Abstract
Mitochondria are important organelles that are responsible for cellular energy metabolism, cellular redox/calcium homeostasis, and cell death regulation in mammalian cells. Mitochondrial dysfunction is involved in various diseases, such as neurodegenerative diseases, cardiovascular diseases, immune disorders, and cancer. Defective mitochondria and metabolism remodeling are common characteristics in cancer cells. Several factors, such as mitochondrial DNA copy number changes, mitochondrial DNA mutations, mitochondrial enzyme defects, and mitochondrial dynamic changes, may contribute to mitochondrial dysfunction in cancer cells. Some lines of evidence have shown that mitochondrial dysfunction may promote cancer progression. Here, several mitochondrial stress responses, including the mitochondrial unfolded protein response and the integrated stress response, and several mitochondrion-derived molecules (reactive oxygen species, calcium, oncometabolites, and others) are reviewed; these pathways and molecules are considered to act as retrograde signaling regulators in the development and progression of cancer. Targeting these components of the mitochondrial stress response may be an important strategy for cancer treatment.
Impact statement
Dysregulated mitochondria often occurred in cancers. Mitochondrial dysfunction might contribute to cancer progression. We reviewed several mitochondrial stresses in cancers. Mitochondrial stress responses might contribute to cancer progression. Several mitochondrion-derived molecules (ROS, Ca2+, oncometabolites, exported mtDNA, mitochondrial double-stranded RNA, humanin, and MOTS-c), integrated stress response, and mitochondrial unfolded protein response act as retrograde signaling pathways and might be critical in the development and progression of cancer. Targeting these mitochondrial stress responses may be an important strategy for cancer treatment.
Keywords: Mitochondria, cancer progression, retrograde signaling, mitochondrial stress response, integrated stress response, unfolded protein response
Introduction
The earliest report of the existence of mitochondria traces back to the 1840s.1In the 1890s, the term mitochondria was introduced by combining Greek terms mitos (thread) and chondros (granule), according to their special morphology during spermatogenesis.1Mitochondria are the major energy-producing organelles in eukaryotic cells, and they are responsible for converting nutrients into usable energy sources, such as adenosine triphosphate (ATP), through oxidative phosphorylation (OXPHOS) in conjunction with the citric acid cycle.2In addition to glucose metabolism, mitochondria carry out fatty acid β-oxidation and amino acid metabolism.3,4Mitochondria also play critical roles in numerous physiological processes such as programmed cell death, innate immunity, autophagy, redox homeostasis, and calcium homeostasis.5–7In addition, mitochondria are essential regulators of stem cell activation and fate decisions.8Reactive oxygen species (ROS) are byproducts of OXPHOS and are linked to several diseases, such as aging, neurodegenerative disease, diabetes, and cancer.9,10Mitochondria have their own genome mitochondrial DNA (mtDNA), that is located in the mitochondrial matrix. The number of mtDNA copies in each mitochondrion usually varies.11Human mtDNA is a double-stranded, circular DNA molecule, approximately 16.6 kb in size, that contains the genes of 2 rRNAs, 22 tRNAs, and 13 subunits of the respiratory enzyme complex for the OXPHOS system.11The biogenesis of mitochondria requires tight coordination between the genomes of the mitochondria and nucleus.12
Deregulation of cellular energetics is proposed as one of cancer hallmarks.13In the 1920s, Otto Warburg14proposed that cancer cells utilize glycolysis instead of mitochondrial OXPHOS for glucose metabolism even in aerobic condition. Normal cells can metabolize glucose through mitochondria under oxygen abundant circumstance, but in the absence of oxygen, glucose will be converted into lactate by glycolysis. Although cancer microenvironment is nutrient and oxygen limited, cancer cells highly utilize glucose in the presence of oxygen and elevate lactate production. This was thus suggested by Dr. Warburg that there were defects in OXPHOS or mitochondrial respiration in cancer, and which forced the cells to revert to glycolysis.14This metabolic characteristic provides the base for clinical use of 18F-fluorodeoxyglucose positron emission tomography (PET) in cancer diagnosis. The Warburg effect has led to identification of the cause factor for mitochondrial dysfunction in cancer, though recent studies showed that cancer cells have intact mitochondrial metabolism.15,16
Tight coordination and communication between mitochondrial and nuclear genomes are essential for the maintenance of mitochondrial function. The nuclear genome can regulate mitochondrial activity depending on cellular needs for proliferation through anterograde regulation. Conversely, mitochondria can further regulate the expression of nuclear genes to modify cellular function and cell metabolism remodeling via retrograde signaling. The mitochondria-to-nucleus signaling pathway was first identified in yeast. The Rtg family proteins are the major regulators of retrograde signaling17–19and are essential for maintaining yeast survival under OXPHOS-deficient circumstances.18,20However, the mammalian orthologs of Rtg proteins have not yet been identified.21In mammalian cells, mitochondrial retrograde signaling was first proposed in skeletal myoblast cells and later confirmed in human lung cancer cells.22,23Several types of mitochondrial retrograde signaling have been identified and largely investigated in cancer cells.24,25Mitochondrial dysfunction can be caused by various mitochondrial stresses, such as mtDNA mutations, mitochondrial enzyme defects, mitochondrial dynamic changes, and mitochondrial unfolded protein accumulation. The mitochondrial stress responses not only reprogram cellular energetic metabolism but also induce signaling for cancer progression by releasing ROS, Ca2+, some metabolites/proteins, or mitochondrion-derived molecules from mitochondria. In this minireview, we discuss what factors may contribute to mitochondrial dysfunction in cancers, how mitochondrial dysfunction promotes cancer progression, and the role of the mitochondrial stress response in cancer progression.
The factors contribute to mitochondrial dysfunction in cancers
Defective mitochondria and increased aerobic glycolysis are frequently observed in cancer cells compared to normal cells. The mechanistic understanding of what factors contribute to mitochondrial dysfunction and how they further regulate cell growth and carcinogenesis is expanding beyond the Warburg effect as an area of research that is underexplored in terms of its significance for clinical application in cancer prevention and treatment.
MtDNA copy number changes and mutations in cancers
In human cancers, several mtDNA alterations have been identified, such as mtDNA copy number changes, point mutations, insertions, and large-scale deletions.26MtDNA mutations may also provide a powerful molecular diagnostic marker for noninvasive detection of cancer because mutated mtDNA can be detected in number of cancers.27
MtDNA copy number alterations can change mitochondrial function.28Great variations in mtDNA copy number are detected across various cancers. An increase or decrease in mtDNA copy number may be tissue specific in different types of cancers. In glioma, endometrial adenocarcinoma, lymphoma, esophageal squamous cell carcinoma, and colorectal cancer, the mtDNA copy number is increased.29–33On the other hand, the mtDNA copy number is decreased in most hepatocellular carcinomas (HCC, 60%), gastric cancers (55%), and breast cancers (63%).34–38
The majority of somatic mutations in mtDNA are located in the D-loop region (51%), followed by the protein coding region (40%), rRNA genes (5%), and tRNA genes (4%).26,39The D-loop is thus thought to be a “hot spot” for the mutation of mtDNA in tumors.27Mutations in the D-loop region may induce mitochondrial dysfunction and subsequently elevate ROS production, which may contribute to cancer initiation.40,41The D-loop region is responsible for the replication and expression of the mitochondrial genome. Somatic mutations in the mtDNA D‐loop coincide with decreased mtDNA copy number in several human cancers.34–36,38
MtDNA mutations in the protein coding region have high potential to induce mitochondrial dysfunction in cancer.39Some of these somatic mtDNA mutations are pathogenic in patients with mitochondrial disorders.42,43Moreover, several somatic mtDNA mutations may result in missense, nonsense, or frame-shift mutations, which potentially lead to mitochondrial dysfunction.42–44These findings support that somatic mutations in the protein coding region of mtDNA can lead to mitochondria defects during tumorigenesis.
Most of the somatic mtDNA mutations are homoplasmic, indicating cancer cells harboring mutated mtDNA become dominant in tumor. The homoplasmic mtDNA mutations in cancer cells might be through selection process during cancer development.45,46Pathogenic mtDNA mutations give an advantage in tumor growth and overcome wild-type mtDNA in the promotion of tumors.
Large-scale deletions of mtDNA, especially the 4977-bp common deletion that result in the loss of 5 tRNA genes and 7 protein-coding genes, have been detected in various cancers.34,35,47–52The mtDNA 4977-bp deletion is considered a pathogenic mutation in human cells. It can lead to completely impaired energy production and subsequently induce mitochondrial dysfunction.53A correlation was found between the 4977-bp deletion and betel quid chewing history in oral cancer patients, which suggests that the accumulation of this deletion may play an important role during the early phase of oral carcinogenesis.54Moreover, NADPH quinone oxidoreductase 1 (NQO1) deficiency-mediated ROS elevation may contribute to mtDNA 4977 deletion in breast cancer patients.55However, lower levels of the mtDNA 4977 deletion in tumors were noted than in nontumor tissue in different kinds of cancers, such as gastric cancer and colorectal cancer.34,56The low accumulation of mtDNA 4977 deletion in the cancerous area might be the consequence of a dilution effect after cancer progression or a selection process that eliminates cancer cells harboring the mtDNA deletion.
In cancers, most somatic point mutations in mtDNA are homoplastic. Large-scale mtDNA deletions accumulate less readily in tumor tissue than in nontumor tissue. The mtDNA copy number decrease alone might not affect the homoplasmic/heteroplasmic level of the point mutation or the accumulation level of large-scale deletions in the mtDNA of cancer cells.57These results suggest that mitochondrial genome instability and reduced mtDNA copy number may be independent of each other in human cancer.
The mitochondrial genome is highly susceptible to oxidative damage and mutation because ROS are byproducts of OXPHOS and mtDNA lacks efficient DNA repair systems.58Mutated mtDNA-mediated mitochondrial dysfunction can increase ROS production.59ROS might be a causing factor for mtDNA mutation. Moreover, ROS may stimulate several signaling pathways to maintain homeostasis. Therefore, ROS play a dual role as an inducer as well as a protector via apoptosis signals against cancer depending upon the development stage of cancer.60,61
In the past decades, the mitochondrial alterations have been studied in detail with the advances of biotechnology. We realize that the cancer cells do not exhibit universal pattern such as mtDNA copy number alterations. This may be due to tumor heterogeneity arising from the heritable causes or the origin of tumor such as regional differences in the tumor (e.g. various structures of blood and lymphatic, different types and amounts of infiltrated normal cells, and different extracellular matrix composition).62
Mitochondrial enzyme defects and mitochondrial dynamic changes in cancer
Mitochondrial enzyme defects and mitochondrial dynamic changes can lead to mitochondrial dysfunction. Although mitochondria contain mtDNA, most proteins are encoded by nuclear DNA.63Mitochondrial stress might also be induced by defects in nuclear-encoded mitochondrial enzymes, such as citric acid cycle enzymes or other mitochondrial proteins, such as sirtuin 3 (SIRT3), which is responsible for the deacetylation of mitochondrial proteins.
Mitochondrial enzyme defects
Mutations in fumarate hydratase (FH), succinate dehydrogenase (SDH), and isocitrate dehydrogenase (IDH), which are nuclear genome-coded mitochondrial enzymes, have been found in cancers.64–66The FH germline mutation might contribute to increased cancer risk in renal cell carcinoma and uterine leiomyosarcoma.67SDH complex subunit A (SDH-A) germline mutations might be a driver of tumorigenesis in neuroblastoma.68Moreover, IDH mutations might promote the development of a number of malignancies, such as glioma, myeloid neoplasia, chondrosarcoma, and cholangiocarcinoma.69,70
SIRT3 (mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase, sirtuin-3) is responsible for protein deacetylation and regulates mitochondrial activity and energy metabolism.71SIRT3 regulates the redox status, stress response, and aging. SIRT3 defects are associated with different cancers, such as oral cancer, breast cancer, and HCC.72–74SIRT3 may act as a tumor suppressor in gastric cancer.75Moreover, SIRT3-mediated deregulation was found to decrease the expression of the mitochondrial DNA repair gene (8-oxoguanine DNA glycosylase, OGG1-2a) and to increase proliferation activity, which may be important factors in the development of head and neck squamous cell carcinoma.76These results suggest that mitochondrial enzyme defects can contribute to tumorigenesis.
Mitochondrial dynamic changes
In mammalian cells, mitochondria are highly dynamic organelles that have tightly coordinated cycles for fission and fusion, which are processes involved in “mitochondrial dynamics.” Changes in mitochondrial dynamics can regulate the shape, distribution, size and function of mitochondria. Mitochondrial dynamics play an important role in many cellular homeostasis, such as cell cycle, immunity, apoptosis, and mitochondrial quality control.77
Mitochondrial dynamics is responsible for the altered extracellular nutrient level.78Moreover, cancer metabolism has its flexibility to the surrounding nutrient availability.79In addition, cancer cells acquire different metabolic remodeling corresponding to their malignant stages, such as rapid proliferating cancer cells have a high glycolytic activity, while metastatic cancer cells have high OXPHOS activity.80–82In general, the fragmented mitochondria (fission state) have less active OXPHOS compared to the tubular mitochondria (fusion state). It can provide glycolytic intermediates as the building blocks for cancer cell proliferation. However, the relationships between mitochondrial dynamics and cellular metabolism are veiled due to complex mechanisms and factors involved such as the cellular environment, cell type, and differences between metabolic cues.78
The fission morphology of mitochondria is often observed in tumor cells, which may be related to tumorigenesis.83While the underlying mechanisms that regulate mitochondrial dynamics in cancer remain unclear, some hyperactivated oncogenic signals, such as Ras, Raf, MYC, CDKN2A and p53, can remodel mitochondrial shape and metabolism during tumorigenesis.84Primary fibroblasts display fused mitochondria and rely on OXPHOS, and B-RAFV600Emutation-driven melanoma cells contain fragmented mitochondria.85Moreover, changes in mitochondrial dynamics/fission status, such as decreased OPA1 expression in HCC, downregulated mitofusin-2 (Mfn2) in human gastric tumors, and upregulated dynamin-related protein 1 (DRP1) in various cancers, have been found in many cancers.86–88These results suggest that mitochondrial dynamic changes may play a potential role during tumorigenesis.
Mitochondrial dysfunction contributes to cancer malignant progression
Different types of mitochondrial dysfunction may contribute to metabolic switch and malignant processes involved in cancer progression, including tumorigenesis, metastasis, and chemoresistance.39The relationship between mitochondrial dysfunction or tumor environment and metabolic switch is complicated. In addition to mitochondrial dysfunction, acquired mutations can remodel the cancer metabolism, and tumor microenvironment is another factor that regulates cancer metabolism and provides metabolic heterogeneity.89Solid tumors encompass highly disorganized normal tissues and numbers of cell types including endothelial cells for blood vessels, stromal fibroblasts, immune cells, and cancer cells. Stromal fibroblasts can recruit immune cells and further affect the development of vascular system.90Leaky vessels inefficiently transport nutrients and eliminate cellular metabolism wastes, such as lactate.91,92In addition to insufficient nutrients and waste accumulation, hypoxia exists in vasculature uncovered by limited oxygen supply. The hypoxia response leads to metabolism remodeling though enhanced glycolysis and additional lactate deposition.93The metabolism remolding of cancer cells by microenvironments and acquired mutations confers a selective advantage for survival and proliferation in the vile tumor microenvironment.94
MtDNA copy number changes
Compared to high mtDNA copy number, low mtDNA copy number was found to be associated with poor prognosis in HCC patients.95Moreover, a reduced mtDNA copy number was observed in malignant gastric cancer phenotypes, such as ulcerated, infiltrating, and diffuse types (Bormann’s type III-IV).34Reduced mtDNA copy number was also correlated with older onset age, higher histological grade, and poorer disease-free survival and overall survival rates in breast cancer patients.96MtDNA-depleted (long term-ethidium bromide EtBr, an mtDNA replication inhibitor treated and adapted) cancer cells were found to be linked to invasiveness and metastasis through induction of the expression of epithelial-to-mesenchymal transition (EMT) proteins and stemness markers.97The mtDNA-depleted prostate cancer cells exhibit cancer stem cell features such as CD44 and ABCG2.98,99A reduced mtDNA copy number (such as mutant mitochondrial polymerase γ- or EtBr-mediated) was found to induce an invasive phenotype.100,101
However, whether decreased mtDNA copy number contributes to cancer progression is still controversial in some cancers, such as head and neck cancer and esophageal squamous cell carcinoma.102,103The alterations of mtDNA copy number required for cancer initiation and progression are tissue specific and complicated. Two ρ0murine cancer cells (B16 melanoma and 4T1 breast carcinoma) formed tumors in vivomore slowly than mtDNA-sufficient (ρ+) parental cells. Moreover, the mtDNA copy number could be recovered in derivatives of the originally ρ0cells at different stages of malignant progression, such as primary cells at the tumor injection site, circulating tumor cells, and lung metastasis cancer cells.104These results suggest that mtDNA-deficient cancer cells can recover mtDNA from host cells, restoring their OXPHOS activities to a level that is sufficient for tumor initiation and progression.
Mitochondrial transcription factor A (TFAM) plays an important role in regulating mtDNA copy number. TFAM is an important mediator of mitochondrial damage-associated molecular patterns and can further regulate inflammation and immunity.105TFAM is an important regulator of mtDNA replication. Recently, it was found that a TFAM-mediated increase in mtDNA copy number is important to promote cancer progression by enhancing OXPHOS in microsatellite-stable colorectal cancer.106In addition, the TFAM-mtDNA-calcium-cilia and flagella-associated protein 65 (CFAP65)-cytoplasmic phosphoenolpyruvate carboxykinase (PCK1) axis, which is connected to mitochondrial retrograde signaling, affects cancer cell differentiation and proliferation and contributes to cancer progression.107Therefore, whether increased or decreased mtDNA copy number contributes to cancer progression is still controversial and may also have tissue-specific trend corresponding to different types of cancers.
Some lines of evidence show that mtDNA-depleted cells (long term-EtBr treated) are resistant to chemotherapeutic agents such as doxorubicin, cisplatin, and etoposide.108–110Some underlying mechanisms linking mtDNA copy number alterations in cancer progression have been proposed, such as an increase of manganese superoxide dismutase (MnSOD) and elimination of the effects of chemotherapeutic agents by developing P-glycoproprotein-mediated multidrug resistance (MDR) phenotype or activation of mitochondria-to-nucleus retrograde signaling to increase the expression of antiapoptotic genes (including B cell lymphoma-2 (Bcl-2) and pro-survival enzymes such as Akt).111–115Moreover, mtDNA copy number alterations (EtBr-treated) may contribute to endocrine therapy resistance in prostate and breast cancer cells.116,117MtDNA-depleted prostate cancer cells and breast cancer cells lose their hormone dependence and exhibit tamoxifen and fulvestrant resistance. However, it was reported that a high copy number of mtDNA could be a potential biomarker for predicting unfavorable efficacy of anthracycline treatments in breast cancer patients.118The exact role of mtDNA copy number alterations in cancer treatment resistance needs further investigation.110,117,119,120
MtDNA mutations
The incidence of somatic mtDNA D-loop mutations is high in advanced staged cancers such as HCC, gastric, lung and colorectal cancers.38In breast cancer patients, the incidence of mtDNA D-loop mutations is associated with old age and lack of hormone receptor (such as estrogen receptor and progesterone receptor) expression. Moreover, mtDNA D-loop mutations are significantly related to poor prognosis in breast cancer patients.35Furthermore, several somatic mtDNA mutations in the coding region were identified in breast cancers. The occurrence of these somatic mtDNA mutations is also associated with old age, late stage, and malignant histological grade.44These findings suggest that mtDNA somatic mutations may be the biomarkers for breast cancer prognosis.
In chronic lymphocytic leukemia, patients which refractory to conventional therapeutic agents have higher rate of cancer mtDNA mutations than good responder patients.121It was also reported that mutant mtDNA (such as mtDNA ATP synthase subunit 6 gene pathogenic point mutation) cybrids confer cisplatin resistance via resistant to apoptosis.122Reduced ATP synthase activity contributes to 5-flurouracil resistance in colon cancer cells.123Reduced mitochondrial Complex I (NADH dehydrogenase) activity was also found to significantly regulate the aggressiveness of human breast cancer cells via NAD+/NADH redox balance, mTORC1 activity, and autophagy.124Moreover, normal mitochondrial transplantation was found to decrease cell growth, ROS levels, and chemoresistance in breast cancer cells. In addition, replacement of normal mitochondria with mtDNA A8344G-mutated dysfunctional mitochondria abolished the original suppression of cancer cell growth via distinct metabolic remodeling such as switches to the energetic and glycolytic phenotypes.125These findings suggest that mtDNA mutation-induced mitochondrial dysfunction or decreased mitochondrial activity may contribute to the malignant progression of various cancers. Interestingly, it was reported that patients with common pathogenic mtDNA mutations and mitochondrial dysfunction do not appear to be at increased risk of cancer compared with the general population.126However, this might not rule out that these mtDNA mutations contribute to a vicious cycle of further malignant transformation.127
Mitochondrial enzyme defects
FH-deficient renal cancers are often highly aggressive and frequently metastasize even when the tumors are small, resulting in poor clinical prognosis.128,129FH deficiency contributes to cancer progression through enhanced invasion and migration in clear cell renal cancers or induced EMT in kidney cancer cells.130–132In clear cell renal cancers, low SDH subunit B (SDH-B)-expressing patients have a poorer prognosis than high-expressing patients.133Moreover, SDH mutations contribute to cancer progression by promoting EMT cell migration, invasion, and angiogenesis.134Low IDH1 expression in breast cancer is significantly correlated with late stage, lymph node metastasis, and poor prognosis.135Accumulation of 2-hydroxyglutarate (2-HG) may contribute to poor prognosis and treatment response in acute myeloid leukemia (AML).136Glioma-derived IDH2 mutations may contribute to chemoresistance through HIF-1α and β-catenin signaling.137Furthermore, low expression of IDH could be observed in doxorubicin-resistant breast cancer cells.138However, it was reported that the chemotherapy response is conversely correlated with FH deficiency in gastric cancers.139The role of mutations in the tricarboxylic acid cycle (TCA) cycle enzymes in cancer therapy resistance has not been fully investigated.
In gastric cancer, low SIRT3 expression was found to be associated with poor prognosis.75Moreover, low SIRT3 expression may contribute to poor prognosis in pancreatic cancers.140p53 and p21 may be mediators of the SIRT3-mediated mitochondrial stress response in lung adenocarcinoma or oral carcinoma cells.141,142In addition to cancer progression, the loss of SIRT3, which leads to the acetylation of MnSOD and other mitochondrial proteins, has a connection with ROS and the development of luminal B breast cancer and may contribute to endocrine therapy resistance.143
Mitochondrial dynamic changes
The inhibition of mitochondrial fragmentation by DRP1 knockdown can increase genomic instability and decrease migration and invasion by cellular stress in breast cancer cells.144,145Impaired mitochondrial fission can cause mtDNA mutation-mediated mitochondrial dysfunction and deregulation of redox homeostasis. Inhibition of mitochondrial fission can be a potential modality for enhancing cancer cell apoptosis and increasing sensitivity to cancer therapy.145,146Some oncogenes are responsible for fragmented mitochondria in cancer cells through the RAS-RAF-MEK-ERK (MAPK) pathway. DRP1 can be phosphorylated by extracellular-signal-regulated kinase 2 (ERK2) on Ser616 and is required for mitochondrial fission and tumor growth in RAS-transformed tumors. This result indicates that MAPK activation and consequent mitochondrial fragmentation are needed in tumors expressing oncogenic RAS.87On the other hand, tissue from tumor metastasis to lymph nodes was found to highly express DRP1 compared to the original tumor or normal/adjacent tissue.144Moreover, hormones such as androgen and estradiol have a strong influence on mitochondrial dynamics. These findings suggest that targeting mitochondrial dynamics for cancer progression in hormone-related malignancies may be a newly effective treatment strategy.147,148
Mitophagy, which is a specific type of autophagy for damaged, dysfunctional or unhealthy mitochondria, can maintain mitochondrial dynamics by the lysosome-mediated pathway. Several canonical mitophagy pathways have been proposed, such as PTEN-induced putative kinase 1 (PINK1)/Parkin, bcl-2/adenovirus E1B protein-interacting protein 3 (BNIP3)/NIX, and FUN14 domain-containing 1 (FUNDC1).149Several lines of evidence have shown that inhibition of mitophagy could contribute to increased efficacy of chemotherapy.150,151Moreover, chemoresistance in some cancer cells may be induced by increased mitophagy.152,153However, the mitochondrial fusion status may contribute to resistance to cisplatin therapy.154The role of mitophagy in cancer progression is controversial.155,156The mitochondrial stress response that occurs in response to changes in mitochondrial dynamics is very complicated, and the detailed regulatory mechanism for cancer progression and/or therapy resistance remains to be further investigated.
Mitochondrion-derived molecules are involved in mitochondrial retrograde signaling pathway for cancer progression
Mitochondrial dysfunction can produce various retrograde signals. Through these signals, cells can regulate cellular homeostasis and protect cells against environmental stresses by retrograde regulation of the expression of nuclear genes.157The nature of retrograde signals can vary depending on their trigger. ROS, Ca2+, and oncometabolites are common mitochondrion-derived molecules.158Other mitochondrion-derived molecules, such as exported mtDNA, exported mitochondrial double-stranded RNA (mt-dsRNA), humanin and MOTS-c, are also proposed to be involved in the retrograde signaling pathway.159–162
ROS and Ca2+
ROS are common byproducts of OXPHOS that are often elevated due to a defective electron transport chain; ROS directly affect redox homeostasis and act as signaling molecules in a number of cellular processes under normal or stress environments.163In mammalian cells, increased ROS activate retrograde signaling to activate detoxification enzymes or increase antioxidant ability by nuclear factor erythroid 2-related factor 2 (NRF2).164In addition, ROS can compensate for increased mitochondrial biogenesis by activating the JNK–PGC1α pathway and promoting mitochondrial Complex II phosphorylation.165,166In cancer cells, mitochondrial ROS contribute to promoting cell growth and survival via the nuclear factor-κB (NF-κB) pathway.167On the other hand, mitochondria are important organelles responsible for calcium storage and homeostasis.168Mitochondrial stressors such as mtDNA mutation, OXPHOS disruption, and mitochondrial membrane potential uncoupling can trigger Ca2+release from mitochondria. Free cytosolic Ca2+can activate the NF-κB, Jun N-terminal kinase (JNK), and p38 MAPK pathways. Moreover, Ca2+can increase the expression of various transcription factors, such as CREB, early growth response protein 1 (EGR1), ATF2, CCAAT/enhancer-binding protein-δ, and CHOP.169,170Calcium retrograde signaling not only contributes to mitochondrial adaptation but is also involved in calcium homeostasis, insulin regulation, glucose metabolism remodeling, and cell proliferation.158
Mitochondrial stress induced by mitochondrial inhibitor (such as oligomycin)-decreased mitochondrial activity enhances the migration of gastric cancer cells via ROS-mediated retrograde signaling.42Mitochondrial inhibitors (such as oligomycin and antimycin A)-induced ROS-β5-integrin retrograde signaling plays an important role in promoting cell migration.171In addition to metastasis, ROS are involved in mitochondrial stress (by mitochondrial inhibitors)-induced chemoresistance in gastric cancer cells.172Moreover, ROS- and calcium-mediated expression of amphiregulin (AR) is important for mitochondrial stress (by mitochondrial inhibitors-decreased mitochondrial activity or interfering mtDNA transcription and translation)-induced chemoresistance and migration in HCC cancer cells.173ROS are also involved in defective SIRT3-mediated cancer progression.141,142Furthermore, mitochondrion-derived ROS mediate the regulation of vascular endothelial growth factor (VEGF), which may be one of the possible mechanisms of tumorigenesis and metastasis regulated by MAPK-mediated mitochondrial fission. These results indicate that mitochondrial dysfunction-mediated ROS and Ca2+changes contribute to cancer progression.
Oncometabolites
The mutations of FH and SDH, two nuclear genome-encoded enzymes of the citric acid cycle, may lead to the accumulation of fumarate and succinate. These two metabolites have been shown to lead to malignant transformation and tumorigenesis.174–1762-HG accumulates in response to defects in NADP-dependent IDH 1 (cytosolic) and 2 (mitochondrial) in different cancers.69,177Moreover, mutations of IDH in cancer strongly implicate metabolism remodeling during tumorigenesis.178Fumarate, succinate, and 2-HG are thus thought to be oncometabolites. Accumulation and subsequent release of fumarate and succinate from mitochondria might lead to HIF1 stabilization and α-ketoglutarate-dependent dioxygenase inhibition-mediated DNA and histone modifications, which promote cancer progression by EMT, angiogenesis, and cellular glucose or energy metabolism remodeling.179–1812-HG accumulation and release from mitochondria contribute to the malignant phenotype by affecting DNA demethylation and promoting epigenetic changes. Furthermore, 2-HG might inhibit the activity of Complex IV/V and subsequently induce the mitochondrial stress response, which is responsible for deregulating cellular energetics.182,183These results indicate that oncometabolites such as fumarate, succinate, and 2-HG are involved in retrograde signaling for cancer progression.
MtDNA and mitochondrial double-stranded RNA
The inflammasome plays an important role in a myriad of acute/chronic inflammatory and degenerative diseases.184The inflammasome is composed of a set of intracellular protein complexes that enable autocatalytic activation of inflammatory caspases and drive some cytokine secretion. Cytosolic oxidized mtDNA was identified to activate the NLRP3 inflammasome complex.162The NLRP3 inflammasome is unique and can be triggered by a number of stresses.184Persistently aberrant NLRP3 signaling contributes to several immune disorders and degenerative diseases, such as autoimmune disorders, gout, osteoarthritis, Alzheimer’s disease, type 2 diabetes, atherosclerosis, lupus, macular degeneration, and cancer.185,186Circulating mtDNA inhibited the production of proinflammatory cytokines, horizontal transfer of mtDNA from tumor cells to surrounding immune cell-activated apoptosis in immune cells, and inappropriate sensing of mtDNA leading to dysfunction of the host immune system, consequently contributing to cancer progression.187Mitochondrial double-stranded RNA exported from mitochondria was recently demonstrated to engage an MDA5-driven antiviral signaling pathway that triggers a type I interferon response with antiviral effects.161Synthetic dsRNA may potentially be a new immunotherapy for cancer treatment.188However, the exact role of mitochondrial double-stranded RNA in cancer progression is still unclear.
Humanin and MOTS-c
Some mitochondrial-derived peptides are encoded from the mitochondrial genome. Humanin is the first reported and better characterized mitochondrial-derived peptide that provides protective effects against various stresses.189Humanin is a 24-amino acid short peptide originally isolated from a cDNA library that was screened for survival factors in a study about Alzheimer’s disease (AD).190It is transcribed by a part of the mitochondrial MT-RNR2 gene, which encodes 16S mitochondrial ribosomal RNA (16S rRNA). Humanin expression can be triggered by mitochondrial stressors such as serum deprivation and chemotherapy or inhibited by steroid hormones such as estrogen. In cancer, humanin was initially proposed to be a potential oncopeptide.191ERK1/2 and STAT3 may be humanin downstream targets in the development of several types of tumors, such as glioblastoma, triple-negative breast cancer, and pituitary tumors.189Moreover, evidence has shown that humanin contributes to chemoresistance and cancer aggressiveness.192,193
Mitochondrial open reading frame of the 12S rRNA type-c (MOTS-c), which is a 16-amino acid peptide encoded by the mitochondrial 12S rRNA gene, is another mitochondrial-derived peptide. It was originally found in the in silicosearch for potential short open reading frames (sORFs) within the human 12S rRNA and was then identified to have a biological function in metabolic homeostasis.159,194The physiological function of MOTS-c can be exhibited by the relief of metabolic syndromes such as obesity, insulin resistance, and Q fever/chronic fatigue syndrome.159,195,196Initially, MOTS-c was demonstrated to target the one-carbon pool and de novopurine synthesis pathways to increase 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) levels and activate AMP-activated protein kinase (AMPK).159It can stimulate glucose utilization and lactate production coupled with reduced mitochondrial oxygen consumption as well as increased fatty acid utilization, which suggests that MOTS-c can maintain metabolic homeostasis for the regulation of obesity, diabetes, exercise, and longevity.159Increased intracellular ROS in response to metabolic stress may mediate the translocation of MOTS-c to the nucleus via AMPK-dependent regulation.197Moreover, MOTS-c is responsive to retrograde signaling via interaction with multiple stress-response transcription factors, including nuclear factor erythroid 2-related factor 2 (NFE2L2/NRF2) and activating transcription factor 1 and 7 (ATF1/ATF7). Metabolic dysfunction and remodeling are characteristics of cancer cells. Mitochondrial dysfunction can down-regulate HIF-1α via the activated AMPK pathway in HCCs.198Hence, this may provide a potential link between MOTS-c and cancer progression. There is still much to be unveiled about metabolic rewiring via these mitochondrion-derived molecules in tumor formation and malignant progression.
Mitochondrial unfolded protein response in cancer progression
The mitochondrial proteome contains more than a thousand mitochondrial proteins that are encoded by nuclear and mitochondrial genomes. Mitochondrial biogenesis and function are dependent on the maintenance of protein import pathways and the protein-folding circumstances. Deregulating mitochondrial proteostasis can induce mitochondrial stress and negatively affect mitochondrial function. Mitochondrial stress in response to accumulation of misfolded mitochondrial proteins in the mitochondrial matrix, impairment of the protein quality control system, mitonuclear imbalance or inhibition of the electron transport chain (ETC) can induce the mitochondrial unfolded protein response (UPRmt).199Cells usually use impaired protein as a sensor for mitochondrial dysfunction to activate the specific UPRmt, a mitochondrial stress response. It can activate an adaptive transcriptional program that promotes mitochondrial function recovery, metabolic adaptations, and innate immunity.200
The UPRmtcan activate the transcription of the CCAAT-enhancer-binding protein homologous protein (CHOP) gene, dimerize with CCAAT/enhancer-binding protein β (C/EBP-β), and bind to the promoters of UPRmt-responsive genes. It can further regulate mitochondrial quality control proteins via molecular chaperones and proteases.201UPRmt-induced mitochondrial chaperones heat shock protein 60 (hsp60) and mthsp70, which promote protein folded and prevent aggregated formation are involved in mitochondrial recovery program.200UPRmt-activated AAA proteases such as Lon and ClpXP are responsible for removing damaged mitochondrial proteins.200Moreover, the UPRmtcan promote mitochondrial biogenesis and function via elevation of iron–sulfur cluster and ubiquinone synthesis which is required for OXPHOS complex biogenesis. Furthermore, UPRmtcan promote clearance of defective mitochondria by mitochondrial dynamic, such as DRP1.200These transcriptional outputs of the UPRmtmediate its recovery of damaged mitochondria.
The UPRmthas been extensively investigated in C. elegansmodel system. Digestion of unfolded or unassembled mitochondrial proteins into peptides by the matrix protease ClpP can activate UPRmtthrough efflux of short peptides to cytoplasm by HAF-1 transporter.202Accumulation of digested short peptides induces a transcriptional response by activating transcription factor associated with stress 1 (ATFS-1) in worms.203ATFS-1 contains mitochondrial and nuclear targeting signals. ATFS-1 is normally imported into mitochondria and degraded by the Lon protease. Under mitochondrial stress, the import of ATFS-1 to mitochondria is attenuated. ATFS-1 will be translocated to the nucleus along with two other factors, DVE-1 and ubiquitin-like 5 (UBL-5) to regulate the gene expressions of mitochondrial chaperones (such as hsp-6, hsp-60, and DNaJ domain 10 (dnj-10)) and proteases (such as ymel-1) for mitochondrial quality control and restoring proteostasis. Moreover, it can positively regulate the DRP-1, glycolytic genes such as gpd-2, detoxification genes such as skn-1, and translocase of the inner membrane 23 (TIM23). Furthermore, it can negatively regulate the expression of other nuclear mitochondrial genes, such as TCA cycle enzymes and ETC subunits.158,204,205In addition to regulating genes involved in mitochondrial proteostasis, the UPRmtalso mediates homeostasis through metabolic remodeling from mitochondrial OXPHOS to cytoplasmic glycolysis.
In mammalian cells, UPRmtwas initially introduced by the overexpression of the mitochondrial matrix-localized misfolded mutant ornithine transcarbamylase.201The transcription factor ATF5 as the mammalian ortholog of ATFS-1 was recently identified.206Both ATF4 and ATF5 have been considered for harboring bZip domain homologous to ATFS-1. In addition, evidence showed that ATF5 have a putative, but relatively weak mitochondrial targeting sequences (MTS). However, multi-omics analysis identifies ATF4 as a key regulator of UPRmt, not through the canonical ATF5 in mammalian cells.207The key regulator of UPRmtin mammalian cells remains controversial.
In addition to the first identified regulation pathway, CHOP-ATF5, a number of mitochondrial chaperones and proteases, such as ClpP, hsp10, and hsp60, are involved in the UPRmt. In addition, several mechanisms have been proposed, such as those involving SIRT7, estrogen receptor α (ERα), and the SIRT3 response pathway.208The UPRmtcontributes to mitoprotective outcomes by regulating antioxidant ability, proteostasis, OXPHOS, mitochondrial biogenesis, mitophagy, and so on. Recently, mitohormesis was introduced as a response to mitochondrial stress in the hormetic zone, which is composed of low-level exposure mitochondrial stress and induces favorable biological responses.209–211Mitohormesis can induce cancer invasion/metastasis and poor clinical outcomes through the UPRmt.211Moreover, UPRmthas been reported to involve the activation of ERα, and the UPR might play an important role in aromatase inhibitor-resistant breast cancer cells.212,213
ATF5 is up-regulated in numbers of cancers and is related to apoptosis resistance.214The synthetic cell-penetrating dominant-negative ATF5 peptide provides the antitumor activity against treatment-resistant cancers by monotherapy or in combination therapy.215This is relevant considering that ATF5-mediated UPRmtmight play an important role in cancer progression. Moreover, targeting to other UPRmtdownstream targets such as LonP1 by obtusilactone A and sesamin compounds from Cinnamomum kotoense, ClpP by genetic or chemical inhibition and hsp60 by genetic ablation are helpful against numbers of cancers.216–218Mitochondrial stress-induced UPRmtis important to cancer progression. In addition, mitochondrial ROS are important for UPRmt-induced mitoprotective pathways. These lines of evidence suggest that UPRmtis one of the mitochondrial stress responses and is involved in retrograde mitonuclear communication for cancer progression.
Integrated stress response is involved in mitochondrial stress response and contributes to cancer progression
Depending on the severity and nature of the stress, the integrated stress response (ISR) modulates various cellular functions to adapt to stress. The core of ISR is the phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) and activation of the activating transcription factor-4 (ATF4) pathway. The eIF2α belongs to the multimeric eIF2 complex and is responsible for cap-dependent protein translation.219Four eIF2α kinases have been identified to be responsible for eIF2α phosphorylation: PKR-like endoplasmic reticulum kinase (PERK), general control nonderepressible 2 (GCN2), protein kinase R, and heme-regulated eIF2α kinase.219–222
Phosphorylated eIF2α inhibits cap-dependent protein translation under stress conditions and allows cells to adapt stress through upregulation of ATF4-translation and subsequent activation of its downstream genes.223The ISR protects against intrinsic/extrinsic cellular stress (such as endoplasmic reticulum stress, hemoglobin deficiency, nutrient deficiency, viral infection, or hypoxia) by regulating transporters, antioxidant systems, chaperones, and so on.223–225The eIF2α-ATF4 pathway not only maintains cellular redox homeostasis but also regulates cellular metabolism and nutrient uptake.226,227On the other hand, it can induce cell death through activation of proapoptotic bc l-2 family proteins or death receptor 5 via the ATF4-CHOP pathway.228–230Nutrient deprivation can also induce cell necrosis through the ATF4-dependent ISR pathway.231ISR encompasses a dual role in cellular homeostasis.232,233
The ISR has been proposed to be involved in mitochondrial-nuclear communication and thus responsible for cellular homeostasis and lifespan.158ATF4 has been identified as a key regulator of the mitochondrial stress response in mammalian cells.207Mitochondrial stress in response to arsenic or doxycycline can decrease mitochondrial function and reprogram gene expression to maintain mitochondrial protein homeostasis through the eIF2α-ATF4 pathway.234–236PERK and GCN2 were recently identified to be involved in the mitochondrial stress response.237–239Moreover, ETC dysfunction, ROS elevation and mitochondrial unfolded protein stress can activate GCN2, PERK, or HRI, depending on clinical situation.158
The ISR is an important way by which tumor cells adapt to environmental stress, and it contributes to tumor growth.240ATF4 expression is higher in tumor tissues compared to normal tissues.240,241Evidence has shown that the eIF2α-ATF4 pathway is critical for tumor cell survival and proliferation in response to nutrient deprivation.242In addition, cancer cells with knockdown of ATF4 formed fewer tumor lesions that were smaller and had slower growth compared with the large burden and rapid growth of control tumor lesions. ATF4 promotes cancer metastasis by induction of heme oxygenase 1-mediated reducing anoikis of cancer cells.243Moreover, ATF4 is involved in c-Myc oncogene-driven malignant progression through uncharged transfer RNA-mediated GCN2 activation.244These results indicate that ISR is critical for the initiation and progression of cancer.
The ATF4 signaling pathway includes several downstream targets, such as apoptotic genes (Bcl-2, NOXA/PUMA, BIM), adaptive genes (such as amino acid transporters, metabolic enzymes, redox balance, endoplasmic reticulum chaperones), and several recycling of cellular material-related genes (such as autophagy genes, REDD1/DDIT4/SESN2, and GADD34).245
SLC7A11 (xCT), a downstream protein in the ATF4 pathway, is involved in the xc−system and is responsible for cysteine uptake and supports cellular glutathione (GSH) synthesis.246Mitochondrial inhibitor-induced mitochondrial stress was found to induce chemoresistance via the ROS-mediated GCN2-ISR-xCT pathway.172Increased antioxidant ability in response to GSH generation is responsible for the ISR-xCT-mediated chemoresistance in gastric cancer cells.247xCT is also important for cystine dependency in triple-negative breast cancer cells.248Moreover, the increased expression of xCT contributes to glucose and glutamine dependency via reduced metabolic inflexibility and imbalance of redox status.249–251Therefore, the ATF4-xCT pathway may play a critical role in the metabolism remodeling and therapy resistance of cancer cells.
On the other hand, cysteine starvation-induced high ROS production and mitochondrial stress were found to induce necroptosis and ferroptosis via the GCN2-ISR-CHAC1 pathway in triple-negative breast cancer cells.252The ISR can result in adaptive or deleterious effects depending on the extent of the mitochondrial changes. Since cancer cells need abundant energy and macromolecular supplies for sustainable cell growth, most cancer cells have tolerable levels of mitochondrial dysfunction and acquire an ability to adapt to cancer microenvironments via the mitochondrial stress response. Therefore, the mitochondrial stress response may be a treatment target for cancer patients.
Until 2011, no reliable biomarker for mitochondrial disorders or mitochondrial dysfunction was identified. The serum fibroblast growth factor 21 (FGF-21) is identified as a useful biomarker for the screening and diagnosis of muscle-manifested mitochondrial disorders.253The diagnostic process of mitochondrial disorders is proposed such as clinical assessment, FGF‐21 level, sequencing of nuclear DNA, and with/without biopsy, which can clinically be used for differential diagnosis of about 70–80% of suspected mitochondrial disorders.254Recently, the evidence showed that ISR is involved in FGF-21 regulation.255,256Although FGF-21 might be used for early diagnosis of liver cancer or renal cancer and might be a biomarker for predicting tumor progression, the understanding the role of FGF-21 in cancer initiation and progression is limited.257–259The gene expression of FGF-21 in tumors part is not increased compared to the normal counterpart in cancers by the gene expression profiling interactive analysis (GEPIA) website (http://gepia.cancer-pku.cn/).
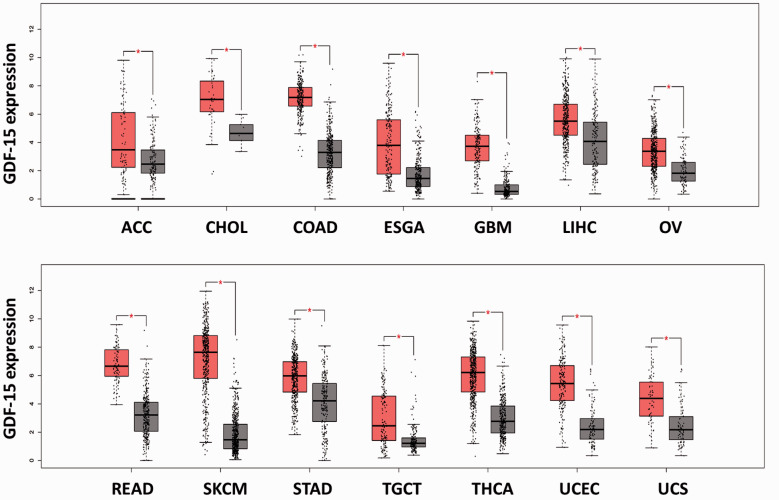
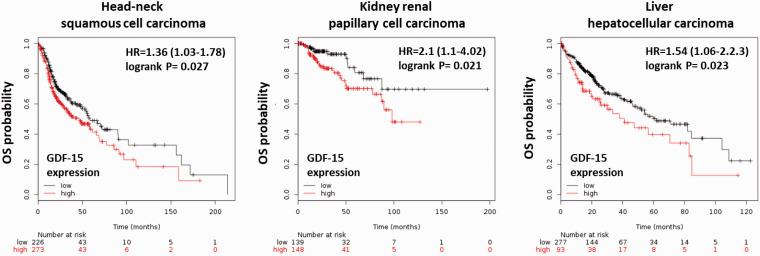
On the other hand, evidence showed that growth differentiation factor 15 (GDF‐15) has greater sensitivity and specificity than FGF‐21 for the diagnosis and/or the monitoring of disease progression of mitochondrial disorders in adults and children.260GDF‐15, a member of the transforming growth factor beta (TGF-β) superfamily, is also regulated by ISR and may be a useful biomarker for mitochondrial dysfunction.255,256,260,261The expression of GDF-15 is often elevated in response to cellular stress such as inflammation, cancer, cardiovascular diseases, obesity, kidney disease, and brain disease.262High gene and protein expression levels of GDF-15 have been identified in several cancers 263(Figure 1), but the findings regarding the function of GDF-15 in cancers are limited and controversial.262High GDF-15 expression is a poor prognostic factor in head-neck, kidney, and liver cancers (Figure 2). In addition, GDF-15 was found to be associated with gastric wall invasion and lymph node metastasis in diffuse-type gastric cancers.264It was also suggested that the GDF-15-activated Akt pathway may contribute to proliferation and migration in cervical and pancreatic cancer cells.265,266Moreover, it was suggested that circulating GDF15 may be a powerful biomarker for bone metastasis in several cancers.267However, GDF15 can inhibit proliferation and bone metastasis in lung adenocarcinoma cancer cells.268Low expression of GDF15 is associated with a poor prognosis in nonsmall-cell lung cancer (NSCLC) patients.269The role of GDF-15 in cancer progression is still unclear and may depend on cell-type specificity.
Figure 1.
The gene expression of GDF-15 in tumors and normal tissues in several cancers. The RNA sequencing expression data were obtained from the TCGA and the GTEx projects. The gene expression of GDF-15 in normal tissues (gray box) and tumor tissues (red box) from several cancers was analyzed by box-plot. The box-plot was generated by the GEPIA website and software (http://gepia.cancer-pku.cn/).270|Log2FC| cutoff: 1; P-value cutoff: 0.01 (ACC: adrenocortical carcinoma, tumor (T) number: 77, normal (N) number: 128; CHOL: cholangiocarcinoma, T number: 36, N number: 9; COAD: colon adenocarcinoma, T number: 275, N number: 349; ESGA: esophageal carcinoma, T number: 182, N number: 286; GBM: glioblastoma multiforme, T number: 163, N number: 207; LIHC: liver hepatocellular carcinoma, T number: 369, N number: 160; OV: ovarian serous cystadenocarcinoma, T number: 426, N number: 88; READ: rectum adenocarcinoma, T number: 92, N number: 318; SKCM: skin cutaneous melanoma, T number: 461, N number: 558; STAD: stomach adenocarcinoma, T number: 408, N number: 211; TGCT: testicular germ cell tumors, T number: 137, N number: 165; THCA: thyroid carcinoma, T number: 512, N number: 337; UCEC: uterine corpus endometrial carcinoma, T number: 174, N number: 91; UCS: uterine carcinosarcoma, T number: 57, N number: 78). (A color version of this figure is available in the online journal.)
Figure 2.
Kaplan–Meier survival analyses for GDF-15 expression on overall survival (OS) in several cancers. The RNA-seq data were collected from several databases, including GEO, EGA, and TCGA. The Kaplan–Meier survival analysis (overall survival) was analyzed by the KM plotter website and software (https://kmplot.com/analysis/).271Kaplan–Meier survival analyses showed that high expression of GDF-15 is a poor prognostic factor in head-neck squamous cell carcinoma, kidney renal papillary cell carcinoma, and liver hepatocellular carcinoma. (A color version of this figure is available in the online journal.)
Conclusion
Numbers of endogenous or exogenous stresses can induce mitochondrial dysfunction. However, not all of mitochondrial dysfunction can produce mitohormesis to adaptations. Mild mitochondrial stress can actually protect cells from detrimental outcomes of subsequent larger stress. Cancer cells with Warburg effects characteristic or mitochondrial dysfunction (in hormesis) might contribute to adaption of tumor microenvironments or supporting for cell proliferation through several stress response pathways.
Herein, we summarize that mitochondrial dysfunction (non-lethal condition)-mediated mitochondrial stresses contribute to tumor malignant phenotype and increased ability of environmental adaption through increasing metastatic/invasion activity and promoting therapy resistance. Mitochondrial dysfunction-mediated stress response might provide the metastatic/invasion activity through EMT, stemness activity remodeling, and increased antioxidant ability for reducing anoikis. Moreover, mitochondrial dysfunction-mediated increasing detoxification enzymes, such as MnSOD, P-glycoprotein-mediated MDR, and xCT-mediated GSH elevation, might contribute to chemoresistance. Furthermore, ISR-activated xCT is important for glucose metabolism remodeling and dependency. This might provide the tumor heterogeneity such as more sensitive to nutrient demand through mitochondrial dysfunction.
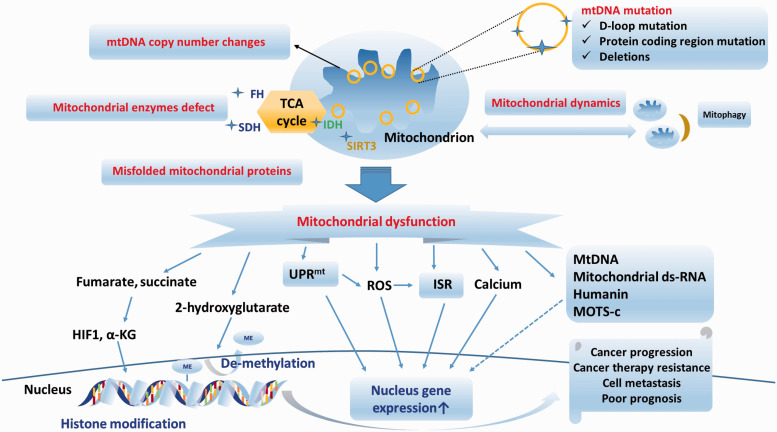
Mitochondria are responsible for cell homeostasis. Several mitochondrial stresses are induced by mtDNA mutation, mitochondrial enzyme defects, mitochondrial dynamic changes, and unfolded mitochondrial proteins in cancers. Mitochondrial stress impairs mitochondrial function and induces cancer progression via various mitochondrial stress responses and retrograde signaling. The UPRmt, the ISR, and mitochondrial-derived molecules have recently been proposed to be involved in the mitochondrial-nuclear signaling pathway. Nondeleterious mitochondrial dysfunction can activate the mitochondrial stress response and play an important role in cancer progression. Several retrograde signaling pathways and mitochondrial stress responses contribute to cancer progression (Figure 3). Therefore, targeting the regulatory pathway of the mitochondrial stress response may be a potential therapeutic strategy for addressing cancer progression or therapy resistance in the future.
Figure 3.
Summary of retrograde signaling pathways and mitochondrial stress responses in mitochondrial stress-induced cancer progression and carcinogenesis. Several mitochondrial alterations, such as mtDNA copy number changes, mtDNA mutations, mitochondrial enzyme defects, and mitochondrial dynamic changes, can induce mitochondrial dysfunction in cancer cells. Several retrograde signaling pathways, such as ROS, calcium, oncometabolites, exported mtDNA/mt-dsRNA, humanin, MOTS-c, UPRmtand ISR, are involved in the mitochondrial stress responses. The retrograde mitochondrial stress response can affect several nuclear gene expressions and plays an important role in cancer progression. (A color version of this figure is available in the online journal.)
Authors’ contributions
SFW reviewed the literature and prepared the first manuscript; SC, LMT, and HCL edited the final manuscript. All authors participated in the writing and discussion of this manuscript.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The present work is supported by study grants (V107A-015) from the Taipei Veterans General Hospital, Taipei, Taiwan; grants from Cheng Hsin General Hospital (CY10707, CY10805); and partly by a grant from the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan; as well as grants MOST 107–2321-B-006–019, MOST 108–2320-B-010–016-MY3 and MOST 108–2314-B-075–052-MY3 from the Ministry of Science and Technology, Taiwan; and the SPROUT Project—Center For Intelligent Drug Systems and Smart Biodevices (IDS2B) of National Chiao Tung University, from the Ministry of Education, Taiwan.
ORCID iD
Hsin-Chen Lee https://orcid.org/0000-0001-7455-9593
References
- 1.Ernster L, Schatz G. Mitochondria: a historical review. J Cell Biol 1981; 91:227s–55s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee HC, Wei YH. Mitochondrial role in life and death of the cell. J Biomed Sci 2000; 7:2–15 [DOI] [PubMed] [Google Scholar]
- 3.Guda P, Guda C, Subramaniam S. Reconstruction of pathways associated with amino acid metabolism in human mitochondria. Genom Proteom Bioinform 2007; 5:166–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bratic I, Trifunovic A. Mitochondrial energy metabolism and ageing. Biochim Biophys Acta 2010; 1797:961–7 [DOI] [PubMed] [Google Scholar]
- 5.Rambold AS, Pearce EL. Mitochondrial dynamics at the interface of immune cell metabolism and function. Trends Immunol 2018; 39:6–18 [DOI] [PubMed] [Google Scholar]
- 6.Nikoletopoulou V, Markaki M, Palikaras K, Tavedallarnarakis N. Crosstalk between apoptosis, necrosis and autophagy. Biochim Biophys Acta 2013; 1833:3448–59 [DOI] [PubMed] [Google Scholar]
- 7.Kamer KJ, Mootha VK. The molecular era of the mitochondrial calcium uniporter. Nat Rev Mol Cell Biol 2015; 16:545–53 [DOI] [PubMed] [Google Scholar]
- 8.Bahat A, Gross A. Mitochondrial plasticity in cell fate regulation. J Biol Chem 2019; 294:13852–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan LB, Chandel NS. Mitochondrial reactive oxygen species and cancer. Cancer Metab 2014; 2:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell 2012; 48:158–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta 1999; 1410:103–23 [DOI] [PubMed] [Google Scholar]
- 12.van den Heuvel L, Smeitink J. The oxidative phosphorylation (OXPHOS) system: nuclear genes and human genetic diseases. Bioessays 2001; 23:518–25 [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646–74 [DOI] [PubMed] [Google Scholar]
- 14.Warburg O. On respiratory impairment in cancer cells. Science 1956; 124:269–70 [PubMed] [Google Scholar]
- 15.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer 2011; 11:85–95 [DOI] [PubMed] [Google Scholar]
- 16.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell 2012; 148:1132–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, Sekito T, Spirek M, Thornton J, Butow RA. Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Mol Cell 2003; 12:401–11 [DOI] [PubMed] [Google Scholar]
- 18.Sekito T, Thornton J, Butow RA. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell 2000; 11:2103–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 1993; 72:61–71 [DOI] [PubMed] [Google Scholar]
- 20.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell 2004; 14:1–15 [DOI] [PubMed] [Google Scholar]
- 21.Arnould T, Michel S, Renard P. Mitochondria retrograde signaling and the UPR mt: where are we in mammals? Int J Mol Sci 2015; 16:18224–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amuthan G, Biswas G, Ananadatheerthavarada HK, Vijayasarathy C, Shephard HM, Avadhani NG. Mitochondrial stress-induced calcium signaling, phenotypic changes and invasive behavior in human lung carcinoma A549 cells. Oncogene 2002; 21:7839–49 [DOI] [PubMed] [Google Scholar]
- 23.Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J 1999; 18:522–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang D, Kim J. Mitochondrial retrograde signalling and metabolic alterations in the tumour microenvironment. Cells 2019; 8:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guha M, Avadhani NG. Mitochondrial retrograde signaling at the crossroads of tumor bioenergetics, genetics and epigenetics. Mitochondrion 2013; 13:577–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HC, Huang KH, Yeh TS, Chi CW. Somatic alterations in mitochondrial DNA and mitochondrial dysfunction in gastric cancer progression. World J Gastroenterol 2014; 20:3950–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science 2000; 287:2017–9 [DOI] [PubMed] [Google Scholar]
- 28.Li H, Slone J, Fei L, Huang T. Mitochondrial DNA variants and common diseases: a mathematical model for the diversity of age-related mtDNA mutations. Cells 2019; 8:608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang BC, Hays L. Mitochondrial DNA copy number changes in human gliomas. Cancer Lett 1996; 105:167–73 [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Liu VW, Xue WC, Tsang PC, Cheung AN, Ngan HY. The increase of mitochondrial DNA content in endometrial adenocarcinoma cells: a quantitative study using laser-captured microdissected tissues. Gynecol Oncol 2005; 98:104–10 [DOI] [PubMed] [Google Scholar]
- 31.Lan Q, Lim U, Liu CS, Weinstein SJ, Chanock S, Bonner MR, Virtamo J, Albanes D, Rothman N. A prospective study of mitochondrial DNA copy number and risk of non-Hodgkin lymphoma. Blood 2008; 112:4247–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin CS, Chang SC, Wang LS, Chou TY, Hsu WH, Wu YC, Wei YH. The role of mitochondrial DNA alterations in esophageal squamous cell carcinomas. J Thorac Cardiovasc Surg 2010; 139:189–97 e4 [DOI] [PubMed] [Google Scholar]
- 33.Feng S, Xiong L, Ji Z, Cheng W, Yang H. Correlation between increased copy number of mitochondrial DNA and clinicopathological stage in colorectal cancer. Oncol Lett 2011; 2:899–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu CW, Yin PH, Hung WY, Li AF, Li SH, Chi CW, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes Cancer 2005; 44:19–28 [DOI] [PubMed] [Google Scholar]
- 35.Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer 2006; 45:629–38 [DOI] [PubMed] [Google Scholar]
- 36.Lee HC, Li SH, Lin JC, Wu CC, Yeh DC, Wei YH. Somatic mutations in the D-loop and decrease in the copy number of mitochondrial DNA in human hepatocellular carcinoma. Mutat Res 2004; 547:71–8 [DOI] [PubMed] [Google Scholar]
- 37.Lee HC, Wei YH. Mitochondrial DNA instability and metabolic shift in human cancers. Int J Mol Sci 2009; 10:674–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee HC, Yin PH, Lin JC, Wu CC, Chen CY, Wu CW, Chi CW, Tam TN, Wei YH. Mitochondrial genome instability and mtDNA depletion in human cancers. Ann N Y Acad Sci 2005; 1042:109–22 [DOI] [PubMed] [Google Scholar]
- 39.Hsu CC, Tseng LM, Lee HC. Role of mitochondrial dysfunction in cancer progression. Exp Biol Med 2016; 241:1281–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gille JJ, Joenje H. Cell culture models for oxidative stress: superoxide and hydrogen peroxide versus normobaric hyperoxia. Mutat Res 1992; 275:405–14 [DOI] [PubMed] [Google Scholar]
- 41.Lievre A, Chapusot C, Bouvier AM, Zinzindohoue F, Piard F, Roignot P, Arnould L, Beaune P, Faivre J, Laurent-Puig P. Clinical value of mitochondrial mutations in colorectal cancer. J Clin Oncol 2005; 23:3517–25 [DOI] [PubMed] [Google Scholar]
- 42.Hung WY, Wu CW, Yin PH, Chang CJ, Li AF, Chi CW, Wei YH, Lee HC. Somatic mutations in mitochondrial genome and their potential roles in the progression of human gastric cancer. Biochim Biophys Acta 2010; 1800:264–70 [DOI] [PubMed] [Google Scholar]
- 43.Yin PH, Wu CC, Lin JC, Chi CW, Wei YH, Lee HC. Somatic mutations of mitochondrial genome in hepatocellular carcinoma. Mitochondrion 2010; 10:174–82 [DOI] [PubMed] [Google Scholar]
- 44.Tseng LM, Yin PH, Yang CW, Tsai YF, Hsu CY, Chi CW, Lee HC. Somatic mutations of the mitochondrial genome in human breast cancers. Genes Chromosomes Cancer 2011; 50:800–11 [DOI] [PubMed] [Google Scholar]
- 45.Ohta S. Contribution of somatic mutations in the mitochondrial genome to the development of cancer and tolerance against anticancer drugs. Oncogene 2006; 25:4768–76 [DOI] [PubMed] [Google Scholar]
- 46.Chinnery PF, Samuels DC, Elson J, Turnbull DM. Accumulation of mitochondrial DNA mutations in ageing, cancer, and mitochondrial disease: is there a common mechanism? Lancet 2002; 360:1323–5 [DOI] [PubMed] [Google Scholar]
- 47.Wallace DC, Shoffner JM, Trounce I, Brown MD, Ballinger SW, Corral-Debrinski M, Horton T, Jun AS, Lott MT. Mitochondrial DNA mutations in human degenerative diseases and aging. Biochim Biophys Acta 1995; 1271:141–51 [DOI] [PubMed] [Google Scholar]
- 48.Hertweck KL, Dasgupta S. The landscape of mtDNA modifications in cancer: a tale of two cities. Front Oncol 2017; 7:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo ZS, Jin CL, Yao ZJ, Wang YM, Xu BT. Analysis of the mitochondrial 4977 bp deletion in patients with hepatocellular carcinoma. Balkan J Med Genet 2017; 20:81–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maximo V, Soares P, Seruca R, Rocha AS, Castro P, Sobrinho-Simoes M. Microsatellite instability, mitochondrial DNA large deletions, and mitochondrial DNA mutations in gastric carcinoma. Genes Chromosomes Cancer 2001; 32:136–43 [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Lu YY. Mitochondrial DNA 4977-bp deletion correlated with reactive oxygen species production and manganese superoxidedismutase expression in gastric tumor cells. Chin Med J 2009; 122:431–6 [PubMed] [Google Scholar]
- 52.Zhu W, Qin W, Sauter ER. Large-scale mitochondrial DNA deletion mutations and nuclear genome instability in human breast cancer. Cancer Detect Prev 2004; 28:119–26 [DOI] [PubMed] [Google Scholar]
- 53.Wei YH, Lee CF, Lee HC, Ma YS, Wang CW, Lu CY, Pang CY. Increases of mitochondrial mass and mitochondrial genome in association with enhanced oxidative stress in human cells harboring 4,977 BP-deleted mitochondrial DNA. Ann N Y Acad Sci 2001; 928:97–112 [DOI] [PubMed] [Google Scholar]
- 54.Lee HC, Yin PH, Yu TN, Chang YD, Hsu WC, Kao SY, Chi CW, Liu TY, Wei YH. Accumulation of mitochondrial DNA deletions in human oral tissues – effects of betel quid chewing and oral cancer. Mutat Res 2001; 493:67–74 [DOI] [PubMed] [Google Scholar]
- 55.Tseng LM, Yin PH, Tsai YF, Chi CW, Wu CW, Lee LM, Lee HC. Association between mitochondrial DNA 4,977 bp deletion and NAD(P)H:quinone oxidoreductase 1 C609T polymorphism in human breast tissues. Oncol Rep 2009; 21:1169–74 [DOI] [PubMed] [Google Scholar]
- 56.Dani MA, Dani SU, Lima SP, Martinez A, Rossi BM, Soares F, Zago MA, Simpson AJ. Less DeltamtDNA4977 than normal in various types of tumors suggests that cancer cells are essentially free of this mutation. Genet Mol Res 2004; 3:395–409 [PubMed] [Google Scholar]
- 57.Lee HC, Hsu LS, Yin PH, Lee LM, Chi CW. Heteroplasmic mutation of mitochondrial DNA D-loop and 4977-bp deletion in human cancer cells during mitochondrial DNA depletion. Mitochondrion 2007; 7:157–63 [DOI] [PubMed] [Google Scholar]
- 58.Croteau DL, Bohr VA. Repair of oxidative damage to nuclear and mitochondrial DNA in mammalian cells. J Biol Chem 1997; 272:25409–12 [DOI] [PubMed] [Google Scholar]
- 59.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A 2005; 102:719–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Porporato PE, Filigheddu N, Pedro JMB, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Res 2018; 28:265–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vyas S, Zaganjor E, Haigis MC. Mitochondria and cancer. Cell 2016; 166:555–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta 2010; 1805:105–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Area-Gomez E, Schon EA. Mitochondrial genetics and disease. J Child Neurol 2014; 29:1208–15 [DOI] [PubMed] [Google Scholar]
- 64.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst 2010; 102:932–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toro JR, Nickerson ML, Wei MH, Warren MB, Glenn GM, Turner ML, Stewart L, Duray P, Tourre O, Sharma N, Choyke P, Stratton P, Merino M, Walther MM, Linehan WM, Schmidt LS, Zbar B. Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North america. Am J Hum Genet 2003; 73:95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bardella C, Pollard PJ, Tomlinson I. SDH mutations in cancer. Biochim Biophys Acta 2011; 1807:1432–43 [DOI] [PubMed] [Google Scholar]
- 67.Lehtonen HJ, Kiuru M, Ylisaukko-Oja SK, Salovaara R, Herva R, Koivisto PA, Vierimaa O, Aittomaki K, Pukkala E, Launonen V, Aaltonen LA. Increased risk of cancer in patients with fumarate hydratase germline mutation. J Med Genet 2006; 43:523–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dubard Gault M, Mandelker D, DeLair D, Stewart CR, Kemel Y, Sheehan MR, Siegel B, Kennedy J, Marcell V, Arnold A, Al-Ahmadie H, Modak S, Robson M, Shukla N, Roberts S, Vijai J, Topka S, Kentsis A, Cadoo K, Carlo M, Latham Schwark A, Reznik E, Dinatale R, Hechtman J, Borras Flores E, Jairam S, Yang C, Li Y, Bayraktar EC, Ceyhan-Birsoy O, Zhang L, Kohlman W, Schiffman J, Stadler Z, Birsoy K, Kung A, Offit K, Walsh MF. Germline SDHA mutations in children and adults with cancer. Cold Spring Harb Mol Case Stud 2018; 4:a002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov 2013; 3:730–41 [DOI] [PubMed] [Google Scholar]
- 70.Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol 2016; 27:599–608 [DOI] [PubMed] [Google Scholar]
- 71.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr., Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 2010; 464:121–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang CZ, Liu L, Cai M, Pan Y, Fu J, Cao Y, Yun J. Low SIRT3 expression correlates with poor differentiation and unfavorable prognosis in primary hepatocellular carcinoma. PLoS One 2012; 7:e51703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ashraf N, Zino S, Macintyre A, Kingsmore D, Payne AP, George WD, Shiels PG. Altered sirtuin expression is associated with node-positive breast cancer. Br J Cancer 2006; 95:1056–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D’Silva NJ, Kapila YL. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer 2011; 117:1670–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang KH, Hsu CC, Fang WL, Chi CW, Sung MT, Kao HL, Li AF, Yin PH, Yang MH, Lee HC. SIRT3 expression as a biomarker for better prognosis in gastric cancer. World J Surg 2014; 38:910–7 [DOI] [PubMed] [Google Scholar]
- 76.Mahjabeen I, Kayani MA. Loss of mitochondrial tumor suppressor genes expression is associated with unfavorable clinical outcome in head and neck squamous cell carcinoma: data from retrospective study. PLoS One 2016; 11:e0146948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tilokani L, Nagashima S, Paupe V, Prudent J. Mitochondrial dynamics: overview of molecular mechanisms. Essays Biochem 2018; 62:341–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen H, Chan DC. Mitochondrial dynamics in regulating the unique phenotypes of cancer and stem cells. Cell Metab 2017; 26:39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Palm W, Thompson CB. Nutrient acquisition strategies of mammalian cells. Nature 2017; 546:234–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang L, Xiao L, Sugiura H, Huang X, Ali A, Kuro-O M, Deberardinis RJ, Boothman DA. Metabolic reprogramming during TGFbeta1-induced epithelial-to-mesenchymal transition. Oncogene 2015; 34:3908–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jiang L, Deberardinis R, Boothman DA. The cancer cell ‘energy grid’: TGF-beta1 signaling coordinates metabolism for migration. Mol Cell Oncol 2015; 2:e981994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang L, Deberardinis RJ. Cancer metabolism: when more is less. Nature 2012; 489:511–2 [DOI] [PubMed] [Google Scholar]
- 83.Corrado M, Scorrano L, Campello S. Mitochondrial dynamics in cancer and neurodegenerative and neuroinflammatory diseases. Int J Cell Biol 2012; 2012:729290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trotta AP, Chipuk JE. Mitochondrial dynamics as regulators of cancer biology. Cell Mol Life Sci 2017; 74:1999–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Serasinghe MN, Wieder SY, Renault TT, Elkholi R, Asciolla JJ, Yao JL, Jabado O, Hoehn K, Kageyama Y, Sesaki H, Chipuk JE. Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol Cell 2015; 57:521–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ferreira-da-Silva A, Valacca C, Rios E, Populo H, Soares P, Sobrinho-Simoes M, Scorrano L, Maximo V, Campello S. Mitochondrial dynamics protein Drp1 is overexpressed in oncocytic thyroid tumors and regulates cancer cell migration. PLoS One 2015; 10:e0122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL, Counter CM, Kashatus DF. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell 2015; 57:537–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wieder SY, Serasinghe MN, Sung JC, Choi DC, Birge MB, Yao JL, Bernstein E, Celebi JT, Chipuk JE. Activation of the mitochondrial fragmentation protein DRP1 correlates with BRAF(V600E) melanoma. J Invest Dermatol 2015; 135:2544–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lyssiotis CA, Kimmelman AC. Metabolic interactions in the tumor microenvironment. Trends Cell Biol 2017; 27:863–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016; 16:582–98 [DOI] [PubMed] [Google Scholar]
- 91.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature 2011; 473:298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jain RK. Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science 2005; 307:58–62 [DOI] [PubMed] [Google Scholar]
- 93.Petrova V, Annicchiarico-Petruzzelli M, Melino G, Amelio I. The hypoxic tumour microenvironment. Oncogenesis 2018; 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell 2008; 134:703–7 [DOI] [PubMed] [Google Scholar]
- 95.Yamada S, Nomoto S, Fujii T, Kaneko T, Takeda S, Inoue S, Kanazumi N, Nakao A. Correlation between copy number of mitochondrial DNA and clinico-pathologic parameters of hepatocellular carcinoma. Eur J Surg Oncol 2006; 32:303–7 [DOI] [PubMed] [Google Scholar]
- 96.Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, Zhang N, Hao X, Niu R. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life 2007; 59:450–7 [DOI] [PubMed] [Google Scholar]
- 97.Cook CC, Higuchi M. The awakening of an advanced malignant cancer: an insult to the mitochondrial genome. Biochim Biophys Acta 2012; 1820:652–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li X, Zhong Y, Lu J, Axcrona K, Eide L, Syljuasen RG, Peng Q, Wang J, Zhang H, Goscinski MA, Kvalheim G, Nesland JM, Suo Z. MtDNA depleted PC3 cells exhibit warburg effect and cancer stem cell features. Oncotarget 2016; 7:40297–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Y, Wu X, Li X, Kvalheim G, Axcrona U, Axcrona K, Suo Z. Blocking mtDNA replication upregulates the expression of stemness-related genes in prostate cancer cell lines. Ultrastruct Pathol 2013; 37:258–66 [DOI] [PubMed] [Google Scholar]
- 100.Singh KK, Ayyasamy V, Owens KM, Koul MS, Vujcic M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J Hum Genet 2009; 54:516–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Naito A, Cook CC, Mizumachi T, Wang M, Xie CH, Evans TT, Kelly T, Higuchi M. Progressive tumor features accompany epithelial-mesenchymal transition induced in mitochondrial DNA-depleted cells. Cancer Sci 2008; 99:1584–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheau-Feng Lin F, Jeng YC, Huang TY, Chi CS, Chou MC, Chin-Shaw Tsai S. Mitochondrial DNA copy number is associated with diagnosis and prognosis of head and neck cancer. Biomarkers 2014; 19:269–74 [DOI] [PubMed] [Google Scholar]
- 103.Masuike Y, Tanaka K, Makino T, Yamasaki M, Miyazaki Y, Takahashi T, Kurokawa Y, Nakajima K, Mori M, Doki Y. Esophageal squamous cell carcinoma with low mitochondrial copy number has mesenchymal and stem-like characteristics, and contributes to poor prognosis. PLoS One 2018; 13:e0193159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, Pesdar EA, Sobol M, Filimonenko A, Stuart S, Vondrusova M, Kluckova K, Sachaphibulkij K, Rohlena J, Hozak P, Truksa J, Eccles D, Haupt LM, Griffiths LR, Neuzil J, Berridge MV. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab 2015; 21:81–94 [DOI] [PubMed] [Google Scholar]
- 105.Hill S, Sataranatarajan K, Van Remmen H. Role of signaling molecules in mitochondrial stress response. Front Genet 2018; 9:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun X, Zhan L, Chen Y, Wang G, He L, Wang Q, Zhou F, Yang F, Wu J, Wu Y, Xing J, He X, Huang Q. Increased mtDNA copy number promotes cancer progression by enhancing mitochondrial oxidative phosphorylation in microsatellite-stable colorectal cancer. Signal Transduct Target Ther 2018; 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee WR, Na H, Lee SW, Lim WJ, Kim N, Lee JE, Kang C. Transcriptomic analysis of mitochondrial TFAM depletion changing cell morphology and proliferation. Sci Rep 2017; 7:17841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Biswas G, Guha M, Avadhani NG. Mitochondria-to-nucleus stress signaling in mammalian cells: nature of nuclear gene targets, transcription regulation, and induced resistance to apoptosis. Gene 2005; 354:132–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Singh KK, Russell J, Sigala B, Zhang Y, Williams J, Keshav KF. Mitochondrial DNA determines the cellular response to cancer therapeutic agents. Oncogene 1999; 18:6641–6 [DOI] [PubMed] [Google Scholar]
- 110.Qian W, Nishikawa M, Haque AM, Hirose M, Mashimo M, Sato E, Inoue M. Mitochondrial density determines the cellular sensitivity to cisplatin-induced cell death. Am J Physiol Cell Physiol 2005; 289:C1466–75 [DOI] [PubMed] [Google Scholar]
- 111.Gonzalez-Sanchez E, Marin JJ, Perez MJ. The expression of genes involved in hepatocellular carcinoma chemoresistance is affected by mitochondrial genome depletion. Mol Pharm 2014; 11:1856–68 [DOI] [PubMed] [Google Scholar]
- 112.Lee W, Choi HI, Kim MJ, Park SY. Depletion of mitochondrial DNA up-regulates the expression of MDR1 gene via an increase in mRNA stability. Exp Mol Med 2008; 40:109–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ferraresi R, Troiano L, Pinti M, Roat E, Lugli E, Quaglino D, Taverna D, Bellizzi D, Passarino G, Cossarizza A. Resistance of mtDNA-depleted cells to apoptosis. Cytometry A 2008; 73:528–37 [DOI] [PubMed] [Google Scholar]
- 114.Park SY, Chang I, Kim JY, Kang SW, Park SH, Singh K, Lee MS. Resistance of mitochondrial DNA-depleted cells against cell death: role of mitochondrial superoxide dismutase. J Biol Chem 2004; 279:7512–20 [DOI] [PubMed] [Google Scholar]
- 115.Srinivasan S, Guha M, Avadhani NG. Mitochondrial respiratory defects promote the Warburg effect and cancer progression. Mol Cell Oncol 2016; 3:e1085120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Higuchi M, Kudo T, Suzuki S, Evans TT, Sasaki R, Wada Y, Shirakawa T, Sawyer JR, Gotoh A. Mitochondrial DNA determines androgen dependence in prostate cancer cell lines. Oncogene 2006; 25:1437–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Naito A, Carcel-Trullols J, Xie CH, Evans TT, Mizumachi T, Higuchi M. Induction of acquired resistance to antiestrogen by reversible mitochondrial DNA depletion in breast cancer cell line. Int J Cancer 2008; 122:1506–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hsu CW, Yin PH, Lee HC, Chi CW, Tseng LM. Mitochondrial DNA content as a potential marker to predict response to anthracycline in breast cancer patients. Breast J 2010; 16:264–70 [DOI] [PubMed] [Google Scholar]
- 119.Liang BC, Ullyatt E. Increased sensitivity to cis-diamminedichloroplatinum induced apoptosis with mitochondrial DNA depletion. Cell Death Differ 1998; 5:694–701 [DOI] [PubMed] [Google Scholar]
- 120.Radde BN, Ivanova MM, Mai HX, Alizadeh-Rad N, Piell K, Van Hoose P, Cole MP, Muluhngwi P, Kalbfleisch TS, Rouchka EC, Hill BG, Klinge CM. Nuclear respiratory factor-1 and bioenergetics in tamoxifen-resistant breast cancer cells. Exp Cell Res 2016; 347:222–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Carew JS, Zhou Y, Albitar M, Carew JD, Keating MJ, Huang P. Mitochondrial DNA mutations in primary leukemia cells after chemotherapy: clinical significance and therapeutic implications. Leukemia 2003; 17:1437–47 [DOI] [PubMed] [Google Scholar]
- 122.Shidara Y, Yamagata K, Kanamori T, Nakano K, Kwong JQ, Manfredi G, Oda H, Ohta S. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res 2005; 65:1655–63 [DOI] [PubMed] [Google Scholar]
- 123.Shin YK, Yoo BC, Chang HJ, Jeon E, Hong SH, Jung MS, Lim SJ, Park JG. Down-regulation of mitochondrial F1F0-ATP synthase in human Colon cancer cells with induced 5-fluorouracil resistance. Cancer Res 2005; 65:3162–70 [DOI] [PubMed] [Google Scholar]
- 124.Santidrian AF, Matsuno-Yagi A, Ritland M, Seo BB, LeBoeuf SE, Gay LJ, Yagi T, Felding-Habermann B. Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. J Clin Invest 2013; 123:1068–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chang JC, Chang HS, Wu YC, Cheng WL, Lin TT, Chang HJ, Kuo SJ, Chen ST, Liu CS. Mitochondrial transplantation regulates antitumour activity, chemoresistance and mitochondrial dynamics in breast cancer. J Exp Clin Cancer Res 2019; 38:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lund M, Melbye M, Diaz LJ, Duno M, Wohlfahrt J, Vissing J. Mitochondrial dysfunction and risk of cancer. Br J Cancer 2015; 112:1134–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria in cancer cells: what is so special about them? Trends Cell Biol 2008; 18:165–73 [DOI] [PubMed] [Google Scholar]
- 128.Schmidt LS, Linehan WM. Hereditary leiomyomatosis and renal cell carcinoma. Int J Nephrol Renovasc Dis 2014; 7:253–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lau HD, Chan E, Fan AC, Kunder CA, Williamson SR, Zhou M, Idrees MT, Maclean FM, Gill AJ, Kao CS. A clinicopathologic and molecular analysis of fumarate hydratase-deficient renal cell carcinoma in 32 patients. Am J Surg Pathol 2020; 44:98–110 [DOI] [PubMed] [Google Scholar]
- 130.Sciacovelli M, Goncalves E, Johnson TI, Zecchini VR, da Costa AS, Gaude E, Drubbel AV, Theobald SJ, Abbo SR, Tran MG, Rajeeve V, Cardaci S, Foster S, Yun H, Cutillas P, Warren A, Gnanapragasam V, Gottlieb E, Franze K, Huntly B, Maher ER, Maxwell PH, Saez-Rodriguez J, Frezza C. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature 2016; 537:544–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sciacovelli M, Frezza C. Fumarate drives EMT in renal cancer. Cell Death Differ 2017; 24:1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sudarshan S, Shanmugasundaram K, Naylor SL, Lin S, Livi CB, O’Neill CF, Parekh DJ, Yeh IT, Sun LZ, Block K. Reduced expression of fumarate hydratase in clear cell renal cancer mediates HIF-2alpha accumulation and promotes migration and invasion. PLoS One 2011; 6:e21037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cornejo KM, Lu M, Yang P, Wu S, Cai C, Zhong WD, Olumi A, Young RH, Wu CL. Succinate dehydrogenase B: a new prognostic biomarker in clear cell renal cell carcinoma. Hum Pathol 2015; 46:820–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dalla Pozza E, Dando I, Pacchiana R, Liboi E, Scupoli MT, Donadelli M, Palmieri M. Regulation of succinate dehydrogenase and role of succinate in cancer. Semin Cell Dev Biol 2020; 98:4–14 [DOI] [PubMed] [Google Scholar]
- 135.Liu WS, Chan SH, Chang HT, Li GC, Tu YT, Tseng HH, Fu TY, Chang HY, Liou HH, Ger LP, Tsai KW. Isocitrate dehydrogenase 1-snail axis dysfunction significantly correlates with breast cancer prognosis and regulates cell invasion ability. Breast Cancer Res 2018; 20:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Medeiros BC, Fathi AT, DiNardo CD, Pollyea DA, Chan SM, Swords R. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia 2017; 31:272–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fu Y, Zheng S, Zheng Y, Huang R, An N, Liang A, Hu C. Glioma derived isocitrate dehydrogenase-2 mutations induced up-regulation of HIF-1alpha and beta-catenin signaling: possible impact on glioma cell metastasis and chemo-resistance. Int J Biochem Cell Biol 2012; 44:770–5 [DOI] [PubMed] [Google Scholar]
- 138.Wang Z, Liang S, Lian X, Liu L, Zhao S, Xuan Q, Guo L, Liu H, Yang Y, Dong T, Liu Y, Liu Z, Zhang Q. Identification of proteins responsible for adriamycin resistance in breast cancer cells using proteomics analysis. Sci Rep 2015; 5:9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yu HE, Wang F, Yu F, Zeng ZL, Wang Y, Lu YX, Jin Y, Wang DS, Qiu MZ, Pu HY, Kang TB, Xie D, Ju HQ, Xu RH, Luo HY. Suppression of fumarate hydratase activity increases the efficacy of cisplatin-mediated chemotherapy in gastric cancer. Cell Death Dis 2019; 10:413. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 140.Huang S, Chen X, Zheng J, Huang Y, Song L, Yin Y, Xiong J. Low SIRT3 expression contributes to tumor progression, development and poor prognosis in human pancreatic carcinoma. Pathol Res Pract 2017; 213:1419–23 [DOI] [PubMed] [Google Scholar]
- 141.Xiao K, Jiang J, Wang W, Cao S, Zhu L, Zeng H, Ouyang R, Zhou R, Chen P. Sirt3 is a tumor suppressor in lung adenocarcinoma cells. Oncol Rep 2013; 30:1323–8 [DOI] [PubMed] [Google Scholar]
- 142.Kao YY, Chou CH, Yeh LY, Chen YF, Chang KW, Liu CJ, Fan Chiang CY, Lin SC. MicroRNA miR-31 targets SIRT3 to disrupt mitochondrial activity and increase oxidative stress in oral carcinoma. Cancer Lett 2019; 456:40–8 [DOI] [PubMed] [Google Scholar]
- 143.Zou X, Santa-Maria CA, O’Brien J, Gius D, Zhu Y. Manganese superoxide dismutase acetylation and dysregulation, due to loss of SIRT3 activity, promote a luminal b-like breast carcinogenic-permissive phenotype. Antioxid Redox Signal 2016; 25:326–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhao J, Zhang J, Yu M, Xie Y, Huang Y, Wolff DW, Abel PW, Tu Y. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene 2013; 32:4814–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qian W, Wang J, Van Houten B. The role of dynamin-related protein 1 in cancer growth: a promising therapeutic target? Expert Opin Ther Targets 2013; 17:997–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, Martinou JC. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS One 2008; 3:e3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sastre-Serra J, Nadal-Serrano M, Pons DG, Roca P, Oliver J. Mitochondrial dynamics is affected by 17beta-estradiol in the MCF-7 breast cancer cell line. Effects on fusion and fission related genes. Int J Biochem Cell Biol 2012; 44:1901–5 [DOI] [PubMed] [Google Scholar]
- 148.Choudhary V, Kaddour-Djebbar I, Lakshmikanthan V, Ghazaly T, Thangjam GS, Sreekumar A, Lewis RW, Mills IG, Bollag WB, Kumar MV. Novel role of androgens in mitochondrial fission and apoptosis. Mol Cancer Res 2011; 9:1067–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vara-Perez M, Felipe-Abrio B, Agostinis P. Mitophagy in cancer: a tale of adaptation. Cells 2019; 8:493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wu HM, Shao LJ, Jiang ZF, Liu RY. Gemcitabine-induced autophagy protects human lung cancer cells from apoptotic death. Lung 2016; 194:959–66 [DOI] [PubMed] [Google Scholar]
- 151.Abdrakhmanov A, Kulikov AV, Luchkina EA, Zhivotovsky B, Gogvadze V. Involvement of mitophagy in cisplatin-induced cell death regulation. Biol Chem 2019; 400:161–70 [DOI] [PubMed] [Google Scholar]
- 152.Yan C, Luo L, Guo CY, Goto S, Urata Y, Shao JH, Li TS. Doxorubicin-induced mitophagy contributes to drug resistance in cancer stem cells from HCT8 human colorectal cancer cells. Cancer Lett 2017; 388:34–42 [DOI] [PubMed] [Google Scholar]
- 153.Villa E, Proics E, Rubio-Patino C, Obba S, Zunino B, Bossowski JP, Rozier RM, Chiche J, Mondragon L, Riley JS, Marchetti S, Verhoeyen E, Tait SWG, Ricci JE. Parkin-independent mitophagy controls chemotherapeutic response in cancer cells. Cell Rep 2017; 20:2846–59 [DOI] [PubMed] [Google Scholar]
- 154.Kong B, Wang Q, Fung E, Xue K, Tsang BK. p53 is required for cisplatin-induced processing of the mitochondrial fusion protein L-Opa1 that is mediated by the mitochondrial metallopeptidase Oma1 in gynecologic cancers. J Biol Chem 2014; 289:27134–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chang JY, Yi HS, Kim HW, Shong M. Dysregulation of mitophagy in carcinogenesis and tumor progression. Biochim Biophys Acta Bioenerg 2017; 1858:633–40 [DOI] [PubMed] [Google Scholar]
- 156.Yan C, Li TS. Dual role of mitophagy in cancer drug resistance. Anticancer Res 2018; 38:617–21 [DOI] [PubMed] [Google Scholar]
- 157.Jazwinski SM. The retrograde response: when mitochondrial quality control is not enough. Biochim Biophys Acta 2013; 1833:400–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Quiros PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol 2016; 17:213–26 [DOI] [PubMed] [Google Scholar]
- 159.Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, Kim SJ, Mehta H, Hevener AL, de Cabo R, Cohen P. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab 2015; 21:443–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC. Humanin peptide suppresses apoptosis by interfering with bax activation. Nature 2003; 423:456–61 [DOI] [PubMed] [Google Scholar]
- 161.Dhir A, Dhir S, Borowski LS, Jimenez L, Teitell M, Rotig A, Crow YJ, Rice GI, Duffy D, Tamby C, Nojima T, Munnich A, Schiff M, de Almeida CR, Rehwinkel J, Dziembowski A, Szczesny RJ, Proudfoot NJ. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 2018; 560:238–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhong Z, Liang S, Sanchez-Lopez E, He F, Shalapour S, Lin XJ, Wong J, Ding S, Seki E, Schnabl B, Hevener AL, Greenberg HB, Kisseleva T, Karin M. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 2018; 560:198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shadel GS, Horvath TL. Mitochondrial ROS signaling in organismal homeostasis. Cell 2015; 163:560–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 2009; 284:13291–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Acin-Perez R, Carrascoso I, Baixauli F, Roche-Molina M, Latorre-Pellicer A, Fernandez-Silva P, Mittelbrunn M, Sanchez-Madrid F, Perez-Martos A, Lowell CA, Manfredi G, Enriquez JA. ROS-triggered phosphorylation of complex II by Fgr kinase regulates cellular adaptation to fuel use. Cell Metab 2014; 19:1020–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chae S, Ahn BY, Byun K, Cho YM, Yu MH, Lee B, Hwang D, Park KS. A systems approach for decoding mitochondrial retrograde signaling pathways. Sci Signal 2013; 6:rs4. [DOI] [PubMed] [Google Scholar]
- 167.Formentini L, Sanchez-Arago M, Sanchez-Cenizo L, Cuezva JM. The mitochondrial ATPase inhibitory factor 1 triggers a ROS-mediated retrograde prosurvival and proliferative response. Mol Cell 2012; 45:731–42 [DOI] [PubMed] [Google Scholar]
- 168.Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol 2012; 13:566–78 [DOI] [PubMed] [Google Scholar]
- 169.Biswas G, Anandatheerthavarada HK, Zaidi M, Avadhani NG. Mitochondria to nucleus stress signaling: a distinctive mechanism of NFkappaB/rel activation through calcineurin-mediated inactivation of IkappaBbeta. J Cell Biol 2003; 161:507–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Arnould T, Vankoningsloo S, Renard P, Houbion A, Ninane N, Demazy C, Remacle J, Raes M. CREB activation induced by mitochondrial dysfunction is a new signaling pathway that impairs cell proliferation. EMBO J 2002; 21:53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hung WY, Huang KH, Wu CW, Chi CW, Kao HL, Li AF, Yin PH, Lee HC. Mitochondrial dysfunction promotes cell migration via reactive oxygen species-enhanced beta5-integrin expression in human gastric cancer SC-M1 cells. Biochim Biophys Acta 2012; 1820:1102–10 [DOI] [PubMed] [Google Scholar]
- 172.Wang SF, Chen MS, Chou YC, Ueng YF, Yin PH, Yeh TS, Lee HC. Mitochondrial dysfunction enhances cisplatin resistance in human gastric cancer cells via the ROS-activated GCN2-eIF2alpha-ATF4-xCT pathway. Oncotarget 2016; 7:74132–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chang CJ, Yin PH, Yang DM, Wang CH, Hung WY, Chi CW, Wei YH, Lee HC. Mitochondrial dysfunction-induced amphiregulin upregulation mediates chemo-resistance and cell migration in HepG2 cells. Cell Mol Life Sci 2009; 66:1755–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Guaragnella N, Giannattasio S, Moro L. Mitochondrial dysfunction in cancer chemoresistance. Biochem Pharmacol 2014; 92:62–72 [DOI] [PubMed] [Google Scholar]
- 175.Alam NA, Olpin S, Leigh IM. Fumarate hydratase mutations and predisposition to cutaneous leiomyomas, uterine leiomyomas and renal cancer. Br J Dermatol 2005; 153:11–7 [DOI] [PubMed] [Google Scholar]
- 176.Zhao T, Mu X, You Q. Succinate: an initiator in tumorigenesis and progression. Oncotarget 2017; 8:53819–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fathi AT, Sadrzadeh H, Comander AH, Higgins MJ, Bardia A, Perry A, Burke M, Silver R, Matulis CR, Straley KS, Yen KE, Agresta S, Kim H, Schenkein DP, Borger DR. Isocitrate dehydrogenase 1 (IDH1) mutation in breast adenocarcinoma is associated with elevated levels of serum and urine 2-hydroxyglutarate. Oncologist 2014; 19:602–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Losman JA, Kaelin WG., Jr., What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev 2013; 27:836–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 2010; 29:625–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, Merino M, Trepel J, Zbar B, Toro J, Ratcliffe PJ, Linehan WM, Neckers L. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell 2005; 8:143–53 [DOI] [PubMed] [Google Scholar]
- 181.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 2005; 7:77–85 [DOI] [PubMed] [Google Scholar]
- 182.Latini A, da Silva CG, Ferreira GC, Schuck PF, Scussiato K, Sarkis JJ, Dutra Filho CS, Wyse AT, Wannmacher CM, Wajner M. Mitochondrial energy metabolism is markedly impaired by D-2-hydroxyglutaric acid in rat tissues. Mol Genet Metab 2005; 86:188–99 [DOI] [PubMed] [Google Scholar]
- 183.Kolker S, Pawlak V, Ahlemeyer B, Okun JG, Horster F, Mayatepek E, Krieglstein J, Hoffmann GF, Kohr G. NMDA receptor activation and respiratory chain complex V inhibition contribute to neurodegeneration in d-2-hydroxyglutaric aciduria. Eur J Neurosci 2002; 16:21–8 [DOI] [PubMed] [Google Scholar]
- 184.Gross O, Thomas CJ, Guarda G, Tschopp J. The inflammasome: an integrated view. Immunol Rev 2011; 243:136–51 [DOI] [PubMed] [Google Scholar]
- 185.Lamkanfi M, Dixit VM. Inflammasomes and their roles in health and disease. Annu Rev Cell Dev Biol 2012; 28:137–61 [DOI] [PubMed] [Google Scholar]
- 186.Heneka MT, Kummer MP, Latz E. Innate immune activation in neurodegenerative disease. Nat Rev Immunol 2014; 14:463–77 [DOI] [PubMed] [Google Scholar]
- 187.Liu S, Feng M, Guan W. Mitochondrial DNA sensing by STING signaling participates in inflammation, cancer and beyond. Int J Cancer 2016; 139:736–41 [DOI] [PubMed] [Google Scholar]
- 188.Braunstein MJ, Kucharczyk J, Adams S. Targeting toll-like receptors for cancer therapy. Target Oncol 2018; 13:583–98 [DOI] [PubMed] [Google Scholar]
- 189.Zuccato CF, Asad AS, Nicola Candia AJ, Gottardo MF, Moreno Ayala MA, Theas MS, Seilicovich A, Candolfi M. Mitochondrial-derived peptide humanin as therapeutic target in cancer and degenerative diseases. Expert Opin Ther Targets 2019; 23:117–26 [DOI] [PubMed] [Google Scholar]
- 190.Hashimoto Y, Niikura T, Tajima H, Yasukawa T, Sudo H, Ito Y, Kita Y, Kawasumi M, Kouyama K, Doyu M, Sobue G, Koide T, Tsuji S, Lang J, Kurokawa K, Nishimoto I. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and abeta. Proc Natl Acad Sci U S A 2001; 98:6336–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Maximov V, Martynenko A, Hunsmann G, Tarantul V. Mitochondrial 16S rRNA gene encodes a functional peptide, a potential drug for alzheimer’s disease and target for cancer therapy. Med Hypoth 2002; 59:670–3 [DOI] [PubMed] [Google Scholar]
- 192.Omar NN, Tash RF, Shoukry Y, ElSaeed KO. Breaking the ritual metabolic cycle in order to save acetyl CoA: a potential role for mitochondrial humanin in T2 bladder cancer aggressiveness. J Egypt Natl Canc Inst 2017; 29:69–76 [DOI] [PubMed] [Google Scholar]
- 193.Mottaghi-Dastjerdi N, Soltany-Rezaee-Rad M, Sepehrizadeh Z, Roshandel G, Ebrahimifard F, Setayesh N. Genome expression analysis by suppression subtractive hybridization identified overexpression of humanin, a target gene in gastric cancer chemoresistance. Daru 2014; 22:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Harhay GP, Sonstegard TS, Keele JW, Heaton MP, Clawson ML, Snelling WM, Wiedmann RT, Van Tassell CP, Smith TP. Characterization of 954 bovine full-CDS cDNA sequences. BMC Genomics 2005; 6:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lu H, Wei M, Zhai Y, Li Q, Ye Z, Wang L, Luo W, Chen J, Lu Z. MOTS-c peptide regulates adipose homeostasis to prevent ovariectomy-induced metabolic dysfunction. J Mol Med 2019; 97:473–85 [DOI] [PubMed] [Google Scholar]
- 196.Raijmakers RPH, Jansen AFM, Keijmel SP, Ter Horst R, Roerink ME, Novakovic B, Joosten LAB, van der Meer JWM, Netea MG. Bleeker-Rovers CP. A possible role for mitochondrial-derived peptides humanin and MOTS-c in patients with Q fever fatigue syndrome and chronic fatigue syndrome. J Transl Med 2019; 17:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kim KH, Son JM, Benayoun BA, Lee C. The mitochondrial-encoded peptide MOTS-c translocates to the nucleus to regulate nuclear gene expression in response to metabolic stress. Cell Metab 2018; 28:516–24 e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hsu CC, Wang CH, Wu LC, Hsia CY, Chi CW, Yin PH, Chang CJ, Sung MT, Wei YH, Lu SH, Lee HC. Mitochondrial dysfunction represses HIF-1alpha protein synthesis through AMPK activation in human hepatoma HepG2 cells. Biochim Biophys Acta 2013; 1830:4743–51 [DOI] [PubMed] [Google Scholar]
- 199.Jovaisaite V, Mouchiroud L, Auwerx J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J Exp Biol 2014; 217:137–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Naresh NU, Haynes CM. Signaling and regulation of the mitochondrial unfolded protein response. Cold Spring Harb Perspect Biol 2019; 11:a033944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J 2002; 21:4411–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell 2010; 37:529–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 2012; 337:587–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Topf U, Wrobel L, Chacinska A. Chatty mitochondria: keeping balance in cellular protein homeostasis. Trends Cell Biol 2016; 26:577–86 [DOI] [PubMed] [Google Scholar]
- 205.Mottis A, Herzig S, Auwerx J. Mitocellular communication: shaping health and disease. Science 2019; 366:827–32 [DOI] [PubMed] [Google Scholar]
- 206.Fiorese CJ, Schulz AM, Lin YF, Rosin N, Pellegrino MW, Haynes CM. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr Biol 2016; 26:2037–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Quiros PM, Prado MA, Zamboni N, D’Amico D, Williams RW, Finley D, Gygi SP, Auwerx J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol 2017; 216:2027–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kenny TC, Manfredi G, Germain D. The mitochondrial unfolded protein response as a non-oncogene addiction to support adaptation to stress during transformation in cancer and beyond. Front Oncol 2017; 7:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mattson MP. Hormesis defined. Ageing Res Rev 2008; 7:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mattson MP. Hormesis and disease resistance: activation of cellular stress response pathways. Hum Exp Toxicol 2008; 27:155–62 [DOI] [PubMed] [Google Scholar]
- 211.Kenny TC, Craig AJ, Villanueva A, Germain D. Mitohormesis primes tumor invasion and metastasis. Cell Rep 2019; 27:2292–303 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Papa L, Germain D. SirT3 regulates the mitochondrial unfolded protein response. Mol Cell Biol 2014; 34:699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wang Y, Zhou D, Phung S, Warden C, Rashid R, Chan N, Chen S. SGK3 sustains ERalpha signaling and drives acquired aromatase inhibitor resistance through maintaining endoplasmic reticulum homeostasis. Proc Natl Acad Sci U S A 2017; 114:E1500–E08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Angelastro JM. Targeting ATF5 in cancer. Trends Cancer 2017; 3:471–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Karpel-Massler G, Horst BA, Shu C, Chau L, Tsujiuchi T, Bruce JN, Canoll P, Greene LA, Angelastro JM, Siegelin MD. A synthetic cell-penetrating dominant-negative ATF5 peptide exerts anticanceractivity against a broad spectrum of treatment-resistant cancers. Clin Cancer Res 2016; 22:4698–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ghosh JC, Dohi T, Kang BH, Altieri DC. Hsp60 regulation of tumor cell apoptosis. J Biol Chem 2008; 283:5188–94 [DOI] [PubMed] [Google Scholar]
- 217.Cole A, Wang Z, Coyaud E, Voisin V, Gronda M, Jitkova Y, Mattson R, Hurren R, Babovic S, Maclean N, Restall I, Wang X, Jeyaraju DV, Sukhai MA, Prabha S, Bashir S, Ramakrishnan A, Leung E, Qia YH, Zhang N, Combes KR, Ketela T, Lin F, Houry WA, Aman A, Al-Awar R, Zheng W, Wienholds E, Xu CJ, Dick J, Wang JC, Moffat J, Minden MD, Eaves CJ, Bader GD, Hao Z, Kornblau SM, Raught B, Schimmer AD. Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 2015; 27:864–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang HM, Cheng KC, Lin CJ, Hsu SW, Fang WC, Hsu TF, Chiu CC, Chang HW, Hsu CH, Lee AY. Obtusilactone a and (-)-sesamin induce apoptosis in human lung cancer cells by inhibiting mitochondrial lon protease and activating DNA damage checkpoints. Cancer Sci 2010; 101:2612–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 2006; 34:7–11 [DOI] [PubMed] [Google Scholar]
- 220.Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie 2007; 89:799–811 [DOI] [PubMed] [Google Scholar]
- 221.Han AP, Yu C, Lu L, Fujiwara Y, Browne C, Chin G, Fleming M, Leboulch P, Orkin SH, Chen JJ. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J 2001; 20:6909–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 2011; 334:1081–6 [DOI] [PubMed] [Google Scholar]
- 223.B’Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, Bruhat A. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res 2013; 41:7683–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 2003; 11:619–33 [DOI] [PubMed] [Google Scholar]
- 225.Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep 2016; 17:1374–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wek RC, Staschke KA. How do tumours adapt to nutrient stress? Embo J 2010; 29:1946–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lewerenz J, Maher P.Basal levels of eIF2alpha phosphorylation determine cellular antioxidant status by regulating ATF4 and xCT expression. J Biol Chem284:1106–15 [DOI] [PMC free article] [PubMed]
- 228.Galehdar Z, Swan P, Fuerth B, Callaghan SM, Park DS, Cregan SP. Neuronal apoptosis induced by endoplasmic reticulum stress is regulated by ATF4-CHOP-mediated induction of the bcl-2 homology 3-only member PUMA. J Neurosci 2010; 30:16938–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Puthalakath H, O’Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, Gotoh T, Akira S, Bouillet P, Strasser A. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 2007; 129:1337–49 [DOI] [PubMed] [Google Scholar]
- 230.Zou W, Yue P, Khuri FR, Sun SY. Coupling of endoplasmic reticulum stress to CDDO-Me-induced up-regulation of death receptor 5 via a CHOP-dependent mechanism involving JNK activation. Cancer Res 2008; 68:7484–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Leon-Annicchiarico CL, Ramirez-Peinado S, Dominguez-Villanueva D, Gonsberg A, Lampidis TJ, Munoz-Pinedo C. ATF4 mediates necrosis induced by glucose deprivation and apoptosis induced by 2-deoxyglucose in the same cells. FEBS J 2015; 282:3647–58 [DOI] [PubMed] [Google Scholar]
- 232.Jiang HY, Wek RC. GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation. Biochem J 2005; 385:371–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Jiang HY, Wek RC. Phosphorylation of the alpha-subunit of the eukaryotic initiation factor-2 (eIF2alpha) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J Biol Chem 2005; 280:14189–202 [DOI] [PubMed] [Google Scholar]
- 234.Rainbolt TK, Atanassova N, Genereux JC, Wiseman RL. Stress-regulated translational attenuation adapts mitochondrial protein import through Tim17A degradation. Cell Metab 2013; 18:908–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 2013; 497:451–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Moullan N, Mouchiroud L, Wang X, Ryu D, Williams EG, Mottis A, Jovaisaite V, Frochaux MV, Quiros PM, Deplancke B, Houtkooper RH, Auwerx J. Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep 2015; 10:1681–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS Genet 2012; 8:e1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Martinez-Reyes I, Sanchez-Arago M, Cuezva JM. AMPK and GCN2-ATF4 signal the repression of mitochondria in Colon cancer cells. Biochem J 2012; 444:249–59 [DOI] [PubMed] [Google Scholar]
- 239.Silva JM, Wong A, Carelli V, Cortopassi GA. Inhibition of mitochondrial function induces an integrated stress response in oligodendroglia. Neurobiol Dis 2009; 34:357–65 [DOI] [PubMed] [Google Scholar]
- 240.Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, Ron D, Wouters BG, Koumenis C. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 2005; 24:3470–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ameri K, Lewis CE, Raida M, Sowter H, Hai T, Harris AL. Anoxic induction of ATF-4 through HIF-1-independent pathways of protein stabilization in human cancer cells. Blood 2004; 103:1876–82 [DOI] [PubMed] [Google Scholar]
- 242.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J 2010; 29:2082–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Dey S, Sayers CM, Verginadis II, Lehman SL, Cheng Y, Cerniglia GJ, Tuttle SW, Feldman MD, Zhang PJ, Fuchs SY, Diehl JA, Koumenis C. ATF4-dependent induction of heme oxygenase 1 prevents anoikis and promotes metastasis. J Clin Invest 2015; 125:2592–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Tameire F, Verginadis II, Leli NM, Polte C, Conn CS, Ojha R, Salas Salinas C, Chinga F, Monroy AM, Fu W, Wang P, Kossenkov A, Ye J, Amaravadi RK, Ignatova Z, Fuchs SY, Diehl JA, Ruggero D, Koumenis C. ATF4 couples MYC-dependent translational activity to bioenergetic demands during tumour progression. Nat Cell Biol 2019; 21:889–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wortel IMN, van der Meer LT, Kilberg MS, van Leeuwen FN. Surviving stress: modulation of ATF4-mediated stress responses in normal and malignant cells. Trends Endocrinol Metab 2017; 28:794–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lo M, Ling V, Wang YZ, Gout PW. The xc- cystine/glutamate antiporter: a mediator of pancreatic cancer growth with a role in drug resistance. Br J Cancer 2008; 99:464–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wang SF, Wung CH, Chen MS, Chen CF, Yin PH, Yeh TS, Chang YL, Chou YC, Hung HH, Lee HC. Activated integrated stress response induced by salubrinal promotes cisplatin resistance in human gastric cancer cells via enhanced xCT expression and glutathione biosynthesis. Int J Mol Sci 2018; 19:3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Timmerman LA, Holton T, Yuneva M, Louie RJ, Padro M, Daemen A, Hu M, Chan DA, Ethier SP, van ‘T Veer LJ, Polyak K, McCormick F, Gray JW. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell 2013; 24:450–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Shin CS, Mishra P, Watrous JD, Carelli V, D’Aurelio M, Jain M, Chan DC. The glutamate/cystine xCT antiporter antagonizes glutamine metabolism and reduces nutrient flexibility. Nat Commun 2017; 8:15074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Goji T, Takahara K, Negishi M, Katoh H. Cystine uptake through the cystine/glutamate antiporter xCT triggers glioblastoma cell death under glucose deprivation. J Biol Chem 2017; 292:19721–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Koppula P, Zhang Y, Zhuang L, Gan B. Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun 2018; 38:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Chen MS, Wang SF, Hsu CY, Yin PH, Yeh TS, Lee HC, Tseng LM. CHAC1 degradation of glutathione enhances cystine-starvation-induced necroptosis and ferroptosis in human triple negative breast cancer cells via the GCN2-eIF2alpha-ATF4 pathway. Oncotarget 2017; 8:114588–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Suomalainen A, Elo JM, Pietilainen KH, Hakonen AH, Sevastianova K, Korpela M, Isohanni P, Marjavaara SK, Tyni T, Kiuru-Enari S, Pihko H, Darin N, Ounap K, Kluijtmans LA, Paetau A, Buzkova J, Bindoff LA, Annunen-Rasila J, Uusimaa J, Rissanen A, Yki-Jarvinen H, Hirano M, Tulinius M, Smeitink J, Tyynismaa H. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol 2011; 10:806–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Liang C, Ahmad K, Sue CM. The broadening spectrum of mitochondrial disease: shifts in the diagnostic paradigm. Biochim Biophys Acta 2014; 1840:1360–7 [DOI] [PubMed] [Google Scholar]
- 255.Forsstrom S, Jackson CB, Carroll CJ, Kuronen M, Pirinen E, Pradhan S, Marmyleva A, Auranen M, Kleine IM, Khan NA, Roivainen A, Marjamaki P, Liljenback H, Wang L, Battersby BJ, Richter U, Velagapudi V, Nikkanen J, Euro L, Suomalainen A. Fibroblast growth factor 21 drives dynamics of local and systemic stress responses in mitochondrial myopathy with mtDNA deletions. Cell Metab 2019; 30:1040–54 e7 [DOI] [PubMed] [Google Scholar]
- 256.Khan NA, Nikkanen J, Yatsuga S, Jackson C, Wang L, Pradhan S, Kivela R, Pessia A, Velagapudi V, Suomalainen A. mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab 2017; 26:419–28 e5 [DOI] [PubMed] [Google Scholar]
- 257.Knott Me Minatta Jn Roulet L, Gueglio G, Pasik L, Ranuncolo SM, Nunez M, Puricelli L, De Lorenzo MS. Circulating fibroblast growth factor 21 (Fgf21) as diagnostic and prognostic biomarker in renal cancer. J Mol Biomark Diagn 2016; 1:015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Kang YE, Kim JT, Lim MA, Oh C, Liu L, Jung SN, Won HR, Lee K, Chang JW, Yi HS, Kim HJ, Ku BJ, Shong M, Koo BS. Association between circulating fibroblast growth factor 21 and aggressiveness in thyroid cancer. Cancers 2019; 11:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Yang C, Lu W, Lin T, You P, Ye M, Huang Y, Jiang X, Wang C, Wang F, Lee MH, Yeung SC, Johnson RL, Wei C, Tsai RY, Frazier ML, McKeehan WL, Luo Y. Activation of liver FGF21 in hepatocarcinogenesis and during hepatic stress. BMC Gastroenterol 2013; 13:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Yatsuga S, Fujita Y, Ishii A, Fukumoto Y, Arahata H, Kakuma T, Kojima T, Ito M, Tanaka M, Saiki R, Koga Y. Growth differentiation factor 15 as a useful biomarker for mitochondrial disorders. Ann Neurol 2015; 78:814–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Fujita Y, Ito M, Kojima T, Yatsuga S, Koga Y, Tanaka M. GDF15 is a novel biomarker to evaluate efficacy of pyruvate therapy for mitochondrial diseases. Mitochondrion 2015; 20:34–42 [DOI] [PubMed] [Google Scholar]
- 262.Emmerson PJ, Duffin KL, Chintharlapalli S, Wu X. GDF15 and growth control. Front Physiol 2018; 9:1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Welsh JB, Sapinoso LM, Kern SG, Brown DA, Liu T, Bauskin AR, Ward RL, Hawkins NJ, Quinn DI, Russell PJ, Sutherland RL, Breit SN, Moskaluk CA, Frierson HF, Jr., Hampton GM. Large-scale delineation of secreted protein biomarkers overexpressed in cancer tissue and serum. Proc Natl Acad Sci U S A 2003; 100:3410–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ishige T, Nishimura M, Satoh M, Fujimoto M, Fukuyo M, Semba T, Kado S, Tsuchida S, Sawai S, Matsushita K, Togawa A, Matsubara H, Kaneda A, Nomura F. Combined secretomics and transcriptomics revealed cancer-derived GDF15 is involved in diffuse-type gastric cancer progression and fibroblast activation. Sci Rep 2016; 6:21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kalli M, Minia A, Pliaka V, Fotis C, Alexopoulos LG, Stylianopoulos T. Solid stress-induced migration is mediated by GDF15 through akt pathway activation in pancreatic cancer cells. Sci Rep 2019; 9:978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Li S, Ma YM, Zheng PS, Zhang P. GDF15 promotes the proliferation of cervical cancer cells by phosphorylating AKT1 and Erk1/2 through the receptor ErbB2. J Exp Clin Cancer Res 2018; 37:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Windrichova J, Fuchsova R, Kucera R, Topolcan O, Fiala O, Finek J, Slipkova D. MIC1/GDF15 as a bone metastatic disease biomarker. Anticancer Res 2017; 37:1501–5 [DOI] [PubMed] [Google Scholar]
- 268.Duan L, Pang HL, Chen WJ, Shen WW, Cao PP, Wang SM, Liu LL, Zhang HL. The role of GDF15 in bone metastasis of lung adenocarcinoma cells. Oncol Rep 2019; 41:2379–88 [DOI] [PubMed] [Google Scholar]
- 269.Lu X, He X, Su J, Wang J, Liu X, Xu K, De W, Zhang E, Guo R, Shi YE. EZH2-Mediated epigenetic suppression of GDF15 predicts a poor prognosis and regulates cell proliferation in Non-Small-Cell lung cancer. Mol Ther Nucleic Acids 2018; 12:309–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017; 45:W98–W102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018; 8:9227. [DOI] [PMC free article] [PubMed] [Google Scholar]