Abscisic acid and gibberellic acid exert opposite effects on root growth and tillering by antagonistically regulating the APC/CTE-mediated degradation of rice SHORT-ROOT1 (OsSHR1) and MONOCULM1 (MOC1).
Abstract
The antagonistic regulation of seed germination by the phytohormones abscisic acid (ABA) and gibberellic acid (GA) has been well-established. However, how these phytohormones antagonistically regulate root growth and branching (tillering in rice, Oryza sativa) remains obscure. Rice TILLER ENHANCER (TE) encodes an activator of the APC/CTE E3 ubiquitin ligase complex that represses tillering but promotes seed germination. In this study, we identified a dual role of GA and APC/CTE in regulating root growth. High GA levels can activate APC/CTE to promote the degradation of rice SHORT-ROOT1 (OsSHR1, a key factor promoting root growth) in the root meristem (RM) or MONOCULM1 (MOC1, a key factor promoting tillering) in the axillary meristem (AM), leading to restricted root growth and tillering, while low GA levels can activate the role of APC/CTE in stimulating RM cell division to promote root growth. In addition, moderate enhancement of ABA signaling helps maintain the RM and AM size, sustaining root growth and tillering by antagonizing the GA-promoted degradation of OsSHR1 and MOC1 through the SnRK2-APC/CTE regulatory module. We conclude that APC/CTE plays a key role in regulating plant architecture by mediating the crosstalk between ABA and GA signaling pathways.
INTRODUCTION
The root system and tillering are two important aspects of plant architecture and critical determinants of crop yield. The development of lateral branches (tillering in cereal crops) includes two steps: Formation of an axil meristem (AM) in the leaf axil and outgrowth of an axil bud to form branches (tillers; Wang and Li, 2008; Xing and Zhang, 2010). The root system is key for plant anchorage and efficient uptake of water and nutrients, and has a major effect on fertilizer usage and yield in crops (Rogers and Benfey, 2015). Continuous root growth and development are sustained by the stable root meristem (RM) composed of multipotent stem cells (Perilli et al., 2012). Above the RM is the transition zone (TZ) where cell division ceases to initiate cell differentiation for further cell expansion in the elongation zone. RM maintenance is strictly regulated by the balance between the rate of RM cell division and the rate of TZ cell differentiation at the TZ (Perilli et al., 2012).
A pair of classic phytohormones, abscisic acid (ABA) and gibberellic acid (GA), display opposite effects on various processes of plant growth and development. For example, a high concentration of ABA acts as a growth inhibitor to repress seed germination and plant stem elongation and as an inducer of axil bud dormancy under adverse environmental conditions (Cutler et al., 2010; Golldack et al., 2013; Yao and Finlayson, 2015; González-Grandío et al., 2017), while GA acts to promote seed germination, plant stem elongation, leaf expansion, and flowering (Yamaguchi, 2008). The signaling pathways for ABA and GA have now been well-established in the model plant species Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). In the ABA signaling pathway, ABA binds to its receptor proteins PYRABACTIN RESISTANCE1 LIKE (PYL)/PYRABACTIN RESISTANCE1/REGULATORY COMPONENTS OF ABA RECEPTOR (RCAR) for further binding to PROTEIN PHOSPHATASES TYPE 2C, thus releasing the SNF1-RELATED KINASES2 (SnRK2s) from the inhibition by PROTEIN PHOSPHATASES TYPE 2Cs. Then the activated SnRK2s phosphorylate the downstream targets to induce ABA responses (Fujii et al., 2009; Ma et al., 2009; Melcher, 2009; Miyazono, 2009; Nishimura et al., 2009; Park, 2009; Santiago et al., 2009; Yin et al., 2009). In the GA signaling pathway, the receptor GA-INSENSITIVE DWARF1 and SKP1-CUL1-F-box type E3 ligase SCFSLEEPY1/GA-INSENSITIVE DWARF2 together promote the degradation of the DELLA repressor proteins in a GA-dependent manner to relieve their repression of GA action (Ueguchi-Tanaka et al., 2005, 2007; Sun, 2011).
Extensive molecular genetic studies have revealed a conserved LATERAL SUPPRESSOR/LATERAL SUPPRESSOR/MONOCULM1 (MOC1) genetic pathway controlling the initiation of AM and inflorescence branch meristem in both dicots and monocots. LATERAL SUPPRESSOR/LATERAL SUPPRESSOR/MOC1 encode homologous GRAS proteins that act as key promoting factors of AM formation and thus branching/tillering (Schumacher et al., 1999; Greb et al., 2003; Komatsu et al., 2003; Li et al., 2003). In addition, previous studies also showed that the root growth regulatory pathway composed of SHORT ROOT (SHR) and SCARECROW (SCR) in Arabidopsis (Di Laurenzio et al., 1996; Helariutta et al., 2000; Wysocka-Diller et al., 2000; Nakajima et al., 2001; Kamiya et al., 2003; Sabatini et al., 2003; Wu et al., 2014; Moreno-Risueno et al., 2015) is likely conserved in rice (Cui et al., 2007). SHR and SCR also encode GRAS proteins that play a key role in regulating RM asymmetric cell division, endodermis identity, and RM maintenance (Di Laurenzio et al., 1996; Helariutta et al., 2000; Wysocka-Diller et al., 2000; Nakajima et al., 2001; Kamiya et al., 2003; Sabatini et al., 2003; Wu et al., 2014; Moreno-Risueno et al., 2015). Recently, it was shown that both ABA and GA participate in the coordination of the middle cortex formation in the RM through the SHR/SCR pathway at the transcriptional level in Arabidopsis (Choi and Lim, 2016; Gong et al., 2016; Lee et al., 2016). However, how ABA and GA antagonistically regulate root and tiller development remains largely unclear at the molecular level.
We previously reported that rice TILLER ENHANCER (TE) encodes an activator of the ANAPHASE-PROMOTING COMPLEX/CYCLOSOME (APC/C) E3 ubiquitin ligase complex that acts to repress tillering (branching) by promoting the degradation of MOC1 (Lin et al., 2012). More recently, we showed that GA can block SnRK2 activity to reduce SnRK2-mediated phosphorylation of TE, thus activating APC/CTE-mediated degradation of ABA receptors to repress ABA signaling (Lin et al., 2015). Here, we show that high GA levels can activate APC/CTE to promote the degradation of rice SHORT-ROOT1 (OsSHR1) in the RM or MOC1 in the AM, leading to restricted root growth and tillering, whereas low GA levels can activate the role of APC/CTE in stimulating RM cell division to promote root growth. Moreover, moderate enhancement of ABA signaling can help to maintain the RM and AM by antagonizing GA-APC/CTE-mediated degradation of OsSHR1 and MOC1. Our results show that GA and APC/CTE have dual roles in regulating root growth dependent on their activity levels and reveal a novel mechanism by which TE regulates root growth and tillering activity through mediating the crosstalk between ABA and GA signaling pathways.
RESULTS
TE Mediates the Antagonistic Effects of ABA and GA on Root Growth
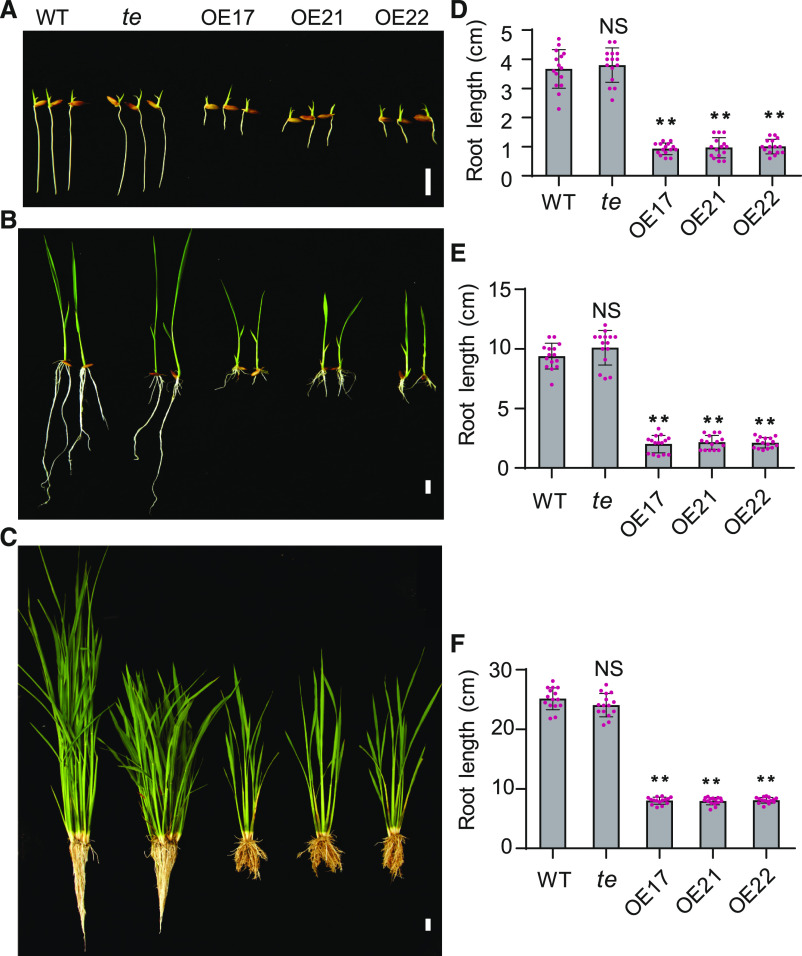
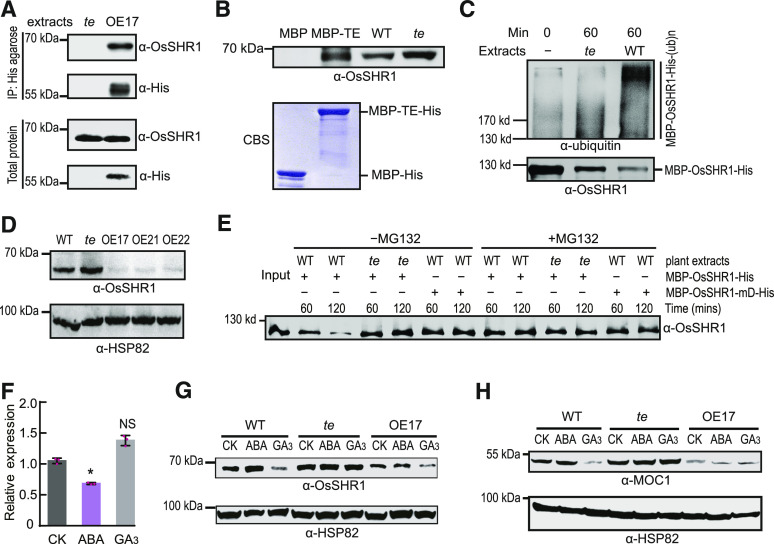
In previous studies, we reported that TE can repress tillering by mediating the degradation of MOC1 (Lin et al., 2012) and promote seed germination by mediating the GA-promoted degradation of ABA receptors (Lin et al., 2015) through the APC/CTE-26S proteasome pathway. During these studies, we noticed that, besides fewer tillers, the TE overexpressing lines (OE17, OE21, and OE22) also displayed an evident short-root phenotype in comparison to the wild type and te mutant (Figures 1A to 1F), suggesting that APC/CTE also regulates root growth.
Figure 1.
Overexpression of TE Represses Rice Root Growth.
(A) to (C) The seedling phenotype of 7-d–old (A), 14-d–old (B), or 3-month–old (C) wild type (WT), te, and OE17, OE21, and OE22. Scale bar, 1 cm in (A) and (B), and 5 cm in (C).
(D) to (F) The root length of 7-d–old (D), 14-d–old (E), or 3-month–old (F) wild-type, te, OE17, OE21, and OE22 plants. Values are means ±sd. (n = 15 seedlings). **P < 0.01, Student’s t test (two-tailed). NS, not significant.
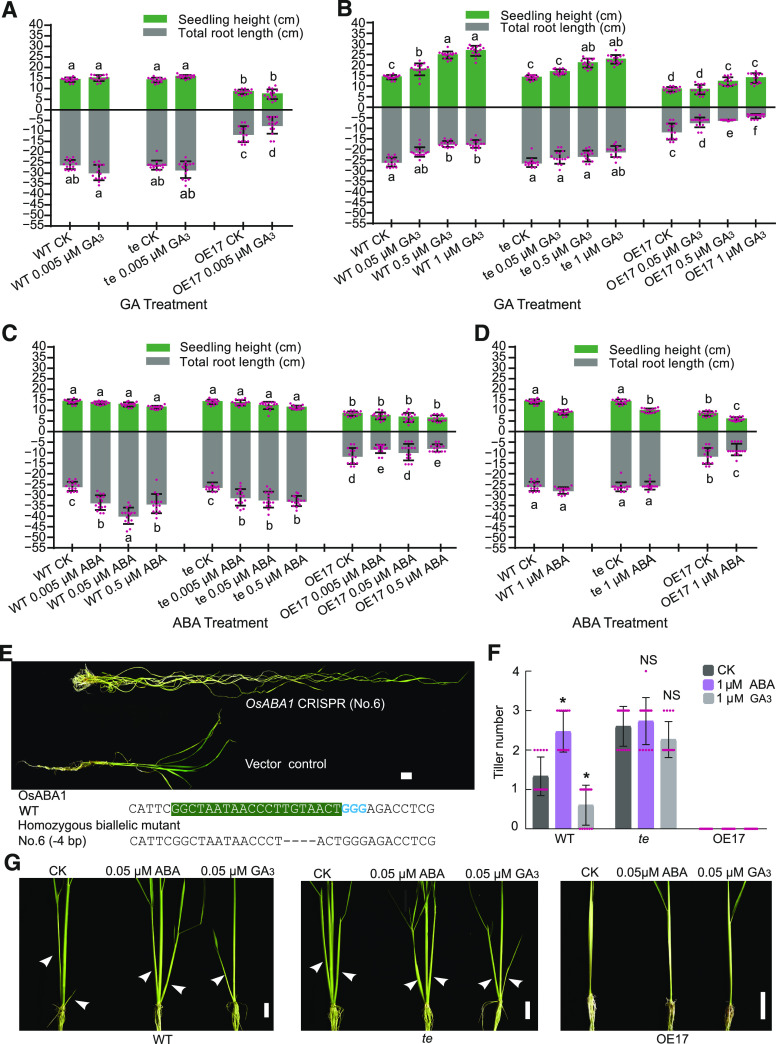
To establish whether ABA and GA are involved in APC/CTE-regulated root growth, we conducted a series of phytohormone treatment experiments. We found that a low level of exogenous GA3 (0.005 µM) slightly promoted root growth of wild type but not te; by contrast, 0.005 µM of GA3 evidently repressed root growth of the TE overexpression line OE17 (Figure 2A; Supplemental Figure 1). However, treatments with higher levels of exogenous GA3 (from 0.05 μM to 1 μM) evidently repressed root growth of both wild type and OE17, but only a higher level of GA3 (1 μM) could slightly repress root growth of te. Notably, the heights of wild-type, OE17, and te seedlings all increased gradually to varying degrees by treatments with increasing concentrations of GA3 (Figure 2B; Supplemental Figure 1). These observations suggest that the promotive effect of weak GA signaling or repressive effect of strong GA signaling on root growth is at least partially mediated by TE. On the contrary, treatment with low concentrations of ABA (from 0.005 μM to 0.5 μM) evidently promoted root growth of the wild-type and te seedlings, but not that of the OE17 seedlings (Figure 2C; Supplemental Figure 1), suggesting that moderate enhancement of ABA signaling can promote root growth and the promotive effect of ABA on root growth was likely counteracted by overdose of TE. However, when treated with a higher concentration of ABA (1 μM), the promoting effect of ABA on root growth of wild-type and te seedlings was compromised while the heights of the treated wild-type, te, and OE17 seedlings were evidently reduced (Figure 2D; Supplemental Figure 1). These results suggest that GA and ABA antagonistically regulate rice root growth in a concentration- and TE-dependent manner.
Figure 2.
APC/CTE Mediates the Opposite Effects of ABA and GA on Root Growth and Tillering.
(A) to (D) Seedling height and total root length of 3-y–old wild-type (WT), te, and OE17 seedlings treated with different doses of GA3 or ABA for 10 d. CK, no phytohormone treatment. Values are means ±sd of 15 seedlings from one of three independent experiments with similar results. The letters a, b, c, d, e, and f indicate significant differences at P < 0.01, and ab at P < 0.05 according to two-way ANOVA test with Tukey correction.
(E) The upper representation shows the phenotype of tissue cultured OsABA1 knockout plant. The lower representation shows the deletion of OsABA1 using CRISPR/CAS9 technology. Shading and colored bases show the single guide DNA site. Scale bar, 2 cm.
(F) and (G) The tiller number (F) and tiller phenotypes (G) of 3-month–old wild-type, te, and OE17 plants treated with 0.05 μM of ABA or 0.05 μM of GA3 for 1.5-month. Values are means ±sd of 15 seedlings from one of three independent experiments with similar results and *P < 0.05, Student’s t test (two-tailed) in (F). Scale bar, 2 cm; arrowheads denote the tillers in (G).
In further support of a role of ABA in regulating root growth in rice, we knocked out a single ABA biosynthesis gene, OsABA1 (Agrawal et al., 2001) using the clustered regularly interspaced short palindromic repeats (CRISPR)/CAS9 genome-editing technology. All four mutants of OsABA1 (Osaba1-6 [No.6], Osaba1-13 [No.13], Osaba1-14 [No.14], and Osaba1-15 [No.15]) displayed a short-root phenotype accompanied with a slender plant phenotype (Figure 2E; Supplemental Figure 2), indicating that ABA is a positive regulator of root growth and a negative regulator of plant height. It is also worth noting that all these plants died in the field without seed setting, indicating that ABA is essential for plants to complete their life cycle under ambient conditions.
TE Mediates the Antagonistic Effects of ABA and GA on Tillering
In a previous study, we reported that TE represses tillering by mediating the degradation of MOC1 (Lin et al., 2012). To investigate the effects of ABA and GA on tillering, we treated wild-type, te, and OE17 seedlings with ABA and GA over a prolonged period. Treatment with 0.05 μM of ABA for 1.5 months promoted tillering in the wild type but not in OE17 (Figures 2F and 2G). On the contrary, treatment with 0.05 μM of GA3 for 1.5 months repressed tillering of the wild-type but not that of the te seedlings (Figures 2F and 2G). These observations suggest that TE is likely also involved in mediating the antagonistic regulation of tillering by ABA and GA. Notably, we found that OE17 did not develop any tillers under all tested conditions, indicating that the TE dosage in OE17 might be high enough to repress the promoting effect of ABA on tillering.
TE Mediates the Antagonistic Effects of ABA and GA on RM Size in a Concentration-Dependent Manner
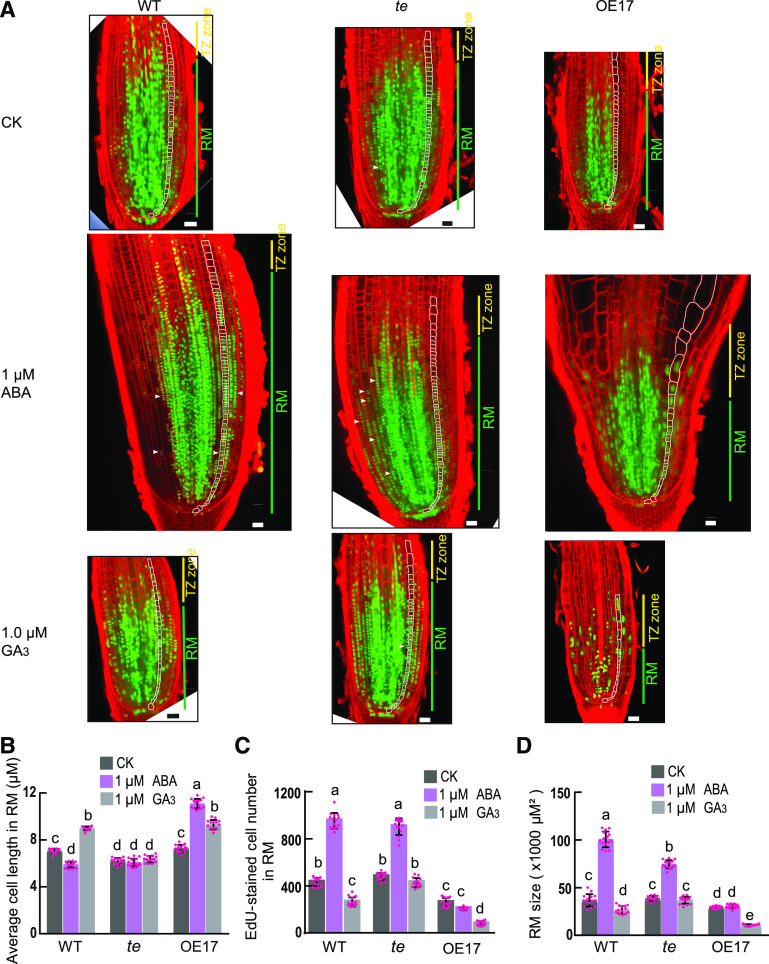
Previous reports have shown that APC/CCdh1(TE/CCS52A) promotes cell cycle progression from mitotic exit to entry into the next cell cycle (Li and Zhang, 2009; Wäsch et al., 2010; Cappell et al., 2018). We previously observed more binucleated cells in the young flag leaves of the rice te mutant compared with wild-type flag leaves (Lin et al., 2012), suggesting that TE is likely required for mitotic exit. Thus, we examined whether APC/CTE regulates the cell cycle progression of RM cells using a combined Edu-(ethynyl deoxyuridine, a marker showing active cell proliferation; Kotogány et al., 2010) and modified pseudo-Schiff-propidium iodide (mPS-PI) staining analysis. In the RM, the te mutant had more binucleated cells than OE17 and the wild type under all tested conditions (Figure 3A; Supplemental Figure 3A), indicating that TE acts to promote cell cycle progression of RM cells. Consistent with this, we found that 0.005 μM of GA3 treatment slightly stimulated RM cell proliferation and increased the RM size in wild type but not in the te mutant (Supplemental Figures 3A to 3C). These results suggest that a very low level of GA can promote root growth by stimulating APC/CTE-mediated progression of RM cell division.
Figure 3.
APC/CTE Mediates the Opposite Effects of ABA and GA on the RM Size.
(A) EdU- and mPS-PI staining analysis shows the proliferating cells (marked by green nucleus) and binucleated cells (marked by white arrowheads) in RMs of 3-d–old wild-type (WT), te, and OE17 seedlings treated with 1 μM of ABA, 1 μM of GA3, or no phytohormone treatment (CK, Control Check) for 3 d. Scale bar, 20 μm.
(B) to (D) Average cell length (B), EdU-stained cell number (C), and RM size (D) of 3-d–old wild-type, te, and OE17 seedlings treated with 1 μM of ABA, 1 μM of GA3, or no phytohormone treatment (CK) for 3 d. Values are means ±sd of 15 root tips and the letters a, b, c, d, and e indicate significant differences at P < 0.01 according to two-way ANOVA test with Tukey correction.
In addition, earlier studies have shown that strong APC/CCdh1(TE/CCS52A) activity can trigger cell elongation and endoreplication to promote cell differentiation (Wäsch et al., 2010; Lin et al., 2012; Su’udi et al., 2012). To test whether APC/CTE can induce RM cell elongation to promote RM cell differentiation, we checked the cell length under the treatment of ABA and GA. Results showed that treatment with 1 μM of GA3 but not 0.005 μM of GA3 increased the longitudinal cell length of RM in wild type and OE17 but not in te (Figure 3B; Supplemental Figure 3D), suggesting that a higher concentration of GA can promote RM cell elongation and differentiation in an APC/CTE-dependent manner. Consistent with this, we found that 1 µM of GA treatment markedly reduced RM cell proliferation and RM size in wild type and OE17, but not in the te mutant, in comparison to the no phytohormone treatment (CK; Figures 3A, 3C, and 3D). On the contrary, treatment with 1 μM of ABA decreased longitudinal cell length of RM in wild type but not in OE17 (Figure 3B), suggesting that ABA may inactivate APC/CTE to repress RM cell elongation and that the repressive effect of ABA on cell elongation in the RM is likely offset by an overdose of TE in OE17. Consistent with this finding, 1 µM of ABA treatment evidently increased the number of EdU-labeled cells and the RM size in the wild type and te mutant but not in OE17 in comparison to the no phytohormone treatment (CK; Figures 3A, 3C, and 3D). Taken together, these findings suggest that high levels of GA likely induce a strong APC/CTE activity to stimulate RM cell elongation, thus promoting RM cell differentiation and reducing RM size; on the contrary, ABA likely represses the APC/CTE activity to restrict RM cell elongation and differentiation, thus maintaining RM size and cell proliferation.
TE Mediates the Antagonistic Effects of ABA and GA on AM Size
To probe the cellular mechanisms of ABA and GA on tillering, we examined the effects of ABA and GA on AM size using an EdU-staining analysis (Kotogány et al., 2010). At the AM initiation stage, 1 μM of ABA treatment increased the AM size in wild-type but not in the OE17 plants, while 1 μM of GA treatment reduced the AM size in wild type and OE17, but not in the te mutant plants (Supplemental Figure 4), indicating that ABA’s promoting effects on AM size is likely counteracted by an overdose of TE in the OE17 plants and that GA’s repressive effects on AM size is dependent on APC/CTE. Therefore, these findings suggest that TE also mediates the antagonistic regulation of AM size by ABA and GA.
OsSHR1 Promotes Root Growth and Is a Substrate of the APC/CTE E3 Ubiquitin Ligase Complex
We have previously characterized MOC1, a key promoter of tillering, as a substrate of APC/CTE in the tillering regulatory pathway (Lin et al., 2012). To investigate the regulatory mechanisms by which TE mediates the antagonistic regulation of RM cell proliferation and size by ABA and GA, we attempted to identify the potential substrates of APC/CTE in the root growth regulatory pathway. Through manually searching for D-box (RxxLxxxxN/D/E), a motif recognized by TE (Lin et al., 2012; Xu et al., 2012), we found that OsSHR1, a rice homologue of Arabidopsis SHR (Helariutta et al., 2000), is a D-box–containing protein (Supplemental Figure 5), suggesting that OsSHR1 might be a substrate of APC/CTE.
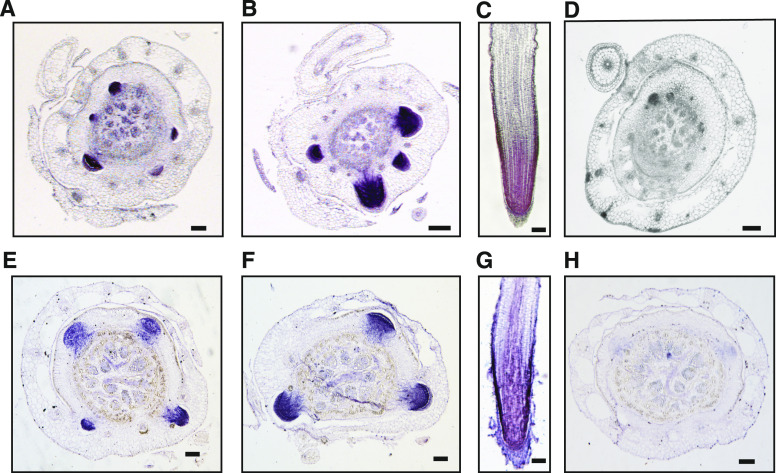
To test whether OsSHR1 is an authentic substrate of TE, we conducted a series of analyses. In situ hybridization showed that from the root primordium to the elongated root, both TE and OsSHR1 were expressed strongly throughout the RM zone (Figure 4). Both co-immunoprecipitation (Co-IP) and in vitro pull-down analyses showed that TE interacted with OsSHR1 (Figures 5A and 5B). Moreover, an in vitro ubiquitination assay showed that MBP-OsSHR1-His was polyubiquitinated more efficiently by wild-type plant extracts than by te plant extracts (Figure 5C). Further, immunoblot analysis showed that evidently more OsSHR1 protein accumulated in the te plants, compared with the TE OE lines and wild-type plants (Figure 5D). A cell-free degradation assay showed that MBP-OsSHR1-His recombinant protein could be effectively degraded by wild-type extracts, but its degradation was compromised in the te mutant extracts (Figure 5E). In addition, the degradation of MBP-OsSHR1-His could be effectively blocked by the proteasome inhibitor MG132 and the mutant MBP-OsSHR1-mD-His protein (with a mutated D-box; RSLL→aSLa) remained relatively stable in wild-type plant extracts compared with the wild-type MBP-OsSHR1-His protein (Figure 5E). These results collectively suggest that OsSHR1 is an authentic substrate of APC/CTE.
Figure 4.
Expression Patterns of TE and OsSHR1 in the Root.
(A) to (H) RNA in situ hybridization showing that from the root primordium to the elongated root, TE (A) to (C) and OsSHR1 (E) to (G) are strongly expressed in the RM zone of them. The sense probe of TE (D) or OsSHR1 (H) was used as the negative control. Scale bar, 100 μm.
Figure 5.
APC/CTE Mediates the Opposite Effects of ABA and GA on the Degradation of OsSHR1 and MOC1.
(A) Co-IP assay shows that His-agarose simultaneously pulls down TE and OsSHR1 from the OE17 but not te plant extracts by specifically binding to the His epitope in the N terminus of TE. “α-His” indicates detected TE proteins by the α-His antibody.
(B) In vitro pull-down assay shows that MBP-TE-His pulls down OsSHR1 from the te plant extracts. WT, wild type.
(C) In vitro ubiquitination assay of MBP-OsSHR1-His by the wild-type and te plant extracts.
(D) Immunoblot analysis shows the levels of OsSHR1 protein in the shoot bases of wild-type, te, OE17, OE21, and OE22 plants. “α-HSP82” indicates that roughly equal amounts of total plant extracts were used.
(E) Cell-free degradation assay shows the stability of MBP-OsSHR1-His protein in wild-type and te plant extracts or MBP-OsSHR1-mD-His protein in wild-type extracts with or without 50 μM of proteasome inhibitor MG132. “Input” shows the initiation amounts of MBP-OsSHR1-His proteins.
(F) and (G) The mRNA levels (F) of OsSHR1 in 6-d–old wild type treated with 1 μM of ABA, 1 μM of GA3 or no phytohormone treatment (CK) for 3 dor protein levels (G) of OsSHR1 in 6-d–old wild type, te and OE17 treated with 1 μM of ABA, 1 μM of GA3 or no phytohormone treatment (CK) for 3 d. Values are means ±sd (n = 3 biological replicates) and *P < 0.001, Student’s t test (two-tailed) in (F). “α-HSP82” indicates that roughly equal amounts of total plant extracts were used in (G). NS, not significant.
(H) Immunoblot analysis showing the quantities of MOC1 proteins in 1-month–old wild-type, te, and OE17 plants treated with 1 μM of ABA, 1 μM of GA3, or no phytohormone treatment (CK) for 3 d. “α-HSP82” indicates that roughly equal amounts of total plant extracts were used.
To test whether OsSHR1 is involved in regulating root growth in rice, we knocked out OsSHR1 using CRISPR/CAS9 technology. The results showed that the monoallelic mutant (Osshr1-1[No. 1]) and a biallelic mutant of OsSHR1 (Osshr1-2 [No. 2]) displayed a short-root phenotype as expected (Supplemental Figure 6). In addition, we generated RNA interference (RNAi) transgenic lines of OsSHR1 and transgenic plants expressing the undegradable OsSHR1-mD-His protein (driven by its endogenous promoter) in the Nipponbare background. The RNAi plants had reduced OsSHR1 expression and exhibited a short-root phenotype with increased plant height, while the transgenic plants expressing OsSHR1-mD-His accumulated more OsSHR1 proteins and displayed a long-root phenotype with reduced plant height, in comparison with Nipponbare (Supplemental Figure 7). Further EdU- and mPS-PI analyses showed that both RM cell proliferation and size were reduced in the OsSHR1 RNAi plants but increased evidently in the OsSHR1-mD-His transgenic plants in comparison to Nipponbare (Supplemental Figure 8). We further used these materials to examine the possible effects of ABA and GA on OsSHR1-mediated root growth. A 0.05 μM of GA3 treatment appeared to repress the root growth of Nip and NipOsSHR1-RNAi but not that of the NipOsSHR1-mD-His transgenic line while a 0.05 μM of ABA treatment appeared to promote the root growth of Nip and NipOsSHR1-mD-His but not that of NipOsSHR1-RNAi (Supplemental Figure 9), suggesting that stabilized OsSHR1-mD-His antagonizes GA-mediated inhibition of root growth, whereas RNAi-destabilized OsSHR1 disrupts ABA-mediated promotion of root growth. Taken together, these results support the notion that OsSHR1 acts as a key positive regulator of root growth in rice by maintaining RM activity and size, and it is subject to antagonistic regulation by ABA and GA.
To further investigate the genetic relationship between TE and OsSHR1, we generated RNAi lines of OsSHR1 in the te background. Phenotypic analyses showed that the RNAi lines with reduced OsSHR1 expression in the te background displayed a short-root phenotype as did the TE overexpression lines (OE17, OE21, and OE22; Supplemental Figure 10), supporting the notion that OsSHR1 acts downstream of TE to promote root growth.
GA and ABA Antagonistically Regulate APC/CTE-Mediated Degradation of OsSHR1 and MOC1
We previously showed that GA promotes APC/CTE-mediated degradation of the ABA receptor protein OsPYL/RCAR10 (R10), while ABA stabilizes R10, thus antagonistically regulating seed germination (Lin et al., 2015) . We thus speculated that ABA and GA may also regulate root growth by influencing APC/CTE-mediated degradation of OsSHR1. To test this possibility, we examined the effects of ABA and GA on the protein levels of OsSHR1. Treatment with 1 μM of ABA stabilized the protein levels of OsSHR1 in wild-type but not in OE17 roots, while treatment with 1 μM of GA3 reduced the protein levels of OsSHR1 in wild-type and OE17 but not in te roots, although the level of OsSHR1 mRNA reduced slightly under ABA treatment and did not change significantly under GA treatment in the wild-type plants (Figures 5F and 5G). These observations suggest that OsSHR1 protein levels were mainly regulated by ABA and GA at the post-transcriptional level. Similarly, treatment with 1 μM of ABA stabilized the MOC1 protein in the shoot bases of wild type but not in these of OE17, while treatment with 1 μM of GA destabilized the MOC1 protein in the shoot bases of wild type and OE17 but not in these of te mutant (Figure 5H). In addition, te plants accumulated high levels of OsSHR1 or MOC1 proteins regardless of the treatment with ABA or GA (Figures 5G and 5H). These results together suggest that ABA and GA antagonistically regulate APC/CTE-mediated degradation of OsSHR1 and MOC1 to regulate root growth and tillering, respectively.
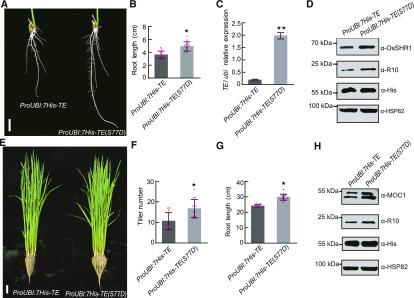
ABA and GA Antagonistically Regulate APC/CTE Activity through Modulating SnRK2s-Mediated Phosphorylation of TE
We previously showed that TE’s binding ability to its substrate R10 protein is impaired by ABA-activated SnRK2s-mediated phosphorylation of TE and that the S77 residue in TE is a key phosphorylation site recognized by SnRK2s (Lin et al., 2015). To test whether SnRK2s-mediated phosphorylation of TE interferes with the interaction between TE with its substrates, we overexpressed 7His-TE or 7His-TE(S77D)—S77D is a mutation mimicking the phosphorylation status of S77, driven by the maize (Zea mays) ubiquitin promoter in the te mutant background. The root length of 1-week–old 7His-TE transgenic seedlings was shorter than that of 7His-TE(S77D) transgenic seedlings (Figures 6A and 6B), although the mRNA level of 7His-TE(S77D) was evidently higher than that of 7His-TE (Figure 6C). Immunoblot analysis showed that both OsSHR1 and R10 proteins were overaccumulated in the roots of the 7His-TE(S77D) plants compared with the 7His-TE plants (Figure 6D). Further, 3-month–old 7His-TE transgenic seedlings had shorter roots and fewer tillers than 3-month–old 7His-TE(S77D) transgenic seedlings (Figures 6E to 6G). In addition, MOC1 and R10 proteins overaccumulated in the shoot bases of 3-month–old 7His-TE(S77D) transgenic plants compared with the 7His-TE transgenic plants (Figure 6H). These results suggest that phosphorylation of TE at S77 by SnRK2s (Lin et al., 2015) can stabilize OsSHR1, MOC1, and R10 proteins in planta likely by impairing the binding ability of TE to its substrates.
Figure 6.
S77D Phosphorylation Mimic of TE Stabilizes Its Substrates In Planta.
(A) to (D) Root phenotypes (A), root lengths (B), relative TE mRNA levels detected by RT-qPCR analysis (C), and the quantities of TE’s substrate proteins (OsSHR1 and R10) detected by immunoblot analysis (D) of 6-d–old ProUBI:7His-TE and ProUBI:7His-TE(S77D) transgenic seedlings. Scale bar, 1 cm in (A). Values are means ±sd and Student’s t test (two-tailed) analysis indicates a significant difference (n = 15 seedlings in (B) and n = 3 biological replicates in (C); *P < 0.05, **P < 0.01). “α-HSP82” indicates that roughly equal amounts of root extracts were used and “α-His” indicates detected 7His-TE and 7His-TE(S77D) proteins by “α-His” antibody in (D).
(E) to (H) Plant phenotypes (E), tiller numbers (F), root lengths (G), and the quantities of TE’s substrates (MOC1 and R10) detected by immunoblot analysis (H) of 3-month–old ProUBI:7His-TE and ProUBI:7His-TE(S77D) transgenic plants in the field. Values are means ±sd of 15 seedlings and *P < 0.05, Student’s t test (two-tailed) in (F) and (G). Scale bar, 10 cm in (E). “α-HSP82” indicates that roughly equal amounts of shoot base extracts were used and “α-His” indicates detected 7His-TE and 7His-TE(S77D) proteins by “α-His” antibody in (H).
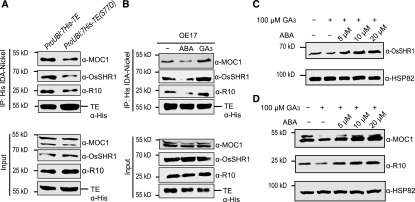
In support of this notion, Co-IP analyses showed that more substrates (OsSHR1, MOC1, and R10) could be pulled down by 7His-TE than by 7His-TE (S77D; Figure 7A), indicating that S77 phosphorylation of TE by SnRK2s did impair the binding ability of TE to its substrates. Consistently, our previous study has shown that ABA repressed the interaction between TE and ABA receptor R10 by activating SnRK2s-mediated phosphorylation of TE, while GA promoted the interaction by inactivating SnRK2s-mediated phosphorylation of TE (Lin et al., 2015). To test if ABA and GA also regulate the affinity of TE with OsSHR1 and MOC1 by a similar mechanism, we performed Co-IP experiments and the results showed that ABA treatment repressed the interaction between TE with OsSHR1 and MOC1 while GA treatment promoted these interactions (Figure 7B), suggesting that ABA repressed the interaction between TE with OsSHR1 and MOC1, thus stabilizing OsSHR1 and MOC1, while GA promoted these interactions, thus destabilizing these proteins. Interestingly, we found that ABA can block GA-promoted degradation of OsSHR1, MOC1, or R10 in a dosage-dependent manner (Figures 7C and 7D). Together, these results suggest that ABA stabilizes while GA destabilizes TE’s substrates and that the GA’s destabilization on TE’s substrates can be blocked by ABA.
Figure 7.
ABA Represses while GA Promotes the Activity of APC/CTE.
(A) and (B) Co-IP assay shows the quantities of TE, R10, OsSHR1, and MOC1 proteins pulled down from the extracts of 1-month–old ProUBI:7His-TE transgenic plants and ProUBI:7His-TE(S77D) transgenic plants (A) or from the extracts of 1-month–old OE17 plants treated with 10 μM of ABA or 100 μM of GA3 for 3 h (B) by equal amounts of His-agarose. “Input” shows the starting quantities of TE, R10, OsSHR1, and MOC1 proteins that were used in the Co-IP assays. “α-His” indicates detected 7His-TE and 7His-TE(S77D) proteins in (A) or TE proteins in (B) by “α-His” antibody.
(C) and (D) GA-promoted degradation of OsSHR1 is repressed gradually in wild-type roots (C) or GA-promoted degradation of MOC1 and R10 is repressed gradually in wild-type shoot bases (D) by application of increased amounts of ABA. “α-HSP82” indicates that roughly equal amounts of total plant extracts were used in (C) and (D).
DISCUSSION
APC/CTE Mediates the Antagonistic Regulation of GA and ABA on Root Growth and Tillering
In this study, we showed that GA (within a concentration range of 0.05 μM to 1 μM) represses root growth and tillering while ABA (within a concentration range of 0.005 μM to 0.5 μM) promotes root growth and tillering (Figure 2). Further, we collected several lines of evidence to demonstrate that the SnRK2s-APC/CTE regulatory module plays a critical role in mediating the antagonistic regulation of root growth and tillering by ABA and GA. First, we showed that TE mediates the opposite effects of ABA and GA on root growth and tillering (ABA promotes while GA represses root growth and tillering; Figures 1 and 2). Second, we showed that TE mediates the opposite effects of ABA and GA on the size of RM and AM (ABA promotes while GA represses the sizes of RM and AM; Figure 3; Supplemental Figure 4). Third, we showed that the APC/CTE E3 ubiquitin ligase complex is responsible for ubiquitination and targeted degradation of OsSHR1 by the 26S proteasome to repress root growth (Figures 4 and 5; Supplemental Figures 5 to 10). Finally, we showed that GA can promote APC/CTE-mediated degradation of OsSHR1 in the RM or MOC1 in the AM, whereas ABA can stabilize these proteins through stimulating SnRK2s-mediated phosphorylation of TE, thus impairing the binding ability of TE to its substrates (Figures 6 and 7).
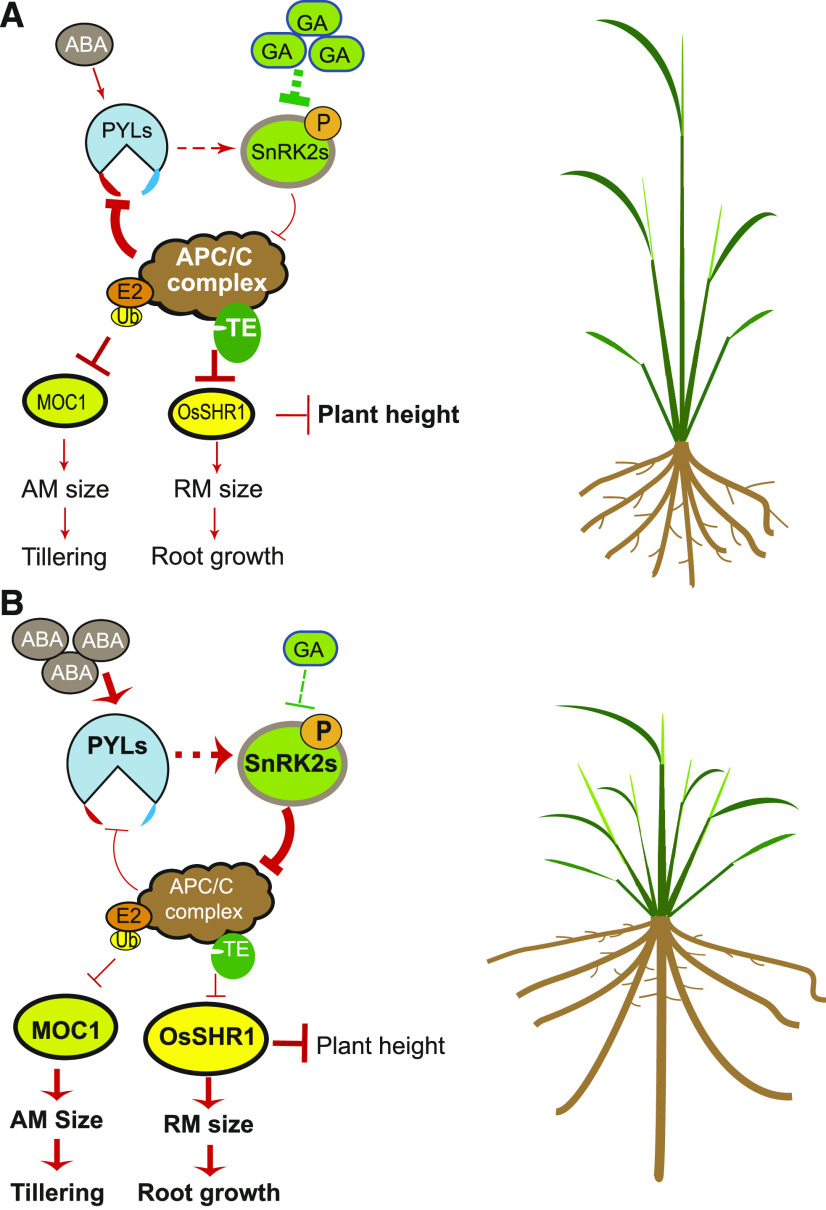
Based on these results, we propose a model in which strong GA signaling can reduce RM size or AM size to restrict root growth or tillering by promoting the APC/CTE-mediated degradation of a key RM promoting factor, OsSHR1, in the RM or a key AM promoting factor, MOC1, in the AM through the SnRK2s-APC/CTE regulatory module (Figure 8A). Conversely, moderate enhancement of ABA signaling can maintain RM or AM sizes to sustain root growth or tillering by antagonizing the GA-APC/CTE-promoted degradation pathway to stabilize OsSHR1 in the RM or MOC1 in the AM through the SnRK2s-APC/CTE regulatory module (Figure 8B). In addition, stabilized OsSHR1 can repress the elongation growth of plant height (Supplemental Figure 7) and GA-inhibited root growth (Supplemental Figure 9). Consistent with our model, a previous study reported that root growth and tillering are reduced due to reduced OsSHR1 and MOC1 activity, respectively, in the strong GA signaling enhancement mutant low tillering and high plant height1 (ltn1, defective in the rice homologue of Arabidopsis o-fucosyltransferase SPINDLY, which activates DELLA to repress GA signaling; Supplemental Figure 11; Guo et al., 2017; Zentella et al., 2017); on the contrary, slight weakening of GA signaling in Zhonghua11’s GA biosynthesis-deficient mutant semi-dwarf1 (sd1, which is defective in OsGA20ox2) can promote root growth by stabilizing OsSHR1 and repressing plant height (Supplemental Figure 12A to 12C) and enhance rice tillering by stabilizing MOC1 (Liao et al., 2019). Further, our model is supported by the previous reports that Gibberellin 2-oxidases (GA2oxs) regulate plant growth by inactivating endogenous bioactive gibberellins (GAs) and that overexpression of these GA2oxs can promotes tillering and adventitious root growth (Lo et al., 2008). However, severe weakening of GA signaling significantly represses both root and plant growth in Kittaka’s GA-deficient mutant dwarf18 (which is defective in OsGA3ox2 required for the biosynthesis of GAs) although in which OsSHR1 is evidently accumulated (Supplemental Figures 12D to 12F), indicating that a certain amount of GA is required for optimal root and plant growth.
Figure 8.
A Work Model Depicting the SnRK2s-APC/CTE Module-Mediated Antagonistic Regulation of Root Growth and Tillering by ABA and GA.
(A) Strong GA signaling can reduce RM size or AM size to restrict root growth or tillering by promoting the APC/CTE-mediated degradation of OsSHR1 (a key RM promoter) in RM or MOC1 (a key AM promoter) in AM through the SnRK2s-APC/CTE regulatory module.
(B) Conversely, moderate enhancement of ABA signaling can maintain RM or AM sizes to sustain root growth or tillering by antagonizing the GA-APC/CTE-promoted degradation pathway to stabilize OsSHR1 in RM or MOC1 in AM through the SnRK2s-APC/CTE regulatory module. In addition, stabilized OsSHR1 can repress the elongation growth of plant height.
APC/CTE Plays a Dual Role in Regulating both RM Maintenance and RM Differentiation
The observation that there is no evident difference between the root lengths of wild type and te was initially confusing to us. Through careful analyses, we found that APC/CTE is involved in regulating both RM maintenance and RM differentiation, and that both ABA and GA regulate root growth in a concentration-dependent manner.
For GA, we found that a very low level of GA (0.005 μM) activates APC/CTE’s role in promoting cell cycle progression to increase RM size and promote root growth (Figure 2A; Supplemental Figures 1 and 3). By contrast, high levels of GA (from 0.05 μM to 1 μM) activate APC/CTE’s role in promoting RM cell elongation (differentiation) to repress root growth (Figures 2B and 3; Supplemental Figure 1). Consistent with this finding, we found that only severe GA-deficient mutants such as dwarf18 and ks1 (which is defective in ent-kaurene synthase1 required for the biosynthesis of GAs; Li et al., 2015) showed reduced root growth while the weak GA-deficient mutant sd1 showed slightly enhanced root growth (Supplemental Figure 12). Moreover, we found that GA’s constitutive response (manifested by the slender plant phenotype) caused by loss of the GA signaling inhibitor SLENDER RICE1 (Ikeda et al., 2001) or strong ABA deficiency (Figure 2E; Supplemental Figure 2) significantly reduced root growth. For ABA, we found that a low concentration range of ABA (from 0.005 µM to 0.5 µM) promotes root growth likely by repressing APC/CTE’s role in promoting RM cell elongation (differentiation), while a higher concentration of ABA (1 µM) has a declined promoting effect on root growth (Figures 2C, 2D, and 3; Supplemental Figure 1).
In addition, we found that in te, the lack of TE has two opposite effects on root growth: On the one hand, the lack of TE activity can repress RM cell division by delaying mitotic exit of RM cell cycle progression (Figure 3A; Supplemental Figure 3A) thus repressing root growth; on the other hand, the lack of TE activity can weaken RM cell elongation (differentiation; Figures 3A and 3B) and stabilize OsSHR1 to promote RM cell division by stimulating the S-progression (DNA replication) of RM cell cycle progression (Figure 5; Supplemental Figures 6 to 8), thus promoting root growth. The dual effect of lacking TE on RM cell cycle progression (Supplemental Figure 13A) may offset each other, thus resulting in similar root lengths in the te mutant as in the wild type.
In summary, these findings support the notion that in rice, APC/CTE plays a dual role in regulating both RM maintenance and RM differentiation dependent on its activity level: Lower APC/CTE activity, which could be induced by weak GA signaling, promotes RM cell cycle progression (cell division) to maintain RM for sustainable root growth; whereas higher APC/CTE activity, which could be induced by strong GA signaling (could be repressed by ABA signaling), promotes RM cell elongation (differentiation) to restrict root growth (Supplemental Figure 13B). This notion is supported by the previous findings that APC/CCdh1(TE/CCS52A) plays a dual role in regulating both cell division and cell differentiation (including cell elongation and cell endoreplication) from yeast to animal and plant dependent on its activity level (Larson-Rabin et al., 2009; Wäsch et al., 2010; Lin et al., 2012; Baloban et al., 2013; Edgar et al., 2014; Cappell et al., 2018).
Improve Crop Plant Architecture via Fine-Tuning the Crosstalk between ABA and GA
Our results showed that intriguingly, although as a major stress response phytohormone, ABA traditionally acts as a growth inhibitor to preferentially confer plant fitness adaptation to various environmental stresses by restricting the growth of the whole plant including shoot elongation, branching, and root growth in a high concentration range (Golldack et al., 2013), lack of ABA severely impairs root growth and plant survival in ambient conditions while moderate levels of ABA can maintain root growth and shoot branching (tillering; Figure 2; Supplemental Figures 1 and 2), suggesting that ABA is essential for plant survival and a certain level of ABA is required for plant growth and development, a notion in agreement with several earlier studies (Mulkey et al., 1983; Humplík et al., 2017; Yoshida et al., 2019). On the other hand, it is known that a moderate amount of GA is also required for plant growth and development (Yamaguchi, 2008). Interestingly, the Osaba1 mutants with severe ABA deficiency (Figure 2E; Supplemental Figure 2) display a similar constitutive GA response (i.e., tall or slender phenotype) as the slr1 mutant (loss of the GA signaling inhibitor SLENDER RICE1; Ikeda et al., 2001; Sasaki et al., 2003), and that both of them eventually die—suggesting that the balance between ABA and GA signaling is very important for fine-tuning plant growth and survival. This notion is supported by the finding that exaggerated GA signaling in the ltn1 rice mutant caused a tall plant stature with a weak root system (Supplemental Figure 11) accompanied with reduced 1,000-grain weight, seed setting rate, and dramatic decline yield per plant (Guo et al., 2017), whereas slight weakening of GA signaling in the rice sd1 mutant shaped a dwarf plant stature with a strong root system (Supplemental Figure 12), thus leading to the successful breeding of lodging-tolerant, semi-dwarf rice and wheat (Triticum aestivum) cultivars that constitute the first “green evolution” (Peng et al., 1999; Sasaki et al., 2002).
In recent years, the efforts to breed rice cultivars with “ideal plant architecture” (less unproductive tillers, larger panicles, and stronger root) started to show promising results with improved yield (Khush, 1995; Wang et al., 2018). Our findings that in a low concentration range, moderate enhancement of ABA signaling can enhance the root system and tillering activity, may offer new strategies for breeding modern crop varieties with enhanced adaptation to worsened environmental conditions due to global warming by optimizing plant architecture via fine-tuning the crosstalk between ABA and GA signaling pathways.
METHODS
Plant Materials and Growth Conditions
The wild-type, te mutant, and TE overexpression transgenic lines (OE17, OE21, and OE22) used in this study were previously described by Lin et al. (2012). For RT-qPCR assays, phytohormone treatments, and Co-IP assays, the seeds of wild-type, te, OE17, and rice (Oryza sativa) Nipponbare were treated for one week at 37°C to break dormancy and the germinated seeds or seedlings were grown in climate chambers (HP1500GS; Ruihua) at 70% humidity, under long-day conditions with a photocycle of 14-h light (30°C) and 10-h darkness (25°C). Light was provided by white-light emitting diode (LED) tubes (400 nm to 700 nm, 250 μmol m−2 s−1).
Vector Construction and Plant Transformation
To generate the overexpression vectors ProUBI:7HIS-TE and ProUBI:7HIS-TE(S77D), the full-length coding sequence (CDS) or partial CDS of TE were amplified using the primers shown in Supplemental Table 1, and the PCR products were cloned into the binary vector pCUBI1390 with the In-Fusion Advantage PCR Cloning Kit (Cat. no. PT4065; Clontech). To generate the CRISPR/CAS9 DNA knockout constructs, pCAS9-OsSHR1 and pCAS9-OsABA1, the guide RNA scaffolds targeting them were synthesized by Life Technology/Thermo Fisher Scientific and cloned into the binary vector pCAS9-single guide RNA-AarI. To generate the OsSHR1-RNAi construct, the OsSHR1 CDS was amplified with the primers shown in Supplemental Table 1, and the PCR products were inserted into the LH-FAD2-1390 RNAi vector. To generate the construct ProOsSHR1:OsSHR1-mD(RSLL→aSLa)-HIS, two fragments were PCR-amplified with primers containing the mutated sites (RSLL→aSLa) in the D-box shown in Supplemental Table 1 and the products were infused into the pCAMBIA 1305 plasmid. The resulting constructs were introduced into the Nipponbare variety or te mutant by Agrobacterium tumefaciens-mediated transformation.
Antibody Preparation and Immunoblot Analysis
The CDS of N-terminal 1 to 287 amino acids of OsSHR1 was amplified using the primers shown in Supplemental Table 2, and the PCR products were inserted into the pMAL-C2× vector to express the MBP-OsSHR1-N287aa-HIS fusion protein. Antibodies against OsSHR1 were prepared by immunizing rabbits with the MBP-OsSHR1-N287aa-HIS fusion protein, and then affinity-purified with corresponding MBP-OsSHR1-N287aa-HIS fusion protein. Immunoblots were performed with the purified antibodies and visualized with enhanced chemiluminescence reagent (ECL; GE Healthcare). The antibody against OsPYL/R10 and MOC1 were previously described by Lin et al. (2012, 2015). The antibodies for detecting His (Cat. no. D291-7, 1:1,000 dilution) were purchased from the company Medical & Biological Laboratories, and the antibodies against Ubiquitin (Cat. no. 3936, 1:1,000 dilution) and HSP82 (Cat. no. AbM51099-31-PU, 1:5,000 dilution) were purchased from Cell Signaling Technology and Beijing Protein Innovation, respectively.
All immunoblot experiments were repeated at least three times, essentially with the same conclusions, and representative results are shown.
Construction of Protein Expression Plasmids
The construction of MBP-His and MBP-TE-His was performed as previously described by Lin et al. (2015). To generate the expression vector of MBP-OsSHR1-His and MBP-OsSHR1-mD(RSLL-aSLa)-His, the CDS of OsSHR1 or OsSHR1-mD were amplified using the primers shown in Supplemental Table 2, and the PCR products were cloned into the pMAL-C2× vector (New England BioLabs) with the In-Fusion Advantage PCR Cloning Kit (Cat. no. PT4065; Clontech) to generate the following plasmids: pMAL-C2x::MBP-OsSHR1-His and pMAL-C2x::MBP-OsSHR1-mD-His.
In Vitro Pull-Down Assays
For the in vitro pull-down assay of OsSHR1, total proteins were extracted from 1-week–old te seedlings in a degradation buffer (containing 25 mM of Tris-HCl at pH 7.5, 10 mM of NaCl, 10 mM of MgCl2, 4 mM of phenylmethylsulfonyl fluoride, 5 mM of DTT, and 10 mM of ATP; Wang et al., 2009). Roughly equal amounts of purified MBP-His and MBP-TE-His fusion proteins (∼1 μg) were affixed to Amylose Resin (New England BioLabs), then incubated in 200-μL te seedling extract (containing 0.8-mg total proteins) for each assay, with 50 μm of MG132, a proteasome inhibitor. The mixture was gently shaken at 28°C for 30 min. The pull-down assay was performed as reported in Tagwerker et al. (2006) and Miernyk and Thelen (2008), using anti-His (Cat. no. D291-7, Medical & Biological Laboratories, 1:1,000 dilution) or anti-OsSHR1 antibodies (1:4,000 dilution).
Cell-Free Degradation Assays
Total protein extracts were prepared from te and wild-type seedlings as previously described by Lin et al. (2012). Then MG132 was selectively added to various in vitro degradation assays, as indicated. For the degradation assays, equal amounts (∼500 ng) of the purified MBP-OsSHR1-His protein or the MBP-OsSHR1-mD-His protein were each incubated in 50 μL of rice total protein extract (containing ∼200-μg total proteins). The mixtures were incubated at 28°C for 60 min or 120 min, and the samples were processed for immunoblot analysis to determine the abundances of MBP-OsSHR1-His protein or MBP-OsSHR1-mD-His protein. The membranes were probed with anti-OsSHR1 antibodies.
RNA In Situ Hybridization
Shoot apexes or root tips of rice seedlings at the third and fourth leaf stage were fixed using a Formalin–acetic acid–alcohol fixative solution (RNase-free) at 4°C overnight followed by dehydration steps (dehydrated successively in 30% [v/v], 50%[v/v], 70% [v/v], 80% [v/v], 90% [v/v], and 100% [v/v] ethanol; and cleared successively in 25% [v/v] xylene +75% [v/v] ethanol, 50% [v/v] xylene +50% [v/v] ethanol, 75% [v/v] xylene +25% [v/v] ethanol, and three times in 100% [v/v] xylene) and then embedded in paraffin (Paraplast Plus; Sigma-Aldrich). RNA in situ hybridization was performed as previously described by Lin et al. (2012).
In Vitro Ubiquitination Assays
Purified MBP-OsSHR1-His protein was bound to Ni-NTA-Agarose (Novagen) and then incubated at 28°C with equal amounts of crude rice seedling extracts in a buffer previously described by Lin et al. (2015). After incubating for the indicated time intervals, the MBP-OsSHR1-His and the polyubiquitinated MBP-OsSHR1-His (MBP-OsSHR1-His-(Ub)n) were added to the SDS-PAGE loading buffer, then loaded onto an SDS-PAGE gel. Immunoblots were performed with antibodies against the polyubiquitin tail or OsSHR1. The upshifted bands of MBP-OsSHR1-His in Figure 5C were confirmed to be the polyubiquitinated MBP-OsSHR1-His by blotting with anti-polyubiquitin antibodies (Cat. no. 3936; Cell Signaling Technology).
RT-qPCR Analyses
RNA was extracted from frozen samples using Direct-zol RNA Kits (Zymo Research) according to the manufacturer’s instructions. RT-qPCRs were performed using the SYBR Premix Ex Taq RT-PCR kit (Takara), according to the manufacturer’s instructions, with the primers listed in Supplemental Table 3.
Effects of ABA and GA Treatment on the Stability of OsSHR1, R10, and MOC1 Proteins
Three-d–old seedlings or three-leaf-stage seedlings of the wild type, te, OE17, and Nipponbare were grown in a growth chamber (Ruihua) under a 14-h/10-h light/dark cycle at 30°C in the light (provided by white-light LED tubes [400 nm to 700 nm, 250 μmol m−2 s−1] photo-cycle) and 25°C in the dark. These plants were subjected to ABA and GA3 treatment to determine their effects on the stability of OsSHR1, R10, and MOC1 proteins. For phytohormone treatments, seedlings were placed in 500-mL tanks containing 450 mL of half-strength Murashige and Skoog (MS) liquid medium supplemented with 1 mM of cycloheximide (Inalco), 4.5 μL of ethanol (control), 4.5 μL of 100 mM ABA, or 100 mM of GA3. Subsequently, the tanks were placed in a growth chamber (Ruihua) under a 14-h/10-h light (provided by white-light LED tubes [400 nm to 700 nm, 250 μmol m−2 s−1]) at 30°C/dark at 25°C/light photocycle for the indicated times. After phytohormone treatments, the seedlings were frozen in liquid nitrogen for further analysis. Immunoblots were performed with antibodies against OsPYL/R10, OsSHR1, MOC1, or HSP82, and signals were visualized with ECL reagent (GE Healthcare).
Co-IP Assays
For experiments represented in Figures 5A and 7A, no phytohormone treatment was done. For those shown in Figure 7B, 1-week–old OE17 plants were washed several times with water. Then, they were treated as described in Lin et al., (2015). After phytohormone treatment, the seedlings were frozen in liquid nitrogen for further analysis.
For Co-IP assays, total proteins were extracted from 1-week–old te and OE17 seedlings in radioimmunoprecipitation assay buffer (Lin et al., 2015). Then 200 μL of His•Bind resin (Novagen) was incubated at 4°C with 800 μL of te or OE17 plant extracts (containing 3-mg total proteins) for each assay. The mixture was gently shaken at 4°C for 15 min and loaded on the His•Bind Columns (Novagen). Then the columns were washed two times with 1 mL of radioimmunoprecipitation assay buffer. The total protein extracts and final resins were resolved in 1×SDS-PAGE sample buffer and immunoblots were conducted with anti-His, anti-R10, anti-MOC1, or anti-OsSHR1 antibodies.
EdU- and mPS-PI Analyses
For EdU- and mPS-PI analysis of root, 3-d–old seedlings were treated in half strength MS liquid medium by ethanol (CK), 1 µM of ABA, and 1 μM of GA3 for 3 d under 10-h 25°C dark/14-h 30°C light (provided by white-light LED tubes [400 nm to 700 nm, 250 μmol m−2 s−1 photocycle) with 70% relative humidity. Then these seedlings were incubated in half strength MS liquid medium supplemented with 5 μM of EdU (Invitrogen) and grown for 6 to 18 h. The roots were cut and pre-fixed for 15 min in 1.6 mL of fixative (45%[v/v] [95% ethanol]+45% [v/v] methanol +10% [v/v] acetic acid) and then in (40% [v/v] double-distilled water +50% [v/v] methanol +10% [v/v] acetic acid)at 4°C for at least 12 h; after that, the samples were washed for ≥15 min each in 70% (v/v) ethanol for three times and cleared for 45 min to 60 min (for these roots treated by ABA), 30 min to 45 min (for these roots treated by ethanol), or 15 min to 20 min (for these roots treated by GA) in 600 μL to 800 μL 0.01% (v/v) 84 disinfectant (effective chlorine content ≥ 5% [w/v]) with gentle shaking at room temperature. Cleared samples were washed for 15 min in water for three times. Then the samples were incubated at 37°C for ≥ 24 h in alpha-amylase (Sigma-Aldrich) and 0.6 mg/mL in phosphate buffer at pH 7.0, containing 2 mM of NaCl and 0.25 mM of CaCl2. Then the samples were stained in solution containing 10 mM of Alexa 488-azide (Life Technologies/Thermo Fisher Scientific), 100 mM of Tris, 1 mM of CuSO4, and 100 mM of ascorbic acid at pH 8.5 according to the manufacturer’s methods. Then mPS-PI staining was performed according to a method reported in Truernit et al. (2008).
For the EdU-staining analysis of the AM and shoot apical meristem, the seedlings with three leaves were treated in 400 mL of half strength MS liquid medium with 4 μL of ethanol (CK), 1 μM of ABA, and 1 μM of GA3 for 6 d under a 10-h 25°C dark/14-h 30°C light photo-cycle. Then these seedlings were incubated in half strength MS liquid medium supplemented with 5 μM of EdU (Invitrogen) and grown for 6 h to 18 h. After the treatments, the outer leaf sheaths of the seedlings were peeled off and the bases of seedlings were fixed in Formalin–acetic acid–alcohol fixative for 24 h. Then the samples were washed three times in water and stained with Alexa 488-azide (Life Technologies/Thermo Fisher Scientific) according to the manufacturer’s methods. After the staining, the samples were dehydrated in 30% (v/v), 50% (v/v), 70% (v/v), 80% (v/v), 90% (v/v), and 100% (v/v) ethanol, then cleared three times in a mixture containing an equal volume of ethanol and methyl salicylate and four times in methyl salicylate. The completely transparent samples were examined using a laser confocal scanning microscope (model no. LSM 700; Zeiss Microsystems).
The AM size, RM size, and EdU-stained cell number in RM were measured using the software ImageJ (http://rsb.info.nih.gov/ij/index.html). Fifteen shoot bases were used for the analysis of AM size and 15 roots were used for analysis of RM size and EdU-stained cell number in RM.
Statistics
Statistical analyses were performed with the software Prism (v.7; GraphPad; https://www.graphpad. com/). Statistical tests, sample sizes, and P-values are described in each figure legend. Detailed descriptions of statistical analyses are presented in the Supplemental File.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: TE (LOC_Os03g03150 or Os03g0123300), OsPYL/R10 (LOC_Os10g42280 or Os10g0573400), OsSHR1 (LOC_Os07g39820 or Os07g0586900), MOC1 (LOC_Os06g40780 or Os06g0610350), OsABA1 (LOC_Os04g37619 or Os04t0448900), LTN1 (LOC_Os08g44510 or Os08g0559300), SD1 (LOC_Os01g66100 or Os01g0883800), D18 (LOC_Os01g08220 or Os01g0177400), and KS1 (LOC_Os04g52230.1 or Os04g0611800).
Supplemental Data
Supplemental Figure 1. Root phenotypes of wild-type, te, and OE17 seedlings treated with different doses of GA3 or ABA (supports Figures 2A to 2D).
Supplemental Figure 2. The mutation of OsABA1 by CRISPR/CAS9 technology impairs root growth (supports Figure 2E).
Supplemental Figure 3. APC/CTE mediates the promotive effect of weaker GA signal on RM size (supports Figure 2A).
Supplemental Figure 4. APC/CTE mediates the opposite effects of ABA and GA on the AM size (supports Figures 2F and 2G).
Supplemental Figure 5. A conserved D-Box in the C-terminal of SHR-like proteins (supports Figures 5A to 5D).
Supplemental Figure 6. The mutation of OsSHR1 by CRISPR/CAS9 technology impairs root growth (supports Figures 5A to 5E).
Supplemental Figure 7. OsSHR1-mD-His with a D-box mutation (RSLL→aSLa) is stable in planta and can increase root growth (supports Figure 5E).
Supplemental Figure 8. OsSHR1 is critical for maintaining RM size (supports Figures 5A to 5E).
Supplemental Figure 9. OsSHR1-mD-His transgenic line is insensitive to GA’s inhibition of root growth (supports Figures 5A to 5G).
Supplemental Figure 10. OsSHR1 RNAi in the te mutant impairs root growth (supports Figures 5A to 5G).
Supplemental Figure 11. Strong GA signaling represses root growth and tillering (supports Figure 8).
Supplemental Figure 12. Moderate weakening of GA signaling promotes root growth but represses plant height while severe weakening of GA signaling represses both root growth and plant height (supports Figure 8).
Supplemental Figure 13. Schematic diagram of a dual role of APC/CTE in regulating RM cell division and cell differentiation (modified from Wäsch et al., 2010 with some modifications; supports Figure 8).
Supplemental Table 1. Primers used for construction of transgenic plasmids.
Supplemental Table 2. Primers used for construction of protein expression plasmids.
Supplemental Table 3. Primers used in RT-PCR.
Supplemental File. Detailed descriptions of statistical analyses.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Tao Zhang (Institute of Rice and Sorghum, Sichuan Academy of Agricultural Sciences, Chengdu Sichuan province, China) for providing the ltn1 mutant seeds and the Core Facility Platform, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, for their assistance with confocal imaging. This work was supported by the National Key Research and Development Program of China (2016YFD0100901), the National Natural Science Foundation (91735304, 31201271, and 31671769), the National Natural Science Foundation of China–Guangdong Province Joint Program (U1701232), and the Chinese Academy of Agricultural Sciences project (CAAS-ZDXT2018001).
AUTHOR CONTRIBUTIONS
J.-M.W. supervised the project; J.-M.W., H.W., and Q.L. designed research and wrote the article; Q.L. and Z.-J.Z. performed most of the experiments; Q.L. and M.F. performed RNA in situ hybridization; F.W., M.F., and J.W. performed some of the RT-qPCR analysis, immunoblotting analysis, and hormone treatment experiments; Y.S., W.C., X.Z., X.G., Y.R., and S.Z. generated the transgenic plants; C.L., J.W., Z.-C.Z., and Z.-J.C. cultivated the transgenic plants in the field.
References
- Agrawal G.K., Yamazaki M., Kobayashi M., Hirochika R., Miyao A., Hirochika H.(2001). Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel ostatc gene. Plant Physiol. 125: 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloban M., Vanstraelen M., Tarayre S., Reuzeau C., Cultrone A., Mergaert P., Kondorosi E.(2013). Complementary and dose-dependent action of AtCCS52A isoforms in endoreduplication and plant size control. New Phytol. 198: 1049–1059. [DOI] [PubMed] [Google Scholar]
- Cappell S.D., Mark K.G., Garbett D., Pack L.R., Rape M., Meyer T.(2018). EMI1 switches from being a substrate to an inhibitor of APC/CCDH1 to start the cell cycle. Nature 558: 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.W., Lim J.(2016). Control of asymmetric cell divisions during root ground tissue maturation. Mol. Cells 39: 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Levesque M.P., Vernoux T., Jung J.W., Paquette A.J., Gallagher K.L., Wang J.Y., Blilou I., Scheres B., Benfey P.N.(2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R.(2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N.(1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433. [DOI] [PubMed] [Google Scholar]
- Edgar B.A., Zielke N., Gutierrez C.(2014). Endocycles: A recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 15: 197–210. [DOI] [PubMed] [Google Scholar]
- Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S.Y., Cutler S.R., Sheen J., Rodriguez P.L., Zhu J.K.(2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D., Li C., Mohan H., Probst N.(2013). Gibberellins and abscisic acid signal crosstalk: Living and developing under unfavorable conditions. Plant Cell Rep. 32: 1007–1016. [DOI] [PubMed] [Google Scholar]
- Gong X., Flores-Vergara M.A., Hong J.H., Chu H., Lim J., Franks R.G., Liu Z., Xu J.(2016). SEUSS integrates gibberellin signaling with transcriptional inputs from the SHR-SCR-SCL3 module to regulate middle cortex formation in the Arabidopsis root. Plant Physiol. 170: 1675–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Grandío E., Pajoro A., Franco-Zorrilla J.M., Tarancón C., Immink R.G.H., Cubas P.(2017). Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. USA 114: E245–E254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schafer E., Muller D., Herrero R., Schmitz G., Theres K.(2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17: 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., et al. (2017). Genetic analysis and gene mapping of a low-tiller number and high plant height mutant in rice. Mol. Plant Breed. 15: 3524–3530. [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N.(2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567. [DOI] [PubMed] [Google Scholar]
- Humplík J.F., Bergougnoux V., van Volkenburgh E.(2017). To stimulate or inhibit? That is the question for the function of abscisic acid. Trends Plant Sci. 22: 830–841. [DOI] [PubMed] [Google Scholar]
- Ikeda A., Ueguchi-Tanaka M., Sonoda Y., Kitano H., Koshioka M., Futsuhara Y., Matsuoka M., Yamaguchi J.(2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N., Itoh J., Morikami A., Nagato Y., Matsuoka M.(2003). The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J. 36: 45–54. [DOI] [PubMed] [Google Scholar]
- Khush G.S.(1995). Breaking the yield frontier of rice. GeoJournal 35: 329–332. [Google Scholar]
- Komatsu K., Maekawa M., Ujiie S., Satake Y., Furutani I., Okamoto H., Shimamoto K., Kyozuka J.(2003). LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 100: 11765–11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotogány E., Dudits D., Horváth G.V., Ayaydin F.(2010). A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Rabin Z., Li Z., Masson P.H., Day C.D.(2009). FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol. 149: 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.A., et al. (2016). Interplay between ABA and GA modulates the timing of asymmetric cell divisions in the Arabidopsis root ground tissue. Mol. Plant 9: 870–884. [DOI] [PubMed] [Google Scholar]
- Li J., Zhao Y., Chu H., Wang L., Fu Y., Liu P., Upadhyaya N., Chen C., Mou T., Feng Y., Kumar P., Xu J.(2015). SHOEBOX modulates root meristem size in rice through dose-dependent effects of gibberellins on cell elongation and proliferation. PLoS Genet. 11: e1005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zhang P.(2009). The function of APC/CCdh1 in cell cycle and beyond. Cell Div. 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., et al. (2003). Control of tillering in rice. Nature 422: 618–621. [DOI] [PubMed] [Google Scholar]
- Liao Z., Yu H., Duan J., Yuan K., Yu C., Meng X., Kou L., Chen M., Jing Y., Liu G., Smith S.M., Li J.(2019). SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat. Commun. 10: 2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., et al. (2012). Rice APC/CTE controls tillering by mediating the degradation of MONOCULM 1. Nat. Commun. 3: 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Wu F., Sheng P., Zhang Z., Zhang X., Guo X., Wang J., Cheng Z., Wang J., Wang H., Wan J.(2015). The SnRK2-APC/CTE regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 6: 7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S.F., Yang S.Y., Chen K.T., Hsing Y.I., Zeevaart J.A., Chen L.J., Yu S.M.(2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E.(2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Melcher K., et al. (2009). A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk J.A., Thelen J.J.(2008). Biochemical approaches for discovering protein–protein interactions. Plant J. 53: 597–609. [DOI] [PubMed] [Google Scholar]
- Miyazono K., et al. (2009). Structural basis of abscisic acid signalling. Nature 462: 609–614. [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno M.A., Sozzani R., Yardımcı G.G., Petricka J.J., Vernoux T., Blilou I., Alonso J., Winter C.M., Ohler U., Scheres B., Benfey P.N.(2015). Transcriptional control of tissue formation throughout root development. Science 350: 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkey T.J., Evans M.L., Kuzmanoff K.M.(1983). The kinetics of abscisic acid action on root growth and gravitropism. Planta 157: 150–157. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P.N.(2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311. [DOI] [PubMed] [Google Scholar]
- Nishimura N., Hitomi K., Arvai A.S., Rambo R.P., Hitomi C., Cutler S.R., Schroeder J.I., Getzoff E.D.(2009). Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326: 1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261. [DOI] [PubMed] [Google Scholar]
- Perilli S., Di Mambro R., Sabatini S.(2012). Growth and development of the root apical meristem. Curr. Opin. Plant Biol. 15: 17–23. [DOI] [PubMed] [Google Scholar]
- Rogers E.D., Benfey P.N.(2015). Regulation of plant root system architecture: Implications for crop advancement. Curr. Opin. Biotechnol. 32: 93–98. [DOI] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B.(2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J., Dupeux F., Round A., Antoni R., Park S.Y., Jamin M., Cutler S.R., Rodriguez P.L., Márquez J.A.(2009). The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668. [DOI] [PubMed] [Google Scholar]
- Sasaki A., Ashikari M., Ueguchi-Tanaka M., Itoh H., Nishimura A., Swapan D., Ishiyama K., Saito T., Kobayashi M., Khush G.S., Kitano H., Matsuoka M.(2002). Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 416: 701–702. [DOI] [PubMed] [Google Scholar]
- Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Jeong D.H., An G., Kitano H., Ashikari M., Matsuoka M.(2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898. [DOI] [PubMed] [Google Scholar]
- Schumacher K., Schmitt T., Rossberg M., Schmitz G., Theres K.(1999). The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 96: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P.(2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21: R338–R345. [DOI] [PubMed] [Google Scholar]
- Su’udi M., Cha J.Y., Jung M.H., Ermawati N., Han C.D., Kim M.G., Woo Y.M., Son D.(2012). Potential role of the rice OsCCS52A gene in endoreduplication. Planta 235: 387–397. [DOI] [PubMed] [Google Scholar]
- Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., Kaiser P.(2006). A tandem affinity tag for two-step purification under fully denaturing conditions: Application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell. Proteomics 5: 737–748. [DOI] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthélémy J., Palauqui J.C.(2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T.Y., Hsing Y.I., Kitano H., Yamaguchi I., Matsuoka M.(2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Motoyuki A., Matsuoka M.(2007). Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58: 183–198. [DOI] [PubMed] [Google Scholar]
- Wang B., Smith S.M., Li J.(2018). Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69: 437–468. [DOI] [PubMed] [Google Scholar]
- Wang F., Zhu D., Huang X., Li S., Gong Y., Yao Q., Fu X., Fan L.M., Deng X.W.(2009). Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J.(2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59: 253–279. [DOI] [PubMed] [Google Scholar]
- Wäsch R., Robbins J.A., Cross F.R.(2010). The emerging role of APC/CCdh1 in controlling differentiation, genomic stability and tumor suppression. Oncogene 29: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Lee C.M., Hayashi T., Price S., Divol F., Henry S., Pauluzzi G., Perin C., Gallagher K.L.(2014). A plausible mechanism, based upon short-root movement, for regulating the number of cortex cell layers in roots. Proc. Natl. Acad. Sci. USA 111: 16184–16189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller J.W., Helariutta Y., Fukaki H., Malamy J.E., Benfey P.N.(2000). Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603.10631180 [Google Scholar]
- Xing Y., Zhang Q.(2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61: 421–442. [DOI] [PubMed] [Google Scholar]
- Xu C., Wang Y., Yu Y., Duan J., Liao Z., Xiong G., Meng X., Liu G., Qian Q., Li J.(2012). Degradation of MONOCULM 1 by APC/CTAD1 regulates rice tillering. Nat. Commun. 3: 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S.(2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251. [DOI] [PubMed] [Google Scholar]
- Yao C., Finlayson S.A.(2015). Abscisic acid is a general negative regulator of Arabidopsis axillary bud growth. Plant Physiol. 169: 611–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P., Fan H., Hao Q., Yuan X., Wu D., Pang Y., Yan C., Li W., Wang J., Yan N.(2009). Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 16: 1230–1236. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Obata T., Feil R., Lunn J.E., Fujita Y., Yamaguchi-Shinozaki K., Fernie A.R.(2019). The role of abscisic acid signaling in maintaining the metabolic balance required for Arabidopsis growth under nonstress conditions. Plant Cell 31: 84–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R., Sui N., Barnhill B., Hsieh W.P., Hu J., Shabanowitz J., Boyce M., Olszewski N.E., Zhou P., Hunt D.F., Sun T.P.(2017). The Arabidopsis o-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Nat. Chem. Biol. 13: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]