Abstract
Deep phenotyping during pregnancy offers an opportunity to define the antecedents of lifelong health and wellness, and to improve pregnancy outcomes.
Systems biology offers a holistic approach to deciphering physiology or disease by deriving and integrating biological information to delineate relevant networks and their dynamics. It enables medicine that is predictive, preventive, personalized, and participatory, with a focus on the two primary domains of healthcare—wellness and disease—using “deep phenotyping” to decipher the complexities of human health. This approach is defined by the longitudinal aggregation of high-throughput information on individuals, including digital health data, clinical laboratory tests, ‘omics data that are based on the genome, proteome, metabolome, microbiome, and exposome, and the deployment of mathematical and statistical models for interrogation of biological networks. The integration of these data enables: i) the identification of actionable possibilities to improve individual health, ii) personalized diagnostics to identify the earliest transitions from wellness to disease, and iii) the ability to reverse pathophysiological changes before disease manifests, using systems-driven preventive measures. This strategy, including deep phenotyping of individuals coupled with data-informed actions to improve health and avoid or mitigate disease trajectories, is defined as “scientific wellness” (1). In this article, we discuss why pregnancy is particularly well-suited for prototyping deep phenotyping, scientific wellness, and related systems biology approaches to predictive and preventive medicine.
The Emergence of Deep Phenotyping
Beyond the basic generation of omics data in isolation, deep phenotyping has been successfully used to track health-related transitions for thousands of individuals in the context of scientific wellness. For example, genetic risk scores coupled with health-risk markers such as high concentrations of ferritin, mercury, lead, or LDL cholesterol, or low vitamin D concentrations motivated low-risk wellness interventions that redirected these harbinger disease trajectories back to a healthy range (1, 2). Similar deep phenotyping, with an emphasis on wearable sensors, has been used to identify changes in inflammatory markers of Lyme disease, diagnose pre-diabetes, and develop a personalized, activity-based framework to direct disease transitions (3). Although this approach has the potential to improve individualized clinical care, it has inherent limitations, such as the requirement for data tracking well before chronic diseases, such as cancer and heart disease manifest, which may escalate cost and reduce patient compliance. For deep phenotyping to gain clinical traction, it must be proven viable, effective, broadly relevant, and cost-effective during a manageable time period. Although not proving causality, deep phenotyping provides means to inform potential causal relationships based on (a) temporality, with measurements captured before and during disease progression; (b) biological plausibility, where potential disease mechanisms can be inferred from comprehensive data, gathered across multiple systems; and (c) specificity and consistency, as distinctive differences are attained by comparing trajectories derived from a large number of well-phenotyped individuals. Pregnancy is uniquely suited to model, deploy, and study this approach because it is critically important to lifelong health, it typically involves frequent engagement with the healthcare system, it requires safe maternal and fetal strategies, and key outcomes can be measured within a short period of time.
Deep Phenotyping of Pregnancy
Multi-omics data collection initiatives have emerged as a popular approach to investigate pregnancy. A few recent manuscripts demonstrated how such studies can bridge knowledge gaps in a major complication of pregnancy, preterm birth (4–6). Several efforts to collect omics data in pregnancy and establish pregnancy-related biobanks are ongoing. Although a review of these pioneering efforts is beyond the scope of this Focus, they include the Danish National Birth cohort, the PRenatal DetermiNants of Children’s hEalth (PRINCE) cohort in Germany, the WHO’s Alliance for Maternal Newborn Health Improvement (AMANHI) consortium, the Environmental Child Health Outcomes (ECHO) study, and the longitudinal cohorts, such as the Stanford cohort, enabled by the March Of Dimes Foundation. With the appropriate longitudinal deep phenotyping data, a systems-driven strategy should illuminate the coordinated molecular and phenotypic trajectories of healthy pregnancies and those complicated by maternal-fetal disorders. As outlined below, we propose that prototyping a broader application of predictive and preventive medicine may build upon these studies by generating and tracking dynamic data for each pregnancy and deploying this information through patient-driven care initiatives. This would allow the identification of the earliest deviation from a normal trajectory to disease and may suggest safe interventions aimed at guiding the perturbed networks back to a healthy state. This should aid women and their providers in identifying at-risk pregnancies, improve care before pregnancy and throughout gestation, and thereby reduce perinatal and postnatal morbidity and mortality and their consequences.
There are three main reasons that explain the particular relevance of deep phenotyping to the 9 months of human pregnancy.
1. Pregnancy is critically important for lifetime health:
Compelling evidence, presented by Barker and colleagues and corroborated by investigators worldwide (known as the developmental origins of adult disease hypothesis (7)), highlighted the central impact of pregnancy and early childhood on the entire lifetime. During the 9 months of human gestation, rapid fetal growth depends on the drastically changing maternal physiology. Perturbations during this vulnerable window may adversely impact placental function and disrupt fetal development. Conditions such as fetal growth restriction, spontaneous preterm birth, and preeclampsia may leave a lifelong mark on the child, with a higher incidence of chronic diseases and neurobehavioral disorders during childhood and beyond (7). Certain pregnancy-related diseases, such as preeclampsia, are associated with maternal cardiovascular diseases long after pregnancy (8). Considering the nearly four million annual deliveries in the US, it is not surprising that perinatal diseases may take a major toll on our healthcare. To best identify the impact of these pathological perturbations, we must first conduct comprehensive mapping of normal dynamic changes during pregnancy, which will serve as a baseline for comparisons to disease trajectories (Fig. 1). Deep phenotyping can not only generate a high-resolution view of the biology of pregnancy, but also provide opportunities to identify the earliest wellness-to-disease transitions that may occur at any point during the 9 months in order to develop means to predict and prevent complications.
Fig. 1: The role of deep phenotyping during pregnancy in informing lifelong health.
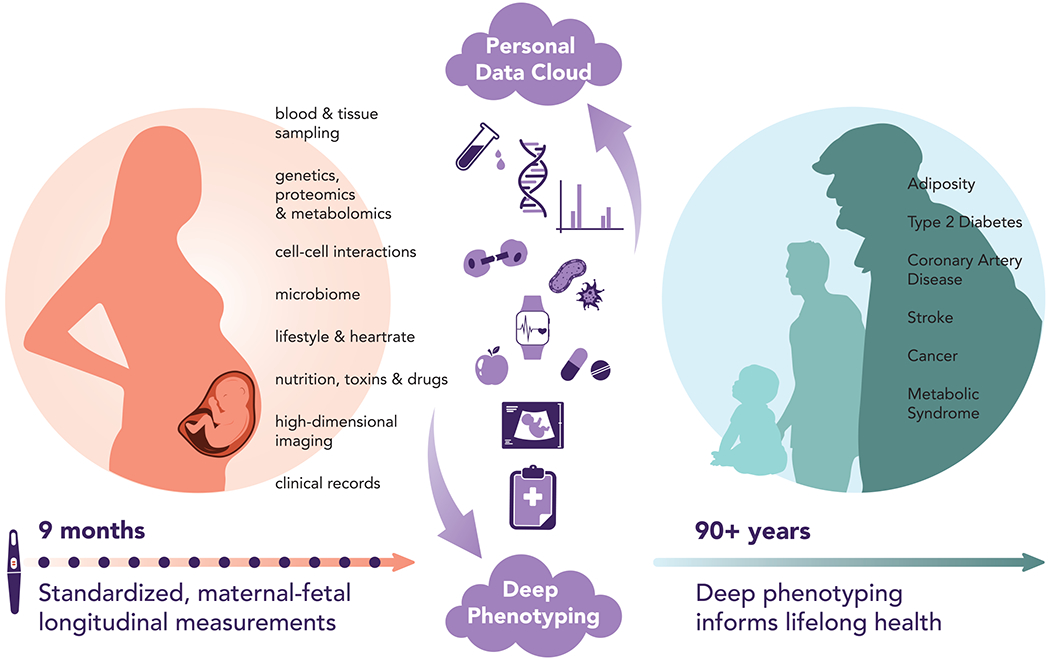
The use of systems biology approaches to defining key early human development parameters and to deploying actionable disease prevention strategies that promote lifelong health and wellness.
2. The complexities and risks of pregnancy motivate a scientific wellness approach:
Our current understanding of the pathobiology underlying pregnancy-related diseases is limited. Most gestational disorders are multifactorial, representing complex interactions among the fetus, placenta, and mother. Disruptions to this interface may result in pregnancy complications, where symbiotic harmony may rapidly evolve into conflicts between the two organisms. Existing therapeutic or preventive strategies are largely deficient, offered late after the disease becomes irreversible, or avoided altogether for compelling safety reasons. Deciphering these inherent complexities may greatly benefit from a deep phenotyping approach combined with integrated data analytics.
3. Pregnancy is a well-suited period to prototype future predictive and preventive medicine:
The relatively short 9-month duration of pregnancy offers many practical opportunities to determine whether deep phenotyping can identify health-to-disease transitions, inform personalized diagnostics, and deploy therapeutic and even preventive strategies. Pregnant women tend to be actively engaged with the healthcare system, and generally undergo routine, regularly recurring prenatal care visits. Characteristically, these visits include the recording of quantifiable physiological parameters, such as weight, blood pressure, and fundal height, documented using prenatal records that are standardized and fairly uniform across low- and high-income countries. Additionally, pregnancy may be a period of particularly high scientific and clinical interest in tracking activity, sleep patterns, and other health measurements, because these behavioral and physiological parameters rapidly change across pregnancy and may affect maternal and fetal health. We propose that active, real-time engagement of pregnant women and their providers – informed by deep phenotyping clinical trials – will allow adjustments to disease trajectories, as recently deployed in other contexts (1, 2). Data-informed and patient-driven behavioral, nutritional, and lifestyle alterations may provide a safe and feasible way to improve pregnancy outcomes, whereas developing pharmaceutical agents for pregnancy is challenging because of safety concerns. For example, a pregnant woman may learn early on that she is deficient in a critical nutrient, which may have long-term health impacts on her or on her fetus. With such information, it may be possible to mitigate maternal-fetal risk in a safe and straightforward way, and before symptoms arise. Many health parameters may be suited for personal health tracking with wearable sensors, which have garnered recent interest for their applications to pregnancy (9). Regular sampling of blood, urine, and other bodily fluids is an integral part of standard prenatal screening tests and can be easily expanded to other specimens. When combined with the numerous tissue specimens that are accessible at delivery (placenta, membranes, cervico-vaginal and rectal microbiome specimens), these materials may serve as biobanked sources for deep phenotyping (10). Relevant pregnancy outcome parameters and predictive biomarkers of later-life health, including the gestational age at delivery, birth weight, and placental histopathology are attainable within 9 months or less, resulting in a relatively rapid, iterative learning cycle between prediction and validation. Lastly, women with preexisting conditions such as diabetes, history of recurrent pregnancy losses, or genetic disorders that classify them as “high-risk” may be interested in pre-conceptional evaluation and counseling to prevent complications and improve health during pregnancy.
We recognize that there may be limitations to this scientific wellness approach due to emotional, social, economic, and logistical challenges that women face during routine prenatal care, which may disproportionately affect certain socioeconomic and ethnic groups or those with inadequate access to care. It is also clear that researchers and providers in the field should remain sensitive to women’s autonomy and independence, particularly during engagement with researchers and providers. As in any study, we hope that the data are collected in a manner that limits the burden placed on women and may involve diverse pregnancy care settings, including mobile clinics. Together, these deep phenotyping studies will not only drive discovery, but may also identify the most valuable measurements of maternal well-being and pregnancy outcomes that can be tested through large-scale clinical trials, and safely and ethically expanded to pregnant women worldwide.
Looking Beyond the 9 Months and Into the Future
Pregnancy is gradually being recognized as a pivotal challenge for medicine, because its outcomes are associated with lifelong consequences for both the mother and child. It provides an opportunity for prototyping early identification and personalized correction of deviations that may negatively impact intrauterine development and long-term maternal and neonatal health. Deep phenotyping of pregnancy, combined with data on neonates and infants (for example, breastfeeding and early life exposures), may usher in a comprehensive view on early developmental perturbations before they manifest as diseases. Realizing the immense potential of these approaches, when deployed during or after pregnancy, should facilitate collaborations for enhancement of pregnancy-focused, systems-oriented transdisciplinary research and personalized clinical interventions that may vastly impact health and wellness during pregnancy and across the human lifespan.
Acknowledgements
We thank Lisa S. Parker, PhD, Professor and Director, Center for Bioethics & Health Law at the University of Pittsburgh, as well as Kalliopi Trachana, Technical Program Manager for Clinical Genomics at the Institute for Systems Biology for their critical review, comments and insights. We also thank Allison Kudla, Associate Director of Communications at the Institute for Systems Biology, for her assistance with generating the figure.
Funding: This work was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development grants R01HD091527 (N.P.) and K99HD096112 (A.P.); R01HD086325 and the Richard King Mellon Foundation (Y.S.).
Footnotes
Competing interests: L.H. and N.P. serve as scientific advisors to Sera Prognostics and hold stock options in the company. Y.S. is an advisory board member for Illumina, Inc.
References and notes:
- 1.Price ND, Magis AT, Earls JC, Glusman G, Levy R, Lausted C, McDonald DT, Kusebauch U, Moss CL, Zhou Y, Qin S, Moritz RL, Brogaard K, Omenn GS, Lovejoy JC, Hood L, A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat. Biotechnol. 35, 747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zubair N, Conomos MP, Hood L, Omenn GS, Price ND, Spring BJ, Magis AT, Lovejoy JC, Genetic predisposition impacts clinical changes in a lifestyle coaching program. Sci. Rep. 9, 6805 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schussler-Fiorenza Rose SM, Contrepois K, Moneghetti KJ, Zhou W, Mishra T, Mataraso S, Dagan-Rosenfeld O, Ganz AB, Dunn J, Hornburg D, Rego S, Perelman D, Ahadi S, Sailani MR, Zhou Y, Leopold SR, Chen J, Ashland M, Christle JW, Avina M, Limcaoco P, Ruiz C, Tan M, Butte AJ, Weinstock GM, Slavich GM, Sodergren E, McLaughlin TL, Haddad F, Snyder MP, A longitudinal big data approach for precision health. Nat Med 25, 792 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang G, Feenstra B, Bacelis J, Liu X, Muglia LM, Juodakis J, Miller DE, Litterman N, Jiang PP, Russell L, Hinds DA, Hu Y, Weirauch MT, Chen X, Chavan AR, Wagner GP, Pavlicev M, Nnamani MC, Maziarz J, Karjalainen MK, Ramet M, Sengpiel V, Geller F, Boyd HA, Palotie A, Momany A, Bedell B, Ryckman KK, Huusko JM, Forney CR, Kottyan LC, Hallman M, Teramo K, Nohr EA, Davey Smith G, Melbye M, Jacobsson B, Muglia LJ, Genetic associations with gestational duration and spontaneous preterm birth. N. Engl. J. Med. 377, 1156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knijnenburg TA, Vockley JG, Chambwe N, Gibbs DL, Humphries C, Huddleston KC, Klein E, Kothiyal P, Tasseff R, Dhankani V, Bodian DL, Wong WSW, Glusman G, Mauldin DE, Miller M, Slagel J, Elasady S, Roach JC, Kramer R, Leinonen K, Linthorst J, Baveja R, Baker R, Solomon BD, Eley G, Iyer RK, Maxwell GL, Bernard B, Shmulevich I, Hood L, Niederhuber JE, Genomic and molecular characterization of preterm birth. Proc. Natl. Acad. Sci. U.S.A 116, 5819 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngo TTM, Moufarrej MN, Rasmussen MH, Camunas-Soler J, Pan W, Okamoto J, Neff NF, Liu K, Wong RJ, Downes K, Tibshirani R, Shaw GM, Skotte L, Stevenson DK, Biggio JR, Elovitz MA, Melbye M, Quake SR, Noninvasive blood tests for fetal development predict gestational age and preterm delivery. Science 360, 1133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gluckman PD, Hanson MA, Cooper C, Thornburg KL, Effect of in utero and early-life conditions on adult health and disease. N Engl J Med 359, 61 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton GJ, Redman CW, Roberts JM, Moffett A, Pre-eclampsia: pathophysiology and clinical implications. BMJ 366, l2381 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Runkle J, Sugg M, Boase D, Galvin SL, C CC, Use of wearable sensors for pregnancy health and environmental monitoring: Descriptive findings from the perspective of patients and providers. Digit Health 5, 2055207619828220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghaemi MS, DiGiulio DB, Contrepois K, Callahan B, Ngo TTM, Lee-McMullen B, Lehallier B, Robaczewska A, McIlwain D, Rosenberg-Hasson Y, Wong RJ, Quaintance C, Culos A, Stanley N, Tanada A, Tsai A, Gaudilliere D, Ganio E, Han X, Ando K, McNeil L, Tingle M, Wise P, Maric I, Sirota M, Wyss-Coray T, Winn VD, Druzin ML, Gibbs R, Darmstadt GL, Lewis DB, Partovi Nia V, Agard B, Tibshirani R, Nolan G, Snyder MP, Relman DA, Quake SR, Shaw GM, Stevenson DK, Angst MS, Gaudilliere B, Aghaeepour N, Multiomics modeling of the immunome, transcriptome, microbiome, proteome and metabolome adaptations during human pregnancy. Bioinformatics 35, 95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


