SUMMARY
Active membrane remodeling during myelination relies on phospholipid synthesis and membrane polarization, both of which are known to depend on inositol phospholipids. Here, we show that sciatic nerves of mice lacking phosphatidylinositol 4-kinase alpha (PI4KA) in Schwann cells (SCs) show substantially reduced myelin thickness with grave consequences on nerve conductivity and motor functions. Surprisingly, prolonged inhibition of PI4KA in immortalized mouse SCs failed to decrease plasma membrane phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) levels or PI 3-kinase (PI3K) activation, in spite of large reductions in plasma membrane PI4P levels. Instead, it caused rearrangements of the actin cytoskeleton, which was also observed in sciatic nerves of knockout animals. PI4KA inactivation disproportionally reduced phosphatidylserine, phosphatidylethanolamine, and sphingomyelin content in mutant nerves, with similar changes observed in SCs treated with a PI4KA inhibitor. These studies define a role for PI4KA in myelin formation primarily affecting metabolism of key phospholipids and the actin cytoskeleton.
Grahical Abstract
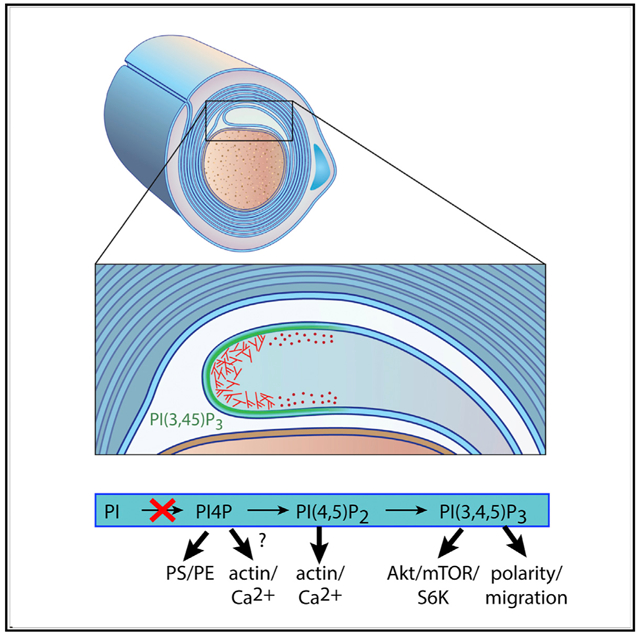
In Brief
Alvarez-Prats et al. show that Schwann cell (SC)-specific inactivation of PI4KA in mice causes myelination defects in peripheral nerves, decreases in key phospholipid myelin components, and gross alterations in actin architecture. The results point to impaired phospholipid and actin defects as the primary cause of the myelination defects.
INTRODUCTION
Myelination of peripheral and central nerves is essential for proper propagation of electrical signals. Several human diseases affect myelination, including multiple sclerosis in the CNS and some forms of Charcot-Marie-Tooth disease in the peripheral nervous system (PNS) (Herbert and Monk, 2017). Myelination of peripheral nerves requires the proper sequence of signaling events that promote the migration, maturation, and differentiation of Schwann cells (SCs) even before the actual myelin wrapping of axons takes place (Herbert and Monk, 2017). The process of axon wrapping uses several signaling pathways to induce membrane polarization, actin remodeling, and the synthesis and delivery of membrane lipids into the membrane sheaths. In many of these events, phosphoinositides play important roles. Phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) is a substrate for phospholipase C (PLC)- or PI 3-kinase (PI3K)-generated second messengers but also controls actin dynamics and ion channel activity (Balla, 2013), all of which are necessary for myelination (Herbert and Monk, 2017). PI(3,4,5)P3 defines membrane polarity but also activates mTOR to increase protein and lipid synthesis. Elevated PI(3,4,5)P3 levels due to PTEN deletion enhance myelination by both oligodendrocytes and SCs (Goebbels et al., 2010). Inactivation of PTEN in cerebellar granule cells initiates myelination of otherwise unmyelinated fibers (Goebbels et al., 2017). Akt and mTORC1, downstream targets of the class I PI3K pathway, are critical for the myelination both in the CNS and the PNS (Kearns et al., 2015; Norrmén et al., 2014). An additional role of phosphoinositides, namely PI3P and PI(3,5)P2 in myelination, is known as mutations in the myotubularin gene family were identified in Charcot-Marie-Tooth disease, CMT 4B1 and −2 (Azzedine et al., 2003; Houlden et al., 2001; Kim et al., 2002).
Phosphatidylinositol 4-phosphate (PI4P) is a precursor of PI(4,5)P2 in the plasma membrane (PM) but is also important in the Golgi and endosomes, where it regulates vesicular trafficking (Balla, 2013). PI4P is generated by four distinct PI 4-kinases (PI4Ks), and each of these enzymes is linked to a different aspect of cell physiology (Boura and Nencka, 2015). PI4KA is the most important source of PI4P in the PM (Bojjireddy et al., 2014; Nakatsu et al., 2012). PI4KA also is an essential host factor for the replication of some RNA viruses (Altan-Bonnet and Balla, 2012), suggesting its more complex role in membrane biogenesis. PI4KA is highly expressed in the brain, yet little is known about its role(s) in the nervous system. Germline inactivation of Pi4ka in mice is embryonic lethal (Nakatsu et al., 2012), and an inactivating mutation has been described as causing major brain developmental abnormalities in human fetuses (Pagnamenta et al., 2015). PI4KA is essential in flies (Tan et al., 2014), and its yeast homolog, Stt4p, is essential for cell wall biogenesis (Audhya and Emr, 2002; Yoshida et al., 1994). PI4KA is recruited to the PM by a protein complex comprised of EFR3, TTC7, and Fam126A (Baskin et al., 2016; Nakatsu et al., 2012), and mutations or depletion of Fam126A destabilizes PI4KA and causes hypomyelination (Baskin et al., 2016; Miyamoto et al., 2014; Traverso et al., 2013).
Recently, PI4KA was implicated in the transport of phosphatidylserine (PS) between various membranes (Chung et al., 2015; Galmes et al., 2016), thereby controlling the level of several lipids (PI4P, PI(4,5)P2, and PS) that define PM identity. In order to test the importance of the enzyme in a biological system that requires the generation of large volumes of PM, such as the SCs, we generated mice with a SC-specific Pi4ka deletion to evaluate the role of this enzyme in the myelination process. These mice display severe myelination defects with substantially reduced myelin thickness and lipid content most severely affecting PS and phosphatidylethanolamine (PE). Surprisingly, mouse SCs kept in culture retain their PM PI(4,5)P2 levels and Akt and mTORC1 responses even after prolonged PI4KA inhibition in spite of the massive reduction in their PM PI4P levels. In contrast, PI4P depletion from the PM causes rearrangements of the actin cytoskeleton both in cultured cells and in nerves of affected animals. Our studies highlight the central role of PI4KA in the myelination process and hint that the role of the enzyme might be closely linked to actin dynamics and PS metabolism.
RESULTS
Inactivation of PI4KA in SCs Causes Neuropathy-like Symptoms
Conditional knockout mice for PI4KA (Figures S1A and S1B; Bojjireddy et al., 2014) were cross-bred with Mpz-Cre mice expressing Cre recombinase under the control of the myelin protein zero (P0 and Mpz) gene promoter (Feltri et al., 1999). P0 is a glycoprotein located in SCs, and its expression is induced at the onset of myelination (Eichberg, 2002). Mice were born at the expected Mendelian ratio. As mice reached day postnatal day 30 (P30), MPZCre/+/PI4KAfl/fl mice (hereafter referred to as “mutants”), started to show abnormal gait compared to controls (MPZCre/+/PI4KAfl/wt and MPZCre/+/PI4Kwt/wt), regardless of their gender. Their gait progressively worsened, mostly affecting hind legs, which they dragged behind with muscles in flaccid extension. Affected animals exhibited a tremor in their hind legs. When suspended by the tail, the hind limbs of mutants either flexed spasmodically or clasped together intermittently (Figure 1A).
Figure 1. Specific Inactivation of PI4KA in SCs Causes Neuropathy-like Symptoms.
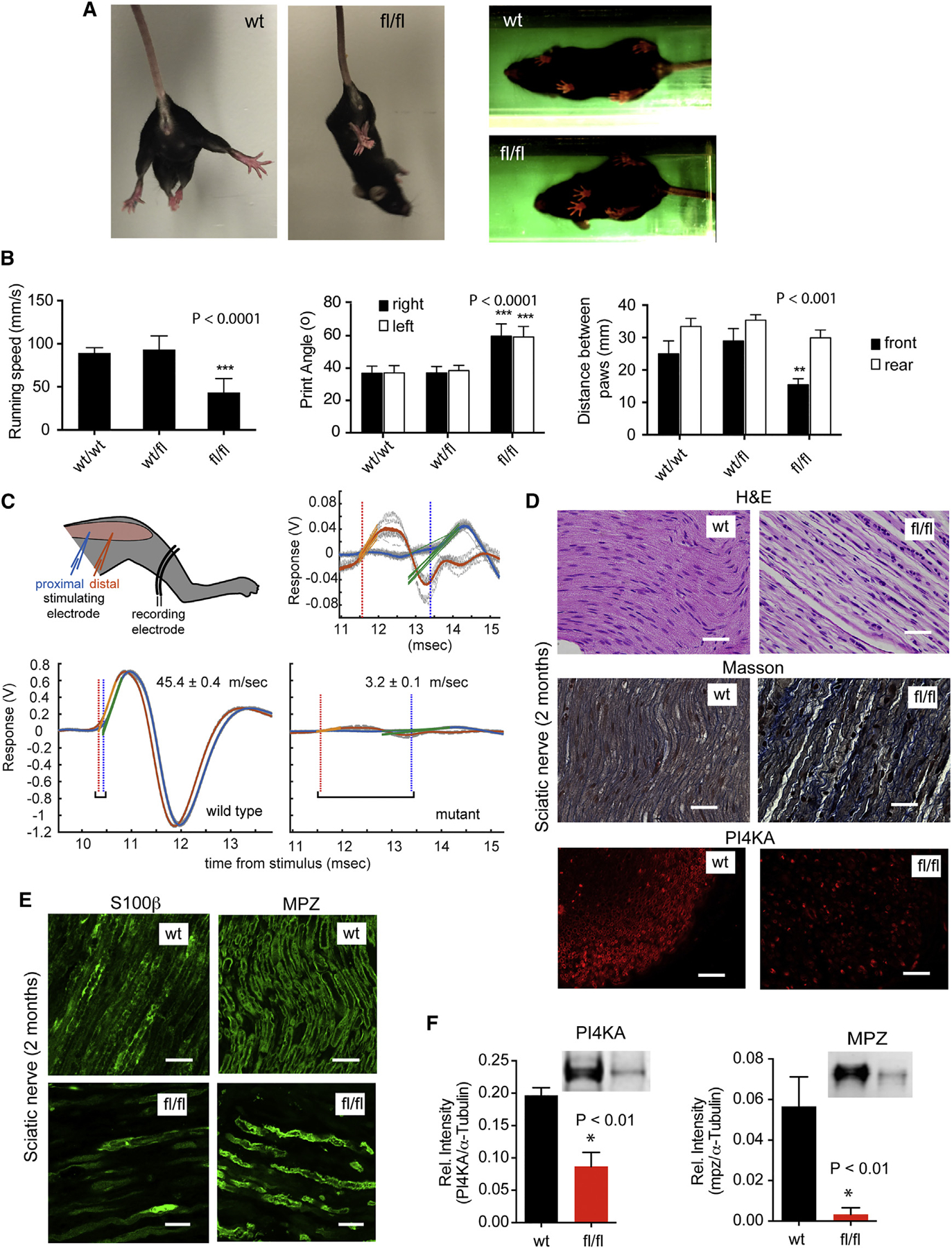
(A) Mutant mice clasp their hind legs together when suspended by the tail, and their stance is different from that of wild-type.
(B) Gait parameters measured with TredScan equipment: running speed of mutant mice is significantly slower than the controls. The print angles (the angle of the hind foot relative to the longitudinal body axis) are significantly larger on both sides of the mutant animals compared to controls. The distance between the front paws is significantly smaller in mutants than in controls. Means ± SEM are shown for n = 5 animals per group. ***p < 0.0001 and **p < 0.001 versus control using one-way ANOVA analysis followed by the Tukey’s multiple comparisons test.
(C) Cartoon and representative traces showing nerve conductance measurements. Electromyography (EMG) recordings are shown from the same animal after stimulating the sciatic nerve from a proximal (blue) and distal (orange) electrode. The time delay and the distance between the two stimulating points allow calculation of the conductivity speed. Note the greatly diminished EMG signal of the mutant mice (enlarged in the insert above).
(D) H&E staining of longitudinal sections of sciatic nerves show substantial structural changes and cellularization in mutant (fl/fl) nerves. Fibers are not tightly packed and not arranged properly compared to control (wt). Masson’s trichrome staining shows high collagen deposition in the sciatic nerves of mutant nerves. Immunohistochemistry (IHC) of cross section of sciatic nerves shows the major loss of PI4KA. The scale bars represent 50 μm.
(E) IHC of both S100b, a marker for mature SCs, and MPZ shows marked decreases in mutant nerves (bars show 20 μm).
(F) Reduced expression of PI4KA and MPZ is shown in western blots (WBs) from sciatic nerve samples. (Means ± SEM, n = 3, *p < 0.01).
Characterization of the phenotype by TreadScan instrument (Beare et al., 2009) showed that mutant mice (P60) walked significantly slower than their control littermates (Figure 1B). Both controls (MPZCre/+/PI4KAfl/wt and MPZCre/+/PI4Kwt/wt) showed a similar stance, whereas mutant animals kept their front paws significantly closer to one another (Figures 1A and 1B). Print angle (PA), which measures the angle between the heel and the linea alba of the body for each rear foot, was also significantly wider in the mutants (Figure 1B). Notably, similar changes in gait parameters have been reported for various forms of CMT models in mice (Bogdanik et al., 2013; Cartoni et al., 2010; Wooley et al., 2005).
Sciatic nerve conductivity was then performed using electromyographic recordings in P60 mice. We measured the time it took for an impulse elicited by a mild electric stimulation delivered at two (distal and proximal) stimulation points on the sciatic nerve to travel a known distance to a reference electrode. Consistent with published reports of healthy animals (Oh et al., 2010; Xia et al., 2010), conductance speed in control animals was 73.0 ± 31.6 m/s (mean ± SD; n = 8 independent measurements). In most mutants, the electromyography (EMG) signal was too small to allow calculating a reliable conductivity speed. In two cases where accurate measurements could be made, conduction velocity was severely reduced (3.2 ± 0.1 and 6.8 ± 0.1 m/s; mean ± SEM calculated from 10 repeated proximal and distal stimulations; Figure 1C).
Histological Alterations in Mutant Nerves
H&E and Masson’s trichrome staining of sciatic nerves of P60 mice showed disorganized fibers with more nuclei and high collagen deposition in mutants compared to controls (Figure 1D). Immunohistochemical and western blot analysis confirmed the dramatic reduction in PI4KA levels and the decrease in the myelin-specific protein, MPZ, in mutant sciatic nerves (Figures 1D–1F). Similar reduction was observed in S100b, a specific marker of mature SCs in peripheral nerves (Figure 1E). Cajal bands were also missing in mutant nerves, and their immunoreactive Drp2 and periaxin signals were greatly diminished (Figure 2A). TUNEL analysis showed the presence of apoptotic nuclei in mutant nerves whereas no apoptosis was observed in controls (Figure 2B). Sciatic nerves of mutant animals showed more nuclei and pronounced staining for the oligodendrocyte marker, neuron-glia 2 (NG2), together with enhanced expression of Krox20, the transcription factor that drives MPZ and MBP transcription (Figure S1C). These changes most likely reflect a reactive process, as proliferation of NG2-positive cells has been described in peripheral nerves in response to injury (Rezajooi et al., 2004).
Figure 2. Histological and EM Analysis of Sciatic Nerves from Control and Mutant Mice.
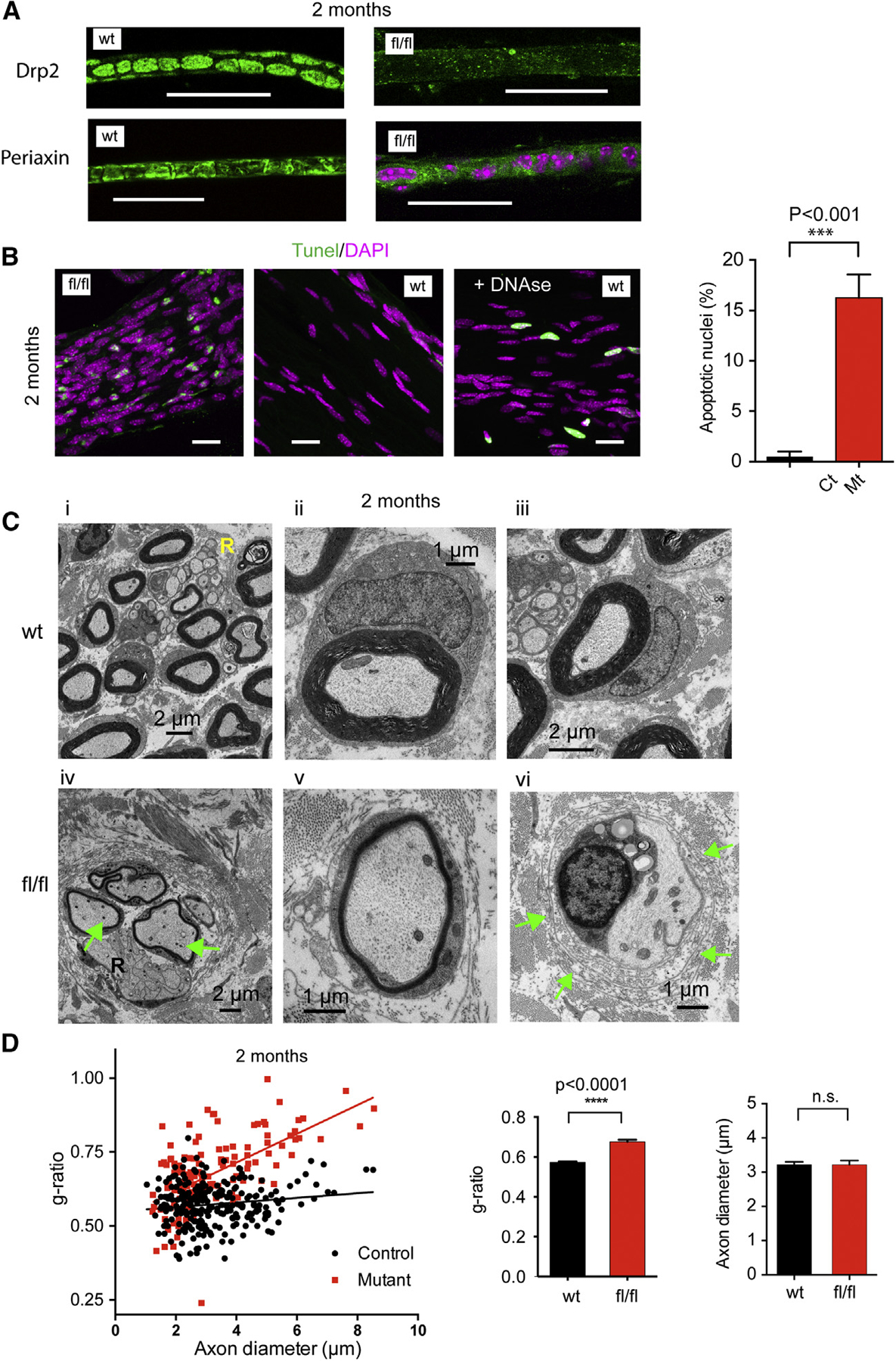
(A) Sciatic nerves from P60 wild-type (wt) or mutant (fl/fl) littermates were teased and immuno-stained for Drp2 and Periaxin. Note the presence of Cajal bands in control nerves and the very low signal and no Cajal bands in mutant nerves (scale bars, 20 μm; pink shows nuclei).
(B) TUNEL staining of mutant nerves shows several apoptotic nuclei (green). Note the more numerous and less elongated nuclei in mutants (scale bars, 20 μm). Statistical significance of difference in the percent of apoptotic nuclei was assessed by unpaired t test (n = 121 and 391 nuclei from control and mutant nerves, respectively, ***p < 0.001).
(C) Electron micrographs of cross sections of sciatic nerves demonstrate significantly reduced myelin thickness in mutant nerves (fl/fl, panels iv–vi). Hallmarks of aberrant myelination, such as myelin outfoldings (see panel iv) and lack of myelin with onion bulbs (see panel vi, arrows), are present in the mutants. No major differences were found in the appearance of Remak bundles (panels i and iv, labeled with “R”). The number of mitochondria within the axons of mutant nerves was higher than those in controls (panel iv arrows show numerous mitochondria).
(D) Scatterplots showing g-ratios as a function of axon diameter. Myelin thickness is lower across the whole spectrum of axon diameters in the mutant nerves, and the slope is significantly steeper in the mutant group. Average g-ratios show reduced myelin thickness but no change in axon diameter. Means ± SEM; ****p < 0.0001 control versus mutant; n = 255 (control) and n = 136 (mutant) from 3 animals for each group assessed by unpaired t test.
To analyze the progression of myelination, we crossed our mouse lines with a reporter line (Gt(ROSA)26Storm4(ACTB-tdTomato-EGFP)), which allows monitoring the Cre activity in the tissues of developing animal as it expresses a PM-targeted tdTomato that is replaced by a PM-targeted GFP, marking the affected cells (Muzumdar et al., 2007). This analysis showed that Cre was already active by P7 and that control mice displayed myelinating SCs in their sciatic nerves at P14. Myelination was complete by 2 months, with clear detection of Schmidt-Lanterman incisures and nodes of Ranvier and showing red fluorescence in the axons (Figure S1E, left). In contrast, mutant nerves showed little signs of myelination at P14 and the lack of organized SC structures even at P60 (Figure S1E, right).
Myelination Is Dramatically Impaired in Mice with SCs Lacking PI4KA
Electron microscopy (EM) analysis of sciatic nerves of P60 mice showed significantly thinner myelin in mutants (Figure 2C, panels iv–vi). This was true for axons of all diameters, but larger axons were more affected as shown by the shift and increased slope of g-ratios compared to control nerves. Importantly, average axon diameters showed no change (Figure 2D). Hallmarks of aberrant myelination, such as myelin outfoldings (Figure 2C, panel iv; none versus 3 or 4/field per mice in wild-type and mutant, respectively) as well as the appearance of onion bulbs (Figure 2C, panel vi, green arrows), were evident in the mutants (none were observed in wild-type nerves versus an average of 19 onion bulbs per field per mice in mutants; n = 3 fields). Notably, we observed more mitochondria in the axons of mutant nerves (1.947 ± 0.1447, n = 57 and 3.943 ± 0.3558, n = 53; mitochondrion per axon in wild-type and mutant, respectively; mean ± SEM; Figure 2C, panel iv, arrows). Importantly, no major alterations were observed in the number and appearance of Remak bundles (Figure 2C, marked “R”), suggesting that radial sorting and early differentiation of the non-myelinating SCs have taken place in mutant nerves.
Inhibition of PI4KA Depletes PI4P, but Not PI(4,5)P2, from the Plasma Membrane of a Mouse SC Cell Line
During myelination, polarized movement of the growing myelin sheet between the existing myelin wraps and the axon requires both polarized PI3K activation (Goebbels et al., 2010, 2017) and reorganization of the actin cytoskeleton (Nawaz et al., 2015; Zuchero et al., 2015; Figure 3A). To investigate the molecular events affected by PI4KA inactivation, we used the mouse immortalized SC line, SW10 (SW10 [ATCC CRL2766]) and A1, a specific inhibitor of PI4KA (Bojjireddy et al., 2014; Hammond et al., 2014). We also analyzed mouse embryonic fibroblast (MEF) cells with inducible inactivation of PI4KA. Specifically, we wanted to evaluate the effects of prolonged inactivation of PI4KA on the PI4P, PI(4,5)P2, and PI(3,4,5)P3 phosphoinositide axis. For PI4P and PI(4,5)P2 imaging, we used the PI4P-specific lipid sensor P4M of the SidM protein of Legionella pneumophila and the PLCδ1-PH domain, respectively (Hammond et al., 2014). A 24-hr treatment with the PI4KA inhibitor completely eliminated PI4P from the PM, causing the sensor to bind to intracellular PI4P containing Golgi and endosomal membranes (Figure 3B, upper panels). Surprisingly, this treatment did not affect the PM localization of the PI(4,5)P2 sensor PH domain (Figure 3B, lower). This finding was corroborated by lipidomic analysis of SW10 cells, which showed a large decrease in PI4P without a significant decrease in total PI(4,5)P2 after 24 hr of PI4KA inhibition (Figure 3C). (Note that the peak values do not reflect the relative amounts among the various PI derivatives). This analysis does not resolve various stereoisomers, but given the relatively small amounts of other isomers, the results can be informative regarding changes in the absolute levels of PI4P and PI(4,5)P2.
Figure 3. PI(4,5)P2 Levels and PI3K/Akt/ mTOR Responses to Serum Stimulation Are Unchanged in a Mouse SC Line after Prolonged Inhibition of PI4KA.
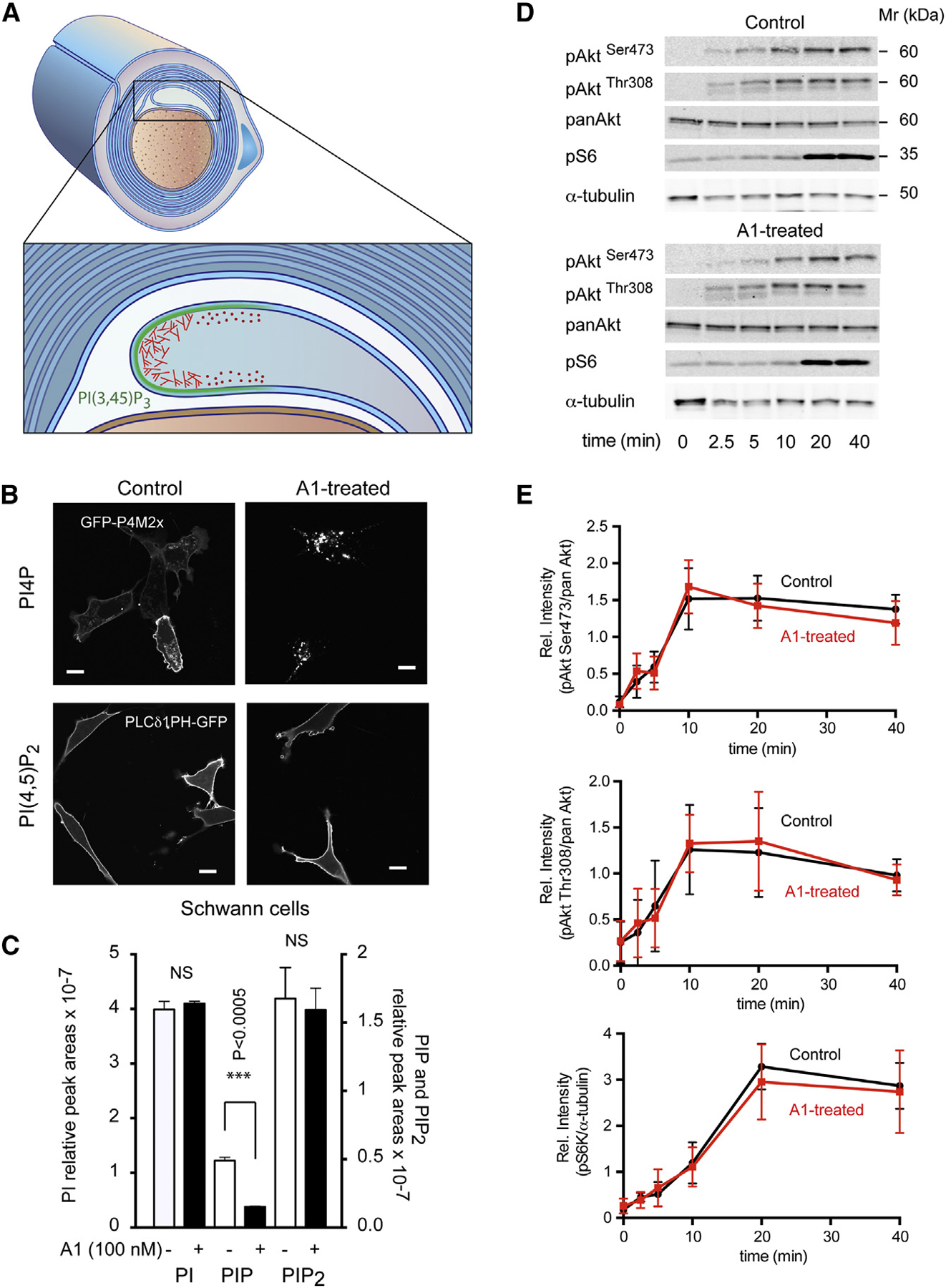
(A) Cartoon showing the growing myelin sheaths around a myelinated axon. This process requires both PI3K activation (green) and actin dynamics with proper spatial control (red).
(B) A mouse SC line (SW10) was treated with the PI4KA inhibitor (A1; 100 nM) for 24 hr, and PI(4,5)P2 and PI4P localization was assessed by expressing the lipid reporters, PLCδ1-PH-GFP and GFP-P4M2x, respectively. A1 treatment causes a major decrease in PM PI4P levels, causing the PI4P reporter probe to re-localize to endosomes and the Golgi, where PI4P levels remain unaffected (lower right panel). The same treatment did not reduce PI(4,5)P2 levels, and the reporter remains on thePM (left-lower panel). The scale bars represent 10 μm.
(C) Lipidomic analysis of phosphoinositides from cells treated with 100 nM A1 or DMSO for 24 hr. Note the massive reduction in the absolute amount of PIP (***p < 0.0005; unpaired t test) and the lack of significant change in PIP2 (means ± SEM; from triplicate samples).
(D) WB analysis of different downstream effectors of the PI3K/Akt/mTOR pathway (AktSer473, AktThr308, and pS6 ribosomal protein) showed similar kinetics of phosphorylation after stimulation with 10% serum following prolonged treatment with the PI4KA inhibitor or DMSO (control).(E) Quantification from three such experiments (means ± SEM; n = 3).
To test the status of the PI3K pathways and their downstream effectors, we stimulated SW10 cells with serum and followed the phosphorylation of Akt (both Ser473 and Thr308) and the S6 ribosomal protein, a substrate of p70 S6-kinase, a target of mTOR. These phosphorylation responses showed that PI3K activation was not significantly affected by prolonged PI4KA inhibition (Figures 3D and 3E).
Inhibition of PI4KA Causes Migration Defects and Actin Rearrangements in SCs
Despite an intact PI3K activation in response to serum stimulation, treatment with the PI4KA inhibitor for 24 hr significantly reduced the migration of SW10 cells toward serum containing media in trans-well chambers (Chen, 2005; Figure 4A). This could be related to the significantly enhanced cortical cytoskeleton relative to stress fibers that we observed both in SW10 cells and MEF cells after prolonged PI4KA inhibition and PI4KA inactivation, respectively (Figures 4B and 4C). Quantitation of the actin rearrangement was performed using line intensity histograms and measuring the ratio of PM intensities divided by the average intensity calculated for the cytoplasm (Figure 4C). Similar changes were observed by Nakatsu et al. (2012) in their study of MEF cells after PI4KA inactivation. Analysis of actin structure of nerves isolated from control animals (P60) showed well-organized actin structures in control nerves with enrichments in nodes of Ranvier and in the Schmidt-Lanterman incisures (SLIs) (Figure 4D). In contrast, actin organization was completely broken down and very strong actin staining was found in the membranes of SCs of the sciatic nerves of the knockout animals (Figure 4D).
Figure 4. PI4KA Inactivation Affects Cell Migration and Actin Distribution.
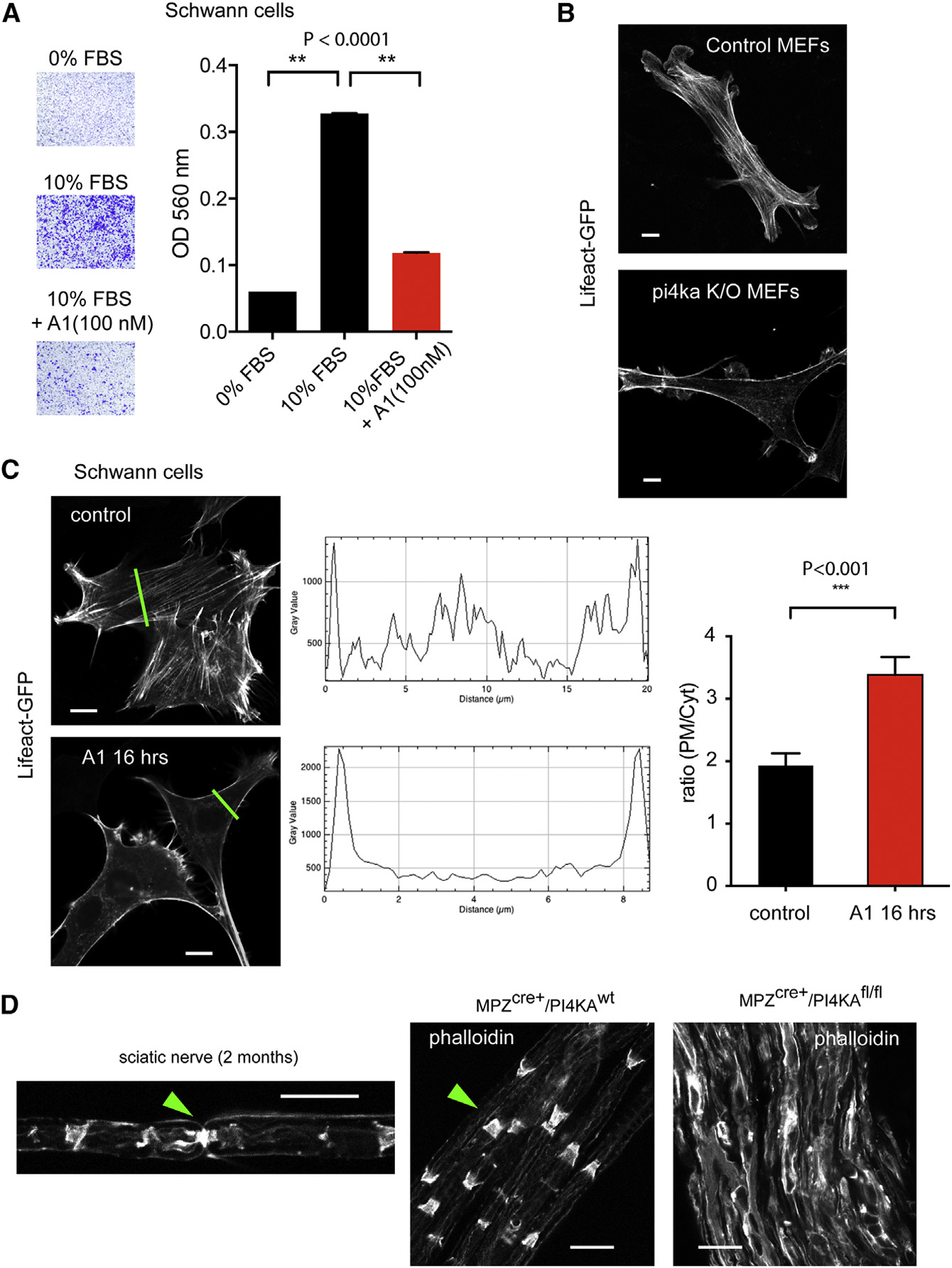
(A) Migration of SW10 cells in Boyden chambers in response to serum. Treatment of cells with A1 (100 nM; during the whole migration time of 24 hr) significantly reduced the number of cells migrating toward the serum-containing medium. Means ± SEM; n = 3 (**p < 0.0001).
(B) Immortalized MEF cells expressing LifeAct-GFP. Immortalized MEF cells prepared from PI4Kfl/fl/R26ERCre+ embryos (Bojjireddy et al., 2014) were treated with tamoxifen or solvent as described under Materials and Methods before transfection with the LifeAct-GFP and confocal microscopy. Representative pictures show the prominent lack of stress fibers and more pronounced cortical actin in the cells with PI4KA deletion compared to controls. The scale bars represent 10 μm.
(C) Prolonged (16-hr) inhibition of PI4KA in SW10 cells shows reduced stress fibers. Line intensity histograms were analyzed to assess the redistribution of F-actin as shown in representative examples (green lines; scale bars: 10 μm). Such analysis of 10 cells showed a significant increase in the plasma membrane intensities relative to the average cytoplasmic signal. ***p < 0.001.
(D) Phalloidin staining of teased sciatic nerves obtained from control or mutant mice. Note the degraded actin organization and strong actin staining in the mutant nerves. Green arrow in the left panel shows the actin structure at a node of Ranvier, whereas in the middle panel, it shows Schmidt-Lanterman incisures (SLIs). The scale bars represent 20 μm.
Inactivation of PI4KA Decreases Levels of Key Phospholipids for Myelination
Sciatic nerves of P60 mice were strikingly different in appearance: mutant nerves were completely transparent and soft, whereas control nerves were white and firm (Figure 5A), indicating loss of lipids from the affected nerves. Lipidomic analysis of nerves from such animals showed more than 50% reduction in the total lipid content of the sciatic nerves in the mutants (Figure 5B, insert). Notably, among the various lipid classes, the reductions were particularly prominent in PS, PE, sphingomyelin (SM), and hexosyl-ceramide (Hex-Cer) (note that the graph in Figure 5B shows mol% of total phospholipids and not absolute amounts). The increase in the relative proportion (not the absolute amount) of phosphatidylcholine (PC) and PI likely reflects a compensatory process. Lipidomic analysis was also performed on SW10 cells treated with the PI4KA inhibitor, A1, for 24 hr, which showed that inhibition of PI4KA in SCs caused reductions in the levels of PS, PE, and SM (Figure 5C). These lipid changes were similar to those observed in the sciatic nerves of the knockout animals and were consistent with the recently recognized close relationship between PI4P and PS metabolism and PS transport (Chung et al., 2015; Sohn et al., 2016).
Figure 5. PI4KA Inactivation Causes Reduction in Total Phospho-and Sphingolipids in Sciatic Nerves and in Cultured SW10 Cells.
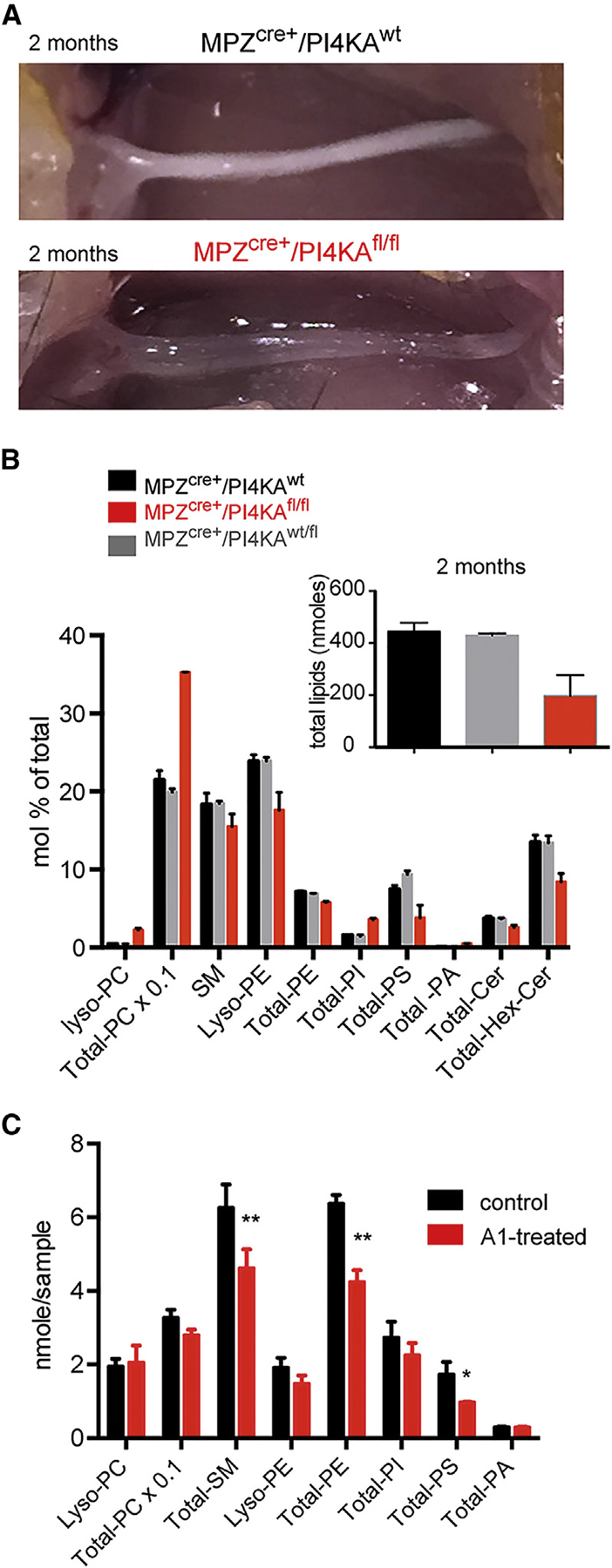
(A) Mutant sciatic nerves of P60 mice are more transparent than those of controls.
(B) Lipidomic analysis of mutant and control nerves shows a disproportional reduction in the major lipid components of the myelin sheath, such as phosphatidylserine (PS), phosphatidylethanolamine (PE), and sphingomyelin (SM) hexose-ceramide (Hex-Cer) in mutants (red) and a relative compensatory increase in phosphatidylcholine (PC) and phosphatidylinositol (PI) (note that mol % and not absolute amounts are plotted). The insert shows the decrease in the total amount of lipids in the mutant nerves from the same experiment. Means and SD of duplicate measurements are shown from a representative experiment that was repeated, showing the same changes.
(C) Lipidomic analysis of SW10 cells treated with 100 nM A1 for 24 hr shows a significant reduction in major lipid components of the myelin sheath, such as PS, PE, and SM. *p < 0.05 and **p < 0.001 versus control using t test; n = 2 for each group. Error bars indicate range. This analysis was repeated in a separate experiment, showing identical results.
DISCUSSION
In the present study, we found that inactivating PI4KA specifically in SCs causes severe myelination defects with pathologies shared by many demyelination diseases. Notably, SCs were not eliminated in the peripheral nerves, although an increased number of apoptotic cells suggested their major functional impairment. Accordingly, we found that the ability of these nerves to myelinate was severely compromised. A critical step in myelination is the generation of a new layer of myelin sheath by a protruding SC membrane that pries between the axon and the already existing myelin (see cartoon in Figure 3A). This process bears several similarities to the PM polarization observed in migrating cells, which requires PI3K activation (presumably in the leading edge) and a concerted actin polymerization-depolymerization sequence. To gain a mechanistic insight in the possible mechanism by which myelination is impaired, we considered three major angles: first, we tested the PI3K angle because of documented roles of PI(3,4,5)P3 (Goebbels et al., 2010) and its downstream target mTOR (Kearns et al., 2015; Mathews and Appel, 2016; Norrmén et al., 2014) in myelination. PI4KA inhibition could, in theory, affect PI(4,5)P2 levels and hence PI(3,4,5)P3 signaling. However, even after prolonged pharmacological inhibition of PI4KA in SCs, PI(4,5)P2 levels were maintained and the PI3K-Akt-mTOR-p70S6K signaling axis in response to serum stimulation was unchanged. This was consistent with previous cellular studies, which showed that PI(4,5)P2 levels did not decrease in cultured cells when PI4KA was inhibited (Bojjireddy et al., 2014) or inactivated (Nakatsu et al., 2012). Despite the unaltered PI3K signaling found in SC after PI4KA inhibition, we would be cautious to discount a contribution from an altered PI3K signaling to the myelination phenotype of the mutant mice. PI3K signaling is activated in a number of ways during myelination through secreted factors as well as integrin engagement at cell-cell contacts (Heller et al., 2014). These highly localized changes could have been impaired in the mutant nerves without being detected in cultured cells. Similarly, mTORC1 plays a different role in the early and late stages of myelination (Figlia et al., 2017), for which the cell culture model cannot be instructive. Analysis of these processes with temporal and spatial details in the intact nerve would require further studies and tools that are not available at this time.
PI4KA-deleted MEF cells showed greatly enhanced cortical actin on the expense of stress fibers, and similar changes were observed in SW10 cells after pharmacological inhibition of PI4KA. Enhancement of cortical actin and lack of stress fibers was also described in MEF cells after PI4KA inactivation in a previous study (Nakatsu et al., 2012). These actin changes could contribute to the migration defect of SC after the PI4KA inhibition. Phalloidin staining of sciatic nerves of mutant mice showed broken down actin organization and greatly enhanced staining. This, together with the data in cultured cells, has indicated that the balance of actin polymerization to depolymerization was affected by the loss of PI4KA activity. Recent studies showed that disassembly of actin is critical for proper myelination (Nawaz et al., 2015; Zuchero et al., 2015). It is notable, however, that actin-polymerization has not been linked to PI4KA activity, and further studies are in progress to explore this angle of PI4P biology in more detail as well as to define its role in the myelination process.
Lastly, a third angle of the myelination defect was explored based on most recent studies that have linked PS metabolism and transport to PI4P production in the PM (Chung et al., 2015; Maeda et al., 2013; Moser von Filseck et al., 2015; Sohn et al., 2016). PS is the main precursor of PE, and these two lipids are major components of the myelin. Lipidomic analysis of the nerves showed that, even beyond the greatly reduced total lipid content of the nerve, PS, PE, lyso-PE, and SM were disproportionally more reduced. Similar lipid changes were found in SCs treated with the PI4KA inhibitor for 24 hr and in HEK293 cells treated the same way (Sohn et al., 2016). These results pointed to a defect in PS metabolism that most likely contributes to the myelination pathology observed in the mutant animals.
In summary, the present study demonstrated a critical role of PI4KA in myelination using a mouse model with genetic inactivation of the kinase selectively in SCs. The myelination defect was not caused by death of SC cells, but changes in actin-polymerization and impaired PS, PE, and SM metabolism were identified as possible contributors to the myelination defect. These studies highlight a critical role of PI4KA in the proper maintenance and definition of the PM, which is especially important during myelination when large PM expansion takes place. The critical role of PI4KA in creating the proper membrane lipid environment for the replication of selected RNA viruses may reflect a broader role of the enzyme in lipid biogenesis and distribution in specific membranes. It is thought provoking that PI4KA may represent a link between certain viral infections and defective myelin formation and/or demyelination.
EXPERIMENTAL PROCEDURES
Generation of Mice
All the animal experiments were performed in compliance with animal protocols approved by the NICHD ACUC Committee. Conditional knockout mice of the Pi4ka gene (Bojjireddy et al., 2014) and B6N.FVB-Tg(Mpz-cre) 26Mes/J transgenic mice (Feltri et al., 1999), from Jackson Laboratories, were bred in order to inactivate PI4KA in SCs. The mice were genotyped, and MPZCre/+/PI4KAfl/fl was referred to as “mutants” or “fl/fl,” whereas MPZCre/+/PI4KAfl/wt and MPZCre/+/PI4Kwt/wt, served as “controls” or “wild-type (WT)”. The MPZ-Cre transgene and the floxed pi4ka allele were also cross-bred into Gt(ROSA)26Storm4(ACTB-tdTomato-EGFP) mice (obtained from Jackson Laboratories; stock no. 007576; Muzumdar et al., 2007), which allows to turn the original tdTomato (red) cells into GFP (green) once the Mpz-cre gets activated. Both male and female mice were used as they showed no apparent phenotypic difference.
MEF Cell Isolation and Immortalization
MEF cells were prepared from 13-day-old (post coitum [p.c.]) embryos from pregnant mice homozygous for the floxed allele of PI4KA and expressing the Cre from the Rosa26 locus under a tamoxifen-inducible promoter (Bojjireddy et al., 2014) as detailed under Supplemental Experimental Procedures.
Gait Analysis
The gait phenotype was analyzed using the TreadScan instrument from Clever Sys (Beare et al., 2009) and the software provided by the manufacturer. Mice were studied at two months of age (n = 5 per genotype), and one-way ANOVA analysis was performed followed by the Tukey’s multiple comparisons test.
Morphometric Analysis of EM Micrographs
Digitalized electron micrographs of semi-thin sciatic nerve cross sections were analyzed for axon diameter and g-ratio. Inner and outer diameters of the myelin sheath were measured from fibers randomly selected from three mice per group by using the GRatio software. The plug-in of this software, available online (http://gratio.efil.de), was implemented for ImageJ (https://imagej.nih.gov/ij), allowing semi-automated analysis.
Cell Culture
Cell studies were carried out in a mouse SC line: SW10 (ATCC CRL2766), which was cultured and maintained in DMEM supplemented with 10% FBS (HyClone) and 1% Pen Strep (Gibco). Transfection of plasmids was performed with Lipofectamine LTX with Plus Reagent (MA, USA) following the manufacturer’s instructions.
Live Cell Imaging by Confocal Microscopy
After 24 hr of transfection on glass-bottom dishes (In Vitro Scientific; cat no. D29-20-1-N), cells were washed and incubated in a modified Krebs-Ringer solution, containing 120 mM NaCl, 4.7 mM KCl, 2 mM CaCl2, 0.7 mM MgSO4, 10 mM glucose, and 10 mM Na-HEPES (pH 7.4). Confocal images were taken with a Zeiss LSM 710 laser confocal microscope (Zeiss, Peabody, MA).
Cell Migration Assay
SW10 cells were plated at 50,000 cells/insert on the top compartment of Boyden chambers (CytoSelect 24-Well Cell Migration Assay; 8 mm; Colorimetric Format from Cell Biolabs, San Diego, CA) in DMEM-high glucose w/o serum and allowed to migrate toward the same medium complemented with 10% fetal bovine serum (FBS) placed in the lower compartment for 24 hr in the presence or absence of A1 (100 nM). Migratory cells on the bottom of the membrane were stained with the “staining solution,” photographed, and quantified by spectrophotometry after elution by the “extraction solution” as provided by the manufacturer.
Statistical Analysis
Most of the analyses were carried out using the two-tailed Student’s t test. When comparisons were performed between more than two groups, one-way ANOVA analysis was performed followed by the Tukey’s multiple comparisons test. The values are expressed as mean ± SEM, and all statistical analyses were performed using GraphPad Prism, version 6.0.
Detailed description of nerve preparation for immunohistochemistry and EM analysis, western Blot and lipidomic analyses, as well as the method of nerve conductance measurements can be found under the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Schwann cell (SC)-specific inactivation of PI4KA causes myelination defects in mice
Inhibition of PI4KA did not decrease PI(4,5)P2 levels or PI(3,4,5)P3 responses in SCs
SC-specific PI4KA inactivation caused major decreases in PS, PE, and sphingomyelin
PI4KA inactivation caused actin disorganization in sciatic nerves and SCs
ACKNOWLEDGMENTS
This work was supported by the intramural research program of the Eunice Kennedy Shriver NICHD at the NIH. Confocal imaging and EM analysis were performed at the NICHD Microscopy and Imaging Core of the NIH, with the kind assistance of Dr. Vincent Schram and Louis (Chip) Dye, respectively. The advice and expertise of Dr. John Heuser in the EM studies is greatly appreciated. We would like to thank Dr. Pietro De Camilli (Yale University) for the PI4KA antibody and Dr. Peter Brophy (University of Edinburgh) for the Drp2 and periaxin antibodies. The lipid analyses were performed at the Kansas Lipidomics Research Center Analytical Laboratory. Instrument acquisition and lipidomics method development was supported by National Science Foundation (EPS 0236913, MCB 1413036, MCB 0920663, DBI 0521587, and DBI1228622), Kansas Technology Enterprise Corporation, K-IDeA Networks of Biomedical Research Excellence (INBRE) of NIH (P20GM103418), and Kansas State University. The lipidomic analysis of PIPs was done by ATK-Analytics Innovation and Discovery (North Bend, WA).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and one figure and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.05.019.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Altan-Bonnet N, and Balla T (2012). Phosphatidylinositol 4-kinases: hostages harnessed to build panviral replication platforms. Trends Biochem. Sci 37, 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, and Emr SD (2002). Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2, 593–605. [DOI] [PubMed] [Google Scholar]
- Azzedine H, Bolino A, Taıëb T, Birouk N, Di Duca M, Bouhouche A, Benamou S, Mrabet A, Hammadouche T, Chkili T, et al. (2003). Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease associated with early-onset glaucoma. Am. J. Hum. Genet 72, 1141–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T (2013). Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev 93, 1019–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin JM, Wu X, Christiano R, Oh MS, Schauder CM, Gazzerro E, Messa M, Baldassari S, Assereto S, Biancheri R, et al. (2016). The leuko-dystrophy protein FAM126A (hyccin) regulates PtdIns(4)P synthesis at the plasma membrane. Nat. Cell Biol 18, 132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare JE, Morehouse JR, DeVries WH, Enzmann GU, Burke DA, Magnuson DS, and Whittemore SR (2009). Gait analysis in normal and spinal contused mice using the TreadScan system. J. Neurotrauma 26, 2045–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanik LP, Sleigh JN, Tian C, Samuels ME, Bedard K, Seburn KL, and Burgess RW (2013). Loss of the E3 ubiquitin ligase LRSAM1 sensitizes peripheral axons to degeneration in a mouse model of Charcot-Marie-Tooth disease. Dis. Model. Mech 6, 780–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojjireddy N, Botyanszki J, Hammond G, Creech D, Peterson R, Kemp DC, Snead M, Brown R, Morrison A, Wilson S, et al. (2014). Pharmacological and genetic targeting of PI4KA reveals its important role in maintaining plasma membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphonate levels. J. Biol. Chem 289, 6120–6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura E, and Nencka R (2015). Phosphatidylinositol 4-kinases: function, structure, and inhibition. Exp. Cell Res 337, 136–145. [DOI] [PubMed] [Google Scholar]
- Cartoni R, Arnaud E, Médard JJ, Poirot O, Courvoisier DS, Chrast R, and Martinou JC (2010). Expression of mitofusin 2(R94Q) in a transgenic mouse leads to Charcot-Marie-Tooth neuropathy type 2A. Brain 133, 1460–1469. [DOI] [PubMed] [Google Scholar]
- Chen HC (2005). Boyden chamber assay. Methods Mol. Biol 294, 15–22. [DOI] [PubMed] [Google Scholar]
- Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F, and De Camilli P (2015). INTRACELLULAR TRANSPORT. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science 349, 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichberg J (2002). Myelin P0: new knowledge and new roles. Neurochem. Res 27, 1331–1340. [DOI] [PubMed] [Google Scholar]
- Feltri ML, D’Antonio M, Previtali S, Fasolini M, Messing A, and Wrabetz L (1999). P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann. N Y Acad. Sci 883, 116–123. [PubMed] [Google Scholar]
- Figlia G, Norrmén C, Pereira JA, Gerber D, and Suter U (2017). Dual function of the PI3K-Akt-mTORC1 axis in myelination of the peripheral nervous system. eLife 6, e29241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmes R, Houcine A, van Vliet AR, Agostinis P, Jackson CL, and Giordano F (2016). ORP5/ORP8 localize to endoplasmic reticulum-mitochondria contacts and are involved in mitochondrial function. EMBO Rep. 17, 800–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebbels S, Oltrogge JH, Kemper R, Heilmann I, Bormuth I, Wolfer S, Wichert SP, Möbius W, Liu X, Lappe-Siefke C, et al. (2010). Elevated phosphatidylinositol 3,4,5-trisphosphate in glia triggers cell-autonomous membrane wrapping and myelination. J. Neurosci 30, 8953–8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebbels S, Wieser GL, Pieper A, Spitzer S, Weege B, Yan K, Edgar JM, Yagensky O, Wichert SP, Agarwal A, et al. (2017). A neuronal PI(3,4,5)P3-dependent program of oligodendrocyte precursor recruitment and myelination. Nat. Neurosci 20, 10–15. [DOI] [PubMed] [Google Scholar]
- Hammond GR, Machner MP, and Balla T (2014). A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol 205, 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller BA, Ghidinelli M, Voelkl J, Einheber S, Smith R, Grund E, Morahan G, Chandler D, Kalaydjieva L, Giancotti F, et al. (2014). Functionally distinct PI 3-kinase pathways regulate myelination in the peripheral nervous system. J. Cell Biol 204, 1219–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert AL, and Monk KR (2017). Advances in myelinating glial cell development. Curr. Opin. Neurobiol 42, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlden H, King RH, Wood NW, Thomas PK, and Reilly MM (2001). Mutations in the 50 region of the myotubularin-related protein 2 (MTMR2) gene in autosomal recessive hereditary neuropathy with focally folded myelin. Brain 124, 907–915. [DOI] [PubMed] [Google Scholar]
- Kearns CA, Ravanelli AM, Cooper K, and Appel B (2015). Fbxw7 limits myelination by inhibiting mTOR signaling. J. Neurosci 35, 14861–14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Taylor GS, Torgersen KM, and Dixon JE (2002). Myotubularin and MTMR2, phosphatidylinositol 3-phosphatases mutated in myotubular myopathy and type 4B Charcot-Marie-Tooth disease. J. Biol. Chem 277, 4526–4531. [DOI] [PubMed] [Google Scholar]
- Maeda K, Anand K, Chiapparino A, Kumar A, Poletto M, Kaksonen M, and Gavin AC (2013). Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature 501, 257–261. [DOI] [PubMed] [Google Scholar]
- Mathews ES, and Appel B (2016). Cholesterol biosynthesis supports myelin gene expression and axon ensheathment through modulation of P13K/Akt/ mTor signaling. J. Neurosci 36, 7628–7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Torii T, Eguchi T, Nakamura K, Tanoue A, and Yamauchi J (2014). Hypomyelinating leukodystrophy-associated missense mutant of FAM126A/hyccin/DRCTNNB1A aggregates in the endoplasmic reticulum.J. Clin. Neurosci 21, 1033–1039. [DOI] [PubMed] [Google Scholar]
- Moser von Filseck J, Čopič A, Delfosse V, Vanni S, Jackson CL, Bourguet W, and Drin G (2015). INTRACELLULAR TRANSPORT. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science 349, 432–436. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, Lee SY, Pirruc-cello M, Hao M, Ingolia NT, Wenk MR, and De Camilli P (2012). PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol 199, 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz S, Sánchez P, Schmitt S, Snaidero N, Mitkovski M, Velte C, Brückner BR, Alexopoulos I, Czopka T, Jung SY, et al. (2015). Actin filament turnover drives leading edge growth during myelin sheath formation in the central nervous system. Dev. Cell 34, 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrmén C, Figlia G, Lebrun-Julien F, Pereira JA, Trötzmüller M, Köfeler HC, Rantanen V, Wessig C, van Deijk AL, Smit AB, et al. (2014). mTORC1 controls PNS myelination along the mTORC1-RXRγ-SREBP-lipid biosynthesis axis in Schwann cells. Cell Rep. 9, 646–660. [DOI] [PubMed] [Google Scholar]
- Oh SS, Hayes JM, Sims-Robinson C, Sullivan KA, and Feldman EL (2010). The effects of anesthesia on measures of nerve conduction velocity in male C57Bl6/J mice. Neurosci. Lett 483, 127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnamenta AT, Howard MF, Wisniewski E, Popitsch N, Knight SJ, Keays DA, Quaghebeur G, Cox H, Cox P, Balla T, et al. (2015). Germline recessive mutations in PI4KA are associated with perisylvian polymicrogyria, cerebellar hypoplasia and arthrogryposis. Hum. Mol. Genet 24, 3732–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezajooi K, Pavlides M, Winterbottom J, Stallcup WB, Hamlyn PJ, Lieberman AR, and Anderson PN (2004). NG2 proteoglycan expression in the peripheral nervous system: upregulation following injury and comparison with CNS lesions. Mol. Cell. Neurosci 25, 572–584. [DOI] [PubMed] [Google Scholar]
- Sohn M, Ivanova P, Brown HA, Toth DJ, Varnai P, Kim YJ, and Balla T (2016). Lenz-Majewski mutations in PTDSS1 affect phosphatidylinositol 4-phosphate metabolism at ER-PM and ER-Golgi junctions. Proc. Natl. Acad. Sci. USA 113, 4314–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Oh K, Burgess J, Hipfner DR, and Brill JA (2014). PI4KIIIa is required for cortical integrity and cell polarity during Drosophila oogenesis. J. Cell Sci 127, 954–966. [DOI] [PubMed] [Google Scholar]
- Traverso M, Assereto S, Gazzerro E, Savasta S, Abdalla EM, Rossi A, Baldassari S, Fruscione F, Ruffinazzi G, Fassad MR, et al. (2013). Novel FAM126A mutations in hypomyelination and congenital cataract disease. Biochem. Biophys. Res. Commun 439, 369–372. [DOI] [PubMed] [Google Scholar]
- Wooley CM, Sher RB, Kale A, Frankel WN, Cox GA, and Seburn KL (2005). Gait analysis detects early changes in transgenic SOD1(G93A) mice. Muscle Nerve 32, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia RH, Yosef N, and Ubogu EE (2010). Dorsal caudal tail and sciatic motor nerve conduction studies in adult mice: technical aspects and normative data. Muscle Nerve 41, 850–856. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Ohya Y, Goebl M, Nakano A, and Anraku Y (1994). A novel gene, STT4, encodes a phosphatidylinositol 4-kinase in the PKC1 protein kinase pathway of Saccharomyces cerevisiae. J. Biol. Chem 269, 1166–1172. [PubMed] [Google Scholar]
- Zuchero JB, Fu MM, Sloan SA, Ibrahim A, Olson A, Zaremba A, Dugas JC, Wienbar S, Caprariello AV, Kantor C, et al. (2015). CNS myelin wrapping is driven by actin disassembly. Dev. Cell 34, 152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


