Abstract
Background:
Clinical evidence suggests that body muscle mass is positively associated with bone mass, of significance for the elderly population at risk of osteoporosis (OP). Furthermore, muscle and bone interact mechanically and functionally, via local interactions as well as remotely via secreted components. Thus, it was of interest to compare muscle transcriptomes in postmenopausal OP and healthy women, and study effects of strength training on the muscle transcriptome, muscle stress proteins and bone mineral density (BMD).
Methods:
Skeletal muscle histological and genetic properties were compared in postmenopausal healthy (n = 18) and OP (n = 17) women before and after heavy-load strength training for 13–15 weeks. The cohorts were of similar age and body mass index without interfering diseases, medication or difference in lifestyle factors. Muscle biopsies obtained before and after intervention were studied histologically, and stress proteins and transcriptomes analyzed.
Results:
The OP women showed distinct muscle transcription profiles when compared with healthy women and had higher levels of the stress proteins HSP70 and α-β-crystalline. A set of 12 muscle transcripts, including ACSS3, FZD4, GNAI1 and IGF1, were differentially expressed before and after intervention (false discovery rate ⩽0.10, p ⩽0.001), and their corresponding bone transcripts were associated with BMD. Experimental data underline and describe the functionality of these genes in bone biology. OP women had 8% (p <0.01) higher proportion of type I fibres, but muscle fibre cross-sectional area did not differ. Muscle strength increased in both groups (p <0.01).
Conclusions:
Postmenopausal healthy and OP women have distinct muscle transcriptomes [messenger ribonucleic acids (mRNAs) and microRNAs] that are modulated by strength training, translating into key protein alterations and muscle fibre changes. The function of common skeletal muscle and bone genes in postmenopausal OP is suggestive of a shared disease trait.
Keywords: heavy-load strength training, muscle biopsies, postmenopausal osteoporosis, transcriptional profiling
Introduction
Muscles, together with the skeleton, represent the human locomotive system; they have a common embryological origin and define an integrated functional entity. In contrast to bone, skeletal muscle is a swiftly adaptable tissue with responses characteristic for exercise type, intensity and duration.1,2 Optimal function of the musculoskeletal system is dependent on regular physical activity, as inactivity leads to muscular atrophy and bone resorption, which may develop into osteoporosis (OP), a condition characterized by low mineral density, bone microstructure deterioration and increased susceptibility to low-energy fractures.
It is estimated that one in three women and one in five men over the age of 50 worldwide will sustain an OP fracture.3 The incidence is highest in the Scandinavian countries, and in Sweden, the remaining lifetime risk of a major OP fracture (clinical spine, hip, forearm or humeral fracture) was found to be 46.4% in women and 22.4% in men.4 In the European Union, 27.5 million people, or 55% of the population were estimated to have OP in 2010, at an estimated yearly cost from incident and prior fragility fractures of EUR 37 billion.3 The most severe complication of OP is femoral neck fracture, 20–30% of the patients die during the first year after surgery, and about 50% of survivors are unable to walk without support 5 years later.5
Muscular asthenia and sarcopenia are associated with postmenopausal OP6 and lead to impaired balance and falls and are thus predisposing factors for fractures.7 OP remains an under-diagnosed disease, often undetected until the first fracture. There is a great need for novel disease insight and effective preventive measures to prevent falls. Improving balance by restoring muscular capacity and performance will reduce the tendency for falls and lead to more active everyday life and healthy ageing. Currently, it is unknown if or how molecular muscle pathology is associated with bone deterioration in postmenopausal OP. Based on the association between muscle asthenia and OP, we hypothesized that the muscle transcriptome might differ between OP and healthy women. Furthermore, strength training, which has been shown to improve BMD,8 might also influence the muscle expression pattern towards the situation in healthy women.8
Our first objective was to compare transcriptional activity [messenger ribonucleic acid (mRNA) and microRNA] and translated key proteins in the skeletal muscle biopsies of women with established OP (T-score ⩽2.5 and at least one fragility fracture9) with that of healthy controls. Unlike mRNAs coding for specific proteins, microRNAs are small RNAs transcribed from individual genes or are products of genes also coding for other RNAs. MicroRNAs have regulatory activity in binding and destabilizing specific mRNAs.10 One microRNA may have multiple mRNA targets, and several microRNAs typically target the same mRNA,10 thus they have general important gene regulatory functions.
Our second objective was to identify muscle characteristics and transcriptomic changes in the cohorts of healthy and OP after a heavy-load strength exercise intervention. Muscle and osteoblastic cells may differentiate from the same stem cells.11 Thus, it was of interest to investigate if the muscle transcripts affected by exercise were also associated to bone mineral density (BMD) and if they were expressed in bone to search for common involved genes.
For validation, we studied whether levels of BMD-associated transcripts in bone, which also changed in thigh muscle with exercise, were correlated with levels of iliac muscle transcripts in a separate cohort of postmenopausal women. Our results show that the skeletal muscle transcriptome and selected key proteins are markedly altered in postmenopausal OP. The heavy-load strength exercise intervention induced transcriptional mRNA and microRNA changes in both groups. A limited set of genes exert pleomorphic functions in both bone and skeletal muscle and may therefore contribute to the clinical picture of postmenopausal OP including sarcopenia.
Materials and methods
Recruitment and ethics
Participants with postmenopausal OP were recruited consecutively through a medical clinic and gave their verbal and written consent as described in Raastad et al.12 The healthy participants were either recruited through advertisements in newspapers and/or included via Lovisenberg Diakonale Hospital outpatient clinic.9 The study was approved by the Norwegian Regional Ethical Committee (REK no: 2010/2539) and conducted according to the Declaration of Helsinki.
Participants
A total of 18 healthy women (T-score over −1.0, no fracture) and 17 women with established OP (T-score no more than −2.5 at the back and hip with at least one fragility fracture; age 55–80 years) were included for the intervention (Table 1 and Supplemental Table S1). Number of previous smokers and the level of physical activity were similar between healthy and patient groups, as were other lifestyle factors and nutrition. The 17 women with established OP had previously donated trans-iliac bone biopsies with muscle attached to the pelvic side as part of a different study including postmenopausal women with varying BMD from OP to healthy.9 The various cohorts and analyses performed are illustrated in Figure 1. Serum and urine biomarkers of all healthy and patient donors of iliac bone biopsies are presented in Supplemental Table S2.
Table 1.
Mean demographic characteristics before intervention of postmenopausal women donating thigh-muscle biopsies.
| Healthy (n = 18), mean (SD) |
Osteoporotic (n = 17), mean (SD) |
t-test p-value |
|
|---|---|---|---|
| Age (years) | 73.9 (5.7) | 68.0 (6.2) | 0.460 |
| Body mass (kg) | 68.1 (11.9) | 63.3 (12.1) | 0.288 |
| BMI (weight/height2) | 25.2 (3.6) | 23.8 (4.8) | 0.358 |
| Femoral neck (T-score) | −0.18 (0.76) | −2.35 (0.54) | p < 0.001 |
| L1–L4 BMD (T-score) | 0.49 (1.17) | −3.46 (0.90) | p < 0.001 |
| Total body BMD (T-score) | 0.24 (1.20) | −2.39 (1.14) | p < 0.001 |
| Lean mass (kg) | 40.3 (4.3) | 39.7 (4.5) | 0.712 |
| Fat mass (kg) | 25.5 (8.2) | 21.3 (8.9) | 0.194 |
| Maximal squat load (kg) | 48.3 (3.9) | 21.2 (3.1) | p < 0.001 |
T-score denotes bone mineral density (g calcium/cm2) calculated in relation to a reference population of young healthy adult White women.
BMD, bone mineral density; BMI, body mass index; L1–L4, average BMD of lumbar vertebra 1–4; SD, standard deviation.
Figure 1.
Overview of cohorts and analyses performed in the current study.
The figure represents the study cohorts and tables and figures containing the various study results. Note that the OP women in cohort III is part of cohort I. These women donated a trans-iliac bone biopsy with pelvic muscle attached, 6–7 years prior to the training intervention as part of a different study.9, 13
OP, osteoporotic women.
Clinical evaluation and laboratory analyses
All participants had a clinical examination and completed detailed interview questionnaires on present and previous diseases, nutrition and lifestyle factors (smoking, alcohol, physical activity). The participants were evaluated by dual-energy X-ray absorptiometry (DXA) and through additional examination of blood biochemistry including measurements of 25(OH) vitamin D, vitamin K and endocrinological analyses covering calcium/parathyroid hormone (PTH) metabolism and thyroid status. No other diseases (kidney, alimentary or inflammatory) or medicine known to affect bone metabolism were allowed (e.g. oestrogen, corticosteroids and psychiatric medication such as selective serotonin reuptake inhibitors). The patients were all on anti-resorptive treatment (peroral bisphosphonates) and two had finished 2 years’ prior treatment with PTH. Patients on bisphosphonate had their medication temporarily withdrawn 3 months prior to and during the training period but continued with daily supplements of vitamin D (1000 IU D3) and calcium (1000 mg). The healthy controls did not take any medication affecting bone metabolism. Bone densitometry was performed by DXA, using a Lunar Prodigy DF+12649 densitometer with the Lunar iDXA encore software, version 12.30 (GE Medical Systems, Madison, USA). Prior to the DXA scan, subjects were requested to avoid training for 24 h and to avoid any ingestion of liquid or food 2 h before the scan. Subjects laid in a standardized position in the machine according to instructions from the manufacturer. The measurements were carried out by skilled bioengineers. Repetitive measurements of the same patients gave a short-term reproducibility of 2.8% at the spine and 2.6% at the hip.
Training protocols
The total duration of the intervention for the OP women was 15 weeks, including 2 weeks to familiarize the participants to the training protocol using lighter-start training loads. The duration of the intervention for the healthy participants was 13 weeks because it was possible to introduce heavy training loads earlier in this group. After the familiarization period in OP women, the training loads were gradually increased to ensure that the following 13 weeks of training was conducted with optimal loading to improve muscle strength and muscle mass.12
Strength training in the last 13 weeks was performed as traditional heavy-load strength training: three times per week with one to three sets involving all major muscle groups. The training protocol consisted of three exercises for leg muscles; squat, leg press and standing toe rise, and three exercises for upper body muscles; chest press, seated rowing and shoulder press. In addition, the participants performed self-selected exercises for abdominals and lower back muscles at the end of each session. The strength-training regime was a mix between linear periodization and daily undulating periodization. The participants started with 8–12 RM (repetition maximum) sets and ended the 13-week protocol with 4–8 RM sets. In two sessions per week, the sets were run until failure, and in the third session, performed between the two maximal sessions, sets were run with a load corresponding to 80–90% of the actual RM load. Total duration of training was about 60 min per session, and the participants exercised in groups of three with a personal instructor present. Training under supervision ensured the quality of the exercise and the safety of the participants. The training load was recorded in a training diary and this log was used to check the progression in all exercises for each participant, as described by Raastad et al.12
The exercises and the training loads were well tolerated by the participants, but one compression fracture in the spine was attained during an accident in the squat exercise. The patient recovered during 3 months of reduced loading and completed the training intervention with excellent results.12
Timeline
Baseline muscle biopsies were performed before participants started the familiarization sessions, and about 1 week after completion of the training intervention, the post-training biopsies were obtained.
Collection of iliac bone biopsies with attached pelvic muscle and RNA purification
As illustrated in Figure 1, a cohort of postmenopausal women donated iliac bone biopsies with muscle attached to the pelvic side. The iliac bone biopsies were taken with Bordier’s trephine, 2 cm from crista iliaca and 2 cm from spina iliaca under local anaesthesia (lidocaine). Pelvic muscle was detached from the bone biopsy and both tissues were immediately frozen in liquid nitrogen and stored at −70°C for later RNA extraction. RNA was isolated from pelvic muscle, thigh muscle and iliac bone by a similar method. In brief, the tissue samples were pulverized with mortar in liquid nitrogen. The pulverized material was added to TRIZOL reagent (Life Technologies, Gaithersburg, USA), homogenized and RNA purified according to the manufacturer’s instructions. RNA was further purified using the miRNeasy micro kit (Qiagen, Oslo, Norway), according to the manufacturer’s protocol. RNA concentration was determined with a spectrophotometer (ND-1000 Nano Drop; Thermo Fisher Scientific, Wilmington, USA). RNA quality was confirmed with Bioanalyzer (2100 System and RNA 6000 Nano Assay; Agilent Technologies, Santa Clara, USA). RNA samples were immediately frozen and stored at −80°C.
Measures: thigh-muscle sampling
Thigh muscle samples were obtained under local anaesthesia (Xylocaine adrenalin, 10 mg/ml + 5 μg/ml; AstraZeneca, London, UK) from the mid portion of musculus vastus lateralis before and after the training intervention, using a modified Bergström technique. Muscle samples were collected at least 2 days after any training or testing, with the post biopsy taken approximately 3 cm distal of the previous biopsy site. A sample for immunohistochemistry (20–30 mg) was embedded in OCT compound (Tissue-Tek, Sakura Finetek, Alphen aan den Rijn, The Netherlands) and frozen in isopentane precooled to approximately −120°C by liquid nitrogen. Another sample (10–20 mg) used for RNA extraction was immediately frozen in in liquid nitrogen (applies to the cohort of OP women, muscle biopsies from healthy were stored in RNA later for one day at 4°C before freezing). Samples were then stored at −80°C until sectioning (immunohistochemistry sample) or RNA extraction. Pieces for immunoassays were rinsed in ice-cold saline (0.9% NaCl, Braun, Melsungen, Germany), and carefully dissected free of visual fat, connective tissue and blood. Pieces of 50 mg were frozen in isopentane on dry ice and stored at −80°C for later homogenization.
Immunohistochemistry; Western blotting; microarray and data analyses
Technical validation by real-time qRT-PCR
To validate the microarray data, we used reverse-transcription polymerase chain reaction (RT-PCR) to test for differentially expressed mRNAs/microRNAs in a select number of muscle genes. Qualitative real-time (qRT)-PCR reactions were performed using the ViiA 7 Real-Time PCR System (Applied Biosystems, Carlsbad, USA) to validate a selected panel of mRNAs. From the samples, 200 ng of total RNA were reverse transcribed in duplicates using the qScript cDNASuperMix (Quanta BioSciences, Beverly, MA, USA). Nine µl complementary deoxyribonucleic acid (cDNA; diluted 1:10 in H2O) and 1 µl of primer/probes (TaqMan Gene Expression Assays, Applied Biosystems) were added to 10 µl universal PCR master mix (TaqMan, Applied Biosystems). Each gene was run in duplicates. MYBPC1, showing a variance of 0.53 in the microarray experiments, served as endogenous control. The following assays were used: MYBPC1_Hs00159451_m1, COL3A1_Hs00943809_m1, FRZB_Hs00173503_m1, IGF2_Hs04188276_m1, ACTN3_Hs00153812_m1, TUBA4A_Hs01081794–g1 (Applied Biosystems). The relative amounts of mRNA for each gene were calculated using the ΔΔCt method which utilizes a formula, ‘fold’-gene expression change = 2−ΔΔCt where ΔΔCt = (Ct gene of interest in the unknown specimen − Ct normalizer in the unknown specimen) − (Ct gene of interest in the calibrator specimen − Ct normalizer in the calibrator specimen). The method is described in Applied Biosystems User Bulletin #2, ‘ABI PRISM 7700 Sequence Detection System’, 11 December 1997 (updated in October 2001). Statistical comparison of qRT-PCR data was performed with the paired Student’s t-test (Excel, Microsoft, Redmond, USA); p <0.05 was considered significant. A good correspondence between RT-PCR and microarray data was found (Supplemental Table S3), as also previously described by Vissing et al.14 The bone expression microarray data have also been validated with very good correspondence.9,13
Statistics
The normality assumption was tested prior to all calculations. Log2 transformed signal values were used in all calculations involving transcript levels. The statistical approach is detailed below:
Cohort and demographic characteristics Healthy versus OP groups were compared using independent sample t-tests (Table 1 and Supplemental Table S4).
Muscle morphology and protein markers of cellular stress before and after strength training
Independent sample t-tests were used for fibre type (Supplemental Table S5); fibre cross-sectional area pre- versus post-strength training [Figure 2(a)]; HSP70 and α-β-crystalline (alpha-B-c) levels [Figure 2(b)]. We also analyzed the data with a two-factor (group × time) analysis of variance (ANOVA) with repeated measures on the time factor. As the test for the interaction was underpowered, we stratified by disease group and calculated paired sample t-tests for fibre type (SupplementalTable S5); fibre cross-sectional area pre- versus post-strength training [Figure 2(a)]; HSP70 and alpha-B-c levels [Figure 2(b)].
Figure 2.
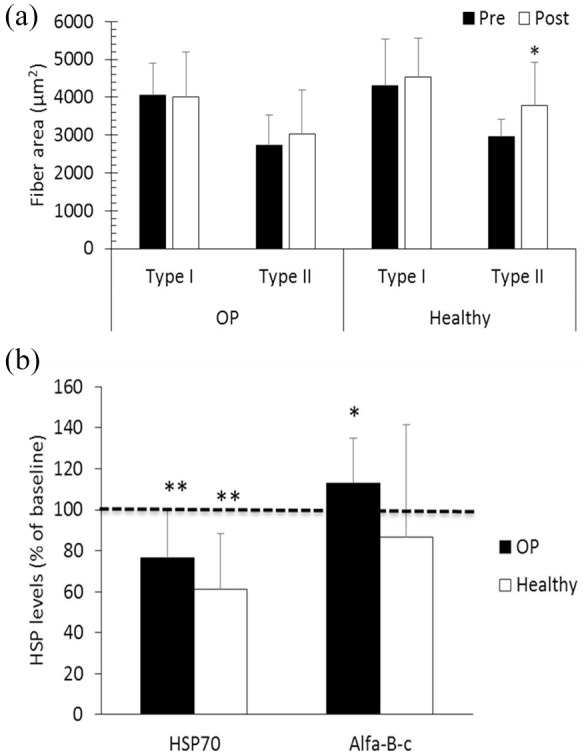
Muscle fibre area and protein expression levels.
(a) Muscle fibre cross-sectional area in postmenopausal osteoporotic (OP) women and healthy controls (Healthy) before (Pre) and after (Post) a heavy-load strength training intervention; (b) HSP70 and α-β-crystalline (alpha-B-c) levels after the strength training intervention in postmenopausal OP women and healthy controls (Healthy). The results are given as percentage of baseline levels (±SD).
*p <0.05, **p <0.01 as evaluated by paired t-test. Significance (*) of pre versus post for type II fibres (panel A) and alpha-B-c (panel B) refer to post hoc analyses.
HSP, heat-shock protein; SD, standard deviation.
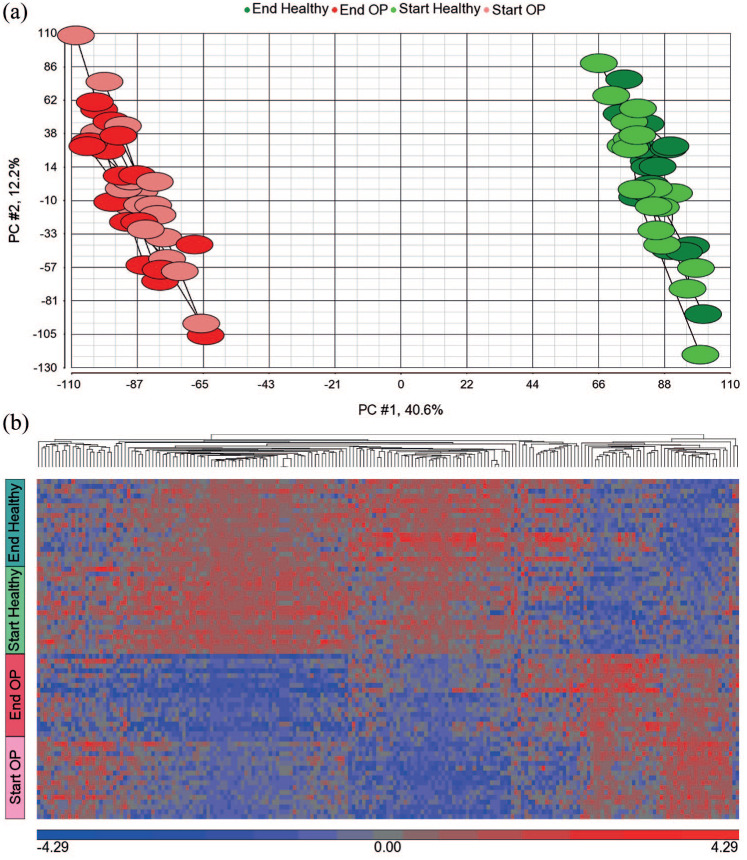
Distinct muscle transcriptomes between healthy and postmenopausal OP women For principal component analysis (PCA) on muscle thigh transcripts [Figure 3(a)], all available transcripts (n = 18,811) were used. In the supervised cluster analysis [Figure 3(b)], 308 transcripts were identified by repeated-measures ANOVA at 5% false discovery rate (FDR) after pooling healthy and OP women, before and after strength training.
Figure 3.
Changes in muscle transcript levels after exercise.
(a) Principal component 1 versus principal component 2 from a PCA using all available (genome-wide) muscle mRNA transcripts (n = 18,811), showing two non-overlapping clusters. Green and light-green colours represent post- and pre-exercise healthy women, respectively. Red and pink colours represent post- and pre-strength-training osteoporotic women, respectively; (b) supervised cluster analysis of muscle transcript levels of 18 healthy and 17 osteoporotic women representing 70 samples, before and after strength training. The heat map shows 308 transcripts with differential expression (FDR < 5%) pre versus post-strength training in healthy and/or osteoporotic women identified by repeated-measures ANOVA ignoring disease status. Separate genes whose transcripts show down- (blue) or up-regulation (red) regulation. The colour bar (blue/red) below the heat map represents gene expression standardized to a mean of zero and standard deviation of one of log2 signal values.
ANOVA, analysis of variance; FDR, false discovery rate; mRNA, messenger ribonucleic acid; OP, osteoporotic; PC, principal component; PCA, principal component analysis.
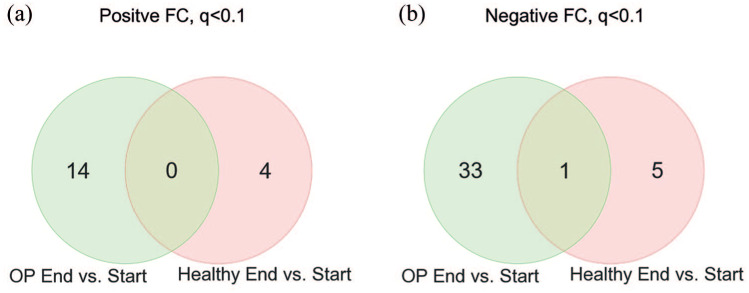
Effect of heavy-load strength training on the muscle transcriptomes We tested a repeated-measures ANOVA, including: group (OP versus healthy); time (before versus after exercise); and group versus time interaction. However, we observed some convergence problems for some transcripts. Therefore, we present the stratified results in the Venn diagrams [Figure 4(a) and 4(b)]. Table 2 contains all muscle transcripts fulfilling p ⩽0.0001 calculated by repeated-measures ANOVA in OP women, after versus before strength training.
Figure 4.
Number of common muscle transcripts in OP and healthy changed after exercise.
(a) Muscle transcripts with positively altered expression upon heavy-load strength training. The Venn diagram depicts 14 and 4 positively changed transcripts (FDR <0.1, repeated-measures ANOVA) upon strength training in healthy (pink) and OP women ( green), respectively; (b) muscle transcripts with negatively altered expression upon heavy-load strength training. The Venn diagram depicts 34 and 6 negatively changed transcripts (FDR <0.1, repeated-measures ANOVA) upon strength training in healthy (pink) and/or OP women (green), respectively.
ANOVA, analysis of variance; FC, fold change; FDR, false discovery rate; OP, osteoporosis.
Table 2.
Shortlist of 15 muscle transcripts in osteoporotic women showing the highest statistical difference after versus before strength training (p ⩽ 0.0001) and fold change.
| Transcript ID | Gene symbol | Gene name | p-value | Fold change (end versus start) |
|---|---|---|---|---|
| 8006229 | RNF135 | Ring finger protein 135 | p < 0.001 | 1.16 |
| 8030128 | PPP1R15A | Protein phosphatase 1, regulatory subunit 15A | p < 0.001 | −1.28 |
| 7924636 | TMEM63A | Transmembrane protein 63A | p < 0.001 | −1.16 |
| 8139330 | CAMK2B | Calcium/calmodulin-dependent protein kinase II beta | p < 0.001 | −1.21 |
| 7899101 | CNKSR1 | Connector enhancer of kinase suppressor of Ras 1 | p < 0.001 | −1.20 |
| 7901256 | CYP4B1 | Cytochrome P450, family 4, subfamily B, polypeptide 1 | p < 0.001 | −1.34 |
| 8148304 | TRIB1 | Tribblespseudokinase 1 | p < 0.001 | −1.23 |
| 8015914 | HDAC5 | Histone deacetylase 5 | p < 0.001 | −1.22 |
| 8063914 | MIR1-1HG | MIR1-1 host gene | p < 0.001 | −1.40 |
| 7963137 | (Not characterized) | p < 0.001 | 1.26 | |
| 8164995 | BRD3 | Bromodomain containing 3 | p < 0.001 | −1.10 |
| 8094056 | HTRA3 | HtrA serine peptidase 3 | p < 0.001 | 1.13 |
| 8124967 | BAG6 | BCL2-associated athanogene 6 | p < 0.001 | −1.14 |
| 8101718 | PIGY | Phosphatidylinositol glycan anchor biosynthesis, class Y | p < 0.001 | 1.14 |
| 7975787 | JDP2 | Jun dimerization protein 2 | p < 0.001 | −1.18 |
p-values were calculated using repeated-measures ANOVA.
ANOVA, analysis of variance.
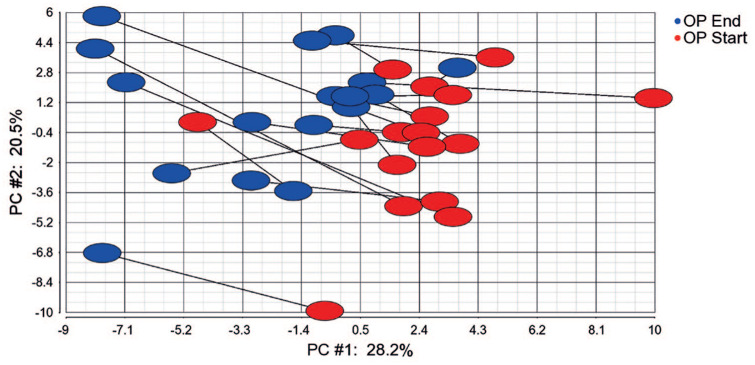
Effect of strength training on mature microRNAs PCA (Figure 5) was based on repeated-measures ANOVA of OP women (p <0.05, 58 microRNAs). The 18 most significantly changed microRNAs (p <0.01) are shown in Supplemental Table S6.
Figure 5.
Changes in miRNA expression after strength training.
Principal component analysis of filtered microRNAs (p <0.05) in 17 patients before and after heavy-load strength training as identified by repeated-measures ANOVA of OP women, end versus start of strength training. Each symbol represents one person depicted as average transcript signal value of 58 microRNAs (p <0.05).
ANOVA, analysis of variance; miRNA, micro ribonucleic acid; OP, osteoporosis; PC, principal component.
Functional gene assignment Top networks (Supplemental Table S7) representing key genes were identified through use of ingenuity pathway analysis (IPA; Ingenuity®Systems, www.ingenuity.com). Briefly, the data sets containing gene identifiers were uploaded into the web-delivered application and each gene identifier was mapped to its corresponding gene object in the IPA software. Fisher’s exact test was performed to calculate a p-value, assigning probability of enrichment to each biological function within the IPA library.
Associations between BMD and the most significantly differentially expressed muscle mRNAs in bone after intervention Independent sample t-tests were used for comparing OP women with healthy donors of iliac bone and pelvic muscle biopsies (Supplemental Table S4). R, p and q values in Table 3 were taken from Supplemental in a previous paper as indicated, and were calculated by regression analysis in R.9 In table 5(b) Pearson correlation r-values and the two tailed p-values were calculated using the linear regression function in GraphPad Prism (GraphPad Software, CA, USA).
Table 3.
Training altered thigh muscle transcripts that have corresponding BMD correlated transcripts in bone; correlation between shared iliac bone and pelvic muscle mRNA levels.
| A |
B |
||||||
|---|---|---|---|---|---|---|---|
| Affymetrix ID | Gene symbol | Gene name | r | p-value | FDR | r |
p-value (unadjusted) |
| 229222_at | ACSS3 | Acyl-CoA synthetase short-chain family member 3 | −0.37 | p <0.001 | 0.100 | 0.136 | 0.526 |
| 218665_at | FZD4 | Frizzled homolog 4 (drosophila) | −0.39 | p <0.001 | 0.078 | −0.164 | 0.444 |
| 209576_at | GNAI1 | Guanine nucleotide-binding protein (G protein), alpha-inhibiting activity polypeptide 1 | −0.38 | p <0.001 | 0.088 | 0.047 | 0.827 |
| 209540_at | IGF1 | Insulin-like growth factor 1 (somatomedin C) | −0.41 | p <0.001 | 0.064 | 0.531 | 0.007 |
| 201505_at | LAMB1 | Laminin, beta 1 | −0.40 | p <0.001 | 0.064 | −0.061 | 0.777 |
| 212274_at | LPIN1 | Lipin 1 | −0.37 | p <0.001 | 0.092 | −0.327 | 0.119 |
| 212636_at | QKI | Quaking homolog, KH domain RNA binding (mouse) | −0.41 | p <0.001 | 0.064 | NA | |
| 227543_at | RNASEH2C | Ribonuclease H2, subunit C | 0.37 | p <0.001 | 0.096 | −0.010 | 0.963 |
| 222669_s_at | SBDS | Shwachman-Bodian-Diamond syndrome/Shwachman-Bodian-Diamond syndrome pseudogene | −0.38 | p <0.001 | 0.088 | −0.407 | 0.048 |
| 201811_x_at | SH3BP5 | SH3-domain binding protein 5 (BTK associated) | −0.44 | p < 0.001 | 0.051 | −0.183 | 0.392 |
| 218077_s_at | ZDHHC3 | Zinc finger, DHHC-type containing 3 | 0.37 | p < 0.001 | 0.097 | −0.052 | 0.809 |
| 210275_s_at | ZFAND5 | Zinc finger, AN1-type domain 5 | −0.38 | p < 0.001 | 0.088 | 0.284 | 0.179 |
Part A is extracted from Supplemental Table S5 in Reppe et al.9 and includes the bone BMD correlated transcripts [with r, p and false discovery rate (FDR)] having corresponding thigh muscle transcripts that changed upon training (pooled healthy and OP). Part B is based on a subset of healthy (n = 12) or OP (n = 12) iliac bone and pelvic muscle donors presented in Table S1, group C and D, and Supplemental Table S4.
BMD, bone mineral density; FDR, false discovery rate; OP, osteoporosis; RNA, ribonucleic acid; mRNA, messenger RNA.
Results
Cohorts and demographic characteristics
Table 1 and Supplemental Table S1 present the cohorts and their demographic characteristics. The study was performed in healthy and OP groups, and thigh-muscle biopsies were taken before and after the strength-training intervention. The healthy women tended to be somewhat older (average 73.9 years) than the patients (average 68.0 years). There was no statistical difference between the groups regarding body mass index (BMI), body mass, lean mass or fat mass. The BMD measurements, measured as T-scores, showed a significant difference between healthy and patients as expected (p < 0.001).
Effects of heavy-load strength training on muscle strength
Recently, Raastad et al.12 showed in the present cohorts (Table 1 and Supplemental Table S1) that even in postmenopausal women with established OP, heavy-load strength training was feasible and effective in improving muscle mass and strength. In fact, the relative increase in strength during the strength-training intervention was comparable in the two groups (31 ± 19% and 32 ± 16%, in OP and healthy, respectively).12
Muscle morphology and protein markers of cellular stress before and after strength training At baseline, OP women had significantly more type I fibres and less type II fibres than the healthy, elderly control group (Supplemental Table S5). No differences were found in fibre cross-sectional area, number of myonuclei per fibre, myonuclear domain, or the number of satellite cells per fibre between healthy and OP women (Supplemental Table S5). In both groups, the percentage of type II fibres was less than type I fibres (≈ 30%, p <0.01).
There were no significant changes in fibre-type distribution after training (not shown), and OP women maintained the higher percentage of type I fibres from before the study (Supplemental Table S5) after the intervention (56% versus 36% type I fibres in OP women and healthy controls, respectively, p <0.01; not shown). Both groups tended to increase the cross-sectional area of type II fibres during the strength-training intervention, but the increase was only statistically significant in healthy [Figure 2(a)]. The interaction between disease status and exercise intervention for cross-sectional area of type I and type II fibres [Figure 2(a)] was not significant (p = 0.11).
There were no significant changes during the training intervention in the number of myonuclei or myonuclear domain. In healthy controls, the number of satellite cells was reduced around type I fibres and increased around type II fibres (−28 ± 29% and +82 ± 97%, respectively), both p <0.05. The same tendency was observed in the OP group but did not reach significance (not shown).
There were no differences between OP women and healthy controls at baseline in levels of cytosolic HSP70 or alpha-B-c. OP women had 110 ± 68%, p = 0.67, and 99 ± 20%, p = 0.84, of levels in healthy controls for HSP70 and alpha-B-c, respectively (students t-test, data not shown). Both groups had significantly reduced HSP70 levels after the intervention (−24 ± 24% and −39 ± 28% in OP and healthy controls, respectively, both p <0.01, [Figure 2(b)]. In OP, alpha-B-c levels increased by 13 ± 22% (p = 0.05), whereas alpha-B-c levels remained unchanged in healthy [Figure 2(b)]. There was no statistically significant interaction between disease status and exercise intervention for HSP70 (p = 0.13) but there was a clear exercise intervention effect (p <0.0001). Alpha-B-c did not reach significance either for interaction (p = 0.11) or group (p = 0.11).
Distinct muscle transcriptomes after exercise and between healthy and postmenopausal OP women
The muscle transcriptomes were markedly altered genome-wide between healthy and postmenopausal OP women, as seen by the two well-separated clusters on the PCA plot [Figure 3(a)], when all muscle mRNA transcripts were included (n = 18,811). mRNA data pooled from all participants (35 women at two time points, 70 samples) revealed 308 transcripts with differential expression (FDR <5%) pre- versus post-strength training [see hierarchical heat map in Figure 3(b)], using repeated-measures ANOVA ignoring disease status. Notably, we can visually distinguish patients and healthy controls from each other in this plot. The patients showed generally lower signal values of the 308 transcripts [Figure 3(a)], than healthy. This tendency persisted in muscle biopsies obtained after exercise.
A shortlist of top 15 transcripts (p <0.0001) showing the lowest p-value in OP comparing end versus start of training is presented in Table 2. The effects of intervention in these 17 patients ranged from −10% (BRD3) to 40% (MIR1-1HG). All except four transcripts showed reduced levels 1 week after exercise.
To search for the most important transcriptome differences and similarities between healthy and OP patients after heavy-load strength training, we included mature microRNAs and visualized the results in two Venn diagrams [Figure 4(a, b)]. Ten transcripts were differentially expressed (FDR <0.1) in healthy women compared with 48 in OP women after intervention. Of these, only one transcript was common and regulated in the same direction in OP and healthy. We found no evidence for group versus time interaction after FDR correction. However, this two-way repeated-measures ANOVA test is very underpowered, and we therefore present the stratified results in Figure 4.
Effect of strength training on mature microRNAs in osteoporotic women
To obtain more complete data on the muscle transcriptome changes in the patients after strength training, we also performed separate analyses of mature microRNAs. The patients were examined at the start and end of the training period, and the mature microRNA transcriptome expression was visualized as PCA (Figure 5), revealing a pattern, but less homogenous than mRNA profiles [Figure 3(a)]; however, a reasonable separation of the results was observed at the start, and strength training resulted in a similar directional shift in the profiles (p <0.05; Figure 5). The 18 muscle microRNAs most differentially expressed (p <0.01) after strength training are shown in Supplemental Table S6.
Functional assignment of genes changed after training in osteoporotic women
Upon pooling of mRNAs and mature microRNAs IPA using p <0.05 and least-fold change >1.2 yielded the following top networks (Supplemental Table S7): organismal development, cardiovascular system development and function, organ morphology, skeletal and muscular system development and function, and tissue morphology.
Common transcripts between thigh and pelvic muscle and iliac bone
For the common muscle, bone transcripts study we used women from cohort I who donated iliac bone biopsies, with muscle attached to the pelvic side (Figure 1). The muscle part was separated from the bone biopsy and both transcriptomes from the same individuals were analyzed and compared. We selected 24 postmenopausal women (12 OP and 12 healthy) who were of similar age and BMI for comparison of pelvic muscle with iliac bone in the same individuals (Figure 1). Table 1 and Supplemental Table S4 summarize these women. The healthy and OP did not differ with regards age, BMI, lean, fat or mean body mass, but were distinct in bone density T-scores (Supplemental Table S4). Except for serum bone-specific alkaline phosphatase and urinary DPD (free deoxypyridinoline), indicating lower bone remodelling activity in healthy, all other biomarkers were similar (not shown).
In the women performing strength training [Figure 1, cohort II (healthy) + cohort III (OP)], we determined that 308 thigh muscle transcripts were differentially expressed before and after strength training at FDR <0.05 [Figure 3(b)]. Twelve corresponding iliac bone mRNAs (Table 3, column A) were correlated also to total hip BMD T-score at FDR ⩽0.10. Furthermore, all the 12 mRNAs, but QKI, were expressed in mechanically unstressed pelvic musculature (Table 3, column B). Interestingly, both pelvic muscle IGF1 and SBDS transcript levels correlated significantly with the corresponding transcript levels in iliac bone from the same donors (Table 3). Most of these transcripts were previously unknown to play a role in bone biology. However, two are recognized as bone-related genes highly associated with BMD:9 FZD4 and IGF1. It is notable that the latter, together with QKI and SH3BP5, had p-values of 9.1−0.000023, FDR ⩽0.064 and r of about −0.4 (Table 3).
Experimentally documented training responsive skeletal muscle genes that are also associated to BMD
A literature search using Gene Cards (www.genecards.org),15 associating the differentially expressed muscle genes in OP women before and after strength training to BMD yielded the nine transcripts presented in Supplemental Table S8. They showed a 1.2–1.43-fold difference in transcript levels after strength training. The difference corresponded with results from transcriptional profiling of the iliac bone biopsies (Supplemental Table S8, column 3)9 and experimental studies in animals (Supplemental Table S8, column 5). Furthermore, all genes in Supplemental Table S8, except IGF1, were expressed in primary cultured osteoblasts from three human donors.16 However, IGF1 is expressed in human bone biopsies9 containing osteoblasts/osteocytes/bone-lining cells at various stages of differentiation, as well as in osteoclasts.17
Discussion
Principal findings, interpretation and comparison to other studies
The present study reports for the first time a molecular pathology in the muscular macromolecular system as documented by transcriptome, protein and histological findings affecting many genes in postmenopausal women with primary OP. We show that the muscle transcriptome differs between OP and healthy women, and that heavy-load strength training alters the muscle expression pattern in both cohorts, apparently making it more similar to the situation in healthy women, providing new insight in muscle genes affected in OP. Nine muscle transcripts affected by training, are also among bone transcripts strongly associated with BMD and bone metabolism, as demonstrated in published studies (Supplemental Table S8). Thus, we present the genetic and molecular background behind the physical improvement in healthy and OP postmenopausal women upon heavy-load strength training, but future research will better connect the expression of gene groups to physical activity.
A recent paper18 strongly supports the present findings where the authors performed weighted gene co-expression network analyses to integrate the largest genome-wide association study (GWAS) set for BMD (GEFOS-2) combined with global bone expression data, and discovered a module containing 44 interacting genes highly enriched within contractile fibres. Moreover, bivariate GWAS revealed the pleiotropic effects of seven BMD-associated loci, including our previously published transcripts with recognized functions in bone [e.g. Wnt-signalling genes, MEPE (matrix extracellular phosphoglycoprotein)].19 Together, these studies underline the close relationship between muscle and bone, describing genes with common functions in these tissues. Thus, available information obtained by previous GWAS18–21 combined with expression data strongly support the present results, and we have been unable to find contradictory results. Therefore, the present study extends the GWAS-based information regarding the macromolecular relationship between skeletal muscle and bone by providing both transcriptome and protein data in general and after strength training in well-defined cohorts of healthy and OP women.
We identify 12 bone-associated muscle mRNAs (Table 3, column A) and document their overlapping functions in the skeleton and muscle. To validate the results, we also used an independent muscle donor group from whom we also obtained BMD and transcriptome data. All muscle transcripts were confirmed, and two showed statistical correlation to BMD (Table 3, column B) despite pelvic muscles having different functions from the weight-carrying thigh muscles. We interpret these findings as muscle and bone sharing common genes and provide the discovered subset of pleiotropic genes.
Strengths and weaknesses
This study hitherto presents the largest unrelated groups of ethnically homogeneous women, including a cohort who has undergone global muscle transcriptome (mRNA, microRNA) analyses with correlations to similar data in bone. The cohorts were well-characterized through extensive clinical and complementary examinations (X-ray, DXA, blood analyses)9 and had similar age and BMI. The microarray data were technically validated using RT-PCR of selected transcripts. In addition to the transcriptome analyses, key stress proteins were measured, and histological evaluation performed before and after heavy-load strength training. The difference in physical levels at the start of training between healthy controls and patients, and the gain in strength and balance in both groups has already been documented.12 Our data are uniformly supported by GWAS combined with expression data,18–21 as described above. In addition, animal experiments show that nine of our bone-associated muscle transcripts play a role in bone remodelling and with functions documented experimentally in the same direction as predicted from the present data. One weakness is the relatively limited number of participants and that the study involved women only, even though primary OP is predominantly a female disease. The study included only one ethnic group and the overall validity of the results for women in general requires confirmation, even if epidemiological studies clearly show that postmenopausal OP is a worldwide disease with common traits. Larger cohorts would probably lead to the detection of additional statistically significant pleomorphic genes and their products and further increase the statistical power. All patients received bisphosphonate treatment, thus, in spite of a 3-month withdrawal before the training started and during this period, we cannot rule out the possibility of a long-term effect on the muscle- and bone-transcriptome.
Meaning of the study and possible explanations
We compared muscle transcriptomes of controls and patients and searched for bone (BMD)-associated mRNAs and microRNAs. The thigh-muscle donors underwent biopsies before and after 13–15 weeks of heavy-load strength training, enabling us to study the training-sensitive genes as expressed in differential transcription profiles and in key protein expression and histology alterations. After heavy-load strength training, most of the changed transcripts showed reduced levels in both groups [Figure 4(a, b)]. The present post-strength-training transcription activity analyses are an instant reflection of gene activity, which most probably changes with time, and a bimodal effect of training would not be unexpected. Interestingly, similar correlations, contrary to what was expected, based on phenotype, have been reported; for example, for traits such as BMI.22 Such unexpected associations increase our insight and awareness of the complex molecular pathology underlying traits and phenotypes.
The plasticity of muscular tissue in adapting to physical activity is well documented.1 In healthy subjects, short-duration, one-leg exercises were accompanied by significant changes in muscle gene expression and DNA methylation,23 suggesting epigenetics as one mode of altering transcription.
It is well known that muscular strain affects the skeleton and is fundamental to bone remodelling.1 The age-dependent loss of muscle mass reflects the decay of fast, high-power glycolytic type II fibres and their conversion to slow, type I fibres with ensuing reduction in power and endurance.24 In the present study, the controls and patients had similar type I and II fibre areas at baseline. Notably, both groups showed the characteristic age-associated atrophy of type II fibres indicated by 30% smaller type II fibres than type I fibres. After the strength training, the cross-sectional area of type II fibres was significantly increased, mostly in the controls. Diminished muscle mass and activity in OP women cannot explain the age-dependent decrease in BMD, nor can OP explain age-associated sarcopenia. In the present study, the positive effect of heavy-load strength training on BMD did not reach statistical significance,12 probably due to the short intervention period. In comparison, Alison et al.25 showed in a randomized control trial in the elderly that 12 months of brief, minutes-long hopping and jumping exercises increased cortical and proximal femur trabecular bone mineral content, more so in the exercised leg, as revealed by clinical computed tomography (CT). These changes were accompanied by improved biomechanical properties of bone strength.
It was surprising to find that baseline fibre-type distribution differed between the OP and healthy women (Supplemental Table S5). As the age difference between the groups was non-significant, we have no good explanation for this finding.
PCA was used to compare controls and patients before and after training, revealing striking and consistent differences between the two groups. Cluster analyses underscored the differences and revealed expression patterns that yielded clear conclusions. Since the patients reported an equal or higher physical activity, inactivity cannot explain the results in this group. We correlated the mRNA and microRNA transcriptomes before and after strength training and present a list of interactive hubs that were statistically different after the intervention. The annotations resulted in maps likely to represent the core important interactions in skeletal muscle. Furthermore, we used the same patients to identify the pleiotropic genes with functions also in bone and found a convincing overlap between how these muscle characteristic genes were predicted to affect bone remodelling in published experimental data (Supplemental Table S8). Many of these genes, in addition to IGF1, have also been linked to bone biology: the Wnt-related transcripts FZD4 and FRZB are both present in Table 3 and Supplemental Table S8, respectively. Frzb-deficient mice have thicker cortical bone, with increased stiffness and higher cortical appositional bone formation after loading.26
The RGS family of proteins regulates G-protein signalling, and Rgs10-deficient mice have severe OP and impaired osteoclast differentiation.27 GNAI1, another G-protein-binding ligand, competes with Ca2+/calmodulin and is affected by heavy-load strength training. In mice, Igf1/Igf2 deficiency exaggerates the negative effect of low calcium on bone accretion and leads to inverse PTH serum levels.28 MGP belongs to a family of mineral-binding GLA proteins that include coagulation factors VII/IX and VIII [VWF (von Willebrand factor); Supplemental Table S8], anti-clotting factors protein S and protein C, and osteocalcin (BGLAP, bone Gla protein), a bone formation inhibitor.29 Mice lacking MGP die shortly after birth because of extensive arterial calcification leading to blood-vessel rupture.29 Decorin (DCN) belongs to the family of small leucine-rich proteoglycans predominantly expressed in skin and bone.30 Dcn-deficient mice develop a phenotype reminiscent of certain Ehlers–Danlos syndromes, affecting collagen type I genes, impairing osteogenesis and causing growth failure and skin fragility. Osteonectin, or SPARC (secreted protein acidic and cysteine-rich), is abundant in bone and is expressed in areas of active remodelling. It binds to collagen and hydroxyapatite and regulates cell proliferation, the production of metalloproteinases, for example, cathepsin K (CTSK), and angiogenesis.31 Sparc-null mice have decreased bone formation due to decreased osteoblast and osteoclast numbers.31 Ctsk-null mice develop OP due to the inability to degrade the bone matrix. Such mice exhibit enhanced compensatory osteoclastogenesis and increased expression of other proteases via an increased RANKL/OPG [tumour necrosis factor (TNF) superfamily member 11/TNF receptor superfamily member 11b] ratio.32
Unanswered questions and further research
A limited set of susceptibility genes associated with BMD in postmenopausal women have been identified in recent years9,13,19 and presently, their putative shared functions in skeletal muscle. It will be essential to study further their molecular functions in bone and muscle cells in order to understand how they interact and influence the cellular phenotypes. Also, the physiological actions of training-sensitive genes and how they translate to physical competence need to be studied further.
Prior knowledge
Postmenopausal OP and muscular asthenia, although present together, are considered as separate entities: as a disease and an accompanying condition co-occurring mainly due to physical inactivity.
Conclusion and added value
The present cohorts represent uniform groups of healthy and OP postmenopausal Norwegian women, as detailed previously,9 providing muscle and bone biopsies for analyses of RNA, key protein and histology changes between healthy controls and patients before and after strength training. Women with postmenopausal OP have distinct transcriptomes compared with healthy controls, translating into muscle pathophysiology and reduced skeletal muscle strength. A set of these disease-correlated genes are also associated with BMD, suggesting a shared genetic susceptibility previously unrecognized.
Our study generates novel insight into a common population disease and should have in the short term, an important impact on preventive measures, and may in the longer perspective lead to discovery of biomarkers to improve diagnosis and treatment for a large group of exposed women.
Supplemental Material
Supplemental material, Revised_Supplementary_040220_2 for Postmenopausal osteoporosis is a musculoskeletal disease with a common genetic trait which responds to strength training: a translational intervention study by Ole Kristoffer Olstad, Vigdis Teig Gautvik, Marissa LeBlanc, Karl Johnny Kvernevik, Tor Paaske Utheim, Anne Runningen, >Håvard Wiig, Camilla Kirkegaard, Truls Raastad, Sjur Reppe and Kaare Morten Gautvik in Therapeutic Advances in Musculoskeletal Disease
Acknowledgments
Sjur Reppe and and Kaare Gautvik contributed to this article equally. We thank Anne Marie Trøseid for technical assistance.
Footnotes
Author contributions: T Raastad and K Gautvik designed research; O Olstad, V Gautvik, K Kvernevik, A Runningen, H Wiig, C Kirkegaard, T Raastad, S Reppe and K Gautvik performed research; O Olstad, V Gautvik, T Utheim, T Raastad and K Gautvik contributed new reagents or analytical tools; O Olstad, M LeBlanc, T Raastad and S Reppe analyzed the data; O Olstad, M LeBlanc, T Utheim, T Raastad, S Reppe and K Gautvik wrote the paper.
Conflict of interest: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Data and materials availability: All primary data are available on request or via Geo Expression Omnibus.
Ethics approval: Verbal and written consents have been obtained from the participants who all were consent competent. Under the clinical interview, they filled out an extensive questionnaire containing all relevant health questions, family history and lifestyle risk factors. All patient information is anonymized and not traceable. We have received formal acceptance from the involved hospitals, health authorities and user committees at Lovisenberg Diakonale Hospital and Oslo University Hospital. The Norwegian Regional Ethical Committee approved the study (REK Sør-Øst C, registration no. 2010/2539 Norway) and commented positively on the patient`s and the societal benefits. The clinical importance of the project has been underlined by REK.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the South East Norway Health Authority under Grant number 52009/8029 to KMG; the 6th EU framework program under Grant number LSHM-CT-2003-502941 to KMG and SR; Oslo University Hospital, Ullevaal under Grant number 52009/8029 to KMG; Lovisenberg Diakonale Hospital to KMG and SR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ORCID iD: Sjur Reppe  https://orcid.org/0000-0002-4330-5379
https://orcid.org/0000-0002-4330-5379
Supplemental material: Supplemental material for this article is available online.
Transparency statement: The manuscript is an honest, accurate, and transparent account of the study, and no important aspect has been omitted.
Contributor Information
Ole Kristoffer Olstad, Department of Medical Biochemistry, Oslo University Hospital, Oslo, Norway.
Vigdis Teig Gautvik, Unger-Vetlesen Institute, Lovisenberg Diaconal Hospital, Oslo, Norway.
Marissa LeBlanc, Oslo Centre for Biostatistics and Epidemiology, Oslo University Hospital, Oslo, Norway.
Karl Johnny Kvernevik, Unger-Vetlesen Institute, Lovisenberg Diaconal Hospital, Oslo, Norway.
Tor Paaske Utheim, Department of Medical Biochemistry, Oslo University Hospital, Oslo, Norway; Department of Plastic and Reconstructive Surgery, Oslo University Hospital, Oslo, Norway; Department of Ophthalmology, Stavanger University Hospital, Stavanger, Norway; Department of Ophthalmology, Sørlandet Hospital Arendal Surgical Unit, Arendal, Norway.
Anne Runningen, Unger-Vetlesen Institute, Lovisenberg Diaconal Hospital, Oslo, Norway.
Håvard Wiig, Department of Physical Performance, Norwegian School of Sports Sciences, Oslo, Norway.
Camilla Kirkegaard, Department of Physical Performance, Norwegian School of Sports Sciences, Oslo, Norway.
Truls Raastad, Department of Physical Performance, Norwegian School of Sports Sciences, Oslo, Norway.
Sjur Reppe, Department of Medical Biochemistry, Oslo University Hospital, Oslo, Norway, Beverly, MA, USA; Unger-Vetlesen Institute, Lovisenberg Diaconal Hospital, Oslo, Norway; Department of Plastic and Reconstructive Surgery, Oslo University Hospital, Oslo, Norway; Department of Molecular Medicine, University of Oslo, Oslo, Norway.
Kaare Morten Gautvik, Lovisenberg Diakonale Sykehus, Unger-Vetlesen Institute, Lovisenberggata 17, Oslo 0456, Norway.
References
- 1. Coffey VG, Hawley JA. The molecular bases of training adaptation. Sports Med 2007; 37: 737–763. [DOI] [PubMed] [Google Scholar]
- 2. Cruz-Jentoft AJ, Baeyens JP, Bauer JM, et al. Sarcopenia: European consensus on definition and diagnosis: report of the European working group on sarcopenia in older people. Age Ageing 2010; 39: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hernlund E, Svedbom A, Ivergard M, et al. Osteoporosis in the European Union: medical management, epidemiology and economic burden. A report prepared in collaboration with the International Osteoporosis Foundation (IOF) and the European Federation of Pharmaceutical Industry Associations (EFPIA). Arch Osteoporos 2013; 8: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kanis JA, Johnell O, Oden A, et al. Long-term risk of osteoporotic fracture in Malmo. Osteoporos Int 2000; 11: 669–674. [DOI] [PubMed] [Google Scholar]
- 5. Johnell O, Kanis J. Epidemiology of osteoporotic fractures. Osteoporos Int 2005; 16(Suppl. 2): S3–S7. [DOI] [PubMed] [Google Scholar]
- 6. Chalhoub D, Cawthon PM, Ensrud KE, et al. Risk of nonspine fractures in older adults with sarcopenia, low bone mass, or both. J Am Geriatr Soc 2015; 63: 1733–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laurent MR, Dubois V, Claessens F, et al. Muscle-bone interactions: from experimental models to the clinic? A critical update. Mol Cell Endocrinol 2016; 432: 24–46. [DOI] [PubMed] [Google Scholar]
- 8. Watson SL, Weeks BK, Weis LJ, et al. High-intensity resistance and impact training improves bone mineral density and physical function in postmenopausal women with osteopenia and osteoporosis: the LIFTMOR randomized controlled trial. J Bone Miner Res 2018; 33: 211–220. [DOI] [PubMed] [Google Scholar]
- 9. Reppe S, Refvem H, Gautvik VT, et al. Eight genes are highly associated with BMD variation in postmenopausal Caucasian women. Bone 2010; 46: 604–612. [DOI] [PubMed] [Google Scholar]
- 10. Schmiedel JM, Klemm SL, Zheng YN, et al. MicroRNA control of protein expression noise. Science 2015; 348: 128–132. [DOI] [PubMed] [Google Scholar]
- 11. Camernik K, Barlic A, Drobnic M, et al. Mesenchymal stem cells in the musculoskeletal system: from animal models to human tissue regeneration? Stem Cell Rev 2018; 14: 346–369. [DOI] [PubMed] [Google Scholar]
- 12. Raastad T, Kvernevik KJ, Johansen M, et al. Marked improvement in physical function through gains in muscle strength and thigh muscle size after heavy-load strength training in women with established postmenopausal osteoporosis. J Osteopor Phys Act 2015; 3: 8. [Google Scholar]
- 13. Jemtland R, Holden M, Reppe S, et al. Molecular disease map of bone characterizing the postmenopausal osteoporosis phenotype. J Bone Miner Res 2011; 26: 1793–1801. [DOI] [PubMed] [Google Scholar]
- 14. Vissing K, Schjerling P. Simplified data access on human skeletal muscle transcriptome responses to differentiated exercise. Sci Data 2014; 1: 140041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stelzer G, Rosen N, Plaschkes I, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics 2016; 54: 1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- 16. Reseland JE, Reppe S, Larsen AM, et al. The effect of enamel matrix derivative on gene expression in osteoblasts. Eur J Oral Sci 2006; 114(Suppl. 1): 205–211; discussion 254–206, 381–202. [DOI] [PubMed] [Google Scholar]
- 17. Cappellen D, Luong-Nguyen NH, Bongiovanni S, et al. Transcriptional program of mouse osteoclast differentiation governed by the macrophage colony-stimulating factor and the ligand for the receptor activator of NFkappa B. J Biol Chem 2002; 277: 21971–21982. [DOI] [PubMed] [Google Scholar]
- 18. Chen YC, Guo YF, He H, et al. Integrative analysis of genomics and transcriptome data to identify potential functional genes of BMDs in females. J Bone Miner Res 2016; 31: 1041–1049. [DOI] [PubMed] [Google Scholar]
- 19. Medina-Gomez C, Kemp JP, Dimou NL, et al. Bivariate genome-wide association meta-analysis of pediatric musculoskeletal traits reveals pleiotropic effects at the SREBF1/TOM1L2 locus. Nat Commun 2017; 8: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Estrada K, Styrkarsdottir U, Evangelou E, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 2012; 44: 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reppe S, Wang Y, Thompson WK, et al. Genetic sharing with cardiovascular disease risk factors and diabetes reveals novel bone mineral density loci. PLoS One 2015; 10: e0144531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015; 518: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lindholm ME, Marabita F, Gomez-Cabrero D, et al. An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics 2014; 9: 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miljkovic N, Lim JY, Miljkovic I, et al. Aging of skeletal muscle fibers. Ann Rehabil Med 2015; 39: 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Allison SJ, Poole KE, Treece GM, et al. The influence of high-impact exercise on cortical and trabecular bone mineral content and 3D distribution across the proximal femur in older men: a randomized controlled unilateral intervention. J Bone Miner Res 2015; 30: 1709–1716. [DOI] [PubMed] [Google Scholar]
- 26. Lories RJ, Peeters J, Bakker A, et al. Articular cartilage and biomechanical properties of the long bones in Frzb-knockout mice. Arthritis Rheum 2007; 56: 4095–4103. [DOI] [PubMed] [Google Scholar]
- 27. Yang S, Li YP. RGS10-null mutation impairs osteoclast differentiation resulting from the loss of [Ca2+]i oscillation regulation. Genes Dev 2007; 21: 1803–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kasukawa Y, Baylink DJ, Wergedal JE, et al. Lack of insulin-like growth factor I exaggerates the effect of calcium deficiency on bone accretion in mice. Endocrinology 2003; 144: 4682–4689. [DOI] [PubMed] [Google Scholar]
- 29. Luo G, Ducy P, McKee MD, et al. Spontaneous calcification of arteries and cartilage in mice lacking matrix GLA protein. Nature 1997; 386: 78–81. [DOI] [PubMed] [Google Scholar]
- 30. Corsi A, Xu T, Chen XD, et al. Phenotypic effects of biglycan deficiency are linked to collagen fibril abnormalities, are synergized by decorin deficiency, and mimic Ehlers-Danlos-like changes in bone and other connective tissues. J Bone Miner Res 2002; 17: 1180–1189. [DOI] [PubMed] [Google Scholar]
- 31. Delany AM, Amling M, Priemel M, et al. Osteopenia and decreased bone formation in osteonectin-deficient mice. J Clin Invest 2000; 105: 915–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kiviranta R, Morko J, Alatalo SL, et al. Impaired bone resorption in cathepsin K-deficient mice is partially compensated for by enhanced osteoclastogenesis and increased expression of other proteases via an increased RANKL/OPG ratio. Bone 2005; 36: 159–172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Revised_Supplementary_040220_2 for Postmenopausal osteoporosis is a musculoskeletal disease with a common genetic trait which responds to strength training: a translational intervention study by Ole Kristoffer Olstad, Vigdis Teig Gautvik, Marissa LeBlanc, Karl Johnny Kvernevik, Tor Paaske Utheim, Anne Runningen, >Håvard Wiig, Camilla Kirkegaard, Truls Raastad, Sjur Reppe and Kaare Morten Gautvik in Therapeutic Advances in Musculoskeletal Disease