Transposable elements (TEs) are a ubiquitous feature of plant genomes. Because of the threat they pose to genome integrity, most TEs are epigenetically silenced. However, even closely related plant species...
Keywords: Ac/Ds, transposon, alternative transposition, inverted duplication, small RNA, transposon silencing
Abstract
Although transposable elements (TEs) comprise a major fraction of many higher eukaryotic genomes, most TEs are silenced by host defense mechanisms. The means by which otherwise active TEs are recognized and silenced remains poorly understood. Here we analyzed two independent cases of spontaneous silencing of the active maize Ac/Ds transposon system. This silencing is initiated by alternative transposition, a type of aberrant transposition event that engages the termini of two nearby separate TEs. Alternative transposition during DNA replication can generate Composite Insertions that contain inverted duplications of the transposon sequences. We show that the inverted duplications of two Composite Insertions are transcribed to produce double-stranded RNAs that trigger the production of two distinct classes of small interfering RNAs: a 24-nt class complementary to the TE terminal inverted repeats and noncoding subterminal regions, and a 21- to 22-nt class corresponding to the TE transcribed regions. Plants containing these small interfering RNA-generating Composite Insertions exhibit decreased levels of Ac transcript and heritable repression of Ac/Ds transposition. Further, we demonstrate that Composite Insertions can heritably silence otherwise active elements in trans. This study documents the first case of transposon silencing induced by alternative transposition and may represent a general initiating mechanism for silencing of DNA transposons.
Introduction
Transposable elements (TEs) are often silenced by their hosts, but how TEs are initially recognized for silencing remains unclear. Here we describe two independent loci that induce de novo heritable silencing of maize Ac/Ds transposons. Plants containing these loci produce double-stranded RNA and Ac-homologous small interfering RNAs, and exhibit decreased levels of Ac transcript and heritable repression of Ac/Ds transposition. We show that these loci comprise inverted duplications of TE sequences generated by alternative transposition coupled with DNA rereplication. This study documents the first case of transposon silencing induced by alternative transposition and may represent a general initiating mechanism for TE silencing.
Transposable elements (TEs) comprise a large proportion of eukaryotic genomes, including those of important crop plants such as rice (>35%, International Rice Genome Sequencing Project, Matsumoto et al. 2005), sorghum (62%) (Paterson et al. 2009), and maize (85%) (Schnable et al. 2009). Over time, multiple sporadic TE proliferations altered the number and distribution of TE sequences, enhancing genome diversity between species and even among varieties of the same species (Tikhonov et al. 1999; Sanmiguel and Vitte 2009). These observations indicate that TEs have been, and continue to be, a major natural force driving genome evolution.
Although uncontrolled TEs can be deleterious to the host (Kidwell 1985), the majority of maize transposons are highly methylated (Regulski et al. 2013; West et al. 2014) and transcriptionally silenced (Anderson et al. 2019), which effectively preserves genome integrity. Small RNA-mediated silencing is an efficient means to initiate and maintain repression of both class 1 (RNA) and class 2 (DNA) TEs. For example, the DNA polymerase IV–RNA-dependent RNA polymerase 2 (Pol IV-RDR2) pathway generates 24-nt small interfering RNAs (siRNAs) to mediate RNA-directed DNA methylation (RdDM) at the homologous DNA targets. Because it relies on transcripts from methylated templates, the Pol IV-RDR2 pathway likely serves to reinforce or maintain silencing of previously silenced TEs [reviewed by Law and Jacobsen (2010), Haag and Pikaard (2011), Castel and Martienssen (2013), and Matzke and Mosher (2014)]. Although the mechanisms of maintenance of TE silencing have been extensively studied, the mechanisms responsible for de novo silencing remain relatively obscure. The first evidence for a locus that can silence an active TE came from analysis of Mu killer (Muk), a locus that can heritably silence MuDR transposons in trans in maize. Muk comprises an inverted duplication of the MuDR 5′ terminal inverted repeat (TIR) and a portion of the mudrA gene, which is required for element excision (Lisch 2002). This long inverted repeat is transcribed into double-stranded RNAs (dsRNAs) which initiate siRNA-mediated silencing of intact MuDR elements (Slotkin et al. 2003, 2005; Li et al. 2010). Recent studies of de novo silencing of active LTR retroelements in Arabidopsis demonstrated the involvement of RdDM (Marí-Ordóñez et al. 2013; McCue et al. 2014; Duan et al. 2015; Panda et al. 2016). Active TEs are transcribed by Pol II, followed by second-strand synthesis by RDR6. The resulting dsRNA is then processed into 21- to 22-nt siRNAs (Nuthikattu et al. 2013); these 21- to 22-nt siRNAs can then induce post-transcriptional silencing as well as transcriptional gene silencing, which is associated with DNA methylation (Pontier et al. 2012; McCue et al. 2014; Matzke and Mosher 2014). Once RdDM is initiated, silencing can then be maintained via the classic RdDM pathway, which involves Pol IV-mediated transcription from previously methylated sequences (Matzke and Mosher 2014).
Maize Ac/Ds TEs were the first transposons discovered and characterized (McClintock 1948, 1949, 1950, 1951). As members of the class 2 hAT transposon superfamily, Ac/Ds elements are less numerous than class 1 retroelements, which are often highly amplified (Sanmiguel and Vitte 2009). However, Ac/Ds elements can strongly affect gene expression because of their preferential insertions into genes (Vollbrecht et al. 2010) and the induction of genome rearrangements via alternative transpositions (Zhang and Peterson 1999). Unlike standard transposition reactions, which act on the 5′ and 3′ termini of a single element, alternative transposition acts on the termini of two separate, usually nearby elements (Gray 2000). During alternative transposition, the termini of two nearby, separate TEs can interact with the transposase and insert into linked or unlinked sites, resulting in various chromosomal rearrangements, including duplications, deletions, inversions, and translocations (Zhang and Peterson 2004; Huang and Dooner 2008; Zhang et al. 2009; Wang et al. 2015). Moreover, alternative transposition during DNA replication can generate novel structures termed Composite Insertions, which are composed of transposon termini surrounding host genome sequences that were copied from the transposon donor site. Because of their complex and heterogeneous structures, Composite Insertions represent an ongoing source of diverse Ac sequence configurations in the genome (Zhang et al. 2014)
In a previous study of Ac/Ds alternative transposition, Zhang and Peterson (1999) identified an allele termed p1-ww-id1 that induced significant repression of Ac/Ds transposition. Because this stock contains only one copy of Ac, the observed Ac/Ds repression could not be explained by the classic Ac negative dosage effect, in which increased Ac copy number results in reduced frequency and developmental delay of Ac/Ds transposition (McClintock 1949; Brink Nilan 1952). Subsequently, we isolated a second allele, p1-ww-id4, that was independently derived by alternative transposition and that exhibits a similar repression of Ac/Ds transposition.
Here, we show that both of these alleles (1) cause de novo and heritable repression of Ac/Ds transposition and Ac mRNA accumulation; (2) contain Composite Insertions with inverted duplications of Ac sequence; (3) produce Ac-homologous dsRNA transcripts driven by either a flanking host gene promoter or the Ac promoter; and (4) accumulate 21-, 22-, and 24-nt siRNAs corresponding to the region of dsRNAs transcribed from each Composite Insertion. The siRNA profile includes two distinct classes: a 24-nt class corresponding to the TIR and subterminal region of Ac/Ds, and a 21- to 22-nt class homologous to portions of the transcribed region of Ac. These data provide the first evidence for heritable siRNA-mediated silencing of Ac/Ds activity. Our results support and extend a previous model of alternative transposition-induced DNA rereplication to generate Composite Insertions (Zhang et al. 2014), and also show that the resulting Composite Insertions can trigger heritable silencing of otherwise active elements in trans. This study is the first demonstration of de novo TE silencing induced by alternative transposition, which may represent a general mechanism of self-repression of class 2 transposons.
Materials and Methods
Maize stocks and screen
The maize p1 gene regulates pigmentation in floral organs, and p1 alleles are identified by a two-letter suffix indicating their expression in kernel pericarp (the maternal tissue surrounding the seed) and cob glumes (e.g., “w” for white, “r” for red, and “v” for variegated). The progenitor p1-vv-9D9A allele (Zhang and Peterson 1999) contains an active Ac element and a fractured Ac element (fAc; a terminally deleted Ac element) inserted into p1 intron 2. The p1-vv-9D9A allele was introgressed into maize inbred B73, which has a p1-wr genotype. To screen for variants in Ac activity derived from p1-vv-9D9A, silks of plants of genotype p1-vv-9D9A/p1-wr were crossed by pollen from an Ac tester line of genotype p1-ww; rm-3::Ds. This tester is recessive for a null allele of p1 and is also homozygous for an Ds insertion allele of r1, which exhibits excisions in the aleurone of Ds in the presence of Ac transposase, resulting in colored sectors (Kermicle 1980). The mature ears were screened for multikernel sectors or individual kernels with developmentally delayed (small) purple spots on kernel aleurone due to repressed Ac activity. These candidate kernels were then planted and backcrossed to B73 for more than six generations to generate the material for study. After introgression, the final genotype of plants carrying these candidates is p1-ww-id/p1-wr; r1/r1 in B73 background.
Genomic PCR and DNA gel blot hybridizations
Total DNA was prepared from seedling shoots by using a modified cetyltrimethylammonium bromide extraction protocol (Allen et al. 2006). HotMaster Taq polymerase (5PRIME) was used in the PCR reactions. PCR reactions were heated at 94° for 2 min; followed by 35 cycles of 94° for 20 sec, 60–68° annealing (depending on primers) for 30 sec, and 65° for 1 min/1 kb expected product length; followed by a final cycle at 65° for 8 min. The sequences of oligonucleotide primers are listed in Supplemental Material, Table S1. For Southern blots, total DNA extracted from seedling shoots was digested with restriction enzymes from Promega and electrophoresed through 0.8% agarose gels. Blotting and hybridization were performed according to standard protocols (Sambrook et al. 1989); blots were washed in stringent conditions (0.5% SDS, 0.5×SSC at 60°).
Plant growth and tissue collection
Seeds were germinated in SB300 Universal Mix soil mix, and grown in a PGW-40 growth chamber (Percival Scientific, Perry, IA) at 25° for 15 hr in the light and 20° for 9 hr in the dark. Both the above-ground tissues (shoot) and below-ground tissues (root) were harvested from seedlings 14 days after sowing. Nine random plants were pooled for each genotype and tissue type.
RNA isolation and complementary DNA preparation
Total RNA was extracted from seedling shoots and roots by two methods: (1) using the RNeasy Plant mini kit (QIAGEN, Valencia, CA) for small-scale RNA isolation, and (2) using the PureLink Plant RNA Reagent (Life Technologies) for large-scale RNA isolation. The RNA extracted by both methods was then treated with DNase I (New England Biolabs) to remove residual genomic DNA. The RNA extracted by RNeasy Plant mini kit was then converted to complementary DNA (cDNA) by Omniscript RT kit (QIAGEN) primed with oligo-dT and was used as template in the RT-PCR and quantitative RT-PCR experiment. The RNA used in experiments involving small RNA-sequencing and detection of dsRNAs was extracted using the PureLink Plant RNA Reagent.
Quantitative RT-PCR
Quantitative RT-PCR was performed using the Stratagene Mx4000 multiplex quantitative PCR system. Total RNA and cDNA were prepared as described above. PCR was catalyzed by SsoFast EvaGreen Supermixes (Bio-Rad, Hercules, CA) with two technical repeats and three biological repeats. Ac transcript levels measured by primers ac10 and ac11 were normalized by comparison to levels of ubiquitin transcript measured by primers Ubi-f and Ubi-r in the same sample as an internal control. The relative expression level of Ac in each sample was then calculated by comparison to levels of Ac transcript in p1-vv-9D9A, which contains a single active Ac element. Standard deviations were calculated among replications. Student’s t-test was then performed to evaluate the differences among samples at significance level of 0.05.
Detection of dsRNAs
Total RNA, prepared as described above, was treated with RNases A/T1 (Thermo Fisher Scientific) with concentrations of 0, 1.5, and 15 units at 37° for 15 min. The treated RNA was precipitated and reverse transcribed into cDNA by SuperScript III Reverse Transcriptase (Life Technologies) primed with random primers (Thermo Fisher Scientific) as the template for PCR. A seminested PCR was used to amplify regions inside the expected dsRNA (primers ac12 + ac6 followed by ac4 + ac6) to detect dsRNA. Genomic DNA contamination was not detected by using primer ac13 + ac14.
Small RNA high-throughput sequencing and data analysis
Total RNA was extracted as described above and previously (Zuo et al. 2016); library preparation and sequencing on Illumina platform HiSeq 2000 were performed by Beijing Genomics Institute. Adapter sequences, contamination, and low-quality reads were filtered from raw data. The small RNA sequences were mapped to Ac full-length DNA sequence (4565 bp) by Bowtie (Langmead et al. 2009). Only perfectly matched short reads were included in the analysis. The mapped reads from each library were normalized to read counts per million reads.
Data availability
The small RNA high-throughput sequencing data analyzed here is deposited in the NCBI under accession number SRP062285. Maize stocks are available upon request. All data necessary for confirming the conclusions of the article were presented within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12150459.
Results
The model of sister chromatid transposition-induced DNA rereplication
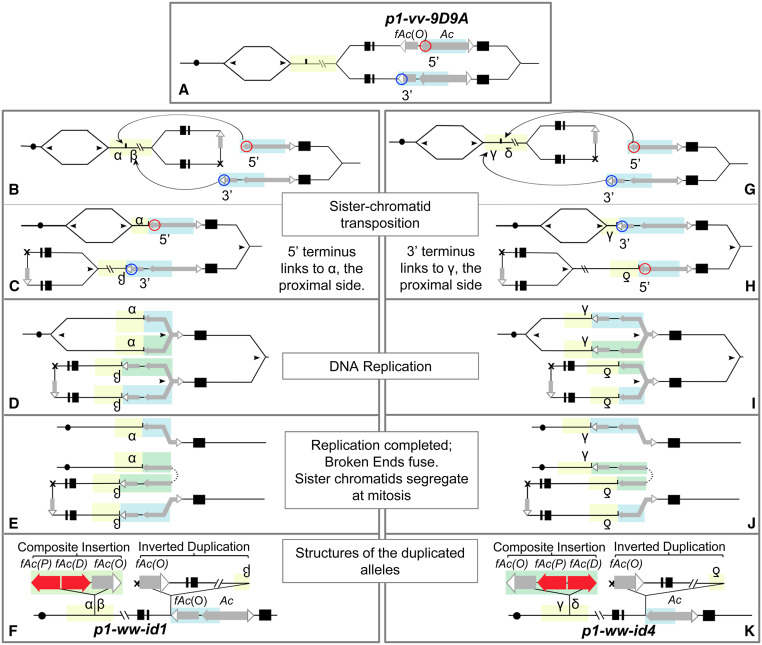
We previously reported that tandem direct duplications and associated Composite Insertions can be produced by reversed end transposition, a type of alternative transposition, followed by DNA rereplication and repair (Zhang and Peterson 2004; Zhang et al. 2014). The Composite Insertions are bordered by partial or full copies of the Ac transposon and may also include sequences flanking the original Ac donor site. Extending the principle of alternative transposition-induced DNA rereplication, we hypothesized that sister chromatid transposition during DNA replication can produce inverted duplications and Composite Insertions (Figure 1 and Supplemental Material Video 1). In sister chromatid transposition, the Ac transposase acts on a pair of directly oriented Ac termini that are present in the progenitor allele p1-vv-9D9A (Figure 1A). This allele contains a complete Ac element inserted 112 bp from a second element termed fAc, which contains only the 3′ half of Ac. The Ac element is known to preferentially transpose during or shortly after S phase (Greenblatt and Brink 1962; Chen et al. 1987), possibly because the Ac transposase preferentially interacts with hemimethylated Ac TIRs (Ros and Kunze 2001). Following replication, the Ac 5′ and fAc 3′ termini located on sister chromatids have strand-specific hemimethylation patterns that are competent for transposition. Excision of the 5′ and 3′ termini followed by religation of the host sequences flanking these termini is expected to produce a sister chromatid fusion with a small sequence footprint at the excision site (Weil and Wessler 1993) (Figure 1B). The excised termini may then reinsert at many possible genomic sites; for example, they can reinsert at a proximal site to generate reciprocal duplication/deletion chromatids (Figure 1C) (Zhang and Peterson 1999). Insertion of the Ac termini into the target site commonly produces 8-bp target site duplications flanking the insertion site, a diagnostic feature of Ac transposition (Döring and Starlinger 1984; Peacock et al. 1984; Pohlman et al. 1984). When sister chromatid transposition from a replicated donor site inserts into an unreplicated target site, previously replicated sequences are joined to unreplicated DNA. As DNA synthesis continues, replication forks progress into the newly inserted Ac termini, rereplicating the Ac/fAc sequences. Replication may continue into the flanking DNA and extend for 10 kb or more (Zhang et al. 2014) (Figure 1D). Ultimately, the DNA rereplication forks spontaneously abort, producing two broken ends that fuse together (McClintock 1951). In some cases, fusion of the two broken ends can generate two fAc elements in an inverted orientation, as shown in Figure 1E. In the ensuing mitosis, the two sister chromatids segregate into two daughter cells: one contains a deletion (p1-ww-def) and the other contains an inverted duplication (p1-ww-id) with a Composite Insertion. In the Composite Insertion, the two new fAcs are both derived from Ac 5′ termini and are in an inverted orientation relative to each other. We term the proximal one fAc(P), and the distal one fAc(D), to distinguish them from the original fAc(O) in the progenitor p1-vv-9D9A allele (Figure 1F). Two possible orientations of insertion are predicted by the model: (1) the 5′ terminus of Ac ligates to the proximal side of the insertion target (Figure 1, B–F), and (2) the 3′ terminus of Ac ligates to the proximal side of the insertion target (Figure 1, G–K). These two orientations are found in the p1-ww-id1 and p1-ww-id4 alleles, respectively.
Figure 1.
Model of sister chromatid transposition-induced DNA rereplication. (A) Maize chromosome 1 and progenitor allele p1-vv-9D9A with DNA replication bubbles. Solid circle indicates centromere. The p1 gene contains 3 exons (black boxes) with Ac and fAc elements located in intron 2 (gray boxes with solid/open arrowheads indicate the 5′ and 3′ Ac/fAc termini, respectively). The 5′ and 3′ Ac/fAc termini involved in sister chromatid transposition are circled in red and blue, respectively. The unreplicated regions are highlighted in yellow, and the replicated regions to be rereplicated are highlighted in blue. (B and C) Sister chromatid transposition, p1-ww-id1 orientation. (B) Excision of the Ac 5′ and fAc 3′ termini results in excision footprint (marked by X) and fusion of the two sister chromatids. Curved arrows indicate insertion of the excised transposon ends into the target site (short vertical line). (C) The 5′ terminus of Ac is ligated to the centromere-proximal side (marked α) and the 3′ terminus of fAc is ligated to the distal side (marked β). This joins the unreplicated sequences at the insertion site to the previously replicated Ac/fAc sequences. The replication fork containing the sister chromatid fusion is flipped. (D) DNA replication. As DNA replication continues, Ac/fAc sequences are rereplicated (highlighted in green). (E) Completion of DNA replication. Rereplication of Ac/fAc sequences aborts, releasing two broken ends that fuse (dotted line). This produces a Composite Insertion between α and β. During mitosis, sister chromatids separate. One sister chromatid (top) has a deletion, and the other sister chromatid (bottom) carries a corresponding inverted duplication and a Composite Insertion between α and β. (F) Maize chromosome 1 with the p1-ww-id1 allele. The Composite Insertion in the α/β site contains two new fAc elements (solid red arrows) containing Ac 5′ sequences in inverted orientation: fAc(P) (proximal) and fAc(D) (distal). P1-ww-id1 also contains an inverted duplication of the segment from fAc(O) (original) to site β. (G and H) Sister chromatid transposition, p1-ww-id4 orientation. (G) Same as B, except the 3′ terminus of fAc will ligate to the centromere-proximal side (marked γ) and the 5′ terminus of Ac will ligate with the distal side (labeled as δ) of the insertion site. (H–J) Same as C–E. (K) Maize chromosome 1 containing the p1-ww-id4 allele. For animation of alternative transposition mechanism, see Supplementary Materials Video 1.
Alleles p1-ww-id1 and p1-ww-id4 contain structural hallmarks expected from the model of sister chromatid transposition-induced DNA rereplication
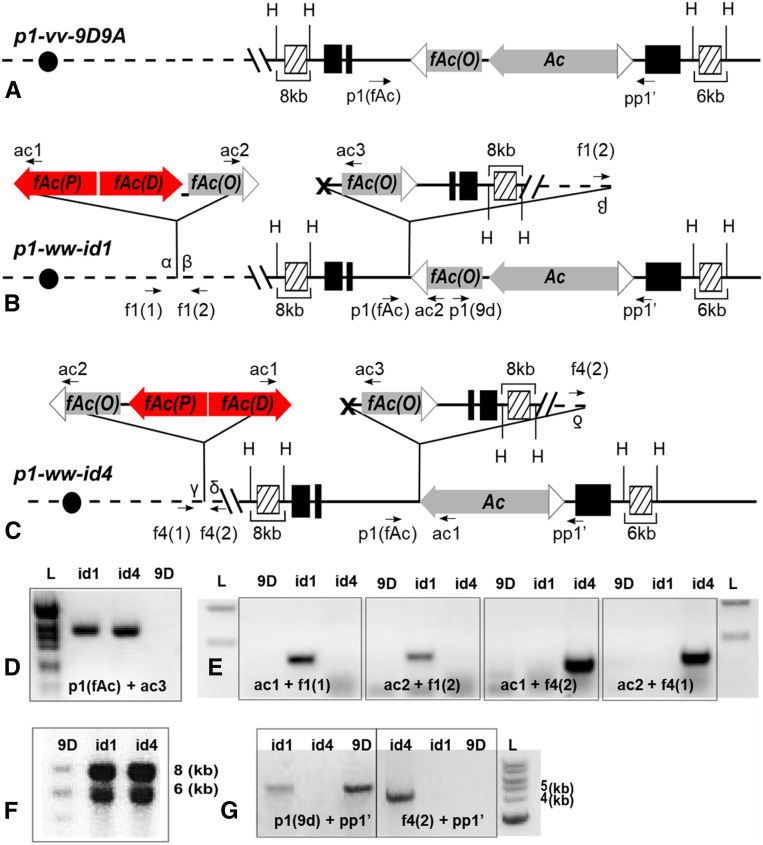
Figure 2 summarizes the structural features of the p1-vv-9D9A, p1-ww-id1, and p1-ww-id4 alleles (Figure 2, A–C.) As predicted by the sister chromatid transposition model, both p1-ww-id1 and p1-ww-id4 contain Composite Insertions and inverted duplications as compared to the progenitor p1-vv-9D9A. To further test the model, we examined the sequences at the junctions of the inverted duplications. As previously reported (Zhang and Peterson 1999), p1-ww-id1 contains a typical Ac excision footprint at the junction of the inverted duplication segments. For p1-ww-id4, we PCR-amplified and sequenced the excision site [using primers ac3 + p1(fAc) in Figure 2C; results shown in Figure 2D]. Similar to the footprint of p1-ww-id1 (Zhang and Peterson 1999), p1-ww-id4 shows changes at the first nucleotides flanking the Ac/fAc excision sites, as is typical for sites of Ac excision (File S1).
Figure 2.
p1-ww-id1 and p1-ww-id4 contain Composite Insertions and inverted duplications. (A–C) Structure of the progenitor allele p1-vv-9D9A (A), and expected structures of p1-ww-id1 (B) and p1-ww-id4 (C) based on the model of sister chromatid transposition-induced DNA rereplication. PCR primers are labeled as arrows, and sequences homologous to hybridization probe 15 are labeled as hatched boxes. H stands for HindIII restriction site. Other symbols are as in Figure 1. (D) Gel analysis of PCR to amplify the sister chromatid transposition footprints in p1-ww-id1 and p1-ww-id4. Bands were excised from the gel and sequenced (File S1). (E) Gel analysis of PCR to amplify the sequences flanking Composite Insertions. Bands were excised from the gel and sequenced to identify the target site duplications flanking each Composite Insertion (File S2). (F) Genomic Southern blot produced by digestion with HindIII and hybridization with probe 15. (G) Gel analysis of PCRs to determine the orientation of sister chromatid transposition (see text for details).
We then isolated the sequences flanking the Composite Insertions in each allele by inverse PCR, and confirmed the insertion junctions by direct PCR using primer pairs f1(1) + ac1 for p1-ww-id1, and f4(2) + ac1 for p1-ww-id4 (Figure 2E). The sequences flanking the Composite Insertions contain perfect 8-bp target site duplications (“GCCTCGCT” in p1-ww-id1 and “GCCCGGAT” in p1-ww-id4; File S2) characteristic of Ac transposition.
The sister chromatid transposition model predicts that the p1-ww-id1 and p1-ww-id4 alleles contain inverted duplications extending from the p1 locus at 48.6 Mb of maize chromosome 1 to the Composite Insertion sites. The sequences flanking the Composite Insertions in p1-ww-id1 and p1-ww-id4 are located at positions 51.8 and 48.9 Mb, respectively, on chromosome 1, maize B73 RefGen_v4 reference genome. These results indicate that the p1-ww-id1 and p1-ww-id4 contain inverted duplications of 3.2 and 0.3 Mb, respectively.
The presence of duplicated segments in p1-ww-id1 and p1-ww-id4 is tested by Southern blot using HindIII digests and probe fragment 15 (Figure 2F). The fragments of 8 and 6 kb originate from the proximal and distal side of p1, respectively. In the progenitor allele p1-vv-9D9A (labeled as 9d), the intensities of the 8- and 6-kb bands are approximately equal, consistent with their single-copy status in p1-vv-9D9A. In the p1-ww-id1 and p1-ww-id4 alleles, the 8-kb band intensity is approximately twice that of the 6-kb band, consistent with a duplication of the 8-kb segment proximal to p1 in those alleles. Hybridization signals in lane 9d are overall weaker due to the fact that there is less DNA loaded on the gel. This experiment confirms the presence of a duplication extending to the proximal side of p1 as predicted by the sister chromatid transposition model.
Finally, we tested the orientations of the Composite Insertions in the p1-ww-id1 and p1-ww-id4 alleles. These orientations depend on whether the excised 5′ Ac and 3′ fAc(O) are joined to the proximal or distal sides of the Composite Insertion site (Figure 1, B and G), and are correlated with the presence or absence of fAc(O) adjacent to Ac in the original p1 donor site (Figure 1, F and K). Therefore, we used PCR primers p1(9d) + pp1’ to amplify from fAc(O) across Ac and into the flanking p1 gene sequence, producing a 6.6-kb product; this 6.6-kb product was observed in p1-ww-id1 and the control p1-vv-9D9A, but not p1-ww-id4 (Figure 2G). This result indicates that in p1-ww-id1, the 3′ terminus of fAc(O) is ligated to the centromere-distal side of the target site (β in Figure 1B). Conversely, primers f4(2) and pp1’ produce a 4.5-kb band from p1-ww-id4, but not p1-ww-id1 and p1-vv-9D9A (Figure 2G). This 4.5-kb band reflects the absence of fAc(O) adjacent to Ac in the p1-ww-id4 allele, indicating that in p1-ww-id4, the 5′ terminus of Ac is ligated to the centromere-distal side of target site (δ in Figure 1G). In summary, these results show that the p1-ww-id1 and p1-ww-id4 alleles contain transposon donor excision footprints, Composite Insertions in two possible orientations flanked by target site duplications, and proximal duplications, all as predicted by sister chromatid transposition with insertion into unreplicated target sites.
p1-ww-id1 and p1-ww-id4 alleles induce de novo and heritable silencing of Ac
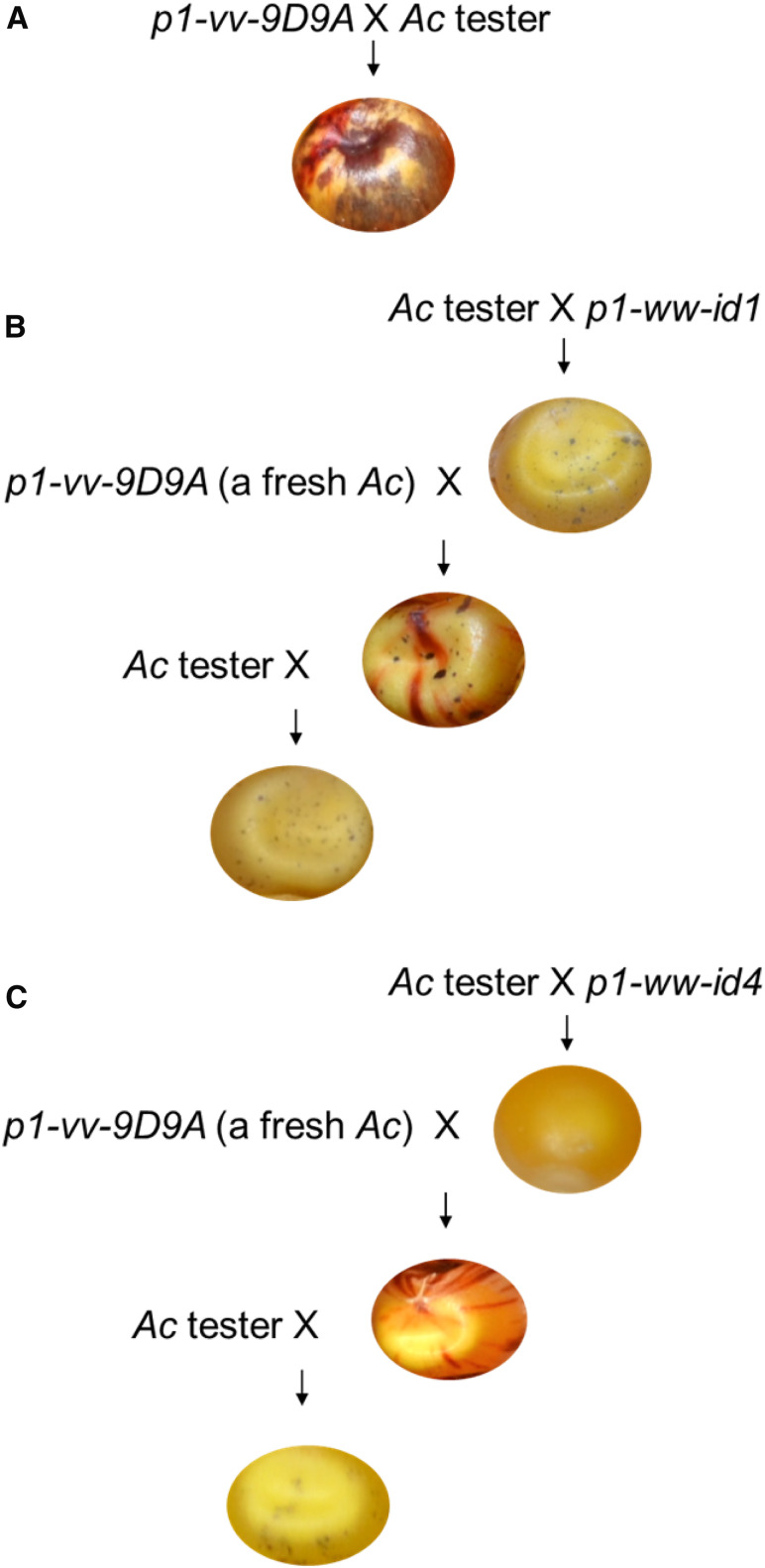
The maize p1 gene is required for kernel pericarp (seed coat) pigmentation (Zhang and Peterson 1999). The p1-vv-9D9A allele contains an Ac element in p1 intron 2 (Figure 2A), which blocks p1 expression and results in colorless pericarp. Somatic excision of Ac can restore p1 function and thus produce red clonal sectors on the kernel pericarp (a variegated pericarp phenotype). The p1-vv-9D9A allele produces frequent kernel pericarp sectors, indicating that the Ac in p1-vv-9D9A is fully active. This is confirmed by crosses of p1-vv-9D9A to an Ac tester line of genotype r1-m3::Ds (Figure 3A). The Ac tester line contains a nonautonomous Ds element inserted in r1 (red1), a gene required for anthocyanin biosynthesis in kernel aleurone. Without an active Ac, the Ds insertion blocks r1 function, resulting in colorless aleurone; however, with an active Ac, Ds can be transposed from r1, producing purple aleurone sectors (Kermicle 1980; Lechelt et al. 1989). The activity of Ac can thus be assessed by the size and frequency of purple spots on the kernel: large spots from early Ds excision events indicate a fully active Ac, while small spots from delayed Ds excisions indicate repressed Ac activity. As shown in Figure 3A, crosses of p1-vv-9D9A to the Ac tester line produce kernels with a coarsely spotted pattern typical of active Ac. In contrast, crosses of p1-ww-id1 with Ac testers produce kernels with fine spots (Figure 3B, top kernel), while p1-ww-id4 gives no spots in test crosses (Figure 3C, top kernel). These results suggest that the Ac elements in p1-ww-id1 and p1-ww-id4 are repressed. Moreover, p1-ww-id1 and p1-ww-id4 can induce trans-dominant repression of a fresh and active Ac. This can be seen in the kernels produced by crossing p1-ww-id alleles to the p1-vv-9D9A allele that contains a single active Ac. Kernels heterozygous for p1-vv-9D9A/p1-ww-id1 show very few and fine aleurone sectors (Figure 3B, second kernel), while kernels produced by crossing p1-vv-9D9A with p1-ww-id4 had no visible aleurone sectors (Figure 3C, second kernel). Note that these kernels exhibit a normal variegated pericarp phenotype because the kernel pericarp is a maternal tissue and thus is not affected by a repressive p1-ww-id allele introduced through the pollen in this cross.
Figure 3.
Genetic crosses indicate the in-trans and heritable repression of Ac/Ds initiated by p1-ww-id1 and p1-ww-id4. (A) Progenitor allele p1-vv-9D9A contains a single active Ac element, shown by the heavily spotted kernel aleurone from the cross of p1-vv-9D9A and Ac tester. Cross: p1-vv-9D9A x r1-m3::Ds. (B) Ac activity is repressed in p1-ww-id1, shown by the fine spots in kernel aleurone in the first cross of p1-ww-id1 and Ac tester (p1-vv-id1 x r1-m3::Ds). The second cross (p1-vv-9D9A x p1-ww-id1) introduces a fresh active Ac in p1-vv-9D9A, which is repressed in trans by p1-ww-id1 (indicated by small aleurone spots). The third cross (r1-m3::Ds x p1-vv-9D9A/p1-ww-id1) shows that silencing of Ac in p1-vv-9D9A is heritable, as small spots are maintained even after p1-vv-9D9A is segregated from p1-ww-id1. (C) Parallel crosses of p1-ww-id4 as in B. Ac activity is repressed in the line of p1-ww-id4, shown as absence of kernel aleurone spots in the first cross between p1-ww-id4 and Ac tester. The repression is trans-dominant, shown by the absence of spots in kernels produced by cross of active Ac from p1-vv-9D9A by p1-ww-id4. Ac repression is heritable, shown by the fine spotting in kernels from the third cross in which p1-vv-9D9A is segregated from p1-ww-id4.
To test whether the repression of Ac induced by p1-ww-id1 and p1-ww-id4 reflects heritable silencing, we crossed p1-vv-9D9A/p1-ww-id plants to the Ac tester line; this cross separates the Ac element in p1-vv-9D9A from each p1-ww-id allele. If the repression of Ac in p1-vv9D9A is relieved following segregation from p1-ww-id, then we would expect to see ∼50% coarsely spotted kernels (containing p1-vv-9D9A) and ∼50% weakly or nonspotted kernels (containing p1-ww-id). However, if Ac is heritably silenced, then most or all progeny kernels would again show few, small Ds excision sectors. The results show that all the kernels produced by crossing p1-vv-9D9A/p1-ww-id1 with the Ac tester show few or zero purple sectors (0% heavily spotted seeds; Figure 3B, third kernel). The corresponding ear for this cross is shown in Figure S1, and kernel count data are provided in Table S2, Cross 6. Parallel genetic tests show that Ac repression induced by p1-ww-id4 is also heritable, but is less stable than that induced by p1-ww-id1. Among 160 progeny kernels from the cross of Ac tester by p1-vv-9D9A/p1-ww-id4, 126 are weakly or nonspotted (Figure 3C, third kernel), and 34 kernels are heavily spotted. Assuming the heavily spotted kernels are derived from a reactivated Ac in the p1-vv-9D9A allele, this indicates that Ac repression is lifted in 42.5% (34/80) of kernels following segregation from p1-ww-id4 (Fisher exact test P < 0.00001, indicating a significant difference from the p1-vv-9D9A ear without the initial silencing; ear for this cross is shown in Figure S2, and kernel count data are provided in Table S2, cross 8). In other words, the silencing of Ac induced by p1-ww-id4 is heritable in only ∼57.5% of the progeny kernels that carry p1-vv-9D9A.
We also assessed the maintenance of Ac silencing in succeeding generations following segregation from the p1-ww-id1 and p1-ww-id4 alleles. Plants of genotype p1-vv-9D9A*/p1-ww (weakly spotted kernels, where * indicates Ac silenced by exposure to p1-ww-id1 or p1-ww-id4 in the prior generation) were crossed again to the Ac tester stock r1-m3::Ds. In these crosses, 50% of the kernels receive p1-ww (no Ac) and are nonspotted, while the remaining 50% of kernels receive p1-vv-9D9A* and their spotting pattern can reflect the stability of Ac silencing. Interestingly, p1-ww-id1 and p1-ww-id4 induce different levels of silencing stability. For p1-vv-9D9A* initiated by p1-ww-id1 and separated from it for two generations, all the kernels are nonspotted or weakly spotted, indicating near complete maintenance of Ac repression (Figure S1; Table S2, cross 7). In contrast, the silencing of Ac initiated by p1-ww-id4 and segregated from it for two generations is not efficiently maintained. Among 144 progeny seeds from the cross of Ac tester by p1-vv-9D9A/p1-ww, 72 seeds are expected to inherit the p1-vv-9D9A allele and thus contain Ac. In the progeny kernels we observed 52 heavily spotted seeds, meaning that Ac is reactivated in 72.2% (52/72) of the kernels (Fisher exact test P-value is 0.0116, indicating a significant difference from the p1-vv-9D9A ear without the initial silencing; Figure S2; Table S2, cross 9). Thus, the repression of Ac initiated by p1-ww-id4 is maintained in only ∼27.8% of progeny kernels after two generations.
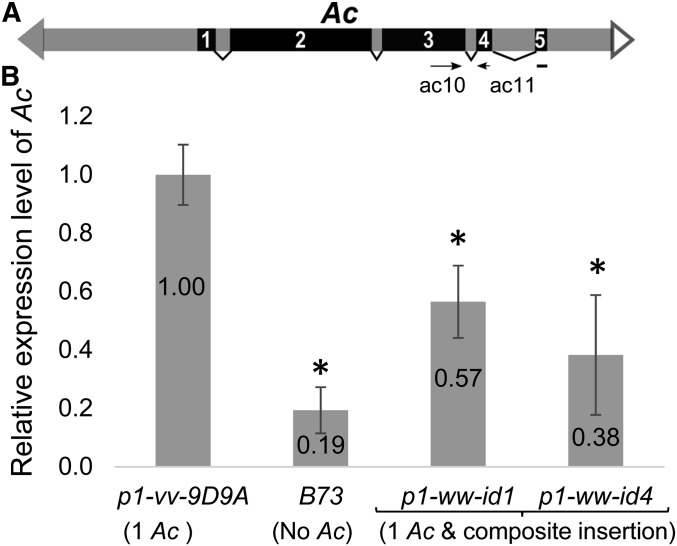
To determine the mechanism of Ac repression, we performed quantitative RT-PCR experiment using Ac-specific primers. Compared to the progenitor allele p1-vv-9D9A, the p1-ww-id1 and p1-ww-id4 alleles express significantly decreased levels of Ac transcript (Student’s t-test P = 0.0015 and 0.0004, respectively) (Figure 4), suggesting that repression occurred via transcriptional and/or post-transcriptional gene silencing.
Figure 4.
The Ac transcript level is decreased in p1-ww-id alleles. (A) Schematic structure of the full-length Ac transposable element. Solid and open triangles indicate Ac 5′ and 3′ termini, respectively. Black boxes with numbers are Ac exons 1–5; boxes with subtending lines indicate introns. Primers used in the quantitative RT PCR (ac10 and ac11) are labeled as arrows; note that primer ac11 spans Ac intron 4. (B) Quantitative RT-PCR measurement of Ac transcript levels. Bars labeled by * indicate transcript levels that are significantly different from p1-vv-9D9A by Student’s unpaired t-test with P < 0.05.
Composite Insertions of alleles p1-ww-id1 and p1-ww-id4 contain inverted repeats of fAc fragments
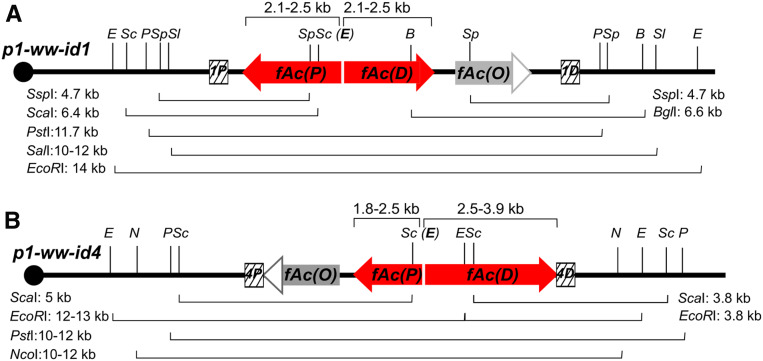
We performed genomic Southern blots to determine the structures of the Composite Insertions in p1-ww-id1 and p1-ww-id4. The results are summarized in Figure 5, and the raw data are shown in Figure S3 and Figure S4. First, the sizes of Composite Insertions are estimated by using restriction enzymes that do not cut within the Ac element. For p1-ww-id1, digestion with SalI and hybridization with probe 1D gives a band of 10–12 kb, indicating a Composite Insertions of ∼6.2–8.2 kb. Consistent with this, enzyme PstI and probe 1P give an estimated size of ∼6.7 kb. We then determined the internal Composite Insertion structure by digesting with a series of endonucleases with known sites in Ac. We observed the expected bands produced by digestion at the SspI and ScaI sites in fAc(P), and the expected bands from digestion at the SspI of fAc(O) and BglI sites of fAc(D). These results indicate that the Composite Insertion of p1-ww-id1 includes the first 1842 bp sequences from the Ac 5′ terminus of fAc(P) and the first 729 bp sequences from fAc(D). If either fAc(P) or fAc(D) contained the EcoRI site located near the center of Ac, then digestion with EcoRI and hybridization with probe 1P would yield a band of size 7.6–9.6 kb. No such band is observed, and instead we detect a larger band of ∼14 kb. This indicates that both fAc(P) and fAc(D) were truncated prior to reaching the EcoRI site at position 2486 bp from the Ac 5′ end. We conclude that the total size of Composite Insertion in p1-ww-id1 is <7.1 kb, consistent with the ∼6.7 kb size estimated by PstI digestion.
Figure 5.
Summary of Southern blot results that elucidate the internal structures of Composite Insertions. Schematic structures of p1-ww-id1 and p1-ww-id4 are shown in A and B, respectively, with restriction sites (vertical lines; E: EcoRI; Sc: ScaI; P: PstI; Sp: SspI; Sl: SalI; B: BglI; N: NcoI) and probes (hatched boxes) shown. Bands observed from Southern blot (Figures S3 and S4) are indicated by braces with actual sizes labeled by each restriction enzyme.
Similar assays were performed on p1-ww-id4 (Figure 5B and Figure S4). PstI and NcoI digestions both show the total size of the Composite Insertion in p1-ww-id4 as ∼6.4–8.4 kb. The Composite Insertion contains the ScaI site in both fAc(P) and fAc(D), indicating that both fAc fragments include at least 1842 bp of Ac 5′ terminal sequences. Additionally, fAc(D) contains the EcoRI site but fAc(P) does not, indicating that fAc(P) is truncated prior to the EcoRI site at position 2486 bp from Ac 5′ terminus. Combining the results from ScaI and EcoRI digests, the size of fAc(P) could be from 1842 to 2486 bp. By subtracting the size of fAc(P) (1.8–2.5 kb) from the total size of Composite Insertion of p1-ww-id4 (6.4–8.4 kb), we infer the size of fAc(D) is 2.5–3.9 kb. Extensive attempts were made to PCR amplify and sequence the exact fusion point connecting fAc(D) and fAc(P) in both Composite Insertions; unfortunately, these attempts failed, most likely due to the long perfectly inverted repeats present in both Composite Insertions, which would give rise to self-annealed hairpins with extensive secondary structures in the template DNA.
The inverted repeats of fAc fragments are transcribed to produce dsRNAs
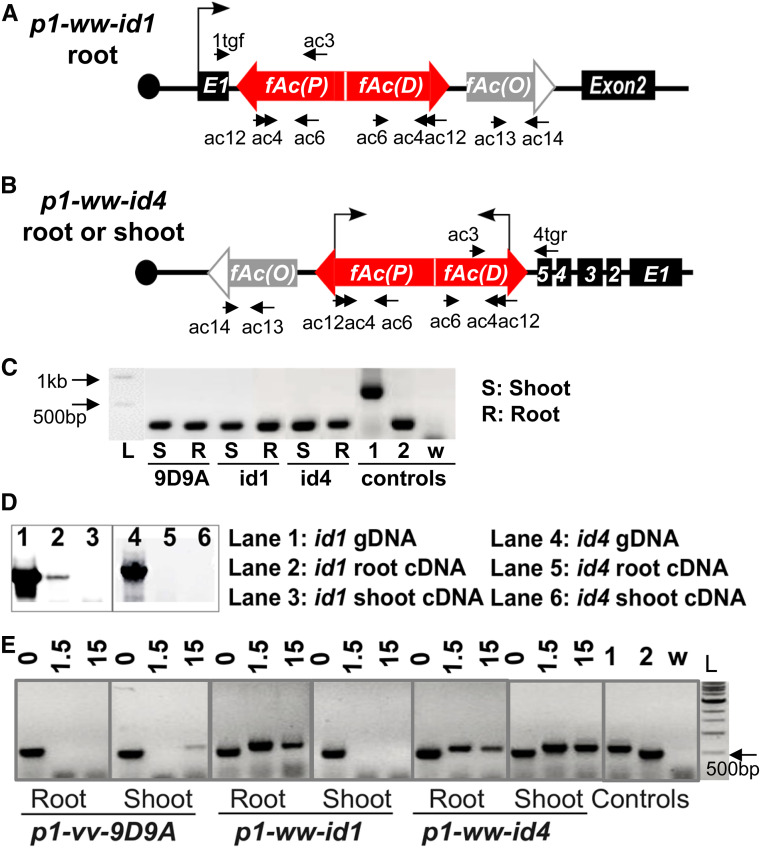
The inverted fAc(P) and fAc(D) fragments in both p1-ww-id1 and p1-ww-id4 retain the Ac promoter. Moreover, consistent with the characteristic preferential insertions of Ac/Ds into linked genes (Vollbrecht et al. 2010), both Composite Insertions are located in the middle of annotated maize genes: the Composite Insertion in p1-ww-id1 is located at intron 1 of Zm00001d028930, while the Composite Insertion in p1-ww-id4 is located at intron 5 of Zm00001d028863 (Figure 6, A and B). The locations of Composite Insertions within genes could allow their expression by read-through transcription from the flanking genes’ promoters.
Figure 6.
Composite Insertions in p1-ww-id1 and p1-ww-id4 produce dsRNA transcripts. (A) Structure of the Composite Insertion in p1-ww-id1, located in intron 1 of gene Zm00001d028930. Bent arrow indicates transcription from the host gene promoter to generate read-through transcripts of the Composite Insertion in root tissues, but not shoot. Labeled arrows indicate primers used in RT-PCR. (B) Structure of the Composite Insertion in p1-ww-id4, located in intron 5 of Zm00001d028863. The promoter of Zm00001d028863 is not active in either root or shoot tissues. Bent arrows indicate transcription from the native Ac promoters. (C) Test of genomic DNA contamination in RNA samples. Primers ac13 + ac14 are located in fAc(O) outside the dsRNA region, in Ac exons 3 and 5 (Figure 6, A and B). Bands of 744 bp and 272 bp are expected from genomic DNA and spliced cDNA, respectively. Control lanes “1” and “2” are from templates of genomic DNA and root cDNA, respectively, of p1-ww-id1; lane “w” is from a water template as a control for PCR contamination. (D) Test for chimeric transcripts by RT-PCR using primers 1tgf + ac3 in p1-ww-id1 (lanes 1–3), and 4tgr + ac3 in p1-ww-id4 (lanes 4–6). Chimeric transcript initiating from flanking gene promoter was detected only in p1-ww-id1 root. No chimeric transcript from flanking gene promoter was detected in p1-ww-id4. (E) Detection of dsRNA by RNase protection assay. Total RNA from the indicated tissues and alleles were treated with DNase1 and three quantities of RNase A/T1 (0, 1.5, and 15 units). Treated RNA samples were then analyzed by a seminested PCR (primers ac12 + ac6; followed by ac4 + ac6). Control lanes “1” and “2” are from templates of genomic DNA and root cDNA, respectively, of p1-ww-id1; lane “w” is from a water template as a control for PCR contamination.
To test for read-through transcription, we performed RT-PCR experiments using total RNA extracted from root and shoot tissues of plants containing p1-ww-id1 and p1-ww-id4. No genomic DNA contamination is detected as illustrated in Figure 6C. For p1-ww-id1, chimeric transcripts are detected in root, but not shoot (Figure 6D); this is the same pattern of expression as the flanking host gene Zm00001d028930 (Wang et al. 2009) (Figure S5). The chimeric transcript includes sequences from exon 1 and intron 1 of the host gene Zm00001d028930, and sequences from the 5′ terminus of Ac (File S3), consistent with the structure expected of read-through transcripts. In contrast, gene Zm00001d028863, the host gene of the Composite Insertion of p1-ww-id4, is not expressed in either root or shoot, and we did not detect chimeric p1-ww-id4 transcripts in those tissues (Figure 6D).
Prompted by the inverted fAc repeat structures in both p1-ww-id1 and p1-ww-id4 Composite Insertions, we searched for Ac-homologous dsRNAs using RNA protection assays. In this experiment, total RNA samples were treated with two concentrations of RNase A/T; linear RNAs can be digested by the RNase, while dsRNAs are resistant. Following RNase treatment, RNAs were reverse transcribed into cDNA, which was used as template for PCR. A seminested PCR was performed to detect sequences protected by dsRNA (primers ac12 + ac6 followed by ac4 + ac6; Figure 6, A and B). We observed bands of the expected size in p1-ww-id1 root, but not shoot. This pattern matches the expression pattern of the host gene Zm00001d028930 and of the presence of chimeric transcripts. We also detected Ac-homologous dsRNAs in both root and shoot of p1-ww-id4 (Figure 6E), suggesting that p1-ww-id4 may be transcribed from its own Ac promoter. Interestingly, both the chimeric transcript and the dsRNAs seem to retain introns (Figure 6, D and E and File S3), suggesting that the hairpin nature of these transcripts may interfere with normal splicing.
We concluded that the inverted repeats of fAc elements in the Composite Insertions of p1-ww-id1 and p1-ww-id4 are transcribed into dsRNAs, either by the flanking gene promoter as in p1-ww-id1, or by the retained Ac promoter as suggested for p1-ww-id4. The accumulation of dsRNAs can thus provide a potential template for production of siRNAs that may directly induce Ac silencing.
Small RNAs derived from Composite Insertions are detected in both p1-ww-id1 and p1-ww-id4 alleles
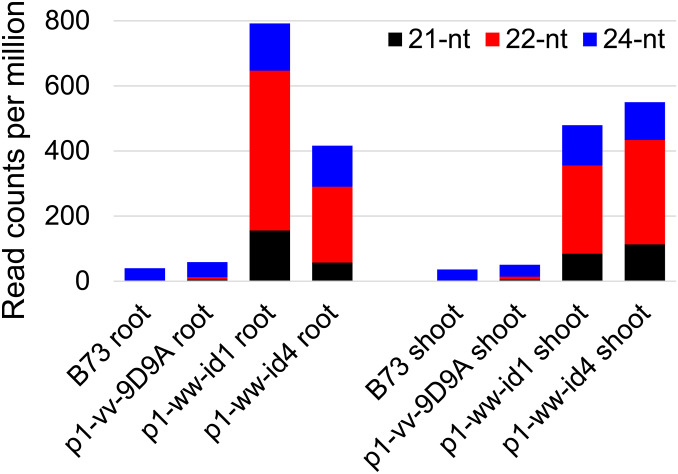
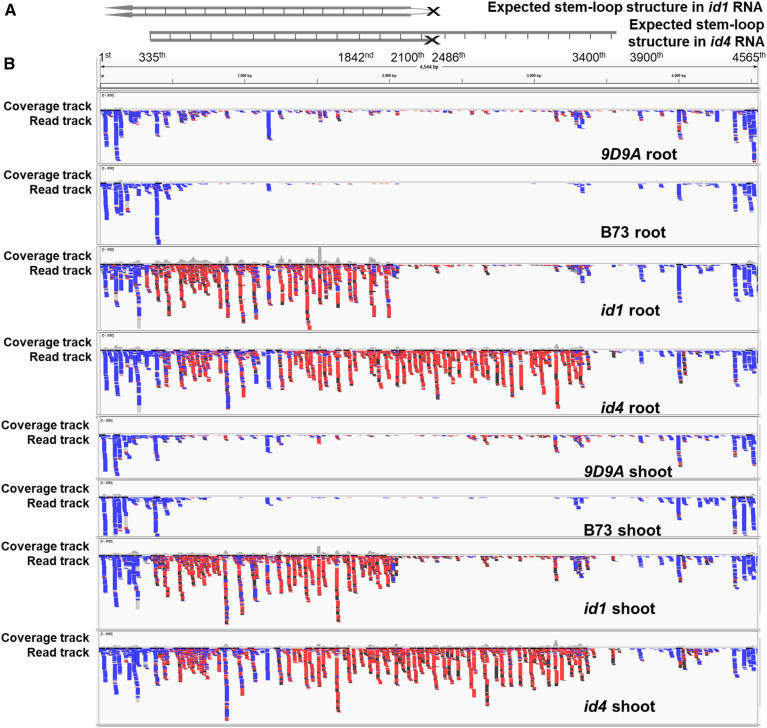
The presence of dsRNAs in p1-ww-id1 and p1-ww-id4 suggests a possible mechanism of Ac silencing by siRNAs. Therefore we performed high-throughput sequencing of small RNAs from root and shoot tissues of p1-ww-id1 and p1-ww-id4 plants. Progenitor allele p1-vv-9D9A and standard maize inbred B73 were used as controls. As shown in Figure 7, siRNAs of 21, 22, and 24 nt that map to Ac are dramatically enriched in both shoot and root from both p1-ww-id1 and p1-ww-id4 plants compared to the same tissues in p1-vv-9D9A and B73. In p1-ww-id1 root and shoot, the enriched small RNAs map to a 1- to 2486-nt region from the Ac 5′ terminus (Figure 8), consistent with the extent of dsRNA predicted by the Composite Insertion inverted repeat structure. In p1-ww-id4, the enriched small RNAs map to the region 1–3400 nt from the Ac 5′ terminus, again matching the expected stem-loop structure with the size range of 2486–3900 nt predicted by the Composite Insertion structure. This finding suggests that small RNAs are synthesized from both the double-strand stem and single-strand loop regions of p1-ww-id4.
Figure 7.
Small RNAs mapped to Ac. Compared to inbred B73 and progenitor allele p1-vv-9D9A, small RNA levels are increased significantly in both p1-ww-id1 and p1-ww-id4. Colored bars indicate the abundance of 21-, 22-, and 24-nt small RNA classes.
Figure 8.
Detection and mapping of small RNAs in p1-ww-id1 and p1-ww-id4. (A) The expected stem-loop structures in p1-ww-id1 and p1-ww-id4. The schematic structures include a ruler with corresponding nucleotide position in Ac. The loop of p1-ww-id4 is open for better presentation of the size. The transcription of Composite Insertion of p1-ww-id1 is driven by the host promoter, and the two inverted fAc fragments in p1-ww-id1 are relatively symmetrical (2100–2486 nt of each indicated by Southern blot), predicting a hairpin structure in the dsRNA contains first ∼ (2100th–2486th) nucleotides of Ac. The transcription of Composite Insertion of p1-ww-id4 is driven by the Ac promoter, and thus the transcript starts from 335th nucleotide of Ac. The inverted fAc fragments in p1-ww-id4 differ in size, with one fAc fragment of 1842–2486 nt and the other fAc of 3400–3900 nt indicated by Southern blot. This asymmetrical structure should give rise to a transcript with a stem containing 335th ∼ (1842nd–2486th) nucleotides of Ac and a loop containing (1843rd–2487th) ∼ (3400th–3900th) nucleotides of Ac in p1-ww-id4. (B) Small RNAs mapped to Ac are visualized by the Integrative Genomics Viewer (Robinson et al. 2011). The y axis of the coverage track was standardized to the same scale for each sample. Colors indicate size of siRNAs: black, 21 nt; red, 22 nt; blue, 24 nt.
Interestingly, the different size-classes of siRNAs map to distinctly different regions of Ac. The 24-nt siRNAs are enriched in the 5′ TIR/sub-TIR regions of Ac (Figure S6, B–D); this region includes the native Ac promoter and the presence of these 24-nt small RNAs is consistent with a Pol IV/RDR2-based maintenance of heritable silencing (Law and Jacobsen 2010; Pikaard 2013; Matzke and Mosher 2014; Li et al. 2015a). In contrast, the 21- to 22-nt siRNAs predominate in the transcribed regions of Ac (Figure S6E). This size of siRNAs have been shown to be associated with the Pol II-RDR6 pathway (Pontier et al. 2012; Marí-Ordóñez et al. 2013; Nuthikattu et al. 2013; Matzke and Mosher 2014; McCue et al. 2014; West et al. 2014; Duan et al. 2015; Panda et al. 2016) and thus may induce de novo silencing of active Ac elements. The 21- to 22-nt siRNAs are also mapped to the single-strand loop region in p1-ww-id4 (Figure 8; the expanded loop region is shown in Figure S7). This is consistent with a transitive process, by which RDR6 converts the single-stranded RNAs in the loop into dsRNAs, followed by the processing of dsRNAs into small RNAs. The ratio of 21- to 22-nt siRNA is increased in the loop region in shoot compared to root tissues (Table S3; chi-squared test gives P < 0.01 in shoot and P = 0.24 in root), suggesting tissue-specific variation in production or stability of siRNA species. Finally, we noted that 21-nt siRNAs in p1-ww-id1 are enriched in Ac intron sequences, consistent with the observation that both the chimeric Ac transcript and dsRNAs retain introns in p1-ww-id1 (Figure S8).
We observed similar profiles of siRNA in both root and shoot tissues of p1-ww-id1, even though no Composite Insertion transcripts or dsRNAs were detected in p1-ww-id1 shoots. Possibly, this might result from long-range transportation of siRNAs from root (where dsRNAs are produced) to shoot (where dsRNAs are absent), as described previously in Arabidopsis (Chitwood and Timmermans 2010; Molnar et al. 2010). In any event, the fact that the siRNAs in p1-ww-id1 and p1-ww-id4 correspond well to the structure of the two Composite Insertions in each allele strongly suggests that these Composite Insertions are the source of the siRNAs.
Discussion
In this study, we characterized two naturally occurring maize alleles (p1-ww-id1 and p1-ww-id4) derived by alternative transposition of the transposable element Ac. Both alleles elicit trans-dominant and heritable repression of Ac-induced transposition, reduce accumulation of Ac mRNA, and contain inverted duplications and novel Composite Insertions derived from sister chromatid transposition-induced DNA rereplication. The inverted duplications are of 3.2 Mb in p1-ww-id1 and 0.3 Mb in p1-ww-id4; both Composite Insertions contain inverted repeats of the Ac 5′ terminal sequences, of 2.1–2.5 kb in p1-ww-id1 and 1.8–3.9 kb in p1-ww-id4. These Ac inverted repeats are transcribed either from a flanking gene promoter in the case of p1-ww-id1, or possibly from the Ac promoter itself in the case of p1-ww-id4. dsRNA transcripts are detected from both alleles, presumably due to fold back pairing of the inverted repeat transcripts.
The alternative transposition-initiated de novo and heritable silencing in the p1-ww-id alleles is consistent with mechanisms of transcriptional gene silencing and post-transcriptional gene silencing reported for class 1 elements in Arabidopsis (de novo silencing: Marí-Ordóñez et al. 2013; Nuthikattu et al. 2013; McCue et al. 2014; Duan et al. 2015; Panda et al. 2016; heritable silencing: Law and Jacobsen 2010; Haag and Pikaard 2011; Castel and Martienssen 2013; Matzke and Mosher 2014). In the p1-ww-id alleles described here, the 21- to 22-nt siRNAs enriched in the transcribed region of the Composite Insertions, and particularly in the loop region of p1-ww-id4, suggests the involvement of RDR6 in the de novo silencing. The reduced levels of Ac transcript can be formally explained by mRNA degradation via post-transcriptional gene silencing, and/or transcriptional gene silencing of the Ac promoter triggered by the Pol II-RDR6 pathway. Transcriptional silencing of Ac seems most likely, as silencing is maintained even after the id1 and id4 alleles are removed by meiotic segregation (Figure 3, B and C). The production of 24-nt siRNAs corresponding to the TIR and subterminal regions of Ac signals the possible involvement of the Pol IV-RDR2 RdDM pathway in the maintenance of transcriptional gene silencing by cytosine methylation in all sequence contexts (Law and Jacobsen 2010; Zemach et al. 2013; Stroud et al. 2014; Li et al. 2015b). The siRNA-independent pathway may also participate in the maintenance of symmetrical methylation (Stroud et al. 2013; Matzke and Mosher 2014).
The structure and effects of the p1-ww-id1 and p1-ww-id4 Composite Insertions are remarkably similar to the Muk allele that represses Mutator transposons in maize (Slotkin et al. 2003, 2005; Li et al. 2010; Burgess et al. 2020), and hence the p1-ww-id1 and p1-ww-id4 alleles can be considered as examples of “killers” targeting a different DNA TE superfamily. Muk and the “Ac killers” share similar features: (1) they are both initiated from naturally occurring inverted duplications of partial DNA transposon sequences; (2) the transcription of inverted duplications in Muk and Ac killer in p1-ww-id1 are both driven by nearby promoters, while p1-ww-id4 is possibly transcribed by the Ac promoter, producing dsRNAs as the precursor of 21-, 22-, and 24-nt siRNAs; and (3) both Muk and Ac killer can trigger in trans and heritable silencing of an active element. These similarities suggest a general mechanism for the heritable silencing of active DNA transposons [see Burgess et al. (2020)]. Active TEs are prone to rearrangements, some of which will be competent to silence otherwise active elements in trans. Thus, it is likely that if an element is active for sufficient time, it will eventually produce transposon killers. To the extent that selection favors a loss of TE activity, these killers would tend to spread within a population, eventually resulting in the heritable epigenetic silencing of homologous elements in that population. In addition to silencing resident active TEs, these killers may also serve as a reservoir of “antigens” that immunize genomes from subsequent invasion by active elements. Further, if expressed in specific cells of the male and female gametophyte, TE killers may serve as sources of small RNAs that reinforce TE silencing in germinal lineages (Martínez and Slotkin 2012).
Our discovery of siRNA-mediated de novo and heritable silencing of Ac/Ds adds another layer of complexity to the regulation of Ac/Ds activity. Previous studies of the regulation of Ac/Ds transposons reveal a complex relationship among Ac dosage, transcript abundance, transposase level, DNA methylation, and transposition frequency. Ac has a GC-rich subterminal region, and hypermethylation of this region is associated with transcriptional silencing (Kunze et al. 1988; Brutnell and Dellaporta 1994; Conrad and Brutnell 2005). It is this region of the p1-ww-id alleles that we see being targeted by 24-nt small RNAs. Given that these small RNAs are also observed in B73, they likely arise from inactive Ac elements present in that genetic background. Additionally, splicing of Ac mRNA is inefficient and inaccurate, producing a large number of aberrant Ac transcripts in Arabidopsis, representing post-transcriptional regulation (Jarvis et al. 1997). The aggregation of Ac transposase protein in tobacco (Kunze et al. 1995), petunia, and maize (Heinlein et al. 1994), may represent a type of post-translational regulation responsible for the Ac negative dosage effect first observed by McClintock (1948, 1949, 1950). Molecular analysis showed that increased dosage of Ac results in increased Ac transposase mRNA and protein levels, at least in the maize wx-m7 allele that was tested (Fußwinkel et al. 1991). In this study, we find that the p1-ww-id1 and p1-ww-id4 alleles derived from alternative transposition-induced DNA rereplication initiate transgenerational silencing of active elements via trans-acting siRNAs.
These results also expand our understanding of the effect of alternative transposition on the structure of the maize genome. Previous studies have described the ability of alternative transposition to induce structural rearrangements such as deletions, inversions, translocations, duplications (Zhang and Peterson 2004; Huang and Dooner 2008; Zhang et al. 2009, 2013), and exon shuffling (Zhang et al. 2006; Wang et al. 2015). Here we show that p1-ww-id alleles derived from sister chromatid transposition contain novel inverted-repeat structures (Composite Insertions) generated by DNA rereplication. The fact that similar structures and mechanisms effect silencing in members of two unrelated transposon superfamilies (hAT and Mu) suggests that inverted repeat-induced siRNA may represent a general mechanism of spontaneous TE silencing.
Acknowledgments
We thank Dr. R. Keith Slotkin for suggestions on the dsRNA detection experiment, Lisa Coffey and Dr. Patrick Schnable for providing the growth chamber, Terry Olson for technical assistance, and Douglas Baker for field assistance. This research is supported by National Science Foundation award 0923826 (to T.P. and J.Z.), US Department of Agriculture National Institute of Food and Agriculture Hatch project number IOW05282, State of Iowa funds, and an Institutional Development Award from the National Institute of General Medical Sciences of the National Institutes of Health (grant P20-GM103476).
Author contributions: D.W., J.Z., and T.P. conceived and designed the experiment; D.W. performed the experiments; T.Z., M.Z., and D.W. analyzed the small RNA-sequencing data; D.W., D.L., and T.P. wrote the paper.
Note added in proof: See Burgess et al. 2020 (pp. 379–391) in this issue for a related work.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12150459.
Communicating editor: J. Birchler
Literature Cited
- Allen G. C., Flores-Vergara M. A., Krasynanski S., Kumar S., and Thompson W. F., 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1: 2320–2325. 10.1038/nprot.2006.384 [DOI] [PubMed] [Google Scholar]
- Anderson S. N., Stitzer M. C., Zhou P., Ross-Ibarra J., Hirsch C. D. et al. , 2019. Dynamic patterns of transcript abundance of transposable element families in maize. G3 (Bethesda) 9: 3673–3682.. 10.1534/g3.119.400431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink R. A., and Nilan R. A., 1952. The relation between light variegated and medium variegated pericarp in maize. Genetics 37: 519–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell T. P., and Dellaporta S. L., 1994. Somatic inactivation and reactivation of Ac associated with changes in cytosine methylation and transposase expression. Genetics 138: 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess D., Li H., Zhao M., Kim S. Y., and Lisch D., 2020. Silencing of Mu elements in maize involves distinct populations of small RNAs and distinct patterns of DNA methylation. Genetics Early online March 30, 2020. 10.1534/genetics.120.303033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel S. E., and Martienssen R. A., 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14: 100–112. 10.1038/nrg3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Greenblatt I. M., and Dellaporta S. L., 1987. Transposition of Ac from the P locus of maize into unreplicated chromosomal sites. Genetics 117: 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D. H., and Timmermans M. C. P., 2010. Small RNAs are on the move. Nature 467: 415–419. 10.1038/nature09351 [DOI] [PubMed] [Google Scholar]
- Conrad L. J., and Brutnell T. P., 2005. Ac-Immobilized, a stable source of Activator transposase that mediates sporophytic and gametophytic excision of Dissociation elements in maize. Genetics 171: 1999–2012. 10.1534/genetics.105.046623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring H. P., and Starlinger P., 1984. Barbara McClintock’s controlling elements: now at the DNA level. Cell 39: 253–259. 10.1016/0092-8674(84)90002-3 [DOI] [PubMed] [Google Scholar]
- Duan C.-G., Zhang H., Tang K., Zhu X., Qian W. et al. , 2015. Specific but interdependent functions for Arabidopsis AGO4 and AGO6 in RNA-directed DNA methylation. EMBO J. 34: 581–592. 10.15252/embj.201489453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Schott G., Himber C., Meyer D., Takeda A. et al. , 2010. Small RNA duplexes function as mobile silencing signals between plant cells. Science 328: 912–916. 10.1126/science.1185880 [DOI] [PubMed] [Google Scholar]
- Fußwinkel H., Schein S., Courage U., Starlinger P., and Kunze R., 1991. Detection and abundance of mRNA and protein encoded by transposable element Activator (Ac) in maize. Mol. Gen. Genet. 225: 186–192. 10.1007/BF00269846 [DOI] [PubMed] [Google Scholar]
- Gray Y. H. M., 2000. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 16: 461–468. 10.1016/S0168-9525(00)02104-1 [DOI] [PubMed] [Google Scholar]
- Greenblatt I., and Brink R., 1962. Twin mutations in medium variegated pericarp maize. Genetics 47: 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag J. R., and Pikaard C. S., 2011. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 12: 483–492. 10.1038/nrm3152 [DOI] [PubMed] [Google Scholar]
- Heinlein M., Brattig T., and Kunze R., 1994. In vivo aggregation of maize Activator (Ac) transposase in nuclei of maize endosperm and petunia protoplasts. Plant J. 5: 705–714. 10.1111/j.1365-313X.1994.00705.x [DOI] [PubMed] [Google Scholar]
- Huang J. T., and Dooner H. K., 2008. Macrotransposition and other complex chromosomal restructuring in maize by closely linked transposons in direct orientation. Plant Cell 20: 2019–2032. 10.1105/tpc.108.060582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P., Belzile F., and Dean C., 1997. Inefficient and incorrect processing of the Ac transposase transcript in iae1 and wild-type Arabidopsis thaliana. Plant J. 11: 921–931. 10.1046/j.1365-313X.1997.11050921.x [DOI] [PubMed] [Google Scholar]
- Kermicle J. L., 1980. Probing the component structure of a maize gene with transposable elements. Science 208: 1457–1459. 10.1126/science.208.4451.1457 [DOI] [PubMed] [Google Scholar]
- Kidwell M. G., 1985. Hybrid dysgenesis in Drosophila melanogaster: nature and inheritance of P element regulation. Genetics 111: 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze R., Starlinger P., and Schwartz D., 1988. DNA methylation of the maize transposable element Ac interferes with its transcription. Mol. Gen. Genet. 214: 325–327. 10.1007/BF00337730 [DOI] [Google Scholar]
- Kunze R., Kuhn S., Jones J. D. G., and Scofield S. R., 1995. Somatic and germinal activities of maize Activator (Ac) transposase mutants in transgenic tobacco. Plant J. 8: 45–54. 10.1046/j.1365-313X.1995.08010045.x [DOI] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J. A., and Jacobsen S. E., 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechelt C., Peterson T., Laird A., Chen J., Dellaporta S. L. et al. , 1989. Isolation and molecular analysis of the maize P locus. Mol. Gen. Genet. 219: 225–234. 10.1007/BF00261181 [DOI] [PubMed] [Google Scholar]
- Li H., Freeling M., and Lisch D., 2010. Epigenetic reprogramming during vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 107: 22184–22189. 10.1073/pnas.1016884108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Gent J. I., Zynda G., Song J., Makarevitch I. et al. , 2015a RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc. Natl. Acad. Sci. USA 112: 14728–14733. 10.1073/pnas.1514680112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Vandivier L. E., Tu B., Gao L., and Won S. Y., 2015b Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 25: 235–245. 10.1101/gr.182238.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D., 2002. Mutator transposons. Trends Plant Sci. 7: 498–504. https://www.ncbi.nlm.nih.gov/pubmed/12417150 [DOI] [PubMed] [Google Scholar]
- Marí-Ordóñez A., Marchais A., Etcheverry M., Martin A., Colot V. et al. , 2013. Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 45: 1029–1039. 10.1038/ng.2703 [DOI] [PubMed] [Google Scholar]
- Martínez G., and Slotkin R. K., 2012. Developmental relaxation of transposable element silencing in plants: functional or byproduct? Curr. Opin. Plant Biol. 15: 496–502. 10.1016/j.pbi.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Matsumoto T., Wu J., Kanamori H., Katayose Y., Fujisawa M. et al. , 2005. The map-based sequence of the rice genome. Nature 436: 793–800. 10.1038/nature03895 [DOI] [PubMed] [Google Scholar]
- Matzke M. A., and Mosher R. A., 2014. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408 (erratum: Nat. Rev. Genet. 15: 570). 10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- Matzke M. A., Kanno T., and Matzke A. J. M., 2015. RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 66: 243–267. 10.1146/annurev-arplant-043014-114633 [DOI] [PubMed] [Google Scholar]
- McClintock B., 1948. Mutable loci in maize. Carnegie Inst. Washingt. Year B. 47: 155–169. [Google Scholar]
- McClintock B., 1949. Mutable loci in maize: the mechanism of transposition of the Ds Locus. Carnegie Inst. Washingt. Year B. 48: 142–154. [Google Scholar]
- McClintock B., 1950. Mutable loci in maize: mode of detection of transpositions of Ds. Carnegie Inst. Washingt. Year B 49: 157–167. [Google Scholar]
- McClintock B., 1951. Chromosome organization and genic expression. Cold Spring Harb. Symp. Quant. Biol. 16: 13–47. 10.1101/SQB.1951.016.01.004 [DOI] [PubMed] [Google Scholar]
- McCue A. D., Panda K., Nuthikattu S., Choudury S. G., Thomas E. N. et al. , 2014. ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 34: 20–35. 10.15252/embj.201489499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A., Melnyk C. W., Bassett A., Hardcastle T. J., Dunn R. et al. , 2010. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875. 10.1126/science.1187959 [DOI] [PubMed] [Google Scholar]
- Nuthikattu S., McCue A. D., Panda K., Fultz D., DeFraia C. et al. , 2013. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21–22 nucleotide small interfering RNAs. Plant Physiol. 162: 116–131. 10.1104/pp.113.216481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda K., Ji L., Neumann D. A., Daron J., Schmitz R. J. et al. , 2016. Full-length autonomous transposable elements are preferentially targeted by expression-dependent forms of RNA-directed DNA methylation. Genome Biol. 17: 170 10.1186/s13059-016-1032-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J. et al. , 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. 10.1038/nature07723 [DOI] [PubMed] [Google Scholar]
- Peacock W. J., Dennis E. S., Gerlach W. L., Sachs M. M., and Schwartz D., 1984. Insertion and excision of Ds controlling elements in maize. Cold Spring Harb. Symp. Quant. Biol. 49: 347–354. 10.1101/SQB.1984.049.01.041 [DOI] [PubMed] [Google Scholar]
- Pikaard C. S., 2013. Methylating the DNA of the most repressed: special access required. Mol. Cell 49: 1021–1022. 10.1016/j.molcel.2013.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlman R. F., Fedoroff N. V., and Messing J., 1984. The nucleotide sequence of the maize controlling element Activator. Cell 37: 635–643. 10.1016/0092-8674(84)90395-7 [DOI] [PubMed] [Google Scholar]
- Pontier D., Picart C., Roudier F., Garcia D., Lahmy S. et al. , 2012. NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin-based pathway in Arabidopsis. Mol. Cell 48: 121–132. 10.1016/j.molcel.2012.07.027 [DOI] [PubMed] [Google Scholar]
- Regulski M., Lu Z., Kendall J., Donoghue M. T., Reinders J. et al. , 2013. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23: 1651–1662. 10.1101/gr.153510.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S. et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros F., and Kunze R., 2001. Regulation of Activator/Dissociation transposition by replication and DNA methylation. Genetics 157: 1723–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J.,F. E. Fritsch., and T. Maniatis, 1989 Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. [Google Scholar]
- Sanmiguel P., and Vitte C., 2009. The LTR-retrotransposons of maize, pp. 307–327 in Maize Handbook - Volume II: Genetics and Genomics, edited by Bennetzen J. L., Hake S. C.. Springer-Verlag, New York. [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., et al. , 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- Slotkin R. K., Freeling M., and Lisch D., 2003. Mu killer causes the heritable inactivation of the Mutator family of transposable elements in Zea mays. Genetics 165: 781–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R. K., Freeling M., and Lisch D., 2005. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat. Genet. 37: 641–644. 10.1038/ng1576 [DOI] [PubMed] [Google Scholar]
- Stroud H., Greenberg M. V. C., Feng S., Bernatavichute Y. V., and Jacobsen S. E., 2013. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364 [corrigenda: Cell 161: 1697–1698 (2015)]. 10.1016/j.cell.2012.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Do T., Du J., Zhong X., Feng S. et al. , 2014. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21: 64–72. 10.1038/nsmb.2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov A. P., SanMiguel P. J., Nakajima Y., Gorenstein N. M., Bennetzen J. L. et al. , 1999. Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96: 7409–7414. 10.1073/pnas.96.13.7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E., Duvick J., Schares J. P., Ahern K. R., Deewatthanawong P. et al. , 2010. Genome-wide distribution of transposed dissociation elements in maize. Plant Cell 22: 1667–1685. 10.1105/tpc.109.073452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Elling A. A., Li X., Li N., Peng Z. et al. , 2009. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069. 10.1105/tpc.109.065714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Yu C., Zuo T., Zhang J., Weber D. F. et al. , 2015. Alternative transposition generates new chimeric genes and segmental duplications at the maize p1 locus. Genetics 201: 925–935. 10.1534/genetics.115.178210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil C. F., and Wessler S. R., 1993. Molecular evidence that chromosome breakage by Ds elements is caused by aberrant transposition. Plant Cell 5: 515–522. 10.1105/tpc.5.5.515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West P. T., Li Q., Ji L., Eichten S. R., Song J. et al. , 2014. Genomic distribution of H3K9me2 and DNA methylation in a maize genome. PLoS One 9: e105267 10.1371/journal.pone.0105267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M. Y., Hsieh P. H., Coleman-Derr D., Eshed-Williams L. et al. , 2013. The arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. 10.1016/j.cell.2013.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., and Peterson T., 1999. Genome rearrangements by nonlinear transposons in maize. Genetics 153: 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., and Peterson T., 2004. Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics 167: 1929–1937. 10.1534/genetics.103.026229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang F., and Peterson T., 2006. Transposition of reversed Ac element ends generates novel chimeric genes in maize. PLoS Genet. 2: e164 10.1371/journal.pgen.0020164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yu C., Pulletikurti V., Lamb J., Danilova T. et al. , 2009. Alternative Ac/Ds transposition induces major chromosomal rearrangements in maize. Genes Dev. 23: 755–765. 10.1101/gad.1776909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zuo T., and Peterson T., 2013. Generation of tandem direct duplications by reversed-ends transposition of maize Ac elements. PLoS Genet. 9: e1003691 10.1371/journal.pgen.1003691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zuo T., Wang D., and Peterson T., 2014. Transposition-mediated DNA re-replication in maize. eLife 3: e03724 10.7554/eLife.03724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T., Zhang J., Lithio A., Dash S., Weber D. F. et al. , 2016. Genes and small RNA transcripts exhibit dosage-dependent expression pattern in maize copy-number alterations. Genetics 203: 1133–1147. 10.1534/genetics.116.188235 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The small RNA high-throughput sequencing data analyzed here is deposited in the NCBI under accession number SRP062285. Maize stocks are available upon request. All data necessary for confirming the conclusions of the article were presented within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12150459.