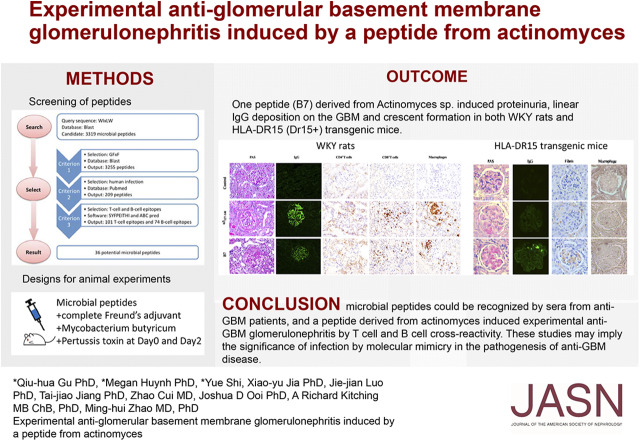
Significance Statement
Antiglomerular basement membrane (anti-GBM) disease is associated with HLA-DRB1*1501 (the major predisposing genetic factor in the disease), with α3127–148 as a nephritogenic T and B cell epitope. Association of infections with anti-GBM disease has been long suspected. In this study, the authors used bioinformatic tools to search for peptides from microbes mimicking the critical motif of a pathogenic epitope for Goodpasture disease (α3127–148). They identified a pathogenic peptide derived from Actinomyces, which was recognized by sera from patients with anti-GBM disease and that induced proteinuria, linear IgG deposition on GBM, and crescent formation in both WKY rats and humanized HLA-DR15 transgenic mice via crossreactivity of lymphocytes. These findings implicate a role for infection and molecular mimicry in the pathogenesis of anti-GBM disease.
Keywords: glomerular disease, pathology, renal injury
Visual Abstract
Abstract
Background
Antiglomerular basement membrane (anti-GBM) disease is associated with HLA-DRB1*1501 (the major predisposing genetic factor in the disease), with α3127–148 as a nephritogenic T and B cell epitope. Although the cause of disease remains unclear, the association of infections with anti-GBM disease has been long suspected.
Methods
To investigate whether microbes might activate autoreactive T and B lymphocytes via molecular mimicry in anti-GBM disease, we used bioinformatic tools, including BLAST, SYFPEITHI, and ABCpred, for peptide searching and epitope prediction. We used sera from patients with anti-GBM disease to assess peptides recognized by antibodies, and immunized WKY rats and a humanized mouse model (HLA-DR15 transgenic mice) with each of the peptide candidates to assess pathogenicity.
Results
On the basis of the critical motif, the bioinformatic approach identified 36 microbial peptides that mimic human α3127–148. Circulating antibodies in sera from patients with anti-GBM recognized nine of them. One peptide, B7, derived from Actinomyces species, induced proteinuria, linear IgG deposition on the GBM, and crescent formation when injected into WKY rats. The antibodies to B7 also targeted human and rat α3127–148. B7 induced T cell activation from human α3127–148-immunized rats. T cell responses to B7 were detected in rats immunized by Actinomyces lysate proteins or recombinant proteins. We confirmed B7’s pathogenicity in HLA-DR15 transgenic mice that developed kidney injury similar to that observed in α3135–145-immunized mice.
Conclusions
Sera from patients with anti-GBM disease recognized microbial peptides identified through a bioinformatic approach, and a peptide from Actinomyces induced experimental anti-GBM GN by T and B cell crossreactivity. These studies demonstrate that anti-GBM disease may be initiated by immunization with a microbial peptide.
Anti-glomerular basement membrane (GBM) disease, also called Goodpasture disease, is a classic autoimmune disease, characterized by circulating autoantibodies against glomerular and/or alveolar basement membranes.1 The noncollagenous domain 1 (NC1) of the α3 chain of type IV collagen [α3(IV)NC1] is the main target antigen of anti-GBM antibodies.2–4 Both autoantibodies and autoreactive T cells against α3(IV)NC1 are pathogenic.5,6
The major predisposing genetic factor (“first hit”) in anti-GBM disease has been identified as HLA-DRB1*15:01,7–10 which with the invariant HLA-DRA1*01:01 forms the “HLA-DR15” allomorph, but little is known about the other etiologic events (“second hit”) that trigger the disease. The association of infections with the onset of anti-GBM disease has been long suspected, with 40%–60% of patients with anti-GBM disease experiencing prodromal infection.1,7,11 Whereas several factors can promote the transition from an immune response to an exogenous agent into an autoimmune response,7 molecular mimicry represents a classic example of this process. In molecular mimicry, autoimmunity is generated when a pathogen that expresses an antigen that is similar in amino acid sequence or structure to a key self-antigen. This foreign antigen activates not only pathogen-specific T or B cells, but also autoreactive lymphocytes. These events may occur even when only a small peptide sequence, or “hot spot,” shared between self and microbial antigens binds to the T cell receptor.7 In anti-GBM nephritis, Arends et al.12 found several microbial peptides on the basis of a critical residue motif of a nephrogenic T cell epitope within rat α3(IV)NC1, Pcol14–26 (all amino acid numbering, including other cited epitopes, converted from the original numbering to that of the reference).13 In this study, a peptide from Clostridium botulinum induced crescentic GN and pulmonary hemorrhage in WKY rats, implicating molecular mimicry in anti-GBM disease.12 However, the corresponding sequence of Pcol14–26 on human α3(IV)NC1 has not been recognized by T cells from patients with anti-GBM disease.14,15
Previous studies in patients with anti-GBM disease have identified a linear human α3(IV)NC1 epitope, α3127–148 (TDIPPCPHGWISLWKGFSFIMF) (Uniprot accession: Q01955 CO4A3_HUMAN1615–1636), that contains both B and T cell epitopes.10,14–16 The critical amino acid residues within human α3127–148 were characterized as tryptophan (W)136, isoleucine (I)137, leucine (L)139, and tryptophan (W)140, which are critical for inducing crescentic GN in WKY rats,17 and glycine (G)142, phenylalanine (F)143, and phenylalanine (F)145, which are crucial for autoantibody binding.18 This epitope and its critical amino acids are broadly consistent with the T cell epitope studies in the context of HLA-DRB1*15:01 restriction, with four critical residues.8 These findings led us to search for mimicking microbial peptides that may participate in anti-GBM disease.
Bioinformatic tools, including SYFPEITHI19 and ABCpred20, are commonly used for T cell and B cell epitope prediction. They have been used for in silico epitope prediction for vaccines against viruses such as HIV and influenza.21–23 In this study, we used bioinformatic tools for peptide alignment and epitope prediction to define 36 candidate microbial peptides, on the basis of the motif WIxLWxGFxF, that may mimic α3127–148. Nine peptides were recognized by the circulating antibodies from patients with anti-GBM disease. One peptide (B7), from the UDP-N-acetylenolpyruvoylglucosamine reductase sequence found in Actinomyces sp, could induce anti-GBM GN after immunization to WKY rats and HLA-DR15 transgenic (DR15+) mice, providing evidence for infections participating in the development of anti-GBM disease through molecular mimicry.
CONCISE METHODS
Searching Microbial Peptides on the Basis of the Critical Motif of α3127–148
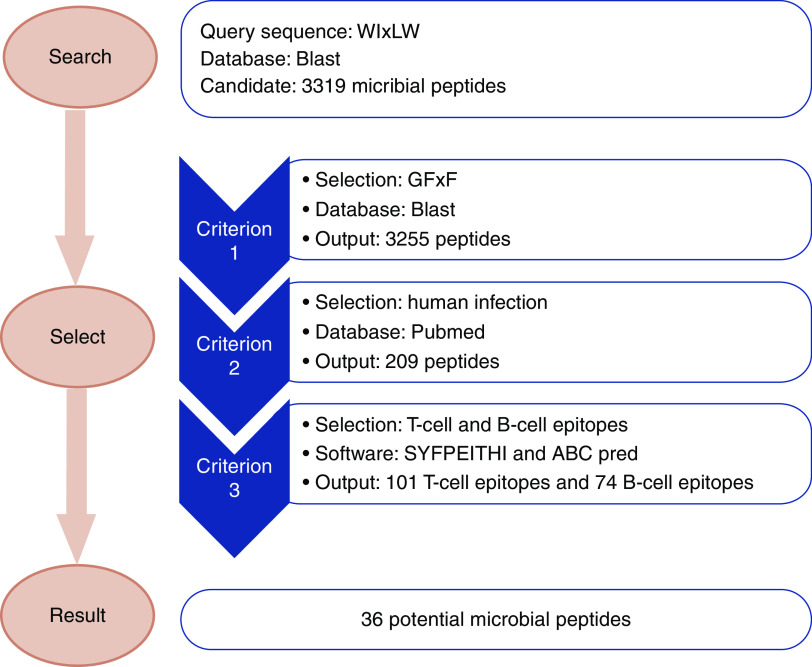
A schematic diagram illustrating the process of searching out the candidate microbial peptides is shown in Figure 1.
The pathogenic motif of human α3127–148 (WIxLW) was applied as the query sequence to search microbial peptides using BLASTp (Protein Basic Local Alignment Search Tool) suite (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The search set was nonredundant protein sequences (nr) and the selected organisms were bacteria (TaxID: 2), fungi (TaxID: 4751), and viruses (TaxID: 10239). The maximum number of target sequences to display was 5000 and other parameters remained on the default settings.
Candidate peptides were selected when they contained residues G and/or F in the form of a GFxF motif, which are crucial for antibody binding.
Candidate peptides were selected when they were derived from probiotics or microbes associated with human infections. We used the keywords “(microbial genus name) AND (human OR human infection)” in PubMed to select microbes that were infectious to human beings, or could be isolated from specimens of humans. Peptides without exact human-related records were eliminated.
Candidate peptides were selected when they had higher probabilities of being T cell and B cell epitopes. B cell epitope prediction was performed using ABCpred (http://crdd.osdd.net/raghava/abcpred/ABC_submission.html). The threshold for the sequence length of B cell epitope prediction was 16 amino acids, which is the default setting of the software. Protein sequences containing peptides within the top 50% of predictions among all peptides were chosen as candidate B cell epitopes. T cell epitope prediction was performed using SYFPEITHI (http://www.syfpeithi.de/bin/MHCServer.dll/EpitopePrediction.htm). HLA-DR15 is the risk allele for patients with anti-GBM disease24 and the structure of rat MHC II molecule RT1-D is similar to that of HLA-DR15,25 thus the HLA-DR15 molecule selected the peptides that could bind to HLA II and be presented to T cells. The threshold for the sequence length of T cell epitope prediction was 15 amino acids, which is a setting for MHC II. Protein sequences containing peptides in the top 50% of predictions were selected as candidate T cell epitopes. Peptides predicted to be both B cell epitopes and T cell epitopes were selected for further evaluation.
Figure 1.
Schematic diagram for the screening of candidate microbial peptides mimicking the critical motif of the anti-GBM epitope. The pathogenic motif of α3127–148 (WIxLW) was applied as the query sequence to search microbial peptides using BLASTp suite. Candidate peptides were selected when they fulfilled the following three criteria. Firstly, the peptide contained at least one residue from GFxF, which is crucial for antibody binding. Secondly, the peptides were derived from probiotics or microbes associated with human infections. Thirdly, the peptides had a relatively high probability of being both T cell and B cell epitopes.
Preparation of Peptides and Proteins
Peptides were synthesized as described previously.17,18 There were 18 peptides that were too hydrophobic to be synthesized and their sequences were adjusted to increase hydrophilicity, as shown in Table 1.
Table 1.
The potential microbial peptide mimicking the critical amino acids of α3127–148
| Peptide | Sequencea | Adjusted Sequenceb | Protein | Species |
|---|---|---|---|---|
| B1 | AGFGWIDLWYGVLFGV | 4-Hydroxybenzoate polyprenyltransferase | Inquilinus limosus | |
| B2 | TWIQLWHGTPFKKMLF | Hypothetical protein | Staphylococcus sp. EGD-HP3 | |
| B3 | ARWISLWLFFMFINQP | ARWISLWLFFMFKK | O-antigen flippase Wzx | Bacillus sp. JCM 19046 |
| B4 | TFPDWIPLWGGESFVF | WIPLWGGESFVFKK | Peptidase A8 | Prevotella multiformis |
| B5 | YMKIQLWICLWFLFDF | QLWICLWFLFDFKK | Potassium channel protein | Bacteroides |
| B6 | YFLFQWILLWKNRSFK | Amino acid racemase | Clostridium sp. CL-2 | |
| B7 | GRWIVLWVEFQFTHAS | GRWIVLWVEFQFKK | UDP-N-acetylenolpyruvoylglucosamine reductase | Actinomyces sp. S4-C9/A. europaeus |
| B8 | WISLWKKAEFLQGLSP | Hypothetical protein | Bacillus sp. J37 | |
| B9 | GLWIGLWSGTTFTFNP | WIGLWSGTTFTFKK | gp240 | Mycobacterium phage ScottMcG |
| B10 | KQFLWIVLSLWTGFTF | WIVLSLWTGFTFKK | Cytochrome C oxidase | Comamonas testosteroni |
| B11 | HFLWVLIALWTGFTFI | WVLIALWTGFTFKK | Cytochrome C oxidase | Ralstonia pickettii |
| B12 | HSAWILIALWTGFTFV | WILIALWTGFTFKK | Cytochrome C oxidase | Ralstonia sp. UNC404CL21Col |
| B13 | MELNIWIALWAGFVSF | IWIALWAGFVSFKK | Cytochrome C biogenesis protein | Paenibacillus |
| B14 | KHFLWIVIALWTGFTF | WIVIALWTGFTFKK | Cytochrome C oxidase | Cupriavidus sp. Amp6 |
| B15 | AVVLWILLWLGAGVSG | WILLWLGAGVSGKK | Envelope glycoprotein C | Human herpesvirus 1 |
| B16 | GRGWITLWKCFKCAGP | Hypothetical protein | Paenibacillus alvei | |
| B17 | DMDGNWIGLWKGKDKW | Hypothetical protein | Proteobacteria bacterium | |
| B18 | LERRWIDLWGKLGFKE | Hypothetical protein | Prevotella sp. MA2016 | |
| B19 | PWIWLWAPNFYFPLSG | Hypothetical protein ×994_4209 | Burkholderia sp. TSV202 | |
| B20 | PFWVWILLWIFLFANF | WILLWIFLFANFKK | Hypothetical protein | Alloscardovia omnicolens |
| B21 | AWIWLWGFEWALLALA | AWIWLWGFEWALKK | Membrane protein | Streptomyces sp. NRRL B-1347 |
| B22 | TEEGWILLWEPKYFDD | Hypothetical protein | Streptococcus sobrinus | |
| B23 | IVIGWIFLWAFVDKLF | WIFLWAFVDKLFKK | Membrane protein | Actinotalea ferrariae CF5–4 |
| B24 | LGQWISLWVYGFSLVL | GQWISLWVYGFSKK | Multispecies: hypothetical protein | Acinetobacter |
| B25 | KVVIWIALWTPLFAFL | Conjugal transfer protein TraG | Acinetobacter baumannii | |
| B26 | YDTWIDLWESGKFPDI | CRISPR-associated protein CasA | Streptococcus oralis | |
| B27 | LAFLNWIRLWKSFKTE | Putative membrane protein | Leptospira sp. B5–022 | |
| B28 | CLTGWIGLWPMAGFLI | TGWIGLWPMAGFKK | Putative uncharacterized protein | Akkermansia sp. CAG:344 |
| B29 | FISNWITLWKGDLIGT | Hypothetical protein FEM08_33490 | Flavobacterium sp. EM1308 | |
| B30 | EVNLWIALWAGVASFI | LWIALWAGVASFKK | Cytochrome C biogenesis protein | Paenibacillus sp. UNC451MF |
| B31 | PDWISLWGFFQSEMKYF | Glycosyl transferase | Proteobacteria bacterium | |
| B32 | NPIQWIKLWSEMELFLF | LysR family transcriptional regulator | Bacillus subtilis | |
| B33 | CLWIGLWGNFPFALGP | Xanthine permease | Oscillospiraceae bacterium | |
| B34 | QWIYLWLKGFESGELR | Hypothetical protein | Clostridiales bacterium | |
| B35 | RGWIMLWGVGFFAVGL | GWIMLWGVGFFAKK | Hypothetical protein | Acinetobacter soli |
| B36 | TGAWIHLWGKGANGGS | gp34 | Mycobacterium phage |
CRISPR, clustered regularly interspaced short palindromic repeats.
The key amino acids are underlined.
Peptides were too hydrophobic to be synthesized and their sequences were adjusted to increase hydrophilicity.
Actinomyces europaeus Funke (American Type Culture Collection 700353) (Actinomyces sp. S4-C9, containing a B7 sequence) was purchased and amplified by the Microorganism Examination Laboratory of Peking University First Hospital (Beijing, China). Actinomyces were collected in radioimmunoprecipitation assay lysis buffer (Plygen Technologies Inc., Beijing, China) supplemented with 1% protease inhibitor cocktail (EMD Millipore Corp., MA) and underwent 1 hour of sonic disruption to obtain whole protein lysates.
Recombinant UDP-N-acetylenolpyruvoylglucosamine reductase protein fused with a His tag was synthesized by GenScript (Genscript Corp., Nanjing, China). Escherichia coli and pET-30a (+) were used as the expression system and vector, respectively. Protein was obtained from supernatants of cell lysates and purified by two-step purification with an Ni column and Superdex 200, and the final purity was around 90%.
Whole protein lysate of Mycobacterium butyricum (Becton, Dickinson and Company) was acquired in the same manner as Actinomyces.
Human Recognition of Microbial Peptides
Sera from 46 patients with anti-GBM disease, diagnosed between 2008 and 2013, were collected before immunosuppressive therapy and plasmapheresis. In addition, 22 sera samples from healthy donors were used as normal controls, and 40 patients with ANCA-associated small vessel vasculitis or lupus nephritis were used as disease controls.
For peptide recognition by human sera, one half of the ELISA plate was coated with peptides at 10 μg/ml and the other half was coated with coating buffer as antigen-free wells. Next, 3% BSA dissolved with PBS was used for blocking at 200 μl/well at 37°C for 1 hour. Human sera were diluted at 1:100 in 1% PBS Tween-20 and incubated at 37°C for 1 hour. Anti-human IgG (γ-chain specific)-alkaline phosphatase antibody (A3187; Sigma-Aldrich, St. Louis, MO) was diluted at 1:5000 in 1% PBS Tween-20 plus 1% BSA and incubated at 37°C for 45 minutes.
Experimental Anti-GBM GN in WKY Rats
Immunization of WKY Rats
WKY rats, 4–6 weeks of age, were immunized with 100 μg/g of each microbial peptide, 1 μg/g of Actinomyces lysate, and 10, 5, 2, and 1 μg/g of recombinant UDP-N-acetylenolpyruvoylglucosamine reductase protein emulsified in CFA (Sigma-Aldrich) in addition to M. butyricum (4.5 mg/ml), by footpad injection, 0.6 ml per each rat, on day 0. Pertussis toxin (Sigma-Aldrich) (500 ng) was injected intraperitoneally on day 0 and day 2. Control rats were immunized with CFA, M. butyricum, and pertussis toxin. Positive control rats were immunized with human α3127–148, as described previously.17 Experiments ended on day 49.
Assessment of Disease
Before and after immunization, 24-hour urine samples and blood samples were collected each week. Clinical manifestations and renal histopathology were evaluated as described previously.17 Glomerular CD4+ T cells, CD8+ T cells, and macrophages were detected by staining frozen sections (for CD4) or paraffin-embedded formalin-fixed sections (CD8 and CD68). All sections were subjected to 3% H2O2 to block endogenous peroxidase, then incubated with mouse anti-rat CD4, CD8, or CD68 monoclonal antibodies (Abcam, Cambridge, MA) at 4°C overnight, followed by anti-mouse Ig conjugated with horseradish peroxidase (ZGSB-BIO, Beijing, China) at 37°C for 30 minutes. Then, the staining was developed with 3,3′diaminobenzidine for 30 seconds. At least 50 consecutive glomeruli were assessed per sample by Image-Pro Plus analysis software (version 6.0; Media Cybernetics, Dallas, TX). CD4+ and CD8+ T cell infiltration was shown as the cells per glomerular cross-section (gcs). Macrophage (CD68+ cell) infiltration was shown as the integrated OD per gcs (IOD/gcs).
Assessment of T and B Cell Responses
Antibodies in kidneys were eluted by glycine (pH 2.7). The antibodies in circulation and in kidney elutes were measured by ELISA.16,18,26 Peptides at 10 μg/ml, and human α3(IV)NC1, Actinomyces lysate protein, recombinant UDP-N-acetylenolpyruvoylglucosamine reductase protein, M. butyricum lysate and pertussis toxin (all at 2 μg/ml) were used to coat plates, then test sera were diluted at 1:100 and elutes were diluted at 1:5, and 100 μl were added to each well.
IFN-γ enzyme-linked immunospot assay (ELISpot) (Mabtech, Stockholm, Sweden) was used to detect cytokine production. Splenocytes were collected 10 days after immunization with 10 μg human α3127–148, B7, Actinomyces lysate protein, or recombinant UDP-N-acetylenolpyruvoylglucosamine reductase protein with CFA, cultured, and restimulated with α3127–148 or other peptides (10 μg/ml) in the plates coated with anti-IFN-γ mAb for 20 hours (37°C and 5% CO2). The culture supernatant was collected, and biotinylated anti-IFN-γ mAb was added and incubated for 2 hours at room temperature. After washing, streptavidin-horseradish peroxidase was added for 1 hour at room temperature. 3,3', 5,5"-Tetramethylbenzidine substrate solution was added and color development was stopped by deionized water. The results were expressed as the mean numbers of spots counted by the ELISpot reader (ELR08; AID, Strasberg, Germany) minus baseline (media alone).
Experimental Anti-GBM GN in HLA-DR15 Transgenic Mice
Immunization of HLA-DR15 Transgenic Mice
HLA-DRB1*15:01 transgenic mice [transgenic for HLA-DRA1*01:01, HLA-DRB1*1501 (DR15+), mouse MHC II−/−, and Fcgr2b−/− or Fcgr2b+/+] were used, with the FcγRIIb-deficient strain being used in the induction of anti-GBM GN.8,10 DR15+.Fcgr2b−/− mice were immunized with 100 µg mouse α3135–145 (Uniprot accession: Q9QZS0 CO4A3_MOUSEM1622–1632), OVA323–339, or B7 in CFA (Sigma-Aldrich) for the first immunization, then with CFA subcutaneously on days 0, 7, and 14. Experiments ended on day 42.
Assessment of Disease
Urinary albumin was quantified by ELISA (Bethyl Laboratories, Montgomery, TX) and expressed as a urinary albumin-creatinine ratio. Serum creatinine and BUN were measured using an autoanalyzer. Histologic injury was determined by assessing glomerular segmental necrosis and crescent formation on periodic acid–Schiff-stained, formalin-fixed paraffin-embedded (FFPE) kidney sections. Linear IgG deposition was analyzed by immunofluorescence staining on frozen tissue using FITC-labeled rabbit anti-mouse IgG antibodies (Thermo Fisher Scientific, Waltham, MA). Inflammatory cell infiltrates were assessed on periodate-lysate paraformaldehyde (PLP)-fixed tissue by immunoperoxidase staining using primary antibodies for CD4+ T cells (clone GK1.5), macrophages (anti-mouse CD68, clone FA/11), and neutrophils (anti-mouse Gr-1, clone RB6–8C5). Fibrin was assessed by immunoperoxidase staining using rabbit anti-mouse fibrinogen antibodies (R-4025; a gift from Dr. J. Degen, Children’s Research Foundation, Cincinnati, OH). At least 30 glomeruli were assessed per sample.
Assessment of T and B Cell Responses in DR15+ Mice
Serum α3(IV)NC1-specific antibodies were measured by ELISA. Plates were coated with 5 µg/ml recombinant α3(IV)NC1,27 then blocked with 2% casein/PBS. Serum samples were diluted 1:100 and detected with horseradish peroxidase-conjugated sheep anti-mouse IgG antibodies (GE Healthcare, Sydney, Australia) and 3,3', 5,5"-tetramethylbenzidine substrate (Sigma-Aldrich). To assess cytokine production, splenocytes were cultured and restimulated with α3135–145, recombinant α3(IV)NC1, and B7 in supplemented RPMI media (10% FCS, 2 mM L-glutamine, 50 µm 2-mercaptoethanol, 100 U/ml penicillin, and 0.1 mg/ml streptomycin; Sigma-Aldrich) for 72 hours (37°C and 5% CO2), after which the culture supernatant was collected. IFN-γ and IL-17A were measured by ELISA.28 To examine initial immune responses, DR15+.Fcgr2b+/+ mice were immunized with 10 µg α3135–145, OVA323–339, or B7 emulsified in CFA, then draining lymph nodes were harvested after 10 days. Immune reactivity to specific peptides was determined by IFN-γ and IL-17A ELISpots.8
Statistical Analyses
Statistical analysis was performed using statistical software SPSS 13.0 (SPSS Inc., Chicago, IL). Differences in quantitative parameters were assessed using the t test or one-way ANOVA for data that were normally distributed, and the Mann–Whitney U test or Kruskal–Wallis test for data that were nonnormally distributed. Differences in qualitative data were compared using the chi-squared or Fisher’s exact tests. Results in the animal experiments were represented as the mean ± SEM. All statistical analyses were two-tailed and a P value of <0.05 was considered to be significant.
Study Approval
The research was conducted in accordance with the Declaration of Helsinki and approved by the ethics committee of Peking University First Hospital. Informed consent was obtained from each patient for sampling blood. All rat experiments were approved by the Experimental Animal Ethics Committee of Peking University First Hospital. Mice were kept in specific pathogen-free conditions at the Monash Medical Centre Animal Facility, and experiments were approved by the Monash University Animal Ethics Committee.
RESULTS
Microbial Peptides Mimicking the Critical Motif WIxLWxGFxF
First, 3319 microbial peptides were identified using the motif WIxLW in the nr database of BLASTp and 3255 peptides were identified containing at least one residue from the GFxF motif. We selected 209 peptides (204 peptides from bacteria, 2 from fungi, and 3 from viruses) associated with human infections. Of the 204 bacteria-derived peptides, 15 peptides came from the genus of Bacillus, 10 peptides from the genus of Paenibacillus, 9 from Clostridium, and 9 from Pseudomonas. The other bacteria-derived peptides were from multiple genera (Figure 1).
All 209 peptides were assessed by B cell and T cell epitope predictive algorithms. Of them, 74 peptides were identified as potential B cell epitopes and 101 as T cell epitopes, of which 36 were common to both the T and B cell list (33 bacterial peptides and 3 viral peptides). The genera of Bacillus, Paenibacillus, and Acinetobacter contained three peptides, respectively. The other peptides came from Actinomyces, Proteobacteria, Prevotella, and 18 other genera, with each genus containing one or two peptides (Table 1).
Circulating Antibodies Against Microbial Peptides in Patients with Anti-GBM Disease
Clinical data of the 46 patients with anti-GBM in this study are detailed in Table 2. Antibodies against the 36 microbial peptides were evaluated in sera from these subjects. Among them, nine peptides were recognized with high frequency: B7 (30.4%), B10 (52.2%), B11 (63.0%), B12 (26.1%), B13 (21.7%), B24 (37.0%), B25 (21.7%), B30 (32.6%), and B35 (45.7%). Table 1 contains the origin of these sequences. The mean levels of antibodies against these nine peptides are shown in Table 3. Very few antibodies against the nine peptides were detected from normal and disease controls, and at very low levels (Table 3). The binding between antibodies and B7 could be inhibited by B7 but not by α3127–148 (Supplemental Figure 1).
Table 2.
The clinical features of patients with anti-GBM disease
| Parameters | n=46 |
|---|---|
| Age (yr) | 47.1 ± 17.6 |
| Men/women | 23:23 |
| Hydrocarbon exposure, n (%) | 8 (17.4) |
| Smoking, n (%) | 18 (39.1) |
| Hemoptysis, n (%) | 15 (32.6) |
| Hemoglobin (g/L) | 88.3 ± 22.8 |
| Oliguria/anuria, n (%) | 22 (47.8) |
| Serum albumin (g/L) | 29.7 ± 4.2 |
| Gross hematuria, n (%) | 17 (37.0) |
| Serum creatinine, μmol/L (upper, lower quartiles) | 756.5 (459.8, 1144.3) |
| Anti-GBM antibody level (relative unit/ml) | 155.3 ± 64.0 |
| Glomerular crescents (%) | 63.7 ± 27.3 |
Table 3.
The recognition frequencies and antibody levels of circulating antibodies against microbial peptides in patients with anti-GBM disease, healthy controls, and patients with other diseases such as ANCA vasculitis and lupus nephritis
| Peptide | Sequencea | Frequency in Anti-GBM Disease, n (%) | Antibody Level (OD Value) | Frequency in Healthy Controls, n (%) | Antibody Level (OD Value) | Frequency in Disease Controls, n (%) | Antibody Level (OD Value) | Cutoff Value (OD Value) |
|---|---|---|---|---|---|---|---|---|
| B7 | GRWIVLWVEFQFKK | 14 (30.4) | 0.57 ± 0.26 | 0 (0) | −0.02 ± 0.13 | 0 (0) | 0.04 ± 0.05 | 0.25 |
| B10 | WIVLSLWTGFTFKK | 24 (52.2) | 0.53 ± 0.29 | 0 (0) | −0.07 ± 0.11 | 1 (2.5) | 0.02 ± 0.04 | 0.14 |
| B11 | WVLIALWTGFTFKK | 29 (63.0) | 0.56 ± 0.37 | 1 (4.5) | 0.06 ± 0.11 | 0 (0) | 0.02 ± 0.04 | 0.17 |
| B12 | WILIALWTGFTFKK | 12 (26.1) | 0.68 ± 0.33 | 1 (4.5) | 0.06 ± 0.13 | 0 (0) | 0.02 ± 0.02 | 0.31 |
| B13 | IWIALWAGFVSFKK | 10 (21.7) | 0.65 ± 0.34 | 2 (9.1) | 0.01 ± 0.07 | 0 (0) | 0.02 ± 0.04 | 0.15 |
| B24 | GQWISLWVYGFSKK | 17 (37.0) | 0.51 ± 0.35 | 0 (0) | −0.07 ± 0.09 | 1 (2.5) | 0.01 ± 0.03 | 0.13 |
| B25 | KVVIWIALWTPLFAFL | 10 (21.7) | 0.60 ± 0.37 | 1 (4.5) | −0.01 ± 0.10 | 0 (0) | 0.02 ± 0.03 | 0.19 |
| B30 | LWIALWAGVASFKK | 15 (32.6) | 0.51 ± 0.28 | 0 (0) | 0.03 ± 0.09 | 0 (0) | 0.02 ± 0.04 | 0.21 |
| B35 | GWIMLWGVGFFAKK | 21 (45.7) | 0.63 ± 0.46 | 0 (0) | −0.001 ± 0.08 | 3 (7.5) | 0.04 ± 0.05 | 0.16 |
The key amino acids are underlined.
Nephritogenicity of Microbial Peptides in WKY Rats
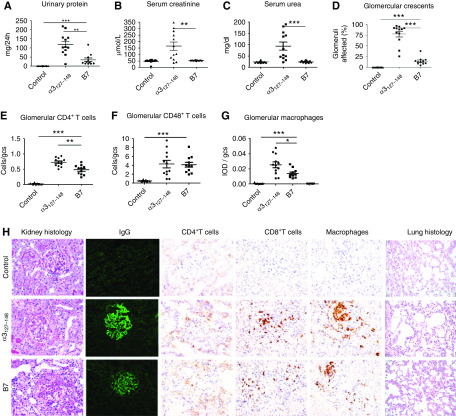
The selected nine peptides were used to immunize WKY rats. Control rats were immunized with either the nephritogenic T cell epitope α3127–148 or CFA. One peptide (B7) induced significant proteinuria in all rats (11/11, 100%) from week 3 after immunization, peaking at week 5 (54.0 ± 11.2 versus 0.9 ± 0.1 mg/24 h in control immunized rats, P<0.001) (Figure 2A), but no increase in serum creatinine or serum urea (Figure 2, B and C). All B7-immunized rats exhibited linear deposits of IgG on the GBM (Figure 2H). The percentage of crescent formation in glomeruli was 14.6% ± 2.7%, predominantly cellular crescents (Figure 2D). Segmental mesangial proliferation, and lymphocyte and monocyte infiltration were also observed in the kidney (Figure 2H). CD4+ T cells, CD8+ T cells, and macrophages were present within glomerular tufts and crescents in all B7-immunized rats (Figure 2H). CD8+ T cells (4.1 ± 0.6 cells/gcs) and macrophages (0.01 ± 0.002 IOD/gcs) were mostly distributed in glomeruli and peri-glomerular regions. They could also be observed in the interstitium. CD4+ T cells were detected in the same areas but with fewer cells observed (0.5 ± 0.05 cells/gcs) (Figure 2H). No pulmonary lesions were observed in any rats (Figure 2H).
Figure 2.
The microbial peptide B7 induced anti-GBM GN in WKY rats. WKY rats were immunized with B7 (n=11), α3127–148 (n=12), or CFA (control, n=12) on day 0, and experiments ended on day 49. Disease was assessed by proteinuria (A), serum creatinine (B), serum urea (C), crescent formation (D), and infiltrating cells (E–G). Illustrative photomicrographs of glomeruli showing crescent formation by periodic acid–Schiff stain, immunofluorescent staining of linear IgG deposits, and cellular infiltrates of CD4+ T cells, CD8+ T cells, and macrophages (H). Infiltrating cells are expressed as cells per gcs for T cells and IOD per gcs for macrophages. No pulmonary lesion was observed (H). *P<0.05, **P<0.01, and *** P<0.001.
All (12/12, 100%) α3127–148-immunized rats developed severe crescentic GN (Figure 2H). Compared with B7-immunized rats, they exhibited more proteinuria (126.4 ± 20.0 versus 34.3 ± 9.6 mg/24 h, P=0.005), higher BUN (94.0 ± 17.9 versus 24.5 ± 1.4 mg/dl, P<0.001), higher serum creatinine levels (165.3 ± 32.0 versus 52.4 ± 0.8 μmol/L, P=0.007), and more crescent formation (78.0 ± 6.7 versus 14.6% ± 2.7%, P<0.001) (Figure 2, A–D). The intensity of linear IgG deposition was stronger (Figure 2H), and the infiltration of CD4+ T cells (0.7 ± 0.04 versus 0.5 ± 0.05 cells/gcs, P=0.001) (Figure 2E) and macrophages (0.03 ± 0.004 versus 0.01 ± 0.002 IOD/gcs, P=0.02) (Figure 2G) was more prominent, although CD8+ T cell numbers were comparable between the B7 and α3127–148 groups (Figure 2F).
None of the rats immunized with the other eight microbial peptides developed proteinuria or kidney injury. No linear IgG staining or crescent formation was observed in the glomeruli (Supplemental Figure 2). No pulmonary lesions were found in the lung tissues.
The lysate proteins of Actinomyces and the recombinant UDP-N-acetylenolpyruvoylglucosamine reductase proteins were used to immunize WKY rats. No linear IgG deposits, crescent formation, or pulmonary lesions were observed (Supplemental Figure 3).
Immune Response Induced by B7 in WKY Rats
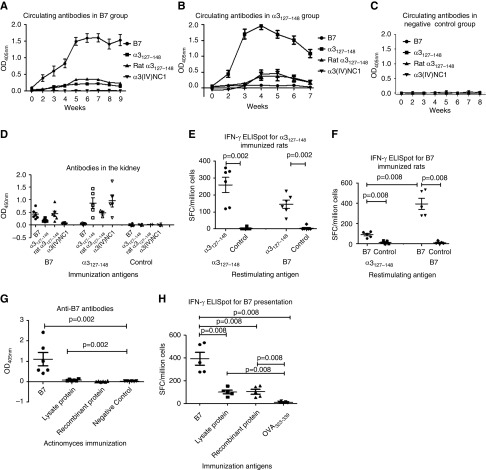
Antigen specificities of the antibodies in sera and kidney eluates were detected by ELISA using B7, human α3127–148, rat α3127–148, and recombinant human α3(IV)NC1 (Figure 3, A–D). Rats immunized with B7 developed a high level of antibodies against B7 by week 2, and lower titers of antibodies toward human and rat α3127–148 week later. Serum antibody responses to B7, human α3127–148, and rat α3127–148 peaked at weeks 5–6 after immunization (Figure 3A). B7-immunized rats did not develop autoantibodies against intact human α3(IV)NC1. However, rats immunized with α3127–148 developed antibodies toward not only human and rat α3127–148, but also human α3(IV)NC1 (Figure 3B). Similarly, IgG eluted from kidneys of B7-immunized rats reacted to the immunogen itself, human α3127–148, and rat α3127–148, but not to human α3(IV)NC1 (Figure 3D). IgG of kidney eluates from the α3127–148 group recognized not only human and rat α3127–148, but also human α3(IV)NC1 (Figure 3D).
Figure 3.
Immune response induced by B7 in WKY rats. Sera (A) and kidney eluates (D) of B7-immunized rats crossreacted with human α3127–148 and rat α3127–148, but not human α3(IV)NC1. Sera (B) and kidney eluates (D) of α3127–148-immunized rats crossreacted with rat α3127–148 and human α3(IV)NC1. No antibody was found in the negative control group (C). Splenocytes from α3127–148- and B7-immunized rats were cultured and restimulated with α3127–148 and B7 for 20 hours; supernatant was collected and measured for IFN-γ by ELISpot. Splenocytes from α3127–148-immunized rats could respond to α3127–148 and B7 (E). Splenocytes from B7-immunized rats could respond to α3127–148 and B7 (F). Antibodies toward B7 were detected in the Actinomyces lysate protein-immunized group (G). Splenocytes from both the Actinomyces lysate protein- and recombinant protein-immunized rats could respond to B7 (H). SFC, spot-forming cells. *P<0.05, **P<0.01, and ***P<0.001.
T cell crossreactivity to B7 and α3127–148 was detected in α3127–148-immunized rats using IFN-γ ELISpot. We found that splenocytes responded to both α3127–148 and B7 (Figure 3E). Splenocytes from B7-immunized rats could also respond to both α3127–148 and B7 (Figure 3F).
Rats immunized with Actinomyces lysate proteins developed antibodies toward B7, although the level of anti-B7 antibodies was low (Figure 3G). No crossrecognition to B7 was found in rats immunized with recombinant UDP-N-acetylenolpyruvoylglucosamine reductase protein. Human antibodies toward the recombinant proteins were detected in sera from 14/46 patients with anti-GBM disease, among them 13 patients had anti-B7 antibodies. No antibody toward Actinomyces lysate protein was found in patients.
Splenocytes from rats immunized with Actinomyces lysate proteins and recombinant UDP-N-acetylenolpyruvoylglucosamine reductase proteins were stimulated with B7 to investigate the presentation and recognition of B7. Results showed that the splenocytes from rats immunized by lysate proteins and recombinant proteins both could respond to B7 (Figure 3H).
Nephritogenicity of B7 in HLA-DR15 Transgenic Mice
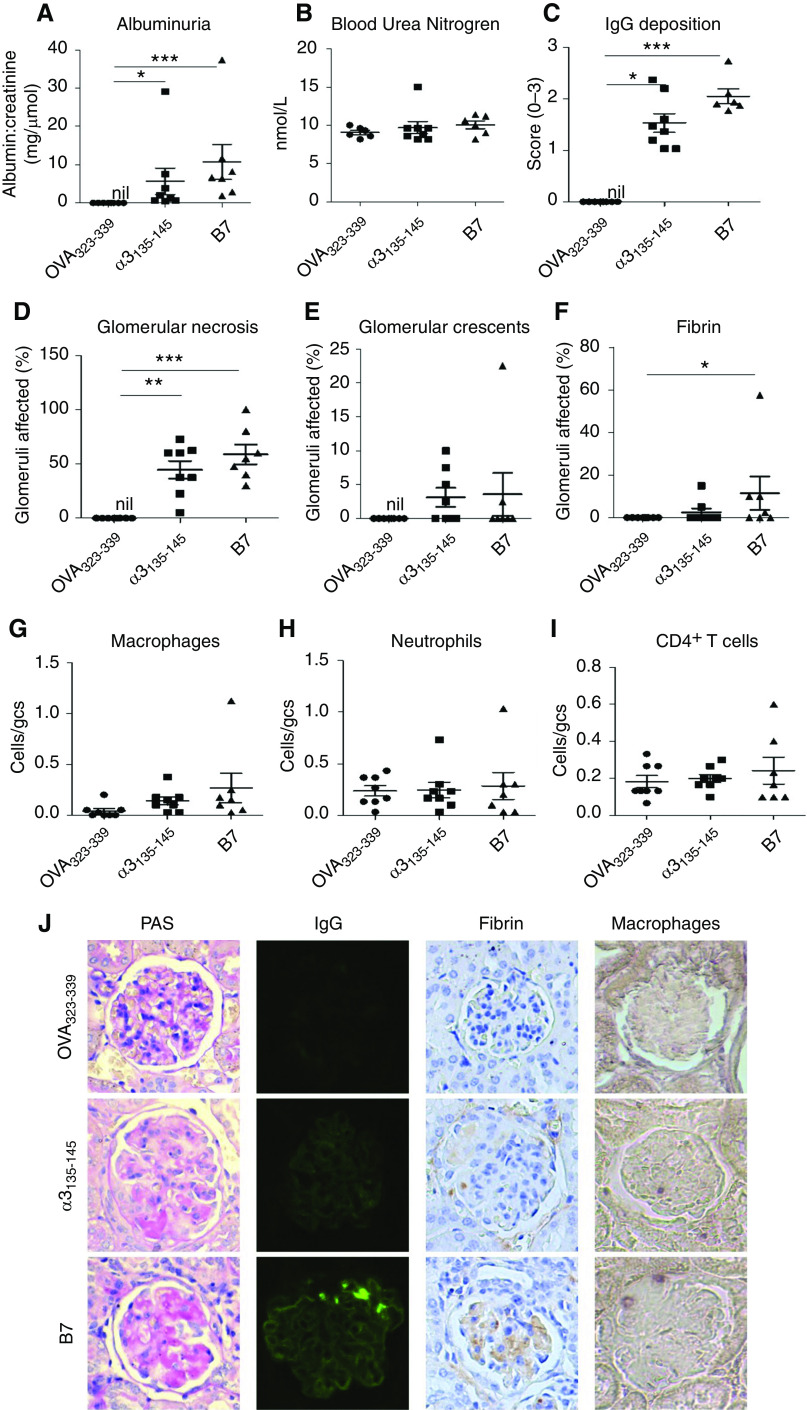
The nephritogenicity of B7 was further confirmed in a humanized mouse model of autoimmune anti-GBM GN.8 HLA-DR15 (DR15+) transgenic mice were immunized with B7. Control mice were immunized with either the nephritogenic T cell epitope α3135–145 or OVA323–339. All seven mice immunized with B7 developed disease, comparable to α3135–145-immunized mice, with albuminuria [10.7 ± 4.6 versus 5.7 ± 3.4 mg/μmol (albumin-creatinine ratio), P=0.08] (Figure 4A) but no increase in BUN (10.1 ± 0.5 versus 9.7 ± 0.8 mmol/L) (Figure 4B).
Figure 4.
HLA-DR15 (Dr15+) transgenic mice immunized with B7 developed anti-GBM GN. DR15+.Fcgr2b−/− mice were immunized with OVA323–339 (n=8), α3135–145 (n=8), or B7 (n=7) on days 0, 7, and 14, and experiments ended on day 42. Disease was assessed by albuminuria (A), BUN (B), glomerular IgG deposition (C), segmental glomerular necrosis (D), crescent formation (E), glomerular fibrin (F), and infiltrating cells (G–I). Illustrative photomicrographs of glomeruli showing necrosis and early crescent formation by periodic acid–Schiff stain, immunofluorescent staining of linear IgG deposits, and immunoperoxidase staining of fibrin deposition and cellular infiltrates with macrophages as an example (J). IgG deposition is expressed semiquantitatively on the basis of intensity; infiltrating cells are expressed as cells per gcs. *P<0.05, **P <0.01, and *** P<0.001.
All mice immunized with B7 or α3135–145 exhibited linear IgG deposits along the GBM (Figure 4J) and focal glomerular necrosis (Figure 4J). Two mice (28.6%) in the B7-immunized group and four α3135–145-immunized mice (50.0%) developed glomerular crescent formation (Figure 4J). The immunofluorescence intensity (2.1 ± 0.1 versus 1.5 ± 0.2, P=0.07) (Figure 4C), percentage of glomeruli affected by necrosis (58.9% ± 9.1% versus 44.7% ± 8.2%, P=0.42) (Figure 4D), and crescents (3.6% ± 3.2% versus 3.1±1.4, P=0.47) (Figure 4E) were comparable in B7- and α3135–145-immunized mice. B7-immunized mice also had increased glomerular fibrin deposits and macrophage infiltration, compared with mice immunized with OVA323–339 that did not develop disease (Figure 4, F–J).
Immune Response Induced by B7 in HLA-DR15 Transgenic Mice
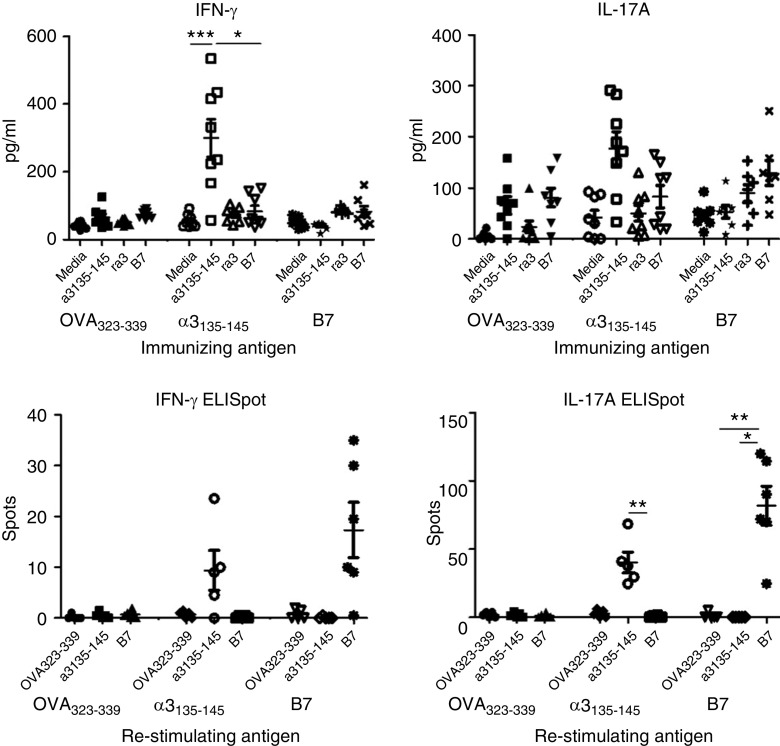
Splenocytes from the B7-immunized group showed a lack of crossreactivity in vitro, with IFN-γ and IL-17A production detectable in cells that were restimulated with B7, but not with α3135–145 (Figure 5A). Similarly, 10 days after immunization, IFN-γ and IL-17A production was detected ex vivo only in cells from mice that were immunized and restimulated with the same peptide (Figure 5B).
Figure 5.
Immune response induced by B7 in HLA-DR15 transgenic mice. B7-specific T cell immune responses do not crossreact with α3135–145. Splenocytes from immunized DR15+Fcgr2b−/− mice were cultured and restimulated with α3135–145, recombinant α3(IV)NC1, and B7 for 72 hours; supernatant was collected and measured for IFN-γ and IL-17A by ELISA (A). Early antigen-specific T cell responses 10 days postimmunization were assessed in DR15+.Fcgr2b+/+ mice by IFN-γ and IL-17A production measured by ELISpot (B). *P<0.05, **P<0.01, and ***P<0.001.
DISCUSSION
Molecular mimicry is a plausible mechanism that might, at least in some instances, explain the links between infection and autoimmunity.29 Reports implicate molecular mimicry in the pathogenesis of a number of autoimmune diseases,30 with microbial peptides inducing experimental autoimmune disease due to sequence homology with the autoantigen.31–33 This study evaluated microbial peptides sharing the critical residues of a nephritogenic α3(IV)NC1 epitope in human anti-GBM disease. Evidence of crossreactivity via the detection of circulating IgG antibodies against nine microbial peptides was demonstrated in 20%–60% of patients with anti-GBM autoantibodies and disease. In particular, one microbial peptide from Actinomyces, B7, could induce crescentic GN in both WKY rats and humanized HLA-DR15 transgenic mice. These results indicate that the recognition of peptides from Actinomyces might be one of the etiologic triggers in anti-GBM disease and provide an example of molecular mimicry in the pathogenesis of this disease.
Although T cell crossreaction has been thought to be dependent on sequence similarities between foreign peptides and self-epitopes, structural analyses found that the motifs of peptides binding to MHC molecules were not strictly confined, which might reduce the sequence-specific requirement to only a few critical amino acids.34–36 After defining the critical residues of the nephrogenic T cell epitope of anti-GBM disease,17 it is rational to search for etiologic microbial peptides on the basis of these critical amino acids. In silico epitope prediction has been widely applied in recent years.21–23 Compared with the traditional laboratory assays, bioinformatics methods are efficient and save both time and cost in searching for new antigens, and antigenic peptides.37 In this study, we evaluated >3000 microbial peptides with similar amino acid sequences to α3127–148 via sequence alignment in BLAST, then screened them by human infection association and by B/T cell epitope prediction, and defined 36 potential peptides for subsequent laboratory experiments. These bioinformatics tools helped us to undertake comprehensive retrieval in big databases and then filter out unlikely candidates, leaving plausible sequences for experimental verification. In a previous study,38 we identified seven human-associated microbial peptides using the motif IWxGF using the Uniprot database; however, none of them induced anti-GBM GN in WKY rats. All of these peptides are different from the 36 peptides identified in this study. This may be due to the more sophisticated bioinformatics strategy used in this study. The BLAST database is more powerful for sequence alignment and may provide more candidate peptides with different amino acids for the nonmatched place “x.” In this study, we added a new criterion that candidate peptides were predicted to be both T cell and B cell epitopes, which might have increased the chance of finding pathogenic peptides.
Among the 36 microbial peptides, 9 peptides could be recognized by the circulating antibodies from 20% to 60% of patients with anti-GBM disease, but not the healthy and disease controls. This indicates that some patients with anti-GBM possess antibodies against some microbial peptides that are similar to the disease epitope. However, more evidence is needed regarding previous infections with the respective microbes in patients with anti-GBM and the production of antibodies during such infections. The nephritogenicity of one peptide (B7) was confirmed in both the WKY rat and humanized mouse model of anti-GBM GN. DR15+ mice immunized with B7 developed kidney disease comparable to that found in mice immunized with the nephritogenic Goodpasture T cell epitope α3135–145, consistent with B7 having a high recognition frequency in patients with anti-GBM disease.
Kidney disease in WKY rats immunized with B7 was less severe than disease in rats immunized with α3127–148. In T cell epitopes, all of the amino acids could potentially affect the structure of the peptide and its interaction with the MHC, which together determine the strength of the signal through the T cell receptor (TCR).39–41 Lang et al.41 found that although an Epstein–Barr virus peptide shared the crucial amino acids for TCR recognition with a peptide derived from myelin basic protein (MBP), it could not activate the Hy.2E11 TCR from patients with multiple sclerosis because of amino acid differences in the “noncrucial” residues. In another study conducted on the basis of the structural requirements for both MHC class II binding and TCR recognition of an immune-dominant MBP peptide, several peptides were identified to activate MBP-specific T cell clones from patients with multiple sclerosis. However, only one peptide was identified as a molecular mimic by sequence alignment.39 Therefore, both sequence alignment and structural similarity are important in the development of molecular mimicry.42,43
Healthy individuals have both α3(IV)NC1-specific T cells and natural anti-GBM antibodies in the circulation. The levels of antibodies are too low to induce autoimmune disease and the autoreactive T cells remain inactive.44,45 Carrizosa et al.46 found that priming with crossreactive peptides contributed to the expansion of autoantigen-specific T and B cells to break immune tolerance. In this study, we identified crossreaction between α3127–148 and the microbial peptide B7 at both the B and T cell level in rats. We propose that this crossreactivity may contribute to the initiation of anti-GBM disease.
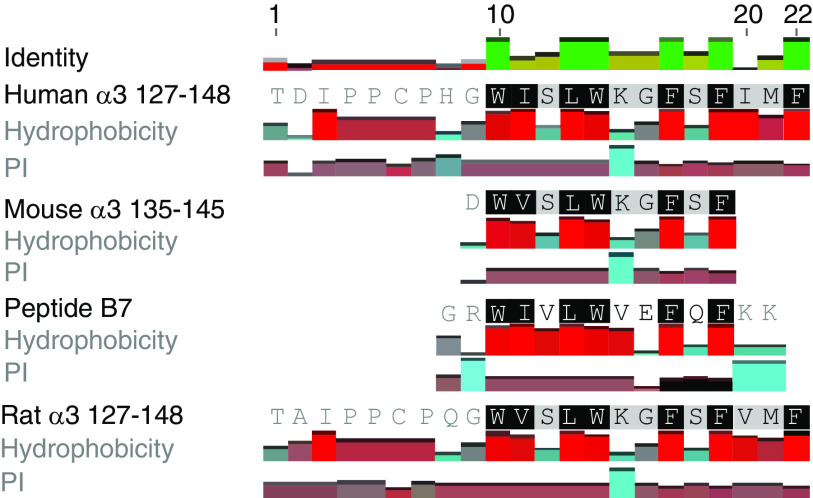
However, no crossreaction of B7 and α3135–145 on T cell responses was shown in DR15+ mice. By amino acid sequence alignment (Figure 6), we found that of the four critical amino acids of mouse α3135–145 (V137, W140, G142, and F143),8 only two residues (W and F) were identical to those of B7. In contrast, six of the seven critical residues of α3127–148 (W136, I137, L139, W140, G142, F143, and F145) were included in B7. That might be one of the reasons for the inability of B7 to crossreact with mouse α3135–145.8,10 There might be other mechanisms involved in the pathogenicity of B7 in DR15+ mice.7 McCoy et al. proposed that virus infection could lead to central nervous system demyelinating diseases including multiple sclerosis, through both molecular mimicry and bystander activation. Viral proteins mimicking self-antigens in the central nervous system can prime genetically susceptible individuals. An subsequent immunologic challenge can initiate the disease through bystander activation mechanisms.47 B7 immunization elicited strong Th1 and Th17 responses in DR15+ mice. This proinflammatory environment might favor the activation of autoreactive T and B cells via bystander effects or by other mechanisms.
Figure 6.
Sequence alignment of B7, mouse α3135–145, and human α3127–148. Two of the four critical amino acids (V137, W140, G142, and F143) of mouse α3135–145 were identical to those of B7. Six of the seven critical residues of α3127–148 (W136, I137, L139, W140, G142, F143, and F145) were identical to those of B7.
B7 is a peptide of the UDP-N-acetylenolpyruvoylglucosamine reductase, derived from Actinomyces. Actinomyces are commensals of the human oropharynx, gastrointestinal tract, and urogenital tract.48 However, when mucosal integrity is breached, they become pathogenic by invading local tissue and organs. Actinomycosis is a chronic granulomatous infectious disease. Whereas there is no report of actinomycosis preceding anti-GBM disease, it is difficult to isolate and culture Actinomyces due to previous antibiotic treatment and overgrowth of other organisms. Furthermore, the diagnosis of actinomycosis depends on visualizing Gram-positive filamentous organisms and sulfur granules on histologic examination,49 which is not commonly performed in routine clinical practice. In this study, immunization by lysate proteins of Actinomyces and by recombinant protein containing B7 both induced T cell responses toward B7. Anti-B7 antibodies could be detected in rats immunized by lysate proteins and in patients recognizing recombinant proteins. These findings indicate that B7 might be presented to and recognized by the immune system during the infection/colocalization of Actinomyces; however, there still exists a gap between the pathogenicity of microorganisms and the immune responses toward microbial peptides.
In conclusion, we found that microbial peptides sharing the critical amino acids of a nephrogenic T cell epitope of anti-GBM disease could be recognized by the sera of patients. Specifically, one microbial peptide derived from Actinomyces could induce crescentic anti-GBM GN in both WKY rats and humanized HLA-DR15 transgenic mice. These results indicate that microbial antigens may initiate anti-GBM disease through molecular mimicry.
Disclosures
All authors have nothing to disclose.
Funding
This work is financially supported by Natural Science Foundation of China grants 81870486 and 81870482. Prof. Kitching is supported by Australian National Health and Medical Research Council Project grant 1008849.
Supplementary Material
Acknowledgments
Dr. Cui, Prof. Kitching, Dr. Ooi, and Prof. Zhao designed the study; Dr. Gu, Dr. Huynh, Dr. Luo, and Ms. Shi carried out experiments; Dr. Gu, Dr. Huynh, and Ms. Shi analyzed the data, completed the figures, and drafted the paper; Dr. Cui, Dr. Jia, Dr. Jiang, Prof. Kitching, Dr. Ooi, and Prof. Zhao revised the paper. The final version of the manuscript was approved by all authors.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2019060619/-/DCSupplemental.
Supplemental Figure 1. The specificity of microbial peptide recognition by patients with anti-GBM.
Supplemental Figure 2. Peptides B10, B11, B12, B13, B24, B25, B30, and B35 could not induce any kidney or pulmonary lesions in WKY rats.
Supplemental Figure 3. Actinomyces lysate protein and recombinant UDP-N-acetylenolpyruvoylglucosamine reductase protein could not induce anti-GBM disease in WKY rats.
References
- 1.Cui Z, Zhao MH: Advances in human antiglomerular basement membrane disease. Nat Rev Nephrol 7: 697–705, 2011. [DOI] [PubMed] [Google Scholar]
- 2.Saus J, Wieslander J, Langeveld JP, Quinones S, Hudson BG: Identification of the Goodpasture antigen as the alpha 3(IV) chain of collagen IV. J Biol Chem 263: 13374–13380, 1988. [PubMed] [Google Scholar]
- 3.Turner N, Mason PJ, Brown R, Fox M, Povey S, Rees A, et al.: Molecular cloning of the human Goodpasture antigen demonstrates it to be the alpha 3 chain of type IV collagen. J Clin Invest 89: 592–601, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedchenko V, Bondar O, Fogo AB, Vanacore R, Voziyan P, Kitching AR, et al.: Molecular architecture of the Goodpasture autoantigen in anti-GBM nephritis. N Engl J Med 363: 343–354, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lerner RA, Glassock RJ, Dixon FJ: The role of anti-glomerular basement membrane antibody in the pathogenesis of human glomerulonephritis. J Exp Med 126: 989–1004, 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J, Hicks J, Borillo J, Glass WF 2nd, Lou YH: CD4(+) T cells specific to a glomerular basement membrane antigen mediate glomerulonephritis. J Clin Invest 109: 517–524, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couser WG, Johnson RJ: The etiology of glomerulonephritis: Roles of infection and autoimmunity. Kidney Int 86: 905–914, 2014. [DOI] [PubMed] [Google Scholar]
- 8.Ooi JD, Chang J, O’Sullivan KM, Pedchenko V, Hudson BG, Vandenbark AA, et al.: The HLA-DRB1*15:01-restricted Goodpasture’s T cell epitope induces GN. J Am Soc Nephrol 24: 419–431, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie LJ, Cui Z, Chen FJ, Pei ZY, Hu SY, Gu QH, et al.: The susceptible HLA class II alleles and their presenting epitope(s) in Goodpasture’s disease. Immunology 151: 395–404, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ooi JD, Petersen J, Tan YH, Huynh M, Willett ZJ, Ramarathinam SH, et al.: Dominant protection from HLA-linked autoimmunity by antigen-specific regulatory T cells. Nature 545: 243–247, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu QH, Xie LJ, Jia XY, Ma R, Liao YH, Cui Z, et al. : Fever and prodromal infections in anti-glomerular basement membrane disease. Nephrology (Carlton) 23: 476–482, 2018 [DOI] [PubMed] [Google Scholar]
- 12.Arends J, Wu J, Borillo J, Troung L, Zhou C, Vigneswaran N, et al.: T cell epitope mimicry in antiglomerular basement membrane disease. J Immunol 176: 1252–1258, 2006. [DOI] [PubMed] [Google Scholar]
- 13.Netzer KO, Leinonen A, Boutaud A, Borza DB, Todd P, Gunwar S, et al.: The goodpasture autoantigen. Mapping the major conformational epitope(s) of alpha3(IV) collagen to residues 17-31 and 127-141 of the NC1 domain. J Biol Chem 274: 11267–11274, 1999. [DOI] [PubMed] [Google Scholar]
- 14.Hu SY, Jia XY, Gu QH, Yu CY, Cheng XY, Jin QZ, et al.: T cell responses to peptides of Goodpasture autoantigen in patients with anti-glomerular basement membrane disease. Nephrology (Carlton) 23: 345–350, 2017 [DOI] [PubMed] [Google Scholar]
- 15.Cairns LS, Phelps RG, Bowie L, Hall AM, Saweirs WW, Rees AJ, et al.: The fine specificity and cytokine profile of T-helper cells responsive to the alpha3 chain of type IV collagen in Goodpasture’s disease. J Am Soc Nephrol 14: 2801–2812, 2003. [DOI] [PubMed] [Google Scholar]
- 16.Jia XY, Cui Z, Yang R, Hu SY, Zhao MH: Antibodies against linear epitopes on the Goodpasture autoantigen and kidney injury. Clin J Am Soc Nephrol 7: 926–933, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu SY, Gu QH, Wang J, Wang M, Jia XY, Cui Z, et al.: The pathogenicity of T cell epitopes on human Goodpasture antigen and its critical amino acid motif. J Cell Mol Med 21: 2117–2128, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia XY, Cui Z, Li JN, Hu SY, Zhao MH: Identification of critical residues of linear B cell epitope on Goodpasture autoantigen. PLoS One 10: e0123277, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanović S: SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics 50: 213–219, 1999. [DOI] [PubMed] [Google Scholar]
- 20.Saha S, Raghava GP: Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins 65: 40–48, 2006. [DOI] [PubMed] [Google Scholar]
- 21.Oyarzún P, Ellis JJ, Bodén M, Kobe B: PREDIVAC: CD4+ T-cell epitope prediction for vaccine design that covers 95% of HLA class II DR protein diversity. BMC Bioinformatics 14: 52, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soria-Guerra RE, Nieto-Gomez R, Govea-Alonso DO, Rosales-Mendoza S: An overview of bioinformatics tools for epitope prediction: Implications on vaccine development. J Biomed Inform 53: 405–414, 2015. [DOI] [PubMed] [Google Scholar]
- 23.Lin HH, Ray S, Tongchusak S, Reinherz EL, Brusic V: Evaluation of MHC class I peptide binding prediction servers: Applications for vaccine research. BMC Immunol 9: 8, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang R, Cui Z, Zhao J, Zhao MH: The role of HLA-DRB1 alleles on susceptibility of Chinese patients with anti-GBM disease. Clin Immunol 133: 245–250, 2009. [DOI] [PubMed] [Google Scholar]
- 25.Duyar H, Dengjel J, de Graaf KL, Wiesmüller KH, Stevanović S, Weissert R: Peptide motif for the rat MHC class II molecule RT1.Da: Similarities to the multiple sclerosis-associated HLA-DRB1*1501 molecule. Immunogenetics 57: 69–76, 2005. [DOI] [PubMed] [Google Scholar]
- 26.Saxena R, Bygren P, Butkowski R, Wieslander J: Specificity of kidney-bound antibodies in Goodpasture’s syndrome. Clin Exp Immunol 78: 31–36, 1989. [PMC free article] [PubMed] [Google Scholar]
- 27.Apostolopoulos J, Ooi JD, Odobasic D, Holdsworth SR, Kitching AR: The isolation and purification of biologically active recombinant and native autoantigens for the study of autoimmune disease. J Immunol Methods 308: 167–178, 2006. [DOI] [PubMed] [Google Scholar]
- 28.Ooi JD, Phoon RK, Holdsworth SR, Kitching AR: IL-23, not IL-12, directs autoimmunity to the Goodpasture antigen. J Am Soc Nephrol 20: 980–989, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steelman AJ: Infection as an environmental trigger of multiple sclerosis disease exacerbation. Front Immunol 6: 520, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cusick MF, Libbey JE, Fujinami RS: Molecular mimicry as a mechanism of autoimmune disease. Clin Rev Allergy Immunol 42: 102–111, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bachmaier K, Neu N, de la Maza LM, Pal S, Hessel A, Penninger JM: Chlamydia infections and heart disease linked through antigenic mimicry. Science 283: 1335–1339, 1999. [DOI] [PubMed] [Google Scholar]
- 32.Olson JK, Croxford JL, Calenoff MA, Dal Canto MC, Miller SD: A virus-induced molecular mimicry model of multiple sclerosis. J Clin Invest 108: 311–318, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kain R, Exner M, Brandes R, Ziebermayr R, Cunningham D, Alderson CA, et al.: Molecular mimicry in pauci-immune focal necrotizing glomerulonephritis. Nat Med 14: 1088–1096, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wucherpfennig KW, Hafler DA, Strominger JL: Structure of human T-cell receptors specific for an immunodominant myelin basic protein peptide: Positioning of T-cell receptors on HLA-DR2/peptide complexes. Proc Natl Acad Sci U S A 92: 8896–8900, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wucherpfennig KW, Sette A, Southwood S, Oseroff C, Matsui M, Strominger JL, et al.: Structural requirements for binding of an immunodominant myelin basic protein peptide to DR2 isotypes and for its recognition by human T cell clones. J Exp Med 179: 279–290, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinigaglia F, Hammer J: Defining rules for the peptide-MHC class II interaction. Curr Opin Immunol 6: 52–56, 1994. [DOI] [PubMed] [Google Scholar]
- 37.Patronov A, Doytchinova I: T-cell epitope vaccine design by immunoinformatics. Open Biol 3: 120139, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li JN, Jia X, Wang Y, Xie C, Jiang T, Cui Z, et al.: Plasma from patients with anti-glomerular basement membrane disease could recognize microbial peptides. PLoS One 12: e0174553, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wucherpfennig KW, Strominger JL: Molecular mimicry in T cell-mediated autoimmunity: Viral peptides activate human T cell clones specific for myelin basic protein. Cell 80: 695–705, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wucherpfennig KW: Structural basis of molecular mimicry. J Autoimmun 16: 293–302, 2001. [DOI] [PubMed] [Google Scholar]
- 41.Lang HL, Jacobsen H, Ikemizu S, Andersson C, Harlos K, Madsen L, et al.: A functional and structural basis for TCR cross-reactivity in multiple sclerosis. Nat Immunol 3: 940–943, 2002. [DOI] [PubMed] [Google Scholar]
- 42.Quaratino S, Thorpe CJ, Travers PJ, Londei M: Similar antigenic surfaces, rather than sequence homology, dictate T-cell epitope molecular mimicry. Proc Natl Acad Sci U S A 92: 10398–10402, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brock R, Wiesmüller KH, Jung G, Walden P: Molecular basis for the recognition of two structurally different major histocompatibility complex/peptide complexes by a single T-cell receptor. Proc Natl Acad Sci U S A 93: 13108–13113, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui Z, Wang HY, Zhao MH: Natural autoantibodies against glomerular basement membrane exist in normal human sera. Kidney Int 69: 894–899, 2006. [DOI] [PubMed] [Google Scholar]
- 45.Zou J, Hannier S, Cairns LS, Barker RN, Rees AJ, Turner AN, et al.: Healthy individuals have Goodpasture autoantigen-reactive T cells. J Am Soc Nephrol 19: 396–404, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carrizosa AM, Nicholson LB, Farzan M, Southwood S, Sette A, Sobel RA, et al.: Expansion by self antigen is necessary for the induction of experimental autoimmune encephalomyelitis by T cells primed with a cross-reactive environmental antigen. J Immunol 161: 3307–3314, 1998. [PubMed] [Google Scholar]
- 47.McCoy L, Tsunoda I, Fujinami RS: Multiple sclerosis and virus induced immune responses: Autoimmunity can be primed by molecular mimicry and augmented by bystander activation. Autoimmunity 39: 9–19, 2006. [DOI] [PubMed] [Google Scholar]
- 48.Könönen E, Wade WG: Actinomyces and related organisms in human infections. Clin Microbiol Rev 28: 419–442, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong VK, Turmezei TD, Weston VC: Actinomycosis. BMJ 343: d6099, 2011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.