Abstract
A set of 1036 U.S. Population Samples were sequenced using the Illumina ForenSeq DNA Signature Prep Kit. This sample set has been highly characterized using a variety of marker systems for human identification. The FASTQ files obtained from a ForenSeq DNA Signature Prep Kit experiment include several STR loci that are not reported in the associated software. These include SE33, DXS8377, DXS10148, DYS456, and DYS461. The sequence variation within the autosomal STR marker SE33 was evaluated using a customized bioinformatic approach to identify and characterize the locus in the 1036 data set. The analysis identified 53 unique alleles by length and 264 by sequence. An additional 10 alleles were detected when selected extended flanking regions were examined to resolve discordances. Allele frequencies and SE33 sequence motif patterns are reported for the 1036 data set. The comparison of numerical allele calls derived from sequence data to the allele calls obtained from commercial capillary electrophoresis-based STR typing kits resulted in 100% concordance, after manual data review and confirmation sequencing of three flanking region deletions. The analysis of this data set involved significant manual sequence curation and information support from length-based genotypes to ensure high confidence in the sequence-based allele calls. The challenges of interpreting the sequence data for SE33 consisted of high sequence noise, allele-size dependent variance in coverage, and heterozygote imbalance. As allele length increased, sequence depth of coverage and quality decreased at the terminal end. Accordingly, heterozygous genotype imbalance increased in proportion to increased distance between alleles.
1. Introduction
SE33 has long been a locus of interest to the human identity community. The highly polymorphic and heterozygotic nature of SE33 provides a more discriminating genotype, which is useful for mixture analysis. However, these advantages are tempered by technical genotyping/separation issues, a complex sequence motif (challenging for nomenclature), and a high mutation rate (challenging for kinship).
SE33 was first reported in Moos and Gallwitz 1983 [1] as an STR marker adjacent to a beta-actin related pseudogene (ACTBP2, also historically used as the STR locus name) on chromosome 6. Warne et al. [2] and Polymeropoulos et al. [3] identified the repeat as [AAAG]n (now considered reverse strand) and generated limited allele frequency data. Urquhart et al. [4] followed in 1993 with expanded sequencing of the region and concluded that variation in SE33 was more extensive than early studies had indicated. The 1998 recommendations of Schneider et al. [5] for assigning numerical allele designations are consistent with current allele typing by electrophoresis methods. In the same year, Rolf et al. [6] published many more sequence variants at this locus.
Figure 1 characterizes the discrete regions that define the landscape of the SE33 locus, and these regions will be referenced throughout this manuscript. The CE-compatible numerical allele designations are generally based on the Repeat Region defined in Figure 1. Recent ISFG STR sequence-based nomenclature guidance [7] orients the locus on the forward strand of the current human genome reference assembly, GRCh38, as [CTTT]n. The Repeat Region aligns with GRCh38, Chromosome 6 (accession number NC_000006.12), base position 88,277,144 to 88,277,245. The primary sequence reported in this manuscript is the forward strand orientation of the extended sequence range reported in Rolf et al. [6], shown in Figure 1 as Repeat with Local Flanks.
Figure 1 –
Landscape of the SE33 region is a representation of the domains for the STR and the surrounding sequence (5’ to 3’) on the forward strand. The three gray bars represent deletions that have been detected in these population sample sequences. The white boxes are the STRait Razor ‘A’nchor sites. The two boxes with black lines are ‘Pre’ and ‘Post’ sequence that are not counted toward the numerical allele designation but contain variable sequences. The terms Repeat Region, Repeat with Local Flanks, and Extended Flanks are used throughout the manuscript and refer to this figure. This schematic is scaled to the GRCh38 reference genome sequence NC_000006.12 base positions 88,277,094 to 88,277,370.
The ForenSeq kit is designed to sequence each locus unidirectionally. In the case of SE33, sequencing occurs in the 5’ to 3’ direction on the forward strand, with the 5’ PCR primer binding site within approximately 15 bp of the start of the Repeat Region. The 3’ PCR primer binding site is located over 200 bp away from the 3’ end of the Repeat Region. The forward strand amplicon is highly thymine rich, 52% based on the reference sequence, followed by 30% cytosine, and 18% total adenine and guanine, which are located primarily at the 3’ end of the sequence. The amplicon length and nucleotide bias are likely factors rendering this locus a challenge to sequence.
Population data generated with the ForenSeq kit has been published by Novroski et al. [8], Devesse et al. [9], and Gettings et al. 2018 (submitted) among others [10, 11, 12]. Although mention of SE33 sequences being present in the FASTQ files [8] can be found, SE33 sequence alleles derived from the ForenSeq kit have not previously been published.
2. Materials and Methods
2.1. Samples
The population samples of self-declared ancestry represented four groups: 342 African American, 97 Asian, 361 Caucasian, and 236 Hispanic for a total of N=1036 samples [13, 14]. All work presented has been reviewed and approved by the NIST Human Subjects Protections Office.
2.2. Sequencing
The samples were sequenced using the ForenSeq DNA Signature Prep Kit on a MiSeq FGx instrument (Illumina, San Diego, CA). Multiplex amplification was performed using DNA Primer Mix B (DPMB), which is intended to sequence 27 autosomal STRs, 7 X chromosome STRs, 24 Y chromosome STRs, and 172 SNPs. Detailed methods and sequencing results for the 27 autosomal STR data set have been submitted for publication (Gettings et al. 2018).
Monoplex SE33 amplification and Sanger sequencing was performed to confirm differences between sequencing and length-based capillary electrophoresis methods [15]. Sanger sequencing and repeated ForenSeq kit sequencing were performed to confirm select occurrences of extended flanking deletions, lower coverage alleles, and motif patterns.
2.3. Bioinformatic methods
FASTQ files for each sample were collected from the MiSeq FGx Universal Analysis Software (UAS) server. The FASTQ files containing all loci were trimmed using BBDuk [https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/bbduk-guide/]. The trimming parameters required a minimum sequence length of 50 bp, a minimum base quality score of 10 at sequence ends and trimming the 3’ end beginning at the Illumina tag sequence onward. The start of the tag sequence (used to bind the index during library preparation) was identified by the 11 bp sequence TGGAATTCTCG (allowing for one base variation), and trimming from this point onward eliminates tag, index, and adapter sequence from the resulting string. An independent analysis of the sequencing data was completed using a modified version of the open source STRait Razor v2.0 software [16]. The required STRait Razor v2.0 configuration information specific to SE33 was added to the configuration file: 1) 5’ and 3’ anchor sites (forward strand TATAGTAACTTG and CTTTTTCTTTCTTTTTCTTC, respectively, and reverse complements); 2) identification of the repeat sequence CTTTCTTT; and 3) length ‘bins’ for all possible SE33 alleles between 1 and 50.3 (200 bins total).
The STRait Razor v2.0 code was additionally modified to retain 75 bp of the 3’ flanking sequence including the 3’ anchor site and provide counts of consensus sequences. The code was also modified to report full sequence strings, identified by the anchor sites, and provide counts of these consensus sequences. The goal of these changes was to maintain useful sequence information, including trimmed and full sequence, associated with the STRait Razor length-based allele call.
The STRait Razor results were further processed to determine the genotypes. This processing generated two report files. The first report included all sequences with a depth of coverage (DoC) greater than 10 reads. The second report was designed to reduce the first report to the true genotypes, exclusive of artifacts, in order to limit manual data review. This report required an allele coverage ratio (ACR) greater than 20% in order to include more than one allele (ACR is calculated by dividing a lower DoC allele by the highest DoC allele, analogous to peak height ratio in CE methods). DoC and ACR metrics were used to identify most of the reported alleles. An additional calculation reported in the results file was non-majority allele (NonMA). NonMA is a high-level representation of the noise present at each allele called. It is the fraction of sequencing reads not identical to the most abundant sequence of the same length for an allele. NonMA was calculated by dividing the number of same-length sequences which differed in base composition from the majority sequence by the total number of same-length sequences.
The allele calls included in the second report file were compared to those obtained from length-based CE methods. Comparing sequence-derived allele calls to CE data identified heterozygotes that were imbalanced beyond the 20% ACR criteria. Alleles which dropped out of the second report due to imbalance were manually recovered from the first report file. Stutter and/or sequence artifacts which exceeded the 20% ACR criteria were identified by the CE comparison and were manually reviewed and removed from the final data set. All sequences reported in this study had a DoC of greater than 30 reads. The annotated sequenced alleles reported in this work can be found in the STRSeq BioProject [19] (GenBank records MH232696 – MH232959). The BioProject ID specific to SE33 is PRJNA380562 [https://www.ncbi.nlm.nih.gov/bioproject/PRJNA380562/].
2.4. Motif Patterns
Reported sequence alleles were categorized into motif patterns by dividing the Repeat and Local Flank region into the Pre, Repeat Region, and Post segments, as identified in Figure 1. Motif Pattern IDs correspond to unique patterns identified in the set and were assigned to assist with describing motifs. Each ID consists a single character (A through D) followed by a single number (0 through 9). Assignments were made based on pattern frequency, starting with the most common pattern being A0, the second most common pattern being A1, and so on.
2.5. Statistical analysis
Allele frequencies and associated population statistic parameters: genetic diversity (GD) or expected heterozygosity, observed heterozygosity (Hobs), polymorphism information content (PIC), match probability (PM), power of discrimination (PD), power of exclusion (PE), typical paternity index (TPI), and Hardy-Weinberg equilibrium (pHW) were performed by the online tool STRAF: STR Analysis for Forensics [17, http://cmpg.unibe.ch/shiny/STRAF/]. Statistical analyses were performed for the global data set and per population group. Statistical parameters were also calculated for the length-based alleles for comparisons [14]. To facilitate the input of sequenced allele information into the STRAF software each unique STR allele sequence was assigned a unique integer value to represent the allele in the program. Random match probabilities were calculated using GenAlEx software [18], and no adjustments were made for theta or minimum allele frequencies.
3. Results
3.1. Concordance
Allele calls from CE methods were compared to the sequence-based alleles which met the criteria for inclusion. Forty-six samples demonstrated dropout of an allele due to imbalance beyond the 20% ACR criteria, all 46 dropped alleles were manually recovered from the first report file generated in the bioinformatic analysis. The average distance between alleles when one allele was manually recovered was approximately 13 repeats. The largest distance was 24 repeats. These recovered alleles were always the larger of the two alleles with a size range between 27.2 and 36 repeats.
After recovery of dropped alleles, there was 93% (1924 / 2072) allele concordance when only evaluating the Repeat Region. An additional 6% (127 / 2072) of allele concordance was recovered when including the Repeat with Local Flanks (Figure 1) region in the evaluation. This difference is caused by ambiguous Repeat Region boundaries, because of sequence variation in the Repeat with Local Flanks [4]. An example of this can be seen in motif pattern A6, shown below with A0 for comparison:
A0 CT [CTTT]3 C [CTTT]n CT [CTTT]3 CT [CTTT]2
A6 CT [CTTT]3 C [CTTT]n [CTTT]3 CT [CTTT]2.
The Repeat Region of the A6 motif could be counted as the uninterrupted stretch of [CTTT], equivalent to n+3, as the CT that is typically between the [CTTT]n and the [CTTT]3 is absent from this motif. However, this results in discordance with the CE-based allele call, which calculates the numerical allele based on the amplicon length. When considering the entire Repeat with Local Flank sequence, it is apparent that one must artificially shift [CTTT]3 to the Post-repeat region, and account for the absence of the intervening CT, to achieve concordance with numerical allele assignment.
The final 1% (21 / 2072) of discordance between sequence and CE derived data was reconciled by manually evaluating the ForenSeq data and Sanger Sequencing in the Extended Flanks. The 21 discordances in this category are representative of three deletions, fully described in Supplementary Table 1 and shown as gray bars in Figure 1. The first is a known four-base deletion in the 3’ Extended Flank, rs369314007 in dbSNP. It was observed eight times in ForenSeq sequences, confirmed with Sanger sequencing, and is associated with the motif pattern A0. The second deletion, also a four-base deletion in the 3’ Extended Flank, was submitted to dbSNP by the authors, and has been assigned rs1371483225. This deletion was observed 12 times in ForenSeq sequences, confirmed with Sanger sequencing, and is associated with motif pattern A4. The third deletion was a single observation of a three-base deletion in the 5’ flank, resulting in a 29.2 allele by sequence and a 28.3 allele by CE. This is the only deletion which was not present in the ForenSeq PCR amplicon. It was confirmed with Sanger sequencing and was previously identified as discordant between commercial CE kits [15]: the AmpFℓSTER® NGM SElect Express CE kit typed the allele as 29.2 whereas the PowerPlex® ESX 17, PowerPlex® ESI 17 and ESSplex SE all typed a 28.3 allele. This deletion is rs1421903850 in dbSNP. By careful evaluation of the sequenced alleles compared to the CE-based allele calls, 100% concordance of length-based genotypes was achieved.
3.2. Increase in number of observed alleles by sequencing
The complete 1036 sample set contained 53 alleles by length-based allele calling [13, 14]. This number increased to 264 unique sequences identified in the Repeat with Local Flanks region (Supplementary Table 2). This represents a fivefold increase in observed alleles by sequencing. An additional ten unique sequences were confirmed in the population set by identifying deletions in the Extended Flanks (Supplementary Table 1).
3.3. Allele frequencies
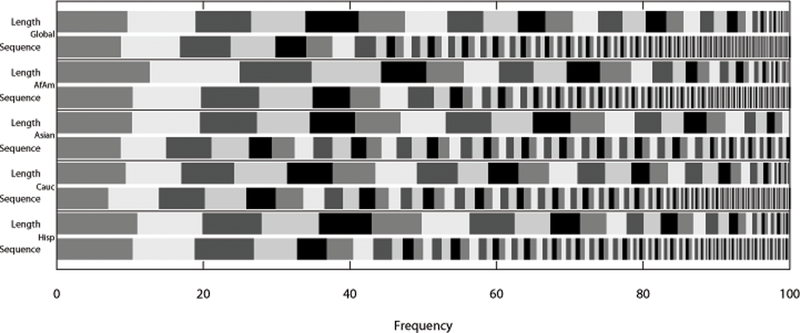
Frequencies for the 264 alleles across the four population groups were calculated and are presented in Supplementary Table 2. Figure 2 represents the number and frequency distribution of length-based and sequence-based alleles. This is illustrated for the global population set as well as each individual population group. Of the 264 alleles, 110 were only observed once in the population set.
Figure 2 -.
SE33 allele frequency distribution by sequence and length plotted in global N = 1036, African American N = 342, Asian N = 97, Caucasian N = 361, and Hispanic N = 236.
3.4. Depth of coverage
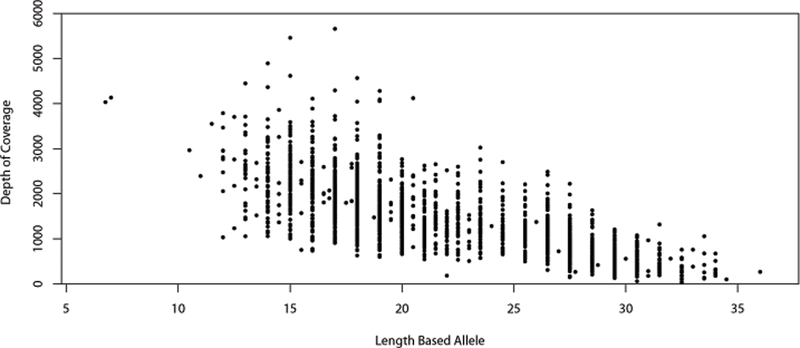
SE33 exhibits one of the larger allele size ranges among STR loci used in human identification. Alleles from 3 to 49 repeats have been observed in the general population [20], corresponding to a difference in length between largest and smallest alleles of 184 bp. The 1036 population sample set contains length-based alleles ranging from 6.3 to 36. Within this data set, the SE33 DoC for the Repeat with Local Flanks region ranged from 35X to 5657X (observed in a 32.2 allele and a 17 allele, respectively). The mean DoC for SE33 was 1436X with a standard deviation of 820X. Figure 3 illustrates the overall trend of DoC versus length-based allele size. As length-based allele size increased the observed DoC decreased. DoC was affected by sample quality as well as the experimental conditions under which the samples were processed and sequenced.
Figure 3 -.
SE33 plot of depth of coverage (DoC) versus length-based allele size. A decrease in DoC was observed as allele size increased. N = 1036
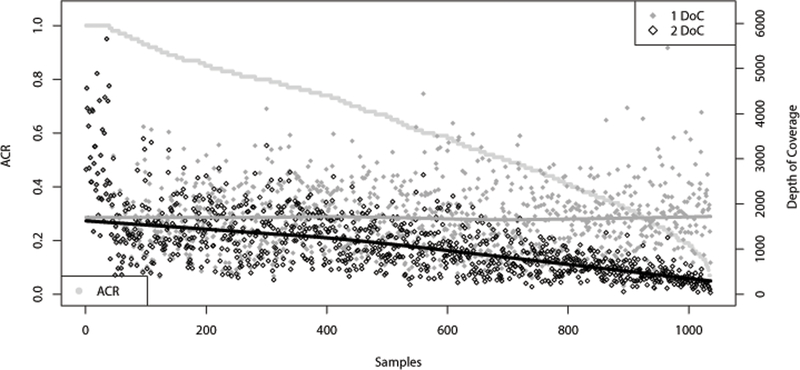
The variation in ACR, DoC, and NonMA are best observed in the aggregate data. Figure 4, which includes results across all 1036 samples, demonstrates that the decline in ACR is correlated with the decline in DoC of the larger length allele in a heterozygous pair, while the DoC of the smaller length allele remains relatively constant. ACR observed in this study ranged from 5% to 100% (genotypes 24.2, 32.2 and 21.2, 26.2, respectively). The sample with 5% ACR had DoC of 701X (24.2 allele) and 35X (32.2 allele).
Figure 4 -.
SE33 depth of coverage (DoC) per allele per sample and allele coverage ratio (ACR) for each allele pair, ordered by decreasing ACR, and including sequence-based homozygous genotypes (ACR=1 and DoC data points overlap). The ACR scale is on the left axis, 0.0 to 1.0 (0% to 100%), and ACR data are represented by light gray filled circles. The DoC scale is on the right axis, and data for allele 1 (smaller allele) are represented by dark gray filled diamonds while data for allele 2 (larger allele) are represented by black outlined diamonds. Trend lines correspond to DoC allele 1, dark gray, and allele 2, black. N = 1036.
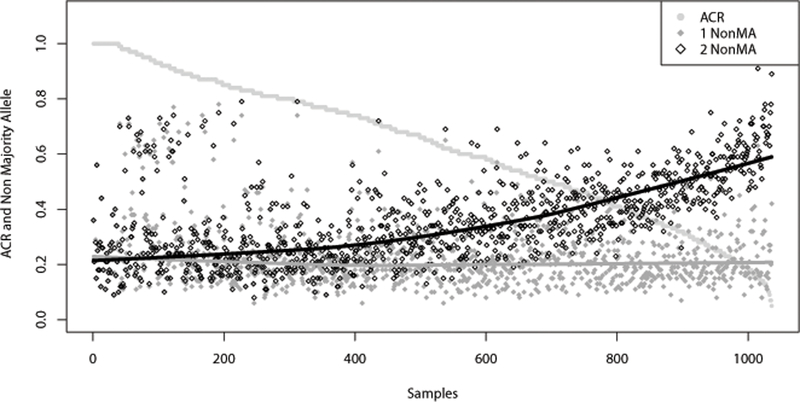
Figure 5 plots the fraction of NonMA for the samples in relation to decreasing ACR across all 1036 samples. In this comparison, a decline in ACR is correlated to an increase in NonMA in the larger length allele of a heterozygous pair (up to a maximum of 91% NonMA, an approximation of noise), while the smaller length allele maintains approximately 20% NonMA. The sample genotype which exhibited the highest NonMA was 27.2 (325X DoC, 63% NonMA), 32.2 (48X DoC, 91% NonMA).
Figure 5 -.
SE33 non-majority allele (NonMA) per allele per sample and allele coverage ratio (ACR) for each allele pair, ordered by decreasing ACR, and including sequence-based homozygous genotypes (ACR=1 and NonMA data points overlap). The ACR and NonMA scale is on the left axis, 0.0 to 1.0 (0% to 100%). ACR data are represented by light gray filled circles. NonMA data for allele 1 (smaller allele) are represented by dark gray filled diamonds while data for allele 2 (larger allele) are represented by black outlined diamonds. Trend lines correspond to NonMA allele 1, dark gray, and allele 2, black. N = 1036.
3.5. SE33 motif patterns
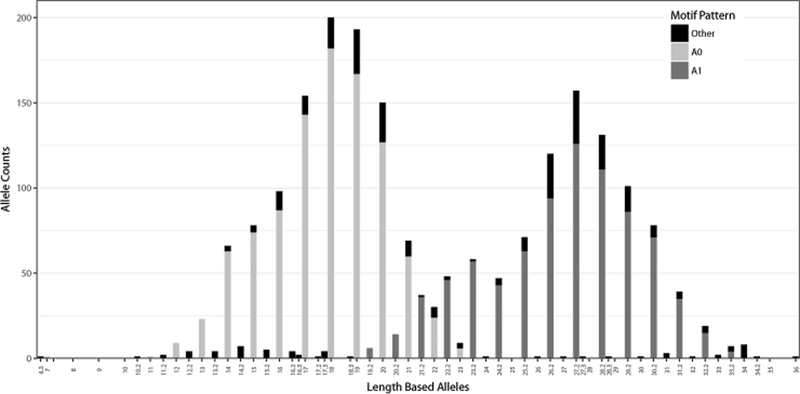
Table 1 contains the 11 most common motif patterns, present at a frequency of greater than 1% in at least one of the four population groups. The table includes the allele range for each motif pattern, the motif pattern, motif pattern ID, and the frequencies of each motif pattern reported for each population. A plot of allele counts versus length-based allele for the population data set appears bimodal (Figure 6). The most common (A0) and second most common (A1) motif patterns represent the majority of the two peaks without any overlapping length alleles.
Table 1 -.
SE33 Sequence-based motif patterns and corresponding frequency by population. N=1036. Motif patterns exceeding 1% in at least one population are represented. Italicized characters indicate ambiguous repeat regions and or bracketing of the repeats to attempt a best visual fit of the Local Flanks and Repeat region.
| Frequency of Motif by Population |
||||||
|---|---|---|---|---|---|---|
| Allele Range | Motif | Motif ID | AfAm | Asian | Cauc | Hisp |
| 7 to 23 | CT [CTTT]3 C [CTTT]n CT [CTTT]3 CT [CTTT]2 | A0 | 54.24% | 36.08% | 42.24% | 46.82% |
| 19.2 to 33.2 | CT [CTTT]2 CCTT C [CTTT]n TT [CTTT]n CT [CTTT]3 CT [CTTT]2 | A1 | 23.98% | 57.73% | 49.86% | 36.23% |
| 15 to 23 | CT [CTTT]2 CCTT C [CTTT]n CT [CTTT]3 CT [CTTT]2 | A2 | 6.58% | 0.14% | 2.33% | |
| 22.2 to 30.2 | CT [CTTT]2 CCTT C [CTTT]n CT [CTTT]n CT [CTTT]3 CT [CTTT]2 | A3 | 0.58% | 4.64% | 1.52% | 6.78% |
| 21.2 to 31.2 | CT [CTTT]2 [CCTT]2 C [CTTT]n TT [CTTT]n CT [CTTT]3 CT [CTTT]2 | A4 | 3.95% | 0.52% | 0.42% | 0.64% |
| 16 to 23 | CT [CTTT]3 CCTT C [CTTT]n CT [CTTT]3 CT [CTTT]2 | A5 | 3.80% | 0.52% | 0.14% | 0.64% |
| 10.2 to 15.2 | CT [CTTT]3 C [CTTT]n [CTTT]3 CT [CTTT]2 | A6 | 1.75% | 0.28% | 1.27% | |
| 30 to 36 | CT [CTTT]2 CCTT C [CTTT]n TT [CTTT]n TT [CTTT]n CT [CTTT]3 CT [CTTT]2 | A7 | 0.44% | 1.39% | 0.42% | |
| 27.2 to 34.2 | CT [CTTT]2 CCTT C [CTTT]n TT [CTTT]n CT [CTTT]3 CT CTTT | A8 | 0.15% | 1.52% | 0.21% | |
| 15 to 20 | CT [CTTT]3 C CCTT [CTTT]n CT [CTTT]3 CT [CTTT]2 | A9 | 1.46% | 0.14% | 0.21% | |
| 26.2 to 32.2 | CT [CTTT]2 [CCTT]2 C [CTTT]n CT [CTTT]3 CT [CTTT]2 | B0 | 0.15% | 1.69% | ||
Figure 6 -.
Distribution of common motif patterns across length-based alleles. Light gray represents the most common A0 motif pattern, dark gray represents the second most common A1 motif pattern, black represents remaining motif patterns. N = 1036.
An inclusive table of all 34 distinct motif patterns is provided in Supplementary Table 3, which also includes a detailed resolution of the motif patterns by pre-repeat, repeat, and post-repeat segments, and serves as a template for determining the numerical allele designation for each motif pattern. Thirteen of these motifs (C1 through D3) are only observed once and therefore are also unique sequences in this data set. The motif C1 was observed in two different samples but the sequences were identical and therefore only one unique sequence was identified for C1.
The pre-repeat had two predominant motifs based on a single base change, SNP rs9362477. The repeat segment also had two predominant motifs which included the basic repeat and the basic repeat interrupted by a pair of Ts. The post-repeat only had one major motif. All three segments varied from the predominant motifs, which increased the number of motif patterns identified.
3.6. Population statistics (by length and sequence)
Supplementary Table 4 contains a summary of the population statistics for the dataset. Statistics calculated from length-and sequence-based alleles are presented for comparisons. By sequencing, a five-fold increase in the number of alleles was observed in the 1036 data set (from 53 to 264 unique alleles) and observed heterozygosity increased by approximately 3% across the four populations.
The observed heterozygosity by sequence was based on the data reported in Supplementary Table 2, which includes the Repeat and Local Flanks region. The 3% increase in heterozygosity can primarily be attributed to 32 samples determined to be sequence-based heterozygotes. One sample was found to be of heterozygous length by CE (27.2, 28.2), homozygous length by sequencing (28.2, 28.2) but a sequence-based heterozygote, and therefore counted as heterozygous by sequence. Upon examining the 3’ Extended Flank, an aforementioned four base deletion, rs1371483225, was detected in the 3’ Extended Flank. Two other samples differed in zygosity by length and sequence due to the presence of the aforementioned rs369314007 four base deletion in the 3’ Extended Flank. One of these samples was homozygous in length by CE (17,17) and heterozygous length by sequence (17,18), while the other sample was heterozygous in length by CE (16, 17) and homozygous length by sequence (17, 17).
Further resolving SE33 through sequencing did not affect Hardy-Weinberg equilibrium, all p-values were greater than 5% (Bonferroni correction was not applied). The median random match probability (RMP) calculated for the 1036 dataset was 0.006787 (1 in 147) and 0.000552 (1 in 1810) using length-and sequence-based allele frequencies, respectively representing a 12-fold increase.
4. Discussion
Sequence-based allele frequencies for the SE33 STR locus were generated for 1036 U.S. population samples, using the FASTQ files from the ForenSeq DNA Signature Prep Kit on a MiSeq FGx instrument. The locus was complex to analyze compared to other loci in the ForenSeq kit, often requiring deliberate evaluation and confirmation of alleles. From the observed variation in DoC and ACR values obtained in this study, it was apparent that CE-based allele calls were needed to provide guidance and confidence in the bioinformatic workflow. Therefore, the sequence-based allele calls were supported by CE-based allele calls for all samples in this set. It would have been far more challenging, and perhaps not possible, to correctly assign all of the SE33 sequence-based alleles in the absence of independently generated allele calls.
In this work 274 unique sequences for the SE33 locus were determined to be present in this population sample set. There are 264 unique sequences present in the Repeat with Local Flanks region analyzed across all samples, and the statistical calculations are based on these alleles. It was not possible to evaluate the Extended Flanks for all 1036 samples with confidence, due to lower DoC for larger SE33 alleles. However, ten additional unique sequences were identified by manually evaluating and Sanger sequencing the 3’ Extended Flank in cases of discordance between CE and sequence allele calls. The presence of three deletions explained the discordance issues, two of which are likely to be observed in other studies (rs369314007 observed nine times in this study and rs1371483225 observed 12 times in this study). Screening dbSNP for additional variants located in the extended flanking regions reveals additional reported deletions which could also affect length-based allele calls.
Once overcoming the technical and informatic challenges of assigning allele calls, the informational gains of sequencing SE33 are obvious. The highly polymorphic nature of the sequence is demonstrated by the five-fold increase in observed alleles by sequence compared to CE (264/53). For the 1036 population samples, the occurrence of rare alleles defined as being observed less than five times in the CE-based allele counts was 1.98% (4½072). An increase to 14.33% (297/2072) was observed in the sequenced-based allele counts.
This work also highlights the fact that CE-concordant assignment of sequenced alleles requires the inclusion of what we refer to as Local Flanks. This includes 15 bases at the 5’ end of the repeat and 24 bases at the 3’ end of the repeat (forward strand), which are both highly variable and ambiguously demarcated from the Repeat Region in some sequences. These two Local Flanks are important to the sequence identification of SE33 and we recommend they be included in the bioinformatic analysis and reporting of sequence data for this locus.
We expect that additional SE33 sequences will be published by laboratories in the future. The evaluation of sequence motifs patterns included herein will help provide a reference for forensic nomenclature and an organized format to evaluate sequence variation at this locus.
Supplementary Material
Supplementary Table 1 – Extended Flank-Represented in this table are ten unique sequences that were observed by analyzing the Extended Flanks and one additional sequence that was unique in both the Repeat with Local Flanks region and unique in the Extended Flank region. Allele call by sequence (column B), CE (column C) and motif ID associated with the sequence (column D). The reported deletion (column E) and the counts observed for the Global, African American, Asian, Caucasian, and Hispanic groupings are in columns F-J. The sequence and the bracketed sequence are in columns K and L.
Supplementary Table 2 – Allele Frequencies-Frequencies for 264 unique SE33 sequences identified in the Repeat with Local Flanks region in the population samples. Listed in order of allele size and global frequency. The sequence counts for Global, African American, Asian, Caucasian, and Hispanic groupings are in columns D-H and the frequencies for these populations are in columns I-M. Sequences corresponding to the Repeat with Local Flanks region are listed in column N with the bracketed sequences in column O. *Italicized characters in column O indicate ambiguous repeat regions and or bracketing of the repeats for compression and simplification of the [CTTT] motif.
Supplementary Table 3-Motif Patterns-34 motif patterns (column A) identified in the data set are listed in order of frequency (column S). These are further separated into the 5’ motifs (pre-repeat) (column B-E), Central Repeat Motifs (column F-J), and 3’ motifs (post-repeat) (column K-N). In each motif the combinations of these three sections create a unique pattern. Italicized characters indicate ambiguous repeat regions and or bracketing of the repeats to attempt a best visual fit of the Local Flanks and Repeat region. Bolded and red font represent non-standard elements within a pattern. The ability to reconcile the CE-based allele call with the sequence is important and for each motif pattern the impact on the numerical allele designation of 5’ motif (column O), repeat (column P), and 3’ motif (column Q) have been determined. The result is the formula in column R that will provide identical numerical allele calls.
Supplementary Table 4-Population Statistics-Values for total number of observed alleles, genetic diversity (GD), polymorphism information content (PIC), match probability (PM), power of discrimination (PD), observed heterozygosity (Hobs), power of exclusion (PE), typical paternity index (TPI), Hardy-Weinberg equilibrium (pHW) and median random match probability (RMP) are listed for the global data set and further separated by population group. Analyses were performed using both the sequenced-based and CE-based alleles for comparisons.
Acknowledgements
We wish to thank Dr. John M. Butler for his review of the manuscript.
Funding – The FBI Biometrics Center of Excellence (BCOE): Forensic DNA Typing as a Biometric tool. NIST Special Programs Office: Forensic DNA.
Glossary
- ACR
Allele Coverage Ratio
- DoC
Depth of Coverage
- DPMB
DNA Primer Mix B
- GD
Genetic Diversity
- Hobs
Observed Heterozygosity
- NGS
Next Generation Sequencing
- NonMA
Non-Majority Allele
- PD
Power of Discrimination
- PE
Power of Exclusion
- pHW
Hardy-Weinberg equilibrium
- PIC
Polymorphism Information Content
- PM
Match Probability
- TPI
Typical Paternity Index
- UAS
Universal Analysis Software
Footnotes
Disclaimer – Points of view in this document are those of the authors and do not necessarily represent the official position or policies of the U.S. Department of Commerce or the Department of Justice. Certain commercial equipment, instruments, and materials are identified in order to specify experimental procedures as completely as possible. In no case does such identification imply a recommendation or endorsement by NIST, nor does it imply that any of the materials, instruments, or equipment identified are necessarily the best available for the purpose. All work presented has been reviewed and approved by the NIST Human Subjects Protections Office.
Conflict of interest – None (each author has to declare conflicts of interest)
All authors declare no conflict of interest
5. References
- [1].Moos M, Gallwitz D, The EMBO Journal. 1983, 2, 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Warne D, Watkins C, Bodfish P, Nyberg K, Spurr NK, Nucleic Acids Research. 1991, 19, 6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Polymeropoulos MH, Rath DS, Xiao H, Merril CR, Nucleic Acids Research. 1992, 20, 1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Urquhart A, Kimpton CP, Gill P, Hum. Genet 1993, 92, 637–638. [DOI] [PubMed] [Google Scholar]
- [5].Schneider HR, Rand S, Schmitter H, Weichhold G, Int. J. Legal Med 1998, 111, 97–100. [DOI] [PubMed] [Google Scholar]
- [6].Rolf B, Schürenkamp M, Junge A, Brinkmann B, Int. J. Legal Med 1998, 110, 69–72. [DOI] [PubMed] [Google Scholar]
- [7].Parson W, Ballard D, Budowle B, Butler JM, Gettings KB, Gill P, Gusmão L, Hares DR, Irwin JA, King JL, de Knijff P, Morling N, Prinz M, Schneider PM, Van Neste C, Willuweit S, Phillips C, Forensic Sci. Int. Genet. 2016, 22, 54–63. [DOI] [PubMed] [Google Scholar]
- [8].Novroski NMM, King JL, Churchill JD, Seah LH, Budowle B, Forensic Sci. Int. Genet 2016, 25, 214–226. [DOI] [PubMed] [Google Scholar]
- [9].Devesse L, Ballard D, Davenport L, Riethorst I, Mason-Buck G, Syndercombe Court D, Forensic Sci. Int. Genet 2017, 34, 57–61. [DOI] [PubMed] [Google Scholar]
- [10].Wendt FR, King JL, Novroski NM, Churchill JD, Ng J, Oldt RF, McCulloh KL, Weise JA, Smith DG, Kanthaswamy S, Budowle B, Forensic Sci. Int. Genet 2017, 28, 146–154. [DOI] [PubMed] [Google Scholar]
- [11].Wendt FR, Churchill JD, Novroski NM, King JL, Ng J, Oldt RF, McCulloh KL, Weise JA, Smith DG, Kanthaswamy S, Budowle B, Forensic Sci. Int. Genet 2016, 24, 18–23. [DOI] [PubMed] [Google Scholar]
- [12].Casals F, Anglada R, Bonet N, Rasal R, van der Gaag KJ, Hoogenboom J, Solé-Morata N, Comas D, Calafell F, Forensic Sci. Int. Genet 2017, 30, 66–70. [DOI] [PubMed] [Google Scholar]
- [13].Hill CR, Duewer DL, Kline MC, Coble MD, Butler JM, Forensic Sci. Int. Genet. 2013, 7, e82–e83. [DOI] [PubMed] [Google Scholar]
- [14].Steffen CR, Coble MD, Gettings KB, Vallone PM, Forensic Sci. Int. Genet 2017, 31, e36–e40. [DOI] [PubMed] [Google Scholar]
- [15].Kline MC, Hill CR, Decker AE, Butler JM, Forensic Sci. Int. Genet. 2011, 5, 329–332. [DOI] [PubMed] [Google Scholar]
- [16].Warshauer DH, King JL, Budowle B, Forensic Sci. Int. Genet 2015, 14, 182–186. [DOI] [PubMed] [Google Scholar]
- [17].Gouy A, Zieger M. Forensic Sci. Int. Genet 2017, 30, 148–151. [DOI] [PubMed] [Google Scholar]
- [18].Peakall R, Smouse PE, Bioinformatics 2012, 28 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gettings KB, Borsuk LA, Ballard D, Bodner M, Budowle B, Devesse L, King J, Parson W, Phillips C, Vallone PM, Forensic Sci. Int. Genet. 2017, 31, 111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Butler JM, Advanced Topics in Forensic DNA Typing: Methodology, Academic Press, San Diego, 2012, pp 590–598. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 – Extended Flank-Represented in this table are ten unique sequences that were observed by analyzing the Extended Flanks and one additional sequence that was unique in both the Repeat with Local Flanks region and unique in the Extended Flank region. Allele call by sequence (column B), CE (column C) and motif ID associated with the sequence (column D). The reported deletion (column E) and the counts observed for the Global, African American, Asian, Caucasian, and Hispanic groupings are in columns F-J. The sequence and the bracketed sequence are in columns K and L.
Supplementary Table 2 – Allele Frequencies-Frequencies for 264 unique SE33 sequences identified in the Repeat with Local Flanks region in the population samples. Listed in order of allele size and global frequency. The sequence counts for Global, African American, Asian, Caucasian, and Hispanic groupings are in columns D-H and the frequencies for these populations are in columns I-M. Sequences corresponding to the Repeat with Local Flanks region are listed in column N with the bracketed sequences in column O. *Italicized characters in column O indicate ambiguous repeat regions and or bracketing of the repeats for compression and simplification of the [CTTT] motif.
Supplementary Table 3-Motif Patterns-34 motif patterns (column A) identified in the data set are listed in order of frequency (column S). These are further separated into the 5’ motifs (pre-repeat) (column B-E), Central Repeat Motifs (column F-J), and 3’ motifs (post-repeat) (column K-N). In each motif the combinations of these three sections create a unique pattern. Italicized characters indicate ambiguous repeat regions and or bracketing of the repeats to attempt a best visual fit of the Local Flanks and Repeat region. Bolded and red font represent non-standard elements within a pattern. The ability to reconcile the CE-based allele call with the sequence is important and for each motif pattern the impact on the numerical allele designation of 5’ motif (column O), repeat (column P), and 3’ motif (column Q) have been determined. The result is the formula in column R that will provide identical numerical allele calls.
Supplementary Table 4-Population Statistics-Values for total number of observed alleles, genetic diversity (GD), polymorphism information content (PIC), match probability (PM), power of discrimination (PD), observed heterozygosity (Hobs), power of exclusion (PE), typical paternity index (TPI), Hardy-Weinberg equilibrium (pHW) and median random match probability (RMP) are listed for the global data set and further separated by population group. Analyses were performed using both the sequenced-based and CE-based alleles for comparisons.