Wounding induces the parallel activation of two signaling cascades: a MKK4/5-MPK3/6 module (rapid) and a jasmonic acid-dependent MAP3K14-MKK3-MPK1/2/7/14 module (slow) to restrict insect feeding.
Abstract
Abiotic and biotic factors cause plant wounding and trigger complex short- and long-term responses at the local and systemic levels. These responses are under the control of complex signaling pathways, which are still poorly understood. Here, we show that the rapid activation of clade-A mitogen-activated protein kinases (MAPKs) MPK3 and MPK6 by wounding depends on the upstream MAPK kinases MKK4 and MKK5 but is independent of jasmonic acid (JA) signaling. In addition, this fast module does not control wound-triggered JA accumulation in Arabidopsis (Arabidopsis thaliana), unlike its orthologs in tobacco. We also demonstrate that a second MAPK module, composed of MKK3 and the clade-C MAPKs MPK1/2/7, is activated by wounding in a MKK4/5-independent manner. We provide evidence that the activation of this MKK3-MPK1/2/7 module occurs mainly through wound-induced JA production via the transcriptional regulation of upstream clade-III MAP3Ks, particularly MAP3K14. We show that mkk3 mutant plants are more susceptible to herbivory from larvae of the generalist lepidopteran herbivore Spodoptera littoralis, indicating that the MKK3-MPK1/2/7 module is involved in counteracting insect feeding.
INTRODUCTION
Wounding is a common stress for plants that can be caused by abiotic factors such as wind, heavy rain, hail, and snow or during biotic interactions, mostly with herbivorous organisms such as insects. Injury may cause severe damage to plant tissues and facilitate the entry of pathogens (Savatin et al., 2014). Plants respond to these challenges by activating several mechanisms to rapidly heal tissues and restrict potential pathogen entry. The cuticle and trichomes are primary constitutive structures that offer a first line of defense involved in wounding prevention. When wounding does occur, intracellular molecules released from dead cells or damaged cell wall components act as signaling molecules: damage-associated molecular patterns (Maffei et al., 2012). In addition, living cells surrounding wounded sites can sense mechanical disturbances through the activation of mechanosensitive channels that activate signaling cascades and local responses (Farmer et al., 2014). Such responses are mediated by efficient and complex intracellular signaling mechanisms involving phosphorylation, lipids, reactive oxygen species (ROS), and calcium (Ca2+) signaling, as well as the biosynthesis of phytohormones leading to gene expression reprogramming and long-distance signaling (Savatin et al., 2014). Of all phytohormones, jasmonates, members of the oxylipin family, play a dominant role (Wasternack and Hause, 2013). Jasmonates, and in particular jasmonic acid (JA), are produced very rapidly by herbivory-induced wounding and regulate over 100 wound-induced genes (Reymond et al., 2004). The isoleucine conjugate of JA, (+)-7-iso-jasmonoyl-L-isoleucine (JA-Ile), is the bioactive form of jasmonates (Fonseca et al., 2009).
Mitogen-activated protein kinase (MAPK) modules are conserved signaling pathways found in all eukaryotes (Colcombet and Hirt, 2008). A MAPK module is minimally composed of three kinases: a MAP3K (or MAP2K kinase), a MAP2K (or MAPK kinase), and a MAPK, which phosphorylate each other, leading to their sequential activation. These kinases are encoded by large gene families, for which functional information is incomplete (Colcombet and Hirt, 2008). Since their discovery in plants in 1993, a number of MAPK components have been described in biotic and abiotic stress signal transduction as well as in developmental processes (reviewed by Rodriguez et al., 2010, and Xu and Zhang, 2015). They have notably been shown to act early during wound signaling. For example, the alfalfa (Medicago sativa) MAPK MMK4 and the tobacco (Nicotiana tabacum) wound-induced protein kinase WIPK are activated within minutes upon leaf wounding (Bogre et al., 1997; Seo et al., 1999). Generally, after an initial peak 15 min after wounding, their activities decline and return to basal levels within 1 h (Seo et al., 1995, 1999; Usami et al., 1995; Bogre et al., 1997). In Arabidopsis (Arabidopsis thaliana), early evidence pointed to the activation of the MAPKs MPK4 and MPK6 in response to wounding (Ichimura et al., 2000). Details have filled in over the years to provide a better picture of a complete module: a pair of MAP2Ks, MKK4 and MKK5, and a pair of MAPKs, MPK3 and MPK6, stimulate ethylene biosynthesis in response to wounding (Li et al., 2018). Several early studies also suggested that this MAPK cascade also controlled JA production (Ahmad et al., 2016). Indeed, tobacco lines with altered WIPK transcript levels adjusted their JA levels up (if WIPK was overexpressed) or down (when it was silenced via RNA interference) after wounding (Seo et al., 1995, 1999, 2007). In addition, JA can modulate MPK6 activity, suggesting a feedback loop that fine tunes JA homeostasis (Takahashi et al., 2007). MAPKs are classified into four clades (A to D); MPK3 and MPK6 belong to the clade A, typically recognized as stress-activated MAPKs. However, data also suggested that less-studied members of the MAPK family can transduce wound signals. For example, wounding activates the clade-D MAPK MPK8, as well as the clade-C MAPKs MPK1 and MPK2 (Ortiz-Masia et al., 2007; Takahashi et al., 2011).
Arabidopsis MPK1 and MPK2, together with MPK7 and MPK14, comprise the entire subgroup of clade-C MAPKs and function downstream of the atypical MAPK kinase MKK3 (Colcombet et al., 2016). For instance, the MKK3-MPK7 module was reported to play a role during plant-pathogen interactions and drought perception. MPK7 is itself activated by ROS, produced when plants are challenged with pathogenic bacteria, or by the drought-induced phytohormone abscisic acid (ABA; Dóczi et al., 2007; Danquah et al., 2015). In agreement, mkk3 mutants are hypersensitive to infection by the Gram-negative bacterium Pseudomonas syringae DC3000 and are more sensitive to water deficit. In the context of ABA signaling, the MKK3-MPK1 module is activated by the clade-III MAP3Ks MAP3K17 and MAP3K18 through transcriptional up-regulation, an additional step that introduces a delay in the activation of the module (Boudsocq et al., 2015; Danquah et al., 2015). In this study, we characterized MAPK-dependent phosphorylation cascades in response to tissue damage. We unveil the coexistence of two independent MAPK modules, which are both activated by wounding but with different kinetics. We also reexamined the relationship between MAPKs and JA biosynthesis and provide evidence that JA production is not controlled by stress-activated MPK3 or MPK6 in Arabidopsis.
RESULTS
Clade-C MAPK Activation by Wounding Depends Strictly on MKK3
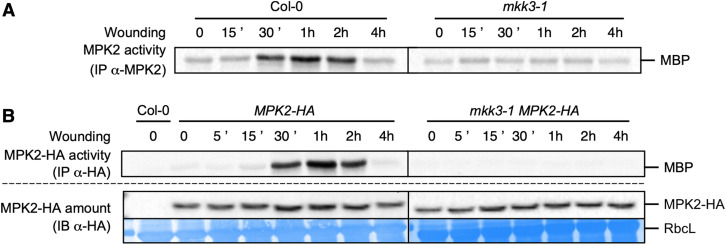
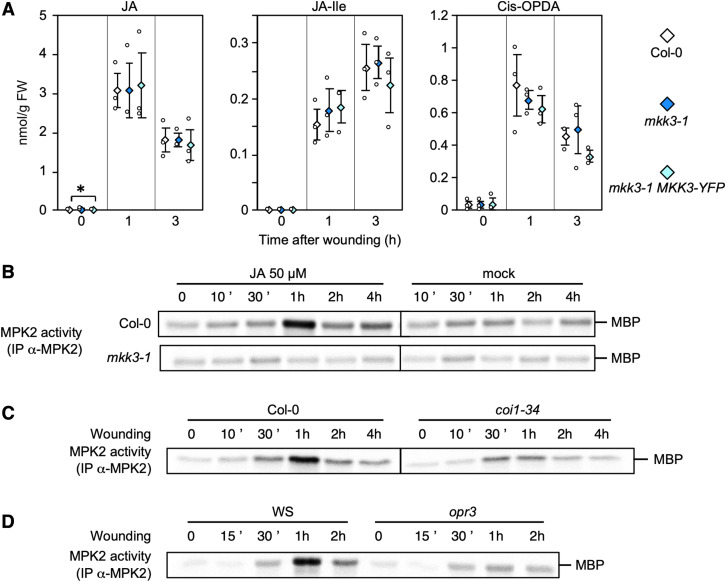
Some clade-C MAPKs were previously shown to be activated by wounding and to act downstream of MKK3 in response to hydrogen peroxide and ABA treatment (Dóczi et al., 2007; Ortiz-Masia et al., 2007; Danquah et al., 2014). To investigate whether MPK2 also acts downstream of MKK3 in the wounding response, we immunoprecipitated MPK2 using a specific antibody from wounded leaves of Arabidopsis wild-type Columbia (Col-0) and mkk3 mutant plants and assayed its activity as the ability to phosphorylate the heterologous substrate myelin basic protein (MBP). As clade-C MAPK activation by ABA occurs slowly, we collected samples over 4 h (Danquah et al., 2015). In Col-0 plants, MPK2 was activated as early as 30 min after wounding and maintained high activity up to 2 h after wounding (Figure 1A). This activation was completely abolished in mkk3-1 and mkk3-2 mutants (Figure 1A; Supplemental Figure 1A) and restored in mkk3-1 plants complemented with the MKK3 gene cloned in-frame with YELLOW FLUORESCENT PROTEIN (YFP; mkk3-1 MKK3-YFP; Supplemental Figure 1B). Because the MPK2-specific antibody is unable to detect MPK2 protein in immunoblots (Ortiz-Masia et al., 2007), we could not exclude the possibility that the increase in MPK2 activity in response to wounding reflects a wound-dependent accumulation of MPK2. To test this hypothesis, we introduced a version of the MPK2 gene cloned in-frame with a HA tag (human influenza hemagglutinin) in both Col-0 and mkk3-1 genetic backgrounds. Immunoprecipitation with an anti-HA antibody revealed a transient increase of MPK2-HA activity with similar kinetics as endogenous MPK2 in Col-0, but not in mkk3-1 wounded leaves (Figure 1B). Importantly, MPK2-HA protein levels remained constant over the time course (Figure 1B), ruling out a contribution from MPK2 translation in the observed rise in MPK2 activity. MPK2 belongs to clade-C of MAPKs together with MPK1, MPK7, and MPK14. In protoplast assays, all four MAPKs (MPK1, MPK2, MPK7, and MPK14, which we will refer to as MPK1/2/7/14 when mentioned collectively) appear to function in a similar module downstream of MKK3 (Danquah et al., 2015). To test whether other clade-C MAPKs are activated by wounding, we generated plants with MPK1 or MPK7 fused in-frame with a HA tag. When challenged by wounding, both MPK1-HA and MPK7-HA displayed a transient activation and followed the same kinetics as MPK2 (Supplemental Figures 2A and 2B). We also introduced the MPK7-HA construct in the mkk3-1 background and confirmed the MKK3 dependence of MPK7-HA activation, which was not detectable in the mkk3-1 MPK7-HA line (Supplemental Figure 2B). In addition, wounding did not affect MPK1-HA or MPK7-HA protein levels (Supplemental Figures 2A and 2B). Together, these results demonstrate that wounding activates clade-C MAPKs in an MKK3-dependent manner with kinetics that are considerably slower than activation of the well-studied MAPKs MPK3 and MPK6 by the flagellin peptide flg22, which occurs within 2 min and peaks around 10 to 15 min (Supplemental Figure 3; Ranf et al., 2011).
Figure 1.
MPK2 Activation by Wounding Depends on MKK3.
(A) Kinase activity of MPK2 after immunoprecipitation with an anti-MPK2-specific antibody from Col-0 and mkk3-1 leaves following wounding at the indicated times. Results were repeated two to five times depending on the time point.
(B) Kinase activity of MPK2 after immunoprecipitation with an anti-HA antibody from leaves of plants expressing an HA-tagged version of MPK2 in Col-0 and mkk3-1 leaves following wounding at the indicated times. Protein amount was monitored by immunoblot using an anti-HA antibody. Equal loading was controlled by Coomassie staining of the membrane. RbcL, RubisCo large subunit. Results were repeated two to five times depending on the time point.
MKK3 and Clade-III MAP3Ks Interact and Form a Functional Module in Arabidopsis Mesophyll Protoplasts
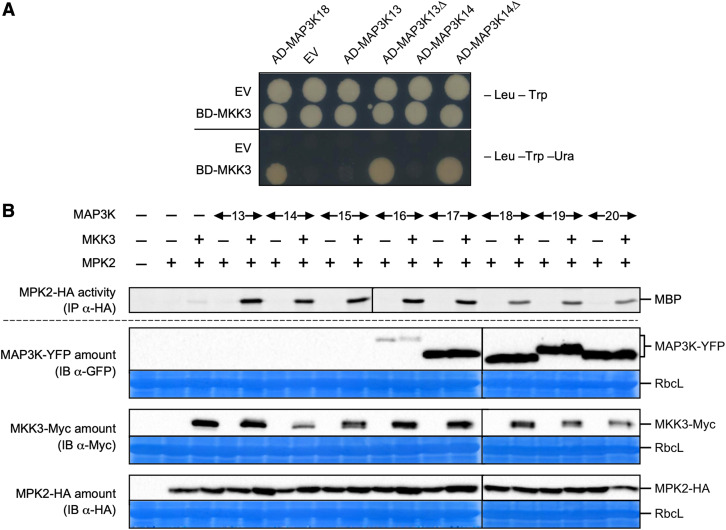
We had previously suggested that the module composed of MKK3 and MPK1/2/7/14 may function downstream of clade-III MAP3Ks, namely MAP3K13 to MAP3K20 (Colcombet et al., 2016). Indeed, MKK3 can interact in yeast two-hybrid (Y2H) assays with a subset of clade-III MAPKs (MAP3K15 to MAP3K20) but not with any of the tested clade-I or clade-II MAP3Ks (Supplemental Figure 4). However, MKK3 did not interact with MAP3K13 or MAP3K14. Among all clade-III MAP3Ks, only MAP3K13 and MAP3K14 are predicted to contain a transmembrane domain at their C terminus (Schwacke et al., 2003). As transmembrane domains may trigger Y2H false negatives, we generated truncations of activation domain (AD)-MAP3K13 and AD-MAP3K14 fusions missing the last 143 and 116 amino acids of the MAP3Ks, respectively, corresponding to the putative transmembrane domain. Only then did MKK3 interact with the truncated forms of MAP3K13 and MAP3K14 in Y2H (Figure 2A).
Figure 2.
Functional Reconstitution of MKK3 Modules.
(A) Yeast two-hybrid analysis of the interaction between MKK3 and full-length or C-terminal truncated forms (Δ) of MAP3K13 and MAP3K14. EV, Empty vector. One typical experiment out of three is shown.
(B) Kinase activity of HA-immunoprecipitated MPK2 transiently expressed in mkk3-1 mesophyll protoplasts in the presence or absence of MKK3-Myc and YFP-tagged clade-III MAP3Ks. Immunoblots show protein expression levels. Equal loading was controlled by Coomassie staining of the membrane. RbcL, RubisCo large subunit. Results were repeated two to five times depending on the MAP3K.
To test whether these MKK3-interacting MAP3Ks can activate the downstream kinases MKK3-MPK2 in planta, we took advantage of a transient expression system in Arabidopsis mesophyll protoplasts. We coexpressed YFP-tagged versions of the eight clade-III MAP3Ks and MPK2-HA in protoplasts isolated from mkk3-1 leaves in the absence or presence of MKK3-Myc as the sole source of MKK3 (Figure 2B). We assayed MPK2 activity after immunoprecipitation with an anti-HA antibody. MPK2-HA was activated only when both MKK3 and one of the tested MAP3Ks were coexpressed together. Surprisingly, the fusion proteins MAP3K13-, MAP3K14-, MAP3K15-, and MAP3K16-YFP were barely detectable at best, despite their ability to activate MPK2 in a MKK3-dependent manner, demonstrating their intact functionality. In the case of MAP3K14, we confirmed that activation of the module depended on kinase activity: the kinase-dead variant MAP3K14D140A could not activate the MKK3-MPK2 module (Supplemental Figure 5). Overall, these results indicate that clade-III MAP3Ks form functional modules with MKK3 and MPK2 (together with other clade-C MAPKs) in planta.
Wounding Induces the Transcriptional Regulation of Clade-III MAP3K
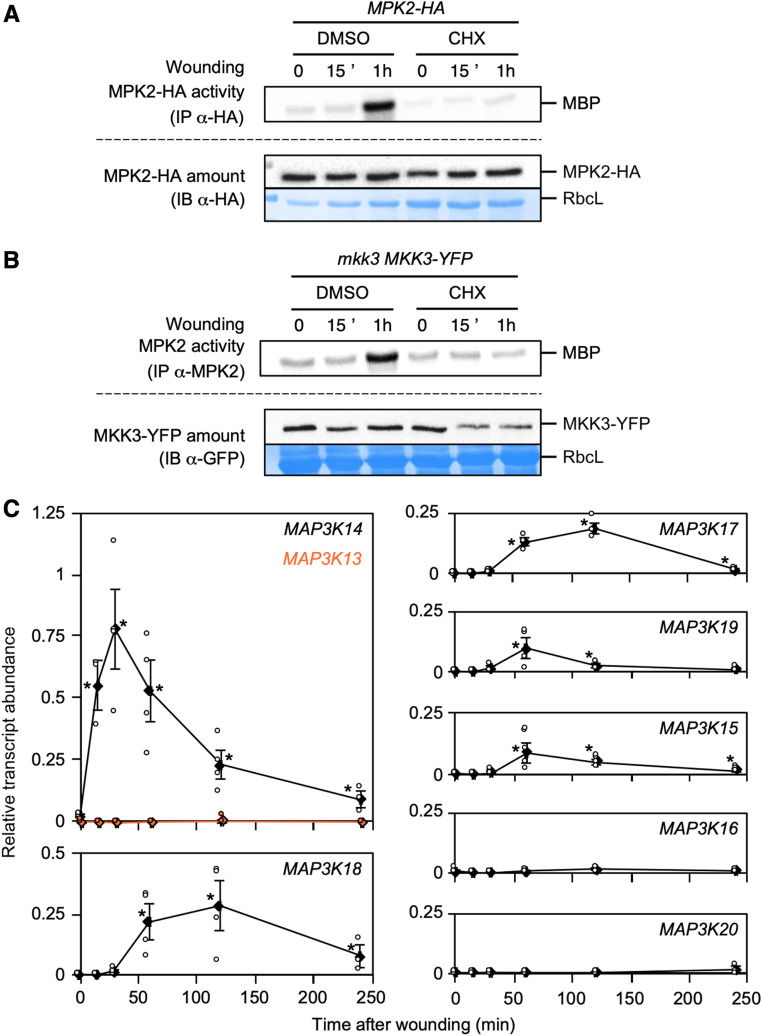
We previously demonstrated that the slow activation of MPK7, a close homologue of MPK2, by ABA was dependent on de novo protein synthesis (Danquah et al., 2015). To determine whether a similar mechanism might be involved in the wound-induced activation of MPK2, we sprayed the protein biosynthesis inhibitor cycloheximide (CHX; 100 µM) onto plants accumulating HA-tagged versions of MAPKs prior to wounding. After immunoprecipitation and kinase assays, we saw no evidence of activation for either MPK1-HA, MPK2-HA, or MPK7-HA in response to wounding, although MAPK protein levels remained unchanged (Figure 3A; Supplemental Figure 6). In addition, MKK3-YFP protein levels only decreased slightly upon CHX spraying prior to wounding, as seen in mkk3-1 MKK3-YFP plants (Figure 3B). These results suggest that the activation of clade-C MAPKs requires the wound-induced synthesis of proteins acting upstream of MKK3. Transcriptional regulation of clade-III MAP3Ks would place a necessary, and conserved, regulatory step to activate clade-C MAPKs. Using RT-qPCR, we measured the expression of clade-III MAP3K genes in Col-0 leaves over a wounding time course (Figure 3C). We observed that MAP3K14 transcript levels rose sharply within 15 min postwounding with peak expression within 30 min. Other MAP3Ks (MAP3K15, MAP3K17, MAP3K18, and MAP3K19) were also induced in response to wounding, although more moderately, while their transcript levels reached their peak later than MAP3K14, 1 to 2 h after wounding. These results suggest that although all clade-III MAP3Ks can activate the MKK3-MPK2 module, only MAP3K14, MAP3K15, MAP3K17, MAP3K18, and MAP3K19 may play a role in its activation upon wounding. We also confirmed that wound-induced transcriptional upregulation of MAP3Ks led to protein accumulation, taking advantage of a previously published map3k17 map3k18 double mutant complemented with a construct bearing a YFP-tagged version of the MAP3K18 locus (Danquah et al., 2015). MAP3K18-YFP protein did accumulate upon wounding, with the same slow kinetics displayed by MAP3K18 transcript accumulation (Supplemental Figure 7). Overall, our expression data are compatible with the slow activation of MPK2 by wounding and suggest that MAP3K14 (as well as other clade-III MAP3Ks) may play a role in the wound-triggered activation of the MKK3-MPK1/2/7/14 module.
Figure 3.
MPK2 Activation by Wounding Requires Protein Synthesis.
(A) and (B) Kinase activity of MPK2 after immunoprecipitation with anti-HA (A) and anti-MPK2 (B) antibodies from leaves of indicated genetic background following 100 µM CHX or mock treatment (0.03% DMSO) spraying prior to wounding. Protein amount was monitored by immunoblot using anti-HA (A) and anti-GFP (B) antibodies. Equal loading was controlled by Coomassie staining of the membrane. RbcL, RubisCo large subunit. Results were repeated three times.
(C) RT-qPCR analysis of clade-III MAP3K genes in response to wounding. Transcript levels are expressed relative to ACTIN2 as reference gene. Values are mean ± SE of three to four biological replicates. Asterisks indicate statistical differences compared to time zero (Mann-Whitney test; α-= 1%).
MAP3K14 Contributes to MPK2 Activation by Wounding
To obtain genetic evidence that MAP3K14 contributes to the activation of the MKK3 module in response to wounding, we first identified homozygous plants carrying a T-DNA insertion in the MAP3K14 gene (Supplemental Figure 8A). Surprisingly, MPK2 activation was higher in the map3k14-1 mutant background relative to Col-0 plants (Supplemental Figure 8B). However, we realized that the map3k14-1 mutant was not a complete loss-of-function but rather a gain-of-function allele. The T-DNA insertion site, located 1041 bp downstream of the start codon, is predicted to interrupt the MAP3K14 coding sequence by removing a large part of the encoded C-terminal tail without affecting the kinase domain. When transfected in mkk3-1 mesophyll protoplasts, the predicted truncated protein MAP3K14-1 activated the MKK3-MPK2 module (Supplemental Figure 8C). Importantly, MAP3K14-1 showed a strong accumulation in immunoblots, in contrast to MAP3K14, potentially explaining the overactivation of MPK2 in map3k14-1 and suggesting that the C terminus of MAP3K14 may contain a domain important for protein stability.
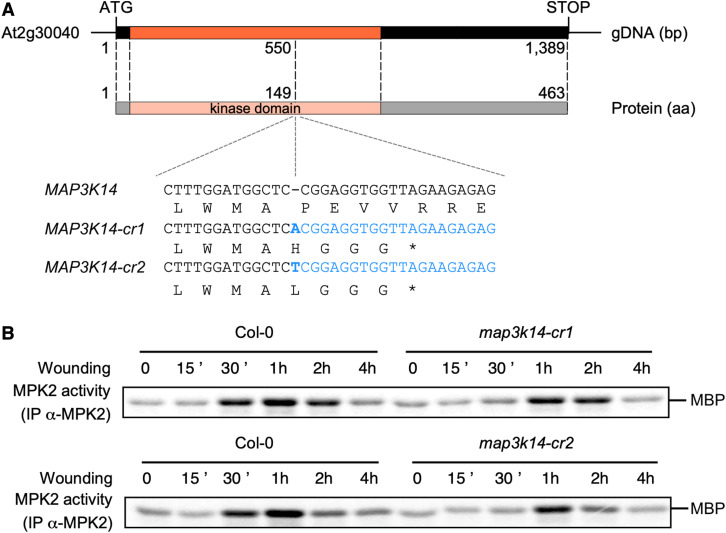
Since no other T-DNA insertion lines besides map3k14-1 were available at this time to determine the phenotype associated with a true loss-of-function in MAP3K14, we turned to clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) technology to generate two independent genome-edited lines. Both lines had a 1-bp insertion at the same position (550 bp downstream of the ATG): the addition of one adenine in map3k14-cr1 and the addition of one thymine in map3k14-cr2. The resulting frame shift introduces an early STOP codon leading to a truncated kinase domain (Figure 4A). map3k14-cr1 and map3k14-cr2 plants challenged by wounding exhibited a delayed and weaker activation of MPK2 (Figure 4B; Supplemental Figure 9). This result is in agreement with MAP3K14 being the only clade-III MAP3K whose transcription is induced at early time points after wounding (Figure 3C). Overall, these data support the hypothesis that MAP3K14 initiates the wound-mediated activation of the MKK3-MPK2 module but also indicates that other MAP3Ks may take over MPK2 activation at later time points.
Figure 4.
MAP3K14 Plays a Role in the Activation of MPK2 by Wounding.
(A) Genomic structure of MAP3K14 and CRISPR lines used in this work.
(B) Kinase activity of MPK2 after immunoprecipitation with an anti-MPK2 antibody from Col-0, map3k14-cr1, and map3k14-cr2 leaves following wounding. Results were repeated four times.
JA Regulates the Activation of the MKK3 Module in Response to Wounding
Wounding rapidly triggers the biosynthesis of the phytohormone JA, its isoleucine conjugate JA-Ile, and its precursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA). Their accumulation appears to be independent of the MKK3 module, as their levels are not affected by the loss of MKK3 function (Figure 5A). We next investigated whether JA plays a role in the wound-triggered activation of MAPKs. JA treatment (50 µM) of 10-d-old Col-0 seedlings caused a slow rise in the activity of endogenous MPK2 (with a peak at 30 to 60 min depending on the experiment) that was absent in mkk3-1 mutant seedlings (Figure 5B). Similarly, JA treatment resulted in the activation of MPK2-HA, MPK1-HA, and MPK7-HA without affecting protein levels (Supplemental Figure 10).
Figure 5.
JA Is Involved in MKK3-MPK2 Activation by Wounding.
(A) JA, JA-isoleucine, and cis-OPDA contents in wounded leaves of indicated genetic backgrounds. Values are mean ± SE of three biological replicates. Asterisks indicate statistical differences compared to Col-0, at each time point (Mann-Whitney test; α-= 5%).
(B) Kinase activity of MPK2 after immunoprecipitation with an anti-MPK2-specific antibody from Col-0 and mkk3-1 in vitro seedlings following 50 µM JA or mock (0.1% ethanol) treatments. Results were repeated two to three times depending on time points.
(C) and (D) Kinase activity of MPK2 after immunoprecipitation with an anti-MPK2-specific antibody from leaves of indicated genetic backgrounds following wounding at indicated times. One typical experiment from two is shown.
JA is perceived by the F-box protein CORONATINE INSENSITIVE1 (COI1); coi1 mutants are insensitive to JA (JA-Ile) treatment (Katsir et al., 2008a, 2008b). To test whether the JA receptor COI1 plays a role in the JA-induced activation of MPK2, we treated coi1 mutant seedlings with JA and monitored subsequent MPK2 activity. We observed a reduction in JA-triggered MPK2 activation in the two mutant alleles tested, coi1-34 and coi1-16 (Supplemental Figure 11). Moreover, MPK2 activation was reduced in response to wounding in both coi1-34 and opr3 (defective in 12-OXOPHYTODIENOATE REDUCTASE 3, one of the enzymes involved in JA biosynthesis; Figures 5C and 5D). Consistent with a role of JA in the signaling pathway, the transcriptional upregulation of clade-III MAP3Ks in response to wounding was also partially compromised in coi1-34 (Supplemental Figure 12). Taken together, these results indicate that JA synthesis and signaling activate the MKK3-MPK2 module upon wounding, likely through the modulation of MAP3K expression.
Rapid Wound-Induced Activation of MPK3 and MPK6 Is Independent of MKK3
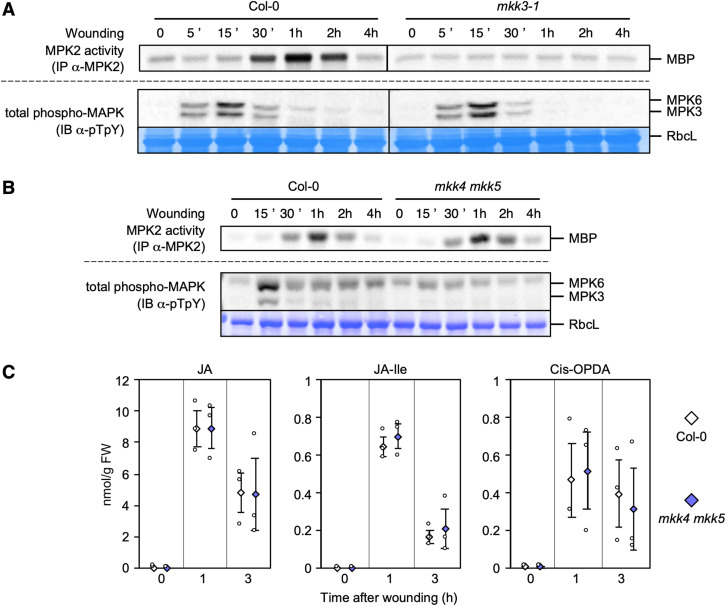
Classical stress-responsive MAPKs, such as Medicago SIMK (STRESS‐INDUCED MAPK) and SAMK (STRESS-ACTIVATED MAPK), tobacco SIPK (SALICYLIC ACID-INDUCED PROTEIN KINASE) and WIPK, and their Arabidopsis homologues MPK3 and MPK6, define functional modules that are activated by wounding (Meskiene et al., 2003; Seo et al., 2007; Li et al., 2018). In our conditions, an antibody raised against the human ERK2 (EXTRACELLULAR SIGNAL-REGULATED KINASE) phospho-motif pT-E-pY (referred to as anti-pTpY here), detected two bands of molecular weights 40 to 45 kD in Arabidopsis leaves rapidly after wounding (Figure 6A; Supplemental Figure 13A). We conclusively assigned the upper band to phosphorylated MPK6, while the lower band corresponds to phosphorylated MPK3, based on leaf samples collected 15 min after wounding in the mpk6-2 and mpk3-1 mutants. These results also confirmed that MPK6 and MPK3 are activated (phosphorylated) in response to wounding (Supplemental Figure 13B). Importantly, the wound-induced activation of MPK3 or MPK6 was not affected in mkk3-1, mkk3-2, map3k14-1, or map3k14-cr1 mutants (Figure 6A; Supplemental Figures 14A to 14C) but strongly impaired in the mkk4 mkk5 double mutant (Figure 6B; Supplemental Figure 13C), as previously described (Li et al., 2018). Moreover, MPK3 and MPK6 were not activated by JA treatment, while their activation by wounding remained intact in the JA receptor mutant coi1-34 (Supplemental Figure 15). Taken together, these results show that two MAPK modules are activated by wounding but with different kinetics: a rapid module involving MKK4/5 and MPK3/6, which is independent of JA, and a slower module comprised of MKK3 and MPK2, which depends on JA signaling.
Figure 6.
MKK4/5-MPK3/6 Are Not Involved in Wound-Induced JA Production.
(A) and (B) Kinase activity of MPK2 after immunoprecipitation with an anti-MPK2-specific antibody from Col-0 (A and B), mkk3-1 (A), and mkk4 mkk5 (B) leaves following wounding at indicated times. MPK3/6 activation was monitored by immunoblot using an antibody raised against the phosphorylated form of ERK2 (anti-pTpY). Equal loading was controlled by Coomassie staining of the membrane. RbcL, RubisCo large subunit. Results were repeated two to three times depending on the time point.
(C) JA, JA-isoleucine, and cis-OPDA contents in wounded leaves of Col-0 and mkk4 mkk5. Values are mean ± SE of three biological replicates. None of the comparisons between Col-0 and mkk4 mkk5 are statistically different (Mann-Whitney test; α-= 5%).
The MKK3-MPK2 and MKK4/5-MPK3/6 Modules Are Independently Activated by Wounding
The fact that the MKK4/5-MPK3/6 module is activated rapidly suggested that it might function upstream of MKK3-MPK2. This hypothesis is supported by the observation that the tobacco homologues of MPK3 and MPK6, SIPK and WIPK, play a role in wound-induced JA biosynthesis (Seo et al., 1995, 1999, 2007; Heinrich et al., 2011). We also showed here that JA is an important signal for the activation of MKK3-MPK2 in response to wounding in Arabidopsis (Figure 5). To test whether the MKK4/5-MPK3/6 module acts upstream of MKK3-MPK2, we compared the activation of MPK2 by wounding in Col-0 and mkk4 mkk5 double mutant plants (Figure 6B). Although we detected little activation of MPK3 and MPK6 by wounding in mkk4 mkk5 leaves, MPK2 activation remained essentially normal. This result indicates that MPK2 activation does not rely on MKK4/5-MPK3/6 function. Although homologues of MPK3/6 in other species were proposed to control JA production, our results suggest that neither MPK3 nor MPK6 are involved in the JA-mediated activation of MPK2 by wounding. To resolve this conundrum, we tested whether tobacco and Arabidopsis regulate JA biosynthesis in a similar fashion. For this purpose, we measured JA levels in Col-0 and mkk4 mkk5 plants at various time points after wounding (Figure 6C; Supplemental Figure 16). Wounding strongly and rapidly increased levels of JA, JA-Ile, and cis-OPDA in both Col-0 and in mkk4 mkk5 mutants, indicating that the MKK4/5-MPK3/6 module is not involved in wound-induced JA accumulation.
The MKK3-MPK2 Module Is Activated by Spodoptera littoralis Feeding
The mechanical wounding we subjected plants to partially mimics attacks by herbivorous insects. To test whether insects can activate the MAPK pathways we described with comparable kinetics, we placed starved S. littoralis fourth-instar larvae on Col-0 leaves and allowed them to feed for 15 min and monitored MAPK activation over 4 h, time zero corresponding to first contact between larvae and leaves (Figure 7A). Under these conditions, MPK2 was activated after 30 min with a peak after 1 h. The activation of MPK3 and MPK6 was weak and restricted to a short time window between 5 and 15 min following contact with larvae (Figure 7A). MPK2 activation was dependent on MKK3 (Figure 7B) and COI1 (Figure 7C), offering a nice parallel to our results in response to wounding and further suggesting that JA plays a prominent role in S. littoralis-triggered MPK2 activation.
Figure 7.
MKK3-MPK2 Module Is Activated by Spodoptera littoralis Feeding.
(A) Kinase activity of MPK2 after immunoprecipitation with an anti-MPK2-specific antibody from leaves on which S. littoralis fed during 15 min before being removed (at T = 15′). MPK3/6 activation was monitored by immunoblot using an antibody raised against the phosphorylated form of ERK2 (anti-pTpY). Equal loading was controlled by Coomassie staining of the membrane. RbcL, RubisCo large subunit. Results were repeated twice.
(B) and (C) Kinase activity of MPK2 after immunoprecipitation with an anti-MPK2-specific antibody from leaves on which S. littoralis fed for 1 to 3 h in Col-0 and coi1-34. One typical experiment out of two is shown.
(D) Weight of S. littoralis caterpillars after feeding for 8 d on Col-0 and mkk3-1 rosettes. Box plot shows distribution of caterpillar weight (n > 120 in five biological replicates). Crosses show averages of biological replicates (39.5 ± 5.2 for Col-0 [n = 5] and 54.4 ± 6.6 mg for mkk3-1 [n = 7]; statistical difference based on Mann-Whitney test; α-= 2.5%).
Lastly, we tested whether the MKK3 module provided a line of defense against insect herbivory. We compared the growth of S. littoralis larvae on Col-0 and mkk3-1 plants by placing first-instar larvae on leaves and allowed them to feed on plants for 8 d. We observed a significant increase in larval weight when they fed on mkk3-1 leaves (Figure 7D). These results demonstrate that the MKK3 module can restrict S. littoralis growth.
DISCUSSION
We report here the activation of two independent MAPK modules by wounding in Arabidopsis plants (Figure 8). The first module is defined by MKK4/5 and MPK3/6. The second module, defined by MKK3 and clade-C MAPKs, was inferred from previous preliminary information (Ortiz-Masia et al., 2007). These two modules are activated independently of each other and with very distinct kinetics. We also demonstrate that JA, which is produced in response to wounding and to herbivore attacks, is a critical mediator for the activation of the MKK3 module.
Figure 8.
Working Model of MAPK Activation by Wounding.
Wounding and insect feeding activate two MAPK modules: (1) a rapid one composed of MKK4/5-MPK3/6, which regulates notably ethylene production, and (2) a slower one composed of clade-III MAP3Ks-MKK3-MPK1/2/7/14, whose activation is under the control of a JA-dependent production of MAP3Ks. Drought also activates MAP3Ks-MKK3-MPK1/2/7/14 through an ABA-dependent step.
Defining General Late Responsive Signaling Modules with Clade-III MAP3Ks, MKK3, and Clade-C MAPKs
Our data suggest that the MKK3-MPK2 module is activated through the transcriptional upregulation of several clade-III MAP3Ks in response to JA and wounding. Among them, MAP3K14 shows the highest and fastest rise in expression upon wounding. Consistent with these observations, we documented a reduction in MPK2 activation upon wounding specifically at early time points in loss-of-function map3k14 genome-edited lines. We also observed a stronger wound-induced MPK2 activation in the gain-of-function allele map3k14-1, which accumulates a truncated and more stable form of MAP3K14. In addition, other members of clade-III MAP3Ks, such as MAP3K17, MAP3K18, MAP3K19, and MAP3K20, which are all induced by wounding but at a later time point and to a lesser extent, may sustain the activation of MPK2 after the initial activation triggered by MAP3K14. Among the eight clade-III MAP3K proteins, only MAP3K13 and MAP3K14 have a carboxyl-terminal extension that may encode a transmembrane domain, possibly suggesting a specific role at a cellular membrane, a location thought to play a role in cellular wound sensing (Farmer et al., 2014). Unfortunately, we failed to detect any fluorescent signal in lines expressing MAP3K14-YFP upon wounding, suggesting either a very low expression level of MAP3K14 or that adding a YFP tag at the C-terminus of the protein reduced MAP3K14 stability. As expected in a process requiring de novo translation, the general protein biosynthesis inhibitor cycloheximide abolished the wound-induced activation of MPK2. De novo synthesis of MAP3Ks has been well documented in the past: for instance, the activation of the MKK3-MPK7 module by drought requires the accumulation of MAP3K18 protein in an ABA-dependent manner (Boudsocq et al., 2015; Danquah et al., 2015). We describe here the activation of the MKK3 module in response to another stress (wounding) by the transcriptional upregulation and subsequent rise in protein synthesis of MAP3Ks. This new result suggests that MAPK signaling pathways with slow kinetics are more general than initially thought. The module may therefore broadly regulate a second layer of events in response to environmental constraints (Colcombet et al., 2016).
MKK3 modules have been far less characterized than the classic stress-activated module MKK4/5-MPK3/6. An obvious explanation lies in its rather atypical slow activation, which was unexpected; textbooks often state that MAPK modules act early and quickly in signaling cascades. In addition, the activity of clade-C MAPKs has thus far never been detected using in-gel kinase assays. Nevertheless, phosphorylated (and thus activated) peptides from clade-C MAPKs do appear in phosphoproteomic data sets, suggesting that they are often activated. For example, phosphorylation of MPK1 and/or MPK2 was reported in response to ABA (Umezawa et al., 2013) and DNA damage-inducing radiation (Roitinger et al., 2015). Interestingly, the anti-pT-E-pY antibody detected an additional phosphorylated protein in map3k14-1, which overactivates MPK2 in response to wounding; the apparent molecular weight of this new band is smaller than MPK3 or MPK6 and follows the expected kinetics fitting clade-C MAPK activation (Supplemental Figure 14B). This suggests that clade-C MAPKs are tightly controlled to avoid full activation. More globally, transcriptional upregulation of clade-III MAP3Ks by stress may occur very often based on expression data sets (Zimmermann et al., 2004; Winter et al., 2007) and suggests crucial roles for MKK3 modules in perception of the environment. Notably, MAP3K13, MAP3K18, and MAP3K20 are induced by multiple environmental signals, including osmotic, salt, and drought stress. While MKK3 constitutes a hub in the module, the related MAP3Ks and MAPKs are encoded by multigenic families. Signal specificity is conferred by the transcriptional regulation of the upstream MAP3K genes, although we cannot exclude that MAP3K activity is also directly modulated by the imposed stress, as suggested in ABA signaling (Matsuoka et al., 2015; Mitula et al., 2015). How signal specificity is achieved is less clear for MAPKs: MPK1/2/7/14 appear to be activated by stress in the same way, based on protoplast experiments and immunoprecipitation from plant tissues (Danquah et al., 2015). We hypothesize that specificity results from cell- or tissue-specific expression of MAPKs or of their target substrates. This should be an important point to address in the future.
Last, MKK3 has also been proposed to function upstream of MPK6 and MPK8 in various contexts such as blue light, dark-to-light transitions, and ROS homeostasis (Takahashi et al., 2007, 2011; Sethi et al., 2014; Lee, 2015). We previously reported on the lack of such functional connections when combining these kinases in a protoplast transient expression system (Danquah et al., 2015). In agreement, this work failed to discover a functional link between MKK3 and MPK6 in the context of wounding. Nonetheless, it is possible that such connections may exist in other physiological contexts and in specific cell types or tissues (Colcombet et al., 2016).
Is Wound-Induced JA Production Connected to MAPK Modules?
The phytohormone JA plays a critical role in wound responses (Wasternack, 2007). It is massively produced within a few minutes after wounding by chloroplast- and peroxisome-localized biosynthetic enzymes and is involved both in local and long-distance signaling (Koo et al., 2009; Wasternack and Hause, 2013). Plants impaired in JA biosynthesis or signaling exhibit weakened responses to wounding and are much more susceptible to chewing insects (Howe and Jander, 2008). The JA core signaling pathway is well described (Chini et al., 2007; Thines et al., 2007): JASMONATE ZIM DOMAIN (JAZ) proteins sequester transcription factors, notably basic helix-loop-helix factors such as MYC2 to prevent their activation of JA-responsive genes. The binding of the bioactive form of JA, JA-Ile, to the COI1 receptor, an F-box protein that forms an E3 ubiquitin ligase Skp, Cullin, F-box (SCF) containing complex with Skp and Cullin proteins, increases the affinity of COI1 for JAZ proteins, triggering their ubiquitination and consequent proteasome-dependent degradation. The released transcription factors are then free to modulate gene expression (Thines et al., 2007). We observed a reduction in the activation of MPK2 triggered by wounding in the opr3 and coi1 mutants, which are impaired in JA biosynthesis and signaling, respectively. We can therefore conclude that the activation of the MKK3-MPK1/2/7/14 module occurs downstream of JA signaling and biosynthesis. The residual activation in these mutants may arise from the existence of JA-independent pathways controlling MAP3K transcriptional regulation in response to wounding. An alternative explanation may build upon the fact that these are not complete loss-of-function mutants. In fact, opr3 mutant still produces some JA-Ile (Chini et al., 2018), while coi1-34 is clearly not a full loss-of-function allele, since it is only partially male-sterile, unlike coi1 null mutants (Acosta et al., 2013). Surprisingly, the up-regulation of MAP3K transcripts triggered by wounding is only mildly blocked in the two JA mutants, although the activation of MAPKs is itself strongly reduced. It is possible that the levels of active MAP3Ks are limiting and titrated by competition between their targets. Alternatively, full activation of MAP3Ks may also require a JA-dependent modification (i.e., phosphorylation) in addition to the transcriptional induction of their encoding genes. Only the inactivation of both transcriptional and posttranslational regulations would lead to a strong reduction of MPK2 activation. This last hypothesis is supported by the fact that MAP3K18 intrinsic activity increases upon ABA treatment (Matsuoka et al., 2015; Mitula et al., 2015).
These results add a new layer of complexity in the chain of events triggered by JA perception. An open question remains: what is controlled by the JA-activated MKK3 module? MKK3-related modules regulate ROS homeostasis in response to wounding, but they were proposed to act upstream of the clade-D MAPK MPK8 (Takahashi et al., 2011). Recently, the MAP2K inhibitor PD98059 was shown to reduce JA-triggered activation of enzymes involved in ascorbate and glutathione metabolism in maize (Zea mays) leaves (Shan and Sun, 2018). Together, these results point to redox homeostasis as a putative target of the MKK3 module in the general response to wounding and JA.
The signaling pathway leading from wound signaling to JA biosynthesis is largely unknown. One known event is a transient increase in cytosolic Ca2+ (Maffei et al., 2004; Kiep et al., 2015) that is then decoded by Ca2+-sensing proteins. Two CALMODULIN-LIKE (CML) proteins, CML37 and CML42, were identified as positive and negative regulators, respectively, of JA-mediated defense in Arabidopsis plants after herbivore attack (Vadassery et al., 2012; Scholz et al., 2014). While CML42 very likely affects the binding of JA-Ile to the receptor, CML37 regulates the expression of JASMONATE RESISTANT1 (JAR1), the enzyme catalyzing JA-Ile formation. Our work shows that JA synthesis is not under the control of MPK3 or MPK6. This is surprising, as the tobacco homologues of MPK3 and MPK6 (WIPK and SIPK) modulate JA levels in response to wounding (Seo et al., 1995, 1999, 2007; Heinrich et al., 2011). This apparent discrepancy might be explained by different experimental conditions used in each study. For example, we took advantage of the mkk4 mkk5 double mutant (Zhao et al., 2014), in which the activation of MPK3 and MPK6 by wounding is strongly reduced, although not fully abolished. It is possible that the residual and weak activation of MPK3 and MPK6 is sufficient to trigger downstream events such as JA biosynthesis. Nevertheless, this possibility is unlikely, since this mutant is clearly compromised in wound-induced ethylene biosynthesis (Li et al., 2018). Alternatively, and more appealingly, these results suggest that homologous MAPKs may not regulate the same responses in various plant species, despite being activated in the same manner by a given stress. This might illustrate diverging adaptation strategies between tobacco and Arabidopsis.
MPK3 and MPK6 are very well characterized stress-responsive MAPKs. Many of their substrates have been identified in the context of microbe-associated molecular pattern signaling (Bigeard et al., 2015; Bigeard and Hirt, 2018; Rayapuram et al., 2018). However, very little is known about their functions in response to other types of stress, including wounding. The first report of a role for MKK4/5-MPK3/6 module in wound signaling came from the observation that they induced the transcription of 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE (ACS) genes and thus ethylene biosynthesis in Arabidopsis (Li et al., 2018). Interestingly, the same module is involved in ethylene biosynthesis in response to flg22 and to infection by the fungus Botrytis (Liu and Zhang, 2004; Li et al., 2012). This regulation occurs through both a direct phosphorylation-dependent stabilization of the ACS6 enzyme and an up-regulation of ACS genes by the phosphorylation-dependent activation of the WRKY33 transcription factor. In rice (Oryza sativa), OsMPK1, the closest homologue of Arabidopsis MPK6, was shown to interact with and phosphorylate WRKYs in vitro (Yoo et al., 2014). It is worth noting that cycloheximide, a potent blocker of MKK3 module activation (Figure 3), is also a strong activator of MPK3 and MPK6 (Supplemental Figure 17) and triggers the transcriptional up-regulation of a large fraction of microbe-associated molecular pattern-regulated genes (Navarro et al., 2004). Overall, these data suggest that the MKK4/5-MPK3/6 module may be involved in the control of gene expression in response to wounding in the same way as other stresses (Frei dit Frey et al., 2014).
Roles of MAPKs in Interactions with Herbivores
MAPKs also play an important role in plant-herbivore interactions (Hettenhausen et al., 2015). Early studies in tobacco showed that WIPK was activated rapidly by wounding and that WIPK-silenced plants had reduced levels of defense-related genes and JA (Seo et al., 1995). In coyote tobacco (Nicotiana attenuata), mechanical wounding and oral secretions of the herbivore Manduca sexta activate SIPK and WIPK within 5 min (Wu et al., 2007). In Arabidopsis, grasshopper oral secretions enhance the wound-induced activation of MPK3 and MPK6 (Schäfer et al., 2011). In tomato (Solanum lycopersicum), M. sexta feeding also activated the WIPK and SIPK homologues SlMPK3 and SlMPK1/2, respectively, while silencing SlMPK1/2 reduces herbivory-induced JA levels, allowing greater growth of larvae (Kandoth et al., 2007). However, other studies in both tobacco species (N. tabacum and N. attenuata) revealed a more complex picture in the JA-MAPK relationship depending on the herbivore. In response to wounding, tobacco MPK4 is activated in minutes, and silencing the encoding gene compromises the induction of JA-responsive genes (Gomi et al., 2005). By contrast, silencing NaMPK4 affects neither JA production nor resistance to the generalist herbivore S. littoralis but increases resistance to M. sexta (Hettenhausen et al., 2015). This indicates that some functions of MPK4 are specific to particular herbivore interactions, and that further studies will be necessary to clarify the role of different MAPK modules in wound- and herbivory-induced JA signaling.
METHODS
Plant Material
mkk3-1, coi1-34 (Acosta et al., 2013), opr3, mpk3-1, mpk6-2 (Galletti et al., 2011), mkk4 mkk5 (Su et al., 2017), and map3k17 map3k18 MAP3K18-YFP (Danquah et al., 2015) were described previously. mkk3-2 (SALK_208528) and map3k14-1 (Gabi_653B01) were identified in public databases and ordered from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/).
To create genome-edited alleles of MAP3K14 by CRISPR/Cas9, we generated CRISPR single guide RNAs targeting MAP3K14 by cloning primers Cs9-3K14-F and Cs9-3K14-R into vector pDGE65 according to Ordon et al. (2017). The resulting construct, pDGE65-MAP3K14CR, was introduced into Agrobacterium tumefaciens (Agrobacterium) strain C58C1 and used to transform Arabidopsis (Arabidopsis thaliana) Col-0 plants by the floral-dipping method (Clough and Bent, 1998). We selected 20 primary transformants resistant to the herbicide BASTA (Bayer) and determined the number of T-DNA insertions using the segregation of the BASTA resistance in T2 plants. To eliminate the pDGE65-MAP3K14CR T-DNA, BASTA-sensitive T2 seedlings were rescued by careful transfer into pots. After recovery, we genotyped them for the presence of mutations in MAP3K14, as well as for the absence of mutations in MAP3K13, its closest homologue. We also confirmed the absence of the pDGE65-MAP3K14CR T-DNA in the genome by PCR. We identified two independent and homozygous lines (map3k14–cr1 and map3k14–cr2).
To generate constructs that add HA or YFP tags to selected kinases, we amplified the full MPK1, MPK2, MPK7, and MKK3 loci (much of the promoter, 5′ UTR, exons, and introns up to the STOP codon) using appropriate primers (Supplemental Table 1) with high-fidelity iProof DNA polymerase (Bio-Rad). We then digested the resulting PCR products with appropriate restriction enzymes and cloned them in-frame with HA or YFP tags using in-house binary vectors derived from pGREEN0229. The final vectors were named pGREEN0229-MPK1-HA, pGREEN0229-MPK2-HA, pGREEN0229-MPK7-HA, and pGREEN0229-MKK3-YFP. Their sequences are provided in Supplemental Table 2. Vectors were introduced into Agrobacterium strain C58C1 containing the pSOUP helper plasmid (Hellens et al., 2000) and then used to transform Col-0 or mkk3-1 plants with the floral-dipping method (Clough and Bent, 1998). We identified homozygous lines by segregation analysis of resistance to BASTA.
ORF Cloning and Protoplast Expression
Constructs for protoplast experiments (MKK3-Myc, MKK3-EE-Myc, MPK2-HA, MAP3K17-YFP, and MAP3K18-YFP) were previously reported by Danquah et al. (2015). Others were generated for this work. In brief, we amplified open reading frames (ORFs) from Col-0 genomic DNA using appropriate primers (Supplemental Table 1) and cloned the PCR products in pDONR207 using Gateway technology (Invitrogen). We amplified ORFs with and without the STOP codon in a single reaction by using reverse degenerate primers (see sequences in Supplemental Table 1). We identified clones lacking the STOP codon by restriction digest (ORFs lacking the STOP codon contain a BamHI restriction site) and by sequencing.
Sequenced ORFs without STOP were then recombined into p2GWF7 to generate MAP3K-YFP C-terminal fusions for protoplast experiments (MAP3K13-YFP, MAP3K14-YFP, MAP3K15-YFP, MAP3K16-YFP, MAP3K19-YFP, MAP3K20-YFP). To generate p2GWF7-MAP3K14-1, we amplified a PCR product with primers p35S and pMAP3K4-1 using p2GWF7-MAP3K14 as a template and then cloned it into p2GWF7-MAP3K14 at the BamHI and HindIII restriction sites. Both vector sequences are provided in Supplemental Table 2. We introduced the D140A mutation into p2GWF7-MAP3K14 and p2GWF7-MAP3K14-1 following a PCR-based mutagenesis. In brief, full vectors p2GWF7-MAP3K14 and p2GWF7-MAP3K14-1 were amplified using high-fidelity iProof DNA polymerase (Bio-Rad) and primers 3K14-D140A-F and 3K14-D140A-R bearing the D140A mutation (Supplemental Table 1). The nonmutated methylated templates p2GWF7-MAP3K14 and p2GWF7-MAP3K14-1 were removed by digestion with DpnI (NEB), and remaining mutated plasmids were transformed in Escherichia coli DH5alpha. Plasmids from individual colonies were verified by sequencing. MAPK-HA immunoprecipitation and kinase assays were performed as previously described (Danquah et al., 2015).
Y2H Assay
Constructs BD-MKK3, AD-MAP3K17, and AD-MAP3K18 were previously reported by Danquah et al. (2015). Additional constructs were generated using gateway technology (Invitrogen) by recombining sequenced ORFs with STOP (see previous section) into pDEST22 to generate AD-MAP3K fusions (AD-MAP3K13, AD-MAP3K14, AD-MAP3K15, AD-MAP3K16, AD-MAP3K19, AD-MAP3K20). We then removed the DNA fragment coding for the putative transmembrane domain from pDEST22-MAP3K13 and pDEST22-MAP3K14 using a PCR-based deletion protocol identical to the one described for site-specific mutations above and appropriate primers (Supplemental Table 1). Y2H assays were performed as previously described, except that the selective medium was generated with 0.2% (w/v) dropout medium lacking Trp, Leu, and Ura (U.S. Biological; Berriri et al., 2012).
Wounding Experiments
Single plants were grown on soil pellets (Jiffy-7 38-mm pellet pack, ref. no. 32204011, Jiffy Products International) in a Percival growth chamber under a 12-h light (80 to 100 μE.m−2.s−1 provided by soft white fluorescent tubes [Philips F17T8/TL830/PLUS/ALTO])/12-h dark photoperiod at 22°C and 70% humidity for 4 weeks before experiments. Genotypes were randomized within the tray to avoid position bias. For each sample, three fully expanded leaves from three Arabidopsis rosettes were wounded three times using tweezers (5-mm round tip with line-serrated surfaces). Wounding was performed at such a time as to harvest samples (3 leaves × 3 plants) 7 to 8 h after lights on (see Supplemental Protocol for details). For cycloheximide treatments, we sprayed a solution containing 100 µM CHX (Sigma-Aldrich, ref. no. C4859) in 0.03% (v/v) DMSO onto rosettes 150 min before collecting samples. We also sprayed control rosettes with 0.03% (v/v) DMSO alone as mock treatment. Samples were frozen in liquid nitrogen, ground using either mortar and pestle or a Tissuelyser II (Qiagen) and kept at −80°C before further investigation. Experiments were typically repeated three times with plants grown independently.
JA Experiments
For JA-treated samples, seeds were sterilized (15 min in 70% [v/v] ethanol) and 20 to 30 seedlings were grown in small Petri dishes (diameter 6 cm) containing 8 mL half-strength Murashige and Skoog medium supplemented with 1% (w/v) Suc for 10 d under a 16-h light/8-h night photoperiod at 22°C. Seedlings were then treated with 0.1% (v/v) ethanol (mock) or 50 µM JA (Sigma-Aldrich, ref. no. J2500). To stop treatments, seedlings were rapidly (<20 s) wiped with a paper towel and frozen in liquid nitrogen.
Jasmonate content in Arabidopsis leaves was analyzed by liquid chromatography-mass spectrometry as previously described by Almeida Trapp et al. (2014).
Insect Experiments
Larvae of the generalist lepidopteran species Spodoptera littoralis were reared as previously published by Vadassery et al. (2012). To monitor MPK2 activation or measure oxylipin production in response to insect, fourth-instar larvae were starved overnight prior to plant feeding for 1 or 3 h. To monitor MPK3/MPK6 activation, larvae were allowed on plants for 15 min before being removed. For long-term feeding assays (8 d), we used first-instar larvae as described by Vadassery et al. (2012). Plant samples were frozen in liquid nitrogen and ground to a fine powder. We kept different plant lines separated from each other when used in the same experiment to avoid plant-to-plant contact; all plant positions were also randomized during the experimental setup.
Kinase Assays and Immunoblots
A detailed kinase assay protocol for plant samples is provided (Supplemental Protocol). Kinase assays using α-MPK2 (Ortiz-Masia et al., 2007), α-HA (Sigma-Aldrich H6908), α-MPK3 (Sigma-Aldrich M8318), and α-MPK6 (Sigma-Aldrich A7104) antibodies were performed as previously described by Danquah et al. (2015) and Ortiz-Masia et al. (2007). It should be noted that α-MPK2 antibodies can specifically immunoprecipitate MPK2, although they fail to detect the protein on immunoblots of total proteins (Ortiz-Masia et al., 2007).
Protein levels were monitored by immunoblotting following Bio-Rad recommendations. Proteins were separated in 10% (w/v) SDS-PAGE gels and transferred onto polyvinylidene fluoride membranes (Bio-Rad). Membranes were blocked in 5% (w/v) nonfat dry milk, except for α-pTpY, for which we used 5% (w/v) bovine serum albumin (Sigma-Aldrich). The following primary antibodies were used: α-HA (Roche 11867431001; 1: 10,000 dilution), α-Myc (Sigma-Aldrich C3956; 1:10,000 dilution), α-GFP (Roche 11814460001; 1:5000 dilution), α-MPK3 (Sigma-Aldrich M8318; 1:5000 dilution), α-MPK6 (Sigma-Aldrich A7104; 1:5000 dilution) and α-pTpY (Cell Signaling 4370S; 1:10,000 dilution). The following horseradish peroxidase-coupled secondary antibodies were used: α-rat (Sigma-Aldrich A9037, 1:10,000), α-rabbit (Sigma-Aldrich A6154, 1/20,000), and α-mouse (Sigma-Aldrich A5906, 1:10,000). Horseradish peroxidase activity was detected with a Clarity western ECL substrate reaction kit (Bio-Rad) and a ChemiDoc Imagers (Bio-Rad). Blots were stained with Coomassie Brilliant Blue for protein visualization.
RT-qPCR Analysis
For gene expression analysis, plants were collected at indicated times and frozen in liquid nitrogen. RNA extraction was performed with Qiagen RNeasy plant mini kit, and first-strand cDNA synthesis was initiated with SuperScript II reverse transcriptase (Invitrogen). RT-qPCR analysis was performed as previously described on a CFX384 Touch real-time PCR detection system (Bio-Rad) with SYBR Green Premix Ex Ta (Tli RNase H Plus; Takara, RR420W). RT-qPCR primers are listed in Supplemental Table 1 (Danquah et al., 2015).
Statistics
The statistical analyses were performed with Mann-Whitney test for qRT-PCR, kinase activity, hormonal content and insect feeding assay. The parameters of the test for each experiment is provided in the Supplemental File.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At1g10210 (MPK1), At1g59580 (MPK2), At3g45640 (MPK3), At2g43790 (MPK6), At2g18170 (MPK7), At5g40440 (MKK3), At1g51660 (MKK4), At3g21220 (MKK5), At1g07150 (MAP3K13), At2g30040 (MAP3K14), At5g55090 (MAP3K15), At4g26890 (MAP3K16), At2g32510 (MAP3K17), At1g05100 (MAP3K18), At5g67080 (MAP3K19), At3g50310 (MAP3K20), and At3g18780 (ACTIN2).
SUPPLEMENTAL DATA
Supplemental Figure 1. MPK2 activation by wounding depends on MKK3.
Supplemental Figure 2. MPK1 and MPK7 are activated by wounding.
Supplemental Figure 3. flg22 rapidly and transiently activates MPK3 and MPK6 in leaf punches.
Supplemental Figure 4. MKK3 and clade-III MAP3Ks interact in a yeast two-hybrid assay.
Supplemental Figure 5. MAP3K14 activity is necessary for MKK3-MPK2 activation.
Supplemental Figure 6. MPK1 and MPK7 activations by wounding require protein synthesis.
Supplemental Figure 7. MAP3K18 accumulates in response to wounding.
Supplemental Figure 8. MPK2 Activation by wounding is higher in map3k14-1.
Supplemental Figure 9. Quantification of MPK2 activation by wounding in map3k14-cr1 and map3k14-cr2 lines.
Supplemental Figure 10. MPK1, MPK2 and MPK7 are activated by JA.
Supplemental Figure 11. JA-triggered MPK2 activation is impaired in mutants of JA receptor.
Supplemental Figure 12. The coi1-34 mutant reduces the expression of some wounding-induced MAP3Ks.
Supplemental Figure 13. MPK3 and MPK6 activation depends on MKK4/MKK5 but not on MKK3: mpk and mkk mutant profiles.
Supplemental Figure 14. MPK3 and MPK6 activation depends on MKK4/MKK5 but not on MKK3: mkk3 and map3k14 genome edited lines.
Supplemental Figure 15. MPK3 and MPK6 activity is independent of JA.
Supplemental Figure 16. Oxylipin contents in mkk4 mkk5.
Supplemental Figure 17. MPK3 and MPK6 are activated by cycloheximide treatment.
Supplemental Table 1. Primers used in this study.
Supplemental Table 2. Sequences of some plasmids used in this work.
Supplemental File. Statistical analysis.
Supplemental Protocol. Typical sample preparation and kinase assay to monitor kinase activity after wounding.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
AUTHOR CONTRIBUTIONS
C.S., H.H., and J.C. designed the research; C.S., S.T.S., M.B., C.C., A.K., M.A.-T., A.M., and J.C. performed research; all the authors contributed to the article.
Acknowledgments
We thank the Stress Signaling group for critical discussion of this work. We also thank Shuqun Zhang and Edward Farmer for providing mkk4 mkk5 and coi1-34 seeds. This work was supported by the French State (LabEx Saclay Plant Sciences-SPS, grant ANR-10-LABX-0040-SPS), managed by the French National Research Agency (“Investments for the Future” program ANR-11-IDEX-0003-02). Support was also provided by Saclay Plant Sciences (PhD fellowships to C.S. and C.C.), by a Capes-Humboldt Research Fellowship (to M.A.T.), and by the AgreenSkills+ fellowship program, which has received funding from the EU’s Seventh Framework Program (grant FP7-609398, AgreenSkills+ contract to S.T.S.).
Footnotes
Articles can be viewed without a subscription.
References
- Acosta I.F., Gasperini D., Chételat A., Stolz S., Santuari L., Farmer E.E.(2013). Role of NINJA in root jasmonate signaling. Proc. Natl. Acad. Sci. USA 110: 15473–15478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad P., Rasool S., Gul A., Sheikh S.A., Akram N.A., Ashraf M., Kazi A.M., Gucel S.(2016). Jasmonates: Multifunctional roles in stress tolerance. Front Plant Sci 7: 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida Trapp M., De Souza G.D., Rodrigues-Filho E., Boland W., Mithöfer A.(2014). Validated method for phytohormone quantification in plants. Front Plant Sci 5: 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriri S., Garcia A.V., Frei dit Frey N., Rozhon W., Pateyron S., Leonhardt N., Montillet J.L., Leung J., Hirt H., Colcombet J.(2012). Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell 24: 4281–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J., Colcombet J., Hirt H.(2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8: 521–539. [DOI] [PubMed] [Google Scholar]
- Bigeard J., Hirt H.(2018). Nuclear signaling of plant MAPKs. Front Plant Sci 9: 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogre L., Ligterink W., Meskiene I., Barker P.J., Heberle-Bors E., Huskisson N.S., Hirt H.(1997). Wounding induces the rapid and transient activation of a specific MAP kinase pathway. Plant Cell 9: 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Danquah A., de Zélicourt A., Hirt H., Colcombet J.(2015). Plant MAPK cascades: Just rapid signaling modules? Plant Signal. Behav. 10: e1062197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Chini A., Monte I., Zamarreño A.M., Hamberg M., Lassueur S., Reymond P., Weiss S., Stintzi A., Schaller A., Porzel A., García-Mina J.M., Solano R.(2018). An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 14: 171–178. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colcombet J., Hirt H.(2008). Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem. J. 413: 217–226. [DOI] [PubMed] [Google Scholar]
- Colcombet J., Sözen C., Hirt H.(2016). Convergence of multiple MAP3Ks on MKK3 identifies a set of novel stress MAPK modules. Front Plant Sci 7: 1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danquah A., et al. (2015). Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 82: 232–244. [DOI] [PubMed] [Google Scholar]
- Danquah A., de Zelicourt A., Colcombet J., Hirt H.(2014). The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32: 40–52. [DOI] [PubMed] [Google Scholar]
- Dóczi R., Brader G., Pettkó-Szandtner A., Rajh I., Djamei A., Pitzschke A., Teige M., Hirt H.(2007). The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 19: 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer E.E., Gasperini D., Acosta I.F.(2014). The squeeze cell hypothesis for the activation of jasmonate synthesis in response to wounding. New Phytol. 204: 282–288. [DOI] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R.(2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350. [DOI] [PubMed] [Google Scholar]
- Frei dit Frey N., et al. (2014). Functional analysis of Arabidopsis immune-related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biol. 15: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti R., Ferrari S., De Lorenzo G.(2011). Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 157: 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi K., Ogawa D., Katou S., Kamada H., Nakajima N., Saji H., Soyano T., Sasabe M., Machida Y., Mitsuhara I., Ohashi Y., Seo S.(2005). A mitogen-activated protein kinase NtMPK4 activated by SIPKK is required for jasmonic acid signaling and involved in ozone tolerance via stomatal movement in tobacco. Plant Cell Physiol. 46: 1902–1914. [DOI] [PubMed] [Google Scholar]
- Heinrich M., Baldwin I.T., Wu J.(2011). Two mitogen-activated protein kinase kinases, MKK1 and MEK2, are involved in wounding- and specialist lepidopteran herbivore Manduca sexta-induced responses in Nicotiana attenuata. J. Exp. Bot. 62: 4355–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M.(2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832. [DOI] [PubMed] [Google Scholar]
- Hettenhausen C., Schuman M.C., Wu J.(2015). MAPK signaling: A key element in plant defense response to insects. Insect Sci. 22: 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A., Jander G.(2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59: 41–66. [DOI] [PubMed] [Google Scholar]
- Ichimura K., Mizoguchi T., Yoshida R., Yuasa T., Shinozaki K.(2000). Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 24: 655–665. [DOI] [PubMed] [Google Scholar]
- Kandoth P.K., Ranf S., Pancholi S.S., Jayanty S., Walla M.D., Miller W., Howe G.A., Lincoln D.E., Stratmann J.W.(2007). Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc. Natl. Acad. Sci. USA 104: 12205–12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Chung H.S., Koo A.J.K., Howe G.A.(2008a). Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11: 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A.(2008b). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiep V., Vadassery J., Lattke J., Maaß J.P., Boland W., Peiter E., Mithöfer A.(2015). Systemic cytosolic Ca(2+) elevation is activated upon wounding and herbivory in Arabidopsis. New Phytol. 207: 996–1004. [DOI] [PubMed] [Google Scholar]
- Koo A.J.K., Gao X., Jones A.D., Howe G.A.(2009). A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 59: 974–986. [DOI] [PubMed] [Google Scholar]
- Lee H.(2015). Mitogen-activated protein kinase kinase 3 is required for regulation during dark-light transition. Mol. Cells 38: 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Meng X., Wang R., Mao G., Han L., Liu Y., Zhang S.(2012). Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 8: e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Han X., Yang L., Deng X., Wu H., Zhang M., Liu Y., Zhang S., Xu J.(2018). Mitogen-activated protein kinases and calcium-dependent protein kinases are involved in wounding-induced ethylene biosynthesis in Arabidopsis thaliana. Plant Cell Environ. 41: 134–147. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang S.(2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei M., Bossi S., Spiteller D., Mithöfer A., Boland W.(2004). Effects of feeding Spodoptera littoralis on lima bean leaves. I. Membrane potentials, intracellular calcium variations, oral secretions, and regurgitate components. Plant Physiol. 134: 1752–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei M.E., Arimura G., Mithöfer A.(2012). Natural elicitors, effectors and modulators of plant responses. Nat. Prod. Rep. 29: 1288–1303. [DOI] [PubMed] [Google Scholar]
- Matsuoka D., Yasufuku T., Furuya T., Nanmori T.(2015). An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol. Biol. 87: 565–575. [DOI] [PubMed] [Google Scholar]
- Meskiene I., Baudouin E., Schweighofer A., Liwosz A., Jonak C., Rodriguez P.L., Jelinek H., Hirt H.(2003). Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. J. Biol. Chem. 278: 18945–18952. [DOI] [PubMed] [Google Scholar]
- Mitula F., Tajdel M., Cieśla A., Kasprowicz-Maluśki A., Kulik A., Babula-Skowrońska D., Michalak M., Dobrowolska G., Sadowski J., Ludwików A.(2015). Arabidopsis ABA-activated kinase MAPKKK18 is regulated by protein phosphatase 2C ABI1 and the ubiquitin-proteasome pathway. Plant Cell Physiol. 56: 2351–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L., Zipfel C., Rowland O., Keller I., Robatzek S., Boller T., Jones J.D.G.(2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135: 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordon J., Gantner J., Kemna J., Schwalgun L., Reschke M., Streubel J., Boch J., Stuttmann J.(2017). Generation of chromosomal deletions in dicotyledonous plants employing a user-friendly genome editing toolkit. Plant J. 89: 155–168. [DOI] [PubMed] [Google Scholar]
- Ortiz-Masia D., Perez-Amador M.A., Carbonell J., Marcote M.J.(2007). Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Lett. 581: 1834–1840. [DOI] [PubMed] [Google Scholar]
- Ranf S., Eschen-Lippold L., Pecher P., Lee J., Scheel D.(2011). Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J. 68: 100–113. [DOI] [PubMed] [Google Scholar]
- Rayapuram N., Bigeard J., Alhoraibi H., Bonhomme L., Hesse A.M., Vinh J., Hirt H., Pflieger D.(2018). Quantitative phosphoproteomic analysis reveals shared and specific targets of Arabidopsis mitogen-activated protein kinases (MAPKs) MPK3, MPK4, and MPK6. Mol. Cell. Proteomics 17: 61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P., Bodenhausen N., Van Poecke R.M.P., Krishnamurthy V., Dicke M., Farmer E.E.(2004). A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16: 3132–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J.(2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649. [DOI] [PubMed] [Google Scholar]
- Roitinger E., Hofer M., Köcher T., Pichler P., Novatchkova M., Yang J., Schlögelhofer P., Mechtler K.(2015). Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol. Cell. Proteomics 14: 556–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatin D.V., Gramegna G., Modesti V., Cervone F.(2014). Wounding in the plant tissue: The defense of a dangerous passage. Front Plant Sci 5: 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer M., Fischer C., Meldau S., Seebald E., Oelmüller R., Baldwin I.T.(2011). Lipase activity in insect oral secretions mediates defense responses in Arabidopsis. Plant Physiol. 156: 1520–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz S.S., Vadassery J., Heyer M., Reichelt M., Bender K.W., Snedden W.A., Boland W., Mithöfer A.(2014). Mutation of the Arabidopsis calmodulin-like protein CML37 deregulates the jasmonate pathway and enhances susceptibility to herbivory. Mol. Plant 7: 1712–1726. [DOI] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W.B., Flügge U.I., Kunze R.(2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Katou S., Seto H., Gomi K., Ohashi Y.(2007). The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic and salicylic acids in wounded tobacco plants. Plant J. 49: 899–909. [DOI] [PubMed] [Google Scholar]
- Seo S., Okamoto M., Seto H., Ishizuka K., Sano H., Ohashi Y.(1995). Tobacco MAP kinase: A possible mediator in wound signal transduction pathways. Science 270: 1988–1992. [DOI] [PubMed] [Google Scholar]
- Seo S., Sano H., Ohashi Y.(1999). Jasmonate-based wound signal transduction requires activation of WIPK, a tobacco mitogen-activated protein kinase. Plant Cell 11: 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi V., Raghuram B., Sinha A.K., Chattopadhyay S.(2014). A mitogen-activated protein kinase cascade module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. Plant Cell 26: 3343–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C., Sun H.(2018). Jasmonic acid-induced NO activates MEK1/2 in regulating the metabolism of ascorbate and glutathione in maize leaves. Protoplasma 255: 977–983. [DOI] [PubMed] [Google Scholar]
- Su J., Zhang M., Zhang L., Sun T., Liu Y., Lukowitz W., Xu J., Zhang S.(2017). Regulation of stomatal immunity by interdependent functions of a pathogen-responsive MPK3/MPK6 cascade and abscisic acid. Plant Cell 29: 526–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F., Mizoguchi T., Yoshida R., Ichimura K., Shinozaki K.(2011). Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Mol. Cell 41: 649–660. [DOI] [PubMed] [Google Scholar]
- Takahashi F., Yoshida R., Ichimura K., Mizoguchi T., Seo S., Yonezawa M., Maruyama K., Yamaguchi-Shinozaki K., Shinozaki K.(2007). The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19: 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J.(2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Takahashi F., Anderson J.C., Ishihama Y., Peck S.C., Shinozaki K.(2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6: rs8. [DOI] [PubMed] [Google Scholar]
- Usami S., Banno H., Ito Y., Nishihama R., Machida Y.(1995). Cutting activates a 46-kilodalton protein kinase in plants. Proc. Natl. Acad. Sci. USA 92: 8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadassery J., Reichelt M., Hause B., Gershenzon J., Boland W., Mithöfer A.(2012). CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 159: 1159–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C.(2007). Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100: 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C., Hause B.(2013). Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J.(2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Hettenhausen C., Meldau S., Baldwin I.T.(2007). Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19: 1096–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang S.(2015). Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 20: 56–64. [DOI] [PubMed] [Google Scholar]
- Yoo S.J., Kim S.H., Kim M.J., Ryu C.M., Kim Y.C., Cho B.H., Yang K.Y.(2014). Involvement of the OsMKK4-OsMPK1 cascade and its downstream transcription factor OsWRKY53 in the wounding response in rice. Plant Pathol. J. 30: 168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Nie H., Shen Q., Zhang S., Lukowitz W., Tang D.(2014). EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. PLoS Genet. 10: e1004389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W.(2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]