Abstract
Background
Organotypic cultures derived from pancreatic ductal adenocarcinoma (PDAC) termed pancreatic ductal cancer organoids (PDOs) recapitulate the primary cancer and can be derived from primary or metastatic biopsies. Although isolation and culture of patient-derived pancreatic organoids were established several years ago, pros and cons for individualized medicine have not been comprehensively investigated to date.
Methods
We conducted a feasibility study, systematically comparing head-to-head patient-derived xenograft tumor (PDX) and PDX-derived organoids by rigorous immunohistochemical and molecular characterization. Subsequently, a drug testing platform was set up and validated in vivo. Patient-derived organoids were investigated as well.
Results
First, PDOs faithfully recapitulated the morphology and marker protein expression patterns of the PDXs. Second, quantitative proteomes from the PDX as well as from corresponding organoid cultures showed high concordance. Third, genomic alterations, as assessed by array-based comparative genomic hybridization, revealed similar results in both groups. Fourth, we established a small-scale pharmacotyping platform adjusted to operate in parallel considering potential obstacles such as culture conditions, timing, drug dosing, and interpretation of the results. In vitro predictions were successfully validated in an in vivo xenograft trial. Translational proof-of-concept is exemplified in a patient with PDAC receiving palliative chemotherapy.
Conclusion
Small-scale drug screening in organoids appears to be a feasible, robust and easy-to-handle disease modeling method to allow response predictions in parallel to daily clinical routine. Therefore, our fast and cost-efficient assay is a reasonable approach in a predictive clinical setting.
Keywords: PDAC, organoids, drug response prediction
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most frequent cancer arising from the pancreas and still has a dismal prognosis. By 2030, predictions indicate that PDAC will become the second leading cause of cancer-related death in Western countries. Despite intensive basic and translational research, the prognosis for PDAC has only slightly improved over the last 20 years, mainly due to late diagnosis, lack of predictive biomarkers, and ineffective treatments. Chemotherapy is still the mainstay for PDAC.1 A better molecular understanding of pancreatic cancer has led to the identification of a variety of potential molecular targets.2 Nevertheless, these increasing options have only had little impact at changing the therapeutic landscape over the last decade. This is mostly due to the dramatic intra- and inter-tumoral heterogeneity in PDAC leading to dozens of subclones escaping treatment.3 Large-scale data sets have helped to better understand the mutagenic landscape of PDAC and allowed subgrouping of this tumor.4 Thus, novel and more tailored treatment algorithms are warranted to eventually reach a higher response rate in a subset of patients with PDAC. However, it is highly questionable that obtaining several omics data layers to design individual treatment regimen in real time is feasible, affordable, and indeed predictive. Instead, a reverse scenario might be better suited to fulfill such needs, namely to directly test all available drugs using an in vitro culture set-up with a validated predictive value for a given tumor.1,5 The advent of organoids is likely to comply with these demands.6 These organotypic cultures grown from single stem cells in a 3D matrix recapitulate organ/tumor structure and function.
Since the first report to establish intestinal organoids in 2009, organoids from primary tissue or tumor have been obtained from virtually all organs including liver, brain, breast, colon, and pancreas.6–10 Their prompt incorporation into the clinical workflow, as a novel diagnostic and predictive tool, has the capacity to improve cancer treatment and open new perspectives, particularly in cancers with dismal prognosis such as PDAC. So far, only three key studies have systematically used pancreatic ductal adenocarcinoma organoids (PDOs) to either (a) develop subtypes based on their WNT-dependence in the culture media, a feature that allowed for the reconstruction of the niche/stromal-cancer cell crosstalk, or (b) to assess the maintenance of a given tumor phenotype in a pancreatic organoid culture.11–13 Deep molecular profiling of mouse PDO proteomes and transcriptomes have uncovered molecular pathways linked to pancreatic cancer progression. One major advantage of organoid cultures over patient-derived xenografts (PDX) might be the capacity to quickly expand tumor-derived cell material to obtain high cell yields for preclinical testing, as PDXs are particularly limited in the clinical setting due to time constrains. However, timing is of key importance in the case of aggressive cancers such as PDAC. Constant improvement in isolation and culture conditions has allowed the establishment and propagation of PDAC organoids from primary tumors either by fine-needle aspiration or true-cut biopsies with a high rate of success.14
Based on culture conditions, two types of PDAC have been identified growing in different combinations, either WNT3A/RSPO1 dependent or independent.13 Interestingly, the grade of PDAC malignant transformation followed a WNT-dependency gradient, with most aggressive tumors being less dependent on WNT or other growth factor substitutions.13 Thus, even the media composition might impact drug testing results; however, up to now, most studies focused on the creation and characterization of organoid biobanks have convincingly shown the recapitulation of primary cancers via organoid cultures. Only two studies reported the use of patient-derived PDAC organoids as an instrument to guide treatment.14,15 These studies reported on PDO-based chemotherapy response predictive gene signatures and also correlated these gene expression data with pharmacotyping results upon drug treatment of the organoids and correlative PDX. Our presented work complements these studies by mimicking a clinical scenario from the establishment of the organoids to the organoid testing using FDA-approved drugs and the validation of the selected drugs in PDXs. This work illustrates the decision-making processes and troubleshooting inherent to guide treatment and to meet the demands of daily clinical routine.
Results
Organoids (PDO) recapitulate core features of the primary tumor (PDX)
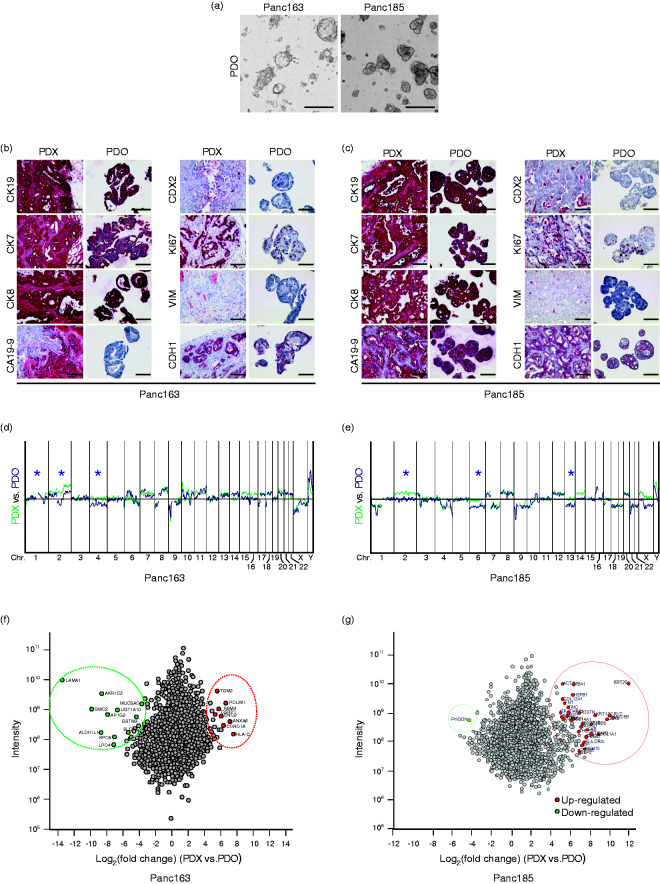
First, we isolated PDO cultures from two PDX pieces (isolated from Panc163 and Panc185 patients). The PDX system was chosen as an in vivo model because it can provide a short-dated and multiplicate in vivo read out of putative drug testing derived from the PDOs. For this we used a set of PDX which has been previously reported to mimic the primary cancer.16–19 PDO163 showed a more dispersed and irregular growth pattern, while PDO185 revealed a glandular growth pattern (Figure 1(a)). Cytokeratin profiles as well as proliferative capacity of PDOs and PDXs were matching (Figure 1(b) and 1(c)). The master transcriptional regulator of intestinal cell differentiation CDX2 is down-regulated during the transformation process from PanIN to PDAC. CDX2-negative PDAC has a better prognosis than scattered CDX2-expressing tumors.13 Interestingly, both Panc163 and Panc185 exhibited more CDX2-positive cells in PDO than in the corresponding PDX (Figure 1(b) and 1(c)). PDOs also successfully classified PDXs as epithelial, non-mesenchymal based on their CDH1-positive and VIM-negative staining pattern (Figure 1(b) and 1(c)). Moreover, array-based comparative genomic hybridization (aCGH) revealed that both PDO163 and PDO185 exhibited the same chromosome rearrangement signature as the PDX with only minor differences (*) indicating some selective genetic gain and loss during the establishment of organoids (Figure 1(d) and 1(e)). Of note, the amount of chromosomal rearrangements was generally higher in PDX185 compared with PDX163 (Figure 1(d) and 1(e)). Finally, we performed quantitative proteome analysis in PDX and their PDO counterparts. Similar to our observations in the immunostaining experiments and aCGH profiling, the proteome analysis revealed only minor differences between organoids and PDX tumors. Indeed, 20% of detected proteins exhibited abundance changes in PDX vs. PDO of more than four-fold in Panc163 (n = 568) and Panc185 (n = 600), respectively. Of these, only 89 proteins showed the same regulation tendency in both samples (online supplementary Figure S1A). To determine significant protein level changes between PDX and PDO, significance was calculated for both tumor entities, resulting in only 19 (Panc163) and 34 (Panc185) regulated proteins (online supplementary Figure S1B). These significantly regulated proteins from Panc163 and Panc185 were compared for regulation tendency (online supplementary Figure S1B), protein interaction network affiliation (online supplementary Figure S1C and S1D) and common GO terms (online supplementary Figure S1E). No overlap of proteins defined as significantly regulated could be observed between samples. Also, the correlation between regulation tendencies was minimal for these proteins. In addition, significantly regulated proteins for the respective samples did not share the same protein interaction networks or molecular function annotations (online supplementary Figure S1E). Overall, proteomes across all analyzed samples were comparable with 99.5% concordance (Figure 1(f) and 1(g)). Altogether PDO reproduces PDX at the protein level, while the few changes resulting upon PDO culture appear to be specific for a given tumor, arguing against a specific pattern caused by the culture condition itself.
Figure 1.
Pancreatic ductal organoid vs. patient-derived xenograft comparison. (a) Representative images of pancreatic ductal organoids (PDO) isolated from human PDAC. Scale bars represent 400 µm. Immunohistochemical characterization of patient-derived xenograft (PDX) and PDO from (b) Panc163 and (c) from Panc185. Scale bars represent 100 µm. Array-Comparative Genomic Hybridization analysis showing genomic rearrangements (*) in PDO vs. PDX of Panc163 (d) and of Panc185 (e). Proteome comparison between PDO vs. PDX of Panc163 (f) and of Panc185 (g).
A drug testing platform for PDOs
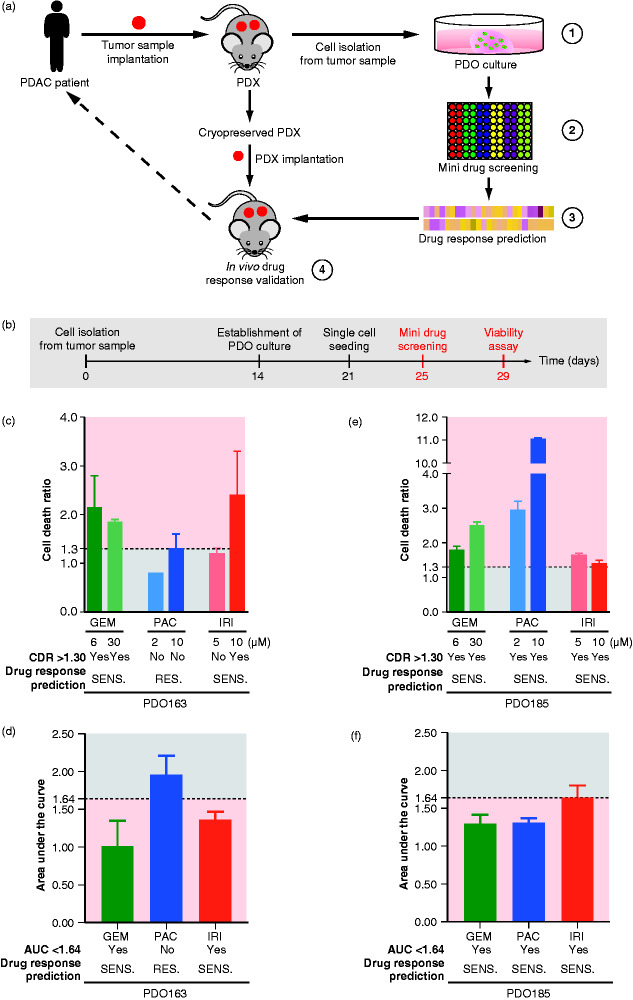
The window to initiate treatment in metastatic PDAC is limited due to diagnosis at an advanced stage in the majority of cases. Therefore, a response prediction scenario has to be rapid, feasible, reproducible, and thus broadly applicable. We tested the predictive value of a drug testing platform operating with only one experimental run (Figure 2(a)). Specifically, we designed a set-up by establishing two PDO lines from two xenografted PDAC PDX cases to mimic the day of a virtual patient’s biopsy in the metastatic setting. The establishment of the PDO culture took approximately 14 days, but the desired yield for multiple drug testing is at least 21 days (Figure 2(b)). Afterward, we performed a small-scale drug testing screen in a potentially scalable format to predict best treatment response in vivo, based on maximum cell death in vitro during 4 days of drug treatment (Figure 2(a) and 2(b)). Based on these results, a limited set of drugs from the screen were validated in a subsequently conducted xenograft trial (Figure 2(a)). After seeding as single cells, we propagated them for 4 days to allow organoid formation, and then applied respective treatments for 4 additional days. To evaluate the drug response (Figure 2(b)), we used a viability assay that sequentially measures dead and living cells, providing a dual readout. To reliably normalize the effect of the treatment across different PDO cultures, we measured the cell death ratio (CDR, dead cell upon drug treatment/dead cell upon vehicle treatment). A threshold was set at 1.30 for CDR analysis reflecting a potent cytotoxic effect of drug.17,18 Moreover, to have a second reliable metric to evaluate the potent drug responsiveness of PDO, we calculated the area under the curve (AUC, following the trapezoidal rule) after a drug-dose response test. This method previously showed one of the best prediction performances of real-life scenarios based on a simulation.20 In our study, we combined CDR and AUC analyses to improve the power of prediction of our drug screening. Therefore, we defined positive drug response based on the association of the following objective criteria: CDR >1.30 and AUC <1.64 (AUC cutoff was defined using the Jenks Natural Breaks classification method). Following these settings, PDO163 was found to be sensitive to gemcitabine (GEM) and irinotecan (IRI) but showed no response upon paclitaxel (PAC) treatment (CDR <1.30 and AUC >1.64) (Figure 2(c) and 2(d)). In contrast, PDO185 showed a broader overall sensitivity to all the drugs tested having a CDR >1.30 (Figure 2(e)) and a positive response after dose escalation experiment with AUC <1.64 (Figure 2(f)). Interestingly, PDO185 displayed a high drug responsiveness to PAC, with a CDR of approximately 3.0 (lower dose) and 11.0 (higher dose). Based on these in vitro experiments conducted on PDOs and our prediction criteria, we hypothesized that PDX163 should significantly respond to GEM or IRI regimen, while it should not be sensitive to PAC treatment. In contrast, based on predictions in PDO185, all three single agents should provide an in vivo response (Figure 2(e) and 2(f)).
Figure 2.
A pancreatic cancer-derived organoids tool to predict drug response. (A) Schematic representation of our pancreatic ductal organoid (PDO)-derived tool. (1) PDO and patient-derived xenograft (PDX) models were generated from cryopreserved xenografts of two patients with pancreatic ductal adenocarcinoma (PDAC). Several FDA-approved drugs were screened either single or in combination (2) and validated on the established organoid cultures (3). Drugs which showed sensitivity in PDO were selected for further small-scale drug screenings and validation in the PDXs (4). (B) Schematic representation of an organoid-based drug screening. (C) Cell death ratio (CDR) analysis of a viability assay and (D) area under the curve (AUC) analysis of a dose-response assay conducted on PDO163. (E) CDR analysis of a viability assay and (F) AUC analysis of a dose-response assay conducted on PDO185. Drug response prediction using CDR and AUC are provided below each bar graph. Yes and No indicate respectively a positive drug response and a lack of drug response. PDO is considered to be sensitive when CDR of one of treatment conditions >1.30 and if AUC < 1.64. GEM (gemcitabine); IRI (irinotecan); PAC (paclitaxel); RES., resistant; SENS., sensitive.
Xenograft validation of PDO predictions
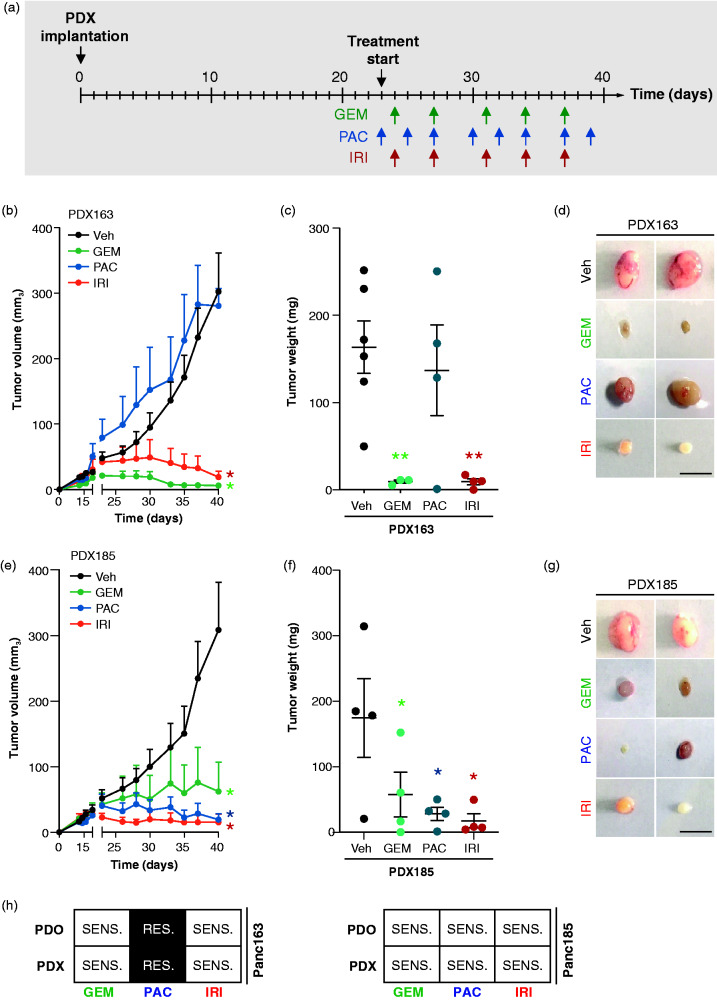
To validate our findings, we set up a state-of-art PDX-based drug testing scenario by implanting and expanding PDX tumor samples for subsequently treatment initiation. As described in Figure 3(a), the PDX approach is time-consuming (approximately 40 days), labor intensive, and technically challenging. Enrolment in a standard mouse treatment regimen (for GEM, PAC, and IRI) was decided when tumors reached an average size of 50 mm3. As shown by its low CDR (<1.30) and high AUC (>1.64), PDX163 was expected to be resistant to PAC. Indeed, there was no significant response in vivo upon PAC treatment (Figure 3(b–d)). In contrast, a strong tumor shrinkage was observed upon treatment with both GEM and IRI, consistently with the PDO drug responsiveness (Figure 3(b–d)). For PDX185, we observed a significant response to all three drugs over time and also at endpoint analysis, again as predicted by our drug screening (Figure 3(e–g)). Altogether, these data underline the predictive power of PDOs and their inherent advantage over effortful and time-consuming PDX models (Figure 2(b) and Figure 4(a) and 4(h)).
Figure 3.
Drug response validation in patient-derived xenograft (PDX). (A) Illustration of the experimental in vivo set-up. (B) Time-dependent development (over the course of 40 days) of subcutaneously engrafted PDX163 treated with GEM (125 mg/kg; n = 3; green lines), PAC (12 mg/kg; n = 4; blue lines), IRI (50 mg/kg; n = 4; red lines), and vehicle (n = 6; black lines). (C) Quantification of tumor weight and (D) representative macroscopic pictures of resected PDXs from subcutaneous assay shown in (B). Scale bars represent 1 cm. (E) Time-dependent development (over the course of 40 days) of subcutaneously engrafted PDX185 treated or not with GEM (125 mg/kg; n = 4; green lines), PAC (12 mg/kg; n = 4; blue lines), IRI (50 mg/kg; n = 4; red lines), and vehicle (n = 4; black lines). (F) Quantification of tumor weight and (G) representative macroscopic pictures of resected PDXs from subcutaneous assay shown in (E). Scale bars represent 1 cm. Error bars indicate SEM. (H) Summary of the prediction efficiency of the mini drug screening on PDOs vs. PDXs. GEM (gemcitabine); IRI (irinotecan); PAC (paclitaxel); Veh, vehicle; RES., resistant; SENS., sensitive. *p < 0.05; **p < 0.001.
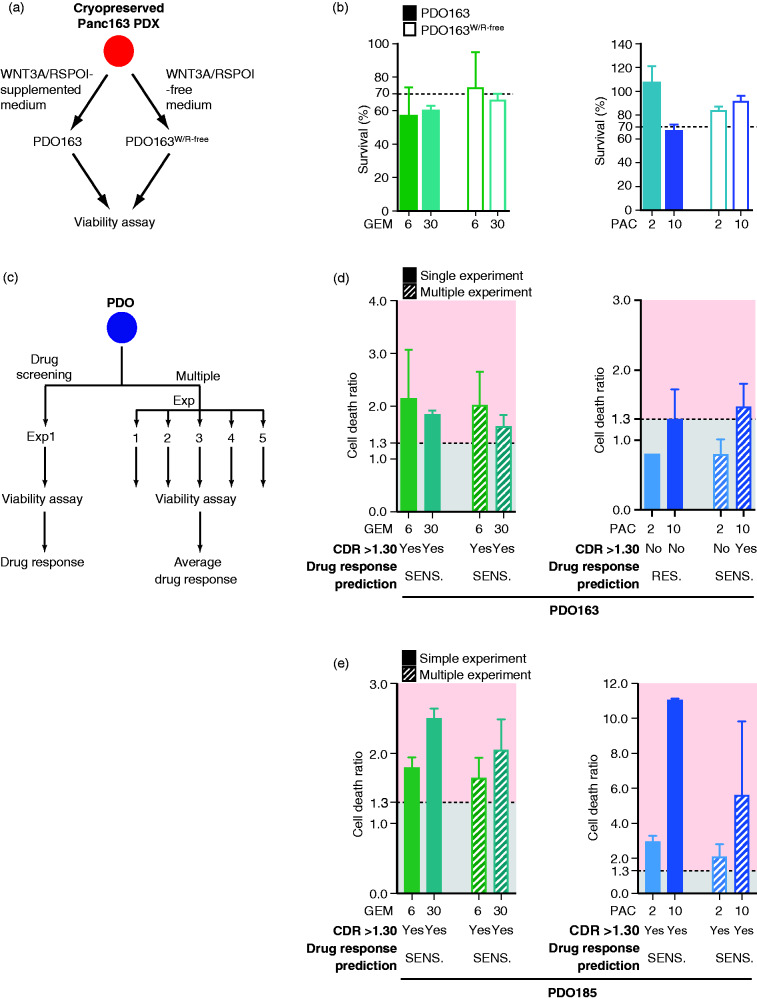
Figure 4.
Reproducibility and power of the assay. (A) Schematic representation of PDO163 and PDO163W/R-free isolation and propagation using pancreatic ductal organoid media respectively supplemented17 or not with WNT3A/RSPOI8. (B) Survival analysis of a viability assay conducted on PDO163 and PDO163W/R-free. (C) Schematic representation of the comparison of multiple independent experiments vs. single experiment of drug screening. Cell death ratio (CDR) analysis of multiple viability assays (n = 5) vs. a single experiment conducted on (D) PDO163 (n = 5) and (E) PDO185 (n = 3). Drug response prediction using CDR is provided below each bar graph. Yes and No indicate respectively a positive drug response and a lack of drug response. PDO is considered to be sensitive when CDR of one of treatment conditions >1.30. GEM (gemcitabine); IRI (irinotecan); PAC (paclitaxel); RES., resistant; SENS., sensitive.
Troubleshooting and power of prediction
PDOs can be isolated and propagated either using WNT3A/RSPO1-supplemented or -free media, both culture conditions reflecting distinct tumor biology.13 Therefore, various clones might be present in a bulk organoid culture being permissive to in vitro culture-driven subclonal selection. The latter might eventually also impact drug prediction results. To test this assumption, two media compositions were used: (a) the originally reported culture conditions including WNT3A/RSPOI11 supplementation and (b) a more minimal media lacking these growth factors12 (Figure 4(a)). We therefore established organoid lines in these two media from the same PDX163 tumor piece isolated. Afterward, drug testing was repeated with GEM and PAC as described above. Overall, the results were similar and would not have modified the drug guidance (Figure 4(b)). Timely initiation of treatment upon diagnosis is critical for most metastatic tumors, and a drug testing platform needs to operate in a robust and quick manner. Hence, we asked whether one experiment is still reliable enough to predict drug response or whether several independently conducted experiments would provide a more robust and stronger response interpretation (Figure 4(c)). As such, we repeated several independent experiments over a period of 2 months and compared them with the initially performed tests using the two most responsive drugs (GEM and PAC). Interestingly, for both PDO163 (n = 5) and PDO185 (n = 3), all results obtained with several independent experiments matched data obtained with the first testing (Figure 4(c–e)), except for one condition, ranging anyhow borderline: indeed, PDO163 responded to the highest dose of PAC (10 µM), with a CDR slightly above 1.30, showing the importance of combining two parameters (CDR plus AUC) for an optimal prediction of drug responsiveness including best dose range.
PDOs derived from primary PDAC and proof of concept
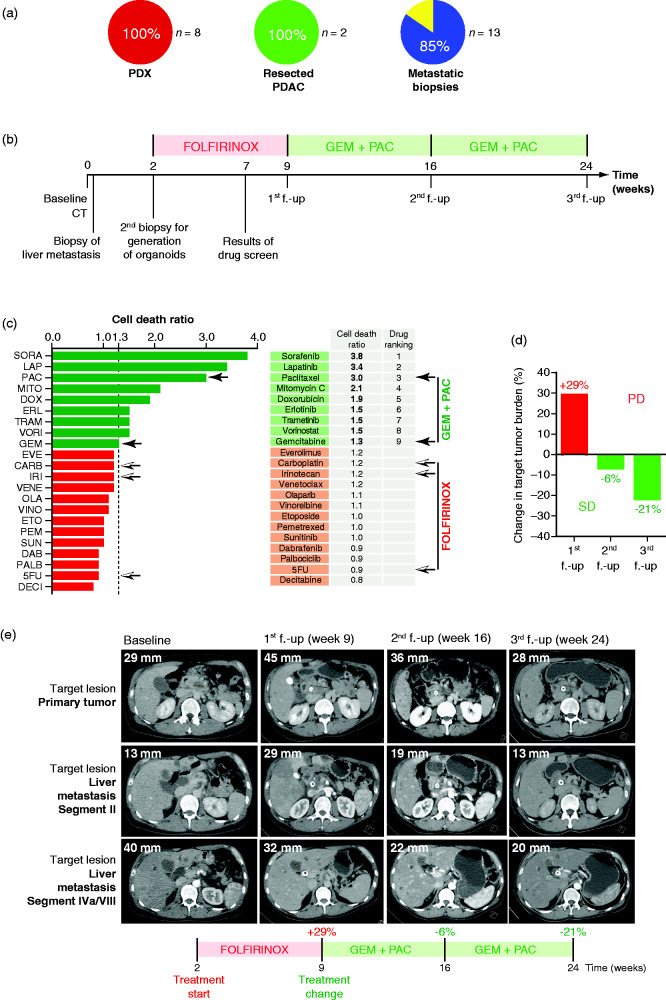
Our comparative attempt using a paired PDX/PDO set-up can be used to standardize and validate the drug response test and extend in vivo validation to a full suite of drugs. However, PDX still harbor disadvantages such as the lack of the appropriate tumor niche to fully recapitulate a PDAC patient. To stress the translational value of our work, we made a head-to-head comparison between the successful PDO culture derivation rate from PDX (100%, 8 from 8 attempts), from primary tumor resectates (100%, 2 from 2 attempts) and, most importantly, from ultrasound-guided biopsy of the primary cancer or liver metastases (85%, 11 from 13 attempts). Indeed, we observed high concordance (Figure 5(a)). To validate the feasibility and the potential value of PDOs to predict sensitivity to chemotherapy in a clinical setting, we conducted a PDO prediction scenario in one patient with synchronous metastatic pancreatic cancer. At an external hospital, a CT scan was performed that suspected an advanced pancreatic malignancy in a 42-year-old male patient. Diagnosis was confirmed histologically by ultrasound-guided biopsy of one liver metastasis (Figure 5(b)). Subsequently, the patient was transferred to our hospital, where a second biopsy of one liver metastasis was taken 2 weeks after primary diagnosis to generate patient-derived organoids. Based on good performance, young age, and current guidelines,21 the patient received first-line palliative chemotherapy with a modified FOLFIRINOX regimen (5FU (fluorouracil) 2,400 mg per m2, leucovorin 400 mg per m2, irinotecan 180 mg per m2, and oxaliplatin 85 mg per m2 every 2 weeks). As early as 5 weeks after initiation of treatment, the results from the organoid drug testing platform were obtained and suggested that 5FU, carboplatin, and irinotecan would not be efficient, showing high survival rates after 4 days of in vitro treatment (126%, 86%, and 89%, respectively) (Figure 5(c)). The PDO was found to be very sensitive to paclitaxel with a survival rate of only 27%, while gemcitabine was predicted to have limited efficiency with a survival rate of 82% (Figure 5(c)). In line with the prediction of resistance to FOLFIRINOX, the first follow-up CT scan (Figure 5(d) and 5(e)) revealed an increase in target lesions of 29% corresponding to progressive disease as determined by RECIST 1.1 (Response Evaluation Criteria In Solid Tumors) following 7 weeks of treatment with four cycles of modified FOLFIRINOX (Figure 5(d) and 5(e)). Subsequently, the patient was treated with second-line therapy gemcitabine/nab-paclitaxel (gemcitabine 1,000 mg per m2, nab-paclitaxel 125 mg per m2 on days 1, 8, 15, every 4 weeks) over a period of 15 weeks according to the results of the PDO pharmacotyping (Figure 5(b), 5(d), and 5(e)). Corresponding to the PDO prediction, the CT scan at the second follow-up after 7 weeks of treatment with second-line therapy showed a decrease of target lesions of 6% and an even more appreciable reduction in target lesions at the third follow-up of 21% under continued treatment with gemcitabine/nab-paclitaxel, although this did not meet RECIST 1.1. criteria for partial response. These findings demonstrate the feasibility and the predictive nature of PDO in one PDAC patient and may open the possibility of personalized therapeutic management in clinical routine.
Figure 5.
Organoid-based tailored therapies. (A) Take rate of pancreatic ductal organoid (PDO) isolation from PDX (n = 8), resected primary pancreatic ductal adenocarcinoma (PDAC) (n = 2) and ultrasound-guided biopsy of the primary cancer or liver metastases (n = 13). (B) Scheme of the course of treatment of the patient. (C) Twenty-two drug viability assay screening performed on patient PDO (one dose per drug). Black arrows show components of GEM + PAC therapy. Black-and-white arrows show components of FOLFIRINOX therapy. (D) Waterfall plot showing percent change in tumor burden determined based on RECIST 1.1. criteria at first follow-up (f.-up) with an increase of 29% corresponding to progressive disease (PD). At second f.-up and third f.-up, there was a decrease of, respectively, 6% and 21%, corresponding to stable disease (SD). (E) Representative pictures of CT scan. Shown is the longest dimension of target lesion of the primary tumor and target lesion of a liver metastasis segment II and segment IVa/VIII at baseline (time of initial diagnosis), at first f.-up (week 9) after treatment with FOLFIRINOX, at second f.-up (week 16) after treatment with gemcitabine/nab-paclitaxel, and at third f.-up (week 24) after continued therapy with gemcitabine/nab-paclitaxel. 5FU: 5-fluorouracil; CARB: carboplatin; DAB: dabrafenib; DECI: decitabine; DOX: doxorubicin; ERL: erlotinib; ETO: etoposide; EVE: everolimus; GEM: gemcitabine; IRI: irinotecan; LAP: lapatinib; MITO: mitomycin C; OLA: olaparib; PAC: paclitaxel; PALB: palbociclib; PEM: pemetrexed; SORA: sorafenib; SUN: sunitinib; TRAM: trametinib; VENE: venetoclax; VINO: vinorelbine tartrate; VORI: vorinostat.
Methods
Isolation and culture of Patient-derived organoids
Briefly, PDX were digested with collagenase/dispase followed by accutase (Sigma-Aldrich) for 30 min at 37℃. Cells were then filtered through a 100 µm filter and seeded on a Matrigel GFR (Corning)-coated plate in organoid culture medium containing WNT-3/RSPOI-conditioned medium11 and 5% Matrigel GFR.
Drug screening
Prior to seeding, organoids were dissociated into single cells using collagenase/dispase and accutase. Organoids were then seeded in 96-well plates (2,000 cells per well) for 4 days before treatment. After a further 4 days of treatment, cell viability was analyzed with the CytoTox-Glo™ Cytotoxicity Assay (Promega) according to the manufacturer’s protocol. Luminescence was measured using a Tecan Infinite M200 Pro. CDR was determined using this formula: CDR = (S1:S2)Drug/(S1:S2)Ctrl. Percentage of survival was determined using this formula: (S2-S1)Drug/(S2-S1)Ctrl. The AUC was estimated using the trapezoidal rule with GraphPad Prism 5 software. AUC cutoff was defined using the Jenks Natural Breaks classification method. The following drugs were used: gemcitabine (LY188011), paclitaxel (NSC 125973), and irinotecan (CPT-11). All the drugs were obtained from the Pharmacy of the University Hospital of Ulm.
Generation of patient-derived xenografts (PDXs)
Six- to eight-week-old female Hsd:Athymic Nude-Foxn1nu mice were purchased from Envigo. Mice were housed and maintained in laminar flow cabinets under a specific pathogen-free environment in accordance with the current standards and regulations of the government of Baden-Württemberg. For subcutaneous implantation, 4 mm3 pieces of Panc163 and Panc185, patient-derived in vivo expanded pancreatic tumors, were embedded in ECM Gel from Engelbreth-Holm-Swarm murine sarcoma (Sigma-Aldrich) and transplanted into the flanks of the mice. Tumor size was measured three times a week with a caliper, and the tumor volumes were determined with the following equation: v = (l × w2)/2 (where v is volume, l is length, and w is width). For each treatment, group of ≥3 tumors were implanted. Tumor-bearing mice received paclitaxel (Taxol; 12 mg/kg i.p. three times per week), gemcitabine (Gemzar; 125 mg/kg i.p. twice per week), or irinotecan (Camptosar; 50 mg/kg i.p. twice per week) provided by the Pharmacy of the University Hospital of Ulm.
Discussion
The discovery of organoids has provided a key tool for next generation disease- and cancer-modeling to analyze disease pathophysiology and to create an innovative platform for drug testing. It is not surprising that “Organoids” have ranked as the Method of the Year 2017.21 The pioneering person in the field is Hans Clevers, who together with his colleagues has foreseen the method’s capacity and clinical value. However, true hope needs to be validated by future studies proving true value for patients care in real time. The establishment of healthy pancreas-specific organoid cultures allowing extensive passaging without deterioration set the stage in the pancreas field.22 Likewise, follow-up work has allowed for the isolation of organoid cultures from pancreatic ductal adenocarcinoma.11–13 Here, therapeutic capacity of PDOs is neither proven nor under consideration. A recent study aimed to develop drug response signatures to provide new perspectives toward PDAC personalized medicine by predicting drug response as wells as tumor evolution based on changes in PDO transcriptomes,14 the latter previously having limited success to be indeed predictive due to high stromal content in PDAC.23 The authors termed their approach “pharmacotyping” and successfully tested a set of PDOs with frequently used drugs. Also, the authors determined in a PDO series, obtained from one patient over several treatment regimens, signs of tumor evolution, which correlated with acquired resistance and a tumor subtype switch.14 Our work puts these findings into a new prospective and clinically relevant context, by demonstrating that compared with xenograft-based approaches PDOs represent a very simple alternative approach to predict drug response.
There are a variety of advantages offered by the proposed testing platform: (a) isolation and culture of PDOs appears feasible and robust albeit our initial head-to-head comparison goes back to PDX source; (b) PDOs indeed recapitulate PDX on a morphological, immunohistochemical, and deep molecular profiling level; (c) PDO testing is less time-consuming and less labor intensive compared with the current preclinical standard prediction tool, the PDX; (d) phenotyping might be even more straightforward than genotype-based in silico predictions, frequently only discovering variants of unknown significance, though costly; (e) the non-responding drugs identified can help to spare toxicity as these could be eventually omitted from poly-chemotherapy regimens, such as FOLFIRINOX. However, this conclusion needs to take synergistic drugs actions into account. Finally, we define clinically relevant pitfalls such as media composition and necessity of at least a limited number of replicate experiments.
Despite the limited number of patient-derived organoids in our study, our work helps to raise hope of implementing these findings into patient care. To support this hope, we present as proof of concept one patient recapitulating our PDX-PDO workflow in a real-life setting, where indeed PDO would have been predictive. Our mini drug testing approach revealed basically two different response groups. A first group represents the most promising drug candidates with a robust response to the treatment shown by a high CDR (>1.30) and a low AUC (<1.64) after the dose-response experiment. For this group, there is a high chance of clinical response. A second group with inefficient drugs, characterized by a low CDR (<1.30) and a high AUC (>1.64), should not provide a significant response in vivo, tumor cells being obviously resistant to their action. False positive results may reduce the therapeutic benefit for the patient. Thus, repetitive experiments with close starting points are desirable to rule out false positive results due to culture quality. This assumption was proved by our head-to-head study comparing results from one pilot study with a follow-up experimental series. Moreover, we realized in the course of this work that the quality of conditioned medium may also play a role on the proliferation of some PDO, and may influence the results from batch to batch. This fact may have to be considered in the standardization and standard operating procedure approach that should be implemented in a clinical routine laboratory. The false negative results are more problematic since they would prevent or delay the use of efficient drugs for the patient, and to exclude them will require additional adjustments. To avoid that, we used in our study two metrics to evaluate potent drug response. When irinotecan was showing a “borderline” response (with two CDRs slightly above 1.30), responsiveness was finally confirmed by a dose-response assay with dose escalation, displaying an AUC below the defined threshold (and ultimately validated by PDX experiment). Combination treatment efficiency as well as synergistic interactions could also be addressed with our PDO-based prediction tool. However, as this further complicates the experimental set-up in a limited time-window (due to dosing strategy adjustments and requirement of a systematic synergy screening), we believe decision making might be better supported by screening a higher number of drugs. Finally, the drug screening efficiency strongly relies on the cellular quality and viability of the patient samples provided to isolate the PDO, rather than the medium used. Establishment of PDOs from primary tumor resections or sonography-guided punctures was reported in several studies with 80% efficiency,14,24 which is consistent with our own findings (Figure 5(a)).
Altogether, our work proposes an approach which might predict PDAC drug response upon future testing and proper standardization using organoids. We also present solutions to interpret the results and rigorously show experimental problems from a real-life setting, improving the decision-making process and opening the possibility of personalized therapeutic management for patients with PDAC.
Supplemental Material
Supplemental material, UEG905183 Supplemental material for Pancreatic cancer-derived organoids – a disease modeling tool to predict drug response by Pierre-Olivier Frappart, Karolin Walter, Johann Gout, Alica K Beutel, Mareen Morawe, Frank Arnold, Markus Breunig, Thomas FE Barth, Ralf Marienfeld, Lucas Schulte, Thomas Ettrich, Thilo Hackert, Michael Svinarenko, Reinhild Rösler, Sebastian Wiese, Heike Wiese, Lukas Perkhofer, Martin Müller, André Lechel, Bruno Sainz Jr Patrick C Hermann, Thomas Seufferlein and Alexander Kleger in United European Gastroenterology Journal
Acknowledgments
We are deeply indebted to Ralf Köhntop and Andrea Wissmann, for excellent technical support. We furthermore gratefully acknowledge Dr. Manuel Hidalgo Medina for donation of patient-derived xenografts.
Declaration of conflicting interests
The authors have no conflicts of interest to declare.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Main funding is provided by the German Cancer Aid grant to A. Kleger (111879). Additional funding came from the Deutsche Forschungsgemeinschaft (DFG, K.L. 2544/1-1, and 1-2; GRK 2254/1 to T. Seufferlein), the BIU fund (Böhringer Ingelheim), the NDIMED-Verbund PancChip, and the Else-Kröner-Fresenius Memorial funding to A. Kleger. AK receives also funding from the DFG within the Heisenberg program and from the Baden-Württemberg Foundation via ExPo Chip. This project was also funded by ANR-DFG collaborative research project (ANR-18-CE92-0031, DFG KL 2544/5-1) to CJ and AK and via additional DFG funding KL 2544/6-1, KL 2544/7-1, KL 2544/1-1, and KL 2544/1-2 to AK. L. Perkhofer is funded by Bausteinprogramm of the Ulm University hospital. This work was supported by a project grant for André Lechel (Deutsche Krebshilfe/111264), for Patrick C. Hermann (Max Eder Fellowship 111746, Projektnummer 316249678 – SFB 1279, and Hector Foundation Cancer Research grant M65.1). Reinhild Rösler and parts of proteomics method development were funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – SFB 1074. Bruno Sainz Jr was funded by a Ramón y Cajal Merit Award from the Ministerio de Economía y Competitividad, Spain and a coordinated grant from the Fundación Asociación Española Contra el Cáncer (AECC).
Ethics approval
All animal care and procedures followed German or Spanish legal regulations and were previously approved by the respective governmental review board of the state of Baden-Württemberg or the Universidad Autónoma de Madrid Ethics Committee (CEI 60-1057-A068) and La Comunidad de Madrid (PROEX 335/14), respectively. All mouse work aspects were carried out following strict guidelines to insure careful, consistent and ethical handling of mice. The biomaterial used was provided either by the PancoBank of the European Pancreas Centre Heidelberg and used in accordance with the regulations of the Biobank and the vote of the Ethics Committee of the University of Heidelberg (resected tumor, Prof. Thilo Hackert) or by the biobank of the University Hospital of Ulm following the regulations of the Biobank and the vote of the Ethics Committee of the University of Ulm (PDAC Liver Metastasis): 72/2019 and 67/2019 (human tissue and blood). The PDAC patient-derived xenografts (PDAC PDX) were obtained from Dr. Manuel Hidalgo under a Material Transfer Agreement with the Spanish National Cancer Centre (CNIO), Madrid, Spain (Reference no. I409181220BSMH and I405271505PHMH).
Informed consent
Written informed consent was obtained from each patient included in the study.
Supplementary material
Supplemental material for this article is available online.
References
- 1.Kleger A, Perkhofer L, Seufferlein T. Smarter drugs emerging in pancreatic cancer therapy. Ann Oncol 2014; 25: 1260–1270. [DOI] [PubMed] [Google Scholar]
- 2.Russell R, Perkhofer L, Liebau S, et al. Loss of ATM accelerates pancreatic cancer formation and epithelial-mesenchymal transition. Nat Commun 2015; 6: 7677–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller S, Engleitner T, Maresch R, et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 2018; 554: 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015; 518: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seufferlein T, Kleger A. Organoidomics – falling star or new galaxy in pancreatic cancer? Nat Rev Gastroenterol Hepatol 2018; 15: 586–587. [DOI] [PubMed] [Google Scholar]
- 6.Perkhofer L, Frappart PO, Müller M, et al. Importance of organoids for personalized medicine. Per Med 2018; 15: 461–465. [DOI] [PubMed] [Google Scholar]
- 7.Hohwieler M, Perkhofer L, Liebau S, et al. Stem cell-derived organoids to model gastrointestinal facets of cystic fibrosis. United European Gastroenterol J 2017; 5: 609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perkhofer L, Illing A, Gout J, et al. Precision medicine meets the DNA damage response in pancreatic cancer. Oncoscience 2018; 5: 6–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seufferlein T, Mayerle J. Pancreatic cancer in 2015: Precision medicine in pancreatic cancer – fact or fiction? Nat Rev Gastroenterol Hepatol 2016; 13: 74–75. [DOI] [PubMed] [Google Scholar]
- 10.Armacki M, Trugenberger AK, Ellwanger AK, et al. Thirty-eight-negative kinase 1 mediates trauma-induced intestinal injury and multi-organ failure. J Clin Invest 2018; 128: 5056–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boj SF, Hwang CI, Baker LA, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015; 160: 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang L, Holtzinger A, Jagan I, et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med 2015; 21: 1364–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seino T, Kawasaki S, Shimokawa M, et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell 2018; 22: 454–467. [DOI] [PubMed] [Google Scholar]
- 14.Tiriac H, Belleau P, Engle DD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov 2018; 8: 1112–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romero-Calvo I, Weber CR, Ray M, et al. Human organoids share structural and genetic features with primary pancreatic adenocarcinoma tumors. Mol Cancer Res 2019; 17: 70–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hidalgo M, Amant F, Biankin AV, et al. Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discov 2014; 4: 998–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter K, Tiwary K, Trajkovic-Arsic M, et al. MEK inhibition targets cancer stem cells and impedes migration of pancreatic cancer cells in vitro and in vivo. Stem Cells Int 2019; 2019: 8475389–8475389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubio-Viqueira B, Jimeno A, Cusatis G, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res 2006; 12: 4652–4661. [DOI] [PubMed] [Google Scholar]
- 19.Izumchenko E, Paz K, Ciznadija D, et al. Patient-derived xenografts effectively capture responses to oncology therapy in a heterogeneous cohort of patients with solid tumors. Ann Oncol 2017; 28: 2595–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang S, Pang L. Comparing statistical methods for quantifying drug sensitivity based on in vitro dose-response assays. Assay Drug Dev Technol 2012; 10: 88–96. [DOI] [PubMed] [Google Scholar]
- 21.Seufferlein T, Porzner M, Becker T, et al. [S3-guideline exocrine pancreatic cancer]. Zeitschrift fur Gastroenterologie 2013; 51: 1395–1440. [DOI] [PubMed] [Google Scholar]
- 22.Huch M, Bonfanti P, Boj SF, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J 2013; 32: 2708–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du Y, Zhao B, Liu Z, et al. Molecular subtyping of pancreatic cancer: Translating genomics and transcriptomics into the clinic. J Cancer 2017; 8: 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aberle MR, Burkhart RA, Tiriac H, et al. Patient-derived organoid models help define personalized management of gastrointestinal cancer. Br J Surg 2018; 105: e48–e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, UEG905183 Supplemental material for Pancreatic cancer-derived organoids – a disease modeling tool to predict drug response by Pierre-Olivier Frappart, Karolin Walter, Johann Gout, Alica K Beutel, Mareen Morawe, Frank Arnold, Markus Breunig, Thomas FE Barth, Ralf Marienfeld, Lucas Schulte, Thomas Ettrich, Thilo Hackert, Michael Svinarenko, Reinhild Rösler, Sebastian Wiese, Heike Wiese, Lukas Perkhofer, Martin Müller, André Lechel, Bruno Sainz Jr Patrick C Hermann, Thomas Seufferlein and Alexander Kleger in United European Gastroenterology Journal