Abstract
Background
In food toxicology, there is growing interest in studying the impacts of foodborne nanoparticles (NPs, originating from food additives, food supplements or food packaging) on the intestinal microbiome due to the important and complex physiological roles of these microbial communities in host health. Biocidal activities, as described over recent years for most inorganic and metal NPs, could favour chronic changes in the composition and/or metabolic activities of commensal bacteria (namely, intestinal dysbiosis) with consequences on immune functions. Reciprocally, direct interactions of NPs with the immune system (e.g., inflammatory responses, adjuvant or immunosuppressive properties) may in turn have effects on the gut microbiota. Many chronic diseases in humans are associated with alterations along the microbiota-immune system axis, such as inflammatory bowel diseases (IBD) (Crohn’s disease and ulcerative colitis), metabolic disorders (e.g., obesity) or colorectal cancer (CRC). This raises the question of whether chronic dietary exposure to inorganic NPs may be viewed as a risk factor facilitating disease onset and/or progression. Deciphering the variety of effects along the microbiota-immune axis may aid the understanding of how daily exposure to inorganic NPs through various foodstuffs may potentially disturb the intricate dialogue between gut commensals and immunity, hence increasing the vulnerability of the host. In animal studies, dose levels and durations of oral treatment are key factors for mimicking exposure conditions to which humans are or may be exposed through the diet on a daily basis, and are needed for hazard identification and risk assessment of foodborne NPs. This review summarizes relevant studies to support the development of predictive toxicological models that account for the gut microbiota-immune axis.
Conclusions
The literature indicates that, in addition to evoking immune dysfunctions in the gut, inorganic NPs exhibit a moderate to extensive impact on intestinal microbiota composition and activity, highlighting a recurrent signature that favours colonization of the intestine by pathobionts at the expense of beneficial bacterial strains, as observed in IBD, CRC and obesity. Considering the long-term exposure via food, the effects of NPs on the gut microbiome should be considered in human health risk assessment, especially when a nanomaterial exhibits antimicrobial properties.
Keywords: Intestinal microbiota, Gut dysbiosis, Nanoparticles, Silver, Titanium dioxide, Zinc oxide, Silicon dioxide, Gut inflammation, Obesity, Colorectal cancer
Background
Nanomaterials are widely used in various industrial manufacturing processes and have applications in everyday consumer products such as foodstuffs, healthcare, clothing, sunscreens and cosmetics. Particles with sizes in the nanoscale range, namely, nanoparticles (NPs; with one dimension between 1 and 100 nm), present unique physical and chemical properties due to their high surface area to volume ratio (e.g., in photocatalysis, mechanical, optical properties), and these properties differ from those of corresponding bulk forms [1–3]. Due to the wide-ranging applications of nanotechnology in everyday products, concerns have been raised regarding the potential health consequences for humans exposed to NPs from different sources and routes (oral, dermal, inhalation). However, the human risk assessment of the oral uptake of NPs is poorly documented compared to that of other routes, although NPs are commonly found in food additives and in food supplements for improving organoleptic properties, shelf life and texture, until used for the nanoencapsulation of specific functional ingredients [4–6].
Among inorganic NPs, titanium dioxide (TiO2), silver (Ag) and silicon dioxide (SiO2) are commonly used as food colouring or anti-caking agents, while others are added as food supplements, such as zinc oxide (ZnO), given that zinc is an essential trace element. Most of these NPs are also used to develop antimicrobial active food packaging or are incorporated into bio-based materials that come in contact with food to act as oxygen, moisture and carbon dioxide barriers. Oral uptake is thought to be one of the major routes of exposure to NPs for the general population, and studies focused on the fate and effects of foodborne NPs have recently gained considerable attention for the assessment of health risks and for regulatory purposes. Following ingestion, NPs interact with a complex gastrointestinal (GI) environment. The non-absorbed fractions of foodborne mineral NPs (or their ionic forms for soluble compounds, such as Ag and ZnO) are accumulated in the intestinal lumen as a result of daily consumption, and can directly interact with the intestinal microbiota colonizing the gut lumen as well as the mucus layer lining the epithelial surface [7–9]. A portion of the NPs then translocate through the epithelial barrier and are possibly captured by the intestinal immune cells (e.g., macrophages and dendritic cells), before reaching systemic circulation. Notably, the gut microbiota (formerly called the intestinal flora) plays important roles in a number of physiological functions as an indispensable substrate for host health. Indeed, the commensal microbial community not only contributes to digestion of dietary fibres leading to the production of key metabolites for host physiology, but also strongly interacts with epithelial cells to maintain an effective gut barrier separating the body from the external environment [10–12]. Intestinal microorganisms also communicate with local immune cells to shape specific responses by balancing tolerance and effector immune functions to various antigens [10, 13, 14]. Beyond the gut, there is also unique coordination between the intestinal microbiota and liver functions, as well as the brain. Indeed, alteration in the microbiota (namely, dysbiosis), in its ecology (microbial population) and/or metabolic functions (production of bacterial metabolites), has been implicated in various chronic GI and metabolic diseases and even neurodevelopmental disorders [15–21]. For general homeostasis of organisms, immune cells of the gut-associated lymphoid tissue (GALT) shape the microbiota from birth to adulthood in terms of composition, quality and activity and allow these microorganisms to be tolerated by the host for its lifetime. In this context, little attention has been paid to the occurrence of intestinal dysbiosis after daily uptake of nanosized and biocidal particles. Consequences of chronic oral exposure to foodborne NPs on GI functions require scrutiny, taking into account all intestinal compartments that are in close contact with the ingested NPs. This includes direct effects on microbiota composition and/or activity and indirect dysbiosis due to NP-mediated immune system dysfunctions, both leading to potential disruption of intestinal homeostasis, affecting the functions of a number of systemic organs, with long-term effects on health.
Here, we review data from in vivo and in vitro studies on the impacts of Ag, TiO2, SiO2 and ZnO along the gut microbiota-immune system axis and examine whether these reports are relevant for health risk assessment in humans in terms of dose levels and effects. Silver (referenced as E174 in the EU) is used to colour the surfaces of some products, such as cakes, ice creams, frozen desserts and chocolates, with exposure levels from children to adults ranging from 0.03 to 2.6 μg/kg of body weight (bw)/day (d) [22]. Food-grade TiO2 (E171) is a white pigment and brightening agent that is used in large amounts in confectionery items, white sauces and icing [3, 23, 24]. Depending on the exposure scenario, the European Food Safety Authority (EFSA) estimated that the daily exposure levels to E171 range from 0.2 to 1.9 mg/kg bw/d in infants, 0.9 to 10.4 mg/kg bw/d in children, and 0.3 to 6.8 mg/kg bw/d in adults [25]. Food-grade SiO2 (E551) is commonly added to powdered food as an anticaking agent, such as in salt, icing sugar, spices, dried milk and dry mixes [26, 27]. The EFSA estimated that daily exposure to E551 ranges between 0.8 and 74.2 mg/kg bw/d in infants, 2.7 and 31.2 mg/kg bw/d in children and 0.9 and 13.2 mg/kg bw/d in adults [28]. In addition, because zinc is important for human health, ZnO-NPs may be found as a nutrient source in supplements and functional foods [29]. To date, the majority of studies evaluating the toxicity of TiO2, SiO2, Ag and ZnO NPs used nanomodels, i.e., composed of strictly nanosized particles and often coated with different compounds to improve their stability and dispersability. Within the scope of food additives, exhibiting size mix in the nano- and submicron range, with particles as aggregates or agglomerates and batch to batch variation, studies should mainly consider oral exposure to food grade NPs originating from E171, E174 and E551 for human health risk assessment, that remains poorly documented. As most animal studies focused on Ag, TiO2, SiO2 and ZnO NPs do not address the potential impacts of these materials on the gut microbiota-immune axis, this review will focus on effects susceptible to affect the delicate balance between these microorganisms and their hosts. In addition, we compared the effect of NPs on the gut microbiome to the intestinal dysbiosis characteristics reported in chronic diseases such as colorectal cancer (CRC), obesity and inflammatory bowel diseases (IBD), where microbiota alteration play important pathogenic roles [15–19]. Understanding the impact of NPs on the crosstalk between the microbiota and the immune system will improve the understanding of the potential effects of chronic exposure to foodborne NPs on host physiology. These effects could lead to a favourable environment for the onset and progression of chronic diseases and/or disruption of the gut-liver and gut-brain axes under the influence of bacterial metabolites.
Main text
The intestinal microbiota-immune axis in host physiology
The human intestinal microbiota is a complex ecosystem mainly composed of bacteria, as well as archaea, viruses, fungi and protozoa. Colonized at birth, the adult human GI tract harbours 100 trillions of bacteria, including at least several hundred species and more than 7000 strains. A majority of these bacteria belong to the phyla Firmicutes and Bacteroidetes, representing approximately 90% of the microbial population, while other species are members of the phyla Proteobacteria, Verrucomicrobia, Actinobacteria, Fusobacteria and Cyanobacteria [30, 31]. The density of these bacterial populations increases from the proximal to the distal end of the intestine, with ~ 102–3 colony forming units (cfu) per gram in the proximal ileum and jejunum, ~ 107–8 cfu in the distal ileum, and ~ 1011–12 cfu in the ascending colon [32]. The biomass of these gut microbes is approximately 1–2 kg, which is similar to the weight of the human brain [33]. The whole genome of intestinal microorganisms (known as the gut microbiome) is 150 times larger than the human genome, providing a diverse range of biochemical and metabolic activities, allowing the microbiota to complement host physiology [31]. Indeed, the gut microbiota and the liver exhibit equal metabolic capacities [34] and due to its beneficial effects for human health, the gut microbiome is now being considered an hidden and indispensable organ of the human body, of which activities need to be integrated as the missing link into host physiology and the development of diseases [35–39].
Local and distant roles of the gut microbiome
The intestinal microbiota plays essential roles in GI functions: these microbes i) facilitate the digestion and fermentation of indigestible polysaccharides and produce vitamins, ii) are essential for the development and differentiation of the intestinal epithelium as well as of the GALT, iii) are involved in host immune defence against pathogens in the luminal content, and, finally, iv) contribute greatly to lifelong maintenance of intestinal homeostasis [40–43]. One of the major biological approaches to examine the importance of the gut microbiota for host physiology is the use of germ-free (GF) mice, which are raised in the absence of any microorganisms [44]. GF mice present an underdeveloped intestinal epithelium, which is consistent with the important role of the microbiota in the maintenance of a functional epithelial surface to prevent the passage of harmful intraluminal entities, including foreign antigens, microorganisms and microbial toxins [45–47]. Specifically, GF mice exhibit decreased brush border differentiation [48] and villus thickness [49] due to reduced cell regeneration [50] and increased cell cycle time [51]. Similar to GF mice, GF piglets show aberrant epithelial surface, notably lengthened villi and shortened crypts, and these effects are reversed by intestinal colonization with faecal microbiota or commensal Escherichia coli strain [52–55]. Studies in Drosophila and mice showed that commensal bacteria promote epithelial development and wound closure through mechanisms that involve reactive oxygen species (ROS) [56, 57]. The intestinal flora also regulates intestinal epithelial cell growth and differentiation indirectly by the production of metabolites such as short-chain fatty acids (SCFAs), and aryl hydrocarbon receptor (AhR) ligands [58–61]. SCFAs (i.e., butyrate, propionate and acetate) are by-products of the fermentation of non-digestible dietary fibres through the action of intestinal bacteria, while AhR ligands are mostly derived from tryptophan metabolism. Moreover, these two metabolite groups are involved in the preservation of intercellular tight junction integrity in the intestinal epithelium, thereby establishing the link between gut microbial activity and the maintenance of effective intestinal barrier function.
Beyond the gut, it is well recognized that the gut microbiome can also modulate the function and/or development of systemic organs, such as the liver and brain, an ongoing dialogue that is enabled by a large variety of bacterial metabolites (SCFAs, polyamines, retinoic acid, AhR ligands). These metabolites can affect distant organs either directly through the bloodstream or indirectly by signalling via nerves or hormones from the gut [20, 62–64]. Both microbiota composition and activity play key roles in such interactions, and dietary alterations may have a significant impact on the gut-liver and gut-brain axes. Consistent with these findings, brain development and social behaviour are impaired in GF mice, and colonization of these mice with a complex microbiota or specific bacterial strains alleviates most of these effects [65–72]. Moreover, GF mice showed an elevated stress response, which was decreased after colonization with wild-type (WT) microbiota or Bifidobacterium infantis, a SCFA producer, whereas colonization with the enteropathogenic proteobacteria E. coli aggravated the stress response [71]. Intestinal microbiota supplementation with Lactobacillus reuteri also restores social behaviours in young mice born to mothers fed a high-fat diet that is known to alter microbiota composition [72]. The social deficit observed in these young mice was not rescued after Lactobacillus johnsonii supplementation, indicating that a specific metabolite produced by L. reuteri is required [72]. This metabolite remains to be determined, but L. reuteri (among other commensals) is able to metabolize the essential amino acid tryptophan to AhR ligands [73, 74] that are known to protect the brain from inflammation [75]. Regarding other bacterial products, the increased blood-brain barrier permeability commonly reported in GF mice was reversed after colonization with the SCFA-producing Clostridium tyrobutyricum or Bacteroides thetaiotaomicron [69]. SCFAs are also involved in the maturation of microglia, which are responsible for immune defence in the brain [70]. Along the gut-liver axis, AhR ligands such as indole are also able to alleviate liver inflammation in mice [76]. Moreover, SCFA administration in animal models of obesity decreases the accumulation of fat in the liver and improves insulin resistance through mechanisms involving liver glycogenesis and lipogenesis [77]. Again, administration of SCFA-producing bacteria in rats helped prevent non-alcoholic fatty liver disease and lower triglyceride levels [78], reinforcing the idea that the SCFA production level is crucial for liver homeostasis and susceptibility to metabolic diseases.
Microbiota and the gut-associated lymphoid tissue
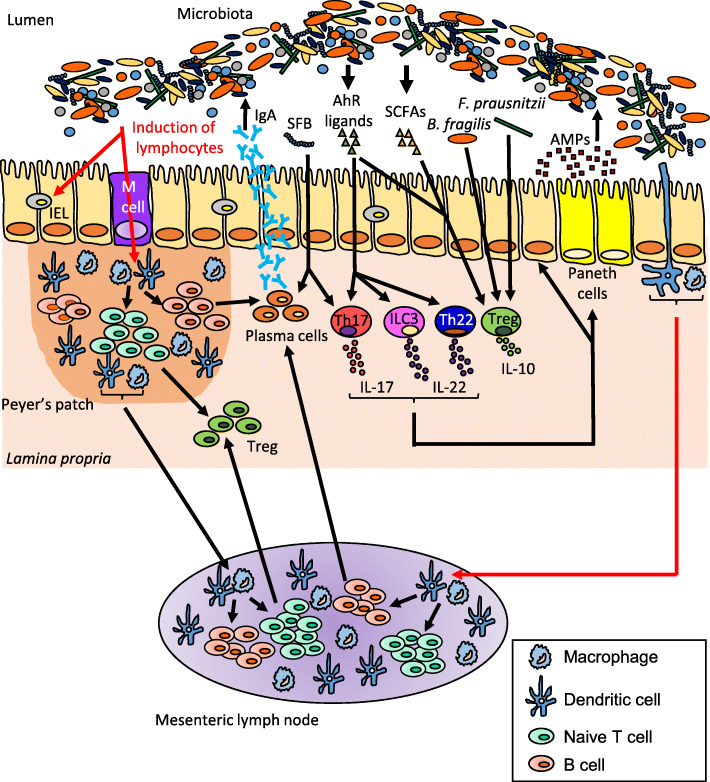
The intestinal mucosa, encompassing the surface epithelium and the GALT, represents a complex interface participating in the homeostatic relationships between the gut microbiota and the host immune system in response to the environment and diet [79–82]. A balanced dialogue at this interface requires collaboration between mucosal immune cells (macrophages, dendritic cells (DCs), B and T lymphocytes) and epithelial cells producing antimicrobial peptides (e.g., defensins, cathelicidins and histatins) and enzymes such as lysozyme produced by the secretory Paneth cells (Fig. 1). Moreover, GALT also consists of isolated or aggregated lymphoid follicles that form Peyer’s patches (PPs) in the small intestine. PPs exhibit a strong ability to sample luminal antigens and bacteria through microfold (M) cells, a class of antigen-presenting cells and immune sensors of the gut located in the follicle-associated epithelium overlying PP domes (Fig. 1). M cells transport bacteria and macromolecules to the underlying immune cells where they play a critical role in gut-oriented immune responses, triggering the induction of immune tolerance to innocuous antigens derived from food and microbiota or activation of immune defence against pathogens (Fig. 1). In the lamina propria (LP), DCs complement these surveillance systems in linking innate immune sensing of the environment to the initiation of adaptive immune responses. By extending transepithelial dendrites into the gut lumen, DCs sample antigens and subsequently migrate to mesenteric lymph nodes (MLNs), where they initiate T-cell responses with an intestinal tropism. Under homeostatic conditions, DCs preferentially promote regulatory T-cell (Treg) responses for oral tolerance [79, 83, 84] while they can also initiate the immunity to pathogens [79, 85] (Fig. 1).
Fig. 1.
The gut microbiota modulates the intestinal immune response. The gut microbiota influences the development of T cell subsets, intraepithelial lymphocytes (IELs) and are critical for the induction of plasma cells which produce immunoglobulin A (IgA). Dendritic cells (DCs) sample microbial antigens that pass through the epithelial barrier via microfold (M) cells or capture antigens from the lumen directly by extending dendrites between the intestinal epithelial cells. Some of these DCs migrate to the mesenteric lymph nodes and induce naïve T cells differentiation into regulatory T-cell (Treg) by production of transforming growth factor β (TGF-β) and retinoic acid. Segmented filamentous bacteria (SFB) exhibit pro-inflammatory effects by inducing IL-17 and IgA production, whereas Bacteroides fragilis, Faecalibacterium prausnitzii and short-chain fatty acids (SCFAs) exhibit anti-inflammatory effects via recruitment of Treg that produce the immunosuppressive cytokine IL-10. The intestinal flora also regulates immune response by the production of aryl hydrocarbon receptor (AhR) ligands able to activate AhR, highly expressed on IELs, Th17, Th22, innate lymphoid cells group 3 (ILC3) that produce IL-17 and/or IL-22. These cytokines induce secretion of antimicrobial peptides (AMPs) from Paneth cells and intestinal epithelial cells. AMPs shape the microbiota and are also involved in colonization resistance against pathogens
The intestinal microbiota is essential for GALT maturation and development, as proven by GF mice exhibiting highly immature immune system. The intestine of these mice exhibit small MLNs and PPs and decreased numbers of immune cells, such as immunoglobulin A (IgA)-producing plasma cells and T lymphocytes, for example, LP CD4+ T cells and intraepithelial αβ CD8+ T cells, leading to a reduced capacity to combat pathogens [40, 86]. All these immune alterations are microbiota dependent and can be reversed by faecal microbiota colonization [87]. Notably, recent studies highlighted the existence of anti- or pro-inflammatory microorganisms in the gut. In mice, segmented filamentous bacteria (SFB) direct the accumulation of pro-inflammatory T helper 17 (Th17) cells in the LP [88, 89] (Fig. 1). This finding is in contrast with the results for other bacteria, such as Bacteroides fragilis [90, 91] and Faecalibacterium prausnitzii [92–94], which exhibit anti-inflammatory activity via metabolite production and/or the recruitment of Treg-producing interleukin (IL)-10, a potent immunosuppressive cytokine (Fig. 1). Indeed, most metabolites derived from commensal bacteria, such as SCFAs and AhR ligands, may regulate immune cell functions via indirect and direct mechanisms [10, 74, 95] (Fig. 1). In the intestine, butyrate is a key regulator of host energy, and the butyrate level in the lumen is correlated with the abundance of Lactobacillus, Allobacterium, Bifidobacterium, Dorea and Blautia. Many cells are affected by SCFAs, such as intestinal epithelial cells (IECs), DCs and T cells, leading to modulation of different aspects of cell development, survival and function and to modulation of enzymes and transcription factors [96, 97]. Gurav and colleagues demonstrated that SCFAs modulate T cell activation and effector response by inducing a tolerogenic profile [98]. This study also showed that DCs stimulated with SCFAs in vitro exhibited increased expression of indoleamine 2,3-dioxygenase 1 [98], the role of which in inflammation is to attenuate the immune response through an immunosuppressive effect on T cell proliferation and activation. On the other hand, AhR ligands produced by the gut microbiota induce AhR activation, with endogenous downstream functions on the cell cycle, immune response, and cell differentiation [99–101] (Fig. 1). IECs have a role in the regulation of AhR ligands availability to GALT, including macrophages, DCs, innate lymphoid cells (ILCs), Th17/Th22 cells and intraepithelial lymphocytes (IELs). All of these immune cell types express AhR, activation of which by microbiota-derived appropriate ligands being pivotal in the regulation of mucosal immune responses in the gut [99].
Interactions of inorganic NPs with the gut microbiome
Despite several studies highlighting the deleterious effects of nanomaterials in soil [102] or plant [103] microbial communities, the impact of inorganic NPs on bacteria that colonize the gut remain poorly documented. Tables 1 and 2 provides a detailed description of data on NPs that exhibit biocidal effects on intestinal bacteria, mainly metal NPs such as Ag, ZnO and TiO2, with additional information for SiO2. Given the very limited information on nanostructured food additives (namely, E174, E171, E551 in EU for food-grade formulations of nano-Ag, TiO2 and SiO2, respectively), it should be noted that this review mostly provides data obtained with NP models. The in vivo studies are categorized according to the duration of treatment and dose levels (Table 1), followed by classification of in vitro data on the human microbiota (Table 2), to provide information relevant for risk assessment of NP exposure through food.
Table 1.
Studies using animal models to determine the impact of NPs on the gut microbiota
| Ag-NP | TiO2-NP | SiO2-NP | ZnO-NP | ||||
|---|---|---|---|---|---|---|---|
| Duration (days) | ≤14 | ≥28 | 7 | ≥28 | 7 | ≤14 | ≥28 |
| Actinobacteria | ↗[104] | ||||||
| Bifidobacterium (g) | ↙[105] | ||||||
| Corynebacterium (g) | ↙[106] | ||||||
| Rhodococcus (g) | ↙[104] | ||||||
| Bacteroidetes | ↗[107] | ↗[105] or ↙[108]♦ | ↙[107]♦ | ↙[104] | ↙[107]♦ | ↙[109] | ↗[110] |
| Bacteroidaceae (f) | ↙[108]♦ | ↗[110] | |||||
| Bacteroides (g) | ↗[105] or ↙[108]♦ | ↙[107]♦ | ↙[107]♦ | ↗[110] | |||
| Bacteroides uniformis | ↙[106] | ||||||
| Odoribacteraceae (f) | ↙[108]♦ | ||||||
| Prevotellaceae (f) | ↙[109] | ||||||
| Prevotella (g) | ↗[107] | ↙[104] | ↗[107]♦ | ↙[109] | |||
| Rikenellaceae (f) | ↗[108]♦ | ↙[110] | |||||
| Alistipes (g) | ↗[107] | ↗[107]♦ | ↙[110] | ||||
| S24–7 (f) | ↙[108]♦ | ↙[109] | |||||
| Barnesiella (g) | ↙[104] | ||||||
| Firmicutes | ↙[107] | ↙[105] or ↗[108]♦ | ↙[104] | ↗[107]♦ | ↗colon, ↙ileum [109] | ↙[110] | |
| Bacillaceae (f) | ↗[109] | ||||||
| Bacillus (g) | ↗[109] | ||||||
| Christensenellaceae (f) | ↙[106] | ||||||
| Erysipelotrichaceae (f) | ↙[104] | ||||||
| Turicibacter (g) | ↙[104] | ↗[111] | |||||
| Lachnospiraceae (f) | ↙[107] | ↗[108]♦ | ↗[107]♦ | ||||
| Blautia (g) | ↗[108]♦ | ||||||
| Coprococcus (g) | ↗[108]♦ | ||||||
| Coprococcus eutatus | ↙[106] | ||||||
| Lactobacillaceae (f) | ↗[108]♦ | ↗colon, ↙ileum [109] | ↙[110, 111] | ||||
| Lactobacillus (g) | ↙[107] | ↙[105] or ↗[108]♦ | ↙[104] | ↙[107]♦ | ↗colon, ↙ileum [109] | ↙[110, 111] | |
| Peptococcaceae (f) | ↙[106] | ||||||
| Ruminococcaceae (f) | ↗[107]♦ | ||||||
| Oscillospira (g) | ↙[106] | ↙[109] | |||||
| Streptococcaceae (f) | ↗[109] | ||||||
| Streptococcus (g) | ↗[109] | ||||||
| Clostridium (g) | ↙[106] | ↗[111] | |||||
| Dehalobacterium (g) | ↙[106] | ||||||
| Oscillibacter (g) | ↗[107]♦ | ||||||
| SMB53 (g) | ↗[111] | ||||||
| Proteobacteria | ↗[104] | ↗[107]♦ | ↗[109] | ↙[111] | |||
| Enterobacteriaceae (f) | ↗[105] | ||||||
| Escherichia/Shigella (g) | ↗[104] | ||||||
| Halomonadaceae (f) | ↗[109] | ||||||
| Halomonas (g) | ↗[109] | ||||||
| Aggregatibacter pneumotropica | ↙[106] | ||||||
♦ Studies using NPs at human relevant doses
Table 2.
In vitro studies exploring the impact of NPs on human microbiota
| Ag-NP | TiO2-NP | |
|---|---|---|
| Duration (days) | 2 | 2 |
| Bacteroidetes | ||
| Bacteroides ovatus | ↙[112] | ↙[113] |
| Firmicutes | ||
| Acidaminococcus intestini | ↗[113] | |
| Clostridium cocleatum | ↗[113] | |
| Eubacterium rectale | ↙[112] | ↗[113] |
| Eubacterium ventriosum | ↗[113] | |
| Faecalibacterium prausnitzii | ↙[112] | |
| Roseburia faecalis | ↙[112] | |
| Roseburia intestinalis | ↙[112] | |
| Ruminococcus torques | ↙[112] | |
| Proteobacteria | ||
| Escherichia coli | ↗[112] | |
| Raoultella (sp) | ↗[112] | |
Impacts of nano-silver and titanium dioxide
Regarding Ag-NPs, several studies were first performed in rats or mice using high concentrations (up to 36 mg/kg bw/d) compared to human dietary levels (0.03 to 0.65 μg/kg bw/d [22]) and with different methods of administration (gavage, addition to drinking water or incorporation into food pellets) and/or exposure times (between 7 and 90 days). Contradictory results were reported, with some studies showing no impact on microbiota ecology and activity and, others revealing profound alterations. Indeed, two studies in rats and mice (9 and 10 mg/kg bw/d, respectively) showed non-significant alterations in the caecal microbiota composition following 28 days of oral exposure to Ag-NPs (stabilized with polyvinylpyrolidone or citrate), regardless of particle size (14, 20 and 110 nm) [114, 115]. In contrast, rats chronically exposed for 13 weeks to Ag-NPs (10, 75 and 110 nm at 9, 18 and 36 mg/kg bw/d) showed a shift in their ileal microbial populations towards increased proportions of Bacteroidetes and pathogenic gram-negative bacteria, and decreased proportions of Firmicutes, Lactobacillus and Bifidobacterium [105]. These effects on Lactobacillus and Bacteroidetes appeared to be more prominent in males than in females, whereas the increased proportion of Enterobacteria was higher in female rats [105]. In addition to the dose of exposure, authors considered NP size as an influencing factor on host gene expression, but the heterogeneity of the results does not allow to fully conclude on a size and/or dose effect of Ag-NPs [105]. The shift in the Firmicutes/Bacteroidetes (F/B) ratio has been confirmed in the faecal microbiota of rats and mice orally given Ag-NPs at 2.5 or 3.6 mg/kg bw/d for 7 days or 2 weeks, respectively [106, 107]. Notably, these authors concluded that microbiota alterations depend on the shapes of Ag-NPs [106], suggesting that NPs structure (which affects the surface-to-volume ratio) could determine the reactivity with gut bacteria. Finally, a shift in the F/B ratio in the faecal microbiota has been reported in mice orally exposed for 28 days to food supplemented with relevant doses of Ag-NPs for human intake [108]. The authors noted that these gut microbiota alterations did not occur when mice were fed aged food pellets due to sulfidation of Ag-NPs in contact with the sulfur-containing food matrix [108]. Because Ag sulfidation limits the release of the toxic Ag+ ions responsible for antimicrobial effects, one may hypothesize that this ageing mechanism could contribute to the aforementioned discrepancies in rodent studies. Some differences in terms of gut microbiota alterations might also be due to differences in NP doses, durations of treatment, microbiota sampling sites along the rodent intestine (i.e., small bowel or more distal regions in the caecum or colon), and the methods and analysis techniques used to determine bacterial composition. Regarding the sampling site, the faecal samples from the distal colon should remain the predominant material for microbial community analysis in toxicity testings with NPs, because this region is characterized by a low transit time making reservoir for NP accumulation after chronic exposure, as reported in rats daily treated with E171 [116], with expected stable impacts on the microbiome.
Although the impacts of Ag-NP ingestion on the rodent microbiota are well documented, their effects on the human microbiota remain to be fully characterized. Only one study has determined, in vitro, the short-term impacts of Ag-NPs on a defined human bacterial community called microbial ecosystem therapeutic-1 (MET-1, consisting of 33 bacterial strains, established from stool obtained from a healthy donor) [112]. The authors observed a reduction in culture-generated gas production and changes in fatty acid methyl ester profiles after 48 h of exposure to several concentrations of Ag-NPs (0–200 mg/L) [112]. Microbiota sequencing confirmed alterations in bacterial composition, characterized by decreased abundances of Bacteroides ovatus, F. prausnitzii, Roseburia faecalis, Roseburia intestinalis, Eubacterium rectale and Ruminococcus torques, and increased proportions of Raoultella sp. and of E. coli, i.e., a negative shift in the microbial community that favours the growth of pathogenic bacteria [112].
The same bacterial community, MET-1, has also been used in a custom colon reactor to determine the impact on the human microbiota of TiO2-NPs and different food-grade TiO2 formulations (the E171 food additive in EU) [113]. A minor effect on bacterial ecology, restricted to a decrease in B. ovatus in favour of Clostridium cocleatum, was reported after 48 h of TiO2 treatment [113]. However, using long exposure times in vitro (5 days) at environmentally relevant concentrations, significant changes in bacterial metabolites were observed in the human colon microbiome, including in SCFA production [117]. To date, whether the effect of TiO2 (on phyla, strains and/or metabolic activity) could occur in vivo at dietary levels for humans has remained poorly studied. Nevertheless, oral bioavailability studies in rodents and humans clearly showed very limited systemic absorption of TiO2 (0.1 to 0.6% of the initial dose, respectively) [118, 119]. This finding indicates that at least 99% of the ingested TiO2 matter accumulates in the lumen of the gut with the commensals in permanent contact with the particles, especially due to repeated exposure, with the potential for alterations in the growth profiles of bacteria as shown in vitro for E171 [8]. Mice exposed for one week to TiO2-NPs at a relevant dose for humans (2.5 mg/kg bw/d) did not reveal any changes in the faecal microbiota composition [107]. However, an increased proportion of potentially harmful Actinobacteria and Proteobacteria and a decrease in the abundance of beneficial Firmicutes and Bacteroidetes were observed in the same region after 28 days of oral treatment, but at higher dosage (100 mg/kg bw/d) [104]. Currently, due to the absence of specific tests for assessing NP-related changes in the composition and activity of the gut microbiota, it is important to elucidate the potential induction of dysbiosis by TiO2-NPs to perform these examinations at appropriate doses and over long periods for relevant dietary exposure models.
Impacts of zinc oxide and silicon dioxide
Zinc oxide NPs exhibit potent antimicrobial activity on various non-intestinal microorganisms. ZnO is currently listed as a generally recognized as safe (GRAS) material by the Food and Drug Administration and is commonly used as food supplement or in food packaging. Due to a widespread use to enhance the bioavailability of zinc in the body, these metal particles are good candidates for NP-induced intestinal dysbiosis, but this aspect has been poorly explored to date. Most of the available studies have been conducted using the microbiota from piglets and hens due to efficient functioning of ZnO to promote growth and relieve diarrhoea in livestock animals [120]. The ingestion of ZnO-NPs at 600 mg/kg for 14 days increased the bacterial richness and diversity in the ileum of piglets, while these parameters decreased in both caecum and colon [109]. Nevertheless, the authors observed different effects on microbiota composition according to the collection sites along the GI tract. An increased abundance of Streptococcus concomitant with a decreased proportion of Lactobacillus was observed in the ileum, while the abundance of Lactobacillus increased and the abundance of Oscillospira and Prevotella decreased in the colon [109]. In hens, gene-sequencing analysis of the 16S rRNA of the ileal digesta microbiota showed that the richness of the bacterial community decreased in a dose-dependent manner (25, 50 and 100 mg/kg for 9 weeks), with increased populations of Bacteroidetes, Fusobacteria and Bacilli and decreased populations of Proteobacteria and Lactobacillus [111]. Moreover, blood metabolite analysis clearly indicated a positive correlation between the richness of the microbiota and choline, lactate and methionine metabolism, suggesting an impact of ZnO-NPs on bacterial metabolic activity [111]. These observations are consistent with those of another study that showed decreased abundance of the SCFA-producing Lactobacillus in the caecum of broiler chickens fed ZnO-NPs at 5 mg/kg of feed for 42 days [110]. One study focused on the human faecal microbiota isolated from a healthy donor confirmed the impacts of ZnO-NPs on the metabolic activity of the gut microbiota, impairing the production capacity of SCFAs as well as extracellular polymeric substances (i.e., polysaccharides, proteins, lipids, and extracellular DNA secreted by bacteria in protective biofilms) [117]. Overall, these studies showed that oral ingestion of ZnO-NPs can lead to changes in both the composition and metabolic activity of the intestinal microbiota, the main feature being decreased abundance of the genus Lactobacillus regardless of the animal model. Finally, with regard to non-metal NPs without well-known biocidal properties, one study in mice exposed to SiO2-NPs for one week at relevant dose for humans (2.5 mg/kg bw/d) reported enrichment and increased diversity of microbial species, with increased populations of Firmicutes and Proteobacteria and decreased proportion of Bacteroidetes and Lactobacillus [107]. Such an unexpected effect should be taken into consideration for risk assessment in the context of the low absorption rate in the human GI tract for precipitated or fumed (amorphous) silicate (E551) [28], leading to gut lumen accumulation.
Microbiota-immune system dysfunction in chronic diseases: could inorganic NPs favour host vulnerability?
NP-related GALT dysfunctions as possible inducers of disease development
Despite several in vitro studies highlighting the variety of immunotoxic effects of TiO2-NPs, SiO2-NPs, Ag-NPs and ZnO-NPs (Table 3) on bone marrow-derived cells and systemic and pulmonary immune cells [121–151], the potential impact of these NPs on GALT functions and consequences for the host remain poorly documented. From a risk perspective, current challenges include the determination of whether NP-related dysbiosis and immune dysfunction may increase the susceptibility to chronic diseases in humans where disruption of the microbiota-GALT crosstalk is central to pathogenesis, such as in CRC, obesity and IBD [13, 152, 153]. IBD, namely Crohn’s disease and ulcerative colitis, are chronic relapsing disorders of multifactorial aetiology characterized by intestinal dysbiosis and severe mucosal inflammation with epithelial injuries. In addition, gut dysbiosis and low-grade inflammation of the intestinal mucosa are often viewed as contributors to CRC and obesity [13, 152, 153]. Notably, micro- and nano-particles have been found in colon biopsies of patients with IBD and CRC, whereas the absence of these particles is consistently reported in the colon of healthy subjects [154]. Again, blood titanium (Ti) levels are high in IBD patients [155], and TiO2 particles of dietary origin were shown to be accumulated in PPs of IBD patients [156], including infants [157]. Such localization is consistent with data from studies in rats and mice that commonly report bioaccumulation of Ti, Ag or silicon (Si) in the ileum (including PPs) and the colon as a result of repeated oral treatment with TiO2-NPs (or E171) [107, 116, 158, 159], nano-Ag [160] or SiO2-NPs [107], respectively. In the colon of rats chronically exposed for 100 days to food-grade TiO2 (E171) at a relevant dose for humans (i.e., 10 mg/kg bw/d), a micro-inflammatory state of the mucosa has been demonstrated [116]. Using a ten-fold higher dosage, another study in mice exposed to TiO2 particle models (nano- and microsized) found increased production of Th1 pro-inflammatory cytokines (tumour necrosis factor alpha (TNF-α), interferon-gamma (IFN-γ), IL-12) and of the Th2 cytokine IL-4 in the mucosa of the small intestine after 10 days of exposure [161]. Inflammation associated with other immune system disturbances has also been observed following the ingestion of SiO2-NPs [107, 162] and Ag-NPs [107, 163], including impairment of oral tolerance mechanisms to food proteins and bacterial antigens present in the gut lumen [162, 163]. Furthermore, IBD-like symptoms, including intestinal upregulation of pro-inflammatory cytokines, were notably induced in mice treated with Ag-NPs at 2.5 mg/kg bw/d for 7 days [107]. Again, oral administration of TiO2-NPs worsened existing gut inflammation in a mouse model of acute colitis through activation of the inflammasome [155]. As an exception, the intestinal expression of pro-inflammatory genes was decreased in piglets exposed orally to ZnO-NPs for 2 weeks [109], which is consistent with ZnO-NPs exhibiting markedly dose-dependent effects on the remission of experimental colitis in mice [164].
Table 3.
In vitro studies on immunological properties of NPs
| NPs | cell response | cell frequence/activity | Potential effect |
|---|---|---|---|
| TiO2 (6-48 h) | ↗ oxidative stress (ROS) [121–125] | ↗ DC, μɸ, lymphocytes [121, 122, 126] | Innate inflammatory response [122, 126–128] |
| ↗ expression of TLR (TLR3, TLR4, TLR7 and TLR10) [126, 128] | ↗ naïve T cells [122] | Cytotoxicity and inflammation [122, 123, 125, 129] | |
| ↗ pro-inflammatory cytokines (IL-6, TNF-α, IL1-β) [121–123, 127, 130–132] | ↗ mature DC [122, 131, 132] | Adjuvant [121, 130–132] | |
| ↗ chemokines secretion (IL-8 and CXCL1) ↙ activity (IL-8) [122, 133] | ↗ mast cell activation [134] | Allergic response [134, 135] | |
| ↗ co-stimulatory molecules (CD80 and CD86) [122, 123, 131, 132] | ↙ neutrophil and μɸ mobility [133] | Tissue damage [124, 133] | |
| ↗ MPO, MMP-9 and NET [124, 136] | ↗ neutrophil activity [124, 136] | NETosis: inflammation, necrosis and apoptosis [124] | |
| ↗ β-hexosaminidase release [134] | |||
| inflammasome activation [123] | |||
| activation of NFkB pathway [128, 131] | |||
| SiO2 (6-48 h) | ↗ apoptose [123, 125, 137, 138] | ↙ DC and lymphocytes [123, 137, 138] | Imbalance of immune response [125, 126, 137–140] |
| ↗ pro inflammatory cytokines (IL1-β, IL-2, TNF-α) [121, 123, 126, 138, 139, 141] | ↗ DC maturation [123, 139] | Immunogenic or adjuvant potential [121, 123, 126, 140, 141] | |
| ↗ CD80, CD86 and MHCII [123, 138, 139] | ↗ M1 μɸ polarization [126, 140] | Inflammation [121, 123, 125, 126, 137–141] | |
| ↙ TLR9 expression [126] | ↗ neutrophil activity [142] | ↗ Susceptibility IBD [123, 126, 140], autoimmune diseases [139, 140] | |
| ↗ oxidative stress (ROS) [125] | ↗ cross-presentation [141] | NETosis: inflammation, necrosis and apoptosis [142] | |
| inflammasome activation [123] | Cytotoxic effect [121, 123, 125, 137, 138, 140] | ||
| NFkB activation [138] | Susceptibility to infection [126] | ||
| ↗ DNA release – NET [142] | Allergic response [135] | ||
| ZnO (6-48 h) | ↗ pro inflammatory cytokines (IL-1β, TNF-α, IL-6, IFN-ɣ) [125, 130, 143, 144] | ↗ DC activity [130, 145] | Cytotoxicity and inflammation [125, 129, 130, 134, 143, 146] |
| ↗ chemokines secretion (IL-8, CXCL9) [130, 144] | ↙ Lymphocytes [146] | Imbalance of immune response [125, 129, 130, 143, 144] | |
| ↗ oxidative stress (ROS) [125, 145] | ↗ neutrophil functions [136] | Chronic pathologies [143] | |
| induces neo-synthesis of polypeptides [144] | ↙ mast cell activation [134] | Allergenic response [134, 135] | |
| ↙ or ↗ apoptose [125, 129, 130, 144] | ↗ eosinophils [144] | Protective effect [134, 144] | |
| ↗ DNA damage [125, 129, 146] | Genomic instability [146] | ||
| ↙ β-hexosaminidase and histamine release [134] | Cell cycle imbalanced [125] | ||
| Ag (6-48 h) | ↗ oxidative stress (ROS) [147–149] | ↙ μɸ, lymphocyte [147–149] | Apoptosis and cytotoxicity [147–149] |
| ↗ DNA damage [148] | ↗ mast cell activation [135, 150, 151] | Inflammation/imbalance of immune response [147–149] | |
| ↗ pro inflammatory cytokines (IL-1β, TNF-α) [147, 149] | Allergic response [135, 150, 151] | ||
| inflammasome activation [147] | |||
| ↗ β-hexosaminidase release [135, 150, 151] |
CD Cluster of differentiation, CXCL1 chemokine ligand 1, DC dendritic cell, IBD Inflammatory bowel disease, Ig immunoglobulin, IL interleukin, μɸ macrophage, MHCII major histocompatibility complex II, MMP-9 matrix metalloproteinase 9, MPO myeloperoxidase, NET Neutrophil extracellular trap, NFkB nuclear factor-kappa B, ROS reactive oxygen species, TLR Toll-like receptor
Taken together, these data highlight the considerable impact of TiO2-NPs, Ag-NPs and SiO2-NPs on the modulation of the immune response in vivo (Table 4) [105, 116, 161–163, 165–170], which may in turn modulate the microbiota. Indeed, recent studies in mice have shown a number of immune deficiencies evoking intestinal dysbiosis in ways that predispose to disease. For example, mice lacking the caspase recruitment domain 9 (Card9−/−, a key adapter protein for innate immunity against a wide range of microorganisms) developed colitis in a microbiota-dependent manner [74]. Remarkably, this colitis phenotype was transmissible to WT mice via transfer of the Card9−/− microbiota and was rescued after inoculation with Lactobacillus strains [74]. Similarly, immune-driven dysbiosis was demonstrated in mice lacking Toll-like receptor 5 (Tlr5−/−), which detects bacterial flagellin. The mice exhibit an altered microbiota associated with metabolic disorders characterized by insulin resistance, hyperlipidaemia and increased fat deposition, and all these markers were also observed in WT mice colonized with the gut microbiota of Tlr5-deficient mice [171]. By describing the key role of the local immune system in controlling the composition and activity of the microbiota, these data support the idea that the strong interactions between NPs and GALT can also shape the microbiota of exposed individuals (Fig. 2).
Table 4.
In vivo studies on immunological properties of NPs
| NPs | Time | Cell response | Cell frequency/activity | Dose range | Potential effect | Ref |
|---|---|---|---|---|---|---|
| TiO2 | 1 -14 d | ↗ IFN-ɣ and IL-17 in spleen | ↗ Th17 and Th1 in spleen | 10 mg/kg bw/d | inflammation | [116] |
| ↗ DC frequency in PP | imbalance immune response | |||||
| ↙ Treg and T helper cells in PP | immunossuppression | |||||
|
↗ inflammatory cytokines in ileum (IL-12, IL-4, IL-23, TNF-α, IFN-γ) |
↗ T cells CD4+ in small intestin | 100 mg/kg bw/d | inflammation | [161] | ||
| 5 g/kg bw (single oral gavage) | no toxic effects | [165] | ||||
| > 28 d | ↗ TNF-α, IL-10 and IL-8 in colon | ↙ Treg and T helper cells in PP | 10 mg/kg bw/d | risk of preneoplasic lesions in colon | [116] | |
|
↗ IL-6, TNF-α, IL1-β in ovary (expression and production) |
↙ white cells and lymphocytes in blood | 2.5; 5; 10 mg/kg bw/d | inflammation and follicule atresia | [166] | ||
| fertility reduction | ||||||
| ↗ glucose in blood of young rats | ↗ mast cells activation in stomach | 10, 50, 200 mg/kg bw/d | alteration of gastrointestinal functions | [167] | ||
| Heart, Liver and Kidney injuries | ||||||
| SiO2 | 7 -14 d |
↙ inflammatory cytokines in blood (IL1-β, TNF-α, IL-12p70, IL-6 and IFN-ɣ) |
↙ Natural Killer acitivity in spleen | 750 mg/kg bw/d | immunossuppression | [168] |
| ↙ T cells and B cells proliferation in spleen | dysregulation of immune response | |||||
| ↙ Lymphocytes population in blood | ||||||
| ↗ IgG and IgE in blood | ↗ splenocyte proliferation | 0.1; 1; 10 mg/mouse/d | imbalance immune response (dose dependent) | [162] | ||
|
↗ inflammatory cytokines in spleen (INF-ɣ, IL-4, IL-5 and IL-17) |
risk of food allergy development | |||||
| ≥ 28 d | slight ↗ lymphocytes in blood | 2.5 mg/mouse/d | no effect | [169] | ||
| Ag | 7-14 d | ↗ TGF-β in blood |
↗ B and Natural Killer cells in blood ↗ T CD8+ population in blood |
1 mg/kg bw/d | imbalance immune response | [170] |
|
↗ IgG in blood, ↗ TGF-β and IFN-ɣ in spleen |
↗ splenocyte proliferation | 0.01; 0.1; 1; 10 mg/kg bw/d | imbalance immune response | [163] | ||
| ≥ 28 d |
↗ inflammatory cytokines in blood (IL1-β, IL-6, IL-4, IL-10, IL-12 and TGF-β) |
↗ T CD8+ population in blood | 0.25; 0.50; 1 mg/kg bw/d | inflammation | [170] | |
| ↗ IgE in blood | ↗ B cell in blood | risk of food allergy development | ||||
|
↙ gene expression in ileum (MUC3, TLR2, TLR4, FOXP3, IL-10, TGF-β) |
9 mg/kg bw/d | immunossuppression | [105] | |||
| sexual dimorphism response |
The relevant doses for humans are indicated in bold. DC dendritic cell, FOXP3 forkhead box P3, GPR43 G-protein coupled receptor 43, IFN-γ interferon-gamma, Ig immunoglobulin, IL interleukin, MUC3 mucin 3, NOD2 nucleotide-binding oligomerization domain 2, TGF-β transforming growth factor beta, Th T helper, TLR Toll-like receptor, TNF-α tumor necrosis factor alpha, Treg regulatory T-cell
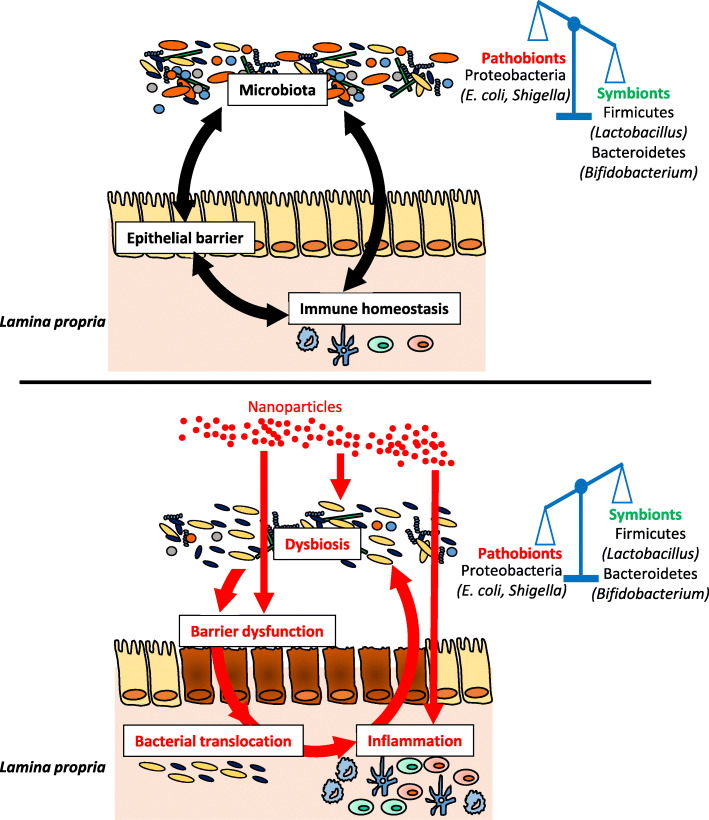
Fig. 2.
Potential impact of NP ingestion on the crosstalk between the microbiota and the immune system. After ingestion, NPs interact with the gastrointestinal environment and can alter the gut microbiota, characterized by an alteration of the F/B ratio, a depletion of Lactobacillus strains and an increase in the abundance of Proteobacteria. NPs exhibit also deleterious effects on the epithelial barrier and the intestinal immune response, which can amplifies the dysbiosis in a vicious circle favouring intestinal inflammation in susceptible individuals
The NP-induced gut microbiota signature resembles that of dysbiosis-associated human diseases
Despite some contradictory studies, the majority of observations (Table 1) [104–113] reveals a microbiota signature for nano-Ag, TiO2, ZnO and SiO2 characterized by alteration of the F/B ratio together with depletion of Lactobacillus and enrichment of Proteobacteria. The F/B ratio is often considered an informative parameter for the general state of the intestinal microbiota. Alteration of the F/B ratio has been observed in diseases associated with dysbiosis [172–175], and helps to predict the decrease in the relative abundance of SCFAs [176, 177]. Furthermore, Lactobacillus confers a health benefit on the host, notably via SCFAs and AhR ligand production [73, 74, 99, 178–180], while pathogenic Proteobacteria (E. coli, Shigella, Listeria, etc.,) are often overrepresented in several intestinal and extra-intestinal diseases with an inflammatory phenotype [181, 182]. Altogether, these data emphasize a negative shift in the microbial community in response to NP exposure, favouring the growth of pathogenic bacteria at the expense of beneficial strains such as Lactobacillus (Fig. 2). Importantly, these reported effects were very similar to those observed in patients suffering from IBD, CRC or chronic metabolic disorders such as obesity (Table 5) [17, 92, 173, 183–222].
Table 5.
Microbiota alteration observed in IBD, CRC and obesity compared to those induced after NP ingestion
| IBD | CRC | Obesity | NPs effects on microbiota | |
|---|---|---|---|---|
| Actinobacteria | ↗[183–185] | ↗[186] | ↗[187] | ↗TiO2 [104] |
| Acidimicrobidae ellin 7143 | ↗[184] | |||
| Actinobacterium GWS-BW-H99 | ↗[184] | |||
| Actinomycinaeae (o) | ↗[184] | |||
| Actinomyces (g) | ↗[188] | |||
| Actinomyces oxydans | ↗[184] | |||
| Bifidobacteriales (o) | ↗[186] | |||
| Bifidobacterium (g) | ↙[189] | ↙[190, 191] | ↙[192, 193] | ↙Ag [105] |
| Bifidobacterium adolescentis | ↙[194, 195] | |||
| Corynebacteriaceae (f) | ↗[184] | ↗[196] | ||
| Nocardioides NS/27 | ↗[184] | |||
| Bacteroidetes | ↙[183, 184, 188] | ↙[17, 197] | ↙[173, 187, 198–200] | ↙Ag, TiO2, SiO2, ZnO [104, 107–109]; ↗Ag, ZnO [105, 107, 110] |
| Bacteroides (g) | ↙[183, 194] | ↙[17, 186, 197, 201] | ↗[202, 203] | ↙Ag, TiO2, SiO2 [107, 108]; ↗Ag, ZnO [105, 110] |
| Bacteroides ovatus | ↙[204, 205] | ↙[206] | ↙Ag, TiO2 [112, 113] | |
| Bacteroides uniformis | ↙[183] | ↙[186] | ↙[206] | ↙Ag [106] |
| Prevotellaceae (f) | ↗[196, 207] | ↙ZnO [109] | ||
| Prevotella | ↙[183] | ↙[201] | ↗[207, 208] | ↙TiO2, ZnO [104, 109]; ↗Ag, SiO2 [107] |
| Rikenellaceae (f) | ↙[209] | ↙[200, 207] | ↙ZnO [110]; ↗Ag [108] | |
| Alistipes (g) | ↙[184, 185, 188] | ↗[210] | ↙[193, 206] | ↙ZnO [110]; ↗Ag, SiO2 [107] |
| Firmicutes | ↙[92, 184, 211, 212] | ↙[197] | ↗[173, 199, 200] | ↙Ag, TiO2, ZnO [104, 105, 107, 109, 110]; ↗Ag, SiO2, ZnO [107–109] |
| Erysipelotrichaceae (f) | ↙[194] | ↗[17, 190] | ↗[196] | ↙TiO2 [104] |
| Turicibacter (g) | ↙[183] | ↙TiO2 [104] | ||
| Lachnospiraceae (f) | ↙[194, 209] | ↙[197, 201] | ↙[207] | ↙Ag [107]; ↗Ag, SiO2 [104, 107] |
| Blautia (g) | ↙[209] | ↙[17] | ↙[207] | ↗Ag [108] |
| Blautia faecis | ↙[183] | |||
| Coprococcus (g) | ↙[209] | ↙[193, 207] | ↗Ag [108] | |
| Roseburia (g) | ↙[185, 209] | ↙[186, 210] | ↙[207] | |
| Roseburia intestinalis | ↙[183, 194] | ↗[206, 213] | ↙Ag [112] | |
| Roseburia inulinivorans | ↙[183] | |||
| Clostridium XIVa and IV groups | ↙[92, 184] | ↗[206] | ||
| Clostridium lavalense | ↙[183] | |||
| Dialister invisus | ↙[195] | |||
| Enterococcus (g) | ↗[186] | |||
| Eubacterium (g) | ↙[186] | ↙[207] | ||
| Eubacterium rectale | ↙[194, 211] | ↗[213] | ↙Ag [112]; ↗TiO2 [113] | |
| Eubacterium ventriosum | ↙[183] | ↗[198, 213] | ↗TiO2 [113] | |
| Faecalibacterium (g) | ↙[185, 209] | ↙[17, 190, 197, 210] | ↙[193] | |
| Faecalibacterium prausnitzii | ↙[92, 183, 194, 211] | ↙[191, 210] | ↙[206, 214] | ↙Ag [112] |
| Lactobacillus (sp) | ↙[183] | ↙[191, 215] | ↙[193, 216] | ↙Ag, TiO2, SiO2, ZnO [104, 105, 107, 109–111]; ↗Ag, ZnO [108, 109] |
| Oscillospira (g) | ↙[207, 213] | ↙Ag, ZnO [106, 109] | ||
| Ruminococcus gnavus | ↗[195, 209, 217] | |||
| Ruminococcus torques | ↙[183, 194] | ↙Ag [112] | ||
| Streptococcus (g) | ↗[186, 218] | ↗ZnO [109] | ||
| Proteobacteria | ↗[184, 185] | ↗[186] | ↗[200, 206, 216] | ↗TiO2, SiO2, ZnO [104, 107, 109]; ↙ZnO [111] |
| Enterobacteriaceae (f) | ↗[209, 219] | ↗[191, 218] | ↗[207] | ↗Ag [105] |
| Escherichia coli | ↗[194, 211, 220–222] | ↗[186, 210] | ↗ Ag, TiO2 [104, 112] | |
| Shigella (sp) | ↗[185, 211] | ↗[186, 210] | ↗ TiO2 [104] | |
| Sutterella (g) | ↗[216] | |||
| Listeria (sp) | ↗[211] |
A deficiency in AhR ligand production by the microbiota was reported in IBD and obese patients compared to healthy subjects [74, 178]. Some commensal bacteria are able to metabolize the essential amino acid tryptophan in AhR ligands such as indole, indole-3-acetic acid, tryptamine, and indole-3-aldehyde. This ability is mainly exhibited by Lactobacillus [73, 74, 99, 178], the abundance of which is decreased in the microbiota of IBD and obese patients [183, 193, 216]. Recent studies in mice demonstrated that the reduced capacity of the microbiota to produce AhR ligands is involved in the pathogenesis of IBD and obesity through a mechanism that involves decreased IL-22 production by intestinal immune cells [74, 178]. In the intestine, IL-22 is involved in mucosal wound healing [223] and production of antimicrobial peptides (AMPs) by IECs, such as regenerating islet-derived 3 gamma (Reg3γ) and Reg3β [224, 225], which can modulate the microbiota composition. Interestingly, defective intestinal production of IL-22 was observed in mice fed a high-fat diet, while administration of exogenous IL-22 reversed many metabolic symptoms, including hyperglycaemia and insulin resistance [226]. IL-22 shows diverse metabolic benefits, as it improves insulin sensitivity, preserves the gut mucosal barrier and endocrine functions, decreases endotoxaemia and chronic inflammation, and regulates lipid metabolism in liver and adipose tissues [226]. Similarly, mice treated with an AhR agonist or with Lactobacillus strains with high AhR-ligand production capacity exhibited improvement in symptoms of colitis and metabolic syndrome [74, 178]. These treatments alleviate inflammatory insults in mice subjected to experimental colitis [74] and reduce glucose dysmetabolism and liver steatosis in both dietary and genetic animal models of metabolic syndrome [178]. Mechanistically, metabolic improvement is linked to the restoration of intestinal barrier function and the production of the intestinal hormone incretin [178].
The proportion of Lactobacillus is also decreased in the microbiota of CRC patients [191, 215], while a number of Lactobacillus species exhibited notable anti-carcinogenic effects via the inactivation of microbial enzymes with procarcinogenic activities, such as β-glucuronidase and nitroreductase [227]. To date, no studies have shown that the ability of some Lactobacillus species to produce AhR ligands contributes to the anti-carcinogenic potential of these bacteria. However, this mechanism cannot be excluded because AhR is an important node for the development of cancer. Indeed, in comparison to APCMin/+ mice that spontaneously developed intestinal tumours, APCMin/+/AhR+/− mice exhibited increased tumour incidence, suggesting a tumour suppressor role for AhR. Moreover, a diet rich in AhR ligands can prevent or decrease CRC in mice: first, by inhibiting the Wnt-β-catenin pathway, which is known to be crucial for the proliferation of intestinal stem cells [228], and second, by regulating components of the DNA damage response (DDR) in epithelial stem cells through a mechanism that involves IL-22 production by intestinal immune cells [229]. DDR culminates in either transient cell-cycle arrest and DNA repair or elimination of damaged cells by apoptosis, thereby inhibiting the development of mutations that can lead to CRC [229].
Collectively, these data strongly support the AhR/IL-22 axis a key regulator of intestinal homeostasis. Hence, whether an NP-induced imbalance in this signalling pathway along the microbiota-immune axis could be a first cause of disease development and/or maintenance needs to be further examined in future studies. As noted above, because an NP-induced depletion in AhR-producing bacterial strains is commonly reported (Tables 1 and 5), one may hypothesize that this change represents the missing mechanistic link for colon cancer development in rodents after long-term treatment with a food-grade form of TiO2 (i.e., the food additive E171). Cancer evolves through a sequential process from normal cells to preneoplastic lesions before tumour development. Among rats exposed for 100 days to a commercial E171 vial (≈45% of TiO2-NPs by number) at a relevant dose for humans, nearly 40% of the animals developed spontaneously preneoplastic lesions in the colon (i.e., initiation of premalignant colorectal lesions), and a steady, low level of inflammation was reported in the colon mucosa [116]. Furthermore, the same study showed increased severity of preneoplastic lesions in E171-treated rats pretreated with dimethylhydrazine to initiate colon carcinogenesis, suggesting the promotion of disease development by TiO2 [116]. This effect was confirmed in mice by using a similar E171 batch and showing exacerbated tumour formation (i.e., colorectal adenoma) in a chemical colitis-associated cancer model [230]. These TiO2 initiator and promoter effects of preneoplastic lesions in the colon could be due to intestinal micro-inflammation [116] and defective expression of genes involved in oxidative stress, immune response and cancer [231, 232] as well as due to alteration of the intestinal microbiota, as reviewed herein.
In the context of dysbiosis as a susceptibility factor for CRC development, a review of human data clearly shows a reduction in the levels of SCFA-producing bacteria in the microbiota of patients, namely, Faecalibacterium, Roseburia and Bifidobacterium, as well as Lactobacillus (Table 5). Decreased abundances of these specific strains was observed in IBD and obese patients, while similar depletion appears in rodents orally exposed to TiO2, Ag, SiO2 and ZnO NPs, as indicated in Tables 1 and 5. The SCFAs, mainly acetate, propionate and butyrate, are absorbed in the intestine. First, butyrate serves as a major source of energy for colonocytes and for epithelial renewal [233–235], whereas propionate and acetate reach the liver via portal circulation. Propionate is primarily used in gluconeogenesis [233, 234, 236], while in the case of acetate, this organic product enters systemic circulation to reach peripheral tissues, where it serves as a substrate for cholesterol synthesis [233, 236]. As previously detailed, SCFAs exhibit anti-inflammatory effects but are also involved in different physiological processes. Acetate participates in the de novo synthesis of lipids in colonic epithelial cells [237], and similar to propionate, butyrate reduces food intake and stimulates the formation of the anorexigenic hormone leptin [234, 236, 238]. Butyrate has been shown to enhance intestinal barrier function and regulate cellular apoptosis, cell proliferation and differentiation [239]. Moreover, in mice, treatment with butyrate improves insulin sensitivity and increases energy expenditure, leading to reduced obesity [240]. In addition to these anti-obesogenic properties, both butyrate and propionate play a protective role against IBD [241–244] and colon carcinogenesis [245–248]. Altogether, these observations suggest that NP-related depletion of bacteria responsible for SCFA production could be viewed as an additional risk factor for the development of these diseases.
Finally, it should be noted that the inflammatory state induced by the absence of a healthy butyrate-producing microbiota leads to increased expression of the gene encoding nitric oxide synthase, Nos2, as well as of nitrate production by the host, a substrate favouring the growth of Enterobacteriaceae belonging to the Proteobacteria [249]. The proportion of Proteobacteria is increased in the gut microbiota of patients suffering from IBD, obesity or CRC (Table 5), and some authors have proposed that the abundance of Proteobacteria as a “microbial signature” of disease progression [182]. The role of Proteobacteria in inflammation has been revealed in various mouse models of obesity, colitis and colitis-associated colorectal cancer. For example, the resistance of GF mice to the development of an obesity phenotype after being fed a high-fat diet was overcome by inoculation of these mice with an Enterobacter population isolated from the obese human gut, highlighting the obesogenic potential of Proteobacteria [250]. In mice that exhibit spontaneous development of colitis, such as those lacking Toll-like receptors-5 (Tlr5−/−), IL-10 (Il-10−/−) or the transcription factor T-bet and the recombinase activating gene Rag (T-bet−/− x Rag2−/−), an increased intestinal level of Proteobacteria was reported [251–255]. Inoculation of WT mice with two Enterobacteriaceae species isolated from faeces of T-bet−/− x Rag2−/− mice induced intestinal inflammation leading to colitis [253]. In addition, monocolonization with the commensal E. coli NC101 of GF Il-10−/− mice treated with the colon-specific carcinogen azoxymethane promoted the development of invasive carcinoma [256, 257]. These data indicate that some Proteobacteria are able to induce intestinal inflammation that may promote IBD and/or CRC development. Notably, in recent studies, microbiota composition analysis concluded on a Proteobacteria bloom in mice orally exposed to inorganic NPs (Tables 1 and 3). However, the shift towards pathogenic bacterial colonization is not specific to NP exposure because such a shift was also observed with other xenobiotics. For example, mice exposed to artificial sweeteners and emulsifiers added to many processed foods or to chlorpyrifos (an organophosphorus pesticide frequently detected in the diet) exhibit a high abundance of Proteobacteria, a scenario that predisposes mice to obesity [258–260], colitis [259, 261] and colon cancer [262]. Despite in vivo studies demonstrating that oral exposure to NP induces inflammatory [107, 116, 155, 161, 230] or metabolic [263–265] effects which could be related to IBD, CRC and obesity, it is not known whether the NP-induced bloom of the pathogenic phylum Proteobacteria may also promote the development of one or more of these diseases. In such a context, it should also be investigated whether NP-related GALT dysfunctions could be a cause or consequence of intestinal dysbiosis and whether specific NPs, alone or in mixture, are associated with different disease phenotypes, rendering the host highly vulnerable to IBD, CRC or metabolic disorders (Fig. 2).
Conclusions
The increasing use of NPs in the food chain, as additives or incorporated into food packaging, has led to concerns regarding the daily exposure of consumers. On the basis of reports presented in this review, one may suggest that the antimicrobial and/or immunotoxic properties of inorganic NPs have the potential to alter the intestinal microbiota and GALT functions, interactions of which are important for many physiological processes in the organism (Fig. 2). This review has summarized the impacts on the gut microbiota-immune axis of the four most common NPs found in the food sector. The existing data highlight a recurrent microbiota signature for nano-Ag, TiO2, ZnO and SiO2, characterized by an alteration of the F/B ratio, a depletion of Lactobacillus strains (SCFA and AhR ligand producers) and an increase in the abundance of Proteobacteria, which may resemble the microbiome shift in IBD, CRC or obesity where gut dysbiosis play a key pathogenic role. These observations raise the need for additional studies for the re-evaluation of NP-containing food additives used for decades in foodstuffs, especially given the uncertainties associated with the long-term effects of these NPs on the gut microbiome as described in this review. In Europe, the new guidance document by EFSA on risk assessment for the application of nanotechnologies in food and the feed chain highlights the need for studies on the composition of the microbiome for nanomaterials, especially for those with antimicrobial properties [266]. This caution appears to be of particular importance given the limited absorption of insoluble particles from the intestine (e.g., TiO2, SiO2), meaning that the non-absorbed fraction of NPs remains in direct contact with the resident bacteria before being excreted. In this context and based on available data, the NP-microbiota interactions appeared to be localization dependent along the gut, probably due to differences in microbial composition and density from the small intestine to the colon. Changes in the physicochemical properties of NPs during gut transit (i.e., differences in pH and influence of the food matrices and biliary acids) or the extent of particle dissolution for soluble materials (i.e., Ag, ZnO), could modify their long term impact on the microbiome. Except for few studies, the effects of foodborne NPs on the metabolic activity of the microbiota remain largely unexplored; however, this parameter is crucial for evaluating biological consequences for the host and potential hazards. Moreover, most studies have been conducted using pure nanoparticulate matter as NP models instead of the food additives that often exhibit mixed submicron- and nanosized particles, and at high doses, i.e., far above human dietary levels for the equivalent food-grade forms; these aspects could elicit different effects on the gut microbiome. It also appears that interpretations of findings from animals to humans are very limited due to differences in gut microbiota composition and activity. However, as 85% of the 16S rRNA sequence dataset for the mouse microbiota represents genera that are not present in humans [199], the use of GF mice inoculated with the human microbiota seems to be a good model for investigation of the chronic impact of foodborne NPs on bacteria that colonize the human intestine. Some studies used a custom colon reactor in this way, but the exposure time of the microbiota to the NPs was short [113, 117] and hence was not representative of chronic exposure conditions. Furthermore, available data are often limited to the impact of NP models/food additives exclusively on the intestinal flora, bypassing the importance of the microbiota-immune system axis for the host health that would require more integrated approaches as microbiota transfer in GF mice.
Another area that requires detailed investigation is the ability of the gut microbiota to recover after the end of NP exposure; such an investigation would help determine whether NP-induced dysbiosis is permanent and has long-lasting consequences for the host. Notably, the period of exposure to NPs during life is rarely taken into account, while exposure when the microbiota and immune system begin to interact, such as the perinatal period, could induce higher alteration of microbiota-GALT crosstalk than a similar exposure experienced during adulthood. One of the other limitations of the current studies reviewed is that the intestinal immune response (GALT) and gut microbiota alterations were explored after exposure to one type of NP. However, humans are continually exposed to different foodborne NPs as well as to a multitude of other xenobiotics that may have synergistic or antagonistic effects on these functions. One of the challenges in the coming years will be to evaluate the effects on humans of this complex exposome, taking into account exposure to foodborne mineral particles with different physicochemical characteristics. For effective risk assessment, an understanding of the effects of this exposome extended to inorganic (nano) particles will be essential for prevention and remediation strategies and for facilitating the design of safe nanomaterials devoid of biocidal activity when used as food additives or food supplements.
Acknowledgements
All contributors who provided help during the research have been listed.
Abbreviations
- Ag
Silver
- AhR
Aryl hydrocarbon receptor
- AMPs
Antimicrobial peptides
- bw
Body weight
- Card9
Caspase recruitment domain 9
- cfu
Colony forming units
- CRC
Colorectal cancer
- d
Day
- DCs
Dendritic cells
- DDR
DNA damage response
- EFSA
European Food Safety Authority
- F/B
Firmicutes/Bacteroidetes
- GALT
Gut-associated lymphoid tissue
- GF
Germ-free
- GI
Gastrointestinal
- GRAS
Generally recognized as safe
- IBD
Inflammatory bowel diseases
- IECs
Intestinal epithelial cells
- IELs
Intraepithelial lymphocytes
- IFN-γ
Interferon-gamma
- IgA
Immunoglobulin A
- IL
Interleukin
- ILCs
Innate lymphoid cells
- LP
Lamina propria
- M cells
Microfold cells
- MET-1
Microbial ecosystem therapeutic-1
- MLNs
Mesenteric lymph nodes
- NPs
Nanoparticles
- PPs
Peyer’s patches
- Reg3
Regenerating islet-derived 3
- ROS
Reactive oxygen species
- SCFAs
Short-chain fatty acids
- SFB
Segmented filamentous bacteria
- Si
Silicon
- SiO2
Silicon dioxide
- Th
T helper
- Ti
Titanium
- TiO2
Titanium dioxide
- Tlr5
Toll-like receptor 5
- TNF-α
Tumour necrosis factor alpha
- Treg
Regulatory T-cell
- WT
Wild-type
- ZnO
Zinc oxide
Authors’ contributions
BL, NMB and EH. Designed the study and contributed to all the sections. All authors read and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bruno Lamas, Email: bruno.lamas@inrae.fr.
Eric Houdeau, Email: eric.houdeau@inrae.fr.
References
- 1.Nel A, Xia T, Mädler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 2.Xia T, Li N, Nel AE. Potential health simpact of nanoparticles. Annu Rev Public Health. 2009;30:137–150. doi: 10.1146/annurev.publhealth.031308.100155. [DOI] [PubMed] [Google Scholar]
- 3.Weir A, Westerhoff P, Fabricius L, Hristovski K, von Goetz N. Titanium dioxide nanoparticles in food and personal care products. Environ Sci Technol. 2012;46:2242–2250. doi: 10.1021/es204168d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaudhry Q, Scotter M, Blackburn J, Ross B, Boxall A, Castle L, et al. Applications and implications of nanotechnologies for the food sector. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2008;25:241–258. doi: 10.1080/02652030701744538. [DOI] [PubMed] [Google Scholar]
- 5.Srinivas PR, Philbert M, Vu TQ, Huang Q, Kokini JL, Saltos E, et al. Nanotechnology research: applications in nutritional sciences. J Nutr. 2010;140:119–124. doi: 10.3945/jn.109.115048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang M, Lee EJ, Kweon SY, Park MS, Jeong JY, Um JH, et al. Risk assessment principle for engineered nanotechnology in food and drug. Toxicol Res. 2012;28:73–79. doi: 10.5487/TR.2012.28.2.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercier-Bonin M, Despax B, Raynaud P, Houdeau E, Thomas M. Mucus and microbiota as emerging players in gut nanotoxicology: The example of dietary silver and titanium dioxide nanoparticles. Crit Rev Food Sci Nutr. 2018;58:1023–1032. doi: 10.1080/10408398.2016.1243088. [DOI] [PubMed] [Google Scholar]
- 8.Radziwill-Bienkowska JM, Talbot P, Kamphuis JBJB, Robert V, Cartier C, Fourquaux I, et al. Toxicity of Food-Grade TiO2 to Commensal Intestinal and Transient Food-Borne Bacteria: New Insights Using Nano-SIMS and Synchrotron UV Fluorescence Imaging. Front Microbiol. 2018;9:794. doi: 10.3389/fmicb.2018.00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talbot P, Radziwill-Bienkowska JM, Kamphuis JBJB, Steenkeste K, Bettini S, Robert V, et al. Food-grade TiO2 is trapped by intestinal mucus in vitro but does not impair mucin O-glycosylation and short-chain fatty acid synthesis in vivo: implications for gut barrier protection. J Nanobiotechnology. 2018;16:53. doi: 10.1186/s12951-018-0379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomas J, Wrzosek L, Bouznad N, Bouet S, Mayeur C, Noordine M-LL, et al. Primocolonization is associated with colonic epithelial maturation during conventionalization. FASEB J. 2013;27:645–655. doi: 10.1096/fj.12-216861. [DOI] [PubMed] [Google Scholar]
- 12.Natividad JMM, Verdu EF. Modulation of intestinal barrier by intestinal microbiota: pathological and therapeutic implications. Pharmacol Res. 2013;69:42–51. doi: 10.1016/j.phrs.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belkaid Y, Harrison OJ. Homeostatic Immunity and the Microbiota. Immunity. 2017;46:562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14:573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verdu EF, Galipeau HJ, Jabri B. Novel players in coeliac disease pathogenesis: role of the gut microbiota. Nat Rev Gastroenterol Hepatol. 2015;12:497–506. doi: 10.1038/nrgastro.2015.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richard ML, Liguori G, Lamas B, Brandi G, da Costa G, Hoffmann TW, et al. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes. 2018;9:131–142. doi: 10.1080/19490976.2017.1379637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meijnikman AS, Gerdes VE, Nieuwdorp M, Herrema H. Evaluating Causality of Gut Microbiota in Obesity and Diabetes in Humans. Endocr Rev. 2018;39:133–153. doi: 10.1210/er.2017-00192. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeld CS. Microbiome Disturbances and Autism Spectrum Disorders. Drug Metab Dispos. 2015;43:1557–1571. doi: 10.1124/dmd.115.063826. [DOI] [PubMed] [Google Scholar]
- 20.Dinan TG, Cryan JF. Gut instincts: microbiota as a key regulator of brain development, ageing and neurodegeneration. J Physiol (Lond). 2017;595:489–503. doi: 10.1113/JP273106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minemura M, Shimizu Y. Gut microbiota and liver diseases. World J Gastroenterol. 2015;21:1691–1702. doi: 10.3748/wjg.v21.i6.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.EFSA ANS Panel. Scientific opinion on the re-evaluation of silver (E 174) as food additive. EFSA J EFSA Journal; 2016;14:4364.
- 23.Peters RJ, van Bemmel G, Herrera-Rivera Z, Helsper HP, Marvin HJ, Weigel S, et al. Characterization of titanium dioxide nanoparticles in food products: analytical methods to define nanoparticles. J Agric Food Chem. 2014;62:6285–6293. doi: 10.1021/jf5011885. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, Doudrick K, Bi X, Hristovski K, Herckes P, Westerhoff P, et al. Characterization of food-grade titanium dioxide: the presence of nanosized particles. Environ Sci Technol. 2014;48:6391–6400. doi: 10.1021/es500436x. [DOI] [PubMed] [Google Scholar]
- 25.EFSA ANS Panel Re-evaluation of titanium dioxide (E 171) as a food additive. EFSA J. 2016;14:4545–4583. [Google Scholar]
- 26.Dekkers S, Krystek P, Peters RJ, Lankveld DPP, Bokkers BG, van Hoeven-Arentzen PH, et al. Presence and risks of nanosilica in food products. Nanotoxicology. 2011;5:393–405. doi: 10.3109/17435390.2010.519836. [DOI] [PubMed] [Google Scholar]
- 27.Peters R, Kramer E, Oomen AG, Rivera ZE, Oegema G, Tromp PC, et al. Presence of nano-sized silica during in vitro digestion of foods containing silica as a food additive. ACS Nano. 2012;6:2441–2451. doi: 10.1021/nn204728k. [DOI] [PubMed] [Google Scholar]
- 28.EFSA ANS Panel Re-evaluation of silicon dioxide (E 551) as food additive. EFSA J. 2018;16:5070–5088. doi: 10.2903/j.efsa.2018.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Yuan L, Yao C, Ding L, Li C, Fang J, et al. A combined toxicity study of zinc oxide nanoparticles and vitamin C in food additives. Nanoscale. 2014;6:15333–15342. doi: 10.1039/c4nr05480f. [DOI] [PubMed] [Google Scholar]
- 30.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger J, Chinwalla A, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stilling RM, Dinan TG, Cryan JF. Microbial genes, brain & behaviour - epigenetic regulation of the gut-brain axis. Genes Brain Behav. 2014;13:69–86. doi: 10.1111/gbb.12109. [DOI] [PubMed] [Google Scholar]
- 34.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amedei A, Boem F. I’ve Gut A Feeling: Microbiota Impacting the Conceptual and Experimental Perspectives of Personalized Medicine. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed]
- 37.Baquero F, Nombela C. The microbiome as a human organ. Clin Microbiol Infect. 2012;18(Suppl 4):2–4. doi: 10.1111/j.1469-0691.2012.03916.x. [DOI] [PubMed] [Google Scholar]
- 38.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, Dinan TG. Minireview: Gut microbiota: the neglected endocrine organ. Mol Endocrinol. 2014;28:1221–1238. doi: 10.1210/me.2014-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 41.Lamas B, Michel M-LL, Waldschmitt N, Pham H-PP, Zacharioudaki V, Dupraz L, et al. Card9 mediates susceptibility to intestinal pathogens through microbiota modulation and control of bacterial virulence. Gut. 2018;67:1836–1844. doi: 10.1136/gutjnl-2017-314195. [DOI] [PubMed] [Google Scholar]
- 42.Moens E, Veldhoen M. Epithelial barrier biology: good fences make good neighbours. Immunology. 2012;135:1–8. doi: 10.1111/j.1365-2567.2011.03506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamada N, Seo S-UU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 44.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki T, Yoshida S, Hara H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br J Nutr. 2008;100:297–305. doi: 10.1017/S0007114508888733. [DOI] [PubMed] [Google Scholar]
- 46.Peng L, Li Z-RR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139:1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han B, Sheng B, Zhang Z, Pu A, Yin J, Wang Q, et al. Aryl Hydrocarbon Receptor Activation in Intestinal Obstruction Ameliorates Intestinal Barrier Dysfunction Via Suppression of MLCK-MLC Phosphorylation Pathway. Shock. 2016;46:319–328. doi: 10.1097/SHK.0000000000000594. [DOI] [PubMed] [Google Scholar]
- 48.ABRAMS GD, BAUER H, SPRINZ H. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab Invest. 1963;12:355–364. [PubMed] [Google Scholar]
- 49.Reinhardt C, Bergentall M, Greiner TU, Schaffner F, Ostergren-Lundén G, Petersen LC, et al. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature. 2012;483:627–631. doi: 10.1038/nature10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banasaz M, Norin E, Holma R, Midtvedt T. Increased enterocyte production in gnotobiotic rats mono-associated with Lactobacillus rhamnosus GG. Appl Environ Microbiol. 2002;68:3031–3034. doi: 10.1128/AEM.68.6.3031-3034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alam M, Midtvedt T, Uribe A. Differential cell kinetics in the ileum and colon of germfree rats. Scand J Gastroenterol. 1994;29:445–451. doi: 10.3109/00365529409096836. [DOI] [PubMed] [Google Scholar]
- 52.Shirkey TW, Siggers RH, Goldade BG, Marshall JK, Drew MD, Laarveld B, et al. Effects of commensal bacteria on intestinal morphology and expression of proinflammatory cytokines in the gnotobiotic pig. Exp Biol Med (Maywood). 2006;231:1333–1345. doi: 10.1177/153537020623100807. [DOI] [PubMed] [Google Scholar]
- 53.Willing BP, Van Kessel AG. Enterocyte proliferation and apoptosis in the caudal small intestine is influenced by the composition of colonizing commensal bacteria in the neonatal gnotobiotic pig. J Anim Sci. 2007;85:3256–3266. doi: 10.2527/jas.2007-0320. [DOI] [PubMed] [Google Scholar]
- 54.Danielsen M, Hornshøj H, Siggers RH, Jensen BB, van Kessel AG, Bendixen E. Effects of bacterial colonization on the porcine intestinal proteome. J Proteome Res. 2007;6:2596–2604. doi: 10.1021/pr070038b. [DOI] [PubMed] [Google Scholar]
- 55.Kozakova H, Kolinska J, Lojda Z, Rehakova Z, Sinkora J, Zakostelecka M, et al. Effect of bacterial monoassociation on brush-border enzyme activities in ex-germ-free piglets: comparison of commensal and pathogenic Escherichia coli strains. Microbes Infect. 2006;8:2629–2639. doi: 10.1016/j.micinf.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 56.Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, et al. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013;32:3017–3028. doi: 10.1038/emboj.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swanson PA, Kumar A, Samarin S, Vijay-Kumar M, Kundu K, Murthy N, et al. Enteric commensal bacteria potentiate epithelial restitution via reactive oxygen species-mediated inactivation of focal adhesion kinase phosphatases. Proc Natl Acad Sci USA. 2011;108:8803–8808. doi: 10.1073/pnas.1010042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stockinger B, Di Meglio P, Gialitakis M, Duarte JHH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 59.Wong JM, de SR, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 60.Comalada M, Bailón E, de Haro O, Lara-Villoslada F, Xaus J, Zarzuelo A, et al. The effects of short-chain fatty acids on colon epithelial proliferation and survival depend on the cellular phenotype. J Cancer Res Clin Oncol. 2006;132:487–497. doi: 10.1007/s00432-006-0092-x. [DOI] [PubMed] [Google Scholar]
- 61.Orchel A, Dzierzewicz Z, Parfiniewicz B, Weglarz L, Wilczok T. Butyrate-induced differentiation of colon cancer cells is PKC and JNK dependent. Dig Dis Sci. 2005;50:490–498. doi: 10.1007/s10620-005-2463-6. [DOI] [PubMed] [Google Scholar]
- 62.Foster JA, Lyte M, Meyer E, Cryan JF. Gut Microbiota and Brain Function: An Evolving Field in Neuroscience. Int J Neuropsychopharmacol. 2016;19. [DOI] [PMC free article] [PubMed]
- 63.Llorente C, Schnabl B. The gut microbiota and liver disease. Cell Mol Gastroenterol Hepatol. 2015;1:275–284. doi: 10.1016/j.jcmgh.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mazagova M, Wang L, Anfora AT, Wissmueller M, Lesley SA, Miyamoto Y, et al. Commensal microbiota is hepatoprotective and prevents liver fibrosis in mice. FASEB J. 2015;29:1043–1055. doi: 10.1096/fj.14-259515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 66.Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Mol Psychiatry. 2014;19:146–148. doi: 10.1038/mp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luczynski P, Whelan SOO, O’Sullivan C, Clarke G, Shanahan F, Dinan TG, et al. Adult microbiota-deficient mice have distinct dendritic morphological changes: differential effects in the amygdala and hippocampus. Eur J Neurosci. 2016;44:2654–2666. doi: 10.1111/ejn.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoban AE, Stilling RM, Ryan FJ, Shanahan F, Dinan TG, Claesson MJ, et al. Regulation of prefrontal cortex myelination by the microbiota. Transl Psychiatry. 2016;6:e774. doi: 10.1038/tp.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Tóth M, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med. 2014;6:263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Erny D, Hrabě de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015;18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu X-NN, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol (Lond). 2004;558:263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. Microbial Reconstitution Reverses Maternal Diet-Induced Social and Synaptic Deficits in Offspring. Cell. 2016;165:1762–1775. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 74.Lamas B, Richard ML, Leducq V, Pham H-PP, Michel M-LL, Da Costa G, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med. 2016;22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beaumont M, Neyrinck AM, Olivares M, Rodriguez J, de RSA, Roumain M, et al. The gut microbiota metabolite indole alleviates liver inflammation in mice. FASEB J. 2018:fj201800544. [DOI] [PMC free article] [PubMed]
- 77.Sakakibara S, Yamauchi T, Oshima Y, Tsukamoto Y, Kadowaki T. Acetic acid activates hepatic AMPK and reduces hyperglycemia in diabetic KK-A(y) mice. Biochem Biophys Res Commun. 2006;344:597–604. doi: 10.1016/j.bbrc.2006.03.176. [DOI] [PubMed] [Google Scholar]
- 78.Endo H, Niioka M, Kobayashi N, Tanaka M, Watanabe T. Butyrate-producing probiotics reduce nonalcoholic fatty liver disease progression in rats: new insight into the probiotics for the gut-liver axis. PLoS One. 2013;8:e63388. doi: 10.1371/journal.pone.0063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Kivit S, Tobin MC, Forsyth CB, Keshavarzian A, Landay AL. Regulation of Intestinal Immune Responses through TLR Activation: Implications for Pro- and Prebiotics. Front Immunol. 2014;5:60. doi: 10.3389/fimmu.2014.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Delgado-Rizo V, Martínez-Guzmán MA, Iñiguez-Gutierrez L, García-Orozco A, Alvarado-Navarro A, Fafutis-Morris M. Neutrophil Extracellular Traps and Its Implications in Inflammation: An Overview. Front Immunol. 2017;8:81. doi: 10.3389/fimmu.2017.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cassard A-MM, Gérard P, Perlemuter G. Microbiota, Liver Diseases, and Alcohol. Microbiol Spectr. 2017;5. [DOI] [PubMed]
- 82.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 83.Coombes JL, Siddiqui KRR, Arancibia-Cárcamo CV, Hall J, Sun C-MM, Belkaid Y, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.LeBien TW, Tedder TF. B lymphocytes: how they develop and function. Blood. 2008;112:1570–1580. doi: 10.1182/blood-2008-02-078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ohno H. Intestinal M cells. J Biochem. 2016;159:151–160. [DOI] [PMC free article] [PubMed]
- 86.Sommer F, Bäckhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 87.Chung H, Pamp SJJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 89.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Telesford KM, Yan W, Ochoa-Reparaz J, Pant A, Kircher C, Christy MA, et al. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39(+)Foxp3(+) T cells and Treg function. Gut Microbes. 2015;6:234–242. doi: 10.1080/19490976.2015.1056973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Quévrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut. 2016;65:415–425. doi: 10.1136/gutjnl-2014-307649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Breyner NM, Michon C, de SCS, Vilas Boas PB, Chain F, Azevedo VA, et al. Microbial Anti-Inflammatory Molecule (MAM) from Faecalibacterium prausnitzii Shows a Protective Effect on DNBS and DSS-Induced Colitis Model in Mice through Inhibition of NF-κB Pathway. Frontiers in microbiology. Front Microbiol. 2017;8:114. doi: 10.3389/fmicb.2017.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Levy M, Thaiss CA, Elinav E. Metabolites: messengers between the microbiota and the immune system. Genes Dev. 2016;30:1589–1597. doi: 10.1101/gad.284091.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Corrêa-Oliveira R, Fachi JLL, Vieira A, Sato FT, Vinolo MA. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunology. 2016;5:e73. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gurav A, Sivaprakasam S, Bhutia YD, Boettger T, Singh N, Ganapathy V. Slc5a8, a Na+−coupled high-affinity transporter for short-chain fatty acids, is a conditional tumour suppressor in colon that protects against colitis and colon cancer under low-fibre dietary conditions. Biochem J. 2015;469:267–278. doi: 10.1042/BJ20150242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lamas B, Natividad JM, Sokol H. Aryl hydrocarbon receptor and intestinal immunity. Mucosal Immunol. 2018;11:1024–1038. doi: 10.1038/s41385-018-0019-2. [DOI] [PubMed] [Google Scholar]
- 100.Agus A, Planchais J, Sokol H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 101.Lee HU, McPherson ZE, Tan B, Korecka A, Pettersson S. Host-microbiome interactions: the aryl hydrocarbon receptor and the central nervous system. J Mol Med. 2017;95:29–39. doi: 10.1007/s00109-016-1486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McGee CF, Storey S, Clipson N, Doyle E. Soil microbial community responses to contamination with silver, aluminium oxide and silicon dioxide nanoparticles. Ecotoxicology. 2017;26:449–458. doi: 10.1007/s10646-017-1776-5. [DOI] [PubMed] [Google Scholar]
- 103.Timmusk S, Seisenbaeva G, Behers L. Titania (TiO2) nanoparticles enhance the performance of growth-promoting rhizobacteria. Sci Rep. 2018;8:617. doi: 10.1038/s41598-017-18939-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li J, Yang S, Lei R, Gu W, Qin Y, Ma S, et al. Oral administration of rutile and anatase TiO 2 nanoparticles shifts mouse gut microbiota structure. Nanoscale. 2018;10:7736–7745. doi: 10.1039/c8nr00386f. [DOI] [PubMed] [Google Scholar]
- 105.Williams K, Milner J, Boudreau MD, Gokulan K, Cerniglia CE, Khare S. Effects of subchronic exposure of silver nanoparticles on intestinal microbiota and gut-associated immune responses in the ileum of Sprague-Dawley rats. Nanotoxicology. 2015;9:279–289. doi: 10.3109/17435390.2014.921346. [DOI] [PubMed] [Google Scholar]
- 106.Javurek AB, Suresh D, Spollen WG, Hart ML, Hansen SA, Ellersieck MR, et al. Gut Dysbiosis and Neurobehavioral Alterations in Rats Exposed to Silver Nanoparticles. Sci Rep. 2017;7:2822. doi: 10.1038/s41598-017-02880-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen H, Zhao R, Wang B, Cai C, Zheng L, Wang H, et al. The effects of orally administered Ag, TiO2 and SiO2 nanoparticles on gut microbiota composition and colitis induction in mice. Nanoimpact. 2017;8:80–88. [Google Scholar]
- 108.van den Brule S, Ambroise J, Lecloux H, Levard C, Soulas R, De Temmerman P-JJ, et al. Dietary silver nanoparticles can disturb the gut microbiota in mice. Part Fibre Toxicol. 2016;13:38. doi: 10.1186/s12989-016-0149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xia T, Lai W, Han M, Han M, Ma X, Zhang L. Dietary ZnO nanoparticles alters intestinal microbiota and inflammation response in weaned piglets. Oncotarget. 2017;8:64878–64891. doi: 10.18632/oncotarget.17612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yausheva Е, Miroshnikov S, Sizova Е. Intestinal microbiome of broiler chickens after use of nanoparticles and metal salts. Environ Sci Pollut R. 2018;25:18109–18120. doi: 10.1007/s11356-018-1991-5. [DOI] [PubMed] [Google Scholar]
- 111.Feng Y, Min L, Zhang W, Liu J, Hou Z, Chu M, et al. Zinc Oxide Nanoparticles Influence Microflora in Ileal Digesta and Correlate Well with Blood Metabolites. Front Microbiol. 2017;8:992. doi: 10.3389/fmicb.2017.00992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Das P, Julie A, Elaine O, Emma A-V, Virginia K. Nanosilver-Mediated Change in Human Intestinal Microbiota. J Nanomed Nanotechnol. 2014:05.
- 113.Dudefoi W, Moniz K, Allen-Vercoe E, Ropers M-HH, Walker VK. Impact of food grade and nano-TiO2 particles on a human intestinal community. Food Chem Toxicol. 2017;106:242–249. doi: 10.1016/j.fct.2017.05.050. [DOI] [PubMed] [Google Scholar]
- 114.Hadrup N, Loeschner K, Bergström A, Wilcks A, Gao X, Vogel U, et al. Subacute oral toxicity investigation of nanoparticulate and ionic silver in rats. Arch Toxicol. 2012;86:543–551. doi: 10.1007/s00204-011-0759-1. [DOI] [PubMed] [Google Scholar]
- 115.Wilding LA, Bassis CM, Walacavage K, Hashway S, Leroueil PR, Morishita M, et al. Repeated dose (28-day) administration of silver nanoparticles of varied size and coating does not significantly alter the indigenous murine gut microbiome. Nanotoxicology. 2016;10:513–520. doi: 10.3109/17435390.2015.1078854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bettini S, Boutet-Robinet E, Cartier C, Coméra C, Gaultier E, Dupuy J, et al. Food-grade TiO2 impairs intestinal and systemic immune homeostasis, initiates preneoplastic lesions and promotes aberrant crypt development in the rat colon. Sci Rep. 2017;7:40373. doi: 10.1038/srep40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Taylor A, Marcus I, Guysi R, Walker S. Metal Oxide Nanoparticles Induce Minimal Phenotypic Changes in a Model Colon Gut Microbiota. Environ Eng Sci. 2015;32:602–612. [Google Scholar]
- 118.Kreyling WG, Holzwarth U, Schleh C, Kozempel J, Wenk A, Haberl N, et al. Quantitative biokinetics of titanium dioxide nanoparticles after oral application in rats: Part 2. Nanotoxicology. 2017;11:443–453. doi: 10.1080/17435390.2017.1306893. [DOI] [PubMed] [Google Scholar]
- 119.Jones K, Morton J, Smith I, Jurkschat K, Harding A-HH, Evans G. Human in vivo and in vitro studies on gastrointestinal absorption of titanium dioxide nanoparticles. Toxicol Lett. 2015;233:95–101. doi: 10.1016/j.toxlet.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 120.Shen J, Chen Y, Wang Z, Zhou A, He M, Mao L, et al. Coated zinc oxide improves intestinal immunity function and regulates microbiota composition in weaned piglets. Br J Nutr. 2014;111:2123–2134. doi: 10.1017/S0007114514000300. [DOI] [PubMed] [Google Scholar]
- 121.Tsugita M, Morimoto N, Nakayama M. SiO2 and TiO2 nanoparticles synergistically trigger macrophage inflammatory responses. Part Fibre Toxicol. 2017;14:11. doi: 10.1186/s12989-017-0192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schanen BC, Karakoti AS, Seal S, Drake DR, Warren WL, Self WT. Exposure to titanium dioxide nanomaterials provokes inflammation of an in vitro human immune construct. ACS Nano. 2009;3:2523–2532. doi: 10.1021/nn900403h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Winter M, Beer H-DD, Hornung V, Krämer U, Schins RP, Förster I. Activation of the inflammasome by amorphous silica and TiO2 nanoparticles in murine dendritic cells. Nanotoxicology. 2011;5:326–340. doi: 10.3109/17435390.2010.506957. [DOI] [PubMed] [Google Scholar]
- 124.Shah SN, Shah Z, Hussain M, Khan M. Hazardous Effects of Titanium Dioxide Nanoparticles in Ecosystem. Bioinorg Chem Appl. 2017;2017:4101735. doi: 10.1155/2017/4101735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Setyawati MI, Tay CY, Leong DT. Mechanistic Investigation of the Biological Effects of SiO2, TiO2, and ZnO Nanoparticles on Intestinal Cells. Small. 2015;11:3458–3468. doi: 10.1002/smll.201403232. [DOI] [PubMed] [Google Scholar]
- 126.Lucarelli M, Gatti AM, Savarino G, Quattroni P, Martinelli L, Monari E, et al. Innate defence functions of macrophages can be biased by nano-sized ceramic and metallic particles. Eur Cytokine Netw. 2004;15:339–346. [PubMed] [Google Scholar]
- 127.Mano SS, Kanehira K, Taniguchi A. Comparison of cellular uptake and inflammatory response via toll-like receptor 4 to lipopolysaccharide and titanium dioxide nanoparticles. Int J Mol Sci. 2013;14:13154–13170. doi: 10.3390/ijms140713154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen P, Kanehira K, Taniguchi A. Role of toll-like receptors 3, 4 and 7 in cellular uptake and response to titanium dioxide nanoparticles. Sci Technol Adv Mater. 2013;14:015008. doi: 10.1088/1468-6996/14/1/015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Andersson-Willman B, Gehrmann U, Cansu Z, Buerki-Thurnherr T, Krug HF, Gabrielsson S, et al. Effects of subtoxic concentrations of TiO2 and ZnO nanoparticles on human lymphocytes, dendritic cells and exosome production. Toxicol Appl Pharmacol. 2012;264:94–103. doi: 10.1016/j.taap.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 130.Palomäki J, Karisola P, Pylkkänen L, Savolainen K, Alenius H. Engineered nanomaterials cause cytotoxicity and activation on mouse antigen presenting cells. Toxicology. 2010;267:125–131. doi: 10.1016/j.tox.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 131.Zhu R, Zhu Y, Zhang M, Xiao Y, Du X, Liu H, et al. The induction of maturation on dendritic cells by TiO2 and Fe (3) O (4)@TiO (2) nanoparticles via NF-κB signaling pathway. Mater Sci Eng C Mater Biol Appl. 2014;39:305–314. doi: 10.1016/j.msec.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 132.Vandebriel RJ, Vermeulen JP, van ELB, de JB, Verhagen LM, de la F-BLJ, et al. The crystal structure of titanium dioxide nanoparticles influences immune activity in vitro and in vivo. Part Fibre Toxicol. 2018;15:9. doi: 10.1186/s12989-018-0245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Batt J, Milward M, Chapple I, Grant M, Roberts H, Addison O. TiO2 nanoparticles can selectively bind CXCL8 impacting on neutrophil chemotaxis. Eur Cell Mater. 2018;35:13–24. doi: 10.22203/eCM.v035a02. [DOI] [PubMed] [Google Scholar]
- 134.Feltis BN, Elbaz A, Wright PF, Mackay GA, Turney TW, Lopata AL. Characterizing the inhibitory action of zinc oxide nanoparticles on allergic-type mast cell activation. Mol Immunol. 2015;66:139–146. doi: 10.1016/j.molimm.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 135.Johnson MM, Mendoza R, Raghavendra AJ, Podila R, Brown JM. Contribution of engineered nanomaterials physicochemical properties to mast cell degranulation. Sci Rep. 2017;7:43570. doi: 10.1038/srep43570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Babin K, Antoine F, Goncalves DM, Girard D. TiO2, CeO2 and ZnO nanoparticles and modulation of the degranulation process in human neutrophils. Toxicol Lett. 2013;221:57–63. doi: 10.1016/j.toxlet.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 137.Sabziparvar N, Saeedi Y, Nouri M, Najafi Bozorgi AS, Alizadeh E, Attar F, et al. Investigating the Interaction of Silicon Dioxide Nanoparticles with Human Hemoglobin and Lymphocyte Cells by Biophysical, Computational, and Cellular Studies. J Phys Chem B. 2018;122:4278–4288. doi: 10.1021/acs.jpcb.8b00193. [DOI] [PubMed] [Google Scholar]
- 138.Kang K, Lim J-S. Induction of Functional Changes of Dendritic Cells by Silica Nanoparticles. Immune Netw. 2012;12:104–112. doi: 10.4110/in.2012.12.3.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Winkler HC, Kornprobst J, Wick P, von Moos LM, Trantakis I, Schraner EM, et al. MyD88-dependent pro-interleukin-1β induction in dendritic cells exposed to food-grade synthetic amorphous silica. Part Fibre Toxicol. 2017;14:21. doi: 10.1186/s12989-017-0202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Herd HL, Bartlett KT, Gustafson JA, McGill LD, Ghandehari H. Macrophage silica nanoparticle response is phenotypically dependent. Biomaterials. 2015;53:574–582. doi: 10.1016/j.biomaterials.2015.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hirai T, Yoshioka Y, Takahashi H, Ichihashi K, Yoshida T, Tochigi S, et al. Amorphous silica nanoparticles enhance cross-presentation in murine dendritic cells. Biochem Biophys Res Commun. 2012;427:553–556. doi: 10.1016/j.bbrc.2012.09.095. [DOI] [PubMed] [Google Scholar]
- 142.Desai J, Foresto-Neto O, Honarpisheh M, Steiger S, Nakazawa D, Popper B, et al. Particles of different sizes and shapes induce neutrophil necroptosis followed by the release of neutrophil extracellular trap-like chromatin. Sci Rep. 2017;7:15003. doi: 10.1038/s41598-017-15106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chang H, Ho C-CC, Yang CS, Chang W-HH, Tsai M-HH, Tsai H-TT, et al. Involvement of MyD88 in zinc oxide nanoparticle-induced lung inflammation. Exp Toxicol Pathol. 2013;65:887–896. doi: 10.1016/j.etp.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 144.Silva LR, Girard D. Human eosinophils are direct targets to nanoparticles: Zinc oxide nanoparticles (ZnO) delay apoptosis and increase the production of the pro-inflammatory cytokines IL-1β and IL-8. Toxicol Lett. 2016;259:11–20. doi: 10.1016/j.toxlet.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 145.Heng BC, Zhao X, Tan EC, Khamis N, Assodani A, Xiong S, et al. Evaluation of the cytotoxic and inflammatory potential of differentially shaped zinc oxide nanoparticles. Arch Toxicol. 2011;85:1517–1528. doi: 10.1007/s00204-011-0722-1. [DOI] [PubMed] [Google Scholar]
- 146.Gümüş D, Berber AA, Ada K, Aksoy H. In vitro genotoxic effects of ZnO nanomaterials in human peripheral lymphocytes. Cytotechnology. 2014;66:317–325. doi: 10.1007/s10616-013-9575-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yang E-JJ, Kim S, Kim JS, Choi I-HH. Inflammasome formation and IL-1β release by human blood monocytes in response to silver nanoparticles. Biomaterials. 2012;33:6858–6867. doi: 10.1016/j.biomaterials.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 148.Ghosh M, Manivannan J, Sinha S, Chakraborty A, Mallick SK, Bandyopadhyay M, et al. In vitro and in vivo genotoxicity of silver nanoparticles. Mutat Res. 2012;749:60–69. doi: 10.1016/j.mrgentox.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 149.Park E-JJ, Yi J, Kim Y, Choi K, Park K. Silver nanoparticles induce cytotoxicity by a Trojan-horse type mechanism. Toxicol In Vitro. 2010;24:872–878. doi: 10.1016/j.tiv.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 150.Aldossari AA, Shannahan JH, Podila R, Brown JM. Influence of physicochemical properties of silver nanoparticles on mast cell activation and degranulation. Toxicol In Vitro. 2015;29:195–203. doi: 10.1016/j.tiv.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Alsaleh NB, Persaud I, Brown JM. Silver Nanoparticle-Directed Mast Cell Degranulation Is Mediated through Calcium and PI3K Signaling Independent of the High Affinity IgE Receptor. PLoS One. 2016;11:e0167366. doi: 10.1371/journal.pone.0167366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Burcelin R. Gut microbiota and immune crosstalk in metabolic disease. Mol Metab. 2016;5:771–781. doi: 10.1016/j.molmet.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lazar V, Ditu L-MM, Pircalabioru GG, Gheorghe I, Curutiu C, Holban AM, et al. Aspects of Gut Microbiota and Immune System Interactions in Infectious Diseases, Immunopathology, and Cancer. Front Immunol. 2018;9:1830. doi: 10.3389/fimmu.2018.01830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gatti AM. Biocompatibility of micro- and nano-particles in the colon. Part II. Biomaterials. 2004;25:385–392. doi: 10.1016/s0142-9612(03)00537-4. [DOI] [PubMed] [Google Scholar]
- 155.Ruiz P, Morón B, Becker H, Lang S, Atrott K, Spalinger M, et al. Titanium dioxide nanoparticles exacerbate DSS-induced colitis: role of the NLRP3 inflammasome. Gut. 2016;66:gutjnl–2015–310297. doi: 10.1136/gutjnl-2015-310297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Powell JJ, Ainley CC, Harvey RS, Mason IM, Kendall MD, Sankey EA, et al. Characterisation of inorganic microparticles in pigment cells of human gut associated lymphoid tissue. Gut. 1996;38:390–395. doi: 10.1136/gut.38.3.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hummel TZ, Kindermann A, Stokkers PC, Benninga MA, ten Kate FJ. Exogenous pigment in Peyer patches of children suspected of having IBD. J Pediatr Gastroenterol Nutr. 2014;58:477–480. doi: 10.1097/MPG.0000000000000221. [DOI] [PubMed] [Google Scholar]
- 158.Janer G, Mas del Molino E, Fernández-Rosas E, Fernández A, Vázquez-Campos S. Cell uptake and oral absorption of titanium dioxide nanoparticles. Toxicol Lett. 2014;228:103–110. doi: 10.1016/j.toxlet.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 159.Brun E, Barreau F, Veronesi G, Fayard B, Sorieul S, Chanéac C, et al. Titanium dioxide nanoparticle impact and translocation through ex vivo, in vivo and in vitro gut epithelia. Part Fibre Toxicol. 2014;11:13. doi: 10.1186/1743-8977-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Loeschner K, Hadrup N, Qvortrup K, Larsen A, Gao X, Vogel U, et al. Distribution of silver in rats following 28 days of repeated oral exposure to silver nanoparticles or silver acetate. Part Fibre Toxicol. 2011;8:18. doi: 10.1186/1743-8977-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nogueira CM, de Azevedo WM, Dagli ML, Toma SHH, Leite AZZ, Lordello ML, et al. Titanium dioxide induced inflammation in the small intestine. World J Gastroenterol. 2012;18:4729–4735. doi: 10.3748/wjg.v18.i34.4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Toda T, Yoshino S. Amorphous nanosilica particles block induction of oral tolerance in mice. J Immunotoxicol. 2016;13:723–728. doi: 10.3109/1547691X.2016.1171266. [DOI] [PubMed] [Google Scholar]
- 163.Xu Y, Tang H, Wang H, Liu Y. Blockade of oral tolerance to ovalbumin in mice by silver nanoparticles. Nanomedicine (Lond). 2015;10:419–431. doi: 10.2217/nnm.14.111. [DOI] [PubMed] [Google Scholar]
- 164.Li J, Chen H, Wang B, Cai C, Yang X, Chai Z, et al. ZnO nanoparticles act as supportive therapy in DSS-induced ulcerative colitis in mice by maintaining gut homeostasis and activating Nrf2 signaling. Sci Rep. 2017;7:43126. doi: 10.1038/srep43126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang J, Zhou G, Chen C, Yu H, Wang T, Ma Y, et al. Acute toxicity and biodistribution of different sized titanium dioxide particles in mice after oral administration. Toxicol Lett. 2007;168:176–185. doi: 10.1016/j.toxlet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 166.Zhao X, Ze Y, Gao G, Sang X, Li B, Gui S, et al. Nanosized TiO2-induced reproductive system dysfunction and its mechanism in female mice. PLoS One. 2013;8:e59378. doi: 10.1371/journal.pone.0059378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 167.Wang Y, Chen Z, Ba T, Pu J, Chen T, Song Y, et al. Susceptibility of young and adult rats to the oral toxicity of titanium dioxide nanoparticles. Small. 2013;9:1742–1752. doi: 10.1002/smll.201201185. [DOI] [PubMed] [Google Scholar]
- 168.Kim J-HH, Kim C-SS, Ignacio RM, Kim D-HH, Sajo ME, Maeng EH, et al. Immunotoxicity of silicon dioxide nanoparticles with different sizes and electrostatic charge. Int J Nanomedicine. 2014;9(Suppl 2):183–193. doi: 10.2147/IJN.S57934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yoshida T, Yoshioka Y, Takahashi H, Misato K, Mori T, Hirai T, et al. Intestinal absorption and biological effects of orally administered amorphous silica particles. Nanoscale Res Lett. 2014;9:532. doi: 10.1186/1556-276X-9-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Park E-JJ, Bae E, Yi J, Kim Y, Choi K, Lee SH, et al. Repeated-dose toxicity and inflammatory responses in mice by oral administration of silver nanoparticles. Environ Toxicol Pharmacol. 2010;30:162–168. doi: 10.1016/j.etap.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 171.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lu Y, Chen J, Zheng J, Hu G, Wang J, Huang C, et al. Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci Rep. 2016;6:26337. doi: 10.1038/srep26337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 174.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 175.Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 176.Mariat D, Firmesse O, Levenez F, Guimarăes V, Sokol H, Doré J, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Voreades N, Kozil A, Weir TL. Diet and the development of the human intestinal microbiome. Front Microbiol. 2014;5:494. doi: 10.3389/fmicb.2014.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Natividad JM, Agus A, Planchais J, Lamas B, Jarry AC, Martin R, et al. Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab. 2018;28:737–749. doi: 10.1016/j.cmet.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 179.LeBlanc JG, Chain F, Martín R, Bermúdez-Humarán LG, Courau S, Langella P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Fact. 2017;16:79. doi: 10.1186/s12934-017-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pessione E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front Cell Infect Microbiol. 2012;2:86. doi: 10.3389/fcimb.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. Proteobacteria: A Common Factor in Human Diseases. Biomed Res Int. 2017;2017:9351507. doi: 10.1155/2017/9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shin N-RR, Whon TW, Bae J-WW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 183.Takahashi K, Nishida A, Fujimoto T, Fujii M, Shioya M, Imaeda H, et al. Reduced Abundance of Butyrate-Producing Bacteria Species in the Fecal Microbial Community in Crohn’s Disease. Digestion. 2016;93:59–65. doi: 10.1159/000441768. [DOI] [PubMed] [Google Scholar]
- 184.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 186.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Forbes JD, Van Domselaar G, Bernstein CN. Microbiome Survey of the Inflamed and Noninflamed Gut at Different Compartments Within the Gastrointestinal Tract of Inflammatory Bowel Disease Patients. Inflamm Bowel Dis. 2016;22:817–825. doi: 10.1097/MIB.0000000000000684. [DOI] [PubMed] [Google Scholar]
- 189.Favier C, Neut C, Mizon C, Cortot A, Colombel JF, Mizon J. Fecal beta-D-galactosidase production and Bifidobacteria are decreased in Crohn’s disease. Dig Dis Sci. 1997;42:817–822. doi: 10.1023/a:1018876400528. [DOI] [PubMed] [Google Scholar]
- 190.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, et al. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2015;50:167–179. doi: 10.1007/s00535-014-0963-x. [DOI] [PubMed] [Google Scholar]
- 192.Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring). 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 193.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 194.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 196.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, Inoue H, et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015;15:100. doi: 10.1186/s12876-015-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen Y-YY, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8:e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ferrer M, Ruiz A, Lanza F, Haange S-BB, Oberbach A, Till H, et al. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environ Microbiol. 2013;15:211–226. doi: 10.1111/j.1462-2920.2012.02845.x. [DOI] [PubMed] [Google Scholar]
- 203.Patil DP, Dhotre DP, Chavan SG, Sultan A, Jain DS, Lanjekar VB, et al. Molecular analysis of gut microbiota in obesity among Indian individuals. J Biosci. 2012;37:647–657. doi: 10.1007/s12038-012-9244-0. [DOI] [PubMed] [Google Scholar]
- 204.Noor SO, Ridgway K, Scovell L, Kemsley EK, Lund EK, Jamieson C, et al. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 2010;10:134. doi: 10.1186/1471-230X-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Takaishi H, Matsuki T, Nakazawa A, Takada T, Kado S, Asahara T, et al. Imbalance in intestinal microflora constitution could be involved in the pathogenesis of inflammatory bowel disease. Int J Med Microbiol. 2008;298:463–472. doi: 10.1016/j.ijmm.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 206.Verdam FJ, Fuentes S, De JC, Zoetendal EG, Erbil R, Greve JW, et al. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (Silver Spring) 2013;21:E607–E615. doi: 10.1002/oby.20466. [DOI] [PubMed] [Google Scholar]
- 207.Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 208.Durbán A, Abellán JJ, Jiménez-Hernández N, Latorre A, Moya A. Daily follow-up of bacterial communities in the human gut reveals stable composition and host-specific patterns of interaction. FEMS Microbiol Ecol. 2012;81:427–437. doi: 10.1111/j.1574-6941.2012.01368.x. [DOI] [PubMed] [Google Scholar]
- 209.Sokol H, Leducq V, Aschard H, Pham H-PP, Jegou S, Landman C, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 211.Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M, et al. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis. 2010;16:2034–2042. doi: 10.1002/ibd.21319. [DOI] [PubMed] [Google Scholar]
- 212.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tims S, Derom C, Jonkers DM, Vlietinck R, Saris WH, Kleerebezem M, et al. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. 2013;7:707–717. doi: 10.1038/ismej.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Furet J-PP, Kong L-CC, Tap J, Poitou C, Basdevant A, Bouillot J-LL, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59:3049–3057. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zhu Q, Jin Z, Wu W, Gao R, Guo B, Gao Z, et al. Analysis of the intestinal lumen microbiota in an animal model of colorectal cancer. PLoS One. 2014;9:e90849. doi: 10.1371/journal.pone.0090849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sen T, Cawthon CR, Ihde BT, Hajnal A, DiLorenzo PM, La Serre CB de, et al. Diet-driven microbiota dysbiosis is associated with vagal remodeling and obesity. Physiol Behav. 2017;173:305–317. [DOI] [PMC free article] [PubMed]
- 217.Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9:103. doi: 10.1186/s13073-017-0490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Geng J, Song Q, Tang X, Liang X, Fan H, Peng H, et al. Co-occurrence of driver and passenger bacteria in human colorectal cancer. Gut Pathog. 2014;6:26. doi: 10.1186/1757-4749-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1720–1728. doi: 10.1053/j.gastro.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 221.Sokol H, Lepage P, Seksik P, Doré J, Marteau P. Temperature gradient gel electrophoresis of fecal 16S rRNA reveals active Escherichia coli in the microbiota of patients with ulcerative colitis. J Clin Microbiol. 2006;44:3172–3177. doi: 10.1128/JCM.02600-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 223.Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 225.Stelter C, Käppeli R, König C, Krah A, Hardt W-DD, Stecher B, et al. Salmonella-induced mucosal lectin RegIIIβ kills competing gut microbiota. PLoS One. 2011;6:e20749. doi: 10.1371/journal.pone.0020749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wang X, Ota N, Manzanillo P, Kates L, Zavala-Solorio J, Eidenschenk C, et al. Interleukin-22 alleviates metabolic disorders and restores mucosal immunity in diabetes. Nature. 2014;514:237–241. doi: 10.1038/nature13564. [DOI] [PubMed] [Google Scholar]
- 227.Geier MS, Butler RN, Howarth GS. Probiotics, prebiotics and synbiotics: a role in chemoprevention for colorectal cancer? Cancer Biol Ther. 2006;5:1265–1269. doi: 10.4161/cbt.5.10.3296. [DOI] [PubMed] [Google Scholar]
- 228.Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E, et al. The Environmental Sensor AHR Protects from Inflammatory Damage by Maintaining Intestinal Stem Cell Homeostasis and Barrier Integrity. Immunity. 2018;49:353–362. doi: 10.1016/j.immuni.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Gronke K, Hernández PP, Zimmermann J, Klose CSNS, Kofoed-Branzk M, Guendel F, et al. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature. 2019. [DOI] [PMC free article] [PubMed]
- 230.Urrutia-Ortega IM, Garduño-Balderas LG, Delgado-Buenrostro NL, Freyre-Fonseca V, Flores-Flores JOO, González-Robles A, et al. Food-grade titanium dioxide exposure exacerbates tumor formation in colitis associated cancer model. Food Chem Toxicol. 2016;93:20–31. doi: 10.1016/j.fct.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 231.Proquin H, Jetten MJ, Jonkhout MCMC, Garduño-Balderas LG, Briedé JJ, de Kok TM, et al. Gene expression profiling in colon of mice exposed to food additive titanium dioxide (E171) Food Chem Toxicol. 2018;111:153–165. doi: 10.1016/j.fct.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 232.Proquin H, Jetten MJ, Jonkhout MCMC, Garduño-Balderas LG, Briedé JJ, de Kok TM, et al. Transcriptomics analysis reveals new insights in E171-induced molecular alterations in a mouse model of colon cancer. Sci Rep. 2018;8:9738. doi: 10.1038/s41598-018-28063-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Shoaie S, Karlsson F, Mardinoglu A, Nookaew I, Bordel S, Nielsen J. Understanding the interactions between bacteria in the human gut through metabolic modeling. Sci Rep. 2013;3:2532. doi: 10.1038/srep02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lin HV, Frassetto A, Kowalik EJ, Nawrocki AR, Lu MM, Kosinski JR, et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One. 2012;7:e35240. doi: 10.1371/journal.pone.0035240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hartstra AV, Bouter KE, Bäckhed F, Nieuwdorp M. Insights into the role of the microbiome in obesity and type 2 diabetes. Diab Care. 2015;38:159–165. doi: 10.2337/dc14-0769. [DOI] [PubMed] [Google Scholar]
- 236.Harris K, Kassis A, Major G, Chou CJ. Is the gut microbiota a new factor contributing to obesity and its metabolic disorders? J Obes. 2012;2012:879151. doi: 10.1155/2012/879151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Zambell KL, Fitch MD, Fleming SE. Acetate and butyrate are the major substrates for de novo lipogenesis in rat colonic epithelial cells. J Nutr. 2003;133:3509–3515. doi: 10.1093/jn/133.11.3509. [DOI] [PubMed] [Google Scholar]
- 238.Xiong Y, Miyamoto N, Shibata K, Valasek MA, Motoike T, Kedzierski RM, et al. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc Natl Acad Sci USA. 2004;101:1045–1050. doi: 10.1073/pnas.2637002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Brahe LK, Astrup A, Larsen LH. Is butyrate the link between diet, intestinal microbiota and obesity-related metabolic diseases? Obes Rev. 2013;14:950–959. doi: 10.1111/obr.12068. [DOI] [PubMed] [Google Scholar]
- 240.Vrieze A, Van NE, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–916. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 241.Scheppach W, Sommer H, Kirchner T, Paganelli GM, Bartram P, Christl S, et al. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology. 1992;103:51–56. doi: 10.1016/0016-5085(92)91094-k. [DOI] [PubMed] [Google Scholar]
- 242.Vernia P, Monteleone G, Grandinetti G, Villotti G, Di Giulio E, Frieri G, et al. Combined oral sodium butyrate and mesalazine treatment compared to oral mesalazine alone in ulcerative colitis: randomized, double-blind, placebo-controlled pilot study. Dig Dis Sci. 2000;45:976–981. doi: 10.1023/a:1005537411244. [DOI] [PubMed] [Google Scholar]
- 243.Di Sabatino A, Morera R, Ciccocioppo R, Cazzola P, Gotti S, Tinozzi FP, et al. Oral butyrate for mildly to moderately active Crohn’s disease. Aliment Pharmacol Ther. 2005;22:789–794. doi: 10.1111/j.1365-2036.2005.02639.x. [DOI] [PubMed] [Google Scholar]
- 244.Vernia P, Marcheggiano A, Caprilli R, Frieri G, Corrao G, Valpiani D, et al. Short-chain fatty acid topical treatment in distal ulcerative colitis. Aliment Pharmacol Ther. 1995;9:309–313. doi: 10.1111/j.1365-2036.1995.tb00386.x. [DOI] [PubMed] [Google Scholar]
- 245.Tang Y, Chen Y, Jiang H, Robbins GT, Nie D. G-protein-coupled receptor for short-chain fatty acids suppresses colon cancer. Int J Cancer. 2011;128:847–856. doi: 10.1002/ijc.25638. [DOI] [PubMed] [Google Scholar]
- 246.Canani RB, Costanzo MD, Leone L, Pedata M, Meli R, Calignano A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol. 2011;17:1519–1528. doi: 10.3748/wjg.v17.i12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Keku TO. Dulal S, Deveaux A, Jovov B, Han X. The gastrointestinal microbiota and colorectal cancer. Am J Physiol Gastrointest Liver Physiol. 2015;308:G351–G363. doi: 10.1152/ajpgi.00360.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Donohoe DR, Holley D, Collins LB, Montgomery SA, Whitmore AC, Hillhouse A, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014;4:1387–1397. doi: 10.1158/2159-8290.CD-14-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, Lokken KL, et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science. 2017;357:570–575. doi: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Fei N, Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013;7:880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Maharshak N, Packey CD, Ellermann M, Manick S, Siddle JP, Huh EY, et al. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes. 2013;4:316–324. doi: 10.4161/gmic.25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan T-JJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Arthur JC, Gharaibeh RZ, Mühlbauer M, Perez-Chanona E, Uronis JM, McCafferty J, et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun. 2014;5:4724. doi: 10.1038/ncomms5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza O, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514:181–186. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- 259.Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Liang Y, Zhan J, Liu D, Luo M, Han J, Liu X, et al. Organophosphorus pesticide chlorpyrifos intake promotes obesity and insulin resistance through impacting gut and gut microbiota. Microbiome. 2019;7:19. doi: 10.1186/s40168-019-0635-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Rodriguez-Palacios A, Harding A, Menghini P, Himmelman C, Retuerto M, Nickerson KP, et al. The Artificial Sweetener Splenda Promotes Gut Proteobacteria, Dysbiosis, and Myeloperoxidase Reactivity in Crohn’s Disease-Like Ileitis. Inflamm Bowel Dis. 2018;24:1005–1020. doi: 10.1093/ibd/izy060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Viennois E, Merlin D, Gewirtz AT, Chassaing B. Dietary Emulsifier-Induced Low-Grade Inflammation Promotes Colon Carcinogenesis. Cancer Res. 2017;77:27–40. doi: 10.1158/0008-5472.CAN-16-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Ebabe Elle R, Gaillet S, Vidé J, Romain C, Lauret C, Rugani N, et al. Dietary exposure to silver nanoparticles in Sprague-Dawley rats: effects on oxidative stress and inflammation. Food Chem Toxicol. 2013;60:297–301. doi: 10.1016/j.fct.2013.07.071. [DOI] [PubMed] [Google Scholar]
- 264.Chen Z, Wang Y, Zhuo L, Chen S, Zhao L, Luan X, et al. Effect of titanium dioxide nanoparticles on the cardiovascular system after oral administration. Toxicol Lett. 2015;239:123–130. doi: 10.1016/j.toxlet.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 265.Hu H, Li L, Guo Q, Jin S, Zhou Y, Oh Y, et al. A mechanistic study to increase understanding of titanium dioxide nanoparticles-increased plasma glucose in mice. Food Chem Toxicol. 2016;95:175–187. doi: 10.1016/j.fct.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 266.EFSA ANS Panel Guidance on risk assessment of the application of nanoscience and nanotechnologies in the food and feed chain: Part 1, human and animal health. EFSA J. 2018;16:5327–5395. doi: 10.2903/j.efsa.2018.5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.