Although analysis of modern and ancient genomes showed that Neanderthals contributed genetic material to the ancestors of extant human populations, when and where Neanderthals interacted with modern human populations remain exciting...
Keywords: Copy number variation, genomic structural variation, haplotype blocks, introgression, selection
Abstract
The time, extent, and genomic effect of the introgressions from archaic humans into ancestors of extant human populations remain some of the most exciting venues of population genetics research in the past decade. Several studies have shown population-specific signatures of introgression events from Neanderthals, Denisovans, and potentially other unknown hominin populations in different human groups. Moreover, it was shown that these introgression events may have contributed to phenotypic variation in extant humans, with biomedical and evolutionary consequences. In this study, we present a comprehensive analysis of the unusually divergent haplotypes in the Eurasian genomes and show that they can be traced back to multiple introgression events. In parallel, we document hundreds of deletion polymorphisms shared with Neanderthals. A locus-specific analysis of one such shared deletion suggests the existence of a direct introgression event from the Altai Neanderthal lineage into the ancestors of extant East Asian populations. Overall, our study is in agreement with the emergent notion that various Neanderthal populations contributed to extant human genetic variation in a population-specific manner.
HUMANS and Neanderthals interbred following the emergence of humans in Eurasia 60,000–50,000 years ago (Green et al. 2010; Prüfer et al. 2014; Moorjani et al. 2016). As a result, all present-day humans from outside of Africa inherit 1–2% Neanderthal DNA in their genomes (Green et al. 2010; Prüfer et al. 2014). The observation that Eurasian genomes harbor similar levels of Neanderthal ancestry was initially interpreted as evidence for a single pulse of introgression that occurred in the Middle East shortly after the migrations of modern humans into Eurasia (Green et al. 2010). Contrary to these initial findings, however, it was subsequently shown that East Asians carry ∼20% more Neanderthal ancestry relative to Europeans (Wall et al. 2013; Sankararaman et al. 2014; Vernot and Akey 2014).
Three scenarios were discussed in the literature to explain this excess Neanderthal ancestry in East Asia. First, it is possible that negative selection has acted in different strengths in East Asian and European populations due to the differences in the effective population sizes of these two human populations. Second, a hypothetical Basal Eurasian population, which has little to no Neanderthal ancestry, may have contributed to present-day Europeans, diluting the overall prevalence of Neanderthal ancestry in this population. Third, an additional pulse of population-specific Neanderthal introgression may have increased the prevalence of Neanderthal ancestry in East Asia.
Having observed a substantial depletion of Neanderthal ancestry in the functional parts of the human genome, Sankararaman et al. (2014) suggested that the excess Neanderthal ancestry in East Asia might be due to the smaller effective population of East Asians and hence less effective purifying selection that has acted against deleterious Neanderthal DNA in this human population. Two independent studies tested this hypothesis and showed that the smaller effective population of East Asians cannot explain the excess Neanderthal ancestry in East Asia (Kim and Lohmueller 2015; Vernot and Akey 2015). Moreover, forward-time simulations indicated that the widespread purifying selection against Neanderthal ancestry in human populations was strongest during the very early few generations following the introgression (Harris and Nielsen 2016; Petr et al. 2019), a time frame largely preceding the split of East Asians and other Eurasians. Although deleterious Neanderthal DNA continued to be purged from human populations, the strength of the selection was diminished after this early phase, effectively not changing the genome-wide Neanderthal ancestry levels from 400 generations following the introgression to the present day (Harris and Nielsen 2016; Petr et al. 2019). Assuming a generation time of 29 years, 400 generations amounts to 11,600 calendar years. Hence, in a conservative estimation, purifying selection has effectively not changed Neanderthal ancestry levels in Eurasia for the past 35,000 years.
A second hypothesis addressing the differential retention of Neanderthal ancestry in East Asia and Europe suggests that the Neanderthal ancestry in Europe was diluted by an ancestry component not carrying Neanderthal introgression. It was previously shown that the proportion of Neanderthal ancestry carried by ancient European and Western Asian human genomes decreases linearly with the proportion of Basal Eurasian ancestry found in the same genomes (Lazaridis et al. 2016). The Basal Eurasian ancestry is derived from a hypothetical population that remained isolated in the Middle East after humans migrated out of Africa and did not admix with Neanderthals as much as other Eurasian populations did. Basal Eurasian ancestry, therefore, carried little or no Neanderthal introgression. This ancestral component later entered the European gene pool via first the expanding agricultural populations during the Neolithic and later the Yamnaya expansion during the Bronze Age (Lazaridis et al. 2016). It is plausible, therefore, that the Basal Eurasian ancestry that present-day Europeans carry might have diluted the Neanderthal ancestry in Europe in the past 10,000 years.
However, Petr et al. (2019) recently showed that there was no significant decrease in the Neanderthal ancestry in Europe during the past 10,000 years. They documented that the inferred decrease in the Neanderthal ancestry in European genomes was due to the design of the F4-ratio test used in Lazaridis et al. (2016). Specifically, the F4-ratio tests used in previous work estimated the proportion of Neanderthal ancestry as the remaining ancestry component after accounting for the sub-Saharan African ancestry found in the test European genomes. Moreover, Lazaridis et al. (2016) used West African populations as the outgroup in their F4-ratio tests, which Petr et al. (2019) and, more recently, Chen et al. (2020) showed to be biased due to the recent gene flow from Eurasia back to Africa and gene flow between West and East African populations. Instead, they developed an alternative model by using the two high-quality Neanderthal genomes (Altai and Vindija Neanderthals; Prüfer et al. 2014, 2017) and designed a new F4-ratio test to avoid the aforementioned bias. Their work showed that there is no significant decrease in the Neanderthal ancestry in Europe in the past 10,000 years (Petr et al. 2019). Combined, these findings indicate that the differential retention of Neanderthal ancestry in different parts of Eurasia is unlikely to explain the excess Neanderthal ancestry in East Asia.
The third hypothesis involving multiple pulses of Neanderthal introgression into modern humans is now gaining more traction. Investigating the joint distribution of the frequencies of introgressed haplotypes in East Asia and Europe, Villanea and Schraiber (2019) showed that Neanderthals contributed genetic material to modern humans in three independent bouts: Once to the common ancestor of East Asians and Europeans, and in two independent bouts to East Asian and European populations following the split of these two human populations. Similarly, Mondal et al. (2019) found evidence for a second pulse of Neanderthal introgression into East Asians from an inferred Neanderthal–Denisovan hybrid population, which likely separated from the parental populations of Neanderthals and Denisovans at an early point following the split of these two hominin populations.
A frequently ignored portion of present-day human genetic diversity in the studies of human population genetics is structural variants; that is, large deletion and duplications, insertions, inversions, and translocations. Yet, structural variants account for a much larger proportion of genetic variation—in the number of affected base-pairs—between any two human genomes (Conrad et al. 2009; Sudmant et al. 2015). It is thus possible to gain additional insights into the admixture history between humans and Neanderthals studying structural variants. In our previous work, we investigated allele sharing involving deletion polymorphisms between present-day humans and ancient human populations and further scrutinized the haplotypic context of these deletions (Lin et al. 2015). We found 38 deletion variants that were likely introgressed from Neanderthals into human genomes (Lin et al. 2015).
Here, we built on these insights and identified thousands of haplotypes introgressed from Neanderthals into Eurasian human genomes. Analyses on these haplotypes indicate introgression from different Neanderthal lineages into Europe as well as East Asia. In parallel, we investigated deletion polymorphisms to identify specific haplotypic variation found in present-day human populations that were potentially descended from different bouts of introgression from ancient hominins. Both haplotype data and deletion allele sharing indicate introgression from different Neanderthals lineages into present-day Europeans and East Asians.
Materials and Methods
S* calculations
To detect haplotypes introgressed from Neanderthals into modern human genomes, we used S* statistics following the framework applied in Taskent et al. (2017). S* uses 20 Eurasian test genomes and 13 Yoruba reference genomes. S* scans each of 20 test genomes in turn for 50-kb windows across the chromosomes (with a step size of 20 kb) and seeks single nucleotide variants (SNVs) where the test genome carries the derived allele and the 13 reference Yoruba genomes carry the ancestral chimpanzee allele. For those set of SNVs, S* follows a dynamic programming algorithm to assess whether SNV pairs segregate together in the remaining 19 Eurasian test genomes and assigns a score (S*-score) to SNV pairs based on a scoring scheme developed in Vernot and Akey (2014). S* then detects the combination of SNVs with the highest score for each test Eurasian genome for each 50-kb window. In this study, we applied S* framework for 200 genomes from each of Western European (Finnish, Great Britain, and CEU populations) and East Asian populations (Han Chinese from Beijing, Han Chinese from South China, and Japanese populations) included in the 1000 Genomes Project, Phase I data set (1000 Genomes Project Consortium et al. 2012).
To derive a null S*-score distribution, we performed coalescent simulations not including introgression by using ms (Hudson 2002). The demographic parameters as well as the recombination rate and number of segregating site parameters for coalescent simulations were used as in Taskent et al. (2017). In particular, we sampled the number of segregating sites from a uniform distribution with a range of 30–350 and a step size of five. Recombination rate, on the other hand, was sampled from a natural-log-transformed uniform distribution ranging from −10.25 to 2.75 cM/Mb with a step size of 0.25 cM/Mb. A total of 20,000 50-kb-long sequences were generated for 13 Africans and 20 East Asians and Western Europeans each by coalescent simulations for each segregating site-recombination rate pair (a total of 68,900,000 simulations for the 3445 recombination rate-number of segregating sites parameter combinations, Supplemental Material, Figure S1). Demographic parameters used in the simulations are as follows:
Divergence of African and non-African populations at 51 KYA.
Divergence of European and East Asian populations at 23 KYA.
Gradual growth of non-African populations from 23 KYA to 5 KYA, to East Asian effective population size (Ne) of 8,879 and European Ne of 9,475. African Ne remained at 14,474 during this period.
Rapid growth of all populations starting at 5 KYA, to a modern-day Ne of 424,000 of Africans, 512,000 of Europeans, and 1,370,990 of East Asians.
Migration rates were fixed as follows: 1.498975 × 10−4 between Africans and the ancestors of Europeans and East Asians, 2.498291 × 10−5 between Africans and Europeans, 7.794668 × 10−6 between Africans and East Asians, and 3.107874 × 10−5 between Europeans and East Asians.
An example ms script is as follows: ms 106 20000 -s 290 -r 2.6679060934e-07 50000 -I 3 26 40 40 0.0 -n 1 58.002735978 -n 2 70.041039672 -n 3 187.55 -eg 0 1 482.46 -eg 0 2 570.18 -eg 0 3 720.23 -em 0 1 2 0.7310 -em 0 2 1 0.7310 -em 0 1 3 0.228072 -em 0 3 1 0.228072 -em 0 2 3 0.909364 -em 0 3 2 0.909364 -eg 0.006997264 1 0 -eg 0.006997264 2 20.89166 -eg 0.006997264 3 30.06376 -en 0.006997264 1 1.98002736 -en 0.031463748 2 0.7774282 -en 0.031463748 3 0.5820793 -ej 0.031463748 3 2 -en 0.031463748 2 0.7774282 -em 0.031463748 1 2 4.386 -em 0.031463748 2 1 4.386 -ej 0.0697674412173913 2 1 -en 0.0697674412173913 1 1.98002736.
S* statistic was then calculated for the Eurasian sequences generated by these simulations and the null S*-score distributions for each segregating site and recombination rate parameter pair were generated.
To compare the empirical S*-scores calculated for the haplotypes detected in the test genomes with the null S*-score distributions, we calculated the total number of segregating sites that S* used for the 33 total modern human genomes (20 Eurasian test genomes , 13 Yoruba reference genomes) within each 50-kb window as well as the average recombination rate for those sites. Recombination rate data for SNVs were obtained from HapMap recombination map data set (International HapMap Consortium et al. 2007). We then compared empirical S*-scores with the null S*-score distributions for sequences generated by coalescent simulations with the number of segregating sites and recombination rate parameters matching the empirical results. Haplotypes detected in human genomes with S*-scores falling above 0.99 quantile value of the null distributions were considered as putatively introgressed haplotypes.
As S* uses genotype information to infer regions where introgressed haplotypes are located, it does not distinguish between phased haplotypes. To detect haplotypes on the phased human genomes, we applied an additional filter by counting the number of derived alleles in each phased chromosome of an individual genome for the SNVs that S* used to detect the putatively introgressed fragments for that individual. Only haplotypes found on the chromosomes carrying more than half of the derived alleles for S* SNVs were retained after this filter. We then merged the overlapping S* haplotypes detected for different present-day Eurasian genomes. A merged haplotype starts from the upstream-most S*-significant SNV to the downstream-most S*-significant SNV for overlapping S* haplotypes and covers the entire overlapping regions. To estimate how closely related the putatively introgressed haplotypes to the archaic genomes, we calculated the average pairwise nucleotide differences (pi-symbol) between the merged, phased S* haplotypes and the two high quality Neanderthal genomes (Altai Neanderthal (Prufer et al. 2014) and Vindija Neanderthal (Prufer et al. 2071)) as well as the Denisovan genome (Meyer et al. 2012).
We used custom python and shell scripts to perform S* statistics, find S*-significant haplotypes, find phased S*-significant haplotypes and compute average pairwise nucleotide differences (π) between the S*-significant putatively introgressed haplotypes and the two Neanderthal genomes. The scripts that we used for these analyses can be found in the following GitHub repository: https://github.com/taskent/Multiple-Neanderthal-Introgression-. We used R scripts for the remaining analyses and to make the figures.
Calculations for residuals for nucleotide difference comparisons:
Given the null hypothesis of one pulse of introgression from the Vindija Neanderthal lineage into the ancestors of Eurasians, we performed a linear regression analysis where π between the S*-significant (putatively introgressed) haplotypes and the Vindija Neanderthal genome was used as the explanatory variable and the corresponding values for the Altai Neanderthal genome were used as the response variable.
Bayesian simulations:
For the Bayesian analysis, we simulated three “populations”: human, Altai, and Vindija. From each population, we have sampled one haploid genome. The reason that we chose one haploid genome per population is that by using just a single genome, the demography becomes irrelevant (since there is no coalescent with only a single genome). In our simulation, the two Neandertal populations join each other in the past within the period 0.1625–0.18125 (in coalescent time units). Assuming Ne is 10,000 and generation time is 20 years, then this period corresponds to 130,000–145,000 years. All three populations join each other at a period between 0.625 and 0.875, i.e., between 500,000 and 700,000 years. Based on the known ages of the actual specimens, we sampled the Altai and Vindija haplotypes at 120,000 and 50,000 years ago, respectively.
Priors for the times of population split (forward in time) or join (backward in time) were uniform (with the values that we specified above). There is migration between the two Neandertal populations for a specified period. This time is distributed uniformly (priors) between the sampling time and the time the two Neandertal populations join each other. Also, there is migration (gene flow) between the Homo sapiens and Altai (in the single introgression model) and between Homo sapiens and the two Neandertals in the double introgression model. Again, the period of introgression is distributed uniformly between the sampling time of Altai (which is older) to the time that the two Neanderthal populations merge. (In the scenario with a single migration event, migration may take place between the sampling time of Altai to the time that all three populations merge). Priors were again uniform. We tried different migration rate (not from a distribution, but distinct values; These values were M = 0.1, 1, 10, 50, i.e., M = 4Nm, where m is the probability of a person being a migrant, i.e., that it has originated in another population than the sampling population). Overall, we simulated the nucleotide differences among these three “populations” for 500 independent fragments, and the total number of simulated data sets is 2000. The Bayes factor values were low (∼1) and we could not conclude if a single or double introgression were preferred.
Simulations to test the probability of allele sharing between Altai Neanderthal and East Asian populations:
We observed that a relatively high allele frequency deletion variant and associated haplotype seen only in the East Asian population (and to a lesser extent in South Asian population) showed clear signatures of introgression specifically from the Altai Neanderthal lineage. To test the probability of this observation, we simulated one pulse of introgression from Vindija-like lineage into the ancestors of Eurasians. We sampled 200 East Asian, 200 European, and 200 African 1-Mb-long haplotypes for present-day modern humans and 2 haplotypes for each of the archaic genomes (Altai-like, Vindija-like, and Denisovan-like). The introgressed SNPs in the Eurasian genomes can be tracked with the msprime code that we used (the code can be found here: github.com/taskent/Multiple-Neanderthal-Introgression-/blob/master/msprime_one_pulse_of_admixture.py). As the deletion located on chromosome 9 is only shared with Altai Neanderthal and East Asians, we have counted number of introgressed derived alleles that are found in the East Asian genomes but not in the European or African genomes (African frequency <0.05) and only shared with Altai Neanderthal (but not with Vindija Neanderthal or Denisovan). We have then divided this number to (1) total number of introgressed SNPs and (2) total number of SNPs where the East Asian genome(s) carry the introgressed segment with the derived allele. Results indicate that observing a locus like chromosome-9 deletion is unlikely under a scenario with only one pulse of introgression from the Vindija lineage into the ancestors of Eurasians. Among the 1087951 total introgressed variants, only at 19 of them is the derived allele shared with East Asians and Altai Neanderthal. This is highly improbable (P = 0.0002). Furthermore, it remains highly improbable even when we consider only those variants where the introgressed variant is the derived allele (N = 96,304) (P = 1.75 × 10−5). The results remain qualitatively unchanged when we added migration between the Neanderthal lineages (point migration rates = 0.1, 0.05, and 0.01) in the simulations.
Shared deletion variants:
Deletion polymorphism data for modern humans were gathered from the 1000 Genomes Project, Phase III (1000 Genomes Project Consortium et al. 2015). The 1000 Genomes Project (Phase III) detected 33,350 large deletions (>50 bp) found polymorphic for 2504 human genomes from across 26 populations. As the identification of deletions in the above-mentioned study was performed following extensive validation efforts, we used this data set in our analyses. The average size of the deletions included in this data set is 12202.65 bp (SD = 36025.85 bp, median size = 3776 bp; Figure S2).
To detect deletion variants shared between ancient hominins and modern humans, we genotyped ancient hominin genomes for the deletion variants found polymorphic in modern humans. Ancient hominin genomes used in this analysis are as follows: high-quality genome of Altai Neanderthal from Siberia (∼50-fold mean coverage; Prüfer et al. 2014), high-quality genome of Vindija Neanderthal from Croatia (∼30-fold mean coverage; Prüfer et al. 2017), low-quality genomes of Goyet Neanderthal from Belgium and Les Cottés Neanderthal from France (2.2-fold and 2.7-fold mean coverages for Goyet and Les Cottés Neanderthals, respectively; Hajdinjak et al. 2018), as well as the high-quality genome of the Denisovan individual from Siberia (∼30-fold mean coverage; Meyer et al. 2012). Although the Altai Neanderthal sample dates back to ∼130,000 years ago (Prüfer et al. 2014), the remaining Neanderthal samples dates to comparably much earlier times, all between 50,000 and 40,000 years ago (Prüfer et al. 2017; Hajdinjak et al. 2018). The lineage ancestral to this latter set of Neanderthals replaced earlier Neanderthal populations in Western Europe, and hence were categorized as late Neanderthals (Hajdinjak et al. 2018).
The .bam files for Neanderthal and Denisovan genomes were downloaded from Max Planck Institute for Evolutionary Anthropology’s internet repositories: Altai Neanderthal, http://cdna.eva.mpg.de/neandertal/altai/AltaiNeandertal/bam/; Vindija Neanderthal, http://cdna.eva.mpg.de/neandertal/Vindija/bam/; Goyet Neanderthal, http://cdna.eva.mpg.de/neandertal/GoyetQ56-1/; Les Cottés Neanderthal, http://cdna.eva.mpg.de/neandertal/LesCottes_Z4-1514/; Denisovan, http://cdna.eva.mpg.de/denisova/alignments/.
We used raw read count data for the ancient hominin genomes. Particularly, we counted the number of raw reads coinciding with the regions where deletion variants were detected in human genomes. Raw read count data for these deletion regions correlates well with the size of the region for all ancient hominin genomes except Goyet Neanderthal genome (Altai Neanderthal, R2 = 0.025, P < 0.001; Vindija Neanderthal, R2 = 0.023, P < 0.001; Denisovan, R2 = 0.013, P = 0.014; Les Cottés Neanderthal, R2 = 0.029, P < 0.001; Goyet Neanderthal, R2 = 0.007, P = 0.22). To detect regions with less than expected number of raw reads, we fitted normal distributions on raw read count data with the mean and SD observed for the ancient hominin genomes (e.g., Figure S3 for Altai and Vindija Neanderthal genomes). Regions with raw read counts below 0.01 quantile value of the normal distribution were considered as deletions for the ancient hominin genome being genotyped. The Goyet Neanderthal genome comprises three .bam files. Thus, we fitted normal distributions on each separate .bam file for Goyet Neanderthal and considered regions with raw read counts below 0.01 quantile values of all three normal distributions as deletions for Goyet Neanderthal genome. Similarly, the Les Cottés Neanderthal genome comprises multiple .bam files. For six out of eight .bam files for the Les Cottés Neanderthal genome, a normal distribution did not seem to be an appropriate approximation to data as below 0.01 quantile values of the normal distributions were <0 or very close to 0. Hence, we removed these .bam files from further analyses. We considered regions with raw read counts below 0.01 quantile values of all remaining normal distributions as deletions for the Les Cottés Neanderthal genome. We detected 296 deletions for Les Cottés Neanderthal, 429 deletions for Goyet Neanderthal, 643 deletions for Vindija Neanderthal, 621 deletions for Altai Neanderthal, and 598 deletions for the Denisovan individual.
As variants introgressed from Neanderthals or Denisovans are not expected to be found in sub-Saharan African genomes, we focused on deletion variants with <0.05 frequency in Yoruba. Intersect function of Bedtools was used to find S*-significant haplotypes overlapping the deletion variants in this filtered data set (Quinlan and Hall 2010). Deletions variants located within S*-significant haplotypes detected for the modern human genomes carrying the same deletion variants were classified as introgressed.
As using only the top 1% of S* haplotypes may underestimate the number of introgressed deletions, we repeated the aforementioned analysis with more permissive criteria. To that end, we used the empirical S*-score distribution and sampled haplotypes with S*-scores above a certain quantile S*-score value. The S*-scores of all haplotypes ranged from 5010 to 1,558,325. The distribution of S*-scores is truncated on the left at 5000 due to intrinsic features of S* statistics. However, a manual investigation of the distribution indicates that a better minimum for the S*-scores would be 10,000 (Figure S1). Thus, we retained only the haplotypes with S*-scores ∼10,000 for further analyses. We fit a normal distribution with the mean and SD of the empirical data, which is truncated on the left at 10,000, and used the quantile values of this distribution. From the 0.01 quantile to the 0.99 quantile and by increasing the quantile threshold by 0.01 at each iteration, we retained only the haplotypes with S*-scores above the quantile threshold value and counted the number of deletions shared and not shared with Neanderthals and found within S* haplotypes (Table S4).
At the 0.8 quantile threshold, S* haplotypes carry >50% of all deletions shared with Neanderthals. Thus, in further analyses, we focused on haplotypes with S*-scores above the 0.8 quantile value. To compare the East Asian and European frequencies of deletions found within this set of S* haplotypes, we used the Mann–Whitney U test (Table S5).
We used the --hap-r2-positions function of vcftools (Danecek et al. 2011) to detect SNVs in linkage disequilibrium with the deletion variants exclusively shared with one Neanderthal lineage.
We focused on one deletion variant found on chromosome 9 and shared exclusively between Asians and Altai Neanderthal. We used VCFtoTree software (Xu et al. 2017) to first align human haplotypes included in the 1000 Genomes Project Phase III data set (1000 Genomes Project Consortium et al. 2015) and the Neanderthal, Denisovan, and chimpanzee haplotypes, and then build a phylogenetic tree with these haplotypes for this deletion variant. We used iTOL (Letunic and Bork 2016) to visualize the tree.
Data availability statement
All data used in this study are available in public domain, the main and supplementary text, or in the supplementary table. Supplemental material available at figshare: https://doi.org/10.25386/genetics.10320743.
Results and Discussion
An estimation of introgressed haplotypes in Eurasia
It has been suggested that the main gene flow from Neanderthals into the ancestors of present-day Eurasian populations originated from the Vindija branch of the Neanderthal phylogeny (Prüfer et al. 2017). It was further suggested that this branch belongs to a late Neanderthal population from which the Vindija Neanderthal descended, which replaced earlier Neanderthal populations, including the population that is represented by the Altai Neanderthal (Hajdinjak et al. 2018). If true, we expect that haplotypes introgressed from Neanderthals into the present-day human populations to be closer to the Vindija Neanderthal genome compared with the Altai Neanderthal genome. To test this hypothesis, we first identified Neanderthal-introgressed haplotypes using S* statistics. This statistic is particularly suitable for our purposes, as it can predict introgressed haplotypes without input from the source of introgression (Vernot and Akey 2014).
We computed S* statistics for 200 individual genomes each of Western European (Finnish, British, and Utah residents with Central and Western European ancestry) and East Asian ancestry (Japanese, Han Chinese from Beijing, and Southern Han Chinese), included in the 1000 Genomes Project, Phase I data set (1000 Genomes Project Consortium et al. 2012). Similar to Vernot and Akey (2014), a null distribution for S*-scores were created with coalescent simulations not including introgression. Haplotypes with S*-scores falling above the 0.99 percentile of the null distribution were considered as S*-significant putatively introgressed haplotypes.
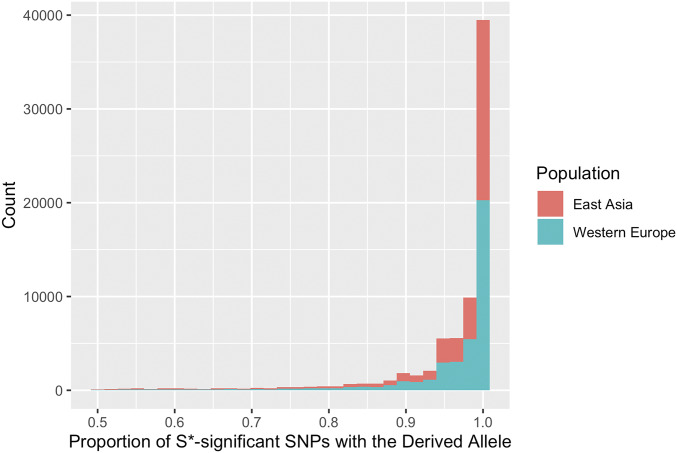
S* uses diploid genotype data to detect introgression at a particular region of the genome. That is, data that S* uses have twos for positions homozygous for the derived allele, ones for heterozygous positions, and zeros for positions homozygous for the ancestral allele. S* treats positions with genotype scores of one and two equally. In other words, for these positions, S* acknowledges that the test genome carries the derived allele. Therefore, it does not estimate on which phased chromosome of the test genome the introgressed haplotype is located. To distinguish the introgressed haplotype from the nonintrogressed human haplotype found on the other phased chromosome of the same genome, we counted the proportion of SNVs that S* used to detect the introgressed haplotype (S*-significant SNVs) using phased data available from the 1000 Genomes Project, Phase I release (1000 Genomes Project Consortium et al. 2012). We found that indeed the vast majority of the S*-significant SNVs reside on a single haplotype (Figure 1). Based on this observation, we considered the haplotype carrying the derived allele for more than half of all S*-significant SNVs as a putatively introgressed phased haplotype. We then merged the overlapping haplotypes. Based on these criteria, we detected in total 9435 and 11,027 phased, putatively introgressed haplotypes for East Asian and Western European genomes, respectively (Table S1).
Figure 1.
Phasing S*′ haplotypes. Distribution of the proportion of S*-significant single nucleotide polymorphisms (SNPs) with the derived alleles on a single phased haplotype per S*-significant region per test genome. Note that only one of the two phased haplotypes per genome, the one which carries the derived allele at >0.5 of all S*-significant SNPs, is shown here. Red and green bars show the frequency clusters of haplotypes detected for East Asian and Western European genomes, respectively, included in the 1000 Genomes Project, Phase I release.
Comparative analysis of introgressed haplotypes suggest additional introgression events in Eurasians
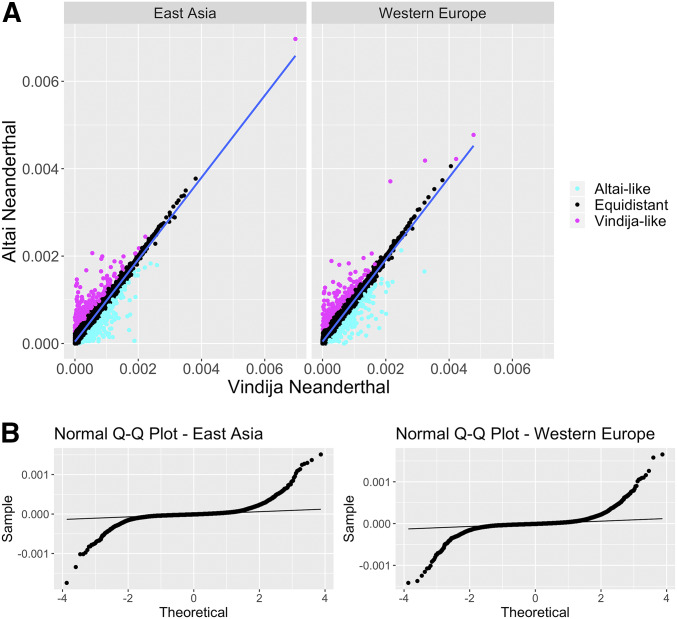
These introgressed haplotypes gave us a suitable framework to compute average pairwise nucleotide differences (π) between the introgressed haplotypes detected in the human genomes and the high-quality Neanderthal genomes (Altai and Vindija Neanderthals) (Figure 2A and Table S2). We found that introgressed haplotypes on average are closer to the Vindija Neanderthal genome than to the Altai Neanderthal genome (Wilcoxon rank-sum test, W = 1.73 × 108, P = 0.0073). This is in agreement with the earlier findings showing that the admixing Neanderthal population was more closely related to the late Neanderthal lineage, which is represented here with Vindija Neanderthal, than to the Altai Neanderthal lineage (Prüfer et al. 2017). However, we also found that a considerable number of haplotypes match one Neanderthal more than the other, creating an upside-down arrow-shaped pattern in Figure 2A. Given the hypothesis of one-pulse of introgression from a lineage closer to Vindija Neanderthal, we expect that the distances of introgressed haplotypes to the Altai Neanderthal genome should be a function of distances to the Vindija Neanderthal genome. To investigate the relationship between these two variables, we performed a linear regression analysis where the distance to the Vindija Neanderthal genome was used to predict the distance to the Altai Neanderthal genome of the form Y = a + bX + ϵ (where X is π between the S* haplotypes and the Vindija Neanderthal genome, Y is π between the S* haplotypes and the Altai Neanderthal genome, and ϵ is residuals).
Figure 2.
Pairwise nucleotide distances between putatively introgressed haplotypes and Neanderthal genomes. (A) Average pairwise nucleotide differences (π) between the Neanderthal-introgressed haplotypes detected for 200 genomes from each of East Asian and European populations included in the 1000 Genomes Project data set (Phase I) and the Vindija (x-axis) and Altai Neanderthal genomes (y-axis). The blue lines show the linear regression of the form Y = 4.3 × 10−5 + 9.4 × 10−1 × X + ϵ, where π between introgressed haplotypes and the Vindija genome (X) is used to predict π between introgressed haplotypes and the Altai Neanderthal genome (Y), and ϵ is residuals. Residuals of these linear regressions are used to detect Vindija-like and Altai-like haplotypes. Haplotypes with residuals below the 0.025 quantile value of the empirical distribution are considered as Altai-like and shown in cyan. Haplotypes with residuals above the 0.975 quantile value of the empirical distribution are considered as Vindija-like and shown in magenta. (B) Normal quantile-quantile (Q-Q) plots for the residuals of the linear regressions between the variables on the x- and y-axes of A.
An analysis of residuals indicate that the residuals deviate from the normal distribution (Kolmogorov–Smirnov test, D = 0.21119, P < 2.2 × 10−16 for East Asia; D = 0.21465, P < 2.2 × 10−16 for Western Europe; Figure 2B). Furthermore, the residuals are on average <0 (−4.862733 × 10−22), suggesting that the distance between the introgressed haplotypes and Vindija Neanderthal genome overestimates the distance to the Altai Neanderthal genome. By calculating the distance from the regression line (Figure 2B), we identified haplotypes that are equidistant to Vindija and Altai Neanderthal genomes in East Asian and Western European populations, as well as those that are significantly closer to one Neanderthal genome than the other (Figure 2A and Table 1). Interestingly, we found a substantial number of haplotypes closer to the Altai Neanderthal genome in both East Asian and European genomes. First, we attempted to estimate the expected number of nucleotide differences (π) between the introgressed haplotypes and the two Neanderthal genomes under one-pulse and two-pulse of introgression scenarios using Bayesian simulations. These estimations would allow us to test whether a single introgression scenario can be rejected. However, the power of these comparisons is greatly hindered by the fact that the Altai Neanderthal individual that is available for analysis was sampled very close in time to the coalescence of late and early Neanderthal lineages. Thus, the expected number of informative, lineage-specific variants to be found in the Altai Neanderthal genome is very low. As a result, our Bayesian approach returned inconclusive results (Bayes factor of ∼1).
Table 1. The number of putatively introgressed haplotypes.
| Equidistant | Altai-like | Vindija-like | |
|---|---|---|---|
| Western Europe | 8766 | 234 | 220 |
| East Asia | 8790 | 229 | 242 |
Haplotypes are categorized based on their distance to early and late Neanderthal genomes.
Next, we conducted a relatively simple, coalescent-based estimation for the π that depends on several assumptions. We found that our observations does not fit with a single introgression model. Instead, we found more than the expected number of Altai-like haplotypes both in European and East asian populations (see Supplementary Materials for details). While our results are in line with the findings of earlier studies inferring a second pulse of Neanderthal introgression into East Asians (Kim and Lohmueller 2015; Vernot and Akey 2015; Vernot et al. 2016), the source of this excess Neanderthal ancestry does not seem to originate from the Altai Neanderthal lineage per se as the proportion of haplotypes showing more than an expected affinity to Altai Neanderthal genome did not differ between East Asians and Europeans (chi-square statistic = 0.08, P = 0.78). Collectively, our results are in line with the findings of Villanea and Schraiber (2019), who found evidence for independent pulses of Neanderthal introgression into both East Asian and European populations following the initial Neanderthal introgression into the ancestors of these two human populations.
Analysis of deletion polymorphisms hint at specific introgression events from the Altai lineage
Once we established that there may be multiple sources of introgression that explain archaic haplotypes in modern human genomes, we widened our search to include large deletion polymorphisms that can be shared by either late or early Neanderthal populations. We are particularly interested in these variants because they are distinctive and so less likely to be false positives. For example, the deletion calls for these regions cannot be attributed to the technical errors due to the nature of ancient DNA (i.e., short sequencing reads are more difficult to be successfully mapped to the reference genome) or the problems associated with calling structural variants in regions enriched for segmental duplications. If these technical confounding effects were present, we would expect them to be present for all Neanderthal genomes. We reasoned that deletion polymorphisms that are introgressed from specific Neanderthal lineages will have the following characteristics. First, they are the derived allele as compared to the chimpanzee allele. Second, they are shared with either the early or late Neanderthal lineages, but not with Denisovans. Third, they are not recurrent in the human and Neanderthal lineages, i.e., they have the same breakpoints.
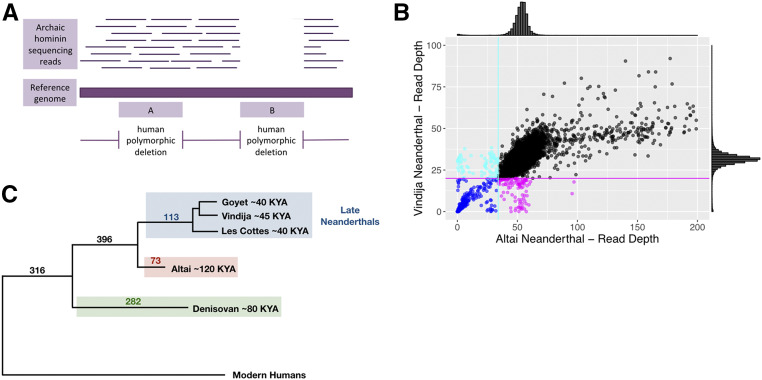
Based on this reasoning, we first genotyped 33,350 polymorphic large deletions (>50 bp) reported for present-day humans in the 1000 Genomes Project, Phase III data set (1000 Genomes Project Consortium et al. 2015) in the genomes of Altai Neanderthal and late Neanderthals (Vindija, Goyet, and Les Cottés Neanderthals) as well as the Denisovan (Figure 3, A and B and Table S3). We found that a total of 32,271 (∼96.8%) large deletions are human-specific and the remaining 1079 (∼3.2%) are shared with at least one ancient hominin (Figure 3C). This value is consistent with our previous estimates of allele sharing (Lin et al. 2015).
Figure 3.
Genotyping deletion variants found polymorphic in modern humans for ancient hominin genomes. (A) Schematic representation of genotyping the Neanderthal genomes for the human-polymorphic deletion variants included in the 1000 Genomes Project data set (Phase III). Horizontal thin lines show the raw sequencing reads for ancient hominin genomes. A horizontal thick line represents the human reference genome on which the raw sequencing reads for the ancient hominin genome are mapped. Regions found deleted in modern humans are shown in the lower part of the figure with pink shaded boxes. Box A represents an example region deleted in some modern human genomes where the ancient hominin genome carries raw sequence reads. Box B represents an example region deleted in some modern human genomes where the ancient hominin genome lacks raw sequence reads. (B) Raw read depth distribution of Neanderthal genomes for the regions found polymorphically deleted in humans. Histograms above the x- and y-axes show the read-depth distribution for the Altai and Vindija Neanderthal genomes. A normal distribution with the mean and SD of the empirical read depth distribution was fitted on the empirical read-depth distribution for the two Neanderthal genomes. Cyan and magenta lines show the 0.01 quantile values of normal distributions (34 and 20 for Altai and Vindija Neanderthal, respectively). Cyan and magenta points below these lines are considered as deleted in the Altai and Vindija Neanderthal genomes, respectively. Blue points show the regions where both Neanderthal genomes are genotyped as deleted. Black points show the regions where both Neanderthal genomes carry the intact sequences. (C) A schematic tree of ancient hominins and modern humans. The numbers of deletion variants shared with each branch of ancient hominins are shown on the corresponding branches.
Among those shared ones, we found 113 and 73 deletions that are shared in a lineage-specific manner with the late and Altai Neanderthal branches, respectively (Figure 3C), including those that may have functional consequences (Figure 6). We considered three scenarios to explain this lineage-specific sharing. First, it is possible that this situation can be explained by incomplete lineage sorting dating back to the common ancestor of humans and Neanderthals, followed by the loss of the deletion in one Neanderthal branch because of drift. These shared deletions may indeed have introgressed from Neanderthals into human populations. Second, they could either represent ancient population structure within the Neanderthal populations and explain our observation without invoking multiple introgressions. Third, it is plausible that these shared deletions are the putative candidates that represent independent introgression events from these specific branches of the Neanderthal phylogeny into modern humans.
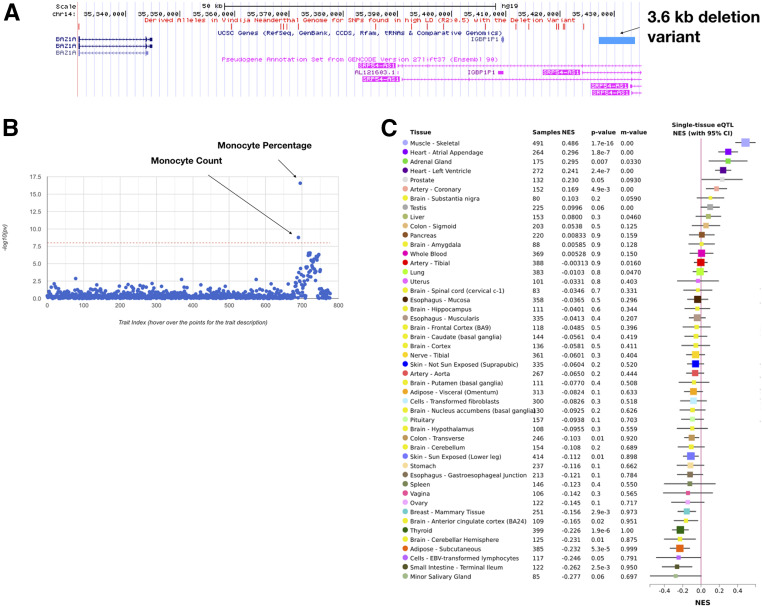
Figure 6.
A deletion variant shared between modern humans and late Neanderthals. (A) UCSC Genome browser snapshot of the genomic region encompassing the deletion variant (esv3634045, position: chr14:35428788-35432438) as well as the SNVs in high LD with the deletion variant (R2 > 0.5). The genotype of Vindija Neanderthal for SNVs in high LD with the deletion variant are shown with red vertical bars above. For all SNVs in high LD with the deletion variant for which genotype data were available for Neanderthals, Vindija Neanderthal carried at least one derived allele, hence the red vertical bars. UCSC genes are shown in dark blue; long noncoding RNAs are shown in pink; the 3.6-kb-long deletion variant is shown in the light blue bar. (B) Haplotype carrying the deletion variant has phenotypic effects in the UK population. An example SNV in high LD with the deletion variant is associated with decreased monocyte count and percentage in the UK population. The x- and y-axes show different traits and the negative log-transformed P-values of the correlation between the trait and the SNV. The dashed red line represents the multiple-hypotheses corrected P-value threshold for significance. The figure was obtained from GeneATLAS data set (Canela-Xandri et al. 2018). (C) GTEx gene expression profile of FAM177A1 under the influence of an SNV in high LD with the deletion variant. SNVs in high LD with the deletion variant affect the expression of FAM177A1 in multiple tissues, including two heart tissues in humans. Tissue names, sample sizes for each tissue, the effect size of the expression quantitative trait locus (eQTL) of FAM177A1, and m-values are shown in the first five columns of the table. Mean effect sizes of FAM177A1 with two SD around the means for each tissue are shown on the sixth column. The figure was obtained from the GTEx Portal on June 10, 2019.
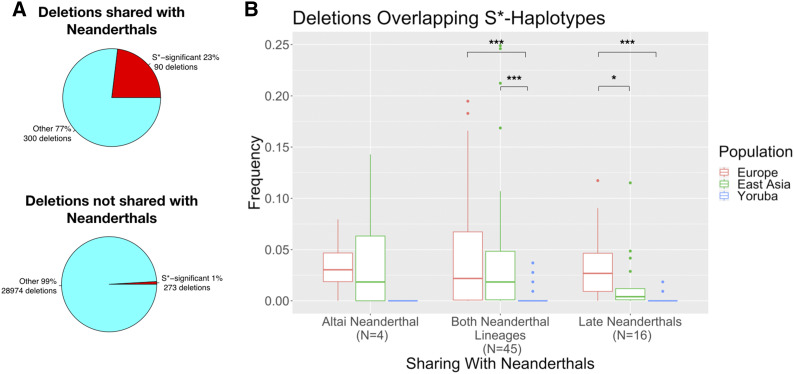
To enrich our data set for the introgressed deletions, we first eliminated all deletion polymorphisms that have >5% allele frequency in Yoruba. Given that Neanderthal introgression affected the ancestral population of present-day Eurasians, we argue that this step eliminated most of the common deletion alleles that are shared because of incomplete lineage sorting. Second, we used the putatively introgressed haplotypes that we identified in the earlier section of this study (top 1% S* haplotypes) and asked how many of the deletions reside in introgressed haplotypes found in the same individuals. We found that 90 out of a total 390 deletions shared with either Neanderthal lineage overlap the region where we detected putatively introgressed haplotypes in the same human genomes carrying the deletion variant, corresponding to 23% of all shared deletions (Figure 4A). The majority of these deletions are not shared with the Denisovan genome (65 out of 90). In a comparative analysis, we found that only ∼0.9% (273 out of 29,247) of the human deletions not shared with Neanderthals are within the putatively introgressed haplotypes (Figure 4A). Compared to each other, these analyses indicate that the deletions that we identified to be shared with Neanderthals clearly reside in putatively introgressed haplotypes more often than expected by chance (chi-square statistic = 1559.9, P < 0.0001).
Figure 4.
Enrichment for introgression and population differentiation of human polymorphic deletion variants shared with Neanderthals. (A) Pie charts show the proportion of deletion variants found (red) and not found (light blue) in S*-significant introgressed haplotypes. Pie charts for deletion variants shared and not shared with Neanderthals are shown above and below, respectively. (B) Frequencies of deletion variants overlapping S*-significant putatively introgressed haplotypes in European, East Asian, and Yoruba populations included in the 1000 Genomes Project data set Phase III release. x-Axis shows with which Neanderthal lineage the deletion variants are shared. Note that the deletion variants shared with Altai and late Neanderthal lineages are exclusively shared with those lineages. Significant frequency differences between populations are shown with asterisks on the top of the plots. * P < 0.05, ** P < 0.01, *** P < 0.001.
As limiting the data set with top 1% S* haplotypes may underestimate the enrichment for introgressed deletions, we progressively loosened the quantile threshold for S* haplotypes. To that end, we used the empirical distribution of S*-scores of all haplotypes detected by S* (see Materials and Methods). From the 0.99 quantile value to 0.01 quantile value and by decreasing the quantile threshold by 0.01 at each iteration, we retained only the S* haplotypes with scores above the corresponding quantile threshold value. We observed that approximately half of all deletions shared with Neanderthals are found within S* haplotypes at 0.8 quantile threshold (Figure S4 and Table S4).
To make sure to eliminate false positives in our data set, we analyzed only those deletion variants that are found within the top 1% S* haplotypes (S*-score quantile >0.99). We identified 4 and 16 deletion variants, which are strong candidates for being exclusively introgressed from Altai Neanderthal and late Neanderthals, respectively. When we examined the allele frequency distribution of these deletion variants, we found that deletion variants exclusively shared with late Neanderthals have significantly higher frequencies in Europe than in East Asia (Wilcoxon rank-sum test, W = 183.5, P = 0.038, Figure 4B). This trend remained unchanged at all quantile thresholds above the 0.8 S*-score quantile value (Table S5). As the size of the putatively introgressed deletion variants does not differ for East Asians and Europeans (average size of putatively introgressed deletions in Europe is 5971.5 bp, average size of putatively introgressed deletions in East Asia is 4857.6 bp; Wilcoxon rank-sum test, W = 24449, P = 0.84), selection occurring in different strengths against introgressed Neanderthal sequences (in this case, the deletion variants) in the two human populations is unlikely to have created the observed result.
This result is especially intriguing given that Neanderthal-introgressed variants on average are found in higher frequencies in East Asia than in Europe both in previous studies (Vernot and Akey 2015) and in our own calculations. One explanation for this observation would be an additional introgression event from a Neanderthal population closer to Vindija Neanderthal into the ancestors of present-day Europeans during their range expansion out of Africa. This scenario was considered in detail and rejected by an excellent simulation-based framework by Currat and Excoffier (2004). However, that study has considered only a single admixing Neanderthal population and did not discriminate between genetic differences among Neanderthal populations. Here, by focusing on only putatively lineage-specific events in an extremely conservative manner, we may have detected a low-level, lineage-specific introgression event into the ancestral European population that coincides with its range expansion. This event was not visible in our broader S* analysis of all introgressed haplotypes possibly because they were shaped by additional introgression events, as well as the potential noise introduced by incomplete lineage sorting and back migration of Western Eurasian groups back in Africa (Mondal et al. 2019; Villanea and Schraiber 2019; Chen et al. 2020). Overall, our study raises interesting questions that may be conclusively answered when additional Neanderthal samples are available for analysis.
The haplotype architecture of deletions shared with specific Neanderthal lineages
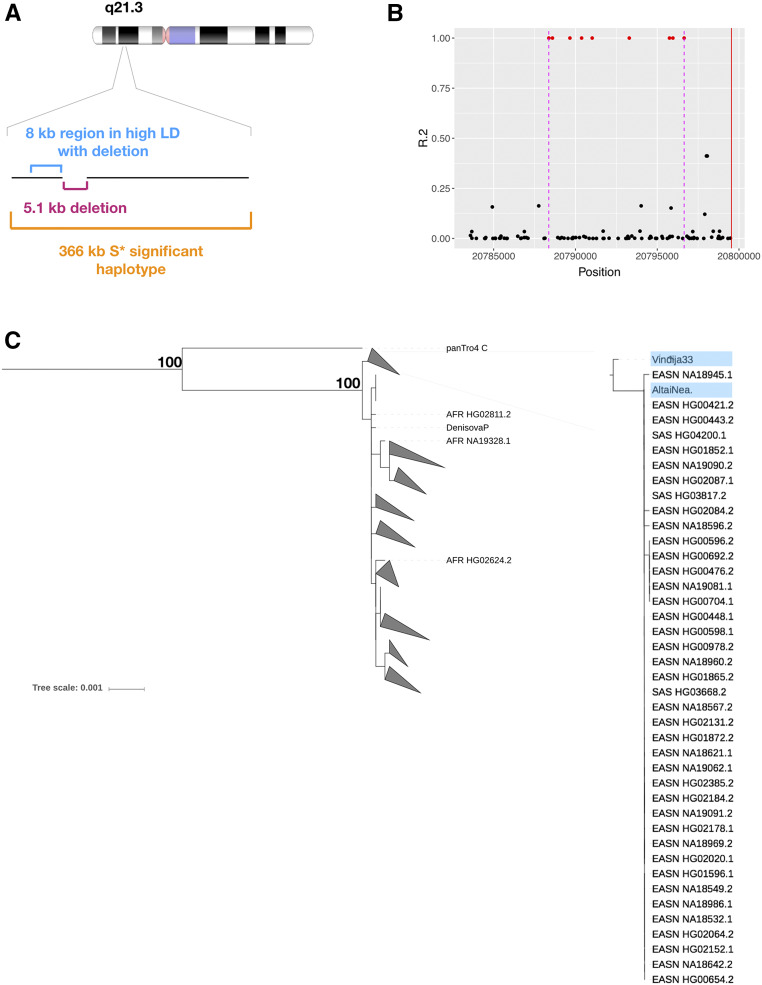
To better understand the haplotypic architecture of the deletions shared with only late or Altai Neanderthal lineages, we calculated the linkage disequilibrium between those deletion variants and the SNVs. We were able to identify SNVs that are in near-perfect LD for the majority of these deletions, allowing us to better resolve their phylogenetic context (Table S6). We were specifically interested in the phylogenetic context of the deletions that are shared only with the Altai Neanderthal lineage. As a case example, we further analyzed the haplotypic variation of one such deletion on chromosome 9 (Figure 5A), for which we were able to identify a “target” region which harbors multiple SNVs in near-perfect linkage disequilibrium with the deletion (Figure 5B). Using data from 5008 present-day human haplotypes, Denisovan, Vindija, and Altai Neanderthal genomes, as well as the chimpanzee reference sequence, we built a maximum likelihood phylogenetic tree of the variation in this target region by using RAxML implemented in VCFtoTree software (Xu et al. 2017) (Figure 5C).
Figure 5.
An example deletion variant shared between East and South East Asians and Altai Neanderthal. (A) The 5.1-kb-long deletion variant is located on the q21.3 segment of chromosome 9. Single nucleotide variants (SNVs) in high LD with the deletion are found in the 8-kb upstream flanking region of the deletion. A 366-kb-long putatively introgressed S* haplotype overlapping the deletion variant was detected for three East Asian chromosomes carrying the deletion. (B) LD between the deletion variant and SNVs found in the upstream flanking region of the deletion. The 8-kb upstream flanking region where SNVs in high LD with the deletion are found is delimited between the purple dashed lines. This region was used to build the phylogenetic tree to understand the evolutionary relationships between the haplotypes carrying and not carrying the deletion. The deletion variant encompasses a 5.1-kb-long region downstream of the starting position shown by the red line. (C) A maximum likelihood phylogenetic tree built with the sequences overlapping the 8-kb upstream flanking region of the deletion variant. All human haplotypes carrying the SNVs in high LD with the deletion variant are found in the same branch of the tree with Neanderthals. This branch is basal to other branches of the tree including other human haplotypes. Significant bootstrap supports (>90) are shown in the tree. The phylogenetic tree was built with RAxML implemented in VCFtoTree software (Xu et al. 2017) and visualized with iTOL (Letunic and Bork 2016).
This tree confirms several of our assumptions with regards to the ancestry of the deletion, which we think was introgressed from a Neanderthal lineage closer to the Altai branch than the Vindija branch. First, as expected, the chimpanzee sequence is an outgroup to all hominin haplotypes. Second, we found that Neanderthal haplotypes diverged from the presumably ancestral branch that includes Denisovans. This ancestral branch harbors all present-day human haplotypes that do not harbor the deletion polymorphism. Third, within the derived branch, Vindija and Altai Neanderthal haplotype further branch into two clusters, where all present-day human haplotypes that harbor the deletion variant reside with the Altai Neanderthal which also has the deletion. To estimate the probability of this observation under a model where we assumed a single introgression from Vindija Neanderthal, we conducted additional, simulation-based analyses with msprime (Kelleher et al. 2016, see Materials and Methods). We found that a single introgression model cannot explain the allele frequency distribution and the introgression patter we observed for this haplotype, even when gene flow from Vindija and Altai Neanderthals were considered (P ≈ 0.0002). These results collectively indicate that this haplotype was introgressed from the Altai lineage, specifically into the ancestors of East Asian populations.
To further investigate this issue, we have conducted a more thorough analysis of the broader haplotype that was detected by S* encompassing the deletion (Figure 5A). Specifically, we mapped the Vindija-matched and Altai-matched derived SNVs across this haplotype block. This analysis revealed an intriguing pattern. We found that Altai Neanderthal-specific alleles homozygously match closely to the introgressed haplotype along the entirety of the ∼366 kb of the introgressed haplotype. In contrast, we found that Vindija Neanderthal matches this haplotype only partially and heterozygously. Although we cannot rule out some sort of structure among Neanderthal lineages that we cannot resolve with the available samples, our results are mostly in line with a model of low-level Altai Neanderthal lineage-specific introgression.
Functional consequences of the introgressed deletions
We then asked whether any of the deletions and associated haplotypes that are shared specifically with either Vindija or Altai Neanderthals have functional consequences. To do this, we first conducted a general function enrichment analysis using GREAT (http://great.stanford.edu/) for the deletion variants that are shared with Neanderthals, but found no significant enrichment. Then, we conducted a phenome-wide association study search in the GWAS Atlas (https://atlas.ctglab.nl/PheWAS), using the “tag” SNVs for these deletions. The tag SNVs are those that are found in high LD with the deletion variants in human populations. We found that out of 20 deletions that are shared with either Vindija or Altai Neanderthals, three of them show associations with traits with P < 10−7 (Table S6). These associations include “Immature fraction of reticulocytes,” “Monocyte percentage of white cells,” and “Diuretics.” Given that the largest variants in each of these haplotypes are deletions, it is highly plausible that they are the causal factor in these associations. We then investigated these particular associations in the UK Biobank data set and were able to confirm one of these associations (“Monocyte percentage of white cells”) with a nominal P < 10−9 in this cohort as well (Figure 6). We further interrogated the haplotype harboring this deletion that is shared with the Vindija but not Altai Neanderthal genome. We found that this haplotype is mostly found in Western Eurasia (∼9% allele frequency) and in lower frequencies in South Asia, but not observed in Eastern Eurasia. This haplotype covers the long noncoding RNA SRP54-AS1 (Figure 6), which has been associated with cardiovascular risk in patients with autoimmune disorders. Further characterizations showed that this noncoding RNA regulates the expression patterns of FAM177A1, which was argued to play a role in vascular inflammation. Indeed, when we searched for expression quantitative trait loci databases, we were able to find that the introgressed haplotypes harboring the deletion were associated with significantly increased expression of FAM177A1 in two different heart tissues (Figure 6). Overall, this haplotype is an example where a Neanderthal-introgressed haplotype has important health consequences through mediating immune response, and in this instance, leading to increased risk of cardiovascular disease.
Summary
Recent studies revealed that admixture between different species of humans was widespread in the ancient past (Gokcumen 2019). In this study, we showed that modern humans share different amounts of single nucleotide as well as large deletion polymorphisms with the two Neanderthal lineages. The Altai Neanderthal lineage, on the one hand, represents the ancestral lineage of Neanderthals and was sampled only in Asia. Late Neanderthals, on the other hand, represents a more derived Neanderthal lineage that replaced the ancestral Neanderthal lineage in Europe ∼50,000 years ago. We showed that although the putatively introgressed haplotypes detected in modern humans genomes from Western Europe and East Asia are on average closer to the Vindija Neanderthal genome (a late Neanderthal genome) than the Altai Neanderthal genome, there are more than expected haplotypes that show excess distances to Vindija Neanderthal genome under a single-pulse introgression model in both East Asia and Western Europe. This indicates that multiple pulses of introgression from different lineages of Neanderthals into modern humans occurred for both East Asians and Western Europeans. In line with these results, we found a deletion variant that is located within a 366 kb introgressed haplotype detected in East Asian genomes and exclusively shared between Altai Neanderthal and extant humans from East and Southeast Asia. Coalescent simulations indicate that the allele sharing observed for this locus is highly unlikely under single-pulse introgression from a lineage closer to Vindija Neanderthal.
Deletion polymorphisms, furthermore, show population differentiation in allele sharing with late Neanderthals for East Asians and Europeans. Specifically, the putatively introgressed deletion variants that are shared with late Neanderthals are found in significantly higher frequencies in Europe than in East Asia. Thus, a second pulse of introgression from a late Neanderthal lineage into the ancestors of Europeans after they split from the East Asians is the most likely scenario. Lastly, we found that a deletion variant that has been introgressed into Europeans from late Neanderthals affects the monocyte count in the UK population and increases the expression of FAM177A1, a gene involved in vascular inflammation, in two heart tissues.
Our results present a more complex admixture history between modern humans and Neanderthals than what was assumed before, and increase our understanding of human evolutionary history. This is in line with the increasing number of studies showing a much more dynamic evolutionary history of humans than previously thought (Xu et al. 2017; Chen et al. 2020; Durvasula and Sankararaman 2020; Rogers et al. 2020). We also provide new questions to be investigated by future studies, which will have more power when more Neanderthal sequences will become available. Finally, we have shown that structural variants such as large deletion polymorphisms, when supplemented with SNVs, provide a powerful tool to study admixture.
Acknowledgments
We thank Joe LaChance and Mehmet Somel for their insightful comments and discussion during the preparation of this manuscript. We acknowledge Justin Bradley for his initial phylogenetic analyses that hinted at multiple introgression events. We thank The National Science Foundation (grant 1714867 to O.G.) for allowing us to explore new avenues in anthropological genomics. We thank the editors and the reviewers for their very helpful and constructive suggestions.
Footnotes
Available freely online through the author-supported open access option.
Supplemental material available at figshare: https://doi.org/10.25386/genetics.10320743.
Communicating editor: R. Nielsen
Literature Cited
- Canela-Xandri O., Rawlik K., and Tenesa A., 2018. An atlas of genetic associations in UK Biobank. Nat. Genet. 50: 1593–1599. 10.1038/s41588-018-0248-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wolf A. B., Fu W., Li L., and Akey J. M., 2020. Identifying and interpreting apparent neanderthal ancestry in african individuals. Cell 180: 677–687.e16. 10.1016/j.cell.2020.01.012 [DOI] [PubMed] [Google Scholar]
- Conrad D. F., Pinto D., Redon R., Feuk L., Gokcumen O. et al. , 2009. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712. 10.1038/nature08516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currat M., and Excoffier L., 2004. Modern humans did not admix with Neanderthals during their range expansion into Europe. PLoS Biol. 2: e421 10.1371/journal.pbio.0020421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E. et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durvasula A., and Sankararaman S., 2020. Recovering signals of ghost archaic introgression in African populations. Sci Adv 6: eaax5097 10.1126/sciadv.aax5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, Abecasis G. R., Auton A., Brooks L. D, DePristo M. A. et al. , 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, Auton A., Brooks L. D., Durbin R. M., Garrison E. P. et al. , 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokcumen O., 2019. Archaic hominin introgression into modern human genomes. Am. J. Phys. Anthropol. (in press). 10.1002/ajpa.23951 [DOI] [PubMed] [Google Scholar]
- Green R. E., Krause J., Briggs A. W., Maricic T., Stenzel U. et al. , 2010. A draft sequence of the neandertal genome. Science 328: 710–722. 10.1126/science.1188021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdinjak M., Fu Q., Hübner A., Petr M., Mafessoni F. et al. , 2018. Reconstructing the genetic history of late Neanderthals. Nature 555: 652–656. 10.1038/nature26151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K., and Nielsen R., 2016. The genetic cost of neanderthal introgression. Genetics 203: 881–891. 10.1534/genetics.116.186890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. 10.1093/bioinformatics/18.2.337 [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium, Frazer K. A., Ballinger D. G., Cox D. R., Hinds D. A. et al. , 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. 10.1038/nature06258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher J., Etheridge A. M., and McVean G., 2016. Efficient coalescent simulation and genealogical analysis for large sample sizes. PLOS Comput. Biol. 12: e1004842 10.1371/journal.pcbi.1004842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. Y., and Lohmueller K. E., 2015. Selection and reduced population size cannot explain higher amounts of Neandertal ancestry in East Asian than in European human populations. Am. J. Hum. Genet. 96: 454–461. 10.1016/j.ajhg.2014.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I., Nadel D., Rollefson G., Merrett D. C., Rohland N. et al. , 2016. Genomic insights into the origin of farming in the ancient Near East. Nature 536: 419–424. 10.1038/nature19310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., and Bork P., 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44: W242–W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-L., Pavlidis P., Karakoc E., Ajay J., and Gokcumen O., 2015. The evolution and functional impact of human deletion variants shared with archaic hominin genomes. Mol. Biol. Evol. 32: 1008–1019. 10.1093/molbev/msu405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Kircher M., Gansauge M.-T., Li H., Racimo F. et al. , 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338: 222–226. 10.1126/science.1224344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal M., Bertranpetit J., and Lao O., 2019. Approximate Bayesian computation with deep learning supports a third archaic introgression in Asia and Oceania. Nat. Commun. 10: 246 10.1038/s41467-018-08089-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorjani P., Sankararaman S., Fu Q., Przeworski M., Patterson N. et al. , 2016. A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years. Proc. Natl. Acad. Sci. USA 113: 5652–5657. 10.1073/pnas.1514696113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petr M., Pääbo S., Kelso J., and Vernot B., 2019. Limits of long-term selection against Neandertal introgression. Proc. Natl. Acad. Sci. USA 116: 1639–1644. 10.1073/pnas.1814338116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S. et al. , 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505: 43–49. 10.1038/nature12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K., de Filippo C., Grote S., Mafessoni F., Korlević P. et al. , 2017. A high-coverage neandertal genome from Vindija cave in Croatia. Science 358: 655–658. 10.1126/science.aao1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., and Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A. R., Harris N. S., and Achenbach A. A., 2020. Neanderthal-Denisovan ancestors interbred with a distantly related hominin. Sci Adv 6: eaay5483 10.1126/sciadv.aay5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S., Mallick S., Dannemann M., Prüfer K., Kelso J. et al. , 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507: 354–357. 10.1038/nature12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant P. H., Rausch T., Gardner E. J., Handsaker R. E., Abyzov A. et al. , 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526: 75–81. 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taskent R. O., Alioglu N. D., Fer E., Melike Donertas H., Somel M. et al. , 2017. Variation and functional impact of neanderthal ancestry in western Asia. Genome Biol. Evol. 9: 3516–3524. 10.1093/gbe/evx216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B., and Akey J. M., 2014. Resurrecting surviving Neandertal lineages from modern human genomes. Science 343: 1017–1021. 10.1126/science.1245938 [DOI] [PubMed] [Google Scholar]
- Vernot B., and Akey J. M., 2015. Complex history of admixture between modern humans and Neanderthals. Am. J. Hum. Genet. 96: 448–453. 10.1016/j.ajhg.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B., Tucci S., Kelso J., Schraiber J. G., Wolf A. B. et al. , 2016. Excavating neandertal and denisovan DNA from the genomes of melanesian individuals. Science 352: 235–239. 10.1126/science.aad9416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanea F. A., and Schraiber J. G., 2019. Multiple episodes of interbreeding between Neanderthal and modern humans. Nat. Ecol. Evol. 3: 39–44. 10.1038/s41559-018-0735-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall J. D., Yang M. A., Jay F., Kim S. K., Durand E. Y. et al. , 2013. Higher levels of neanderthal ancestry in East Asians than in Europeans. Genetics 194: 199–209. 10.1534/genetics.112.148213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Pavlidis P., Taskent R. O., Alachiotis N., Flanagan C. et al. , 2017. Archaic hominin introgression in Africa contributes to functional salivary MUC7 genetic variation. Mol. Biol. Evol. 34: 2704–2715. 10.1093/molbev/msx206 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used in this study are available in public domain, the main and supplementary text, or in the supplementary table. Supplemental material available at figshare: https://doi.org/10.25386/genetics.10320743.