Abstract
Background
While the prognostic relevance of lymphovascular invasion (LVI) in breast cancer is well known, its molecular biology is poorly understood. We hypothesized that pathologically determined LVI reflects molecular features of tumors and can be discerned from their genomic and transcriptomic profiles.
Methods
LVI status and Nottingham histological scores of primary breast tumors of The Cancer Genome Atlas (TCGA) project were assessed from pathology reports; other clinical and molecular data were obtained from TCGA data portals and publications. Two independent datasets (GSE5460 and GSE7849) were combined and used for validation.
Results
LVI status was determinable for 639 and 196 cases of the TCGA and validation cohorts, among whom LVI incidence was 37.8% and 37.2%, respectively. LVI was associated with high tumor Ki67 expression, advanced pathologic stage, and high Nottingham scores. LVI-positive cases had worse overall and progression-free survival regardless of cancer subtype. Surprisingly, in both cohorts, LVI was not associated with lymphangiogenesis or lymphatic vessel density as estimated from tumor expression of lymphatic endothelium-associated genes. LVI-positive tumors had higher genome copy number aberrations, aneuploidy, and homologous recombination defects, but not single nucleotide variations or intra-tumor genome heterogeneity. Tumor immune cell composition and cytolytic activity was not associated with LVI status. On the other hand, expression of cell proliferation-related genes was significantly increased in LVI-positive tumors.
Conclusions
Our study suggests that breast cancer with LVI is a highly proliferative cancer, and it does not correlate with gene expression markers for lymphangiogenesis or immune response.
Keywords: breast cancer, cell proliferation, lymphatic invasion, lymphovascular invasion, transcriptome
Background
The association of pathological features of cancer and tumor microenvironment, a battlefield between cancer cells and host stroma including immune system, has been attracting a lot of attention. Lymphovascular invasion (LVI) is a pathological determination that cancer cells have infiltrated into blood or lymphatic vessels in or around tumors. LVI is thought to lead to cancer dissemination through lymphatic vessels [1,2]. It is associated with worse clinical and pathological features [3,4], and is predictive of worse survival.
LVI was reported to have strong correlation with increased lymphatic vessel density (LVD) in a recent meta-analysis of 12 studies [5]. LVD, whose determination requires immunohistochemical staining of lymphatic vessels, has been the gold standard to quantify lymphangiogenesis, the generation of new lymphatic vessels. Lymphatic endothelial cell (LEC) markers such as prospero homeobox protein 1 (PROX1), podoplanin (PDPN/D2–40), vascular endothelial growth factor C (VEGFC) [6,7], and lymphatic vessel endothelial hyaluronan receptor 1 (LYVE1) [8] have been used for immunohistochemical staining of lymphatic vessels [9]. Increased LVD and expression of LEC markers have been shown to correlate with metastasis and poor prognosis [2,10,11]. It has therefore been thought that lymphangiogenesis associates with LVI, and spreading of cancer cells through the lymphatic vessels contributes to distant metastasis or tumor recurrence and worse outcome [12]. In agreement, our group has reported that sphingosine-1-phosphate (S1P) generated by sphingosine kinase 1 (SPHK1) is a mediator of lymphangiogenesis [13] and linked to inflammation-related metastasis in breast cancer [13], and demonstrated that S1P-high breast cancer is associated with increased lymph node metastasis [14]. Lymphatic vessels also have an important role in conveying tumor antigens and immune cells from peripheral tissues to lymph nodes. The anti-cancer immune response includes transportation of tumor antigens to lymph nodes by dendritic cells, expansion of antigen-recognizing naïve T cells, and infiltration of the matured T cells into tumors [1]. In this regard, LVI may be expected to enhance tumor-associated immunity.
While the clinical relevance of LVI in breast cancer has been studied well, the molecular biology of LVI is still largely undefined. We hypothesized that pathologically determined LVI in breast cancer may reflect an aggressive cancer biology characterized by features such as enhanced lymphangiogenesis, suppressed cancer immunity, and accelerated cell proliferation, and that such features can be discerned from the genomic and transcriptomic profiles of tumors. We examined treatment-naïve primary breast tumors of The Cancer Genome Atlas (TCGA) project for this purpose. Data from two independent, previously published studies was combined and used for validation.
Materials and Methods
Examination of pathology reports of the TCGA-BRCA project
As detailed in supplementary Text S1, the Text Information Extraction System (TIES) Cancer Research Network [15] was used to obtain LVI status and Nottingham histological system scores from pathology reports of hematoxylin-eosin (H&E)-stained tissue sections of 1,046 cases. A determinable LVI status could be recorded for 639 cases, with LVI present in 242 and absent in 397. The groups of LVI-determinable and non-determinable cases did not significantly differ for age, lymph node status, or positivity for estrogen receptor (ER), human epidermal growth factor receptor 2 (HER2), or progesterone receptor (PR) expression (Table S1). However, the two groups had modest but significant differences for overall surviving fraction (88.7% vs. 82.4%) and tumor size distribution (both Fisher exact test p < 0.05; Table S1). At least two types of histological scores could be recorded for 625 cases. Collected LVI status and histological scores are in supplementary Table S2.
Data of the TCGA-BRCA cohort
Tumor gene expression data was generated from mapped RNA sequencing counts obtained from Genome Data Commons portal of National Cancer Institute, USA (Text S1). Survival data was from the TCGA PanCancer Clinical Data Resource [16]. Other clinical information and gene mutation status were obtained from the cBio Cancer Genomics Portal [17] in late 2017. Tumor aneuploidy, copy number alteration (CNA), genome heterogeneity, and homologous recombination defect (HRD) values were from Thorsson et al [18]. All and APOBEC-mediated single nucleotide mutation counts of tumors were estimated using TCGA’s public MAF (mutation annotation format) data [19] with Helmsman [20] software (1.1.0) and deconstructSigs [21] Bioconductor package (1.8.0). Immune-related measurements like T cell receptor diversity and relative immune cell fractions estimated from tumor gene expression by CIBSERSORT method [22] were from Thorsson et al [18]. Immune cytolytic activity was calculated from tumor gene expression as CYT score [23].
Data of the validation cohort
We identified two studies that collected information on both LVI status and gene expression of primary breast cancer tumors. Both studies, with cohorts of 128 [24] and 68 [25] patients, used H&E-stained tissue sections for LVI assessment. All samples of the former cohort and about half of the latter were therapy naïve. Clinical and gene expression data were obtained from Gene Expression Omnibus (accession numbers GSE5460 and GSE7849). Gene expression data of the two studies were merged using ComBat (3.30.1) [26] as detailed in Text S1.
Curation of endothelium-specific gene-sets
A set of 88 genes whose expression was determined to be > 20x higher in lymphatic compared to vascular endothelia was collected from six studies [27–32] (Text S3). A 45-gene lymphatic-specific gene-set was generated by combining the six lymphatic endothelial cell signatures defined in the xCell cell subset enumeration method [33]. General endothelium and microvascular endothelium-specific sets with 174 and 359 genes were similarly generated using nine and 11 xCell signatures, respectively. Supplementary Table S3 lists the gene-set members.
Analyses of gene expression data
Unless noted otherwise, log2-transformed normalized gene expression measurements were used. For t-distributed stochastic neighbor embedding (tSNE), the t-SNE R package (0.15) with perplexity of 30 was used. GSEA (3.0) and GSVA Bioconductor package (1.30.0) with the gene-set variation analysis (GSVA) method [34] were used for gene set enrichment analysis [35] of Hallmark [36] (50 sets) and C2 CP:Reactome [37] (674 sets) gene set collections of the Molecular Signatures Database (mSigDB 6.2). Categories of Reactome sets were the top-level pathway node in the Reactome pathway hierarchy [37]. Raw, gene-level, mapped sequencing read count values and Poisson distribution were used for GSVA analysis of TCGA tumors; log2-transformed normalized values and the Gaussian distribution were used for others. Two-group comparison of tumor GSVA scores as well as of gene expression were performed using an empirical Bayes-moderated t test as implemented in limma Bioconductor package (3.38.2) [38], with the Benjamini-Hochberg method used for false discovery control.
Statistical analyses
Statistical analyses and data plotting were performed using R (3.6) and Prism (7.0d; GraphPad Software®, San Diego, USA). Unless noted otherwise, Fisher’s exact and equal-variance t tests were respectively used in group comparisons for categorical and continuous variables, and a threshold of 0.05 was used to deem significance from p values of statistical tests. For survival analyses, performed with survival (2.44) and greyzoneSurv (1.0) packages in R, Cox proportional hazard model and Kaplan-Meier method with log-rank test were used. The glmer function in lme4 package for R (1.1.21) was used for multivariate analysis with age as random effect.
Results
LVI associates with worse survival in both TCGA and validation cohorts
Among patients with determinable tumor LVI status, LVI was present in 242 (37.8 %) and 73 (37.2 %) cases of the TCGA and validation cohorts, respectively, and absent in the others. The incidence of LVI was not associated with age (< 50 vs. ≥ 50 years) or ethnicity (Caucasian vs. other). TCGA patients with LVI-positive tumors had shorter progression-free interval (p = 0.044, hazard ratio [HR] = 2.3), as well as worse overall survival (OS; p = 0.002, HR = 3.2; Fig. 1a). Incidence of LVI had no association with tumor status for ER, HER2, or PR expression as determined by immunohistochemical staining. On the other hand, LVI was about 2-fold more frequent among HER2-positive compared to -negative tumors in the validation cohort (p = 0.01; Fig. 1b). OS was consistently worse for LVI-positive TCGA patients regardless of ER, HER2, or PR status in TCGA cohort (Fig. 1c), which is in agreement with the previous reports [39,40]. Survival analyses by receptor subtype was not performed for the validation cohort because of limited availability of survival data.
Figure 1.
Lymphovascular invasion (LVI) in primary tumors of TCGA-BRCA cohort. a Kaplan-Meier survival plots along with log-rank test p and hazard ratio (HR) values are shown for association of LVI with progression-free and overall survival. b LVI incidence rates (fractions) as per tumor status for estrogen (ER), HER2, and progesterone (PR) receptors are depicted with barplots. P values were determined with Fisher’s exact test. c Kaplan-Meier survival plots along with log-rank test p values are shown for overall survival among LVI-positive and -negative cases of the cohort sub-grouped by ER, HER2, and PR status.
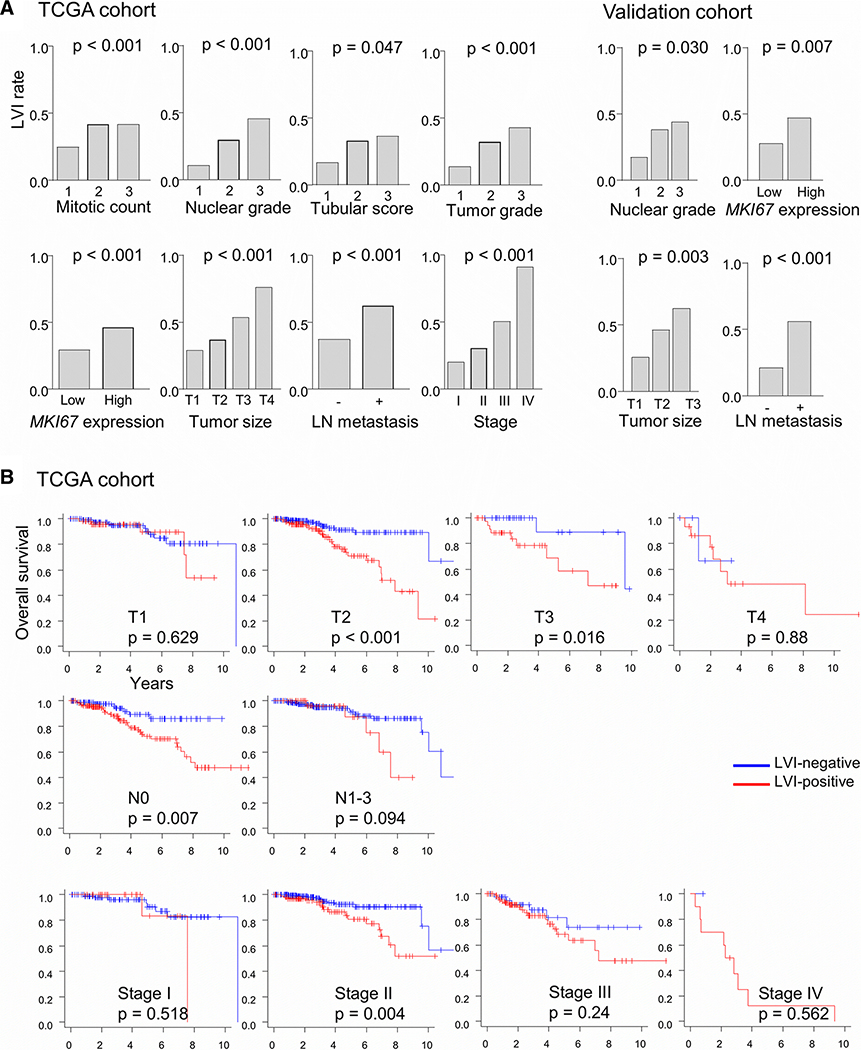
LVI associates with aggressive clinical features in both TCGA and validation cohorts
Tumors with high histological grade, high scores for mitotic rate, nuclear grade, or tubular formation, and increased MKI67 gene expression had higher frequency of LVI in the TCGA cohort. Similar observations were obtained with the validation cohort (Fig. 2a). Interestingly, the association of LVI with significantly worse OS was seen only in patients with T2 or T3 (p < 0.001 and 0.016, respectively), N0 (p = 0.007), and stage II (p = 0.004) disease, but not with the other clinical stages (Fig. 2b). These results suggest that the survival impact of LVI is largest for cancer of intermediate aggressiveness.
Figure 2.
Lymphovascular invasion (LVI) and clinical and pathological features. a LVI incidence rates (fractions) in the TCGA-BRCA and validation cohorts are plotted for different features. P values were determined with Fisher’s exact test. Median value was used to classify tumors into two groups by their MKI67 gene expression. LN, lymph node. b Kaplan-Meier survival plots along with log-rank test p values are shown for overall survival among LVI-positive and -negative cases of the TCGA cohort sub-grouped by TNM T and N status and pathologic stage.
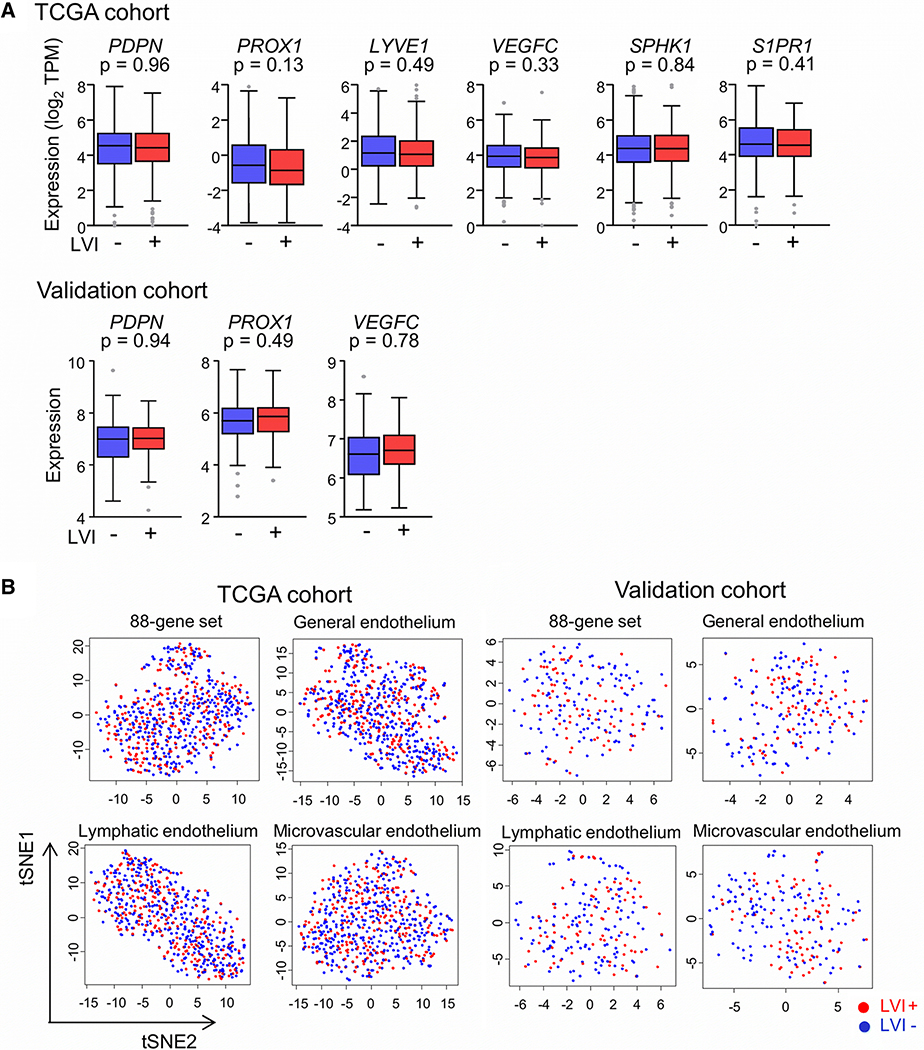
LVI is not associated with tumor expression of lymphatic endothelial cell (LEC)-specific genes
It has been reported that LVI has a strong positive correlation with intratumoral LVD [2,5]. We therefore expected that tumors with LVI will have a higher expression of LEC marker genes. However, the expression levels of four well-known marker genes, PDPN, PROX1, LYVE1 and VEGFC, or of the S1P-related genes SPHK1 and S1PR1 were not statistically different between tumors with and without LVI in any cohort (Fig. 3a). In order to further clarify the association between LVI and LVD, we obtained a set of 88 genes with high specificity for expression in LECs through a review of previously published studies. A second such set was generated using 45 genes that constitute LEC-specific gene signatures used with the xCell method to enumerate cell subsets from transcriptomes [33]. Examination of the TCGA tumors for expression of these two LEC-specific gene sets did not reveal a difference between LVI-positive and -negative cases. Of the 88-gene set, only one gene (STK26) was identified as differentially expressed, with FDR < 0.05 and fold-change > 1.2. For the 45-gene xCell set, no gene was identified as differentially expressed. Clustering analyses of the TCGA tumors by their gene expression for the two sets also did not reveal any difference between the LVI-positive and -negative cases (Fig. 3b).We also examined the tumors for clustering by expression of 174 general endothelium-specific or 359 microvascular endothelium-specific genes of the xCell signatures. As observed for two LEC-specific gene sets, clustering by LVI status was also absent for these two gene sets in both cohorts (Fig. 3b). These findings indicate that the density of lymphatic or blood microvessels in tumor is not associated with LVI.
Figure 3.
Association of lymphovascular invasion (LVI) with tumor expression of lymphatics-associated genes. a Tukey boxplots of expression of six lymphatics-associated genes in groups of LVI-positive and -negative tumors are shown for the TCGA-BRCA and validation cohorts. P values in two-group comparison with standard t test are shown. Expression measurements were unavailable for the validation cohort for three genes. TPM, transcripts per million. b Clustering of LVI-positive and -negative cases of the TCGA-BRCA and validation cohorts by expression of genes of the 88-gene or the three xCell endothelium-specific gene sets. T-distributed stochastic neighborhood embedding (tSNE) was used to generate the two dimension values that are plotted.
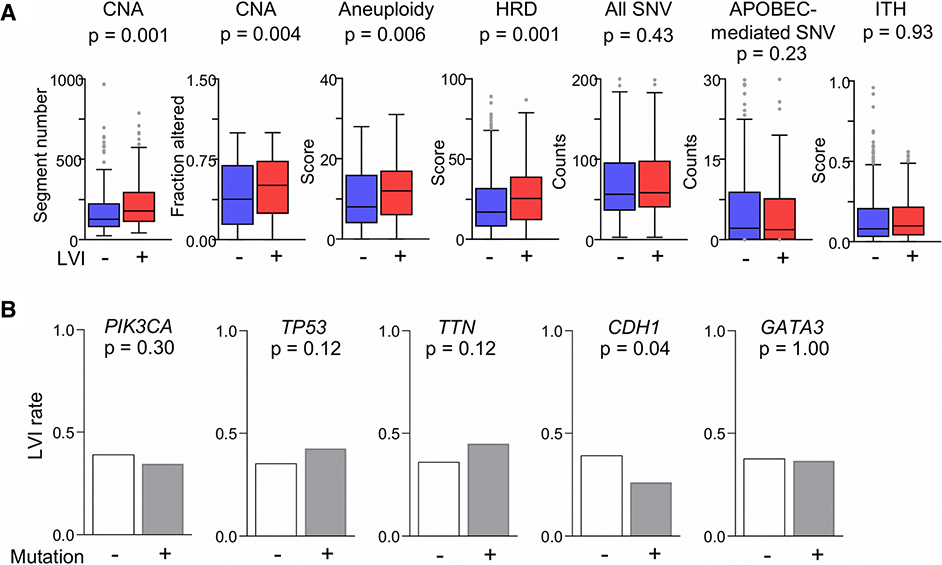
LVI is more prevalent in tumors with higher genome instability and tumors without CDH1 mutation
Given the association of LVI with clinically aggressive features, we expected that LVI will associate with higher genome instability. Copy number alterations (both segment numbers and fractions altered [18]) and aneuploidy were more prevalent in LVI-positive compared to -negative tumors in the TCGA cohort (Fig. 4a). Defects in homologous recombination, a mechanism to repair double-strand breaks in DNA during cell cycle [41], were also higher in tumors with LVI. Burden of single nucleotide variation, either all or those likely caused by APOBEC DNA deaminases, and intra-tumor clonal heterogeneity were not different between LVI-positive and -negative tumors (Fig. 4a). We also examined the association between LVI and the ten most commonly mutated genes in TCGA-BRCA primary tumors (Table S4). Other genes were excluded because of low mutation frequencies. LVI rate was significantly lower in tumors with CDH1 mutation (p = 0.04; Fig. 4b). Interestingly, CDH1 mRNA levels in LVI-positive tumors were significantly higher (∼1.4x) compared to -negative tumors in both TCGA and validation cohorts (data not shown).
Figure 4.
Association of lymphovascular invasion (LVI) with genomic alterations in primary tumors of TCGA-BRCA cohort. a Tukey boxplots of values for the LVI-positive and -negative cases are shown for various cancer genome characteristics. P values in two-group comparisons were calculated with standard t tests. CNA, copy number alteration; HRD, homologous recombinant defect; ITH, intra-tumor heterogeneity; SNV, single nucleotide variation. b The barplots show LVI incidence rates among tumors with and without mutations in five genes that are most frequently mutated in the cohort. Fisher’s exact test was used to determine the p values.
Immune microenvironments of tumors with and without LVI are similar
There was no significant difference between the tumors for not only innate immune cells such as dendritic cells and macrophages but also for adaptive immune cells like memory B and regulatory T cells. Only the activated dendritic cell fraction was significantly higher among LVI-positive tumors in the TCGA cohort (p = 0.02; Fig. 5a). However, this was not observed for the validation cohort (Fig. 5b). Tumor immune cytolytic activity (CYT score) as well as B or T cell receptor diversity (Shannon’s score), were also not different between LVI-positive and -negative tumors (Fig. 5a). Our findings suggest that the immune microenvironment is not associated with LVI.
Figure 5.
Association of lymphovascular invasion (LVI) with immune characteristics of primary tumors of a TCGA-BRCA and b validation cohorts. Tukey boxplots of values for relative fractions of tumor-infiltrating immune cells of different types, tumor immune cytolytic activity, and B (BCR) and T cell receptor (TCR) diversity in the tumor microenvironment are shown for the LVI-positive and -negative cases of the two cohorts, respectively. Measurements of some features were unavailable for the validation cohort. The p values in two-group comparisons were calculated with standard t test. NK, natural killer.
Tumors with LVI have enriched expression of genes associated with cell proliferation
Due to the association of aggressive clinical characteristics with LVI, we hypothesized that cellular processes of cancer progression such as cell proliferation, cell invasion, angiogenesis, and epithelial-mesenchymal transition are enhanced in tumors with LVI. To test this, we compared LVI-positive and -negative tumors for enriched expression of mSigDb Hallmark and Reactome gene sets using both classical GSEA and GSVA methods. In GSEA analysis of the TCGA cohort, 12 Hallmark sets were significantly enriched among LVI-positive tumors (FDR < 0.25, normalized enrichment score > 2). Four of these sets are related to cell proliferation: E2F targets, G2M checkpoint, mTORC1 signaling, and Myc targets V1. Enrichment plots for these gene sets are shown in Fig. 6a. For Reactome, enrichment for LVI-positive tumors was observed for 107 sets in the TCGA cohort and 16 sets in the validation cohort. Respectively, 44 (41.1%) and 7 (43.8%) of the enriched gene sets belong to categories of either cell cycle or DNA replication (Fig. 6b). GSEA results were validated with the GSVA method. Enrichment for LVI-positive tumors was seen in the TCGA and validation cohorts, respectively, for 17 and 6 Hallmark sets, and 201 and 38 Reactome sets. Of the Reactome sets, 56 (27.8 %) and 15 (39.4%) respectively are of cell cycle or DNA replication category. Examples of ridgeline plots of GSVA scores for some of the Hallmark gene sets with enrichment in the TCGA cohort are shown in Fig. 6c. Gene sets with significant enrichment in LVI-positive tumors as per the GSVA method and for both cohorts are listed in Tables S5 and S6, and categories of the Reactome sets are shown in Fig. 6d. Enrichment for gene sets related to cancer development processes such as angiogenesis or epithelial mesenchymal transition, or enrichment for the four endothelium-related gene sets of this study was not observed for any cohort with either GSEA or GSVA methods. In a multivariate analysis with a logit-linked generalized linear mixed model using the various cell proliferation-related variables that we had thus far identified to be associated with LVI, significance was seen only for tumor size (p = 0.01) and Nottingham mitotic score (p = 0.02) but not MKI67 gene expression (p = 0.79) or GSVA scores for proliferation-related gene sets such as E2F targets (p = 0.19). This indicates that the gene expression indicators for proliferation are secondary to tumor size and mitoses.
Figure 6.
Gene set enrichment comparison of primary tumors with and without lymphovascular invasion (LVI). a Examples of enrichment plots obtained with the classical gene set enrichment analysis (GSEA) method for mSigDb Hallmark gene sets for the TCGA-BRCA cohort. NES, normalized enrichment score. b Pie charts show the categories of mSigDb Reactome gene sets for which significant enrichment was observed in LVI-positive compared to -negative tumors in analysis with GSEA method for TCGA-BRCA and validation cohorts. c Ridgeline plots of gene set variation analysis (GSVA) scores of LVI-positive and -negative tumors for the mSigDb Hallmark gene sets are shown for the five sets with highest absolute fold-change values in LVI-positive vs. -negative comparison. EMT, epithelial mesenchymal transition. d Pie charts show the categories of mSigDb Reactome gene sets for which significant enrichment was observed in LVI-positive compared to -negative tumors in analysis with GSVA method for TCGA-BRCA and validation cohorts.
Given the association of LVI with cell cycle- and DNA replication-related gene sets, we compared the transcriptomes of LVI-positive and -negative tumors to identify individual differentially expressed genes. Expression of 152 and 60 genes respectively was up- and down-regulated by > 1.2x in LVI-positive tumors in the TCGA cohort at FDR < 0.05 (Table S7). Fifty and 20 genes respectively were similarly up- and down-regulated in both the TCGA and validation cohorts with p < 0.05 and fold-change > 1.2. Genes related to cell proliferation, like CDC20, CDC45, and FOXM1 were up-regulated the most in LVI-positive tumors. Overall, these findings support our hypothesis that LVI-positive tumors have increased cell proliferation.
Discussion
In this study, we investigated primary breast cancers from TCGA and two independent and published datasets to understand the molecular biology of LVI-positive human breast cancer. The incidence of LVI in the TCGA (37.8 %) and validation (37.2 %) cohorts as well as its association with worse prognosis (progression-free and/or overall survival) are consistent with previous studies [42–47]. LVI incidence rates reported for different subtypes of breast cancer have been inconsistent [47,48]. In the current study also, HER2 subtype had significantly higher rate of LVI in the validation cohort but not in the TCGA cohort.
Digital images of H&E-stained tumor section slides are available for all TCGA-BRCA cases, and this has been utilized in novel ways including assessment of LVI [49–51]. An advantage of using the TCGA image collection is that it allows for a uniform, standardized LVI assessment. A possible disadvantage is that the number and the area of images/slides is less than what is commonly used by the pathologists who assessed LVI for the reports that we used in our study, though we cannot truly assert this without a proper comparison. We did not utilize TCGA image collection to include more cases in our study because we did not want to introduce variability in our LVI assessments.
The level of diagnostic agreement for LVI in invasive breast cancer with H&E staining has been reported to be 0.86 for LVI-positive and 0.93 for LVI-negative tumors [40]. The incidence rate of LVI and the associations of LVI with various clinical and pathological factors that we observed for our cohorts are similar to that reported in multiple previously published studies. This indicates that the LVI determinations in our study were reliable.
The impact of LVI on OS was not observed for either very early (stage I) or advanced (stage III/IV) disease. We interpret these findings as that a tumor in early-stage such as T1 or stage I is not malignant enough for LVI to affect breast cancer mortality. For instance, cancer cells of such tumors may have a slow cell proliferation, or a limited ability to establish metastasis. Conversely, an advanced cancer such as T4, lymph node-positive, or stage III or IV has such high metastatic potential that LVI has no consequence on survival. In our study, LVI was more frequent in tumors with higher tumor grade or MKI67 gene expression. These factors are widely accepted as indicators of rapid cancer growth. These associations between LVI and clinical and pathological features such as larger tumor size and Ki67 expression that are found in our study are consistent with previous studies [e.g., 52,53,54].
LVI in breast cancer has been associated with tumor LVD [5], and positive correlation of LVD with classical LEC markers such as PDPN and LYVE1 has been reported in many studies [55,9]. However, there was no difference between LVI-positive and -negative tumors for the expression of such LEC marker genes in either cohort of our study. We also did not observe any association of LVI with tumor expression for a larger set of LEC markers. These observations suggest that LVI is independent of LVD, a disagreement with the reported correlation of LVI and LVD. This discrepancy can mean that our hypothesis was faulty. We had speculated that tumors that have more lymphatic vessels, i.e., a higher LVD, should have a greater LEC marker content in their bulk gene expression data, which we failed to observe. This may be because many of the LEC marker genes that we used are not highly specific for lymphatic vessels. Furthermore, because LECs constitute only a small fraction of tumors, their contribution to the whole tumor gene expression profile may have been minor. Another reason for our failure to observe an LVI-LVD association could be that different regions of the tumors were used for LVI and gene expression assessment. It is well known that gene expression level as well as both LVD and LVI vary across different regions of tumors [9,55]. Finally, it is possible that the association of LVI with LVD is strong only for LVD in the peri-tumoral region. The LVD that we inferred from bulk tumor gene expression data is intra-tumoral and not peri-tumoral. Peri-tumoral LVD is typically greater than intra-tumoral LVD [56], and while both are associated with LVI as well as lymph node metastasis in breast cancer, the associations for peri-tumoral LVD are stronger than for intra-tumoral LVD [5,57,58].
We found that CNA, aneuploidy and HRDs were significantly higher in LVI-positive tumors. It is known that homologous recombination repair during the cell cycle is more prevalent in tumors with higher cell proliferation [59,60]. LVI-positive and -negative tumors had similar burden of SNVs, and they did not differ in intra-tumor genomic heterogeneity. Incidence of LVI was about 40% less in tumors that had a mutation in CDH1 gene. CDH1 encodes the E-cadherin epithelial cell adhesion molecule and CDH1 mutations generally result in a reduction of E-cadherin protein level [61,62]. LVI incidence was less among tumors with reduced CDH1 gene expression. Moreover, it was significantly less in invasive lobular carcinoma compared to invasive ductal carcinoma in TCGA cohort (24 % vs. 44 %, p < 0.05). Additionally, expression of other cell adhesion-related genes, such as CEACAM5 and CNTNAP2, was also significantly higher in LVI-positive tumors (supplementary Table S6). These findings suggest that LVI can be driven by cell adhesion mediated by proteins like E-cadherin.
Because of the association of LVI with tumor aggressiveness, we expected the immune microenvironment of LVI-positive tumors to be different from LVI-negative ones. However, we found that the two types of tumors had similar anti-cancer immune responses, as assessed from the relative fractions of tumor-infiltrating CD8 T cells, M1 macrophages, and NK cells, as well as tumor immune cytolytic activity and B and T cell receptor diversity. Pro-cancerous immune features such as abundance of M2 macrophages and regulatory T cells were also not different between LVI-positive and -negative tumors. These results implicate that LVI has no significant association with the tumor immune microenvironment. An important caveat for this conclusion is that our quantification of lymphangiogenesis and immune features were based on gene expression signatures of the same sample, which is most likely not taken from the region where lymphangiogenesis is commonly measured by immunohistochemistry [9,63]. Nonetheless, a lack of association of LVI with various tumor immune features such as intra-tumoral total lymphocyte, T cell, and macrophage infiltration has been noted in many studies [56,64,65].
While enrichment of genes related to angiogenesis or epithelial-mesenchymal transition was not observed, a higher expression of cell proliferation-related genes was noticeable among LVI-positive tumors. This agrees with the observations on positive association of LVI incidence with high MKI67 gene expression, and Nottingham mitotic count and nuclear grade scores, which all indicate increased cell proliferation. Previously, Kurozumi et al. identified 99 genes whose expression was significantly different between LVI-positive and -negative cases; they included cell proliferation-related genes such as CDCA5 [49]. Increased cell proliferation in breast cancer tumors has been associated with greater genomic aberrations like CNAs and HRDs [66], which too were associated with presence of LVI in our study. These results imply that highly proliferative cancer cells have a greater tendency to invade lymphatic vessels.
A limitation of our study is that the LVI determinations made by pathologists for both the TCGA and validation cohorts were neither centralized nor standardized, and lymphatic vessel and blood vessel invasion were not distinguished for determining LVI status. This may not be important because invasion of lymphatic vessels is the predominant form of LVI in breast cancer compared to blood vascular invasion [47,67]. The portion of tumor used for LVI assessment may have been different from that used for genome or transcriptome profiling. Another limitation is that some patients with LVI may have received additional radiation therapy, and that could affect survival data. Indeed, radiation after mastectomy has been noted to improve survival in only LVI-positive patients [68]. However, we were unable to assess any treatment association with LVI in our study because of data unavailability in neither of the cohorts.
In conclusion, our assessment of LVI in the TCGA and validation cohorts were comparable with previous studies in terms of incidence and association with survival. We found that LVI in breast cancer is associated with gene expression signatures of cell proliferation, but not that of lymphangiogenesis or tumor immune microenvironment features. We thus demonstrate that LVI, a pathological finding, can reflect breast cancer biology.
Supplementary Material
Text S1
Details of methods.
Table S1
Characteristics of TCGA-BRCA tumors for which LVI status was determinable or not.
Table S2
Nottingham histologic scores and LVI status for TCGA-BRCA tumors examined in this study.
Table S3
Members of the endothelium-specific 88-gene and xCell gene sets.
Table S4
Mutation frequency among LVI-positive and -negative tumors of TCGA-BRCA cohort for ten most commonly mutated genes.
Table S5
mSigDb Hallmark and Reactome gene sets with significant enrichment in LVI-positive vs. -negative tumor comparison of TCGA-BRCA cohort by GSVA method.
Table S6
mSigDb Hallmark and Reactome gene sets with significant enrichment in LVI-positive vs. -negative tumor comparison of validation cohort by GSVA method.
Table S7
Top 20 each of genes whose expression is significantly down- or up-regulated in LVI-positive compared to -negative tumors of TCGA-BRCA cohort.
Acknowledgments
Funding
This study was supported by National Institutes of Health (NIH), USA grants R01CA160688 to K.T. and R25CA181003 to Roswell Park Comprehensive Cancer Center in support of F.Z.’s research internship.
Abbreviations
- CNA
copy number alteration
- ER
estrogen receptor
- GSEA
gene set enrichment analysis
- GSVA
gene set variation analysis
- HER2
human epidermal growth factor receptor 2
- HRD
homologous recombination defect
- LEC
lymphatic endothelial cell
- LVD
lymphatic vessel density
- LVI
lymphovascular invasion
- PR
progesterone receptor
- S1P
sphingosine-1-phosphate
- SNV
single nucleotide variation
- TCGA
The Cancer Genome Atlas
- TIES
Text Information Extraction System
- TPM
transcripts per million
- tSNE
t-distributed stochastic neighbor embedding
Footnotes
Data availability
The GEO datasets used in this study are available at https://www.ncbi.nlm.nih.gov/geo with accession numbers GSE5460 and GSE7849. Clinical, gene-level mapped read counts of RNA sequencing data, and MAF files of TCGA-BRCA cases are available at Genome Data Commons portal of National Cancer Institute, USA at https://gdc.cancer.gov. Information on Nottingham scores and LVI status that was collated from pathology reports of the TCGA cases is in Table S2. Other data used in this study is available from sources cited in the Materials and Methods section.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
This study examined human data that had been generated in the past by other studies. Informed consent was therefore not obtained in this study.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Ma Q, Dieterich LC, Detmar M (2018) Multiple roles of lymphatic vessels in tumor progression. Current opinion in immunology 53:7–12. doi: 10.1016/j.coi.2018.03.018 [DOI] [PubMed] [Google Scholar]
- 2.Zhang S, Zhang D, Gong M, Wen L, Liao C, Zou L (2017) High lymphatic vessel density and presence of lymphovascular invasion both predict poor prognosis in breast cancer. BMC cancer 17 (1):335. doi: 10.1186/s12885-017-3338-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rakha EA, Martin S, Lee AH, Morgan D, Pharoah PD, Hodi Z, Macmillan D, Ellis IO (2012) The prognostic significance of lymphovascular invasion in invasive breast carcinoma. Cancer 118 (15):3670–3680. doi: 10.1002/cncr.26711 [DOI] [PubMed] [Google Scholar]
- 4.Zhang ZQ, Han YZ, Nian Q, Chen G, Cui SQ, Wang XY (2015) Tumor Invasiveness, Not Lymphangiogenesis, Is Correlated with Lymph Node Metastasis and Unfavorable Prognosis in Young Breast Cancer Patients (</=35 Years). PloS one 10 (12):e0144376. doi: 10.1371/journal.pone.0144376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang S, Zhang D, Yi S, Gong M, Lu C, Cai Y, Tang X, Zou L (2017) The relationship of lymphatic vessel density, lymphovascular invasion, and lymph node metastasis in breast cancer: a systematic review and meta-analysis. Oncotarget 8 (2):2863–2873. doi: 10.18632/oncotarget.13752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enholm B, Karpanen T, Jeltsch M, Kubo H, Stenback F, Prevo R, Jackson DG, Yla-Herttuala S, Alitalo K (2001) Adenoviral expression of vascular endothelial growth factor-C induces lymphangiogenesis in the skin. Circulation research 88 (6):623–629 [DOI] [PubMed] [Google Scholar]
- 7.Oh SJ, Jeltsch MM, Birkenhager R, McCarthy JE, Weich HA, Christ B, Alitalo K, Wilting J (1997) VEGF and VEGF-C: specific induction of angiogenesis and lymphangiogenesis in the differentiated avian chorioallantoic membrane. Developmental biology 188 (1):96–109. doi: 10.1006/dbio.1997.8639 [DOI] [PubMed] [Google Scholar]
- 8.Du Y, Liu H, He Y, Liu Y, Yang C, Zhou M, Wang W, Cui L, Hu J, Gao F (2013) The interaction between LYVE-1 with hyaluronan on the cell surface may play a role in the diversity of adhesion to cancer cells. PloS one 8 (5):e63463. doi: 10.1371/journal.pone.0063463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagahashi M, Ramachandran S, Rashid OM, Takabe K (2010) Lymphangiogenesis: a new player in cancer progression. World journal of gastroenterology 16 (32):4003–4012. doi: 10.3748/wjg.v16.i32.4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tammela T, Alitalo K (2010) Lymphangiogenesis: Molecular mechanisms and future promise. Cell 140 (4):460–476. doi: 10.1016/j.cell.2010.01.045 [DOI] [PubMed] [Google Scholar]
- 11.Klahan S, Wong HS, Tu SH, Chou WH, Zhang YF, Ho TF, Liu CY, Yih SY, Lu HF, Chen SC, Huang CC, Chang WC (2017) Identification of genes and pathways related to lymphovascular invasion in breast cancer patients: A bioinformatics analysis of gene expression profiles. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine 39 (6):1010428317705573. doi: 10.1177/1010428317705573 [DOI] [PubMed] [Google Scholar]
- 12.Paduch R (2016) The role of lymphangiogenesis and angiogenesis in tumor metastasis. Cellular oncology (Dordrecht) 39 (5):397–410. doi: 10.1007/s13402-016-0281-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagahashi M, Abe M, Sakimura K, Takabe K, Wakai T (2018) The role of sphingosine-1-phosphate in inflammation and cancer progression. Cancer science 109 (12):3671–3678. doi: 10.1111/cas.13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsuchida J, Nagahashi M, Nakajima M, Moro K, Tatsuda K, Ramanathan R, Takabe K, Wakai T (2016) Breast cancer sphingosine-1-phosphate is associated with phospho-sphingosine kinase 1 and lymphatic metastasis. The Journal of surgical research 205 (1):85–94. doi: 10.1016/j.jss.2016.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobson RS, Becich MJ, Bollag RJ, Chavan G, Corrigan J, Dhir R, Feldman MD, Gaudioso C, Legowski E, Maihle NJ, Mitchell K, Murphy M, Sakthivel M, Tseytlin E, Weaver J (2015) A Federated Network for Translational Cancer Research Using Clinical Data and Biospecimens. Cancer research 75 (24):5194–5201. doi: 10.1158/0008-5472.Can-15-1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Cancer Genome Atlas Research N, Hu H (2018) An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 173 (2):400–416 e411. doi: 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2 (5):401–404. doi: 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CE, Cancer Genome Atlas Research N, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich L (2018) The Immune Landscape of Cancer. Immunity 48 (4):812–830 e814. doi: 10.1016/j.immuni.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, Hess J, Ma S, Chiotti KE, McLellan M, Sofia HJ, Hutter C, Getz G, Wheeler D, Ding L, Group MCW, Cancer Genome Atlas Research N (2018) Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst 6 (3):271–281 e277. doi: 10.1016/j.cels.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson J, Li JZ, Zollner S (2018) Helmsman: fast and efficient mutation signature analysis for massive sequencing datasets. BMC Genomics 19 (1):845. doi: 10.1186/s12864-018-5264-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C (2016) DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 17:31. doi: 10.1186/s13059-016-0893-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA (2018) Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol 1711:243–259. doi: 10.1007/978-1-4939-7493-1_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N (2015) Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160 (1–2):48–61. doi: 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu X, Lu X, Wang ZC, Iglehart JD, Zhang X, Richardson AL (2008) Predicting features of breast cancer with gene expression patterns. Breast cancer research and treatment 108 (2):191–201. doi: 10.1007/s10549-007-9596-6 [DOI] [PubMed] [Google Scholar]
- 25.Anders CK, Acharya CR, Hsu DS, Broadwater G, Garman K, Foekens JA, Zhang Y, Wang Y, Marcom K, Marks JR, Mukherjee S, Nevins JR, Blackwell KL, Potti A (2008) Age-specific differences in oncogenic pathway deregulation seen in human breast tumors. PloS one 3 (1):e1373. doi: 10.1371/journal.pone.0001373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics (Oxford, England) 28 (6):882–883. doi: 10.1093/bioinformatics/bts034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Podgrabinska S, Braun P, Velasco P, Kloos B, Pepper MS, Skobe M (2002) Molecular characterization of lymphatic endothelial cells. Proc Natl Acad Sci U S A 99 (25):16069–16074. doi: 10.1073/pnas.242401399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wick N, Saharinen P, Saharinen J, Gurnhofer E, Steiner CW, Raab I, Stokic D, Giovanoli P, Buchsbaum S, Burchard A, Thurner S, Alitalo K, Kerjaschki D (2007) Transcriptomal comparison of human dermal lymphatic endothelial cells ex vivo and in vitro. Physiol Genomics 28 (2):179–192. doi: 10.1152/physiolgenomics.00037.2006 [DOI] [PubMed] [Google Scholar]
- 29.Hirakawa S, Hong YK, Harvey N, Schacht V, Matsuda K, Libermann T, Detmar M (2003) Identification of vascular lineage-specific genes by transcriptional profiling of isolated blood vascular and lymphatic endothelial cells. Am J Pathol 162 (2):575–586. doi: 10.1016/S0002-9440(10)63851-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amatschek S, Kriehuber E, Bauer W, Reininger B, Meraner P, Wolpl A, Schweifer N, Haslinger C, Stingl G, Maurer D (2007) Blood and lymphatic endothelial cell-specific differentiation programs are stringently controlled by the tissue environment. Blood 109 (11):4777–4785. doi: 10.1182/blood-2006-10-053280 [DOI] [PubMed] [Google Scholar]
- 31.DiMaio TA, Wentz BL, Lagunoff M (2016) Isolation and characterization of circulating lymphatic endothelial colony forming cells. Exp Cell Res 340 (1):159–169. doi: 10.1016/j.yexcr.2015.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrova TV, Makinen T, Makela TP, Saarela J, Virtanen I, Ferrell RE, Finegold DN, Kerjaschki D, Yla-Herttuala S, Alitalo K (2002) Lymphatic endothelial reprogramming of vascular endothelial cells by the Prox-1 homeobox transcription factor. The EMBO journal 21 (17):4593–4599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aran D, Hu Z, Butte AJ (2017) xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 18 (1):220. doi: 10.1186/s13059-017-1349-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14:7. doi: 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102 (43):15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P (2015) The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1 (6):417–425. doi: 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, Milacic M, Roca CD, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Viteri G, Weiser J, Wu G, Stein L, Hermjakob H, D’Eustachio P (2018) The Reactome Pathway Knowledgebase. Nucleic acids research 46 (D1):D649–D655. doi: 10.1093/nar/gkx1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smyth G (2005) Limma: linear models for microarray data In: Gentleman R, Carey VJ, Huber W, Dudoit S, Irizarry RA (eds) Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer, New York, pp 397–420 [Google Scholar]
- 39.Ryu YJ, Kang SJ, Cho JS, Yoon JH, Park MH (2018) Lymphovascular invasion can be better than pathologic complete response to predict prognosis in breast cancer treated with neoadjuvant chemotherapy. Medicine 97 (30):e11647. doi: 10.1097/md.0000000000011647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rakha EA, Abbas A, Pinto Ahumada P, ElSayed ME, Colman D, Pinder SE, Ellis IO (2018) Diagnostic concordance of reporting lymphovascular invasion in breast cancer. Journal of clinical pathology 71 (9):802–805. doi: 10.1136/jclinpath-2017-204981 [DOI] [PubMed] [Google Scholar]
- 41.Moynahan ME, Jasin M (2010) Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature reviews Molecular cell biology 11 (3):196–207. doi: 10.1038/nrm2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee AH, Pinder SE, Macmillan RD, Mitchell M, Ellis IO, Elston CW, Blamey RW (2006) Prognostic value of lymphovascular invasion in women with lymph node negative invasive breast carcinoma. European journal of cancer (Oxford, England : 1990) 42 (3):357–362. doi: 10.1016/j.ejca.2005.10.021 [DOI] [PubMed] [Google Scholar]
- 43.Lauria R, Perrone F, Carlomagno C, De Laurentiis M, Morabito A, Gallo C, Varriale E, Pettinato G, Panico L, Petrella G, et al. (1995) The prognostic value of lymphatic and blood vessel invasion in operable breast cancer. Cancer 76 (10):1772–1778 [DOI] [PubMed] [Google Scholar]
- 44.Schoppmann SF, Bayer G, Aumayr K, Taucher S, Geleff S, Rudas M, Kubista E, Hausmaninger H, Samonigg H, Gnant M, Jakesz R, Horvat R (2004) Prognostic value of lymphangiogenesis and lymphovascular invasion in invasive breast cancer. Annals of surgery 240 (2):306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ejlertsen B, Jensen MB, Rank F, Rasmussen BB, Christiansen P, Kroman N, Kvistgaard ME, Overgaard M, Toftdahl DB, Mouridsen HT (2009) Population-based study of peritumoral lymphovascular invasion and outcome among patients with operable breast cancer. Journal of the National Cancer Institute 101 (10):729–735. doi: 10.1093/jnci/djp090 [DOI] [PubMed] [Google Scholar]
- 46.Bettelheim R, Penman HG, Thornton-Jones H, Neville AM (1984) Prognostic significance of peritumoral vascular invasion in breast cancer. British journal of cancer 50 (6):771–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohammed RA, Martin SG, Mahmmod AM, Macmillan RD, Green AR, Paish EC, Ellis IO (2011) Objective assessment of lymphatic and blood vascular invasion in lymph node-negative breast carcinoma: findings from a large case series with long-term follow-up. The Journal of pathology 223 (3):358–365. doi: 10.1002/path.2810 [DOI] [PubMed] [Google Scholar]
- 48.Mohammed RA, Menon S, Martin SG, Green AR, Paish EC, Ellis IO (2014) Prognostic significance of lymphatic invasion in lymph node-positive breast carcinoma: findings from a large case series with long-term follow-up using immunohistochemical endothelial marker. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 27 (12):1568–1577. doi: 10.1038/modpathol.2014.60 [DOI] [PubMed] [Google Scholar]
- 49.Kurozumi S, Joseph C, Sonbul S, Alsaeed S, Kariri Y, Aljohani A, Raafat S, Alsaleem M, Ogden A, Johnston SJ, Aleskandarany MA, Fujii T, Shirabe K, Caldas C, Ashankyty I, Dalton L, Ellis IO, Desmedt C, Green AR, Mongan NP, Rakha EA (2019) A key genomic subtype associated with lymphovascular invasion in invasive breast cancer. British journal of cancer 120 (12):1129–1136. doi: 10.1038/s41416-019-0486-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper LA, Demicco EG, Saltz JH, Powell RT, Rao A, Lazar AJ (2018) PanCancer insights from The Cancer Genome Atlas: the pathologist’s perspective. The Journal of pathology 244 (5):512–524. doi: 10.1002/path.5028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, Samaras D, Shroyer KR, Zhao T, Batiste R, Van Arnam J, Cancer Genome Atlas Research N, Shmulevich I, Rao AUK, Lazar AJ, Sharma A, Thorsson V (2018) Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep 23 (1):181–193 e187. doi: 10.1016/j.celrep.2018.03.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen S, Wu G, Xiao G, Du R, Hu N, Xia X, Zhou H (2018) Prediction model of lymphovascular invasion based on clinicopathological factors in Chinese patients with invasive breast cancer. Medicine 97 (43):e12973. doi: 10.1097/md.0000000000012973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanyilmaz G, Yavuz BB, Aktan M, Karaagac M, Uyar M, Findik S (2019) Prognostic Importance of Ki-67 in Breast Cancer and Its Relationship with Other Prognostic Factors. Eur J Breast Health 15 (4):256–261. doi: 10.5152/ejbh.2019.4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He KW, Sun JJ, Liu ZB, Zhuo PY, Ma QH, Liu ZY, Yu ZY (2017) Prognostic significance of lymphatic vessel invasion diagnosed by D2–40 in Chinese invasive breast cancers. Medicine 96 (44):e8490. doi: 10.1097/MD.0000000000008490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng Y, Wang W, Hu J, Ma J, Zhang Y, Zhang J (2010) Expression of VEGF-C and VEGF-D as significant markers for assessment of lymphangiogenesis and lymph node metastasis in non-small cell lung cancer. Anatomical record (Hoboken, NJ : 2007) 293 (5):802–812. doi: 10.1002/ar.21096 [DOI] [PubMed] [Google Scholar]
- 56.Ding M, Fu X, Tan H, Wang R, Chen Z, Ding S (2012) The effect of vascular endothelial growth factor C expression in tumor-associated macrophages on lymphangiogenesis and lymphatic metastasis in breast cancer. Mol Med Rep 6 (5):1023–1029. doi: 10.3892/mmr.2012.1043 [DOI] [PubMed] [Google Scholar]
- 57.Zhang S, Yi S, Zhang D, Gong M, Cai Y, Zou L (2017) Intratumoral and peritumoral lymphatic vessel density both correlate with lymph node metastasis in breast cancer. Sci Rep 7:40364. doi: 10.1038/srep40364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Agarwal S, Singh A, Bagga PK (2018) Immunohistochemical evaluation of lymphovascular invasion in carcinoma breast with CD34 and D2–40 and its correlation with other prognostic markers. Indian J Pathol Microbiol 61 (1):39–44. doi: 10.4103/IJPM.IJPM_791_16 [DOI] [PubMed] [Google Scholar]
- 59.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434 (7035):917–921. doi: 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- 60.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434 (7035):913–917. doi: 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- 61.Rakha EA, Abd El Rehim D, Pinder SE, Lewis SA, Ellis IO (2005) E-cadherin expression in invasive non-lobular carcinoma of the breast and its prognostic significance. Histopathology 46 (6):685–693. doi: 10.1111/j.1365-2559.2005.02156.x [DOI] [PubMed] [Google Scholar]
- 62.Corso G, Intra M, Trentin C, Veronesi P, Galimberti V (2016) CDH1 germline mutations and hereditary lobular breast cancer. Familial cancer 15 (2):215–219. doi: 10.1007/s10689-016-9869-5 [DOI] [PubMed] [Google Scholar]
- 63.Nagahashi M, Ramachandran S, Kim EY, Allegood JC, Rashid OM, Yamada A, Zhao R, Milstien S, Zhou H, Spiegel S, Takabe K (2012) Sphingosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Cancer research 72 (3):726–735. doi: 10.1158/0008-5472.can-11-2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyan M, Schmidt-Mende J, Kiessling R, Poschke I, de Boniface J (2016) Differential tumor infiltration by T-cells characterizes intrinsic molecular subtypes in breast cancer. J Transl Med 14 (1):227. doi: 10.1186/s12967-016-0983-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eryilmaz MK, Mutlu H, Unal B, Salim DK, Musri FY, Coskun HS (2018) The importance of stromal and intratumoral tumor lymphocyte infiltration for pathologic complete response in patients with locally advanced breast cancer. J Cancer Res Ther 14 (3):619–624. doi: 10.4103/0973-1482.174550 [DOI] [PubMed] [Google Scholar]
- 66.Duijf PHG, Nanayakkara D, Nones K, Srihari S, Kalimutho M, Khanna KK (2019) Mechanisms of Genomic Instability in Breast Cancer. Trends Mol Med 25 (7):595–611. doi: 10.1016/j.molmed.2019.04.004 [DOI] [PubMed] [Google Scholar]
- 67.Mohammed RA, Martin SG, Gill MS, Green AR, Paish EC, Ellis IO (2007) Improved methods of detection of lymphovascular invasion demonstrate that it is the predominant method of vascular invasion in breast cancer and has important clinical consequences. The American journal of surgical pathology 31 (12):1825–1833. doi: 10.1097/PAS.0b013e31806841f6 [DOI] [PubMed] [Google Scholar]
- 68.Su YL, Li SH, Chen YY, Chen HC, Tang Y, Huang CH, Chou FF, Wu SC, Rau KM (2014) Post-mastectomy radiotherapy benefits subgroups of breast cancer patients with T1–2 tumor and 1–3 axillary lymph node(s) metastasis. Radiology and oncology 48 (3):314–322. doi: 10.2478/raon-2013-0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Text S1
Details of methods.
Table S1
Characteristics of TCGA-BRCA tumors for which LVI status was determinable or not.
Table S2
Nottingham histologic scores and LVI status for TCGA-BRCA tumors examined in this study.
Table S3
Members of the endothelium-specific 88-gene and xCell gene sets.
Table S4
Mutation frequency among LVI-positive and -negative tumors of TCGA-BRCA cohort for ten most commonly mutated genes.
Table S5
mSigDb Hallmark and Reactome gene sets with significant enrichment in LVI-positive vs. -negative tumor comparison of TCGA-BRCA cohort by GSVA method.
Table S6
mSigDb Hallmark and Reactome gene sets with significant enrichment in LVI-positive vs. -negative tumor comparison of validation cohort by GSVA method.
Table S7
Top 20 each of genes whose expression is significantly down- or up-regulated in LVI-positive compared to -negative tumors of TCGA-BRCA cohort.