Abstract
Chemotherapy is one of the fundamental methods of cancer treatment. However, drug resistance remains the main cause of clinical treatment failure. We comprehensively review the newly identified roles of long noncoding RNAs (lncRNAs) in oncobiology that are associated with drug resistance. The expression of lncRNAs is tissue-specific and often dysregulated in human cancers. Accumulating evidence suggests that lncRNAs are involved in chemoresistance of cancer cells. The main lncRNA-driven mechanisms of chemoresistance include regulation of drug efflux, DNA damage repair, cell cycle, apoptosis, epithelial-mesenchymal transition (EMT), induction of signaling pathways, and angiogenesis. LncRNA-driven mechanisms of resistance to various antineoplastic agents have been studied extensively. There are unique mechanisms of resistance against different types of drugs, and each mechanism may have more than one contributing factor. We summarize the emerging strategies that can be used to overcome the technical challenges in studying and addressing lncRNA-mediated drug resistance.
Keywords: cancer, drug resistance, EMT, long noncoding RNAs, mechanisms
Introduction
The discovery and development of antineoplastic treatment modalities have significantly improved the survival and quality of life of cancer patients. Although surgery is regarded as the main and optimal treatment option for early-stage primary cancers, nonsurgical approaches based on small molecules, monoclonal antibodies, and humanized cell transplants are minimally invasive treatment modalities that increase survival of patients with late-stage and metastatic cancers.1–3 However, patients receiving pharmacological antineoplastic treatments often develop pronounced drug resistance, which leads to reduced therapeutic benefit, recurrent tumor growth, metastasis, and, ultimately, treatment failure.4,5
Thus, drug resistance remains a major challenge in cancer treatment. The mechanisms underlying drug resistance of tumors involve both intrinsic and extrinsic factors. Drug inactivation mediated through increased extracellular release of the drugs or reduced intracellular drug absorption is frequently observed in cancer cells.6,7 Moreover, cancer cells can induce changes in drug metabolism. Regulatory mechanisms such as mutation, epigenetic regulation, and posttranslational modification that lead to changes in enzyme expression and activity can impact drug efficacy and resistance, thereby affecting enzymatic reactions, which are classified into two complementary phases: phase I (oxidation, reduction, and hydrolysis) and phase II reactions (consumption and conversion). In addition, tumor cells may develop enhanced DNA repair systems, which directly counteract the efficacy of antitumor drugs in antineoplastic treatments that generally cause DNA damage (directly and/or indirectly) in tumor cells, leading to cell apoptosis.8,9 Furthermore, gene amplifications increase the number of copies of oncogenes per cell up to a 100-fold, which leads to drug resistance of tumors.10 Notably, the populations of tumor cells are heterogeneous, comprising cells with specific changes such as mutations, deletions, and chromosomal rearrangements. Therefore, only drug-sensitive cells are killed during the therapeutic course, while drug-resistant cells survive.7 Additionally, normal stromal cells contribute to acquired non-cell-autonomous drug resistance; this has been comprehensively reviewed by us elsewhere.11
Although several novel therapeutic strategies have been developed to overcome drug resistance and the short-term effects of the new treatments appear promising, rapid adaptation of the tumor cells and their surrounding microenvironment has resulted in unfavorable therapeutic outcomes.12
Noncoding RNAs (ncRNAs) are RNA products that do not encode proteins.13,14 Based on size, ncRNAs are classified into small ncRNAs (<200 nucleotides, nt) and long ncRNAs (lncRNA).15–18 LncRNAs are noncoding RNA molecules longer than 200 nts (coding potential of < 100 amino acids), which lack a defined open reading frame.18–21 LncRNA genes can be classified into five categories based on their position relative to the nearest protein-coding gene: (1) sense lncRNAs, which are superimposed on the exons of a protein-coding gene on the coding strand of the gene; (2) antisense lncRNAs, which are superimposed on exons of a protein-coding gene on the noncoding strand of the gene; (3) bidirectional lncRNAs, which are mirror-transcribed from the start site of another transcript; (4) intronic lncRNAs, which are completely located in the intron of another transcript; and (5) long intergenic ncRNAs (lincRNAs), which are located between two protein-coding genes.22 This heterogeneity of the lncRNAs allows them to control a broad spectrum of molecular and cellular functions through differential gene expression. Several lncRNAs in the nucleus participate in important transcriptional and posttranscriptional regulatory processes of gene expression. In certain cases, a single lncRNA regulates the expression of multiple genes by adjusting the expression of genes located in close proximity and controlling the expression of local and distal genes. At the same time, lncRNAs can also regulate the expression of proteins associated with specific pathways and biological processes, leading to the establishment of an lncRNAs-mediated regulatory network of specific cellular pathways.23,24
Differential expression of lncRNAs is generally observed in different tissues, diseases, and even in specific stages of a disease, highlighting the potential of these molecules as attractive therapeutic targets for the disease.25–28 Whereas the roles of small ncRNAs, including small interfering RNAs (siRNAs), microRNAs (miRNAs), and PIWI-interacting RNAs (piRNAs), have been reviewed extensively and critically elsewhere,29–36 it has only been recognized recently that a variety of lncRNAs play crucial functions in the initiation and development of human cancers.37–39 However, the role of lncRNAs in regulating the sensitivity of cancer cells to antineoplastic treatments is yet to be summarized. Given that lncRNAs play an essential role in regulating the level of gene expression through mechanisms such as chromatin modification and transcriptional and posttranscriptional regulation,28,40 which concomitantly occur during the development of drug resistance, it is important to critically summarize the recent advances regarding the functional roles of lncRNAs in the development of drug resistance and outline potential solutions for lncRNA-induced drug resistance during cancer treatment.
Mechanism of lncRNAs in chemotherapeutic resistance of cancer
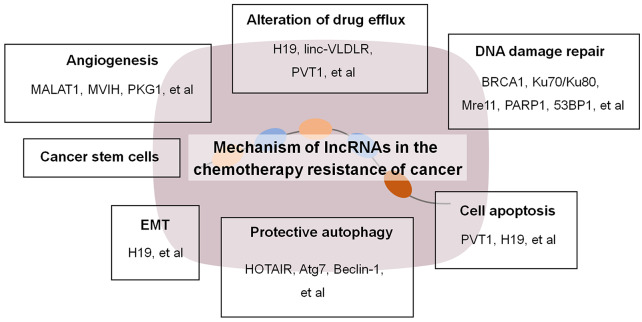
Increasing numbers of studies have explored the mechanisms underlying lncRNA-mediated drug resistance in human cancers. MALAT1 is the primary lncRNA reported to exert cancer-promoting effects in multiple myeloma (MM) through the enhancement of cell survival, mainly by regulating the expression of the proteasome mechanism. MALAT1 regulates the expression of Keap1 and Nrf1/2, which are key transcription factors of the genes encoding the proteasome subunits. Moreover, MALAT1 activates DNA repair in MM cells by acting as a scaffold molecule in the formation of PARP1/LIG3 complexes.41,42 NEAT1 is also highly expressed in MM, and regulates cellular stress response mechanisms by modulating UPR and p53 pathways.43 Moreover, lncRNAs regulate the expression of genes related to various pathways, including cellular metabolism of drugs, cell repair, cell death, cell transformation, and stemness, which can directly or indirectly lead to the initiation of drug resistance in human cancers (Figure 1).
Figure 1.
Mechanism of lncRNAs in chemotherapeutic resistance of cancer.
EMT, epithelial-mesenchymal transition; lncRNAs, long noncoding RNAs.
LncRNA-mediated alteration of drug efflux
In humans, the ATP-binding cassette (ABC) family of proteins comprises 49 known transporters that are involved in regulating the absorption, distribution, and excretion of pharmacological agents. ABC overexpression leads to enhanced drug efflux, which makes it challenging to maintain the therapeutic intracellular drug concentration and results in failure of antineoplastic therapy.44 Among these ABC proteins, P-glycoprotein (P-gp), adenosine triphosphate-binding cassette, superfamily G, member 2 (ABCG2), and multidrug resistance-associated protein (MRP) are frequently overexpressed in many types of cancers such as breast cancer (BC) and ovarian cancer (OC).45,46 This may be attributed to the activation of multiple kinases, including H-Ras, Raf-1, MEK1, and MEK2, that mediate the upregulation of P-gp level in tumor cells. In addition, cellular environmental stress conditions can also lead to the overexpression of these proteins.47 For example, increased expression of the lncRNAs H19 and linc-VLDLR induced the expression of MDR1/P-gp and ABCG2 in hepatocellular carcinoma (HCC); overexpression of the lncRNAs PVT1 and MRL led to increased expression of MDR1 and ATP-binding cassette, subfamily B, member 1 (ABCB1), respectively, in gastric cancer (GC); increased level of the lncRNA AK022798 led to the upregulation of MRP1 and P-gp in drug resistant cells.48–51
LncRNA-mediated DNA damage repair
Genomic instability is an important cause of chemotherapeutic resistance in cancers.48 Antineoplastic chemotherapeutics generally trigger DNA damage to kill tumor cells; however, tumor cells develop enhanced DNA repair pathways to maintain chromosome stability and avoid apoptosis. Cancer cells are able to repair DNA damage, thereby resisting the cytotoxicity of chemotherapy.47,52 DNA damage results in cell cycle arrest mediated by the activation of checkpoint pathways and suppression of cyclin-dependent kinase (CDK) activities.47,48,53–55 In tumor cells, lncRNAs regulate the activity of transcription factors (such as p53) that respond to DNA damage, mediate the repair of the damaged site, isolate the factors hindering DNA repair, interact with DNA repair proteins (such as BRCA1, Ku70/Ku80, Mre11, PARP1, 53BP1), and sequester miRNAs (regulators of DNA repair protein stability) to modulate mRNA expression levels.56
LncRNA-mediated regulation of cell apoptosis
Deregulation of apoptosis also affects drug resistance, as most chemotherapeutic agents inhibit the proliferation of cancer cells by promoting apoptosis.57 The upregulation of various survival factors such as nuclear factor inhibitors of apoptotic proteins, Bcl-2 family proteins, and nuclear factor κB (NF-κB) inhibit apoptotic cell death. Zhang et al. reported overexpression of PVT1 in cisplatin-resistant cancer cells, which results in reduced apoptosis of the cells following cisplatin treatment.58 In cisplatin-resistant lung cancer cells, the expression of p53 and Bcl-XL was regulated by the mitochondrial apoptotic pathway to reduce the expression of the lncRNA MEG3 and restore the sensitivity of cells to cisplatin treatment.59
The Bcl-2 family of proteins includes both antiapoptotic proteins such as Bcl-XL and Bcl-2, and proapoptotic proteins such as BAK, BAX, BAD, and BIK, which are essential components of the mitochondrial apoptotic pathway.60 The lncRNA H19 facilitates the induction of cisplatin resistance in lung adenocarcinoma by compromising the expression of the proapoptotic proteins BAX, BAK, and FAS.61 In contrast, the lncRNA ENST00000457645 significantly attenuates cisplatin resistance of CD70 cells by promoting BAX-associated cell apoptosis.62 In general, it is suggested that lncRNAs can influence the proliferation and apoptosis of cancer cells to affect radiosensitivity, mainly by regulating the relevant signal transduction pathways, such as the Wnt/β-catenin pathway and PI3K/AKT pathway, and acting as miRNA sponges.63
LncRNA-mediated protective autophagy
Autophagy is a catabolic process that plays an important role in some diseases, including cancer and neurodegenerative diseases, and has been defined as an adaptive pathway that maintains cell homeostasis. The role of autophagy in promoting survival or death mechanisms depends on several factors. Interestingly, a close correlation between lncRNAs and autophagy has been reported. Moreover, several studies have shown that lncRNAs play a role in regulating autophagy and further promote the development and progression of cancer.64,65 In addition, autophagy promotes drug resistance by mediating tumor hypoxia, cancer stem cells (CSCs), and DNA damage repair. In pancreatic cancer cells, HOTAIR promotes autophagy by stimulating the expression of Atg7, which mediates drug resistance. Similarly, HOTAIR mediates autophagy in human endometrial cancer cells by regulating the expression of Beclin-1, which results in drug resistance. XIST is highly expressed in non-small cell lung cancer (NSCLC) and regulates autophagy through the miR-17/ATG7 pathway to enhance the resistance of NSCLC cells to treatment.63,66 However, the precise mechanisms involved in these processes remains unclear and further studies are needed to clarify the underlying molecular mechanisms.64
Modulation of EMT
In the EMT process, epithelial cells lose several epithelial characteristics while gaining various mesenchymal characteristics. In addition, epithelial cells transform into a mesenchymal phenotype and lose their plasticity and intercellular adhesion, thereby becoming drug resistant.67 EMT is frequently observed in stem cell-like cells in cancers, which are generally characterized by resistance to antineoplastic agents, high expression of ABC transporters, non-responsiveness to induction of apoptosis, and enhanced capability for DNA repair.68 During the EMT process, epithelial cells lose their polarity and cell–cell contacts such as desmosomes, adhesion junctions, and tight junctions, leading to their separation from the epithelial layer, and acquire mesenchymal properties, including enhanced motility, invasiveness, resistance to apoptosis, and increased production of extracellular matrix components.69–71 In support of the role of lncRNAs in tumor progression and metastasis through EMT, a study reported that lncRNAs could function as either promoters or suppressors of EMT.72
Resistance/sensitivity to cisplatin and other chemotherapeutic agents has been associated with EMT-related lncRNAs. EMT is closely associated with reduced response to EGFR-TKIs and has been shown to mediate drug resistance by upregulating the PI3K-AKT pathway and reducing dependence on the MAPK/Erk pathway.73 The lncRNA H19 has been reported to regulate the expression of multiple EMT-associated genes in cancer cells. Overexpression of H19 decreases the epithelial marker E-cadherin and induces the mesenchymal marker vimentin, resulting in an elongated mesenchymal-like morphological change in the cells.74 Mechanistically, this function of lncRNA could be mediated through the epigenetic silencing of EMT-related genes and posttranscriptional regulation through binding of competing miRNA to target genes related to EMT regulation.73
LncRNA-mediated angiogenesis
Growth and metastasis of tumors depend on the establishment of a tumor vasculature that provides nutrition, oxygen, and other necessary factors. Tumor angiogenesis is a key sign of tumor development. Research into the potential mechanisms of angiogenesis can provide novel anti-angiogenic therapies for solid tumors. Angiogenesis is a vital process in the progression of cancer because it provides the tumor with nutrients and oxygen, as well as promotes proliferation and migration of tumor cells. Vascular endothelial growth factor (VEGF), the most effective activator of angiogenesis, is regulated by lncRNAs.75,76 Abnormal expression of lncRNAs can drive angiogenesis in cancer by activating carcinogenic signaling pathways such as STAT3, NF-κB, AKT, mTOR, and WNT in tumor cells. PVT1, which is highly expressed in GC tissues and cells, binds to STAT3 in the nucleus, and forms a complex that promotes angiogenesis in tumors through the STAT3/VEGFA axis. High expression of MALAT1 activates the mTOR/HIF-1α pathway to promote angiogenesis in osteosarcoma, and, through a feedback mechanism, TOR/HIF-1α mediates the expression of MALAT1 to maintain the existence of the cancer. LncRNAs also bind to various angiogenic proteins within tumor cells, thus affecting protein secretion, protein stability, enzyme activity, and the recruitment of proteins to specific genes. Moreover, lncRNAs sequester transcription factors away from target gene promoters, and, thus, regulate their expression. MVIH was the first lncRNA identified to induce angiogenesis, mainly by binding to PKG1 protein and inhibiting its secretion. Moreover, lncRNAs can regulate the crosstalk with miRNAs that may be involved in tumor angiogenesis.77
Cancer stem cells
CSCs are a small group of heterogeneous cells with the capacity of self-renewal and ability of directional differentiation. CSCs participate in tumorigenesis, tumor progression, distant metastasis, and chemoresistance. Emerging evidence suggests that lncRNAs play an essential role in maintaining CSCs and increasing the resistance of tumor cells to chemotherapy.78 LncRNAs are a new type of gene regulators in CSCs that mediate their effects through different mechanisms of regulating epigenetic changes and transcription levels, such as through chromatin modification and regulation and transcriptional activation or repression, respectively.79
Antitumor drug resistance driven by lncRNAs
Treatment with antineoplastic agents induces a series of changes in gene expression, not only in coding genes but also in noncoding genes such as lncRNAs. LncRNA-driven mechanisms of resistance to various types of antineoplastic agents have been well studied. Different types of drugs trigger unique drug resistance mechanisms, which may have more than one contributing factor (Table 1).
Table 1.
A summary of the lncRNA related to resistance of cancer.
| LncRNA | Type of cancer | Type of drug | Drug | Factors | Signaling pathway | Reference |
|---|---|---|---|---|---|---|
| PVT1 | GC | DNA-targeted drugs | Cisplatin | MRP1 | Apoptosis, mTOR/HIF-1α/P-gp | Zhang et al.58 |
| OC | DNA-targeted drugs | Cisplatin or carboplatin-Docetaxel | TGF-beta 1, p-Smad4, Caspase-3 | Apoptosis | Liu et al.80; Worku et al.81 | |
| HOTAIR | OC | DNA-targeted drugs | Cisplatin | XAV939 | Apoptosis, Wnt/β-catenin | Li et al.82 |
| NSCLC | DNA-targeted drugs | Cisplatin | Klf4, Iκ-Bα | NF-κB, CSC | Ozes et al.83; Janssens et al.84; Liu et al.85 | |
| OC | DNA-targeted drugs | Carboplatin | – | DNA methylation | Wu et al.86 | |
| BC | Antihormone therapy | Tamoxifen | – | ER-mediated transcription inhibition | Xue et al.87 | |
| AL | Tyrosine kinase inhibitors | imatinib | PI3K, Akt | Apoptosis, PI3K/Akt | Cruz-Miranda et al.88; Wang et al.89 | |
| AK022798 | GC | DNA-targeted drugs | Cisplatin | MRP1 and P-gp | – | Hang et al.90 |
| UCA1 | Bladder cancer | DNA-targeted drugs | Cisplatin | srpk1 | Wnt, apoptosis, | Fan et al.91; Wang et al.92 |
| NSCLC | Tyrosine kinase inhibitors | Gefitinib | – | – | Cheng et al.93 | |
| CRC | Antimetabolite therapy | 5-FU | miR-204-5p | Apoptosis | Luo et al.94 | |
| BC | Antihormone therapy | Tamoxifen | – | Wnt/β-catenin | Liu et al.95 | |
| CML | Tyrosine kinase inhibitors | imatinib | MDR1, miR-16 | MDR1/miR-16 | Xiao et al.96 | |
| H19 | OC | DNA-targeted drugs | Cisplatin | MEG3, miR-214 | Nrf2 | Wang et al.61; Zhan et al.97 |
| CRC | Antimetabolite therapy | Methotrexate | – | WNT/β-catenin | Luo et al.94 | |
| ANRIL | GC | DNA-targeted drugs | cisplatin | – | – | Lan et al.98 |
| GC | Antimetabolite therapy | Cisplatin and 5-FU | – | Apoptosis | Lan et al.98 | |
| LAD | Cytotoxic drugs | Paclitaxel | cleaved-PARP, Bcl-2 | Apoptosis | Xu et al.99 | |
| ROR | NSCLC | DNA-targeted drugs | Cisplatin | – | PI3K/Akt/mTOR | Shi et al.100 |
| Zinc | OC | DNA-targeted drugs | Cisplatin | miRNA-150-5p, SP1 | – | Xia et al.101 |
| LINC00161 | osteosarcoma | DNA-targeted drugs | Cisplatin | miR-645, IFIT2 | Apoptosis | Wang et al.102 |
| ENST00000457645 | OC | DNA-targeted drugs | Cisplatin | – | Apoptosis | Yan et al.62 |
| BLACAT1 | GC | DNA-targeted drugs | Oxaliplatin | miR-361, ABCB1 | – | Wu et al.103 |
| MALAT-1 | Pancreatic cancer | DNA-targeted drugs | Gemcitabine | miR-200c, miR-145, Sox2 | CSC | Jiao et al.104 |
| UCA1 | GC | Cytotoxic drugs | Adriamycin | PARP, Bcl-2 | Apoptosis | Shang et al.105 |
| TUG1 | BUC | Cytotoxic drugs | adriamycin | – | Wnt/β-catenin | Xie et al.106 |
| HANR | HCC | Cytotoxic drugs | adriamycin | GSK3β | GSK3β | Xiao et al.107 |
| ENST00000500843 | LAD | Cytotoxic drugs | Paclitaxel | – | Apoptosis | Tian et al.108 |
| NEAT1 | PC | Cytotoxic drugs | Docetaxel | miR-34a | – | Tian et al.109 |
| lnc-ATB | BC | Tyrosine kinase inhibitors | Trastuzumab | ZEB1, ZNF-217, miR-200c | TGF-β | Gajria et al.110 |
| GAS5 | BC | Tyrosine kinase inhibitors | Trastuzumab | mTOR, PTEN | mTOR | Li et al.111 |
| BC087858 | NSCLC | Tyrosine kinase inhibitors | Gefitinib | – | PI3K/AKT, MEK/ERK | Pan et al.112 |
| SNHG5 | LAD | Tyrosine kinase inhibitors | Gefitinib | miR-377 | – | Wang et al.113 |
| linc-VLDLR | HCC | Tyrosine kinase inhibitors | Sorafenib | ABC-G2 | EVs | Takahashi et al.114 |
| ARSR | RCC | Tyrosine kinase inhibitors | Sunitinib | AXL, c-MET, miR-34/miR-449 | – | Qu et al.115 |
| LINC00152 | CRC | Antimetabolite therapy | 5-FU | miR-139-5p | Apoptosis | Bian et al.116 |
| LUCAT1 | osteosarcoma | Antimetabolite therapy | Methotrexate | ABCB1, miR-200c | – | Han and Shi117 |
| BCAR4 | BC | Antihormone therapy | Tamoxifen | ERBB2, ERBB3, AKT | ERBB, apoptosis | van Agthoven et al.118 |
| FENDRR | CML | Cytotoxic drugs | adriamycin | MDR1 | Apoptosis | Zhang et al.119 |
BC, breast cancer; BUC, bladder urothelial carcinoma; CML, chronic myeloid leukemia; CRC, colorectal cancer; CSC, cancer stem cells; 5-FU, 5-fluorouracil; GC, gastric cancer; HCC, hepatocellular carcinoma; LAD, lung adenocarcinoma; lncRNA, long noncoding RNA; NFκB, nuclear factor κB’; NSCLC, non-small cell lung cancer; OC, ovarian cancer; PC, prostate cancer; RCC, renal cell carcinoma; TGF-β, transforming growth factor beta.
DNA-targeted drugs
Platinum-based chemotherapy
Cisplatin or cis-diamminedichloroplatinum (II) (CDDP), a commonly used platinum-based chemotherapy drug, was originally approved by the Food and Drug Administration (FDA) in 1978 for the treatment of testicular and bladder cancer, and its clinical use has now been expanded to ovarian, colorectal, lung, and head and neck cancers. In the early 1980s, a second-generation platinum compound was developed with the specific purpose of reducing its side effects without jeopardizing its anticancer properties.120 Cisplatin is one of the most potent drugs used in cancer chemotherapy that triggers cell apoptosis by cross-linking DNA.121
PVT1 is a lncRNA located adjacent to the MYC gene on human chromosome 8q24. Several researchers have reported that abnormal expression of PVT1 promotes cisplatin resistance in cancer. Notably, the overexpression of PVT1 is associated with cisplatin-resistance in GC cell lines due to the inhibition of apoptosis. Overexpression of PVT1 activates mTOR/HIF-1α/P-gp and MRP1 signaling pathways, which, in turn, induce the expression of drug resistance-related genes, and, consequently, lead to the development of multidrug resistance in GC.58 Moreover, PVT1 may differentially regulate the expression of proteins related to the apoptotic pathway in OC, and result in resistance to cisplatin treatment.80 Overexpression of PVT1 promotes drug resistance in OC cells by regulating downstream apoptotic proteins.80,81 In addition, increased expression of PVT1 and its negative function in cell apoptosis leads to cisplatin or carboplatin-docetaxel resistance in OC. Overexpression of HOTAIR was found in the cisplatin-resistant SKOV-3CDDP/R ovarian carcinoma cell line model, and the sensitivity to cisplatin was altered in an siRNA-induced HOTAIR knockout cell line. The above phenomena are also associated with increased apoptosis and cytotoxicity.122 Liu et al. showed that high expression of HOTAIR in OC cells activates Wnt/β-catenin signaling pathway and promotes cell cycle progression, which contributes to the induction of cisplatin resistance; suppressing HOTAIR leads to inactivation of Wnt/β-catenin signaling pathway as well as cell cycle arrest at G1 phase, and renders OC cells susceptible to cisplatin treatment.82 On the other hand, overexpression of HOTAIR activates nuclear factor-κB (NF-κB) signal transduction-associated DNA damage response, promoting cell repair against cisplatin-induced DNA damage. This effect of HOTAIR may be due to the downregulation of the endogenous NF-κB inhibitor, Iκ-Bα.83,84 In addition, HOTAIR promotes cisplatin resistance by maintaining the CSCs in NSCLC patients.85 HOTAIR promotes the ability of tumor sphere formation through the upregulation of tumor stem cell-related Klf4 expression in lung CSCs, which, in turn, contributes to the development of cisplatin resistance of NSCLC.85
LncRNA AK022798, whose expression is regulated by Notch 1, was found to promote the development of resistance to cisplatin treatment in GC cells. Silencing of AK022798 expression significantly decreased the expression of MRP1 and P-gp, leading to increased cellular uptake of cisplatin by the resistant GC cells.90
UCA1, a member of the human endogenous retrovirus H (HERV-H) family, facilitates cisplatin resistance in bladder cancer through mediating the enhanced expression of Wnt6.91 At the same time, overexpression of UCA1 leads to reduced apoptosis in cells following cisplatin treatment. Upregulation of UCA1 leads to overexpression of the anti-apoptotic protein serine-arginine protein kinase 1 (srpk1); however, the sensitivity to cisplatin is partially restored following the downregulation of sprk1.92 This suggests the existence of multiple regulatory mechanisms of UCA1 in platinum resistance of cancer cells.
The overexpression of lncRNA H19 promotes the acquisition of cisplatin resistance in lung cancer cells. Dysregulation of H19 expression was found in cisplatin-resistant lung cancer cells.61 Upregulation of H19 in cisplatin-resistant OC cell lines promoted the expression of Nrf2 and its target genes, which are associated with the regulation of glutathione metabolism and glutathione expression, leading to the inactivation of cisplatin and reduced intracellular free radicals. After treating epithelial OC cell lines with curcumin and 5-AZA-dC, demethylation of MEG3 weakened the ability of extracellular vesicles (EVs) to enhance cisplatin resistance through binding with miR-214.97
In GC patients with cisplatin resistance, the expression of ANRIL was significantly upregulated, and transfection with si-ANRIL led to a significant decrease in the tumor growth rate. A similar effect was observed in 5-fluorouracil-resistant patients.98 LncRNA regulator of reprogramming (ROR) activates the PI3K/Akt/mTOR signaling pathway during the initiation of drug resistance in NSCLC cells, whereas knockdown of lncRNA ROR renders NSCLC cells susceptible to cisplatin treatment.100 LncRNA Zinc finger antisense 1 (ZFAS1) interacts with miRNA-150-5p, promotes expression of the specific protein 1 (SP1) gene, and induces resistance of OC cells to cisplatin and paclitaxel.101
In contrast, lncRNA long intergenic noncoding RNA 161 (LINC00161) is induced by cisplatin in osteosarcoma cells, and subsequently sponges miR-645 to attenuate its binding to the 3′ UTR of IFIT2 mRNA. Consequently, IFIT2 is upregulated and promotes cell apoptosis following cisplatin treatment. LINC00161 thereby reverses the cisplatin-resistant phenotype of osteosarcoma cells.102 LncRNA ENST00000457645 is suppressed in cisplatin-resistant OC cells. Transfection of ENST00000457645 upregulates apoptosis-related proteins to abrogate cisplatin resistance in these cells.62
Overexpression of lncRNA HOTAIR in carboplatin resistant primary OC cells results in DNA methylation that contributes to drug resistance.86 Downregulation of another lncRNA, BC200, in OC tissues leads to increased cell viability and chemoresistance, although the mechanism remains unknown.123
Oxaliplatin is a third-generation platinum compound and is widely used as a first-line treatment drug for advanced GC. The expression of the lncRNA, bladder cancer-associated transcript-1 (BLACAT1, also known as linc-UBC1), is upregulated in oxaliplatin-resistant GC tissue and cells compared with oxaliplatin-sensitive tissue and cells. BLACAT1 antagonizes miR-361 and restores ABCB1 protein expression, which results in the oxaliplatin resistance of GC. BLACAT1 knockdown abolished oxaliplatin resistance and suppresses the growth of GC tumor.103
Nucleotide analogs and precursor analogs
Nucleoside analogs have been used clinically for nearly 50 years as antineoplastic agents for various tumors, such as in certain types of lymphomas and prostate cancer.124 Upregulation of the lncRNA, MALAT-1, is observed in CSCs which increases the CSCs population in pancreatic cancer. MALAT-1 sponges miR-200c and miR-145 to restore the expression of Sox2, a gene associated with stemness of CSCs. As a result, MALAT-1 decreases the sensitivity of pancreatic cancer cells to gemcitabine.104
Cytotoxic drugs
The development of cytotoxic agents has revolutionized cancer treatment, making it possible to cure certain tumors such as childhood acute leukemia, Hodgkin’s disease, non-Hodgkin’s lymphoma, gestational trophoblastic disease, and germ cell tumors. In addition to surgical treatment alone, adjuvant therapy using cytotoxic drugs has obvious therapeutic benefits in many types of cancers.125–127 Adriamycin/doxorubicin is a cytotoxic drug (an anthracycline antibiotic) that is commonly used in chemotherapy.
Anthracyclines
Suppression of lncRNA UCA1 reduced adriamycin-resistance in GC. This was associated with the upregulation of PARP and downregulation of Bcl-2 due to silencing of UCA1 in adriamycin resistant GC cells.105
The expression of lncRNA ODRUL is increased in adriamycin resistant osteosarcoma cell lines, and the knockdown of ODRUL leads to suppression of the ABCB1 gene.128 Zhou et al. reported that high expression of SNHG12 mediated adriamycin-resistance in osteosarcoma through the miR-320a/MCL1 axis.129
The lncRNA taurine upregulated 1 (TUG1) gene is highly expressed in several malignant tumors, including bladder cancer. Xie et al. discovered overexpression of TUG1 in bladder urothelial carcinoma (BUC) tissues and cell lines that are resistant to adriamycin. Knockdown of TUG1 restored adriamycin cytotoxicity, which was associated with the inhibition of the Wnt/β-catenin pathway. Reversal of adriamycin drug resistance by TUG1 knockdown could be achieved by the partial activation of the Wnt/β-catenin pathway. Thus, lncRNA TUG1 results in the non-responsiveness of BUC patients to adriamycin through the Wnt/β-catenin pathway.106
HANR was found to be upregulated in HCC patients and cell lines and plays an essential role in the development of HCC; this was further proven using clinical specimens, HCC cell lines in vitro, and xenograft/orthotopic mouse models in vivo. HANR regulates the phosphorylation of GSK3β in HCC, resulting in a decrease in its sensitivity to Adriamycin.107
Other cytotoxic drugs
Paclitaxel is one of the most effective plant-derived anticancer drugs for lung adenocarcinoma (LAD) treatment; it functions mainly by causing cell cycle arrest at the G2/M phase and subsequently inducing apoptosis.130 ANRIL was initially found in familial melanoma patients; its expression is upregulated in LAD and affects the therapeutic effect of paclitaxel. ANRIL acts as a potential oncogene and promotes the acquisition of chemoresistance to paclitaxel by regulating the expression of cleaved-PARP and Bcl-2 among the apoptosis-related proteins.99
The expression of the lncRNA ENST00000500843 is downregulated in LAD tissues and A549/paclitaxel cells. The knockdown of lncRNA ENST00000500843 decreased apoptosis in A549 cells exposed to paclitaxel, which led to paclitaxel-resistance in the LAD cells.108
The lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) is upregulated in human prostate cancer (PC) tissues and docetaxel-resistant PC cells, whereas miR-34a-5p is downregulated. NEAT1 knockdown re-sensitizes the resistant PC cells to docetaxel, which may be associated with the sponging of miR-34a-5p.109,131
Tyrosine kinase inhibitors
A variety of protein kinases, including receptor tyrosine kinases such as VEGF receptors (VEGFRs) engage in tumor progression. A series of tyrosine kinase inhibitors (TKIs) have been developed for the systematic treatment of various types of cancers. TKIs such as sunitinib, sorafenib, and pazopanib bind to different types of tyrosine kinases with different affinities and have been approved for the treatment of advanced cancers such as renal cell cancer, gastrointestinal stromal tumors, and HCC.132
Human epidermal growth factor receptor (HER2) inhibitors
Trastuzumab, currently used for early-stage and metastatic HER2-positive BC and GC, was designed to block the extracellular domain of HER2.110 Shi et al. found that lncRNA activated by TGF-beta (lnc-ATB) was the most significantly upregulated lncRNA in trastuzumab-resistant SKBR-3 cells and trastuzumab-resistant BC patient tissues.110 LncRNA-ATB also promotes trastuzumab resistance in BC by competitively binding with miR-200c, upregulating ZEB1 and ZNF-217, and then inducing EMT. In addition, a high level of lncRNA-ATB is correlated with trastuzumab resistance in BC patients, which acts as a vital regulatory factor of TGF-β signaling pathways and trastuzumab resistance in BC.110,111
GAS5 is another lncRNA related to trastuzumab resistance; reduced expression of GAS5 activates mTOR and suppresses PTEN.111
Epidermal growth factor receptor (EGFR) inhibitors
Gefitinib is a selective inhibitor of EGF and is used for the treatment of NSCLC patients. It acts by controlling cell growth, inducing apoptosis, and suppressing angiogenesis.133
Pan et al. found that overexpression of the lncRNA, BC087858, was associated with acquired resistance of NSCLC cell lines and patients to EGFR-TKIs. LncRNA, BC087858, activates PI3K/AKT and MEK/ERK pathways, and initiates EMT of NSCLC cells, which in turn results in acquired resistance to EGFR-TKIs in EGFR T790M-mutant NSCLC cells.112 Others studies also revealed that UCA1 had a similar effect on mediating the development of acquired resistance of EGFR-mutant NSCLC patients to EGFR-TKIs.93
The expression of SNHG5 was significantly downregulated in gefitinib-resistant LAD patients and LAD cell lines. Overexpression of SNHG5 suppressed the expression of miR-377, and knockdown of SNHG5 increased miR-377 expression. The overexpression of SNHG5 rendered LAD cells sensitive to gefitinib therapy in vitro and in vivo.112 An EGFR-TKI-resistant HCC827-8-1 cell line was generated and analyzed for expression patterns by lncRNA microarray and these patterns were compared with those of its parental HCC827 cell line. The results showed that lncRNAs may be used as a new candidate biomarkers and potential targets for EGFR-TKI therapy in LAD.134
B-Raf inhibitors
HCC cell-derived EVs contain and transfer lncRNAs. Takahashi et al. found that the transfer of lincRNA-VLDLR (linc-VLDLR), which is enriched in HCC-derived EVs, promotes drug resistance in HCC. During chemotherapeutic stress, exposure of HCC cells to sorafenib increased linc-VLDLR expression in cells, and within the EVs released from these cells, to promote chemoresistance. Moreover, the expression of ABC subfamily G member 2 (ABC-G2) was induced in tumor cells by incubation with tumor cell-derived EVs, suggesting lncRNA-containing EVs may modulate chemoresistance through the ABC transporters.114
Other TKIs
LncARSR is highly expressed in renal cell carcinoma (RCC) cells with sunitinib resistance, which remains a major challenge in RCC treatment. LncARSR, identified by Qu et al., correlated with sunitinib resistance in RCC in the clinic. LncARSR promoted the expression of AXL and c-MET by competitive binding with miR-34/miR-449 leading to sunitinib resistance. LncARSR could be transmitted to sensitive cells through the incorporation into exosomes, resulting in acquired resistance to sunitinib. Treatment of sunitinib-resistant RCC with locked nucleic acids targeting lncARSR or an AXL/c-MET inhibitor restored sunitinib response and improved the therapeutic response.115
In a study by Yan et al., chip technology was used to analyze the expression of lncRNAs in paired normal gastric tissues, primary gastrointestinal stromal tumor (GIST) tissues and GIST tissues resistant to imatinib mesylate. Expression of these lncRNAs may be associated with GIST and mediate the secondary resistance of imatinib mesylate.135
Antimetabolite therapy
Radiosensitization of antimetabolites improves the clinical outcome for malignant solid tumors, such as gastrointestinal tract cancer, cervical cancer, and head and neck cancer.136 LINC00152 plays an oncogenic role in gastric, liver, gallbladder, and lung cancer and is involved in regulating cell proliferation, cell cycle and other biological processes. In CRC, overexpression of LINC00152 was negatively correlated with the survival rate of patients. LINC00152 enhanced the migration and invasion of cells and promoted cell proliferation and inhibited apoptosis; thus, causing therapeutic resistance of CRC cells to 5-fluorouracil (5-FU). The expression of NOTCH1 is frequently upregulated in CRC, and miR-139-5p targets NOTCH1 to regulate CRC growth. Further studies have shown that LINC00152 sponges miR-139-5p to regulate the expression and activity of NOTCH1, which promotes the progression of CRC.116
The expression of LEIGC in GC tissue is higher than that in noncancerous tissue, whereas its reduction causes 5-FU resistance in MGC-803 cells.137 High expression of ANRIL was observed in the GC tissues of patients with cisplatin and 5-FU resistance, as well as in resistant GC cell lines. Knockdown of ANRIL suppresses proliferation and invasion, and promotes apoptosis of GC cells upon treatment of cisplatin and 5-FU, suggesting a functional role of ANRIL in the development of drug resistance in GC.98
UCA1 causes 5-FU-resistance in CRC by preventing miR-204-5p from inhibiting apoptosis. In CRC, methotrexate resistance is mediated through the activation of the WNT/β-catenin pathway by lncRNA H19.94
LncRNA LUCAT1 and ABCB1 protein are upregulated in methotrexate-resistant osteosarcoma cells (MG63/MTX and HOS/MTX) compared with parental cells. Bioinformatics tools and luciferase assays were used to verify that the 3′-UTR of both LUCAT1 and ABCB1 mRNA competed for binding with miR-200c, and sponging of miR-200c by LICAT1 modulated ABCB1 expression, thereby contributing to the development of chemoresistance.117
Antihormone therapy
Antihormone therapy is widely used in the treatment of cancer, and research has focused mainly on the development of effective and selective estrogen receptor modulators for BC therapies. Antihormone therapy can be used for the prevention or treatment of women with BC and at high risk for BC.138
Breast cancer antiestrogen resistance 4 (BCAR4), defined as a lncRNA, transforms BC cells into an state independent to estrogen and antiestrogen, and was also associated with tumor aggressiveness and tamoxifen resistance.118 Godinho et al. analyzed the expression pattern of BCAR4 in 280 estrogen receptor α (ERα)-positive primary BC patients with advanced disease and found that 29% of the patients showed high expression levels of BCAR4. High expression of BCAR4 led to robust phosphorylation of v-erb-b2 erythroblastic leukemia viral oncogene homolog (ERBB)2, and ERBB3, resulting in activation of the critical downstream mediators of ERBB signaling such as AKT and extracellular signal-regulated kinase 1/2 (ERK 1/2) expression, leading to the inhibition of apoptosis in the cancer cells.139 Knockdown of BCAR4 suppressed cell proliferation of these BC cells, which was associated with the expression and activity of ERBB2/3.
LncRNA HOTAIR also plays an essential role in tamoxifen resistance. In their study, Xue et al. found that, compared with primary BC tissues, expression of HOTAIR was increased in tamoxifen-resistant BC tissues. HOTAIR is a direct target of ER-mediated transcriptional inhibition, and its overexpression promotes the growth of BC and contributes to tamoxifen resistance.87
Liu et al. studied the role and clinical significance of UCA1 in BC drug resistance using tamoxifen-resistant BC cell lines that they established in vitro and in mouse xenograft models, and found that the expression of UCA1 was positively correlated with mortality in patients with BC, and the expression of UCA1 was significantly increased in tamoxifen-resistant cell lines compared with the wild-type parental cells.135 In addition, hyper-activation of the Wnt/β-catenin pathway was found in the resistant BC cells compared with their sensitive counterparts. UCA1 depletion significantly reduced the activity of the Wnt/β-catenin pathway and the tumorigenicity of the tamoxifen-resistant BC cells, suggesting that UCA1/Wnt signaling plays a pivotal role in mediating the resistance of BC cells to tamoxifen.95
LncRNA-mediated drug resistance in leukemia
Acute lymphoblastic leukemia (ALL), chronic lymphoblastic leukemia (CLL), acute myeloid leukemia (AML), and chronic myeloid leukemia (CML) are subsets of leukemia, which belong to hematological malignancies.88
Imatinib plays an extremely important role in the treatment of CML. However, some patients who are in an accelerated phase develop imatinib resistance. The main cause of imatinib resistance in CML is MDR1-mediated external drug efflux. LncRNA UCA1 is an important regulator of MDR1 and increased expression of UCA1 leads to increased expression of MDR1. Overexpressed UCA1 in CML cells binds to miR-16, functioning as a competitive endogenous RNA (ceRNA) of MDR1, leading to imatinib resistance.96 HOTAIR is one of the lncRNAs found to be overexpressed in acute leukemia (AL). HOTAIR promotes the proliferation of leukemic blast and leukemic stem cells, and its high expression is associated with a low survival rate in AL, and with imatinib resistance.88 High levels of HOTAIR can induce apoptosis of human CML cell line and reduce cell sensitivity to imatinib through the PI3K/AKT pathway, resulting in drug resistance.89 The reduced expression of FENDRR reduces the sensitivity of CML cells to adriamycin, which leads to reduced apoptosis. FENDRR and MDR1 are negatively correlated. FENDRR inhibits the binding of MDR1 and miR-184 to reverse adriamycin-resistance.119
Therapeutic prospects
Accumulating evidence suggests that expression of lncRNA is globally altered in various cancers and is involved in all aspects of tumorigenesis by inactivating tumor inhibitors or activating oncogenes. Due to their ubiquitous expression and relative stability, lncRNAs are suitable as biomarkers for the early diagnosis of cancers. PVT1 is overexpressed in tissues of NSCLC and lung squamous cell carcinoma, and overexpression of PVT1 is significantly associated with poor prognosis. High expression of UCA1 is associated with decreased survival rate of patients, suggesting that UCA1 is a predictive biomarker in OC.140 HOTAIR level and upregulation of HOTAIR expression in patients with lung cancer are associated with the pathological stage and poor prognosis of lung cancer. Plasma HOTAIR expression level may be a biomarker for the diagnosis and monitoring of NSCLC patients.141–143 Further, overexpression and methylation of HOTAIR mediates therapeutic resistance of OC cells. As a prognostic biomarker for mediating DNA repair, HOTAIR could be used as a reliable classifier to distinguish patients for personalized treatment.123 Overexpression of HOTAIR is also associated with short survival time in patients with esophageal carcinoma, suggesting that increased HOTAIR level may be a prognostic factor for esophageal cancer.144 ANRIL is recruited and associates with PRC2, and upregulated in GC tissues.145
The diverse expression and functions of lncRNA in cancers, as discussed above, make them an attractive target for developing therapeutic target. A number of lncRNAs are not only biomarkers for drug resistance in the clinical application of chemotherapeutic drugs, but also therapeutic targets to overcome drug resistance. MALAT1 is expressed extensively in various human solid and hematological malignancies and promotes cell proliferation and metastasis. Moreover, due to the biological signature of MALAT1, selective strategies using synthetic oligonucleotides have been developed to inhibit MALAT1.146 A feedback loop was identified between MALAT1 and Nrf1, which is activated in resistant cells to affect the proteasome machinery.147 The inhibitor of AXL/c-MET is the downstream effector of the lncRNAs, restoring lncRNA-mediated resistance to sunitinib in orthotopic xenograft models.115 VEGFR, AXL, and c-MET are targets of cabozantinib, a RTK inhibitor, revealed in a randomized phase III trial that prolonged progression-free survival of RCC patients following VEGFR-targeted treatment.148 These results reinforce the idea that understanding specific biological mechanisms of resistance can help overcome the resistance to chemotherapeutic agents which remains a critical goal for anticancer therapy.
Conclusion
Recent studies have identified several lncRNAs that mediate drug resistance, a critical mechanism that causes refractory cancer and antineoplastic treatment failure. In the present review, the important biological functions of some lncRNAs and the mechanism of action underlying the regulation of key signaling pathways are discussed. As a precise tool for regulating cell homeostasis, lncRNAs are involved in vital biological processes. Their functions are diverse and often altered in cancer. Accumulating evidence shows that lncRNAs are involved in the resistance of cancer cells to various therapeutic drugs, and the main mechanisms include regulation of drug efflux, DNA damage repair, cell cycle, apoptosis, EMT, and induction of signaling pathways and angiogenesis. Several lncRNAs have confirmed to mediate chemoresistance in cancer cells. Rapid advances have been made in research relating to lncRNA and its diverse biological processes, and many studies have shown that lncRNA are candidates for cancer treatment. However, the application of lncRNAs as a therapeutic target in the development of new treatments to overcome chemotherapeutic resistance requires more in-depth studies. Further research is also needed to identify more lncRNAs associated with resistance, and to understand their role in cancer and chemotherapy resistance. In conclusion, the roles and mechanisms of action of lncRNAs in mediating resistance to anticancer treatments summarized in this review provide new ideas for the development of more effective treatment strategies.
Footnotes
Author contributions: YDQ, HYT, YTC and HBJ retrieved the data and draft the manuscript. NW and DW revised the manuscript and initiated the idea and draft the manuscript.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the National Key Research and Development Program of China (Project code: No. 2018YFE0107800), the Special Projects of Cooperation between Jilin University and Jilin Province in China (Project code: SXGJSF2017-1), Research Grant Council, HKSAR (Project code: RGC GRF 17152116), Commissioner for Innovation Technology, HKSAR (Project code: ITS/091/16FX) and Health and Medical Research Fund (Project code: 15162961).
Contributor Information
Yidi Qu, School of Life Sciences, Jilin University, Changchun, China.
Hor-Yue Tan, School of Chinese Medicine, The University of Hong Kong, Pokfulam, Hong Kong S.A.R., P.R. China.
Yau-Tuen Chan, School of Chinese Medicine, The University of Hong Kong, Pokfulam, Hong Kong S.A.R., P.R. China.
Hongbo Jiang, School of Life Sciences, Jilin University, Changchun, China.
Ning Wang, School of Chinese Medicine, The University of Hong Kong, Pokfulam, Hong Kong S.A.R., P.R. China.
Di Wang, School of Life Sciences, Jilin University, Changchun, 130012, China.
References
- 1. Reghupaty SC, Sarkar D. Current status of gene therapy in hepatocellular carcinoma. Cancers (Basel) 2019; 11: 1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cruz E, Kayser V. Monoclonal antibody therapy of solid tumors: clinical limitations and novel strategies to enhance treatment efficacy. Biologics 2019; 13: 33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dugger SA, Platt A, Goldstein DB. Drug development in the era of precision medicine. Nat Rev Drug Discov 2018; 17: 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garraway LA, Jänne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov 2012; 2: 214–226. [DOI] [PubMed] [Google Scholar]
- 5. Corso S, Giordano S. Cell-autonomous and non-cell-autonomous mechanisms of HGF/MET-driven resistance to targeted therapies: from basic research to a clinical perspective. Cancer Discov 2013; 3: 978–992. [DOI] [PubMed] [Google Scholar]
- 6. Zahreddine H, Borden KL. Mechanisms and insights into drug resistance in cancer. Front Pharmacol 2013; 4: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Housman G, Byler S, Heerboth S, et al. Drug resistance in cancer: an overview. Cancers (Basel) 2014; 6: 1769–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borst P, Evers R, Kool M, et al. A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst 2000; 92: 1295–1302. [DOI] [PubMed] [Google Scholar]
- 9. Laura B, Adolfo F, Rafael R. Platinum drugs and DNA repair mechanisms in lung cancer. Anticancer Res 2014; 34: 493–501. [PubMed] [Google Scholar]
- 10. Mansoori B, Mohammadi A, Davudian S, et al. The different mechanisms of cancer drug resistance: a brief review. Adv Pharm Bull 2017; 7: 339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qu Y, Dou B, Tan H, et al. Tumor microenvironment-driven non-cell-autonomous resistance to antineoplastic treatment. Mol Cancer 2019; 18: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holohan C, Van Schaeybroeck S, Longley DB, et al. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 2013; 13: 714–726. [DOI] [PubMed] [Google Scholar]
- 13. Sakamoto N, Honma R, Sekino Y, et al. Non-coding RNAs are promising targets for stem cell-based cancer therapy. Noncoding RNA Res 2017; 2: 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poller W, Dimmeler S, Heymans S, et al. Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur Heart J 2018; 39: 2704–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6: 857–866. [DOI] [PubMed] [Google Scholar]
- 16. Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene 2008; 27 Suppl 2: S52–S57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Winter J, Jung S, Keller S, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 2009; 11: 228–234. [DOI] [PubMed] [Google Scholar]
- 18. Chen LL, Carmichael GG. Long noncoding RNAs in mammalian cells: what, where, and why? Wiley Interdiscip Rev RNA 2010; 1: 2–21. [DOI] [PubMed] [Google Scholar]
- 19. Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta 2010; 1799: 597–615. [DOI] [PubMed] [Google Scholar]
- 20. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009; 136: 629–641. [DOI] [PubMed] [Google Scholar]
- 21. Dinger ME, Pang KC, Mercer TR, et al. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 2008; 4: e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gutschner T, Diederichs S. The hallmarks of cancer A long non-coding RNA point of view. RNA Biol 2012; 9: 703–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun QY, Hao QY, Prasanth KV. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet 2018; 34: 142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kornienko AE, Guenzl PM, Barlow DP, et al. Gene regulation by the act of long non-coding RNA transcription. BMC Biol 2013; 11: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Amaral PP, Neyt C, Wilkins SJ, et al. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA 2009; 15: 2013–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fu XQ, Ravindranath L, Tran N, et al. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol 2006; 25: 135–141. [DOI] [PubMed] [Google Scholar]
- 27. Ravasi T, Suzuki H, Pang KC, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 2006; 16: 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009; 23: 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He YQ, Lin JJ, Kong DL, et al. Current state of circulating microRNAs as cancer biomarkers. Clin Chem 2015; 61: 1138–1155. [DOI] [PubMed] [Google Scholar]
- 30. Ohtsuka M, Ling H, Doki Y, et al. MicroRNA processing and human cancer. J Clin Med 2015; 4: 1651–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017; 16: 203–221. [DOI] [PubMed] [Google Scholar]
- 32. Oh YK, Park TG. siRNA delivery systems for cancer treatment. Adv Drug Deliv Rev 2009; 61: 850–862. [DOI] [PubMed] [Google Scholar]
- 33. He CB, Lu KD, Liu DM, et al. Nanoscale metal-organic frameworks for the co-delivery of cisplatin and pooled siRNAs to enhance therapeutic efficacy in drug-resistant ovarian cancer cells. J Am Chem Soc 2014; 136: 5181–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan LP, Mai DM, Zhang BL, et al. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol Cancer 2019; 18: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen Z, Krishnamachary B, Pachecho-Torres J, et al. Theranostic small interfering RNA nanoparticles in cancer precision nanomedicine. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2019: e1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Romano G, Veneziano D, Acunzo M, et al. Small non-coding RNA and cancer. Carcinogenesis 2017; 38: 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martens-Uzunova ES, Bottcher R, Croce CM, et al. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol 2014; 65: 1140–1151. [DOI] [PubMed] [Google Scholar]
- 38. Zhang H, Chen ZH, Wang XX, et al. Long non-coding RNA: a new player in cancer. J Hematol Oncol 2013; 6: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci 2016; 73: 2491–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10: 155–159. [DOI] [PubMed] [Google Scholar]
- 41. Morelli E, Gulla A, Rocca R, et al. The non-coding RNA landscape of plasma cell dyscrasias. Cancers (Basel) 2020; 12: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taiana E, Favasuli V, Ronchetti D, et al. Long non-coding RNA NEAT1 targeting impairs the DNA repair machinery and triggers anti-tumor activity in multiple myeloma. Leukemia 2020; 34: 234–244. [DOI] [PubMed] [Google Scholar]
- 43. Taiana E, Ronchetti D, Favasuli V, et al. Long non-coding RNA NEAT1 shows high expression unrelated to molecular features and clinical outcome in multiple myeloma. Haematologica 2019; 104: e72–e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Breier A, Gibalova L, Seres M, et al. New insight into P-Glycoprotein as a drug target. Anti-Cancer Agents Med Chem 2013; 13: 159–170. [PubMed] [Google Scholar]
- 45. Cui HG, Zhang AJ, Chen MW, et al. ABC transporter inhibitors in reversing multidrug resistance to chemotherapy. Curr Drug Targets 2015; 16: 1356–1371. [DOI] [PubMed] [Google Scholar]
- 46. Langcharoen P, Punfa W, Yodkeeree S, et al. Anti-P-glycoprotein conjugated nanoparticles for targeting drug delivery in cancer treatment. Arch Pharm Res 2011; 34: 1679–1689. [DOI] [PubMed] [Google Scholar]
- 47. Chen QN, Wei CC, Wang ZX, et al. Long non-coding RNAs in anti-cancer drug resistance. Oncotarget 2017; 8: 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Majidinia M, Yousefi B. Long non-coding RNAs in cancer drug resistance development. DNA Repair 2016; 45: 25–33. [DOI] [PubMed] [Google Scholar]
- 49. Doyle LA, Yang W, Rishi AK, et al. H19 gene overexpression in atypical multidrug-resistant cells associated with expression of a 95-kilodalton membrane glycoprotein. Cancer Res 1996; 56: 2904–2907. [PubMed] [Google Scholar]
- 50. Tsang WP, Kwok TT. Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene 2007; 26: 4877–4881. [DOI] [PubMed] [Google Scholar]
- 51. Kong JH, Qiu YJ, Li Y, et al. TGF-β1 elevates P-gp and BCRP in hepatocellular carcinoma through HOTAIR/miR-145 axis. Biopharm Drug Dispos 2019; 40: 70–80. [DOI] [PubMed] [Google Scholar]
- 52. Yousefi B, Zarghami N, Samadi N, et al. Peroxisome proliferator-activated receptors and their ligands in cancer drug-resistance: opportunity or challenge. Anti-Cancer Agents Med Chem 2016; 16: 1541–1548. [DOI] [PubMed] [Google Scholar]
- 53. Salehan MR, Morse HR. DNA damage repair and tolerance: a role in chemotherapeutic drug resistance. Br J Biomed Sci 2013; 70: 31–40. [DOI] [PubMed] [Google Scholar]
- 54. Shah MA, Schwartz GK. Cell cycle-mediated drug resistance: an emerging concept in cancer therapy. Clinical Cancer Res 2001; 7: 2168–2181. [PubMed] [Google Scholar]
- 55. Liu ZL, Sun M, Lu KH, et al. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One 2013; 8: e77293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thapar R. Regulation of DNA double-strand break repair by non-coding RNAs. Molecules 2018; 23: pii: E2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tsuruo T, Naito M, Tomida A, et al. Molecular targeting therapy of cancer: drug resistance, apoptosis and survival signal. Cancer Sci 2003; 94: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang XW, Bu P, Liu L, et al. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun 2015; 462: 227–232. [DOI] [PubMed] [Google Scholar]
- 59. Liu J, Wan L, Lu KH, et al. The long noncoding RNA MEG3 contributes to cisplatin resistance of human lung adenocarcinoma. PLoS One 2015; 10: e0114586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 2008; 9: 47–59. [DOI] [PubMed] [Google Scholar]
- 61. Wang Q, Cheng NN, Li XF, et al. Correlation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinoma. Oncotarget 2017; 8: 2558–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yan H, Xia JY, Feng FZ. Long non-coding RNA ENST00000457645 reverses cisplatin resistance in CP70 ovarian cancer cells. Genet Mol Res. Epub ahead of print 23 January 2017. DOI: 10.4238/gmr16019411. [DOI] [PubMed] [Google Scholar]
- 63. Zhu JM, Chen SS, Yang BX, et al. Molecular mechanisms of lncRNAs in regulating cancer cell radiosensitivity. Biosci Rep 2019; 39: pii: BSR20190590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sun T. Long noncoding RNAs act as regulators of autophagy in cancer. Pharmacol Res 2018; 129: 151–155. [DOI] [PubMed] [Google Scholar]
- 65. Yang LX, Wang HY, Shen Q, et al. Long non-coding RNAs involved in autophagy regulation. Cell Death Dis 2017; 8: e3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hu Y, Zhu QN, Deng JL, et al. Emerging role of long non-coding RNAs in cisplatin resistance. OncoTargets Ther 2018; 11: 3185–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Radisky DC. Epithelial-mesenchymal transition. J Cell Sci 2005; 118: 4325–4326. [DOI] [PubMed] [Google Scholar]
- 68. Xia H, Hui KM. MicroRNAs involved in regulating epithelial-mesenchymal transition and cancer stem cells as molecular targets for cancer therapeutics. Cancer Gene Ther 2012; 19: 723–730. [DOI] [PubMed] [Google Scholar]
- 69. Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest 2003; 112: 1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009; 119: 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee JM, Dedhar S, Kalluri R, et al. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol 2006; 172: 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gugnoni M, Ciarrocchi A. Long noncoding RNA and epithelial mesenchymal transition in cancer. Int J Mol Sci 2019; 20: pii: E1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Heery R, Finn SP, Cuffe S, et al. Long non-coding RNAs: key regulators of epithelial-mesenchymal transition, tumour drug resistance and cancer stem cells. Cancers (Basel) 2017; 9: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 2015; 6: 22513–22525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fantozzi A, Gruber DC, Pisarsky L, et al. VEGF-mediated angiogenesis links EMT-induced cancer stemness to tumor initiation. Cancer Res 2014; 74: 1566–1575. [DOI] [PubMed] [Google Scholar]
- 76. Barzi A, Lenz HJ. Angiogenesis-related agents in esophageal cancer. Expert Opin Biol Ther 2012; 12: 1335–1345. [DOI] [PubMed] [Google Scholar]
- 77. Zhao J, Li L, Han ZY, et al. Long noncoding RNAs, emerging and versatile regulators of tumor-induced angiogenesis. Am J Cancer Res 2019; 9: 1367–1381. [PMC free article] [PubMed] [Google Scholar]
- 78. Yan HW, Bu PC. Non-coding RNAs in cancer stem cells. Cancer Lett 2018; 421: 121–126. [DOI] [PubMed] [Google Scholar]
- 79. Ma ZW, Wang YY, Xin HW, et al. The expanding roles of long non-coding RNAs in the regulation of cancer stem cells. Int J Biochem Cell Biol 2019; 108: 17–20. [DOI] [PubMed] [Google Scholar]
- 80. Liu EL, Liu Z, Zhou YX, et al. Overexpression of long non-coding RNA PVT1 in ovarian cancer cells promotes cisplatin resistance by regulating apoptotic pathways. Int J Clin Exp Med 2015; 8: 20565–20572. [PMC free article] [PubMed] [Google Scholar]
- 81. Worku T, Bhattarai D, Ayers D, et al. Long non-coding RNAs: the new horizon of gene regulation in ovarian cancer. Cell Physiol Biochem 2017; 44: 948–966. [DOI] [PubMed] [Google Scholar]
- 82. Li J, Yang SQ, Su N, et al. Overexpression of long non-coding RNA HOTAIR leads to chemoresistance by activating the Wnt/β-catenin pathway in human ovarian cancer. Tumor Biol 2016; 37: 2057–2065. [DOI] [PubMed] [Google Scholar]
- 83. Ozes AR, Miller DF, Ozes ON, et al. NF-κB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene 2016; 35: 5350–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Janssens S, Tinel A, Lippens S, et al. PIDD mediates NF-kappa B activation in response to DNA damage. Cell 2005; 123: 1079–1092. [DOI] [PubMed] [Google Scholar]
- 85. Liu MY, Li XQ, Gao TH, et al. Elevated HOTAIR expression associated with cisplatin resistance in non-small cell lung cancer patients. J Thorac Dis 2016; 8: 3314–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wu D, Wang TZ, Ren CC, et al. Downregulation of BC200 in ovarian cancer contributes to cancer cell proliferation and chemoresistance to carboplatin. Oncol Lett 2016; 11: 1189–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xue X, Yang YA, Zhang A, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 2016; 35: 2746–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cruz-Miranda GM, Hidalgo-Miranda A, Barcenas-Lopez DA, et al. Long non-coding RNA and acute leukemia. Int J Mol Sci 2019; 20: 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wang HY, Li Q, Tang SS, et al. The role of long noncoding RNA HOTAIR in the acquired multidrug resistance to imatinib in chronic myeloid leukemia cells. Hematology 2017; 22: 208–216. [DOI] [PubMed] [Google Scholar]
- 90. Hang Q, Sun RC, Jiang CQ, et al. Notch 1 promotes cisplatin-resistant gastric cancer formation by upregulating lncRNA AK022798 expression. Anticancer Drugs 2015; 26: 632–640. [DOI] [PubMed] [Google Scholar]
- 91. Fan Y, Shen B, Tan MY, et al. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J 2014; 281: 1750–1758. [DOI] [PubMed] [Google Scholar]
- 92. Wang F, Zhou J, Xie X, et al. Involvement of SRPK1 in cisplatin resistance related to long non-coding RNA UCA1 in human ovarian cancer cells. Neoplasma 2015; 62: 432–438. [PubMed] [Google Scholar]
- 93. Cheng NN, Cai WJ, Ren SX, et al. Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget 2015; 6: 23582–23593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Luo J, Qu J, Wu DK, et al. Long non-coding RNAs: a rising biotarget in colorectal cancer. Oncotarget 2017; 8: 22187–22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Liu HY, Wang G, Yang LL, et al. Knockdown of long non-coding RNA UCA1 increases the tamoxifen sensitivity of breast cancer cells through inhibition of Wnt/β-catenin pathway. PLoS One 2016; 11: e0168406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xiao Y, Jiao CJ, Lin YQ, et al. lncRNA UCA1 contributes to imatinib resistance by acting as a ceRNA against miR-16 in chronic myeloid leukemia cells. DNA Cell Biol 2017; 36: 18–25. [DOI] [PubMed] [Google Scholar]
- 97. Zhan L, Li J, Wei B. Long non-coding RNAs in ovarian cancer. J Exp Clin Cancer Res 2018; 37: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lan WG, Xu DH, Xu C, et al. Silencing of long non-coding RNA ANRIL inhibits the development of multidrug resistance in gastric cancer cells. Oncol Rep 2016; 36: 263–270. [DOI] [PubMed] [Google Scholar]
- 99. Xu R, Mao YQ, Chen KB, et al. The long noncoding RNA ANRIL acts as an oncogene and contributes to paclitaxel resistance of lung adenocarcinoma A549 cells. Oncotarget 2017; 8: 39177–39184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Shi H, Pu J, Zhou XL, et al. Silencing long non-coding RNA ROR improves sensitivity of non-small-cell lung cancer to cisplatin resistance by inhibiting PI3K/Akt/mTOR signaling pathway. Tumor Biol 2017; 39: 1010428317697568. [DOI] [PubMed] [Google Scholar]
- 101. Xia BR, Hou Y, Chen H, et al. Long non-coding RNA ZFAS1 interacts with miR-150-5p to regulate Sp1 expression and ovarian cancer cell malignancy. Oncotarget 2017; 8: 19534–19546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wang Y, Zhang L, Zheng XF, et al. Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett 2016; 382: 137–146. [DOI] [PubMed] [Google Scholar]
- 103. Wu X, Zheng YZ, Han B, et al. Long noncoding RNA BLACAT1 modulates ABCB1 to promote oxaliplatin resistance of gastric cancer via sponging miR-361. Biomed Pharmacother 2018; 99: 832–838. [DOI] [PubMed] [Google Scholar]
- 104. Jiao F, Hu H, Han T, et al. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int J Mol Sci 2015; 16: 6677–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Shang C, Guo Y, Zhang JX, et al. Silence of long noncoding RNA UCA1 inhibits malignant proliferation and chemotherapy resistance to adriamycin in gastric cancer. Cancer Chemother Pharmacol 2016; 77: 1061–1067. [DOI] [PubMed] [Google Scholar]
- 106. Xie DL, Zhang H, Hu XH, et al. Knockdown of long non-coding RNA taurine up-regulated 1 inhibited doxorubicin resistance of bladder urothelial carcinoma via Wnt/β-catenin pathway. Oncotarget 2017; 8: 88689–88696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Xiao J, Lv Y, Jin FJ, et al. LncRNA HANR promotes tumorigenesis and increase of chemoresistance in hepatocellular carcinoma. Cell Physiol Biochem 2017; 43: 1926–1938. [DOI] [PubMed] [Google Scholar]
- 108. Tian X, Gao S, Liu Y, et al. Long non-coding RNA ENST00000500843 is downregulated and promotes chemoresistance to paclitaxel in lung adenocarcinoma. Oncol Lett 2019; 18: 3716–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Tian X, Zhang GW, Zhao H, et al. Long non-coding RNA NEAT1 contributes to docetaxel resistance of prostate cancer through inducing RET expression by sponging miR-34a. RSC Adv 2017; 7: 42986–42996. [Google Scholar]
- 110. Gajria D, Chandarlapaty S. HER2-amplified breast cancer: mechanisms of trastuzumab resistance and novel targeted therapies. Expert Rev Anticancer Ther 2011; 11: 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Li WT, Zhai LM, Wang H, et al. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget 2016; 7: 27778–27786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Pan H, Jiang T, Cheng NN, et al. Long non-coding RNA BC087858 induces non-T790M mutation acquired resistance to EGFR-TKIs by activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell lung cancer. Oncotarget 2016; 7: 49948–49960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang ZX, Pan LM, Yu HX, et al. The long non-coding RNA SNHG5 regulates gefitinib resistance in lung adenocarcinoma cells by targetting miR-377/CASP1 axis. Biosci Rep 2018; 38: pii: BSR20180400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Takahashi K, Yan IK, Wood J, et al. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol Cancer Res 2014; 12: 1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Qu L, Ding J, Chen C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 2016; 29: 653–668. [DOI] [PubMed] [Google Scholar]
- 116. Bian ZH, Zhang JW, Li M, et al. Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis 2017; 6: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Han Z, Shi LY. Long non-coding RNA LUCAT1 modulates methotrexate resistance in osteosarcoma via miR-200c/ABCB1 axis. Biochem Biophys Res Commun 2018; 495: 947–953. [DOI] [PubMed] [Google Scholar]
- 118. van Agthoven T, Dorssers LCJ, Lehmann U, et al. Breast cancer anti-estrogen resistance 4 (BCAR4) drives proliferation of IPH-926 lobular carcinoma cells. PLoS One 2015; 10: e0136845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Zhang F, Ni HW, Li XM, et al. LncRNA FENDRR attenuates adriamycin resistance via suppressing MDR1 expression through sponging HuR and miR-184 in chronic myelogenous leukaemia cells. FEBS Lett 2019; 593: 1993–2007. [DOI] [PubMed] [Google Scholar]
- 120. Galluzzi L, Senovilla L, Vitale I, et al. Molecular mechanisms of cisplatin resistance. Oncogene 2012; 31: 1869–1883. [DOI] [PubMed] [Google Scholar]
- 121. Xu W, Chen Q, Wang Q, et al. JWA reverses cisplatin resistance via the CK2-XRCC1 pathway in human gastric cancer cells. Cell Death Dis 2014; 5: e1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wang Y, Wang HL, Song TF, et al. HOTAIR is a potential target for the treatment of cisplatin-resistant ovarian cancer. Mol Med Rep 2015; 12: 2211–2216. [DOI] [PubMed] [Google Scholar]
- 123. Teschendorff AE, Lee SH, Jones A, et al. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med 2015; 7: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Jordheim LP, Durantel D, Zoulim F, et al. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat Rev Drug Discov 2013; 12: 447–464. [DOI] [PubMed] [Google Scholar]
- 125. Ismae GF, Rosa DD, Mano MS, et al. Novel cytotoxic drugs: old challenges, new solutions. Cancer Treat Rev 2008; 34: 81–91. [DOI] [PubMed] [Google Scholar]
- 126. Ribeiro JT, Macedo LT, Curigliano G, et al. Cytotoxic drugs for patients with breast cancer in the era of targeted treatment: back to the future? Ann Oncol 2012; 23: 547–555. [DOI] [PubMed] [Google Scholar]
- 127. Morgan G, Ward R, Barton M. The contribution of cytotoxic chemotherapy to 5-year survival in adult malignancies. Clin Oncol 2004; 16: 549–560. [DOI] [PubMed] [Google Scholar]
- 128. Zhang CL, Zhu KP, Shen GQ, et al. A long non-coding RNA contributes to doxorubicin resistance of osteosarcoma. Tumor Biol 2016; 37: 2737–2748. [DOI] [PubMed] [Google Scholar]
- 129. Zhou BH, Li LJ, Li YJ, et al. Long noncoding RNA SNHG12 mediates doxorubicin resistance of osteosarcoma via miR-320a/MCL1 axis. Biomed Pharmacother 2018; 106: 850–857. [DOI] [PubMed] [Google Scholar]
- 130. Mekhail TM, Markman M. Paclitaxel in cancer therapy. Expert Opin Pharmacother 2002; 3: 755–766. [DOI] [PubMed] [Google Scholar]
- 131. Majidinia M, Alizadeh E, Yousefi B, et al. Downregulation of notch signaling pathway as an effective chemosensitizer for cancer treatment. Drug Res 2016; 66: 571–579. [DOI] [PubMed] [Google Scholar]
- 132. Gotink KJ, Verheul HMW. Anti-angiogenic tyrosine kinase inhibitors: what is their mechanism of action? Angiogenesis 2010; 13: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Lin YX, Wang X, Jin HC. EGFR-TKI resistance in NSCLC patients: mechanisms and strategies. Am J Cancer Res 2014; 4: 411–435. [PMC free article] [PubMed] [Google Scholar]
- 134. Wu Y, Yu DD, Hu Y, et al. Genome-wide profiling of long non-coding RNA expression patterns in the EGFR-TKI resistance of lung adenocarcinoma by microarray. Oncol Rep 2016; 35: 3371–3386. [DOI] [PubMed] [Google Scholar]
- 135. Yan JY, Chen DD, Chen XL, et al. Identification of imatinib-resistant long non-coding RNAs in gastrointestinal stromal tumors. Oncol Lett 2019; 17: 2283–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Shewach DS, Lawrence TS. Antimetabolite radiosensitizers. J Clin Oncol 2007; 25: 4043–4050. [DOI] [PubMed] [Google Scholar]
- 137. Han YH, Ye J, Wu D, et al. LEIGC long non-coding RNA acts as a tumor suppressor in gastric carcinoma by inhibiting the epithelial-to-mesenchymal transition. BMC Cancer 2014; 14: 932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Miller WR. Biological rationale for endocrine therapy in breast cancer. Best Pract Res Clin Endoc Metab 2004; 18: 1–32. [DOI] [PubMed] [Google Scholar]
- 139. Godinho MF, Sieuwerts AM, Look MP, et al. Relevance of BCAR4 in tamoxifen resistance and tumour aggressiveness of human breast cancer. Br J Cancer 2010; 103: 1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Hong HH, Hou LK, Pan X, et al. Long non-coding RNA UCA1 is a predictive biomarker of cancer. Oncotarget 2016; 7: 44442–44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Loewen G, Jayawickramarajah J, Zhuo Y, et al. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol 2014; 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Nakagawa T, Endo H, Yokoyama M, et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun 2013; 436: 319–324. [DOI] [PubMed] [Google Scholar]
- 143. Li ND, Wang YC, Liu XF, et al. Identification of circulating long noncoding RNA HOTAIR as a novel biomarker for diagnosis and monitoring of non-small cell lung cancer. Technol Cancer Res Treat 2017; 1533034617723754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ma GX, Wang QY, Lv CY, et al. The prognostic significance of HOTAIR for predicting clinical outcome in patients with digestive system tumors. J Cancer Res Clin Oncol 2015; 141: 2139–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Yang Q, Zhang RW, Sui PC, et al. Dysregulation of non-coding RNAs in gastric cancer. World J Gastroenterol 2015; 21: 10956–10981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Amodio N, Raimondi L, Juli G, et al. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol 2018; 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Amodio N, Stamato MA, Juli G, et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia 2018; 32: 1948–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Choueiri TK, Escudier B, Powles T, et al. Cabozantinib versus everolimus in advanced renal-cell carcinoma. N Engl J Med 2015; 373: 1814–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]