Abstract
Protein-protein interactions (PPIs) are important in human disease. Developing and refining tools to understand physical contacts between signaling proteins is crucial. This unit describes a split luciferase complementation (SLC) method designed to discover inhibitors for PPI. Different fusion proteins with split luciferase are constructed, expressed, and purified to determine the best pair which can generate the strongest luminescence. SLC specificity and affinity are further confirmed. We also describe step-by-step instructions to perform these assays using the NS2B-NS3 interaction as an example. NS2B is an essential cofactor for flaviviral NS3 protease function. Advantages and disadvantages of these assays are further discussed.
Keywords: Protein-protein interaction, flaviviral protease, NS2B-NS3, split luciferase complementation
INTRODUCTION
Human diseases usually involve protein-protein interactions (PPIs). Some of these interactions, such as envelope glycoprotein GP120 of the human immunodeficiency virus (HIV) and C-C chemokine receptor type 5 (CCR5) (Farzan et al., 1998), can facilitate pathogen invasion of human cells. Viruses usually use this receptor binding trick to initiate the first step of cell entry. PPIs can affect protein functions as well, such as NS2B and NS3 of the flaviviral protease (Li et al., 2017; Li et al., 2018). Along with host proteases, the flaviviral protease can cleave the viral polyprotein precursor, which is a critical step for virus replication. In cell signaling pathways, PPIs involving cytokines are indispensable for regulating immune and inflammatory responses (Panga and Raghunathan, 2018). “Cytokine storm,” an aberrant immune response induced by infection, is related to the pandemic 1918–19 Spanish H1N1 influenza, the pandemic H1N1 influenza of 2009, and H5N1 avian influenza (Tisoncik et al., 2012). Therefore, it is critical to develop and refine tools to study these proteins’ physical contacts.
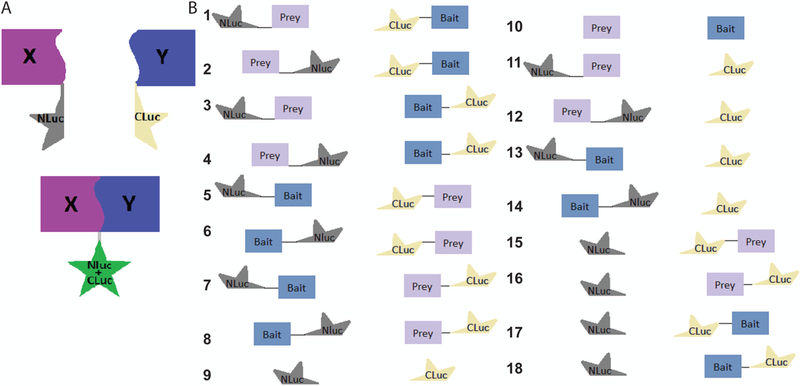
To better study the interactions between two proteins, luciferase is widely used as a probe for visualization and quantification. This unit describes protocols using a split luciferase complementation (SLC) method to measure PPI which were initially established to identify novel flavivirus NS2B-NS3 inhibitors (Li et al., 2017). Firefly luciferase (FLuc) can be split into two fragments: N-terminal (NLuc) and C-terminal (CLuc) fragments. Given two binding proteins which have NLuc/CLuc fused with them, the PPI should bring the NLuc and CLuc in close proximity and subsequently reconstitute a functional FLuc, leading to detectable luminescence. Otherwise, non-reactive proteins with fused NLuc/CLuc should not generate signals (Porter et al., 2008). Figure 1A is a schematic diagram displaying the firefly SLC strategy to monitor PPIs. In the Commentary section, advantages and disadvantages of this protocol are discussed.
Figure 1.
(A) Schematic diagram displaying the firefly SLC strategy to monitor PPIs. (B). Schematic illustrations of all possible interaction pairs and controls.
BASIC PROTOCOL 1
EXPRESSION AND PURIFICATION OF DIFFERENT FUSION PROTEINS
The aim of this assay is to obtain functional target proteins (NS3 and NS2B) with NLuc/CLuc fusions. The FLuc is composed of 550 amino acids (aa) that can be split to NLuc (aa 1–416) and CLuc (aa 398–550), which can be fused to the PPI pairs, known as the bait and prey proteins. Because the positions of luciferase fragments may affect the intensity of luminescence (Li et al., 2017), testing many or all possible combinations of the bait and prey fusion proteins may be required, listed here: NLuc-prey/CLuc-bait, prey-Nluc/CLuc-bait, NLuc-prey/bait-CLuc, prey-NLuc/bait-CLuc, CLuc-prey/NLuc-bait, prey-CLuc/NLuc-bait, CLuc-prey/bait-NLuc, and prey-CLuc/bait-NLuc (Figure 1B). In addition, controls without prey or bait (NLuc and CLuc only) should also be included. Moreover, to demonstrate interaction specificity, “cold” prey and bait not fused to NLuc or CLuc, and/or “fused” mutants of prey or bait that are known to abolish prey-bait interactions should also be prepared. Then, the pair with the strongest luminescence signal after incubating with luciferin substrate should be further analyzed to confirm the specificity and affinity of the SLC (see Basic Protocol 2). Standard molecular cloning techniques or gene synthesis through commercial vendors may be employed to generate these constructs. As an example, gene cloning, overlapping PCR, and PCR mutagenesis are employed to generate correct constructions for fusion proteins, as reported previously (Li et al., 2017). All proteins are expressed in E. coli and purified via protein purification columns. In this Basic Protocol 1, we will use the NLuc-NS2B-E66stop and GST-CLuc-NS3 (GCN) as examples to describe protein expression and purification.
Materials
Expression vectors: the NLuc-NS2B-E66stop in the pET28a vector (Sigma-Aldrich, cat. No. 69864-3) and the GST-CLuc-NS3 in the pGEX-6P-1 vector (GE HealthCare, cat. No. 28954648).
SOC Medium
Super broth medium containing the appropriate antibiotic for the vector used
10 cm LB agar plate containing the appropriate antibiotic for the vector used
E. coli strain Rosetta 2(DE3) (Sigma-Aldrich, cat. No. 71400-3)
DNase 1 (Worthington Biochemical, cat. No. LS002139)
Nickel-nitrilotriacetic acid (Ni-NTA) column (Qiagen, cat. No. 30210)
Glutathione sepharose 4B (GE HealthCare, cat. No. 17075601)
Econo-Column® Chromatography Columns (Bio-rad, cat. No. 7372507)
Buffer A: 50 mM Tris, pH 7.5, 150 mM NaCl
Buffer B: 50 mM Tris, pH 8.0, 500 mM NaCl, 20 mM imidazole
Buffer C: 50 mM Tris, pH 8.0, 500 mM NaCl, 250 mM imidazole
Buffer D: 50 mM Tris, pH 7.5, 150 mM NaCl, 10 mM reduced glutathione
Buffer E: 20 mM Tris, pH 8.0, 150 mM NaCl
Buffer F: 1x PBS + 0.05% Chaps + 0.1% BSA + 5mM DTT (freshly added)
Buffer G: PBS+2mM MgCl2 + 4mM EGTA
Buffer H: Buffer G + D-luciferin (1 mg/ml) + 4 mM ATP (freshly added)
Stock D-luciferin, 10 mg/ml (100x) in DMSO
Stock ATP 0.4 M in H2O
IPTG solution (1 M)
Refolding buffer: 0.4M Arginine; 100mM Tris PH8.0; 5mM reduced glutathione; 0.5mM oxidized glutathione; 2mM EDTA; 5% glycerol
100 mM phenylmethylsulfonyl fluoride (PMSF) stock solution in Isopropanol
Guanidine stock solution: 6M Guanidine-HCl; 10mM NaAc; 10mM EDTA PH 8.0. Adjust to PH 4.2 at room temperature (RT).
Water bath
Refrigerated shaker incubator
Spectrophotometer
Floor centrifuge
Microcentrifuge
Sonicator
NanoDrop UV visible spectrophotometer (ThermoFisher Scientific, cat. No. ND-2000)
Amicon stirred cell Model (Sigma-Aldrich, cat. No. UFSC05001 and 5121)
Filtration membranes with appropriate molecular weight cutoff (Sigma-Aldrich, cat. No. PLGC04310 and PLGC02510)
16/60 Superdex 200 column (GE HealthCare, cat. No. 28989335)
AKTA protein purification instrument (GE HealthCare, cat. No. 29022094)
Protein expression
-
1
To disinfect the working site, spray the work bench with 70% ethanol, allowing it to dry before use, and prepare a bucket of ice. Turn on a water bath and set the temperature at 42°C. Bring SOC Medium to RT.
-
2
Thaw one tube of chemically competent E. coli strain Rosetta 2(DE3) cells, which should contain 20–50 µl of cells, on ice. Mix gently to ensure that the cells are distributed evenly.
As the competent cells are fragile, always leave them on ice and avoid high-speed vertexing. Fingertip-tap the tube 3–5 times to gently mix. Depending on the number of constructs, transformation of several tubes of cells with different plasmids at once is also feasible. To reduce error, we do not recommend transforming more than 10 tubes of cells at the same time. Other competent cells such as BL21(DE3) can be considered as well.
-
3
Add 1 µl of the purified plasmid DNA solution (usually 10 pg – 100 ng) directly to the cells. Fingertip-tap the tube 3–5 times to gently mix. Then place the tubes on ice for 20–30 min.
Gentle mix should be done quickly and cells put back into ice promptly.
-
4
Heat the tubes for 30–60 s in a 42 °C water bath without shaking and immediately place the tubes on ice for 2 min.
Keep the ice bucket beside the water bath to cool down the cells immediately.
-
5
Add 250–1000 µl of RT SOC Medium to each tube and shake at 37 °C for 1–2 h.
Avoid using conical bottom tubes to shake the cells because cells cannot be suspended evenly via shaking. Round bottom sterilized tubes are preferred.
-
6
Plate some or all of the transformation onto a 10 cm LB agar plate containing the appropriate antibiotic.
Distribute the culture cells evenly on the plates so that single colonies can be picked up from each plate later. Because the transformation rate is unknown, we suggest spreading two plates with different volumes of cells. Put 100 µl of the cell culture on one plate and spread the remaining cell culture on the other plate.
-
7
Label plates clearly and incubate them upside down into a 37 °C incubator overnight.
-
8
Pick up a single colony from each plate with a 10 µl sterile pipette tip (or a sterilized toothpick) and put it into an Erlenmeyer flask containing 20 mL cell culture media with an appropriate antibiotic (depending on the vector used). Shake flasks in a shaker at 220 rpm at 37°C overnight.
-
9
Expand the cell culture by adding the 20 mL overnight cell culture into 2 L fresh super broth medium with an appropriate antibiotic.
-
10
Shake cells in a shaker at 220 rpm at 37 °C.
-
11
Monitor cell culture at OD600 using a spectrophotometer. When the OD600 reaches 0.8–1.0, add 1 ml stock IPTG solution (1 M) to a final 0.5 mM IPTG concentration to induce protein expression.
-
12
Shake cells in a refrigerated shaker at 220 rpm at 16 °C overnight.
-
13
Centrifuge the cell cultures at 2800 x g for 30 min at 4 °C and discard the supernatant.
-
14
Continue to protein purification directly or store cells at −80 °C for further purification.
Protein purification
-
15
Put 1 L of refolding buffer at 4 °C overnight and pre-chill the floor centrifuge to 4 °C.
-
16
Resuspend cell pellets on ice with buffer A for GST-tagged protein (GST-CLuc-NS3) and buffer B for His-tagged protein (NLuc-E66stop) (10 mL/g cell pellet), add appropriate amount of DNase 1 stock solution (1 mg/mL) to a final concentration of 10 µg/mL, and add fresh 2-Mercaptoethanol (2-Me) to 10 mM final concentration. Incubate the mixture on ice for 10–15 min.
-
17
Sonicate using a tip sonicator at 70% power with ice bath (3.0 s sonication and 9.0 s break for 10 times to avoid overheating) and centrifuge the cell suspension at 45000 x g for 30 min at 4 °C.
To prevent overheating during sonication, put the container into ice during this process. Sonicate the cells thoroughly and carefully. Before centrifuge, centrifuge tubes containing cells must be balanced first.
-
18
Carefully pool supernatants together for each sample and discard the pellets.
Purification of His-tagged NLuc-E66stop
-
19
To purify NLuc-E66stop, transfer Ni-NTA resin into a clean Econo-Column with a final settled bed volume of 2 mL resin/L cell culture. Wash the resin with 10 bed volumes of buffer B containing 10 mM 2-Me.
Do not allow the resin bed to run dry at any point. Make sure that the column is always stored in liquid. Avoid introducing bubbles to the column by adding solutions carefully against the wall of the column.
-
20
Apply the pooled supernatant to the column with a speed < 1 ml/min. Collect flow-through in a container.
-
21
Wash the column with 10 bed volumes of buffer B containing 10 mM 2-Me. Collect flow-through in a container.
-
22
Elute NLuc-E66stop with buffer C (5–10 column volumes) containing 10 mM 2-Me. Collect eluted samples with 5 ml per fraction in test tubes. Measure the OD280 value of each fraction until OD280 < 0.1, using the Nanodrop spectrophotometer.
-
23
Run SDS-PAGE of eluted protein to determine the protein quality.
-
24
Concentrate the pooled protein to 5–10 ml using an EMD Millipore Amicon stirred cell Model at 4 °C.
Depending on the target protein molecular weight, choose the right size of protein filter membrane. Do not discard the flow-through immediately after concentration. Check the protein concentration with NanoDrop UV visible spectrophotometer and use the flow-through as blank. If the concentrated protein concentration is much higher than the flow-through, discard the flow-through and keep the concentrated protein. Otherwise, check the membrane and reset the Amicon device. Repeat step 23 with the flow-through.
Purification of GST-tagged GST-CLuc-NS3
-
25
To purify GST-CLuc-NS3, transfer Glutathione sepharose 4B resin into a clean Econo-Column with a final settled bed volume of 2 mL resin/L cell culture. Wash the resin with 10 bed volumes of buffer B with 10 mM 2-Me.
-
26
Apply the pooled supernatant to the column with a speed < 1 ml/min. Collect flow-through in a container.
-
27
Wash the column with 30 bed volumes of buffer B with 10 mM 2-Me. Collect flow-through in a container. Stop column flow using a 2-way stopcock.
-
28
Elute GST-CLuc-NS3 by addition of 1 ml of buffer D (with 10 mM 2-Me) per ml bed volume. Incubate the column at room temperature (22–25°C) for 10 minutes. Open the stopcock to elute GST-CLuc-NS3.
-
29
Repeat the elution and collection for 5–10 times, or until OD280 < 0.1.
-
30
Run SDS-PAGE of eluted protein to determine the protein quality.
-
31
Concentrate the pooled protein to 5–10 ml using an EMD Millipore Amicon stirred cell Model at 4 °C.
Purification using size-exclusion chromatography
-
32
Equilibrate a HiLoad Superdex 200 column with 1 column volume of buffer E with freshly-added 1 mM DTT.
-
33
Inject 5 ml of concentrated samples (NLuc-E66stop or GST-CLuc-NS3). Run size exclusion chromatography (0.5 ml/min) on an AKTA protein purification instrument.
Avoid bubbles when loading protein samples to columns.
-
34
Pool fractions containing the target protein by analyzing the peak fractions using SDS-PAGE.
-
35
Concentrate the pooled factions using an EMD Millipore Amicon stirred cell Model at 4 °C. Use a NanoDrop UV visible spectrophotometer to measure the protein concentration (> 1 mg/ml). Aliquot the concentrated proteins and store them in −80 °C freezer.
Purification of all other alternative constructs (such as His-tagged NLuc-NS2B, NS2B-NLuc, E66stop-NLuc, NLuc-E66stop mutants, “cold” MBP-NS3, “cold” NS2B, and MBP, or GST-NS3-CLuc (GNC)) will be carried out using protocols similar to the ones stated above.
BASIC PROTOCOL 2
ANALYSIS OF SLC-BASED NS2B-NS3 INTERACTION ASSAY
Although we obtain different constructions of NS2B (NLuc-NS2B, NLuc-E66stop, NS2B-NLuc, and E66stop-NLuc) and NS3 (GCN, GNC) proteins from the Basic Protocol 1 described above, the best pair needs to be identified via SLC-based NS2B-NS3 interaction assay. The pair with the strongest luminescent signal after adding luciferin substrate should be selected for further testing. Interaction specificity, dose-response, competition binding, and mutations with residues related to protease function should be studied to confirm that the detectable signal is due to the PPI (Li et al., 2017; Li et al., 2018).
Materials
NLuc-NS2B
NLuc-E66stop
NS2B-NLuc
E66stop-NLuc
GST-CLuc-NS3 (GCN)
GST-NS3-CLuc (GNC)
96-well white plates
8-channel pipette (10–100 µL)
8-channel pipette (0.1–10 µL)
12-channel pipette (0.1–10 µL)
12-channel pipette (10–100 µL)
10–100 µL pipette
Orbital shaker
Microplate Luminometer
SLC-based NS2B-NS3 interaction assay
Prepare serial 5x dilutions as stocks for each protein at the following concentrations, using buffer F as diluent: 4 µM, 2 µM, 1 µM, 0.5 µM, and 0.25 µM.
In a 96-well white plate (Figure 2A), dispense 50 µl buffer F into each well using an 8- or 12-channel pipette.
Use an 8-channel pipette, transfer 40 µl buffer E with fresh 1 mM DTT to ONLY columns 1 and 12 as illustrated.
Use an 8-channel pipette, transfer 20 µl of each stock GCN or GNC proteins (5x concentrated, 4 µM, 2 µM, 1 µM, 0.5 µM, and 0.25 µM) to columns 2–11 as illustrated to make a final concentration series of 0.8 µM, 0.4 µM, 0.2 µM, 0.1 µM, and 0.05 µM.
Use a 10–100 µL pipette, transfer 20 µl of each concentration series of stock proteins of NLuc-NS2B (rows AB), NS2B-NLuc (rows CD), E66stop-NLuc (rows EF), NLuc-E66stop (rows GH) in duplicates into columns 2–11 to match the GCN or GNC concentrations.
Incubate the mixture with gentle agitation at room temperature for 30 min.
Prepare substrate buffer H by diluting stock D-luciferin (10 mg/ml) 10-fold using buffer G and adding ATP stock (100x) to 4 mM final concentration.
Using an 8- or 12-channel pipette or injector, transfer 10 µl of buffer H to each well.
Read luminescence using a luminometer every 5 min for 2–3 hours or until the reading is maximized and plateaued.
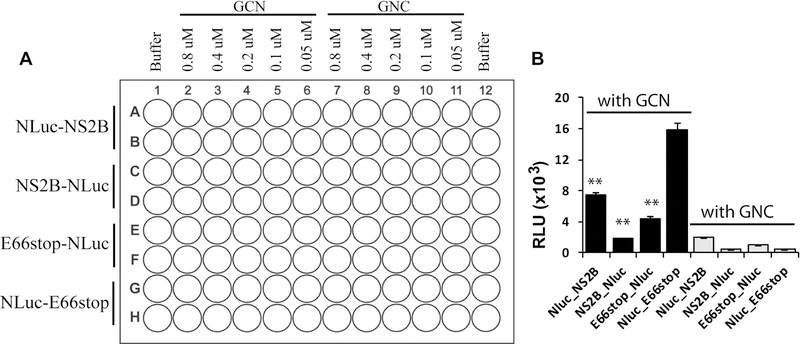
Figure 2. The DENV2 NS2B/NS3 SLC assays.
(A) Experimental design using 96-well plate. (B) Relative luminescence unit (RLU) generated by each pair of binding partners. Equal concentrations (100 nM) of each pair of NS2B/NS3 constructs were mixed and incubated with luciferin substrate. **P < 0.001. In all bar graphs, means and SD from triplicate experimental data were shown, unless otherwise specified. Figure is modified from Figure 2 in Li et al. (Li et al., 2017).
From this assay, a pair of interacting partners can be selected. First, determine the time when the readings are plateaued or maximized and stabilized. Second, choose a minimum concentration required to give readings in the range of 103 to 104 relative luminescence unit (RLU) at the maximum reading. Third, plot a figure to show the readings for different pairs at that concentration (Figure 2B). Fourth, choose the pair that shows the highest reading for further development. For example, in our case, the NLuc-E66stop and GCN pair gave the highest signal at 100 nM. Therefore, they were chosen for further analysis.
Support Protocol 1
INTERACTION SPECIFICITY ASSAY
For any PPI assay, it is important to determine the interaction specificity. As a general guide, a signal/noise ratio larger than 3 is required for any high throughput screening assay (https://ncats.nih.gov/preclinical/drugdev/assay#criteria). Although there is no specific guideline in terms of assay specificity, a 5 to 10-fold difference between signals for specific vs. non-specific bindings may be required. To achieve these, NLuc-only and CLuc-only controls should be used.
Additional Materials:
NLuc-control
GST-CLuc-control
Assay Procedure
Prepare 5x dilutions (0.5 µM) as stocks for NLuc-E66stop, GCN, NLuc-only, and GST-CLuc-only, using buffer F as diluent.
In a 96-well white plate (Figure 3A), dispense 50 µl buffer F into columns 1–6 using an 8-channel pipette.
Use an 8-channel pipette, transfer 20 µl of buffer E with fresh 1 mM DTT to columns 1–2 or each stock proteins (5x concentrated, 0.5 µM), NLuc-E66stop to columns 3–4, or NLuc-only to columns 5–6, to make final concentration of 100 nM.
Use an 8-channel pipette, transfer 20 µl of buffer E with fresh 1 mM DTT to rows A-B or each stock proteins (5x concentrated, 0.4 µM), GCN to rows C to D, and GST-CLuc-only to rows E to F, to make final concentration of 100 nM.
Incubate the mixture with gentle agitation at room temperature for 30 min.
Prepare substrate buffer H by diluting stock D-luciferin (10 mg/ml) 10-fold using buffer G and adding ATP stock (100x) to 4 mM final concentration.
Using an 8- or 12-channel pipette or injector, transfer 10 µl of buffer H to each well.
Read luminescence using a luminometer every 5 min for 2–3 hours or until the reading is maximized and plateaued.
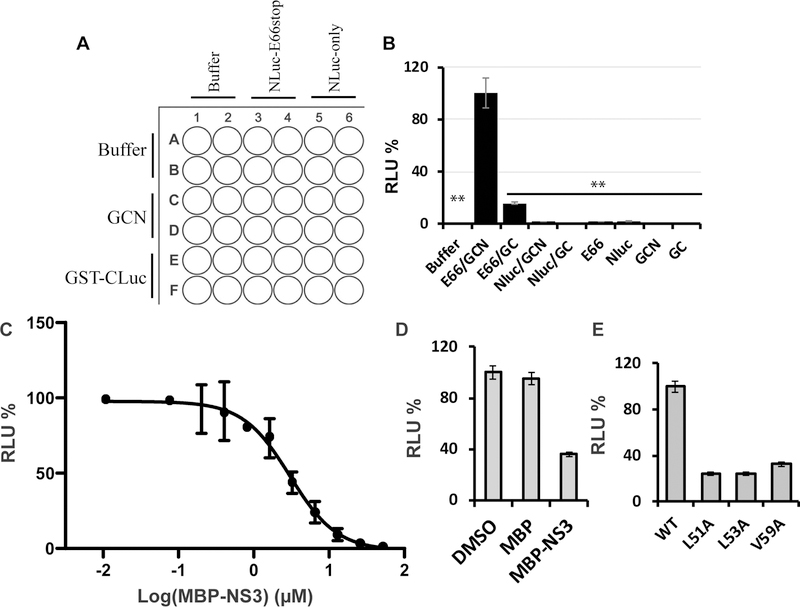
Figure 3. SLC Assay specificity.
(A) Experimental design using 96-well plate. (B) Relative luminescence unit (RLU) generated by each pair of binding partners or controls. Equal concentrations (100 nM) of each pair of NS2B/NS3 constructs or controls (or alone) were mixed and incubated with luciferin substrate. **P < 0.001. In all bar graphs, means and SDs from triplicate experimental data were shown, unless otherwise specified. (C) Dose-response inhibition of the SLC signals from NLuc-E66stop and GCN by “cold” MBP-NS3. Experimental data were fitted using the sigmoidal function with Prism GraphPad 8.1 software. (D) The DENV2 MBP-NS3 fusion protein specifically inhibited the SLC by NLuc-E66stop and GCN (80 nM each). MBP-NS3 or MBP were at 3.25 μM each. **P < 0.001. (E) NS2B mutations greatly reduced SLC. GCN was paired with equal molar of NLuc-E66stop or NLuc-E66stop mutants (L51A, L53A, and V59A). **P < 0.001. Figure is modified from Figure 2 in Li et al. (Li et al., 2017).
Average the readings for each pair and plot a figure to show interaction specificity (Figure 3B). As shown, individual fusion proteins and any pairs with control NLuc-only and GST-CLuc-only did not show significant luminescence, whereas NLuc-E66stop and GCN interaction led to robust luminescence, indicating that the luminescence signal seen from the NLuc-E666stop and GCN is specific to the interactions between NS2B and NS3.
Support Protocol 2
COMPETITION BINDING ASSAY: DOSE-RESPONSE INHIBITION USING “COLD” PREY OR BAIT
To further demonstrate interaction specificity in SLC, a competition binding assay can be performed using “cold” prey or bait proteins that are not fused to NLuc or CLuc. Here we use unfused “cold” MBP-NS3 and MBP-tag as examples. We first conduct a dose-response competition assay using “cold” MBP-NS3 as a competitor to determine the competition efficacy with IC50 defined as the concentration of the “cold” competitor at which the luminescence signal is reduced 50% (Support protocol 2). Then we compare the inhibition efficacy using an irrelevant protein such as the MBP tag at a concentration to achieve 50% inhibition (Support protocol 3).
Additional Materials:
MBP-NS3
Assay Procedure
-
1
Prepare 5x dilutions (0.4 µM) stocks for NLuc-E66stop and GCN, using buffer F as diluent.
-
2
Prepare a 10x dilution (0.8 µM) stock of Nluc-E66stop, using buffer F as diluent.
-
3
Prepare a 4x dilution (200 µM) of MBP-NS3, using buffer F as diluent.
-
4
In a 96-well white plate, dispense buffer F 30 µl into columns 2–12 (rows A-C) and 10 µl into column 1 (rows A-C) using a 12- channel pipette.
-
5
Use a 12-channel pipette, transfer 5x stock NLuc-E66stop protein (5x concentrated, 0.4 µM) 20 µl to columns 2–12 (rows A-C); transfer 10x stock NLuc-E66stop protein 40 µl to column 1 (rows A-C).
-
6
Use an 8-channel pipette, transfer 50 µl of 4x dilution (200 µM) of MBP-NS3 into column 1 (rows A-C); mix the solutions by pipetting up and down a few times.
-
7
Using an 8-channel pipet with 3 tips, transfer 50 µl column 1 mixture solutions (rows A-C) into column 2 (rows A-C); mix the solutions by pipetting up and down a few times.
-
8
Repeat 7 by transferring and mixing column 2 solutions to column 3, 3 to 4, etc., until 9 to 10, to generate 2-fold serial dilutions of the “cold” MBP-NS3 competitor; remove and discard 50 µl of mixture from the final dilution in column 10.
-
9
Incubate the mixture with gentle agitation at room temperature for 1 hour.
-
10
Using an 8- or 12-channel pipette, transfer 20 µl of buffer F into column 1–12 (rows A-C).
-
11
Using an 8- or 12-channel pipette, transfer 20 µl of stock GCN protein (5x concentrated, 0.4 µM).
-
12
Incubate the mixture with gentle agitation at room temperature for 30 min.
-
13
Prepare substrate buffer H by diluting stock D-luciferin (10 mg/ml) 10-fold using buffer G and adding ATP stock (100x) to 4 mM final concentration.
-
14
Using an 8- or 12-channel pipette or injector, transfer 10 µl of buffer H to each well (rows A-C).
-
15
Read luminescence using a luminometer every 5 min for 2–3 hours or until the reading is maximized and plateaued.
Using Prism GraphPad or Origin to process the dose-response data to determine the inhibition IC50 (Figure 3C). As shown, MBP-NS3 inhibited the NS2B-NS3 interactions in a dose-dependent manner with an IC50 value of about 2.5 µM.
Support Protocol 3
COMPETITION BINDING ASSAY: COMPARE INHIBITION BY MBP-NS3 AND IRRELEVANT MBP-TAG
To determine the specificity of the competition binding inhibition assay, we compared the decrease in luminescence from incubation with MBP-NS3 to the MBP tag alone. To simplify this assay, we compared the inhibition by MBP-NS3 and MBP-tag at a fixed concentration, at which 50% or more inhibition could be seen for the specific binder. We chose a concentration of about 3.25 µM, which will lead to approximately 50% inhibition by MBP-NS3.
Additional Materials:
MBP
Assay Procedure
-
16
Prepare 5x dilutions (0.4 µM) as stocks for NLuc-E66stop and GCN, using buffer F as diluent.
-
17
Prepare a 5x dilution (16.25 µM) of MBP-NS3 and MBP, using buffer F as diluent.
-
18
In a 96-well white plate, dispense buffer F 30 µl into columns 1–3 (rows A-C), using an 8-channel pipette.
-
19
Use an 8-channel pipette, transfer 5x stock NLuc-E66stop protein (5x concentrated, 0.4 µM), 20 µl to columns 1–3 (rows A-C).
-
20
Use an 8-channel pipette, transfer 20 µl of buffer F into column 1 (rows A-C), or 20 µl of 5x dilution (16.25 µM) of MBP-NS3 into column 2 (rows A-C), or 20 µl of 5x dilution (16.25 µM) of MBP into column 3 (rows A-C); mix the solutions by pipetting up and down a few times.
-
21
Incubate the mixture with gentle agitation at room temperature for 1 hour.
-
22
Using an 8-channel pipette, transfer 20 µl of stock GCN protein (5x concentrated, 0.4 µM).
-
23
Incubate the mixture with gentle agitation at room temperature for 30 min.
-
24
Prepare substrate buffer H by diluting stock D-luciferin (10 mg/ml) 10-fold using buffer G and adding ATP stock (100x) to 4 mM final concentration.
-
25
Using an 8-channel pipette or injector, transfer 10 µl of buffer H to each well (rows A-C).
-
26
Read luminescence using a luminometer every 5 min for 2–3 hours or until the reading is maximized and plateaued.
Average the readings for each triplicate and plot a figure to show interaction specificity (Figure 3D). As shown, MBP-NS3 at 3.25 µM significantly inhibited the NS2B-NS3 interaction, whereas the maltose-binding protein (MBP) fusion tag did not show any inhibition.
Support Protocol 4
SLC-BASED NS2B-NS3 INTERACTION ASSAY WITH MUTATIONS IN NS2B KNOWN TO DISRUPT NS2B-NS3 INTERACTIONS
To further demonstrate interaction specificity in SLC, fusion proteins with mutations known to disrupt the prey-bait interactions can be used. In our NS2B-NS3 system, a few NS2B mutants (L51A, L53A, and V59A) are known to be essential for both NS2B-NS3 interactions and NS2B-NS3 protease activities (Chappell et al., 2008). Here, we will use these mutant fusion proteins to further demonstrate the SLC specificity.
Additional Materials:
NLuc-E66stop-L51A
NLuc-E66stop-L53A
NLuc-E66stop-V59A
Assay Procedure
Prepare 5x dilutions (0.4 µM) as stocks for NLuc-E66stop, NLuc-E66stop-L51A, NLuc-E66stop-L53A, NLuc-E66stop-V59A, GCN, using buffer F as diluent.
In a 96-well white plate (Figure 1), dispense 50 µl buffer F into columns 1–4 (rows A-C to make triplicates) using an 8-channel pipette.
Use an 8-channel pipette, transfer 20 µl of each of the stock proteins (5x concentrated, 0.4 µM) NLuc-E66stop, NLuc-E66stop-L51A, NLuc-E66stop-L53A, and NLuc-E66stop-V59A into columns 1 to 4 (rows A-C), respectively. The final concentration for these proteins will be 80 nM.
Use an 8-channel pipette, transfer 20 µl of stock GCN protein (5x concentrated, 0.4 µM) into rows A to C to make final concentration of 80 nM.
Incubate the mixture with gentle agitation at room temperature for 30 min.
Prepare substrate buffer H by diluting stock D-luciferin (10 mg/ml) 10-fold using buffer G and adding ATP stock (100x) to 4 mM final concentration.
Using an 8-channel pipette or injector, transfer 10 µl of buffer H to each well.
Read luminescence using a luminometer every 5 min for 2–3 hours or until the reading is maximized and plateaued.
Average the readings for each pair and plot a figure to show interaction specificity (Figure 3E). As shown, mutations of the NS2B residues L51, L53, and V59, known to be essential for protease function (Chappell et al., 2008), significantly reduced the SLC (Figure 3E). All these data indicate that the SLC signal from NLuc-E66stop/GCN pair is specific to the interactions between NS2B and NS3.
Reagents and Solutions
LB broth per liter:
10 g tryptone
5 g yeast extract
5 g NaCl
1 ml 1 N NaOH
Autoclave
Store up to 1 month at room temperature
LB plates Per liter:
10 g tryptone
5 g yeast extract
5 g NaCl
1 ml 1 N NaOH
15 g agar Autoclave
Store up to 1 month at 4°C
COMMENTARY
Background Information
The SLC assay was first used as an optical probe to detect PPI in mammalian cells and in live animals (Ozawa et al., 2001; Paulmurugan et al., 2002). Since then, this assay has been used widely to visualize and quantify PPI in biological sciences, including botany, microbiology, proteomics, pathology, immunology, etc. (Chen et al., 2008; Hashimoto et al., 2011; Luker et al., 2003; Sasaki et al., 2018). Most of the SLC assays as described in previous literatures were performed in living cells where the PPI naturally happens. However, some PPIs may affect cell cycle or even worse, poison cells. Therefore, cell toxicity needs to be considered when designing the PPI assay. Currently, five luciferase proteins have been applied to SLC assays: firefly (Photinus pyralis) (Chen et al., 2008), Renilla (Renilla reniforms) (Fujikawa and Kato, 2007), Click beetle (Pyrophorus plagiophthalamus) (Misawa et al., 2010), NanoLuc (Sasaki et al., 2018), and Gaussia (Gaussia Princeps) (Hashimoto et al., 2011) luciferase. Firefly (61kDa) luciferase, which catalyzes oxidation of luciferin and emits light of 560 nm, has been used in SLC assays for more than 15 years in different areas of biology (Ozawa et al., 2001).
There are four main advantages to using an SLC assay. First, this assay can be used in a large-scale format. In our study, the SLC assay for NS3 and NS2B protein pair was initially established to discover inhibitors via high throughput screening (Li, Brecher, Deng, Zhang, Sakamuru, Liu, Huang, Koetzner, Allen and Jones, 2017). Second, SLC assays generate low background signals because they do not require external light. Unlike fluorescent-based protein complementation assays, in which excitation light (lower wavelength) is required to generate emission light (higher wavelength), luciferase in SLC assay oxidizes its chemical substrate and subsequently emits detectable light known as bioluminescence. Third, the sensitivity of SLC assay is high. As the light signal is emitted only when PPI happens, this assay possesses the binary ‘on or off’ characteristic which increases the detection sensitivity. Last, SLC assays can detect both association and dissociation of protein pairs (Paulmurugan et al., 2004),while bimolecular fluorescence complementation assays can only detect protein association but not dissociation because the complemented fluorescent protein can only stably emit a detectable signal once the PPI happens (Fujikawa and Kato, 2007).
As any other assays, SLC assay has several limitations as well. First, an exogenous substrate is required to generate the bioluminescence. Therefore, there is a small chance of obtaining false negative results in the presence of a luciferase inhibitor (Braeuning, 2015; Leitão et al., 2010). Second, unlike the stable fluorescent protein, which can produce detectable signal stably, the bioluminescence signals in SLC assay gradually reduce since the luciferase and luciferin reaction is a chemical reaction. Therefore, this assay is not suitable to monitor PPI for long time. Last, as the minimum physical distance between the split luciferase pair that emits an interaction signal is unknown, currently this assay cannot be used to measure physical distances between a PPI protein pair. Instead, a fluorescence resonance energy transfer assay is suitable to achieve this goal.
Critical Parameters
The key to perform the SLC assay is obtaining functional high-quality protein pair with a split luciferase tag that has not degraded, precipitated, oxidized, or otherwise been damaged. To achieve this goal, different protein constructions should be meticulously designed before starting these experiments. After proteins have been expressed, quality must be monitored via SDS-PAGE. As the bioluminescence signals might vary among experiments, it is important to use the same positive and negative controls in all experiments.
Troubleshooting
To perform the transformation successfully, always keep an ice bucket beside the reaction. To obtain the proteins expressed, sonicate the E. Coli cells thoroughly without overheating. After sonication, always put the protein mixture on ice between centrifugations to minimize protein degradation. To monitor the protein quality, SDS-PAGE must be performed. After proteins have been expressed and purified, quantify with nanodrop and freeze in −80 °C after aliquot.
One common problem in split luciferase complementation assay is that high background signal can be detected in some cases (Ataei et al., 2013; Luker et al., 2004; Misawa et al., 2010). It has been suggested that the high background signal arises from overlapping the luciferase fragments between NLuc and CLuc (Ataei et al., 2013; Luker et al., 2004; Misawa et al., 2010). To minimize background noise, constructs with different linkers and luciferase splitting sites should be constructed.
In addition, the proper controls are an important part of reporter assays. The background issue is related to negative control. It is also important to include a positive control to develop split luciferase complementation-based assay to detect protein-protein interactions. Essentially, any known binding pairs could be served as positive controls.
Statistical Analyses
All experiments were performed in triplicates. One-way analysis of variance (ANOVA) was used to do statistical analyses with Prism software.
Understanding Results
These assays generate a stable method to detect PPI between a protein pair that can be used as a tool to find PPI inhibitors in a large-scale way. Herein, we used NS3 and NS2B proteins as an example to study PPI. For the SLC assay, we expect the data is comparable among experiments. However, the bioluminescence signals may vary among different experiments. Therefore, same negative and positive controls should be included in each experiment so data can be normalized to the same scale based on these controls.
Time Considerations
E. Coli transformation takes at least 2 days. Protein expression and purification takes at least 2 weeks, depending on the number of constructions. Each SLC-based interaction assay, SLC-based dose-response test, and competition binding assay can take up to 2 h, respectively, depending on the number of samples.
ACKNOWLEDGEMENTS
(mandatory for NIH, optional for all others)
This work was partially supported by NIH grants AI131669, AI140726, AI141178, AI140491, AI133219, and AI134568.
LITERATURE CITED
- Ataei F, Torkzadeh-Mahani M, Hosseinkhani S 2013. A novel luminescent biosensor for rapid monitoring of IP3 by split-luciferase complementary assay. Biosens Bioelectron, 41: 642–648. [DOI] [PubMed] [Google Scholar]
- Braeuning A, 2015. Firefly luciferase inhibition: a widely neglected problem. Arch Toxicol, 89: 141–142. [DOI] [PubMed] [Google Scholar]
- Chappell KJ, Stoermer MJ, Fairlie DP and Young PR 2008. Mutagenesis of the West Nile virus NS2B cofactor domain reveals two regions essential for protease activity. J. Gen. Virol, 89: 1010–1014. [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X and Zhou JM 2008. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol, 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardins E, Ruffing N, Wu L, Wyatt R, Gerard N, Gerard C, and Sodroski J 1998. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J. Virol, 72: 1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa Y and Kato N 2007. TECHNICAL ADVANCE: Split luciferase complementation assay to study protein–protein interactions in Arabidopsis protoplasts. Plant J, 52: 185–195. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Adams KW, Fan Z, McLean PJ and Hyman BT, 2011. Characterization of oligomer formation of amyloid-β peptide using a split-luciferase complementation assay. J. Biol. Chem, 286: 27081–27091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitão JM and da Silva JCGE 2010. Firefly luciferase inhibition. J. Photochem. Photobiol. B: Biol, 101: 1–8. [DOI] [PubMed] [Google Scholar]
- Li Z, Brecher M, Deng Y, Zhang J, Sakamuru S, Liu B, Huang R, Koetzner CA, Allen CA, Jones SA, Chen H, Zhang N, Tian M, Gao F, Lin Q, Banavali N, Zhou J, Boles N, Xia M, Kramer LD, Qin C and Li H 2017. Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction. Cell Res, 27: 1046–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang J and Li H 2018. Flavivirus NS2B/NS3 Protease: Structure, Function, and Inhibition, In Viral Proteases and Their Inhibitors (Eds Gupta SP). pp. 163–188. Elsevier (Academic Press), San Diego, CA, USA. [Google Scholar]
- Luker GD, Sharma V and Piwnica-Worms D 2003. Noninvasive imaging of protein-protein interactions in living animals, In Handbook of Proteomic Methods (Eds Conn PM). pp. 283–298. Humana Press, Totowa, NJ, USA. [Google Scholar]
- Luker KE1, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. 2004. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci U S A, 101:12288–12293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa N, Kafi A, Hattori M, Miura K, Masuda K and Ozawa TJ 2010. Rapid and high-sensitivity cell-based assays of protein− protein interactions using split click beetle luciferase complementation: an approach to the study of G-protein-coupled receptors. Anal. Chem, 82: 2552–2560. [DOI] [PubMed] [Google Scholar]
- Ozawa T, Kaihara A, Sato M, Tachihara K and Umezawa YJ 2001. Split Luciferase as an Optical Probe for Detecting Protein− Protein Interactions in Mammalian Cells Based on Protein Splicing. Anal. Chem, 73: 2516–2521. [DOI] [PubMed] [Google Scholar]
- Panga V and Raghunathan S 2018. A cytokine protein-protein interaction network for identifying key molecules in rheumatoid arthritis. PLos One, 13: e0199530 10.1371/journal.pone.0199530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulmurugan R, Massoud TF, Huang J and Gambhir SS 2004. Molecular imaging of drug-modulated protein-protein interactions in living subjects. Cancer Res, 64: 2113–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulmurugan R, Umezawa Y and Gambhir SJ 2002. Noninvasive imaging of protein–protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc. Natl. Acad. Sci. U.S.A, 99: 15608–15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JR, Stains CI, Jester BW and Ghosh IJ 2008. A general and rapid cell-free approach for the interrogation of protein− protein, protein− DNA, and protein− RNA interactions and their antagonists utilizing split-protein reporters. J. Am. Chem. Soc, 130: 6488–6497. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Anindita PD, Phongphaew W, Carr M, Kobayashi S, Orba Y and Sawa HJ 2018. Development of a rapid and quantitative method for the analysis of viral entry and release using a NanoLuc luciferase complementation assay. Virus Res, 243: 69–74. [DOI] [PubMed] [Google Scholar]
- Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR and Katze MG 2012. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev, 76: 16–32. [DOI] [PMC free article] [PubMed] [Google Scholar]