Abstract
Tumor microenvironment is a complex niche consisting of cancer cells and stromal cells in a network of extracellular matrix proteins and various soluble factors. Dynamic interactions among cellular and non-cellular components of the tumor microenvironment regulate tumor initiation and progression. Fibroblasts are the most abundant stromal cell type and dynamically interact with cancer cells both in primary tumors and in metastases. Cancer cells activate resident fibroblasts to produce and secrete soluble signaling molecules that support proliferation, migration, matrix invasion, and drug resistance of cancer cell and tumor angiogenesis. In recent years, various forms of three-dimensional tumor models have been developed to study tumor–stromal interactions and to identify anti-cancer drugs that block these interactions. There is currently a technological gap in development of tumor models that are physiologically relevant, scalable, and allow convenient, on-demand addition of desired components of the tumor microenvironment. In this review, we discuss three studies from our group that focus on developing bioengineered models to study tumor-stromal signaling. We will present these studies chronologically and based on their increasing complexity. We will discuss the validation of the models using a CXCL12-CXCR4 chemokine-receptor signaling present among activated fibroblasts and breast cancer cells in solid tumors, highlight the advantages and shortcomings of the models, and conclude with our perspectives on their applications.
Impact statement
Tumor stroma plays an important role in progression of cancers to a fatal metastatic disease. Modern treatment strategies are considering targeting tumor stroma to improve outcomes for cancer patients. A current challenge to develop stroma-targeting therapeutics is the lack of preclinical physiologic tumor models. Animal models widely used in cancer research lack human stroma and are not amenable to screening of chemical compounds for cancer drug discovery. In this review, we outline in vitro three-dimensional tumor models that we have developed to study the interactions among cancer cells and stromal cells. We describe development of the tumor models in a modular fashion, from a spheroid model to a sophisticated organotypic model, and discuss the importance of using correct physiologic models to recapitulate tumor-stromal signaling. These biomimetic tumor models will facilitate understanding of tumor-stromal signaling biology and provide a scalable approach for testing and discovery of cancer drugs.
Keywords: Tumor microenvironment, tumor model, cancer-associated fibroblasts, breast cancer
Introduction
Tumor microenvironment
Solid tumors consist of a mass of neoplastic cells surrounded by stromal cells including fibroblasts, immune cells, and endothelial cells embedded in an extracellular matrix (ECM).1,2 These cohorts of cells, their secreted factors, and the ECM are referred to as the tumor microenvironment (TME).3 Contact-mediated or soluble factor-dependent signaling among the cellular and non-cellular components of the TME lead to mechanical and biochemical interactions among these components. While interactions of epithelial cells with their environment in normal tissues are critical to homeostasis, interactions of epithelial cancer cells with the TME facilitate tumor growth and subsequent processes that very often lead to metastatic dissemination of cancer cells. Due to the role of interactions among various components of the TME in tumor growth and progression, the TME is considered a key regulator of tumorigenesis.4 Studies show that during tumor progression, genetic profiles of both tumor and stromal cells change.5–7 These genetic instabilities can drive abnormal paracrine interactions between tumor and stromal cells, causing tumor cells to proliferate, invade the stroma, and metastasize.
Stromal cells in solid tumors
During tumor progression, cancer cells recruit stromal cells such as fibroblasts, immune cells, pericytes, mesenchymal stem cells, and endothelial cells into the TME and increase its cellular heterogeneity.8 These recruited stromal cells change their phenotypes to provide a tumorigenic niche for cancer progression. For example, macrophages convert into tumor-associated macrophages (TAMs) that promote tumor cell dissemination through angiogenesis, invasion, and metastasis.9,10 It is important to note that TAMs also have tumor killing properties through presenting tumor antigens to cytotoxic T-cells.11 Endothelial cells in the TME also transform into fibroblast-like cells with increased expression of alpha smooth muscle actin (αSMA).12 Transformed endothelial cells may express a normal endothelial cell marker, CD31, but contain genetic abnormalities with aneuploid cells and abnormal centrosomes.13 These cells have been found to release from primary tumors and co-migrate with tumor cells in the blood stream to distal sites.14,15
Among different stromal cells, fibroblasts play pivotal roles in initiation and progression of epithelial tumors. In normal tissues, fibroblasts are present in the stroma, deposit ECM proteins such as collagen and fibronectin,16 and degrade the ECM by producing matrix metalloproteinases (MMPs).17 Balanced remodeling of the ECM is necessary for normal functioning of tissues, whereas disruption in the remodeling process is implicated in pathologies such as fibrosis and cancer.18 Fibroblasts are also important in wound healing. Fibroblasts migrate into the lesions and accelerate ECM deposition by transforming into myofibroblasts. Myofibroblasts are activated fibroblasts with a unique spindle shape and express αSMA and fibroblast activation protein (FAP).19,20 After the wound healing is complete, the myofibroblasts may revert back to a normal phenotype or undergo apoptosis.21 In tumors, normal fibroblasts are recruited in large numbers to convert the TME into a reactive stroma.22 Fibroblasts are transformed into an activated phenotype by various stimuli such as transforming growth factor-β (TGF-β) and epidermal growth factor (EGF). The activated fibroblasts produce significantly large amounts of ECM proteins that result in the stiffening of the tumor stroma. In the past, ECM was considered to be a passive component of the TME. However, research in the past two decades has shown that ECM is a dynamic component of the TME and provides biochemical and biophysical signaling cues to cancer cells to facilitate tumor progression.23 Various studies have shown that a stiffer ECM correlates with invasiveness and malignant behavior of cancer cells.24–26 During cancer progression, ECM undergoes continuous remodeling that results in changes in its structural and mechanical properties. This reorganization of the ECM promotes tumor progression by disrupting cell polarity, altering integrin adhesions, and inducing focal adhesions.24,27,28 Tumor stiffness is considered a prognostic factor and stiffer tumors often have a poorer prognosis.29,30 Activated fibroblasts in the TME are called cancer-associated fibroblasts (CAFs). Unlike in normal processes, activated fibroblasts in the TME do not revert to a normal phenotype nor do they undergo apoptosis.31
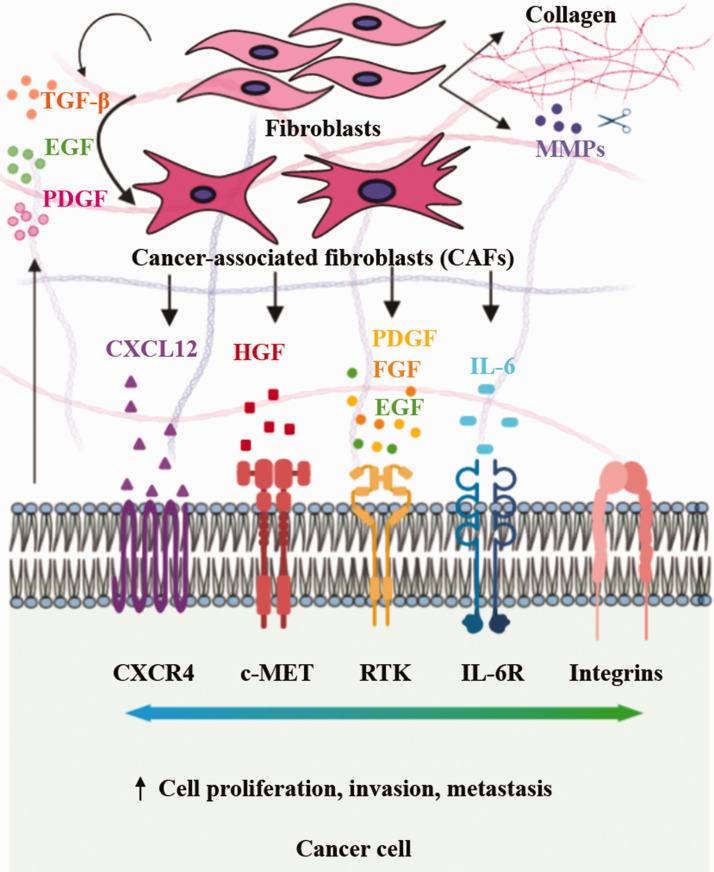
CAFs are the most abundant stromal cells in solid tumors and associate with cancer cells during tumor initiation, progression, invasion, and metastasis.32 Immunohistochemical analysis of human tumors showed that abundance of CAFs correlate with a poor prognosis.33 CAFs are derived from various cell types such as resident fibroblasts, vascular smooth muscle cells, endothelial cells, and pericytes, and hence are heterogeneous in origin.34,35 CAFs support tumor growth through soluble signaling of various CAFs-derived factors such as TGF-β,36 hepatocyte growth factor (HGF),37,38 fibroblast growth factor (FGF),39 interleukin-6 (IL-6),40,41 stromal derived factor-1α (SDF-1α or CXCL12),42,43 and epidermal growth factor (EGF),44 as highlighted in Figure 1. This has prompted a therapeutic strategy using antagonists of the corresponding receptors, i.e. EGFR,45,46 c-MET,47 FGFR,48 VEGFR,49 and CXCR4,50,51 expressed on cancer cells to inhibit the corresponding signaling pathways in cancer cells.
Figure 1.
Cancer cells secrete soluble factors to activate fibroblasts into cancer-associated fibroblasts (CAFs), which in turn remodel the tumor microenvironment and promote tumor growth, invasion, and metastasis. (A color version of this figure is available in the online journal.)
Models of tumor–stromal interactions
Considering the importance of stromal cells in tumor progression, a mechanistic understanding of interactions among components of the TME is critical to develop novel therapeutics to target tumor–stromal interactions. Typically, animal models and cell cultures are used to study signaling among cancer cells and the tumor stroma. Animal models, in particular mouse models of human cancers, have been a useful research tool due to their anatomical and physiological similarities with humans. Mouse models have played a significant role in understanding mechanisms of tumor development, angiogenesis, invasion, and metastasis, and identifying novel biomarkers for cancer drug discovery. Nevertheless, limitations of mouse models including significant failure rates in developing tumors in mice, differences in the tumor stroma and immune system between mouse and human, difficulty of handling a large number of animals and analysis of responses to treatments, low throughput for drug screening applications, expense, and ethical issues remain major obstacles to accelerate new discoveries. In vitro co-cultures of cancer cells and stromal cells such as fibroblasts in a monolayer have also been widely used to study stroma effect on phenotypes and functions of cancer cells. Monolayer cultures (2D) are convenient to use, adaptable with robotic instruments used in the pharmaceutical industry, allow high throughput screening of chemical compounds, and enable straightforward analysis of responses of cells to drug compounds. However, they lack the three-dimensional (3D) geometry and architecture of human tumors. To address the need for in vitro tumor models, 3D co-cultures of cancer cells and fibroblasts as spheroids have been used. Studies show that spheroids reproduce certain aspects of solid tumors including close cell–cell contacts, gradients of nutrients and oxygen that may lead to hypoxia and necrosis, expression of pro-angiogenic proteins, and upregulation of ABC transporter efflux pumps implicated in multidrug resistance (MDR) of cancers.52,53 Despite these advantages, co-culture spheroids do not precisely mimic the architecture of solid tumors in terms of spatial distribution of cancer cells, stromal cells, and the ECM.34 This emphasizes a need for more advanced and sophisticated in vitro tumor models to study tumor–stromal interactions and testing the efficacy of therapeutic compounds. In addition to resembling the tumor architecture, the ease-of-use and scalability are other important features that in vitro models should offer to enable high throughput testing of drug libraries or arrays of combinations of drugs, and molecular analysis of drug responses of cells.
Scope of this review
In this review, we will focus on engineered 3D tumor models to study the interactions of fibroblasts and triple negative breast cancer (TNBC) cells. We will mainly review research from our laboratory on developing in vitro models of increasing complexity, i.e. from intermixed co-culture spheroids of cancer and fibroblast cells to an organotypic model consisting of a cancer cell mass, fibroblast cells, and ECM. To establish effects of fibroblasts on cancer cells throughout these studies, we used a CXCL12-CXCR4 chemokine-receptor signaling axis. CXCL12 is a major paracrine signaling molecule produced by activated fibroblasts, allowing us to model activity of CAFs in the TME. CXCL12 signals through its cognate CXCR4 receptor, which is often overexpressed on TNBC cells. The focus on TNBC in our research is due to its aggressive biology and higher mortality rate than other subtypes of breast cancer. Due to the lack of estrogen and progesterone receptors and HER2 amplification, commonly used hormonal and targeted therapies with other breast cancers are not feasible with TNBC. Availability of physiologic in vitro tumor models that help elucidate mechanisms of tumor–stromal interactions is critical to develop novel therapeutics especially for cancers such as TNBC that currently lack effective treatments.
Free-floating co-culture spheroids
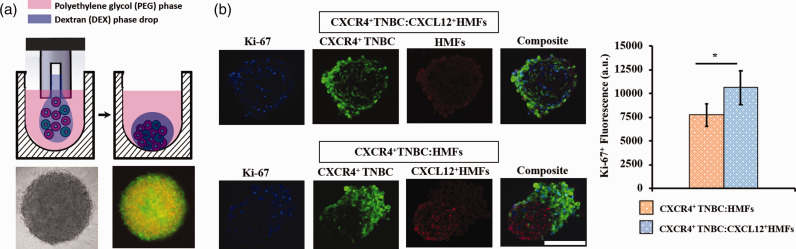
To overcome the drawbacks of monolayer cell cultures, Ham et al.54 developed a tumor spheroid printing technology to model cancer cell–stromal cell interactions. The authors formed an array of intermixed co-culture spheroids of TNBC and activated fibroblasts to examine the role of chemokine-receptor signaling on TNBC cell growth. Tumor spheroids were developed using an aqueous two-phase system (ATPS) technology with polyethylene glycol (PEG) and dextran (DEX) as phase-forming polymers (Figure 2(a)).55–58 Both polymers are soluble in water but their aqueous solutions phase-separate when used above specific concentrations determined by a unique phase diagram.59 Briefly, a nanoliter-volume DEX phase drop containing intermixed TNBC cells and CXCL12-secreting fibroblasts was dispensed into the immersion PEG phase solution in each well of a microwell plate. Due to an ultralow interfacial tension between the aqueous PEG and DEX phases,60 cells remained partitioned to the denser aqueous DEX phase drop that resided at the bottom of each well.61 Cells aggregated to form a co-culture spheroid inside each drop within 24 h. This technology was previously adapted to robotic liquid handling to enable forming large numbers of consistently sized spheroids that reproduced several key biological properties of solid tumors including non-uniform spatial distribution of proliferative cells within spheroids and a cell density-dependent hypoxic core.62,63 The co-culture spheroids contained a 2:1 ratio of fibroblast:TNBC cells to mimic high stromal cell content present in advanced human breast tumors.64–67
Figure 2.
(a) Free-floating co-culture spheroid of cancer cells and fibroblasts generated using the ATPS technology. (b) Proliferation of TNBC cells in co-culture spheroids containing normal human mammary fibroblasts (HMF) or activated human mammary fibroblasts (CXCL12+HMFs) by quantifying Ki-67+ TNBC cells. *p < 0.05. Panels a and b are reproduced with permission from Oncotarget, 2018;9:249. (A color version of this figure is available in the online journal.)
Next, they examined effects of CXCL12-CXCR4 and CXCL12-CXCR7 chemokine-receptor signaling between fibroblasts and TNBC cells on growth of spheroids characterized through measurements of biochemical activity of cells. Using engineered cancer cells that expressed or lacked either receptor, engineered fibroblasts that secreted CXCL12, and normal human mammary fibroblasts, this systematic study used eight different co-culture spheroid models. Results showed that the co-culture spheroids with active CXCL12-CXCR4 signaling had the greatest metabolic activity over time. This was consistent with studies from mouse models that showed the impact of the CXCL12-CXCR4 axis on proliferation of breast cancer cells.68–71 Overexpression of CXCR7 receptors did not confer proliferative advantage to TNBC cells.
To test the hypothesis that CXCL12-CXCR4 signaling increased the proliferative activity of the spheroids, Ham et al. performed inhibition and stimulation experiments. Treating spheroids with AMD3100, which is an antagonist of CXCR4 receptors to block signaling of CXCL12 chemokine to TNBC cells, significantly reduced the proliferation of spheroids. Additionally, treating the co-culture spheroids of CXCR4+ TNBC cells and normal human mammary fibroblasts lacking CXCL12 production with conditioned medium of CXCL12-secreting fibroblasts stimulated the proliferation of the spheroids to the level of co-culture spheroids containing active CXCL12-CXCR4 signaling. These experiments validated that CXCL12-CXCR4 axis is responsible for the elevated proliferation of TNBC cells in spheroids. Ham et al. also confirmed higher cell proliferation of spheroids by immunohistochemistry (IHC) staining of the cells with a Ki-67 proliferative cell marker (Figure 2(b)). Again, the co-culture spheroids containing active CXCL12-CXCR4 axis showed statistically significantly higher proliferation than the spheroids without this signaling. In separate experiments, measuring the fluorescent signal of green fluorescent protein (GFP)-expressing TNBC cells in the co-culture spheroids confirmed that proliferation of cancer cells in the spheroids significantly increases due to this signaling. Molecular analysis using both Western blotting and IHC showed significantly higher activities of p-ERK and p-AKT in co-culture spheroids with CXCL12-CXCR4 signaling than spheroids lacking this chemokine-receptor signaling. In addition, this signaling conferred resistance to paclitaxel, which is a clinically used drug for TNBC patients. Paclitaxel-treated co-culture spheroids had higher activity of MAPK/ERK and PI3K/AKT pathways, indicating that activation of these oncogenic pathways due to interactions among cancer cells and activated fibroblasts renders cancer cells chemotherapy resistant. This was consistent with studies that showed increased activation of MAPK and PI3K/AKT pathways in paclitaxel-treated breast cancers.72–74 Additionally, inhibiting MAPK/ERK and PI3K/AKT pathways using specific molecular inhibitors significantly blocked TNBC cell proliferation and compromised their viability.75
Overall, Ham et al. generated scalable intermixed co-culture spheroids of cancer cells and fibroblasts to study tumor–stromal interactions through a specific signaling axis. This study indicated the importance of understanding molecular effects of cancer cell–stromal cell interactions and the feasibility of targeting these interactions to prevent growth of cancer cells and using activated oncogenic pathways as therapy targets.
ECM-embedded co-culture spheroids
Although the intermixed co-culture spheroids helped elucidate the role of activated fibroblasts on proliferation and drug resistance of TNBC cells,54 this model lacked ECM and the resulting cell-ECM interactions implicated in tumor progression.76 In a recent study, Plaster et al.77 further developed this model by embedding the intermixed co-culture spheroids in a type I collagen matrix to study the interactions between TNBC cells and fibroblasts in presence of the ECM. This study also used CXCL12-CXCR4 signaling to model tumor–stromal interactions and showed that it significantly enhances proliferation of cancer cells while blocking it normalizes the effect. The CXCL12-CXCR4 signaling promoted matrix invasion of TNBC cells that showed an elongated, mesenchymal morphology. Additionally, blocking this signaling pathway inhibited matrix invasion of TNBC cells. This study developed a more complex tumor model than that developed by Ham et al. to study the interactions among components of the TME, i.e. cancer cells and fibroblasts embedded in a collagen ECM.
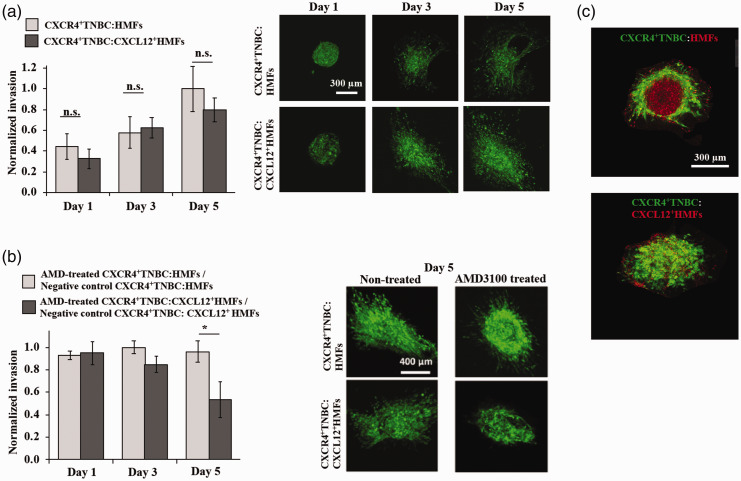
They prepared co-culture spheroids of TNBC cells with either normal fibroblasts or CXCL12-secreting fibroblasts using the ATPS technology and encapsulated the spheroids in a collagen hydrogel with a stiffness that approximated that of human breast tumors. Unexpectedly, no significant difference in 3D spreading of cancer cells in the collagen matrix between the co-culture models containing normal mammary fibroblasts and CXCL12-secreting mammary fibroblasts was observed (Figure 3(a)). To understand this phenomenon, both ECM-embedded co-culture models were treated with a CXCR4 antagonist, AMD3100. This treatment significantly reduced the matrix invasion of cancer cells only in the co-culture model containing CXCL12-producing fibroblasts (Figure 3(b)). The treatment did not inhibit the spreading of cancer cells into the matrix in the co-culture model containing normal mammary fibroblasts. Fluorescence microscopy of normal fibroblasts-TNBC cell co-culture spheroids without embedding them in ECM showed that the normal fibroblasts tend to separate out from the TNBC cells, and this was attributed to the incompatibility of cell adhesion molecules of the two cell types (Figure 3(c)). Therefore, the dispersion of TNBC cells from the co-culture with normal fibroblasts into the ECM was not due to invasiveness of the TNBC cells. On the other hand, imaging showed that CXCL12-producing fibroblasts remained mixed with cancer cells in the co-culture spheroids and during matrix invasion of the cells (Figure 3(c)). Overall, this study demonstrated the role of signaling of a fibroblast-derived chemokine receptor and TNBC cells on invasiveness of cancer cells. In addition, it underscored that intermixed co-cultures are not a physiologic model and their use may result in misleading experimental results.
Figure 3.
(a) Invasion of TNBC cells from co-culture spheroids into the collagen matrix. (b) Matrix invasion of TNBC cells inhibited by using AMD3100. *p < 0.05. (c) Normal human mammary fibroblasts (red) segregate from TNBC cells (green), while CXCL12-secreting fibroblasts (red) remain mixed with TNBC cells. Panels a, b, and c are reproduced with permission from Advanced Therapeutics, 2019; 2:1900121. (A color version of this figure is available in the online journal.)
Organotypic tumor model
Intermixed co-culture spheroids embedded in an ECM do not truly represent the spatial distribution of the components of a solid tumor. In native tumors, stromal cells such as fibroblasts are dispersed within the matrix bordering the mass of cancer cells,34 and they communicate with cancer cells primarily through soluble factors signaling. To overcome this technological barrier and develop a more realistic tumor model, Singh et al.78 robotically encapsulated a breast cancer spheroid within a collagen matrix containing dispersed fibroblasts. They also used the CXCL12-CXCR4 signaling to validate the tumor model.
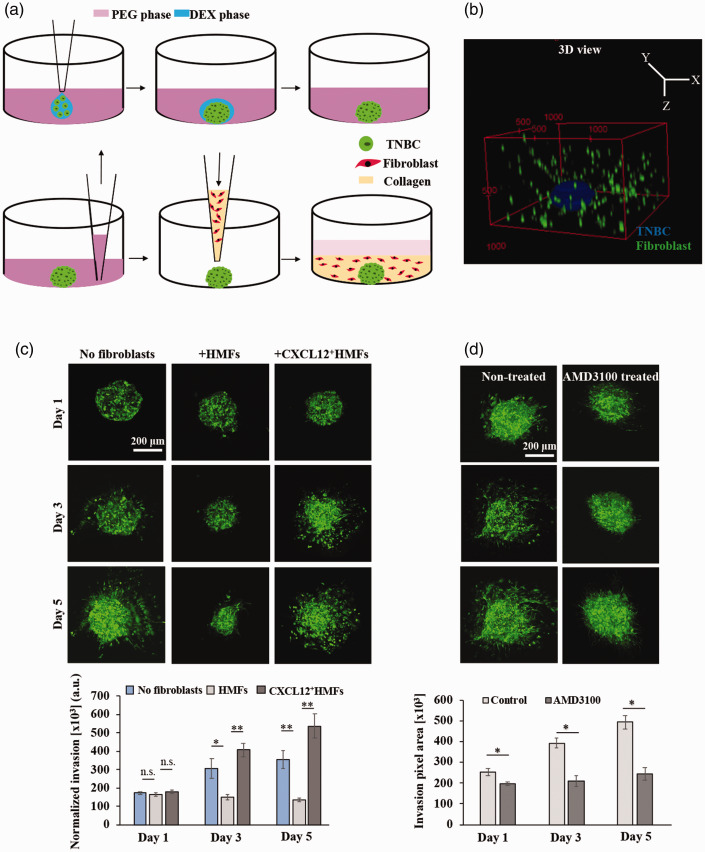
Singh et al. generated the high throughput organotypic tumor model in two convenient steps: first forming TNBC spheroid using the ATPS technology followed by overlaying the spheroid with a type I collagen solution containing dispersed fibroblasts (Figure 4). The entire operation was performed at low temperatures to avoid premature collagen gelation.79 The elastic modulus of the matrix was adjusted to ∼2.53 kPa to mimic the stiffness of human breast tumors and a 2:1 ratio of fibroblasts:cancer cells was used to mimic cellular composition in advanced breast tumors that are stroma-rich. The organotypic cultures detached from the walls of the microwells and shrank over time. Excluding either fibroblasts or TNBC cells from the cultures established that dispersed fibroblasts were responsible for shrinking of collagen, while TNBC spheroids were not able to induce matrix contraction at all. Cultures with only fibroblasts, either normal or CXCL12-secreting fibroblasts, dispersed in collagen were prepared to study collagen contraction, considering effects of elastic modulus of collagen, fibroblast cell density, and fibroblast type. Keeping the elastic modulus of collagen hydrogel and fibroblast cell density constant, activated fibroblasts contracted the collagen matrix significantly more than normal fibroblasts did. Stiffer collagen matrices showed less contraction at a constant cell density, and larger cell densities caused a greater matrix contraction at a constant matrix elastic modulus. Whole gel immunofluorescence of fibroblasts showed that CXCL12-secreting fibroblasts displayed spindle-like and elongated morphology resembling myofibroblasts and cancer-associated fibroblasts. Molecular analysis showed that higher collagen contraction by CXCL12-secreting fibroblasts was due to the activated RhoA/ROCK/myosin light chain-2 signaling pathway in these cells.
Figure 4.
(a) Schematics of formation of organotypic tumor model in two convenient steps: first forming spheroids using the ATPS technology and then embedding it in a collagen hydrogel containing dispersed fibroblasts. (b) 3D reconstructed confocal image of the tumor model. Note that collagen is not shown. (c) Confocal z-projected images of TNBC cells and quantified matrix invasion of TNBC cells in the tumor models with and without fibroblasts. (d) Confocal images of TNBC cells with and without AMD3100 treatment and the quantified invasion results. *p < 0.05, **p < 0.001. This figure is reproduced with permission from Biomaterials, 2020; 238:119853. (A color version of this figure is available in the online journal.)
Next, Singh et al. generated organotypic cultures by embedding TNBC spheroids in a collagen matrix containing dispersed fibroblasts or without fibroblasts to study the role of fibroblasts on cancer cell invasiveness. The matrix invasion of TNBC cells depended on the type of fibroblasts. Confocal imaging showed that normal fibroblasts suppressed matrix invasion of cancer cells, while CXCL12-secreting fibroblasts significantly increased matrix invasion of TNBC cells (Figure 4(c)). Molecular analysis showed a significantly higher activity of p-ERK1/2 in the culture with CXCL12-secreting fibroblasts than that with normal fibroblasts. Treating the cultures containing activated fibroblasts with AMD3100 inhibited matrix invasion of TNBC cells through downregulating ERK1/2 activity (Figure 4(d)). This indicated that CXCL12-CXCR4 signaling between fibroblasts and TNBC cells causes cancer cell invasiveness and that blocking this pathway is a potential strategy against tumor–stromal signaling to prevent matrix invasion of cancer cells.
Overall, Singh et al. developed a scalable organotypic breast tumor model that mimics the architecture of solid tumors in terms of spatial distribution of tumor and stromal cells and mechanical properties of breast tumors. The study clearly demonstrated the role of activated fibroblasts in promoting cancer cell invasiveness. Unlike the study by Plaster et al. that used collagen-embedded intermixed co-cultures of fibroblasts and TNBC cells, this organotypic model clearly distinguished the invasion-promoting role of activated fibroblasts compared to normal fibroblasts, indicating the importance of developing and using physiologic models to elucidate effects of stroma on functional characteristics of cancer cells.
Perspectives
Due to the major role of cell-based models in preclinical cancer drug discovery, incorporating physiologically relevant tumor models that replicate the complexity of solid tumors is expected to expedite the development of effective therapies for cancer. In the bioengineering community, there has recently been major efforts to develop 3D tumor models that recapitulate certain aspects of tumor–stromal interactions by incorporating key cellular and non-cellular elements of the TME. Fibroblasts are the most prominent cell type in solid tumors and play significant roles in therapeutic responses of tumors. Therefore, to understand mechanisms of signaling between fibroblasts and cancer cells and develop therapeutics that target these interactions, it is essential to use tumor models that contain both fibroblasts and cancer cells and replicate the spatial distribution of these cells in tumors. Research, including from our group, has shown that including cells in a physiologic context is important to correctly model intercellular interactions in the TME and to screen for effective therapeutic compounds. Here, we reviewed three separate studies from our group that were designed to study interactions among breast cancer cells and normal or activated fibroblasts in models of increasing complexity.
Ham et al. generated the simplest 3D model that included intermixed co-culture spheroids of TNBC cells and fibroblasts. Tumor–stromal interactions via CXCL12-CXCR4 axis enhanced the proliferation of breast cancer cells. Although fibroblasts in the co-culture spheroids produced collagen and allowed limited interactions of cancer cells and fibroblasts with the ECM, a shortcoming of this model was its lack of ECM containing cells as in the native TME. To address this problem, Plaster et al. embedded the intermixed co-culture spheroids in a collagen matrix. In addition to tumor–stromal interactions, this tumor model allowed drug testing to inhibit cancer cell proliferation and matrix invasion that are two important processes in tumor progression. The shortcoming of this study was the relative positioning of fibroblasts and cancer cells. Singh et al. addressed this problem and further developed the tumor model to replicate the spatial distribution of fibroblasts and cancer cells in solid tumors. This organotypic tumor model enabled evaluating biological signaling between activated fibroblasts and cancer cells on processes such as cancer cell invasion that facilitates tumor metastasis. This model is convenient to use and scalable for high throughput drug testing against tumor–stromal interactions. This model enables modular addition of different stromal cells such as immune cells for cancer immunotherapy applications and endothelial cells for tumor angiogenesis. From a viewpoint of developing the models and readouts, this may require optimization and validation of techniques that work in a simpler system, but they have not been used in more complex systems. For example, we previously showed that biochemical assays of metabolic activity originally developed for monolayer cultures had to be optimized for use with 3D cultures.56 Or, while measuring the fluorescence signal of fluorescently labeled cancer cells in free-floating spheroids using standard plate readers reliably reflects the cell population, this approach fails when spheroids are embedded in the ECM due to the interference of the protein network with the light beams. As such, other methods such as confocal imaging or flow cytometry may be required for quantitative analysis.
Due to the heterogeneity of tumors and significant differences in therapy responses among patients of the same type of cancer, therapies in future will shift toward personalized medicine where patient-specific tumor models containing patient-derived cancer and stromal cells are developed. Although maintaining primary tumor cells in vitro has proved challenging, the use of techniques such as conditional reprogramming of primary cells will allow expanding them in sufficient numbers that accommodate tumor models in high throughput for testing the efficacy of arrays of cancer drugs and identify effective therapies for specific patients.80 The use of analytical techniques such as next generation sequencing will help identify activated signaling pathways to target therapeutically in the tumor models. Availability of such information will aid clinicians with selecting treatments that are tailored toward each patient. We envision that incorporating bioengineered tumor models of patients in the pipeline will dramatically improve personalized cancer therapies in future. Increasing investments by federal funding agencies and private enterprises have accelerated new scientific discoveries to help realize this important goal.
Authors’ contributions
All authors participated in the design and writing of the manuscript.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
This work was supported by grants CA216225549 and CA216413 from National Institutes of Health and 1801591 from National Science Foundation.
ORCID iD
Hossein Tavana https://orcid.org/0000-0003-3872-1869
References
- 1.Balkwill FR, Capasso M, Hagemann T. The tumor microenvironment at a glance. J Cell Sci 2012; 125:5591–6 [DOI] [PubMed] [Google Scholar]
- 2.Ham SL, Joshi R, Thakuri PS, Tavana H. Liquid-based three-dimensional tumor models for cancer research and drug discovery. Exp Biol Med 2016; 241:939–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thakuri PS, Liu C, Luker GD, Tavana H. Biomaterials-based approaches to tumor spheroid and organoid modeling. Adv Healthc Mater 2018; 7:e1700980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646–74 [DOI] [PubMed] [Google Scholar]
- 5.Finak G, Sadekova S, Pepin F, Hallett M, Meterissian S, Halwani F, Khetani K, Souleimanova M, Zabolotny B, Omeroglu A, Park M. Gene expression signatures of morphologically normal breast tissue identify basal-like tumors. Breast Cancer Res 2006; 8:R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma X-J, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res 2009; 11:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polyak K, Hu M. Molecular chacarterization of the tumor microenvironment in breast cancer. Eur J Cancer 2008; 44:2760–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong Y. Hematopoietic stem cell-derived adipocytes and fibroblasts in the tumor microenvironment. World J Stem Cells 2015; 7:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell 2006; 124:263–6 [DOI] [PubMed] [Google Scholar]
- 10.Linde N, Casanova-Acebes M, Sosa MS, Mortha A, Rahman A, Farias E, Harper K, Tardio E, Reyes Torres I, Jones J, Condeelis J, Merad M, Aguirre-Ghiso JA. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat Commun 2018; 9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leek RD, Harris AL. Tumor-associated macrophages in breast cancer. J Mammary Gland Biol Neoplasia 2002; 7:177–89 [DOI] [PubMed] [Google Scholar]
- 12.Bussard KM, Mutkus L, Stumpf K, Gomez-Manzano C, Marini FC. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res 2016; 18:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hida K, Hida Y, Amin DN, Flint AF, Panigrahy D, Morton CC, Klagsbrun M. Tumor-associated endothelial cells with cytogenetic abnormalities. Cancer Res 2004; 64:8249–55 [DOI] [PubMed] [Google Scholar]
- 14.Beerepoot LV, Mehra N, Vermaat JSP, Zonnenberg BA, Gebbink M, Voest EE. Increased levels of viable circulating endothelial cells are an indicator of progressive disease in cancer patients. Ann Oncol 2004; 15:139–45 [DOI] [PubMed] [Google Scholar]
- 15.Yadav A, Kumar B, Yu J-G, Old M, Teknos TN, Kumar P. Tumor-associated endothelial cells promote tumor metastasis by chaperoning circulating tumor cells and protecting them from anoikis. PLoS One 2015; 10:e0141602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grinnell F, Billingham RE, Burgess L. Distribution of fibronectin during wound healing in vivo. J Invest Dermatol 1981; 76:181–9 [DOI] [PubMed] [Google Scholar]
- 17.Simian M, Hirai Y, Navre M, Werb Z, Lochter A, Bissell MJ. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development 2001; 128:3117–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oxford JT, Reeck JC, Hardy MJ. Extracellular matrix in development and disease. Int J Mol Sci 2019; 20:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garin-Chesa P, Old LJ, Rettig WJ. Cell surface glycoprotein of reactive stromal fibroblasts as a potential antibody target in human epithelial cancers. Proc Natl Acad Sci U S A 1990; 87:7235–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B, Wang J. Fibroblasts and myofibroblasts in wound healing: force generation and measurement. J Tissue Viability 2011; 20:108–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xing F, Saidou J, Watabe K. Cancer associated fibroblasts (CAFs) in tumor microenvironment. Front Biosci 2010; 15:166–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flier JS, Underhill LH, Dvorak HF. Tumors: wounds that do not heal. N Engl J Med 1986; 315:1650–9 [DOI] [PubMed] [Google Scholar]
- 23.Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol 2010; 22:697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell 2005; 8:241–54 [DOI] [PubMed] [Google Scholar]
- 25.Provenzano PP, Inman DR, Eliceiri KW, Keely PJ. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene 2009; 28:4326–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer 2009; 9:108–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang M, Yuan J, Peng C, Li Y. Collagen as a double-edged sword in tumor progression. Tumour Biol 2014; 35:2871–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paszek MJ, Weaver VM. The tension mounts: mechanics meets morphogenesis and malignancy. J Mammary Gland Biol Neoplasia 2004; 9:325–42 [DOI] [PubMed] [Google Scholar]
- 29.Boyd NF, Li Q, Melnichouk O, Huszti E, Martin LJ, Gunasekara A, Mawdsley G, Yaffe MJ, Minkin S. Evidence that breast tissue stiffness is associated with risk of breast cancer. PLoS One 2014; 9:e100937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayashi M, Yamamoto Y, Ibusuki M, Fujiwara S, Yamamoto S, Tomita S, Nakano M, Murakami K, Iyama KI, Iwase H. Evaluation of tumor stiffness by elastography is predictive for pathologic complete response to neoadjuvant chemotherapy in patients with breast cancer. Ann Surg Oncol 2012; 19:3042–9 [DOI] [PubMed] [Google Scholar]
- 31.Littlepage LE, Egeblad M, Werb Z. Coevolution of cancer and stromal cellular responses. Cancer Cell 2005; 7:499–500 [DOI] [PubMed] [Google Scholar]
- 32.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature 2004; 432:3323–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amornsupak K, Jamjuntra P, Warnnissorn M, O-Charoenrat P, Sa-Nguanraksa D, Thuwajit P, Eccles SA, Thuwajit C. High ASMA+ fibroblasts and low cytoplasmic HMGB1+ breast cancer cells predict poor prognosis. Clin Breast Cancer 2017; 17:441–52.e2 [DOI] [PubMed] [Google Scholar]
- 34.Ishii G, Ochiai A, Neri S. Phenotypic and functional heterogeneity of cancer-associated fibroblast within the tumor microenvironment. Adv Drug Deliv Rev 2016; 99:186–96 [DOI] [PubMed] [Google Scholar]
- 35.Rønnov-Jessen L, Petersen OW, Koteliansky VE, Bissell MJ. The origin of the myofibroblasts in breast cancer: recapitulation of tumor environment in culture unravels diversity and implicates converted fibroblasts and recruited smooth muscle cells. J Clin Invest 1995; 95:859–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen WJ, Ho CC, Chang YL, Chen HY, Lin CA, Ling TY, Yu SL, Yuan SS, Louisa Chen YJ, Lin CY, Pan SH, Elizabeth Chou HY, Chang GC, Chu WC, Lee YM, Lee JY, Lee PJ, Li KC, Chen HW, Yang PC. Cancer-associated fibroblasts regulate the plasticity of lung cancer stemness via paracrine signalling. Nat Commun 2014; 5:3472. [DOI] [PubMed] [Google Scholar]
- 37.Tyan SW, Kuo WH, Huang CK, Pan CC, Shew JY, Chang KJ, Lee E, Lee WH. Breast cancer cells induce cancer-associated fibroblasts to secrete hepatocyte growth factor to enhance breast tumorigenesis. PLoS One 2011; 6:e15313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X, Chen X, Zhou Q, Li P, Yu B, Li J, Qu Y, Yan J, Yu Y, Yan M, Zhu Z, Liu B, Su L. Hepatocyte growth factor activates tumor stromal fibroblasts to promote tumorigenesis in gastric cancer. Cancer Lett 2013; 335:128–35 [DOI] [PubMed] [Google Scholar]
- 39.Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr Relat Cancer 2000; 7:165–97 [DOI] [PubMed] [Google Scholar]
- 40.Sun X, Mao Y, Wang J, Zu L, Hao M, Cheng G, Qu Q, Cui D, Keller ET, Chen X, Shen K, Wang J. IL-6 secreted by cancer-associated fibroblasts induces tamoxifen resistance in luminal breast cancer. Oncogene 2014; 33:4450. [DOI] [PubMed] [Google Scholar]
- 41.Wu X, Tao P, Zhou Q, Li J, Yu Z, Wang X, Li J, Li C, Yan M, Zhu Z, Liu B, Su L. IL-6 secreted by cancer-associated fibroblasts promotes epithelial-mesenchymal transition and metastasis of gastric cancer via JAK2/STAT3 signaling pathway. Oncotarget 2017; 8:20741–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005; 121:335–48 [DOI] [PubMed] [Google Scholar]
- 43.Ray P, Stacer AC, Fenner J, Cavnar SP, Meguiar K, Brown M, Luker KE, Luker GD. CXCL12-γ in primary tumors drives breast cancer metastasis. Oncogene 2014; 34:2043–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao L, Huang G, Song H, Chen Y, Chen L. Cancer associated fibroblasts: an essential role in the tumor microenvironment (review). Oncol Lett 2017; 14:2611–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Addison CL, Pond GR, Cochrane B, Zhao H, Chia SK, Levine MN, Clemons M. Correlation of baseline biomarkers with clinical outcomes and response to fulvestrant with vandetanib or placebo in patients with bone predominant metastatic breast cancer: an OCOG ZAMBONEY Sub-study. J Bone Oncol 2015; 4:47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, Moasser MM. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature 2007; 445:437–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du Y, Yamaguchi H, Wei Y, Hsu JL, Wang HL, Hsu YH, Lin WC, Yu WH, Leonard PG, Lee GR, Chen MK, Nakai K, Hsu MC, Chen CT, Sun Y, Wu Y, Chang WC, Huang WC, Liu CL, Chang YC, Chen CH, Park M, Jones P, Hortobagyi GN, Hung MC. Blocking c-Met-mediated PARP1 phosphorylation enhances anti-tumor effects of PARP inhibitors. Nat Med 2016; 22:194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharpe R, Pearson A, Herrera-Abreu MT, Johnson D, Mackay A, Welti JC, Natrajan R, Reynolds AR, Reis-Filho JS, Ashworth A, Turner NC. FGFR signaling promotes the growth of triple-negative and basal-like breast cancer cell lines both in vitro and in vivo. Clin Cancer Res 2011; 17:5275–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sitohy B, Nagy JA, Dvorak HF. Anti-VEGF/VEGFR therapy for cancer: reassessing the target. Cancer Res 2012; 72:1909–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feig C, Jones JO, Kraman M, Wells RJ, Deonarine A, Chan DS, Connell CM, Roberts EW, Zhao Q, Caballero OL, Teichmann SA, Janowitz T, Jodrell DI, Tuveson DA, Fearon DT. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci U S A 2013; 110:20212–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang S, Peng X, Li X, Yang P, Xie L, Li Y, Du C, Zhang G. Silencing of CXCR4 sensitizes triple-negative breast cancer cells to cisplatin. Oncotarget 2015; 6:1020–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Modok S, Mellor HR, Callaghan R. Modulation of multidrug resistance efflux pump activity to overcome chemoresistance in cancer. Curr Opin Pharmacol 2006; 6:350–4 [DOI] [PubMed] [Google Scholar]
- 53.Zanoni M, Piccinini F, Arienti C, Zamagni A, Santi S, Polico R, Bevilacqua A, Tesei A. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci Rep 2016; 6:19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ham SL, Thakuri PS, Plaster M, Li J, Luker KE, Luker GD, Tavana H. Three-dimensional tumor model mimics stromal-breast cancer cells signaling. Oncotarget 2018; 9:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Atefi E, Lemmo S, Fyffe D, Luker GD, Tavana H. High throughput, polymeric aqueous two-phase printing of tumor spheroids. Adv Funct Mater 2014; 24:6509–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lemmo S, Atefi E, Luker GD, Tavana H. Optimization of aqueous biphasic tumor spheroid microtechnology for anti-cancer drug testing in 3D culture. Cell Mol Bioeng 2014; 7:344–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tavana H, Jovic A, Mosadegh B, Lee QY, Liu X, Luker KE, Luker GD, Weiss SJ, Takayama S. Nanolitre liquid patterning in aqueous environments for spatially defined reagent delivery to mammalian cells. Nat Mater 2009; 8:736–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shahi Thakuri P, Ham SL, Luker GD, Tavana H. Multiparametric analysis of oncology drug screening with aqueous two-phase tumor spheroids. Mol Pharm 2016; 13:3724–35 [DOI] [PubMed] [Google Scholar]
- 59.Atefi E, Fyffe D, Kaylan KB, Tavana H. Characterization of aqueous two-phase systems from volume and density measurements. J Chem Eng Data 2016; 61:1531–39 [Google Scholar]
- 60.Atefi E, Mann JA, Tavana H. Ultralow interfacial tensions of aqueous two-phase systems measured using drop shape. Langmuir 2014; 30:9691–99 [DOI] [PubMed] [Google Scholar]
- 61.Atefi E, Joshi R, Mann JA, Tavana H. Interfacial tension effect on cell partition in aqueous two-phase systems. ACS Appl Mater Interfaces 2015; 7:21305–14 [DOI] [PubMed] [Google Scholar]
- 62.Ham SL, Atefi E, Fyffe D, Tavana H. Robotic production of cancer cell spheroids with an aqueous two-phase system for drug testing. J Vis Exp 2015; 98:e52754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ham SL, Joshi R, Luker GD, Tavana H. Engineered breast cancer cell spheroids reproduce biologic properties of solid tumors. Adv Healthc Mater 2016; 5:2788–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Kruijf EM, Van Nes JGH, Van De Velde CJH, Putter H, Smit V, Liefers GJ, Kuppen PJK, Tollenaar R, Mesker WE. Tumor-stroma ratio in the primary tumor is a prognostic factor in early breast cancer patients, especially in triple-negative carcinoma patients. Breast Cancer Res Treat 2011; 125:687–96 [DOI] [PubMed] [Google Scholar]
- 65.Dekker TJA, Van De Velde CJH, Van Pelt GW, Kroep JR, Julien JP, Smit V, Tollenaar R, Mesker WE. Prognostic significance of the tumor-stroma ratio: validation study in node-negative premenopausal breast cancer patients from the EORTC perioperative chemotherapy (POP) trial (10854). Breast Cancer Res Treat 2013; 139:371–79 [DOI] [PubMed] [Google Scholar]
- 66.Gujam FJA, Edwards J, Mohammed ZMA, Going JJ, McMillan DC. The relationship between the tumour stroma percentage, clinicopathological characteristics and outcome in patients with operable ductal breast cancer. Br J Cancer 2014; 111:157–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moorman AM, Vink R, Heijmans HJ, Van Der Palen J, Kouwenhoven EA. The prognostic value of tumour-stroma ratio in triple-negative breast cancer. Eur J Surg Oncol 2012; 38:307–13 [DOI] [PubMed] [Google Scholar]
- 68.Dewan MZ, Ahmed S, Iwasaki Y, Ohba K, Toi M, Yamamoto N. Stromal cell-derived factor-1 and CXCR4 receptor interaction in tumor growth and metastasis of breast cancer. Biomed Pharmacother 2006; 60:273–76 [DOI] [PubMed] [Google Scholar]
- 69.Liang Z, Wu T, Lou H, Yu X, Taichman RS, Lau SK, Nie S, Umbreit J, Shim H. Inhibition of breast cancer metastasis by selective synthetic polypeptide against CXCR4. Cancer Res 2004; 64:4302–08 [DOI] [PubMed] [Google Scholar]
- 70.Liang Z, Yoon Y, Votaw J, Goodman MM, Williams L, Shim H. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res 2005; 65:967–71 [PMC free article] [PubMed] [Google Scholar]
- 71.Smith MCP, Luker KE, Garbow JR, Prior JL, Jackson E, Piwnica-Worms D, Luker GD. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res 2004; 64:8604–12 [DOI] [PubMed] [Google Scholar]
- 72.López-Knowles E, O’Toole SA, McNeil CM, Millar EKA, Qiu MR, Crea P, Daly RJ, Musgrove EA, Sutherland RL. PI3K pathway activation in breast cancer is associated with the basal-like phenotype and cancer-specific mortality. Int J Cancer 2010; 126:1121–31 [DOI] [PubMed] [Google Scholar]
- 73.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Franklin RA, Montalto G, Cervello M, Libra M, Candido S, Malaponte G, Mazzarino MC. Ras/raf/MEK/ERK and PI3K/PTEN/akt/mTOR Cascade inhibitors: how mutations can result in therapy resistance and how to overcome resistance. Oncotarget 2012; 3:1068–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, Stivala F. Roles of the raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta 2007; 1773:1263–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saini KS, Loi S, de Azambuja E, Metzger-Filho O, Saini ML, Ignatiadis M, Dancey JE, Piccart-Gebhart MJ. Targeting the PI3K/AKT/mTOR and raf/MEK/ERK pathways in the treatment of breast cancer. Cancer Treat Rev 2013; 39:935–46 [DOI] [PubMed] [Google Scholar]
- 76.Plodinec M, Loparic M, Monnier CA, Obermann EC, Zanetti-Dallenbach R, Oertle P, Hyotyla JT, Aebi U, Bentires-Alj M, Lim RY, Schoenenberger CA. The nanomechanical signature of breast cancer. Nat Nanotechnol 2012; 7:757–65 [DOI] [PubMed] [Google Scholar]
- 77.Plaster M, Singh S, Tavana H. Fibroblasts promote proliferation and matrix invasion of breast cancer cells in co-culture models. Adv Ther 2019; 2:1900121 [Google Scholar]
- 78.Singh S, Ray LA, Shahi Thakuri P, Tran S, Konopka MC, Luker GD, Tavana H. Organotypic breast tumor model elucidates dynamic remodeling of tumor microenvironment. Biomaterials 2020; 238:119853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh S, Tavana H. Collagen partition in polymeric aqueous two-phase systems for tissue engineering. Front Chem 2018; 6:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuan H, Myers S, Wang J, Zhou D, Woo JA, Kallakury B, Ju A, Bazylewicz M, Carter YM, Albanese C, Grant N, Shad A, Dritschilo A, Liu X, Schlegel R. Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N Engl J Med 2012; 367:1220–27 [DOI] [PMC free article] [PubMed] [Google Scholar]