Abstract
Desmoplastic infantile ganglioglioma (DIG) and desmoplastic infantile astrocytoma (DIA) are extremely rare tumors that typically arise in infancy; however, these entities have not been well characterized in terms of genetic alterations or clinical outcomes. Here, through a multi-institutional collaboration, the largest cohort of DIG/DIA to date is examined using advanced laboratory and data processing techniques. Targeted DNA exome sequencing and DNA methylation profiling were performed on tumor specimens obtained from different patients (n = 8) diagnosed histologically as DIG/DIGA. Two of these cases clustered with other tumor entities, and were excluded from analysis. The remaining 16 cases were confirmed to be DIG/DIA by histology and by DNA methylation profiling. Somatic BRAF gene mutations were discovered in 7 instances (43.8%); 4 were BRAFV600E mutations, and 3 were BRAFV600D mutations. Three instances of malignant transformation were found, and sequencing of the recurrence demonstrated a new TP53 mutation in one case, new ATRX deletion in one case, and in the third case, the original tumor harbored an EML4–ALK fusion, also present at recurrence. DIG/DIA are distinct pathologic entities that frequently harbor BRAFV600 mutations. Complete surgical resection is the ideal treatment, and overall prognosis is excellent. While, the small sample size and incomplete surgical records limit a definitive conclusion about the risk of tumor recurrence, the risk appears quite low. In rare cases with wild-type BRAF, malignant progression can be observed, frequently with the acquisition of other genetic alterations.
Implications:
DIG/DIA are a distinct molecular entity, with a subset frequently harboring either BRAFV600E or BRAFV600D mutations.
Introduction
The World Health Organization’s classification of central nervous system (CNS) neoplasms categorizes desmoplastic infantile astrocytoma (DIA) and desmoplastic infantile ganglioglioma (DIG) together as one diagnosis, among grade I “neuronal and mixed neuronal-glial” entities (1). In 1978, Friede reported a desmoplastic glial tumor of the medulla with leptomeningeal metastases, proposing the terms “gliofibroma” or “desmoplastic glioma” (2). DIA was introduced by Taratuto, describing 6 infants with “superficial cerebral astrocytoma attached to dura” (3). Vandenberg reported 11 infants with DIG in 1987, noting mixed glial and neuronal histology (4). The typically large size, nodular contrast enhancement pattern, cellular pleomorphism and atypia, frequent mitoses, and undifferentiated small-cell component, all conjure the visage of an aggressive malignancy. Yet, complete resection is expected to yield an excellent prognosis (5). DIG/DIA most frequently occur in patients under 24 months of age, and accounts for a significant proportion of intracranial neoplasms seen in the first year of life (6, 7). Mitotic figures, primitive-appearing cells, and cellular pleomorphism are common histologic features of DIGs and DIAs. Yet, such high-grade characteristics have repeatedly failed to correlate with worse clinical outcomes in DIGs and DIAs, regardless of location (8, 9).
DIG/DIAs commonly present in the periphery of the supratentorial compartment, consisting of a large cystic component with a contrast-enhancing solid nodule attached to the overlying dura. Even without a significant cystic portion at presentation, this can develop over time (Fig. 1A; ref. 10). Rarely, DIG/DIA presents as an intraventricular lesion (Fig. 1B), or a mostly solid sellar/hypothalamic mass (Fig. 1C). Histologically, desmoplasia is prominent within dense stroma, with fibroblastic and neuroepithelial elements. Neoplastic cells are limited to the solid nodule and adjacent leptomeninges (5, 10–13). Neuroepithelial elements vary in proportions of astrocytic and neuronal cells, and as the names suggest, presence of neuronal differentiation distinguishes DIG from DIA. DIGs, but not DIAs, commonly involve rests of primitive ganglion cells suggesting anaplasia, giving them an appearance such that the term “desmoplastic neuroblastoma” was previously used to described this entity (14). Presence of primitive neuronal components does not seem to imply more aggressive biology in DIG/DIA (9, 15–17). Craniospinal seeding or metastasis (Fig. 1D) has been reported (15–19), as have few instances of malignant transformation (20–22).
Figure 1:
A, Solid dura-based DIG. B, Multicystic intraventricular DIG. C, Suprasellar DIA with leptomeningeal metastases. D, Suprasellar and cisternal DIA
A common origin of DIG/DIA has been suggested, as has a relationship with PXA and GG (23–28). The argument for divergence along a common ancestral lineage accounting for the similarities and differences between these tumor types has been made entirely on histopathologic findings thus far (29). PXA shares many features with DIA in particular—both are frequently desmoplastic and involve the leptomeninges, are often cystic, and have a generally benign prognosis. PXA tends to present at an older age, and the pleomorphic, xanthomatous appearance of its astrocytes differentiate the two tumor types. Vandenberg notes a basal lamina seen in DIG/DIA that is also typical of PXA (9).
Little in the way of genetic and molecular characterization of DIG/DIA has been published, and no clonal chromosomal abnormalities have been found in DIA or DIG (22, 30–33). While various nonclonal aberrations have been reported, no specific locus has consistently appeared. Gessi and colleagues, performed a genome-wide DNA copy number analysis in combination with a multiplex analysis of candidate genes in 4 DIAs and 10 DIGs, observing inconsistent focal recurrent genomic losses in chromosome regions 5q13.3, 21q22.11, and 10q21.3 in both DIA and DIG (34). Principal component analysis did not show any significant differences; however, in 6 of these cases, gain of genomic material at 7q31, which corresponds to the MET gene, was found. A total of 7 cases of BRAFV600E mutations have been found previously in DIG/DIA (34–38). Hypermethylated alleles in the P14ARF gene were seen in one case (32). Lonnrot and colleagues, identified MYCN amplifications in 2 cases and EGFR amplifications in 3 cases (39). Several negative TP53 analyses in patients with DIG/DIA have been performed (7, 31, 32, 39, 40), with 1 case report of a TP53 SNV found in both a primary DIG and in its subsequent malignant transformation (22).
With this study, we aimed to more precisely understand the genetic underpinnings of DIG and DIA among glial–neuronal CNS tumors.
Materials and Methods
SCH cohort
Institutional review board approval was obtained prior to retrieval of pathologic tissue samples from Seattle Children’s Hospital (SCH) and contributing sites. All tissue samples were re-reviewed by independent neuropathology faculty of the Department of Pathology at SCH for diagnosis, tumor content, and adequacy for DNA extraction. UW-OncoPlex assay version 5, a targeted, massively parallel gene sequencing assay that detects mutations in 262 cancer-related genes (http://tests.labmed.washington.edu/UW-OncoPlex, last accessed May 5, 2017), was performed as described previously (41). The test uses next-generation “deep” sequencing to detect mutations including single nucleotide variants (SNV), small insertions and deletions (InDel), copy number alterations including gene amplifications, and selected gene fusions.
Briefly, DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) solid tumor tissue samples using the Gentra Puregene DNA Isolation Kit (catalog #158489; Qiagen). H&E-stained slides were reviewed before DNA extraction for all FFPE samples, and when feasible, macrodissection of tumor-containing regions was performed to enrich tumor cellularity. Tumor cellularity was estimated by review of H&E-stained slides. DNA sequencing was performed on a HiSeq2500 sequencing system (Illumina) with 2 × 101 bp, paired-end reads, and on a NextSeq 500 (Illumina) with 2 × 150 bp, paired end reads according to the manufacturer’s instructions. The average coverage across the capture region was 500×, minimum gene-level average coverage was set to 50×, with genes below this threshold reported as failed. The minimum acceptable average coverage for the entire panel was set at 150×, and the minimum library complexity (the fraction of unique DNA fragments sequenced) was set at 20%.
HD cohort
Tissue samples were retrieved with Ethics Committee approval from the archives of the Department of Neuropathology of the German Cancer Research Center/Heidelberg University (HD), and the N.N. Burdenko Neurosurgical Institute (Moscow, Russia). All included cases were reviewed by faculty in the Department of Neuropathology at HD. DNA from FFPE tissue was extracted on the Promega Maxwell device (Promega) following manufacturer’s instructions. Targeted exome sequencing was performed as described previously (42).
DNA methylation analysis
DNA was extracted from tumors and analyzed for genome-wide DNA methylation patterns using the Illumina Infinium Methylation EPIC BeadChip (850k) array. Processing of DNA methylation data was performed with custom approaches as described previously (43, 44). Analysis of tumor subgroups was performed using a t-SNE-based approach taking the 5,000 most variably methylated probes (45).
Results and Discussion
Individual molecular and genetic characterization of pediatric tumors may yield unexpected avenues for treatment
Ten primary tumor samples from SCH were examined using targeted DNA exome sequencing. In 5 tumors (50%), we found no genetic aberrations involving the 262 genes included in UW-OncoPlex. Within the other five tumors, we discovered 2 (20%) BRAFV600E mutations, 2 (20%) BRAFV600D mutations, and one EML4-ALK fusion (Table 1A). No other gene fusions were detected in either BRAF-mutant or wild-type tumors, including the KIAA1549–BRAF fusion. CDKN2A (p16) was intact at the DNA level in all of our tested samples. CDK4 and CDK6 are also at a normal copy number state. Detailed clinical and sequencing information are listed in the Supplementary Data.
A replication cohort of 8 samples histologically diagnosed as DIG/DIA was collected at HD. Analysis of DNA methylation profiles through a molecular classification platform (www.molecularneuropathology.org) indicated that 2 of these cases clustered with PA, which were therefore excluded from subsequent analysis. Targeted DNA exome sequencing in the remaining 6 cases revealed two V600E and one V600D mutations (Table 1B). Of note, the 2 excluded cases did not harbor BRAF gene mutations. The clinical implications of our findings, in terms of potential therapeutic targets, argue for incorporation of broad molecular and genetic characterization in the diagnostic process for pediatric CNS tumors. This is particularly true of lower grade tumors likely driven by few oncogenic aberrations, and in patients for whom radiation sparing is paramount.
BRAF mutations are common in DIG/DIA
Seven of 16 (43.8%) patients in our series harbored somatic nonsynonymous single nucleotide variant substitutions at codon 600 of the BRAF gene. Four were the canonical V600E valine-to-glutamic acid; interestingly, 3 of 4 were found in histologically diagnosed DIAs. The other 3 were exceptionally rare V600D valine-to-aspartic acid point mutations, which account for less than 1% of all V600 BRAF mutations (http://cancer.sanger.ac.uk, last accessed January 8, 2018; refs. 46, 47). All three BRAFV600D mutations occurred in patients with DIG diagnosed at less than 1 year of age. BRAF mutations were thus discovered in 4 of 12 (25%) DIGs, but in 3 of 4 (75%) DIAs. None of the patients harboring BRAF mutations experienced recurrence.
Prior to our study, eight instances of BRAF mutations had been described in DIG/DIA, seven V600E substitutions, and one V600D (48). Dougherty and colleagues, reported a BRAFV600E mutation in 1 of 2 cases of DIG (35). Karabagli reported finding a BRAFV600E mutation in a skull base DIA (37). Prabowo and colleagues, found BRAFV600E mutations in 2 of 4 DIGs (38). Gessi and colleagues, discovered a single case of a BRAFV600E mutation in a DIA, among 14 DIG/DIAs (34). Koelsche and colleagues, found 2 cases of BRAFV600E mutations among 18 cases of DIG/DIA, 1 in a DIG and 1 in a suprasellar DIA (36). Of note, in several of these series, VE1 IHC staining was performed as screening, and molecular analysis was then performed only on positively staining cases to confirm or disprove the presence of the mutation. As a screen for BRAFV600E mutations, VE1 staining accurately identified these mutations in only 67%–75% of DIG (36, 38); moreover, it does not appear to detect other BRAFV600 mutations, including the V600D mutation.
BRAF mutations have been sparsely seen among adult low-grade glial–neuronal tumors (49–51). TCGA low-grade glioma (LGG) analysis revealed only two BRAF mutations among 283 patients, including one V600E mutation and one D594G mutation, while the UCSF LGG analysis included one V600E mutation in 23 patients. (http://www.cbioportal.org, last accessed January 8, 2017; refs. 52, 53) Certain typically pediatric tumors are far more likely to show BRAF alterations; however, BRAFV600E mutations have been found in an estimated 66% of pleomorphic xanthoastrocytomas (PXA; ref. 42), 18%–58% of gangliogliomas (38, 42, 54), 30% of dysembryoplastic neuroepithelial tumors (DNET; ref. 38), and 9% of pilocytic astrocytomas (42). Bergthold and colleagues, found that BRAFV600E mutations clustered among supratentorial pediatric LGG, a group comprised primarily of gangliogliomas, diffuse astrocytoma, DNET, and a small proportion of pilocytic astrocytomas, also demonstrating that this cluster was significantly enriched for a significant number of gene sets that together suggested a neuronal signature (55). Furthermore, they did not detect any alteration in gene expression profile among BRAF-altered tumors when compared with wild-type BRAF in LGG.
The effect of these two types of BRAF mutations on RAS/MAPK function is thought to be equivalent; however, implications for treatment and diagnostic options are potentially confounding. While a small number of targeted molecular therapies have demonstrated efficacy in tumors harboring V600E and V600K mutations, V600D mutations have not been specifically evaluated in a clinical setting. However, preclinical studies suggest that BRAFV600D mutations are likely to be sensitive to vemurafenib (56) and dabrafenib (57), and as alternative proven options are lacking, BRAF inhibitors are likely to play an important role in the management of patients with V600D mutations, even in infants (58).
Nosology of DIG and DIA is distinct, and likely involves a primitive precursor capable of glial and neuronal differentiation
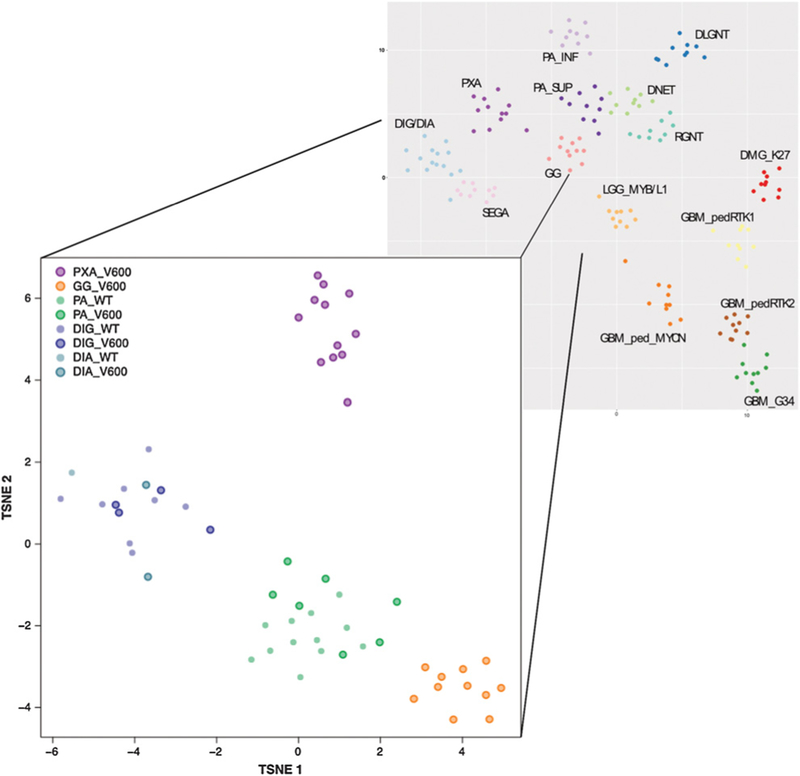
To further examine the relationship between BRAFV600-mutant and wild-type DIG/DIA and other pediatric glioneuronal tumors, including other entities that harbor BRAFV600E mutations, we performed a global DNA methylation analysis using Illumina Infinium (850k) arrays. This analysis included 9 primary DIG/DIA from SCH, 6 primary DIG/DIA from the HD series, and reference samples of other entities collected in HD (11 GG BRAFV600E mutant, 12 PXA BRAFV600E mutant, 7 PA BRAFV600E mutant, and 12 PA with other MAPK pathway alterations). Similarities in methylation profiles were visualized using a t-distributed stochastic neighbor embedding (TSNE) approach, as described previously (59).
DIG/DIA clearly formed a distinct molecular group regardless of BRAF mutation status (Fig. 2). No overlap with other BRAFV600E-enriched pediatric brain tumor subtypes (gangliogliomas and PXA) was observed, demonstrating that DIG/DIA are molecularly distinct from these other entities. Of note, no discernible separation was observed between DIG and DIA samples, indicating that these may rather be morphologic variants of a single molecular entity.
Figure 2:
DNA methylation profiling of DIG/DIA, relative to other pediatric glioneuronal tumors frequently harboring BRAF gene mutations. This t-distributed stochastic neighbor embedding plot represents DNA methylation assay data of the 5,000 most variable CpG sites across a larger cohort of pediatric glioneuronal CNS tumors. PXA with BRAFV600E mutations and GG with BRAFV600E mutations are clearly distinct from DIG/DIA. PA forms two other distinct groups, with BRAFV600E mutant and wild-type cases intermixed. Similarly, DIG/DIA form a distinct group with the BRAF mutant and wild-type cases intermixed, with no clear differentiation between DIA and DIG. These data suggest that DIG/DIA is one distinct pathologic entity, with a significant proportion of cases harboring BRAF mutations.
On an autopsy of a patient with suprasellar/intraventricular DIG, Komori and colleagues, noted that, within the optic nerves, tumor infiltrates consisted of neuroepithelial cells of varying levels of differentiation, only occasional staining for GFAP or neurofilament, and unassociated with desmoplasia (60). Tumor astrocytes, primitive-appearing neuroepithelial cells, and Schwann-like cells were all intimately coexistent within the reticulin-rich basal lamina typical of DIG/DIA, as has been repeatedly observed in DIG/DIA (9, 28, 60). While astrocytes are neural-tube derivatives, Schwann cells originate from the neural crest, findings such as these indicate a more primitive derivation, perhaps related to the neural plate, to be capable of neuronal, astrocytic, and Schwannian differentiation.
Koelsche and colleagues, found BRAFV600E mutations in 41 of 71 (58%) of gangliogliomas, further localizing the mutant BRAFV600E protein product predominantly to the neuronal compartments within these tumors using VE1 IHC, indicating that BRAF mutations occur in cells that have the capacity to differentiate into ganglionic cells (54). A positive association has been found between VE1 positivity and synaptophysin-positive clusters, while a separate study made the similar observation of colocalization of BRAFV600E mutation with the neuronal markers synaptophysin and NeuN (38). In addition, BRAFV600E mutation was found to be tightly associated with pS6 (a marker for mTOR activation) and CD34 immunoreactivity within only the neuronal components of glial–neuronal tumors, but not within the glial components, where pS6 and VE1 staining was sparse. Although the number of tumors studied in this fashion is too small to draw conclusions regarding nosology, the frequent observation of both neural tube and neural crest derivatives within these tumors suggests a derivation more primitive than both. Case-by-case correlation between the BRAF mutation allele frequency and ganglion cell content would provide crucial understanding of the derivation of this tumor.
Acquired genetic alterations drive malignant transformation of DIG and DIA
Four (40%) patients with SCH experienced tumor recurrence/progression; 3 (30%) suffered malignant transformation of their tumors, and so UW-Oncoplex next-generation sequencing and DNA methylation profiling were performed on the recurrent tumors as well. One patient (Fig. 3A) presented at 4 months of age with a DIG negative for somatic variants on sequencing of the native tumor, underwent subtotal resection (STR) and received carboplatin and vincristine according to the Children’s Oncology Group (COG) 9952 protocol for growth of the residual tumor almost immediately after surgery. The tumor further progressed 2 years later, at which time she again underwent STR, and sequencing of this sample showed a new frameshift insertion affecting TP53. The patient then underwent gross total resection for tumor progression 1 year later, and sequencing of this tumor held the same TP53 mutation. She survived over 3 years from her presentation, before dying from her disease. Somatic TP53 gene mutations are some of the most frequently observed in human cancers, including malignant CNS tumors (61).
Figure 3.
A, Malignant transformation driven by TP53 mutation. B, Malignant transformation driven by ATRX mutation. C, Malignant transformation driven by EML4–ALK fusion.
Another patient (Fig. 3B), who presented at 7 months of age with a DIA negative for somatic variants on sequencing of the native tumor, underwent STR. Because of leptomeningeal involvement at presentation, she received COG 99703 protocol chemotherapy with a 25% dose reduction for age. Over 10 years passed before local recurrence was discovered, for which she again underwent STR of the recurrent tumor. Sequencing of this tumor showed a frameshift deletion affecting ATRX, as well as a nonsynonymous single-nucleotide substitution in BCORL1. Histopathology revealed high-grade malignant transformation of the native tumor, and 5,400 cGy of focal fractionated radiotherapy was administered. She continues to be followed, now 13 years after her initial presentation.
The ATRX gene is a core component of a chromatin remodeling complex active in telomere function, the alteration of which results in alternative lengthening of telomeres, a presumed precursor to genomic instability, and a recognized contributor to gliomagenesis (62). The additional finding of a BCORL1 point mutation is of uncertain significance. Both ATRX and BCORL1 are on the X chromosome, and BCOR and BCORL1 alterations are known to be involved in acute myelogenous leukemia (63). They are also rarely observed in CNS malignancies, including medulloblastoma, glioblastoma, and supratentorial CNS embryonal tumors (59, 64, 65). While 1 case of concurrent ATRX and BCOR mutations was discovered in a pediatric malignant glioma, ATRX has not been consistently described to coincide with BCOR (L1) specifically (65).
A 3rd patient (Fig. 3C) presented at 8 months of age with a DIG, and underwent GTR. Sequencing of this native tumor revealed an EML4–ALK gene fusion event. Tumor recurrence within 4 months was treated according to COG 9952 regimen A protocol with carboplatin and vincristine. She subsequently developed acute lower extremity paresis and bulky leptomeningeal disease progression, and sequencing of this sample showed the same EML4–ALK gene fusion. Of note, the native tumor sequencing also showed a premature stop mutation in CREBBP, which was not detected on the recurrent tumor. She was treated at this time according to CCG 99703 protocol. At her most recent follow-up, the patient was 7 years removed from her initial presentation, with complete response to treatment, and no signs of disease recurrence.
Conclusion
DIG/DIA are a distinct molecular entity, without overlap with other BRAFV600E-enriched pediatric brain tumor subtypes (gangliogliomas and PXA). We have identified a subset of DIG/DIAs harboring BRAF mutations with approximately a 43.8% frequency. While BRAFV600D mutations have, to this point, proven exceedingly rare in primary CNS tumors, our cohorts revealed 3 among only 16 total DIG/DIAs. No other oncogenic mutation was consistently identified in the wild-type or mutant BRAF DIG/DIAs. BRAFV600E mutations were seen in 3 of the 4 DIAs, whereas the BRAFV600D mutations were all found in DIGs, perhaps suggesting a tendency for the former to arise in cells of astrocytic, rather than neuronal, lineage.
Malignant transformation of these tumors might be more common than previously recognized. Malignant transformation was identified in 33.3% of the patients followed long-term at SCH, with 2 of these 3 patients acquiring a new oncogenic mutation, not present in the native tumor, when the malignant tumor was sequenced. Each incidence of malignant transformation was driven by an identifiable genetic aberration other than a BRAF mutation.
Maximal safe resection remains the mainstay of treatment for these tumors. The need for chemotherapy and radiation in cases where complete resection is unable to be obtained does not appear to predict a higher mortality rate, and these remain optional in cases of tumor progression. However, in a small percentage of BRAF wild-type cases, we identified malignant transformation with the acquisition of other genetic alterations. Our findings in this regard serve to highlight the need to test all DIG/DIA for BRAF and other gene mutations including gene fusions, which might allow for the use of targeted molecular therapies, or dictate the need for chemotherapy or radiation in refractory or recurrent cases.
Supplementary Material
Acknowledgments
This work was supported by grants from the Richard G. Ellenbogen Chair of Pediatric Neurological Surgery.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
D. Capper has ownership interest (including stock, patents, etc.) in DIANOVA GmbH. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. WHO classification of tumours of the central nervous system. Lyon, France: World Health Organization; 2016. [DOI] [PubMed] [Google Scholar]
- 2.Friede RL. Gliofibroma. A peculiar neoplasia of collagen forming glia-like cells. J Neuropathol Exp Neurol 1978;37:300–13. [DOI] [PubMed] [Google Scholar]
- 3.Taratuto AL, Monges J, Lylyk P, Leiguarda R. Superficial cerebral astrocytoma attached to dura. Report of six cases in infants. Cancer 1984;54: 2505–12. [DOI] [PubMed] [Google Scholar]
- 4.VandenBerg SR, May EE, Rubinstein LJ, Herman MM, Perentes E, Vinores SA, et al. Desmoplastic supratentorial neuroepithelial tumors of infancy with divergent differentiation potential (“desmoplastic infantile gangliogliomas”). Report on 11 cases of a distinctive embryonal tumor with favorable prognosis. J Neurosurg 1987;66:58–71. [DOI] [PubMed] [Google Scholar]
- 5.Sugiyama K, Arita K, Shima T, Nakaoka M, Matsuoka T, Taniguchi E, et al. Good clinical course in infants with desmoplastic cerebral neuroepithelial tumor treated by surgery alone. J Neurooncol 2002;59:63–9. [DOI] [PubMed] [Google Scholar]
- 6.Zuccaro G, Taratuto AL, Monges J. Intracranial neoplasms during the first year of life. Surg Neurol 1986;26:29–36. [DOI] [PubMed] [Google Scholar]
- 7.Taratuto AL, VandenBerg SR, Rorke LB. Desmoplastic infantile astrocytoma and ganglioglioma In: Kleihues P, Cavenee WK, editor. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of the Nervous System. Lyon: IARC Press; 2000. pp. 99–102. [Google Scholar]
- 8.Bachli H, Avoledo P, Gratzl O, Tolnay M. Therapeutic strategies and management of desmoplastic infantile ganglioglioma: two case reports and literature overview. Childs Ner Syst 2003;19:359–66. [DOI] [PubMed] [Google Scholar]
- 9.VandenBerg SR. Desmoplastic infantile ganglioglioma and desmoplastic cerebral astrocytoma of infancy. Brain Pathol 1993;3:275–81. [DOI] [PubMed] [Google Scholar]
- 10.Tseng JH, Tseng MY, Kuo MF, Tseng CL, Chang YL. Chronological changes on magnetic resonance images in a case of desmoplastic infantile ganglioglioma. Pediatr Neurosurg 2002;36:29–32. [DOI] [PubMed] [Google Scholar]
- 11.Kurose A, Beppu T, Miura Y, Suzuki M, Ogawa A, Arai H, et al. Desmoplastic cerebral astrocytoma of infancy intermingling with atypical glial cells. Pathol Int 2000;50:744–9. [DOI] [PubMed] [Google Scholar]
- 12.Kim JH, Kim IO, Kim WS, Kim KH, Park CM, Yeon KM. MR findings of desmoplastic cerebral astrocytoma of infancy. Acta Radiol 2003;44: 688–90. [DOI] [PubMed] [Google Scholar]
- 13.Beppu T, Sato Y, Uesugi N, Kuzu Y, Ogasawara K, Ogawa A. Desmoplastic infantile astrocytoma and characteristics of the accompanying cyst. Case report. J Neurosurg Pediatr 2008;1:148–51. [DOI] [PubMed] [Google Scholar]
- 14.Horten BC, Rubinstein LJ. Primary cerebral neuroblastoma. A clinicopathological study of 35 cases. Brain 1976;99:735–56. [DOI] [PubMed] [Google Scholar]
- 15.De Munnynck K, Van Gool S, Van Calenbergh F, Demaerel P, Uyttebroeck A, Buyse G, et al. Desmoplastic infantile ganglioglioma: a potentially malignant tumor? Am J Surg Pathol 2002;26:1515–22. [DOI] [PubMed] [Google Scholar]
- 16.Darwish B, Arbuckle S, Kellie S, Besser M, Chaseling R. Desmoplastic infantile ganglioglioma/astrocytoma with cerebrospinal metastasis. J Clin Neurosci 2007;14:498–501. [DOI] [PubMed] [Google Scholar]
- 17.Setty SN, Miller DC, Camras L, Charbel F, Schmidt ML. Desmoplastic infantile astrocytoma with metastases at presentation. Mod Pathol 1997;10:945–51. [PubMed] [Google Scholar]
- 18.Taranath A, Lam A, Wong CK. Desmoplastic infantile ganglioglioma: a questionably benign tumour. Australas Radiol 2005;49:433–7. [DOI] [PubMed] [Google Scholar]
- 19.Milanaccio C, Nozza P, Ravegnani M, Rossi A, Raso A, Gambini C, et al. Cervico-medullary desmoplastic infantile ganglioglioma: an unusual case with diffuse leptomeningeal dissemination at diagnosis. Pediatr Blood Cancer 2005;45:986–90. [DOI] [PubMed] [Google Scholar]
- 20.Loh JK, Lieu AS, Chai CY, Howng SL. Malignant transformation of a desmoplastic infantile ganglioglioma. Pediatr Neurol 2011;45: 135–7. [DOI] [PubMed] [Google Scholar]
- 21.Phi JH, Koh EJ, Kim SK, Park SH, Cho BK, Wang KC. Desmoplastic infantile astrocytoma: recurrence with malignant transformation into glioblastoma: a case report. Childs Ner Syst 2011;27:2177–81. [DOI] [PubMed] [Google Scholar]
- 22.Prakash V, Batanian JR, Guzman MA, Duncavage EJ, Geller TJ. Malignant transformation of a desmoplastic infantile ganglioglioma in an infant carrier of a nonsynonymous TP53 mutation. Pediatr Neurol 2014;51: 138–43. [DOI] [PubMed] [Google Scholar]
- 23.Gambarelli D, Hassoun J, Choux M, Toga M. Complex cerebral tumor with evidence of neuronal, glial and Schwann cell differentiation: a histologic, immunocytochemical and ultrastructural study. Cancer 1982;49:1420–8. [DOI] [PubMed] [Google Scholar]
- 24.Ng TH, Fung CF, Ma LT. The pathological spectrum of desmoplastic infantile gangliogliomas. Histopathology 1990;16:235–41. [DOI] [PubMed] [Google Scholar]
- 25.Kordek R, Biernat W, Alwasiak J, Liberski PP. Pleomorphic xanthoastrocytoma and desmoplastic infantile ganglioglioma–have these neoplasms a common origin? Folia Neuropathol 1994;32:237–9. [PubMed] [Google Scholar]
- 26.Paulus W, Schlote W, Perentes E, Jacobi G, Warmuth-Metz M, Roggendorf W. Desmoplastic supratentorial neuroepithelial tumours of infancy. Histopathology 1992;21:43–9. [DOI] [PubMed] [Google Scholar]
- 27.Aydin F, Ghatak NR, Salvant J, Muizelaar P. Desmoplastic cerebral astrocytoma of infancy. A case report with immunohistochemical, ultrastructural and proliferation studies. Acta Neuropathol 1993;86:666–70. [DOI] [PubMed] [Google Scholar]
- 28.Mallucci C, Lellouch-Tubiana A, Salazar C, Cinalli G, Renier D, Sainte-Rose C, et al. The management of desmoplastic neuroepithelial tumours in childhood. Childs Nerv Syst 2000;16:8–14. [DOI] [PubMed] [Google Scholar]
- 29.de Chadarevian JP, Pattisapu JV, Faerber EN. Desmoplastic cerebral astrocytoma of infancy. Light microscopy, immunocytochemistry, and ultra-structure. Cancer 1990;66:173–9. [DOI] [PubMed] [Google Scholar]
- 30.Park JP, Dossu JR, Rhodes CH. Telomere associations in desmoplastic infantile ganglioglioma. Cancer Genet Cytogenet 1996;92:4–7. [DOI] [PubMed] [Google Scholar]
- 31.Kros JM, Delwel EJ, de Jong TH, Tanghe HL, van Run PR, Vissers K, et al. Desmoplastic infantile astrocytoma and ganglioglioma: a search for genomic characteristics. Acta Neuropathol 2002;104:144–8. [DOI] [PubMed] [Google Scholar]
- 32.Cerda-Nicolas M, Lopez-Gines C, Gil-Benso R, Donat J, Fernandez-Delgado R, Pellin A, et al. Desmoplastic infantile ganglioglioma. Morphological, immunohistochemical and genetic features. Histopathology 2006;48:617–21. [DOI] [PubMed] [Google Scholar]
- 33.Alghamdi S, Castellano-Sanchez A, Brathwaite C, Shimizu T, Khatib Z, Bhatia S. Strong desmin expression in a congenital desmoplastic infantile ganglioglioma mimicking pleomorphic rhadomyosarcoma: a case report including ultrastructural and cytogenetic evaluation and review of the literature. Childs Nerv Syst 2012;28:2157–62. [DOI] [PubMed] [Google Scholar]
- 34.Gessi M, Zur Muhlen A, Hammes J, Waha A, Denkhaus D, Pietsch T. Genome-wide DNA copy number analysis of desmoplastic infantile astrocytomas and desmoplastic infantile gangliogliomas. J Neuropathol Exp Neurol 2013;72:807–15. [DOI] [PubMed] [Google Scholar]
- 35.Dougherty MJ, Santi M, Brose MS, Ma C, Resnick AC, Sievert AJ, et al. Activating mutations in BRAF characterize a spectrum of pediatric low-grade gliomas. Neuro Oncol 2010;12:621–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koelsche C, Sahm F, Paulus W, Mittelbronn M, Giangaspero F, Antonelli M, et al. BRAF V600E expression and distribution in desmoplastic infantile astrocytoma/ganglioglioma. Neuropathol Appl Neurobiol 2014;40:337–44. [DOI] [PubMed] [Google Scholar]
- 37.Karabagli P, Karabagli H, Kose D, Kocak N, Etus V, Koksal Y. Desmoplastic non-infantile astrocytic tumor with BRAF V600E mutation. Brain Tumor Pathol 2014;31:282–8. [DOI] [PubMed] [Google Scholar]
- 38.Prabowo AS, Iyer AM, Veersema TJ, Anink JJ, Schouten-van Meeteren AY, Spliet WG, et al. BRAF V600E mutation is associated with mTOR signaling activation in glioneuronal tumors. Brain Pathol 2014;24:52–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lonnrot K, Terho M, Kahara V, Haapasalo H, Helen P. Desmoplastic infantile ganglioglioma: novel aspects in clinical presentation and genetics. Surg Neurol 2007;68:304–8. [DOI] [PubMed] [Google Scholar]
- 40.Louis DN, von Deimling A, Dickersin GR, Dooling EC, Seizinger BR. Desmoplastic cerebral astrocytomas of infancy: a histopathologic, immunohistochemical, ultrastructural, and molecular genetic study. Hum Pathol 1992;23:1402–9. [DOI] [PubMed] [Google Scholar]
- 41.Pritchard CC, Salipante SJ, Koehler K, Smith C, Scroggins S, Wood B, et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn 2014;16:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schindler G, Capper D, Meyer J, Janzarik W, Omran H, Herold-Mende C, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol 2011;121:397–405. [DOI] [PubMed] [Google Scholar]
- 43.Hovestadt V, Remke M, Kool M, Pietsch T, Northcott PA, Fischer R, et al. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol 2013;125:913–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012;22:425–37. [DOI] [PubMed] [Google Scholar]
- 45.van der Maarten L, Hinton G. Visualizing high-dimensional data using t-SNE. J Mach Learn Res 2008;9:2579–605. [Google Scholar]
- 46.Rubinstein JC, Sznol M, Pavlick AC, Ariyan S, Cheng E, Bacchiocchi A, et al. Incidence of the V600K mutation among melanoma patients with BRAF mutations, and potential therapeutic response to the specific BRAF inhibitor PLX4032. J Translat Med 2010;8:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015;43:D805–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greer A, Foreman NK, Donson A, Davies KD, Kleinschmidt-DeMasters BK. Desmoplastic infantile astrocytoma/ganglioglioma with rare BRAF V600D mutation. Pediatr Blood Cancer 2017;64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, et al. Wholegenome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet 2013;45:602–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chappe C, Padovani L, Scavarda D, Forest F, Nanni-Metellus I, Loundou A, et al. Dysembryoplastic neuroepithelial tumors share with pleomorphic xanthoastrocytomas and gangliogliomas BRAF (V600E) mutation and expression. Brain Pathol 2013;23:574–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014;343:189–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koelsche C, Wohrer A, Jeibmann A, Schittenhelm J, Schindler G, Preusser M, et al. Mutant BRAF V600E protein in ganglioglioma is predominantly expressed by neuronal tumor cells. Acta Neuropathol 2013;125:891–900. [DOI] [PubMed] [Google Scholar]
- 55.Bergthold G, Bandopadhayay P, Hoshida Y, Ramkissoon S, Ramkissoon L, Rich B, et al. Expression profiles of 151 pediatric low-grade gliomas reveal molecular differences associated with location and histological subtype. Neuro Oncol 2015;17:1486–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang H, Higgins B, Kolinsky K, Packman K, Go Z, Iyer R, et al. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res 2010;70:5518–27. [DOI] [PubMed] [Google Scholar]
- 57.Gentilcore G, Madonna G, Mozzillo N, Ribas A, Cossu A, Palmieri G, et al. Effect of dabrafenib on melanoma cell lines harbouring the BRAF (V600D/R) mutations. BMC Cancer 2013;13:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lassaletta A, Guerreiro Stucklin A, Ramaswamy V, Zapotocky M, McKeown T, Hawkins C, et al. Profound clinical and radiological response to BRAF inhibition in a 2-month-old diencephalic child with hypothalamic/chiasmatic glioma. Pediatr Blood Cancer 2016;63:2038–41. [DOI] [PubMed] [Google Scholar]
- 59.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DT, Capper D, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 2016;164:1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Komori T, Scheithauer BW, Parisi JE, Watterson J, Priest JR. Mixed conventional and desmoplastic infantile ganglioglioma: an autopsied case with 6-year follow-up. Mod Pathol 2001;14:720–6. [DOI] [PubMed] [Google Scholar]
- 61.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 2007;28:622–9. [DOI] [PubMed] [Google Scholar]
- 62.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482:226–31. [DOI] [PubMed] [Google Scholar]
- 63.Li M, Collins R, Jiao Y, Ouillette P, Bixby D, Erba H, et al. Somatic mutations in the transcriptional corepressor gene BCORL1 in adult acute myelogenous leukemia. Blood 2011;118:5914–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ, et al. Medulloblastomics: the end of the beginning. Nat Rev Cancer 2012;12: 818–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 2014;46:444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.