Abstract
Background
Tuberculosis is a leading cause of death worldwide. BCG is an effective vaccine, but not widely used in many parts of the world due to a variety of issues. Mycobacterium vaccae (M. vaccae) is another vaccine used in human subjects to prevent tuberculosis. In the current study, we investigated the potential mechanisms of M. vaccae vaccination by determining differentially expressed genes in mice infected with M. tuberculosis before and after M. vaccae vaccination.
Methods
Three days after exposure to M. tuberculosis H37Rv strain (5 × 105 CFU), adult BALB/c mice randomly received either M. vaccae vaccine (22.5 μg) or vehicle via intramuscular injection (n = 8). Booster immunization was conducted 14 and 28 days after the primary immunization. Differentially expressed genes were identified by microarray followed by standard bioinformatics analysis.
Results
M. vaccae vaccination provided protection against M. tuberculosis infection (most prominent in the lungs). We identified 2326 upregulated and 2221 downregulated genes in vaccinated mice. These changes could be mapped to a total of 123 signaling pathways (68 upregulated and 55 downregulated). Further analysis pinpointed to the MyD88-dependent TLR signaling pathway and PI3K-Akt signaling pathway as most likely to be functional.
Conclusions
M. vaccae vaccine provided good protection in mice against M. tuberculosis infection, via a highly complex set of molecular changes. Our findings may provide clue to guide development of more effective vaccine against tuberculosis.
Keywords: Mycobacterium tuberculosis, Immunotherapeutic effect, Immunotherapy, Vaccae vaccine, Differentially expressed genes, Signaling pathway
Background
Since the discovery of Mycobacterium tuberculosis by Robert Koch over a century ago [1], human beings have made significant achievements in the fight against tuberculosis (TB). However, with the increase of multidrug-resistant (MDR) strains, human immunodeficiency virus (HIV) co-infection, and lack of effective TB vaccines, TB remains a major threat to human health [2].
Bacillus Calmette–Guérin (BCG), the first vaccine used against TB, is prepared from a strain of the attenuated live Mycobacterium bovis. A major limitation of BCG is the variable efficacy across ethnicity and population [2–4]. Vaccae™ vaccine is one of the most promising vaccines against TB. It is a non-cell Mycobacterium vaccae vaccine produced by Anhui Zhifei Longcom [5]. Our previous study suggested that it played an important role in improving immunity, promoting phagocytosis, regulating bidirectional immunoreaction, and reducing pathological damage [2]. At present, this vaccine has been given a Chinese new drug certificate and approved by the China Food and Drug Administration (CFDA) for the adjuvant treatment of TB. Currently, a large double-blind Phase III trial has been completed to evaluate the efficacy and safety of the Vaccae™ vaccine in 10,000 cases whose skin tests of PPD (purified protein derivative) were strongly positive in Guangxi province in China [6], and the results have not yet been published.
M. vaccae is a nonpathogenic species of the Mycobacteriaceae family and belongs to the same genus as M. tuberculosis. This bacterium contains many protective antigens with immunomodulating effects [2, 7]. Previous studies in animal models have demonstrated that M. vaccae vaccine had a good immunotherapeutic effect by stimulating T lymphocytes producing high-level cytokines such as interferon-gamma (IFN-γ) [8], interleukin 12 (IL-12) [9], IL-4 delta 2 [10], and tumor necrosis factor-alpha (TNF-α) [11]. Furthermore, this vaccine also has been used as an immunotherapeutic adjunct to treat TB [12–14], MDR-TB [15], surgery-elicited neuroinflammation and cognitive dysfunction [16], metastatic malignant melanoma [17], and neuroimmune processes [18].
Although the potential mechanisms of M. vaccae vaccine immunotherapy have been studied from the immunological and proteomic levels [19–21], the molecular mechanism of the immunotherapeutic effect of this vaccine is still unclear. Herein, we assessed the immunotherapeutic effect of the M. vaccae vaccine in mouse animal model and identified the differential expression (DE) genes of mice before and after M. vaccae vaccine treatment for the first time by using DNA (deoxyribonucleic acid) microarray. Based on these data, we hope to identify possible target molecules and signaling pathways of M. vaccae vaccine, which will give a new perspective for the molecular mechanism of its immunotherapy.
Methods
Mice and ethics statement
Female BALB/c mice (6–8 weeks of age) were purchased from the Institute of Military Medicine, Academy of Military Sciences of Chinese PLA (People’s Liberation Army) (Beijing, China). Experimental protocol was approved by the Animal Ethical Committee of the 8th Medical Center of Chinese PLA General Hospital, and conducted in compliance with the Experimental Animal Regulation Ordinances of the China National Science and Technology Commission.
Mycobacterial strains and M. vaccae vaccine
Mycobacterium tuberculosis (H37Rv strain) was cultured and purified as previously described [22, 23]. M. vaccae vaccine (Vaccae™) was purchased from Anhui Zhifei Longcom Co., Ltd. (Anhui, China).
Immunization and challenge
General experimental design is shown in Fig. 1. The schedule of immunization and challenge is shown in Fig. 2. Mice received 5 × 105 colony formation units (CFUs) of M. tuberculosis H37Rv strain via the caudal vein. Three days later, mice were randomly divided to receive intramuscular injection of either M. vaccae vaccine (22.5 μg in 100-μl distilled water) or vehicle (n = 8). Booster immunization was conducted 14 and 28 days after the primary immunization.
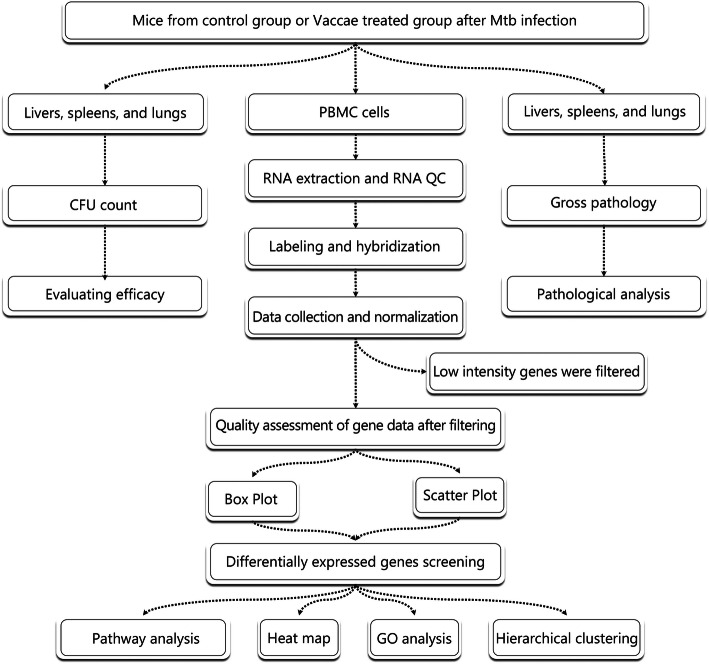
Fig. 1.
The flow chart of all experiments. Eight mice of M. vaccae group or control group were challenged with M. tuberculosis H37Rv strain. After 3 times immunization, mice were sacrificed and their lungs, livers, and spleens were collected to efficacy evaluation and pathological observation. The PBMCs of 3 mice of each group were isolated to extract total RNA. Hierarchical Clustering was performed to show the distinguishable gene expression profiling between samples. DE genes with statistical significance were identified through Volcano Plot filtering. Finally, pathway analysis and GO Analysis were applied to determine the roles of these DE genes played in these biological pathways or GO terms. GO, Gene Ontology; PBMCs, peripheral blood mononuclear cells; CFUs, colony formation units
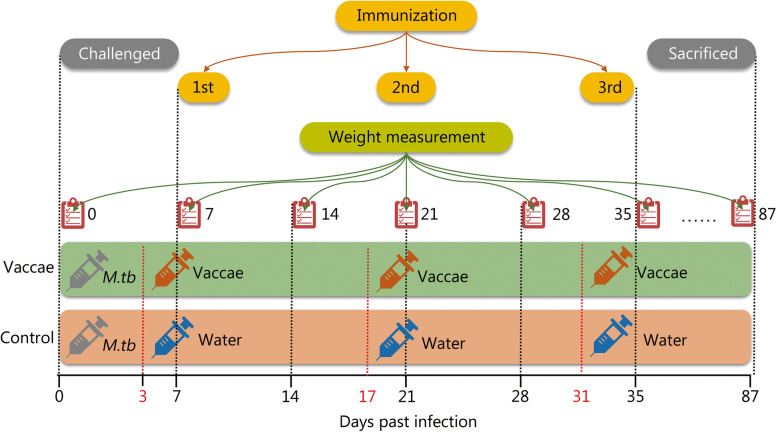
Fig. 2.
The schedule of immunotherapy evaluation. Each mouse in the control group (showed as an orange belt) and M. vaccae group (showed as a green belt) was challenged with M. tuberculosis H37Rv strain at day 0. Three days after challenge, mice were immunized intramuscularly 3 times at 2-week intervals with water (orange belt) and M. vaccae vaccine (green belt), respectively. Eighty-seven days after challenge, each mouse was sacrificed and their organs were collected for protective evaluation and histopathological analysis. Additionally, the weight of each mouse was measured per week (showed as a red notebook)
Infection severity assessment
Mouse body weight was measured once per week. Eight weeks after the last immunization, the mice per group were killed and the lungs, liver, and spleen were collected for gross pathological observation, histopathological examination, and CFU counting. Firstly, the organ coefficients were evaluated by the ratio of organ weight to body weight, the average areas of lesions in the liver, the number of the tubercular nodules in the lung, and the size of spleen were observed following the standards listed in Table 1. The spleen and the left lobe of lung were homogenized in 3-ml saline, serially diluted (10-fold in each step), inoculated in duplicate on Lowenstein-Jensen medium plate (100 μl) and cultured at 37 °C for 4 weeks. Colonies on the medium were counted and the results are showed as CFUs per organ. The right lung was fixed in 10% (vol/vol) formalin overnight and embedded in paraffin. Sections (3 μm) thickness were stained with hematoxylin and eosin (H&E) for histopathological examination as previously described [22–28].
Table 1.
The standard of identifying gross pathological lesion indexes of organs by pathological observation
| Organs | Lesion indexes | |||
|---|---|---|---|---|
| – | 1+ | 2+ | 3+ | |
| Lung | Without TB nodules and caseous necrosis | The number of TB nodules ≤10 or the area of caseous necrosis up to 20% | The number of TB nodules ≥10 or the area of caseous necrosis up to 40% | The number of TB nodules ≥20 or the area of caseous necrosis up to 40% |
| Liver | Normal size | Slight swell a | Moderate swell b | Severe swell c |
| Spleen | Normal size | Slight swell a | Moderate swell b | Severe swell c |
Note: Relative to normal liver and spleen, a swell was less than 20%; b swell was more than 20% and less than 40%; c swell was more than 40%
PBMCs isolation and total RNA extraction
On days 87 after challenge, 3 mice of each group were sacrificed. PBMCs (peripheral blood mononuclear cells) were prepared using a Mouse PBMCs Isolation Kit (TBDscience, Tianjin, China). Total RNA was extracted using a kit from Solarbio Life Science (Beijing, China). The integrity of RNA was assessed by electrophoresis on a denaturing agarose gel. Sharp 28S and 18S rRNA bands at a ratio of 2:1 are used as the hallmark for intact RNA.
Sample RNA purity and concentration
The NanoDrop ND-1000 was used to measure RNA concentration (OD260), protein contamination (ratio of OD260/OD280) and organic compound impureness (ratio OD260/OD230). The OD260/OD280 ratio should be > 1.8.
DNA microarray
DNA microarray experiment was conducted using a Mouse 4x44K Gene Expression Array (Agilent) with 39,000+ mouse genes and transcripts, all with public domain annotations.
RNA labeling and array hybridization
Sample labeling and array hybridization were conducted according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology). Briefly, total RNA from each sample was amplified and labeled with Cy3-UTP. Labeled cRNAs were purified by RNeasy Mini Kit (Qiagen), and NanoDrop ND-1000 was used to measure the concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA). One microgram of each labeled cRNA was fragmented by adding 2.2-μl 25 × fragmentation buffer and 11-μl 10 × blocking agents, heated at 60 °C for 30 min, and diluted by adding 55- μl 2 × GE hybridization buffer. Then, 100 μl of hybridization solution was added into the gasket slide and assembled to the gene expression microarray slide. The slides were incubated for 17 h at 65 °C in an Agilent Hybridization Oven. The hybridized arrays were washed, fixed and scanned using the Agilent DNA Microarray Scanner (part number G2505C).
Data analysis
Microarray images were analyzed using Agilent Feature Extraction software (version 11.0.1.1). Quantile normalization and subsequent data processing were performed using GeneSpring GX v11.5.1 software package (Agilent Technologies, USA). DE genes were identified through volcano plot filtering. Hierarchical clustering was performed using the Agilent GeneSpring GX software (version 11.5.1). GO analysis and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis were performed using a standard enrichment computation method.
Statistical analysis
Statistical analyses were conducted using SAS (version 9.1, SAS Institute, Cary, NC). The sample size was estimated according to our previous studies [24–27]. The results of M. vaccae protective experiments, gross pathological observation, histopathological examination, and CFU count were compared with Student’s t-test or Wilcoxon Two-Sample test according to data normality and homogeneity of variances. Differential expression was defined as fold-change ≥2. P < 0.05 was considered statistically significant.
Results
Efficacy of the vaccine
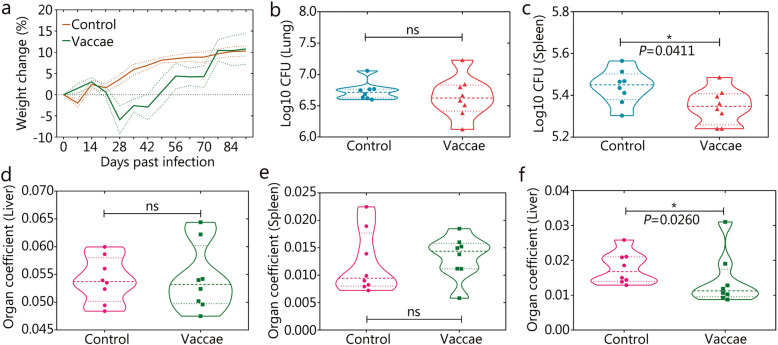
With the exception of temporary reduction in the first week, body weight in mice receiving the vehicle increased over the entire experimental period as expected (Fig. 3a). In mice receiving the M. vaccae vaccine, body weight started to decrease on day 14 day, reached a nadir on day 28, and then increased gradually back to the control level on day 77 (Fig. 3a).
Fig. 3.
Immunotherapeutic efficacy of M. vaccae vaccine. After the challenge, the weight change of each mouse was measured weekly (a, the error bar is represented by dotted lines). Ninety-one days after challenge, all of the mice were killed and their left lobe of the lung b and spleen c was collected for CFU counting. Additionally, organ coefficient of the lung d, spleen e, or liver f was performed. All data are presented as means + S.E.M. (n = 8). Differences were considered statistically significant at P < 0.05. *, P < 0.05; ns. Not significant
In comparison to the control, there was a statistically non-significant trend for decreased CFUs in the lungs in the M. vaccae group (Fig. 3b). The CFUs in the spleen was lower in the M. vaccae group (P = 0.041 vs. control, Fig. 3c). In comparison to the control, mice in the M. vaccae group had similar organ coefficient of the liver (Fig. 3d) and spleen (Fig. 3e), but significantly lower organ coefficient of the lungs (P = 0.026, Fig. 3f).
Histopathological and gross pathological analyses
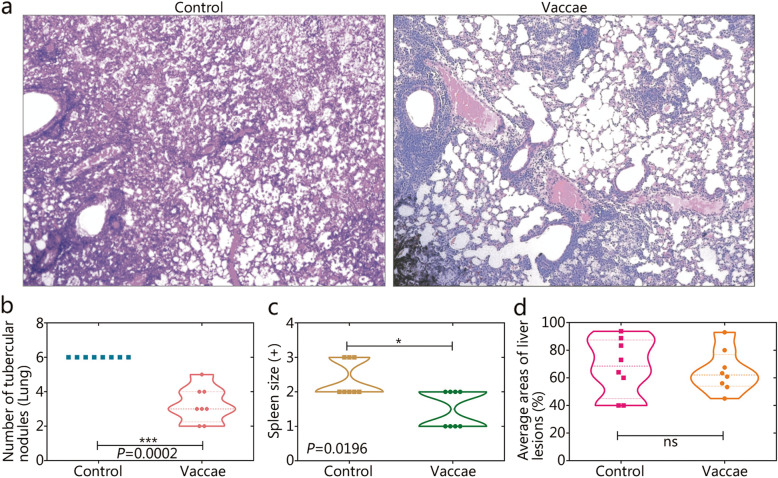
The structure of alveoli was damaged severely in the control group (Fig. 4a). Inflammatory cell infiltration of the lungs was apparent in the M. vaccae group, but the alveolar wall was intact, with no thickening. Gross pathological analysis showed fewer tubercular nodules in the lungs (P = 0.0002, Fig. 4b) and decreased spleen size (P = 0.0196, Fig. 4c) in the M. vaccae group. The average area of the lesions in the liver did not differ significantly between the two groups (Fig. 4d).
Fig. 4.
Gross pathology and histopatological analysis. The right lobe of lungs collected from the mice in the control group (a, left) and M. vaccae group (a, right) were used to undergo histopathological examination (H&E). The gross pathology of organs was also observed, including the number of the tubercular nodules in the lung b, spleen size average c, and areas of lesions in the liver d. Original magnification times: ×100. All data are presented as means + S.E.M. (n = 8). Differences were considered statistically significant at P < 0.05. *, P < 0.05; ***, P < 0.001; ns. Not significant
DE genes
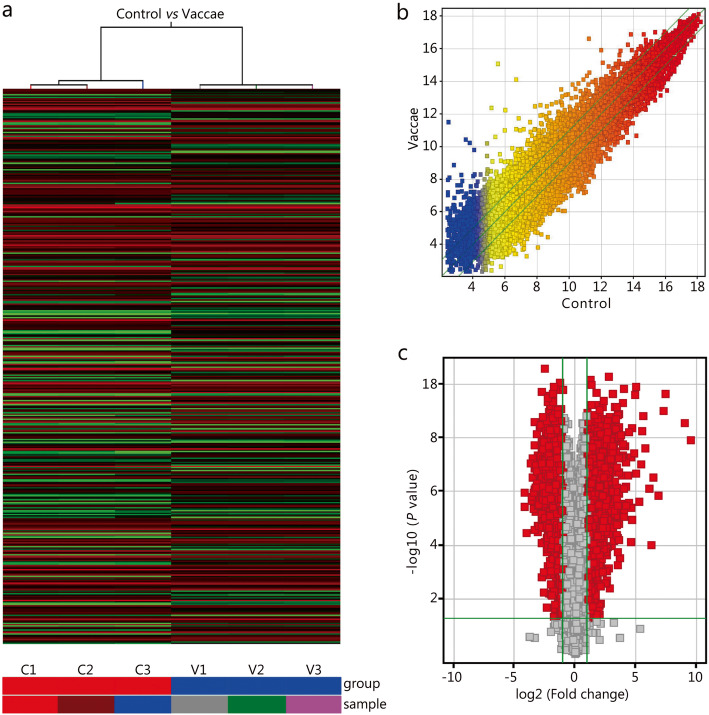
Agilent Mouse 4x44K Gene Expression Microarrays v2 was used to identify DE genes. The array image of each sample was obtained (Fig. S1), and the intensity data was extracted. After quantile normalization of the raw data, genes listed in Table S1 were chosen for DE gene screening. Differential gene expression is shown using a heat map, hierarchical cluster (Fig. 5a) and scatter plot (Fig. 5b). The DE genes were screened with volcano plot (Fig. 5c). We identified 2326 upregulated genes and 2221 downregulated genes in the M. vaccae group. The top 20 upregulated genes were Retnlg, Tiprl, Gyg, Ptgs2, Zfp281, Gbp2, Cxcl2, Ear6, Ighv1–77, Slpi, Azin1, S100a8, Ccrl2, Igj, Tnf, Prkcd, Marcksl1, Prss34, Cct6a, and Il1a (Table 2). The top 20 downregulated genes were Afp, Pcdhga9, Cdc42ep5, Hrsp12, Rnasek, Nprl3, 4932443I19Rik, Ly6g6c, Kdr, 2810416G20Rik, Tubb2a, Triqk, Slc6a16, Cxx1c, Fez2, 1810058I24Rik, Egfbp2, Efna5, Cd151, and 2210013O21Rik (Table 2). More detailed information (all DE genes) are shown in Table S2.
Fig. 5.
Heat map, hierarchical clustering presentation, scatter plot, and volcano plot of the expression profile of genes in control and M. vaccae groups. a. Heat map and hierarchical clustering showed the relationships among gene expression patterns of samples (n = 3). Red indicates high relative expression, and green indicates low relative expression. b. Scatter plot, the values of X and Y axes in the scatter plot are the normalized signal values of the samples (log2 scaled) or the averaged normalized signal values of the groups (log2 scaled). The green lines are Fold Change Lines (the default fold change value given is 2.0). The genes above the top green line and below the bottom green line indicated more than a 2-fold change of genes between two samples or groups (n = 3). c. DE genes with statistical significance were identified through the volcano plot, and the red diamonds represented DE genes with fold change ≥2.0, P ≤ 0.05 (n = 3). C1 – C3, sample 1 to sample 3 in control group; V1-V3, sample 1 to sample 3 in Vaccae group
Table 2.
Top 20 DE genes between control and M. vaccae groups
| Gene name | P-value fold change and regulation | GenBank accession | Relationship with TB | |||
|---|---|---|---|---|---|---|
| P-value a | FDR b | FCAbsolute c | Regulation | |||
| Retnlg | 5.2646E-11 | 3.00721E-07 | 6.762246 | Up | NM_181596 | Unknown |
| Tiprl | 6.7691E-11 | 3.00721E-07 | 2.3434467 | Up | NM_145513 | Unknown |
| Gyg | 1.0686E-10 | 3.00721E-07 | 2.590874 | Up | NM_013755 | Unknown |
| Ptgs2 | 1.1782E-10 | 3.00721E-07 | 33.16254 | Up | NM_011198 | Decreased transcription of PTGS2 was beneficial to the survival of M. tuberculosis |
| Zfp281 | 1.9767E-10 | 3.00721E-07 | 3.3773346 | Up | NM_177643 | Unknown |
| Gbp2 | 2.162E-10 | 3.00721E-07 | 8.493201 | Up | NM_010260 | One of prominent hubs in a highly active common core in TB |
| Cxcl2 | 2.2714E-10 | 3.00721E-07 | 176.0849 | Up | NM_009140 | Blocking of CXCL2 can significantly reduce the M. tuberculosis-induced IL-1β production |
| Ear6 | 2.4307E-10 | 3.00721E-07 | 30.299973 | Up | NM_053111 | Unknown |
| Ighv1–77 | 4.1282E-10 | 4.21673E-07 | 4.998427 | Up | AF045501 | Unknown |
| Slpi | 5.6974E-10 | 4.97498E-07 | 3.1604574 | Up | NM_011414 | Exposure of murine peritoneal macrophages to M. tuberculosis led to an increase in SLPI secretion accelerating both the phagocytosis and killing of the pathogen |
| Azin1 | 5.9447E-10 | 4.97498E-07 | 2.5270393 | Up | NM_018745 | Unknown |
| S100a8 | 7.0447E-10 | 4.97498E-07 | 9.958652 | Up | NM_013650 | A major pathologic role for S100A8/A9 proteins in decreasing lung tissue damage without impacting protective immunity against TB |
| Ccrl2 | 7.0963E-10 | 4.97498E-07 | 10.672567 | Up | NM_017466 | Unknown |
| Igj | 8.5416E-10 | 5.25022E-07 | 4.127778 | Up | NM_152839 | Unknown |
| Tnf | 9.694E-10 | 5.66345E-07 | 156.13841 | Up | NM_013693 | TNF-α has a prominent role in defense and pathological responses to TB and its production in TB patients was higher than that in the control group |
| Prkcd | 1.1807E-09 | 6.28255E-07 | 2.054293 | Up | NM_011103 | Unknown |
| Marcksl1 | 1.2759E-09 | 6.28255E-07 | 11.7962 | Up | NM_010807 | Unknown |
| Prss34 | 1.3143E-09 | 6.28255E-07 | 11.188594 | Up | NM_178372 | Unknown |
| Cct6a | 1.4218E-09 | 6.40172E-07 | 2.9756305 | Up | NM_009838 | Unknown |
| Il1a | 1.5183E-09 | 6.40172E-07 | 48.50243 | Up | NM_010554 | The expression of the IL1A gene was increased in both the TB-infected and the healthy cattle to M. bovis stimulation |
| Afp | 2.4759E-11 | 3.00721E-07 | 5.7038527 | Down | NM_007423 | Only a few literatures reported that AFP was normal in TB patients, but increased significantly in TB patients with hepatocellular carcinoma |
| Pcdhga9 | 8.4144E-11 | 3.00721E-07 | 2.3555577 | Down | NM_033592 | Unknown |
| Cdc42ep5 | 1.2007E-10 | 3.00721E-07 | 3.3431478 | Down | NM_021454 | Unknown |
| Hrsp12 | 1.5283E-10 | 3.00721E-07 | 2.9907024 | Down | NM_008287 | Unknown |
| Rnasek | 1.5763E-10 | 3.00721E-07 | 2.0300574 | Down | NM_173742 | Unknown |
| Nprl3 | 1.6254E-10 | 3.00721E-07 | 2.5463195 | Down | NM_001284359 | Unknown |
| 4932443I19Rik | 1.9509E-10 | 3.00721E-07 | 6.6056542 | Down | NM_001101519 | Unknown |
| Ly6g6c | 2.3228E-10 | 3.00721E-07 | 3.6154532 | Down | NM_023463 | Unknown |
| Kdr | 2.6218E-10 | 3.06341E-07 | 4.521906 | Down | NM_010612 | Unknown |
| 2810416G20Rik | 3.0664E-10 | 3.39439E-07 | 4.7047515 | Down | XM_003945668 | Unknown |
| Tubb2a | 4.3967E-10 | 4.21673E-07 | 3.6115813 | Down | NM_009450 | Unknown |
| Triqk | 4.4108E-10 | 4.21673E-07 | 4.918867 | Down | NM_173746 | Unknown |
| Slc6a16 | 6.2508E-10 | 4.97498E-07 | 4.1382565 | Down | XM_355900 | Unknown |
| Cxx1c | 8.1855E-10 | 5.25022E-07 | 3.0826645 | Down | NM_028375 | Unknown |
| Fez2 | 8.6019E-10 | 5.25022E-07 | 2.6090815 | Down | NM_001285940 | Unknown |
| 1810058I24Rik | 8.7371E-10 | 5.25022E-07 | 4.307958 | Down | NR_027875 | Unknown |
| Egfbp2 | 1.0983E-09 | 6.07878E-07 | 4.641702 | Down | NM_010115 | Unknown |
| Efna5 | 1.2336E-09 | 6.28255E-07 | 2.744601 | Down | NM_207654 | Unknown |
| Cd151 | 1.2898E-09 | 6.28255E-07 | 3.2171504 | Down | NM_009842 | CD9High classical monocytes expressed higher levels of tetraspanin CD151 compared to CD9Low classical monocytes |
| 2210013O21Rik | 1.4572E-09 | 6.40172E-07 | 3.4838674 | Down | NM_027327 | Unknown |
a, P-value calculated from t-test; b, FDR calculated from Benjamini Hochberg FDR; c, FCAbsolute, the absolute ratio (no log scale) of normalized intensities between two groups. Notes: mRNA with expression fold change > 2 and with FDR adjusted P-value < 0.05 was considered statistically significant. Here we show the expression fold change > 10
GO analysis
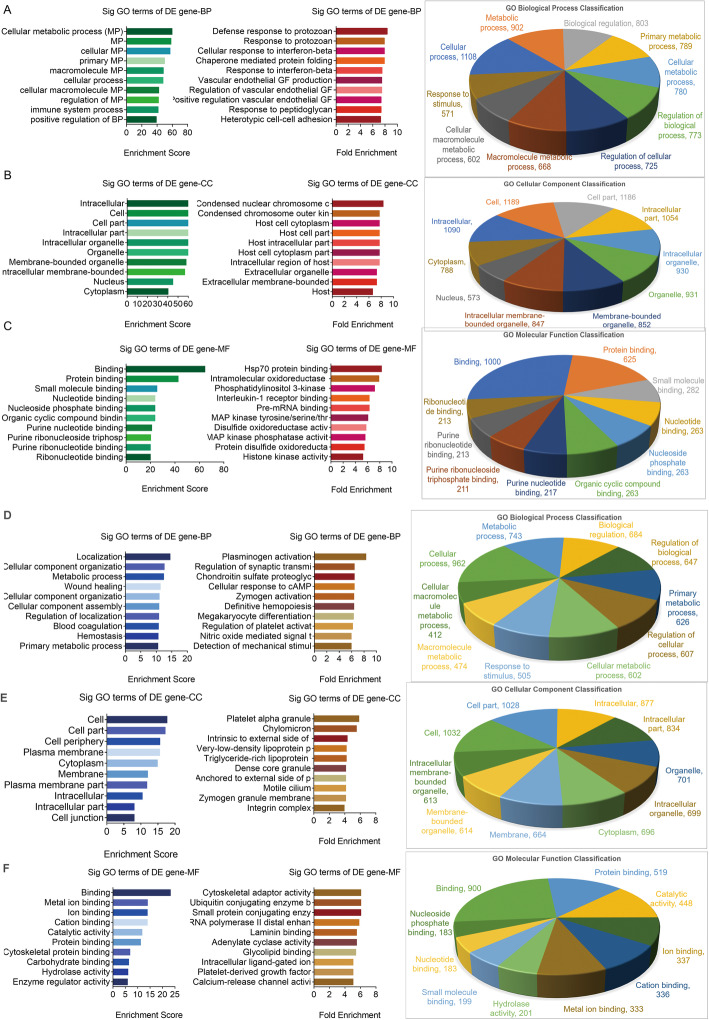
GO analysis showed that, in comparison with the control group, the upregulated genes involve 1672 terms in biological process (BP, P < 0.05, Table S3 BP sheet), 137 terms in cellular component (CC, P < 0.05, Table S3 CC sheet), and 231 terms in molecular function (MF, P < 0.05, Table S3 MF sheet). The downregulated genes involved 1080 terms in BP (P < 0.05, Table S4 BP sheet), 134 terms in CC (P < 0.05, Table S4 CC sheet), and 195 terms in MF (P < 0.05, Table S4 MF sheet).
The top 10 GO terms of the upregulated genes sorted by enrichment score (left lane in Fig. 6), fold enrichment (middle lane in Fig. 6), and classification (right lane in Fig. 6) in BP, CC, and MF are shown in Fig. 6a, Fig. 6b, and Fig. 6c, respectively. The top 10 GO terms of the downregulated genes are showed in Fig. 6d/E/F. Briefly, the upregulated genes in the M. vaccae group are mainly related to metabolic process, cellular metabolic process, primary metabolic process, intracellular, and binding. The downregulated genes are mainly associated with localization, cellular component organization, metabolic process, cell part, cell periphery, and binding.
Fig. 6.
GO analysis of DE genes between two groups. Significant GO terms of TOP 10 upregulated DE genes involved in biological process a, cellular component b, and molecular function c or that of TOP 10 downregulated DE genes involved in biological process d, cellular component e, and molecular function f were identified, respectively. Left lane, Enrichment Score; Middle lane, Fold Enrichment; Right lane, Classification. BP, biological process; CC, cellular component; MF, molecular function. The P-value denotes the significance of GO Term enrichment in the DE gene list. The less the P-value is, the more significant of the GO Term is (P ≤ 0.05 is recommended). DE, differential expression; GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function
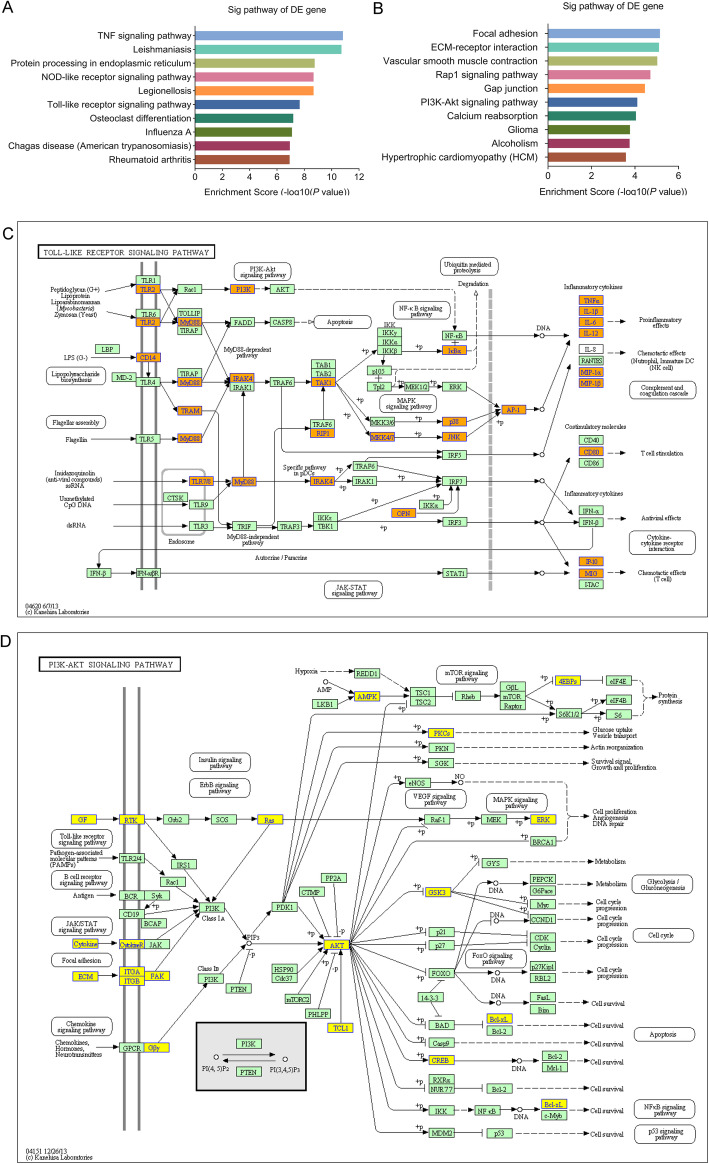
Pathway analysis
KEGG analysis showed that, in comparison with the control group, 68 pathways were unregulated in the M. vaccae group (Table S5); the top 10 were mmu04668- tumor necrosis factor (TNF) signaling pathway, mmu05140 Leishmaniasis, mmu04141 protein processing in endoplasmic reticulum, mmu04621 nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway, mmu05134 Legionellosis, mmu04620 Toll-like receptor (TLR) signaling pathway, mmu04380 osteoclast differentiation, mmu05164 Influenza A, mmu05142 Chagas disease (American trypanosomiasis), and mmu05323 rheumatoid arthritis (Fig. 7a). There were 55 down regulated pathways in the M. vaccae group (Table S6); the top 10 were mmu04510 focal adhesion, mmu04512 extracellular matrix (ECM)-receptor interaction, mmu04270 vascular smooth muscle contraction, mmu04015 Rap1 signaling pathway, mmu04540 gap junction, mmu04151 PI3K (phosphatidylinositol-4,5-bisphosphate 3-kinase)-Akt (protein kinase B) signaling pathway, mmu04961 endocrine and other factor-regulated calcium reabsorption, mmu05214-glioma, mmu05034-alcoholism, and mmu05410-hypertrophic cardiomyopathy (Fig. 7b). The upregulated and downregulated pathway mostly associated with M. vaccae vaccine was MyD88-dependent TLR signaling pathway (Fig. 7c, P = 2.193097 × 10− 8) and PI3K-Akt signaling pathway (Fig. 7d, P = 7.834627 × 10− 5), respectively.
Fig. 7.
Pathway bar plot explanation and pathway map explanation. The bar plot showed the top 10 enrichment score [−log10 (P-value)] value of the significantly upregulated pathways a and downregulated pathways b. The significantly upregulated signaling pathway c and downregulated pathway d mostly associated with M. vaccae vaccine treatment were selected to show here. Yellow marked nodes are associated with downregulated genes, orange marked nodes are associated with upregulated or only whole dataset genes, green nodes have no significance
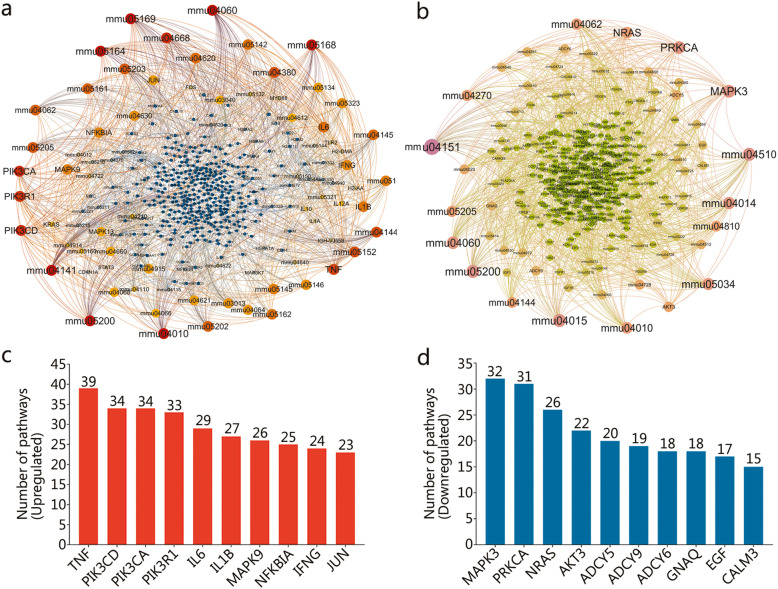
The relationship among DE genes associated with upregulated pathways (Fig. 8a) and downregulated pathways (Fig. 8b) were determined by using Gehpi software. Subsequently, we analyzed the number of upregulated or downregulated pathways involved in a DE gene. The top 10 DE genes associated with upregulated pathways were TNF, Pik3cd, Pik3ca, Pik3r1, Il6, Il1b, Mapk9, Nfkbia, Ifng, and Jun (Fig. 8c). The top 10 DE genes associated with downregulated pathways were Mapk3, Prkca, Nras, Akt3, Adcy5, Adcy9, Adcy6, Gnaq, Egf, and Calm3 (Fig. 8d).
Fig. 8.
Network map of DE genes associated with upregulated or downregulated pathways. The network map of DE genes associated with upregulated a or downregulated b pathways was made by Gehpi software. The pathways were showed as pathway ID such as mmu05168. The color and diameter of each circle represent the degree of enrichment of the genes or pathways, the larger diameter, and redder or pinker color indicate the more upregulated or downregulated pathways the gene involved. In addition, the top 10 genes of upregulated c or downregulated d pathways were also identified
Discussion
The current study showed that the M. vaccae vaccine could decrease the CFUs of M. tuberculosis in mice. Such effect was most robust in the spleen, and statistically significant in the lungs. The organ coefficient of the lungs was decreased. The vaccination attenuated the pulmonary lesion and splenomegaly. These results are generally consistent with the lower M. tuberculosis CFUs, pathological change index, and organ weight index in previous studies [20, 29], and indicated that M. vaccae vaccine had a significant immunotherapeutic effect on TB.
Previous studies suggested that the effects of M. vaccae vaccine immunotherapy mainly depend on enhanced recall IFN-γ responses [8], CD3+CD4+ T cells, IFN-γ+CD4+ T cells, natural killer (NK) cells, and reduced IL-4+CD4+ T cells [29]. However, a systematic review of clinical trials conducted suggested no benefit of M. vaccae vaccine immunotherapy [30]. One study showed that smooth type of M. vaccae could interfere with the production of helper T lymphocytes-1 (Th-1) cytokines, and rough type of M. vaccae could induce the production of Th-1 cytokines [31] by splenocytes, suggesting that the different colonial morphology (smooth type or rough type) of M. vaccae might affect the immunomodulatory effects of M. vaccae preparations. Such discrepancy could have contributed to varying results across different vaccines made with M. vaccae in clinical trials.
We speculated that there are significant differences in gene expression profiles before and after M. vaccae vaccine treatment, and identifying these changes could help to understand the regulatory mechanism of the M. vaccae vaccine. We identified 2326 upregulated genes and 2221 downregulated genes in M. vaccae group, suggesting that M. vaccae vaccine induce more complex and specific gene regulation activities in individuals infected with M. tuberculosis. The top 1 upregulated gene was Retnlg (also known as Xcp1, Fizz3, and Relmg), which encodes the resistin-like gamma protein (alternative names, RELM-γ or XCP1). This protein was first identified as a novel member of the resistin-like molecule/found in inflammatory zone (RELM/FIZZ) family in mice and rats [32]. A subsequent study showed marked increase of Retnlg expression in spontaneously hypertensive hyperlipidemic rats [33], suggesting that RELM-γ has cytokine-like effect and plays a role in promyelocytic differentiation [32, 34]. In addition to RELM-γ, several other upregulated genes identified in our study have been previously reported to be associated with TB. For example, decreased transcription of PTGS2 has been shown to confer a survival benefit to M. tuberculosis [35]. GBP2 is one of the prominent hubs in a highly active common core in TB [36]. Blocking CXCL2 could reduce the M. tuberculosis-induced IL-1β production [37, 38]. Exposure of murine peritoneal macrophages to M. tuberculosis increases SLPI secretion and accelerates both the phagocytosis and killing of the pathogen [39–41], possibly by interacting with S100A8/A9 proteins to decrease lung tissue damage without affecting protective immunity against TB [42]. TNF-α has a prominent role in defense and pathological responses to TB and its production in TB patients has been shown to be increased by the M. vaccae vaccine [43–45]. Expression of the IL1A gene is increased in both the TB-infected and the healthy cattle to M. bovis stimulation [46].
The top 1 downregulated gene was Afp encoding alpha-fetoprotein (AFP). AFP is a shuttle protein that transports nutrients to embryonic cells through receptor-mediated endocytosis and converts drugs into AFP-positive bone marrow-derived inhibitors in adults. Previous studies have implicated AFP in the regulation of cell growth, differentiation, apoptosis, angiogenesis, and immune regulation [47]. A few previous studies reported normal AFP in TB patients, but increased AFP in TB patients with hepatocellular carcinoma [48, 49]. Monocytes can undergo homotypic fusion to produce different types of multinucleated giant cells in response to M. tuberculosis infection. In comparison to CD9Low classical monocytes, CD9High classical monocytes expressed higher levels of tetraspanin CD151, but the role of these cells in immunity remains unknown [50]. Taken together, we identified a number of new downregulated genes in the current study, including Pcdhga9, Cdc42ep5, Hrsp12, Rnasek, Nprl3, 4932443I19Rik, Ly6g6c, Kdr, 2810416G20Rik, Tubb2a, Triqk, Slc6a16, Cxx1c, Fez2, 1810058I24Rik, Egfbp2, Efna5, and 2210013O21Rik. Whether these genes participate in the immune response needs to be investigated in the future.
The upregulated and downregulated genes in the current study are associated with 1672 and 1080 terms in the biological process, 137 or 134 terms in the cellular component, and 231 or 195 terms in the molecular function, indicating the importance of biological process in the regulatory mechanisms of M. vaccae vaccine. Interestingly, GO analysis demonstrated that the most significant GO term of upregulated genes in the biological process is the metabolic process. In contrast, the most significant GO term for downregulated genes in the biological process is localization. It is well known that the immune responses depend on energy. Maintaining adequate energy supply is the basis for immunocytes to attack M. tuberculosis. It has also been shown that M. tuberculosis can adhere to and taken up by alveolar epithelial cells [51, 52]. The interactions between M. tuberculosis and host molecules within the alveolar certainly play a key role in determining whether M. tuberculosis could successfully invade the host [53]. These findings suggested that M. vaccae vaccine activate more immunocytes to participate in the elimination of M. tuberculosis by enhancing metabolism, and antagonize the invasion of M. tuberculosis by downregulating the molecules involved in recognition, adhesion, and invasion.
KEGG pathway analysis in the current study identified 68 upregulated and 55 downregulated pathways by M. vaccae vaccination. The upregulated pathways most associated with M. vaccae vaccine treatment were TNF signaling pathway, NOD-like receptor signaling pathway, TLR signaling pathway, and mitogen-activated protein kinase (MAPK) signaling pathway. M. tuberculosis is primarily recognized by macrophages via TLR2/4 signaling pathways, but the TLR2 and TLR4 signal can be inhibited by the antigens secreted by bacteria [54], which makes it possible to inhibit autophagy, and allow the long-term presence of M. tuberculosis in macrophages [55]. After M. vaccae vaccination, the expression of TLR2 was significantly enhanced to induce upregulation of inflammatory cytokines (TNF-α, IL-6, IL-12, IL-18, and IL-1) and chemokines (CXCL, MCP-1) via MyD88-dependent TLR signaling pathway, NOD-like receptor signaling pathway, and subsequently activating two downstream pathways NF-κB and MAPK to accelerate the killing and elimination of M. tuberculosis [56–58]. MyD88 is one of the most extensively investigated adaptor proteins in the TLR signaling cascade, and plays a critical role in immune response to M. tuberculosis infection [59]. Our study determined that the expression of MyD88 is significantly upregulated in response to M. vaccae vaccine. However, MyD88-independent pathway also participates in the host defense against mycobacterial infection [57]. We speculate that M. vaccae vaccination could induce the transition of the TLR signaling pathway from MyD88-independent to MyD88-dependent.
Downregulated pathways associated with M. vaccae vaccination in the current study included focal adhesion, ECM-receptor interaction, Rap1 signaling pathway, and PI3K-Akt signaling pathway. Focal adhesions are integrin-containing, multi-protein structures that form mechanical links between intracellular actin bundles and the extracellular substrate in many cell types [60]. ECM is a highly dynamic structure that provides structural and biochemical support of surrounding cells [61, 62]. Both play a dominant role in the control of cell-cell and cell-matrix interactions by regulating the function of integrins and other adhesion molecules in various cell types. In addition, growth factor (GF) is a naturally occurring substance capable of stimulating cellular growth, proliferation, healing, and cellular differentiation [63]. In the present study, we found reduced expression of GF and ECM by the M. vaccae vaccine. Recognition of both molecules and their receptors on cell membrane could induce the activation of PI3K and FAK, thus triggering the downstream signaling events, including PI3K-Akt signaling pathway, Wnt signaling pathway, and Rap1 signaling pathway. These pathways have been implicated in macrophage invasion, M. tuberculosis survival, and impaired immune response [64, 65].
There are several limitations to this study. Firstly, the number of mice used to identify DE genes is relatively small (n = 3/group), and therefore must be considered preliminary. Secondly, the study was conducted in BALB/c mice; extrapolation to other animal species, and particularly human beings, must be cautious. Third, the changes induced by the M. vaccae vaccine were not compared to the BCG vaccine. Finally, the upregulated and downregulated signaling pathways were identified by bioinformatics based on microarray data; validation with more quantitative measures and at the protein levels is required.
Conclusions
M. vaccae vaccine produces fairly robust protection against M. tuberculosis. The vaccination resulted in 2326 upregulated and 2221 downregulated genes and 68 upregulated and 55 downregulated pathways. Enhanced release of pro-inflammatory factors via MyD88-dependent TLR signaling pathway might be a key component of the action. Accelerated apoptosis of host cells due to downregulated PI3K-Akt signaling pathway could be another important mechanism.
Supplementary information
Additional file 1: Figure S1. Array image of each sample. c1-c3, the serial number of mice in the control group; v1-v3, the serial number of mice in the M. vaccae group.
Additional file 2: Table S1. Raw and log2 value of the normalized intensity of each sample in the control group and the M. vaccae group.
Additional file 3: Table S2. Detail information of 2326 upregulated genes and 2221 downregulated genes.
Additional file 4: Table S3. GO analysis for upregulated DE genes in control and M. vaccae groups in terms of biological process (BP sheet), cellular component (CC sheet), and molecular function (MF sheet).
Additional file 5: Table S4. GO analysis for downregulated DE genes in control and M. vaccae groups in terms of biological process (BP sheet), cellular component (CC sheet), and molecular function (MF sheet).
Additional file 6: Table S5. The whole analysis results of upregulated pathways in control and M. vaccae groups.
Additional file 7: Table S6. The whole analysis results of downregulated pathways in control and M. vaccae groups.
Acknowledgments
We thank all funding agencies for supporting this study.
Abbreviations
- AFP
Alpha-fetoprotein
- BCG
Bacillus Calmette–Guérin
- BP
Biological process
- CC
Cellular component
- CFDA
China Food and Drug Administration
- CFUs
Colony formation units
- DE
Differential expression
- ECM
Extracellular matrix
- GF
Growth factor
- GO
Gene Ontology
- HIV
Human immunodeficiency virus
- IFN-γ
Interferon-gamma
- IL
Interleukin
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- M. tuberculosis
Mycobacterium tuberculosis
- M. vaccae
Mycobacterium vaccae
- MAPK
Mitogen-activated protein kinase
- MDR
Multidrug-resistant
- MF
Molecular function
- NK
Natural killer
- NOD
Nucleotide-binding oligomerization domain
- PBMCs
Peripheral blood mononuclear cells
- PI3K
Phosphatidylinositol-4,5-bisphosphate 3-kinase
- PLA
People’s Liberation Army
- PPD
Purified protein derivative
- RELM/FIZZ
Resistin-like molecule/found in inflammatory zone
- TB
Tuberculosis
- Th-1
Helper T lymphocytes-1
- TLR
Toll-like receptor
- TNF-α
Tumor necrosis factor-alpha
- WHO
World Health Organization
Authors’ contributions
WPG collected and analyzed the data and was a major contributor to the writing of the manuscript. YL and YBL analyzed the data and performed the experiments of mice infection and immunization. JXZ, LW, and JW performed the experiments of PBMCs isolation and total RNA extraction, YCS conducted mice immunization and challenge. XQW contributed to the study design, data analysis, reviewed the manuscript. All authors read and approved the final manuscript.
Authors’ information
Army Tuberculosis Prevention and Control Key Laboratory/Beijing Key Laboratory of New Techniques of Tuberculosis Diagnosis and Treatment, Institute for Tuberculosis Research, The 8th Medical Center of Chinese PLA General Hospital.
Funding
This study was supported by Grants from the National Natural Science Foundation of China (81801643), the National Key Program for Infectious Disease of China (2018ZX10731301–005), Beijing Municipal Science & Technology Commission (Z181100001718005), and the Medical Science and Technology Youth Cultivation Program of PLA (16QNP075).
Availability of data and materials
The datasets used during the current study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
The experiments involving animals were approved by the Animal Ethical Committee of the 8th Medical Center of Chinese PLA General Hospital, and conducted in compliance to the standards of Experimental Animal Regulation Ordinances defined by the China National Science and Technology Commission.
Consent for publication
Not applicable.
Competing interests
All authors have read and approved the final manuscript and declare that they have no competing interests.
Footnotes
Wen-Ping Gong, Yan Liang and Yan-Bo Ling contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40779-020-00258-4.
References
- 1.Gordon SV, Parish T. Microbe Profile: Mycobacterium tuberculosis: Humanity's deadly microbial foe. Microbiology (Reading, Engl) 2018;164(4):437–439. doi: 10.1099/mic.0.000601. [DOI] [PubMed] [Google Scholar]
- 2.Gong W, Liang Y, Wu X. The current status, challenges, and future developments of new tuberculosis vaccines[J] Hum Vaccin Immunother. 2018;14(7):1697–1716. doi: 10.1080/21645515.2018.1458806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colditz GA, Brewer TF, Berkey CS, Wilson ME, Burdick E, Fineberg HV, et al. Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. JAMA. 1994;271(9):698–702. [PubMed] [Google Scholar]
- 4.Fine PE. Variation in protection by BCG: implications of and for heterologous immunity. Lancet. 1995;346(8986):1339–1345. doi: 10.1016/s0140-6736(95)92348-9. [DOI] [PubMed] [Google Scholar]
- 5.Huang CY, Hsieh WY. Efficacy of Mycobacterium vaccae immunotherapy for patients with tuberculosis: a systematic review and meta-analysis. Hum Vaccin Immunother. 2017;13(9):1960–1971. doi: 10.1080/21645515.2017.1335374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO. Global tuberculosis report 2017. Geneva: World Health Organization; 2017. Report No.: 978–92–4-156551-6 Contract No.: 20.
- 7.Tsukamura M, Mizuno S, Tsukamura S. Classification of rapidly growing mycobacteria. Jpn J Microbiol. 1968;12(2):151–166. doi: 10.1111/j.1348-0421.1968.tb00379.x. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Guell E, Agusti G, Corominas M, Cardona PJ, Luquin M, Julian E. Mice with pulmonary tuberculosis treated with Mycobacterium vaccae develop strikingly enhanced recall gamma interferon responses to M. vaccae cell wall skeleton. Clin Vaccine Immunol. 2008;15(5):893–896. doi: 10.1128/CVI.00477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skinner MA, Yuan S, Prestidge R, Chuk D, Watson JD, Tan PL. Immunization with heat-killed Mycobacterium vaccae stimulates CD8+ cytotoxic T cells specific for macrophages infected with Mycobacterium tuberculosis[J] Infect Immun. 1997;65(11):4525–4530. doi: 10.1128/iai.65.11.4525-4530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez-Pando R, Aguilar D, Orozco H, Cortez Y, Brunet LR, Rook GA. Orally administered Mycobacterium vaccae modulates expression of immunoregulatory molecules in BALB/c mice with pulmonary tuberculosis. Clin Vaccine Immunol. 2008;15(11):1730–1736. doi: 10.1128/CVI.00286-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Jiang Y, Cui Z, Yang W, Yue L, Ma Y, et al. Mycobacterium vaccae induces a strong Th1 response that subsequently declines in C57BL/6 mice[J] J Vet Sci. 2016;17(4):505–513. doi: 10.4142/jvs.2016.17.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanford JL, Bahr GM, Rook GA, Shaaban MA, Chugh TD, Gabriel M, et al. Immunotherapy with Mycobacterium vaccae as an adjunct to chemotherapy in the treatment of pulmonary tuberculosis. Tubercle. 1990;71(2):87–93. doi: 10.1016/0041-3879(90)90002-p. [DOI] [PubMed] [Google Scholar]
- 13.Weng H, Huang JY, Meng XY, Li S, Zhang GQ. Adjunctive therapy of Mycobacterium vaccae vaccine in the treatment of multidrug-resistant tuberculosis: a systematic review and meta-analysis. Biomed Rep. 2016;4(5):595–600. doi: 10.3892/br.2016.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang XY, Chen QF, Li YP, Wu SM. Mycobacterium vaccae as adjuvant therapy to anti-tuberculosis chemotherapy in never-treated tuberculosis patients: a meta-analysis. PLoS One. 2011;6(9):e23826. doi: 10.1371/journal.pone.0023826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo Y, Lu S, Guo S. Immunotherapeutic effect of Mycobacterium vaccae on multi-drug resistant pulmonary tuberculosis. Zhonghua Jie He He Hu Xi Za Zhi. 2000;23(2):85–88. [Article in China]. [PubMed]
- 16.Fonken LK, Frank MG, D'Angelo HM, Heinze JD, Watkins LR, Lowry CA, et al. Mycobacterium vaccae immunization protects aged rats from surgery-elicited neuroinflammation and cognitive dysfunction. Neurobiol Aging. 2018;71:105–114. doi: 10.1016/j.neurobiolaging.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cananzi FC, Mudan S, Dunne M, Belonwu N, Dalgleish AG. Long-term survival and outcome of patients originally given Mycobacterium vaccae for metastatic malignant melanoma. Hum Vaccin Immunother. 2013;9(11):2427–2433. doi: 10.4161/hv.25618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank MG, Fonken LK, Dolzani SD, Annis JL, Siebler PH, Schmidt D, et al. Immunization with Mycobacterium vaccae induces an anti-inflammatory milieu in the CNS: attenuation of stress-induced microglial priming, alarmins and anxiety-like behavior. Brain Behav Immun. 2018;73:352–363. doi: 10.1016/j.bbi.2018.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng J, Chen L, Liu L, Li H, Liu B, Zheng D, et al. Proteogenomic analysis and discovery of immune antigens in Mycobacterium vaccae. Mol Cell Proteomics. 2017;16(9):1578–1590. doi: 10.1074/mcp.M116.065813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, Jiang X, Luo M, Feng G, Sun Q, Chen Y. Mycobacterium vaccae nebulization can protect against asthma in BALB/c mice by regulating Th9 expression[J] PLoS One. 2016;11(8):e0161164. doi: 10.1371/journal.pone.0161164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fowler DW, Copier J, Wilson N, Dalgleish AG, Bodman-Smith MD. Mycobacteria activate gammadelta T-cell anti-tumour responses via cytokines from type 1 myeloid dendritic cells: a mechanism of action for cancer immunotherapy. Cancer Immunol Immunother. 2012;61(4):535–547. doi: 10.1007/s00262-011-1121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang Y, Zhang J, Yang Y, Bai X, Yu Q, Li N, et al. Immunogenicity and therapeutic effects of recombinant Ag85AB fusion protein vaccines in mice infected with Mycobacterium tuberculosis. Vaccine. 2017;35(32):3995–4001. doi: 10.1016/j.vaccine.2017.05.083. [DOI] [PubMed] [Google Scholar]
- 23.Liang Y, Zhang X, Bai X, Xiao L, Wang X, Zhang J, et al. Immunogenicity and therapeutic effects of a Mycobacterium tuberculosis rv2190c DNA vaccine in mice. BMC Immunol. 2017;18(1):11. doi: 10.1186/s12865-017-0196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong W, Xiong X, Qi Y, Jiao J, Duan C, Wen B. Surface protein Adr2 of Rickettsia rickettsii induced protective immunity against Rocky Mountain spotted fever in C3H/HeN mice. Vaccine. 2014;32(18):2027–2033. doi: 10.1016/j.vaccine.2014.02.057. [DOI] [PubMed] [Google Scholar]
- 25.Gong WP, Wang PC, Xiong XL, Jiao J, Yang XM, Wen BH. Chloroform-methanol residue of Coxiella burnetii markedly potentiated the specific Immunoprotection elicited by a recombinant protein fragment rOmpB-4 derived from outer membrane protein B of rickettsia rickettsii in C3H/HeN mice. PLoS One. 2015;10(4):e0124664. doi: 10.1371/journal.pone.0124664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong WP, Wang PC, Xiong XL, Jiao J, Yang XM, Wen BH. Enhanced protection against Rickettsia rickettsii infection in C3H/HeN mice by immunization with a combination of a recombinant adhesin rAdr2 and a protein fragment rOmpB-4 derived from outer membrane protein B. Vaccine. 2015;33(8):985–992. doi: 10.1016/j.vaccine.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 27.Gong W, Qi Y, Xiong X, Jiao J, Duan C, Wen B. Rickettsia rickettsii outer membrane protein YbgF induces protective immunity in C3H/HeN mice. Hum Vaccin Immunother. 2015;11(3):642–649. doi: 10.1080/21645515.2015.1011572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang PC, Xiong XL, Jiao J, Yang XM, Jiang YQ, Wen BH, et al. Th1 epitope peptides induce protective immunity against rickettsia rickettsii infection in C3H/HeN mice. Vaccine. 2017;35(51):7204–7212. doi: 10.1016/j.vaccine.2017.09.068. [DOI] [PubMed] [Google Scholar]
- 29.Xu LJ, Wang YY, Zheng XD, Gui XD, Tao LF, Wei HM. Immunotherapeutical potential of Mycobacterium vaccae on M. tuberculosis infection in mice. Cell Mol Immunol. 2009;6(1):67–72. doi: 10.1038/cmi.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groschel MI, Prabowo SA, Cardona PJ, Stanford JL, van der Werf TS. Therapeutic vaccines for tuberculosis--a systematic review. Vaccine. 2014;32(26):3162–3168. doi: 10.1016/j.vaccine.2014.03.047. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez-Guell E, Agusti G, Corominas M, Cardona PJ, Casals I, Parella T, et al. The production of a new extracellular putative long-chain saturated polyester by smooth variants of Mycobacterium vaccae interferes with Th1-cytokine production. Antonie Van Leeuwenhoek. 2006;90(1):93–108. doi: 10.1007/s10482-006-9062-1. [DOI] [PubMed] [Google Scholar]
- 32.Gerstmayer B, Kusters D, Gebel S, Muller T, Van Miert E, Hofmann K, et al. Identification of RELMgamma, a novel resistin-like molecule with a distinct expression pattern. Genomics. 2003;81(6):588–595. doi: 10.1016/s0888-7543(03)00070-3. [DOI] [PubMed] [Google Scholar]
- 33.Koizumi G, Kumai T, Egawa S, Yatomi K, Hayashi T, Oda G, et al. Gene expression in the vascular wall of the aortic arch in spontaneously hypertensive hyperlipidemic model rats using DNA microarray analysis. Life Sci. 2013;93(15):495–502. doi: 10.1016/j.lfs.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Schinke T, Haberland M, Jamshidi A, Nollau P, Rueger JM, Amling M. Cloning and functional characterization of resistin-like molecule gamma. Biochem Biophys Res Commun. 2004;314(2):356–362. doi: 10.1016/j.bbrc.2003.12.100. [DOI] [PubMed] [Google Scholar]
- 35.Gleeson LE, Sheedy FJ, Palsson-McDermott EM, Triglia D, O'Leary SM, O'Sullivan MP, et al. Cutting edge: Mycobacterium tuberculosis induces aerobic glycolysis in human alveolar macrophages that is required for control of intracellular bacillary replication. J Immunol. 2016;196(6):2444–2449. doi: 10.4049/jimmunol.1501612. [DOI] [PubMed] [Google Scholar]
- 36.Sambarey A, Devaprasad A, Baloni P, Mishra M, Mohan A, Tyagi P, et al. Meta-analysis of host response networks identifies a common core in tuberculosis. NPJ Syst Biol Appl. 2017;3:4. doi: 10.1038/s41540-017-0005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boro M, Balaji KN. CXCL1 and CXCL2 regulate NLRP3 Inflammasome activation via G-protein-coupled receptor CXCR2. J Immunol. 2017;199(5):1660–1671. doi: 10.4049/jimmunol.1700129. [DOI] [PubMed] [Google Scholar]
- 38.Yu EA, John SH, Tablante EC, King CA, Kenneth J, Russell DG, et al. Host transcriptional responses following ex vivo re-challenge with Mycobacterium tuberculosis vary with disease status. PLoS One. 2017;12(10):e0185640. doi: 10.1371/journal.pone.0185640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimura J, Saiga H, Sato S, Okuyama M, Kayama H, Kuwata H, et al. Potent antimycobacterial activity of mouse secretory leukocyte protease inhibitor. J Immunol. 2008;180(6):4032–4039. doi: 10.4049/jimmunol.180.6.4032. [DOI] [PubMed] [Google Scholar]
- 40.Tateosian NL, Pasquinelli V, Hernandez Del Pino RE, Ambrosi N, Guerrieri D, Pedraza-Sanchez S, et al. The impact of IFN-gamma receptor on SLPI expression in active tuberculosis: association with disease severity. Am J Pathol. 2014;184(5):1268–1273. doi: 10.1016/j.ajpath.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomez SA, Arguelles CL, Guerrieri D, Tateosian NL, Amiano NO, Slimovich R, et al. Secretory leukocyte protease inhibitor: a secreted pattern recognition receptor for mycobacteria. Am J Respir Crit Care Med. 2009;179(3):247–253. doi: 10.1164/rccm.200804-615OC. [DOI] [PubMed] [Google Scholar]
- 42.Gopal R, Monin L, Torres D, Slight S, Mehra S, McKenna KC, et al. S100A8/A9 proteins mediate neutrophilic inflammation and lung pathology during tuberculosis. Am J Respir Crit Care Med. 2013;188(9):1137–1146. doi: 10.1164/rccm.201304-0803OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirzaei A, Mahmoudi H. Evaluation of TNF-alpha cytokine production in patients with tuberculosis compared to healthy people. GMS Hyg Infect Control. 2018;13:Doc09. doi: 10.3205/dgkh000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh JH, Yang CS, Noh YK, Kweon YM, Jung SS, Son JW, et al. Polymorphisms of interleukin-10 and tumour necrosis factor-alpha genes are associated with newly diagnosed and recurrent pulmonary tuberculosis. Respirology. 2007;12(4):594–598. doi: 10.1111/j.1440-1843.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- 45.Basingnaa A, Antwi-Baffour S, Nkansah DO, Afutu E, Owusu E. Plasma levels of cytokines (IL-10, IFN-γ and TNF-α) in multidrug resistant tuberculosis and drug responsive tuberculosis patients in Ghana. Diseases. 2018;7(1). pii: E2. doi: 10.3390/diseases7010002. [DOI] [PMC free article] [PubMed]
- 46.Wang Y, Zhou X, Lin J, Yin F, Xu L, Huang Y, et al. Effects of Mycobacterium bovis on monocyte-derived macrophages from bovine tuberculosis infection and healthy cattle. FEMS Microbiol Lett. 2011;321(1):30–36. doi: 10.1111/j.1574-6968.2011.02304.x. [DOI] [PubMed] [Google Scholar]
- 47.Meng W, Bai B, Bai Z, Li Y, Yue P, Li X, et al. The immunosuppression role of alpha-fetoprotein in human hepatocellular carcinoma. Discov Med. 2016;21(118):489–494. [PubMed] [Google Scholar]
- 48.Corapcioglu F, Guvenc BH, Sarper N, Aydogan A, Akansel G, Arisoy ES. Peritoneal tuberculosis with elevated serum CA 125 level mimicking advanced ovarian carcinoma in an adolescent. Turk J Pediatr. 2006;48(1):69–72. [PubMed] [Google Scholar]
- 49.Limaiem F, Gargouri F, Bouraoui S, Lahmar A, Mzabi S. Co-existence of hepatocellular carcinoma and hepatic tuberculosis. Surg Infect. 2014;15(4):437–440. doi: 10.1089/sur.2012.177. [DOI] [PubMed] [Google Scholar]
- 50.Champion TC, Partridge LJ, Ong SM, Malleret B, Wong SC, Monk PN. Monocyte subsets have distinct patterns of tetraspanin expression and different capacities to form multinucleate giant cells. Front Immunol. 2018;9:1247. doi: 10.3389/fimmu.2018.01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bermudez LE, Goodman J. Mycobacterium tuberculosis invades and replicates within type II alveolar cells. Infect Immun. 1996;64(4):1400–1406. doi: 10.1128/iai.64.4.1400-1406.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bermudez LE, Sangari FJ, Kolonoski P, Petrofsky M, Goodman J. The efficiency of the translocation of Mycobacterium tuberculosis across a bilayer of epithelial and endothelial cells as a model of the alveolar wall is a consequence of transport within mononuclear phagocytes and invasion of alveolar epithelial cells. Infect Immun. 2002;70(1):140–146. doi: 10.1128/IAI.70.1.140-146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hall-Stoodley L, Watts G, Crowther JE, Balagopal A, Torrelles JB, Robison-Cox J, et al. Mycobacterium tuberculosis binding to human surfactant proteins a and D, fibronectin, and small airway epithelial cells under shear conditions. Infect Immun. 2006;74(6):3587–3596. doi: 10.1128/IAI.01644-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fortune SM, Solache A, Jaeger A, Hill PJ, Belisle JT, Bloom BR, et al. Mycobacterium tuberculosis inhibits macrophage responses to IFN-gamma through myeloid differentiation factor 88-dependent and -independent mechanisms. J Immunol. 2004;172(10):6272–6280. doi: 10.4049/jimmunol.172.10.6272. [DOI] [PubMed] [Google Scholar]
- 55.Doz E, Rose S, Court N, Front S, Vasseur V, Charron S, et al. Mycobacterial phosphatidylinositol mannosides negatively regulate host toll-like receptor 4, MyD88-dependent proinflammatory cytokines, and TRIF-dependent co-stimulatory molecule expression. J Biol Chem. 2009;284(35):23187–23196. doi: 10.1074/jbc.M109.037846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giacomini E, Iona E, Ferroni L, Miettinen M, Fattorini L, Orefici G, et al. Infection of human macrophages and dendritic cells with Mycobacterium tuberculosis induces a differential cytokine gene expression that modulates T cell response. J Immunol. 2001;166(12):7033–7041. doi: 10.4049/jimmunol.166.12.7033. [DOI] [PubMed] [Google Scholar]
- 57.Sugawara I, Yamada H, Mizuno S, Takeda K, Akira S. Mycobacterial infection in MyD88-deficient mice. Microbiol Immunol. 2003;47(11):841–847. doi: 10.1111/j.1348-0421.2003.tb03450.x. [DOI] [PubMed] [Google Scholar]
- 58.Wright KM, Friedland JS. Regulation of monocyte chemokine and MMP-9 secretion by proinflammatory cytokines in tuberculous osteomyelitis. J Leukoc Biol. 2004;75(6):1086–1092. doi: 10.1189/jlb.0903433. [DOI] [PubMed] [Google Scholar]
- 59.Gu X, Gao Y, Mu DG, Fu EQ. MiR-23a-5p modulates mycobacterial survival and autophagy during Mycobacterium tuberculosis infection through TLR2/MyD88/NF-kappaB pathway by targeting TLR2. Exp Cell Res. 2017;354(2):71–77. doi: 10.1016/j.yexcr.2017.03.039. [DOI] [PubMed] [Google Scholar]
- 60.Chen CS, Alonso JL, Ostuni E, Whitesides GM, Ingber DE. Cell shape provides global control of focal adhesion assembly. Biochem Biophys Res Commun. 2003;307(2):355–361. doi: 10.1016/s0006-291x(03)01165-3. [DOI] [PubMed] [Google Scholar]
- 61.Theocharis AD, Skandalis SS, Gialeli C, Karamanos NK. Extracellular matrix structure. Adv Drug Deliv Rev. 2016;97:4–27. doi: 10.1016/j.addr.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol. 2014;15(12):786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Del Angel-Mosqueda C, Gutierrez-Puente Y, Lopez-Lozano AP, Romero-Zavaleta RE, Mendiola-Jimenez A, Medina-De la Garza CE, et al. Epidermal growth factor enhances osteogenic differentiation of dental pulp stem cells in vitro. Head Face Med. 2015;11:29. doi: 10.1186/s13005-015-0086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Villasenor T, Madrid-Paulino E, Maldonado-Bravo R, Urban-Aragon A, Perez-Martinez L, Pedraza-Alva G. Activation of the Wnt pathway by Mycobacterium tuberculosis: a Wnt-Wnt situation. Front Immunol. 2017;8:50. doi: 10.3389/fimmu.2017.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X, Huang T, Wu Y, Peng W, Xie H, Pan M, et al. Inhibition of the PI3K-Akt-mTOR signaling pathway in T lymphocytes in patients with active tuberculosis. Int J Infect Dis. 2017;59:110–117. doi: 10.1016/j.ijid.2017.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Array image of each sample. c1-c3, the serial number of mice in the control group; v1-v3, the serial number of mice in the M. vaccae group.
Additional file 2: Table S1. Raw and log2 value of the normalized intensity of each sample in the control group and the M. vaccae group.
Additional file 3: Table S2. Detail information of 2326 upregulated genes and 2221 downregulated genes.
Additional file 4: Table S3. GO analysis for upregulated DE genes in control and M. vaccae groups in terms of biological process (BP sheet), cellular component (CC sheet), and molecular function (MF sheet).
Additional file 5: Table S4. GO analysis for downregulated DE genes in control and M. vaccae groups in terms of biological process (BP sheet), cellular component (CC sheet), and molecular function (MF sheet).
Additional file 6: Table S5. The whole analysis results of upregulated pathways in control and M. vaccae groups.
Additional file 7: Table S6. The whole analysis results of downregulated pathways in control and M. vaccae groups.
Data Availability Statement
The datasets used during the current study are available from the corresponding author upon reasonable request.