Abstract
Transposable elements (TEs) are a ubiquitous feature of plant genomes. Because of the threat they post to genome integrity, most TEs are epigenetically silenced. However, even closely related plant species often have dramatically different populations of TEs, suggesting periodic rounds of activity and silencing. Here, we show that the process of de novo methylation of an active element in maize involves two distinct pathways, one of which is directly implicated in causing epigenetic silencing and one of which is the result of that silencing. Epigenetic changes involve changes in gene expression that can be heritably transmitted to daughter cells in the absence of changes in DNA sequence. Epigenetics has been implicated in phenomena as diverse as development, stress response, and carcinogenesis. A significant challenge facing those interested in investigating epigenetic phenomena is determining causal relationships between DNA methylation, specific classes of small RNAs, and associated changes in gene expression. Because they are the primary targets of epigenetic silencing in plants and, when active, are often targeted for de novo silencing, TEs represent a valuable source of information about these relationships. We use a naturally occurring system in which a single TE can be heritably silenced by a single derivative of that TE. By using this system it is possible to unravel causal relationships between different size classes of small RNAs, patterns of DNA methylation, and heritable silencing. Here, we show that the long terminal inverted repeats within Zea mays MuDR transposons are targeted by distinct classes of small RNAs during epigenetic silencing that are dependent on distinct silencing pathways, only one of which is associated with transcriptional silencing of the transposon. Further, these small RNAs target distinct regions of the terminal inverted repeats, resulting in different patterns of cytosine methylation with different functional consequences with respect to epigenetic silencing and the heritability of that silencing.
Keywords: maize, Mutator transposon, epigenetic, methylation
PLANT genomes are host to large numbers of potentially deleterious endogenous mutagens known as transposable elements (TEs). Because of the activity of a sophisticated regulatory system, the vast majority of TEs are epigenetically silenced (Slotkin and Martienssen 2007; Lisch 2009; Law and Jacobsen 2010; Bucher et al. 2012; Sigman and Slotkin 2016). This silencing is associated with DNA methylation and modification of histones, and can be propagated over many generations (Lisch 2009; Saze and Kakutani 2011).
Because most transposons are silenced most of the time, much of what we know about TE silencing involves maintenance, rather than initiation of silencing. However, recent work suggests that aberrant RNAs can trigger silencing of otherwise active TEs via a pathway that involves the production of trans-acting 21–22 nt small RNAs via the activity components of both the post-transcriptional gene silencing and the transcriptional gene silencing pathways (Li et al. 2010; Marí-Ordóñez et al. 2013; Nuthikattu et al. 2013; Fultz et al. 2015; Cuerda-Gil and Slotkin 2016). The available data suggests that Polymerase II (Pol II) transcripts from active TEs are recognized by small RNAs that then act as triggers for RDR6/SGS3-mediated production of double-stranded RNAs (dsRNAs) using RNA-directed RNA Polymerase 6 (RDR6) in conjunction with Suppressor of Gene Silencing 3 (SGS3). The resulting dsRNA is then processed into 21–22 nt trans-acting small RNAs by Dicer like 2 (DCL2) or Dicer like 4 (DCL4). These small RNAs are then incorporated into a complex that includes Argonaute 6 (AGO6), which is then competent to trigger de novo and heritable transcriptional gene silencing using Pol V transcript arising from the active elements as a scaffold (Fultz et al. 2015; McCue et al. 2015; Cuerda-Gil and Slotkin 2016; Fultz and Slotkin 2017). The initial triggers for silencing are not always well understood, but it appears that they may be small RNAs derived from the Pol II transcript itself, or from an unlinked aberrant version of the TE (Slotkin et al. 2005; Marí-Ordóñez et al. 2013; Creasey et al. 2014).
Following the initiation of silencing via trans-acting small RNAs, TE silencing can be maintained by stable propagation of CG and CHG methylation, as well as reinforcement via 24 nt small RNAs derived from Pol IV transcripts that are tethered to the target gene via a Pol V transcript, which is required for de novo methylation in the CHH context (Matzke and Mosher 2014; Holoch and Moazed 2015). Data from maize shoot apical meristems, meiocytes, endosperm, and nurse cells of gametophytes suggest that silencing of TEs is enhanced in cells lineages that are, or are likely to be, inherited in the next generation because of the production of small RNA in cells and tissues that are adjacent to, but distinct from these cells (Martínez and Slotkin 2012). This results in a recapitulation of the initial silencing event, in which expression of otherwise inactive elements triggers production of trans-acting small RNAs, which are then thought to be transported to the germinal lineage (Slotkin et al. 2009; Li et al. 2010; Creasey et al. 2014). The net effect of this process is that active TEs can be recognized and silenced by previously silenced versions of the element, and potentially active TEs can be kept in a stably silenced state over long periods of time.
The initiation and maintenance of TE silencing is particularly well characterized in the Mutator system in maize, primarily because MuDR, the autonomous regulator of the system, can be heritably silenced by a trans-acting locus called Mu killer (Muk), a naturally occurring derivative of MuDR that expresses a long hairpin transcript (Slotkin et al. 2003; Slotkin et al. 2005). This transposon system is composed of several related classes of cut-and-paste elements, all of which share similar, ∼200 bp terminal inverted repeats (TIRs), but each of which carries unique internal sequences (Lisch 2002). The system is regulated by autonomous MuDR elements, which carry two genes: mudrA, which encodes the putative transposase MURA; and mudrB, which encodes the helper protein MURB (Figure 1) (Hershberger et al. 1995; Lisch et al. 1999). Expression of mudrA and mudrB is convergent, with transcripts from each gene originating from within the 220 bp TIRs adjacent to each gene and extending toward the middle of the element (Figure 1). In our lines, activity of MuDR is monitored by a reporter element, a nonautonomous Mu1 element located in the a1-mum2 allele of the A1 gene, whose function is required for color expression in both the plant and the seed (Chomet et al. 1991). In the seed, Mu1 excises from a1-mum2 somatically in the presence of a functional MuDR element, giving rise to spotted kernels. In the absence of functional MuDR elements, the kernels are uniformly pale. All of the plants used in these experiments are homozygous for a1-mum2.
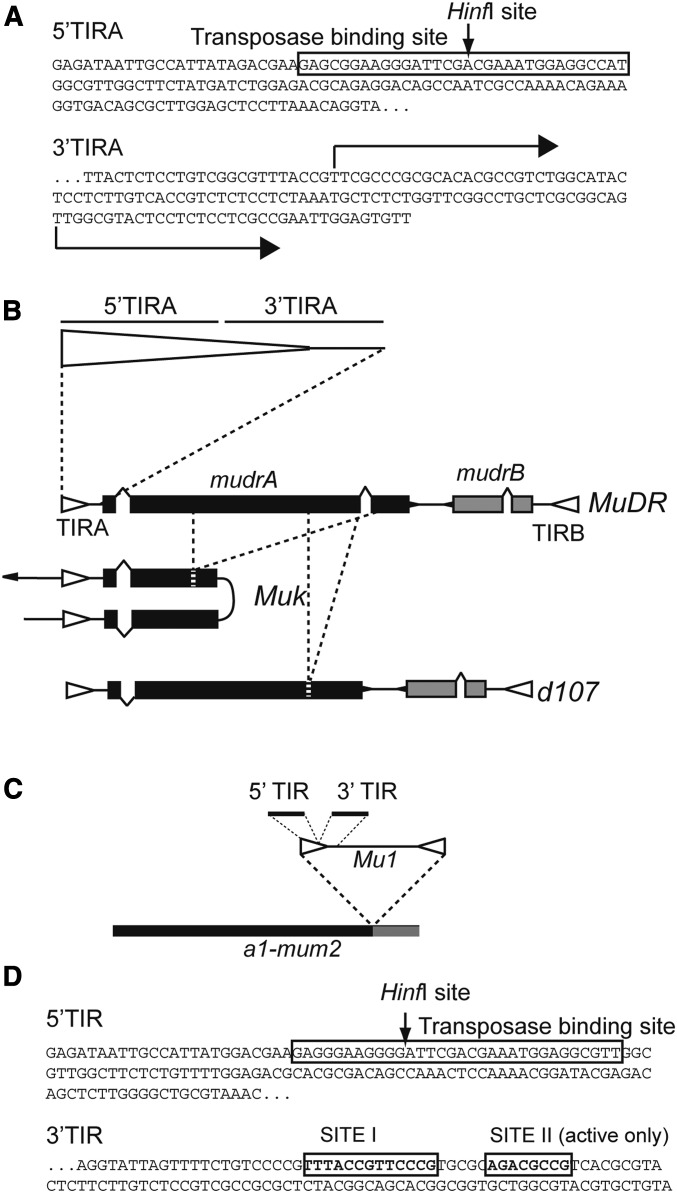
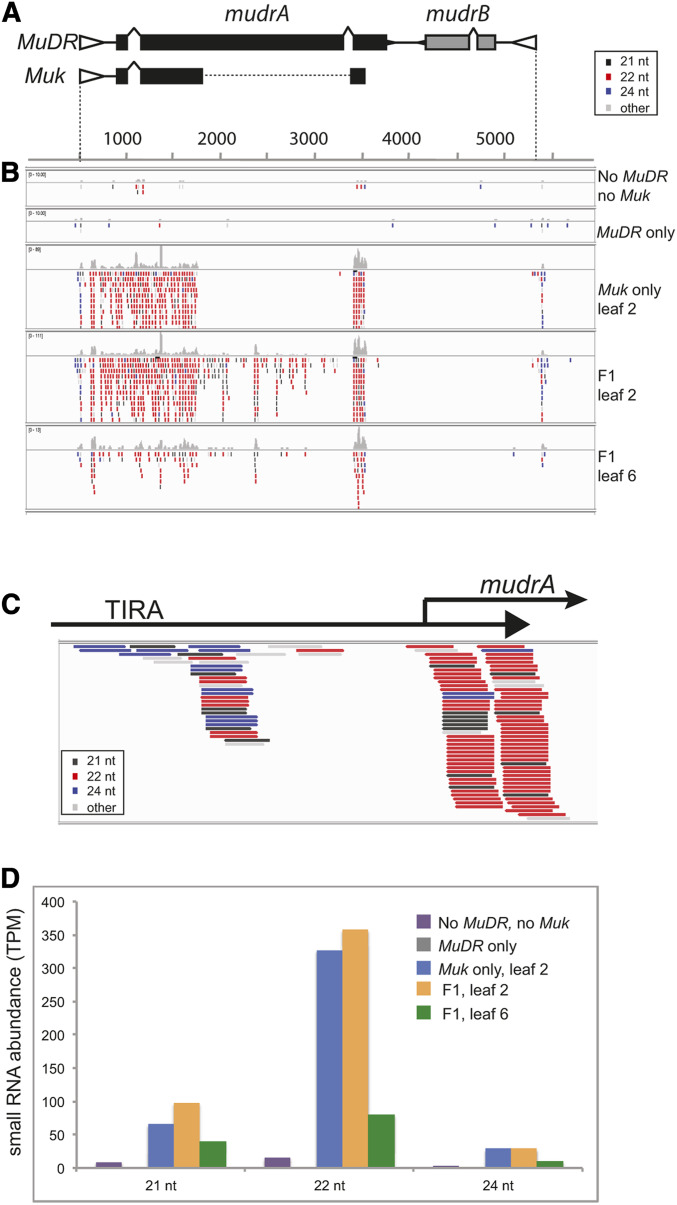
Figure 1.
The structures of the Mu elements examined. (A) The DNA sequence of the terminal inverted repeat (TIR) adjacent to the mudrA gene in MuDR (TIRA). 5′TIRA refers to the first 144 bp of the TIR and includes the known binding site for the MuDR transposase. The Hinf I site has long been diagnostic for methylation in Mu TIRs. The adjacent 3′TIRA includes the last 75 bp of the TIR along with 56 bp of internal sequences corresponding to a portion of the mudrA 5′ UTR, indicated by arrows. This region includes both of the alternative transcriptional start sites for mudrA. (B) A diagram of the structure of MuDR, Muk, and d107. TIRs are indicated as open triangles and exons as shaded boxes. The regions missing in Muk and d107 are indicated by dashed lines. (C) The structure of the Mu1 insertion at a1-mum2. (D) The sequence of the Mu1 TIR, divided into its 5′ and 3′ parts. The transposase binding site is as indicated, as are additional protected sites within the 3′ portion of the Mu1 TIR.
Also present in this and likely all maize lines are MuDR derivatives called hMuDR elements (Rudenko and Walbot 2001). Although nearly identical to portions of MuDR, none of these elements are intact and they do not appear to contribute to Mutator activity, either positively or negatively (Lisch and Jiang 2008). They are, however, a source of nuclear localized transcript and are thus the likely source of the abundant MuDR-similar small RNAs that have been observed in immature ears and embryos (Rudenko et al. 2003; Woodhouse et al. 2006a; Nobuta et al. 2008). Finally, the reference maize genome contains a limited number of nonautonomous Mu elements with high homology to known active Mu elements within their TIRs (Lisch 2015). The available data suggests that both hMuDRs and nonautonomous elements are targeted by 24–26 nt small RNAs that are dependent on the RNA-directed DNA methylation pathway (Nobuta et al. 2008; Hale et al. 2009).
When Mutator activity is lost due to silencing or genetic segregation of autonomous MuDR elements, methyl-sensitive sites within the TIRs of nonautonomous Mu transposons such as Mu1 at a1-mum2 become methylated (Chandler and Walbot 1986; Chomet et al. 1991). This methylation is fully reversible when a source of transposase is added, and invariably occurs when transposase is lost. Indeed, this can occur in somatic sectors in developing plants when spontaneous deletions within MuDR elements occur, suggesting that the RNA-directed DNA methylation pathway is competent to trigger de novo methylation of Mu elements during somatic development (Chomet et al. 1991; Lisch and Jiang 2008). Recent work has demonstrated that this default methylation requires a component of the RNA-directed DNA methylation pathway, MOP1, a protein that is homologous to Arabidopsis RNA-dependent RNA Pol II (Lisch et al. 2002; Alleman et al. 2006; Woodhouse et al. 2006a). This has led to the conclusion that the TIRs of nonautonomous elements are subject to a default methylation pathway that operates in the absence of the transposase, but that can be blocked and even reversed by the presence of the transposase (Hershberger et al. 1995; Lisch et al. 1995; Benito and Walbot 1997).
Active MuDR elements can be heritably and reliably silenced by genetically combining them with Muk, whose transcript is identical to a portion of the mudrA gene, as well as its associated TIR (TIRA) (Figure 1). In active MuDR elements, TIRA is devoid of DNA methylation. In contrast, in plants carrying both Muk and MuDR, TIRA sequences are densely methylated in all three sequence contexts, CG, CHG, and CHH (where H represents any nucleotide but guanine) (Li et al. 2010). This methylation and the associated silencing of the MuDR element can be heritably propagated over many generations, even in the absence of Muk (Slotkin et al. 2003; Lisch 2013).
Interestingly, we have found that silencing of MuDR by Muk is sensitive to changes in components of the trans-acting silencing (tasiRNA) pathway. Specifically, we found that a transient loss of expression of Leafbladeless1 (lbl1), the maize homolog of SGS3, in leaves that emerge during the transition from juvenile to adult growth is associated with an alleviation of transcriptional silencing of the mudrA gene (Li et al. 2010). Since SGS3 works in conjunction with RDR6 to produce secondary dsRNAs (Kumakura et al. 2009), this suggests that silencing of MuDR elements in leaves requires production of secondary dsRNAs triggered by small RNAs produced by the hairpin Muk transcript.
Here, we show that cytosine methylation of different regions within the MuDR TIR has distinct causes and consequences, and corresponds to distinct populations of small RNAs derived from the Muk hairpin, the mudrA transcript, and other Mu elements in the maize genome. In addition, we demonstrate that although active MuDR elements can reverse methylation at one end of the TIR of a silenced MuDR element, they do not heritably reactivate that silenced element, nor does the silenced element inactivate the active element. Finally, we demonstrate that the previously described transient relaxation of Muk-induced silencing of MuDR during vegetative change (Li et al. 2010) is associated with a dramatic reduction in small RNAs targeting that element. Based on these observations, we present a model wherein processed mudrA transcript and the resulting 3′ TIRA methylation (directed by the small RNA products of the processed transcripts) are the cause of the transcriptional silencing, and 5′ TIRA methylation is a consequence of that silencing.
Materials and Methods
Plant materials
The minimal Mutator line consists of one fully active MuDR element at position 1 (p1) on chromosome 3L, MuDR(p1) (Chomet et al. 1991). Muk is a derivative version of MuDR, as described previously (Figure 1) (Slotkin et al. 2005). Activity is monitored in seeds via excisions of a nonautonomous Mu1 element inserted into the a1-mum2 allele of the A1 color gene (Chomet et al. 1991). All plants used in these experiments are homozygous for a1-mum2. “Test crosses” refers to crosses to plants that are homozygous for a1-mum2 but that lack MuDR or Muk.
The crosses used to generate the genotypes examined are depicted in Figure 3A and Figure 4A. To construct lines containing silenced MuDR(p1) elements with and without active elements, plants carrying MuDR(p1) were crossed to plants heterozygous for Muk. Progeny plants that were heterozygous for Muk and that carried MuDR(p1) were then crossed to plants that carried MuDR at a second position, MuDR(p5). Progeny that lacked Muk and that carried silenced MuDR(p1), designated MuDR(p1)*, with or without MuDR(p5), were then compared. Plants carrying MuDR(p1)* and MuDR(p5) were then test crossed to a1-mum2 testers, and progeny plants carrying only MuDR(p1)* were examined. Tests of the ability of an active element to activate a silenced element were performed by crossing a plant carrying an active element at a third position, MuDR(p4), to a plant carrying a previously silenced MuDR(p1)* element and then test crossing plants carrying both MuDR(p1)* and MuDR(p4). Progeny of this cross were then separated into spotted (exhibiting somatic excisions of the reporter element) and pale kernels, genotyped for MuDR(p1)* and MuDR(p4), and then test crossed. Genotyping for MuDR(p1)* employed primers RLTIR2 and Ex1. Genotyping for MuDR(p4) employed primers RLTIR2 and p4flankB. Genotyping for MuDR(p5) employed primers RLTIR2 and p5flankB (all primers used in experiments described in this manuscript are supplied in Supplemental Material, Table S1).
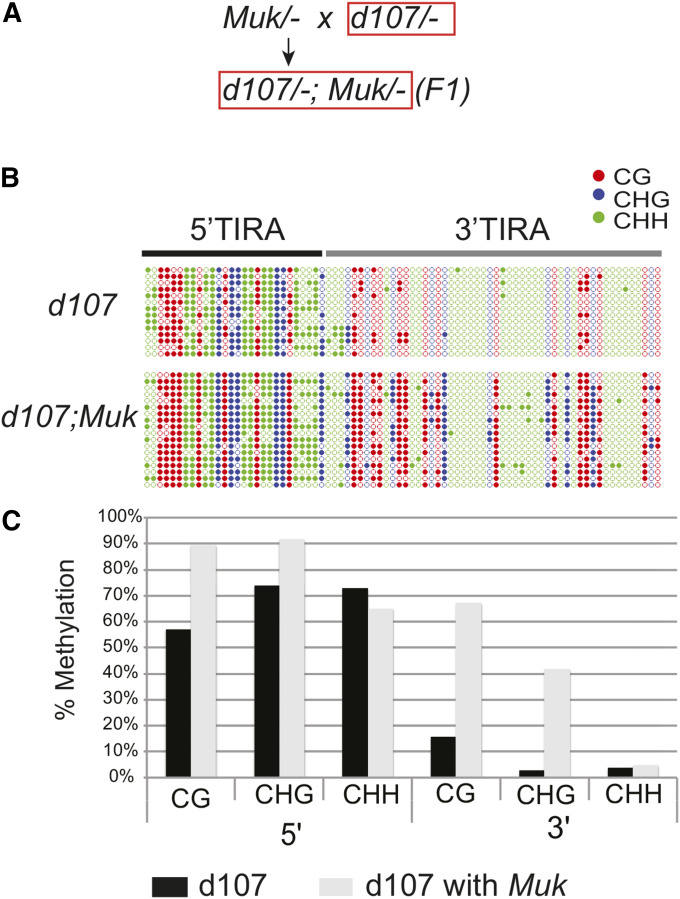
Figure 3.
Depiction of patterns of methylation in TIRA of the deletion derivative, d107. (A) The cross used to generate the individuals examined (one of each genotype) (red box). d107 represents MuDR(d107) and Muk represents Mu killer. (B) A graphic representation of the patterns of methylation at d107 TIRA. Samples include a transcriptionally active d107 and d107 in the presence of Muk. Cytosines in different sequence contexts (CG, CHG, and CHH) are as indicated. (C) Percent methylation of cytosines in each sequence context in the same samples depicted graphically, with the results separated into 5′TIRA and 3′TIRA.
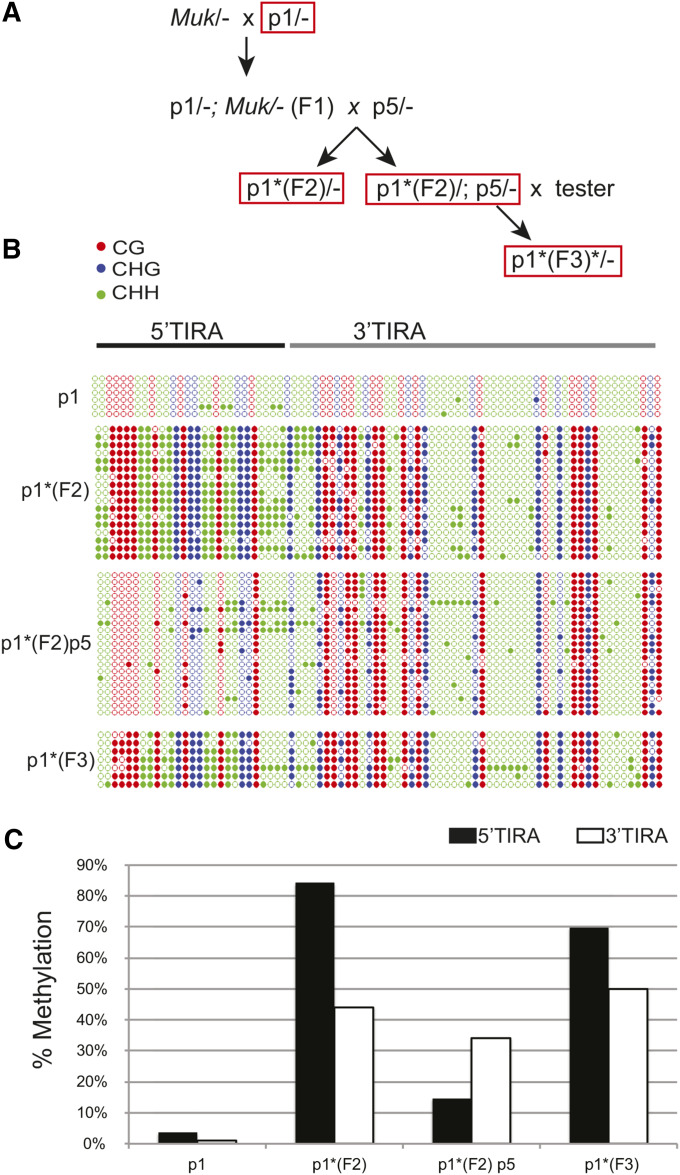
Figure 4.
The effect of an active element on methylation of a silenced element. (A) The crosses used to generate the samples subject to bisulfite sequencing (red box) (B) A graphic representation of patterns of cytosine methylation within TIRA in plants of the indicated genotypes. Cytosines in different sequence contexts (CG, CHG, and CHH) are as indicated. (C) Percent methylation of cytosines within 5′ TIRA and 3′TIRA in plants of the indicated genotype. F2, MuDR(p1)* in the generation after exposure to Muk after Muk has been segregated away; F3, MuDR(p1)* in the following generation after MuDR(p5) has segregated away; p1, MuDR(p1); p1*, silenced MuDR(p1); p5, MuDR(p5).
Tissue sampling
All plants used in bisulfite sequencing experiments were genotyped individually. Immature ears, ∼10 cm long, were collected from each individual plant. To check for variation in patterns of methylation, fully mature leaves (the third leaf from the top of each plant) were also examined (Figure S1). For small RNA analysis, the distal 10 cm of emerging leaves two and six were collected as previously described (Li et al. 2010).
Genomic bisulfite sequencing
Genomic DNA was isolated as previously described (Lisch et al. 1995). Two micrograms of genomic DNA from the appropriate genotype were digested with restriction enzymes (XhoI and BamHI) that cut just outside of the region of interest. Bisulfite conversion was performed using an EpiTect Bisulfite kit (Qiagen). PCR fragments from TIRA were amplified using p1bis2f and TIRAbis2R, and reamplified using TIRAbis2R and the nested primer TIRAmF6 or p1bis7Fmed. In addition, all samples were also amplified using a different set of primers (TIRAMF6 and Autr1R, followed by reamplification with Autr1R and the nested primers TIRAF1 or p1bis7Fmed). The sequences from each amplification were then compared. This was done to ensure that biases had not been introduced due to the selection of a particular pair of primers. No substantive differences were detected and thus clones from each set of primers were combined (Figure S1). In addition, for many of the plants examined, both fully mature leaves (the third from the top leaf) and immature ears were examined. The results in all cases (different primers or different tissues) were substantially equivalent. To ensure that duplicate clones resulting from amplification did not skew the analysis, sequences with zero mismatches were only counted once for each sample. The resulting sequences were analyzed using kismeth (http://katahdin.mssm.edu/kismeth/revpage.pl) (Gruntman et al. 2008).
For Mu1 methylation analysis, a similar strategy was employed, but the initial use of restriction enzymes was not necessary. Following bisulfite conversion, the DNA was amplified using primers Mu1bis1 (located in the Mu1 element) and either a1mum2bis1 or a1mum2bis2 (located in the a1-mum2 allele, flanking the Mu1 insertion).
Small RNA sequencing
Plants were grown in a greenhouse with a 12 hr light/dark cycle. Young leaf tissue for small RNA samples was obtained from two closely related families segregating for MuDR and Muk. Each sample class contained at least three pooled individuals, each one of which was genotyped. RNA was independently extracted from two separate sets of individuals on different days, and each set is referred to as an experimental replicate, the results of which are presented separately because of large overall differences in relative abundance of normalized MuDR-identical small RNAs. The classes included MuDR by itself, Muk by itself, MuDR with Muk, and neither MuDR nor Muk. The small RNA extraction and enrichment protocol was adapted from Dalmay et al. (2000). Total RNA was extracted using an SDS-based extraction solution and precipitated using ethanol. The pellet was dissolved in water, heated to 65° for 5 min, and then placed on ice. Polyethylene glycol (molecular weight 8000; Sigma, St. Louis, MO) was added to a final concentration of 5% and NaCl to a final concentration of 0.5 M. After an hour’s incubation on ice, the RNA was centrifuged at 10,000 × g for 10 min. Three volumes of ethanol were added to the supernatant, and the RNA was precipitated at −20° for at least 2 hr. The low-molecular-weight RNAs were pelleted by centrifugation for 10 min at 10,000 × g. Small RNAs were detected in PAGE gel and purified by ZR small-RNA PAGE Recovery Kit. Small RNA library was prepared by ScriptMiner Small RNA-Seq Library Preparation Kit for Illumina sequencing.
Small RNA data analysis
The small RNA sequencing data from different libraries was trimmed and filtered for low-quality reads, adapter sequences, and reads matching structural noncoding RNAs (total, ribosomal, small nuclear, and small nucleolar RNAs) collected from Rfam (http://rfam.xfam.org). The kept reads with a length of 18–26 nt were mapped to MuDR(p1) and Muk, and their flanking 500 bp upstream and downstream sequences using Bowtie only, allowing perfect matches (Langmead et al. 2009). Small RNA abundance was normalized to reads per million. The data were viewed using Integrative Genomics Viewer (Robinson et al. 2011). Small RNAs for the mop1 and lbl1 mutants were downloaded from previous studies (Nobuta et al. 2008; Dotto et al. 2014), trimmed, and mapped to MuDR sequences.
Reagent and data availability
All small RNA data generated for this work is freely and publicly available. The small RNA sequencing data have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE103833 Is correct. Maize lines used for these analyses are also freely available for noncommercial purposes. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12009360.
Results
The absence of transposase results in default methylation of cytosines in all sequence contexts at TIRs of nonautonomous elements
Data concerning methylation of Mu elements has been largely restricted to methyl-sensitive restriction sites. We wanted to understand the distribution and nature of this methylation more fully, so we examined methylation of the TIR of the nonautonomous Mu1 element inserted into the A1 gene in the a1-mum2 allele (O’Reilly et al. 1985; Chomet et al. 1991), using bisulfite sequencing.
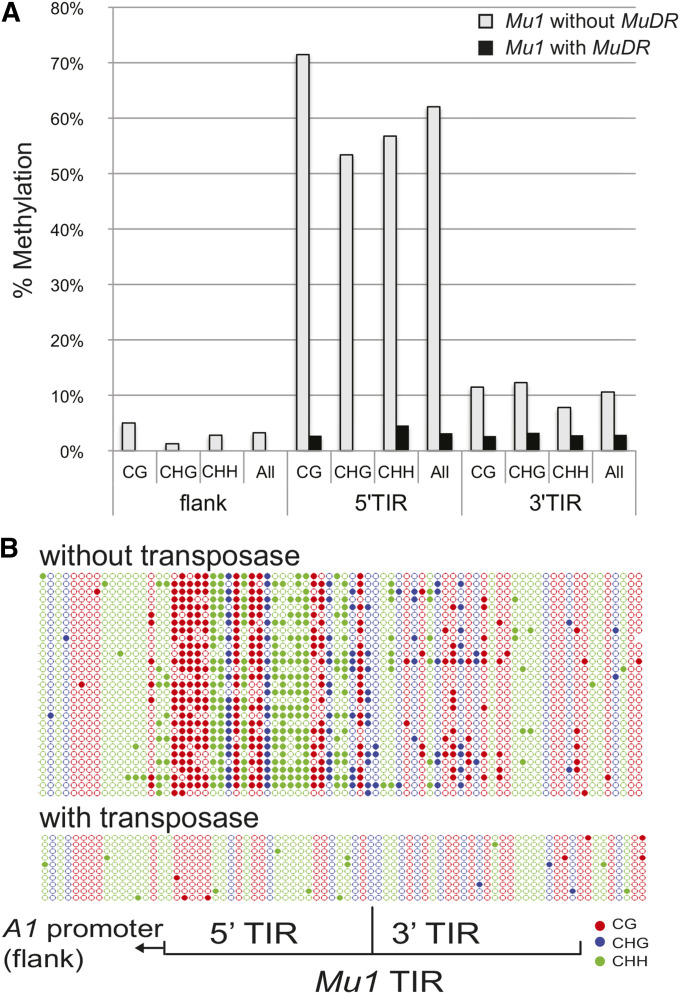
The results were entirely consistent with previous observations. In the absence of transposase, the Mu1 TIR is extensively methylated in all three sequence contexts (CG, CHG, and CHH), although it is interesting to note that methylation of this TIR is much higher in the 5′ end of the TIR (62% methylated cytosines in the first 110 nt) than the 3′ end of the TIR (11% methylated cytosines in the second 110 nt), only marginally more than the 3% observed in the presence of the transposase (Figure 2). The 5′ end of the TIR contains sequences known to bind the MURA protein (Benito and Walbot 1997); the 3′ end of the TIR has two binding sites for unknown proteins that were previously identified (Figure 1D) (Zhao and Sundaresan 1991). Methylation was restricted to the Mu1 TIR and did not extend into adjacent A1 promoter sequences. In plants carrying an active MuDR element, nearly all of the Mu1 methylation was lost, indicating that the presence of the transposase is sufficient to remove that methylation.
Figure 2.
Methylation of the nonautonomous Mu1 element in the presence and absence of an active MuDR element. (A) Percent methylated cytosines in sequences flanking the Mu1 insertion, as well as the 5′ and 3′ portions of the Mu1 TIR. Cytosines are classified by sequence context, with “H” meaning any nucleotide except guanine. (B) A graphic depiction of the same patterns of cytosine methylation. For this and all subsequent depictions of cytosine methylation, filled circles indicate methylated cytosines in the CG (red), CHG (blue), and CHH (green) sequence contexts. Open circles represent unmethylated cytosines. Each line represents one clone from a PCR amplification of a bisulfite-treated DNA sample.
Because the Mu1 TIR is not identical to that of MuDR (roughly 83% identity over 200 bp and 88% identity within the first 100 bp.), we wanted to examine the effects of the transposase on a nonautonomous element with TIRs that are identical to those of MuDR. Fortunately, we had a direct derivative of MuDR (MuDR-d107, or d107) that is identical to the autonomous element, with the exception of a 700-bp deletion within a conserved portion of the mudrA transposase gene (Lisch 1995; Lisch and Jiang 2008) (Figure 1B). As is the case for other MuDR deletion derivatives, sequence analysis of the deletion in d107 suggests that it arose as a consequence of strand slippage during gap repair following excision of MuDR(p1) (Figure S1) (Hsia and Schnable 1996). This derivative cannot cause excision of the reporter element, nor can it trigger hypomethylation of nonautonomous elements, suggesting that it does not make a functional transposase. However, it is transcriptionally active, producing a full-length mudrB transcript and a polyadenylated but internally deleted mudrA transcript. d107 also has the advantage of being at the same chromosomal location as the originally cloned MuDR element at position 1 on chromosome 3L [MuDR(p1)] (Chomet et al. 1991) and can be efficiently silenced by Muk (Slotkin 2005). Thus, the only difference between d107 and the functional MuDR from which it was derived is the presence of the deletion.
Examination of the TIR of d107 revealed that the default methylation that we observed at Mu1 also occurs within the d107 TIR. As in the case of Mu1, methylation was largely restricted to the 5′ end of the TIR (Figure 3B). With this in mind, we have split the analysis of this TIR into 5′ (5′TIRA) and 3′ (3′TIRA) portions (Figure 1A). 5′TIRA includes the first 144 bp of TIRA. This region of the TIR includes the binding site for the transposase (MURA) (Benito and Walbot 1997) and is the most highly conserved region among Mu elements (Bennetzen 1996). 3′TIRA includes the last 75 bp of the TIR along with 69 bp of internal sequences corresponding to a portion of the mudrA 5′ UTR. This region includes both of the alternative transcriptional start sites for mudrA (Hershberger et al. 1995).
Within transcriptionally active d107, 68% of the cytosines in 5′TIRA were methylated. In contrast, only 7% of the cytosines in 3′TIRA were methylated (Figure 3B; P < 0.0001, Fisher’s exact test). These data indicate that the default methylation observed at Mu1 is also observed at d107. However, because d107 is transcriptionally active, we can conclude that the dense methylation we observe in 5′TIRA in all sequence contexts in this derivative is not sufficient to trigger transcriptional silencing.
To determine the effects of Muk on d107 a plant carrying d107 was crossed to a plant carrying Muk, and the pattern of methylation at TIRA was examined in a progeny plant carrying both d107 and Muk (Figure 3A). In this plant, the level of methylation of 5′TIRA was quite similar to that observed in d107 not exposed to Muk (78% and 69%, respectively; P = 0.2327, Fisher’s exact test). In contrast, exposure to Muk caused extensive CG and CHG methylation of 3′TIRA of d107 (30%), significantly higher than that observed in d107 by itself (7%) (P < 0.0001, Fisher’s exact test). This pattern of methylation is nearly identical to that seen in TIRA of MuDR elements as they are being silenced by Muk in immature ears (Li et al. 2010).
Interestingly, we find that the sequence context of the methylated cytosines is quite different between 5′TIRA and 3′TIRA of d107. In both d107 and F1 d107;Muk plants, the cytosines are densely methylated in all three sequence contexts in 5′TIRA. In contrast, in 3′TIRA, cytosine methylation in the CHH context was uniformly low (4% for d107 and 5% for d107;Muk) (P = 0.6210, Fisher’s exact test), but CG and CHG was considerably enriched in d107;Muk relative to d107 plants in 3′TIRA (67% vs. 16% for CG P < 0.0001, Fisher’s exact test), and 42% vs. 3% for CHG, respectively (P < 0.0001, Fisher’s exact test). Collectively, from the examination of d107, we conclude that methylation of 5′TIRA in all three sequence contexts is independent of transcriptional silencing, and that exposure of transcriptionally active d107 to Muk induces new CG and CHG methylation to 3′TIRA, the region that includes both alternative transcriptional start sites. These data suggest that 5′TIRA and 3′TIRA methylation may involve distinct molecular mechanisms that result in distinct patterns of cytosine methylation.
Active elements remove default methylation within silenced elements but do not heritably reactivate them
Having established the nature of default methylation, and the distinction between default methylation and methylation induced by Muk at d107, we sought to examine the effects of an active element on a previously silenced element. To do this, we crossed a plant carrying MuDR at position 1 [MuDR(p1)] to a plant carrying Muk. Previous analysis had determined that MuDR TIRA is densely methylated in the mature leaves and immature ears of F1 [MuDR(p1)/-;Muk/−] plants (Li et al. 2010). Plants carrying MuDR(p1) and Muk were crossed to plants carrying an active MuDR element at a second chromosomal position [referred to as MuDR(p5)] (Figure 4A) (Singh et al. 2008). A progeny plant carrying MuDR(p1) that had been silenced in the previous generation but that no longer carried Muk [MuDR(p1)*] was then compared to a sibling carrying both MuDR(p1)* and MuDR(p5) to examine the heritability of silencing and the effects of an active element on a silenced element. Consistent with the fact that Muk induces heritable silencing of MuDR elements, MuDR(p1)* by itself was extensively methylated. Overall, 5′TIRA of MuDR(p1)* had 81% methylated cytosines and 3′TIRA had 48%, similar to the levels and distribution of methylation in F1 [MuDR(p1)/-;Muk/−] plants (Li et al. 2010). These data confirm our previous observation that methylation at 5′TIRA established due to the presence of Muk is maintained in subsequent generations in its absence (Li et al. 2010). Analysis of a sibling plant that carried both MuDR(p1)* and MuDR(p5) revealed extensive changes in the pattern of methylation at MuDR(p1)* TIRA. In the 5′TIRA of these plants, only 15% of cytosines were methylated, suggesting that the methylation established in this region due to the activity of Muk was largely lost in a manner similar to what is observed at nonautonomous elements when exposed to the transposase. In contrast, within 3′TIRA, 34% of the cytosines remained methylated in this region, somewhat less than the 48% methylation observed in 3′TIRA in siblings that carried only MuDR(p1)* and the F1 MuDR(p1);Muk parent in the previous generation (44%) (P = 0.0032, Fisher’s exact test).
Having established that an active element can reverse at least some of the methylation associated with Muk-induced silencing of MuDR(p1), we wanted to determine whether or not this has a heritable effect on the silenced element. To do this, a plant carrying MuDR(p1)* by itself and a sibling carrying both MuDR(p1)* and MuDR(p5) were test crossed to a plant that lacked both MuDR and Muk, referred to as an a1-mum2 tester (Figure 4A). When crossed to the a1-mum2 tester, the plant carrying only MuDR(p1)* gave rise to an ear where all of its kernels were uniformly pale. The plant carrying both MuDR(p5) and MuDR(p1)* yielded an ear that segregated for a single active element (44 spotted to 41 pale kernels), suggesting that MuDR(p5) had not been silenced by MuDR(p1)*, and MuDR(p1)* had not been heritably activated as a consequence of exposure to MuDR(p5). Analysis of methylation of TIRA in a progeny plant that carried only MuDR(p1)* but that had been exposed to MuDR(p5) in the previous generation [p1*( F3) in Figure 4A] revealed that exposure to MuDR(p5) had no obvious heritable effect on the silenced element. A total of 70% of the cytosines in 5′TIRA were methylated, as were 50% of the cytosines in 3′TIRA (Figure 4C). Further, when this plant was test crossed, none (0 of 137) of its progeny kernels were spotted.
To extend this observation, we performed a cross between a plant carrying a previously silenced MuDR(p1) element [MuDR(p1)*] and a plant that carried an active MuDR element at a third position, MuDR(p4) (Singh et al. 2008). The resulting ear segregated for a single active MuDR element (52% spotted progeny kernels). Spotted and pale progeny kernels were genotyped for MuDR(p1) and MuDR(p4) and then test crossed to plants that lacked either element. Of the plants grown from spotted progeny kernels, six of 14 carried MuDR(p1)* and all of them contained MuDR(p4). When test crossed, these 14 plants gave rise to an average of 52% spotted progeny (Table S2), consistent with the segregation of a single active element. Siblings grown from spotted kernels that lacked MuDR(p1)* gave rise to an average of 50% spotted progeny. Of the pale progeny, eight of 17 carried MuDR(p1)*. When test crossed, none of these gave rise to any spotted progeny. From these experiments, we conclude that the presence of the transposase from the active element had no heritable effect on the silenced element, nor did the silenced elements affect the active element.
Association of 22 nt mudrA-specific small RNAs with silencing of MuDR by Muk
Previously, we had demonstrated that small RNAs are associated with silencing of MuDR by Muk (Slotkin et al. 2005). This conclusion was based on gel hybridization of RNAs from young juvenile leaves. To more comprehensively characterize these small RNAs, small RNAs were sequenced from plants lacking both Muk and MuDR, plants carrying a single active MuDR element, plants carrying only Muk, and plants carrying both MuDR and Muk (F1 plants). In each of these cases, tissue was collected from young leaf 2. Leaf 2 was chosen because it is the tissue that had previously shown ample evidence of an accumulation of Muk-specific small RNAs, and because TIRA is heavily methylated in all three sequence contexts in F1 plants in this leaf (Li et al. 2010). Further, in contrast to immature ears, this leaf also lacks the ubiquitous MuDR-homologous heterochromatic small interfering RNAs (hc-siRNAs) present in all genotypes regardless of activity present in that tissue (Woodhouse et al. 2006a). Small RNAs of leaf 6 from F1 plants were also analyzed because previous work in our laboratory had demonstrated that this leaf exhibits a striking loss of TIRA methylation, concomitant with a loss of expression of lbl1, an important component of the tasiRNA silencing pathway in maize (Li et al. 2010; Dotto et al. 2014).
Consistent with previous results, plants with Muk by itself contained large numbers of MuDR-identical small RNAs, the vast majority of which were 21 nt and (particularly) 22 nt in length (Figure 5 and Tables S3 and S4). These small RNAs were oriented in both sense and antisense orientation relative to mudrA, and were restricted to the portion of the Muk transcript that can form an inverted repeat, consistent with the hypothesis that the small RNAs are a product of processing and/or amplification of the hairpin transcript produced by Muk. In contrast, plants carrying only MuDR and those that lacked both MuDR and Muk had very few MuDR-identical small RNAs.
Figure 5.
Small RNAs associated with silencing of MuDR by Muk. (A) An illustration of MuDR and Muk showing the regions shared by both. The gap in Muk indicates the region of MuDR that is missing in Muk. Genes mudrA and mudrB are as indicated. These models are aligned with the small RNAs illustrated in B and C. (B) A representation of perfectly matched small RNAs from tissues of various genotypes mapped onto MuDR. Small RNAs are color coded for size, as indicated. Note that small RNA samples from plants containing only MuDR or neither MuDR nor Muk have very few small RNAs matching MuDR. F1 leaf 2 and F1 leaf 6 refer to leaves 2 and 6 of plants that carry both MuDR(p1) and Muk in the first generation. (C) Small RNAs with exact matches to the first 250 bp of MuDR (data are taken from F1 plants). (D) Numbers of perfect matches to MuDR of the indicated size classes in the indicated genotypes.
The presence of a deletion within Muk relative to MuDR resulted in a junction that is unique to Muk. We found one prominent group of 22 nt small RNAs that spanned this junction and thus can only be derived from Muk (Figure S2). Interestingly, this particular class of unique, Muk-specific small RNAs are largely in the sense orientation relative to the mudrA transcript, suggesting that processing of the Muk hairpin transcript at this site favors retention of small RNAs in this orientation.
The small RNAs sequenced from plants carrying both MuDR and Muk are quite similar to those observed in plants carrying only Muk, with one important exception. In these plants there are many 21 and 22 nt small RNAs corresponding to regions of MuDR that map exclusively to the mudrA transcript that are not in present in Muk (Figure 5B and Table S4). The presence of small RNAs unique to mudrA data suggest a transitive process by which small RNAs produced by one locus can trigger conversion of a target transcript into dsRNA, which is then cleaved into 21 and 22 nt small RNAs (Choudhary et al. 2019). Interestingly, the ratio of 22 nt to 21 nt small RNAs differs between the regions shared by both MuDR and Muk and the regions only present in MuDR. In the former, the ratio of 22 nt to 21 nt small RNAs is 4.8:1. In the latter it is 1.2:1 (P < 0.0001, χ2 test) (Table S4). Given that these distinct size classes require distinct DCL proteins, generally DCL4 for 21 nt trans-acting small RNAs and DCL2 for the 22 nt size class (Choudhary et al. 2019), this observation suggests the relative contribution of these two proteins may differ in these two distinct regions.
Interestingly, the vast majority of MuDR-identical small RNAs in plants that carry only Muk or that carry both MuDR and Muk correspond to transcribed mudrA sequences. This is most apparent at the transcriptional start site of mudrA, where there is a prominent cluster of 22 nt small RNAs, most of which are in an antisense orientation relative to mudrA (Figure 5C). There are also a smaller number of small RNAs corresponding to sequences upstream of the start of transcription within TIRA. However, many of them are sizes other than 22 nt. Further, when two mismatches are permitted, roughly half of them have sequences that match a nonautonomous Mu element present elsewhere in the maize reference genome rather than MuDR (Figure S3) (Tan et al. 2011).
The distribution of these two distinct small RNA populations in all of the plants carrying Muk matches the distribution of DNA methylation within TIRA of both d107 and silenced MuDR elements. The first of these two distinct small RNA populations, the variably sized and polymorphic small RNAs correspond to the 5′ end of TIRA that is default methylated in all sequence contexts in the absence of the transposase in d107 (Figure 3). The second, more numerous, population of 22 nt small RNAs correspond to the expressed portion of TIRA whose stable CG and CHG methylation is triggered in response to Muk (Figure 3 and Figure 4). Interestingly, analysis of large populations of small RNAs from mop1 mutant and wild-type immature ears, as well as lbl1 mutant and wild type leaf apexes provides similar evidence (Table S5). In these libraries, derived from plants that did not carry MuDR or Muk, there are very few small RNAs of any size class perfectly matching either 5′TIRA or 3′TIRA. When two mismatches are permitted, there are a substantial number of small RNAs matching TIRA, but the vast majority of them are in 5′TIRA. In combination with analysis of our samples, these data suggest that de novo methylation of 5′TIRA, but not 3′TIRA, is mediated by “background” small RNAs with one or more mismatch, but de novo methylation of 3′TIRA requires 21 and 22 nt small RNAs derived from the Muk hairpin and processed Pol-II-derived mudrA transcript.
Given the involvement of LBL in the production of dsRNA, and the requirement for LBL in methylation of TIRA in leaf tissue, we hypothesized that there would be a reduction of small RNAs in leaves that are transitional between juvenile and adult, which exhibit a loss of both TIRA methylation and a reduction of lbl1 expression (Li et al. 2010). In fact, we observed a dramatic reduction in the number of small RNAs of all size classes in transition leaves (Figure 5D and Table S3).
TIRB silencing is not associated with small RNAs
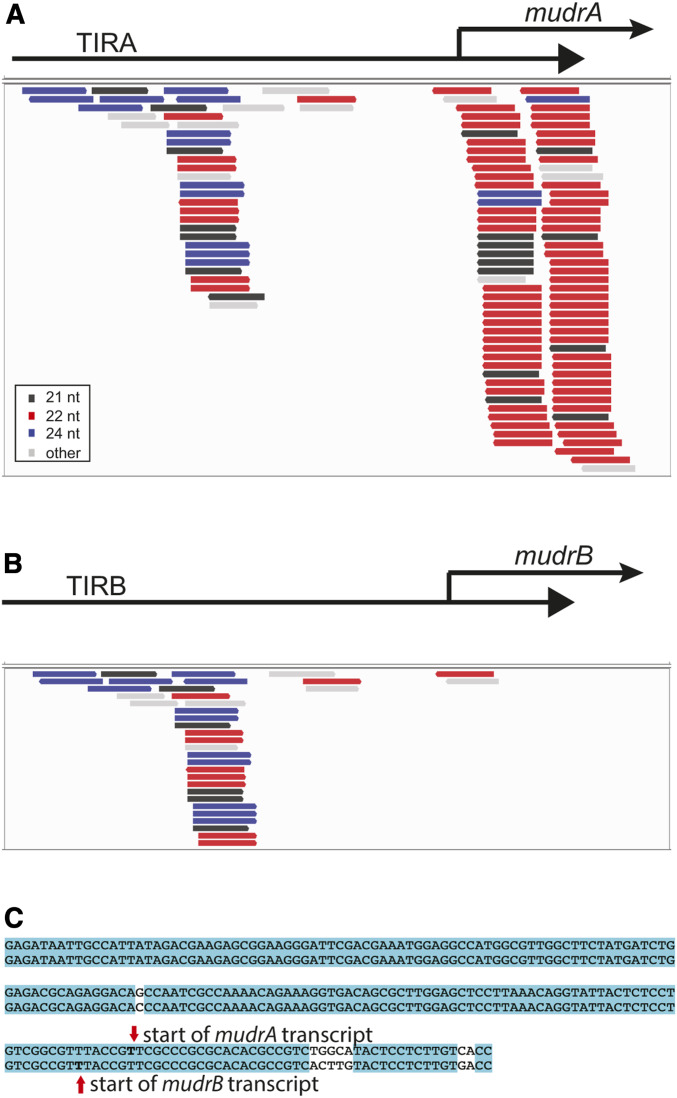
Muk does not include sequences from mudrB. However, it does include TIRA, which is 99% identical with TIRB over the first 180 bp, suggesting that small RNAs from the Muk hairpin transcript would be hypothetically competent to direct methylation of TIRB. Although mudrB is eventually silenced by Muk, the trajectory of mudrB silencing is distinct. Unlike mudrA, which is transcriptionally silenced by the immature ear stage in p1;Muk F1 plants, mudrB is not transcriptionally silenced by this stage (Slotkin et al. 2003). Instead, in these plants transcript from mudrB is readily detectable, but it does not appear to be polyadenylated. By the next generation, however, plants that carry only MuDR(p1)* do not have detectable mudrB transcript. Further, Muk can only heritably silence mudrB in this way when this gene is in cis to mudrA (on the same transposon); it is not silenced when it is in trans to a mudrA gene that is being silenced (Slotkin et al. 2005).
Previous work using RNA gel hybridization showed that small RNAs similar to mudrB did not accumulate to high levels in F1 MuDR;Muk plants (Slotkin et al. 2005). In some ways, this was surprising given that TIRB is identical over much of its length to TIRA, and the hairpin formed by the Muk transcript includes all of TIRA (and thus TIRB as well). A closer examination of the small RNA population in F1 leaf 2 tissues provides a possible explanation for this discrepancy. Although the two TIRs are quite similar to each other, they are more diverged near the internal portion of each TIR, within 3′TIRA and 3′TIRB (Figure 6C). The mudrA transcript initiates 168 bp from the end of the element and the mudrB transcript initiates 163 bp from the other end of the element (Hershberger et al. 1995). Since this is the region in which TIRA and TIRB begin to diverge in sequence, there are very few 22 nt small RNAs that perfectly match TIRB, particularly in the transcribed portion of TIRB (Figure 6, A and B). If silencing requires the presence of both the target transcript and small RNAs from the Muk hairpin, this distribution of small RNAs may explain why Muk acts only indirectly on mudrB via a distinct mechanism that does not involve direct interaction between Muk small RNAs and mudrB transcript.
Figure 6.
Small RNAs in F1 leaf 2 plants with perfect matches to TIRA (A) and TIRB (B). (C) An alignment of TIRA and TIRB with the transcriptional start sites for mudrA and mudrB as indicated.
Discussion
The evidence presented here suggests that two distinct silencing pathways operate on MuDR TIRA as it is being silenced. One pathway, dependent on MOP1, appears to involve a number of heterogeneous small RNAs likely derived from other Mu TIRs in the genome that target 5′TIRA for DNA methylation in the absence of the transposase (Woodhouse et al. 2006a,b; Nobuta et al. 2008). They do not, however, trigger transcriptional silencing, as is evidenced by d107, which accumulates dense methylation in 5′TIRA but is transcriptionally active. In contrast, when d107 or MuDR is silenced by Muk, methylation spreads from 5′TIRA into the transcribed portion of the mudrA (Figure 3; Li et al. 2010). Once MuDR(p1) is silenced, a source of transposase can reverse the methylation at 5′TIRA but does not reverse the heritably silenced state of MuDR(p1), nor does it reverse methylation within 3′TIRA (Figure 4). Thus, one can gain methylation at 5′TIRA and not gain transcriptional silencing, and one can lose 5′TIRA methylation and not lose heritable transcriptional silencing. Interestingly, these data also suggest that the methylation required for a heritably silenced state is restricted to the 3′TIRA, and is largely composed of CG and CHG methylation.
The second pathway involves 21 and 22 nt small RNAs specifically associated with sequences within 3′TIRA corresponding to the transcribed portion of mudrA. These small RNAs correspond to cytosines within 3′TIRA that are stably methylated in the CG and CHG sequence contexts, and are only observed in leaves carrying Muk, either by itself or in combination with MuDR. Plants carrying both MuDR and Muk also have many small RNAs that are derived from portions of mudrA that are not present in Muk. This is consistent with the activity of a transitive process that is triggered by small RNAs processed from the Muk hairpin. This hypothesis is supported by the observation that in leaf 6 in which lbl1 expression is reduced, the number of these small RNAs is also reduced (Figure 5 and Table S3) (Li et al. 2010). The fact that the 22 nt small RNAs do not extend upstream of the transcriptional start site of mudrA suggests that these small RNAs are processed almost entirely from transcript initiated from within TIRA rather than the dsRNA hairpin formed by Muk, which includes 5′TIRA. Since LBL1 works in conjunction with RDR6 to produce a variety of trans-acting small RNAs, these data suggest that the Muk triggers silencing in leaves via a pathway that interacts with Pol II TE transcripts in a manner that is similar to that described in Arabidopsis (Wu et al. 2012; Nuthikattu et al. 2013; Panda and Slotkin 2013; Panda et al. 2016). In this context, an interaction between small RNAs and Pol II transcript represents a natural and expected means by which otherwise active TEs are recognized and silenced. The key here is the source of small RNAs that can trigger silencing. For low-copy-number elements such as MuDR in the Minimal line, it may be necessary for rearrangements such as those observed in Muk to occur for effective silencing to take place. In maize lines containing larger numbers of Mu elements that are prone to spontaneous silencing (Robertson 1986; Skibbe et al. 2012), we speculate that this silencing could be due to the accumulation of aberrant MuDR elements that collectively trigger silencing of active elements in these lines.
One important conclusion from our analysis is that default methylation such as that observed at d107 and Mu1 is not sufficient to trigger silencing. d107 expresses a transcript and Mu1 element has a functional outward reading promoter even when it is methylated (Barkan and Martienssen 1991). Because hypomethylated Mu1 elements can be remethylated due to genetic segregation of MuDR, or even in somatic sectors in which the transposase is lost, it is apparent that this pathway is competent enough to trigger de novo methylation during plant development (Lisch et al. 1995; Lisch and Jiang 2008). However, this methylation is readily reversed when transposase is reintroduced. This may be because binding of the transposase to the TIR directly blocks methyltransferase activity or due to demethylation activity on the part of the transposase, as has been proposed for the Spm transposase (Cui and Fedoroff 2002).
The default methylation at 5′TIRA is dependent on the maize homolog of RDR2, MOP1, which is required for the vast majority of 24 nt hc-siRNAs (Lisch et al. 2002; Alleman et al. 2006; Nobuta et al. 2008). Previous work in our laboratory has demonstrated that heritable silencing of MuDR by Muk occurs efficiently in a mop1 mutant background in the absence of those hc-siRNAs, and that Muk-specific small RNAs are retained in this mutant (Woodhouse et al. 2006a). Further, in immature ears, the presence of the hc-siRNAs, likely derived from hMuDR elements in this genetic background, has no effect on an otherwise active MuDR element. Thus, it would appear that de novo silencing of MuDR elements requires small RNAs derived from Pol II transcript, but likely not those derived from processed Pol IV transcripts such as those that are produced from previously silenced elements. This would explain why silenced MuDR elements do not silence active elements, as the silenced elements would be expected to produce only the Pol IV transcript, which would not be expected to produce trans-acting small RNAs.
There is evidence that a transient loss of transposon silencing in the vegetative nucleus in pollen can serve to reinforce silencing in sperm cells (Slotkin et al. 2009; Martínez et al. 2016), and it has been shown that silenced MuDR elements are reactivated in pollen (Slotkin et al. 2009). However, our results demonstrate that silenced MuDR elements are not competent to silence active elements in trans, and similar results have been obtained in Arabidopsis (Kato et al. 2004). Inactive elements would only be expected to act in trans to silence active elements via this pathways if, when reactivated, they produced a Pol II transcript that, like Muk, could be processed into trans-acting small RNAs. This seems to be the case for epigenetically activated small interfering RNAs in Arabidopsis, which are produced from hairpin micro-RNA-like transcripts that are expressed when they are epigenetically reactivated during reprogramming of the germ line and that target transposons (Creasey et al. 2014).
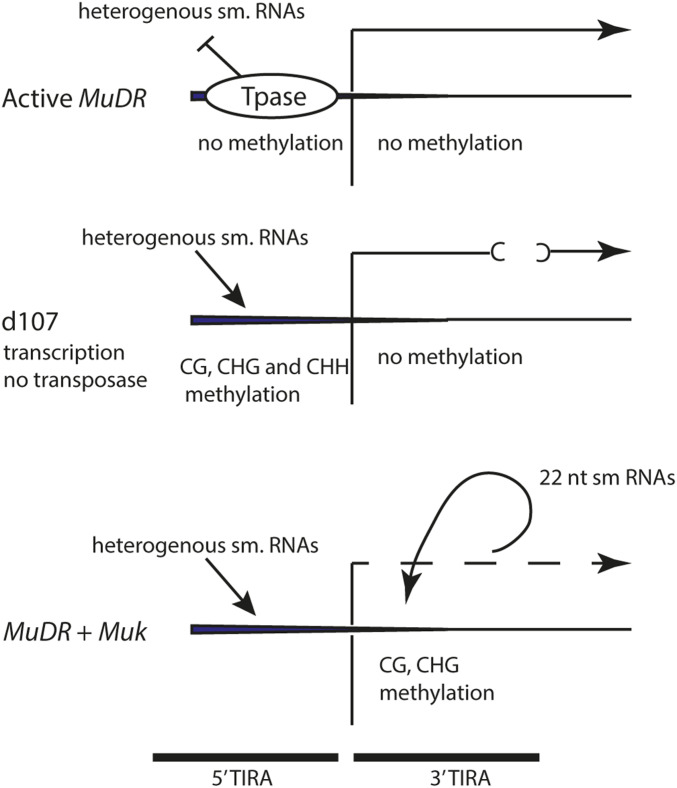
Our observations are consistent with a relatively simple model (Figure 7). TIRA methylation is absent whenever a functional transposase is present, presumably because the transposase blocks methyltransferase activity. When 22 nt small RNAs are introduced from the Muk hairpin, they trigger RDR6-dependent production of dsRNA using the Pol-II-derived mudrA transcript as a template. This dsRNA is cleaved by a DICER (presumably DCL2 and/or DCL4) and the resulting small RNAs are then used to direct DNA methylation, largely in the CG and CHG sequence contexts in 3′TIRA. Meanwhile, because the transposase is no longer present, hc-siRNAs derived from other silenced Mu elements present in the genome can direct de novo methylation of 5′TIRA, exactly as they do at d107 and Mu1 in the absence of functional transposase (Figure 2 and Figure 3). According to this model, processed mudrA transcript and the resulting 3′TIRA methylation is the cause of transcriptional silencing, and 5′TIRA methylation is a consequence of that silencing. Based on this model, we would predict that a MuDR derivative that had a genetic lesion that prevented it from expressing Pol II transcript would accumulate methylation in 5′TIRA, but would not accumulate methylation in 3′TIRA when combined with Muk.
Figure 7.
A model for the interaction between different classes of small RNAs and DNA methylation in 5′TIRA and 3′TIRA.
The involvement of two parallel pathways interacting with MuDR TIRA raises some interesting questions. The default methylation pathway suggests that there are small RNAs that are competent to trigger de novo methylation that are present even when the elements are active. However, active MuDR elements retain that activity in the face of this “soup” of hc-siRNAs, and the same is true for some other active transposons, such as CACTA elements in Arabidopsis and Mu elements in maize (Kato et al. 2004; Woodhouse et al. 2006a). In contrast, others, such as ONSEN and Tos17, are rapidly resilenced after periods of activity, a process that requires Pol IV (Hirochika et al. 1996; Ito et al. 2011; Matsunaga et al. 2015). Since the Pol IV transcript is thought to be derived from previously methylated elements, this would suggest that at some threshold, silenced elements can contribute to de novo silencing of active elements. In contrast, Muk silencing of MuDR does not appear to involve 24 nt Pol-IV-dependent small RNAs, presumably because processing of the Muk hairpin can produce siRNAs in the absence of hc-siRNAs (Woodhouse et al. 2006a). Further, those hc-siRNAs that are present in the background largely target a portion of active MuDR elements (5′TIRA) that is irrelevant to transcriptional gene silencing (Figures 5 and Figure S3). This may be due to random sequence divergence within the relevant portions of TIRA relative to other Mu elements, or due to selection in favor of differences between autonomous and nonautonomous elements at the mudrA and mudrB transcriptional start sites. Indeed, few hc-siRNAs specifically target the transcribed portion of TIRA (Table S5).
Collectively, our data suggests that active elements in maize remain active because of an absence of small RNAs that are competent to trigger heritable silencing, and that small RNAs competent to trigger silencing are most likely those that directly target the Pol II transcript arising from those active elements, rather than the small RNAs derived from previously silenced elements. Further, our data suggest that heritable silencing information is contained within 3′TIRA in the form of CG and CHG methylation. Finally, our analysis of small RNAs in transition leaves reveals that the loss of LBL1 in these leaves results in a reduction of small RNAs targeting mudrA, suggesting that the accumulation of these small RNAs is dependent on the tasiRNA pathway.
Acknowledgments
This work was supported by the National Science Foundation (grants DBI-1237931and DBI-0820828 to D.L.) as well as the Purdue Center for Plant Biology. We thank Xinyan Zhang and Thomas Peterson for providing useful comments.
Author contributions: D.B. performed the bisulfite analysis and contributed to experimental design. H.L. performed the small RNA sequencing analysis contributed to experimental design. S.Y.K. performed bisulfite analysis on the MuDR active individual. M.Z. performed analysis of the small RNA data set. D.L. wrote the manuscript, performed the genetic analysis, and analyzed the small RNA and deletion junction.
Note added in proof: See Wang et al. 2020 (pp. 393–406) in this issue for a related work.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12009360.
Communicating editor: J. Birchler
Literature Cited
- Alleman M., Sidorenko L., McGinnis K., Seshadri V., Dorweiler J. E. et al. , 2006. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442: 295–298. 10.1038/nature04884 [DOI] [PubMed] [Google Scholar]
- Barkan A., and Martienssen R. A., 1991. Inactivation of maize transposon Mu suppresses a mutant phenotype by activating an outward-reading promoter near the end of Mu1. Proc. Natl. Acad. Sci. USA 88: 3502–3506. 10.1073/pnas.88.8.3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito M. I., and Walbot V., 1997. Characterization of the maize Mutator transposable element MURA transposase as a DNA-binding protein. Mol. Cell. Biol. 17: 5165–5175. 10.1128/MCB.17.9.5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., 1996. The Mutator transposable element system of maize. Curr. Top. Microbiol. Immunol. 204: 195–229. [DOI] [PubMed] [Google Scholar]
- Bucher E., Reinders J., and Mirouze M., 2012. Epigenetic control of transposon transcription and mobility in Arabidopsis. Curr. Opin. Plant Biol. 15: 503–510. 10.1016/j.pbi.2012.08.006 [DOI] [PubMed] [Google Scholar]
- Chandler V. L., and Walbot V., 1986. DNA modification of a maize transposable element correlates with loss of activity. Proc. Natl. Acad. Sci. USA 83: 1767–1771. 10.1073/pnas.83.6.1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomet P., Lisch D., Hardeman K. J., Chandler V. L., and Freeling M., 1991. Identification of a regulatory transposon that controls the Mutator transposable element system in maize. Genetics 129: 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary S., Thakur S., and Bhardwaj P., 2019. Molecular basis of transitivity in plant RNA silencing. Mol. Biol. Rep. 46: 4645–4660. 10.1007/s11033-019-04866-9 [DOI] [PubMed] [Google Scholar]
- Creasey K. M., Zhai J., Borges F., Van Ex F., Regulski M. et al. , 2014. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415. 10.1038/nature13069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuerda-Gil D., and Slotkin R. K., 2016. Non-canonical RNA-directed DNA methylation. Nat. Plants 2: 16163 (erratum: Nat. Plants 3: 16211). 10.1038/nplants.2016.163 [DOI] [PubMed] [Google Scholar]
- Cui H., and Fedoroff N. V., 2002. Inducible DNA demethylation mediated by the maize Suppressor-mutator transposon-encoded TnpA protein. Plant Cell 14: 2883–2899. 10.1105/tpc.006163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Hamilton A., Mueller E., and Baulcombe D. C., 2000. Potato virus X amplicons in arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell 12: 369–379. 10.1105/tpc.12.3.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto M. C., Petsch K. A., Aukerman M. J., Beatty M., Hammell M. et al. , 2014. Genome-wide analysis of leafbladeless1-regulated and phased small RNAs underscores the importance of the TAS3 ta-siRNA pathway to maize development. PLoS Genet. 10: e1004826 10.1371/journal.pgen.1004826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fultz D., and Slotkin R. K., 2017. Exogenous transposable elements circumvent identity-based silencing, permitting the dissection of expression-dependent silencing. Plant Cell 29: 360–376. 10.1105/tpc.16.00718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fultz D., Choudury S. G., and Slotkin R. K., 2015. Silencing of active transposable elements in plants. Curr. Opin. Plant Biol. 27: 67–76. 10.1016/j.pbi.2015.05.027 [DOI] [PubMed] [Google Scholar]
- Gruntman E., Qi Y., Slotkin R. K., Roeder T., Martienssen R. A. et al. , 2008. Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371 10.1186/1471-2105-9-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C. J., Erhard K. F. Jr., Lisch D., and Hollick J. B., 2009. Production and processing of siRNA precursor transcripts from the highly repetitive maize genome. PLoS Genet. 5: e1000598 10.1371/journal.pgen.1000598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger R. J., Benito M. I., Hardeman K. J., Warren C., Chandler V. L. et al. , 1995. Characterization of the major transcripts encoded by the regulatory MuDR transposable element of maize. Genetics 140: 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H., Sugimoto K., Otsuki Y., Tsugawa H., and Kanda M., 1996. Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93: 7783–7788. 10.1073/pnas.93.15.7783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D., and Moazed D., 2015. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16: 71–84. 10.1038/nrg3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia A. P., and Schnable P. S., 1996. DNA sequence analyses support the role of interrupted gap repair in the origin of internal deletions of the maize transposon, MuDR. Genetics 142: 603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Gaubert H., Bucher E., Mirouze M., Vaillant I. et al. , 2011. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472: 115–119. 10.1038/nature09861 [DOI] [PubMed] [Google Scholar]
- Kato M., Takashima K., and Kakutani T., 2004. Epigenetic control of CACTA transposon mobility in Arabidopsis thaliana. Genetics 168: 961–969. 10.1534/genetics.104.029637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumakura N., Takeda A., Fujioka Y., Motose H., Takano R. et al. , 2009. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 583: 1261–1266. 10.1016/j.febslet.2009.03.055 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J. A., and Jacobsen S. E., 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Freeling M., and Lisch D., 2010. Epigenetic reprogramming during vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 107: 22184–22189. 10.1073/pnas.1016884108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D., 1995. Genetic and molecular characterization of the mutator system in Mmize, pp. 408 in Plant and Microbial Biology, University of California, Berkeley, CA. [Google Scholar]
- Lisch D., 2002. Mutator transposons. Trends Plant Sci. 7: 498–504. 10.1016/S1360-1385(02)02347-6 [DOI] [PubMed] [Google Scholar]
- Lisch D., 2009. Epigenetic regulation of transposable elements in plants. Annu. Rev. Plant Biol. 60: 43–66. 10.1146/annurev.arplant.59.032607.092744 [DOI] [PubMed] [Google Scholar]
- Lisch D., 2013. Regulation of the Mutator system of transposons in maize. Methods Mol. Biol. 1057: 123–142. 10.1007/978-1-62703-568-2_9 [DOI] [PubMed] [Google Scholar]
- Lisch D., 2015. Mutator and MULE transposons, Mobile DNA III, edited by Craig P. R. N. L., Lambowitz A., Chandler M., Gellert M., and Sandmeyer S.. American Society for Microbiology Press, Washington, D.C. 10.1128/9781555819217.ch36 [DOI] [Google Scholar]
- Lisch D., and Jiang N., 2008. Mutator and pack-MULEs, pp. 277–306 in The Maize Handbook - Volume II: Domestication, Genetics and Genomics of Maize, edited by Bennetzen J. L. and Hake S.. Springer, NY. [Google Scholar]
- Lisch D., Chomet P., and Freeling M., 1995. Genetic characterization of the Mutator system in maize: behavior and regulation of Mu transposons in a minimal line. Genetics 139: 1777–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D., Girard L., Donlin M., and Freeling M., 1999. Functional analysis of deletion derivatives of the maize transposon MuDR delineates roles for the MURA and MURB proteins. Genetics 151: 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D., Carey C. C., Dorweiler J. E., and Chandler V. L., 2002. A mutation that prevents paramutation in maize also reverses Mutator transposon methylation and silencing. Proc. Natl. Acad. Sci. USA 99: 6130–6135. 10.1073/pnas.052152199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí-Ordóñez A., Marchais A., Etcheverry M., Martin A., Colot V. et al. , 2013. Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 45: 1029–1039. 10.1038/ng.2703 [DOI] [PubMed] [Google Scholar]
- Martínez G., and Slotkin R. K., 2012. Developmental relaxation of transposable element silencing in plants: functional or byproduct? Curr. Opin. Plant Biol. 15: 496–502. 10.1016/j.pbi.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Martínez G., Panda K., Kohler C., and Slotkin R. K., 2016. Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat. Plants 2: 16030 10.1038/nplants.2016.30 [DOI] [PubMed] [Google Scholar]
- Matsunaga W., Ohama N., Tanabe N., Masuta Y., Masuda S. et al. , 2015. A small RNA mediated regulation of a stress-activated retrotransposon and the tissue specific transposition during the reproductive period in Arabidopsis. Front Plant Sci 6: 48 10.3389/fpls.2015.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M. A., and Mosher R. A., 2014. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408 (erratum: Nat. Rev. Genet. 15: 570). 10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- McCue A. D., Panda K., Nuthikattu S., Choudury S. G., Thomas E. N. et al. , 2015. ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 34: 20–35. 10.15252/embj.201489499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K., Lu C., Shrivastava R., Pillay M., De Paoli E. et al. , 2008. Distinct size distribution of endogeneous siRNAs in maize: evidence from deep sequencing in the mop1–1 mutant. Proc. Natl. Acad. Sci. USA 105: 14958–14963. 10.1073/pnas.0808066105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthikattu S., McCue A. D., Panda K., Fultz D., DeFraia C. et al. , 2013. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21–22 nucleotide small interfering RNAs. Plant Physiol. 162: 116–131. 10.1104/pp.113.216481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly C., Shepherd N. S., Pereira A., Schwarz-Sommer Z., Bertram I. et al. , 1985. Molecular cloning of the a1 locus of Zea mays using the transposable elements En and Mu1. EMBO J. 4: 877–882. 10.1002/j.1460-2075.1985.tb03713.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda K., and Slotkin R. K., 2013. Proposed mechanism for the initiation of transposable element silencing by the RDR6-directed DNA methylation pathway. Plant Signal. Behav. 8: e25206. 10.4161/psb.25206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda K., Ji L., Neumann D. A., Daron J., Schmitz R. J. et al. , 2016. Full-length autonomous transposable elements are preferentially targeted by expression-dependent forms of RNA-directed DNA methylation. Genome Biol. 17: 170 10.1186/s13059-016-1032-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S. et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. S., 1986. Genetic studies on the loss of mu mutator activity in maize. Genetics 113: 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko G. N., and Walbot V., 2001. Expression and post-transcriptional regulation of maize transposable element MuDR and its derivatives. Plant Cell 13: 553–570. 10.1105/tpc.13.3.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko G. N., Ono A., and Walbot V., 2003. Initiation of silencing of maize MuDR/Mu transposable elements. Plant J. 33: 1013–1025. 10.1046/j.1365-313X.2003.01683.x [DOI] [PubMed] [Google Scholar]
- Saze H., and Kakutani T., 2011. Differentiation of epigenetic modifications between transposons and genes. Curr. Opin. Plant Biol. 14: 81–87. 10.1016/j.pbi.2010.08.017 [DOI] [PubMed] [Google Scholar]
- Sigman M. J., and Slotkin R. K., 2016. The first rule of plant transposable element silencing: location, location, location. Plant Cell 28: 304–313. 10.1105/tpc.15.00869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Freeling M., and Lisch D., 2008. A position effect on the heritability of epigenetic silencing. PLoS Genet. 4: e1000216 [corrigenda: PLoS Genet. 5 (2009)]. 10.1371/journal.pgen.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbe D. S., Fernandes J. F., and Walbot V., 2012. Mu killer-mediated and spontaneous silencing of Zea mays mutator family transposable elements define distinctive paths of epigenetic inactivation. Front Plant Sci 3: 212 10.3389/fpls.2012.00212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R. K., 2005. The heritable silencing of mutator transposons by Mu killer, pp. 223 in Plant and Microbial Biology, University of California, Berkeley, CA. [Google Scholar]
- Slotkin R. K., and Martienssen R., 2007. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8: 272–285. 10.1038/nrg2072 [DOI] [PubMed] [Google Scholar]
- Slotkin R. K., Freeling M., and Lisch D., 2003. Mu killer causes the heritable inactivation of the Mutator family of transposable elements in Zea mays. Genetics 165: 781–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R. K., Freeling M., and Lisch D., 2005. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat. Genet. 37: 641–644. 10.1038/ng1576 [DOI] [PubMed] [Google Scholar]
- Slotkin R. K., Vaughn M., Borges F., Tanurdzic M., Becker J. D. et al. , 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. 10.1016/j.cell.2008.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B. C., Chen Z., Shen Y., Zhang Y., Lai J. et al. , 2011. Identification of an active new mutator transposable element in maize. G3 (Bethesda) 1: 293–302. 10.1534/g3.111.000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse M. R., Freeling M., and Lisch D., 2006a Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 4: e339 10.1371/journal.pbio.0040339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse M. R., Freeling M., and Lisch D., 2006b The mop1 (mediator of paramutation1) mutant progressively reactivates one of the two genes encoded by the MuDR transposon in maize. Genetics 172: 579–592. 10.1534/genetics.105.051383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Mao L., and Qi Y., 2012. Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant Physiol. 160: 990–999. 10.1104/pp.112.200279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z. Y., and Sundaresan V., 1991. Binding sites for maize nuclear proteins in the terminal inverted repeats of the Mu1 transposable element. Mol. Gen. Genet. 229: 17–26. 10.1007/BF00264208 [DOI] [PubMed] [Google Scholar]