ODR1 negatively controls seed dormancy by interacting with the transcription factor bHLH57 and preventing its induction of NCED6 and NCED9 expression and ABA biosynthesis.
Abstract
The control of seed dormancy by abscisic acid (ABA) has been extensively studied, but the underlying mechanism is not fully understood. Here, we report the characterization of two ABA-related seed dormancy regulators in Arabidopsis (Arabidopsis thaliana): ODR1 (for reversal of rdo5), an ortholog of the rice (Oryza sativa) Seed dormancy4 (Sdr4), and the basic helix-loop-helix transcription factor bHLH57. ODR1, whose transcript levels are directly suppressed by the transcription factor ABA INSENSITIVE3 (ABI3), negatively regulates seed dormancy by affecting ABA biosynthesis and ABA signaling. By contrast, bHLH57 positively regulates seed dormancy by inducing the expression of the genes 9-CIS-EPOXYCAROTENOID DIOXYGENASE6 (NCED6) and NCED9, which encode ABA biosynthetic enzymes, and thus leads to higher ABA levels. ODR1 interacts with bHLH57 and inhibits bHLH57-modulated NCED6 and NCED9 expression in the nucleus. bhlh57 loss-of-function alleles can partially counteract the enhanced NCED6 and NCED9 expression seen in odr1 mutants and can therefore rescue their associated hyper-dormancy phenotype. Thus, we identified a novel ABI3-ODR1-bHLH57-NCED6/9 network that provides insights into the regulation of seed dormancy by ABA biosynthesis and signaling.
INTRODUCTION
Dormancy prevents germination when seeds are exposed to short but temporarily favorable periods before the return of adverse conditions and thus delays seedling establishment until the start of the growing season. Dormancy therefore plays a vital role in plant survival and evolution (Linkies et al., 2010; Née et al., 2017b). Seed dormancy is imposed during seed maturation and released during after-ripening or by hydration at specific temperatures. Precise control of seed dormancy in crops results in fast and uniform germination after sowing and prevents preharvest sprouting, which would otherwise negatively impact agricultural production (Gubler et al., 2005).
Seed dormancy is a complex trait influenced by genetic and environmental factors (Graeber et al., 2012; Penfield and MacGregor, 2017). Previous studies revealed that phytohormones, including abscisic acid (ABA), gibberellins (GAs), ethylene, strigolactones, and brassinosteroids, all play important roles in the control of seed dormancy (Seo et al., 2006; Shu et al., 2016a). Among these hormones, ABA and GA have central and antagonistic roles: ABA enhances dormancy, whereas GA stimulates germination. The roles of ABA and GA biosynthesis and signal transduction in the control of seed dormancy and germination have been intensively studied in the past decades (Gubler et al., 2005; Lefebvre et al., 2006; Née et al., 2017b).
During seed maturation, endogenous ABA gradually accumulates to enforce dormancy (Kanno et al., 2010). Cleavage of the ABA precursors 9-cis-violaxanthin and 9-cis-neoxanthin into the intermediate xanthoxin by 9-cis-epoxycarotenoid dioxygenase (NCED) is considered the key rate-limiting step in ABA biosynthesis (Nambara and Marion-Poll, 2005). Of the nine Arabidopsis (Arabidopsis thaliana) NCED genes, seed-specific NCED6 and NCED9 significantly contribute to ABA biosynthesis during seed development. The corresponding nced6 nced9 double mutant shows a significant decrease of seed ABA content and concomitant reduced seed dormancy (Lefebvre et al., 2006), suggesting that both NCED6 and NCED9 are important for the establishment and maintenance of seed dormancy. A tight control of NCED6 and NCED9 expression is therefore vital for seed dormancy. Previous studies have reported that several transcription factors (TFs) control the expression of NCED6 and NCED9 during seed development, such as ABSCISIC ACID INSENSITIVE4 (ABI4), DEHYDRATION-RESPONSIVE ELEMENT BINDING FACTOR2C (DREB2C), and MYB96. These TFs activate NCED6 and NCED9 expression by binding to their promoters (Je et al., 2014; Lee et al., 2015; Shu et al., 2016b). Aside from biosynthesis, ABA degradation also plays an important role in determining endogenous ABA content and release of dormancy. Among four cytochrome P450 CYP707A family (CYP707A1 to CYP707A4) members in Arabidopsis, CYP707A2 is considered to be the major factor that executes ABA degradation in mature and hydrated seeds (Kushiro et al., 2004). The cyp707a2 mutant over-accumulates ABA and shows stronger seed dormancy (Okamoto et al., 2006).
ABA signal transduction also influences seed dormancy, acting primarily through a cascade that comprises ABA receptors (encoded by PYRABACTIN RESISTANCE1 [PYR1]/PYR1-like [PYL] 1-13, also known as REGULATORY COMPONENTS OF ABA RECEPTOR [RCAR]), type 2C protein phosphatases (PP2Cs), and sucrose non-fermenting 1 related protein kinase (SnRK; Ma et al., 2009; Park et al., 2009; Cutler et al., 2010; Miyakawa et al., 2013). Loss-of-function mutants in ABA signaling show decreased ABA sensitivity, resulting in early release of seed dormancy (Park et al., 2009; Fuchs et al., 2014). ABI3 is a vital ABA-responsive TF that plays a central role in seed maturation and primary dormancy establishment in Arabidopsis. Mutations in ABI3 lead to reduced dormancy and premature seed germination (Nambara et al., 1995). ABI3 contains a B3 domain that can physically associate with the RY motif found in the promoter of downstream genes, such as SOMNUS (SOM) and STAY-GREEN1 (SGR1) and SGR2 (Ezcurra et al., 2000; Mönke et al., 2004; Park et al., 2011; Delmas et al., 2013). In addition, ABI3 interacts with the important ABA-responsive TFs ABI4 and ABI5 to establish seed dormancy (Söderman et al., 2000; Lopez-Molina et al., 2002).
Apart from phytohormones, seed dormancy is also controlled by several genes originally identified as major quantitative trait loci (QTLs) such as DELAY OF GERMINATION1 (DOG1), DOG18/REDUCED DORMANCY5 (RDO5), and DOG6 (Bentsink et al., 2010). DOG1 is a major seed dormancy factor in Arabidopsis (Bentsink et al., 2006). DOG1 protein levels are tightly correlated with dormancy levels in freshly harvested seeds, with higher DOG1 levels causing delayed germination (Nakabayashi et al., 2012). The basic Leu zipper TFs bZIP67 and Ethylene Response Factor12 (ERF12) bind to the promoter of and control the expression of DOG1 and seed dormancy in response to cool temperatures and ethylene exposure, respectively (Bryant et al., 2019; Li et al., 2019). DOG1 transcript levels are also modulated by its antisense transcript asDOG1 (Fedak et al., 2016). The DOG1 protein interacts with ABA-HYPERSENSITIVE GERMINATION1 (AHG1) and AHG3, which are PP2Cs that belong to the same clade as those interacting with the ABA receptors. Therefore, these PP2Cs are considered a converging point for DOG1- and ABA-dependent dormancy control pathways (Née et al., 2017a; Nishimura et al., 2018). Another seed dormancy factor, RDO5/DOG18, was identified both in a mutagenesis screen and by QTL mapping. RDO5 was shown to control seed dormancy independently of ABA, and a transcriptome analysis suggested that RDO5 does so by controlling the expression of the genes encoding the PUMILIO RNA binding proteins APUM9 and homologs, revealing a posttranscriptional dormancy pathway (Xiang et al., 2014, 2016). RDO5 belongs to the PP2C family of protein phosphatases but is found in a clade distinct from AHG1 and AHG3. In fact, RDO5 functions as a pseudo-phosphatase because it lacks phosphatase activity (Xiang et al., 2016). Interestingly, the RDO5 and DOG1 proteins interact in Arabidopsis seeds (Née et al., 2017a), although the reason or the role of such interaction in seed dormancy is unknown.
In order to dissect the RDO5-mediated seed dormancy network, we performed a suppressor mutagenesis screen with the low dormancy mutant rdo5-2. Here, we report the identification of one such suppressor (globally called odr, for reversal of the rdo phenotype): odr1. Mutations in ODR1 cause stronger seed dormancy. Expression of ODR1 is repressed by ABI3, and ODR1 negatively affects the expression of NCED6 and NCED9 and ABA content in freshly harvested seeds. Furthermore, ODR1 interacts with bHLH57 and prevents bHLH57-mediated induction of NCED6 and NCED9 expression. In agreement, the odr1-2 bhlh57 double mutant decreased the expression of NCED6 and NCED9 compared to bhlh57 single mutants, while rescuing the hyper-dormancy phenotype of odr1-2. We therefore discovered a new seed dormancy pathway that includes ABI3, ODR1, bHLH57, NCED6, and NCED9. Because the rice (Oryza sativa) ortholog of ODR1 was previously identified as a QTL in rice, our work also provides a molecular link between presprouting research in rice and seed dormancy in Arabidopsis.
RESULTS
The Seed-Specific Protein ODR1 Negatively Controls Seed Dormancy
We had previously reported the cloning and initial characterization of the positive dormancy factor RDO5, which controls seed dormancy without influencing ABA metabolism or signal transduction. Loss-of-function rdo5 mutant alleles cause strongly reduced seed dormancy (Xiang et al., 2014, 2016). Seeking to uncover the function of RDO5 in seed dormancy regulation, we performed a γ-ray mutagenesis screen with rdo5-2, a T-DNA insertion mutant with substantially reduced dormancy duration (Xiang et al., 2014). We identified six mutants that suppressed the rdo5-2 dormancy phenotype and named them odr1 to odr6 (for reversal of the rdo phenotype). The odr1 mutant exerted the most significant effect on seed dormancy and was therefore selected for further characterization. We used bulked segregant analysis–based sequencing to identify mutations that might be responsible for the phenotype and identified an 8-bp deletion in At1g27461 that caused a reading frame shift leading to a premature stop codon (Supplemental Figures 1A and 1B). To confirm the identity of At1g27461 as ODR1, we obtained an independent mutant in the gene, the homozygous T-DNA insertion mutant SALK_022729, which we named odr1-2. This mutant carried a T-DNA insertion in the single ODR1 exon and lacked full-length ODR1 transcript (Supplemental Figures 1C and 1D). The odr1-2 allele is therefore presumed to be a null allele. Germination assays showed that odr1-2 had reduced germination compared to the wild-type Columbia (Col-0; Figures 1A and 1B). We also introduced the odr1-2 T-DNA insertion into rdo5-2 via crossing and conducted germination assays with the double mutant: it showed lower germination percentage than rdo5-2, indicating that odr1-2 also effectively suppressed the dormancy phenotype seen in rdo5-2 (Figure 1A). Additionally, to confirm the repressive role of ODR1 in seed dormancy, we generated overexpression lines in the odr1-2 background by placing a copy of ODR1 under the control of the cauliflower mosaic virus 35S promoter (Supplemental Figure 1D). Germination assays showed that homozygous 35Spro:ODR1/odr1-2 seeds had similar dormancy rates as Col-0 (Figure 1B). These results established that constitutive expression of ODR1 rescues the enhanced dormancy phenotype of odr1-2 and that ODR1 negatively impacts seed dormancy.
Figure 1.
ODR1 Is a Nucleoprotein That Reduces Seed Dormancy and Is Predominantly Expressed in Mature Dry Seeds.
(A) Germination percentages of Col-0, rdo5-2, odr1-1 rdo5-2, odr1-2, and rdo5-2 odr1-2 seeds after 1 week of storage. Values are means ± sd of three independent batches of seeds per genotype. **P < 0.01 by ANOVA analysis.
(B) Germination percentages after different periods of dry storage of Col-0, odr1-2, and two overexpression lines containing a 35Spro:ODR1 insertion in the odr1-2 background. Values are means ± sd of three independent batches of seeds per genotype.
(C) Subcellular localization of ODR1-YFP in N. benthamiana leaves. YFP is used as the control. Bars = 50 µm.
(D) RT-qPCR analysis of ODR1 transcript levels in various organs and developing siliques and mature seeds in Arabidopsis. The expression values were normalized to PP2A. Values are means ± sd of three biological repeats. DAP, days after pollination; HAH, hours after hydration.
The ODR1 gene consists of a single exon of 1065 bp, encoding a 355–amino acid protein. Protein sequence alignment and phylogenetic analysis demonstrated that ODR1 and its homologues are conserved within angiosperms (Supplemental Figures 2A and 2B). ODR1 was previously described as DROUGHT RESPONSIVE GENE, a seed-specific gene induced under drought conditions (Moon et al., 2016). The drg mutant was shown to be more sensitive to a number of abiotic stresses, including osmotic stress, drought, and freezing. The putative rice ortholog of DRG, the zinc finger protein Sdr4, is a positive factor of seed dormancy (Sugimoto et al., 2010). In another study, germination of freshly harvested drg/SEED DORMANCY4-LIKE (Atsdr4L) mutant seeds were found to be insensitive to exogenous GA (Cao et al., 2020). SDR is already assigned to designate short-chain dehydrogenase reductases in Arabidopsis, while DRG can also stand for developmentally regulated GTP binding proteins. We therefore propose to rename DRG/AtSDR4 as ODR1 to avoid confusion and to adopt a single gene identifier representative of its underlying function.
We analyzed the subcellular localization of ODR1 by transient expression in tobacco (Nicotiana benthamiana) leaves. The ODR1-yellow fluorescent protein (YFP) fusion protein was exclusively localized to the nucleus (Figure 1C). We next evaluated ODR1 expression levels in different Arabidopsis tissues by RT-qPCR: ODR1 transcript was undetectable in roots, stems, leaves, or flowers, but it gradually increased during seed maturation and reached its highest level in seeds 20 d after pollination, while it sharply declined in hydrated seed (Figure 1D). This pattern of expression is consistent with the expression data for ODR1 in the Arabidopsis eFP Browser (Supplemental Figure 3). Taken together, ODR1 is specifically expressed in seeds and encodes a nuclear protein that is involved in the control of dormancy.
ODR1 Strongly Impacts ABA-Mediated Dormancy
ABA plays a crucial role in the control of seed dormancy and germination. To test whether the enhanced dormancy of odr1-2 was associated with ABA, we first measured the ABA content in freshly harvested seeds from different genotypes, including Col-0, odr1-2, 35Spro:ODR1/odr1-2, rdo5-2, and rdo5-2 odr1-2. ABA levels in odr1-2 freshly harvested seeds were ∼40% higher compared to those in Col-0 and the overexpression lines (Figure 2A), which is consistent with the stronger dormancy seen in odr1-2 seeds. The rdo5-2 odr1-2 double mutant also showed higher ABA levels compared to the rdo5-2 single mutant, which itself was similar to Col-0 in freshly harvested seeds (Figure 2B), as previously reported by Xiang et al. (2014). These results suggest that ODR1 might influence ABA metabolism. Next, we evaluated the germination behavior of freshly harvested seeds of Col-0 and odr1-2 in the presence of the ABA biosynthesis inhibitor fluridone. The odr1-2 mutant responded more strongly to fluridone compared to Col-0 by exhibiting lower germination percentage, suggestive of heightened ABA sensitivity (Figure 2C). Overall, these results demonstrate that the stronger seed dormancy in odr1-2 is largely caused by a high level of endogenous ABA in dry seeds and de novo ABA synthesis during seed hydration. Finally, we evaluated the ABA sensitivity of Col-0 and odr1-2 after-ripened seeds. Seeds of both genotypes germinated fully and synchronously after stratification on half-strength Murashige and Skoog (MS) growth medium in the absence of ABA. However, odr1-2 seeds were more sensitive to ABA as shown by reduced germination percentage when the medium was supplemented with 0.5 μM ABA. This suggests that loss of function of ODR1 affects ABA sensitivity of seeds (Figure 2D).
Figure 2.
ODR1 Affects ABA Content and Seed ABA Sensitivity.
(A) ABA content in Col-0, odr1-2, and 35Spro:ODR1/odr1-2-2 freshly harvested dry seeds. Values are means ± sd of three biological repeats. *P < 0.05 or **P < 0.01 by Student’s t test.
(B) ABA content in Col-0, rdo5-2, and rdo5-2 odr1-2 freshly harvested seeds. Values are means ± sd of three biological repeats. **P < 0.01 by Student’s t test.
(C) Germination percentages of Col-0 and odr1-2 seeds after 1 week of storage, tested in the presence of 0.05% ethanol (control) or 0.05% ethanol plus the indicated concentrations of fluridone. Values are means ± sd of three independent batches of seeds per genotype.
(D) Germination percentages of Col-0 and odr1-2 freshly harvested seeds on 1/2 MS medium supplemented with 0 or 0.5 µM ABA. Values are means ± sd of three independent batches of seeds per genotype. Significance analysis was performed between odr1-2 and Col-0 with 0 µM ABA treatment, and 0.5 µM ABA treatment. *P < 0.05 or **P < 0.01 by Student’s t test.
(E) RT-qPCR analysis of NCEDs and CYP707As transcript levels in Col-0 and odr1-2 freshly harvested dry seeds and seeds hydrated for 6 h. The expression values were normalized to PP2A. Values are means ± sd of three biological repeats. *P < 0.05 or **P < 0.01 by Student’s t test.
(F) Germination percentages of Col-0, rdo5-2, cyp707a2-1, and rdo5-2 cyp707a2-1 freshly harvested seeds. Values are means ± sd of three independent batches of seeds per genotype.
Since ABA content was higher in odr1-2 seeds compared to Col-0, we evaluated the expression levels of pivotal ABA metabolism genes (Supplemental Figure 4), including NCEDs (NCED2, NCED3, NCED5, NCED6, and NCED9, involved in ABA biosynthesis) and CYP707As (CYP707A1 to CYP707A4, linked to ABA degradation) in Col-0 and odr1-2 freshly harvested and in seeds that had been hydrated for 6 h. Nearly all of the NCEDs (except NCED5) and CYP707A2 were significantly upregulated in dry and/or hydrated seeds in odr1-2 compared with Col-0 (Figure 2E). Differential expression of NCEDs could well explain the increased ABA content of odr1-2 freshly harvested seeds, while the elevated expression levels of CYP707A2 might indicate a feedback reaction to the increased ABA content in odr1-2 seeds. However, ODR1 does not activate gene expression by direct binding to promoters, as we failed to detect ODR1 binding ability to any of the NCEDs and CYP707As promoters in yeast one-hybrid assays (Supplemental Figure 5). We also found that ODR1 indirectly regulated DREB2C and ABI4 gene expression in dry and/or hydrated seeds but did not physically interact with the encoded proteins (Supplemental Figure 6). Taken together, these results suggest that ODR1 controls seed dormancy by affecting the expression of genes involved in ABA metabolism but is unlikely to behave as a TF.
We had shown previously that RDO5 controls seed dormancy in an ABA-independent manner. However, loss-of-function mutants of odr1-1 and odr1-2 both suppressed the weak dormancy phenotype of rdo5-2 in double mutant combinations, which indicated that the ODR1-mediated ABA metabolism pathway was epistatic to RDO5 for seed dormancy. To further confirm this relationship, we generated the rdo5-2 cyp707a2 double mutant by crossing. Germination assays showed that the rdo5-2 cyp707a2 double mutant had a stronger dormancy compared to rdo5-2 (Figure 2F). This result demonstrated that the cyp707a2 mutant, which causes an over-accumulation of ABA in seeds, is epistatic to rdo5-mediated reduced seed dormancy. Surprisingly, the dormancy level of rdo5-2 cyp707a2 was even higher than that of the cyp707a2 single mutant. This might be due to a feedback response to the rdo5 mutation in order to keep seeds in a dormant status, which becomes noticeable in the cyp707a2 mutant background. This is supported by increased transcript levels following 6 h of seed hydration in the rdo5-2 background for two PYR/PYL/RCAR genes (At4g17870 and At2g38310) and an SnRK2 gene (At1g78290) involved in ABA signaling (Xiang et al., 2014). We also found that DOG1, the core factor controlling seed dormancy, was upregulated in odr1-2, but again without direct interaction between the encoded protein and ODR1. Higher DOG1 expression might also contribute to the hyper-dormancy phenotype of the odr1-2 mutant (Supplemental Figure 7).
Overall, ODR1 controls seed dormancy at least partially through modulation of ABA biosynthesis, which acts downstream of the RDO5-mediated dormancy pathway.
ODR1 Is a Direct Target of ABI3
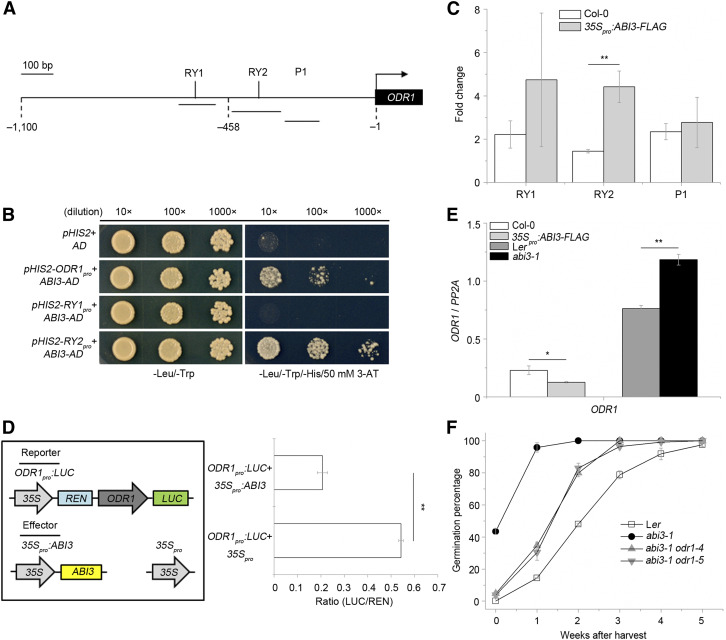
A previous chromatin immunoprecipitation (ChIP) followed by hybridization to tiling arrays (ChIP-chip) and transcriptome analysis predicted that ODR1 was one of the 98 direct targets of ABI3 (Mönke et al., 2012). Based on this finding, we analyzed the ODR1 promoter sequence and found two potential RY motifs (CATGCA) known to be binding sites for ABI3 (Figure 3A). To validate the control of ODR1 expression by ABI3, we performed a yeast one-hybrid assay, whose results indicated that ABI3 specifically binds to the ODR1 promoter at the proximal RY motif (CATGCA-363), but not at the more distal motif (CATGCA-700) (Figure 3B). Furthermore, we performed ChIP followed by quantification of immunoprecipitated chromatin by qPCR with chromatin extracted from seedlings overexpressing ABI3 fused to a FLAG tag (35Spro:ABI3-FLAG; Supplemental Figure 8; Park et al., 2011). We found that the DNA fragment containing the proximal RY motif (CATGCA-363) of the ODR1 promoter was highly enriched compared to other DNA fragments (Figure 3C). To investigate how ABI3 controlled ODR1 expression, we then performed a transient expression assay using N. benthamiana leaves, and it showed that ABI3 repressed the expression of ODR1 (Figure 3D). In addition, expression levels of ODR1 in abi3-1 and 35Spro:ABI3-FLAG seeds demonstrated that overexpression of ABI3 reduced ODR1 expression, while the weak allele abi3-1 showed enhanced ODR1 expression during seed hydration compared to the wild-type control (Figure 3E). Finally, we used the genome editing technique clustered regularly interspaced short palindromic repeats and CRISPR-associated nuclease (CRISPR/Cas9) to inactivate ODR1 in an abi3-1 background. After genomic PCR verification and Sanger sequencing, three lines with different editing scars in ODR1 (containing a 1-bp insertion, a 32-bp deletion, and a 55-bp deletion, respectively) were identified and named odr1-3, odr1-4, and odr1-5 (Supplemental Figure 9). Germination assays of the wild-type Landsberg erecta (Ler), abi3-1, abi3-1 odr1-4, and abi3-1 odr1-5 showed that removal of ODR1 activity could largely suppress the weak dormancy rates of abi3-1 (Figure 3F), suggesting that ODR1 genetically acts downstream of ABI3 and that ABI3 requires functional ODR1 to control seed dormancy. Taken together, we found that ABI3, which is an essential factor of ABA signaling transduction and seed dormancy, represses ODR1 expression by binding to its promoter.
Figure 3.
ABI3 Represses ODR1 Expression.
(A) Schematic diagram of the ODR1 promoter. Three fragments were amplified for yeast one-hybrid assay: the –1- to –457-bp fragment (containing RY2 element [RY2pro]), the –458- to –1100-bp fragment (containing RY1 element [RY1pro]), and the –1- to –1100-bp fragment (ODR1pro). Three fragments for ChIP-qPCR detection (named RY1, RY2, and P1) are depicted with parallel black lines below the promoter. Bar = 100 bp.
(B) Yeast one-hybrid assay: ABI3 binds to the ODR1 promoter through the RY2-containing fragment. 3-AT (50 mM) was added in the -Leu/-Trp/-His medium. Dilution 10×, 100×, and 1000× corresponds to an OD600 of 0.1, 0.01, and 0.001, respectively.
(C) ChIP-qPCR assay: ABI3 mainly binds to the RY2 motif-containing fragment of the ODR1 promoter. Immunoprecipitation was performed using seeds hydrated for 6 h from Col-0 and 35Spro:ABI3-FLAG with anti-FLAG antibody or anti-lgG. qPCR was performed with specific primers listed in Supplemental Data Set 1. The comparison is between the nonimmune control (anti-IgG) and the immunoprecipitation (anti-FLAG). Values are means ± sd of three biological replicates. **P < 0.01 by Student’s t test.
(D) Transient expression assay: ABI3 represses ODR1 expression in N. benthamiana leaves. Firefly LUC activity was normalized to REN activity (as an internal control). Schematic representation of various constructs used in the assay is shown on the left panel. Values are means ± sd from three independent transformants for each construct. **P < 0.01 by Student’s t test.
(E) RT-qPCR analysis of ODR1 transcript levels in 35Spro:ABI3-FLAG seeds (Col-0 background) and abi3-1 mutant seeds (Ler background). Values represent the ODR1 expression levels change after 6 h of seed hydration. Values are means ± sd of three biological replicates. *P < 0.05 or **P < 0.01 by Student’s t test.
(F) Germination percentages after different periods of dry storage of the wild-type Ler, abi3-1, abi3-1 odr1-4, and abi3-1 odr1-5 seeds. Values are means ± sd of three independent batches of seeds per genotype.
ODR1 Interacts with the bHLH-Type Protein bHLH57 in the Nucleus
We performed a yeast two-hybrid screen using ODR1 as bait to identify potential interacting partners and identified bHLH57 (At4g01460), a putative bHLH-type TF. The direct physical interaction between ODR1 and bHLH57 was confirmed using a yeast two-hybrid assay using their full-length coding sequences. A yeast strain AH109 (Saccharomyces cerevisiae) containing both ODR1 and bHLH57 was able to grow on synthetic medium lacking His, Leu, and Trp and supplemented with 2 mM 3-amino-1,2,4-triazole (3-AT), in contrast to the negative controls (Figure 4A). We also performed bimolecular fluorescence complementation (BiFC) assays by cotransfecting different combinations of proteins fused to fragments of YFP into N. benthamiana leaves. Yellow fluorescence signal was detected in the nuclei of N. benthamiana cells coexpressing ODR1-YFPn and bHLH57-YFPc, while the expression of either fusion protein alone did not result in measurable YFP signal (Figure 4B). Finally, we confirmed the interaction between ODR1 and bHLH57 with pull-down experiments using total protein extracts from Escherichia coli producing bHLH57-glutathione S-transferase (GST) and ODR1-6×His. The bHLH57-GST fusion protein was able to pull down ODR1-6×His, indicating that bHLH57 interacts with ODR1 in vitro (Figure 4C). These results therefore confirm that ODR1 physically interacts with bHLH57, suggesting that they may modulate seed dormancy as a complex.
Figure 4.
ODR1 Interacts with bHLH57.
(A) Yeast two-hybrid assay: ODR1 interacts with bHLH57. 3-AT (2 mM) was added in the -Leu/-Trp/-His/5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-Gal) medium. Interaction between TTG1 and MYB5 was used as positive control. bHLH57ΔN represents a truncated bHLH57 (N terminus [1 to 111 amino acids] of bHLH57 is removed). Dilution 10×, 100×, and 1000× corresponds to an OD600 of 0.1, 0.01, and 0.001, respectively.
(B) BiFC assay: interaction between ODR1 and bHLH57 in N. benthamiana leaves. 4′,6-Diamidino-2-phenylindole (DAPI)–labeled nuclei. Bars = 20 µm.
(C) GST-bHLH57 pull-down assays: in vitro interaction between bHLH57 and ODR1. GST and GST-bHLH57 served as bait. ODR1-6×His served as prey.
bHLH57 Imposes NCED6- and NCED9-Mediated Seed Dormancy
To assess whether bHLH57 controls seed dormancy, we obtained the homozygous T-DNA insertion mutants bhlh57-1 (SALK_027604) and bhlh57-2 (SAIL_84_E04). The bhlh57-1 allele carries the insertion ∼50 bp upstream of the ATG and reduced bHLH57 transcript levels to 10% of Col-0. The T-DNA is inserted within the first exon in bhlh57-2 and abrogates bHLH57 transcript accumulation (Figure 5A; Supplemental Figure 10A). Germination assays showed that both mutants had weaker dormancy compared to Col-0, indicating that bHLH57 positively controls seed dormancy (Figure 5B). bHLH57 localizes to the nucleus and is expressed in various plant organs, including roots, stems, and leaves, but is very low in flowers (Figures 5C and 5D). bHLH57 transcript levels gradually decreased during seed maturation and declined after seed hydration (Figure 5D).
Figure 5.
bHLH57 Imposes Seed Dormancy and Induces ABA Biosynthesis.
(A) RT-qPCR analysis of bHLH57 transcript levels in Col-0, bhlh57-1, and bhlh57-2 freshly harvested seeds. The expression values were normalized to PP2A. Values are means ± sd from three biological repeats. ND, not detected.
(B) Germination percentages after different periods of dry storage of Col-0, bhlh57-1, and bhlh57-2 freshly harvested seeds. Values are means ± sd of three independent batches of seeds per genotype.
(C) Subcellular localization of bHLH57-YFP in N. benthamiana leaves. Bars = 50 µm.
(D) RT-qPCR analysis of bHLH57 transcript levels in various organs and developing siliques and mature seeds in Arabidopsis. The expression values were normalized to PP2A. Values are means ± sd from three biological repeats. DAP, days after pollination; HAH, hours after hydration.
(E) ABA content in Col-0 and bhlh57-2 freshly harvested seeds. Values are means ± sd of three independent batches of seeds per genotype. *P < 0.05 by Student’s t test.
(F) RT-qPCR analysis of NCEDs and CYP707As transcript levels in Col-0 and bhlh57-2 freshly harvested dry seeds. The expression values were normalized to PP2A. Values are means ± sd from three biological repeats. *P < 0.05 by Student’s t test.
Considering the reduced seed dormancy phenotype of bhlh57 mutants and the interaction between ODR1 and bHLH57, we speculated that the ABA content and ABA sensitivity of bhlh57-2 seeds might also be affected. Indeed, ABA content in bhlh57-2 freshly harvested seeds was significantly reduced compared to Col-0 seeds (Figure 5E). ABA sensitivity of bhlh57-2 seeds was however similar to that of Col-0 (Supplemental Figure 10B), suggesting that the reduced dormancy of bhlh57-2 seeds was related to ABA metabolism. Next, we analyzed the expression of NCEDs and CYP707As in bhlh57-2 freshly harvested seeds. NCED6 and NCED9 transcripts were both significantly downregulated in bhlh57-2 compared with Col-0 (Figure 5F), indicating that bHLH57 normally induces the expression of NCED6 and NCED9. In addition, CYP707A2 expression was elevated in bhlh57-2, collectively contributing to the decreased ABA content in mutant seeds. By contrast, both CYP707A3 and CYP707A4 were downregulated, which could be due to an ABA metabolic feedback in bhlh57-2 (Figure 5F). These results, in combination with the interaction between ODR1 and bHLH57 and the altered transcript levels of NCED6 and NCED9 in the odr1-2 mutant, suggest that bHLH57 may directly control NCED6 and NCED9 expression.
bHLH TFs generally modulate the expression of their target genes through binding to E-box (CANNTG) motifs in their promoters (Atchley and Fitch 1997). The 1400-bp promoter region upstream of NCED6 contains five E-box motifs, and an E-box and a G-box motif (CACGTG) were also found to be present in the 600-bp promoter region upstream of NCED9 (Figure 6A). To determine whether bHLH57 could bind to the promoters of NCED6 and NCED9, we performed yeast one-hybrid and ChIP-qPCR assays. The yeast one-hybrid assays showed that bHLH57 did bind to the NCED6 and NCED9 promoters, but it could not bind to the promoters of NCED2, NCED3, CYP707A2 to CYP707A4, or DOG1 (Figure 6B; Supplemental Figure 11A). Furthermore, we determined that bHLH57 bound to the first, fourth, and fifth E-box motifs in the NCED6 promoter and to the G-box of the NCED9 promoter (Supplemental Figure 11B). ChIP-qPCR assays with 35Spro:bHLH57-GFP transgenic seeds (Supplemental Figure 12) further confirmed the binding between bHLH57 and the promoters of NCED6 and NCED9: the bHLH57 protein preferentially bound to the first, third, fourth, and fifth E-box motifs in the NCED6 promoter and to both the E-box and G-box motifs in the NCED9 promoter (Figure 6C). We also performed a transient expression assay in N. benthamiana leaves to confirm the positive control of bHLH57 in NCED6 and NCED9 expression in plants. bHLH57 activated both NCED6 and NCED9 expression relative to negative controls (Figure 6D). Collectively, our data demonstrate that bHLH57 induces NCED6 and NCED9 expression through binding to specific E/G-boxes within their promoters, thereby controlling seed dormancy by influencing ABA metabolism.
Figure 6.
bHLH57 Binds to the NCED6 and NCED9 Promoters and Induces Their Expression.
(A) Schematic diagrams of the NCED6 and NCED9 promoters. Fragments (from –1 to –266 bp [E-box5pro], –267 to –619 bp [E-box4pro], –620 to –949 bp [E-box3pro], –950 to –1169 bp [E-box2pro], and –1170 to –1400 bp [E-box1pro]) of NCED6 promoter and fragments (from –1 to –224 bp [G-boxpro] and –225 to –600 bp [E-boxpro]) of the NCED9 promoter were amplified for a yeast one-hybrid assay. The relative positions of the ChIP-qPCR-amplified fragments in the NCED6 promoter (named E-box1, E-box2, E-box3, F1, E-box4, E-box5) and in the NCED9 promoter (named E-box, F2, G-box) are depicted with parallel black lines below the promoter. Bars = 100 bp.
(B) Yeast one-hybrid assay: direct binding of bHLH57 to the NCED6 and NCED9 promoters. 3-AT (50 mM) was added in the -Leu/-Trp/-His medium. ABI4 binding to NCED6 promoter was used as a positive control. Dilution 10×, 100×, and 1000× corresponds to an OD600 of 0.1, 0.01, and 0.001, respectively.
(C) ChIP-qPCR assay: bHLH57 binds to the E/G-box motifs of the NCED6 (left) and NCED9 (right) promoters. Immunoprecipitation was performed using seeds hydrated for 6 h from Col-0 and 35Spro:bHLH57-GFP with anti-GFP or anti-IgG antibody. qPCR was performed with specific primers listed in Supplemental Data Set 1. The comparison is between the nonimmune control (anti-IgG) and the immunoprecipitation (anti-GFP). Values are means ± sd of three biological replicates. *P < 0.05 or **P < 0.01 by Student’s t test.
(D) Transient expression assay: bHLH57 induces the expression of NCED6pro:LUC and NCED9pro:LUC in N. benthamiana leaves. The pGREENII 62-SK empty vector (35Spro) was used as control. LUC activity was normalized to REN activity. Schematic representation of various constructs used in assay is shown on the left panel. Values are means ± sd of three independent transformants for each construct. *P < 0.05 or **P < 0.01 by Student’s t test.
ODR1 Inhibits the Regulation of NCED6 and NCED9 by bHLH57
Our results above demonstrated an interaction between ODR1 and bHLH57. We also documented the opposite phenotypes on seed dormancy, ABA content, and NCED6 and NCED9 expression exhibited by the odr1-2 and bhlh57-2 mutants. These observations prompted us to investigate whether ODR1 may prevent the induction of NCED6 and NCED9 expression by bHLH57. To test this hypothesis, we performed a transient dual-luciferase (LUC) assay by coexpressing 35Spro:ODR1 and/or 35Spro:bHLH57 with the LUC reporters NCED6pro:LUC or NCED9pro:LUC in N. benthamiana leaves. Coexpression of ODR1 and bHLH57 indeed resulted in a significant decrease in LUC activity compared with expression of bHLH57 alone (Figure 7A), suggesting that the upregulation of NCED6 and NCED9 by bHLH57 is blocked by ODR1. Furthermore, we generated the odr1-2 bhlh57-2 double mutant (Supplemental Figure 13) to evaluate its germination behavior and pattern of NCED6 and NCED9 expression. Transcript levels for NCED6 and NCED9 were both upregulated in odr1-2 seeds, but this was largely counteracted when bHLH57 was also inactivated in the odr1-2 bhlh57-2 double mutant (Figure 7B). Germination percentages indicated that the dormancy level of the odr1-2 bhlh57-2 double mutant was significantly lower than that of odr1-2 (Figure 7C), demonstrating that ODR1 and bHLH57 had opposite roles in the control of dormancy and that ODR1 requires bHLH57 for its full function in seed dormancy. Overall, these results suggest that ODR1 inhibits the upregulation of NCED6 and NCED9 by bHLH57 in seeds. Interestingly, the odr1-2 bhlh57-2 double mutant was still more dormant than bhlh57-2 (Figure 7C) and also showed increased expression of NCED6 and NCED9 compared to the wild type (Figure 7B). This indicates that ODR1 also controls NCED6 and NCED9 expression through other factors. Collectively, these results demonstrate that ODR1 negatively controls NCED6 and NCED9 expression partly through direct interaction and inhibition of bHLH57, which induces NCED6 and NCED9 expression.
Figure 7.
ODR1 Prevents the Induction of NCED6 and NCED9 Expression by bHLH57.
(A) Transient expression assay: ODR1 represses the promotion of bHLH57 on luciferase activity driven by the NCED6 and NCED9 promoters in N. benthamiana leaves. The pGREENII 62-SK empty vector (35Spro) was used as the control. LUC activity was normalized to REN activity. A schematic representation of various constructs used in assay is listed on the left panel. Values are means ± sd of three independent transformants for each construct. *P < 0.05 or **P < 0.01 by Student’s t test.
(B) RT-qPCR analysis of NCED6 and NCED9 transcript levels in Col-0, odr1-2, bhlh57-2, and odr1-2 bhlh57-2 freshly harvested seeds. The expression values were normalized to PP2A. Values are means ± sd of three biological repeats. **P < 0.01 by Student’s t test.
(C) Germination percentages of Col-0, odr1-2, bhlh57-2, and odr1-2 bhlh57-2 after-ripened seeds after 1 week of storage. Values are means ± sd of three independent batches of seeds per genotype. *P < 0.05 or **P < 0.01 by Student’s t test.
DISCUSSION
The identification and characterization of seed dormancy genes and understanding their roles in the dormancy mechanism are important for crop genetic improvement. In this study, we isolated two new dormancy factors, ODR1 and bHLH57, and provide genetic, biochemical, and molecular evidence to support the signaling network in which they work alongside ABI3 and NCED6/9. We propose that this network functions in a positive feedback loop with ABA: higher levels of ABA lead to the upregulation of ABI3 during seed maturation. ABI3 then binds physically to the ODR1 promoter and represses its expression. In the absence of the ODR1-imposed inhibition, bHLH57 can bind to the promoters of NCED6 and NCED9 and elevate their expression levels, resulting in enhanced ABA biosynthesis and stronger seed dormancy. This control may be gradually amplified via a positive feedback loop involving the further induction of ABI3 by ABA (Figure 8).
Figure 8.
Proposed Working Model for the Control of Seed Dormancy and Germination by ODR1/AtSdr4L.
Over the course of seed maturation, increasing levels of ABA lead to the upregulation of ABI3. ABI3 binds to the promoter of ODR1 and represses its expression, resulting in the alleviation of ODR1-imposed inhibition of bHLH57. bHLH57 will then bind to the promoter of NCED6 and NCED9 and elevate their expression levels, leading to higher ABA biosynthesis and seed dormancy. This regulation may be amplified by the stimulation of ABI3 by ABA. Recently, Cao et al. (2020) demonstrated that ODR1/AtSdr4L also controls seed germination through GA biosynthesis in hydrated seeds. Taken together, we propose that ODR1/AtSdr4L plays a negative role in seed dormancy by adjusting the ABA and GA balance during seed maturation and germination. Our data also support an indirect function for ODR1 on ABA biosynthesis by decreasing the transcription of ABI4 and DREB2C as well as that of the core dormancy gene DOG1. This indicates that there are still other pathways in ODR1-mediated dormancy that need further investigation.
Cao et al. (2020) recently reported a role for ODR1/AtSdr4L in seed dormancy. Based on a physiological and genetic analysis, the authors concluded that this role was mainly mediated by the GA biosynthesis and signaling pathways. Indeed, odr1/atsdr4l loss of function was associated with higher expression of the GA biosynthesis genes GA20-OXIDASE1 (GA20OX1) and GA20OX2 (Cao et al., 2020). Taken together, we propose that ODR1/AtSdr4L, a homolog of OsSdr4, plays a negative role in seed dormancy by adjusting the ABA and GA balance during seed maturation and germination (Figure 8).
ODR1 Interacts with bHLH57 and Inhibits bHLH57 Upregulation of NCED6 and NCED9 Expression
We identified ODR1 as a suppressor of the low dormant rdo5-2 mutant during a γ-ray mutagenesis screen. Considering the germination behavior and ABA content of rdo5-2, odr1-2 single mutants, and rdo5-2 odr1-2 double mutant, we hypothesized that ODR1 controls seed dormancy by modulating ABA biosynthesis, which is independent of RDO5-mediated seed dormancy. ODR1 interacts with bHLH57 and represses the induction of NCED6 and NCED9 expression and ABA biosynthesis directly (Figures 4 to 7). Therefore, ODR1 and bHLH57 control ABA metabolism and dormancy in opposite directions. The mechanisms underlying these control pathways are consistent with the reported role of ABA in seed dormancy (Lefebvre et al., 2006; Seo et al., 2006). In Arabidopsis, NCED6 and NCED9 are the main contributors to ABA biosynthesis during seed maturation; in agreement, the nced6 nced9 double mutant has significantly reduced ABA levels and weaker seed dormancy (Lefebvre et al., 2006). By contrast, induction of NCED6 during seed development increased seed dormancy (Martínez-Andújar et al., 2011).
Studies have reported that the expression of NCED6 and NCED9 is directly promoted by TFs, including ABI4, DREB2C, and MYB96 (Je et al., 2014; Lee et al., 2015; Shu et al., 2016b). Therefore, it is interesting to note that our data also support an indirect function for ODR1 on ABA biosynthesis by decreasing the transcription of ABI4 and DREB2, as well as that of the core dormancy gene DOG1 (Supplemental Figures 6 and 7). This indicates that there are still other pathways in ODR1-mediated dormancy that need further investigation.
It was previously reported that two bHLH TFs, SPATULA (SPT) and PHYTOCHROME INTERACTING FACTOR1 (PIF1), control seed germination and GA3OX expression in response to light and temperature signaling in Arabidopsis (Penfield et al., 2005; Oh et al., 2006). bHLH TFs also function in seed dormancy in other plant species (Gao et al., 2018; Zhao et al., 2019). These findings collectively demonstrate the conservation and diversity of the bHLH gene family in the control of dormancy. Our study extends a role for the TF bHLH57 in seed dormancy by promoting NCED6 and NCED9 expression.
It is noteworthy that our genetic analysis demonstrated that the loss of bHLH57 function in the odr1-2 background was only partially able to counteract the higher NCED6 and NCED9 expression levels and associated hyper-dormancy phenotype brought on by the loss of ODR1 (Figures 7B and 7C). These results indicate that ODR1 probably also controls NCED6/9 by factors other than bHLH57, like the above-mentioned ABI4 and DREB2C. Furthermore, other dormancy factors such as GA and DOG1 have recently also been reported to be involved in ODR1/AtSdr4L-mediated seed dormancy (Figure 8; Cao et al., 2020).
ODR1 Is Negatively Controlled by ABI3 during Seed Dormancy Establishment
ABI3 is a vital ABA-responsive TF that directly binds to the RY motif in the promoter of its target genes via a B3 domain, and it plays a central role in seed maturation and the establishment of primary seed dormancy (Ezcurra et al., 2000; Mönke et al., 2004). Mutations in ABI3 lead to reduced dormancy and premature seeds germination (Nambara et al., 1995). ChIP-chip and transcriptome analysis in a previous study identified a set of 98 genes (including ODR1) involved in seed development and seed protein and lipid accumulation as direct targets of ABI3 (Mönke et al., 2012). In our study, we confirmed the binding activity of ABI3 to the proximal RY motif in the promoter of ODR1 by yeast one-hybrid and ChIP-qPCR assays (Figures 3B and 3C). Furthermore, we demonstrated that expression of ODR1 is repressed by ABI3 using gene expression analysis and transient expression assays (Figures 3D and 3E). Finally, inactivation of ODR1 in the abi3-1 background by CRISPR/Cas9 genome editing supported the position of ABI3 upstream of ODR1 in the control of seed dormancy (Figure 3F).
AtODR1 and Its Rice Homolog OsSdr4 Have Opposite Roles in Seed Dormancy
Seed dormancy is a key decision point in plant development. Hence, maintaining suitable levels of dormancy is of great importance and has been under strong natural selection during plant evolution. In our study, we found that ODR1 and its homologs are conserved in angiosperms (Supplemental Figure 2), indicating that they might have a conserved function in seed dormancy. Sdr4 is a putative ODR1 ortholog in rice, with ∼35% amino acid identity (and 49% similarity). However, Sdr4 positively affects seed dormancy in several rice cultivars (Sugimoto et al., 2010), which is the opposite of ODR1 in Arabidopsis. We speculate that such divergence in function might be related to their associated ABA-responsive factors: ABI3 and OsVP1. In rice, OsVP1 induces the expression of Sdr4 during seed development and maturation (Sugimoto et al., 2010), whereas ABI3 represses ODR1 expression in Arabidopsis seeds. These opposite effects confer a deeper dormancy in odr1-2 and preharvest sprouting and nondormant phenotypes in sdr4. Furthermore, the expression levels of the two closest rice homologues of Arabidopsis DOG1 were lower in the rice sdr4 mutant compared to the wild type, while our results showed that DOG1 was upregulated in odr1-2 freshly harvested and hydrated Arabidopsis seeds (Supplemental Figure 7). The different expression behavior of DOG1 and its rice homologues may contribute to the distinct mutant phenotypes. We hypothesize that the relationship between ODR1 and DOG1 might be conserved between dicots and monocots. Given the opposite modes of action displayed by ABI3 and OsVP1 on ODR1 and Sdr4 in seed dormancy, it will be of interest to assess the effects of Sdr4 on ABA metabolism and dormancy in rice. Moreover, considering the relatively low identity between ODR1 and Sdr4, we speculate that the opposite dormancy phenotypes of ODR1 and sdr4 in dormancy may also be attributed to sequence variation in their coding sequences, which could lead to a divergence in protein function.
METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) genotypes used in this study were either in the Col-0 or Ler background. T-DNA insertion lines odr1-2 (SALK_022729), bhlh57-1 (SALK_027604), bhlh57-2 (SAIL_84_E04), and cyp707a2-1 (SALK_072410) were ordered from Arabidopsis Biological Resource Center stock center and are in Col-0. Homozygous lines were confirmed by PCR with gene-specific primers (Supplemental Data Set 1). abi3-1 is in Ler and was a kind gift from Yongxiu Liu. 35Spro:ABI3-FLAG is in the Col-0 background and was a kind gift from Giltsu Choi (Park et al., 2011).
Seeds were sown in soil in soil mixture (potting soil:vermiculite [v/v] = 3:1) and grown in the greenhouse or growth chamber with 16-h-light, under 80 to 90 μmol m−2 s−1 white light intensity/8-h-dark cycle at 24°C/22°C. For germination assays on medium, seeds were sterilized with 10% (v/v) NaClO and 75% (v/v) ethanol and then washed with sterile water and sown on plates containing half-strength MS medium containing 1% (w/v) Suc and 0.8% (w/v) plant agar. Plates were kept in the dark at 4°C for 3 d before transfer to a growth chamber.
Mapping of ODR1
To clone the ODR1 gene, we followed a next-generation sequencing–based bulked-segregant analysis approach. The odr-1 was backcrossed with its wild-type rdo5-2 four times to minimize background effects. Next, a segregating population was produced by outcrossing ODR1-1 with Ler. F2 progeny with a homozygous rdo5-2 background were selected for phenotyping. Bulk DNA was in two groups based on their dormant or nondormant phenotype. After sequencing of bulk DNA, we performed linkage analysis based on whole genome–distributed single-nucleotide polymorphism markers and detailed genome sequence analyses in the candidate region.
Constructs and Plant Transformation
To generate 35Spro:ODR1/odr1-2 plants, the full-length coding sequence (CDS) of ODR1 was amplified and cloned into the pFAST-R01 vector (Shimada et al., 2010) and transformed into odr1-2 via Agrobacterium (Agrobacterium tumefasciens) strain GV3101. To generate 35Spro:bHLH57-GFP–overexpressing plants, the bHLH57 CDS was amplified and cloned into the pFAST-R05 vector (Shimada et al., 2010) and transformed into Col-0 with Agrobacterium strain GV3101. Single copy insertions and homozygous lines were identified based on red fluorescence screening (the red fluorescent protein being encoded by the binary vector).
The ODR1 genome-edited lines in the abi3-1 background were generated using the pYAO-based CRISPR/Cas9 system according to the instructions from Yan et al. (2015). Briefly, two 20-nucleotide single guide RNAs (sgRNA; 5′-CAAGCGAGGCCGAGGCGGGA-3′ and 5′-AAGAGACCAAAGTCGTCATG-3′) with specific recognition for ODR1 were synthesized and cloned into the vector pAtU6-26-sgRNA-SK and digested with BsaI. The fragment harboring AtU6-26-sgRNAODR1 was digested with SpeI and cloned into the pCAMBIA1300-pYAO-Cas9 vector. Finally, the plasmid pCAMBIA1300-pYAO-Cas9-AtU6-26-sgRNAODR1 was transformed into abi3-1 with Agrobacterium strain GV3101. Homozygous lines were identified by genome PCR and sequencing.
Seed Germination Assays
Seed germination assays were performed as previously described by Xiang et al. (2014) and Née et al. (2017). For ABA response assays, freshly harvested seeds were sterilized and plated on half-strength MS medium containing 1% (w/v) Suc and 0.8% (w/v) plant agar supplemented with 0 or 0.5 μM ABA. After 3 d of stratification at 4°C in the dark, plates were moved into a growth chamber at 25°C/20°C (day/night) under a 12-h-light/12-h-dark photoperiod. Germination percentages were calculated every day. For fluridone treatments, freshly harvested seeds were plated onto a filter paper soaked with 0, 5, or 10 μM fluridone in Petri dishes and incubated in a growth chamber at 25°C/20°C (day/night) under a 12-h-light/12-h-dark photoperiod. Germination percentages were calculated after 7 d. Stock solutions of ABA and fluridone were dissolved in ethanol.
Subcellular Localization and BiFC Assay
To generate constructs for subcellular localization analysis, the CDSs of ODR1 and bHLH57 were amplified and cloned in frame with enhanced YFP driven by the 35S promoter in pEarleyGate101(Earley et al., 2006). Vectors were transiently transfected into Nicotiana benthamiana leaves as previously described by Liu et al. (2010). Yellow fluorescence signal was detected using a confocal laser scanning microscope (Zeiss) at the excitation wavelength of 513 nm. For BiFC assays, the CDSs of ODR1 and bHLH57 and truncated bHLH57 (bHLH57ΔN) were amplified and inserted into pBatTL-B sYFPn (YFPn) and pBatTL-B sYFPc (YFPc) vectors, respectively. The BiFC assay was performed as described by Née et al. (2017a).
RT-qPCR Assay
Total RNA was extracted from various organs using RNAqueous columns and RNA Isolation Aid (Invitrogen) as previously described by Kushiro et al. (2004). First-strand cDNA was synthesized using All-In-One RT MasterMix (Applied Biological Materials) according to the product instructions. qPCR was then performed with PowerUp SYBR Green Master Mix (Life Technologies) with gene-specific primers (Supplemental Data Set 1). PP2A (At1g69960) was used as the internal control. Three independent biological replicates with three technological repeats each were performed.
Yeast Two-Hybrid and One-Hybrid Assays
The yeast two-hybrid library screen was performed with the Arabidopsis Mate and Plate Library (Clontech) according to the user’s manual. For confirmation of interaction between ODR1 and bHLH57, bHLH57 CDS was amplified and inserted into the pGADT7 vector and cotransformed into yeast strain AH109 harboring pGBKT7-ODR1 and confirmed with the method described previously by Liu et al., 2016). Interaction between TTG1 and MYB5 was used as positive control (Gonzalez et al., 2009). Yeast one-hybrid assays were performed as described previously by Li et al. (2011). The respective promoter fragments (http://www.cbs.dtu.dk/services/Promoter/) for ODR1 (1100 bp upstream of ATG), NCEDs (NCED2 [1800 bp upstream of ATG], NCED3 [1000 bp upstream of ATG], NCED5 [1100 bp upstream of ATG], NCED6 [1400 bp upstream of ATG], NCED9 [600 bp upstream of ATG]), and CYP707As (CYP707A1 [1800 bp upstream of ATG], CYP707A2 [1800 bp upstream of ATG], CYP707A3 [2000 bp upstream of ATG], and CYP707A4 [1800 bp upstream of ATG]) were amplified and inserted into the pHIS2 vector digested with EcoRI and SacI individually. The CDSs of ABI3, ODR1, bHLH57, and ABI4 were amplified and individually fused in frame with the GAL4 activation domain in the pGADT7-Rec2 vector digested with SmaI. The various pairs of recombined pHIS2 and pGADT7-Rec2 plasmids were cotransformed into the yeast strain AH109 individually and grown on synthetic defined (SD)/-Leu/-Trp/-His/-Ade medium with 50 mM 3-AT. Binding of ABI4 to the promoters of NCED6 and CYP707A1 was used as positive controls (Shu et al., 2016b).
GST Pull-Down Assay
To generate constructs for GST pull-down assays, the CDS of ODR1 and bHLH57 were amplified and fused in frame with a His tag or GST in the vectors pET-28a and modified pGEX, to generate vectors expressing ODR1-6×His and GST-bHLH57 proteins in Escherichia coli (BL21). GST pull-down assays were performed with the Pierce GST Protein Interaction Pull-Down Kit (Thermo Fisher Scientific) according to the user’s manual. Briefly, isolated bait lysate GST or GST-bHLH57 was incubated with glutathione agarose resin in Tris-buffered saline (TBS) solution (25 mM Tris-HCl and 150 mM NaCl, pH 7.2) for 1 h at 4°C with gentle rocking. After immobilization of the bait protein, the prey lysate was incubated with the glutathione agarose resin-bait protein complex in TBS solution for 1 h at 4°C. The resin was washed several times with wash solution (TBS solution: lysis buffer [v/v] =1:1), and proteins bound to the resin were eluted with 10 mM glutathione elution buffer, pH 8.0, and analyzed by immunoblotting using anti-GST (at a 1:1000 dilution; catalog no. BE2013, EASYBIO) and anti-6×His (at a 1:10,000 dilution; catalog no. ab184607, Abcam).
Transient Expression Assay
To generate plasmids for the transient expression assays, the respective promoters of ODR1, NCED6, and NCED9 (same promoter fragments used in yeast one-hybrid assays) were amplified and inserted into pGreenII-0800-LUC vector to produce ODR1pro:LUC, NCED6pro:LUC, and NCED9pro:LUC. The coding sequences of ABI3, bHLH57, and ODR1 were amplified and inserted into pGreenII 62-SK vector to generate 35Spro:ABI3, 35Spro:bHLH57, and 35Spro:ODR1, respectively The various pairs of reporter and effector plasmids were cotransfected into 4-week-old N. benthamiana leaves with Agrobacterium strain GV3101. Transfected plants were maintained under continuous white light for 4 d. Firefly LUC and Renilla luciferase (REN) activity were assayed using the Dual-Luciferase assay reagents (Promega) and a GLOMAX 20/20 luminometer (Ye et al., 2017). Three biological replicates were measured for each sample.
ChIP-qPCR Assay
ChIP assays were performed following the procedure described previously (Wang et al., 2016). Briefly, ∼1 g of hydrated seeds was cross-linked with 1% (w/v) formaldehyde solution for 15 min under vacuum, and the fixation reaction was then terminated by adding 2 M Gly to a final concentration of 0.125 M. After grinding all the samples into a powder, chromatin was isolated and sheared by sonication to 300 to 1000 bp. The genomic DNA fragments were immunoprecipitated by the addition of anti-FLAG (catalog no. F1804, Sigma-Aldrich) or anti-GFP (catalog no. ab290, Abcam) antibodies. The precipitated DNA was recovered with the EpiQuik Plant ChIP Kit (Epigentek, P-2014-24) and analyzed by qPCR with specific primers (Supplemental Data Set 1). Three biological replicates were measured for each sample.
Quantification of ABA
The measurement of ABA was performed as described previously, with small modifications (Müller and Munné-Bosch 2011), Briefly, 20 mg of seeds was homogenized in a precooled methanol:isopropanol (20:80 [v/v]) solution containing 0.2% (v/v) formic acid using a TissueLyser (JX-24) with zirconia beads for 3 min at 30 Hz. ABA was extracted at –20°C overnight. The supernatant was collected after 14,000g centrifugation at 4°C for 15 min and dried with a flow of nitrogen. The residue was dissolved with 100 μL of cold methanol solution containing internal standard d6-ABA (CDN Isotopes). Quantification of ABA was performed with an ultra-performance liquid chromatography–tandem mass spectrometry system consisting of an ultra-performance liquid chromatography system (Waters) and a triple quadruple tandem mass spectrometer (AB Sciex). Three independent biological replicates were performed for each sample.
Accession Numbers
Sequence data from this investigation can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ODR1 (At1g27461), ABI3 (At3g24650), bHLH57 (At4g01460), NCED2 (At4g18350) NCED3 (At3g14440), NCED5 (At1g30100), NCED6 (At3g24220), NCED9 (At1g78390), CYP707A1 (At4g19230), CYP707A2 (At2g29090), CYP707A3 (At5g45340), CYP707A4 (At3g19270), ABI4 (At2g40220), DREB2C (At2g40340), TTG1 (At5g24520), MYB5 (At3g13540), DOG1 (At5g45830), RDO5 (At4g11040), and PP2A (At1g13320).
Supplemental Data
Supplemental Figure 1. Mapping of ODR1 and Identification of ODR1 mutants.
Supplemental Figure 2. Sequence alignments and phylogenetic analysis of ODR1 and ODR1-related proteins.
Supplemental Figure 3. Expression profile of ODR1.
Supplemental Figure 4. Expression profiles of NCEDs and CYP707As in seed.
Supplemental Figure 5. Yeast one-hybrid assay between ODR1 and predicted promoters of NCEDs and CYP707As.
Supplemental Figure 6. The relation of ODR1 with DREB2C and ABI4.
Supplemental Figure 7. The relation of ODR1 with DOG1.
Supplemental Figure 8. Expression levels of ABI3 in freshly harvested dry and imbibed seeds of 35Spro:ABI3-FLAG overexpression lines.
Supplemental Figure 9. Construction of ODR1 knock-out lines in the abi3-1 background through CRISPR-Cas9 genome editing.
Supplemental Figure 10. Identification of bhlh57 and sensitivity of bhlh57-2 to exogenous ABA treatment.
Supplemental Figure 11. Yeast one-hybrid between bHLH57 and predicted promoters of NCEDs, CYP707As and DOG1.
Supplemental Figure 12. Expression levels of bHLH57 in 35Spro:bHLH57-GFP overexpression lines.
Supplemental Figure 13. Identification of the odr1-2 bhlh57-2 double mutant with genomic PCR.
Supplemental Data Set 1. Primers used in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Yongxiu Liu (Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences) for kindly providing abi3-1 homozygous seeds. We thank Giltsu Choi (Department of Biological Sciences, Korea Advanced Institute of Science and Technology) for kindly providing 35Spro:ABI3-FLAG overexpression homozygous seeds. We also thank the Arabidopsis Biological Resource Center for providing T-DNA insertion lines used in this study. This work was supported by the National Natural Science Foundation of China (grants 31670323 and 31000123), Shenzhen City Science and Technology Program (grants JCYJ20160530192105268 and JCYJ20180306173702268), and Dapeng New District Science and Technology Program (grants KY20160205 and RCTD20180102).
AUTHOR CONTRIBUTIONS
Y.X. and F.L. designed the experiments; F.L., Y.X., L.D., and H.Z. performed the experiments; F.L. analyzed the data; F.L., Y.X., and W.J.J.S. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Atchley W.R., Fitch W.M.(1997). A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. USA 94: 5172–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L., et al. (2010). Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc. Natl. Acad. Sci. USA 107: 4264–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L., Jowett J., Hanhart C.J., Koornneef M.(2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant F.M., Hughes D., Hassani-Pak K., Eastmond P.J.(2019). Basic LEUCINE ZIPPER TRANSCRIPTION FACTOR 67 transactivates DELAY OF GERMINATION 1 to establish primary seed dormancy in Arabidopsis. Plant Cell 31: 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Han Y., Li J., Ding M., Li Y., Li X., Chen F., Soppe W.J., Liu Y.(2020). Arabidopsis thaliana SEED DORMANCY 4-LIKE regulates dormancy and germination by mediating the gibberellin pathway. J. Exp. Bot. 71: 919–933. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R.(2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Delmas F., Sankaranarayanan S., Deb S., Widdup E., Bournonville C., Bollier N., Northey J.G., McCourt P., Samuel M.A.(2013). ABI3 controls embryo degreening through Mendel’s I locus. Proc. Natl. Acad. Sci. USA 110: E3888–E3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S.(2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Ezcurra I., Wycliffe P., Nehlin L., Ellerström M., Rask L.(2000). Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 24: 57–66. [DOI] [PubMed] [Google Scholar]
- Fedak H., Palusinska M., Krzyczmonik K., Brzezniak L., Yatusevich R., Pietras Z., Kaczanowski S., Swiezewski S.(2016). Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript. Proc. Natl. Acad. Sci. USA 113: E7846–E7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S., Tischer S.V., Wunschel C., Christmann A., Grill E.(2014). Abscisic acid sensor RCAR7/PYL13, specific regulator of protein phosphatase coreceptors. Proc. Natl. Acad. Sci. USA 111: 5741–5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Liu J., Chen Y., Tang H., Wang Y., He Y., Ou Y., Sun X., Wang S., Yao Y.(2018). Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins. Hortic. Res. 5: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A., Mendenhall J., Huo Y., Lloyd A.(2009). TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 325: 412–421. [DOI] [PubMed] [Google Scholar]
- Graeber K., Nakabayashi K., Miatton E., Leubner-Metzger G., Soppe W.J.(2012). Molecular mechanisms of seed dormancy. Plant Cell Environ. 35: 1769–1786. [DOI] [PubMed] [Google Scholar]
- Gubler F., Millar A.A., Jacobsen J.V.(2005). Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 8: 183–187. [DOI] [PubMed] [Google Scholar]
- Je J., Chen H., Song C., Lim C.O.(2014). Arabidopsis DREB2C modulates ABA biosynthesis during germination. Biochem. Biophys. Res. Commun. 452: 91–98. [DOI] [PubMed] [Google Scholar]
- Kanno Y., Jikumaru Y., Hanada A., Nambara E., Abrams S.R., Kamiya Y., Seo M.(2010). Comprehensive hormone profiling in developing Arabidopsis seeds: Examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 51: 1988–2001. [DOI] [PubMed] [Google Scholar]
- Kushiro T., Okamoto M., Nakabayashi K., Yamagishi K., Kitamura S., Asami T., Hirai N., Koshiba T., Kamiya Y., Nambara E.(2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 23: 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.G., Lee K., Seo P.J.(2015). The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol. Biol. 87: 371–381. [DOI] [PubMed] [Google Scholar]
- Lefebvre V., North H., Frey A., Sotta B., Seo M., Okamoto M., Nambara E., Marion-Poll A.(2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45: 309–319. [DOI] [PubMed] [Google Scholar]
- Li X., Chen T., Li Y., Wang Z., Cao H., Chen F., Li Y., Soppe W.J.J., Li W., Liu Y.(2019). ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-TPL complex on DELAY OF GERMINATION1 expression. Plant Cell 31: 832–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Gao X., Wei Y., Deng L., Ouyang Y., Chen G., Li X., Zhang Q., Wu C.(2011). Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5′-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell 23: 1416–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A., Graeber K., Knight C., Leubner-Metzger G.(2010). The evolution of seeds. New Phytol. 186: 817–831. [DOI] [PubMed] [Google Scholar]
- Liu F., Zhang L., Luo Y., Xu M., Fan Y., Wang L.(2016). Interactions of Oryza sativa OsCONTINUOUS VASCULAR RING-LIKE 1 (OsCOLE1) and OsCOLE1-INTERACTING PROTEIN reveal a novel intracellular auxin transport mechanism. New Phytol. 212: 96–107. [DOI] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q.(2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D.T., Chait B.T., Chua N.H.(2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32: 317–328. [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E.(2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Martínez-Andújar C., Ordiz M.I., Huang Z., Nonogaki M., Beachy R.N., Nonogaki H.(2011). Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. USA 108: 17225–17229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa T., Fujita Y., Yamaguchi-Shinozaki K., Tanokura M.(2013). Structure and function of abscisic acid receptors. Trends Plant Sci. 18: 259–266. [DOI] [PubMed] [Google Scholar]
- Mönke G., Altschmied L., Tewes A., Reidt W., Mock H.P., Bäumlein H., Conrad U.(2004). Seed-specific transcription factors ABI3 and FUS3: Molecular interaction with DNA. Planta 219: 158–166. [DOI] [PubMed] [Google Scholar]
- Mönke G., Seifert M., Keilwagen J., Mohr M., Grosse I., Hähnel U., Junker A., Weisshaar B., Conrad U., Bäumlein H., Altschmied L.(2012). Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res. 40: 8240–8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H.D., Lee M.S., Kim S.H., Jeong W.J., Choi D.W.(2016). Identification of a drought responsive gene encoding a nuclear protein involved in drought and freezing stress tolerance in Arabidopsis. Biol. Plant. 60: 105–112. [Google Scholar]
- Müller M., Munné-Bosch S.(2011). Rapid and sensitive hormonal profiling of complex plant samples by liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Plant Methods 7: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K., Bartsch M., Xiang Y., Miatton E., Pellengahr S., Yano R., Seo M., Soppe W.J.J.(2012). The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 24: 2826–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Keith K., McCourt P., Naito S.(1995). A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121: 629–636. [Google Scholar]
- Nambara E., Marion-Poll A.(2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56: 165–185. [DOI] [PubMed] [Google Scholar]
- Née G., Kramer K., Nakabayashi K., Yuan B., Xiang Y., Miatton E., Finkemeier I., Soppe W.J.J.(2017a). DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 8: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Née G., Xiang Y., Soppe W.J.(2017b). The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 35: 8–14. [DOI] [PubMed] [Google Scholar]
- Nishimura N., Tsuchiya W., Moresco J.J., Hayashi Y., Satoh K., Kaiwa N., Irisa T., Kinoshita T., Schroeder J.I., Yates J.R.I.I.I. III, Hirayama T., Yamazaki T.(2018). Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat. Commun. 9: 2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W.I., Choi G.(2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47: 124–139. [DOI] [PubMed] [Google Scholar]
- Okamoto M., Kuwahara A., Seo M., Kushiro T., Asami T., Hirai N., Kamiya Y., Koshiba T., Nambara E.(2006). CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141: 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee N., Kim W., Lim S., Choi G.(2011). ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 23: 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Josse E.M., Kannangara R., Gilday A.D., Halliday K.J., Graham I.A.(2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15: 1998–2006. [DOI] [PubMed] [Google Scholar]
- Penfield S., MacGregor D.R.(2017). Effects of environmental variation during seed production on seed dormancy and germination. J. Exp. Bot. 68: 819–825. [DOI] [PubMed] [Google Scholar]
- Seo M., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48: 354–366. [DOI] [PubMed] [Google Scholar]
- Shimada T.L., Shimada T., Hara-Nishimura I.(2010). A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61: 519–528. [DOI] [PubMed] [Google Scholar]
- Shu K., et al. (2016a). ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 85: 348–361. [DOI] [PubMed] [Google Scholar]
- Shu K., Liu X.D., Xie Q., He Z.H.(2016b). Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 9: 34–45. [DOI] [PubMed] [Google Scholar]
- Söderman E.M., Brocard I.M., Lynch T.J., Finkelstein R.R.(2000). Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 124: 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Takeuchi Y., Ebana K., Miyao A., Hirochika H., Hara N., Ishiyama K., Kobayashi M., Ban Y., Hattori T., Yano M.(2010). Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA 107: 5792–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., et al. (2016). Arabidopsis seed germination speed is controlled by SNL histone deacetylase-binding factor-mediated regulation of AUX1. Nat. Commun. 7: 13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Nakabayashi K., Ding J., He F., Bentsink L., Soppe W.J.J.(2014). Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 26: 4362–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Song B., Née G., Kramer K., Finkemeier I., Soppe W.J.J.(2016). Sequence polymorphisms at the REDUCED DORMANCY5 pseudophosphatase underlie natural variation in Arabidopsis dormancy. Plant Physiol. 171: 2659–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Wei S., Wu Y., Hu R., Li H., Yang W., Xie Q.(2015). High-efficiency genome editing in arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8: 1820–1823. [DOI] [PubMed] [Google Scholar]
- Ye J., Wang X., Hu T., Zhang F., Wang B., Li C., Yang T., Li H., Lu Y., Giovannoni J.J., Zhang Y., Ye Z.(2017). An indel in the Promoter of Al-ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell 29: 2249–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Li X., Jia J., Yuan G., Chen S., Qi D., Cheng L., Liu G.(2019). bHLH92 from sheepgrass acts as a negative regulator of anthocyanin/proanthocyandin accumulation and influences seed dormancy. J. Exp. Bot. 70: 269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]