Abstract
Background
Fanconi anemia (FA) is the most common inherited bone marrow failure (BMF) syndrome with 22 related genes identified. The ALDH2 rs671variant has been proved related to accelerate the progression of BMF in FA patients. The phenotype and genetic basis of Chinese FA patients have not been investigated yet.
Methods
We analyzed the 22 FA-related genes of 63 BMF patients suspected to be FA. Clinical manifestations, morphological and cytogenetic feathers, ALDH2 genotypes, treatment, and outcomes of the definite cases were retrospectively studied.
Results
A total of 21 patients were confirmed the diagnosis of FA with the median age of BMF onset was 4-year-old. The number of patients manifested as congenital malformations and growth retardation were 20/21 and 14/21, respectively. BM dysplasia and cytogenetic abnormalities were found in 13/20 and 8/19 patients. All the patients with abnormal karyotypes also manifested as BM dysplasia or had evident blasts. Thirty-five different mutations were identified involving six genes and including twenty novel mutations. FANCA mutations contributed to 66.67% of cases. Eight patients harboring ALDH2-G/A genotype have a significantly younger age of BMF onset (p = 0.025). Within the 19 patients adhering to continuous follow-up, 15 patients underwent hematopoietic stem cell transplantations (HSCTs). During the 29 months of follow-up, 8/19 patients died, seven of which were HSCT-related, and one patient who did not receive HSCT died from severe infection.
Conclusions
The phenotypic and genetic spectrum of Chinese FA patients is broad. Bone marrow dysplasia and cytogenetic abnormalities are prevalent and highly consistent. The overall outcome of HSCTs is disappointing. Nationwide multicenter studies are needed for the rarity and adverse outcome of this disease.
Keywords: Fanconi anemia, Bone marrow failure, Aldehyde dehydrogenase, Hematopoietic stem cell transplantation
Background
Fanconi anemia (FA) is a rare genetic disease highly heterogeneous in clinical manifestations and genetics. Clinical features primarily include congenital malformations, progressive bone marrow failure (BMF), and predisposition to hematopoietic and solid malignancies [1, 2]. The most common congenital abnormalities include skin pigmentation, café au lait spots, short stature, and hypoplastic of radii and/or thumbs [2]. The time of BMF onset is variable but usually at pre-school age, with the cumulative incidence of 90% by the age of 40 [3]. The malignancy risk in FA patients is mounting, especially the risks of myelodysplastic syndrome/acute myeloid leukemia (MDS/AML), which are several hundred folds higher than those of the general population [3–6].
Twenty-two genes have been identified related to FA (Table S1) to date. Products of the 22 genes participate in FA-BRCA pathway, which is responsible for correcting interstrand crosslinks (ICLs) and other DNA damage events induced by genotoxic agents. Endogenous aldehyde is a genotoxic agent and is detoxicated by aldehyde dehydrogenases (ALDHs) in vivo [7]. Previous studies have suggested aldehydes are highly toxic in FA deficient cells and could speed up the development of BMF and leukemia in FA deficient mice models [8–10]. The mitochondrial ALDH2 isoform is the most efficient acetaldehyde-detoxifying enzyme in humans [11]. Inactivating ALDH2 variant (rs671 c.1510G > A/p.E504L) is highly prevalent in east Asia and can abolish ALDH2 activity by a dominant-negative effect [12]. ALDH2-A/A and ALDH2-G/A genotypes have been proved related to accelerated progression of BMF and malignant transformation in FA patients [13, 14].
Although the genetic basis, pathological mechanisms, and epidemiology of FA have been extensively studied, few researches focus on Chinese patients [15]. In the present study, we report 21 Chinese FA patients aiming to depict their genetic basis and clinical characteristics.
Methods
Patient enrollment
We retrospectively analyzed 63 BMF patients who were suspected to be inherited BMF in Hebei Yanda Lu Daopei Hospital from May 2012 to Dec. 2017. Detailed disease histories and examination files were retrieved from the electronic medical record system of our institute. BMF is considered with one or more lineages decreased in whole blood cell counts and reduced hematopoiesis with routine Wright-Giemsa staining bone marrow morphological analysis and Hematoxylin-Eosin staining pathological analysis. Categorization of hematopoietic cells, blast percentage, hematopoietic grade, dysplasia, and diagnosis of MDS were according to the 2008 edition of the World Health Organization Classification of Tumors of Hematopoietic and Lymphoid Tissues [16]. All patients enrolled were confirmed BMF and should meet at least two following inclusive criteria: 1) growth retardation; 2) congenital physical malformations; 3) early onset of BMF (≤ 6 years old); 4) chronic onset of BMF with a progressive course (disease course > 6 months); 5) suggestive family history (consanguinity or family history of cancer or hematological disorders); 6) positive for chromosome breakage test (Supplementary methods) (Table S2). Other inherited syndromes manifested as BMF and malformations such as dyskeratosis congenita, Diamond-Blackfan anemia, and Neurofibromatosis-Noonan syndrome diagnosed based on syndromic presentations combined with genetic tests were excluded. The follow-up duration was defined as the time from referral to the last follow-up or loss of follow-up/death.
Written informed consents were obtained from the patients or their statutory guardians and all tested family members in accordance with the Declaration of Helsinki. The study was approved by the ethics committee of the Hebei Yanda Lu Daopei hospital.
Nucleic acid extraction
Peripheral blood (PB), bone marrow (BM), or cryopreserved DNA samples of the patients and their parents were obtained. Genomic DNA was extracted from PB/BM nucleated cells using silica gel column method.
High throughput sequencing, variant calling, and ALDH2 genotyping
We carried out Sanger sequencing on the entire coding exons and flank regions of FANCA, FANCC, and FANCG in patients suspected to be inherited BMF from Apr. 2012 to May 2016. Targeted high-throughput sequencing (THS) has been applied since May 2016, FANCD2 and BRCA2 were added in the panel. Whole genomic sequencing (WGS) was carried out using cryopreserved samples for the enrolled cases where the panel test could not find the pathogenic mutations, and all the 22 FA genes were analyzed.
The THS process has been described previously [17]. For the WGS, libraries were constructed with NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, US), followed by sequencing on Illumina HiSeq X Ten platform (Illumina, US) using HiSeq X Ten Reagent Kit v2.5 (Illumina, US) running on paired-end 150 bp mode.
Reads yielded by the two kinds of sequencing were all aligned to the human reference genome (hg19) with the Burrow-Wheeler Aligner (BWA) mem. Variants were called according to Genome Analysis Toolkit (GATK) best practices using bam files. Final confident variants were annotated using annovar and oncotator. Variants with minimal allele frequency (MAF) ≥ 1% in general population were filtered out according to 1000 Genomes, EXAC, and gnomAD databases. The pathogenicity of the germline missense mutations was assessed by in silico prediction algorithms, and the putative causal variants were classified according to the standards and guidelines recommended by the American College of Medical Genetics and Genomics (ACMG) [18]. Only pathogenic, likely pathogenic, or uncertain significance variants were considered causative in the present study. The process of CNV analysis based on WGS has been described elsewhere [17].
ALDH2 genotyping was based on WGS data or Sanger sequencing with primers 5′- TGCTATGATGTGTTTGGAGCC-3′ (forward) and 5′-ATTTAGGGTCTCTGCTGGGCG-3′ (reverse).
Validation by sanger sequencing
Polymerase chain reaction (PCR) and Sanger sequencing performed on the ABI 3500xL Genetic Analyzer (Thermo Fisher, US) were adopted to confirm all the mutations reported in this study. Single nucleotide variants (SNVs) and small insertions/deletions (InDels) were validated by PCR and Sanger sequencing using pedigree’s samples when accessible. For the validation of CNVs, the breakpoints were confirmed by Sanger sequencing using patients’ DNA, and the parental origins were verified through PCR and agarose gel electrophoresis.
Results
Demography and clinical characteristics
A total of 21 patients (six females and fifteen males) from non-related families were finally diagnosed as FA, including one who has already been reported (Case 8) [17]. The median referral age of this cohort was 7 years old, and the median age of BMF onset was 4 years old (range, 1–13 years old). There were 20 Han Chinese and one Uyghur Chinese, and the geographical distribution spread nationwide though over half of the patients came from the south or southwest of China. All patients were referred to our institute because of severe cytopenia except a thirty-year-old boy (Case 10) who was initially diagnosed as MDS for the myeloid dysplasia and increased myeloblasts indicated by BM morphology. Five patients had an indicative family history with two patients had family members died from anemia (Case 3, Case 15), two patients were from consanguineous families (Case 16, Case 21), and one patient was an in vitro fertilize baby whose paternal grandmother died from pancreatic cancer (Case 4) (Table 1).
Table 1.
Clinical features
| Case No. | Gender | Age of referral (years) | Age of clinical BMF onset (years) | Congenital malformations | Growth retardation | Family history |
|---|---|---|---|---|---|---|
| 1 | M | 12 | 2 | S, C, M | Yes | Negative |
| 2 | M | 7 | 2 | S, C, G | Yes | Negative |
| 3 | M | 10 | 5 | S, G | Yes | One sibling manifested as polydactyly and died from anemia |
| 4 | M | 11 | 5 | C, G, M, | Yes |
One sibling manifested as polydactyly and died from anemia IVF and paternal grandmother died from pancreatic cancer |
| 5 | F | 11 | 10 | C, H | Yes |
IVF and paternal grandmother died from pancreatic cancer Negative |
| 6 | M | 7 | 5 | S, M | No | Negative |
| 7 | M | 17 | 10 | S, M | No | Negative |
| 8 | M | 7 | 7 | S, C, M, H | No | Negative |
| 9 | M | 5 | 4 | S, M | No | Negative |
| 10 | M | 13 | 13 | E | Yes | Negative |
| 11 | M | 9 | 6 | S | Yes | Negative |
| 12 | F | 7 | 7 | M | Yes | Negative |
| 13 | M | 7 | 1 | S, M | Yes | Negative |
| 14 | M | 5 | 2 | S | No | Negative |
| 15 | M | 14 | 5 | C, G | Yes | Two family members died from anemia |
| 16a | M | 6 | 4 | S, C, M | Yes | 2nd degree consanguinity |
| 17 | M | 4 | 3 | None | No | Negative |
| 18 | F | 9 | 4 | S, M | No | Negative |
| 19 | F | 7 | 4 | S, G, N | Yes | Negative |
| 20 | F | 6 | 3 | S, C, G | Yes | Negative |
| 21 | F | 6 | 4 | S, M, H, E, G | Yes | 2nd degree consanguinity |
Skin and annex abnormalities include skin pigmentation, café au lait spots, excess hair; craniofacial anomalies include microcephalus, ptosis, hypertelorism, hypotelorism, flat nose bridge; malformations in musculoskeletal system include polydactyly, deformity of thumbs, absence of thumbs, hypoplasia of thenar eminence, and scoliosis; genitourinary system malformations include kidney malformation, hydronephrosis, indirect inguinal hernia, cryptorchidism, ovary absence, and uterine malformation/absence; cardiovascular system defects include patent ductus arteriosus and ventricular septal defect; nervous system abnormalities include encephalatrophy and moyamoya disease; endocrine system defects include hypothyroidism, primary adrenocortical insufficiency, and obesity
F female, M male, S skin and annex, C craniofacial anomalies, M musculoskeletal system, G genitourinary system, H cardiovascular system, E endocrine system, N nervous system, IVF in vitro fertilized
aCase 16 is of Chinese Uyghur ancestry
Fourteen (66.67%) patients were growth-retarded, and 20 (95.24%) patients manifested as congenital malformations. Congenital abnormalities in our cohort included skin pigmentation (13/21), café au lait spots (5/21), spin and limbs deformation (11/21), craniofacial malformations (8/21), genitourinary system malformations (7/21), cardiovascular system defects (2/21), nervous system diseases (2/21), and endocrine system defects (2/21) (Table 1).
Thoroughly evaluation of the hematologic phenotype is crucial to FA patients since BM dysplasia or pathological cytogenetics relate to disease progression and adverse hematopoietic stem cell transplantations (HSCT) outcomes [5, 14]. Twenty patients’ morphologic test results and nineteen patients’ cytogenetics test results before pre-HSCT conditioning regimen and/or chemotherapy were available. BM dysplasia was found in 13/20 (65%) patients, including one AML with the myeloblast count of 41% (Case 5) and one myelodysplasia with the blast count of 6% (Case 10). Karyotypes were described according to the International System for Human Cytogenetic Nomenclature 2013 [15]; at least 20 metaphases were analyzed for each assay. Cytogenetic abnormalities were found in 8/19 (42.11%) patients with clonality found in five patients, and half of the abnormal karyotypes involved chromosome 7 (− 7, 7q-, or der(7)t(1;7)) (Table 2). The cytogenetic result of Case 5 who was diagnosed as AML was 46, XX, der(7)t(1;7)(q21;q36) [19], which was confirmed to be non-constitutional by matched peripheral blood, and the karyotype of patient Case 10 was highly complex (Table 2). All the patients with abnormal karyotypes also manifested as dysplasia on bone marrow smear or had evident blasts, suggesting the initiation of clonal evolution in hematopoietic system.
Table 2.
Bone marrow morphology, karyotype, chromosome breakage tests, and ALDH2 genotypes
| Case No. | BM morphology | BM karyotype | Chromosome breakage test | ALDH2 genotype |
|---|---|---|---|---|
| 1 | Dysplasia | NA | Positive | G/A |
| 2 | Dysplasia | 47,XY,+ 15[1]/46,XY[20] | Positive | G/A |
| 3 | Hypoplasia | normal | Positive | G/A |
| 4 | NA | NA | Positive | G/G |
| 5a | AML | 46,XX,der(7)t(1;7)(q21;q36)[20] | Positive | G/G |
| 6 | Hypoplasia | Normal | Positive | G/A |
| 7 | Dysplasia | 46,XY,-7,+ 21[5]/46,XY[16] | Positive | G/G |
| 8 | Hypoplasia | Normal | Positive | G/G |
| 9 | Hypoplasia | Normal | Positive | G/A |
| 10b | MDS | Complex | Positive | G/G |
| 11 | Hypoplasia | Normal | Positive | G/A |
| 12 | Dysplasia | Normal | Positive | G/G |
| 13 | Dysplasia | Normal | Positive | G/G |
| 14 | Dysplasia | Normal | Positive | G/A |
| 15 | Dysplasia | 46,XY,del(7)(p13)[13]/46,XY[7] | Positive | G/A |
| 16 | Hypoplasia | Normal | Positive | G/A |
| 17 | Dysplasia | Normal | Positive | G/A |
| 18 | Dysplasia | 46,XX,t(1;5)(p36.1;q13)[1]/46,XX[19] | Positive | G/G |
| 19 | Dysplasia | 46,XX,del(14)(q24)[1]/46,XX[20] | Positive | G/G |
| 20 | Dysplasia | 46,XX,del(7)(q22)[8]/46,XX,del(5)(p11)/46,XX[19] | Positive | G/A |
| 21 | Hypoplasia | Normal | Positive | G/A |
Chromosome breakage tests were induced by mitomycin C
NA not available, AML acute myeloid leukemia, MDS myelodysplastic syndrome
aCase 5 is diagnosed as acute myeloid leukemia. The myeloblasts count 41% of the nucleated cells according to morphologic test of bone marrow smears
bCase 10 is diagnosed as myelodysplastic syndrome. His bone marrow morphology shows dysplasia was observed in his granulocytic lineage and megakaryocytic lineage with the myeloblasts count 6% of the nucleated cells. The result of his karyotype is:46,XY,dup(1)(q21q23),add(2)(p11.2),add(3)(q27),der(5)t(1;5)(q21;q35),add(20)(p12)[17]/45,XY,der(1)(?::1q42- > 1q21::1p36.3- > 1q32::1q21- > 1q44::?),add(2)(p11.2),add(3)(q27),add(4)(p16),der(5)t(1;5)(q21;q35),-18,add(20)(p12),ace [2]/46,XY [1]
Characteristics of mutations
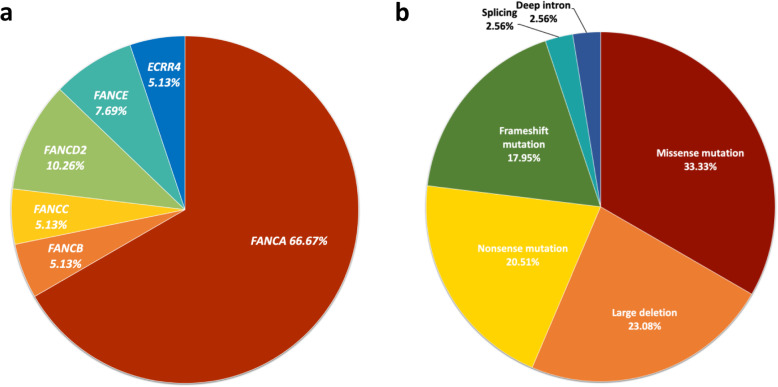
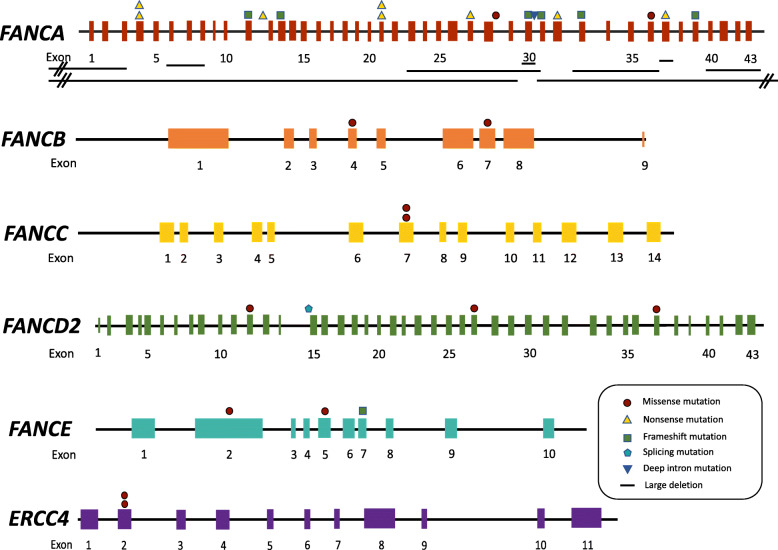
A total of 39 mutations were identified involving six different FA genes and composed of 13 missense mutations, nine large deletions, eight nonsense mutations, seven frameshift mutations, one splicing mutations, and one deep intron mutation (Fig. 1, Table 3). All the large deletions were found within the FANCA gene. 20 (47.73%) mutations identified in our cohort were novel and the majority of mutations were private except FANCA c.367C > T, which was shared by two patients. (Fig. 2, Table 3). We did not find FANCA c.2546delC in our cohort, which accounts for over 30% FANCA mutations in Japanese and Korean patients [22, 24].
Fig. 1.
Distribution and composition of the 39 mutations. a. mutation distribution. b. mutation composition
Table 3.
Mutation details
| Case No. | Gene | Mutation 1 (maternal) | Mutation 2 (paternal) | ||||
|---|---|---|---|---|---|---|---|
| Genomic location | cDNA/Protein | Ref./Com. | Genomic location | cDNA/Protein | Ref./Com. | ||
| 1 | FANCA | chr16:89833593 | c.2557C > T/p.R853X | [20] | chr16:89877396 | c.367C > T/p.Q123X | NA |
| 2 | FANCA | chr16:89815145–89,815,146 | c.3270_3271delCT/p.C1090RfsX25 | Novel | chr16:89868906–89,875,410 | c.792 + 761_c.523-635del | Novel |
| 3 | FANCA | chr16:89877396 | c.367C > T/p.Q123X | NA | chr16:89818822 | c.2982-192A > G | [21] |
| 4a | FANCA | chr16:89842183 | c.1867C > T/p.Q623X | Novel | chr16:89842183 | c.1867C > T/p.Q623X | Novel |
| 5 | FANCA | chr16:89804935–89,806,139 | c.3935-178_4368 + 74del | Novel | chr16:89819567–89,839,134 | c.c.2014 + 545_2982-937del | Novel |
| 6 | FANCA | chr16:89811185–89,815,741 | c.3239 + 397_3626 + 202del | Novel | chr16:89858887 | c.1074_1075delGT/p.Y359PfsX49 | NA |
| 7 | FANCA | chr16:89826812–89,919,023 | FANCA c.2852 + 1545_SPIRE2 c.646-1671del | Novel | chr16:89825071 | c.2894_2895delCT/p.P965RfsX9 | Novel |
| 8 | FANCA | chr16:89780001–89,822,000 | VPS9D1 c.432-877_FANCA c.2981 + 2985del | [16] | chr16: 89808940–89,809,954 | c.3627-607_3765 + 268del | [16] |
| 9 | FANCA | chr16:89823177–89,825,446 | c.2853-333_2981 + 1808del | Novel | chr16:89809270 | c.3703C > T/p.Q1235X | Novel |
| 10 | FANCA | chr16:89818619 | c.2990_2993delGTTA/p.S997MfsX28 | NA | chr16:89862229 | c.987_990delTCAC/p.H330AfsX4 | [22, 23] |
| 11 | FANCA | chr16:89816286 | c.3091C > T/p.Q1031X | NA | chr16:89792569–89,821,767 | ZNF276 c.1007-1118_FANCA c.2982-3137del | Novel |
| 12 | FANCA | chr16:89806417 | c.3918dupT/p.Q1307SfsX6 | [24] | chr16:89831438 | c.2638C > G/p.R880G | NA |
| 13 | FANCA | chr16:89858941 | c.1021C > T/p.Q341X | Novel | chr16:89811412 | c.3581C > T/p.P1194L | [24] |
| 14 | FANCB | chrX:14868651 | c.1472 T > A/p.V491E | Novel | – | – | – |
| 15 | FANCB | chrX:14877390 | c.1018C > A/p.Q340K | Novel | – | – | – |
| 16a | FANCC | chr9:97912346 | c.545C > A/p.S182Y | Novel | chr9:97912346 | c.545C > A/p.S182Y | Novel |
| 17 | FANCD2 | chr3:10084828 | c.983G > A/p.R328Q | NA | chr3:10114634 | c.2574 T > G/p.I858M | Novel |
| 18 | FANCD2 | chr3:10132005 | c.3713 T > A/p.M1238K | NA | chr3:10089599 | c.1279-2A > T | Novel |
| 19 | FANCE | – | – | NA | chr6:35426215 | c.1111C > T/p.R371W | [21, 22, 25] |
| 20 | FANCE | chr6:35423547 | c.272C > T/p.P91L | Novel | chr6:35427467–35,427,470 | c.1246_1249delCAAA/p.T417SfsX7 | NA |
| 21a | ERCC4 | chr16:14015937 | c.257G > A/p.R86H | NA | chr16:14015937 | c.257G > A/p.R86H | NA |
NA not available
a Case 4, Case 16, and Case 21 carry homozygous variants
Fig. 2.
Locations, frequencies, and types of mutations in FANCA, FANCB, FANCC, FANCD2, FANCE, and ERCC4 genes. Exons represent by colored rectangles; mutation types are represented by colored patterns; large deletions are represented by the black horizontal bars
Among the 21 patients,15 patients carried compound heterozygous mutations, three patients carried homozygous mutations, two patients harbored hemizygous FANCB mutations, and one patient with a heterozygous FANCE mutation were identified. Biallelic FANCA mutations caused 61.90% (13/21) of the cases, followed by monoallelic FANCB mutations and FANCD2 mutations, which both constituted 9.52% (2/21) of the cases; and FANCC, FANCE, and ERCC4 mutations caused one case each (Table 3). We did not find any case attributed to FANCG mutations, which is the second most prevalent responsible gene in East Asian according to Japanese and Korean studies [22, 24]. Despite the limited size of this cohort, we identified two FANCB mutations, making it rank one of the most common causative genes in line with the Japanese study [22]. There were three homozygous mutations, FANCA c.1867C > T, FANCC c.545C > A, and ERCC4 c.257G > A; the latter two mutations were carried by patients both came from consanguineous families, and the FANCC c.545C > A was carried by the only Uyghur patient in our cohort.
Rigorous criteria were adopted in the process of criminal variant identification (Table 3, Table S3). Majority of the patients were assigned with compelling mutations with two exceptions. All mutations were classified as pathogenic or likely pathogenic according to the guideline of ACMG. Case 19 carried compound heterozygous FANCE c.1111C > T mutation and FANCE c.1317-237C > G mutation. The c.1111C > T mutation was considered pathogenic, but the c.1317-237C > G mutation is an intron variant and classified as uncertain significance, therefore it was excluded in statistics.
ADLH2 rs671 genotype
12/21 (58.33%) patients in our cohort carried ALDH2-G/A genotype, and the other patients were all ALDH2-G/G genotype. There was no ALDH2-A/A genotype identified (Table 2). The age of BMF onset of ALDH2-G/A patients was significantly younger than that of the ALDH2-G/G patients (p = 0.025, t-test).
Treatment and outcome
Within the 21 patients, continuous medical records of 19 patients can be retrieved except Case 8 and Case 21, who only came to us once and were excluded in this section. All the 21 patients were eligible for HSCT for they were all transfusion-dependent, and HSCT was performed on 15 patients (71.43%). The numbers of patients accepted HSCT from HLA-matched unrelated donors (MUD), HLA-unmatched unrelated donors (UUD), HLA-haploidentical related (sibling or parental) donors (HRD), and HLA-matched related donors (MRD) were three, four, six, and one, respectively. Another patient accepted HLA-unmatched unrelated cord blood (UUC) HSCT. The other four patients who did not undergo HSCT accepted androgen, cytokine, and/or intermittent transfusion support. All the patients with abnormal karyotype underwent HSCTs. In the HSCT subgroup, 9/15 (60%) were ALDH2-G/A genotype. The median follow-up duration was 29 months ranged from 1 month to 68 months. By the end of the study, eight patients (38.10%) have been dead. Seven of them were HSCT-related, mainly severe acute graft-versus-host disease (aGVHD) and/or infections, accounting for 46.67% of the subgroup. One patient who did not receive HSCT died from severe infection (Table 4).
Table 4.
Treatment and outcomes
| Case No. | Therapeutics | Donor & HLA matching | Pre-HSCT conditioning regimen | Outcomes |
|---|---|---|---|---|
| 1 | HSCT | UUD; 8/10 | Bu + CTX + Flu+Alemtuzumab | Dead (aGVHD, infections) |
| 2 | HSCT | UUD; 9/10 | Bu + CTX + Flu+ATG + Me-CCNU | Alive |
| 3 | HSCT | MUD; 10/10 | Bu + Flu+CTX + ATG | Alive |
| 4 | HSCT | HRD; 8/10 | Bu + CTX + Flu+ATG + Me-CCNU | Alive |
| 5 | HSCT | HRD; 6/10 | Dead (aGVHD, drug-induced encephalopathy) | |
| Decitabine+Ara-C + Bu + Flu+ATG + Me-CCNU | ||||
| 6 | Androgen and transfusion | – | – | Alive |
| 7 | HSCT | HRD; 7/10 | Dead (aGVHD, MODS) | |
| Decitabine+Ara-C + Bu + Flu+ATG + Me-CCNU | ||||
| 8 | Lost follow-up | – | – | – |
| 9 | HSCT | MRD; 10/12 | Bu + Flu+CTX + ATG | Dead (aGVHD, septic shock) |
| 10 | HSCT | HRD; 5/10 | Dead (aGVHD, septic shock) | |
| Decitabine+Ara-C + Bu + Flu+ATG + Me-CCNU | ||||
| 11 | HSCT | UUC; 5/8 | Bu + Flu+CTX + ATG | Dead (aGVHD, pulmonary infection, CMV infection) |
| 12 | Androgen, cytokine, transfusion | – | – | Alive |
| 13 | Androgen and cytokine | – | – | Alive |
| 14 | HSCT | HRD; 7/10 | Bu + Flu+CTX + ATG | Dead (aGVHD, TMA, pulmonary infection) |
| 15 | HSCT | HRD; 7/10 | Bu + Flu+CTX + ATG | Alive |
| 16 | Androgen and transfusion | – | – | Dead (pulmonary infection, septic shock) |
| 17 | HSCT | MUD; 10/10 | Bu + Flu+CTX + ATG | Alive |
| 18 | HSCT | UUD; 8/10 | Bu + Flu+CTX + ATG | Alive |
| 19 | HSCT | UUD; 9/10 | TBI + CTX + Flu+ATG | Alive |
| 20 | HSCT | MUD; 10/10 | Bu + Flu+CTX + ATG | Alive |
| 21 | Loss to follow-up | – | – | – |
HSCT hematologic stem cell transplantation, UUD HLA-unmatched unrelated donor, MUD HLA-matched unrelated donor, HRD HLA-haploidentical related donor, MRD HLA-matched related donors, UUC HLA-unmatched unrelated cord blood, Bu Busulfan, CTX cyclophosphamide, Flu Fludarabine, ATG antithymocyte globulin, Me-CCNU Semustine, TBI total body irradiation, aGVHD acute graft-versus-host disease, CMV cytomegalovirus
Discussion
The 21 patients displayed a wide range of clinical phenotype and genetic variation spectrum that all physiological systems were involved (Table 1), and the responsible mutations were detected in six different genes (Fig. 1, Table 3). In keeping with other studies, bone marrow dysplasia and abnormal karyotypes were prevailing (65 and 42.11%, respectively) and highly consistent [14, 26], denoting the risk of hematologic malignant transformation, especially the ones with aberration in chromosome 7, which is the most prevalent cytogenetic abnormality in pediatric MDS and indicates an adverse long-term outcome even after HSCTs in MDS/AML patients [27]. ALDH2-G/A and ALDH-A/A genotypes are confirmed to be associated with more severe hematologic phenotype and more adverse outcomes of FA in Asian patients [13, 14]. The same tendency was observed in our cohort, despite there was no patient of ALDH2-AA genotype.
All patients in our cohort presented with a more severe hematologic manifestation and the proportion of patients who received HSCTs was higher than that of most studies [3–6, 14, 28]. Although BMF is the typical and most prevalent feature, our data may not reflect the actual behavior of FA since all the patients were referred to our institute seeking for HSCTs. Studies suggest the high HSCT-related mortality in FA patients, of which infection and aGVHD were the two leading causes [5, 28]. In our cohort, 46.67% of HSCT patients died from HSCT-related acute complications. Studies also suggest the overall dismal outcome that 10 years cumulative risk of death was over 22% and the overall survival after 30 years of diagnosis dropped to below 40%; besides, the long-term survival of HSCT patients and non-HSCT patients were comparable [5, 26, 28, 29], partly because HSCT in the context of FA is explicitly challenging. Therefore, even with the optimized pre-HSCT conditioning regimens like the reduced intensity and the combination of fludarabine, meticulousness is needed in decision-making. Whether HSCT is the best treatment strategy depends much on the severity of cytopenia and the hematologic adverse events of a given patient and the type of donor he/she could get.
The cumulative incidence of leukemia and solid tumors in the middle age of FA patients was reported to be ~ 20% and ~ 30%, respectively [4–6, 30]. In our cohort, no patient developed hematologic or solid malignancies during the follow-up up to date except the ones initially diagnosed as AML (Case 5) and MDS (Case 10), but the longest follow-up in our cohort was only 5.5 years, which may not be long enough for the malignant phenotype to emerge.
Conclusions
Although this study is limited by its cohort size, it is still informative and enriches the knowledge on Chinese FA patients which was nearly a barren. Here we thoroughly investigated the clinical manifestations, morphologic and cytogenetic changes, genetic basis, and outcomes of 21 Chinese FA patients. Our data displayed a broad phenotypic and genetic variant spectrum of Chinese FA patients, the disappointing outcomes which need improving, and highlighted the urgency of nationwide multicenter studies to reveal the mask of Chinese FA patients and optimize the clinical management.
Supplementary information
Additional file 1. Supplementary methods.
Additional file 2: Table S1. Details of 22 FA-related genes.
Additional file 3. Data of hromosome breakage test.
Additional file 4: Table S3. Mutation interpretations.
Acknowledgments
The authors would like to thank the patients and their families for participating in the study.
Abbreviations
- ACMG
American College of Medical Genetics and Genomics
- AGE
Agarose gel electrophoresis
- aGVHD
Acute graft-versus-host disease
- ALDHs
Aldehyde dehydrogenases
- AML
Acute myeloid leukemia
- BM
Bone marrow
- BMF
Bone marrow failure
- BWA
Burrow-Wheeler Aligner
- CNVs
Copy number variants
- FA
Fanconi anemia
- HLA
Human leukocyte antigen
- HRD
HLA-haploidentical related donors
- HSCT
Hematopoietic stem cell transplantations
- ICLs
Interstrand crosslinks
- InDels
Insertions/deletions
- MAF
Minimal allele frequency
- MDS
Myelodysplastic syndrome
- MRD
HLA-matched related donors
- MUD
HLA-matched unrelated donors
- PB
Peripheral blood
- PCR
Polymerase chain reaction
- SNVs
Single nucleotide variants
- THS
Targeted high-throughput sequencing
- UUC
HLA-unmatched unrelated cord blood
- UUD
HLA-unmatched unrelated donors
- WGS
Whole genomic sequencing
Authors’ contributions
DN reviewed the medical history of the patients, analyzed sequencing data, and wrote the manuscript. JZ, FW, WZ, XM, and LL performed the sequencing process, analyzed the data, and wrote the manuscript. XC and YZ analyzed the morphology and karyotype results, PC designed the bioinformatic analysis process. MX analyzed and interpreted the clinical significance of the genetic variation, guided the clinical diagnosis and treatment. TW and PW carried out the morphologic and cytogenetic study. WT, MW, and KC analyzed the clinical data and supervised the study. HL designed and supervised the study. All authors have read and approved the manuscript.
Funding
Retrospective WGS, like cost of reagent, was supported by grants from the Shandong Nature Science Fund (ZR2016HP02) and Peking University Medicine Seed Fund for Interdisciplinary Research (BMU2018ME002, supported by the Fundamental Research Funds for the Central Universities).
Availability of data and materials
The datasets generated during the current study are available in the Sequence Read Archive (SRA) repository of the National Center for Biotechnology Information [accession number: PRJNA631475; https://www.ncbi.nlm.nih.gov/bioproject/631475]. The reference datasets used in this study is human reference genome (hg19).
Ethics approval and consent to participate
The study was approved by the ethics committee of the Hebei Yanda Lu Daopei hospital. Written informed consent to genetic analysis and participate in this study was obtained from the all of the participants or parents or legal guardians of any participant under the age of 16.
Consent for publication
Written informed consent for publication of clinical details was obtained from the parents or legal guardians of any participant under the age of 18.
Competing interests
The authors declare that no competing interests in this study.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12881-020-01057-3.
References
- 1.Kimble DC, Lach FP, Gregg SQ, Donovan FX, Flynn EK, Kamat A, Young A, Vemulapalli M, Thomas JW, Mullikin JC, Auerbach AD, Smogorzewska A, Chandrasekharappa SC. A comprehensive approach to identification of pathogenic FANCA variants in Fanconi anemia patients and their families. Hum Mutat. 2018;39(2):237–254. doi: 10.1002/humu.23366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frohnmayer D, Frohnmayer L, Guinan E, Kennedy T, Larsen K. Fanconi anemia: guidelines for diagnosis and management. 4. New York: Fanconi Anemia Research Fund, Inc; 2014. [Google Scholar]
- 3.Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, Hanenberg H, Auerbach AD. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101(4):1249–1256. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- 4.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in the National Cancer Institute inherited bone marrow failure syndrome cohort after fifteen years of follow-up. Haematologica. 2018;103(1):30–39. doi: 10.3324/haematol.2017.178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Risitano AM, Marotta S, Calzone R, Grimaldi F, Zatterale A, Contributors R. Twenty years of the Italian Fanconi Anemia Registry: where we stand and what remains to be learned. Haematologica. 2016;101(3):319–327. doi: 10.3324/haematol.2015.133520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alter BP, Giri N, Savage SA, Peters JA, Loud JT, Leathwood L, Carr AG, Greene MH, Rosenberg PS. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol. 2010;150(2):179–188. doi: 10.1111/j.1365-2141.2010.08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang QS, Tang W, Deater M, Phan N, Marcogliese AN, Li H, Al-Dhalimy M, Major A, Olson S, Monnat RJ, Jr, Grompe M. Metformin improves defective hematopoiesis and delays tumor formation in Fanconi anemia mice. Blood. 2016;128(24):2774–2784. doi: 10.1182/blood-2015-11-683490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, Arakawa H, Buerstedde JM, Gillespie DAF, Sale JE, Yamazoe M, Bishop DK, Takata M, Takeda S, Watanabe M, Swenberg JA, Nakamura J. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67(23):11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- 9.Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475(7354):53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 10.Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489(7417):571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- 11.Klyosov AA, Rashkovetsky LG, Tahir MK, Keung WM. Possible role of liver cytosolic and mitochondrial aldehyde dehydrogenases in acetaldehyde metabolism. Biochemistry. 1996;35(16):4445–4456. doi: 10.1021/bi9521093. [DOI] [PubMed] [Google Scholar]
- 12.Crabb DW, Edenberg HJ, Bosron WF, Li TK. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest. 1989;83(1):314–316. doi: 10.1172/JCI113875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hira A, Yabe H, Yoshida K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Nakamura J, Kojima S, Ogawa S, Matsuo K, Takata M, Yabe M. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood. 2013;122(18):2306–2309. doi: 10.1182/blood-2013-06-507962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yabe M, Koike T, Ohtsubo K, Imai E, Morimoto T, Takakura H, Koh K, Yoshida K, Ogawa S, Ito E, Okuno Y, Muramatsu H, Kojima S, Matsuo K, Mori M, Hira A, Takata M, Yabe H. Associations of complementation group, ALDH2 genotype, and clonal abnormalities with hematological outcome in Japanese patients with Fanconi anemia. Ann Hematol. 2019;98(2):271–280. doi: 10.1007/s00277-018-3517-0. [DOI] [PubMed] [Google Scholar]
- 15.Li N, Ding L, Li B, Wang J, D'Andrea AD, Chen J. Functional analysis of Fanconi anemia mutations in China. Exp Hematol. 2018;66:32–41. doi: 10.1016/j.exphem.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellström-Lindberg E, Tefferi A, Bloomfield CD. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 17.Nie D, Cao P, Wang F, Zhang J, Liu M, Zhang W, Liu L, Zhao H, Teng W, Tian W, Chen X, Zhang Y, Nan H, Wei Z, Wang T, Liu H. Analysis of overlapping heterozygous novel submicroscopic CNVs and FANCA-VPS9D1 fusion transcripts in a Fanconi anemia patient. J Hum Genet. 2019;64(9):899–909. doi: 10.1038/s10038-019-0629-x. [DOI] [PubMed] [Google Scholar]
- 18.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL. Standards and guidelines for the interpretation of sequence variants a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaffer L, Mcgowan J, Schmid M, Schmid M, Schmidt M. ISCN 2013: an international system for human cytogenetic nomenclature. Basel: S. Karger; 2013. [Google Scholar]
- 20.Chandra S, Levran O, Jurickova I, Maas C, Kapur R, Schindler D, Henry R, Milton K, Batish SD, Cancelas JA, Hanenberg H, Auerbach AD, Williams DA. A rapid method for retrovirus-mediated identification of complementation groups in Fanconi anemia patients. Mol Ther. 2005;12(5):976–984. doi: 10.1016/j.ymthe.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 21.Gille JJ, Floor K, Kerkhoven L, Ameziane N, Joenje H, De Winter JP. Diagnosis of Fanconi anemia: mutation analysis by multiplex ligation-dependent probe amplification and PCR-based Sanger sequencing. Anemia 603253. 10.1155/2012/603253. [DOI] [PMC free article] [PubMed]
- 22.Ameziane N, Errami A, Leveille F, Fontaine C, de Vries Y, van Spaendonk RM, de Winter JP, Pals G, Joenje H. Genetic subtyping of Fanconi anemia by comprehensive mutation screening. Hum Mutat. 2008;29(1):159–166. doi: 10.1002/humu.20625. [DOI] [PubMed] [Google Scholar]
- 23.De Rocco D, Bottega R, Cappelli E, Cavani S, Criscuolo M, Nicchia E, Corsolini F, Greco C, Borriello A, Svahn J, Pillon M, Mecucci C, Casazza G, Verzegnassi F, Cugno C, Locasciulli A, Farruggia P, Longoni D, Ramenghi U, Barberi W, Tucci F, Perrotta S, Grammatico P, Hanenberg H, Della Ragione F, Dufour C, Savoia A, Bone Marrow Failure Study Group of the Italian Association of Pediatric O-H Molecular analysis of Fanconi anemia: the experience of the Bone Marrow Failure Study Group of the Italian Association of Pediatric Onco-Hematology. Haematologica. 2014;99(6):1022–1031. doi: 10.3324/haematol.2014.104224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yagasaki H, Hamanoue S, Oda T, Nakahata T, Asano S, Yamashita T. Identification and characterization of novel mutations of the major Fanconi anemia gene FANCA in the Japanese population. Hum Mutat. 2004;24(6):481–490. doi: 10.1002/humu.20099. [DOI] [PubMed] [Google Scholar]
- 25.Nookala RK, Hussain S, Pellegrini L. Insights into Fanconi Anaemia from the structure of human FANCE. Nucleic Acids Res. 2007;35(5):1638–1648. doi: 10.1093/nar/gkm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayas M, Saber W, Davies SM, Harris RE, Hale GA, Socie G, LeRademacher J, Thakar M, Deeg HJ, Al-Seraihy A, Battiwalla M, Camitta BM, Olsson R, Bajwa RS, Bonfim CM, Pasquini R, Macmillan ML, George B, Copelan EA, Wirk B, Al Jefri A, Fasth AL, Guinan EC, Horn BN, Lewis VA, Slavin S, Stepensky P, Bierings M, Gale RP. Allogeneic hematopoietic cell transplantation for fanconi anemia in patients with pretransplantation cytogenetic abnormalities, myelodysplastic syndrome, or acute leukemia. J Clin Oncol. 2013;31(13):1669–1676. doi: 10.1200/JCO.2012.45.9719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luna-Fineman S, Shannon MK, Lange JB. Childhood Monosomy 7 epidemiology, biology, and mechanistic implications. Blood. 1995;85(8):1985–1999. doi: 10.1182/blood.V85.8.1985.bloodjournal8581985. [DOI] [PubMed] [Google Scholar]
- 28.Svahn J, Bagnasco F, Cappelli E, Onofrillo D, Caruso S, Corsolini F, De Rocco D, Savoia A, Longoni D, Pillon M, Marra N, Ramenghi U, Farruggia P, Locasciulli A, Addari C, Cerri C, Mastrodicasa E, Casazza G, Verzegnassi F, Riccardi F, Haupt R, Barone A, Cesaro S, Cugno C, Dufour C. Somatic, hematologic phenotype, long-term outcome, and effect of hematopoietic stem cell transplantation. An analysis of 97 Fanconi anemia patients from the Italian national database on behalf of the Marrow Failure Study Group of the AIEOP (Italian Association of Pediatric Hematology-Oncology) Am J Hematol. 2016;91(7):666–671. doi: 10.1002/ajh.24373. [DOI] [PubMed] [Google Scholar]
- 29.Mori M, Hira A, Yoshida K, Muramatsu H, Okuno Y, Shiraishi Y, Anmae M, Yasuda J, Tadaka S, Kinoshita K, Osumi T, Noguchi Y, Adachi S, Kobayashi R, Kawabata H, Imai K, Morio T, Tamura K, Takaori-Kondo A, Yamamoto M, Miyano S, Kojima S, Ito E, Ogawa S, Matsuo K, Yabe H, Yabe M, Takata M. Pathogenic mutations identified by a multimodality approach in 117 Japanese Fanconi anemia patients. Haematologica. 2019. 10.3324/haematol.2018.207241.
- 30.Rosenberg PS, Alter BP, Ebell W. Cancer risks in Fanconi anemia: findings from the German Fanconi Anemia Registry. Haematologica. 2008;93(4):511–517. doi: 10.3324/haematol.12234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary methods.
Additional file 2: Table S1. Details of 22 FA-related genes.
Additional file 3. Data of hromosome breakage test.
Additional file 4: Table S3. Mutation interpretations.
Data Availability Statement
The datasets generated during the current study are available in the Sequence Read Archive (SRA) repository of the National Center for Biotechnology Information [accession number: PRJNA631475; https://www.ncbi.nlm.nih.gov/bioproject/631475]. The reference datasets used in this study is human reference genome (hg19).