The LARGE1/LARGE1/Mei2-Like protein4 (OML4), which is phosphorylated by GSK2, negatively regulates grain size and weight in rice.
Abstract
Regulation of grain size is crucial for improving crop yield and is also a basic aspect in developmental biology. However, the genetic and molecular mechanisms underlying grain size control in crops remain largely unknown despite their central importance. Here, we report that the MEI2-LIKE PROTEIN4 (OML4) encoded by the LARGE1 gene is phosphorylated by GLYCOGEN SYNTHASE KINASE2 (GSK2) and negatively controls grain size and weight in rice (Oryza sativa). Loss of function of OML4 leads to large and heavy grains, while overexpression of OML4 causes small and light grains. OML4 regulates grain size by restricting cell expansion in the spikelet hull. OML4 is expressed in developing panicles and grains, and the GFP-OML4 fusion protein is localized in the nuclei. Biochemical analyses show that the GSK2 physically interacts with OML4 and phosphorylates it, thereby possibly influencing the stability of OML4. Genetic analyses support that GSK2 and OML4 act, at least in part, in a common pathway to control grain size in rice. These results reveal the genetic and molecular mechanism of a GSK2-OML4 regulatory module in grain size control, suggesting that this pathway is a suitable target for improving seed size and weight in crops.
INTRODUCTION
The world population continues to increase rapidly, and this increase has led to a growing demand for rice (Oryza sativa; Lee et al., 2015). Grain yield is determined by tiller number, grain number, and grain weight. As grain size is a key component of grain weight, regulation of grain size is a crucial strategy to increase grain production. Grain growth is restricted by spikelet hulls, which influence final grain size in rice (Li and Li, 2016; Li et al. 2019). The growth of the spikelet hull is determined by cell proliferation and cell expansion processes. Several genes that regulate the grain size by influencing cell proliferation in the spikelet hull have been described in rice, such as GRAIN WIDTH AND WEIGHT2 (GW2; Song et al., 2007), GRAIN SIZE ON CHROMOSOME5 (GW5/GSE5; Duan et al., 2017; Liu et al., 2017), GRAIN WIDTH ON CHROMOSOME8 (GW8/OsSPL16; Wang et al., 2012), GRAIN SIZE ON CHROMOSOME3 (GS3; Fan et al., 2006; Li et al., 2011), GRAIN SHAPE ON CHROMOSOME9 (GS9; Zhao et al., 2018), OsMKKK10-OsMKK4-OsMPK6 (Duan et al., 2014; Xu et al. 2018a, 2018b), and Mitogen-activated protein Kinase Phosphatase1 (MKP1; Guo et al., 2018; Xu et al., 2018b). In addition, several genes that control grain size by influencing cell expansion in the spikelet hulls have been reported in rice, such as GRAIN SIZE ON CHROMOSOME2 (GS2/OsGRF4; Che et al., 2015; Duan et al., 2015; Hu et al., 2015), GSK3/SHAGGY-like kinase5 (OsGSK5; Hu et al., 2018; Xia et al., 2018; Ying et al., 2018), GRAIN LENGTH AND WEIGHT ON CHROMOSOME7 (GLW7/SPL13; Si et al., 2016), GRAIN LENGTH ON CHROMOSOME7 (GL7/GW7/SLG7; Wang et al., 2015a, 2015b; Zhou, 2015), POSITIVE REGULATOR OF GRAIN LENGTH1/2 (PGL1/PGL2; Heang and Sassa, 2012a, 2012b), and ANTAGONIST OF PGL1 (APG; Heang and Sassa, 2012a, 2012b). However, the genetic and molecular relationships between these factors remain largely unknown.
SHAGGY-LIKE KINASE2 (GSK2), a homologue of the Arabidopsis (Arabidopsis thaliana) BRASSINOSTEROID INSENSITIVE2 (BIN2) kinase, has been reported to influence grain size and also other growth processes in rice (Tong et al., 2012). Overexpression of GSK2 leads to small grains and short plants, whereas downregulation of GSK2 produces long grains. GSK2 can associate with and phosphorylate DWARF AND LOW-TILLERING (DLT). Interestingly, downregulation of DLT/GS6 causes multiple growth defects as well as wide, short, and heavy grains due to increased cell proliferation in the grain-width direction (Sun et al., 2013). GSK2 also interacts with the transcription factor OsGRF4/GS2 described to influence grain growth predominantly by promoting cell expansion in the spikelet hull (Che et al., 2015; Duan et al., 2015; Hu et al., 2015). A recent study has shown that GW5/GSE5, a calmodulin binding protein, can associate with GSK2 and repress its kinase activity (Duan et al., 2017; Liu et al., 2017). GW5/GSE5 regulates grain width by limiting cell proliferation in the spikelet hull (Duan et al., 2017; Liu et al., 2017). However, genetic interactions between GSK2 and these grain size regulators are not clear so far.
To further reveal the mechanisms of grain size determination, we have identified several grain size genes whose loss and gain of function lead to opposite effects on grain size in rice. Here, we report that LARGE1, which encodes MEI2-LIKE PROTEIN4 (OML4) with three RNA recognition motif (RRM) domains, regulates grain size and weight by restricting cell expansion in spikelet hulls in rice. Homologs of OML4 have been reported to influence meiosis, the plastochron, and leaf maturation in plant species (Veit et al., 1998; Kaur et al., 2006; Kawakatsu et al., 2006; Xiong et al., 2006), but their function in seed/grain size control has not been characterized so far. The large1-1 mutant forms large and heavy grains, while overexpression of OML4 causes small and light grains. OML4 interacts with GSK2 and can be phosphorylated by GSK2. Genetic analyses indicate that GSK2 and OML4 function, at least in part, in a common pathway to control grain size. Therefore, our findings reveal a significant genetic and molecular mechanism of the GSK2-OML4 regulatory module in grain size control, suggesting that it is useful for grain size and weight improvement in crops.
RESULTS
The large1 Mutant Forms Large and Heavy Grains
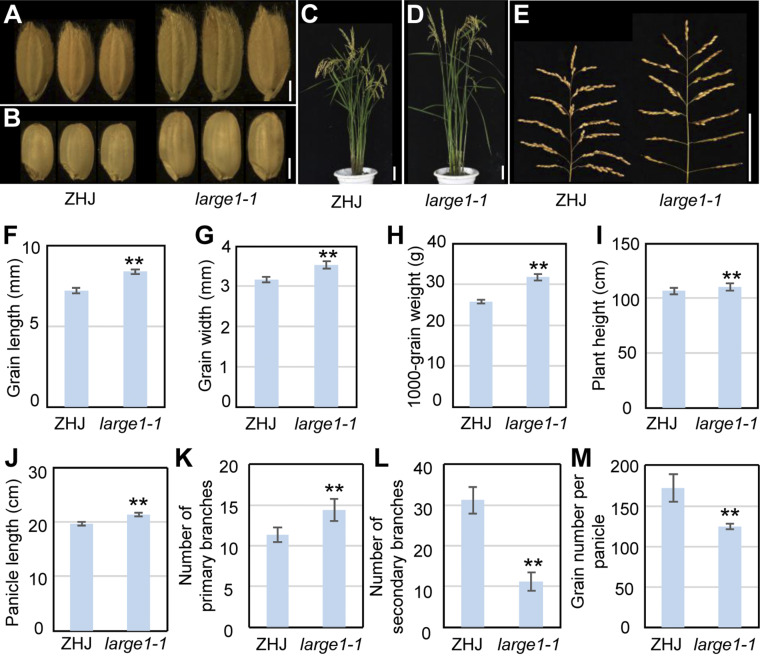
We have previously identified a number of grain size mutants in rice. The large1-1 mutant was isolated from γ-ray–treated M2 populations of the japonica var Zhonghuajing (ZHJ). The large1-1 mutant displayed large grains and tall plants (Figures 1A to 1E). The length of large1-1 grains was increased by 16.24% compared with that of ZHJ grains (Figure 1F). Similarly, the width of large1-1 grains was increased by 11.54% compared with that of ZHJ grains (Figure 1G). The large1-1 grains were also significantly heavier than ZHJ grains (Figure 1H). The weight of large1-1 grains was increased by 23.11% compared with that of ZHJ grains. These results indicate that LARGE1 negatively regulates grain size and weight in rice.
Figure 1.
LARGE1 Influences Grain Size and Plant Morphology.
(A) Mature paddy grains of ZHJ and large1-1.
(B) Brown rice grains of ZHJ and large1-1.
(C) and (D) ZHJ (C) and large1-1 (D) plants at mature stage.
(E) ZHJ (left) and large1-1 (right) panicles.
(F) and (G) Grain length (F) and width (G) of ZHJ and large1-1.
(H) The 1000-grain weight of ZHJ and large1-1.
(I) Plant height of ZHJ and large1-1.
(J) Panicle length of ZHJ and large1-1.
(K) Number of ZHJ and large1-1 primary panicle branches.
(L) Number of ZHJ and large1-1 secondary panicle branches.
(M) Grain number per panicle of ZHJ and large1-1.
Values ( [F] to [H]) are given as mean ± sd (n ≥ 50). Values ( [I] to [M]) are given as means ± sd (n = 20). Asterisks indicate significant differences between ZHJ and large1-1. **, P < 0.01 compared with the wild type (ZHJ) by Student’s t test. Bar in (A) and (B) = 2 mm; bar in (C) to (E) = 10 cm.
Mature large1-1 plants were significantly higher than ZHJ plants (Figure 1I). The large1-1 panicles were long and loose in comparison with the wild-type panicles (Figure 1J), indicating that LARGE1 also negatively influences panicle length. As panicle structure and shape are determined by panicle branches, we investigated ZHJ and large1-1 panicle branches. The primary branches of large1-1 panicles were more than those of ZHJ (Figure 1K), and the large1-1 had fewer secondary branches than ZHJ (Figure 1L). We also investigated the number of grains per panicle in ZHJ and large1-1. The number of grains per panicle in large1-1 was decreased in comparison with that in ZHJ (Figure 1M). These results suggest that LARGE1 influences the balance between grain number and grain size in rice.
LARGE1 Regulates Cell Expansion in Spikelet Hulls
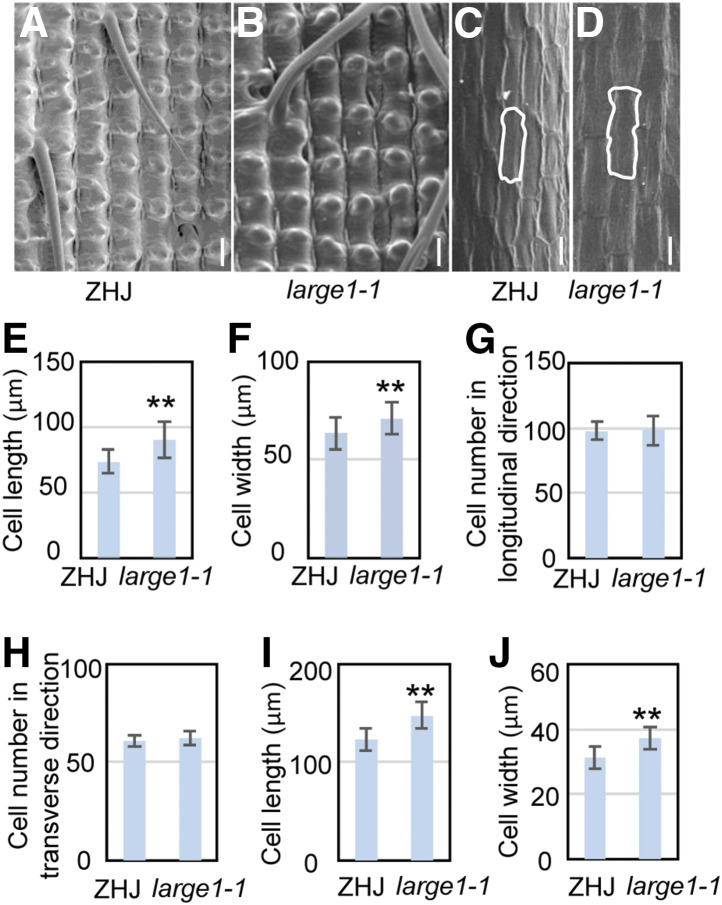
Grain growth is limited by spikelet hulls, and spikelet hull growth is determined by cell proliferation and cell expansion processes (Li and Li, 2016). To uncover the cellular mechanism for LARGE1 in grain growth, we investigated cells in ZHJ and large1-1 spikelet hulls. As shown in Figure 2, the outer epidermal cells in large1-1 lemmas were longer and wider cells than those of ZHJ lemmas, while cell numbers in large1-1 lemmas were similar to that in the wild-type lemmas in both longitudinal and transverse directions (Figures 2A, 2B, and 2E to 2H). Similarly, the inner epidermal cells of large1-1 were longer and wider than those of ZHJ (Figures 2C, 2D, 2I, and 2J). These results indicate that the long and wide grain phenotypes of large1-1 result from the long and wide cells in spikelet hulls. Thus, LARGE1 regulates grain size by limiting cell expansion in spikelet hulls.
Figure 2.
large1 Forms Large Grains Due to Increased Cell Expansion in the Spikelet Hull.
(A) and (B) Scanning electron microscopy analysis of the outer surface of ZHJ (A) and large1-1 (B) lemmas.
(C) and (D) Scanning electron microscopy analysis of the inner surface of ZHJ (C) and large1-1 (D) lemmas.
(E) and (F) Average length (E) and width (F) of outer epidermal cells in ZHJ and large1-1 lemmas.
(G) Outer epidermal cell number in the longitudinal direction in ZHJ and large1-1 lemmas.
(H) Outer epidermal cell number in the transverse direction in ZHJ and large1-1 lemmas.
(I) and (J) Average length (I) and width (J) of inner epidermal cells in the longitudinal direction in ZHJ and large1-1 lemmas.
Values ( [E] to [J]) are given as the means ± sd (n ≥ 50). **, P < 0.01 compared with the wild type by Student’s t test.
Bar in (A) to (D) = 50 µm.
As several genes were reported to regulate grain size by influencing cell expansion in spikelet hulls, we investigated their expression levels in the wild-type and large1-1 panicles (Supplemental Figure 1). SPL13/GWL7, a transcription factor, positively influences grain length by increasing cell expansion (Si et al., 2016). Higher expression of SPL13 in large1-1 panicles was observed. GL7 promotes cell elongation in spikelet hulls, resulting in long grains (Wang et al., 2015a, 2015b; Zhou et al., 2015), although GL7/GW7/SLG7 is also proposed to increase grain length by influencing cell proliferation (Wang et al., 2015b). Expression of GL7 was obviously increased in large1-1 compared with ZHJ (Supplemental Figure 1). The putative Ser carboxypeptidase GS5 and the transcription factor GS2 affect grain growth by increasing both cell expansion and cell proliferation (Li et al., 2011; Duan et al., 2015; Hu et al., 2015). Expression levels of GS5 and GS2 in large1-1 were significantly higher than those in ZHJ (Supplemental Figure 1). The basic helix-loop-helix transcription factor PGL1 controls grain length by increasing cell expansion (Heang and Sassa, 2012a, 2012b). APG, another basic helix-loop-helix transcription factor, regulates grain length by restricting cell expansion in spikelet hulls (Heang and Sassa, 2012a, 2012b). Expression of APG and PGL1 in large1-1 was lower and higher than that in ZHJ, respectively (Supplemental Figure 1). These data indicate that LARGE1 influences the expression of several grain size genes that regulate cell expansion.
LARGE1 Encodes the Mei2-Like Protein OML4
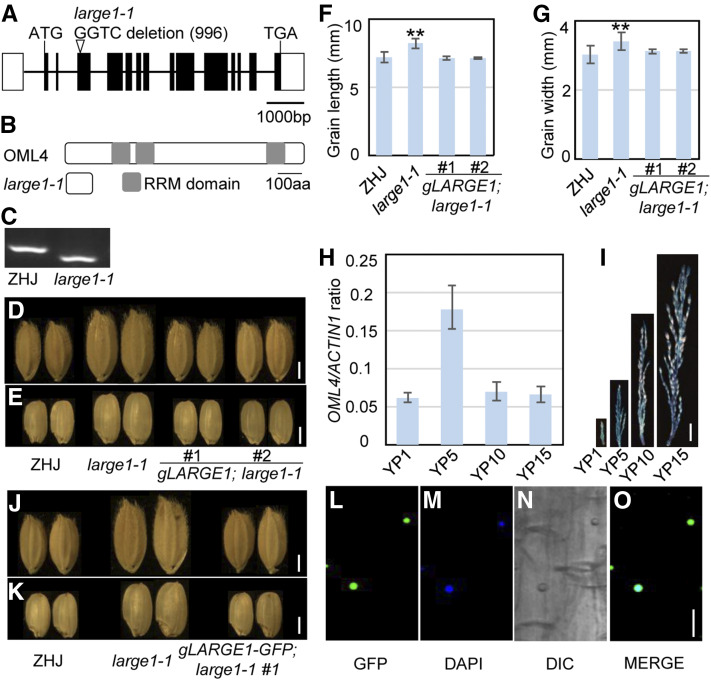
The MutMap approach was used to identify the large1-1 mutation (Abe et al., 2012; Fang et al., 2016; Huang et al., 2017). We crossed ZHJ with large1-1 and generated an F2 population. In the F2 population, the progeny segregation showed that the single recessive mutation determines the large-grain phenotype of large1-1. The genomic DNAs from F2 plants with the large-grain phenotype were pooled and applied for whole-genome resequencing. The wild-type ZHJ was also sequenced as a control. Single-nucleotide polymorphism (SNP) analyses were performed as described previously (Fang et al., 2016; Huang et al., 2017). We detected 3913 SNPs and 1280 small insertions and deletions (INDELs) between ZHJ and the pooled F2 plants with large1-1 phenotypes. The SNP:INDEL ratio in the pooled F2 plants was calculated in the whole genome (Supplemental Figure 2). Among them, only one INDEL in the coding region had an SNP:INDEL ratio = 1. This INDEL contained a 4-bp deletion in large1-1 in the gene (LOC_Os02g31290; Figure 3A; Supplemental Figure 3; Supplemental Table 1), which leads to a premature stop codon (Figure 3B). We further confirmed this deletion in LOC_Os02g31290 by developing a derived cleaved-amplified polymorphic sequence marker (Figure 3C). These results indicate that LOC_Os02g31290 is the candidate gene for LARGE1.
Figure 3.
LARGE1 Encodes the Mei2-Like Protein OML4.
(A) LARGE1/OML4 gene structure. The coding sequence is shown as the black box, and introns are indicated using black lines. ATG and TGA represent the start codon and the stop codon, respectively.
(B) OML4 and mutated protein encoded by large1. The OML4 protein contains three RRM domains. The mutation results in a premature termination codon in OML4, causing a truncated protein.
(C) Derived cleaved-amplified polymorphic sequence marker was developed according to the large1-1 mutation. The PCR products were digested by the restriction enzyme HphI.
(D) and (E) Mature paddy (D) and brown (E) rice grains of ZHJ, large1-1, gLARGE1;large1-1 #1, and gLARGE1;large1-1 #2.
(F) and (G) Grain length (F) and width (G) of ZHJ, large1-1, gLARGE1;large1-1 #1, and gLARGE1;large1-1 #2. Asterisks indicate significant differences between ZHJ and large1-1. **, P < 0.01 compared with the wild type by Student’s t test.
(H) Relative OML4 gene expression in young panicles of 1 cm (YP1) to 15 cm (YP15) in ZHJ. RT-qPCR was used to measure expression levels of OML4 in panicles. Values are given as mean ± sd. Three biological replicates were used (n = 3).
(I) OML4 expression activity was monitored by proOML4:GUS transgene expression. Histochemical analysis of GUS activity in panicles at different developmental stages.
(J) and (K) Mature paddy (J) and brown (K) rice grains of ZHJ, large1-1, and gLARGE1-GFP;large1-1 #1.
(L) to (O) Subcellular location of OML4-GFP in gLARGE1-GFP;large1-1 #1 root cells. GFP fluorescence of GFP-OML4 (L), 4′,6-diamidino-2-phenylindole staining (DAPI; see [M]), differential interference contrast (DIC; see [N]), and merged (O) images are shown.
Bar in (D), (E), (J), and (K) = 2 mm; bar in (I) = 1 cm; bar in (L) to (O) = 10 µm.
A genetic complementation test was conducted to confirm whether the deletion in LOC_Os02g31290 was responsible for the large1-1 phenotypes. The genomic fragment of LOC_Os02g31290 (gLARGE1) was transformed into the large1-1 mutant, and 11 transgenic lines were generated. The gLARGE1 construct complemented the large-grain phenotypes of the large1-1 mutant (Figures 3D and 3E). The grain length and width of gLARGE1;large1-1 transgenic plants were similar to those of ZHJ (Figures 3F and 3G). Genomic complementation plants also recovered to the wild type in plant height and morphology (Supplemental Figure 4). Therefore, the complementation test supported that the LARGE1 gene is LOC_Os02g31290.
LARGE1/LOC_Os02g31290 encodes the OML4 with three RRMs (Figure 3B; Supplemental Figure 5). Homologs of OML4 were found in crops and mammals (Supplemental Figures 5 and 6; Supplemental Data Set). Homologs of OML4 were reported to influence meiosis, the plastochron, and leaf maturation in plants (Veit et al., 1998; Kaur et al., 2006; Kawakatsu et al., 2006; Xiong et al., 2006), but the roles of OML4 and its homologs in grain size control are totally unknown so far. The mutation in large1-1 resulted in a premature stop codon. The proteins encoded by large1-1 (OML4large1-1) lacked RRM motifs (Figure 3B), which indicated that large1-1 is a loss-of-function allele.
Expression and Subcellular Localization of OML4
We investigated the expression of OML4 in developing panicles using RT-qPCR analysis. The OML4 gene expression was detected and was also variable during panicle development (Figure 3H). We further generated the OML4 promoter:GUS transgenic plants (proOML4:GUS) and examined the expression patterns of OML4 in developing panicles. During panicle development, β-glucuronidase (GUS) activity was detected in the panicles with ∼1 cm of length. The strongest GUS activity was observed in the panicles with ∼5 cm of length. The GUS activity then gradually decreased during panicle development (Figure 3I). Similarly, RT-qPCR analysis indicate that expression of OML4 was relatively high in the panicles with ∼5 cm of length (Figure 3H).
To investigate the subcellular localization of OML4 in rice, we generated gLARGE1-GFP transgenic plants. As shown in Figures 3J and 3K, the gLARGE1-GFP construct rescued the phenotypes of the large1-1 mutant, indicating that the LARGE1-GFP fusion protein is functional. GFP signal in gLARGE1-GFP;large1-1 roots was predominantly detected in nuclei (Figures 3L to 3O). Thus, this finding indicated that OML4 is localized in nuclei in rice.
Overexpression of OML4 Results in Short Grains Due to Short Cells in Spikelet Hulls
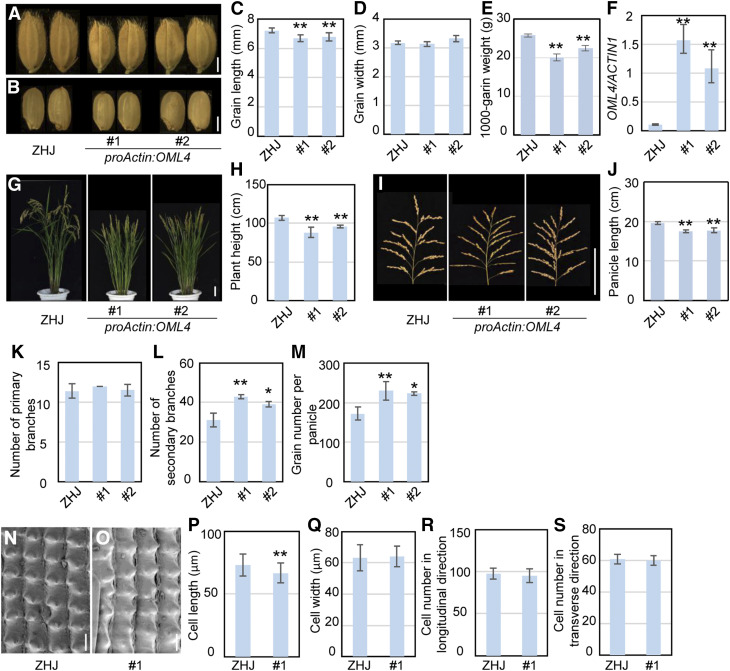
To further explore the functions of OML4 in grain growth, we generated the proActin:OML4 construct, transformed it into ZHJ, and isolated 14 transgenic lines. The proActin:OML4 transgenic plants had short grains compared with ZHJ (Figures 4A to 4C), while the width of proActin:OML4 grains was similar to that of ZHJ (Figure 4D). The grains were also significantly lighter than those of ZHJ (Figure 4E). Grain length of proActin:OML4 transgenic lines was associated with the expression levels of OML4 (Figure 4F). These data reveal that OML4 functions to restrict grain growth in rice.
Figure 4.
Overexpression of OML4 Results in Smaller Grains.
(A) and (B) Mature paddy (A) and brown rice (B) grains of ZHJ and proActin:OML4.
(C) and (D) Grain length (C) and width (D) of ZHJ and proActin:OML4 transgenic lines.
(E) The 1000-grain weight of ZHJ and proActin:OML4 transgenic lines.
(F) Expression level of OML4/LARGE1 in ZHJ and proActin:OML4 transgenic lines. RT-qPCR was used to measure expression levels of OML4/LARGE1. Three biological replicates were used (n = 3). ACTIN1 was used to normalize expression.
(G) ZHJ and proActin:OML4 plants.
(H) Plant height of ZHJ and proActin:OML4 transgenic lines.
(I) ZHJ and proActin:OML4 panicles.
(J) Panicle length of ZHJ and proActin:OML4 transgenic lines.
(K) and (L) Primary (K) and secondary (L) panicle branch number of ZHJ and proActin:OML4 transgenic lines.
(M) Total grain number per panicle of ZHJ and proActin:OML4 transgenic lines.
(N) and (O) Scanning electron microscopy analysis of the outer surface of ZHJ (N) and proActin:OML4 #1 (O) lemmas.
(P) and (Q) Average length (P) and width (Q) of outer epidermal cells in the longitudinal direction in ZHJ and proActin:OML4 #1 lemmas.
(R) and (S) Number of outer epidermal cells in the longitudinal (R) and transverse (S) direction in ZHJ and proActin:OML4 #1 lemmas.
Values (see [C] to [E] and [P] to [S]) are given as the means ± sd (n ≥ 50). Values (see [F]) are given as the mean ± sd. Values (see [H] and [J] to [M] are given as the means ± sd (n = 20). Asterisks indicate significant differences between ZHJ and proActin:OML4 transgenic lines. *, P < 0.05; **, P < 0.01 compared with the wild type by Student’s t test.
Bar in (A) and (B) = 2 mm; bar in (G) and (I) = 10 cm; bar in (N) and (O) = 50 µm.
Mature proActin:OML4 transgenic plants were shorter than ZHJ (Figures 4G and 4H). The average length of proActin:OML4 panicles was significantly decreased compared with that of ZHJ panicles (Figures 4I and 4J). The primary panicle branches of proActin:OML4 were comparable to those of ZHJ, while the secondary panicle branches of proActin:OML4 were obviously increased in comparison to those of ZHJ (Figures 4K and 4L), resulting in the increased grain number per panicle (Figure 4M).
As proActin:OML4 transgenic lines produced short grains, we tested whether overexpression of OML4 could decrease cell length in spikelet hulls. We examined the size of outer epidermal cells in the wild-type and proActin:OML4 spikelet hulls (Figures 4N and 4O). Outer epidermal cells in proActin:OML4 spikelet hulls were shorter than those of ZHJ spikelet hulls (Figures 4P and 4Q). By contrast, the number of epidermal cells in the longitudinal and transverse direction in proActin:OML4 spikelet hulls was similar to that in ZHJ spikelet hulls (Figures 4R and 4S). These results further revealed that OML4 affects grain growth by limiting cell expansion in spikelet hulls.
OML4 Interacts with GSK2
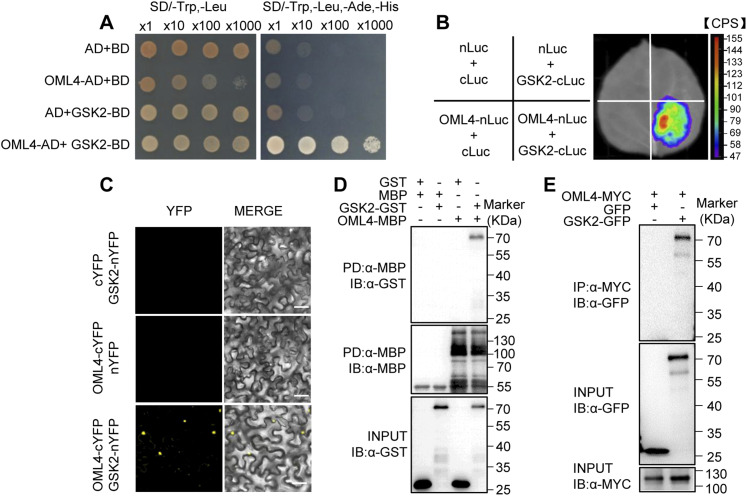
To further understand the molecular role of OML4 in grain growth control, we identified its interacting partners through a yeast two-hybrid assay (Supplemental Table 2). The OML4 full-length protein was used as the bait. Among several interacting proteins, six different clones corresponding to GSK2 were found in this screen. GSK2 has been reported to restrict grain growth in rice (Tong et al., 2012), suggesting that GSK2 is a candidate OML4-interacting partner. We further confirmed the interaction of OML4 with the full-length GSK2 in yeast cells (Figure 5A).
Figure 5.
OML4 Physically Interacts with GSK2 in Vitro and in Vivo.
(A) OML4 interacts with GSK2 in yeast cells. Yeast cells were cultured on SD/-Trp-Leu or SD/-Trp-Leu-His-Ade media. AD, GAL4 activation domain; BD, GAL4 DNA-binding domain; SD, synthetic defined.
(B) OML4 associates with GSK2 in N. benthamiana. OML4-nLUC and GSK2-cLUC were coexpressed in N. benthamiana leaves. LUC activity was observed 48 h after infiltration. The range of luminescence intensity is indicated by the pseudocolor scale bar.
(C) BiFC assays showing that OML4 interacts with GSK2 in N. benthamiana. OML4-cYFP was coexpressed with GSK2-nYFP in leaves of N. benthamiana. Bar = 50 µm.
(D) OML4 binds GSK2 in vitro. GSK2-GST was incubated with OML4-MBP and pulled down by OML4-MBP and detected by immunoblot with anti-GST antibody. IB, immunoblot.
(E) Interaction between OML4 and GSK2 in the coimmunoprecipitation assays. Anti-MYC beads were used to immunoprecipitate GSK2-GFP proteins. Gel blots were probed with anti-MYC or anti-GFP antibody. IB, immunoblot.
We next verified the interaction between OML4 and GSK2 in plant cells using the firefly luciferase (LUC) complementation imaging assay (Figure 5B). The OML4-nLUC and GSK2-cLUC constructs (nLUC, N-terminal luciferase; cLUC, C-terminal luciferase) were transformed and coexpressed in Nicotiana benthamiana leaves. The LUC activity was detected when we coexpressed OML4-nLUC and GSK2-cLUC, while no signal was observed in both combinations of OML4-nLUC/cLUC and nLUC/GSK2-cLUC. We then performed a bimolecular fluorescence complementation (BiFC) assay to test the interaction between OML4 and GSK2 in plant cells (Figure 5C). OML4 was fused with the C terminus of the yellow fluorescent protein (OML4-cYFP), and GSK2 was fused with the N terminus of the yellow fluorescent protein (GSK2-nYFP). Confocal laser-scanning microscopy observation showed strong YFP fluorescence in nuclei when we coexpressed OML4-cYFP and GSK2-nYFP in N. benthamiana leaves. These results indicate that OML4 associates with GSK2 in plant cells.
To investigate whether OML4 could directly interact with GSK2, we performed an in vitro pull-down assay (Figure 5D). We expressed maltose binding protein (MBP)–fused OML4 (OML4-MBP) and glutathione S-transferase (GST) tag–fused GSK2 (GSK2-GST) proteins in Escherichia coli cells. As shown in Figure 5D, OML4-MBP physically interacted with GSK2-GST, but not the negative control (GST) in vitro. The coimmunoprecipitation analyses were used to examine the association of GSK2 and OML4 in N. benthamiana. We coexpressed GSK2-GFP and OML4-MYC in N. benthamiana leaves (Figure 5E). Total proteins were isolated and incubated with MYC beads to immunoprecipitate OML4-MYC. The anti-MYC and anti-GFP antibodies were used to detect immunoprecipitated proteins. GSK2-GFP proteins were detected in the immunoprecipitated OML4-MYC complexes (Figure 5E), indicating that GSK2 associated with OML4 in vivo. These results reveal that OML4 can directly interact with GSK2 in vitro and in vivo.
GSK2 Phosphorylates OML4 and Modulates Its Protein Level
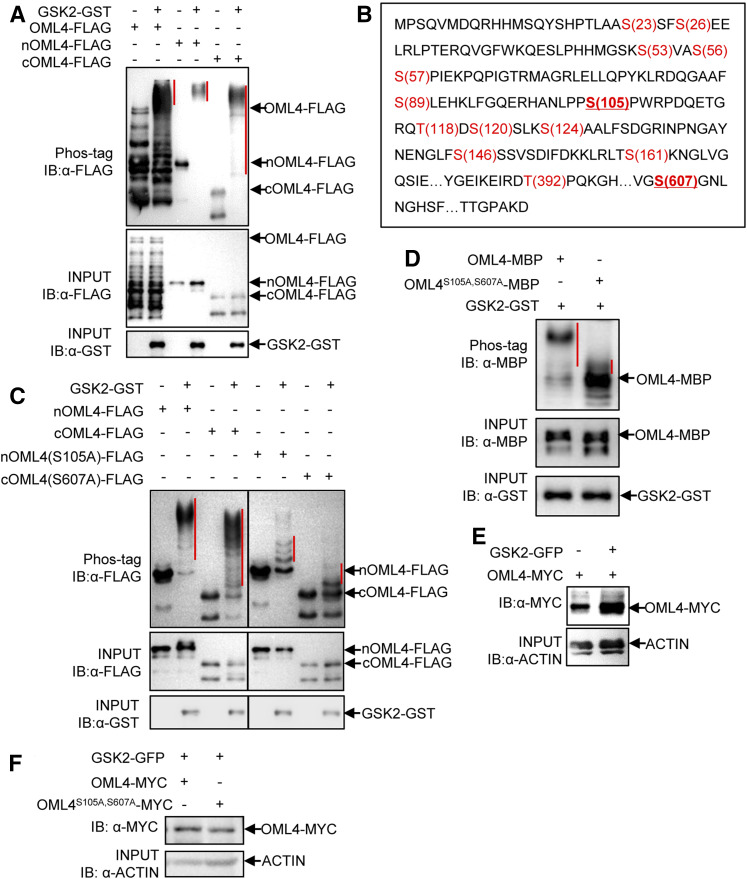
As GSK2 possesses kinase activity and interacts with OML4, we examined whether GSK2 could phosphorylate OML4. To test this, we performed an in vitro kinase assay. GST-fused GSK2 (GSK2-GST) proteins were incubated with OML4-Flag, the N-terminal region of OML4-fused Flag (nOML4-Flag), and the C-terminal region of OML4-fused Flag (cOML4-Flag) in an in vitro kinase assay buffer. Phosphorylated OML4-Flag, nOML4-Flag, and cOML4-Flag were detected in the presence of GSK2-GST, while the phosphorylated OML4-Flag, nOML4-Flag, and cOML4-Flag proteins were not found in the absence of GSK2-GST (Figure 6A). These results show that GSK2 can phosphorylate OML4 in vitro.
Figure 6.
GSK2 Is Required for the Phosphorylation of OML4.
(A) GSK2 phosphorylates OML4 in vitro. The phosphorylated OML4-FLAG, nOML4-FLAG (the N terminus of OML4) and cOML4-FLAG (the C terminus of OML4) were separated by phos-tag SDS-PAGE. The phosphorylated protein is marked with the red vertical line. IB, immunoblot.
(B) Detection of phosphorylation sites of OML4 by LC-MS/MS after in vitro phosphorylation. OML4 contains 1001 residues. The phosphorylate residues detected by LC-MS/MS are shown in red. Two important residues (underlined) were substituted with phosphor-dead residues.
(C) Ser-105 and Ser-607 partially influence the phosphorylation of OML4. The phosphorylated nOML4-FLAG, nOML4(S105A)-FLAG, cOML4-FLAG, and cOML4(S607A)-FLAG were separated by phos-tag SDS-PAGE. The phosphorylated protein is marked with the red vertical line.
(D) Ser-105 and Ser-607 partially influence the phosphorylation of OML4. The phosphorylated OML4-MBP, OML4S105A,S607A-MBP and GSK2-GST were separated by phos-tag SDS-PAGE. The phosphorylated protein is marked with red vertical line. IB, immunoblot.
(E) GSK2 influences the abundance of OML4. GSK2-GFP and OML4-MYC were coexpressed in N. benthamiana leaves, and protein levels were detected by immunoblotting (IB). This experiment was repeated three times, with similar results.
(F) Ser-105 and Ser-607 partially influence the abundance of OML4. GSK2-GFP and OML4-MYC or OML4S105A,S607A-MYC were coexpressed in N. benthamiana leaves and protein levels were detected by immunoblotting (IB). This experiment was repeated three times, with similar results.
To further verify that GSK2 can phosphorylate OML4, we investigated phosphorylation sites of OML4. To identify the phosphorylation sites in OML4, the recombinant OML4 was incubated with the recombinant GSK2 in an in vitro kinase assay buffer, separated by SDS-PAGE electrophoresis, and then subjected to liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis for phosphopeptides. We identified 18 phosphopeptides of OML4, which corresponded to 14 phosphosites (Figure 6B; Supplemental Figure 7; Supplemental Table 3). Among 14 phosphorylation sites of OML4, we observed that Ser-105, Ser-146, and Ser-607 are Ser/Thr, Ser, and Ser in its closest homologs in different plant species, respectively (Supplemental Figure 8), suggesting that these three amino acids are possible conserved phosphorylation sites. We then mutated two amino acids into phosphor-dead Ala (OML4S105A,S607A) and detected their phosphorylation levels by GSK2. Mutations of the two aforementioned Ser residues to Ala reduced the phosphorylation level of OML4, although OML4S105A,S607A was still phosphorylated by GSK2 (Figures 6C and 6D), indicating that Ser-105 and Ser-607 partially contribute to its phosphorylation by GSK2. This result further supports that GSK2 can phosphorylate OML4 in vitro.
Considering that GSK2 can interact with and phosphorylate OML4 in vitro, we asked whether the protein level of OML4 could be affected by GSK2. As shown in Figure 6E, we found that the level of OML4-MYC was increased when GSK2-GFP was coexpressed in leaves of N. benthamiana. Considering that the phosphorylation level of OML4S105A,S607A was lower than that of OML4 in vitro, we asked whether mutations in Ser-105 and Ser-607 could influence the protein level of OML4. As shown in Figure 6F, the level of OML4S105A,S607A was obviously lower than that of OML4 when we transiently overexpressed GSK2-GFP with OML4-MYC or OML4S105A,S607A-MYC in leaves of N. benthamiana. These results indicate that GSK2 affects the level of OML4, possibly by influencing its phosphorylation.
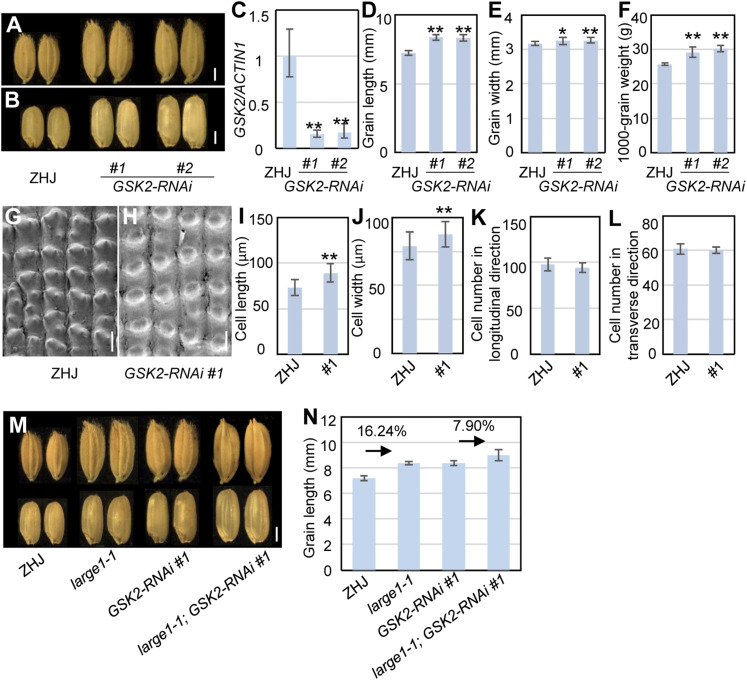
GSK2 Acts Genetically with OML4 to Regulate Grain Size
Although GSK2 has been described to affect grain size, the function of GSK2 in grain size control has not been characterized in detail. To investigate in detail the role of GSK2 in grain size control, we downregulated the expression of GSK2 using RNA interference (RNAi; GSK2-RNAi), as described previously (Tong et al., 2012). GSK2-RNAi lines showed longer and slightly wider grains than ZHJ (Figures 7A to 7E), indicating that GSK2 predominantly regulates grain length in rice. The grain weight of GSK2-RNAi transgenic lines was also significantly increased in comparison with that of ZHJ (Figure 7F). We then observed epidermal cells in ZHJ and GSK2-RNAi spikelet hulls. GSK2-RNAi spikelet hulls contained longer and slightly wider epidermal cells than ZHJ spikelet hulls (Figures 7G to 7J). By contrast, cell number in the grain-length and grain-width directions in GSK2-RNAi lemmas was similar to that in the wild-type lemmas (Figures 7K and 7L). These results demonstrate that GSK2 controls grain growth by limiting cell elongation in spikelet hulls.
Figure 7.
GSK2 Acts Genetically with OML4 to Regulate Seed Size.
(A) Mature paddy grains of ZHJ and GSK2-RNAi.
(B) Brown rice grains of ZHJ and GSK2-RNAi.
(C) Expression level of GSK2 in ZHJ and GSK2-RNAi transgenic lines. RT-qPCR was used to measure expression levels of GSK2. Three biological replicates were used (n = 3). ACTIN1 was used to normalize expression.
(D) and (E) Grain length (D) and width (E) of ZHJ and GSK2-RNAi transgenic lines.
(F) The 1000-grain weight of ZHJ and GSK2-RNAi transgenic lines.
(G) and (H) Scanning electron microscopy analysis of the outer surface of ZHJ (G) and GSK2-RNAi #1 (H) lemmas.
(I) and (J) Average length (I) and width (J) of outer epidermal cells in the longitudinal direction in ZHJ and GSK2-RNAi #1 lemmas.
(K) and (L) Number of outer epidermal cells in the longitudinal (K) and transverse (L) direction in ZHJ and GSK2-RNAi #1 lemmas.
(M) Grains of ZHJ, large1-1, GSK2-RNAi#1, and large1-1; GSK2-RNAi#1.
(N) Grain length of ZHJ, large1-1, GSK2-RNAi#1 and large1-1; GSK2-RNAi#1.
Values ([D] to [F], [I] to [L], and [N]) are given as the means ± sd (n ≥ 50). *, P < 0.05; **, P < 0.01 compared with the wild type by Student’s t test.
Bar in (A), (B), and (M) = 2 mm; bar in (G) and (H) = 50 µm.
GSK2-RNAi produced long grains, like those observed in the large1-1 mutant, and GSK2 and OML4 restrict cell elongation in spikelet hulls (Figures 2 and 7). In addition, GSK2 can phosphorylate OML4 in vitro. We therefore speculated that GSK2 and OML4 could function in a common pathway to regulate grain length in rice. To test this hypothesis, we crossed large1-1 with GSK2-RNAi and isolated large1-1;GSK2-RNAi plants (Figure 7M). As shown in Figure 7N, the length of large1-1 grains was increased by 16.24% in comparison with that of ZHJ, while the length of large1-1;GSK2-RNAi grains was increased by 7.90% compared with GSK2-RNAi. The results suggest that GSK2 acts, at least in part, in a common genetic pathway with OML4 to control grain length.
As GSK2 acts as a negative regulator of brassinosteroid (BR) signaling, and GSK2-RNAi lines are sensitive to BR (Tong et al., 2012), we tested whether the large1-1 mutant could be involved in BR responses. Previous studies revealed that the roots of the wild-type plants become curly after BR treatment. We therefore examined the BR response of the large1-1 mutant by treatment with different concentrations of epi-brassinolide (epi-BL). In half-strength Murashige and Skoog medium without epi-BL, the roots of large1-1 were similar to those of ZHJ (Supplemental Figure 9A). However, in half-strength Murashige and Skoog medium with epi-BL, the roots of large1-1 seedlings were more curved than those of ZHJ (Supplemental Figure 9B). We also performed a lamina joint bending assay for BR sensitivity. The large1-1 mutant showed more bending than ZHJ in response to BR treatment (Supplemental Figures 9C to 9E). These findings indicated that large1-1 is more sensitive to epi-BL. Expression levels of BR-related genes, including DWARF, D2, BZR1, and D61, in large1-1 were significantly higher than those in ZHJ (Supplemental Figure 9F). Thus, it is plausible that the GSK2-OML4 module is involved in BR responses.
Nucleotide Diversity and Selection Signatures
Previous studies showed that the chromosome region covering OML4 contains multiple quantitative-trait loci for grain size and weight (Gao et al., 2004; Marri et al., 2005; Wan et al., 2005), suggesting that this region could be selected by breeders. To test this, we investigated the nucleotide diversity of a 10,200-bp genomic fragment containing the OML4 gene in wild and cultivated rice using 446 Oryza rufipogon and 1083 O. sativa varieties (Huang et al., 2012). We measured the ratio of the genetic diversity in wild rice to that in cultivated rice (πW:πC). The nucleotide diversity of this region was reduced in cultivated rice in comparison with wild rice (Supplemental Figure 10A). The ratio of the genetic diversity πO. rufipogon:πO. sativa was 3.145 (Supplemental Figure 10B), which was higher than the genome-wide threshold of selection signals (πW:πC > 3; Huang et al., 2012). These results suggest that this region could have selective signatures in the full population.
The population structures of the 446 O. rufipogon accessions were classified into three types, simply designated as Or-I, Or-II, and Or-III (Huang et al., 2012). A previous study showed that O. s. indica and japonica are descended from Or-I and Or-III, respectively (Huang et al., 2012). We then investigated the nucleotide diversity between japonica and Or-III and that between indica and Or-I. The ratio of the genetic diversity πOr-III:πjaponica was 13.046 (Supplemental Figure 10B), which was lower than the genome-wide threshold of selection signals (πW:πC < 14; Huang et al., 2012). The ratio of the genetic diversity πOr-I:πindica was 2.610 in this whole region and 4.127 in the 5′-flanking region, respectively (Supplemental Figure 10B), while the genome-wide threshold of selection signals was below 3 (πW:πC < 3). These results suggest that a part of this region could have selective signatures in the indica population, but not in the japonica population.
DISCUSSION
Grain size and weight are critical determinants of grain yield, but the genetic and molecular mechanisms of grain size control in rice are still unlcear. In this study, we identify OML4 as a regulator of grain size and weight. GSK2 interacts with and phosphorylates OML4. GSK2 and OML4 function, at least in part, in a common pathway to control grain length in rice. These findings reveal an important genetic and molecular mechanism of the GSK2-OML4 regulatory module in grain size control.
The large1-1 mutant produced long, wide, and heavy grains in comparison to the wild type. By contrast, overexpression of LARGE1 caused short and light grains. Thus, LARGE1 is a negative regulator of grain size and weight. Cellular analyses support that LARGE1 controls grain size by restricting cell expansion. Consistent with this, expression of several genes (e.g., SPL13, GS2, GS5, and GL7; Li et al., 2011; Che et al., 2015; Duan et al., 2015; Hu et al., 2015; Zhou et al., 2015; Si et al., 2016), which control grain size by regulating cell expansion, was altered in large1-1. The large1-1 plants also formed long panicles with increased primary panicle branches and decreased secondary panicle branches, resulting in a reduction in grain number per panicle (Figure 1M). This result suggests that LARGE1 influences the balance between grain size and grain number per panicle. Consistent with this idea, several genes have been shown to affect the trade-off between grain size and grain number per panicle. For example, mutations in MKP1 resulted in an increase in grain size and a decrease in grain number per panicle (Guo et al., 2018; Xu et al., 2018). Similarly, overexpression of constitutively active OsMKKK10 produced long panicles with large grains and reduced grain number per panicle (Xu et al., 2018). It is an important challenge to understand how LARGE1 regulates the trade-off between grain size and grain number per panicle in the future. In addition, the LARGE1 influences plant height. These results indicate that LARGE1 regulates both vegetative and reproductive organ growth in rice.
LARGE1 encodes a Mei2-like protein (OML4) in rice. There are many Mei2-like proteins in plants that have the conserved RRMs but appear to have taken on distinct functions in plant development (Jeffares et al., 2004). The Arabidopsis-Mei2-Like (AML) genes comprise a five-member gene family that plays a role in meiosis and vegetative growth (Kaur et al., 2006). In maize (Zea mays), terminal ear1 (te1), encoding a Mei2-like protein, plays a role in regulating leaf initiation (Veit et al., 1998). In rice, PLASTOCHRON2 (PLA2)/LEAFY HEAD2 (LHD2) encodes a Mei2-like protein (OML1; Kawakatsu et al., 2006). The pla2 mutant exhibited precocious maturation of leaves, a shortened plastochron, and ectopic shoot formation during the reproductive phase (Kawakatsu et al., 2006). However, a function for Mei2-like proteins in seed/grain size control has not been reported in plants. In this study, we identify OML4 as a negative regulator of grain size in rice.
The LARGE1/OML4 protein contains three RRM domains. RRMs are found in a variety of RNA binding proteins. For example, Arabidopsis FCA, which contains two RRMs, controls flowering time by binding the cold-induced antisense transcript COOLAIR (Tian et al., 2019). In rice, PigmR-INTERACTING and BLAST RESISTANCE PROTEIN1 (PIBP1), an RRM protein, functions as a transcription factor to regulate the defense genes expression (Zhai et al., 2019). In our study, expression of several grain size genes and BR-related genes was altered in the large1-1 mutant. It will be worthwhile to test whether LARGE1/OML4 could bind mRNAs, miRNAs, or noncoding RNAs and associate with the promoters to regulate their expression in the future.
We further identified OML4-interacting proteins. Interestingly, one of them is the GSK2, a homologue of Arabidopsis BIN2 kinase, which has been reported to influence grain size and multiple growth processes in rice (Tong et al., 2012). Previous studies showed that GSK2 interacts with several grain size regulators. However, the effect of GSK2 on cell proliferation and/or cell expansion in spikelet hulls has not been characterized in detail. In this study, we found that downregulation of GSK2 formed large grains as a result of large cells in spikelet hulls. These results indicate that GSK2 restricts cell expansion rather than cell proliferation in spikelet hulls. Consistent with this, it has been proposed that GSK2 regulates grain size by interacting with GS2 that predominately promotes cell expansion in spikelet hulls (Che et al., 2015). GSK5, a homolog of GSK2, has been reported to control grain size by restricting cell expansion in spikelet hulls (Hu et al., 2018). Considering that GSK2 is a functional protein kinase, we presumed that GSK2 could phosphorylate OML4. Consistent with this idea, we found that GSK2 can interact with and phosphorylate OML4. We further observed that GSK2 influences the level of OML4. It is possible that GSK2 might phosphorylate OML4 and prevent the degradation of OML4. Supporting this, we observed that mutations in Ser-105 and Ser-607 partially influence the abundance of OML4. Our genetic analyses suggest that GSK2 and OML4 function, at least in part, in a common pathway to control grain length in rice. In addition, we found that large1-1 is hypersensitive to epi-BL, like GSK2-RNAi plants. The large1-1 mutation also influences the expression of several BR-related genes. It is possible that the GSK2-OML4 module may provide a link between BRs and grain growth. Therefore, our findings reveal an important genetic and molecular mechanism of grain size control involving the GSK2-OML4 regulatory module in rice, suggesting this module is a promising target for grain size improvement in crops.
METHODS
Plant Materials and Growth Conditions
γ-Rays were used to irradiate the grains of the wild-type ZHJ, and the large1-1 mutant was isolated from the M2 population. Rice (Oryza sativa) plants were grown in the field according to a previous report (Huang et al., 2017). Rice plants were cultivated at Lingshui from December 2016 to April 2017 and from December 2017 to April 2018 and at Zhejiang (Hangzhou) from July 2017 to November 2017 and from July 2018 to November 2018.
Phenotypic Evaluation and Cellular Analysis
The ZHJ and large1-1 plants grown in the paddy fields were taken photographs after completing ripe. A Scan Marker i560 (MICROTEK) was used to scan mature seeds. We use the Rice Test System (WSeen) to measure the grain length and width. We also measured the 1000-grain weight with three replicates (separate samples from the same experiment; Huang et al., 2017). We used a scanning electron microscope to observe the cell size and cell number. Observation via scanning electron microscopy was performed as previously described by Duan et al. (2015). ImageJ software was used to measure cell length and width.
RNA Extraction and RT-qPCR Analysis
Total RNA of seedlings of ZHJ, proActin:OML4 transgenic lines, and GSK2-RNAi transgenic lines or young panicles of 1, 5, 10, and 15 cm from ZHJ were extracted using an RNA Pre Pure Plant Kit (Tiangen). cDNAs were synthesized according to a previous study (Duan et al., 2015). qPCR was conducted on an ABI7500 real-time PCR system using a SYBR Green Mix Kit (Bio-Rad). Rice ACTIN1 was used as an internal control. Primers for RT-qPCR are shown in Supplemental Table 4.
Identification of the LARGE1 Gene
We crossed large1-1 with the wild-type ZHJ to produce F2 populations. We cloned the LARGE1 gene using the F2 population. The whole genomes of the wild-type ZHJ and a mixed pool of 50 individual plants with mutant phenotypes were resequenced using a NextSeq 500 system (Illumina). MutMap was used to isolate LARGE1 as previously described by Abe et al. (2012), and the SNP:INDEL ratio was analyzed as previously described by Fang et al. (2016).
Constructs and Plant Transformation
The genomic sequence of OML4, which contained a 2049-bp 5′-flanking region, the whole gene region, and a 1259-bp 3′-flanking region, was amplified using the primers gOML4-99-F and gOML4-99-R. We used the GBclonart Seamless Cloning Kit to fuse the OML4 genomic sequence to the pMDC99 vector and generated the gOML4 recombinant construct. The latter series of the recombinant vectors was constructed using the same kit and similar methods. The related vectors we used in this study were pIPKB003 (containing the ACTIN promoter and fused with the coding sequence of the OML4 gene), pMDC107 (constructing the gOML4-GFP plasmid), and pMDC164 (constructing the proOML4:GUS vector). Primers used for constructs are listed in Supplemental Table 4. The plasmids gOML4, proACTIN:OML4, gOML4-GFP, and proOML4:GUS were introduced into the Agrobacterium strain GV3101. The gOML4 and gOML4-GFP constructs were transferred into large1-1, and other plasmids were transferred into the wild type according to a previous report by Hiei et al. (1994).
GUS Staining and Subcellular Localization of OML4
GUS staining of panicles in different developmental stages was performed as previously described by Fang et al. (2016). The GFP fluorescence of gOML4-GFP transgenic seedlings was observed using the Zeiss LSM 710 confocal microscope. 4′,6-Diamidino-2-phenylindole (1 μg/mL) was used to stain cell nuclei.
Yeast Two-Hybrid Assays
The cDNA sequences of GSK2 and OML4 were amplified using gene-specific primers (Supplemental Table 4), and the products were fused into the linearized pGADT7 and pGBKT7 vectors, respectively. Yeast two-hybrid analysis was conducted according to the manufacturer’s instructions (Clontech).
BiFC Assay
Full-length cDNA fragments of OML4 and GSK2 were recombined into the pGBW414-cYFP and pGBW414-nYFP vectors. The constructs were transformed into Nicotiana benthamiana mesophyll cells with acetosyringone for transient expression. Confocal imaging analysis was performed using a LSM 710 confocal microscope (Zeiss).
Pull-Down Assay
Recombinant proteins (OML4-MBP and MBP) and prey proteins (GSK2-GST and GST) were incubated in TGH buffer (50 mM Hepes, pH 7.5, 10% glycerol, 150 mM NaCl, Triton X-100, 1.5 mM MgCl2, 1 mM EGTA, and Protease Inhibitor cocktail tablet) for 0.5 h at 4°C with 20 μL of MBP-beads per tube. After centrifugation at 23g for 2 min, the supernatant was discarded to stop the reaction. The beads were washed with ice-cold TGH buffer five times, and then 50 μL SDS-loading buffer was added. The samples were denatured at 98°C for 5 min and subjected to the SDS-PAGE analysis. We used anti-MBP (E8032, lot no. 10,009,443; New England Biolabs) and anti-GST (M20007, lot no. 294175; Abmart) to detect the input and the pull-down samples, respectively. Signals were detected using eECL Western Blot Kit (CW0049, Cwbiotech), and images were scanned using Tanon-4500 gel-imaging system according to the manufacturer’s instructions.
Coimmunoprecipitation
The OML4 coding sequence was cloned into the KpnI and BamHI sites of the pCAMBIA1300-221-Myc vector to generate the 35S:Myc-OML4 plasmid. The GSK2 coding sequence was cloned into the AscI and PacI sites of pMDC43 to generate the 35:GFP-GSK2 plasmid. The constructs were transformed into N. benthamiana mesophyll cells with acetosyringone for transient expression. Total protein was isolated with extraction buffer (150 mM NaCl, 50 mM Tris-HCl, 2% Triton X-100, 20% glycerol, 1 mM EDTA, 1× Complete Protease Inhibitor cocktail, and 1 mM phenylmethylsulfonyl fluoride) and incubated with GFP-beads for 40 min at 4°C. After incubation, wash buffer (150 mM NaCl, 50 mM Tris-HCl, 0.1% Triton X-100, 20% glycerol, 1 mM EDTA, and 1× Complete Protease Inhibitor cocktail) was used to wash the beads, and then 50 μL of SDS-loading buffer was added. The samples were denatured at 98°C for 10 min and finally subjected to the SDS-PAGE analysis. We used anti-GFP (M20004, lot no. 313769; Abmart) and anti-MYC (M20002, lot no. 314082; Abmart) to detect the input and immunoprecipitates, respectively. Signals were detected using eECL Western Blot Kit, and images were scanned using a Tanon-4500 gel-imaging system according to the manufacturer’s instructions.
Phylogenetic Analysis
To build a phylogenetic tree, Mei2-like proteins were aligned by ClustalX2. The phylogenetic tree was built using this alignment output based on a neighbor-joining method in MEGA7. The parameters used were as follows: complete deletion and bootstrap (1000 replicates).
Phosphorylation Analysis
The coding sequences of OML4, nOML4, and cOML4 were amplified using the specific primers (OML4-FLAG-F/R, nOML4-FLAG-F/R, and cOML4-FLAG-F/R) in Supplemental Table 4. The products were cloned to the vector pETnT to construct OML4-FLAG, nOML4-FLAG, and cOML4-FLAG plasmids. The GSK2 coding sequence was amplified using the primers GSK2-GST-F/R and subcloned to the vector pGEX4T-1 to construct GSK2-GST plasmid.
These plasmids were transformed into Escherichia coli (host strain BL21). Induction, isolation, and purification of OML4-FLAG, nOML4-FLAG, cOML4-FLAG, and GSK2-GST proteins were done as described previously (Xia et al., 2013). GSK2-GST (10 μL) was incubated with 5 μL of OML4-FLAG, nOML4-FLAG, and cOML4-FLAG in 20 μL of reaction buffer (25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, and 50 mM ATP) for 2 h. Phosphorylated products were analyzed by phos-tag SDS-PAGE. Anti-GST (M20007, lot no. 294175; Abmart) and anti-FLAG (M20008, lot no. 314059; Abmart) antibodies were utilized to detect the phosphorylated products and the input. Signals were detected using eECL Western Blot Kit, and images were scanned using Tanon-4500 gel-imaging system according to the manufacturer’s instructions. To detect the phosphorylation sites of OML4, phosphorylated proteins were cut from the SDS-PAGE and digested using trypsin (Sigma-Aldrich) at 37°C overnight. LC-MS/MS was used to analyze the tryptic peptides. Next, these peptides were identified by searching the UniProt database. The Proteome Discoverer software (version 1.3 was used to perform the phosphosite assignment.
BR Treatment
NaClO (30%) was used to sterilize grains. The sterilized grains were grown in 1/2MS medium (Duchefa Biochemie BV) without or with 1 μM epi-BL (Sigma-Aldrich), respectively. After 7 d in a growth incubator (12-h-light/12-h-dark cycle, 3000 lux), the roots were observed for the root waving analysis. For the lamina joint test, the second leaf laminas of 7-d-old seedings were submerged in the solutions containing different epi-BL concentrations.
Domestication Analysis
A sliding window approach was used to analyze the polymorphism levels (π, pairwise nucleotide variation as a measure of variability; Huang et al., 2012). Our analysis was performed for 200-bp windows sliding with 100-bp steps. The 1529 worldwide accessions, including 446 of the wild rice species Oryza rufipogon and 1083 of cultivated rice species, were used to analyze the domestication of OML4 by VCFtools software. To do the genetic analysis, we selected six groups as follows: 446 wild rice species O. rufipogon; 155 wild_Or-I (the wild type corresponding to indica); 170 wild_Or-III (the wild type corresponding to japonica); 484 japonica species; 520 indica species; and 1083 cultivated rice species.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: OML4 (Os02g0517531), GSK2 (Os05g0207500).
Supplemental Data
Supplemental Figure 1. Expression level of the indicated genes in ZHJ and large1-1 panicles.
Supplemental Figure 2. Δ(SNP/INDEL-index) and G’ value plots of the whole genome generated from MutMap analysis of large1-1 mutant alleles.
Supplemental Figure 3. CDS and protein sequence of OML4.
Supplemental Figure 4. The plant height, panicle size and grain number per panicle of gLARGE1;large1-1.
Supplemental Figure 5. Structural features and phylogenetic tree of OML4.
Supplemental Figure 6. Phylogenetic tree of MEI2-LIKE proteins in plants.
Supplemental Figure 7. b and y ions peptides map under the mass spectrometric analysis.
Supplemental Figure 8. Alignment of OML4 and its homologs in different plant species.
Supplemental Figure 9. The large1-1 mutant is more sensitive to brassinolide (BL).
Supplemental Figure 10. Nucleotide diversity analysis of OML4.
Supplemental Table 1. Identification of the large1-1 mutation using the MutMap approach.
Supplemental Table 2. Y2H screening interaction proteins of OML4.
Supplemental Table 3. Phosphopeptides of OML4.
Supplemental Table 4. Primers used in this study.
Supplemental Data Set. Multiple sequence alignment for Supplemental Figures 5B and 6.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Chengcai Chu for the GSK2-RNAi vector. This work is supported by grants from the National Natural Science Foundation of China (91735302, 91735304, 31425004, 91535203, 31771340, 31571742, and 31871219), the National Basic Research Program of China (2016YFD0100501, 2016YFD0100400, 2016YFD0100402, 2018 YFD1000700-706-10, and 2017YFD0101701), the National Special Project (2016ZX08009003-003), the Youth Innovation Promotion Association CAS (2019102), and the strategic priority research program of the Chinese Academy of Sciences (XDB27010102).
AUTHOR CONTRIBUTIONS
J.L., D.W., P.D., and Y.H.L. designed experiments. Y.H.L. supervised this project. J.L., D.W., P.D., Y.P.L., D.Z., L.Z., G.D., B.Z., N.L., K.H., Y.J.L., Y.W., and X.H. performed the experiments. J.L., P.D., Y.P.L., Q.Q., and Y.H.L. analyzed data. J.L., P.D., and Y.H.L. prepared figures and wrote the article.
References
- Abe A., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30: 174–178. [DOI] [PubMed] [Google Scholar]
- Che R., Tong H., Shi B., Liu Y., Fang S., Liu D., Xiao Y., Hu B., Liu L., Wang H., Zhao M., Chu C.(2015). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2: 15195. [DOI] [PubMed] [Google Scholar]
- Duan P., Ni S., Wang J., Zhang B., Xu R., Wang Y., Chen H., Zhu X., Li Y.(2015). Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2: 15203. [DOI] [PubMed] [Google Scholar]
- Duan P., Rao Y., Zeng D., Yang Y., Xu R., Zhang B., Dong G., Qian Q., Li Y.(2014). SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J. 77: 547–557. [DOI] [PubMed] [Google Scholar]
- Duan P., Xu J., Zeng D., Zhang B., Geng M., Zhang G., Huang K., Huang L., Xu R., Ge S., Qian Q., Li Y.(2017). Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 10: 685–694. [DOI] [PubMed] [Google Scholar]
- Fan C., Xing Y., Mao H., Lu T., Han B., Xu C., Li X., Zhang Q.(2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112: 1164–1171. [DOI] [PubMed] [Google Scholar]
- Fang N., Xu R., Huang L., Zhang B., Duan P., Li N., Luo Y., Li Y.(2016). SMALL GRAIN 11 controls grain size, grain number and grain yield in rice. Rice (N. Y.) 9: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.M., Zhu J., Song Y.S., He C.X., Shi C.H., Xing Y.Z.(2004). Analysis of digenic epistatic effects and QE interaction effects QTL controlling grain weight in rice. J. Zhejiang Univ. Sci. 5: 371–377. [DOI] [PubMed] [Google Scholar]
- Guo T., Chen K., Dong N.Q., Shi C.L., Ye W.W., Gao J.P., Shan J.X., Lin H.X.(2018). GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30: 871–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heang D., Sassa H.(2012a). Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS One 7: e31325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heang D., Sassa H.(2012b). An atypical bHLH protein encoded by POSITIVE REGULATOR OF GRAIN LENGTH 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breed. Sci. 62: 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T.(1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Hu J., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8: 1455–1465. [DOI] [PubMed] [Google Scholar]
- Hu Z., et al. (2018). A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 11: 736–749. [DOI] [PubMed] [Google Scholar]
- Huang K., Wang D., Duan P., Zhang B., Xu R., Li N., Li Y.(2017). WIDE AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size and shape in rice. Plant J. 91: 849–860. [DOI] [PubMed] [Google Scholar]
- Huang X., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares D.C., Phillips M.J., Moore S., Veit B.(2004). A description of the Mei2-like protein family; structure, phylogenetic distribution and biological context. Dev. Genes Evol. 214: 149–158. [DOI] [PubMed] [Google Scholar]
- Kaur J., Sebastian J., Siddiqi I.(2006). The Arabidopsis-mei2-like genes play a role in meiosis and vegetative growth in Arabidopsis. Plant Cell 18: 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T., Itoh J., Miyoshi K., Kurata N., Alvarez N., Veit B., Nagato Y.(2006). PLASTOCHRON2 regulates leaf initiation and maturation in rice. Plant Cell 18: 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.M., Park J., Kim B., Seo J., Lee G., Jang S., Koh H.J.(2015). Influence of multi-gene allele combinations on grain size of rice and development of a regression equation model to predict grain parameters. Rice (N. Y.) 8: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Li Y.(2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33: 23–32. [DOI] [PubMed] [Google Scholar]
- Li N., Xu R., Li Y.(2019). Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 70: 11.1-11.30. [DOI] [PubMed] [Google Scholar]
- Li Y., Fan C., Xing Y., Jiang Y., Luo L., Sun L., Shao D., Xu C., Li X., Xiao J., He Y., Zhang Q.(2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43: 1266–1269. [DOI] [PubMed] [Google Scholar]
- Liu J., et al. (2017). GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 3: 17043. [DOI] [PubMed] [Google Scholar]
- Marri P.R., Sarla N., Reddy L.V., Siddiq E.A.(2005). Identification and mapping of yield and yield related QTLs from an Indian accession of Oryza rufipogon. BMC Genet. 6: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si L., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48: 447–456. [DOI] [PubMed] [Google Scholar]
- Song X.J., Huang W., Shi M., Zhu M.Z., Lin H.X.(2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630. [DOI] [PubMed] [Google Scholar]
- Sun L., Li X., Fu Y., Zhu Z., Tan L., Liu F., Sun X., Sun X., Sun C.(2013). GS6, a member of the GRAS gene family, negatively regulates grain size in rice. J. Integr. Plant Biol. 55: 938–949. [DOI] [PubMed] [Google Scholar]
- Tian Y., Zheng H., Zhang F., Wang S., Ji X., Xu C., He Y., Ding Y.(2019). PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci. Adv. 5: eaau7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Liu L., Jin Y., Du L., Yin Y., Qian Q., Zhu L., Chu C.(2012). DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24: 2562–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit B., Briggs S.P., Schmidt R.J., Yanofsky M.F., Hake S.(1998). Regulation of leaf initiation by the terminal ear 1 gene of maize. Nature 393: 166–168. [DOI] [PubMed] [Google Scholar]
- Wan X.Y., Wan J.M., Weng J.F., Jiang L., Bi J.C., Wang C.M., Zhai H.Q.(2005). Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theor. Appl. Genet. 110: 1334–1346. [DOI] [PubMed] [Google Scholar]
- Wang S., Wu K., Yuan Q., Liu X., Liu Z., Lin X., Zeng R., Zhu H., Dong G., Qian Q., Zhang G., Fu X.(2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44: 950–954. [DOI] [PubMed] [Google Scholar]
- Wang S., et al. (2015a). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47: 949–954. [DOI] [PubMed] [Google Scholar]
- Wang Y., et al. (2015b). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47: 944–948. [DOI] [PubMed] [Google Scholar]
- Xia D., Zhou H., Liu R., Dan W., Li P., Wu B., Chen J., Wang L., Gao G., Zhang Q., He Y.(2018). GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to produce extra-long grains in rice. Mol. Plant 11: 754–756. [DOI] [PubMed] [Google Scholar]
- Xia T., Li N., Dumenil J., Li J., Kamenski A., Bevan M.W., Gao F., Li Y.(2013). The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25: 3347–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G.S., Hu X.M., Jiao Y.Q., Yu Y.C., Chu C.C., Li J.Y., Qian Q., Wang Y.H.(2006). Leafy head2, which encodes a putative RNA-binding protein, regulates shoot development of rice. Cell Res. 16: 267–276. [DOI] [PubMed] [Google Scholar]
- Xu R., et al. (2018a). Control of grain size and weight by the OsMKKK10-OsMKK4-OsMAPK6 signaling pathway in rice. Mol. Plant 11: 860–873. [DOI] [PubMed] [Google Scholar]
- Xu R., Yu H., Wang J., Duan P., Zhang B., Li J., Li Y., Xu J., Lyu J., Li N., Chai T., Li Y.(2018b). A mitogen-activated protein kinase phosphatase influences grain size and weight in rice. Plant J. 95: 937–946. [DOI] [PubMed] [Google Scholar]
- Ying J.Z., Ma M., Bai C., Huang X.H., Liu J.L., Fan Y.Y., Song X.J.(2018). TGW3, a major QTL that negatively modulates grain length and weight in rice. Mol. Plant 11: 750–753. [DOI] [PubMed] [Google Scholar]
- Zhai K., et al. (2019). RRM transcription factors interact with NLRs and regulate broad-spectrum blast resistance in rice. Mol. Cell 74: 996–1009.e7. [DOI] [PubMed] [Google Scholar]
- Zhao D.S., Li Q.F., Zhang C.Q., Zhang C., Yang Q.Q., Pan L.X., Ren X.Y., Lu J., Gu M.H., Liu Q.Q.(2018). GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 9: 1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., et al. (2015). Natural variations in SLG7 regulate grain shape in rice. Genetics 201: 1591–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]