Abstract
Advanced-generation multiparent populations (MPPs) are a valuable tool for dissecting complex traits, having more power than genome-wide association studies to detect rare variants and higher resolution than F2 linkage mapping. To extend the advantages of MPPs in budding yeast, we describe the creation and characterization of two outbred MPPs derived from 18 genetically diverse founding strains. We carried out de novo assemblies of the genomes of the 18 founder strains, such that virtually all variation segregating between these strains is known, and represented those assemblies as Santa Cruz Genome Browser tracks. We discovered complex patterns of structural variation segregating among the founders, including a large deletion within the vacuolar ATPase VMA1, several different deletions within the osmosensor MSB2, a series of deletions and insertions at PRM7 and the adjacent BSC1, as well as copy number variation at the dehydrogenase ALD2. Resequenced haploid recombinant clones from the two MPPs have a median unrecombined block size of 66 kb, demonstrating that the population is highly recombined. We pool-sequenced the two MPPs to 3270× and 2226× coverage and demonstrated that we can accurately estimate local haplotype frequencies using pooled data. We further downsampled the pool-sequenced data to ∼20–40× and showed that local haplotype frequency estimates remained accurate, with median error rates 0.8 and 0.6% at 20× and 40×, respectively. Haplotypes frequencies are estimated much more accurately than SNP frequencies obtained directly from the same data. Deep sequencing of the two populations revealed that 10 or more founders are present at a detectable frequency for > 98% of the genome, validating the utility of this resource for the exploration of the role of standing variation in the architecture of complex traits.
Keywords: budding yeast, de novo assembly, haplotype inference, multiparental populations, Multiparent Advanced Generation Inter-Cross (MAGIC), MPP
A complete understanding of the genetic basis of complex traits is a goal shared by many disciplines. Although much progress has been made in dissecting the genetic architecture of complex traits such as adaptation, disease susceptibility, human height, and crop performance, a major fraction of standing variation for most traits has remained recalcitrant to dissection (Manolio et al. 2009) . This is often referred to as the “missing” heritability problem. Rapid progress in addressing the missing heritability problem seems most likely in model systems that can be genetically and experimentally manipulated in a controlled setting. In contrast to humans, in model genetic systems variants of subtle effect can be validated via allele replacement experiments.
One of the mainstays of modern genetic mapping studies has been the use of pairwise crosses between genetically diverged founder strains. Large segregating populations can then be used to map phenotypes to genotypes. This approach, laid out in its modern form for complex traits, was initially described by Lander and Botstein (1989) and is reviewed in Flint and Mott (2001), Mackay (2001), and Liti and Louis (2012), and has proven to be especially fruitful in budding yeast, such that mapped QTL tend to explain > 70% of the narrow-sense heritability of most traits (Ehrenreich et al. 2010; Bloom et al. 2013, 2015, 2019; Märtens et al. 2016). However, QTL mapping has suffered both from a lack of resolution and a severe undersampling of the functional variation potentially segregating in natural populations. In this regard, association studies enjoy much finer mapping resolution and sample a larger proportion of the variation present in a natural population (Wellcome Trust Case Control Consortium 2007; Visscher 2008). However, large-scale association studies are often underpowered to detect rare alleles (Spencer et al. 2009), regions that harbor multiple causal sites in weak linkage disequilibrium (LD) with one another (Pritchard 2001; Thornton et al. 2013), rare or poorly tagged structural variants (Hehir-Kwa et al. 2016), or variants that are poorly tagged more generally. Furthermore, as genome-wide association studies grow to include tens of thousands of individuals, they can suffer from false positives from population stratification (Berg et al. 2019) or other experimental block artifacts associated with large-scale projects (Sebastiani et al. 2011; Chen et al. 2017).
Advanced-generation multiparent populations (MPPs) consisting of recombinants derived from several founder individuals have been proposed as a bridge between pairwise linkage mapping and association studies in outbred populations (Churchill et al. 2004; Macdonald and Long 2007). MPPs are created by crossing several (inbred or isogenic) founder strains to one another to maximize diversity, and then intercrossing the resulting population for several additional generations to increase the number of recombination events in the population. In many model systems, recombinant inbred lines (RILs) are derived from the MPP via inbreeding. The resulting homozygous RILs are fine-grained mosaics of the original founding strains that have been successfully used to dissect complex traits in Arabidopsis thaliana (Kover et al. 2009; Huang et al. 2011), Drosophila melanogaster (Macdonald and Long 2007; King et al. 2012a,b), Mus musculus (Aylor et al. 2011; Threadgill and Churchill 2012), Saccharomyces cerevisiae (Cubillos et al. 2013), Zea mays (McMullen et al. 2009), Caenorhabditis elegans (Noble et al. 2019 preprint), and several other systems (de Koning and McIntyre 2017). MPP RILs are a powerful resource for dissecting complex traits due to increased mapping resolution relative to F2 populations and increased natural variation sampled by the founders. Furthermore, unlike association studies, both rare alleles of large effect segregating among the founders as well as allelic heterogeneity can be detected in MPP RILs (Long et al. 2014). Although the majority of studies to date have studied RILs derived from MPPs, it is possible to dispense with the creation and maintenance of RILs and sample the MPP directly (Mott et al. 2000; Macdonald and Long 2007), and indeed early MPP efforts did not employ RILs.
Despite the clear advantages of MPPs, only a single MPP has been described in budding yeast (Cubillos et al. 2013), which is surprising as this species is ideally suited in many other ways for the dissection of complex traits. Large population sizes can be maintained in a controlled environment and a few rounds of meiosis result in recombination events spaced at near-genic resolution. The potential of MPPs in budding yeast was demonstrated by Cubillos et al., who crossed four genetically highly diverged strains and intercrossed the resulting population for 12 generations to generate a highly recombined population that has been shown to be capable of mapping complex traits to high resolution (Cubillos et al. 2013, 2017). To expand the potential of budding yeast to contribute to our understanding of complex traits we have developed two large, highly outbred populations of budding yeast derived from a cross of 18 genetically diverged founders. Like previous work, populations were intercrossed for 12 generations to produce highly recombined mosaic populations that capture a large amount of the standing variation present in S. cerevisiae. Here, we describe the derivation of the founders that allows the 18-way cross to be carried out, de novo PacBio (Pacific Biosciences) assemblies of each founder such that all variation segregating in the population is known, and the characterization of 10 haploid recombinant clones from each population to estimate the size distribution of haplotype blocks in the MPPs. We further carry out deep short-read resequencing of the MPPs, estimate founder haplotype frequencies as a function of location in the genome, and show that at Illumina sequencing coverages as low as ∼20–40× haplotype frequencies can be accurately estimated. The MPPs and tools we derive have great utility for dissecting complex traits in yeast.
Materials and Methods
Strains and media
All yeast strains used in this study came from heterothallic, haploid derivatives of a subset of the SGRP yeast strain collection kindly provided by Gianni Liti (Cubillos et al. 2009). A list of strains used, relevant genotypes (before and after our modifications), and their geographical origins is shown in Table 1. Additionally, two mating-type testing yeast strains were used (kindly provided by Ian Ehrenreich) that are selectively killed by the presence of either MATa or MATα haploids, but not by diploids. For propagating plasmids, Escherichia coli strain DH5α was used according to the manufacturer’s recommendations (Invitrogen, Carlsbad, CA). Bacterial transformants were selected on LB agar supplemented with 100 μg/ml ampicillin (LB Amp) (Fisher). Nonselective media for growth and maintenance of all yeast strains included rich media consisting of 1% yeast extract, 2% peptone, and 2% dextrose (YPD) (Fisher). For solid media, 2% agar was added. Additionally, media consisting of 1% yeast extract, 2% peptone, 2% glycerol, and 2.5% ethanol (YPEG) was used to prevent the growth of petite mutants. For selecting yeast transformants, when Ura3MX was the marker, SC drop-out uracil (SC -Ura) plates were used (Sunrise Scientific). When KanMX, HphMX, or NatMX were the markers used, transformants were selected on YPD plates supplemented with 200 μg/ml of G418, 300 μg/ml of hygromycin B (“hyg”), or 100 μg/ml nourseothricin sulfate (“cloNAT”), respectively. For counterselection of yeast that lost the Ura3MX marker, SC media supplemented with 1 mg/ml 5-FOA was used. Two types of sporulation media were used in this study. Type 1 consisted of 1% potassium acetate, 0.1% yeast extract, and 0.05% dextrose (“PYD”) to which ampicillin was added to a final concentration of 50 μg/ml, while type 2 consisted of 1% potassium acetate and a 1× dilution of a 10× amino acid stock [composed of 3.7 g of CSM -lysine (Sunrise Scientific) supplemented with 10 ml of 10 mg/ml lysine in 1 liter total volume], pH adjusted to 7 (“PA7”). Just before use, ampicillin was added to PA7 to a final concentration of 100 μg/ml.
Table 1. An overview of the strains used in this study.
| ADLa | NCYC | Isolate | Origin | Original genotype | Modified genotypeb |
|---|---|---|---|---|---|
| A1 | 3597 | DBVPG6765 | Europe | MATa, ho::HygMX,ura3::KanMX-Barcode | MATa, hoΔ, ura3::KanMX-Barcode, ygr043C::NatMX |
| A2 | 3600 | DBVPG6044 | West Africa; wine | MATa, ho::HygMX,ura3::KanMX-Barcode | MATa, hoΔ, ura3::KanMX-Barcode, ygr043C::NatMX |
| A3 | 3607 | YPS128 | USA; soil beneath Quercus alba | MATa, ho::HygMX,ura3::KanMX-Barcode | MATa, hoΔ, ura3::KanMX-Barcode, ygr043C::NatMX |
| A4 | 3605 | Y12 | Japan; sake | MATa, ho::HygMX,ura3::KanMX-Barcode | MATa, hoΔ, ura3::KanMX-Barcode, ygr043C::NatMX |
| A5 | 3586 | YIIc17_E5 | France; wine | MATa, ho::HygMX,ura3::KanMX-Barcode | MATa, hoΔ, ura3::KanMX-Barcode, ygr043C::NatMX |
| A6 | 3591 | BC187 | USA; wine | MATa, ho::HygMX,ura3::KanMX-Barcode | MATa, hoΔ, ura3::KanMX-Barcode, ygr043C::NatMX |
| A7 | 3590 | SK1 | USA; soil | MATa, ho::HygMX,ura3::KanMX-Barcode | MATa, hoΔ, ura3::KanMX-Barcode, ygr043C::NatMX |
| A8 | 3598 | L_1374 | Chile; wine | MATa, ho::HygMX,ura3::KanMX-Barcode | MATa, hoΔ, ura3::KanMX-Barcode, ygr043C::NatMX |
| A9 | 3602 | UWOPS03_461_4 | Malaysia; nectar, Bertram palm | MATa, ho::HygMX,ura3::KanMX-Barcode | MATa, hoΔ, ura3::KanMX-Barcode, ygr043C::NatMX |
| A10c | 3604 | UWOPS05_227_2 | Malaysia; stingless bee, near Bertram palm | MATa, ho::HygMX,ura3::KanMX-Barcode | MATa, hoΔ, ura3::KanMX-Barcode, ygr043C::NatMX |
| A11 | 3592 | YJM978 | Italy; vagina, clinical isolate | MATa, ho::HygMX,ura3::KanMX-Barcode | MATa, hoΔ, ura3::KanMX-Barcode, ygr043C::NatMX |
| A12 | 3594 | YJM975 | Italy; vagina, clinical isolate | MATa, ho::HygMX,ura3::KanMX-Barcode | MATa, hoΔ, ura3::KanMX-Barcode, ygr043C::NatMX |
| B1 | 3622 | DBVPG6765 | Europe | MATα, ho::HygMX,ura3::KanMX-Barcode | MATα, hoΔ, ura3::KanMX-Barcode, ygr043C::HygMX |
| B2 | 3625 | DBVPG6044 | West Africa; wine | MATα, ho::HygMX,ura3::KanMX-Barcode | MATα, hoΔ, ura3::KanMX-Barcode, ygr043C::HygMX |
| B3 | 3632 | YPS128 | USA; soil beneath Q. alba | MATα, ho::HygMX,ura3::KanMX-Barcode | MATα, hoΔ, ura3::KanMX-Barcode, ygr043C::HygMX |
| B4 | 3630 | Y12 | Japan; sake | MATα, ho::HygMX,ura3::KanMX-Barcode | MATα, hoΔ, ura3::KanMX-Barcode, ygr043C::HygMX |
| B5 | 3611 | 273614N | UK; fecal sample, clinical isolate | MATα, ho::HygMX,ura3::KanMX-Barcode | MATα, hoΔ, ura3::KanMX-Barcode, ygr043C::HygMX |
| B6 | 3631 | YPS606 | USA; bark of Q. rubra | MATα, ho::HygMX,ura3::KanMX-Barcode | MATα, hoΔ, ura3::KanMX-Barcode, ygr043C::HygMX |
| B7 | 3624 | L_1528 | Chile; wine | MATα, ho::HygMX,ura3::KanMX-Barcode | MATα, hoΔ, ura3::KanMX-Barcode, ygr043C::HygMX |
| B8 | 3614 | UWOPS83_787_3 | Bahamas; fruit, Opuntia megacantha | MATα, ho::HygMX,ura3::KanMX-Barcode | MATα, hoΔ, ura3::KanMX-Barcode, ygr043C::HygMX |
| B9 | 3609 | UWOPS87_2421 | USA; cladode, O. megacantha | MATα, ho::HygMX,ura3::KanMX-Barcode | MATα, hoΔ, ura3::KanMX-Barcode, ygr043C::HygMX |
| B10c | 3628 | UWOPS05_217_3 | Malaysia; nectar, Bertram palm | Mat α, ho::HygMX,ura3::KanMX-Barcode | MATα, hoΔ, ura3::KanMX-Barcode, ygr043C::HygMX |
| B11 | 3618 | YJM981 | Italy; vagina, clinical isolate | MATα, ho::HygMX,ura3::KanMX-Barcode | MATα, hoΔ, ura3::KanMX-Barcode, ygr043C::HygMX |
| B12 | 3613 | Y55 | France; grape | MATα, ho::HygMX,ura3::KanMX-Barcode | MATα, hoΔ, ura3::KanMX-Barcode, ygr043C::HygMX |
ADL, [Anthony D. Long]; NCYC, [National Collection of Yeast Cultures].
These are the abbreviated names used throughout this manuscript. Note that all A strains are MATa and all B strains MATα.
Bold text indicates changes made from the original strain genotypes.
These two strains were excluded from subsequent experiments as they mate poorly with the other strains.
Modification of 24 haploid budding yeast strains to create founders for the synthetic population
The strains used in this study were modified by generating clean deletions of the HO gene to recover the HphMX marker, followed by replacement of a pseudogene, YCR043C, which is closely linked to the mating type locus, with either a NatMX cassette in MATa haploids or a HphMX cassette in MATα haploids. This manipulation was carried out to enable high-throughput selection of diploids.
The HphMX marker in HO was recovered via transformation with a URA3 cassette flanked with direct repeats and selection on URA plates followed by selection on 5-FOA plates to recover URA3. The URA3 cassette was assembled from four fragments: a pBluescript II KS(+) backbone linearized with EcoRV and gel purified (for propagation in E. coli), the URA3 gene from Candida albicans with flanking 500-bp direct repeats from Aschbya gossypii (pAG61, #35129; Addgene), and a 450-bp region directly upstream of the HO gene and a 390 bp region directly downstream of the HO gene. Primers pAG61_HO-F/R were used to amplify URA3 and the flanking direct repeats, while primers HO-US F/R and HO-DS F/R were used to amplify the regions flanking the HO gene from strain DBVPG6765. Primers used in this study are listed in Supplemental Material, Table S1 and included overhangs to allow for HiFi assembly. The four fragments were assembled using the New England Biolabs (Beverly, MA) HiFi Assembly Master Mix according to the manufacturer’s recommendations, transformed into chemically competent DH5α (Invitrogen), and recovered on LB Amp plates. Recovered URA plasmid cassettes with HO flanking sequences were PCR amplified from the plasmid template using primers HO-US-F and HO-DS-R, transformed into all 24 haploid strains using a standard lithium acetate protocol, and plated onto SC -Ura plates. Single colonies were restreaked onto SC -Ura (X2), and final colonies were tested for the presence of the KanMX4 marker and absence of HphMX4 marker via G418 and hyg plating, respectively. Overnight cultures of successfully knocked-out transformants were spread onto 5-FOA plates and grown for 2 day at 30° to select for cells that had “popped out” the Ura3 cassette. Single colonies were restreaked onto 5-FOA plates (2×). DNA was extracted (adapted from Cold Spring Harbor handbook, p. 116) from the resulting colonies, and DNA amplicons spanning the HO locus were obtained and Sanger sequenced to confirm the clean deletion of the HO gene.
To delete YGR043C in the 24 newly generated haploid hoΔ Ura3::KanMX4 strains, oligos were ordered from Integrated DNA Technologies that amplify the entire MX4 cassette, including the promoter and terminator regions, and were tailed with 100 base pairs of homology to the regions immediately upstream and downstream of the YGR043C coding sequence. Either pAG32 (#35122; Addgene) or pAG25 (#35121; Addgene) were used as a template to generate knockout constructs that incorporated the HphMX4 or NatMX4 cassettes, respectively. PCR reactions were cleaned up to remove unamplified circular plasmid template by gel extraction followed by digestion with DpnI and a PCR cleanup reaction (PCR purification kit; QIAGEN, Valencia, CA). MATa yeast were then transformed with the cloNAT resistance cassette, while MATα yeast were transformed with the hyg resistance cassette using the standard lithium acetate protocol, and selecting on YPD supplemented with cloNAT and G418, or hyg and G418, respectively. This double selection with G418 was done to ensure that cassette swapping had not occurred. To ensure YGR043C had been correctly replaced in each strain, the region was amplified and Sanger sequenced. All 24 newly generated strains were checked again for HO deletion using the HO-big-flank-F/R primers. The strains were also checked to ensure they had maintained the correct barcodes originally inserted (Cubillos et al. 2009) throughout all the manipulation steps by amplifying the barcodes using the barcode-check-F/R primer pair and Sanger sequencing the amplicons using the M13(-47)F primer. As a final check, all 24 haploid strains were streaked onto YPD supplemented with hyg and cloNAT to ensure that none of the strains could grow on both antibiotics.
The 18-way crossing scheme, version 1
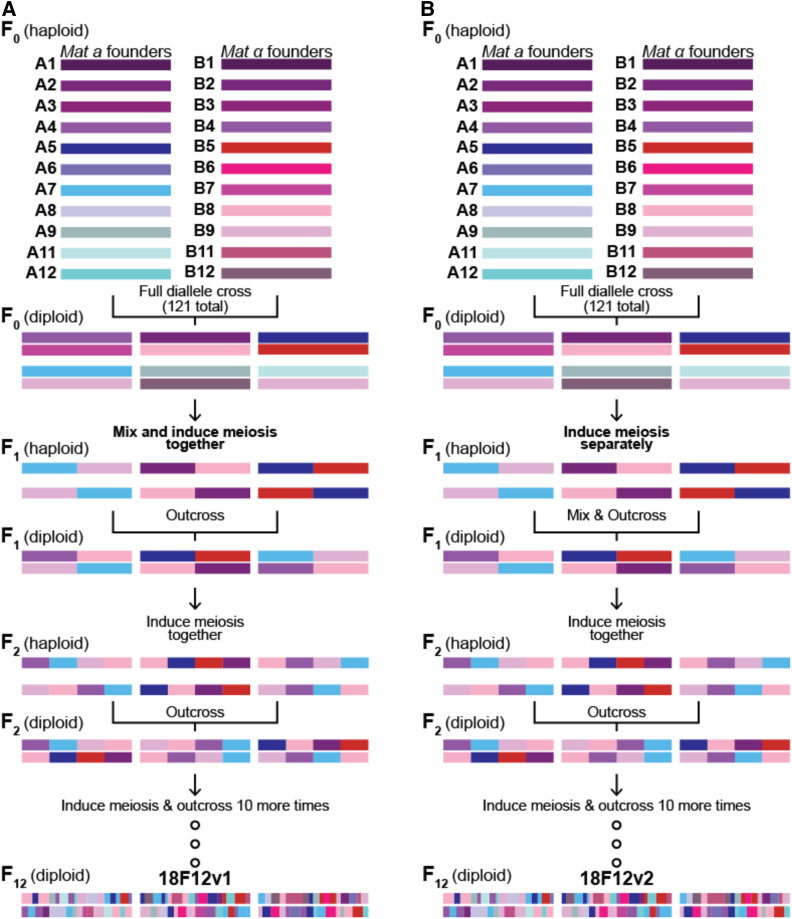
A full diallele cross of 11 MATa and 11 MATα strains (excluding strains A10 and B10) was carried out (with four strains in common). A schematic of the mating scheme is shown in Figure 1A, while Table 1 lists the strains used in this study. Strains A1–A5 and A6–A12 (excluding A10) were struck in horizontal rows onto two YPD plates each (total of four YPD plates), then strains B1–B5 and B6–B12 (excluding B10) were each struck in vertical rows onto two of the YPD plates such that each B strain intersected with each A strain. All 121 pairwise combinations of the A and B strains were thus represented across the four YPD mating plates. Mating occurred overnight at 30° after which diploids were selected by replica plating onto YPD plates with hyg and cloNAT. A single colony from each of the 121 crosses was then incubated overnight in YPD with hyg and cloNAT at 30° at 180 rpm. Equal volumes of each culture and 30% glycerol were used to make frozen stock that were then archived at −80°. An equal volume from each diploid culture was then combined to make the 18-way population, which was washed twice with PYD + ampicillin then split into two 1-liter flasks with 200 ml total PYD + ampicillin each. Sporulation was carried out for 5 days en masse at 30° at 180 rpm to complete the first round of outcrossing.
Figure 1.
Schematic of the outcrossing process used to make the two 18F12 diploid populations. Both populations were established by a full diallele cross of all 22 isogenic haploid founder strains. A1/B1, A2/B2, A3/B3, and A4/B4 are different mating types of the same strains and are the same strains used in Cubillos et al. (2013). In (A), all pairwise crosses were mixed before the first round of sporulation. This is in contrast to (B), in which mixing did not occur until after an initial sporulation event. In both cases, mixed populations were taken through additional rounds of sporulation and random mating for a total of 12 meiotic generations.
Additional outcrossing in the 18-way cross, version 1
Eleven additional cycles of mass sporulation followed by random mating were carried out for a total of 12 rounds of outcrossing for both replicates (Table S3). After sporulation, 50 ml of culture was spun down at 2000 × g for 2 min and resuspended in 1 ml of Yeast Protein Extraction Reagent. Samples were transferred to a 1.5-ml centrifuge tube and vortexed. Cells were washed twice and resuspended in 500 μl of ddH2O with 5 μl of 5 U/μl zymolyase. The tubes were shaken vigorously in a Geno Grinder 2000 at 750 shakes/min for 45 min. Next, 500 μl of 400-μm silica beads were added to the samples, which were again put in the Geno Grinder 2000 for 5 min at 1500 shakes/minute. The supernatant was transferred to a fresh 1.5-ml centrifuge tube, washed once in YPD, resuspended in 500 μl of YPD, and transferred into 50 ml of YPD in a 1-liter flask. Mating was carried out overnight at 30° at 40 rpm. The next day, mated cells were harvested, transferred to YPD with cloNAT, hyg, and amp, and incubated overnight at 30° at 180 rpm. The next day, 7 ml from the overnight culture was used to make glycerol stock while 5 ml was harvested, washed twice, and resuspended in 200 ml of PYD + ampicillin. Sporulation was carried out for 5 days at 30° at 180 rpm (see Table S2). If the experiment had to be paused, 5 ml of glycerol stock from the most recently completed cycle was used to begin the next cycle of sporulation.
The 18-way crossing scheme, version 2
A full diallele cross of the same 11 MATa and 11 MATα founder strains used to create 18F12v1 was again carried out (Figure 1B). To initiate this process, equal volumes from cultures containing each MATa founder strain were mixed with each MATα founder strain in all 121 possible pairwise combinations in 24-well deep-well plates (hereafter “24DWPs”) in a total volume of 1 ml of YPD (no ampicillin added) (see Note S1). Mating was carried out in liquid culture for 4–5 hr at 30° at 50 rpm, after which mating was verified by checking for the presence of zygotes and/or shmooing under a microscope. At this point, 1 ml of YPD supplemented with 200 μg/ml cloNAT and 600 μg/ml hyg was added to each culture (the final concentrations of cloNAT and hyg were 100 μg/ml and 300 μg/ml, respectively) to select for successfully mated diploids, and incubated overnight at 200 rpm at 30°. After overnight selection, 140 μl from each culture was combined with 140 μl of 30% glycerol to make frozen stock of each cross. The remaining cultures were harvested at 1500 rpm for 5 min, and the pellets were washed and then resuspended in 4 ml of PA7 + ampicillin. Sporulation was carried out for 6 day at 30° at 275 rpm in the 24DWPs.
All 121 sporulating cultures were checked using a microscope to determine the amount of sporulation that occurred; cultures were graded on a scale of 0–5, with 0 being no sporulation and 5 being almost complete sporulation. Crosses that did not sporulate were excluded from subsequent steps (see Table S2). After checking for sporulation, cultures were harvested, washed, and then resuspended in 500 μl of spore isolation solution (hereafter “SIS”: 25 U zymolyase, 10 mM DTT, 50 mM EDTA, and 100 mM Tris-HCl, pH 7.2, made up to 500 μl) and incubated for 1 hr at 30° at 250 rpm to spheroplast cells. Cultures were then harvested and resuspended in 1% Tween 20 to selectively lyse unsporulated cells. Following this, cultures were again harvested and resuspended in 500 μl of spore dispersal solution (hereafter “SDS”: 1 mg lysozyme, 5 U zymolyase, 1% Triton X-100, 2% dextrose, and 100 mM PBS, pH 7.2, made up to 500 μl). Cultures were transferred to Eppendorf tubes with 500 μl of 400 μm beads and bead milled using a Geno Grinder 2000 at 1500 strokes per minute for 5 min to break up tetrads, after which all cultures were placed at 4° overnight. The next day, all tubes were vortexed at high speed for 30 sec, the supernatant was transferred to a 24DWP, 500 μl of 100 mM PBS, pH 7.2 was added back to the beads, followed by vortexing for 30 sec and transferring to the same wells of a 24DWP to maximize recovery of spores from the beads. Cultures were washed once in PBS, then resuspended in 500 μl of 100-mM PBS, pH 7.2, and 100 μl was transferred to a 96-well clear plate to measure the OD630 of each culture in duplicate using a BioTek Synergy HT plate reader. OD630 measurements were then used to normalize the densities of spores from each cross that were pooled together (see Note S2). The spore pool was washed twice with 5 ml of YPD, then resuspended in 12.5 ml of YPD. This culture was split in half and transferred to two 250-ml flasks, each with 6.25 ml of YPD, to establish two replicate populations. Mating was carried out overnight at 30° with gentle shaking at 40 rpm. The next day, 12.5 ml of YPDach (a 2× mix of ampicillin, cloNAT, and hyg) was added to each culture to select for diploids. Cultures were incubated overnight at 200 rpm at 30°. This established replicate F2 populations of the 18-way cross, version 2 (hereafter “18F2v2”). The following day, 7 ml of the replicate populations were frozen down at −80° with an equal volume of 30% glycerol. The remaining volume was spun down and used to initiate a second round of outcrossing.
Additional outcrossing in the 18-way cross, version 2
Eleven additional cycles of mass sporulation followed by random mating were carried out for a total of 12 rounds of outcrossing for both replicates. As replicate 2 was treated differently during a couple of cycles, replicate 1 was the population chosen for subsequent analyses and, as such, will be the only replicate of version 2 described further. Each cycle consisted of 3–6 days of sporulation after which diploids were randomly mated for 3–4 hr. This was followed by an overnight selection step in YPDach to enrich for mated diploids. After selection, an aliquot of each population was frozen down at −80° with the remaining culture used to initiate the next cycle of outcrossing. Table S3 enumerates the days of sporulation for each cycle as well as additional details regarding the culturing conditions for both versions of the 18-way population. After each round of sporulation, cultures were processed as detailed above with the following modifications: 5 ml of SIS, 10 ml of 1% Tween 20, and 5 ml of SDS were used to kill vegetative cells. Tetrads were disrupted by bead milling at 1500 strokes/min using a Geno Grinder 2000 for 25–45 min. The contents of the tubes were mixed thoroughly with a pipette to ensure maximal recovery of cells from the bead slurry. The supernatant was then transferred to 50-ml Falcon tubes, after which 500 μl of YPD + ampicillin was added back to the tubes, which were then briefly vortexed at the highest setting. The supernatant was transferred to the same 50-ml Falcon tube. Cultures were harvested, washed, and then resuspended in 5 ml of YPD + ampicillin. At this point, cells were carefully mixed by pipetting and then transferred to a 250-ml Erlenmeyer flask with 7.5 ml of YPD + ampicillin. Spores were mated for 3–4 hr at 30° at 40 rpm, after which the presence of shmoos and/or zygotes was checked under the microscope. Next, 12.5 ml of YPDach was added to the mated cells, which were incubated overnight at 200 rpm at 30°. The next day, cells were transferred to 50-ml Falcon tubes, and 7 ml of culture was mixed with an equal volume of 30% glycerol to make frozen stock, while the rest of the culture was spun down at 3000 rpm for 5 min. Cultures were washed twice, resuspended in 25 ml of PA7, then transferred to a 250-ml flask with 25 ml of PA7 and 50 μl of 100 mg/ml ampicillin, and sporulated at 30° at 275 rpm to initiate the next cycle of outcrossing. Following the twelfth cycle of sporulation followed by random mating, cells were transferred to 1-liter flasks with 187.5 ml of YPDach and incubated overnight at 30° at 200 rpm. The following day, all 200 ml of culture was mixed with an equal volume of 40% glycerol and frozen down at −80° in a combination of 2-ml cryotubes and 15-ml Falcon tubes.
Whole-genome sequencing of the haploid founder strains
All 18 founder strains were sequenced using a combination of PacBio long-read and Illumina short-read technology. PacBio sequencing data were available from a previous study for 6 of the 18 strains (founders AB1–4, A7, and A9) (Yue et al. 2017), which were downloaded and reassembled using our pipeline so that all assemblies were directly comparable. The remaining strains were struck out onto YPD plates for 3 days at 30°, after which a single colony was inoculated into 50 ml of YPD + ampicillin and incubated at 30° at 200 rpm overnight. DNA was extracted using the QIAGEN G-tip DNA extraction kit. Purified genomic DNA (gDNA) was sheared using 24-gauge blunt needles. The resulting sheared gDNA samples were quality checked by a Field-inversion gel electrophoresis run at 134 V overnight and concentrations were measured using Qubit. Sample were considered acceptable if the majority of gDNA was sheared to between 20 and 100 kb. In our hands, carefully controlling the gDNA size distribution results in longer N50 PacBio reads, which gives better de novo assemblies with less data. SMRTbell libraries were prepared and sequenced at the University of California, Irvine Genomics High-Throughput Facility using a PacBio RSII machine. The details of PacBio library creation for the purpose of de novo genome assembly are described in Chakraborty et al. (2016). The average per-site coverage of the six previously sequenced strains was 365× as compared with 59× for the 12 strains sequenced in our hands, while the average PacBio read N50 for the previously sequenced strains was 5.73 kb as compared with 11.65 kb for the strains sequenced by our laboratory.
Libraries for Illumina sequencing were made for all 18 founder strains. The same gDNA that had been used to prep the SMRTbell libraries was used to make Illumina libraries. The gDNA from the six remaining strains was prepared using the QIAGEN G-tip kit as above. All gDNA was sheared to ∼300–400 bp using a Covaris S220 Focused Acoustic Shearer with the following settings: peak incident power (W) of 140, duty factor of 10%, cycles per burst of 200, treatment time of 65 sec, temperature of 4°, and water 12. Illumina compatible libraries were prepared using the NEBNext Ultra II DNA Library Prep kit along with the NEBNext Multiplex Oligos for Illumina (Index Primer Set 1), as per the manufacturer’s recommendations. Adaptor-ligated DNA was size-selected and PCR-enriched for five cycles, followed by cleanup of the PCR reaction using AMPure XP Beads as per the NEBNext Ultra II DNA Library Prep protocol. Sequencing was carried out using the Illumina HiSeq4000 with PE100 or PE150 reads (see Note S3). The average per-site coverage of the 18 founder strains was 290×, with the lowest coverage being 186× (founder B11) and the highest coverage at 374× (founder AB4).
Genome assembly
We assembled the PacBio reads using canu v1.7 (commit r8700; options: corMhapSensitivity = high, corOutCoverage = 500, minReadLength = 500, corMinCoverage = 0, and correctedErrorRate = 0.105) (Koren et al. 2017). We generated hybrid assemblies using the PacBio and Illumina reads for the 12 strains for which we generated the PacBio reads. The PacBio reads from the six strains from Yue et al. (2017) were too short to assemble with DBG2OLC, the hybrid assembler we used (Ye et al. 2016). The DBG2OLC hybrid assemblies were used to fill gaps in the corresponding canu assemblies using quickmerge, following the two-step merging approach (Chakraborty et al. 2016; Solares et al. 2018). The PacBio reads from Yue et al. were sequenced using an older chemistry of PacBio (P4-C2) than our PacBio reads (P6-C4), so they required a different algorithm for optimal polishing than the assemblies created with the P6-C4 reads. Hence, we polished the P4-C2-based assemblies twice using Quiver and the P6-C4-based assemblies twice using Arrow (smrtanalysis v5.2.1). Finally, we polished all assemblies twice with the paired-end Illumina reads using Pilon (Walker et al. 2014).
Benchmarking Universal Single-Copy Orthologs assessment
We estimated the number of fungi Benchmarking Universal Single-Copy Orthologs (BUSCOs) (n = 290) in each polished assembly using BUSCO v3.0.2 (Waterhouse et al. 2018) (Table 2). For the augustus gene prediction step in BUSCO we used “saccharomyces_cerevisiae_S288C” as the species option.
Table 2. Assembly statistics for the 18 sequenced founder strains.
| Strain | Assembly size (Mb) | Assembly N50 (kb) | Assembly Quality value | Assembly BUSCO (complete) | Total PacBio data (Mb) | PacBio read N50 (kb) | Total Illumina data (Mb) | Type of Illumina reads | × PacBio coverage | × Illumina coverage |
|---|---|---|---|---|---|---|---|---|---|---|
| A5 | 12.13 | 757 | 48.8 | 0.990 | 737 | 11.10 | 3396 | PE100 | 61.4 | 283.0 |
| A6 | 11.98 | 913 | 46.6 | 0.986 | 620 | 11.68 | 4079 | PE150 | 51.7 | 340.0 |
| A7 | 12.62 | 901 | 66.2 | 0.990 | 3979 | 5.97 | 2775 | PE100 | 331.6 | 231.3 |
| A8 | 12.03 | 917 | 55.5 | 0.993 | 622 | 11.79 | 3420 | PE100 | 51.9 | 285.0 |
| A9 | 12.53 | 571 | 53.0 | 0.993 | 5845 | 5.10 | 2769 | PE100 | 487.1 | 230.7 |
| A11 | 12.10 | 702 | 49.3 | 0.986 | 644 | 12.03 | 2758 | PE100 | 53.7 | 229.8 |
| A12 | 12.00 | 795 | 45.5 | 0.993 | 613 | 11.83 | 3563 | PE100 | 51.1 | 296.9 |
| B5 | 12.32 | 738 | 46.5 | 0.990 | 741 | 10.97 | 3861 | PE100 | 61.7 | 321.8 |
| B6 | 12.10 | 772 | 44.9 | 0.990 | 401 | 11.67 | 2906 | PE100 | 33.4 | 242.2 |
| B7 | 11.91 | 765 | 39.5 | 0.986 | 719 | 11.79 | 4266 | PE100 | 59.9 | 355.5 |
| B8 | 11.99 | 802 | 66.0 | 0.990 | 741 | 12.00 | 3054 | PE100 | 61.8 | 254.5 |
| B9 | 12.40 | 856 | 54.6 | 0.993 | 1083 | 12.05 | 3419 | PE100 | 90.2 | 284.9 |
| B11 | 12.12 | 789 | 55.0 | 0.990 | 765 | 12.13 | 2236 | PE100 | 63.8 | 186.3 |
| B12 | 12.04 | 790 | 48.7 | 0.986 | 804 | 10.79 | 3930 | PE100 | 67.0 | 327.5 |
| AB1 | 12.35 | 901 | 47.9 | 0.993 | 3315 | 5.84 | 3509 | PE150 | 276.3 | 292.4 |
| AB2 | 12.83 | 741 | 66.2 | 0.990 | 5230 | 4.77 | 4230 | PE150 | 435.8 | 352.5 |
| AB3 | 12.44 | 809 | 55.8 | 0.990 | 3254 | 6.21 | 3891 | PE150 | 271.2 | 324.2 |
| AB4 | 12.32 | 800 | 55.7 | 0.990 | 4649 | 6.43 | 4490 | PE150 | 387.4 | 374.2 |
Bold text represents founder strains that were previously sequenced using PacBio technology in Yue et al. (2017). BUSCO, Benchmarking Universal Single-Copy Orthologs; PacBio, Pacific Biosciences.
Quality value (qv) estimate
To estimate assembly error rate, paired-end Illumina reads used in assembly polishing were mapped to the final assembly using bowtie2 (Langmead and Salzberg 2012). SNPs and small insertion/deletions (indels) were identified using freebayes v0.9.21 (-C 10 -0 -O -q 20 -z 0.10 -E 0 -X -u -p 1 -F 0.75) (Garrison and Marth 2012 preprint). To estimate the error rate, total bases due to SNPs and small indels (e) and the total number of assembly bases (b) with read coverage ≥ 3 were counted, and qv was calculated as –10 × log(e/b) (Koren et al. 2017) (Table 2).
Santa Cruz browser tracks
Assembled genomes were aligned to one another and the SacCer3 reference genome using ProgressiveCactus (https://github.com/ComparativeGenomicsToolkit/cactus) (Paten et al. 2011a,b). Santa Cruz Browser Track Hubs were created using the hal2assemblyhub script that is part of the ProgressiveCactus software (https://github.com/ComparativeGenomicsToolkit/Comparative-Annotation-Toolkit). The resulting SNAKE tracks are viewable at http://bit.ly/2ZrreUd. SNPs were identified between the founder strains using a generic GATK pipeline, with SNPs functionally annotated using SNPeff (Cingolani et al. 2012). Scripts to align the genomes and call SNPs in the founders are available here: https://github.com/tdlong/yeast_resource.
Analysis of structural variants
We aligned each founder genome assembly to the s288c reference genome (GCA_000146055.2) using MUMmer v4.0 (Marçais et al. 2018) (nucmer–maxmatch–prefix founder ref.fasta founder.fasta). To annotate the structural variants, the δ alignment file for each strain was then processed with SVMU (commit e9c0ea1) (Chakraborty et al. 2019).
Whole-genome sequencing of the two base populations
The two base populations were deeply sequenced using Illumina technology. In total, 4 ml of the 18F12v1 frozen stock was thawed at room temperature, pelleted at 3000 rpm for 5 min, and resuspended in 20 ml of YPD + ampicillin. This was followed by incubation at 30° for 3.5 hr at 275 rpm. Next, gDNA was extracted using the QIAGEN DNeasy kit. The gDNA was sheared using the Covaris S220 as above and Illumina-compatible libraries were prepared using the NEBNext Ultra II DNA Library Prep kit as above. The NEBNext libraries were pooled and sequenced on the HiSeq4000 using PE100 reads. The NEBNext libraries were sequenced at a mean per-site coverage of 3270×.
For 18F12v2, similarly to 18F12v1, 4 ml of frozen stock was thawed at room temperature, pelleted at 3000 rpm for 5 min, and resuspended in 20 ml of YPD + ampicillin, followed by incubation at 30° for 3.5 hr at 275 rpm. The gDNA was extracted using the QIAGEN G-tip kit. Nextera libraries were prepped for 18F12v2 by following the standard Nextera protocol with slight modifications. Tagmentation reactions were carried out in 2.5-μl reactions for 10 min at 55°. Reactions were stopped by adding SDS to a final concentration of 0.02% followed by incubation at 55° for 7 min. Samples were immediately transferred to ice. Limited-cycle PCR was carried out to add two unique barcodes to each library to enable dual-index sequencing. This avoids the problem of barcode switching when N i7 and M i5 barcodes are used to create MN combinations. The KAPA HiFi Ready Mix (2×) was used in conjunction with the KAPA forward and reverse primers to amplify tagmented libraries in 25 μl total volume. Thermocycling parameters consisted of 3 min at 72°, 5 min at 98°, followed by 15 cycles of 10 sec at 98°, 30 sec at 63°, and 30 sec at 72°, with a hold of 72° for 5 min at the end. PCR reactions were cleaned up using AMPure XP Beads (Beckman, Fullerton, CA) and libraries quantified by Qubit. Sequencing was performed on the HiSeq4000 using PE150 reads. 18F12v2 libraries received 2226× coverage.
Whole-genome sequencing of the first two meiotic generations of the second base population
For 18F1v2 and 18F2v2, ∼1 ml of frozen stock was thawed at room temperature, spun down at 7500 rpm for 5 min in microcentrifuge tubes, resuspended in 1 ml of YPD + ampicillin, transferred to a 250-ml flask with 19 ml of YPD + ampicillin, and incubated at 30° for 3.5 hr at 275 rpm. The gDNA from both samples was extracted using the QIAGEN G-tip kit. Nextera libraries were prepped by following the Nextera flex protocol using one-fifth reactions with slight modifications. Limited-cycle PCR was carried out using the KAPA HiFi Ready Mix (2×) as detailed above to add barcoded Illumina-compatible adapters in 12.5-μl reactions. Thermocycling parameters consisted of 3 min at 72°, 3 min at 98°, followed by 12 cycles of 45 sec at 98°, 30 sec at 62°, and 2 min at 72°, with a hold of 72° for 1 min at the end. Proteinase K was added to each reaction (50 μg/ml final concentration) to digest the polymerase. Samples were incubated for 30 min at 37° and 10 min at 68°. Reactions were cleaned up using the sample purification beads (SPB) beads provided with the Nextera flex kit. Sequencing was performed as above using PE100 reads. 18F1v2 received 98× coverage while 18F2v2 received 73× coverage.
Whole-genome resequencing of recombinant haploid clones
Ten haploid recombinant clones (five of each mating type) were isolated from each of the two base populations. 18F12v1-derived haploids were generated by sporulating an overnight culture of the 18F12v1 population in 2 ml of PA7 in a 10 ml-culture tube at 30° for 3 day. Spore isolation and dispersal were carried out as detailed above for the creation of 18F12v2 with 15 min of bead milling to disperse spores. Spores were plated at low density onto YPD plates and incubated for 2 days at 30°. One of the YPD plates was then replica plated onto four different plates: YPD with hyg, YPD with cloNAT, YPD with mating-type tester 1, and YPD with mating-type tester 2. Five haploids of each mating type were inoculated into YPD overnight. The gDNA was extracted using the QIAGEN DNeasy kit and Nextera libraries prepared as above. Libraries were sequenced on a HiSeq4000 using PE100 reads to a mean per-site coverage of ∼32×.
18F12v2-derived haploids were generated by sporulating an overnight culture of the 18F12v2 population in 4 ml of PA7 in a 24DWP at 30° at 275 rpm for 3 days. Spore isolation and dispersal were carried out as detailed above for the creation of 18F12v2 with 20 min of bead milling to disperse spores. Spores were plated at low density onto YPD plates and incubated at 30° for 3 days. Next, 96 single colonies were transferred into a 96-well deep-well plate with YPD + ampicillin using sterile toothpicks. After overnight incubation at 30°, 200 μl of culture from each well was transferred to a 96-well shallow plate and pinned YPD plates with either cloNAT, hyg, mating-type tester 1, or mating-type tester 2 using a 48-well replicator tool. The source plate was covered with an adhesive membrane and stored at 4°. The mating-type plates were incubated at 30° for 2 days, after which five haploids of each mating type were transferred from the original source plate to 1.5-ml Eppendorfs and gDNA was extracted using a QIAGEN DNeasy kit. Nextera libraries were prepared as above. Libraries were sequenced on a HiSeq4000 using PE150 reads to a mean per-site coverage of ∼60x.
Haplotype calling in Illumina resequenced MPPs and recombinant haploid clones
Demultiplexed fastq files were used in analyses. Detailed scripts/software versions to reproduce our analysis are located at https://github.com/tdlong/yeast_resource.git. Briefly, reads were aligned to the sacCer reference genome using bwa-mem and default parameters (Li and Durbin 2009; Li 2013). We maintain two SNP lists, a set of known SNPs in the strains obtained from a GATK pipeline that only considers the isogenic founders, and a subset of those SNPs that are well behaved (i.e., frequency of the Reference (REF) allele close to zero or one in all founder lines, pass GATK qualify filters, etc.). The list of well-behaved SNPs that are polymorphic in the founders can be used to speed up subsequent steps, where we sometimes examine hundreds of samples, since only variants polymorphic among the founders need be considered when working with samples from a synthetic population (except when calling newly arising mutations). samtools mpileup (Li et al. 2009; Li 2011) and bcftools (Narasimhan et al. 2016) are used to query well-behaved known SNPs. We have no interest in calling genotypes, but instead simply output the frequency of the REF allele in each sample at each location (output = SNPtable). In a separate analysis, freebayes (Garrison and Marth 2012 preprint), vcfallelicprimitives (https://github.com/vcflib/vcflib), and vt normalize (Tan et al. 2015) were used to call all SNPs and the SNPs not in our list of known SNPs considered candidate new mutations.
We have developed custom software to infer the frequency of each founder haplotype at each location in the genome in pooled samples using the SNPtable as input and the haplotyper.limSolve.code.R script in the GitHub archive. This same algorithm can also be used without modification to infer genotypes in recombinant haploid clones. Briefly, we slide through the genome in 1-kb steps considering a 60-kb window for each step. For all SNPs in the window we calculate a Gaussian weight such that the 50 SNPs closest to the window center account for 50% of the sum of the weights. We then consider F founders and use the lsei function of the limSolve package (limSolve: Solving Linear Inverse Models, R package 1.5.1) (Van den Meersche et al. 2009) in R to identify a set of F mixing proportions (each greater than zero and summing to one) that minimize the sum of the weighted squared differences between founder haplotypes and the observed frequency of each SNP in a pooled sample. That is, for an N SNP window we call lsei with the following parameters: A = N*F matrix of founder genotypes, B = N*1 vector of SNP frequencies in a pooled sample, E = F*1 vector of 1’s, F = 1, G = F*F identity matrix, H= F*1 vector of 0’s, and Wa = N*1 vector of weights. Finally, for windows where the ith and jth founders have near indistinguishable haplotypes, implying the sum of the two mixing proportions are correct, but not individual estimates, we estimate the haplotype frequency as one-half the sum of the two mixing proportions. This method of accounting for indistinguishable haplotypes is regional and is generalized to > 2 near identical founders and multiple such sets.
Validation of the haplotype caller
To validate our haplotype-calling algorithm, we identified 70,478 SNPs private to a single founder strain (excluding those present in founders merged due to high sequence similarity). The haplotype caller was run on 18F12v2 using the full coverage data (i.e., 2230×) or downsampled 18F12v2 to simulate a more typical pool-sequenced (poolseq) resequencing depth (typical applications using the MPPs are likely to sequence hundreds of experimental units to ∼20–60×). For each private SNP, the frequency of the SNP in the full coverage data was estimated and the founder harboring that SNP identified. Since the sequence depth of the nondownsampled population is 2230×, the frequency of each private SNP is measured very accurately. We then infer the frequency of the founder haplotype harboring the private SNP at the position closest to the private SNP, in both the full-coverage and each downsampled population. The error rate associated with the haplotype caller is the absolute difference between the frequency of each private SNP and the founder haplotype harboring it.
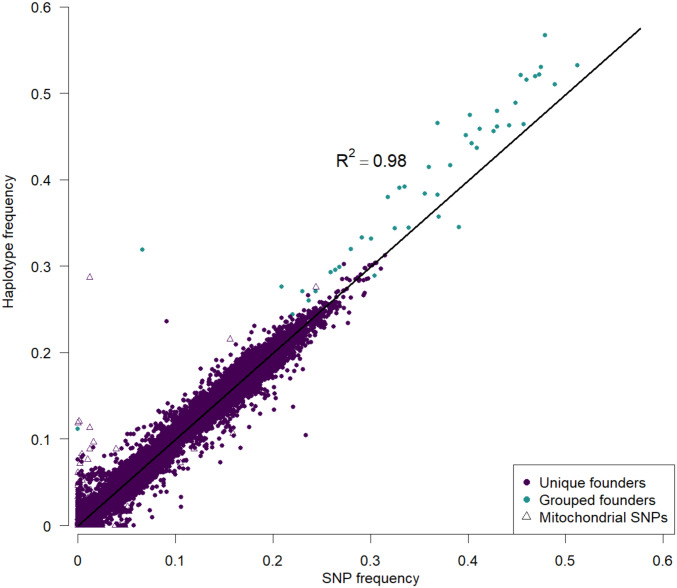
In our examination of the relationship between haplotype and SNP frequency estimates (Figure 4) we identified and removed 91 outlier SNPs among the 71,301 private SNPs. These SNPs were identified as private SNPs whose frequency was > 5% different from the frequency of the haplotype harboring it in the full data set, while exhibiting flanking private SNPs in the same founder whose frequencies agreed with the founder frequency. We believe these outlier SNPs are cases where that particular SNP in a pooled sample cannot be aligned to the reference genome very accurately. It is noteworthy that it is more difficult to identify poorly performing SNPs that are not private to a single founder, and such SNPs likely hurt haplotype inference methods.
Figure 4.
The frequency of SNPs private to a single founder are highly correlated with the estimated haplotype frequencies at these SNPs in 18F12v2. As the frequency of a private SNP should be equal to the corresponding haplotype frequency, this measure provides a benchmark with which the accuracy of our haplotype caller can be measured. Cyan points represent founders that were pooled when estimating haplotype frequencies (“grouped founders”) due to the high degree of sequence similarity between their genomes. Triangles represent mitochondrial SNPs, which, together with SNPs private to pooled founders, represent the bulk of the major outliers. The coefficient of determination was calculated by regressing haplotype frequency onto SNP frequency, excluding SNPs from grouped founders and mitochondrial SNPs.
Delineating haplotype blocks in recombinant haploid clones
The haplotype caller is primarily used to estimate the frequency of founder haplotypes at different positions in the genome in a DNA pool from a segregating population, but it can also be run on DNA obtained from a haploid or diploid clone. In a haploid clone the haplotype caller should return a haplotype frequency of close to 100% for one of the founder haplotypes for much of the genome, with sharp transitions between founder states near recombination breakpoints. In depicting the haplotypic structure of haploid clones we classify genomic regions at which the inferred haplotype frequency of a single founder (or multiple indistinguishable founders) is < 95% as having an “unknown” haplotype (these unknown intervals typically being associated with state transitions). We also observe intervals in which several founders are indistinguishable from one another (due to insufficient SNP divergence between the founders in these window). We could sometimes resolve these intervals to a single founding haplotype when flanking haplotypes were unambiguously called as derived from the same single founder. Custom R scripts were used for these analyses as well as to calculate the length of haplotype blocks in haploid clones. Haplotype block sizes were inferred by finding the positional differences between the beginnings and ends of runs of the same haplotype.
Data availability
Strains and plasmids are available upon request. All genome sequencing data and assemblies have been deposited into public repositories. Sequence data generated for the two base populations (18F12v1, 18F12v2, 18F1v2, and 18F2v2) as well as the recombinant haploid clones are available in the Short Reads Archive under bioproject PRJNA551443 in accessions SRX6465384 to SRX6465405 and SRX6983898 to SRX6983899. All PacBio and Illumina data generated for the 18 founding strains are also available in the Short Reads Archive under the bioproject PRJNA552112 in accessions SRX6380915 to SRX6380944. Detailed scripts/software versions to reproduce our analysis are located at https://github.com/tdlong/yeast_resource.git. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12061659
Results and Discussion
Recovery of hygr and insertion of dominant selectable markers for high-throughput diploid selection
We further engineered a subset of the yeast SGRP resource strains (Cubillos et al. 2009) to serve as founders for an 18-way synthetic population. We first recovered the hygr marker used to delete the HO gene in the haploid SGRP strains. Previous work (McDonald et al. 2016) replaced YGR043C, a pseudogene that is physically close to the mating-type locus, with dominant selectable markers to facilitate high-throughput selection of diploids after mating. We echoed that approach here by replacing YGR043C with NatMX4 in 12 MATa (A) founders and with HphMX4 in 12 MATα (B founders). The presence of these cassettes confers resistance to the antibiotics cloNAT and hyg, respectively, enabling the selection of doubly resistant diploids. All newly engineered strains are given in Table 1.
De novo assembly of high-quality reference genomes for the 18 founding strains
We generated de novo genome assemblies for the founders used to create our MPPs using a hybrid sequencing strategy detailed in Chakraborty et al. (2016) that involves using a combination of long-read (PacBio) and short-read (Illumina paired-end) sequencing technology. The de novo assemblies allow us to reliably identify structural variants while the overall assembly has a low per-base pair error rate. We assembled 58.9× PacBio reads on average (33–90×) for 12 of the founder strains, and reassembled the other 6 strains using publicly available shorter-length 364.9× PacBio reads on average. Despite the different numbers and chemistries of PacBio reads used in assembling the genomes, all of our assemblies are highly accurate (average quality value (qv) = 52.5) and show comparable contiguity. For example, the average contig N50 of our assemblies is ∼800 kb (N50 = 50% of the assembly is contained within sequences of this length or longer), indicating that the majority of the chromosomes are represented as single contigs (Table 2). Examination of 290 conserved fungal single-copy orthologs (BUSCO) shows that completeness (∼99%) of all our assembled genomes is comparable to the reference S288C assembly (99%).
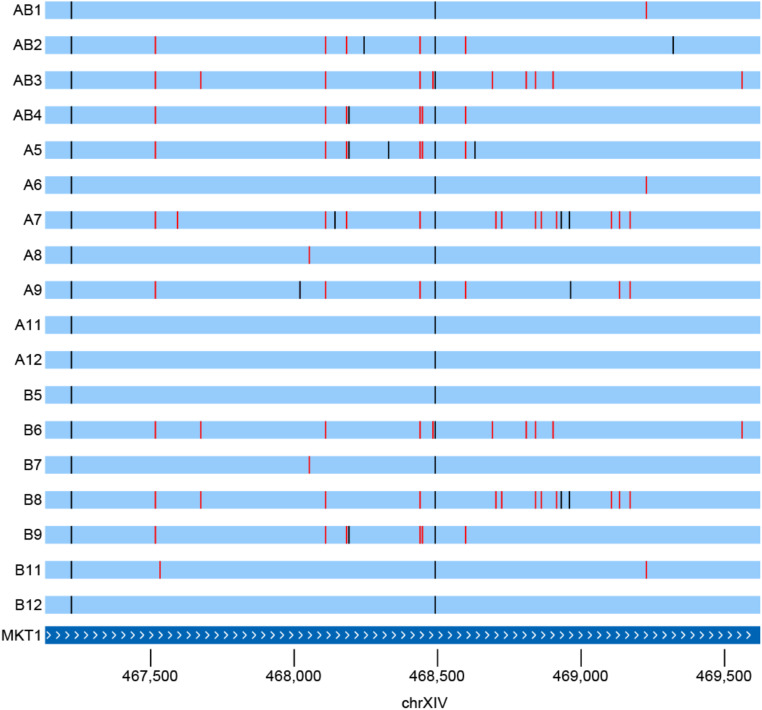
We aligned the assemblies to one another and represent them as Santa Cruz genome browser tracks (http://bit.ly/2ZrreUd). These tracks have utility when looking for candidate causative variants in small regions of genetic interest. The large amount of genetic diversity sampled by the founders can be illustrated by zooming in on regions such as that shown in Figure 2, which highlights the numerous alleles segregating at a gene implicated in many genetic mapping studies in budding yeast, the highly pleiotropic MKT1. MKT1 influences several cellular processes including the DNA damage response, mitochondrial genome stability, drug resistance, and post-transcriptional regulation of HO (Dimitrov et al. 2009; Ehrenreich et al. 2010; Tkach et al. 2012; Kowalec et al. 2015). Studies have found that different alleles of MKT1 can differentially affect several phenotypes, including mitochondrial genome stability and drug resistance. Variation at this gene among our founders includes 10 nonsynonymous SNPs and 34 synonymous SNPs. Of the 10 nonsynonymous SNPs, six are predicted to change the secondary structure of the protein. Taking into account only nonsynonymous SNPs, there are seven different alleles segregating among the founders (all segregating in our 18F12v2 MPP).
Figure 2.
Many alleles of the highly pleiotropic MKT1 gene are segregating among the founder strains, highlighting the potential of uncovering complex allelic series using populations derived from these strains. Seven of these alleles are differentiated by nonsynonymous SNPs, of which six are predicted to be segregating in 18F12v2. Vertical red lines are synonymous SNP differences from the reference S288C strain and black bars are nonsynonymous SNPs.
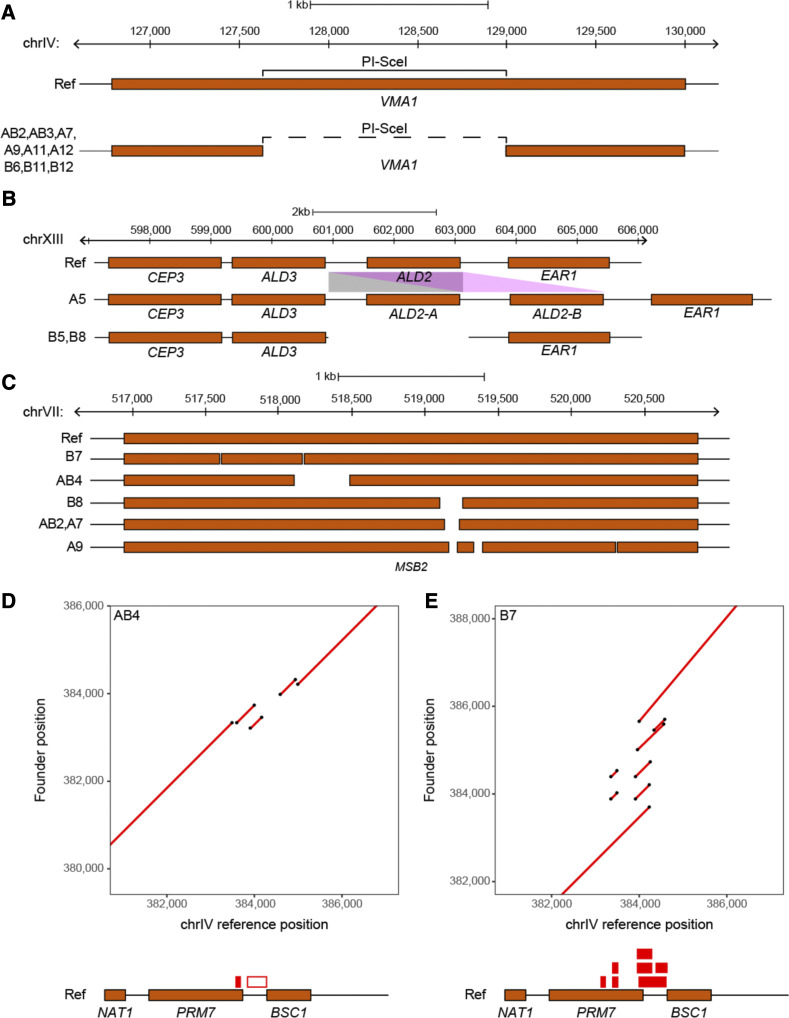
The genome browser tracks are also useful for visualizing structural variants such as those shown in Figure 3, which highlights a large (>1 kb) deletion in the vacuolar ATPase VMA1 (Figure 3A) present in half of the founders. Previous work has shown that the deleted region encodes a self-splicing intein, PI-SceI, a site-specific homing endonuclease that catalyzes its’ own integration into inteinless alleles of VMA1 during meiosis (Gimble and Thorner 1992). This selfish genetic element has been shown to persist in populations solely through horizontal gene transfer and is present in many species of yeast. Perturbation of VMA1 itself has been shown to influence both replicative and chronological life span, resistance to metals, as well as oxidative stress tolerance (Kane 2007; Ruckenstuhl et al. 2014).
Figure 3.
Combining contiguous long-read sequencing with accurate short-read data enables the detection of structural variants such as those depicted to the left. In (A), a large (> 1 kb) deletion within a vacuolar ATPase (VMA1) is present in one-half of the strains used in this study. This deletion directly overlaps the self-splicing intein PI-SceI. Copy number variants of ALD2, an aldehyde dehydrogenase, were detected (B) and include a duplication of this gene in founder A5 (represented as ALD2-A and ALD2-B), as well as its deletion in founders B5 and B8. In (C), multiple deletions of different lengths in the osmosensor MSB2 were detected in multiple founder strains. Dotplots of a structurally complex region on chr IV are shown for founders AB4 (D) and B7 (E). These plots show alignments of regions from the founder strains (depicted on the y-axis) with the corresponding region from the S288C reference strain (depicted on the x-axis). The red boxes present above the genes in the reference strain map duplications (solid boxes) and deletions (empty boxes) detected in each founder strain to the corresponding reference sequence. In all panels, the Ref is used to highlight the various arrangements of structural variants present in the founder strains. chr, chromosome; Ref, reference strain S288C.
In addition to large deletions, copy number variants (CNVs) can also be found, such as that shown in Figure 3B, in which the cytoplasmic aldehyde dehydrogenase, ALD2, is duplicated in founder A5. Conversely, this gene has been deleted in founders B5 and B8. ALD2 has been shown to be involved in the osmotic stress response as well as the response to glucose exhaustion (Navarro-Aviño et al. 1999). A more structurally complex region was identified on chromosome VII (Figure 3C), at which multiple different deletions (ranging from ∼50 bp to > 300 bp) were found to occur at MSB2, an osmosensor involved in the establishment of cell polarity (O’Rourke and Herskowitz 2002; Cullen et al. 2004). Null alleles of MSB2 have been shown to have decreased chemical resistance.
One of the most structurally complex regions we identified contains ∼2 kb of repetitive sequence and is present on chromosome IV (Figure 3, D and E and Figure S1), at which multiple different deletions (ranging from ∼80 bp to > 500 bp) as well as duplications occur in multiple founders within the PRM7 and BSC1 genes. Due to the highly complex nature of the variation present, this region is represented as a series of dot plots, with two founders highlighted in the main text (Figure 3, D and E). Dot plots of this region in all founder strains are shown in Figure S1. A previous study demonstrated that although two distinct genes (PRM7 and BSC1) are present in S288C, a combination of small deletions and point mutations in another yeast strain (W303) have caused the STOP codon to be absent from BSC1, leading to the readthrough transcription of a new gene that encompasses sequence from both PRM7 and BSC1, as well as the intergenic region between them (Kowalec et al. 2015). This gene, IMI1, was shown to affect mitochondrial DNA stability as well as intracellular levels of reduced glutathione.
The above regions highlight the utility of our de novo genome assembly approach, as deletions and CNVs of this scale would be difficult to detect via the usual method of aligning short reads to a reference genome. But if an investigator mapped a QTL to one of these genes, they would certainly want to know about the existence of the segregating structural variation.
Despite the large amount of natural variation present among the founders in general, some of the founders were found to be genetically very similar to one another (AB3/B6) (Figure S2 and shown in Table S4), having < 200 pairwise SNP differences. This lack of divergence makes this set of founders difficult to distinguish from one another for much of the genome and, as a result, we collapsed them for subsequent analyses (despite that fact that a subset of these 200 differences could be functional). Three additional founders (A11/A12/B11) were also found to be highly genetically similar to one another, with, on average, < 2000 pairwise SNP differences. These differences were concentrated in a small number of regions, making these three founders distinguishable for these regions (but indistinguishable for much of the remainder of the genome). We kept these strains separate for downstream analyses.
Creation of two 18-way highly outcrossed populations
MPPs created using multiple rounds of recombination can significantly increase the resolution of genetic mapping studies by virtue of haplotypes sampled from these populations having a greater number of genetic breakpoints. Furthermore, multiple founders result in high levels of standing variation present in the MPP. These two features result in populations that more realistically mimic natural outbred diploid populations, and sample more functional alleles and haplotypes from the species as a whole than a two-way cross. With these goals in mind, we constructed a large, genetically heterogenous population by crossing 18 different founder strains [each strain being derived from the SGRP (Cubillos et al. 2009)]. The 18 founder strains were chosen to represent a broad swathe of the natural diversity of the species and belong to diverse phylogenies, including: Wine/European, West African, North American, Sake, and Malaysian (see Table 1). It is also noteworthy that founder strains A1–4 and B1–4 are the same four strains used in Cubillos et al. (2013), and were introduced into the population as both MATa and MATα mating types. We created two versions of our 18-way MPP. In both cases, a full diallele cross was used to create all 121 unique diploid genotypes from 11 MATa and 11 MATα strains (see Figure 1, A and B). All 121 diploid genotypes were combined and the resulting population was taken through 12 rounds of sporulation followed by random mating to break up LD. Previous work has shown that 12 rounds of random recombination breaks up haplotype blocks to the point where additional outcrossing does not significantly decrease LD (Parts et al. 2011). For brevity, the two different outcrossed populations will be referred to as 18F12v1 and 18F12v2, respectively, throughout the rest of this manuscript. The version 1 MPP differed primarily from version 2 in that the 121 diploid genotypes obtained from the diallele were directly combined and sporulated en masse (version 1; Figure 1A) to create the MPP, as opposed to being individually carried through sporulation and spore disruption before being combined (version 2; Figure 1B). Furthermore, due to a technical artifact during the 12 rounds of outcrossing, 18F12v1 was cross-contaminated with the four-way F12 population from Cubillos et al. (2013), which contains a functional URA3 gene. As a result, 18F12v1 MPP is of limited utility for experiments that require uracil auxotrophy, and 18F12v2 is the current primary focus of work in our laboratory.
Development of an algorithm for accurately inferring haplotype frequencies
In QTL mapping experiments using MPPs, it is often advantageous to map QTL back to founder haplotypes. In experiments derived from a two-way cross between isogenic founders, genotyping SNPs accomplishes this, but with multiple founders parental haplotypes have to be inferred in recombinant offspring (Mott et al. 2000). In a similar manner, when MPPs are used as a base population and genetic changes detected following an experimental treatment, it is often of value to examine changes in haplotype frequency [as done in Burke et al. (2014) and reviewed in Barghi and Schlötterer (2019)]. We developed a sliding-window haplotype caller that can be used in situations when the founder haplotypes are known and applied it to both single haploid clones and pools consisting of millions of diploid individuals. This haplotype caller differs from other widely used callers (Long et al. 2011; Kessner et al. 2013) in that it acknowledges that in some windows pairs of founders are poorly resolved or indistinguishable, and relies solely on read counts at known SNP positions in both founders and recombinant populations.
To benchmark the haplotype-calling algorithm, we compared the frequency of SNPs private to a single founder to the haplotype frequency of the same founder for the interval closest to the SNP location in the 18F12v2 base population (Figure 4). Since this base population is sequenced to 2226×, we initially wished to look at the error in the haplotype estimate at full coverage where the sampling variation on the SNP frequency estimate was quite low [proportional to 1/sqrt(2226) or < 2%]. For the high-coverage base population regions showing large differences between SNPs and haplotype frequencies, estimates likely represent instances where the haplotype caller breaks down, since we attempted to remove SNPs whose frequencies were poorly estimated. Figure S3, depicting the absolute difference in SNP vs. haplotype frequency differences, shows that haplotype and SNP frequencies generally agree with one another with average and median error rates of 0.4% and 0.2%, respectively (below the sampling error of SNP frequency).
Of course, typical experiments employing these base populations will sample the population following some treatment, and compare haplotype frequencies in control vs. treated samples. Although the 18F12v2 base population is sequenced to 2226×, it would be cost-effective if we could infer haplotype frequencies from pooled samples sequenced to much lower coverage. To determine the accuracy of our haplotype estimates as a function of sequencing coverage, the 18F12v2 was downsampled 50- and 100-fold, which corresponded to poolseq data sets of ∼40× and ∼20×, respectively. We then estimated relative haplotype frequency error rates as a function of sequence coverage (Figure S3B), and absolute error rates as a function of coverage and genomic location (Figure S4). It is apparent that the error rate is an increasing function of decreasing coverage, but for much of the genome the absolute error in the haplotype frequency estimate is actually lower than the binomial sampling errors associated with directly estimating SNP frequencies at the same coverage (i.e., at 20–40× coverage binomial sampling errors on frequency are > 10%). It is also apparent that the average error rate is likely driven by a few regions where the haplotype caller struggles; these are presumably regions with poor divergence between founders in the window examined. Overall the mean (median) error rates on haplotype frequency estimates are low, 1.3% (0.8%) at 20× and 1% (0.6%) at 40×, respectively.
Characterization of 18F12v1 and 18F12v2 base populations
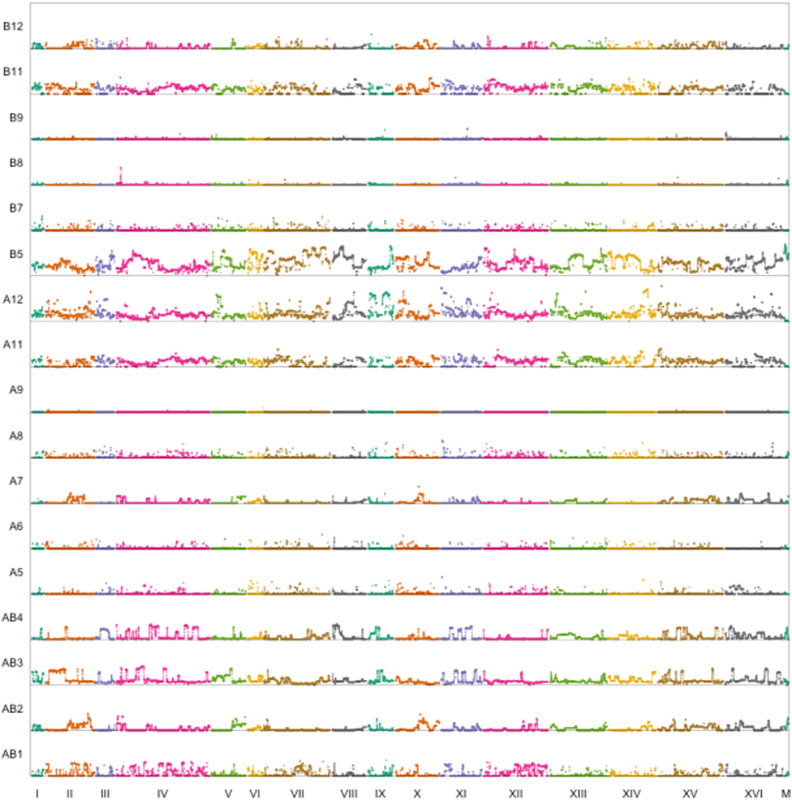
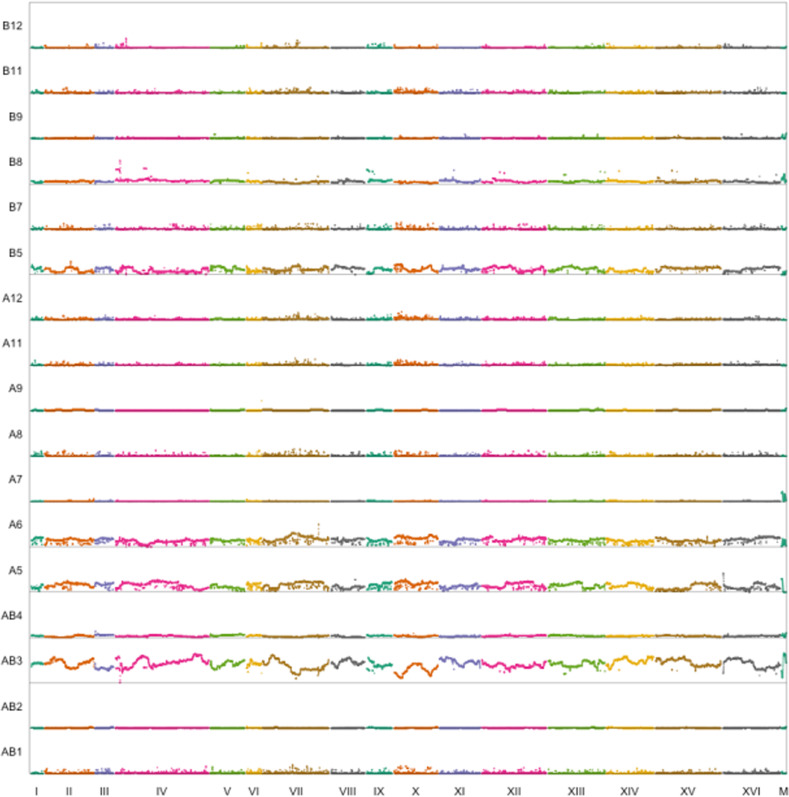
18F12v1 and 18F12v2 were subjected to high-coverage whole-genome sequencing to both characterize their population structure and to establish a baseline for future mapping studies. Figure 5 and Figure 6 show the inferred sliding-window haplotype frequencies for 18F12v1 and 18F12v2, respectively, while Table 3 shows the mean per-founder haplotype frequencies genome-wide. One trend that is evident is that in both the 18F12v1 and 18F12v2 MPPs a small number of founders are overrepresented. To identify the origin of this bias, at least for 18F12v2, the first two meiotic generations of 18F12v2 were sequenced (Figures S5 and S6, and Table S5). Despite having an initially more balanced population after the first round of random mating, a few strains quickly became disproportionately overrepresented. One possible explanation for this is that a few founding haplotypes were selected for early in the 12 rounds of intercrossing. Figure S7 provides suggestive evidence that this may have been the case, as the frequency of haplotypes derived from founder A5 increases genome-wide after the second round of meiosis. The latter one-half of chromosome XIII (from founder A5) emphasizes this point as it was very highly selected for initially. Another potential source of bias was the pooling strategy, which was done using optical density as a proxy for cell numbers. This may have resulted in an uneven distribution of founders in the initial pool. Nonetheless, after 12 rounds of random mating, deep sequencing of 18F12v2 revealed that haplotypes from all founding strains were present in the population at a detectable frequency (Table 3). Specifically, haplotypes from ≥ 10 founders were detected as segregating in > 99% of the genome in 18F12v2 and close to 98% of the genome in 18F12v1. Furthermore, 18F12v2 was verified as being auxotrophic for uracil, facilitating future manipulations for downstream analyses.
Figure 5.
Genome-wide haplotype frequencies for 18F12v1.
Figure 6.
Genome-wide haplotype frequencies for 18F12v2.
Table 3. Mean haplotype frequencies in 18F12v1 and 18F12v2.
| Founder | 18F12v1_frequency (%) | 18F12v2_frequency (%) |
|---|---|---|
| AB1 | 4.4 | 1.2 |
| AB2 | 3.8 | 0.5 |
| AB3 | 10.6 | 41.4 |
| AB4 | 5.8 | 3.3 |
| A5 | 1.2 | 14.0 |
| A6 | 0.8 | 14.3 |
| A7 | 2.4 | 0.4 |
| A8 | 1.2 | 0.7 |
| A9 | 0.1 | 0.5 |
| A11 | 9.9 | 1.2 |
| A12 | 18.1 | 1.7 |
| B5 | 27.1 | 11.3 |
| B7 | 1.1 | 1.0 |
| B8 | 0.5 | 5.7 |
| B9 | 0.9 | 0.8 |
| B11 | 9.8 | 1.3 |
| B12 | 2.4 | 0.6 |
Characterizing the recombination landscape of 18F12v1- and 18F12v2-derived segregants
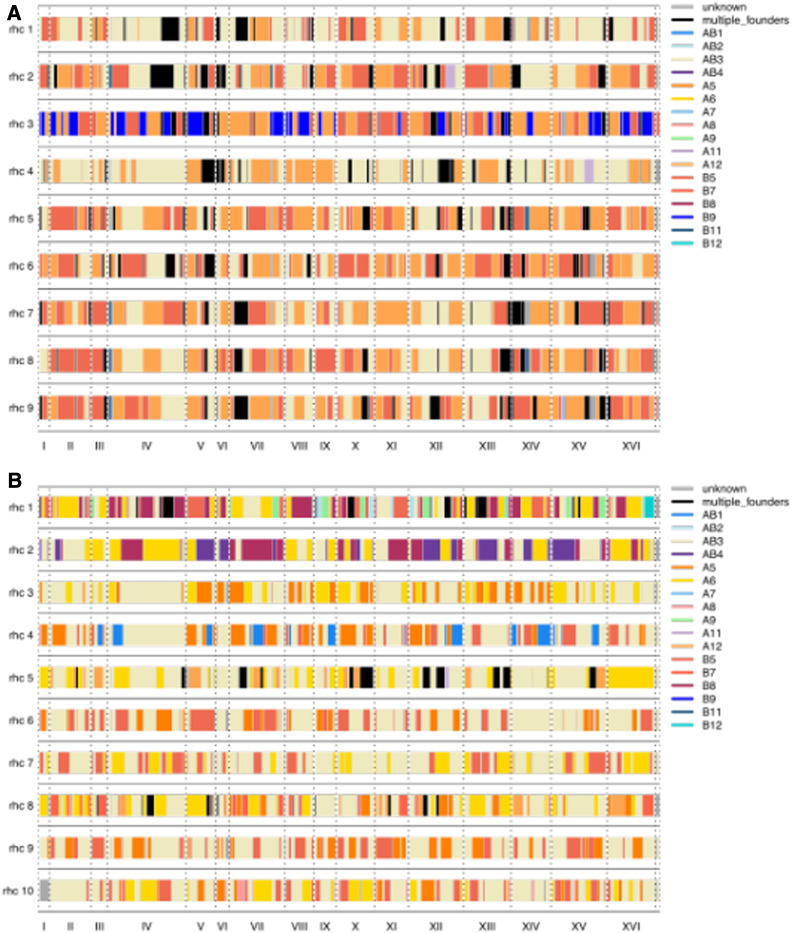
To further characterize 18F12v1 and 18F12v2, 10 haploid segregants were generated from each diploid population and subjected to whole-genome sequencing. The complex structure of these populations is highlighted in Figure 7. The mean (median) size of haplotype blocks in 18F12v1-generated segregants was 103 kb (66 kb) while the mean (median) size of haplotype blocks in 18F12v2-generated segregants was 106 kb (66 kb) (Figure S8). The mean number of discrete haplotype blocks in 18F12v1-generated segregants was 106 as compared with 104 in 18F12v2-generated segregants. A previous study (Cubillos et al. 2013) found that 12 rounds of meiosis in a yeast four-way cross resulted in a median block size of 23 kb with 374 discrete haplotype blocks. Some of the failure to obtain the smaller block sizes and more numerous discrete blocks of this previous study may be due to undetectable recombination events occurring within haplotypes overrepresented in our populations. Another possibility is that, due to the large number of founding haplotypes, recombination events were missed in regions at which multiple founding strains were highly genetically similar. It is also possible that some of the founding strains used in this study have relatively low natural recombination rates.
Figure 7.
Haploids derived from 18F12v1 (A) and 18F12v2 (B) were isolated and sequenced, providing a glimpse into the recombinogenic landscape and haplotype diversity present within these populations.
To highlight the diversity present in the two outbred populations, a close-up view of inferred haplotypes in segregants derived from each population at chromosome X is shown in Figure S9. Regions in which the founding haplotype is unknown tended to occur at the transitions between haplotypes (see Note S4), and are a mean (median) length of 7.8 kb (6 kb) in 18F12v1-derived segregants and 7.5 kb (6 kb) in 18F12v2-derived segregants. Also noticeable is, at least for this chromosome, the larger amount of variation segregating in 18F12v2 (Figure S9B).
Conclusions
The paradigm of utilizing pairwise crosses to dissect the genetic basis of complex traits has enjoyed much success in diverse model organisms. However, such studies typically underestimate the standing variation present in natural populations and often lack the resolution to pinpoint causal variants to a small number of genes. Conversely, association studies are typically underpowered to detect rare alleles, poorly tagged variants, and regions with multiple causal sites in weak LD with one another. MPPs have been proposed to bridge the gap between the above two approaches. Although MPPs have been created in several model systems, only a single MPP has thus far been described in budding yeast. By generating two large, highly outcrossed and genetically heterogeneous populations of S. cerevisiae derived from 18 different founder strains, we have created a powerful resource that can be used in a variety of experimental settings. For instance, these populations can be used in large-scale Bulk segregant analysis (X-QTL) mapping experiments (Ehrenreich et al. 2010) to comprehensively dissect the genetic architecture of complex traits, as well as large-scale evolve-and-resequence experiments (Lang et al. 2011; Parts et al. 2011; Burke et al. 2014) to determine the mechanisms and course of adaptation to diverse stimuli. Large numbers of recombinant haploid clones generated from these populations can be used in complementary large-scale Individual segregant analysis (I-QTL) studies (Bloom et al. 2013; Wilkening et al. 2014). Due to the high levels of standing variation present, these populations should also prove to be a powerful resource in evolutionary engineering applications, as they are presumably capable of being evolved to carry out a plethora of useful tasks.
The haplotype-calling software generated in this study represents a useful resource for the MPP community in general, as it enables highly accurate haplotype calling in poolseq data at reduced coverage. The ability of the algorithm to deal with windows where all founder haplotypes cannot be resolved will have utility in a subset of systems, including our yeast populations. Candidate causal regions can be identified by comparing haplotype frequencies at discrete intervals across the genome in control vs. treatment populations. Candidate regions can then be examined in the University of California, Santa Cruz genome browser, where genome-wide alignments of all founder strains have been posted. Structural variants can be easily visualized in the browser as can nonsynonymous SNPs, thus pointing investigators to potentially causal genes.
In conclusion, the populations generated in this study represent a novel resource that brings together the power of QTL mapping, the resolution of association studies, and a large amount of natural variation to a model system capable of teasing apart and directly testing the molecular underpinnings of complex traits.
Acknowledgments
We thank the University of California, Irvine Genomics High-Throughput Facility for the quick turnaround and efficient processing of libraries for sequencing, and for help with figuring out the parameters for the Covaris S220 Focused Acoustic Shearer. We also acknowledge our funding source: National Institutes of Health grant FG18445 to A.D.L.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12061659.
Communicating editor: P. Wittkopp
Literature Cited
- Aylor D. L., Valdar W., Foulds-Mathes W., Buus R. J., Verdugo R. A. et al. , 2011. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 21: 1213–1222. 10.1101/gr.111310.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barghi N., and Schlötterer C., 2019. Shifting the paradigm in Evolve and Resequence studies: from analysis of single nucleotide polymorphisms to selected haplotype blocks. Mol. Ecol. 28: 521–524. 10.1111/mec.14992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. J., Harpak A., Sinnott-Armstrong N., Joergensen A. M., Mostafavi H. et al. , 2019. Reduced signal for polygenic adaptation of height in UK Biobank. Elife 8: e39725. 10.7554/eLife.39725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. S., Ehrenreich I. M., Loo W., Lite T.-L. V., and Kruglyak L., 2013. Finding the sources of missing heritability in a yeast cross. Nature 494: 234–237. 10.1038/nature11867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. S., Kotenko I., Sadhu M. J., Treusch S., Albert F. W. et al. , 2015. Genetic interactions contribute less than additive effects to quantitative trait variation in yeast. Nat. Commun. 6: 8712 10.1038/ncomms9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. S., Boocock J., Treusch S., Sadhu M. J., Day L. et al. , 2019. Rare variants contribute disproportionately to quantitative trait variation in yeast. Elife 8: e49212. 10.7554/eLife.49212.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke M. K., Liti G., and Long A. D., 2014. Standing genetic variation drives repeatable experimental evolution in outcrossing populations of Saccharomyces cerevisiae. Mol. Biol. Evol. 31: 3228–3239. 10.1093/molbev/msu256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M., Baldwin-Brown J. G., Long A. D., and Emerson J. J., 2016. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 44: e147 10.1093/nar/gkw654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M., Emerson J. J., Macdonald S. J., and Long A. D., 2019. Structural variants exhibit widespread allelic heterogeneity and shape variation in complex traits. Nat. Commun. 10: 4872 10.1038/s41467-019-12884-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.-B., Lee S. H., Robinson M. R., Trzaskowski M., Zhu Z.-X. et al. , 2016. Across-cohort QC analyses of GWAS summary statistics from complex traits. Eur. J. Hum. Genet. 25: 137–146. 10.1038/ejhg.2016.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D. et al. , 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137. 10.1038/ng1104-1133 [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T. et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos F. A., Louis E. J., and Liti G., 2009. Generation of a large set of genetically tractable haploid and diploid Saccharomyces strains. FEMS Yeast Res. 9: 1217–1225. 10.1111/j.1567-1364.2009.00583.x [DOI] [PubMed] [Google Scholar]
- Cubillos F. A., Parts L., Salinas F., Bergström A., Scovacricchi E. et al. , 2013. High-resolution mapping of complex traits with a four-parent advanced intercross yeast population. Genetics 195: 1141–1155. 10.1534/genetics.113.155515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos F. A., Brice C., Molinet J., Tisné S., Abarca V. et al. , 2017. Identification of nitrogen consumption genetic variants in yeast through QTL mapping and bulk segregant RNA-seq analyses. G3 (Bethesda) 7: 1693–1705. 10.1534/g3.117.042127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. J., Sabbagh W., Graham E., Irick M. M., Van Olden E. K. et al. , 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 18: 1695–1708. 10.1101/gad.1178604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning D.-J., and McIntyre L. M., 2017. Back to the future: multiparent populations provide the key to unlocking the genetic basis of complex traits. Genetics 206: 527–529. 10.1534/genetics.117.203265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov L. N., Brem R. B., Kruglyak L., and Gottschling D. E., 2009. Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics 183: 365–383. 10.1534/genetics.109.104497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I. M., Torabi N., Jia Y., Kent J., Martis S. et al. , 2010. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464: 1039–1042. 10.1038/nature08923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., and Mott R., 2001. Finding the molecular basis of quantitative traits: successes and pitfalls. Nat. Rev. Genet. 2: 437–445. 10.1038/35076585 [DOI] [PubMed] [Google Scholar]
- Garrison, E., and G. Marth, 2012 Haplotype-based variant detection from short-read sequencing. arXiv: 1207.3907v2 [q-bio.GN] (Preprint posted July 20, 2012).
- Gimble F. S., and Thorner J., 1992. Homing of a DNA endonuclease gene by meiotic gene conversion in Saccharomyces cerevisiae. Nature 357: 301–306. 10.1038/357301a0 [DOI] [PubMed] [Google Scholar]
- Hehir-Kwa J. Y., Marschall T., Kloosterman W. P., Francioli L. C., Baaijens J. A. et al. , 2016. A high-quality human reference panel reveals the complexity and distribution of genomic structural variants. Nat. Commun. 7: 12989 10.1038/ncomms12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Paulo M.-J., Boer M., Effgen S., Keizer P. et al. , 2011. Analysis of natural allelic variation in Arabidopsis using a multiparent recombinant inbred line population. Proc. Natl. Acad. Sci. USA 108: 4488–4493. 10.1073/pnas.1100465108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane P. M., 2007. The long physiological reach of the yeast vacuolar H+-ATPase. J. Bioenerg. Biomembr. 39: 415–421. 10.1007/s10863-007-9112-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessner D., Turner T. L., and Novembre J., 2013. Maximum likelihood estimation of frequencies of known haplotypes from pooled sequence data. Mol. Biol. Evol. 30: 1145–1158. 10.1093/molbev/mst016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Macdonald S. J., and Long A. D., 2012a Properties and power of the Drosophila Synthetic Population Resource for the routine dissection of complex traits. Genetics 191: 935–949. 10.1534/genetics.112.138537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Merkes C. M., McNeil C. L., Hoofer S. R., Sen S. et al. , 2012b Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Res. 22: 1558–1566. 10.1101/gr.134031.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H. et al. , 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27: 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M. et al. , 2009. A Multiparent Advanced Generation Inter-Cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet 5: e1000551. 10.1371/journal.pgen.1000551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalec P., Grynberg M., Pajak B., Socha A., Winiarska K. et al. , 2015. Newly identified protein Imi1 affects mitochondrial integrity and glutathione homeostasis in Saccharomyces cerevisiae. FEMS Yeast Res. 15: fov048. 10.1093/femsyr/fov048 [DOI] [PubMed] [Google Scholar]
- Lander E. S., and Botstein D., 1989. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [corrigenda: Genetics 136: 705 (1994)]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Botstein D., and Desai M. M., 2011. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics 188: 647–661. 10.1534/genetics.111.128942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997.
- Li H., and Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., and Louis E. J., 2012. Advances in quantitative trait analysis in yeast. PLoS Genet. 8: e1002912 10.1371/journal.pgen.1002912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q., Jeffares D. C., Zhang Q., Ye K., Nizhynska V. et al. , 2011. PoolHap: inferring haplotype frequencies from pooled samples by next generation sequencing. PLoS One 6: e15292 10.1371/journal.pone.0015292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A. D., Macdonald S. J., and King E. G., 2014. Dissecting complex traits using the Drosophila synthetic population resource. Trends Genet. 30: 488–495. 10.1016/j.tig.2014.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald S. J., and Long A. D., 2007. Joint estimates of quantitative trait locus effect and frequency using synthetic recombinant populations of Drosophila melanogaster. Genetics 176: 1261–1281. 10.1534/genetics.106.069641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. 10.1146/annurev.genet.35.102401.090633 [DOI] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A. et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753. 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Märtens K., Hallin J., Warringer J., Liti G., and Parts L., 2016. Predicting quantitative traits from genome and phenome with near perfect accuracy. Nat. Commun. 7: 11512 10.1038/ncomms11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Delcher A. L., Phillippy A. M., Coston R., Salzberg S. L. et al. , 2018. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 14: e1005944 10.1371/journal.pcbi.1005944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald M. J., Rice D. P., and Desai M. M., 2016. Sex speeds adaptation by altering the dynamics of molecular evolution. Nature 531: 233–236. 10.1038/nature17143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen M. D., Kresovich S., Villeda H. S., Bradbury P., Li H. et al. , 2009. Genetic properties of the maize nested association mapping population. Science 325: 737–740. 10.1126/science.1174320 [DOI] [PubMed] [Google Scholar]
- Mott R., Talbot C. J., Turri M. G., Collins A. C., and Flint J., 2000. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 97: 12649–12654. 10.1073/pnas.230304397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan V., Danecek P., Scally A., Xue Y., Tyler-Smith C. et al. , 2016. BCFtools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 32: 1749–1751. 10.1093/bioinformatics/btw044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Aviño J. P., Prasad R., Miralles V. J., Benito R. M., and Serrano R., 1999. A proposal for nomenclature of aldehyde dehydrogenases in Saccharomyces cerevisiae and characterization of the stress-inducible ALD2 and ALD3 genes. Yeast 15: 829–842. [DOI] [PubMed] [Google Scholar]
- Noble L. M., Rockman M. V., and Teotónio H., 2019. Gene-level quantitative trait mapping in an expanded C. elegans multiparent experimental evolution panel. bioRxiv. doi: 10.1101/589432 (Preprint posted March 26, 2019). 10.1101/589432 [DOI] [Google Scholar]
- O’Rourke S. M., and Herskowitz I., 2002. A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch. Mol. Cell. Biol. 22: 4739–4749. 10.1128/MCB.22.13.4739-4749.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parts L., Cubillos F. A., Warringer J., Jain K., Salinas F. et al. , 2011. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 21: 1131–1138. 10.1101/gr.116731.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten, B., M. Diekhans, D. Earl, J. S. John, J. Ma et al., 2011a Cactus graphs for genome comparisons. J. Comput. Biol. 18: 469–481. [DOI] [PMC free article] [PubMed]
- Paten B., Earl D., Nguyen N., Diekhans M., Zerbino D. et al. , 2011b Cactus: algorithms for genome multiple sequence alignment. Genome Res. 21: 1512–1528. 10.1101/gr.123356.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., 2001. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 69: 124–137. 10.1086/321272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckenstuhl C., Netzberger C., Entfellner I., Carmona-Gutierrez D., Kickenweiz T. et al. , 2014. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet. 10: e1004347 10.1371/journal.pgen.1004347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P., Solovieff N., Puca A., Hartley S. W., Melista E. et al. , 2011. Retraction. Science 333: 404 10.1126/science.333.6041.404-a [DOI] [PubMed] [Google Scholar]
- Solares E. A., Chakraborty M., Miller D. E., Kalsow S., Hall K. et al. , 2018. Rapid low-cost assembly of the Drosophila melanogaster reference genome using low-coverage, long-read sequencing. G3 (Bethesda) 8: 3143–3154. 10.1534/g3.118.200162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer C. C. A., Su Z., Donnelly P., and Marchini J., 2009. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 5: e1000477 10.1371/journal.pgen.1000477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A., Abecasis G. R., and Kang H. M., 2015. Unified representation of genetic variants. Bioinformatics 31: 2202–2204. 10.1093/bioinformatics/btv112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium , 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678. 10.1038/nature05911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K. R., Foran A. J., and Long A. D., 2013. Properties and modeling of GWAS when complex disease risk is due to non-complementing, deleterious mutations in genes of large effect. PLoS Genet. 9: e1003258 10.1371/journal.pgen.1003258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill D. W., and Churchill G. A., 2012. Ten years of the collaborative cross. Genetics 190: 291–294. 10.1534/genetics.111.138032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach J. M., Yimit A., Lee A. Y., Riffle M., Costanzo M. et al. , 2012. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 14: 966–976. 10.1038/ncb2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Meersche K., Soetaert K., and Van Oevelen D., 2009. xsample() : an R function for sampling linear inverse problems. J. Stat. Softw. 30: 1–15. [Google Scholar]
- Visscher P. M., 2008. Sizing up human height variation. Nat. Genet. 40: 489–490. 10.1038/ng0508-489 [DOI] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A. et al. , 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse R. M., Seppey M., Simão F. A., Manni M., Ioannidis P. et al. , 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35: 543–548. 10.1093/molbev/msx319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkening S., Lin G., Fritsch E. S., Tekkedil M. M., Anders S. et al. , 2014. An evaluation of high-throughput approaches to QTL mapping in Saccharomyces cerevisiae. Genetics 196: 853–865. 10.1534/genetics.113.160291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Hill C. M., Wu S., Ruan J., and Ma Z., 2016. DBG2OLC: efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci. Rep. 6: 31900 10.1038/srep31900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J.-X., Li J., Aigrain L., Hallin J., Persson K. et al. , 2017. Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat. Genet. 49: 913–924. 10.1038/ng.3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. All genome sequencing data and assemblies have been deposited into public repositories. Sequence data generated for the two base populations (18F12v1, 18F12v2, 18F1v2, and 18F2v2) as well as the recombinant haploid clones are available in the Short Reads Archive under bioproject PRJNA551443 in accessions SRX6465384 to SRX6465405 and SRX6983898 to SRX6983899. All PacBio and Illumina data generated for the 18 founding strains are also available in the Short Reads Archive under the bioproject PRJNA552112 in accessions SRX6380915 to SRX6380944. Detailed scripts/software versions to reproduce our analysis are located at https://github.com/tdlong/yeast_resource.git. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12061659