared.
References
- 1.Biasiotta A, D'Arcangelo D, Passarelli F, Nicodemi EM, Facchiano A. Ion channels expression and function are strongly modified in solid tumors and vascular malformations. J Transl Med. 2016 Oct 04;14(1):285. doi: 10.1186/s12967-016-1038-y. https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-016-1038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Arcangelo D, Giampietri C, Muscio M, Scatozza F, Facchiano F, Facchiano A. WIPI1, BAG1, and PEX3 Autophagy-Related Genes Are Relevant Melanoma Markers. Oxid Med Cell Longev. 2018;2018:1471682. doi: 10.1155/2018/1471682. doi: 10.1155/2018/1471682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Arcangelo D, Facchiano F, Nassa G, Stancato A, Antonini A, Rossi S, Senatore C, Cordella M, Tabolacci C, Salvati A, Tarallo R, Weisz A, Facchiano AM, Facchiano A. PDGFR-alpha inhibits melanoma growth via CXCL10/IP-10: a multi-omics approach. Oncotarget. 2016 Nov 22;7(47):77257–77275. doi: 10.18632/oncotarget.12629. http://www.impactjournals.com/oncotarget/misc/linkedout.php?pii=12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Arcangelo D, Scatozza F, Giampietri C, Marchetti P, Facchiano F, Facchiano A. Ion Channel Expression in Human Melanoma Samples: In Silico Identification and Experimental Validation of Molecular Targets. Cancers (Basel) 2019 Mar 29;11(4) doi: 10.3390/cancers11040446. http://www.mdpi.com/resolver?pii=cancers11040446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallagher EJ, LeRoith D. Obesity and Diabetes: The Increased Risk of Cancer and Cancer-Related Mortality. Physiol Rev. 2015 Jul;95(3):727–48. doi: 10.1152/physrev.00030.2014. http://www.physiology.org/doi/full/10.1152/physrev.00030.2014?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dpubmed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat Rev Cancer. 2012 Feb 16;12(3):159–69. doi: 10.1038/nrc3215. [DOI] [PubMed] [Google Scholar]
- 7.Gu Z, Shan K, Chen H, Chen YQ. n-3 Polyunsaturated Fatty Acids and their Role in Cancer Chemoprevention. Curr Pharmacol Rep. 2015 Oct;1(5):283–294. doi: 10.1007/s40495-015-0043-9. http://europepmc.org/abstract/MED/26457243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Illingworth DR, Harris WS, Connor WE. Inhibition of low density lipoprotein synthesis by dietary omega-3 fatty acids in humans. Arteriosclerosis. 1984;4(3):270–5. doi: 10.1161/01.atv.4.3.270. [DOI] [PubMed] [Google Scholar]
- 9.Warner AB, McQuade JL. Modifiable Host Factors in Melanoma: Emerging Evidence for Obesity, Diet, Exercise, and the Microbiome. Curr Oncol Rep. 2019 Jul 01;21(8):72. doi: 10.1007/s11912-019-0814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauby-Secretan B, Scoccianti C, Loomis D, Grosse Y, Bianchini F, Straif K, International Agency for Research on Cancer Handbook Working Group Body Fatness and Cancer--Viewpoint of the IARC Working Group. N Engl J Med. 2016 Aug 25;375(8):794–8. doi: 10.1056/NEJMsr1606602. http://europepmc.org/abstract/MED/27557308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sergentanis TN, Antoniadis AG, Gogas HJ, Antonopoulos CN, Adami H, Ekbom A, Petridou ET. Obesity and risk of malignant melanoma: a meta-analysis of cohort and case-control studies. Eur J Cancer. 2013 Feb;49(3):642–57. doi: 10.1016/j.ejca.2012.08.028. [DOI] [PubMed] [Google Scholar]
- 12.Tomaipitinca L, Mandatori S, Mancinelli R, Giulitti F, Petrungaro S, Moresi V, Facchiano A, Ziparo E, Gaudio E, Giampietri C. The Role of Autophagy in Liver Epithelial Cells and Its Impact on Systemic Homeostasis. Nutrients. 2019 Apr 11;11(4) doi: 10.3390/nu11040827. http://www.mdpi.com/resolver?pii=nu11040827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Guo S, Li J, Bao W, Zhang P, Huang Y, Ling P, Wang Y, Zhao Q. Effects of high-fat diet-induced adipokines and cytokines on colorectal cancer development. FEBS Open Bio. 2019 Oct 30; doi: 10.1002/2211-5463.12751. doi: 10.1002/2211-5463.12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgoudaki A, Prokopec KE, Boura VF, Hellqvist E, Sohn S, Östling J, Dahan R, Harris RA, Rantalainen M, Klevebring D, Sund M, Brage SE, Fuxe J, Rolny C, Li F, Ravetch JV, Karlsson MCI. Reprogramming Tumor-Associated Macrophages by Antibody Targeting Inhibits Cancer Progression and Metastasis. Cell Rep. 2016 May 31;15(9):2000–11. doi: 10.1016/j.celrep.2016.04.084. https://linkinghub.elsevier.com/retrieve/pii/S2211-1247(16)30529-0. [DOI] [PubMed] [Google Scholar]
- 15.Calvo D, Gómez-Coronado D, Suárez Y, Lasunción MA, Vega MA. Human CD36 is a high affinity receptor for the native lipoproteins HDL, LDL, and VLDL. J Lipid Res. 1998 Apr;39(4):777–88. http://www.jlr.org/cgi/pmidlookup?view=long&pmid=9555943. [PubMed] [Google Scholar]
- 16.Vargas T, Moreno-Rubio J, Herranz J, Cejas P, Molina S, González-Vallinas M, Mendiola M, Burgos E, Aguayo C, Custodio AB, Machado I, Ramos D, Gironella M, Espinosa-Salinas I, Ramos R, Martín-Hernández R, Risueño A, De Las Rivas J, Reglero G, Yaya R, Fernández-Martos C, Aparicio J, Maurel J, Feliu J, Ramírez de Molina A. ColoLipidGene: signature of lipid metabolism-related genes to predict prognosis in stage-II colon cancer patients. Oncotarget. 2015 Mar 30;6(9):7348–63. doi: 10.18632/oncotarget.3130. http://www.impactjournals.com/oncotarget/misc/linkedout.php?pii=3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.IST database. [2020-04-14]. https://ist.medisapiens.com/
- 18.GEO database. [2020-04-14]. https://www.ncbi.nlm.nih.gov/gds.
- 19.Oncomine database. [2020-04-14]. https://www.oncomine.org/resource/login.html.
- 20.The Human Protein Atlas. [2020-04-14]. https://www.proteinatlas.org/
- 21.Tirinato L, Pagliari F, Limongi T, Marini M, Falqui A, Seco J, Candeloro P, Liberale C, Di Fabrizio E. An Overview of Lipid Droplets in Cancer and Cancer Stem Cells. Stem Cells Int. 2017;2017:1656053. doi: 10.1155/2017/1656053. doi: 10.1155/2017/1656053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi M, Li J, Chen S, Cai J, Ban Y, Peng Q, Zhou Y, Zeng Z, Peng S, Li X, Xiong W, Li G, Xiang B. Emerging role of lipid metabolism alterations in cancer stem cells. J Exp Clin Cancer Res. 2018 Jun 15;37(1):118. doi: 10.1186/s13046-018-0784-5. https://jeccr.biomedcentral.com/articles/10.1186/s13046-018-0784-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pascual G, Avgustinova A, Mejetta S, Martín M, Castellanos A, Attolini CS, Berenguer A, Prats N, Toll A, Hueto JA, Bescós C, Di Croce L, Benitah SA. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017 Jan 05;541(7635):41–45. doi: 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 24.Giampietri C, Petrungaro S, Cordella M, Tabolacci C, Tomaipitinca L, Facchiano A, Eramo A, Filippini A, Facchiano F, Ziparo E. Lipid Storage and Autophagy in Melanoma Cancer Cells. Int J Mol Sci. 2017 Jun 15;18(6) doi: 10.3390/ijms18061271. http://www.mdpi.com/resolver?pii=ijms18061271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moghaddam AA, Woodward M, Huxley R. Obesity and risk of colorectal cancer: a meta-analysis of 31 studies with 70,000 events. Cancer Epidemiol Biomarkers Prev. 2007 Dec;16(12):2533–47. doi: 10.1158/1055-9965.EPI-07-0708. http://cebp.aacrjournals.org/cgi/pmidlookup?view=long&pmid=18086756. [DOI] [PubMed] [Google Scholar]
- 26.Sinicrope FA, Foster NR, Sargent DJ, O'Connell MJ, Rankin C. Obesity is an independent prognostic variable in colon cancer survivors. Clin Cancer Res. 2010 Mar 15;16(6):1884–93. doi: 10.1158/1078-0432.CCR-09-2636. http://clincancerres.aacrjournals.org/cgi/pmidlookup?view=long&pmid=20215553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thygesen LC, Grønbaek M, Johansen C, Fuchs CS, Willett WC, Giovannucci E. Prospective weight change and colon cancer risk in male US health professionals. Int J Cancer. 2008 Sep 01;123(5):1160–5. doi: 10.1002/ijc.23612. doi: 10.1002/ijc.23612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bjørge T, Engeland A, Tverdal A, Smith GD. Body mass index in adolescence in relation to cause-specific mortality: a follow-up of 230,000 Norwegian adolescents. Am J Epidemiol. 2008 Jul 01;168(1):30–7. doi: 10.1093/aje/kwn096. [DOI] [PubMed] [Google Scholar]
- 29.Naik GS, Waikar SS, Johnson AEW, Buchbinder EI, Haq R, Hodi FS, Schoenfeld JD, Ott PA. Complex inter-relationship of body mass index, gender and serum creatinine on survival: exploring the obesity paradox in melanoma patients treated with checkpoint inhibition. J Immunother Cancer. 2019 Mar 29;7(1):89. doi: 10.1186/s40425-019-0512-5. https://jitc.biomedcentral.com/articles/10.1186/s40425-019-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donnelly D, Bajaj S, Yu J, Hsu M, Balar A, Pavlick A, Weber J, Osman I, Zhong J. The complex relationship between body mass index and response to immune checkpoint inhibition in metastatic melanoma patients. J Immunother Cancer. 2019 Aug 19;7(1):222. doi: 10.1186/s40425-019-0699-5. https://jitc.biomedcentral.com/articles/10.1186/s40425-019-0699-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuivenhoven JA, Hegele RA. Mining the genome for lipid genes. Biochim Biophys Acta. 2014 Oct;1842(10):1993–2009. doi: 10.1016/j.bbadis.2014.04.028. https://linkinghub.elsevier.com/retrieve/pii/S0925-4439(14)00115-X. [DOI] [PubMed] [Google Scholar]
- 32.Marini ES, Giampietri C, Petrungaro S, Conti S, Filippini A, Scorrano L, Ziparo E. The endogenous caspase-8 inhibitor c-FLIPL regulates ER morphology and crosstalk with mitochondria. Cell Death Differ. 2015 Jul;22(7):1131–43. doi: 10.1038/cdd.2014.197. doi: 10.1038/cdd.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giampietri C, Petrungaro S, Conti S, Facchiano A, Filippini A, Ziparo E. c-Flip KO fibroblasts display lipid accumulation associated with endoplasmic reticulum stress. Biochim Biophys Acta. 2015 Jul;1851(7):929–36. doi: 10.1016/j.bbalip.2015.02.018. https://linkinghub.elsevier.com/retrieve/pii/S1388-1981(15)00066-9. [DOI] [PubMed] [Google Scholar]


