Abstract
Despite enthusiasm about the potential for using fMRI‐based functional connectomes in the development of biomarkers for autism spectrum disorder (ASD), the literature is full of negative findings—failures to distinguish ASD functional connectomes from those of typically developing controls (TD)—and positive findings that are inconsistent across studies. Here, we report on a new study designed to either better differentiate ASD from TD functional connectomes—or, alternatively, to refine our understanding of the factors underlying the current state of affairs. We scanned individuals with ASD and controls both at rest and while watching videos with social content. Using multiband fMRI across repeat sessions, we improved both data quantity and scanning duration by collecting up to 2 hr of data per individual. This is about 50 times the typical number of temporal samples per individual in ASD fcMRI studies. We obtained functional connectomes that were discriminable, allowing for near‐perfect individual identification regardless of diagnosis, and equally reliable in both groups. However, contrary to what one might expect, we did not consistently or robustly observe in the ASD group either reductions in similarity to TD functional connectivity (FC) patterns or shared atypical FC patterns. Accordingly, FC‐based predictions of diagnosis group achieved accuracy levels around chance. However, using the same approaches to predict scan type (rest vs. video) achieved near‐perfect accuracy. Our findings suggest that neither the limitations of resting state as a “task,” data resolution, data quantity, or scan duration can be considered solely responsible for failures to differentiate ASD from TD functional connectomes.
Keywords: autism, fcMRI, functional connectivity, individual differences, naturalistic viewing, resting state
1. INTRODUCTION
Recent years have seen widespread interest in the potential for functional connectivity MRI (fcMRI) to serve as a biomarker or neuroendophenotype for autism spectrum disorders (ASDs). However, despite this enthusiasm, existing studies have failed to differentiate individuals with ASD from typically developing controls (TD) with sufficiently high in‐sample or out‐of‐sample accuracy to meet biomarker standards (Abraham et al., 2017; Chen et al., 2015; Heinsfeld, Franco, Craddock, Buchweitz, & Meneguzzi, 2018; Nielsen et al., 2013; Plitt, Barnes, & Martin, 2015; Yahata et al., 2016), and findings of group‐level differences between ASD and TD groups have been inconsistent across studies and include many negative results (Tyszka, Kennedy, Paul, & Adolphs, 2014; Dajani et al., 2019; He, Byrge, & Kennedy, 2020; for review see Hull, Jacokes, Torgerson, Irimia, & Van Horn, 2016).
Many factors could underlie this overall pattern of results, including (but not limited to) the following. First, most such studies have examined fcMRI data collected during the resting state, but such a task‐free state may not be sufficiently constrained or, alternatively, not sufficiently related to the behavioral presentation of ASD, to give rise to shared atypical patterns of brain functioning. Second, while data that are collected over longer scans and with more data points (Birn et al., 2013; Horien et al., 2018; Laumann et al., 2015) permit the most precise estimates of functional connectivity at the individual level, and accordingly many scanning initiatives are collecting more and more data points per individual with higher temporal resolution (Gordon et al., 2017; Laumann et al., 2015; Smith et al., 2013), existing studies of fcMRI in ASD typically use lower‐resolution imaging protocols and short scan durations, suggesting that estimates of functional connectivity in the literature may not be sufficiently precise to accurately discriminate diagnostic groups. Further, given the likelihood of lower data quality in clinical populations, and thus less usable data, precision and consistency of individual FC estimates may vary between groups, potentially giving rise to less accurate estimates of connectivity in the ASD group.
Here, we report on a new study of individuals with ASD and controls with several key advances designed to address these potential factors. In contrast to the existing literature, here we collect data not only during the resting state but also while individuals watch videos with varied social content in the scanner, which should constrain evoked brain activity and may relate more to atypical social behavior than the resting state. Also in contrast to the existing literature, which predominantly examines a single low temporal resolution (TR 2–3 s) 5–6 min resting‐state fcMRI scan per individual, here we collect for each individual up to eight high‐resolution multiband scans (TR 0.813 s), 13–16 min each, for up to 2 hr of data over two sessions—about 20 times more scanning time and 50 times more data points per individual than most existing studies of ASD. Our overarching goal was to either better differentiate functional connectomes at the group level or, alternatively, refine our understanding of the factors that are more or less likely to be responsible for inconsistency and negative results in the existing literature. In so doing, we asked: (a) are functional connectomes individually discriminable and consistent in both diagnosis groups; (b) are functional connectomes more similar within groups and do they show common group‐specific patterns; and (c) can groups be differentiated, and if so, does video watching or rest allow for better separation of the groups?
Functional imaging in special populations always requires difficult tradeoffs. Analyzing high‐quality data is paramount (see, e.g., Deen & Pelphrey, 2012), yet frequently requires excluding more scans and more subjects than is desirable, limiting generalizability and discarding expensive and unique data. Retaining more subjects may increase generalizability, but at the cost of increased difficulty in disentangling potential effects of diagnosis from those of data quality. Here, we have attempted to bridge that trade‐off by reporting results in multiple ways. In the main text, we report results from a small sample with the highest quality data, using the strictest data inclusion thresholds. In this sample, some reliable group differences in head motion (as indexed using filtered framewise displacement or FDfilt, see Methods) remained. In the Supporting Information, we further address residual effects of data quality by downsampling each rest scan such that no group differences in FDfilt remained, yielding the equivalent of about 5 min of low‐temporal‐resolution data (comparable to the existing ASD fcMRI literature) of the highest quality. We also report in the Supporting Information results from a larger, more inclusive sample with a more relaxed censoring threshold. Finally, we also report results under several additional preprocessing alternatives. Results were generally consistent across approaches.
To preview our results, we obtained functional connectivity estimates that were highly discriminable, allowing for near‐perfect “fingerprinting” or individual identification regardless of diagnosis group, and these functional connectomes were consistent within individuals and equally so in both groups. However, while it might have been expected that dissimilarities to TD functional connectivity patterns would be observable in the ASD group, we did not observe any such reductions in similarity to TD FC patterns robustly or consistently at the group level, although very small reductions were observed in a few isolated preprocessing approaches. We found no consistent atypical FC patterns shared across the ASD group. Prediction of diagnosis group from functional connectomes, using the complete connectome as well as a targeted selection of edges, achieved accuracy not far from chance. However, these same prediction approaches achieved near perfect accuracy when applied to scan type (rest vs. video) irrespective of group. Overall, our findings suggest that none of the potential factors we identified and addressed can be considered fully responsible for the failures in the literature to robustly and consistently differentiate ASD and TD functional connectomes. Despite adding a constrained and socially relevant task and increasing scan resolution, duration, and temporal samples, which resulted in discriminable and consistent FC estimates and near‐perfect prediction of scan type, successful discrimination between ASD and TD connectomes was not achieved.
2. MATERIALS AND METHODS
2.1. Participants
Participants included 25 high‐functioning adults with an autism spectrum disorder (ASD; mean age 24.2; range 17–54; 6 female) and 29 age‐ and full‐scale IQ‐matched controls (TD; mean age 24.5; range 19–37; 5 female), recruited from Bloomington, IN and surrounding areas. All ASD participants had previously received DSM‐IV community diagnoses of autism, Asperger's Syndrome, or Pervasive Developmental Disorder‐Not Otherwise Specified (PDD‐NOS), and the Autism Diagnostic Observation Schedule, Second Edition (ADOS‐2; Lord et al., 2000) was administered and scored by research reliable administrators at the time of study recruitment. All participants were administered Module 4 (the version appropriate for verbally fluent adults). We used the revised scoring algorithm that has improved diagnostic specificity over the previously used scoring algorithms (Hus & Lord, 2014). All participants exceeded the cutoff of 8 (combined social affect [SA] and restricted and repetitive behaviors [RRB] domains), with a mean score of 13.25 (SD = 3.08) (mean SA = 10.5 [2.84]; mean RRB = 2.75 [1.45]). We also administered the Autism‐Spectrum Quotient (AQ; Baron‐Cohen, Wheelwright, Skinner, Martin, & Clubley, 2001) and the Wechsler Abbreviated Scale of Intelligence, Second Edition (Wechsler, 2011). All subjects provided written informed consent; all experimental procedures were approved by the Indiana University Institutional Review Board.
After initial subject‐level exclusions due to consistently poor data quality (N = 4 ASD subjects) and then scan‐level exclusions due to excessive censoring (more than 30% of scan censored at time points with filtered framewise displacement [FDfilt] exceeding 0.2 mm) and insufficient (fewer than 3) scans of each type remaining after censoring, the remaining primary sample included 16 individuals with ASD (14 with at least 3 rest and at least 3 video scans; 1 with 3 rest scans only; 1 with 3 video scans only) and 28 TD individuals (27 with at least 3 rest and at least 3 video scans; 1 with 3 video scans only). In the analyzed sample, there were no group differences in age at time of scanning (ASD group: Mdn 298.5 months (interquartile range [IQR]: 110.5); TD group: Mdn: 279.5 months [IQR: 63.5]; U = 197, z = .65, p > .5) or IQ (ASD group: Mdn: 118.5 [IQR: 16.5]; TD group Mdn: 115 [IQR: 14.5]; U = 196, z = .67, p > .5). As expected, AQ differed between groups (ASD: Mdn 33 [IQR 14], TD: Mdn 17 [IQR 5.5], U = 27.5, z = 4.8, p < .001) and the median ADOS score in the ASD group was 8.5 (IQR 3) (or 11 [IQR 3] using the revised scoring algorithm).
We also repeated the primary analyses with a more relaxed censoring threshold (FDfilt > 0.5 mm) in order to include more data particularly from ASD subjects. This sample included 20 individuals with ASD (19 with at least 3 rest and at least 3 video scans and 1 with 3 video scans only) and 29 TD individuals (25 with at least 3 rest and at least 3 video scans, 3 with 4 video scans only, and 1 with 4 video scans only; see Supporting Information for further detail). The remaining supplemental analyses used the primary sample of individuals.
2.2. Design
The study design consisted of two scanning sessions separated by approximately 1 week (mean 9.3 days between scan sessions, SD 6). Both scanning sessions consisted of two resting‐state scans and two video‐watching scans, interleaved starting with rest. All rest scans were approximately 16 min long (1,200 TRs); video scans were approximately 13–14 min long (1,000, 952, 1,026, and 977 TRs). Subjects were instructed to move as little as possible and remain awake with eyes open.
No visual stimulus was provided during resting state scans. Stimuli for video scans were concatenated short video trailers, with no repetition of specific trailers across sessions. All trailers were obtained from Vimeo (https://vimeo.com). They were selected based on multiple criteria. First, to ensure that videos represented novel stimuli, we excluded any trailer that had a wide theatrical release. Second, we excluded videos with potentially objectionable content (i.e., nudity, swearing, and drug use). Finally, we excluded videos with intentionally startling events that could lead to excessive in‐scanner movement. Each trailer lasted between 45 and 285 s (approximately 1–5 min). Each video scan comprised between four and six trailers with genres that included documentaries, dramas, comedies, sports, mystery, and adventure. Video was back‐projected onto a screen that was visible to subjects via a mirror attached to the head coil. Audio was provided via MR‐compatible headphones.
Wakefulness was monitored via an MR‐compatible video camera; scans in which participants fell asleep were excluded from analysis (10 scans total; 5 from 2 ASD participants and 4 from 4 TD participants). One additional TD scan was excluded due to a technical issue. The final primary sample analyzed included multiple scans for most participants (304 scans analyzed, mean 3.4 [SD 1.28] resting‐state runs and mean 3.5 [SD 1.17] video runs per participant). (For the supplemental analyses, less strictly censored sample: 361 scans analyzed with mean 3.71 [SD = 0.84] rest scans and mean 3.65 [SD = 0.99] video scans per participant). Anatomical images were acquired after functional runs, during which participants were able to watch a video of their choosing.
2.3. Data acquisition and preprocessing
Acquisition and preprocessing were detailed in Byrge and Kennedy (2018) and repeated here. MRI images were acquired using a 3 Tesla whole‐body MRI system (Magnetom Tim Trio, Siemens Medical Solutions, Natick, MA) with a 32‐channel head receive array, using a protocol designed to be similar (but not identical due in large part to hardware constraints) to the Human Connectome Project (Smith et al., 2013). Both raw and prescan‐normalized images were acquired; raw images were used at all preprocessing stages and in all analyses unless specifically noted. During functional scans, T2*‐weighted multiband echo planar imaging (EPI) data were acquired using the following parameters: TR/TE = 813/28 ms; flip angle = 60°; 3.4 mm isotropic voxels; 42 slices acquired with interleaved order covering the whole brain; multi‐band acceleration factor of 3. The number of volumes varied: rest functional scans were 1,200 volumes; video functional scans were 1,000, 952, 1,026, and 977 volumes. Preceding the first functional scan, gradient‐echo EPI images were acquired in opposite phase‐encoding directions (10 images each with P‐A and A‐P phase encoding) with identical geometry to the EPI data (TR/TE = 1175/39.2 ms, flip angle = 60°) to be used to generate a fieldmap to correct EPI distortions, similar to the approach used by the Human Connectome Project (Smith et al., 2013). High‐resolution T1‐weighted images of the whole brain (MPRAGE, 0.7 mm isotropic voxel size; TR/TE/TI = 2499/2.3/1000 ms) were acquired as anatomical references.
Data were preprocessed with an in‐house pipeline using FEAT (v6.00) and MELODIC (v3.14) within FSL (v. 5.0.8; FMRIB's Software Library, http://www.fmrib.ox.ac.uk/fsl), Advanced Normalization Tools (ANTs; v2.1.0) (Avants et al., 2011), and Matlab_R2014b.
Individual anatomical images were bias‐corrected and skull‐stripped using Advanced Normalization Tools (ANTs), and segmented into gray matter, white matter, and CSF partial volume estimates using FSL FAST. From 20 randomly selected anatomical images (10 TD; 10 ASD), a midspace template was constructed using ANTs' buildtemplateparallel tool and then skull‐stripped. Composite (affine and diffeomorphic) transforms warping each individual anatomical image to this midspace template and warping the midspace template to the Montreal Neurological Institute MNI152 1 mm reference template were obtained using ANTs.
The initial five volumes (~4 s) of each functional run were discarded to minimize magnetization equilibration effects. We conducted rigid‐body motion correction, field map‐based geometric distortion correction, and nonbrain removal but not slice‐timing correction (due to the fast TR; Smith et al., 2013). Initial preprocessing also included weak high‐pass temporal filtering (>2,000 s FWHM) to remove slow drifts (as in Smith et al., 2013) but no spatial smoothing. Off‐resonance geometric distortions in the EPI data were corrected using a fieldmap derived from two gradient‐echo EPI images collected in opposite phase‐encoding directions (P‐A and A‐P) using FSL topup (similar to the study by Smith et al., 2013).
As reported in earlier work (Byrge & Kennedy, 2018), we evaluated a number of different preprocessing approaches on this data set and report here for the primary results the preprocessing approach that best removed lagged residual noise: FIX+MGTR, or FIX followed by mean cortical signal regression (equivalently, mean grayordinate time series regression or “MGTR,” following Burgess et al., 2016) in a second step. First, we employed FSL‐FIX (Salimi‐Khorshidi et al., 2014) to regress out independent components classified as noise by a classifier trained on independent but similar data and validated on hand‐classified functional runs. Then, the mean cortical signal (the mean BOLD signal across the individuals' gray matter partial volume estimate obtained from FSL FAST) was regressed from the residuals following FIX in a second step (as in Burgess et al., 2016), and the resulting residuals were analyzed as the cleaned data. Due to the controversy surrounding global signal regression (Murphy & Fox, 2017; Power, Plitt, Laumann, & Martin, 2017), we also repeated the primary analyses using FIX without MGTR in the Supporting Information).
Registration of cleaned functional data occurred as follows. An affine transformation matrix registering the mean prescan‐normalized functional image to each subject's skull‐stripped T1‐weighted anatomical image was obtained using Boundary‐Based Registration (BBR) via epi_reg within FSL. (The mean prescan‐normalized images yielded more accurate alignment than the raw functional images, due to the reduced contrast in fast‐TR EPI data; see also Smith et al., 2013). Next, each subject's functional images were transformed to the MNI152 reference all in one step, using ANTS to apply a concatenation of this affine transformation matrix with the composite (affine + diffeomorphic) transforms mapping between the subject's anatomical image, the midspace template, and the MNI152 reference.
We obtained region of interest time series using a 114‐region‐of‐interest (ROI) cortical parcellation anatomically subdividing the Yeo 17 functional networks (Yeo et al., 2011) and described more fully in the study by Betzel et al. (2014). (We also repeated the primary analyses using a second ROI set, the cortical Harvard‐Oxford parcellation distributed with FSL, subdivided by hemisphere into 96 ROIs, as reported in the Supporting Information). Individual ROI masks were created for each subject from the product of the individuals' gray matter partial volume estimate and the region of interest mask (as in the study by Tyszka et al., 2014). The weighted mean signal across each individual region of interest mask was then extracted from the cleaned BOLD signal for each functional run. These time courses together were used to construct functional connectivity matrices.
2.4. Data analysis
2.4.1. Functional connectome construction
Motion censoring/scrubbing prior to connectome construction was performed in all analyses (Power, Barnes, Snyder, Schlaggar, & Petersen, 2012). Earlier work using slower‐TR fMRI data employed framewise displacement traces to index head movement and identify which data points to censor; however, in faster‐TR data such as the current data set, framewise displacements as originally computed reflect a combination of motion and physiological noise (Byrge & Kennedy, 2018; Power et al., 2018; Power et al., 2019). Following Power et al. (2019), we attempted to better isolate head movements by computing filtered framewise displacement traces (FDfilt) from the backwards differential of the filtered raw head movement traces (using the Butterworth filter reported in the study by Power et al., 2019, with a differential of three TRs rather than four, to adjust for the small difference in sampling rates). We censored time points where FDfilt exceeded three different thresholds: in the primary analyses, a strict threshold of FDfilt > 0.2 mm, and in supplemental analyses, a more relaxed and inclusive threshold of FDfilt > 0.5 mm as well as a moderate threshold of FDfilt > 0.3 mm that permitted randomly downsampling each rest scan prior to FC construction such that no group differences in FDfilt remained (see Supplemental Methods). In all cases, the remaining (uncensored) TRs were used to construct the FC matrices, and scans with more than 30% of time points censored were excluded from analysis. We generated functional connectivity matrices by computing the Fisher‐z transformed pairwise Pearson correlations among all ROI timeseries. Symmetric 114 × 114 FC matrices were reduced to 6,441 × 1 FC vectors for use in subsequent analyses.
2.4.2. Group differences in data quality
Reliable group differences in FDfilt persisted after censoring under the strict and relaxed censoring thresholds (strict [0.2 mm]: for two scans, both rest scans, U = 76, p = .0028; U = 85, p = .0125; all other U > 95 and p > .19); relaxed (0.5 mm); four scans, three rest, U < 134, p < .0175, all other U > 173, p > .055). To address these group differences, we included mean FDfilt as a covariate in analyses where appropriate, and we also repeated the primary analyses upon FC matrices constructed from data randomly downsampled to eliminate group differences in FDfilt prior to FC construction (for rest data only; see Supporting Information).
2.4.3. Statistical reporting
We report nonparametric statistics throughout this article (except in select cases where parametric approaches are clearly appropriate) due to the relatively small number of individuals in the autism group and to reduce the influence of outliers upon group‐level conclusions. When not explicitly indicated, values reported in parentheses following medians are interquartile ranges. Where relevant, we conservatively report the minimum test statistic (i.e., for Mann–Whitney U tests, the smaller of the U statistic for the ranks of the ASD group and for the ranks of the TD group). For group comparisons, positive z‐values reflect higher ranks in the ASD group; for scan‐type comparisons, positive z‐values reflect higher ranks for video scans. There are several methods for reporting effect sizes of nonparametric tests; here we report effect size as r, the absolute value of the corresponding z‐statistic divided by the square root of the total sample size. Correlations were Fisher‐z transformed before analysis and inverse‐Fisher‐z transformed for visualization and reporting. Given the absence of reliable group differences, for ease of comparison, all statistics are reported at α = .05 without correcting for multiple comparisons.
2.4.4. Discriminability analyses (fingerprinting)
We predicted individual subject identity from functional connectomes in two different ways. In both cases, we kept rest scans and video scans separate, using rest scans to predict identity for rest scans but not using rest scans to predict identity for video scans. First, we used an approach (Byrge & Kennedy, 2019) based on the method originally introduced by Finn et al. (2015, 2017) and adapted for comparing four scans in one pass (vs. comparing two scans as in the study by Finn et al., 2015, 2017). In brief, we first computed the similarity between all pairs of scans as the Pearson correlation between the corresponding pair of FC vectors. Next, we predicted subject identity for a given scan by (a) identifying the scan with maximal similarity to the given scan and (b) taking the subject identity corresponding to the maximally similar scan as the predicted subject identity for the given scan. Accuracy was computed as the percentage of scans for which the predicted subject identity was equal to the actual subject identity. Next, we used the original pairwise fingerprinting approach introduced by Finn et al. (2015)—which uses pairs of scans (i.e., rest1 and rest2) to predict identity rather than all four scans of each type but is otherwise the same—then averaged together accuracy for each pair of scans for each subject.
2.4.5. Consistency and similarity analyses
The “fingerprinting” approach compares within‐subject and across‐subject similarity of functional connectomes and succeeds when within‐subject similarity exceeds across‐subject similarity. We also examined these quantities separately. We computed within‐subject similarity—or consistency—as the median of the Fisher‐z transformed Pearson correlations between all pairs of vectorized FC of the same scan type for a given subject. We computed across‐subject similarity analogously, as follows. For each scan, we computed the median of the Fisher‐z transformed Pearson correlations between vectorized FC for that scan and vectorized FC for all scans of the same type from all other subjects in the reference group. We then analyzed these values at the subject level, specifically the median across all scans for the same subject. We computed across‐subject similarity to two reference groups, always using a leave‐one‐subject‐out approach: all (other) TD subjects, and all (other) ASD subjects. As before, rest and video scans were kept separate; that is, rest scans were compared to rest scans but not to video scans.
2.4.6. Extension of maximal similarity matching (fingerprinting) procedure to diagnosis and scan‐type prediction
We extended the fingerprinting or maximal similarity matching procedure for comparing multiple scans in one pass (Byrge & Kennedy, 2019) to predict scan type and diagnosis, rather than individual identity. To do this, we took the predicted diagnosis (or scan type) for a given scan as the diagnosis (or scan type) of the scan, it is most similar to. For these analyses, we excluded scans contributed from the same individual; that is, we compared a given scan only to scans from other individuals.
2.4.7. Connectome‐based predictive modeling
Following Shen et al. (2017), we attempted to predict diagnosis group using functional connectomes. This procedure was conducted separately for rest and for video runs, using leave‐one‐subject‐out cross validation, and using average FC matrices for each subject constructed from the median FC values at each edge across all rest or all video scans. 250 iterations of the following procedure were conducted. First, one subject was left out. Second, a randomly selected subset of TD subjects was selected, to match the size of the remaining ASD group (Spronk et al., 2018). Third, for each FC edge, a GLM was fit, modeling FC in this subset of subjects as a function of diagnosis and mean for each subject of the mean FDfilt for all scans of this type. We included the latter as a proxy for data quality after noticing that many of the edges that differed between groups also differed between scans with low and high movement; we chose to use means here because they are more susceptible to the extreme values reflecting poorer data quality. Fourth, we masked each functional connectome, selecting only those FC edges that covaried with diagnosis above and beyond any associations with data quality at a threshold of p < .01 (note that results did not differ substantially at different thresholds; see Supporting Information). Fifth, we computed two summary values for each included subject: the sum of the FC edges that positively covaried with diagnosis and the sum of the edges that negatively covaried with diagnosis, above and beyond associations with FDfilt. Sixth, we fit a logistic regression modeling diagnosis as a function of two variables, the sums of the positively and the negatively covarying edges (note that including mean FDfilt as a third variable in this model did not change the pattern of results). Seventh, we computed the sums of the positively and negatively covarying edges for the left‐out‐subject and predicted diagnosis using this last logistic regression model. Finally, after all iterations were completed, we binarized the model predictions using a threshold of 0.5, computed the accuracy, sensitivity, and specificity for each iteration, and report the medians across iterations.
We also used the connectome‐based predictive modeling (CPM) procedure to attempt to predict AQ, a continuous scale designed to capture traits associated with the autism spectrum (Baron‐Cohen et al., 2001). The procedure was the same as described above, except the logistic model in Step 6 was replaced with a linear model, and rather than accuracy, we used correlations between actual and predicted AQ to assess model performance.
Finally, we also conducted a parallel CPM procedure in which we attempted to predict scan type rather than diagnosis. For this procedure, the median FC matrix across all rest scans and the median across all video scans for each subject was used as input data, both scans from the left‐out‐subject were excluded from the training sample, and only subjects that had both usable rest and video scans were included. Otherwise, the procedure was identical to that described above.
3. RESULTS
3.1. Functional connectomes are individually discriminable in both TD and ASD groups
We began by examining whether functional connectomes were individually discriminable using the “fingerprinting” approach (Finn et al., 2015), which captures whether within‐subject similarity of functional connectivity exceeds across‐subject similarity. We took two different approaches to “fingerprinting”: one that asks whether a given scan is discriminable relative to any of the other two to three scans of the same type contributed by that same individual (Byrge & Kennedy, 2018), and the other that asks whether a given scan is discriminable relative to each of those other two to three scans, evaluated pairwise (Finn et al., 2015). In both cases, we compared like scans (i.e., rest scans to rest scans but not rest scans to video scans).
Functional connectomes were indeed individually discriminable in both groups. Relative to all available scans of the same type (Byrge & Kennedy, 2018), 100% of rest scans and 100% of video scans were discriminable. Across all subjects, average pairwise discriminability (Finn et al., 2015) across all pairs of like scans was also high (for both rest and video scans, Mdn Acc = 100% [0%]; i.e., median = 100% with interquartile range = 0%, see Statistical Reporting). Average pairwise discriminability was reduced in the ASD group for rest scans (Mdn AccASD = 100% [33.3%], Mdn AccTD = 100% [0%], U = 109, z = −2.77, p = .0056, r = .44) but did not differ between groups for video scans (Video: Mdn AccASD = 100% [0%], Mdn AccTD = 100% [0%], U = 178.5, z = −0.36, p = .72, ns).
Further examination revealed that average pairwise discriminability was strongly linked with the number of scans available across all subjects (Rest: Mdn Acc = 66.6% (0%) across individuals with three scans; Mdn 100% (0%) across individuals with four scans; U = 3, z = −5.06, p < .0001, r = .81; video: Mdn Acc = 66.6% (0%) for 3 scans; Mdn Acc = 100% (0%) with four scans; U = 0, z = −5.8, p < .0001, r = .92). When fewer scans are available for a given individual, the reason is nearly always exclusion due to data quality, making it impossible to decouple whether ASD per se or data quality might underlie the initial group difference in pairwise discriminability for rest scans. Thus, we attempted to further control for potential effects of data quality by regressing mean filtered framewise displacement (FDfilt) from average pairwise discriminability before assessing potential group differences in the residuals. No reliable group differences in pairwise discriminability remained after this analysis (Rest: U = 132, z = −1.37, p > .17, ns; video: U = 147, z = 1.12, p > .26, ns).
Finally, discriminability did not differ reliably between the different scan tasks (mean pairwise discriminability across all subjects, W = 11, p > .3, ns) or separately for either group (ASD: W = 0, p > 0.058, ns; TD: W = 4, p > .7, ns). Thus, the current acquisition and preprocessing choices capture individual distinctness in functional connectivity.
3.2. Functional connectomes are consistent in both groups
After establishing that functional connectomes are individually discriminable in both groups, we next decomposed the fingerprinting approach to examine the consistency or within‐subject similarity of functional connectomes. Median within‐subject similarity of functional connectivity did not differ between groups for rest scans (Mdn r ASD = .84 [.2], Mdn r TD = .86 [.21], U = 152, z = −.79, p > .42, ns) or for video scans (Mdn r ASD = .85 [.16], Mdn r TD = .87 [.18], W = 147, z = −1.11, p > .26, ns). Note that in the larger alternative sample with a more relaxed censoring threshold, there were group differences for rest and marginally so for video scans (rest: Mdn r ASD = .82 [.25], Mdn r TD = .26 [.15], U = 173, z = −2, p = .045, r = .29; video: Mdn r ASD = .84 [.21], Mdn r TD = .87 [.17], U = 175, z = −1.87, p = .061, r = .28); however, these were attenuated when mean FDfilt was first regressed from the data (rest: U = 227, z = −.83, p > .4, ns; video: U = 214, z = −1, p > .3, ns).
Consistency within individuals did not reliably differ between rest and video scans, although it tended to be slightly higher for video scans (Mdn r = .87 [.17] vs. Mdn r = .86 [.18] for rest scans, W = 217, z = 1.6, p = .11).
3.3. Similarity to control functional connectomes did not consistently or robustly differ between groups
Next, we examined the other component of individual discriminability used by the fingerprinting approach: across‐subject similarity. Of most interest in studies of clinical populations is similarity to control subjects. We computed similarity to (other) control subjects as the median of the correlations between an individuals' functional connectivity matrix and those of all (other) controls for the same scan, averaged separately across rest and across video scans. Similarity to controls was marginally reduced in the ASD group (rest: Mdn r ASD = .58 [.04], Mdn r TD = .6 [.05], U = 118, z = −1.78, p = .08, r = .28; video: Mdn r ASD = .6 [0.07], Mdn r TD = 0.61 [.06], U = 125, z = −1.73, p = .08, r = .27). However, once again, regressing mean FDfilt from the data before comparing the groups resulted in the effect becoming no longer reliable (rest: U = 153, z = −0.77, p = .44, r = .12, ns; video: U = 140, z = −1.31, p > .18, r = 0.21, ns). As detailed in the Supporting Information (Supplemental Table S1, Row 8), this analysis was more sensitive to preprocessing differences than the previous analyses, with marginal group differences persisting in some cases and one small significant effect (uncorrected). However, taken in aggregate across the samples and preprocessing choices analyzed, there were no strong, consistent, robust group differences in this measure.
Overall, subjects were more similar to (other) controls during video scans (Mdn r = .6 [.05]) than during rest scans (Mdn r = .59 [.05], W = 88, z = 3.72, p < .001, r = .42), as might be expected given the constrained nature of the video stimuli.
3.4. No shared ASD‐specific FC patterns were evident
In parallel to the previous analysis, we also examined across‐subject similarity to the ASD group. Higher similarity among ASD individuals would be suggestive of the shared, ASD‐specific pattern of functional connectivity that is the goal of many clinical functional connectivity studies. However, median similarity to (other) individuals with ASD was not reliably higher in the ASD group (rest: Mdn r ASD = .56 [.05], Mdn r TD = 0.58 [.06], U = 121, z = −1.69, p = .09, r = .27, ns; video: Mdn r ASD = 0.59 [0.047], Mdn r TD = 0.59 [.06], U = 134, z = −1.48, p = .14, ns), and once again, the marginal group difference for the rest scans is eliminated when mean FDfilt is first regressed from the data (W = 155, z = −.7, p = .48, r = .11, ns). As with the previous similarity to TD analysis, this analysis was somewhat sensitive to preprocessing choices (see Supplemental Table S1, Row 9), with marginal group differences and one small significant effect (uncorrected) persisting after regressing mean FDfilt for a few preprocessing approaches. However, regardless of statistical threshold, z‐values were negative across all preprocessing approaches, indicating that the TD group, rather than the ASD group, was the group that demonstrated numerically higher similarity to (other) ASD individuals. This is somewhat counterintuitive, as the opposite pattern would be expected if there were a shared ASD‐specific FC pattern.
To gain more insight into this pattern of findings, we used multidimensional scaling (MDS) to visualize similarity among pairs of scans, separately for rest (Figure 1, top row) and for video scans (Figure 1, bottom row). ASD and TD subjects were included together in the MDS computation (Figure 1, left column) and also plotted separately for visualization purposes (Figure 1, middle column, ASD subjects; right column, TD subjects), with scans from the same subject plotted in the same color. If there were a shared pattern of functional connectivity among all or some ASD subjects, the MDS plots in Figure 1 (left column) would reveal clusters where ASD scans (in red) are closer together than they are to TD scans (in blue). While there are a few small clusters of ASD data points in the left column, those clusters turn out to reflect individual‐level similarity (c.f. Figure 1, top left, upper right and lower right red clusters to those same clusters in Figure 1, top middle, see also Figure 1, bottom left, rightmost red cluster to that same cluster in Figure 1, bottom middle) rather than increased similarity among different individuals with ASD. Overall, the ASD scans that are most dissimilar (those plotted furthest from the center) tend also to be distant from scans from other subjects from both groups (plotted along the periphery), indicating a lack of shared FC pattern. Indeed, for both rest and video scans, MDS visualization intermixes the majority of TD and ASD scans, indicating comparable similarity at this level of analysis.
Figure 1.
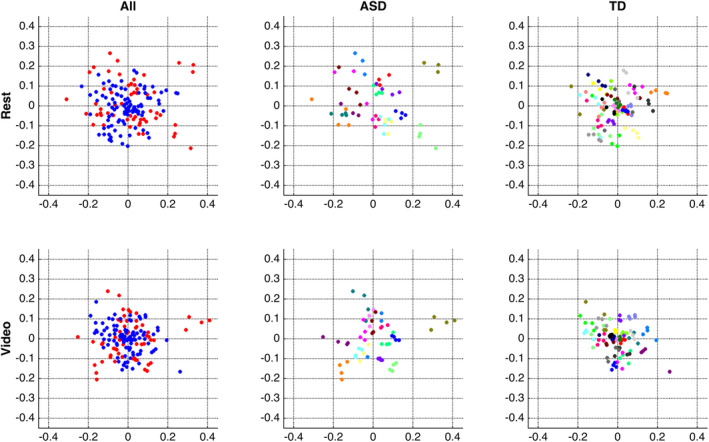
Multidimensional scaling (MDS) visualizations of pairwise similarity among all rest scans (top row) and among all video scans (bottom row). Both MDS computations were conducted using all subjects (left column); results are also re‐plotted separately for each group (ASD, middle column; TD, right column) for closer inspection. The middle and right columns plot scans from the same subject in the same color (note that colors are recycled across groups, i.e., the orange points in the middle and in the right plots represent different subjects). Note that these visualizations present pairwise similarity before regressing out FDfilt
We note that in general, in both groups, it can be seen in Figure 1 that scans from the same subject (plotted in the same color) are plotted near to one another, reflecting the high within‐subject similarity reported above, and also more close to one another than to those of other subjects, reflecting the high discriminability or “fingerprinting” accuracy reported above.
These visualizations also suggest the possibility of a different pattern. Some subjects whose scans have reduced within‐subject similarity—as indicated by greater distance among data points in the same color, such as the green data points in Figure 1 (top middle)—may also have reduced across‐subject similarity, as indicated by greater distance from the center of the plot. Indeed, for both rest and video scans, there was an increasing relationship between within‐subject similarity and across‐subject similarity, such that subjects whose scans were dissimilar to others also tended to be less consistent relative to themselves. These associations hold across all subjects and separately within both groups and hold using both metrics of across‐subject similarity: similarity to only (other) control subjects (rest: Spearman's ⍴ All = .59, p < .001, ⍴ ASD = .65, p = .01, ⍴ TD = .54, p < .007; video: ⍴ All = .45, p < .004, ⍴ ASD = .58, p < .03, ⍴ TD = .38, p < .06) and similarity to only (other) ASD subjects (rest: Spearman's ⍴All = .55, p < .001, ⍴ ASD = .61, p < .02, ⍴ TD = .47, p < .03; video: ⍴ All = .52, p < .001, ⍴ ASD = .68, p < .006, ⍴ TD = .45, p < .03). In other words, this is a general relationship rather than an ASD‐specific one. This association could reflect a number of underlying mechanisms, but it is suggestive of data quality, and indeed, both within‐subject similarity and similarity to controls are associated with mean FDfilt across all subjects (rest: ⍴ All = −.42, p < .008 and ⍴All = −.4, p < .02; video: ⍴ All = −0.57, p < .001 and ⍴All = −.4, p < 0.02), although this relationship held in the control group (⍴ TD ranging from −0.38 to −0.63; p < .0012 to .065) but not the ASD group (⍴ ASD ranging from −0.08 to −0.39; p > .15 to .75, ns).
3.5. Neither rest scans nor video scans permitted differentiating the ASD and TD groups
The MDS visualizations, and specifically the interleaving of the majority of scans from ASD and TD subjects, do not suggest that a clear boundary between the groups can be drawn using full functional connectomes as we have done thus far. Indeed, attempts to predict the diagnosis group for all available scans using maximal similarity matching, in parallel to the fingerprinting procedure used previously, result in accuracy around chance, as most scans are predicted to be those of controls (Rest: 62%, 3.6% sensitivity, 95.8% specificity; video: 66.9%, 10.5% sensitivity, 100% specificity).
Nonetheless, it remains possible that a more targeted selection of connectome edges might permit better prediction of group membership. Therefore, we conducted connectome‐based predictive modeling (CPM; Shen et al., 2017) separately for rest and video scans, iterating over randomly downsampled subsamples of the control group such that the same number of ASD and TD scans were available. This procedure resulted in group prediction accuracy that was still quite low (Rest: Mdn accuracy 51.3% [7.7%], sensitivity 58.3% [8.3%], specificity 46.7% [20%] across iterations; video: Mdn accuracy 57.5% [7.5%], sensitivity 60% [12%], specificity 60% [13.3%]). CPM accuracy for video scans exceeded rest scans (U = 14,171, z = −10.6, p < < .0001), and this was the case in all but one preprocessing approach examined (see Supplemental Table S1). However, such a difference is difficult to interpret in the context of approximately chance‐level accuracy for both scan tasks.
Finally, in an exploratory follow‐up analysis, we also ran CPM models attempting to predict AQ score, a continuous scale designed to capture traits associated with the autism spectrum (Baron‐Cohen et al., 2001), again separately for rest and for movie scans. Results for rest scans looked similar to the results for diagnostic predictions, with the median correlation between actual AQ scores and predicted AQ scores across iterations being r = .035 (IQR = 0.13), p = .69 (IQR = 0.36). However, the distribution of correlations was different for movie scans, and positively skewed, with Mdn r = .26 (IQR = 0.11), Mdn p = .10 (IQR = 0.16), and statistically higher than for rest scans (U = 460, z = −11.09, p < < .001)—despite being predominantly not statistically significant. We consider the differences between these movie and rest results to be an interesting hint deserving follow‐up in future work but not a robust enough result to be considered further in the present work, especially considering the low proportions of variance involved.
3.6. Rest and video scans can themselves be near‐perfectly differentiated
Figure 2 presents the median FC matrices for each scan task, across all subjects (top row), and across each group (middle rows). Median differences between the groups (bottom row) are small, consistent with the overall indistinguishability of scans from the different groups. Contrary to these small median group differences, Figure 2 also shows that differences between FC matrices at rest and during the video are evident by eye, across all subjects (top right) and separately within each group (middle right). Indeed, another MDS visualization of pairwise similarity among scans, this time including all scans together rather than examining rest and video scans separately, reveals that while once again ASD and TD scans are intermingled (Figure 3, left), rest and video scans form visually distinct groups (Figure 3, right).
Figure 2.
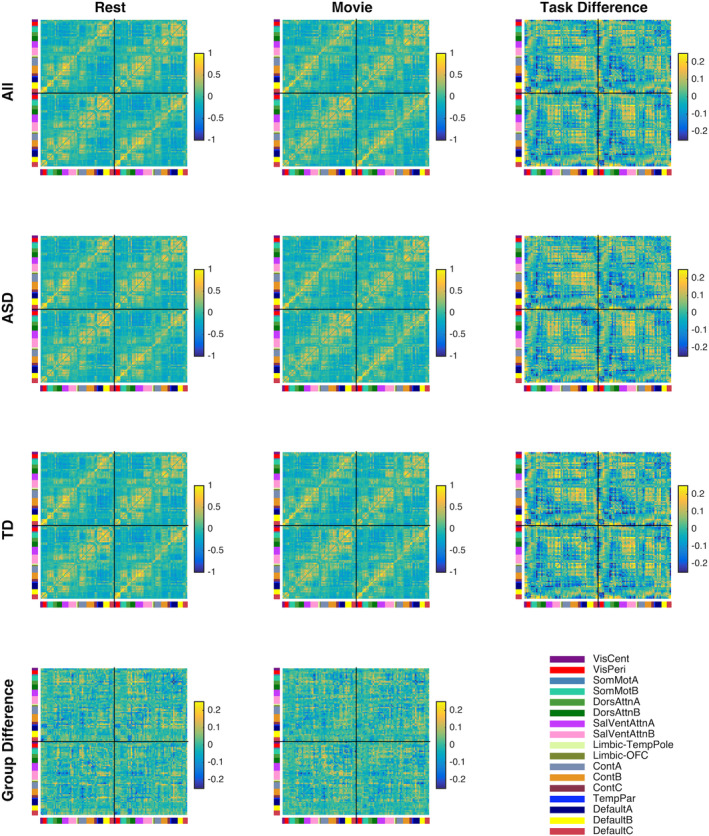
Median and difference functional connectivity matrices for each scan task across all subjects (Row 1), for each diagnosis group (Rows 2 and 3), and differences between diagnosis groups for each task (Row 4). Task differences (Column 3) are more apparent upon visual inspection than group differences (Row 4). Differences are ordered as ASD‐TD (Row 4) and Video‐Rest (Column 3). Left hemisphere ROIs are plotted to the left and above right hemisphere ROIs
Figure 3.
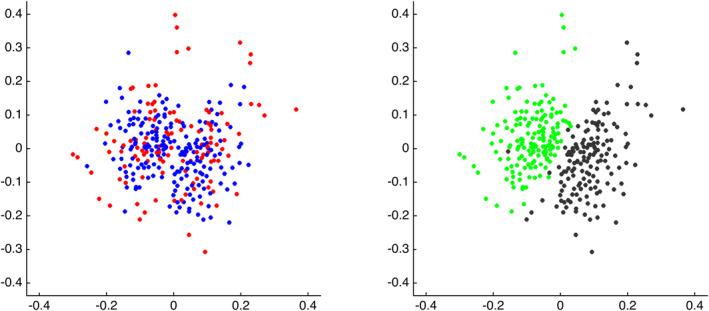
MDS visualizations of pairwise similarity among all scans. Left: scans color‐coded according to diagnosis (red = ASD, blue = TD). Right: scans color‐coded according to scan type (green = video; black = rest). Note that these visualizations present pairwise similarity before regressing out FDfilt
These results suggest that it may be possible to predict scan type based on functional connectivity, using the same analytic techniques that did not differentiate groups. Using maximal similarity matching (parallel to fingerprinting), scan‐type prediction accuracy was 98.4% across all scans, with comparable accuracy using CPM upon both median rest and median video connectomes together (Mdn accuracy = 98.6% (0%) across iterations). Taken all together, scan task and individual identity were stronger drivers of similarity and dissimilarity among scans than diagnosis group membership.
4. DISCUSSION
We found no consistent or robust differences in functional connectomes between the ASD group and the TD group, despite a densely sampled, high‐resolution acquisition that allowed us to predict individual identity and scan task with near‐perfect accuracy. Functional connectomes from individuals with ASD were equally discriminable as those of controls, and equally consistent across different scans. Although a few specific preprocessing choices gave rise to small effects of reduced similarity to controls in the ASD group, the overarching conclusion from these analyses is that ASD functional connectomes in this data set were consistently indistinguishable from those of controls (see also Byrge, Dubois, Tyszka, Adolphs, & Kennedy, 2015; Dajani et al., 2019). Collecting more data per individual and including naturalistic viewing conditions were thus not sufficient to robustly pull apart the diagnosis groups, and whatever shared ASD‐specific signals may ultimately exist in this sample must thus be smaller than those associated with scan task and individual identity.
The success observed in discriminating individual functional connectomes is in line with the existing literature (Finn et al., 2015; Finn et al., 2017; Horien et al., 2018; Vanderwal et al., 2017) and the effectively perfect accuracy in our densely sampled dataset is consistent with work showing that increasing temporal sampling improves discriminability and reliability of FC (Birn et al., 2013; Horien et al., 2018; Laumann et al., 2015). Ours is the first study, to our knowledge, to evaluate functional connectome fingerprinting in ASD and we found that given our acquisition parameters, preprocessing choices, and sample, discriminability of functional connectomes in ASD did not differ from those of controls so long as data quality was controlled in analyses. The high accuracy obtained from using functional connectomes to predict scan task is consistent with prior work demonstrating successful prediction of task from connectivity measures (Kaufmann et al., 2017; Pallares et al., 2018; Richiardi, Eryilmaz, Schwartz, Vuilleumier, & Van De Ville, 2011; Xie et al., 2018). What is most striking, at least superficially, is that a recognizable diagnosis defined by shared clinical features across individuals was not associated with a shared pattern of brain function, in this data set, using these analytic approaches.
Whether common patterns of brain function associated with ASD do ultimately exist in this dataset and in this particular sample of individuals is not something that can be conclusively answered at this time. Several possibilities exist. One is that shared patterns of brain function associated with ASD do exist. Given that an ASD diagnosis is associated with common behavior patterns, and behavior comes from the brain, this possibility is likely to be what most people would expect. In that case, either such shared neurofunctional differences occur at a different temporal or spatial scale than the current fMRI acquisition can measure, and thus detecting them requires technological advances, or perhaps instead such shared differences do exist at the current resolution and detecting them requires instead analytic or denoising advancements. Another possibility recognizes that shared patterns of behavior need not be associated with shared patterns of brain function (Edelman & Gally, 2001; Price & Friston, 2002; Seghier & Price, 2018). Although this principle applies broadly (e.g., to reading, in adult controls; Richardson, Seghier, Leff, Thomas, & Price, 2011), in ASD research, it is typically considered under the umbrella of “heterogeneity”—the idea that because ASD is thought to be associated with multiple distinct etiologies and developmental histories (Fountain, Winter, & Bearman, 2012; Geschwind, 2009; Landa, Gross, Stuart, & Bauman, 2012; Levy, Mandell, & Schultz, 2009), the search for shared patterns of brain function associated with ASD may be more fruitful within ASD subgroups that reflect shared etiological and developmental factors (e.g., Lombardo et al., 2015). Detecting shared patterns of brain function associated with ASD subgroups requires a larger sample size and more extensive phenotyping and/or genotyping than the current dataset. However, at least at the level of full cortical functional connectomes, our analyses revealed no hints of distinct shared patterns among a subset of ASD individuals: similarity to (other) ASD individuals was never higher in the ASD group, and visual inspection of the MDS plots suggested no clusters comprised of more than one individual with ASD.
Our results reveal that across diagnosis groups, and consistent with the literature in controls (Byrge & Kennedy, 2019; Finn et al., 2015; Finn et al., 2017; Horien et al., 2018; Vanderwal et al., 2017), individuals are more similar to themselves across scans than to others, an observation that the fingerprinting approach leverages for predicting identity. Visual inspection of the MDS plots (Figure 1) also reveals that scans from select individuals with ASD seem to form their own cluster, away from the other scans. Whether such examples reflect the individual variability seen across all subjects—perhaps in a more exaggerated form—or instead perhaps a member of a real ASD subgroup that happens to be represented by N = 1 in our particular sample (e.g., perhaps one specific, rare, etiology)—cannot be determined without a larger sample. This is also related to discussions of “idiosyncrasy” in ASD—although definitions vary, numerous studies have observed a form of exaggerated individual variability, in which some individuals with ASD appear both distinct from controls and distinct from other individuals with ASD (Byrge et al., 2015; Hahamy, Behrmann, & Malach, 2015; Hasson et al., 2009; Nunes, Peatfield, Vakorin, & Doesburg, 2019). This pattern is also observed in some individuals with ASD in this sample, although associations between within‐subject similarity, across‐subject similarity, and data quality across all subjects make these dimensions difficult to disentangle.
It is worth noting that residual associations between our measures of interest and indices of data quality persisted in our sample despite our rigorous data inclusion thresholds and state‐of‐the‐art preprocessing approaches. While it has long been known that effects of head motion persist longer than the motion itself (Power et al., 2012), the existing literature would suggest that such residual motion effects should be mitigated by our use of global signal regression (Byrge & Kennedy, 2018; Power et al., 2014; Power, Schlaggar, & Petersen, 2015). A priori, the residual associations with FDfilt that we observed could reflect effects of data quantity—higher FDfilt corresponds with more censoring and thus fewer data points included, potentially leading to less precise FC estimates and reduced similarity to others. However, this association persisted in the downsampled supplemental preprocessing approach (Supplemental Table S1, Column 4), in which data quantity is equated across all individuals. We also cannot rule out the possibility that what we are calling residual associations with motion might be due to some other individual difference, potentially ASD related, that also happens to covary with FDfilt. It is certainly plausible, for instance, that more severely affected members of a clinical group may also move more in the scanner. However, the associations between FDfilt and within‐ and across‐subject similarity in the control group seem to undermine this possibility while not ruling it out. It is also worth recognizing that our understanding of how nuisance factors influence the fMRI signal and fcMRI measures is incomplete and continually evolving. Our indices of data quality are far from perfect; while framewise displacement is a good proxy for data quality, it is not the only measure that is informative (Byrge & Kennedy, 2018; Power et al., 2017, 2018, 2019). Regardless of the underlying causes, these associations point to the need to carefully examine potential residual effects of data quality even after following best practices.
One initial hypothesis was that adding a video stimulus with social content to the commonly used resting state fcMRI paradigms might improve detection of ASD‐specific patterns of brain function, because processing social content may be more related to the behavioral domains in which ASD symptoms are observed, and because driving the brain with a common stimulus might better permit detection of individual and group differences that the unconstrained resting state might obscure. We found instead that patterns of results were generally comparable between rest and video scans. We did find that prediction accuracy was reliably higher using video scans, but in the context of overall very low accuracy for both scan types, it is not prudent to interpret this further. Whether video scans provide a benefit to diagnosis group prediction in a sample where resting state scans achieve above‐chance accuracy remains an open question. Contrary to other work showing improvements in discriminability and reliability during video watching in the scanner (Vanderwal et al., 2017; Wang et al., 2017), we found no differences in those measures in the current study. We speculate that this may be due to the temporal sampling in the current data set and ceiling‐level discriminability, and that such an advantage for video watching could potentially still be observable across shorter FC epochs such as in a dynamic FC paradigm. These are important questions for future study.
Limitations of this study predominantly include the relatively small sample size and limited phenotyping, which, as noted previously, prevent us from addressing pressing questions about heterogeneity in ASD. Due to our relatively small sample size and interest in group‐level effects, we used nonparametric statistical tests for their reduced susceptibility to outliers. While the pattern of results did not materially change under parametric tests, it is possible that the conservative tests we used under‐emphasized real small effects in our data that could become more robust in a larger sample. It is important to make clear that while we do not have strong evidence for ASD‐specific differences, we also do not have strong evidence for a lack of differences. We note that we the current study included a relatively large age range and unbalanced ratio of males and females; we balanced age and sex between our groups in an attempt to best mitigate any effects of these variables on our results. However, complex interactions between age, sex, and diagnosis, together with a whole host of other variables, could still be present, and our sample size is underpowered to detect these interactions. Future studies might examine similar questions within a more narrow age range or separately within males or females. Finally, we used video stimuli with social content to drive brains in a coordinated manner, in a way potentially more related to ASD symptomology than the resting state. However, it is possible that scanning individuals in a manner even more closely approaching the behavioral domains of ASD—such as during more complex and subtle social interactions (as in, e.g., Byrge et al., 2015) than the movie trailers included here, or even during actual social interactions (Redcay et al., 2010, 2013)—may serve to better differentiate the groups. Finally, there are of course existing analytic approaches we have not yet applied to this data set and that could be promising directions for differentiating diagnosis groups, including time‐varying functional connectivity (see, e.g., Falahpour et al., 2016; Mash et al., 2019) and inter‐subject functional correlation (Simony et al., 2016) approaches. Future work will explore these directions.
5. CONCLUSIONS
It was not possible to predict ASD diagnosis much above chance in a high quality and extensively sampled data set including rest and video scans; a data set that permitted predicting individual identity and scan type with effectively perfect accuracy. We found no conclusively ASD‐associated effects for which data quality was not also implicated. Individuals from both groups contributed high quality, discriminable, consistent data, and the overarching picture was that scans from the ASD group were indistinguishable from the control group. Inconsistent and null results in the ASD fcMRI literature cannot be solely attributed to insufficiently sampled scans and limited scan tasks.
CONFLICT OF INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
Both authors developed the study concept, L.B. analyzed the data with input from D.P.K., L.B. drafted the initial version of the manuscript, and both authors revised the manuscript critically and approved the final version. Both authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Supporting information
Appendix S1. Supporting Information.
ACKNOWLEDGMENTS
This work was supported by the NIH (R01MH110630 and R00MH094409 to D.P.K. and T32HD007475 Postdoctoral Traineeship to L.B.). We also acknowledge the Indiana University Pervasive Technology Institute for providing HPC (Big Red II, Karst, Carbonate), visualization, and storage resources, which were supported in part by Lilly Endowment, Inc., the Indiana METACyt Initiative, and based upon work supported by the National Science Foundation under Grant No. CNS‐ 0521433. The authors thank Hu Cheng for MRI protocol development, Soyoung Park for training the FIX classifier, and Brad Caron and Susannah Burkholder for data collection.
Byrge L, Kennedy DP. Accurate prediction of individual subject identity and task, but not autism diagnosis, from functional connectomes. Hum Brain Mapp. 2020;41:2249–2262. 10.1002/hbm.24943
Data Availability Statement:We have made a subset of this data publicly available as part of the Autism Brain Imaging Data Exchange II (ABIDE II) initiative (Indiana University site; Di Martino et al., 2017; http://fcon_1000.projects.nitrc.org/indi/abide/abide_II.html). Data and code are available from the authors upon reasonable request.
Funding information National Institutes of Health, Grant/Award Numbers: R01MH110630, R00MH094409, T32HD007475
DATA AVAILABILITY STATEMENT
We have made a subset of this data publicly available as part of the Autism Brain Imaging Data Exchange II (ABIDE II) initiative (Indiana University site; Di Martino et al., 2017; http://fcon_1000.projects.nitrc.org/indi/abide/abide_II.html). Data and code are available from the authors upon reasonable request.
REFERENCES
- Abraham, A. , Milham, M. P. , Di Martino, A. , Craddock, R. C. , Samaras, D. , Thirion, B. , & Varoquaux, G. (2017). Deriving reproducible biomarkers from multi‐site resting‐state data: An Autism‐based example. NeuroImage, 147, 736–745. [DOI] [PubMed] [Google Scholar]
- Avants, B. B. , Tustison, N. J. , Song, G. , Cook, P. A. , Klein, A. , & Gee, J. C. (2011). A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage, 54(3), 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron‐Cohen, S. , Wheelwright, S. , Skinner, R. , Martin, J. , & Clubley, E. (2001). The autism‐spectrum quotient (AQ): Evidence from asperger syndrome/high‐functioning autism, malesand females, scientists and mathematicians. Journal of Autism and Developmental Disorders, 31(1), 5–17. [DOI] [PubMed] [Google Scholar]
- Betzel, R. F. , Byrge, L., He, Y., Goñi, J., Zuo, X. N., & Sporns, O. (2014). Changes in structural and functional connectivity among resting‐state networks across the human lifespan. Neuroimage, 102, 345–357. [DOI] [PubMed] [Google Scholar]
- Birn, R. M. , Molloy, E. K. , Patriat, R. , Parker, T. , Meier, T. B. , Kirk, G. R. , … Prabhakaran, V. (2013). The effect of scan length on the reliability of resting‐state fMRI connectivity estimates. NeuroImage, 83, 550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, G. C. , Kandala, S. , Nolan, D. , Laumann, T. O. , Power, J. D. , Adeyemo, B. , … Barch, D. M. (2016). Evaluation of denoising strategies to address motion‐correlated artifacts in resting‐state functional magnetic resonance imaging data from the Human Connectome Project. Brain Connectivity, 6(9), 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrge, L. , Dubois, J. , Tyszka, J. M. , Adolphs, R. , & Kennedy, D. P. (2015). Idiosyncratic brain activation patterns are associated with poor social comprehension in autism. Journal of Neuroscience, 35(14), 5837–5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrge, L. , & Kennedy, D. P. (2018). Identifying and characterizing systematic temporally‐lagged BOLD artifacts. NeuroImage, 171, 376–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrge, L. , & Kennedy, D. P. (2019). High‐accuracy individual identification using a “thin slice” of the functional connectome. Network Neuroscience, 3(2), 363–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. P. , Keown, C. L. , Jahedi, A. , Nair, A. , Pflieger, M. E. , Bailey, B. A. , & Müller, R. A. (2015). Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. NeuroImage: Clinical, 8, 238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajani, D. R. , Burrows, C. A. , Odriozola, P. , Baez, A. , Nebel, M. B. , Mostofsky, S. H. , & Uddin, L. Q. (2019). Investigating functional brain network integrity using a traditional and novel categorical scheme for neurodevelopmental disorders. NeuroImage: Clinical, 21, 101678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deen, B. , & Pelphrey, K. (2012). Perspective: Brain scans need a rethink. Nature, 491(7422), S20. [DOI] [PubMed] [Google Scholar]
- Di Martino, A. , O'Connor, D. , Chen, B. , Alaerts, K. , Anderson, J. S. , Assaf, M. , … Blanken, L. M. (2017). Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Scientific Data, 4, 170010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman, G. M. , & Gally, J. A. (2001). Degeneracy and complexity in biological systems. Proceedings of the National Academy of Sciences, 98(24), 13763–13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahpour, M. , Thompson, W. K. , Abbott, A. E. , Jahedi, A. , Mulvey, M. E. , Datko, M. , … Müller, R. A. (2016). Underconnected, but not broken? Dynamic functional connectivity MRI shows underconnectivity in autism is linked to increased intra‐individual variability across time. Brain Connectivity, 6(5), 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, E. S. , Scheinost, D. , Finn, D. M. , Shen, X. , Papademetris, X. , & Constable, R. T. (2017). Can brain state be manipulated to emphasize individual differences in functional connectivity? NeuroImage, 160, 140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, E. S. , Shen, X. , Scheinost, D. , Rosenberg, M. D. , Huang, J. , Chun, M. M. , … Constable, R. T. (2015). Functional connectome fingerprinting: Identifying individuals using patterns of brain connectivity. Nature Neuroscience, 18(11), 1664–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain, C. , Winter, A. S. , & Bearman, P. S. (2012). Six developmental trajectories characterize children with autism. Pediatrics, 129(5), e1112–e1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind, D. H. (2009). Advances in autism. Annual Review of Medicine, 60, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, E. M. , Laumann, T. O. , Gilmore, A. W. , Newbold, D. J. , Greene, D. J. , Berg, J. J. , … Hampton, J. M. (2017). Precision functional mapping of individual human brains. Neuron, 95(4), 791–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahamy, A. , Behrmann, M. , & Malach, R. (2015). The idiosyncratic brain: Distortion of spontaneous connectivity patterns in autism spectrum disorder. Nature Neuroscience, 18(2), 302–309. [DOI] [PubMed] [Google Scholar]
- Hasson, U. , Avidan, G. , Gelbard, H. , Vallines, I. , Harel, M. , Minshew, N. , & Behrmann, M. (2009). Shared and idiosyncratic cortical activation patterns in autism revealed under continuous real‐life viewing conditions. Autism Research, 2(4), 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y. , Byrge, L. , & Kennedy, D. P. (2020). Non‐replication of functional connectivity differences in autism spectrum disorder across multiple sites and denoising strategies. Human Brain Mapping, 10.1002/hbm.24879. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinsfeld, A. S. , Franco, A. R. , Craddock, R. C. , Buchweitz, A. , & Meneguzzi, F. (2018). Identification of autism spectrum disorder using deep learning and the ABIDE dataset. NeuroImage: Clinical, 17, 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horien, C. , Noble, S. , Finn, E. S. , Shen, X. , Scheinost, D. , & Constable, R. T. (2018). Considering factors affecting the connectome‐based identification process: Comment on Waller et al. NeuroImage, 169, 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, J. V. , Jacokes, Z. J. , Torgerson, C. M. , Irimia, A. , & Van Horn, J. D. (2016). Resting‐state functional connectivity in autism spectrum disorders: A review. Frontiers in Psychiatry, 7(205), 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hus, V. , & Lord, C. (2014). The autism diagnostic observation schedule, module 4: Revised algorithm and standardized severity scores. Journal of Autism and Developmental Disorders, 44, 1996–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, T. , Alnæs, D. , Brandt, C. L. , Doan, N. T. , Kauppi, K. , Bettella, F. , … Melle, I. S. (2017). Task modulations and clinical manifestations in the brain functional connectome in 1615 fMRI datasets. NeuroImage, 147, 243–252. [DOI] [PubMed] [Google Scholar]
- Landa, R. J. , Gross, A. L. , Stuart, E. A. , & Bauman, M. (2012). Latent class analysis of early developmental trajectory in baby siblings of children with autism. Journal of Child Psychology and Psychiatry, 53(9), 986–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann, T. O. , Gordon, E. M. , Adeyemo, B. , Snyder, A. Z. , Joo, S. J. , Chen, M. Y. , … Schlaggar, B. L. (2015). Functional system and areal organization of a highly sampled individual human brain. Neuron, 87(3), 657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, S. E. , Mandell, D. S. , & Schultz, R. T. (2009). Autism. Lancet, 374, 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo, M. V. , Pierce, K. , Eyler, L. T. , Barnes, C. C. , Ahrens‐Barbeau, C. , Solso, S. , … Courchesne, E. (2015). Different functional neural substrates for good and poor language outcome in autism. Neuron, 86(2), 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, C. , Risi, S. , Lambrecht, L. , Cook, E. H., Jr. , Leventhal, B. L. , DiLavore, P. C. , … Rutter, M. (2000). A standard measure of social and communication deficits associated with the spectrum of autism. The Autism Diagnostic Observation Schedule‐Generic, 30, 205–223. [PubMed] [Google Scholar]
- Mash, L. E. , Linke, A. C. , Olson, L. A. , Fishman, I. , Liu, T. T. , & Müller, R. A. (2019). Transient states of network connectivity are atypical in autism: A dynamic functional connectivity study. Human Brain Mapping, 40(8), 2377–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, K. , & Fox, M. D. (2017). Towards a consensus regarding global signal regression for resting state functional connectivity MRI. NeuroImage, 154, 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, J. A. , Zielinski, B. A. , Fletcher, P. T. , Alexander, A. L. , Lange, N. , Bigler, E. D. , … Anderson, J. S. (2013). Multisite functional connectivity MRI classification of autism: ABIDE results. Frontiers in Human Neuroscience, 7, 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes, A. S. , Peatfield, N. , Vakorin, V. , & Doesburg, S. M. (2019). Idiosyncratic organization of cortical networks in autism spectrum disorder. NeuroImage, 190, 182–190. [DOI] [PubMed] [Google Scholar]
- Pallares, V. , Insabato, A. , Sanjuan, A. , Kuehn, S. , Mantini, D. , Deco, G. , & Gilson, M. (2018). Extracting orthogonal subject‐and condition‐specific signatures from fMRI data using whole‐brain effective connectivity. NeuroImage, 178, 238–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plitt, M. , Barnes, K. A. , & Martin, A. (2015). Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage: Clinical, 7, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J. D. , Barnes, K. A. , Snyder, A. Z. , Schlaggar, B. L. , & Petersen, S. E. (2012). Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage, 59(3), 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J. D. , Lynch, C. J. , Silver, B. M. , Dubin, M. J. , Martin, A. , & Jones, R. M. (2019). Distinctions among real and apparent respiratory motions in human fMRI data. NeuroImage, 201,116041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J. D. , Mitra, A. , Laumann, T. O. , Snyder, A. Z. , Schlaggar, B. L. , & Petersen, S. E. (2014). Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage, 84, 320–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J. D. , Plitt, M. , Gotts, S. J. , Kundu, P. , Voon, V. , Bandettini, P. A. , & Martin, A. (2018). Ridding fMRI data of motion‐related influences: Removal of signals with distinct spatial and physical bases in multiecho data. Proceedings of the National Academy of Sciences, 115(9), E2105–E2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J. D. , Plitt, M. , Laumann, T. O. , & Martin, A. (2017). Sources and implications of whole‐brain fMRI signals in humans. NeuroImage, 146, 609–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J. D. , Schlaggar, B. L. , & Petersen, S. E. (2015). Recent progress and outstanding issues in motion correction in resting state fMRI. NeuroImage, 105, 536–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, C. J. , & Friston, K. J. (2002). Degeneracy and cognitive anatomy. Trends in Cognitive Sciences, 6(10), 416–421. [DOI] [PubMed] [Google Scholar]
- Redcay, E. , Dodell‐Feder, D. , Mavros, P. L. , Kleiner, M. , Pearrow, M. J. , Triantafyllou, C. , … Saxe, R. (2013). Atypical brain activation patterns during a face‐to‐face joint attention game in adults with autism spectrum disorder. Human Brain Mapping, 34(10), 2511–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redcay, E. , Dodell‐Feder, D. , Pearrow, M. J. , Mavros, P. L. , Kleiner, M. , Gabrieli, J. D. E. , & Saxe, R. (2010). Live face‐to‐face interaction during fMRI: A new tool for social cognitive neuroscience. NeuroImage, 50, 1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, F. M. , Seghier, M. L. , Leff, A. P. , Thomas, M. S. , & Price, C. J. (2011). Multiple routes from occipital to temporal cortices during reading. Journal of Neuroscience, 31(22), 8239–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richiardi, J. , Eryilmaz, H. , Schwartz, S. , Vuilleumier, P. , & Van De Ville, D. (2011). Decoding brain states from fMRI connectivity graphs. NeuroImage, 56(2), 616–626. [DOI] [PubMed] [Google Scholar]
- Salimi‐Khorshidi, G. , Douaud, G. , Beckmann, C. F. , Glasser, M. F. , Griffanti, L. , & Smith, S. M. (2014). Automatic denoising of functional MRI data: Combining independent component analysis and hierarchical fusion of classifiers. NeuroImage, 90, 449–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seghier, M. L. , & Price, C. J. (2018). Interpreting and utilising intersubject variability in brain function. Trends in Cognitive Sciences, 22(6), 517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X. , Finn, E. S. , Scheinost, D. , Rosenberg, M. D. , Chun, M. M. , Papademetris, X. , & Constable, R. T. (2017). Using connectome‐based predictive modeling to predict individual behavior from brain connectivity. Nature Protocols, 12(3), 506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simony, E. , Honey, C. J. , Chen, J. , Lositsky, O. , Yeshurun, Y. , Wiesel, A. , & Hasson, U. (2016). Dynamic reconfiguration of the default mode network during narrative comprehension. Nature Communications, 7, 12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. M. , Beckmann, C. F. , Andersson, J. , Auerbach, E. J. , Bijsterbosch, J. , Douaud, G. , … Kelly, M. (2013). Resting‐state fMRI in the human connectome project. NeuroImage, 80, 144–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spronk, M. , Kulkarni, K. , Ji, J. L. , Keane, B. , Anticevic, A. , & Cole, M. W. (2018). A whole‐brain and cross‐diagnostic perspective on functional brain network dysfunction. bioRxiv, 326728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas Yeo, B. T. , Krienen, F. M., Sepulcre, J., Sabuncu, M. R., Lashkari, D., Hollinshead, M., … & Fischl, B. (2011). The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of Neurophysiology, 106(3), 1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyszka, J. M. , Kennedy, D. P. , Paul, L. K. , & Adolphs, R. (2014). Largely typical patterns of resting‐state functional connectivity in high‐functioning adults with autism. Cerebral Cortex, 24(7), 1894–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderwal, T. , Eilbott, J. , Finn, E. S. , Craddock, R. C. , Turnbull, A. , & Castellanos, F. X. (2017). Individual differences in functional connectivity during naturalistic viewing conditions. NeuroImage, 157, 521–530. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Ren, Y. , Hu, X. , Nguyen, V. T. , Guo, L. , Han, J. , & Guo, C. C. (2017). Test–retest reliability of functional connectivity networks during naturalistic fMRI paradigms. Human Brain Mapping, 38(4), 2226–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler, D. (2011). Wechsler abbreviated scale of intelligence, second edition (WASI‐II). San Antonio: NCS Pearson. [Google Scholar]
- Xie, H. , Calhoun, V. D. , Gonzalez‐Castillo, J. , Damaraju, E. , Miller, R. , Bandettini, P. A. , & Mitra, S. (2018). Whole‐brain connectivity dynamics reflect both task‐specific and individual‐specific modulation: A multitask study. NeuroImage, 180, 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahata, N. , Morimoto, J. , Hashimoto, R. , Lisi, G. , Shibata, K. , Kawakubo, Y. , … Imamizu, H. (2016). A small number of abnormal brain connections predicts adult autism spectrum disorder. Nature Communications, 7, 11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information.
Data Availability Statement
We have made a subset of this data publicly available as part of the Autism Brain Imaging Data Exchange II (ABIDE II) initiative (Indiana University site; Di Martino et al., 2017; http://fcon_1000.projects.nitrc.org/indi/abide/abide_II.html). Data and code are available from the authors upon reasonable request.


