A catalog of large copy number variations (CNVs), CNV-overlapping genes, and their genotypes in 1060 Arabidopsis accessions will stimulate studies on the genetic basis of phenotypic variation.
Abstract
Copy number variations (CNVs) greatly contribute to intraspecies genetic polymorphism and phenotypic diversity. Recent analyses of sequencing data for >1000 Arabidopsis (Arabidopsis thaliana) accessions focused on small variations and did not include CNVs. Here, we performed genome-wide analysis and identified large indels (50 to 499 bp) and CNVs (500 bp and larger) in these accessions. The CNVs fully overlap with 18.3% of protein-coding genes, with enrichment for evolutionarily young genes and genes involved in stress and defense. By combining analysis of both genes and transposable elements (TEs) affected by CNVs, we revealed that the variation statuses of genes and TEs are tightly linked and jointly contribute to the unequal distribution of these elements in the genome. We also determined the gene copy numbers in a set of 1060 accessions and experimentally validated the accuracy of our predictions by multiplex ligation-dependent probe amplification assays. We then successfully used the CNVs as markers to analyze population structure and migration patterns. Finally, we examined the impact of gene dosage variation triggered by a CNV spanning the SEC10 gene on SEC10 expression at both the transcript and protein levels. The catalog of CNVs, CNV-overlapping genes, and their genotypes in a top model dicot will stimulate the exploration of the genetic basis of phenotypic variation.
INTRODUCTION
The frequent occurrence of duplications and deletions in eukaryotic genomes is among the most crucial factors that affect adaptation, evolution, and speciation (Kondrashov, 2012; Panchy et al., 2016). There are numerous lines of evidence that at an intraspecies level, these DNA copy number changes contribute to the phenotypic variation of humans, animals, and plants (McHale et al., 2012; Handsaker et al., 2015; Xu et al., 2016). Accordingly, efforts toward developing tools to detect copy number variations (CNVs) and map polymorphic regions have recently intensified. A good example of this trend is the latest advance in CNV discovery in the human genome, which has been empowered by the consecutive release of data from three phases of the 1000 Genomes Project. Remarkably, 60% of CNVs identified in phase 3 of this project (Sudmant et al., 2015) were novel compared to those identified in previous reports by Mills et al. (2011) and the 1000 Genomes Project Consortium et al. (2012), reflecting the methodological improvements and the importance of using large, diversified data sets.
The number of plant species for which CNV regions have been identified at the genome-wide scale has grown rapidly within the last decade (Swanson-Wagner et al., 2010; Chia et al., 2012; Muñoz-Amatriaín et al., 2013; Duitama et al., 2015; Hardigan et al., 2016; Fuentes et al., 2019). However, for Arabidopsis (Arabidopsis thaliana), an important model plant (Alonso-Blanco and Koornneef, 2000) with more than 1000 accessions whose genomes have been sequenced with coverage between 5× and 118× (1001 Genomes Consortium et al., 2016), comprehensive genome-wide CNV analysis is still required. Previous CNV analyses in Arabidopsis have been limited to individual lines or small populations and most often focused on characterizing presence-absence variation only. One of the earliest studies of this type combined the results of array-based hybridization and short read–based whole-genome sequencing (WGS) to identify ≥100-bp deletions in the genomes of four Arabidopsis accessions: Eil-0, Lc-0, Sav-0, and Tsu-1 (Santuari et al., 2010). These deletions overlapped with 987 to 1344 protein-coding genes (for simplicity, we refer to them as genes hereafter), and many of them were shared by at least two accessions. A larger study that focused on comparing the genomes of 17 accessions that were sequenced and assembled de novo from WGS data revealed multiple polymorphic regions that could not be mapped to the reference genome (Gan et al., 2011). Based on the same WGS data, Bush et al. (2014) identified numerous exon-overlapping regions in the Arabidopsis genome that were absent from at least one accession. These regions overlapped with 411 genes. A wider study that, in addition to detecting large deletions, also identified duplications and multiallelic CNVs included WGS data from 80 accessions from Europe, Asia, and North Africa (Cao et al., 2011). The identified CNVs covered 1.8% of the reference genome and overlapped with nearly 500 genes. Subsequent copy number genotyping of several genes performed by our group using these 80 accessions indicated, however, that the number of genes affected by CNVs may in fact be much higher (Samelak-Czajka et al., 2017). Another study involved the detection of regions of deletions and duplications among 180 accessions, but these accessions represented a narrow local population from Sweden (Long et al., 2013). In these accessions, more than 7700 regions with duplications of a fixed size (3 kb) were identified. A read depth–based approach for CNV detection was used by both Cao et al. (2011) and Long et al. (2013), without further refinement of the CNV breakpoints.
Recently, WGS data from a global collection of 1135 Arabidopsis accessions were released by the 1001 Genomes Consortium et al. (2016), and a catalog of single-nucleotide polymorphisms (SNPs) as well as insertions and deletions shorter than 50 bp (short indels) was created based on these data. Here, we extended the spectrum of characterized genetic variations in these accessions by calling and analyzing large indels and CNVs. We determined the distribution and genomic content of CNV regions and performed population-scale copy number analysis of genes overlapping with CNVs. We investigated the variation in and relative distributions of genes and transposable elements (TEs). We then successfully used gene copy number estimates as markers to reconstruct the genetic structure of the Arabidopsis population. We also demonstrated that natural changes in gene dosage may lead to variations in transcript and protein levels. The CNV map and copy number genotyping data generated in this study provide a background for further studies on the genetic bases of phenotypic variation in Arabidopsis.
RESULTS
Identification of CNVs and Large Indels
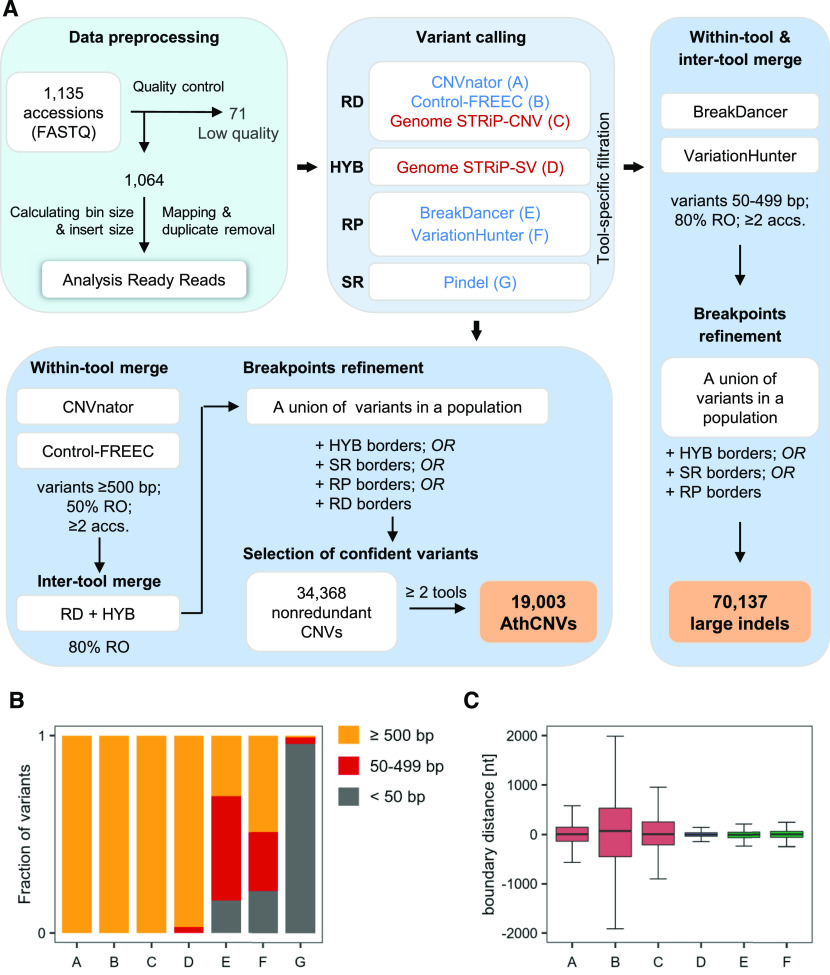
We selected 1064 high-quality WGS data sets from the 1135 data sets available in the 1001 Genomes Project collection and performed an integrated CNV analysis (Figure 1A). To this end, we set up a pipeline that combined the three main types of read signatures that can be used for CNV identification (Alkan et al., 2011). We used three read depth–based tools, namely, CNVnator (Abyzov et al., 2011), Control-FREEC (Boeva et al., 2011), and the Genome STRiP-CNV module (Handsaker et al., 2015); two discordant read pair–based tools, namely, BreakDancer (Chen et al., 2009) and VariationHunter (Hormozdiari et al., 2009); the split read–based tool Pindel (Ye et al., 2009); and a hybrid approach implemented in the Genome STRiP-SV module (Handsaker et al., 2015). Methods relying on read depth signatures are the most sensitive in detecting large size variations (Figure 1B) and are more successful when analyzing regions with segmental duplications (Yoon et al., 2009; Teo et al., 2012). However, their accuracy in estimating CNV breakpoints is low (Figure 1C) and depends on the window size used during the calling step. Tools based on discordant read pair mappings are more precise in setting CNV breakpoints but are unable to detect large variants (Supplemental Figure 1) or to identify highly duplicated regions. Pindel, which is based on split reads, reports variants at a single-nucleotide resolution but is more sensitive to short indels than to CNVs; additionally, it generates a very large number of predictions with a high false-positive rate (Li et al., 2013). To handle these constraints, we separately processed CNVs (defined here as unbalanced variations at least 0.5 kb in length) and large indels (variants 50 to 499 bp in length).
Figure 1.
Genome-Wide Structural Variant Discovery in an Arabidopsis Population.
(A) Variant identification pipeline. The analysis involved three main stages: data preprocessing, variant calling, and merging and filtering. Variants were called with seven different tools, based on read depth (RD), read pair (RP), split read (SR), or hybrid (HYB) approach, in individual samples (blue labels) or in the entire population (red labels). The last stage depended on variant length. RO, reciprocally overlapping each other.
(B) Fraction of variants of different size ranges identified by individual callers.
(C) Comparison of the boundaries set by the callers for variants ≥500 bp reciprocally overlapping each other by 80%. Pindel-derived coordinates served as a reference since this tool reports variants at single-nucleotide resolution. Boxplots show median (inner line) and inner quartiles (box). Whiskers extend to the highest and lowest values no greater than 1.5 times the inner quartile range. nt, nucleotides.
For CNVs, we selected variants that were detected by at least one read depth–based or hybrid approach (Supplemental Tables 1 and 2). In the next step, whenever possible, we further refined the CNV borders with the additional support of the remaining callers to improve the accuracy of CNV breakpoint predictions. Finally, we included only variants supported by at least two of the seven callers that were used in the list of high-confidence regions that are copy number variable in the Arabidopsis genome, hereafter referred to as the AthCNV data set. This data set consists of 19,003 CNVs that vary in length from 500 to 984,676 bp, 92.1% of which are shorter than 20 kb. These variants are listed in Supplemental Data Set 1, along with 15,365 low-confidence CNVs, which were supported by only one caller and were not further investigated.
We identified large indels by combining 50- to 499-bp-long variants from the read pair–based callers only, followed by redundancy removal, and set boundaries with the support from hybrid- and split read–based callers. As a result, we obtained 70,137 variants (Supplemental Data Set 2). Of these, 4149 exceeded the upper size limit defined in our pipeline as a result of merging and breakpoint refinement. We did not remove them from the final large indel data set since they were identified using a different approach from AthCNVs. Overall, large indels had 56% overlap with AthCNVs.
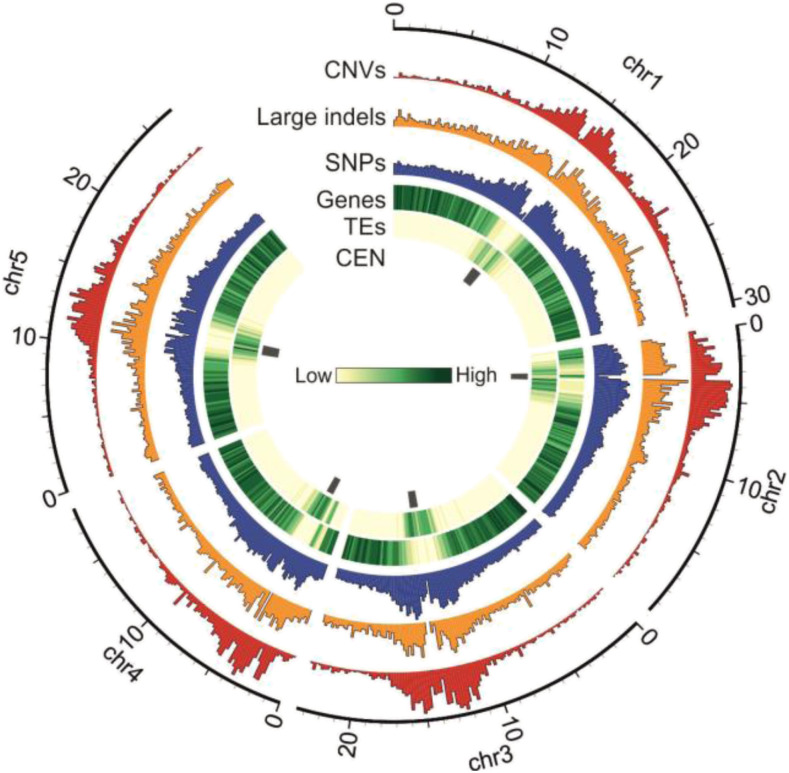
We then compared the genomic distribution of the newly identified variants with that of the previously identified short indels (1001 Genomes Consortium et al., 2016). All types of variants (short indels, large indels, and AthCNVs) were most abundant in the pericentromeric regions and less abundant in the chromosome arms (Figure 2). However, short indels had moderate overlap with AthCNVs (46%) and very little overlap with large indels (8%). Thus, our results substantially complement the existing catalog of known structural variations present in the Arabidopsis genome.
Figure 2.
Genomic Distribution of CNVs, Large Indels, and Short Variants in the Arabidopsis Genome.
Histograms are scaled for equal height. Tracks present: CEN, pericentromeric regions; CNVs, confident CNVs discovered in this study; Genes, protein coding genes; Large indels, variants 50 to 499 bp discovered in this study; SNPs, SNPs and short indels from 1001 Genomes Project; TEs, annotated TEs.
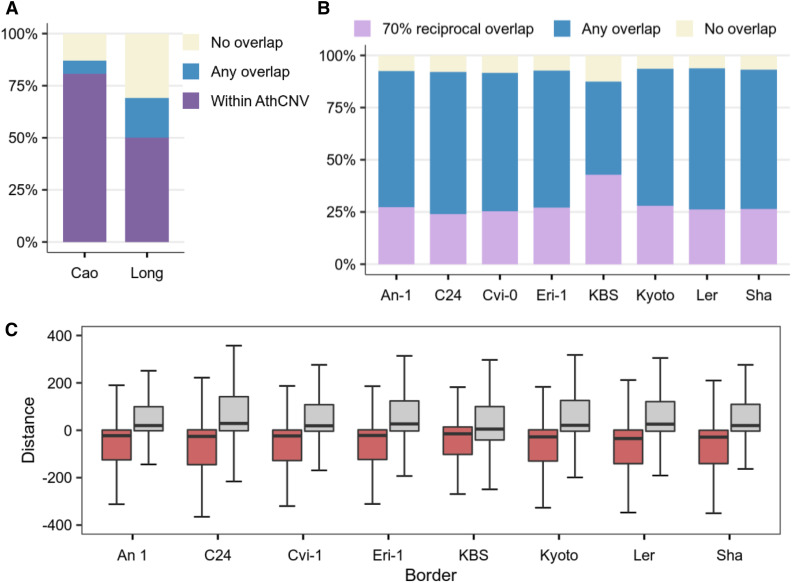
In the subsequent analysis, we focused on CNVs since this class of variants—due to their size—may directly influence the copy number and dosages of entire functional loci, including genes. Since our data analysis pipeline involved two CNV merging steps (between samples and between tools) that preceded the breakpoint refinement step, we attempted to verify the sensitivity and accuracy of our approach at three levels: species, geographically related accessions, and individual genomes (Figure 3A and 3B). For species-level verification, we used CNVs previously identified in a population of 80 accessions that represented a similar geographic range and were not included in our data set (Cao et al., 2011). Of the 1059 CNVs identified in that study, 87% overlapped with AthCNV regions and 81% were positioned entirely within them. This result was in line with our expectations, since the previously identified CNVs were much shorter.
Figure 3.
Overlap of the AthCNV Data Set with Variants Identified in Small Populations and Individual Genomes.
(A) Fractions of CNVs identified previously in a small, worldwide population of 80 accessions (Cao data set) and a narrow population of Swedish accessions (Long data set) that overlap with AthCNVs.
(B) Fractions of CNVs detected in the genomes of individual accessions assembled de novo from long reads that overlap with AthCNVs.
(C) Relative distances between the breakpoints in the AthCNVs and the breakpoints in CNVs in eight accessions (each used as a reference for AthCNV distance calculation). Boxplots depict data for pairs of variants with ≥70% reciprocal overlap. Boxplots show median (inner line) and inner quartiles (box). Whiskers extend to the highest and lowest values no greater than 1.5 times the inner quartile range.
For verification at the level of geographically related accessions, we evaluated the overlap of the AthCNV data set with the duplications and deletions previously detected in 180 Swedish accessions (Long et al., 2013), 174 of which were also included in our analysis. After merging directly adjacent regions with duplications and removing private variants (since they were also filtered out by our CNV discovery pipeline; see Methods), we obtained 235 deletions and 1487 duplications ≥0.5 kb in length in the Swedish samples. We observed that 76% of deletion regions overlapped with the AthCNVs, and 51% were positioned entirely within them. Likewise, 68% of duplication regions overlapped with AthCNVs, and 50% were located entirely within them.
Finally, we investigated how well the AthCNV data set fit the variants identified in eight genomes representing individual accessions. One genome (KBS-Mac-74 accession) has been assembled to the contig level from Nanopore ultralong reads (Michael et al., 2018). We used the Assemblytics tool (Nattestad and Schatz, 2016) to identify CNVs in this genome (Supplemental Data Set 3). The seven remaining genomes (An-1, C24, Cvi-0, Eri-1, Kyoto, Ler, and Sha accessions) were assembled into five chromosome-level scaffolds from PacBio ultralong reads, and structural variants were identified with the SyRI tool (Jiao and Schneeberger, 2020). Note that both SyRI and Assemblytics rely on the same genome aligner, MUMmer. We selected CNVs ≥0.5 kb in length (the reference genome coordinates were considered in the size evaluation) and compared them with our data set.
In each accession, the majority of CNVs (91 to 99%) were shorter than 20 kb, similar to the AthCNVs. From 88 to 94% of the CNVs in each accession overlapped with the AthCNVs by at least 1 bp. As many as 63 to 77% deletions, but only 22 to 25% duplications overlapped with individual AthCNVs by at least 70% and therefore had similar lengths and breakpoint locations (Supplemental Figure 2). We also observed that the AthCNVs for which the breakpoints best fit the breakpoints of variants found in individual genomes, that is, the localization of one of their borders (left or right) differed by no more than ±10 bp (Figure 3C), were mostly refined using the split reads and hybrid approach (91 to 94%) or the discordant read pair approach (7 to 9%). These observations validate the approach we used to assess CNV borders (the highest priority was given to the information provided by the callers based on discordant read pairs and split reads) and explained the lower accuracy of assessing duplication breakpoints. Taking the above-mentioned information into account, the AthCNV map reliably represents variants present in individual accessions.
We also performed literature mining and found 106 genes for which complete or partial duplications/deletions have been reported and—as an obligatory criterion—experimentally confirmed in Arabidopsis (Supplemental Data Set 4; Grant et al., 1995; Stahl et al., 1999; Xiao et al., 2001; Kroymann et al., 2003; Werner et al., 2005; Balasubramanian et al., 2006; Clark et al., 2007; Staal et al., 2008; Vlad et al., 2010; Smith et al., 2011; Bloomer et al., 2012; Cole and Diener, 2013; Karasov et al., 2014; Vukašinović et al., 2014; Pucker et al., 2016; Zmienko et al., 2016; Samelak-Czajka et al., 2017; Michael et al., 2018). We found that 100 genes overlapped with AthCNVs (Supplemental Figure 3). Four additional genes overlapped with low-confidence variants, which were also detected by our CNV discovery pipeline. Thus, our data are highly consistent with the existing experimental evidence on the distribution of CNVs in the Arabidopsis genome.
Genomic Content in CNV Regions
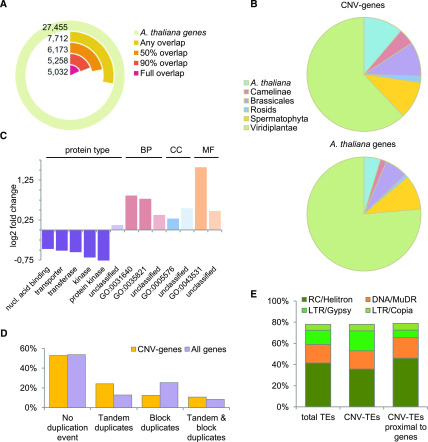
We observed uneven genome coverage by CNVs (Table 1). From 84 to 99% of the centromeric regions were covered by CNVs, with multiple CNVs of various lengths overlapping with each other (Supplemental Figure 4). In Arabidopsis, the centromeres are rich in 178- to 180-bp repeats and TEs (Minoru, 2013). Additionally, in the noncentromeric parts of the genome, the distribution of CNVs was positively correlated with the distribution of TEs and negatively correlated with the distribution of the genes. Nevertheless, a very large number of genes (7712) overlapped with CNV regions. We hereafter refer to genes and TEs covered by AthCNVs by at least 1 bp as CNV-genes and CNV-TEs, respectively, to distinguish them from NONVAR-genes and NONVAR-TEs, which did not overlap with any CNVs. We then investigated more deeply the fraction of CNV-genes that were covered by CNVs for ≥90% of their length (Figure 4A). These genes were highly represented by orphan genes, that is, genes with no detectable homologues in any other species (497 of the 1170 orphan genes present in the Arabidopsis genome) and species-specific gene families (49 of the 55 families found only in this species; Figure 4B; Supplemental Table 3). They were also significantly overrepresented in genes encoding proteins of an unclassified type (binomial test with Bonferroni-corrected P-value < 0.01; Figure 4C). Similarly, we observed significant overrepresentation in CNV-genes that are unclassified based on the Molecular Function, Biological Process, and Cellular Component Gene Ontology (GO) terms. In addition, terms related to plant interactions with other organisms, defense, and stress responses were overrepresented in each category. There were no significantly depleted GO terms, but genes encoding nucleic acid binding proteins, transporters, transferases, and protein kinases were significantly underrepresented in the CNV-genes data set.
Table 1. Arabidopsis Genome Coverage by the Identified CNVs.
| Region Type | No. of Variants | Mean Coverage (%) of the Given Region Typea | Average No. of Variants in Overlapping Segmentsb | |
|---|---|---|---|---|
| Genome | 19,003 | 35.7 | 3.8 | |
| Centromeres | 6,584 | 93.5 | 7.2 | |
| Outside centromeres | 12,419 | 28.0 | 2.4 | |
| Overlapping protein-coding genes | 6,326 | 18.5 | 1.7 | |
| Overlapping pseudogenes | 943 | 59.6 | 2.6 | |
| Overlapping TEs | 8,548 | 94.0 | 3.7 | |
Calculated from the following formula: coverage in individual region of a given type = number of bases overlapped by any CNV/number of all bases in this region × 100%; average value is reported in the table.
Calculated as the number of CNVs overlapping each region in 1-bp windows. Average number is reported in the table. To remove the bias resulting from different overall coverage of various region types, only the positions with nonzero overlap were counted, for example, for a 1000-bp pseudogene overlapped by several CNVs in a total of 46% of its length; the number of overlapping variants was counted for 460 1-bp intervals covered by any CNV and averaged.
Figure 4.
Genomic Content in Regions Overlapped by AthCNVs.
(A) Fractions of annotated Arabidopsis genes with various degrees of overlap with AthCNV variants.
(B) Enrichment of CNV-genes that are overlapped by AthCNVs by at least 90% in the fractions of species-specific and clade-specific genes compared to that of all annotated Arabidopsis genes.
(C) Over- and underrepresented protein types and GO terms among the CNV-genes, in the Biological Process (BP), Cellular Component (CC), and Molecular Function (MF) categories. All terms are either significantly enriched or depleted (binomial test with Bonferroni-corrected P-value < 0.01). The GO terms shown in the chart are killing of cells of other organism (GO:0031640), modification of morphology or physiology of other organism (GO:0035821), extracellular region (GO:0005576), and ADP binding (GO:0043531). nucl., nucleic.
(D) Locations of CNV-genes in regions of tandem and block duplications in the genome compared to those of all genes.
(E) Superfamily composition of Arabidopsis TEs and its comparison with all CNV-TEs and gene-proximal CNV-TEs (located within ±2-kb distance). Top-four most abundant superfamilies are presented. Class I TEs are depicted in orange; class II TEs are in different shades of green. All families are listed in Supplemental Table 4. LTR, long terminal repeat. RC, rolling cycle.
A recent comparative study of seven Arabidopsis genomes assembled de novo from long reads revealed multiple regions with strongly decreased collinearity and multiple haplotypes (Jiao and Schneeberger, 2020). These regions were referred to as hotspots of rearrangements and were enriched in TEs and depleted in genes, similar to the CNVs identified in our study. Additionally, similar to our CNV-genes, the genes within the hotspots of rearrangements were enriched for functions related to biotic stress response. In addition, they displayed high CNV and high mutation frequency among the seven accessions. We therefore expected them to be identified as population-level CNVs in our study. Indeed, we found that 98.6% of rearrangement hotspots overlapped with AthCNVs (73.6% were entirely within CNV regions). Of the eight regions without overlap, two were near AthCNVs (less than 250 bp), and four formed a large cluster with numerous adjacent hotspots of rearrangements, which extended for over 212 kb and was flanked by multiple CNVs on both sides. Many hotspots of rearrangements shared a common pattern of almost exclusively forward tandem gene duplications and large indels (Jiao and Schneeberger, 2020), which prompted us to investigate whether AthCNVs were also enriched in tandem duplications. According to the Plaza 4.0 database (Van Bel et al., 2018), 25.3% of genes in the Arabidopsis genome are located in regions of segmental duplications, while 12.8% arose through tandem duplication events (additionally, 8.3% are located in regions with both segmental and tandem duplications). These proportions were reversed among CNV-genes, with 12.4% of these genes localized in regions of segmental duplications and 24.1% in regions of tandem duplications (additionally, 10.7% underwent both segmental and tandem duplications; Figure 4D). Altogether, these observations indicate that the regions of tandem duplications are sites that accumulate rearrangements and, consequently, show high structural diversity.
In the next step, we analyzed CNV-TEs, which constituted 67.5% of all TEs. These TEs were slightly depleted in RC/Helitron TEs and enriched in long terminal repeat/Gypsy TEs (Figure 4E); however, the composition of CNV-TE superfamilies did not change much compared to all TEs (Supplemental Table 4). We also investigated how many CNV-TEs were proximal to genes, that is, overlapped with genes or were located within 2-kb regions flanking the genes. Only 36.2% of CNV-TEs were proximal to genes, and they were slightly enriched in RC/Helitron TEs but severely depleted in long terminal repeat/Gypsy TEs compared to both all CNV-TEs and the entire genome. They were also moderately enriched in DNA/MuDR elements. In contrast to CNV-TEs, genes with TEs in their proximity constituted the majority (64.4%) of the CNV-gene data set.
Interplay between the Copy Number Polymorphism of Genes and TEs
To investigate the relationship between the copy number polymorphism of genes and TEs, we compared the genomic distributions of CNV-genes and CNV-TEs. Both CNV-genes and CNV-TEs were, on average, located closer to the chromosome centromeres than were their NONVAR counterparts, and this tendency was much stronger for TEs than for genes (Figure 5A). However, the average distance between CNV-genes and the nearest TEs was smaller than the average distance between NONVAR-genes and the nearest TEs. The reverse was observed for CNV-TEs, which were, on average, farther from the nearest gene than were NONVAR-TEs (Supplemental Figure 5). Our observations indicated that some selective forces have opposite effects on shaping the relative distribution patterns of CNV-genes and CNV-TEs. The cut-insert and copy-insert mechanisms underlying TE mobility may affect adjacent genes, usually in a negative manner, for example, by interrupting gene coding or regulatory sequences, by gene rearrangement and duplication, or by altering their DNA methylation status (Quadrana et al., 2016; Bourque et al., 2018). Gene proximity may therefore be considered a negative force acting against nearby TE transposition, especially in the case of genes involved in crucial metabolic processes. On the other hand, TE proximity may contribute to increased copy number polymorphism of nearby genes by inducing DNA breaks and genomic instability.
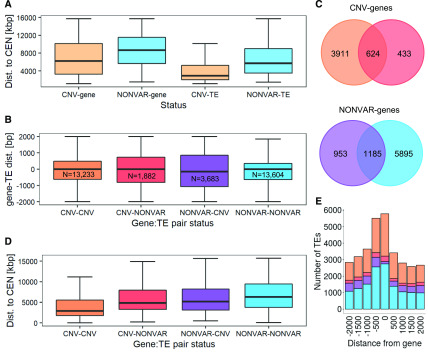
Figure 5.
Links between Genes and TE Variation and Localization.
(A) Distance to centromeres of genes and TEs grouped by variation status (determined based on their overlap with AthCNVs). The groups were significantly different (Wilcoxon rank sum test with continuity correction, P < 0.0001). Genetic elements localized in the pericentromeric regions were not included. dist., distance.
(B) Relative distances between genes and their proximal TEs, grouped by variation status. For each gene, a proximal TE was defined as each TE overlapping with this gene (distance = 0) or overlapping region located within 2 kb upstream from the gene’s 5′ untranslated region (distance < 0) or overlapping region located within 2 kb downstream from 3′ untranslated region (distance > 0). N, number of pairs with a given variation status. dist., distance.
(C) Number of unique CNV-genes and NONVAR-genes with proximal CNV-TEs and NONVAR-TEs and their overlap.
(D) Gene distances to centromeres presented for gene-TE pairs differing by variation status. dist., distance.
(E) Number of proximal TEs within and around genes. Colors in (B) to (E) are identical for the same groups. Boxplots in (A), (B), and (D) show median (inner line) and inner quartiles (box). Whiskers extend to the highest and lowest values no greater than 1.5 times the inner quartile range.
To extend our observations to all genes, we analyzed the distances and compared the CNV statuses of genes and their proximal TEs. We found strong enrichment in pairs where proximal TEs and genes had the same variation statuses (Figure 5B), regardless of whether they were both polymorphic (40% pairs) or invariable (42% pairs). Furthermore, 3911 of 4968 unique CNV-genes (79%) had only CNV-TEs in their proximity and 5895 of 8033 unique NONVAR-genes (73%) had only proximal NONVAR-TEs (Figure 5C). Additionally, the gene-TE pairs with the same variation statuses were located closer to each other than pairs with the opposite statuses. Combining the information about the genomic distribution and relative distances of genes and TEs clearly revealed that the localization of polymorphic gene–TE pairs was biased toward centromeres, while the localization of invariable gene–TE pairs was biased toward chromosome ends (Wilcoxon rank sum test with continuity correction for the difference between CNV-CNV and NONVAR-NONVAR groups, P-value < 0.0001; Figure 5D). Moreover, CNV-genes with proximal CNV-TEs were enriched in extracellular proteins and proteins involved in cell disruption, defense responses, and nucleic acid catabolism (Supplemental Data Set 5). At the same time, NONVAR-genes with proximal NONVAR-TEs were enriched in nuclear proteins and proteins involved in nucleic acid metabolism, regulation of fertilization, and transcription factor activity. There was no difference in the chromosomal distribution of pairs displaying opposite variation statuses, and no or few GO terms were enriched in these two groups.
Interestingly, the combined variation status of gene–TE pairs was also apparently related to the position of TEs relative to nearby genes (Figure 5E). All TEs localized in proximity to genes were 1.2 to 1.4 times more often inserted in their upstream flanking regions compared to downstream flanking regions. CNV-TEs very rarely overlapped with NONVAR-genes (3.8% cases) compared to CNV-genes (19.4%) or NONVAR-TEs, which overlapped with both NONVAR-genes and CNV-genes at similar frequencies (20.2 and 17.4%, respectively). The four groups had similar TE family compositions, which indicated that these differences were not caused by insertion bias of any specific TEs. Altogether, our observations confirmed the presence of selective constraints reciprocally imposed on genes and TEs, which is an important factor contributing to their present variation and genomic distribution patterns.
Copy Number Genotyping and Experimental Evaluation of CNV-Genes
After we identified the genomic regions showing copy number polymorphism in Arabidopsis, we used the Genome STRiP SVGenotyper module (Handsaker et al., 2015) to evaluate the copy number statuses of CNV-genes in individual accessions based on read depth estimates. Based on our earlier observations, we decided to directly evaluate the copy numbers of the genes covered by AthCNVs (using the gene coordinates as the input) instead of the AthCNVs themselves. Our motivation was to simplify the subsequent application of the copy number genotyping data in functional analyses. AthCNVs overlapping with each other may have been formed by different molecular mechanisms and may be present in different accessions (Zmienko et al., 2016); however, at the population scale, they collectively contributed to the copy number diversity of the CNV-genes that they covered (Supplemental Figure 6). Accordingly, we observed that the direct genotyping of CNV-genes provided the most accurate information about their copy number statuses in individual accessions. We ultimately genotyped 7324 CNV-genes as well as—for comparison purposes—5060 genes overlapped by low-confidence CNVs and 14,661 NONVAR-genes in 1060 accessions. These data can be accessed through the web interface at http://athcnv.ibch.poznan.pl in the form of user-generated plots.
Genome STRiP SVGenotyper is capable of assigning integer copy numbers to genotyped regions. We found, however, that it frequently assigned the copy number classes to intervals of only one copy; because Arabidopsis is a predominately selfing species, the expected differences between copy number alleles were multiples of two (Supplemental Figure 7). The integer copy number assignment by Genome STRiP SVGenotyper was also disturbed by the presence of CNV-genes that did not form clear, discrete copy number classes or for which the reported copy number was very high (up to many thousands of copies) in most accessions, including Arabidopsis ecotype Columbia (Col-0), which was expected to have the reference diploid copy number (two copies) for each gene. Such problems were commonly encountered when genotyping complex CNVs and CNVs that were mapped to segmental duplications (Conrad et al., 2010; Campbell et al., 2011; Handsaker et al., 2015). For these reasons, we reported unrounded rather than integer copy number outputs. Additionally, we filtered the genotyping data by excluding genes with extreme copy numbers in Col-0 separately for each of the three data sets. In this step, we removed 451 genes from the analysis.
The global distributions of the copy number estimates obtained for CNV-genes significantly differed from those obtained for NONVAR-genes, which were more uniform (interquartile range for NONVAR-genes was 0.23 versus 0.30 for CNV-genes) and much more concentrated around the reference diploid copy number value (kurtosis = 13 for NONVAR-genes versus 120 for CNV-genes). Moreover, CNV-genes had significantly higher copy number variance, larger copy number ranges, and more extreme maximum and minimum copy number values than did NONVAR-genes (Figure 6). Genes covered by low-confidence CNVs had intermediate values, but overall, they were more similar to NONVAR-genes than to CNV-genes.
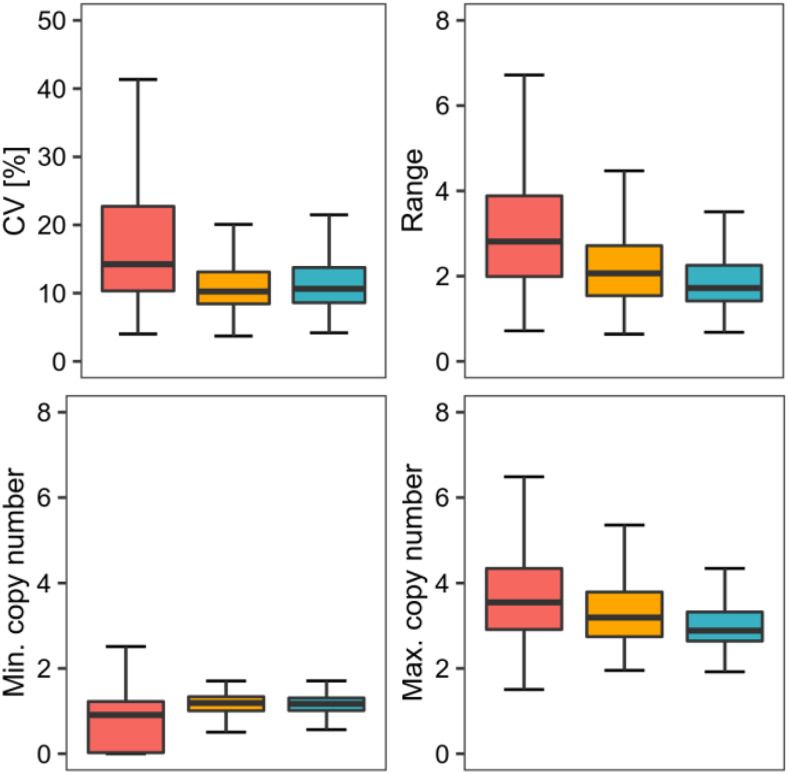
Figure 6.
Differences between CNV-Genes, NONVAR-Genes, and Genes Covered by Low-Confidence CNVs in Terms of the Read Depth–Based Copy Number Genotypes.
The genotyping data for 7031 CNV-genes (red), 4482 low-confidence CNV-genes (orange), and 14,877 NONVAR-genes (blue) were compared for four attributes: the coefficient of the CNV (CV; top left), the copy number range in a population represented by 1060 accessions (top right), and the minimum (min.) and maximum (max.) copy number values (bottom left and bottom right, respectively). For each attribute tested, CNV-genes significantly differed from the other groups (Kruskal–Wallis test, P < 0.0001, Dunn–Bonferroni post hoc method P-value < 0.0001). Boxplots show median (inner line) and inner quartiles (box). Whiskers extend to the highest and lowest values no greater than 1.5 times the inner quartile range.
For 1777 (25.3%) CNV-genes, we observed an unexpectedly small level of variation: for these genes, the copy number difference between any two accessions in the population was <2. One reason for the low level of variation in these CNV-genes was their partial overlap with AthCNVs. In these cases, the reads that mapped to the invariable gene segments contributed to read depth estimates, reducing the observed differences between the accessions with distinct copy number statuses (Supplemental Figure 8). Therefore, for all subsequent analyses, we selected only the 5517 CNV-genes that had ≥50% overlap with AthCNVs. This reduced the percentage of CNV-genes with low variation to 17.7%. To further investigate the possible reasons for their low variation, we assigned each CNV-gene to its longest overlapping AthCNV and found that all CNV-genes with little variation in copy number were contained in only 332 AthCNVs. Moreover, 228 of these AthCNVs also encompassed CNV-genes with high CNV (Supplemental Figure 9). This result suggested that some AthCNVs included small nonvariable subregions, presumably not identified during the segmentation step. We further observed that the presence of this mosaicism was related to AthCNV size—CNV-genes with little variation in copy number were covered by very long AthCNVs, with a median size of 183.4 kb. For comparison, the median size of AthCNVs covering CNV-genes with high CNV was 19.9 kb.
We further verified the accuracy of our read depth–based copy number estimates by performing multiplex ligation-dependent probe amplification (MLPA) assays using 314 accessions (i.e., 30% of the genomes genotyped with Genome STRiP). The experiment involved CNV-genes located in 45 nonoverlapping AthCNVs (Supplemental Figure 10) and four NONVAR-genes. While read depth–based genotyping provided copy number estimates for entire CNV-genes, by disregarding factors such as incomplete overlap with AthCNVs, the fine-scale MLPA approach focused on small (<75-nucleotide) target regions within the assayed genes, which made it more precise but also more sensitive to the presence of local sequence variations such as SNPs and indels. After taking these factors into account, we were able to explain most of the discordant results observed in our experiment by the presence of sequence variation in MLPA probe binding sites in the assayed accessions (Supplemental Figures 11 and 12). Overall, the MLPA-based genotyping results were in agreement with the read depth–based estimates for all assayed genes (Supplemental Figures 13 to 15). For numerous multiallelic CNV-genes, the clusters of samples with the same copy number could be clearly distinguished by plotting the read depth–based data against the MLPA data (Figure 7).
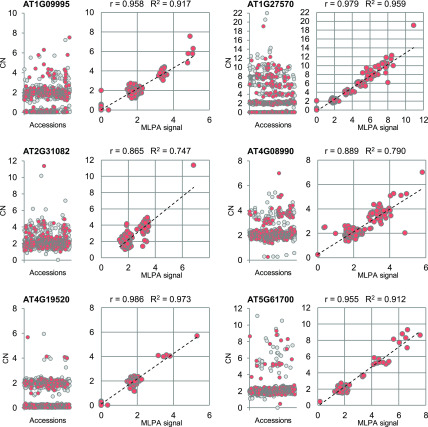
Figure 7.
Experimental Validation of Read Depth–Based Copy Number Genotyping Results.
For each CNV-gene, two scatterplots are presented: read depth–based copy numbers (CN) for 1060 accessions (left) and the correlation of the genotype data with the MLPA results for 314 accessions (right). The same set of accessions was used in all MLPA experiments, which are labeled in red in the plots on the left. The MLPA results were scaled for each CNV-gene using Col-0 signal as a reference value (CN = 2). R, Pearson correlation coefficient; R2, coefficient of determination of linear regression.
Interestingly, the MLPA analysis provided another, although unexpected, piece of evidence supporting the accuracy of our read depth–based genotyping results. Initially, we included 346 accessions in the MLPA assays. However, 32 of them were recently reported as potentially mislabeled in public seed repositories (from which we acquired our seed collection) based on resequencing and SNP analyses, which failed to assign these stocks to the expected strains (Pisupati et al., 2017). In agreement with these findings, we observed a very strong negative effect of these 32 samples on the correlation between the read depth–based and MLPA results (Supplemental Figure 16; Supplemental Table 5). Consequently, we removed them from the MLPA analysis.
Arabidopsis Population Structure Revealed by CNV Markers
The analysis of SNP markers in the 1001 Genomes Project accessions revealed that 95% of Arabidopsis accessions belong to one genetic group composed of several subgroups of accessions sharing a similar geographic origin (Platt et al., 2010; 1001 Genomes Consortium et al., 2016). The remaining 5% of accessions (referred to as relicts) form a few groups that are genetically distant from each other and from the nonrelicts (Lee et al., 2017). We aimed to infer Arabidopsis population structure from CNV markers and verify its consistency with the structure derived from SNP markers. We selected 1050 AthCNVs of various types (deletions, duplications, and multiallelic CNVs) distributed across the genome and used the copy numbers of the representative CNV-genes (one gene per AthCNV) as input for principal component analysis (PCA). We then compared our results to population structure derived from 1001 Genomes Project SNP markers. The first two principal components (PCs) revealed that the population is highly structured and that the accession groupings reflect their geographical distribution (Figure 8A), which is consistent with the SNP-based groupings (Cao et al., 2011; Horton et al., 2012). SNPs better distinguished the genetic subgroups than did the CNVs, which was an expected result, as the subgroups were defined based on SNP variation, and SNPs substantially outnumbered CNV markers (1001 Genomes Consortium et al., 2016). However, the CNV-based analysis better reflected the global distribution of the accessions (the directions of the accessions’ separation were consistent with geographical directions, north to south for PC1 and east to west for PC2, after removing clearly unique U.S. accessions; Figure 8B).
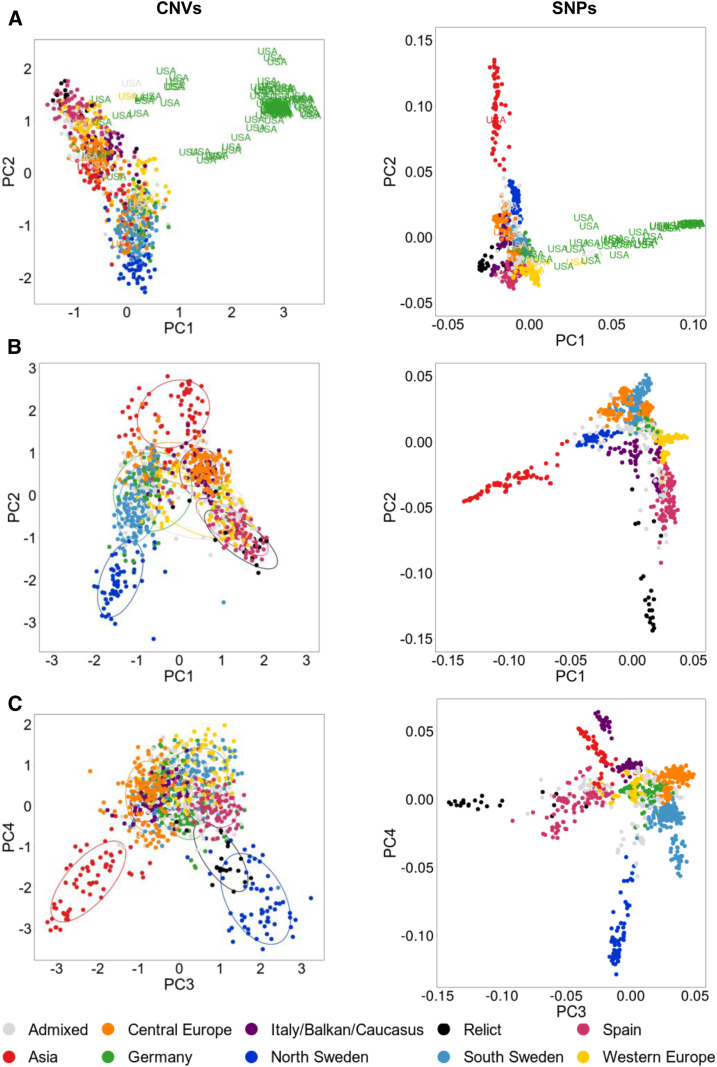
Figure 8.
Arabidopsis Population Structure Based on the Analysis of CNV Genotypes.
PCA was performed on 1060 accessions and on genotyping data from 1050 CNV-PCGs (left). For comparison, another PCA was performed on the same set of accessions and 117,232 SNPs from the 1001 Genomes Project (right).
(A) PC1 and PC2 components; all accessions were included. U.S. accessions assigned to the Germany subgroup were distinguished from the other samples.
(B) PC1 and PC2 components; U.S. accessions from the Germany subgroup were excluded from the analysis.
(C) PC3 and PC4 components; all accessions were included. The accessions in PCA plots are colored based on their 1001 Genomes Project grouping.
Interestingly, CNV-based PCA revealed some similarities between the accessions that were not captured by SNP-based grouping. The third and fourth PCs distinguished the groups from the edges of the natural species range and highlighted the genetic similarity of the northern Sweden accessions to the relict genomes from southern Europe (Figure 8C). Remarkably, this observation is in agreement with the recently proposed two-wave expansion model of Arabidopsis across Eurasia, derived from the analysis of the extent of relict introgression in the nonrelict genomes (Lee et al., 2017). According to this model, the populations from different glacial refugia (relicts) expanded from the south of Europe northward at the end of the last ice age. Subsequently, the ancestors of today’s nonrelicts expanded along the east–west axis, probably from the Balkans or the Black Sea area, and replaced the local accessions, except in the north and south of the species range, where large introgressions from the relict genomes (locally adapted) might have helped the nonrelicts colonize the habitats with more severe climatic conditions.
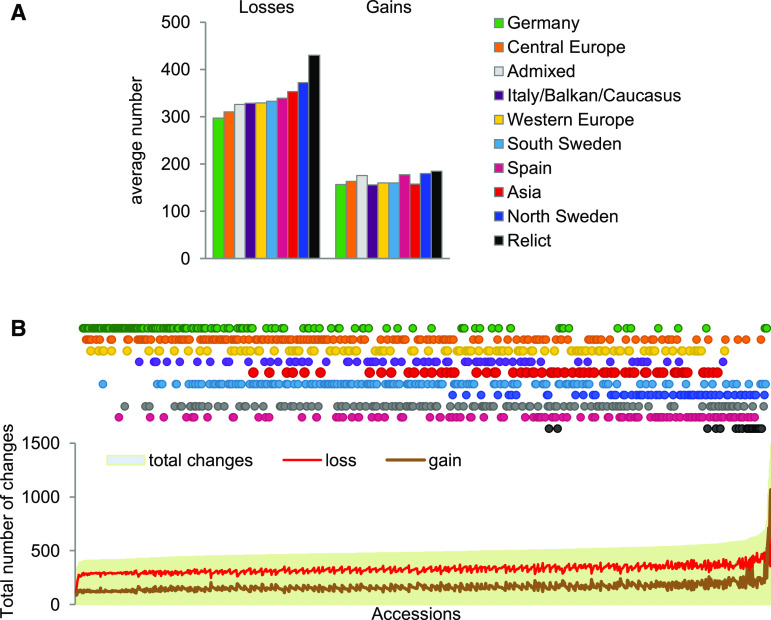
We then compared the extent of CNV-gene copy number changes between 1059 accessions (Col-0 was excluded from this analysis). To this end, we treated all copy number genotypes ≤1 as losses, all copy number genotypes >3 as gains, and all the remaining genotypes as unchanged. These thresholds were justified because the median copy number value for all accessions and all CNV-genes analyzed was 1.98. On average, copy number losses were more frequent in all subgroups (the mean gain-to-loss ratio was 0.5), and their amount differed among the subgroups to a greater extent than did that of copy number gains (Figure 9A). The subgroups least affected by CNVs were Germany (8.2%) and Central Europe (8.6%), while the relicts (11.2%) and northern Sweden (10.0%) subgroups were most affected. This order was in good agreement with the general similarity of the subgroups to the reference genome (the Col-0 accession was assigned to the Germany group) but also confirmed the general rule that the choice of a reference genome is a crucial step that determines the range of variation that may be identified by a mapping-based approach.
Figure 9.
Losses and Gains in Gene Copy Number in Arabidopsis Subgroups.
(A) Average number of gene copy number gains and losses in the subgroups.
(B) Total number of gene copy number changes in individual accessions.
In individual accessions, 3.9 to 26.9% of CNV-genes were affected by copy number changes (Figure 9B), and this broad range was mostly caused by the differences in the number of gains (ranging from 88 to 1068) and, to a lesser extent, by the losses (ranging from 114 to 660). The top five accessions in terms of total copy number changes were also the top five in terms of the number of gains and had a gain-to-loss ratio ranging from 0.93 to 2.77. Two of the accessions were from Sweden (Ull2-5 and Sanna-2), while the remaining accessions were U.S. accessions (KBS-Mac-74, KBS-Mac-68, and BRR57).
Gene Dosage, Gene Expression, and Missing Duplications in the Reference Genome: SEC10 Example
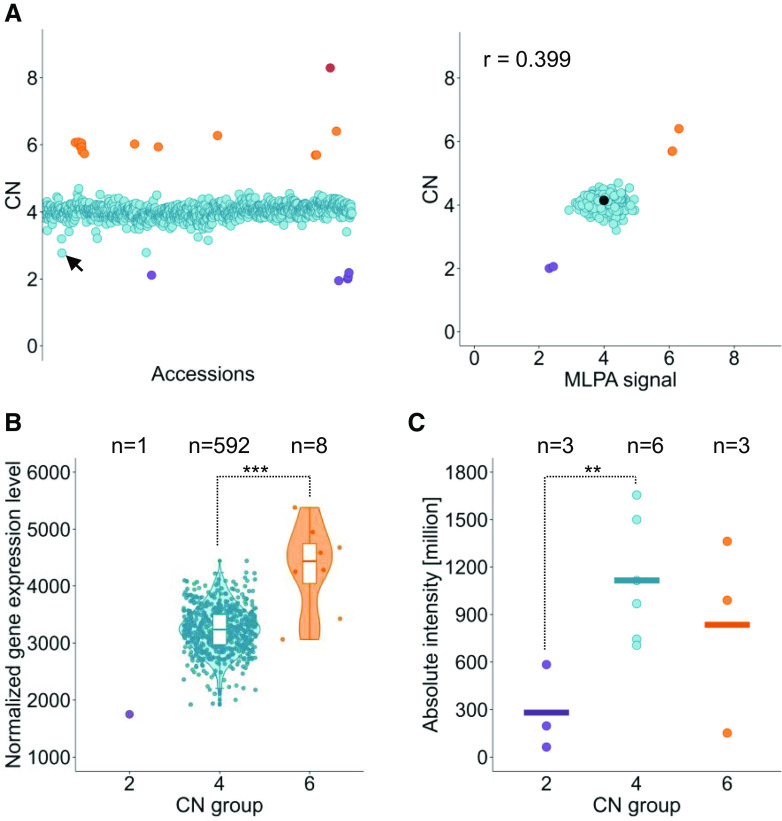
Duplication of AT5G12370, encoding the SEC10 protein involved in exocytotic vesicle fusion, was recently discovered in the Col-0 accession (Vukašinović et al., 2014). The SEC10 duplication is absent from TAIR10 version of the Arabidopsis reference genome (the reference sequence is a chimera of both copies). To determine whether other gene duplications occur in Col-0, we manually searched the genotyping results for the CNV-genes excluded by our interquartile range–based filter. As a result, we identified eight candidates that were possibly duplicated in Col-0, including SEC10 (Supplemental Figure 17). Our genotyping results indicated that the SEC10 duplication was prevalent in the Arabidopsis population, as four, six, and eight copies were detected in the diploid genomes of 1039 accessions, 14 accessions, and 1 accession, respectively, while two copies were detected in only 6 accessions (0.56%; Figure 10A). We also evaluated SEC10 expression in 601 accessions using available RNA sequencing (RNA-seq) data (Kawakatsu et al., 2016) and observed that the transcript levels increased in samples with elevated SEC10 copy numbers (Figure 10B). To determine whether these differences were also reflected at the protein level, we analyzed the SEC10 protein content in 12 accessions representing genotypes with two, four, or six copies of SEC10. Indeed, the mean protein level was significantly higher in accessions with four SEC10 gene copies than in those with two copies (Figure 10C; Supplemental Figure 18). It was also elevated in two of three accessions with six copies compared to samples with no SEC10 duplication.
Figure 10.
Prevalence of the Duplication of the SEC10 Gene and Its Effects on Transcript and Protein Levels.
(A) SEC10 gene copy number in the Arabidopsis population. (Left) Read depth–based copy number (CN) genotypes plotted for 1060 accessions. (Right) Verification of the genotyping data with MLPA assays for 314 accessions. The MLPA signal was scaled to that of the Col-0 accession (marked in black, CN = 4). R, Pearson correlation coefficient.
(B) Distribution of RNA-seq normalized transcript levels among accessions grouped by the copy number class. White boxplots show median (inner line) and inner quartiles (box). Whiskers extend to the highest and lowest values no greater than 1.5 times the inner quartile range, and dots represent the measurements in individual accessions. Asterisks indicate significant differences based on Welch’s t test (***, P < 0.01). Significance was not calculated for the copy number (CN) = 2 group, which included only one sample.
(C) SEC10 protein levels in 3-week-old plants grouped by copy number class. Horizontal lines represent the mean protein level in each group, and the dots represent the measurements in individual accessions. Asterisks indicate significant differences based on Student’s t test (**, P < 0.05). The data were averaged from the measurements of four SEC10 peptide fragments identified by mass spectrometry. The quantification results for individual peptides are presented in Supplemental Figure 18. In each plot, the accessions are colored according to the copy number (CN) classes manually assigned based on the genotyping data: CN = 2 (purple), CN = 4 (blue), CN = 6 (orange), and CN = 8 (red). The accession with the lowest unrounded copy number assigned to the CN = 4 group is KBS-Mac-74 (marked by a black arrow in the left plot); for this accession, the presence of a tandem duplication was confirmed by a BLAST search of the SEC10 nucleotide sequence against a nanopore-based genomic assembly, confirming the correct group assignment.
Genome-Wide Association Study of CNVs
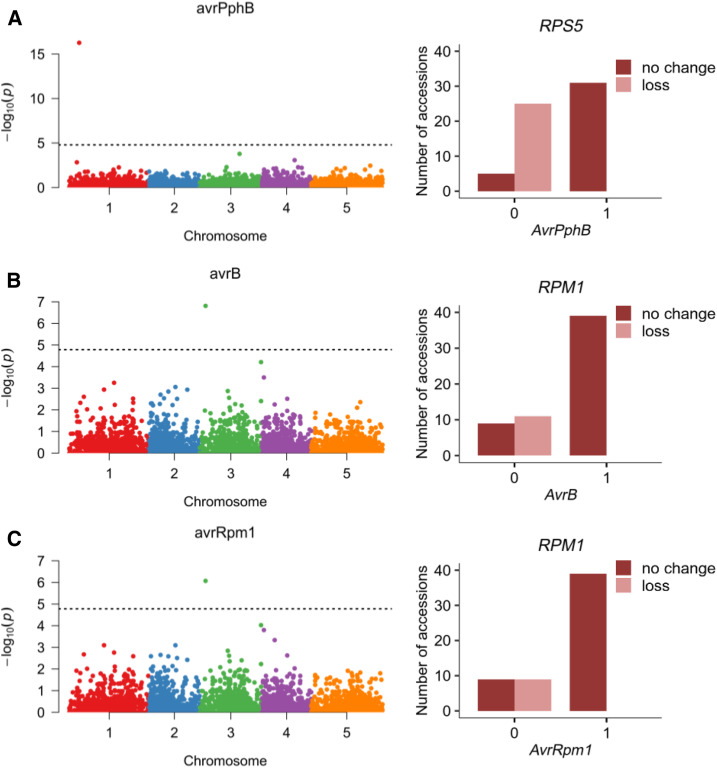
Several studies have provided evidence that CNVs account for a substantial amount of phenotypic variation. In particular, presence-absence polymorphism of resistance genes that are involved in race-specific recognition of pathogen avirulence determinants (McHale et al., 2006) contributes to plant resistance phenotypes. In Arabidopsis, CNVs affect numerous loci related to biotic responses, including RPM1, RPS5, RLM1, RLM3, RPP1, RPP5, and RPP7 (Grant et al., 1998; Henk et al., 1999; Yi and Richards, 2009; Roux and Bergelson, 2016). A previous genome-wide association study revealed strong SNP associations for four hypersensitive response phenotypes to Pseudomonas elicitor proteins: AvrPphB, AvrB, AvrRpm1, and AvrRpt2 (Atwell et al., 2010). Single candidate loci encoding known resistance genes could be associated with these SNPs: RPS5 for AvrPphB, RPM1 for AvrB and AvrRpm1, and RPS2 for AvrRpt2. According to our results, RPS2 is not a CNV-gene; therefore, the association for this gene likely resulted from small-scale variation. We wanted to find out, however, whether the remaining two genes, for which the impact of gene deletion on pathogen resistance has been confirmed previously (Grant et al., 1998; Stahl et al., 1999, Karasov et al., 2014), could be directly distinguished in an association analysis using our genotyping data. To test this possibility, we selected 23 defense-related phenotypes from the Atwell et al. (2010) study, including the four hypersensitive response phenotypes mentioned above (Supplemental Data Set 6). This medium-sized data set consisted of 76 to 175 accessions per phenotype, 51 to 117 of which were shared with our study. Using CNV-gene statuses (gain, loss, or no change) as genetic markers, we filtered the CNV-genes using a 1% minor allele frequency threshold, which left only 2519 CNV-genes. We then evaluated their association with each phenotype using a linear mixed model correcting for population structure (efficient mixed-model association expedited). For eight phenotypes, we obtained significant associations with one to eight CNV-genes (Supplemental Figure 19). Among these, the strongest were single-gene associations with three phenotypes of interest: avrPphB (RPS5 gene, –log10 P-value = 16.27), avrB (Rpm1 gene, –log10 P-value = 6.81), and avrRpm1 (Rpm1 gene, –log10 P-value = 6.07). These results are in perfect agreement with previous results (Figure 11). This serves as a proof of concept that CNVs can serve as powerful and informative markers for traits where copy number polymorphism is a causative agent of the observed phenotypic variation.
Figure 11.
Association of Gene Copy Number Losses in Arabidopsis with Defense Phenotypes.
(A) AvrPphB phenotype.
(B) AvrB phenotype.
(C) AvrRpm1 phenotype. Left panels show Bonferroni-corrected P-values from association analysis; right panels show copy number allele distribution for significantly associated CNV-genes.
DISCUSSION
Analysis using SNP patterns combined with transcriptomic, proteomic, and phenotypic data has led to the efficient discovery of gene function. However, within the last decade, it has become increasingly clear that variation in gene dosage may also lead to phenotypic diversity within a species. Therefore, copy number genotypes must also be considered when attempting to uncover the genetic basis of many traits (Stankiewicz and Lupski, 2010; Żmieńko et al., 2014). To date, unlike our knowledge about SNPs, our inadequate understanding of CNV locations and frequencies in the Arabidopsis 1001 Genomes collection has limited our ability to identify links between genotype and phenotype in this model dicot. Here, we performed an integrative study involving detailed characterization of CNVs in the Arabidopsis genome and their impact on gene dosages. Our map, based on the WGS data for 1064 accessions, substantially extends the list of identified regions with structural variation in this plant obtained from previous studies (Cao et al., 2011; Long et al., 2013). We also performed extensive experimental verification of the genotyping results: we assayed 45 CNV-genes, all in the same set of 314 randomly selected accessions, which guaranteed that the results were not biased toward presenting only a subset of data with the strongest correlations for each CNV. We obtained high concordance between the read depth–based copy numbers and the MLPA signals not only for deletions but also for rare duplications and multiallelic CNV-genes, which is worth noting since experimental verification of duplications has been performed occasionally in large-scale CNV discovery studies in plants (Springer et al., 2009; Swanson-Wagner et al., 2010; Saintenac et al., 2011; Zheng et al., 2011; McHale et al., 2012; Muñoz-Amatriaín et al., 2013; Yu et al., 2013).
Similar to studies involving other plant species (Chia et al., 2012; Muñoz-Amatriaín et al., 2013; Hardigan et al., 2016), we reported high but uneven genome coverage by CNVs in Arabidopsis. We hypothesize that the distribution of CNVs in the genome results from structural and functional constraints on their formation and preservation. The structural constraints may be reflected by the increased representation of tandem duplicates among the CNV-genes identified in our study, which is consistent with the previous finding that CNV regions are hotspots of both past and present large-scale variations (Schuster-Böckler et al., 2010; Jiao and Schneeberger, 2020). The functional constraints might cause highly conserved genes and genes encoding proteins involved in numerous interactions within the cell to be underrepresented in CNV regions due to the usually negative effect of changes in their dosages (Krylov et al., 2003; Platt et al., 2010). In line with this observation, the CNV-genes detected in our study were enriched for less conserved genes, that is, Arabidopsis-specific genes and genes of unknown function. The changes in gene dosage may also provide immediate benefits, for example, a rapid increase in the amount of the enzyme providing drug or herbicide resistance. Indeed, there are several examples highlighting the dynamics of CNV-based adaptation (Harms et al., 1992; Jones et al., 1994; Caretto et al., 1995; Gaines et al., 2010; Kondrashov, 2012). Drawing from nature, processes that induce local changes in DNA copy might therefore be adopted to breed plants with desired traits. However, deeper knowledge about the mechanisms of CNV formation as well as the function of yet-uncharacterized genes is needed to achieve this goal.
AthCNV regions were highly enriched in class I and class II TEs and, similar to the TEs, were unequally distributed across the genome. Indeed, TEs are overrepresented in regions with structural variation (Huang et al., 2008; Cao et al., 2011; Gan et al., 2011; Niu et al., 2019). There is no bias in the localization of newly inserted TEs; however, the deletion of TEs is an ongoing, active, selective process that is largely responsible for the TE distribution pattern in the Arabidopsis genome (Quadrana et al., 2016). A comparison of the genomes of three Arabidopsis accessions, Col-0, Bur-0, and C24, revealed multiple polymorphic TEs for which large deletions were the most common type of variation (93%; Wang et al., 2013). TEs proximal to genes were less variable than distal TEs, suggesting that nearby genes have a negative effect on TE divergence, probably due to stronger selective constraints in these regions. By contrast, TE proximity was positively correlated with the level of small-scale mutations (SNPs and 1- to 3-bp indels) in the genes, pointing to a link between TEs and gene sequence variation. Our observations are in agreement with previous results, and they demonstrate that the variation statuses of genes and TEs are tightly linked and jointly contribute to the unequal distribution of these elements in the genome.
Early studies indicated that the genomes of individual Arabidopsis accessions contain segments not present in the reference genome. The total length of the new sequences in these genomes ranges from 1.3 to 3.3 Mbp (Ossowski et al., 2008; Gan et al., 2011). A recent analysis of the de novo assemblies of seven accessions showed that duplications are the most prevalent type of large CNV (Jiao and Schneeberger, 2020). Because of the limitations of short read–based sequencing (Alkan et al., 2011), we did not use de novo assembly-based approaches for CNV discovery; therefore, our study focused exclusively on regions that were present in the reference genome. Consequently, we detected copy number losses more frequently than copy number gains in most accessions. Nevertheless, by applying population-scale genotyping, we were also able to identify regions missing from the reference genome in our analysis represented by the Col-0 accession, including the recently described duplication of the SEC10 gene (Vukašinović et al., 2014). Homozygous mutant lines with T-DNA insertions in only one SEC10 gene had no obvious mutant phenotype; by contrast, introducing mutations in SEC6 or SEC8, which also encode components of the multiprotein exocyst complex, led to defects in pollen-specific transmission. SEC10 and its duplicate, which share 99% sequence identity, are thought to be functional and complementary (Vukašinović et al., 2014). Here, we showed that the natural duplication of the SEC10 gene is correlated with the increased transcription and production of SEC10 protein. Thus, our results strongly support the opinion of Vukašinović et al. (2014) on the role of SEC10 duplication in the Arabidopsis Col-0 accession. This example also highlights the importance of carefully considering the genetic background in functional and comparative studies. Therefore, we believe that the AthCNV map and the patterns of gene CNV resulting from our study will provide a valuable resource to the Arabidopsis community. They may, for example, guide the selection of the most appropriate sets of accessions for downstream analyses when investigating individual regions in the genome, regardless of whether the presence or lack of variation between these accessions is the main point of interest. As we demonstrated for hypersensitive response phenotypes in Arabidopsis, the copy number data may also complement SNP markers in genome-wide association studies (Fuentes et al., 2019), or to some extent supplement the small number of appropriate plant mutants in comparative functional analyses.
Because of their repetitive nature and the abundance of TE elements, CNV hotspots may accumulate duplications, deletions, and other rearrangements. These rearrangements may be triggered by various mechanisms (Gu et al., 2008; Gabur et al., 2019; Krasileva, 2019). Except for nonallelic homologous recombination events, which lead to recurrent copy number changes with nearly identical breakpoints, the CNV breakpoints in a given region may vary among individuals/accessions. The increasing availability and improvement of the accuracy of long-read DNA sequencing may facilitate more detailed characterizations of such complex CNVs (Michael et al., 2018; Jiao and Schneeberger, 2020). However, the use of population genetics based on chromosome-level sequence assemblies for large numbers of individuals is still a future goal. We observed high consistency between AthCNVs placed at our map, which is a map of merged CNVs and is therefore representative of the entire population rather than individuals, and the variants detected in individual accessions. Thus, we believe that the AthCNV map showing common CNVs in the Arabidopsis genome, combined with the CNV-gene genotyping data, will serve as a useful reference for future studies on variation in Arabidopsis at multiple levels.
METHODS
Data Preprocessing for CNV Discovery and Analysis
The raw reads for 1001 Genomes Project whole-genome shotgun sequence data were downloaded from the National Center for Biotechnology Information Sequence Read Archive repository (PRJNA273563; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA273563). Processed RNA-seq data (normalized counts) for 728 accessions were downloaded from the Gene Expression Omnibus repository (PRJNA319904; https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA319904). The CNV and large indels discovery pipeline was set up based on freely available published tools.
Data Filtering and Quality Analysis
FastQC v.0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) and Trimmomatic v.0.36 (Bolger et al., 2014) were used for read quality analysis and preprocessing. Briefly, the Illumina/Nextera adapter sequences were removed, and the leading and trailing sequences with low base quality (<15) were trimmed. Reads with <30 bases and an average quality score <20 were removed. Finally, reads with a local drop in base quality (average quality <15 measured with a four-base sliding window) were removed. For 45 accessions, fewer than 50% reads or 5,000,000 reads remained following the quality-based filtering, and these accessions were excluded from further analysis (Supplemental Data Set 7). The sequencing data for most rejected accessions were generated during the early stage of the 1001 Genomes Project (Cao et al., 2011), and we decided to remove all data generated at that stage (26 additional accessions) due to their overall lower quality and variable read lengths. The final data set for 1064 accessions was further processed with mapping and CNV detection tools following program-specific parameter optimization, as described below. For 23 accessions, we were unable to extract information about read pairs from the downloaded files; therefore, they were analyzed with read depth–based methods only.
Read Mapping and Marking Duplicates
The genomic reads were mapped to the TAIR10 reference genome assembly using BWA-MEM v.07.15 (Li and Durbin, 2010) and mrsFAST v.3.3.0 (Hach et al., 2014) with default parameters. For mrsFAST mapping, all reads within one sample were first trimmed to obtain a uniform length, and the final read length was calculated separately for each sample based on the largest value that allowed at least 80% of the reads to be kept after trimming. Picard Tools v.2.7.1 (http://broadinstitute.github.io/picard/) and SAMTools v.1.3.1 (Li et al., 2009) were used for data sorting and duplicate removal, respectively. For Genome STRiP analysis, the duplicates were marked, but not removed, to ensure that no unpaired reads remained after the duplicate removal step, since Genome STRiP requires the availability of only paired reads in the input data.
Calculating the Window Size for Read Depth–Based Methods
The number and lengths of the CNV calls when read depth–based methods are used depend on the window/bin size selected for the data-partitioning step. The bin size is a function of coverage, read length, and data quality. To account for all these variables, a bin size evaluation step was performed before the CNV calling step. For CNVnator, the suggested optimal bin size was that for which the ratio of the average read depth signal to its sd was ∼4 to 5. We calculated statistics for a wide range of bin sizes (100 to 1500 bases, with 100-base increments) for all samples (Supplemental Data Set 8). The selection of a very small bin size (100 bases) to ensure the highest sensitivity and resolution was justified for multiple samples, but not for all. Because large discrepancies in the CNV lengths and number between the samples might interfere with the subsequent merging process, we narrowed the acceptable bin size range to 400 to 800 bases. The final bin size was then selected for each sample within this range by determining the smallest value for which the ratio of the average read depth to its sd would be at least 4. For 174 samples, the ratio did not reach the threshold, and they were analyzed with a maximal bin size (800 bases). For Control-FREEC, to evaluate the optimal window size, the coefficient of variation for the read depth data was calculated for a wide range of window sizes, as suggested in a previous report (Boeva et al., 2011). For the final analysis, an overlapping sliding window of 800 bases with a step size of 400 bases was chosen. When this window size was used, the coefficient of variation was below 0.1 for 1025 of 1064 samples (the suggested threshold was 0.05 to 0.1; Supplemental Table 6). We noticed that the optimal window size was similar to the CNVnator bin size parameter, therefore enabling the subsequent comparison and merging of the outputs of the two programs.
Calculating the Insert Size Distributions for the Methods Relying on Paired-End Reads
BreakDancer, VariationHunter, and Pindel require insert size range thresholds as input parameters. The insert size distribution in each sequencing library was therefore evaluated with Picard Tools. At this step, 44 accessions were removed from analyses with these callers due to the bimodal distribution of the insert sizes (Supplemental Figure 20; Chen et al., 2009). The upper and lower threshold cutoffs were then calculated for the remaining libraries using two alternative approaches based on either the mean insert size ± 4 sd or the median insert size ± 5 median absolute deviation, and the maximum result of the two approaches was chosen.
CNV and Large Indels Discovery Pipeline
Variants were called by three read depth–based callers (CNVnator, Control-FREEC, and Genome STRiP-CNV pipeline), two discordant read pair–based tools (BreakDancer and VariationHunter), a split read–based tool (Pindel), and a combination of the above-mentioned approaches (the Genome STRiP-SV pipeline). CNV calling was performed with each tool as specified below. Subsequently, a common filter based on size (50 to 499 bp for large indels and at least 0.5 kb for CNVs) and genomic location was applied to the outputs of each caller. Specifically, variants overlapping with assembly gaps larger than 50 bp (with 50-bp borders) or regions close to the chromosome ends (<1 kb) were discarded. Additional filters specific for each CNV calling algorithm are described below.
CNVnator
BWA-MEM alignments were used to call duplications and deletions with CNVnator v.0.33 (Abyzov et al., 2011) based on read mapping density, separately for each accession, with nonoverlapping windows. The read depth signals were corrected for GC bias with a script implemented in the tool. The raw duplication and deletion calls were filtered based on variant size and genomic location. Additionally, to select the calls with the highest confidence, we applied a q0 filter (q0 describes the fraction of reads with a mapping quality of 0 in the called CNV; a high q0 indicates mapping uncertainty due to a lack of uniqueness in the region). Calls with a q0 ≥ 0.5 were removed. Finally, the read depth threshold was applied to remove uncertain calls (i.e., deletion calls with a normalized read depth >0.5 and duplication calls with a normalized read depth <1.5).
Control-FREEC
Aligned BWA-MEM BAM files for each sample were used to detect regions with gains and losses with Control-FREEC v.9.3 (Boeva et al., 2011) using sliding windows. The average GC content of the Arabidopsis genome varies from 32% in the noncoding regions to 44% in the coding regions; therefore, we set the parameters for GC normalization as follows: minExpectedGC = 0.3 and maxExpectedGC = 0.45. The telocentromeric parameter was set to 0 because it was included in our common filter. The breakPointThreshold value for the segmentation of normalized profiles was set to 0.6 (default is 0.8) to increase sensitivity and obtain more segments (and thus more predicted CNVs). The normalized read depth thresholds for CNV detection were ≤0.25 for loss and ≥1.75 for gain.
BreakDancer
The BreakDancerMax program from the BreakDancer package v.1.3.6 (Chen et al., 2009) was used to detect CNVs in each of 997 samples with paired-end data. Calls were made separately for each sample and each chromosome. The raw results that were indicative of CNVs (deletions or insertions) were filtered by a method-specific filter based on the number of supporting read pairs and the confidence score value. Calls with five or more supporting read pairs and confidence scores >30 were retained. For calls supported by less than five read pairs, the confidence score threshold was raised to 90.
VariationHunter
DIVET files with mrsFAST read alignments were used as the input data (for each sample separately) for VariationHunter v.0.04 (Hormozdiari et al., 2009). The analysis consisted of two main steps: the first step involved the clustering of discordant paired-end read mappings. This was performed with the default parameter values, which resulted in read pairs with more than 500 alternative mapping positions being discarded (-x 500) and low-quality ambiguous mapping alternatives being removed with a pruning parameter (-p 0.001). The required genome.satellite.bed and genome.gap.bed files were prepared with in-house scripts from the RepeatMasker v.4.0.7 output. The second step of VariationHunter analysis was the selection of variants from the created clusters. This was performed with a mismatch score (-ms 0.1) to increase the penalty for reads that were not mapping perfectly; additionally, a heuristic algorithm (-wh) was used with the conflict resolution version (-cr) instead of the greedy algorithm, since this algorithm preferred calls that had reads with decreased multiple mapping, and for reads that had multiple mapping, the mapping with a lower edit distance was preferred. A high number of calls were produced as an initial output (-t 10,000) that were subsequently pruned based on the supporting reads information. Additionally, only regions with an average edit distance (AvgEditDits) ≤ 3 were retained. Eventually, all insertion calls were removed after applying the common filter (Supplemental Table 1) because they were shorter than the lower size threshold.
Pindel
Pindel v.0.2.5b8 (Ye et al., 2009) was used for CNV detection (deletions, insertions, and tandem duplications) in individual samples from BWA-MEM alignments of paired-end reads with the following parameters. The maximum size of the structural variations and the window size were set to the default values (-x 5 -w 10), the balance cutoff was set to 0 (-B 0), and the median of the insert size was calculated for each sample (see above). All insertion calls were shorter than 500 bp, and they were eventually removed with the common filter (Supplemental Table 1).
Genome STRiP
BWA-MEM alignments of all 1064 samples were used as input for Genome STRiP v.2.00.1774 (Handsaker et al., 2015). The software required the precomputing of reference metadata based on the ArabidopsisTAIR10 genome sequence, as described in the software documentation (http://software.broadinstitute.org/software/genomestrip/node_ReferenceMetadata.html). All required information was generated according to this documentation except for the lcmask.fasta file (low-complexity mask), where the regions marked as Low complexity, Satellites, and Simple repeat were obtained from RepeatMasker results. Additionally, the TAIR10 reference sequence contained ambiguous nucleotides, which were not permitted by the CNVDiscoveryPipeline script. Therefore, the positions with nucleotides other than A, C, G, T, or N were changed to N and masked in the genome alignability mask (svmask file) by our own scripts. CNV discovery in Genome STRiP was performed with two separate modes, both of which were preceded by summary metadata computations (SVPreprocess script). This step was run with the default values. Large deletions were then identified in the entire population using the SVDiscovery script with the minimum (-minimumSize) and maximum (-maximumSize) event sizes set to 500 and 1,000,000, respectively. The SVDiscovery pipeline scanned the genome for polymorphic sites with large deletions only. The method was initially seeded with aberrantly spaced read pairs and used the read depth as secondary support for the variant sites. All types of CNVs (biallelic duplications, biallelic deletions, and multiallelic variants) were detected separately with the CNVDiscoveryPipeline script in the entire population with the following parameters: -tilingWindowSize 1000, -tilingWindowOverlap 500, -maximumReferenceGapLength 1000, -boundaryPrecision 100, and -minimumRefinedLength 500. The CNVDiscoveryPipeline script implemented a pipeline for discovering CNVs by seeding based on the read depth of the coverage. CNVs that passed through all read signature filters were retained. The outputs of both pipelines were treated as separate data sets.
Variant Merging and Breakpoint Refinement for CNV Discovery
The CNVs were merged, and the breakpoints were refined as follows. (1) Within-tool merge. Variants ≥0.5 kb detected in individual samples by CNVnator and Control-FREEC were merged separately for each caller and for each CNV type (gains and losses) with 50% reciprocal overlap as a criterion. CNVs detected in fewer than two accessions were subsequently discarded. This step eliminated the initial data redundancy and enabled the subsequent comparison of population-based and sample-based CNV calls. (2) Inter-tool merge. A union of all CNVs detected with read depth and hybrid approaches was created by combining the merged-CNVnator, merged-Control-FREEC, Genome STRiP-CNV pipeline, and Genome STRiP-SV pipeline outputs. To remove redundancy, the variants were merged using reciprocal overlap ≥80% as a criterion, which resulted in 34,366 CNVs. (3) CNV breakpoint refinement. The breakpoints of the merged variants were refined by prioritizing the information obtained from the most accurate methods. Individual variants from BreakDancer, VariationHunter, and Pindel that reciprocally overlapped the merged CNVs by at least 80% were used in this step (Supplemental Table 2). If any variants called by the hybrid method (which combines information from the split reads and discordant read pairs at the population level) supported the merge, the maximal coordinates of these variants were used. For the remaining CNVs, if the split read–based variants supported the merge, the maximal coordinates of these variants were used. For any CNVs remaining after this step, if any discordant read pair–based variants supported the merge, the maximal coordinates of these variants were used. Finally, for the CNVs that still remained, the averaged boundaries of the variants predicted by read depth–based methods were set. (4) CNV selection. We selected 19,003 high-confidence CNVs (supported by two or more different callers) for the final AthCNV data set (Supplemental Table 1). Unless otherwise indicated, these CNVs were analyzed further.
Variant Merging and Breakpoint Refinement for Large Indel Discovery
Large indels were merged, and the breakpoints were refined as follows. (1) Within-tool merge. Variants 50 bp to 499 bp detected in individual samples by BreakDancer and VariationHunter were merged separately for each caller with 80% reciprocal overlap as a criterion. Variants detected in fewer than two accessions were subsequently discarded. This step eliminated the initial data redundancy. (2) Inter-tool merge and breakpoints refinement. Variants overlapping each other by at least 80% were merged and their breakpoints were set by prioritizing the information obtained from the most accurate methods, in the same manner as for CNVs. As a result, we obtained 70,137 variants.
Detection of CNVs in the KBS-Mac-74 Genome Assembly
The KBS-Mac-74 genomic assembly based on Oxford Nanopore long reads was downloaded from the European Nucleotide Archive Genome Assembly Database (PRJEB21270; https://www.ebi.ac.uk/ena/data/view/PRJEB21270). We aligned this assembly to the reference genome (TAIR10) with the nucmer aligner in the MUMmer package (Marçais et al., 2018), followed by variant detection with Assemblytics (Nattestad and Schatz, 2016). For comparison with the AthCNV data set, 1551 KBS-Mac-74 variants that were at least 500 bp long were selected and paired with the best matching AthCNVs.
CNV Genotyping with Genome STRiP SVGenotyper
The genome STRiP SVGenotyper module was used to genotype genes in each accession. Prior to genotyping, the nonunique segments in the reference genome were identified by creating subsequence strings with 40-bp sliding windows and a 1-bp step and aligning them with the reference genome; the nonunique segments were masked. This approach was shown to be successful for distinguishing between highly similar paralogs and resulted in more accurate genotyping (Handsaker et al., 2015). All variants in the input vcf files were marked with a SVTYPE tag specifying a general copy number variant (“CNV”). The genotyping failed for 4 of 1064 accessions, and these data were removed. We ultimately obtained the genotyping data for 26,845 genes. A comparison of the unrounded copy numbers and integer copy number genotypes with the results of the MLPA assays for a subset of CNV-genes indicated that the copy number genotypes were frequently not correctly assigned by the SVGenotyper. Therefore, we did not use the genotype confidence filter integrated into the software. Instead, a custom filter based on the unrounded copy number distribution in the Col-0 accession was used to mark and remove outliers, defined as genes falling below (lower quartile minus 3* sd) value or above (upper quartile plus 3* sd) the value of the copy number range distribution in this accession. The threshold values were calculated separately for CNV-genes, genes overlapped by low-confidence CNVs, and NONVAR-genes. This step resulted in 7031 CNV-genes (5517 of them had at least 50% overlap with the CNVs), 4482 genes overlapped by low-confidence CNVs (2874 overlapped by at least 50%), and 14,877 genes not overlapped by any CNVs in the genotyping data.
Annotation and Analysis of CNV-Genes
The centromere positions were defined as described previously (Clark et al., 2007). The genes and noncoding elements in the CNV regions were located using Araport 11 annotations (Cheng et al., 2017). GO analysis was performed with Panther Tools (Panther database v.13.1; Mi et al., 2013). The classification of the gene duplication types (tandem versus block) and gene family specificity analysis were conducted based on information retrieved from the Plaza v.4.0 database (Van Bel et al., 2018). For PCA, 1050 CNV-genes were manually selected based on the distribution of the copy number genotypes (at least two visibly distinguishable copy number classes) and the genomic location (one CNV-gene represented one AthCNV variant; selected AthCNVs were located throughout the entire genome: 390 in chromosome 1 [Chr1], 153 in Chr2, 203 in Chr3, 129 in Chr4, and 175 in Chr5). The analyses were performed with the R-3.5.0 package prcomp(). Graphical representations of CNVs and genes in the genome were prepared with IGV v.2.3.90 (Robinson et al., 2011), circos-0.69.6 (Krzywinski et al., 2009), and TAIR Chromosome Map Tool (https://www.arabidopsis.org/jsp/ChromosomeMap/tool.jsp).
SNP Analysis
SNP data (1001genomes_snp-short-indel_only_ACGTN_v3.1.vcompared withsnpeff file) were downloaded from the 1001 Genomes Project server. PLINK v.1.90b3w program (https://www.cog-genomics.org/plink2) was used for data preprocessing. Only SNP data for 1060 accessions for which we also had CNV genotyping data were used. Variants with missing call rates exceeding value 0.5 as well as variants with minor allele frequency below 3% were filtered out. The LD parameter for linkage disequilibrium-based filtration was set as follows: –indep-pairwise 200'kb' 25 0.3. The resulting 117,232 SNPs were used for PCA analysis with EIGENSOFT v.7.2.1 (Price et al., 2006). The ggbiplot and ggplot2 packages were used for data visualization in the R version 3.6.1 environment.
Genome-Wide Association Study of CNV Data
Defense-related phenotypes (Atwell et al., 2010) were downloaded from the Arapheno database (Togninalli et al., 2019). For the genome-wide association study, we treated all copy number genotypes ≤1 as losses, all copy number genotypes >3 as gains, and all the remaining genotypes as unchanged. After filtering the CNV-gene data set with a 1% minor allele frequency threshold, 2519 CNV-genes remained in the analysis. Input files were preprocessed with PLINK v.1.90b3w. The IBS kinship matrix was calculated using SNPs for 1060 accessions. Association analysis was performed for each phenotype using a mixed model correcting for population structure using Efficient Mixed-Model Association eXpedited, version emmax-beta-07Mar2010 (Kang et al., 2010). To declare the threshold for significant association, we used Bonferroni correction. Results were further processed using the qqman package in R.
Experimental Procedures
Plant Materials and Growth Conditions
Arabidopsis seeds were obtained from The Nottingham Arabidopsis Stock Centre. The seeds were surface-sterilized, vernalized for 3 d, and grown on Jiffy pellets in ARASYSTEM containers (BETATECH) in a growth chamber (Percival Scientific). A light intensity of 175 µmol m−2 s−1 with proportional blue, red, and far red light was provided by a combination of fluorescent lamps (Philips) and GroLEDs red/far red LED Strips (CLF PlantClimatics). Plants were grown for 3 weeks under a 16-h light (22°C)/8-h dark (18°C) cycle, at 70% RH, with nourishment from Murashige and Skoog medium, 0.5× (Serva). A list of accessions used in the experiments is available in Supplemental Data Set 7.
DNA Extraction and MLPA Assays
DNA was extracted from leaves with a DNeasy Plant Mini Kit (Qiagen). The MLPA assays were performed as described previously (Samelak-Czajka et al., 2017) using 5 ng of DNA template with the SALSA MLPA reagent kit FAM (MRC-Holland). The MLPA products were separated by capillary electrophoresis in an ABI Prism 3130XL analyzer at the Molecular Biology Techniques Facility in the Department of Biology at Adam Mickiewicz University, Poznan, Poland. The results were analyzed with GeneMarker v.2.4.2 (SoftGenetics). Whenever possible, to minimize the risk of incorporating SNPs and indels that might affect the probe hybridization step for some accessions, the MLPA probes were designed within regions of minimal sequence variation, as verified by examining vcf files for 1135 accessions obtained from the 1001 Genomes Project website (1001 Genomes Consortium et al., 2016). The genomic target sequence coordinates for the MLPA probes are provided in Supplemental Table 7.
Protein Extraction and Quantification
Proteins were extracted using the phenol method (Hurkman and Tanaka, 1986). The protein pellet was solubilized in 100 mM ammonium bicarbonate for 2 h with three cycles of sonication using a sonic bath every 0.5 h. The protein concentration was determined using a bicinchoninic acid assay (Pierce). For quantification, 10 µg of total protein was reduced, alkylated, and digested with trypsin (Luczak et al., 2016). Each sample was prepared for digestion in duplicate. For each run, 1.5 μg of protein digest was subjected to nano-liquid chromatography–tandem mass spectrometry analysis using a Dionex UltiMate 3000 chromatograph and a Q-Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) as described previously (Luczak et al., 2016). After each liquid chromatography–tandem mass spectrometry run, the raw files were analyzed by MaxQuant (Cox and Mann, 2008). Quantitative analysis of the experimental groups was based on the label-free quantification intensities. The statistical analyses were performed using Perseus v.1.6.1.3.
Accession Numbers
A detailed list of the accessions and individual data sets used for CNV discovery is provided in Supplemental Data Set 7. The genomic coordinates of CNVs identified in the current study are listed in Supplemental Table 2. The genotyping results for the genes can be accessed through the web interface at http://athcnv.ibch.poznan.pl as user-generated scatterplots that present the copy number values and their distribution across the different genetic groups.
Supplemental Data
Supplemental Figure 1. Comparison of the variants generated by the callers prior to data merging.
Supplemental Figure 2. Fractions of large duplications and deletions detected in the genomes of individual accessions assembled de novo from long reads that overlap with AthCNVs.
Supplemental Figure 3. Chromosome map of 100 genes with evidence for duplication/deletion in A. thaliana that overlap with AthCNVs.
Supplemental Figure 4. Differences in the number of CNVs overlapping with various genetic elements in the A. thaliana genome.
Supplemental Figure 5. Relative distances between genes and TEs and their relationship with CNV status.
Supplemental Figure 6. The accuracy of gene copy number estimates in a complex CNV region calculated for CNV-gene intervals versus AthCNV intervals.
Supplemental Figure 7. Differences between automatic and manual assignment of CNV-gene integer copy numbers from sequencing data.
Supplemental Figure 8. Read depth-based copy number estimates for CNV-genes partially overlapping with CNV regions.
Supplemental Figure 9. Example of a long CNV with a non-uniform pattern of variation of CNV-genes overlapped by this variant.
Supplemental Figure 10. Chromosome map of CNV-genes subjected to experimental verification with MLPA.
Supplemental Figure 11. Intermediate copy number values reported by Genome STRiP for a gene partially covered by CNV.
Supplemental Figure 12. The influence of small-scale sequence variations on oligonucleotide MLPA probe signal and concordance with read depth-based data.
Supplemental Figure 13. Experimental validation of copy number genotypes for NONVAR-genes.
Supplemental Figure 14. Experimental validation of copy number genotypes for CNV-genes with rare duplications (<1%).
Supplemental Figure 15. Experimental validation of copy number genotypes for CNV-genes with common (≥1%) copy number polymorphism.
Supplemental Figure 16. The effect of stock misidentification on the correlation of sequencing-based (source data from the 1001 Genomes Project) and in-house experimental genotyping results.
Supplemental Figure 17. Histograms of gene copy number distribution for CNV-genes that are likely duplicated in the Col-0 accession.
Supplemental Figure 18. Results of mass spectrometry-based identification of SEC10 peptides.
Supplemental Figure 19. Results from GWAS of defense-related phenotypes and CNV-gene data.
Supplemental Figure 20. Insert size distributions in paired-end libraries.
Supplemental Table 1. Variants >0.5 kb in size considered to be copy number changes discovered by each caller in the A. thaliana population.
Supplemental Table 2. CNVs resulting from the inter-tool merging of variants (80% RO) and their support by individual callers.
Supplemental Table 3. Gene family specificity of CNV-genes.
Supplemental Table 4. Superfamily composition of A. thaliana TEs and its comparison with CNV-TEs and CNV-TEs located within +/− 2 kb distance from the genes.
Supplemental Table 5. Effect of excluding suspicious stocks on the correlation of read depth-based and MLPA-based genotyping results.
Supplemental Table 6. Coefficients of variation (CVs) of read depth values in Control-FREEC analysis.
Supplemental Table 7. List of genomic regions targeted by MLPA probes.
Supplemental Data Set 1. CNVs detected in the A. thaliana genome.
Supplemental Data Set 2. Large indels detected in the A. thaliana genome.
Supplemental Data Set 3. CNVs at least 0.5 kb long identified in the KBS-Mac-74 genome assembly.
Supplemental Data Set 4. Genes with previous experimental evidence of CNV among A. thaliana ecotypes and their overlap with AthCNV variants.
Supplemental Data Set 5. Gene Ontology terms enrichment and protein domain enrichment among groups of genes with proximal TEs depending on their variation status.
Supplemental Data Set 6. List of defense-related phenotypes and identified associations with CNV-genes from GWAS.
Supplemental Data Set 7. List of samples and sequencing data used in this study.
Supplemental Data Set 8. Read depth statistics and bin size selection for CNVnator.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Michal Zenczak for help with drawing Circos plots and Ireneusz Stolarek for valuable comments and discussions. The computations were performed using PL Grid infrastructure resources. This work was supported by the Polish National Centre of Science (grants 2014/13/B/NZ2/03837 to M.F., 2017/01/X/NZ2/00144 to A.Z., and 2017/26/D/NZ2/01079 to A.Z.).
AUTHOR CONTRIBUTIONS
A.Z. and M.F. conceived the study. A.Z., P.W., M.M.-Z., and W.M.K. performed methods optimization tests. A.Z., M.M.-Z., and P.W. performed bioinformatics analyses and analyzed the data. A.Z., A.S.-C., and M.L. performed experiments and analyzed the data. P.K. and M.F. contributed to the critical interpretation of the results. P.W. prepared the web interface for data visualization. A.Z. wrote the article. M.M.-Z., P.K., and M.F. revised the article. A.Z. and M.M.-Z. prepared the figures. M.F. supervised the study.
Footnotes
Articles can be viewed without a subscription.
References
- 1000 Genomes Project Consortium (2012). An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1001 Genomes Consortium (2016). 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abyzov A., Urban A.E., Snyder M., Gerstein M.(2011). CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 21: 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C., Coe B.P., Eichler E.E.(2011). Genome structural variation discovery and genotyping. Nat. Rev. Genet. 12: 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C., Koornneef M.(2000). Naturally occurring variation in Arabidopsis: An underexploited resource for plant genetics. Trends Plant Sci. 5: 22–29. [DOI] [PubMed] [Google Scholar]
- Atwell S., et al. (2010). Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Sureshkumar S., Lempe J., Weigel D.(2006). Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomer R.H., Juenger T.E., Symonds V.V.(2012). Natural variation in GL1 and its effects on trichome density in Arabidopsis thaliana. Mol. Ecol. 21: 3501–3515. [DOI] [PubMed] [Google Scholar]
- Boeva V., Zinovyev A., Bleakley K., Vert J.-P., Janoueix-Lerosey I., Delattre O., Barillot E.(2011). Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization. Bioinformatics 27: 268–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B.(2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G., Burns K.H., Gehring M., Gorbunova V., Seluanov A., Hammell M., Imbeault M., Izsvák Z., Levin H.L., Macfarlan T.S., Mager D.L., Feschotte C.(2018). Ten things you should know about transposable elements. Genome Biol. 19: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush S.J., Castillo-Morales A., Tovar-Corona J.M., Chen L., Kover P.X., Urrutia A.O.(2014). Presence-absence variation in A. thaliana is primarily associated with genomic signatures consistent with relaxed selective constraints. Mol. Biol. Evol. 31: 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C.D., Sampas N., Tsalenko A., Sudmant P.H., Kidd J.M., Malig M., Vu T.H., Vives L., Tsang P., Bruhn L., Eichler E.E.(2011). Population-genetic properties of differentiated human copy-number polymorphisms. Am. J. Hum. Genet. 88: 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., et al. (2011). Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963. [DOI] [PubMed] [Google Scholar]
- Caretto S., Giardina M.C., Nicolodi C., Mariotti D. (1995). Acetohydroxyacid synthase GENE amplification induces clorsulfuron resistance in Daucus carota L. In Current Issues in Plant Molecular and Cellular Biology. Current Plant Science and Biotechnology in Agriculture, Terzi M., Cella R., Falavigna A., eds (Dordrecht: Springer; ), Vol. 22: pp. 235–240. [Google Scholar]
- Chen K., et al. (2009). BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6: 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.-Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D.(2017). Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89: 789–804. [DOI] [PubMed] [Google Scholar]
- Chia J.M., et al. (2012). Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44: 803–807. [DOI] [PubMed] [Google Scholar]
- Clark R.M., et al. (2007). Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342. [DOI] [PubMed] [Google Scholar]
- Cole S.J., Diener A.C.(2013). Diversity in receptor-like kinase genes is a major determinant of quantitative resistance to Fusarium oxysporum f.sp. matthioli. New Phytol. 200: 172–184. [DOI] [PubMed] [Google Scholar]
- Conrad D.F., et al. ; Wellcome Trust Case Control Consortium (2010). Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M.(2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26: 1367–1372. [DOI] [PubMed] [Google Scholar]
- Duitama J., Silva A., Sanabria Y., Cruz D.F., Quintero C., Ballen C., Lorieux M., Scheffler B., Farmer A., Torres E., Oard J., Tohme J.(2015). Whole genome sequencing of elite rice cultivars as a comprehensive information resource for marker assisted selection. PLoS One 10: e0124617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes R.R., Chebotarov D., Duitama J., Smith S., De la Hoz J.F., Mohiyuddin M., Wing R.A., McNally K.L., Tatarinova T., Grigoriev A., Mauleon R., Alexandrov N.(2019). Structural variants in 3000 rice genomes. Genome Res. 29: 870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabur I., Chawla H.S., Snowdon R.J., Parkin I.A.P.(2019). Connecting genome structural variation with complex traits in crop plants. Theor. Appl. Genet. 132: 733–750. [DOI] [PubMed] [Google Scholar]
- Gaines T.A., et al. (2010). Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc. Natl. Acad. Sci. USA 107: 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X., et al. (2011). Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M.R., Godiard L., Straube E., Ashfield T., Lewald J., Sattler A., Innes R.W., Dangl J.L.(1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846. [DOI] [PubMed] [Google Scholar]
- Grant M.R., McDowell J.M., Sharpe A.G., de Torres Zabala M., Lydiate D.J., Dangl J.L.(1998). Independent deletions of a pathogen-resistance gene in Brassica and Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 15843–15848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Zhang F., Lupski J.R.(2008). Mechanisms for human genomic rearrangements. PathoGenetics 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hach F., Sarrafi I., Hormozdiari F., Alkan C., Eichler E.E., Sahinalp S.C.(2014). mrsFAST-Ultra: A compact, SNP-aware mapper for high performance sequencing applications. Nucleic Acids Res. 42: W494–W500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handsaker R.E., Van Doren V., Berman J.R., Genovese G., Kashin S., Boettger L.M., McCarroll S.A.(2015). Large multiallelic copy number variations in humans. Nat. Genet. 47: 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardigan M.A., et al. (2016). Genome reduction uncovers a large dispensable genome and adaptive role for copy number variation in asexually propagated Solanum tuberosum. Plant Cell 28: 388–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms C.T., et al. (1992). Herbicide resistance due to amplification of a mutant acetohydroxyacid synthase gene. Mol. Gen. Genet. 233: 427–435. [DOI] [PubMed] [Google Scholar]
- Henk A.D., Warren R.F., Innes R.W.(1999). A new Ac-like transposon of Arabidopsis is associated with a deletion of the RPS5 disease resistance gene. Genetics 151: 1581–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F., Alkan C., Eichler E.E., Sahinalp S.C.(2009). Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res. 19: 1270–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton M.W., et al. (2012). Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 44: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Lu G., Zhao Q., Liu X., Han B.(2008). Genome-wide analysis of transposon insertion polymorphisms reveals intraspecific variation in cultivated rice. Plant Physiol. 148: 25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurkman W.J., Tanaka C.K.(1986). Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 81: 802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao W.-B., Schneeberger K.(2020). Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nat. Commun. 11: 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Weller S.C., Goldsbrough P.B.(1994). Selection for kanamycin resistance in transformed petunia cells leads to the co-amplification of a linked gene. Plant Mol. Biol. 24: 505–514. [DOI] [PubMed] [Google Scholar]
- Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.Y., Freimer N.B., Sabatti C., Eskin E.(2010). Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov T.L., et al. (2014). The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature 512: 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T., et al. ; 1001 Genomes Consortium (2016). Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov F.A.(2012). Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. Biol. Sci. 279: 5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva K.V.(2019). The role of transposable elements and DNA damage repair mechanisms in gene duplications and gene fusions in plant genomes. Curr. Opin. Plant Biol. 48: 18–25. [DOI] [PubMed] [Google Scholar]
- Kroymann J., Donnerhacke S., Schnabelrauch D., Mitchell-Olds T.(2003). Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc. Natl. Acad. Sci. USA 100 (Suppl 2): 14587–14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov D.M., Wolf Y.I., Rogozin I.B., Koonin E.V.(2003). Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 13: 2229–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A.(2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-R., Svardal H., Farlow A., Exposito-Alonso M., Ding W., Novikova P., Alonso-Blanco C., Weigel D., Nordborg M.(2017). On the post-glacial spread of human commensal Arabidopsis thaliana. Nat. Commun. 8: 14458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R.(2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li R., Li H., Lu J., Li Y., Bolund L., Schierup M.H., Wang J.(2013). SOAPindel: Efficient identification of indels from short paired reads. Genome Res. 23: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q., et al. (2013). Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 45: 884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczak M., Suszynska-Zajczyk J., Marczak L., Formanowicz D., Pawliczak E., Wanic-Kossowska M., Stobiecki M.(2016). Label-free quantitative proteomics reveals differences in molecular mechanism of atherosclerosis related and non-related to chronic kidney disease. Int. J. Mol. Sci. 17: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Delcher A.L., Phillippy A.M., Coston R., Salzberg S.L., Zimin A.(2018). MUMmer4: A fast and versatile genome alignment system. PLOS Comput. Biol. 14: e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale L.K., Haun W.J., Xu W.W., Bhaskar P.B., Anderson J.E., Hyten D.L., Gerhardt D.J., Jeddeloh J.A., Stupar R.M.(2012). Structural variants in the soybean genome localize to clusters of biotic stress-response genes. Plant Physiol. 159: 1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale L., Tan X., Koehl P., Michelmore R.W.(2006). Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 7: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Casagrande J.T., Thomas P.D.(2013). Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8: 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., Jupe F., Bemm F., Motley S.T., Sandoval J.P., Lanz C., Loudet O., Weigel D., Ecker J.R.(2018). High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Nat. Commun. 9: 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.E., et al. ; 1000 Genomes Project (2011). Mapping copy number variation by population-scale genome sequencing. Nature 470: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoru M.(2013). Arabidopsis centromeres In Plant Centromere Biology, Jiang J., and Birchler J.A., eds (New York: Wiley; ), pp. 1–14. [Google Scholar]
- Muñoz-Amatriaín M., et al. (2013). Distribution, functional impact, and origin mechanisms of copy number variation in the barley genome. Genome Biol. 14: R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattestad M., Schatz M.C.(2016). Assemblytics: A web analytics tool for the detection of variants from an assembly. Bioinformatics 32: 3021–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X.M., Xu Y.C., Li Z.W., Bian Y.T., Hou X.H., Chen J.F., Zou Y.P., Jiang J., Wu Q., Ge S., Balasubramanian S., Guo Y.L.(2019). Transposable elements drive rapid phenotypic variation in Capsella rubella. Proc. Natl. Acad. Sci. USA 116: 6908–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Clark R.M., Lanz C., Warthmann N., Weigel D.(2008). Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18: 2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N., Lehti-Shiu M., Shiu S.-H.(2016). Evolution of gene duplication in plants. Plant Physiol. 171: 2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisupati R., Reichardt I., Seren Ü., Korte P., Nizhynska V., Kerdaffrec E., Uzunova K., Rabanal F.A., Filiault D.L., Nordborg M.(2017). Verification of Arabidopsis stock collections using SNPmatch, a tool for genotyping high-plexed samples. Sci. Data 4: 170184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A., et al. (2010). The scale of population structure in Arabidopsis thaliana. PLoS Genet. 6: e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D.(2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. [DOI] [PubMed] [Google Scholar]
- Pucker B., Holtgräwe D., Rosleff Sörensen T., Stracke R., Viehöver P., Weisshaar B.(2016). A de novo genome sequence assembly of the Arabidopsis thaliana accession Niederzenz-1 displays presence/absence variation and strong synteny. PLoS One 11: e0164321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L., Bortolini Silveira A., Mayhew G.F., LeBlanc C., Martienssen R.A., Jeddeloh J.A., Colot V.(2016). The Arabidopsis thaliana mobilome and its impact at the species level. eLife 5: e15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.(2011). Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux F., Bergelson J.(2016). The genetics underlying natural variation in the biotic interactions of Arabidopsis thaliana: The challenges of linking evolutionary genetics and community ecology. Curr. Top. Dev. Biol. 119: 111–156. [DOI] [PubMed] [Google Scholar]
- Saintenac C., Jiang D., Akhunov E.D.(2011). Targeted analysis of nucleotide and copy number variation by exon capture in allotetraploid wheat genome. Genome Biol. 12: R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samelak-Czajka A., Marszalek-Zenczak M., Marcinkowska-Swojak M., Kozlowski P., Figlerowicz M., Zmienko A.(2017). MLPA-based analysis of copy number variation in plant populations. Front Plant Sci 8: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santuari L., Pradervand S., Amiguet-Vercher A.-M., Thomas J., Dorcey E., Harshman K., Xenarios I., Juenger T.E., Hardtke C.S.(2010). Substantial deletion overlap among divergent Arabidopsis genomes revealed by intersection of short reads and tiling arrays. Genome Biol. 11: R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Böckler B., Conrad D., Bateman A.(2010). Dosage sensitivity shapes the evolution of copy-number varied regions. PLoS One 5: e9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.M., Bomblies K., Weigel D.(2011). Complex evolutionary events at a tandem cluster of Arabidopsis thaliana genes resulting in a single-locus genetic incompatibility. PLoS Genet. 7: e1002164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N.M., et al. (2009). Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5: e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal J., Kaliff M., Dewaele E., Persson M., Dixelius C.(2008). RLM3, a TIR domain encoding gene involved in broad-range immunity of Arabidopsis to necrotrophic fungal pathogens. Plant J. 55: 188–200. [DOI] [PubMed] [Google Scholar]
- Stahl E.A., Dwyer G., Mauricio R., Kreitman M., Bergelson J.(1999). Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400: 667–671. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P., Lupski J.R.(2010). Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61: 437–455. [DOI] [PubMed] [Google Scholar]
- Sudmant P.H., et al. (2015). An integrated map of structural variation in 2,504 human genomes. Nature 526: 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner R.A., Eichten S.R., Kumari S., Tiffin P., Stein J.C., Ware D., Springer N.M.(2010). Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 20: 1689–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo S.M., Pawitan Y., Ku C.S., Chia K.S., Salim A.(2012). Statistical challenges associated with detecting copy number variations with next-generation sequencing. Bioinformatics 28: 2711–2718. [DOI] [PubMed] [Google Scholar]
- Togninalli M., Seren Ü., Freudenthal J.A., Monroe J.G., Meng D., Nordborg M., Weigel D., Borgwardt K., Korte A., Grimm D.G.(2019). AraPheno and the AraGWAS catalog 2020: A major database update including RNA-seq and knockout mutation data for Arabidopsis thaliana. Nucleic Acids Res. 23: gkz925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M., Diels T., Vancaester E., Kreft L., Botzki A., Van de Peer Y., Coppens F., Vandepoele K.(2018). PLAZA 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 46 (D1): D1190–D1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad D., Rappaport F., Simon M., Loudet O.(2010). Gene transposition causing natural variation for growth in Arabidopsis thaliana. PLoS Genet. 6: e1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukašinović N., Cvrčková F., Eliáš M., Cole R., Fowler J.E., Žárský V., Synek L.(2014). Dissecting a hidden gene duplication: The Arabidopsis thaliana SEC10 locus. PLoS One 9: e94077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Weigel D., Smith L.M.(2013). Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet. 9: e1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner J.D., Borevitz J.O., Warthmann N., Trainer G.T., Ecker J.R., Chory J., Weigel D.(2005). Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc. Natl. Acad. Sci. USA 102: 2460–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Ellwood S., Calis O., Patrick E., Li T., Coleman M., Turner J.G.(2001). Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291: 118–120. [DOI] [PubMed] [Google Scholar]
- Xu L., Hou Y., Bickhart D.M., Zhou Y., Hay H.A., Song J., Sonstegard T.S., Van Tassell C.P., Liu G.E.(2016). Population-genetic properties of differentiated copy number variations in cattle. Sci. Rep. 6: 23161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z.(2009). Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Richards E.J.(2009). Gene duplication and hypermutation of the pathogen resistance gene SNC1 in the Arabidopsis bal variant. Genetics 183: 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Xuan Z., Makarov V., Ye K., Sebat J.(2009). Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 19: 1586–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Wang C.H., Xu Q., Feng Y., Yuan X.P., Yu H.Y., Wang Y.P., Tang S.X., Wei X.H.(2013). Genome-wide copy number variations in Oryza sativa L. BMC Genomics 14: 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L.-Y., Guo X.-S., He B., Sun L.-J., Peng Y., Dong S.-S., Liu T.-F., Jiang S., Ramachandran S., Liu C.-M., Jing H.-C.(2011). Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor). Genome Biol. 12: R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żmieńko A., Samelak A., Kozłowski P., Figlerowicz M.(2014). Copy number polymorphism in plant genomes. Theor. Appl. Genet. 127: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmienko A., Samelak-Czajka A., Kozlowski P., Szymanska M., Figlerowicz M.(2016). Arabidopsis thaliana population analysis reveals high plasticity of the genomic region spanning MSH2, AT3G18530 and AT3G18535 genes and provides evidence for NAHR-driven recurrent CNV events occurring in this location. BMC Genomics 17: 893. [DOI] [PMC free article] [PubMed] [Google Scholar]