Abstract
BACKGROUND
Neurodegenerative diseases feature stereotypical deposits of protein aggregates that selectively accumulate in vulnerable cells. The ability to simultaneously localize multiple targets in situ is critical to facilitate discovery and validation of pathogenic molecular pathways. Immunostaining methods enable in situ detection of specific targets. Effective stripping of antibodies, allowing successive rounds of staining while maintaining tissue adhesion and antigen integrity, is the main roadblock for enabling multiplex immunostaining in standard labs. Furthermore, stripping techniques require antibody-specific optimization, validation, and quality control steps.
NEW METHOD
Aiming to create protocols for multiplex localization of neurodegenerative-related processes, without the need for specialized equipment, we evaluated several antibody stripping techniques. We also recommend quality control steps to validate stripping efficacy and ameliorate concerns of cross-reactivity and false positives based on extensive testing.
RESULTS
A protocol using β-mercaptoethanol and SDS consistently enables reliable antibody stripping across multiple rounds of staining and minimizes the odds of cross-reactivity while preserving tissue adhesion and antigen integrity in human postmortem tissue.
COMPARISON WITH EXISTING METHODS
Our proposed method is optimal for standard lab settings and shows consistent efficacy despite the intricacies of suboptimal human postmortem tissue and the need to strip markers bound to highly aggregated proteins. Additionally, it incorporates quality control steps to validate antibody stripping.
CONCLUSIONS
Multiplex immunofluorescence methods for studying neurodegenerative diseases in human postmortem tissue are feasible even in standard laboratories. Nevertheless, evaluation of stripping parameters during optimization and validation phases of experiments is prudent.
Keywords: Multiplex histology, Immunohistochemistry, Immunofluorescence, Neuropathology, Postmortem human brain tissue
1. Introduction
High-throughput genomic and proteomic methods have enabled probing of the milieu of molecular biology underlying complex diseases. However, these methods destroy tissue integrity precluding insight on topography or co-localization, which are critical in conditions such as neurodegenerative diseases where different cell populations show a gradient of vulnerability and changes (Fu et al., 2018; Seeley, 2008; Seeley et al., 2009). Recent developments in single-cell -omics methods allow more granular probing of the selective changes in individual cells. Available single-cell methods inevitably require a tissue dissociation step that compromises cellular membranes causing the results to reflect the cell nuclear rather than cytoplasmic composition states. As most proteinaceous inclusions associated with neurodegenerative disease localize to the cytoplasm or in the extracellular space, it is challenging to sort out results coming from affected versus unaffected cells just by using single-cell -omics.
Immunostaining remains an attractive method to investigate topographical cellular vulnerabilities in neurodegenerative diseases and to validate the how disease-relevant are the pathways pinpointed by – omics because immunostaining has the potential to unlock insights into molecular cascades while preserving tissue, providing a spatial overview with subcellular resolution. Still, potentially valuable information often gets locked up in a tissue section due to the inherent low-throughput nature of immunostaining. Most immunohistochemistry and immunofluorescent methods rely on visualizing antigens by biding them to a specific (primary) antibody, which in turn, is bound to a secondary antibody. The positive reaction is visualized using a detection system containing and chromogen or a fluorescent dye. Although this approach works well for detecting single antigens, several factors restrict multiple antigen labeling, including the fact that only a few species are used to raise antibodies, and limitations in the number of visualization channels that do not overlap. Pre-staining conjugation of primary and secondary antibodies eliminates the risk of cross-reaction between conspecific antibodies. However, direct antibody conjugation requires large amounts of primary antibody, which makes the protocols too expensive for standard labs and makes it challenging to amplify the signal. The advent of tyramide signal amplification (TSA) brought options for combining conspecific antibodies in the same sample while providing signal amplification (Buchwalow et al., 2018; Chao et al., 1996; Dixon et al., 2015; Lim et al., 2018; Mansfield, 2017; Pirici et al., 2009; Roy et al., 2019; Sorrelle et al., 2019; Stack et al., 2014; Wang et al., 1999; Zhang et al., 2017). In short, TSA uses a peroxidase-mediated reaction to covalently deposit fluorophores or chromogens to tyrosine side chains proximal to the target epitope (Lim et al., 2018; Wang et al., 1999). Because of the covalent bond, stripping of primary and secondary antibodies should not affect tissue-bound tyramide-fluorophores or chromogens.
Nevertheless, even if eliminating the risk of cross-reactivity of conspecific antibodies, the number of spectrally separated detection channels precludes multiplex antibody detection. Signal of chromogenic reporters such as 3,3’-Diaminobenzidine, get detected by the same channel in brightfield microscopy. Thus they get blended, making it challenging to probe more than two targets simultaneously, especially when targets are topographically close (Dixon et al., 2015; Gown et al., 1986; Ilie et al., 2018; Lan et al., 1995; Nakane, 1968; Tramu et al., 1978). Fluorescence microscopy offers the possibility to detect more probes simultaneously by utilizing molecules with specific excitation and emission spectra as reporters, allowing selective visualization of different probes in separate channels, which are subsequently overlapped digitally (Coons, 1961; Coons et al., 1941; Coons et al., 1942; Coons and Kaplan, 1950). Still, because of limitations in a camera’s detectable wavelengths and presence of spectral overlap between fluorophores, most fluorescent microscopes are still limited to detecting three to five markers, depending on the camera. More recently, alternative methods for in situ detection were proposed, such as those replacing fluorescent-based reporters for quantum dots that are directly conjugated in primary antibodies. Quantum dots allow for a higher number of probes to be detected at the same tissue because of reduced spectral overlap (Byers and Hitchman, 2011; Chan et al., 2005; Chen et al., 2013; Dixon et al., 2015; Krenacs et al., 2010; Mansfield, 2017; Prost et al., 2016; Sweeney et al., 2008; Tholouli et al., 2008; Wu et al., 2003; Xu et al., 2013). However, quantum dots and other similar techniques require expensive, specialized equipment.
The ability to strip antibodies after staining allows for using an antibody raised on a species previously used in a second round of staining and frees detecting channels for new antibodies. Several methods for stripping antibodies have been proposed. Still, almost none have been optimized to accommodate the intricacies of human postmortem tissue (that usually show suboptimal quality), or to strip antibodies bound to highly aggregated proteins that may be more difficult to remove, like those used for detecting neurodegenerative pathology. Moreover, most stripping methods tend to affect tissue integrity or slide adhesion, precluding additional rounds of staining. Here, we evaluate different multiplex histology protocols to identify a reliable, low-cost multiplex immunostaining method for routine use in the setting of a standard neuropathology laboratory. Conceptually, our technique of choice would result in a complete, efficient, and reliable stripping of a broad array of antibodies while preserving tissue adhesion on the slide and maintain similar levels of antigenicity in FFPE samples, across multiple rounds of stripping and staining. We chose to use fluorescence detection because immunofluorescence microscopes are widely available and accommodate more detection channels than brightfield microscopy. Next, we make recommendations on limitations, appropriate use, and necessary steps to incorporate techniques into experimental setups with emphasis on quality control steps.
2. Material and Methods
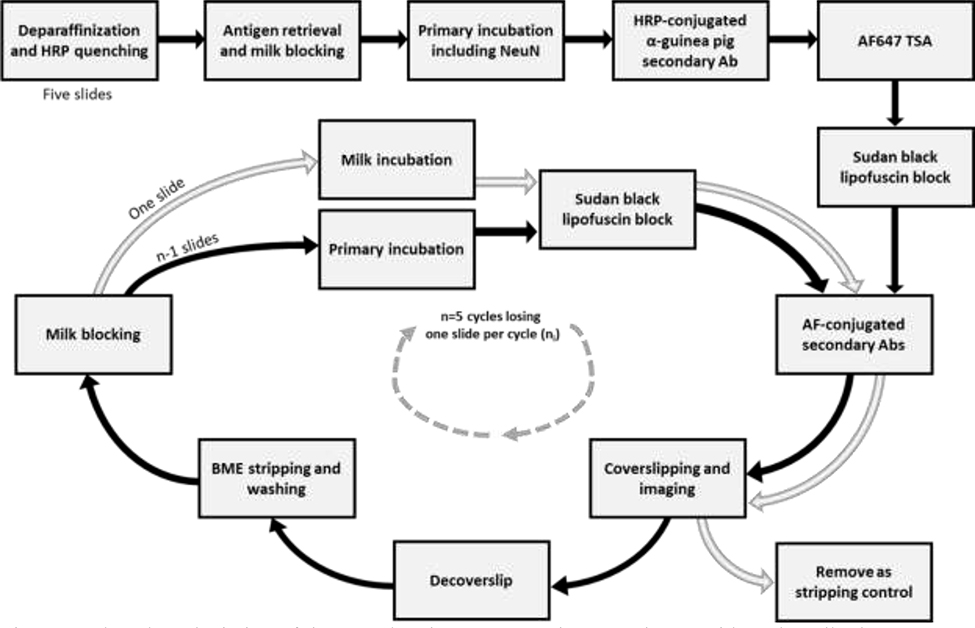
Towards establishing our protocol, we first created a list of principles and published approaches proposed to strip antibodies based on a literature survey. Next, we tested these protocols in two experimental stages. In the first stage, we tested the most promising stripping techniques representing seven different approaches. We use each of these seven approaches to strip three antibodies: one used to label neurons, another to stain astroglia, and most importantly, one used to label phospho-Ser202-tau (phospho-tau), a common component of highly aggregated inclusions in neurodegenerative diseases. Only one stripping protocol met our success criteria. In the second stage, we tested the only successful stripping method to evaluate its efficacy in a larger number of antibodies and its impact on tissue adhesion and antigen integrity across multiple rounds. Multiple rounds of staining are crucial in multiplex protocols relying on immunofluorescence because every round is limited to three to five channels. Moreover, the signal of at least one marker must remain present throughout the multiple rounds to allow digital co-registration of all image sets. Thus we performed five rounds of staining and stripping, using a panel of six antibodies (Figure 1).
Figure 1:
Flowchart depiction of tissue and antigen preservation experiment with each antibody set starting with five slides (stage 2). One slide was removed each round for checking stripping efficiency and then kept in storage.
2.1. Literature review and selection of candidate stripping techniques
We searched PubMed for the terms “antibody elution histology” and “multiplex immunohistochemistry.” “Antibody elution histology” yielded 462 results, and “multiplex immunohistochemistry” yielded 2021 results. In total, these combined searches yielded 2480 unique results ranging from November 1964 to May 2019. These results were manually filtered to exclude articles unrelated to histology. From the remaining, we identified seven main stripping strategies previously used in multiplex immunohistochemistry protocols. Although stripping techniques may either elute or denature antibodies, both terms are used interchangeably in the literature. These seven strategies are (1) heat-based, particularly common in multiplex protocols using TSA (Ilie et al., 2018; Jufas et al., 2008; Lan et al., 1995; Lim et al., 2018; Mansfield, 2017; Parra et al., 2017; Roy et al., 2019; Saylor et al., 2018; Sorrelle et al., 2019; Stack et al., 2014; Toth and Mezey, 2007; Wegner et al., 2017; Zhang et al., 2017); (2) a glycine/SDS solution at a low pH (Bolognesi et al., 2017; Gut et al., 2018; Nakane, 1968; Narhi et al., 1997a; Pirici et al., 2009; Sorrelle et al., 2019); (3) commercially available denaturing solutions for IHC with proprietary formulations (Buchwalow et al., 2018); (4) β-mercaptoethanol/Sodium dodecyl sulfate (BME/SDS) denaturation (Bolognesi et al., 2017; Gendusa et al., 2014; Kim et al., 2012; Mansfield, 2017; van den Brand et al., 2014) which reduces the disulfide bonds present in antibodies, thus breaking down their tertiary structure (Capel et al., 1980; Crivianu-Gaita et al., 2015); (5) a low-pH oxidizing solution of KMnO4 and H2SO4 (Glass et al., 2009; Tramu et al., 1978); (6) chaotropic salts (Bolognesi et al., 2017; Gut et al., 2018; Narhi et al., 1997a; Narhi et al., 1997b); (7) combining antibodies with drastically different abundances (Wang et al., 1999).
From these seven candidate approaches, we initially excluded three. Techniques based on a low-pH oxidizing solution (Glass et al., 2009; Tramu et al., 1978) tend to inactivate fluorophores and damage the tissue. Chaotropic salts, such as guanidinium, are not effective at stripping antibodies bound to highly aggregated, spatially organized targets and require steps to recover antigen conformations following the denaturation step (Bolognesi et al., 2017). Finally, techniques that rely on making a priori assumption that some targets are less abundant than others (Wang et al., 1999) are lack rigor as the use implicit assumptions regarding the sensitivity of microscope cameras and variation in biological samples. Further, we omitted heat-based methods with Tris-EDTA (Cappi et al., 2019), as exposure to EDTA has a tendency to decrease antigenicity for our targets of interest (Cappi et al., 2019). Table 1 depicts the four stripping techniques tested in Stage 1.
Table 1:
Candidate stripping techniques tested on Stage 1.
| Strategy | Description | Reference for protocol |
|---|---|---|
| Heat | Microwave in citrate buffer with 0.05% tween. High setting until boiling then 15 minutes at 20% (Note: referenced paper suggests 5 minutes at 50%). | Toth and Mezey (2007) |
| Glycine/SDS at low pH | Glycine mixed with SDS, at pH 2.0 and heated to 50°C for 30 minutes. | Pirici et al. (2009) |
| Denaturing solution | One part of solution A to two parts of solution B from the Denaturing Solution Kit from Biocare Medical | Manufacturer instructions (part DNS001L) |
| BME | Tris-HCl mixed with SDS and BME, heated to 56C and incubated for 30 minutes. Washed in TBS. | Gendusa et al. (2014) |
2.2. Experimental Stage 1: evaluating stripping techniques
2.2.1. Rationale
We tested stripping efficiency and effects on immunogenicity for each of the four candidate stripping techniques using three different primary antibodies, either labeling tau protein aggregates in Alzheimer’s disease or two cell-type markers, all three raised in different species. Each primary antibody was tested in four serial histological slides (total of 12), one for each stripping technique. We incubated the slides with one of the three primary antibodies, followed by incubation with a host species-specific HRP-conjugated secondary antibody developed with TSA using a tyramide-conjugated to Alexa Fluor 647 (AF647). For this reason, AF647 signal represents the distribution of antibody reactivity. Thus, the signal in this channel (Cy5) is set as the standard. Next, we applied each stripping technique. Because TSA covalently deposits fluorophores to the tissue, the fluorescent signal should remain even after the primary and secondary antibodies are stripped (Lim et al., 2018; Wang et al., 1999), meaning that signal in channel Cy5 should remain intact after stripping. Next, we incubated the same slides again with the same species-specific HRP-conjugated secondary used before, but this time, we developed the reaction with tyramide-conjugated with Alexa Fluor 546 (AF546) to test whether the primary antibodies were successfully stripped. We then imaged the Cy5 and DsRed channels. If the stripping was successful, the reaction would yield no signal in the DsRed channel. Any detected signal in the DsRed channel would indicate a failure in the stripping. Finally, to test whether the stripping treatment damaged the target antigens, we re-incubated the same slides with the same primary antibodies followed by HRP-conjugated secondary antibodies and developed the reaction with tyramide conjugated to Alexa Fluor 488 (AF488) and imaged the GFP channel. Signal overlap between AF488 (visualized in the GFP channel) and AF647 (visualized in the Cy5 channel) would suggest preserved integrity of the target antigen.
2.2.2. Tissue source
De-identified human brain tissue was supplied by the Neurodegenerative Disease Brain Bank from the University of California, San Francisco’s Memory and Aging Center. Post-mortem interval of the cases varied from 7 to 30 hours. Brain samples were fixed in 10% neutral buffered formalin for 72 hours and transferred to PBS-azide at 4°C for long-term storage. Tissue was embedded in paraffin, then 8μm thick FFPE sections were mounted on Ultra Bond adhesive slides (SL6023–1, Avantik BioGroup) and incubated in an oven at 65°C for at least 18 hours before further processing. Tissue collection and processing were approved by the institutional review boards from the University of California, San Francisco, and this experiment was considered to belong to the non-human category (postmortem and de-identified) by the same IRB.
2.2.3. Primary antibodies
We chose a commonly used monoclonal antibody that targets pathologic aggregates of hyperphosphorylated tau protein (CP13, phospho-Ser202 tau, 1:800; gift of Peter Davies, New York). We also used antibodies against Fox-3 (RBFOX3 NeuN, 1:600; #266004, Synaptic Systems, Goettingen, Germany) and glial fibrillary acidic protein (GFAP, 1:1800; #ab68428, Abcam) that label neurons and astroglia, respectively. Table 2 depicts the host species, vendor information, and dilution used for all primary antibodies used in this study, including those used in Experimental Stage 2.
Table 2:
Primary antibodies used in this study
| Antibody | Host | Label | Source | Working dilution |
|---|---|---|---|---|
| CP13 | Ms, monoclonal IgG2b | Phospho-tau (Ser 202) | Gift from Peter Davies, New York, NY | 1:800 |
| TMEM119 | Rb, polyclonal | Yolk-sac derived microglia | Sigma Aldrich (HPA051870) | 1:500 |
| PHF-1 | Ms, monoclonal IgG1 | Phospho-tau (Ser 396/404) | Gift from Peter Davies, New York, NY | 1:9000 |
| GFAP | Rb, monoclonal EPR1034Y | astroglia | Abcam (ab68428) | 1:1800 |
| MC1 | Ms, monoclonal IgG1 | Conformational tau | Gift from Peter Davies, New York, NY | 1:1000 |
| T18 | Rb, polyclonal | Oligomeric tau | Rakez Kayed, Galveston, TX | 1:1500 |
| NeuN | GP, polyclonal | Neurons | Synaptic Systems (266004) | 1:600 |
2.2.4. Immunostaining protocol
Except for the stripping step, all steps of the immunostaining protocol are routine in histology labs. Slides underwent serial deparaffinization steps as follows: three ten-minute immersions in xylene, two two-minute immersions in 100% ethanol, two two-minute immersions in 96% ethanol, and one two-minute immersion in 80% ethanol. Slides were then immersed in a solution of 3% hydrogen peroxide (H2O2) and 80% methanol for 30 minutes to quench any endogenous peroxidase. Sections underwent three two-minute washes in distilled water (dH2O) before being transferred into a 10% solution of 0.1M citrate buffer pH 6.0 with 0.05% tween-20 for antigen retrieval. In the antigen retrieval solution, sections were cycled through an autoclave set at 121oC for five-minutes. Following antigen retrieval, sections were left to cool to room temperature (RT) for approximately 60 minutes. After thorough washing in a solution of 1x PBS and 0.05% tween (PBST), sections were immersed in a solution of 5% milk with 0.05% tween (herein referred to as milk) for 30 minutes to block unspecific binding. Sections were then incubated in CP13 (1:800, gift of Peter Davies), NeuN (1:600; #266004, Synaptic Systems), or GFAP (1:1800; #ab68428, Abcam) diluted in 5% milk PBS solution for 16 hours overnight at RT.
After washes in PBST, sections were incubated in a 1:400 concentration of species-specific HRP-conjugated secondary antibody (goat-α-mouse IgG(H+L)-R-05071, Advansta; goat-anti-guinea pig IgG (H+L)-R-05076, Advansta; or goat-anti-Rabbit IgG (H+L)-R-05072, Advansta) diluted in PBST for 60 minutes. Following additional PBST washes, the antibodies were developed with TSA following the manufacturer’s instructions with a solution of 1:100 Alexa Fluor 647 (AF647) Tyramide (B40958, Thermo Fisher) and 1:100 of 100x H2O2 in 1x tris-buffered saline for 15 minutes and was followed by PBST washes. Following TSA, one of several possible stripping techniques was employed according to the protocols given by the references noted in Table 1.
Next, sections were re-incubated in a 1:400 concentration of species-specific HRP-conjugated secondary antibody for 60 minutes, followed by PBST washes, and an additional TSA step with Alexa Fluor 546 (AF546) Tyramide (B40954, Thermo Fisher) for 15 minutes. The TSA step was ended with a PBST wash. Finally, the sections were re-incubated with the original primary and secondary antibodies as previously described, followed by an additional TSA step according to the manufacturer’s instructions with a solution of 1:100 Alexa Fluor 488 (AF488) Tyramide (B40953, Thermo Fisher) and 1:100 of 100x H2O2 in 1x tris-buffered saline. This amplification occurred for 15 minutes and was followed by PBST washes.
After a transfer through 70% ethanol, the sections were treated with a solution of 0.8% Sudan Black-B in 70% ethanol for 35 minutes for blocking lipofuscin autofluorescence, followed by two ten-second washes in 70% ethanol to remove excess Sudan black-B. Sections were re-hydrated in PBS then coverslipped with Prolong Glass Antifade Mountant with NucBlue (P36981, Thermo Fisher).
2.2.5. Image acquisition
Slides were imaged on a Zeiss AxioImager.A2 microscope equipped with a Zeiss Colibri 7:Type FR-R[G/Y]CBV-UC 7-channel fluorescence light source. NucBlue (Hoechst 33342) was visualized with a DAPI filter set, AF488 visualized with a GFP filter set, AF546 with a DsRed filter set, and AF647 with a Cy5 filter set. Exposure times and visualization parameters were kept consistent for each antibody group.
2.3. Stage 2: Evaluation of tissue and antigen preservation through multiple rounds
2.3.1. Rationale
β-mercaptoethanol/SDS-based (BME/SDS) stripping was the only technique that successfully stripped all antibodies and did not cause the tissue to detach from the slide (Figs. 2 and Supplemental Figs. 1–2). In stage 2, we investigated whether BME/SDS-based stripping could accommodate up to five rounds of staining, coverslipping, imaging, decoverslipping, stripping, and re-staining in six different antibodies. We combined the primary antibodies in groups of three and stained five slides per group. Importantly, all antibody combinations included NeuN, which was selected as an optimal reporter for allowing co-registering the images obtained across different rounds of staining. Thus, immunostained for NeuN developed with TSA always occurred in round one, and the corresponding detection channel would be blocked for other antibodies. The other two antibodies in each group were always developed with species-specific secondary antibodies conjugated with AF488 or AF546 to allow eliminating the fluorescent signal associated with the antibody being stripped.
Figure 2:
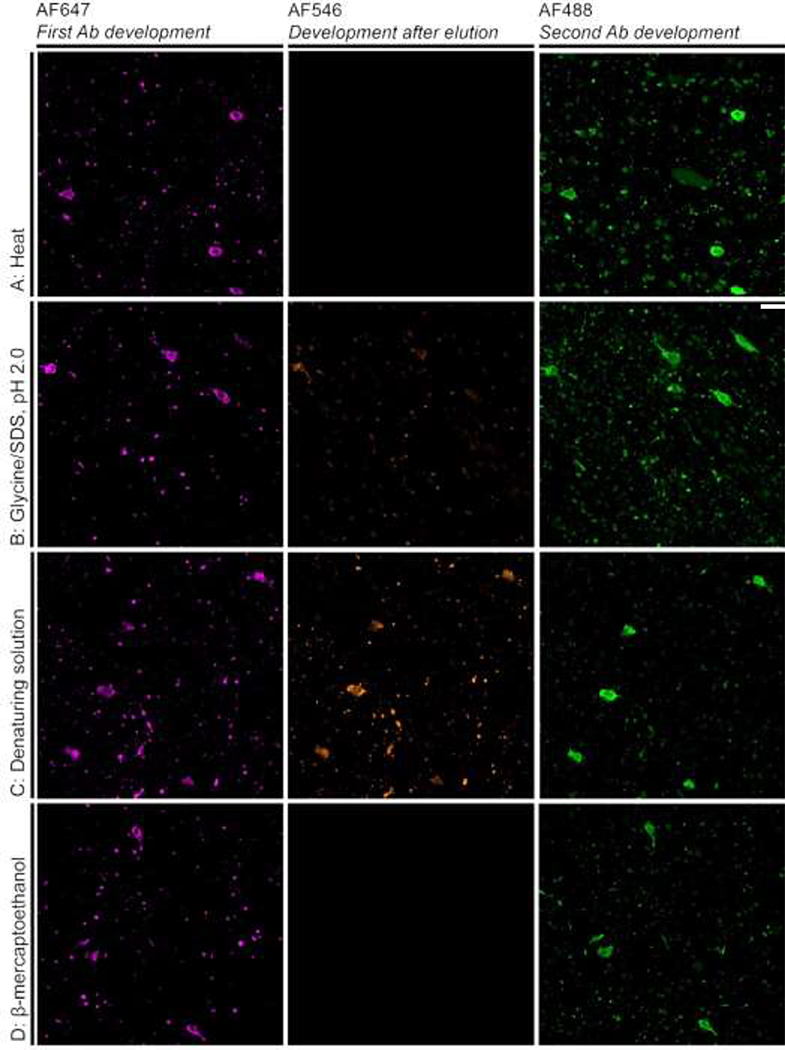
Photomicrographs of the validation experiments for antibody CP13. Overlapping AF647 and AF488 signals represents antigen preservation. Lack of AF546 signal represents successful stripping, which was achieved only for heat and β-mercaptoethanol/SDS strategies (A and D) strategies. The Glycine/SDS, pH 2.0 strategy (B) resulted in a reduced, but noticeable AF546 signal, and denaturing solution strategy (C) resulted in a significant AF546 signal. Scale bar: 10 μm
In the first round of staining, all five slides of each set underwent the same procedure. In the subsequent rounds, one of the slides of each set was set aside and incubated with 5% milk PBS instead of the primary antibodies as quality control for the stripping step. Thus, in rounds 2, 3, 4, and 5, only 4, 3, 2 and 1 slides of the set underwent primary antibodies incubation (Fig. 1). For each group of antibodies, we tested the maximum number of rounds accommodated by the protocol (up to 5), determining at which round, if any, the tissue started to disappear from the slide (either by dissociation or detachment or both) and/or the signal (number of cells labeled and signal intensity) for a given antibody was markedly changed compared to the 1st round.
2.3.1. Tissue selection and primary antibodies
We tested this protocol successfully in different cases. In this study, we used serial sections from the inferior temporal gyrus of a case with a neuropathological diagnosis of Alzheimer’s disease (Braak stage 5) and diffuse neocortical Lewy Body disease. The case had a PMI of 12 hours and was fixed for 72 hours. Immunostaining protocol steps were similar to those described for stage 1 up through the incubation of primary antibodies step. Table 2 provides information on the primary antibodies and grouping. Notably, we included the MC1 antibody against conformationally altered tau proteins because it labels highly aggregated inclusions (Eckermann et al., 2007).
2.3.2. Cycle of staining and stripping
After incubating the slides in a cocktail of the three primary antibodies, slides were washed thoroughly in PBST then incubated in 1:400 goat-anti-guinea pig IgG (H+L) HRP conjugated-secondary antibody (R-05076, Advansta) diluted in PBST for 60 minutes. Following additional PBST washes, the reaction was developed with TSA following the manufacturer’s instructions with a solution of 1:100 AF647 Tyramide (B40958, Thermo Fisher) and 1:100 of 100x H2O2 in 1x tris-buffered saline. The reaction was stopped with a PBST wash.
Following PBST washes and a transfer through 70% ethanol, the sections were treated with a solution of 0.8% Sudan Black-B in 70% ethanol for 35 minutes to block for lipofuscin. After 35 minutes, two 10-second washes in 70% ethanol was used to remove excess Sudan black-B. Sections were incubated in Sudan black-B between TSA development of NeuN and the fluorescent-conjugated secondary antibodies to be consistent with the subsequent test rounds that omitted NeuN incubation and development.
After rehydration in PBS, sections were incubated in a cocktail of 1:400 Goat anti-Rabbit IgG (H+L) (A-11008, Life Technologies) and 1:400 AF546 Goat anti-Mouse IgG (H+L) (A-11003, Life Technologies) in PBS for 90 minutes. Following washing in PBS, the slides were coverslipped with Prolong Glass Antifade Mountant with NucBlue (P36981, Thermo Fisher) then imaged at 10x magnification. After imaging, the slides were submerged in PBST overnight and agitated in order to remove the coverslip. Sections were washed in additional PBST to remove any excess mounting media.
BME/SDS stripping solution was prepared according to Gendusa et al. (2014). Under a fume hood, 20ml of 10% SDS was mixed with 12.5ml of 0.5M Tris-HCl (pH 6.8), 67.5ml of deionized (Millipore) water, and 0.8ml of BME. The solution was heated to 56oC before use. Sections were incubated in the heated BME solution for 30 minutes. Sections were washed in four 15-minute dH2O immersions and then washed in TBST for five minutes, followed by immersion in 5% milk PBS solution for 30 minutes. One slide of each set was left in the milk solution overnight. All other slides were re-incubated with the original primary antibodies of each set (except NeuN) followed by the appropriate incubation in 1:400 Goat anti-Rabbit IgG (H+L) and 1:400 AF546 Goat anti-Mouse IgG (H+L) for 90 minutes, Sudan black treatment, mounting and imaging. The procedure was repeated up to 3 more times (Fig. 1).
2.3.3. Imaging acquisition and quantification
See the imaging parameters described for Stage 1 (see 2.2.5).
Inter-round agreement of antibody signal and TSA signal integrity over the multiple rounds were measured after each round in the same ROIs (coordinates) through all five rounds. For each ROI, the number of cells was counted manually, and the percentage of ROI with signal was measured after thresholding the grayscale image for each channel. The same thresholding parameters were applied for each round. The inter-round agreement of antibody signal represents how each stripping and re-staining step affects antigen integrity. The stripping efficacy was measured in each stripping control slide by probing the signal in a given ROI before and after the stripping step.
3. Results
3.1. Stage 1: Evaluation of stripping techniques
3.1.1. Phospho-tau antibody CP13
Only the heat-based and BME/SDS-based stripping methods were efficient in stripping this antibody, as noted by the lack of AF546 signal (Figure 2A & D). However, tissue partially dissapeared from the slide after the heat treatment. AF546 signal overlapping with AF647 signal on slides treated with glycine/SDS at low pH-based and commercial denaturing solution-based stripping techniques indicated a partial stripping of the antibodies (Figure 2B & C). In all stripping techniques, we observe high colocalization between AF488 and AF647 signals, indicating that antigenicity of the phospho-Ser202 Tau epitope detected by CP13 was preserved through the one round of stripping. Curiously, we noted higher AF488 than AF647 signal in the slides treated with heat-based and glycine/SDS at low pH-based methods, suggesting that these two strategies may expose additional epitopes for antibody reactivity. Table 3 summarizes these results. In a separate set of experiments, we tested if cutting the concentration of BME/SDS by half would be effective in stripping antibodies, and the results were negative (thus data not shown).
Table 3:
Summary of outcomes of various stripping techniques from Stage 1.
| Strategy | Effective stripping | Antigen preservation | Tissue preservation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CP13 | NeuN | GFAP | CP13 | NeuN | GFAP | CP13 | NeuN | GFAP | |
| Heat | + | + | + | + | + | + | - | - | - |
| Glycine/SDS at low pH | - | + | + | + | + | + | + | - | - |
| Denaturing solution | - | + | + | + | + | + | + | + | + |
| BME | + | + | + | + | + | + | + | + | + |
+ represents lack of signal on channel AF546
3.1.2. Antibodies to label cell-types
All the stripping strategies were capable of stripping antibodies, as shown by absent AF546 signal (Supplemental Figures 1–2). Further, we observed high colocalization between AF488 and AF647 in all slides, indicating that the antigenicity of the epitopes for these antibodies was preserved after one round of stripping. In fact, higher AF488 than AF647 signal was noted in all strategies tested, suggesting that either the stripping reagents may expose additional epitopes for antibody reactivity, or, just reflect that most cameras are more sensitive to GFP than to Cy5. Once again, sections treated with a heat-based method partially dissapeared from slides. Table 3 depicts a summary of these results.
Overall, the BME/SDS stripping strategy was the only one meeting our criteria, although other techniques worked for some of the antibodies.
3.2. Stage 2 results
3.2.1. Stripping efficacy
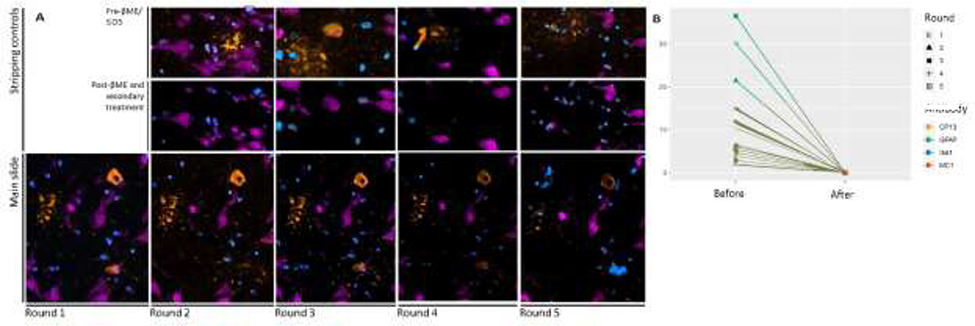
We evaluated stripping efficacy after each round by imaging the slide set aside as the quality control (Fig. 3a illustrates antibody MC1). For all antibodies and in all rounds, cell counts and area fraction with signal went down to zero following BME/SDS-based stripping (Fig. 3b).
Figure 3:
(A) MC1 signal (orange) along with TSA-developed NeuN (pink) and Hoechst 33342 (blue) across five rounds of staining, stripping, and re-staining using βME/SDS-based stripping. (Upper and middle rows) Stripping controls for each round, confirming the lack of signal after stipping and treated with AF546-conjugated secondary antibody. (Lower row) Signal from MC1 (orange) following staining, stripping, and re-staining across five rounds. AF647-TSA-developed NeuN antibody (pink) only occurred in round 1. Hoechst 33342 was stained in each round, as it is contained in the Prolong mounting media. MC1 Signal appeared in all rounds. However a decrease in signal intensity is noted by Round 4. (B) Percentage of ROI area with fluorescent signal, following thresholding, before and after the stripping step for the stripping control slides. Each line connects the pre- and post-stripping signal for a single ROI for a single slide. In all rounds and for all antibodies, the BME/SDS-based stripping technique successfully removed all signal. Scale bar: 10 μm
3.2.2. Tissue adhesion to slide across rounds
For all antibody sets tested, tissue sections had begun to dissapear (either by dissociation or detachment or both) from the glass after the fourth round. Thus, for the experimental conditions described here, if the whole tissue section is required for analysis, we recommend limiting protocols to four rounds, meaning that up to 11 antibodies could be probed in the same section of postmortem human brain tissue when using a fluorescent microscope with 3 channels.
3.2.3. Evaluating antigen integrity across rounds
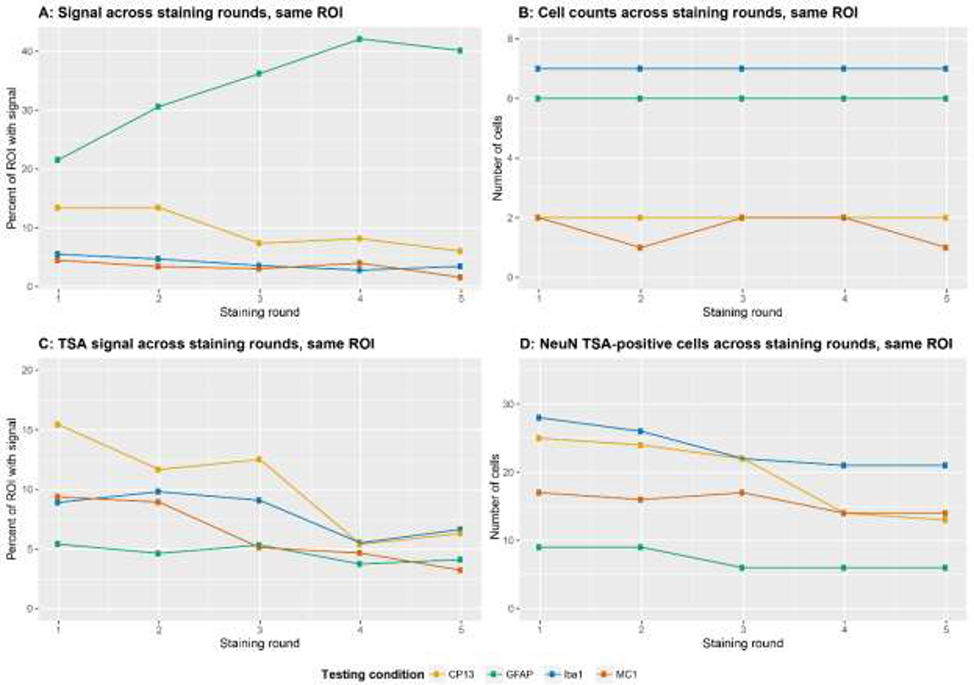
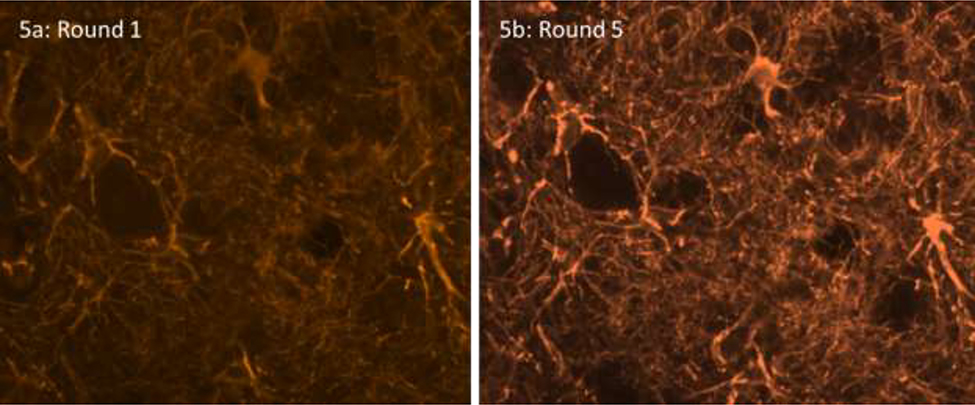
The number of cells positive for each antibody remained constant between rounds (Fig. 4a). However, there were changes in the percent area with a signal (Fig. 4b). Interestingly, GFAP had a net increase in percent area with signal because of increased visibility of distal astrocytic processes (Fig. 5), whereas CP13 and Iba1 had a net decrease in percent area with a signal.
Fig. 4:
In Stage 2, we used a set of 5 slides for each experimental setting. In the one slide per experiment that underwent five rounds of staining and stripping, we measured the percent ROI area with fluorescent signal (A) and number of positive cells (B). Also, in each experimental set, we used TSA-developed NeuN as a reporter. NeuN was only used on the first round of staining and we measured changes in TSA signal after each round of staining, in the one slide per experiment that underwent five rounds. C/D show, respectively, the percent ROI with TSA signal and number of positive cells. Different line colors represent the experimental set.
Fig. 5:
GFAP signal after the first round (5a) and the fifth round (5b) of staining. The number of astrocytes detected remained the same, but the signal in the processes got stronger.
3.2.4. Evaluating TSA signal intensity across rounds
We proposed to reserve one reporter for facilitating co-registering images from these rounds. Here, we used a NeuN antibody. We noticed a decrease in both cell number and percent area with signal for the TSA-deposited fluorophore associated with NeuN (Fig. 4c–d). Still, we saw enough signal to enable co-registration. As a less attractive alternative, Hoechst 33342 signal from each round of staining could be used for co-registration, although it requires more computational power. Here, we detected Hoechst 33342 signal in each round after coverslipping using Prolong with NucBlue, as expected (Fig. 3a – lower row).
3.2.4. Composite image
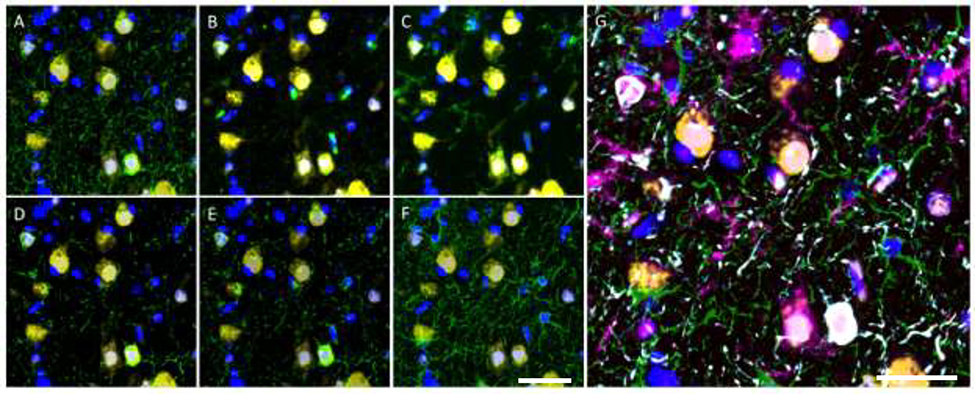
To further validate this pipeline in practice, we applied this same protocol to a single section but with a total of four different phospho-tau antibodies, three antibodies for specific cell types, and Hoechst33342 (Table 2) in three rounds of staining. We used TSA-deposited AF647 to label NeuN in the first round and Hoechst33342 after each round to facilitate co-registering images from each round (Fig. 6).
Figure 6:
Multi-round immunofluorescence from three rounds of staining, separated each by a BME elution. Each round included an anti-Ms and anti-Rb primary antibody and a stripping control. Round 1 also included NeuN (yellow) developed with AF647. After each round, slides were coverslipped with Prolong mounting media containing Hoechst33342 (blue). In panels A-G, NeuN is yellow and Hoechst33342 is blue. In green, panel A has CP13, B has MC1, C has TMEM119, D has T18, E has PHF-1, and F has GFAP. In panel G, blue is Hoechst33342, green is GFAP, pink is TMEM119, yellow is NeuN, red is MC1, gray is T18, cyan is PHF-1, and CP13 is orange. Because of the high overlap between tau inclusions, CP13, MC1, T18, and PHF-1 cannot be distinguished in the composite and appear as white. Scale bar: 10 μm
4. Discussion
In this study aiming to establish a standardized protocol for multiplex immunofluorescence in postmortem human brain tissue for investigating neurodegenerative diseases with the resources of standard histopathology labs, we examined the efficacy of several previously reported techniques for antibody stripping. We chose to focus on antibody stripping methods because a successful protocol would mitigate the most common issues associated with multiplex immunostaining protocols, which includes a restricted availability of detection channels for simultaneous detection in standard fluorescence microscopy and risk of cross-linking among conspecific antibodies. Successful stripping protocols have been published before, but none was optimized explicitly for postmortem human brain tissue with neurodegenerative pathology. From the seven main stripping strategies identified in the literature, we found that the BME/SDS-based stripping was the most reliable technique for stripping antibodies, including those labeling highly aggregated proteins while being relatively gentle to the tissue and antigen integrity. In Stage 2, we tested tissue adhesion and antigen integrity across five rounds of BME/SDS-based stripping in a variety of antibodies. MC1 was of particular interest, as it targets a conformation-specific epitope of tau known to occur in neurofibrillary tangles. The potential for the stripping technique to alter the target confirmation was of concern. Although the signal intensity decreased across the rounds, the number of cells positive for MC1 remained the same (Figs. 3 and 4).
While this study does not provide novelty regarding antibody stripping, we tested several methods using similar conditions in the same tissue, thus providing a roadmap to enable multiple lab to implement multiplex immunostaining protocols that includes “difficult-to-strip” antibodies in postmortem human brain tissue, which is prone to detaching, without the need to invest in expensive equipment. The fact that postmortem brain tissue has a higher tendency to detach from histological slides may explain why in our hands, microwave heating of samples in citrate buffer for antibody stripping, the recommended universal strategy for stripping antibodies in TSA-based multiplex immunostaining protocols from ThermoFisher Scientific (Company publication #MAN0015834) and PerkinElmer (“Opal Multiplex IHC Assay Development Guide”), caused tissue to detach from the slide.
The pipeline requires multiple rounds of staining, but the images of each round can be easily co-registered using open-source or proprietary imaging analysis tools (Alegro et al., 2017; Camp et al., 2002; Marrero et al., 2016; McCabe et al., 2005). Our experiments showed that either the use of Hoechst 33342 in each round of staining or TSA reporting of a neuronal target are good options as landmarks for co-registering images obtained from different rounds of staining.
Using our suggested approach, our protocol may accommodate up to four rounds of staining, representing at least 11 markers. Use of fluorescent microscopes equipped with more than four channels may enable testing of a larger number of markers. Also, in our lab, Ultra Bond adhesive slides proved to be the best cost-effective solution to avoid tissue detachment. Still, other labs may achieve more than four rounds of staining by using other coating methods on histological slides.
Our proposed approach for controlling striping effectiveness and antigen integrity may be valuable to increase the rigor of experiments in easy steps. TSA reporters (in here, a combination of signal from AF546 and AF488) are extremely sensitive to residual antibodies left after stripping steps that might otherwise be missed if using traditional immunofluorescence (Chao et al., 1996; Wang et al., 1999).
Despite being easy to implement and incorporating quality control steps, some precautions are key to guarantee the success of our protocol. As a quality control step, a “stripping control slide” for each antibody should be used following the design proposed on our Stage 1. Further, as the signal captured from an immunohistochemical experiment is not stoichiometric, particularly in a multiplex setup, this protocol is not suitable for experiments having as an outcome signal intensity. However, if the primary outcome is cell or particle counts, this technique is appropriate. The signal of TSA-deposited fluorophores decreased after each round of imaging. Thus, the TSA signal should be re-evaluated prior to starting a new round of staining to ensure that images can be co-registered. Although BME/SDS-based stripping method has been effective in our experiments, we titrated each primary antibody to the optimal signal to noise level. Use of overconcentrated primary antibody may result in poor stripping results. Further, we have noted that BME/SDS solution older than one week leads to worse results.
In conclusion, we present a multiplex immunostaining protocol that can be easily implemented in standard labs, is appropriate for use in suboptimal tissue, and can incorporate a large array of antibodies. We detailed the protocols and the proposed quality control steps to validate antibody stripping. With proper validation, optimization, and implementation of controls, use of multiplex immunostaining protocols is realistic in standard histopathology labs and will likely be key for elucidating the molecular fundamentals of neurodegenerative diseases.
Supplementary Material
Highlights.
Multiplex immunostaining method that does not require specialized equipment
Antibody elution step strip antibodies bound to highly aggregated protein
Elution step is gentle to suboptimal tissue enabling multiple staining rounds
Quality control step to check the elution efficiency in easy steps
5. Acknowledgments
The authors thank the donors to the Neurodegenerative Disease Brain Bank of the University of California, San Francisco Memory and Aging Center and their families for their support of this work. The authors also thank Celica Cosme, Rana Eser, and Mackenzie Hepker for their technical assistance and laboratory support.
5.1 Financial support
Support for this study comes from NIH grants U54NS100717 and K24AG053435, NIH institutional grants P50AG023501 and P01AG019724, and the Tau Consortium.
Footnotes
Declarations of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alegro M, Theofilas P, Nguy A, Castruita PA, Seeley W, Heinsen H, Ushizima DM, Grinberg LT. Automating cell detection and classification in human brain fluorescent microscopy images using dictionary learning and sparse coding. J Neurosci Methods, 2017; 282: 20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi MM, Manzoni M, Scalia CR, Zannella S, Bosisio FM, Faretta M, Cattoretti G. Multiplex Staining by Sequential Immunostaining and Antibody Removal on Routine Tissue Sections. J Histochem Cytochem, 2017; 65: 431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalow I, Samoilova V, Boecker W, Tiemann M. Multiple immunolabeling with antibodies from the same host species in combination with tyramide signal amplification. Acta Histochem, 2018; 120: 405–11. [DOI] [PubMed] [Google Scholar]
- Byers RJ, Hitchman ER. Quantum dots brighten biological imaging. Prog Histochem Cytochem, 2011; 45: 201–37. [DOI] [PubMed] [Google Scholar]
- Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med, 2002; 8: 1323–7. [DOI] [PubMed] [Google Scholar]
- Capel PJ, Gerlag PG, Hagemann JF, Koene RA. The effect of 2-mercaptoethanol on IgM and IgG antibody activity. J Immunol Methods, 1980; 36: 77–80. [DOI] [PubMed] [Google Scholar]
- Cappi G, Dupouy DG, Comino MA, Ciftlik AT. Ultra-fast and automated immunohistofluorescent multistaining using a microfluidic tissue processor. Sci Rep, 2019; 9: 4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P, Yuen T, Ruf F, Gonzalez-Maeso J, Sealfon SC. Method for multiplex cellular detection of mRNAs using quantum dot fluorescent in situ hybridization. Nucleic Acids Res, 2005; 33: e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J, DeBiasio R, Zhu Z, Giuliano KA, Schmidt BF. Immunofluorescence signal amplification by the enzyme-catalyzed deposition of a fluorescent reporter substrate (CARD). Cytometry, 1996; 23: 48–53. [DOI] [PubMed] [Google Scholar]
- Chen ZH, Wu YS, Chen MJ, Hou JY, Ren ZQ, Sun D, Liu TC. A novel homogeneous time-resolved fluoroimmunoassay for carcinoembryonic antigen based on water-soluble quantum dots. J Fluoresc, 2013; 23: 649–57. [DOI] [PubMed] [Google Scholar]
- Coons AH. The beginnings of immunofluorescence. J Immunol, 1961; 87: 499–503. [PubMed] [Google Scholar]
- Coons AH, Creech HJ, Jones RN. Immunological Properties of an Antibody Containing a Fluorescent Group. Proceedings of the Society for Experimental Biology and Medicine, 1941; 47: 200–2. [Google Scholar]
- Coons AH, Creech HJ, Jones RN, Berliner E. The Demonstration of Pneumococcal Antigen in Tissues by the Use of Fluorescent Antibody. The Journal of Immunology, 1942; 45: 159–70. [Google Scholar]
- Coons AH, Kaplan MH. Localization of antigen in tissue cells; improvements in a method for the detection of antigen by means of fluorescent antibody. J Exp Med, 1950; 91: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivianu-Gaita V, Romaschin A, Thompson M. High efficiency reduction capability for the formation of Fab׳ antibody fragments from F(ab)2 units. Biochemistry and Biophysics Reports, 2015; 2: 23–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AR, Bathany C, Tsuei M, White J, Barald KF, Takayama S. Recent developments in multiplexing techniques for immunohistochemistry. Expert Rev Mol Diagn, 2015; 15: 1171–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckermann K, Mocanu MM, Khlistunova I, Biernat J, Nissen A, Hofmann A, Schonig K, Bujard H, Haemisch A, Mandelkow E, Zhou L, Rune G, Mandelkow EM. The beta-propensity of Tau determines aggregation and synaptic loss in inducible mouse models of tauopathy. J Biol Chem, 2007; 282: 31755–65. [DOI] [PubMed] [Google Scholar]
- Fu H, Hardy J, Duff KE. Selective vulnerability in neurodegenerative diseases. Nat Neurosci, 2018; 21: 1350–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendusa R, Scalia CR, Buscone S, Cattoretti G. Elution of High-affinity (>10–9 KD) Antibodies from Tissue Sections: Clues to the Molecular Mechanism and Use in Sequential Immunostaining. J Histochem Cytochem, 2014; 62: 519–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass G, Papin JA, Mandell JW. SIMPLE: a sequential immunoperoxidase labeling and erasing method. J Histochem Cytochem, 2009; 57: 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gown AM, Garcia R, Ferguson M, Yamanaka E, Tippens D. Avidin-biotin-immunoglucose oxidase: use in single and double labeling procedures. J Histochem Cytochem, 1986; 34: 403–9. [DOI] [PubMed] [Google Scholar]
- Gut G, Herrmann MD, Pelkmans L. Multiplexed protein maps link subcellular organization to cellular states. Science, 2018; 361. [DOI] [PubMed] [Google Scholar]
- Ilie M, Beaulande M, Ben Hadj S, Chamorey E, Schiappa R, Long-Mira E, Lassalle S, Butori C, Cohen C, Leroy S, Guerin O, Mouroux J, Marquette CH, Pomerol JF, Erb G, Hofman V, Hofman P. Chromogenic Multiplex Immunohistochemistry Reveals Modulation of the Immune Microenvironment Associated with Survival in Elderly Patients with Lung Adenocarcinoma. Cancers (Basel), 2018; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jufas NE, Roediger B, Armati PJ. A microwave technique for double indirect immunostaining of human brain tissue cultures with mouse monoclonal antibodies. Appl Immunohistochem Mol Morphol, 2008; 16: 83–6. [DOI] [PubMed] [Google Scholar]
- Kim M, Soontornniyomkij V, Ji B, Zhou X. System-wide immunohistochemical analysis of protein co-localization. PLoS One, 2012; 7: e32043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenacs T, Krenacs L, Raffeld M. Multiple antigen immunostaining procedures. Methods Mol Biol, 2010; 588: 281–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan HY, Mu W, Nikolic-Paterson DJ, Atkins RC. A novel, simple, reliable, and sensitive method for multiple immunoenzyme staining: use of microwave oven heating to block antibody crossreactivity and retrieve antigens. J Histochem Cytochem, 1995; 43: 97–102. [DOI] [PubMed] [Google Scholar]
- Lim JCT, Yeong JPS, Lim CJ, Ong CCH, Wong SC, Chew VSP, Ahmed SS, Tan PH, Iqbal J. An automated staining protocol for seven-colour immunofluorescence of human tissue sections for diagnostic and prognostic use. Pathology, 2018; 50: 333–41. [DOI] [PubMed] [Google Scholar]
- Mansfield JR. Phenotyping Multiple Subsets of Immune Cells In Situ in FFPE Tissue Sections: An Overview of Methodologies. Methods Mol Biol, 2017; 1546: 75–99. [DOI] [PubMed] [Google Scholar]
- Marrero A, Lawrence S, Wilsker D, Voth AR, Kinders RJ. Translating pharmacodynamic biomarkers from bench to bedside: analytical validation and fit-for-purpose studies to qualify multiplex immunofluorescent assays for use on clinical core biopsy specimens. Semin Oncol, 2016; 43: 453–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe A, Dolled-Filhart M, Camp RL, Rimm DL. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst, 2005; 97: 1808–15. [DOI] [PubMed] [Google Scholar]
- Nakane PK. Simultaneous localization of multiple tissue antigens using the peroxidase-labeled antibody method: a study on pituitary glands of the rat. J Histochem Cytochem, 1968; 16: 557–60. [DOI] [PubMed] [Google Scholar]
- Narhi LO, Caughey DJ, Horan T, Kita Y, Chang D, Arakawa T. Effect of three elution buffers on the recovery and structure of monoclonal antibodies. Anal Biochem, 1997a; 253: 236–45. [DOI] [PubMed] [Google Scholar]
- Narhi LO, Caughey DJ, Horan TP, Kita Y, Chang D, Arakawa T. Fractionation and characterization of polyclonal antibodies using three progressively more chaotropic solvents. Anal Biochem, 1997b; 253: 246–52. [DOI] [PubMed] [Google Scholar]
- Parra ER, Uraoka N, Jiang M, Cook P, Gibbons D, Forget MA, Bernatchez C, Haymaker C, Wistuba II, Rodriguez-Canales J Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Sci Rep, 2017; 7: 13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirici D, Mogoanta L, Kumar-Singh S, Pirici I, Margaritescu C, Simionescu C, Stanescu R. Antibody elution method for multiple immunohistochemistry on primary antibodies raised in the same species and of the same subtype. J Histochem Cytochem, 2009; 57: 567–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost S, Kishen RE, Kluth DC, Bellamy CO. Working with Commercially Available Quantum Dots for Immunofluorescence on Tissue Sections. PLoS One, 2016; 11: e0163856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Axelrod HD, Valkenburg KC, Amend S, Pienta KJ. Optimization of prostate cancer cell detection using multiplex tyramide signal amplification. J Cell Biochem, 2019; 120: 4804–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saylor J, Ma Z, Goodridge HS, Huang F, Cress AE, Pandol SJ, Shiao SL, Vidal AC, Wu L, Nickols NG, Gertych A, Knudsen BS. Spatial Mapping of Myeloid Cells and Macrophages by Multiplexed Tissue Staining. Front Immunol, 2018; 9: 2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW. Selective functional, regional, and neuronal vulnerability in frontotemporal dementia. Curr Opin Neurol, 2008; 21: 701–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron, 2009; 62: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrelle N, Ganguly D, Dominguez ATA, Zhang Y, Huang H, Dahal LN, Burton N, Ziemys A, Brekken RA. Improved Multiplex Immunohistochemistry for Immune Microenvironment Evaluation of Mouse Formalin-Fixed, Paraffin-Embedded Tissues. J Immunol, 2019; 202: 292–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods, 2014; 70: 46–58. [DOI] [PubMed] [Google Scholar]
- Sweeney E, Ward TH, Gray N, Womack C, Jayson G, Hughes A, Dive C, Byers R. Quantitative multiplexed quantum dot immunohistochemistry. Biochem Biophys Res Commun, 2008; 374: 181–6. [DOI] [PubMed] [Google Scholar]
- Tholouli E, Sweeney E, Barrow E, Clay V, Hoyland JA, Byers RJ. Quantum dots light up pathology. J Pathol, 2008; 216: 275–85. [DOI] [PubMed] [Google Scholar]
- Toth ZE, Mezey E. Simultaneous visualization of multiple antigens with tyramide signal amplification using antibodies from the same species. J Histochem Cytochem, 2007; 55: 545–54. [DOI] [PubMed] [Google Scholar]
- Tramu G, Pillez A, Leonardelli J. An efficient method of antibody elution for the successive or simultaneous localization of two antigens by immunocytochemistry. J Histochem Cytochem, 1978; 26: 322–4. [DOI] [PubMed] [Google Scholar]
- van den Brand M, Hoevenaars BM, Sigmans JH, Meijer JW, van Cleef PH, Groenen PJ, Hebeda KM, van Krieken JH. Sequential immunohistochemistry: a promising new tool for the pathology laboratory. Histopathology, 2014; 65: 651–7. [DOI] [PubMed] [Google Scholar]
- Wang G, Achim CL, Hamilton RL, Wiley CA, Soontornniyomkij V. Tyramide signal amplification method in multiple-label immunofluorescence confocal microscopy. Methods, 1999; 18: 459–64. [DOI] [PubMed] [Google Scholar]
- Wegner KA, Keikhosravi A, Eliceiri KW, Vezina CM. Fluorescence of Picrosirius Red Multiplexed With Immunohistochemistry for the Quantitative Assessment of Collagen in Tissue Sections. J Histochem Cytochem, 2017; 65: 479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, Ge N, Peale F, Bruchez MP. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol, 2003; 21: 41–6. [DOI] [PubMed] [Google Scholar]
- Xu H, Xu J, Wang X, Wu D, Chen ZG, Wang AY. Quantum dot-based, quantitative, and multiplexed assay for tissue staining. ACS Appl Mater Interfaces, 2013; 5: 2901–7. [DOI] [PubMed] [Google Scholar]
- Zhang W, Hubbard A, Jones T, Racolta A, Bhaumik S, Cummins N, Zhang L, Garsha K, Ventura F, Lefever MR, Lu Z, Hurley JK, Day WA, Pestic-Dragovich L, Morrison LE, Tang L. Fully automated 5-plex fluorescent immunohistochemistry with tyramide signal amplification and same species antibodies. Lab Invest, 2017; 97: 873–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.