Significance Statement
Inverted formin 2 (INF2) is the key regulator of a stress response—calcium-mediated actin reset, or CaAR—that reorganizes the actin cytoskeleton of mammalian cells in response to calcium influx. INF2 has been linked to the podocytic kidney disease focal segemental glomerulosclerosis (FSGS) and to cases of the neurologic disorder Charcot–Marie–Tooth disease that are accompanied by FSGS. The authors show that >50 disease-associated INF2 variants lead to deregulation of CaAR in cell lines, in Drosophila nephrocytes, and in cells from patient cells with these disorders. Their findings suggest that CaAR can be used as a sensitive assay for INF2 function and for robust evaluation of disease-linked variants of this formin. This work also highlights the use of quantitative cellular assays in assessing effects of disease-associated mutations to better understand complex disease phenotypes.
Keywords: actin, INF2, CaAR, focal segmental glomerulosclerosis, imaging
Abstract
Background
Monogenic diseases provide favorable opportunities to elucidate the molecular mechanisms of disease progression and improve medical diagnostics. However, the complex interplay between genetic and environmental factors in disease etiologies makes it difficult to discern the mechanistic links between different alleles of a single locus and their associated pathophysiologies. Inverted formin 2 (INF2), an actin regulator, mediates a stress response—calcium mediated actin reset, or CaAR—that reorganizes the actin cytoskeleton of mammalian cells in response to calcium influx. It has been linked to the podocytic kidney disease focal segemental glomerulosclerosis (FSGS), as well as to cases of the neurologic disorder Charcot–Marie–Tooth disease that are accompanied by nephropathy, mostly FSGS.
Methods
We used a combination of quantitative live cell imaging and validation in primary patient cells and Drosophila nephrocytes to systematically characterize a large panel of >50 autosomal dominant INF2 mutants that have been reported to cause either FSGS alone or with Charcot–Marie–Tooth disease.
Results
We found that INF2 mutations lead to deregulated activation of formin and a constitutive stress response in cultured cells, primary patient cells, and Drosophila nephrocytes. We were able to clearly distinguish between INF2 mutations that were linked exclusively to FSGS from those that caused a combination of FSGS and Charcot–Marie–Tooth disease. Furthermore, we were able to identify distinct subsets of INF2 variants that exhibit varying degrees of activation.
Conclusions
Our results suggest that CaAR can be used as a sensitive assay for INF2 function and for robust evaluation of diseased-linked variants of formin. More broadly, these findings indicate that cellular profiling of disease-associated mutations has potential to contribute substantially to sequence-based phenotype predictions.
The increasing availability of human genotypes and sequence variation data can yield insights into the genetic basis of many pathologic conditions. Monogenic diseases offer a particularly amenable basis for the elucidation of the mechanistic and physiologic drivers of disease progression. Charcot–Marie–Tooth disease (CMT) is a hereditary motor and sensory neuropathy characterized by progressive loss of muscle tissue and touch sensation. FSGS exhibits several functionally ill-defined pathologic phenotypes of glomeruli, the filtration units of the kidney. FSGS is one of the most common types of adult glomerular injury1 and has been linked to a combination of genetic and environmental factors.2 Several autosomal dominant genetic lesions have been linked to FSGS, most of which affect proteins expressed in the podocytes, a specialized renal cell type that forms the epithelial layer of the glomerular filtration barrier. These include genetically determined aberrations in the calcium channel TRPC6, as well as defects in proteins linking the slit diaphragm to the actin cytoskeleton, such as α-actinin (ACTN4) and inverted formin 2 (INF2).3 How exactly actin influences podocyte function is not clear.
We have previously shown that INF2 globally reorganizes the actin cytoskeleton of mammalian cells in response to calcium influx (calcium-mediated actin reset; CaAR).4 CaAR acts as a general stress response that influences diverse physiologic processes including transcriptional programs, wound repair, and cellular morphogenesis. To date, >50 individual mutations in INF2 have been linked to either FSGS alone or to a combination of FSGS and Charcot–Marie–Tooth disease (FSGS/CMT) (Table 1).
Table 1.
Summary of INF2 variants included in this study
| INF2 Variant/AA Position | Disease: No of Mutations/Affected | Reference for Individual Variance | PolyPhen: Score; Sensitivity; Specificity | CADD Score | CADD Class | Cellular Phenotype | Actin Reorg/Value | Filopodia | CaAR Reacting | CaAR max(R) | Difference Max(R) to wt | Difference Max(R) to ΔDID | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ± | Mean±SD; n | (±) | (% Cells with Max(R)>14.9) | (Mean ±SD; n) | Sign. | P Value | Sign. | P Value | |||||||
| INF2 | Control | wt | − | 1.03±0.07; 24 | − | 98 | 29±09; 46 | 1 | 7.44×10−69 | ||||||
| ΔDID | Control | Strong | + | 1.21±0.11; 22 | + | 0 | 08±02; 43 | 1 | 7.44×10−69 | ||||||
| A13T | FSGS (1/2)/natural | 5 | 0.999; 0.14; 0.99 | 28.0 | Disease | wt | − | 1.02±0.08; 25 | − | 100 | 28±11; 29 | 0 | 0.419 | 1 | 2.98×10−50 |
| K15R | Natural | 0.711; 0.86; 0.92 | wt | − | ND | − | 97 | 29±13; 29 | 0 | 0.006 | 1 | 2.88×10−56 | |||
| A33T | Natural | 1.000; 0.00; 1.00 | Impaired | − | 1.03±0.09; 29 | − | 82 | 20±07; 34 | 1 | 6.61×10−13 | 1 | 3.47×10−22 | |||
| L42P | FSGS (3/3) | 5 | 1.000; 0.00; 1.00 | 24.8 | Disease | Strong | + | 1.17±0.09; 17 | + | 26 | 12±05; 27 | 1 | 1.36×10−36 | 0 | 0.002 |
| L53P | FSGS/CMT (1/1) | 1.000; 0.00; 1.00 | Strong | + | ND | + | 0 | 07±01; 20 | 1 | 1.07×10−46 | 0 | 0.8 | |||
| L57P | FSGS/CMT (1/1) | 6 | 1.000; 0.00; 1.00 | 26.6 | Disease | Strong | + | 1.14±0.06; 21 | + | 0 | 08±02; 20 | 1 | 1.81×10−45 | 0 | 0.97 |
| L57R | FSGS/CMT (1/1) | 7 | 1.000; 0.00; 1.00 | 27.5 | Disease | Strong | + | ND | + | 0 | 08±02; 17 | 1 | 2.08×10−39 | 0 | 0.81 |
| F68S | FSGS/CMT (1/1) | 7 | 1.000; 0.00; 1.00 | 27.1 | Disease | Strong | + | 1.15±0.08; 24 | + | 4 | 09±06; 26 | 1 | 4.27×10−48 | 0 | 0.41 |
| L69P | FSGS/CMT (2/2) | 8 | 1.000; 0.00; 1.00 | 26.9 | Disease | Strong | + | ND | + | 0 | 08±02; 19 | 1 | 5.48×10−43 | 0 | 0.89 |
| Δ69–72 | FSGS/CMT (1/1) | 9 | Strong | + | ND | + | 0 | 09±02; 24 | 1 | 2.34×10−45 | 0 | 0.37 | |||
| G73S | FSGS (4/4) | 10 | 1.000; 0.00; 1.00 | 32.0 | Disease | Strong | + | ND | + | 0 | 08±02; 27 | 1 | 3.57×10−52 | 0 | 0.75 |
| L76P | FSGS (5/5) | 6 | 1.000; 0.00; 1.00 | 26.5 | Disease | Strong | + | 1.21±0.09; 21 | − | 14 | 10±04; 36 | 1 | 5.97×10−53 | 0 | 0.11 |
| L77P | FSGS/CMT (2/2) | 11 | 1.000; 0.00; 1.00 | 26.2 | Disease | Strong | + | 1.17±0.08; 22 | + | 0 | 09±01; 20 | 1 | 3.62×10−41 | 0 | 0.47 |
| L77R | FSGS/CMT (2/2) | 9 | 1.000; 0.00; 1.00 | 27.0 | Disease | Strong | + | ND | + | 0 | 10±02; 19 | 1 | 7.35×10−38 | 0 | 0.28 |
| L81P | FSGS (6/6) | 10 | 1.000; 0.00; 1.00 | 26.0 | Disease | Strong | + | ND | − | 9 | 09±04; 47 | 1 | 1.21×10−63 | 0 | 0.24 |
| C104F | FSGS/CMT (1/1) | 12 | 1.000; 0.00; 1.00 | 26.4 | Disease | Strong | + | ND | + | 6 | 09±03; 52 | 1 | 3.23×10−67 | 0 | 0.27 |
| C104R | FSGS/CMT (1/1) | 12 | 1.000; 0.00; 1.00 | 24.6 | Disease | Strong | + | 1.26±0.12; 22 | + | 3 | 08±04; 34 | 1 | 3.82×10−59 | 0 | 0.73 |
| C104W | FSGS/CMT (3/3) | 12 | 1.000; 0.00; 1.00 | 26.3 | Disease | Strong | + | ND | + | 0 | 08±02; 26 | 1 | 1.00×10−53 | 0 | 0.91 |
| V105G | FSGS/CMT (1/1) | 13 | 1.000; 0.00; 1.00 | 25.3 | Disease | Strong | + | ND | + | 0 | 08±03; 30 | 1 | 1.70×10−55 | 0 | 0.76 |
| R106P | FSGS/CMT (1/1) | 12 | 1.000; 0.00; 1.00 | 28.5 | Disease | Strong | + | ND | + | 0 | 08±02; 26 | 1 | 4.15×10−54 | 0 | 0.86 |
| V108D | FSGS/CMT (1/1) | 8 | 1.000; 0.00; 1.00 | 25.9 | Disease | Strong | + | ND | + | 5 | 08±03; 22 | 1 | 3.00×10−47 | 0 | 0.9 |
| G114D | FSGS/CMT (2/2) | 9,14 | 1.000; 0.00; 1.00 | 27.,3 | Disease | Strong | + | ND | + | 0 | 08±02; 28 | 1 | 2.93×10−54 | 0 | 0.87 |
| L128P | FSGS/CMT (2/2) | 12 | 1.000; 0.00; 1.00 | 26.7 | Disease | Strong | + | ND | + | 0 | 08±03; 25 | 1 | 1.23×10−51 | 0 | 0.98 |
| S129P | FSGS (1/1) | 10 | 1.000; 0.00; 1.00 | 23.6 | Disease | Strong | + | ND | + | 4 | 09±03; 23 | 1 | 6.04×10−44 | 0 | 0.36 |
| L132P | FSGS/CMT (2/2) | 15 | 1.000; 0.00; 1.00 | 26.6 | Disease | Strong | + | 1.16±0.10; 26 | + | 10 | 10±07; 21 | 1 | 4.57×10−39 | 0 | 0.18 |
| L132R | FSGS/CMT (3/3) | 12 | 1.000; 0.00; 1.00 | 27.5 | Disease | Strong | + | 1.12±0.06; 20 | + | 7 | 10±04; 27 | 1 | 2.54×10−46 | 0 | 0.19 |
| C151R | FSGS (5/5) | 10,14 | 0.999;0.14; 0.99 | 23.8 | Disease | Strong | + | 1.16±0.08; 19 | + | 4 | 08±03; 26 | 1 | 4.74×10−53 | 0 | >0.99 |
| H158D | FSGS (9/9) | 10 | 0.999;0.14; 0.99 | 25.8 | Disease | Strong | + | 1.16±0.08; 31 | + | 20 | 11±04; 41 | 1 | 7.22×10−49 | 0 | 0.004 |
| H158N | Natural | 0.997; 0.41; 0.98 | Impaired | − | ND | − | 79 | 21±08; 42 | 1 | 1.67×10−11 | 1 | 1.76×10−28 | |||
| L162P | FSGS (6/6) | 16 | 1.000; 0.00; 1.00 | 25.9 | Disease | Strong | + | 1.16±0.08; 24 | + | 9 | 09±04; 22 | 1 | 8.00×10−42 | 0 | 0.28 |
| L162R | FSGS (3/3) | 13 | 1.000; 0.00; 1.00 | 26.6 | Disease | Strong | + | ND | − | 27 | 13±06; 33 | 1 | 1.13×10−38 | 0 | 2.46×10−4 |
| A164P | FSGS (7/7) | 17 | 1.000; 0.00; 1.00 | 28.8 | Disease | Strong | + | ND | + | 0 | 08±02; 39 | 1 | 1.22×10−63 | 0 | 0.75 |
| L165P | FSGS/CMT (1+1/2) | 12 | 1.000; 0.00; 1.00 | 27.2 | Disease | Strong | + | ND | + | 4 | 09±02; 25 | 1 | 2.15×10−48 | 0 | 0.57 |
| Δ168 | FSGS (3/3) | 13 | Intermediate | + | ND | − | 41 | 15±07; 44 | 1 | 2.19×10−34 | 1 | 5.64×10−9 | |||
| R177C | FSGS (4/4) | 10,18 | 1.000; 0.00; 1.00 | 34.0 | Disease | Strong | + | ND | − | 16 | 13±07; 25 | 1 | 2.88×10−31 | 0 | 1.81×10−4 |
| R177H | FSGS (9/11) | 6,14,19,20 | 1.000; 0.00; 1.00 | 31.0 | Disease | Strong | + | ND | + | 11 | 11±04; 44 | 1 | 6.93×10−54 | 0 | 0.02 |
| V181G | FSGS/CMT (3+1/4) | 10,21 | 0.998; 0.27; 0.99 | 24.9 | Disease | Strong | + | 1.19±0.07; 20 | + | 0 | 09±02; 25 | 1 | 1.24×10−48 | 0 | 0.6 |
| E184K | FSGS/CMT (12+3/15) | 5,7,10,13 | 1.000; 0.00; 1.00 | 28.5 | Disease | Strong | + | 1.18±0.09; 20 | + | 0 | 07±02; 23 | 1 | 7.22×10−51 | 0 | 0.79 |
| E184Q | FSGS (8/8) | 20 | 1.000; 0.00; 1.00 | 25.1 | Disease | Intermediate | + | 1.10±0.09; 20 | − | 87 | 21±07; 30 | 1 | 4.17×10−11 | 1 | 3.89×10−22 |
| S186P | FSGS (18/28) | 5,10 | 0.998; 0.73; 0.96 | 18.5 | Polymorphism | Intermediate | + | 1.15±0.12; 20 | − | 75 | 17±07; 28 | 1 | 1.17×10−20 | 1 | 2.65×10−11 |
| Y193H | FSGS (1/1) | 6,12 | 1.000; 0.00; 1.00 | 24.1 | Disease | Intermediate | + | 1.17±0.11; 21 | − | 66 | 18±06; 44 | 1 | 2.58×10−23 | 1 | 3.87×10−16 |
| V194M | Natural | 0.708; 0.86; 0.92 | 0.9 | Polymorphism | wt | − | ND | − | 96 | 26±07; 27 | 0 | 0.026 | 1 | 2.41×10−40 | |
| L198R | FSGS (5/5) | 5,6,10 | 1.000; 0.00; 1.00 | 25.3 | Disease | Strong | + | 1.18±0.11; 31 | + | 4 | 10±02; 26 | 1 | 2.39×10−42 | 0 | 0.07 |
| N202D | FSGS (2/2) | 20 | 1.000; 0.00; 1.00 | 24.9 | Disease | Strong | + | ND | + | 5 | 09±03; 21 | 1 | 3.19×10−43 | 0 | 0.55 |
| N202S | FSGS (5/5) | 22 | 1.000; 0.00; 1.00 | 23.,8 | Disease | Intermediate | + | 1.19±0.08; 35 | − | 49 | 15±05; 35 | 1 | 5.25×10−31 | 1 | 5.47×10−8 |
| A203D | FSGS (2/2) | 20 | 1.000; 0.00; 1.00 | 27.0 | Disease | Strong | + | ND | + | 10 | 09±04; 39 | 1 | 6.07×10−61 | 0 | 0.47 |
| R212C | Natural | 1.000; 0.00; 1.00 | 24.7 | Disease | wt | − | ND | − | 88 | 27±11; 26 | 0 | 0.120 | 1 | 5.70×10−43 | |
| R212H | Natural | 0.867; 0.83; 0.93 | 21.6 | Polymorphism | wt | − | ND | − | 92 | 24±10; 25 | 0 | 1.14×10−4 | 1 | 2.95×10−30 | |
| R214C | FSGS (10/13) | 6,10,13,20 | 1.000; 0.00; 1.00 | 28.4 | Disease | Strong | + | 1.13±0.08; 23 | − | 20 | 11±06; 35 | 1 | 8.00×10−45 | 0 | 0.004 |
| R214H | FSGS (20/26) | 5,10,20 | 1.000; 0.00; 1.00 | 27.5 | Disease | Strong | + | 1.18±0.08; 21 | − | 9 | 10±04; 46 | 1 | 2.42×10−58 | 0 | 0.06 |
| R218Q | FSGS (27/36) | 5,6,10,13,20,23 | 1.000; 0.00; 1.00 | 29.5 | Disease | Strong | + | 1.19±0.08; 27 | + | 0 | 09±02; 27 | 1 | 6.20×10−51 | 0 | 0.6 |
| R218W | FSGS (11/11) | 5,10,13 | 1.000; 0.00; 1.00 | 30.0 | Disease | Strong | + | 1.24±0.13; 31 | + | 0 | 08±02; 33 | 1 | 1.23×10−58 | 0 | 0.78 |
| E220K | FSGS (14/14) | 5,6,10,13,24 | 1.000; 0.00; 1.00 | 27.5 | Disease | Strong | + | 1.18±0.08; 26 | − | 33 | 13±05; 27 | 1 | 1.43×10−31 | 0 | 4.84×10−5 |
| L245P | FSGS (3/3) | 25 | 0.998; 0.27; 0.99 | 27.9 | Disease | Strong | + | 1.24±0.13; 24 | + | 0 | 08±02; 24 | 1 | 6.74×10−52 | 0 | 0.83 |
| R261G | Natural | 0.9998; 0.27; 0.99 | 23.6 | Polymorphism | wt | − | ND | − | 89 | 25±09; 27 | 0 | 0.005 | 1 | 2.97×10−37 | |
| R261Q | Natural | 0.929; 0.81; 0.94 | 16.7 | Polymorphism | Impaired | − | ND | − | 58 | 18±07; 24 | 1 | 4.27×10−16 | 1 | 1.20×10−12 | |
| P350L | Natural | 0.985; 0.74; 0.96 | 23.2 | Polymorphism | Impaired | − | ND | − | 87 | 22±07; 23 | 1 | 3.43×10−7 | 1 | 6.02×10−23 | |
| I685V | FSGS (1/1)/var. | 26 | 0.058; 0.94; 0.84 | ND | ND | wt | − | 1.06±0.06; 23 | − | 91 | 25±08; 22 | 0 | 0.006 | 1 | 2.02×10−32 |
| R877Q | FSGS (1/1)/var. | 26 | 0.113; 0.93; 0.86 | ND | ND | Impaired | − | 1.06±0.05; 23 | − | 96 | 23±06; 25 | 1 | 6.44×10−6 | 1 | 2.91×10−27 |
CADD, combined annotation dependent depletion; Reorg, reorganization; CaAR reacting, percentage of cells with Max(R) value >14.9, corresponding to mean+2 SD (ΔDID); Sign., significantly different at significance level 0.01 (1: yes, 0: no; P values given for one-way ANOVA); −, no; +, yes; ND, not determined.
Nearly all mutations identified so far map to the N-terminal diaphanous inhibitory domain (DID) of INF2, a region that interacts with the diaphanous autoregulatory domain located in the C-terminal half of the protein, and might thereby contribute to autoinhibition of the formin.27 Release of autoinhibition would also explain the dominant character of INF2 mutations. Importantly, actin organization in the podocytes of FSGS glomeruli28,29 has been shown to be aberrant, and leads to the effacement of foot processes.5
Previous studies have identified a range of interaction partners, post-translational modifications, and potential biologic functions of INF2.5,6,30,31 However, although these reports suggest many intriguing links between INF2 mutations and podocyte pathologies, no systematic analysis of the phenotypic spectrum of known mutations has been performed. Here, we characterize a large panel (>50) of autosomal dominant INF2 mutants that have been reported to cause either FSGS alone or FSGS/CMT (Table 1). Our results indicate that cellular profiling of disease-associated mutations can substantially contribute to sequence-based phenotype predictions.
Methods
Isolation of Primary Podocytes
Primary mouse podocytes were isolated from a double-fluorescent-reporter mouse line,32,33 in which podocytes are specifically labeled by podocin-Cre-driven expression of GFP and all other glomerular cells express tdTomato. This mouse line facilitates the separation of podocytes from other glomerular cell types. After cervical dislocation of donor mice, kidneys were removed and washed in HBSS containing calcium chloride and magnesium sulfate (Merck). Kidneys were then cut into small pieces and further dissociated by incubation for 30 minutes at 37°C in HBSS with 1 mg/ml collagenase (type 2255 U/mg; Worthington). To isolate glomeruli, the suspension was then passed consecutively through 100-µm, 70-µm, and 40-µm nylon filters (Falcon Cell Strainer; Corning Glass). The final glomerular fraction was eluted from the filter in 20 ml HBSS, centrifuged at 2000×g for 10 minutes, and resuspended in 25 ml RPMI-1640 medium (Merck) supplemented with 10% FCS (FBS; Gibco), 1% penicillin/streptomycin (Merck), 1% nonessential amino acids (Gibco), and 1% sodium pyruvate (HyClone). Glomeruli were then cultured for 7 days at 37°C in the presence of 5% CO2 in Petri dishes (145/20 mm, Cell Star; Greiner Bio-One) coated with collagen type 1 (Merck). During this time, GFP-positive (podocytes) and Tomato-positive cells grew out of the glomeruli. To sort fluorescent populations, cells were washed with PBS−2 (Merck), trypsinized, and passed through a 40-µm nylon filter (20 ml). The flow-through was centrifuged for 5 minutes at 1000×g and the pellet was resuspended in 2 ml medium. GFP-expressing cells were sorted by FACS and subsequently cultivated for 3 days before use in experiments.
Isolation of Urine-Derived Epithelial Cells
To isolate urine-derived epithelial cells, total urine was collected, aliquoted into 50-ml tubes and centrifuged at 400×g for 10 minutes. Pellets were washed once in 20 ml PBS containing 1% penicillin/streptomycin, and resuspended in DMEM/F12 (Gibco) supplemented with 10% FCS, 1% penicillin/streptomycin, 1% nonessential amino acids, 1 mM GlutaMAX (Gibco), 0.1 mM 2-mercaptoethanol (Gibco), and mixed with Renal Epithelial Growth Medium supplements (CC-4127 REGM; Lonza). Cells were seeded into 12-well plates coated with 0.1% gelatin and cultivated for 4 days at 37°C under 5% CO2. When confluent (60%–80%) colonies became visible, cells were trypsinized for 2 minutes, centrifuged for 5 minutes at 400×g, resuspended in fresh medium, and transferred to a fresh six-well plate coated with 0.1% gelatin. Urine-derived cells were cultivated for up to five passages.
Cell Culture and Transfection
HeLa cells were grown at 37°C and 5% CO2 in DMEM (DMEM-GlutaMAX-I; Gibco) supplemented with 10% FBS (FBS; Gibco). Routinely, 2×104 cells/ml were seeded into four- or eight-well glass-bottomed dishes (Sarstedt) and incubated for 24 hours. On the next day, cells were transiently transfected with INF2 constructs using Fugene6 or lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions, and incubated for another 24 hours. Live-cell imaging was performed with cells in HBSS supplemented with 10 mM HEPES (pH 7.4). The following cell lines were used in this study: HeLa (ECACC 93021013), HeLa cells stably expressing Lifeact-mCherry,4 and HeLa INF2 knockout (KO) cells stably expressing Lifeact-mCherry.4 The identities of all cell lines were checked by visual inspection of morphologies, and all tested negative for mycoplasma.
DNA Constructs and Molecular Biology
Full-length GFP-INF2 constructs were derived from the previously described pGFP-INF2-CAAX vectors that contain siRNA-insensitive coding sequences.4 The EGFP coding sequence in pGFP-INF2-CAAX was replaced by that of mCherry after digestion with NheI and BglII to obtain pRFP-INF2-CAAX. GFP-DID constructs contain INF2 fragments coding for amino acids 1–420 cloned in the pEGFP-C1 backbone (Invitrogen). All INF2 mutants were generated by site-directed mutagenesis (Stratagene Quickchange; Agilent) using either pGFP-INF2-CAAX or pEGFP-INF2DID as template. For yeast two-hybrid assays, pDEST22 was used as the backbone for prey constructs and pDEST32 for bait. The yeast strain PJ69–4A was cotransformed with both plasmids, selected on SC-Leu-Trp plates and plated onto SC-Leu-Trp-His plates to detect putative interactions. Variants of the INF2 DID domain (aa 1–420, see above) were tested with INF2 FFW (formin homology 1, formin homology 2 [FH2], WH2; aa 421–1008). Quantitative LacZ analysis was carried out in liquid culture using the o-nitrophenyl-β-D-galactoside as described previously.34
Fluorescence Microscopy
Epifluorescence imaging was performed on a fully automated iMIC-based microscope (FEI/Till Photonics), equipped with an Olympus ×100 1.4 NA objective and solid state lasers at 488 nm (Cobolt Calypso, 75 mW) and 561 nm (Cobolt Jive, 150 mW) as light sources. Lasers were selected through an AOTF and directed through a broadband fiber to the microscope. A galvanometer-driven two-axis scan-head was used to adjust laser incidence angles. Images were collected using an Imago-QE Sensicam camera. Acquisition was controlled by LiveAcquisition software (Till Photonics). Ablation experiments were carried out on an iMIC set-up equipped with a pulsed 355 nm picosecond ultraviolet laser (Sepia, PicoQuant; Rapp) as previously described.4 Confocal microscopy was performed on an iMIC42 set-up equipped with a spinning disk unit (Andromeda) using a ×60 oil immersion (1.49 NA) objective. Images were recorded with an EMCCD camera (Andor iXon Ultra 897).
Immunofluorescence and Cell Labeling
Primary mouse podocytes and urine-derived epithelial cells were grown on glass coverslips, fixed with 4% paraformaldehyde in PBS for 20 minutes, washed with PBS and permeabilized with 0.1% Triton X-100 for 10 minutes before to incubation with Alexa Fluor 350 phalloidin (Thermo Fisher) or DAPI for 1 h in PBS. The coverslips were washed and subsequently mounted on slides in Mowiol/Dabco (Roth). Lysosomes in HeLa cells were labeled with LysoTracker Red (Thermo Fisher) according to the manufacturer’s directions.
Image Analysis
Images were processed with Fiji before data analysis. Linear contrast adjustment and zoom were used for purposes of presentation in the figures only. To measure initial actin reorganization, we acquired z-stacks of cells. For HeLa cell experiments, polygon sections were used as regions of interest (ROIs) to define the area around the nucleus and at the basal plane of the cell to measure actin intensities around the nucleus and at the cell cortex, respectively. For nephrocytes, ROIs were selected at the cell outlines for actin intensity measurements at the cell cortex. The initial actin reorganization (a0) was then calculated as follows: a0=(a [nucleus]+background)/(a [cell cortex]−background).
For analysis of actin reorganization after calcium stimulation, we acquired image series at 1 frame per second. Image series were bleach corrected and the maximum intensity projection of eight frames before stimulation subtracted from all images (reference). We then identified the time point of maximal difference in the series and thresholded this image according to Otsu to generate a binary mask. Max(R) was calculated as relative intensity change (with respect to reference) within the mask at time point of maximal difference. To measure lysosome mobility image series of 60 frames (1 frame per second) were background corrected (rolling ball, five-pixel diameter), filtered (median, 2), and thresholded to obtain binary images. Mobility was then calculated as total area covered by lysosomes (diff=max [t0–60]−t0) relative to the area covered at the start of acquisition: lateral displacement=diff/t0. To quantify colocalization between GFP-DID and Lifeact-RFP images were processed using the Coloc2 plugin of Fiji. Cells of interest were selected by manual ROIs and Pearson correlation coefficients calculated for each time point in a time series. To analyze the density of Sns distribution in fly nephrocytes immunofluorescence images were thresholded and Sns surface coverage as percentage of total area (using manually selected ROIs) was determined for each cell after background subtraction (rolling ball, 20).
Statistical Analyses
Means and SDs are given throughout for quantifications. To test for statistical significance of differences in CaAR max(R) values, one-way ANOVA was performed with the Holm–Bonferroni post hoc correction.
Characterization of INF2 Mutant Phenotypes in Drosophila Nephrocytes
Fly strains were cultured on standard cornmeal agar food and maintained at 25°C. UASt::Myc-INF2 transgenes were established using the Phi-C31 Integrase system,35 with attP40 as landing site. For overexpression of Myc-hINF2, virgin females of the nephrocyte-specific driver line sns::GAL436 were crossed with UASt::Myc-hINF2 males. Garland nephrocytes were isolated from wandering third-instar larvae by dissection, heat fixed, and stained as described previously,37 using a chicken anti-Sns antibody (1:1000). For phalloidin staining of actin, garland nephrocytes were dissected in PBS (pH 7) as described above, and fixed in PBS containing 4% paraformaldehyde for 10 minutes. After washing twice with PBS for 3 minutes each, nephrocytes were permeabilized in PBS with Triton X-100 (0.3% Triton X-100) for 30 minutes and incubated with Alexa Fluor 647 phalloidin (1:100) in PBS for 2 hours; nuclei were then stained with DAPI (diluted 1:1000 in PBS) for 15 minutes. After two further washes in PBS, nephrocytes were mounted in Mowiol. Images were acquired on an SP8 confocal microscope (Leica).
Coimmunoprecipitation Experiments
Coimmunoprecipitation experiments were performed to detect interaction of GFP-INF2 with calmodulin using the GFP-Trap affinity resin (Chromotek). All steps were carried out on ice with centrifugations at 4°C. HEK293T cells were seeded on fibronectin-coated (5 μg/ml in PBS) 10-cm2 dishes, transfected with INF2 constructs, and harvested by scraping into 1 ml PBS after 24–48 hours. After two washing steps with PBS at 500×g for 3 minutes, cells were resuspended in 200 μl lysis buffer (10 mM Tris/Cl, pH 7.5; 150 mM NaCl; 500 µM Ca2+, protease inhibitors [Roche], 2% NP-40). The suspension was left on ice for 30 minutes and pipetted up and down several times every 10 minutes. In the meantime, 25 μl of GFP-Trap beads were cleared by washing three times in lysis buffer without NP-40. Lysed cells were centrifuged for 10 minutes at 20,000×g, the supernatant was mixed with 300 μl buffer, and a sample taken for immunoblotting (“total protein”). The rest of the solution was incubated with GFP-Trap beads by tumbling for 1.5–2 hours at 4°C. Afterward, the suspension was centrifuged at 2500×g for 2 minutes, washed twice in dilution buffer, and bound protein was eluted from the beads by boiling in 50 μl of two-fold concentrated Laemmli buffer for 5 minutes. The protein-containing supernatant was analyzed by Western blotting.
Quantification of INF2 Expression Levels
To analyze expression levels for INF2 variants, HeLa KO cells were seeded in six-well plates, transfected with GFP-INF2 constructs, and grown for 24 hours. Cells were then fixed with 4% paraformaldehyde for 10 minutes. Nuclei were stained with DAPI, and plates were screened using a ScanR High-Content Screening microscope (Olympus). The object-based autofocus was set on the DAPI channel. For cell selection and automated image analysis, a cut-off was set for nuclear size between 1000 and 5000 pixels. For measurement of the GFP signal, a donut-shaped mask (20-pixel width) was drawn around the nuclear periphery. After subtraction of background signal (14 gray values), mean GFP intensities for all cells in a single experiment (n>1000) were calculated. The data reported are means of three experiments (n=3) with the SEM.
Results
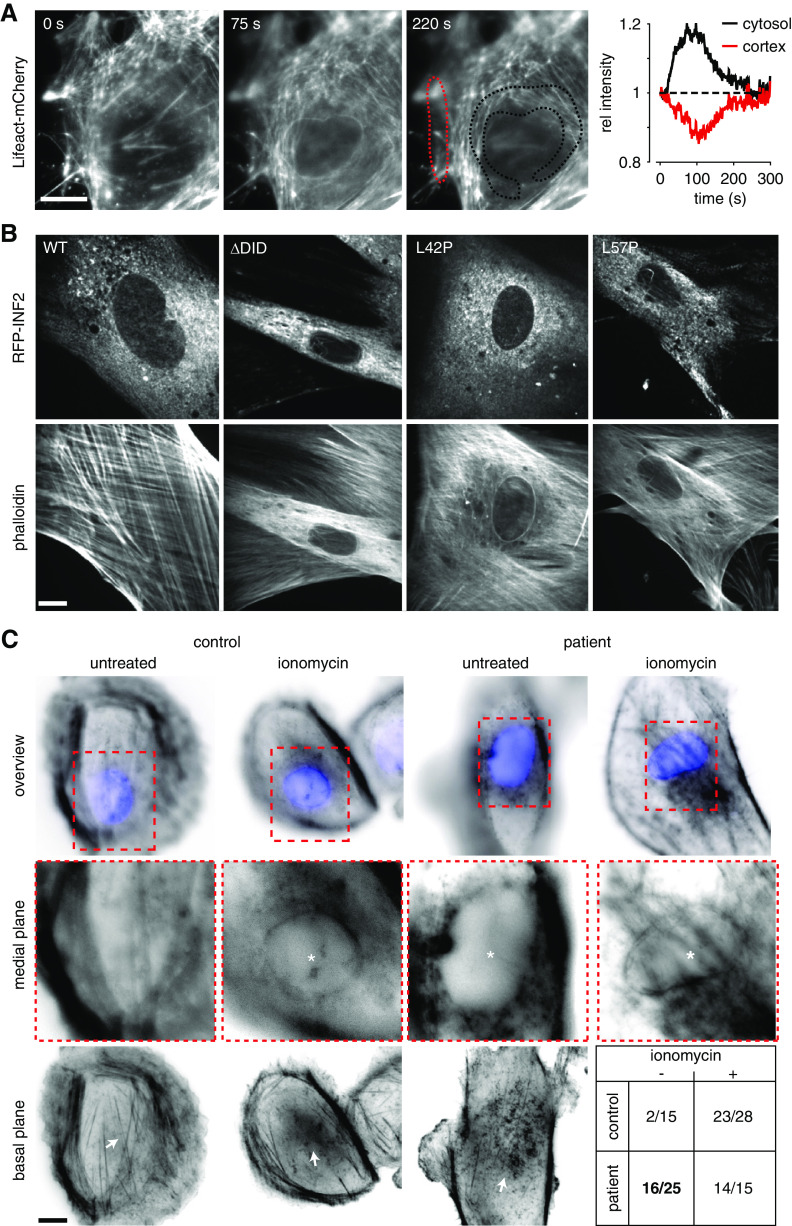
CaAR is a calcium-dependent cellular actin reorganization process that occurs in cultured mammalian cells, including human podocytes. The CaAR stress response is specifically mediated by the formin INF24,38 and osteosarcoma cells expressing disease-linked INF2 variants exhibit intracellular accumulation of actin.30 To assess the effects of INF2 mutations on CaAR in physiologically relevant cell types, we first asked whether the stress response can be induced in primary podocytes. We isolated GFP-labeled podocytes from mouse glomeruli33 and transiently transfected them with Lifeact-mCherry to monitor actin dynamics. Upon treatment with 500 nM ionomycin, we observed transient redistribution of actin from the cell cortex to the perinuclear area38 (Figure 1A, Supplemental Video 1). This phenomenon resembles CaAR, and occurred with kinetics comparable with those observed in immortalized cells4 (Figure 1A). To evaluate the effect of INF2 mutations on cellular actin organization, we expressed different variants of the endoplasmic reticulum (ER)-localized INF2-CAAX isoform39 in isolated primary podocytes. All INF2 variants were N-terminally fused to RFP to track subcellular localization of the formin. We compared INF2 localization and actin organization for wild-type (wt) INF2, a truncated and dominant active form of INF2 (ΔDID) and INF2 point mutants linked to FSGS (L42P) or FSGS/CMT (L57P) (Figure 1B). All INF2 fusions were expressed and localized to reticular tubular networks, as previously described (Figure 1B).4,40 Transient expression of wt INF2 did not alter the normal cellular organization of actin, which is dominated by extensive stress-fiber arrays (Figure 1B, Supplemental Video 2). Truncation of the autoinhibitory DID region led to extensive polymerization of actin in interior regions of the cells (Figure 1B). This is consistent with the reported constitutive activity of this mutant.41 All disease-linked mutations also induced intracellular actin polymerization, most prominently manifested by the formation of a perinuclear actin ring (Figure 1B). Similar actin accumulations have previously been reported for U2OS cells expressing FSGS-linked INF2 variants.30
Figure 1.
INF2 mediates actin reorganization in primary cells. (A) Time-lapse imaging of isolated primary mouse podocytes transfected with Lifeact-mCherry and treated with 500 nM ionomycin at t=0. Dotted lines delineate regions used for intensity plots. (B) Images of primary mouse podocytes transfected with indicated RFP-INF2 constructs, PFA-fixed and stained with Alexa Fluor 350-labeled phalloidin. (C) Cells isolated from urine samples obtained from a healthy control and a patient with FSGS (INF2 L162P), treated with 500 nM ionomycin for 1 minute, PFA-fixed and stained with Alexa Fluor 594-labeled phalloidin and DAPI. Images were taken at the focal plane of the nucleus. Asterisks indicate cells with intracellular actin polymerization. Arrows indicate changes in actin distribution on the basal surface of the cells (note that fewer stress fibers are present in both ionomycin-treated cells and in patient cells before the application of ionomycin). Table in lower right corner indicates number of cells with perinuclear actin (per total number) in indicated conditions. Scale bars, 10 µm.
For further validation of these findings, we isolated cells from the urine of a healthy control person and a patient with FSGS carrying a dominant INF2 mutation (L162P).16 In control cells treated with 500 nM ionomycin, a perinuclear actin ring was detected in 82% of cells (Figure 1C). Strikingly, 64% of unstimulated patient cells already exhibited perinuclear actin localization (Figure 1C, compared with 6% of control cells). This fraction increased to 91% after ionomycin treatment.
Our results show that INF2-dependent actin reorganization occurs in primary podocytes and in urine-derived cells from patients with FSGS. Disease-linked mutations of INF2 induce intracellular actin accumulation similar to that observed after calcium stimulation. Actin reorganization could therefore serve as sensitive readout for the assessment of disease-linked INF2 mutations.
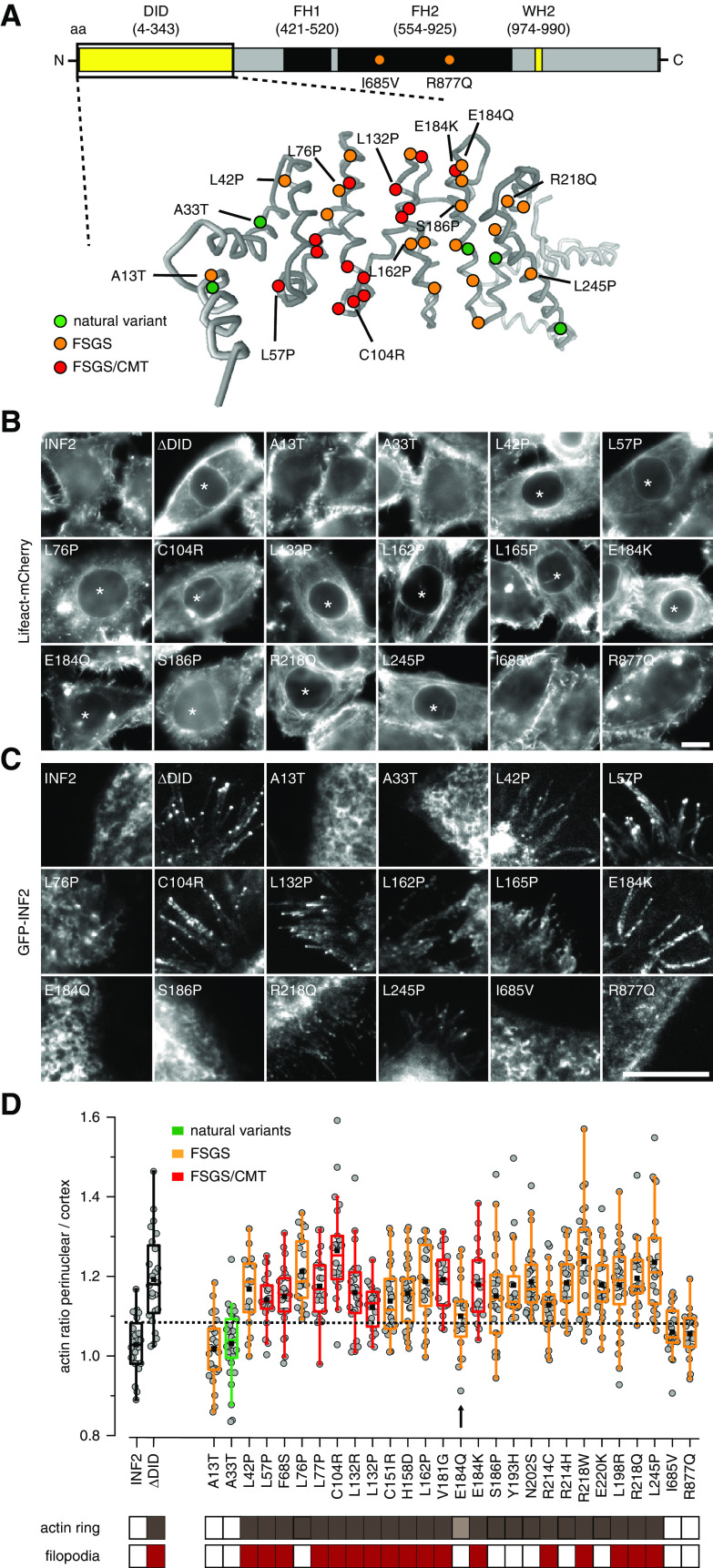
The majority of FSGS- and FSGS/CMT-related mutations have been mapped to the N-terminal DID of INF2. The only exceptions are two missense variants found in the FH2 domain from two sporadic patients26 (Figure 2A). We generated an array of GFP-INF2 fusions, including all proposed FSGS/CMT-linked mutations, as well as a representative set of benign variants from sequence database (Figure 2A, Table 1). We included wt INF2 and ΔDID as positive and negative controls, respectively. We expressed all variants in HeLa INF2 KO cells4 to rule out CaAR reactions mediated by wt INF2. All tested INF2 variants were correctly expressed and localized to ER membranes (Supplemental Figure 1). Interestingly, all amino acid exchanges that lie within the core of the DID domain led to perinuclear actin polymerization (Figure 2B, asterisk). These effects were also observed when variants were expressed in HeLa wt cells (Supplemental Figure 2). Notably, the benign INF2 variant A33T, as well as the two FH2 variants did not induce intracellular actin accumulation (Figure 2B). The A13T variant also behaved similarly to wt (Figure 2B).
Figure 2.
Systematic evaluation of the effects of INF2 mutations on actin organization. (A) Overview of INF2 domains and structural model of the INF2 DID domain (amino acids 1–343) generated by Phyre.2 Amino acid substitutions caused by missense mutations are indicated in green, orange, and red for benign variants, FSGS-only, and FSGS/CMT-linked mutations, respectively. (B and C) Epifluorescence images of HeLa INF2 KO cells stably expressing Lifeact-mCherry after transfection with the indicated GFP-tagged INF2 constructs. Images in (C) were acquired by total internal reflection fluoresecence (TIRF) microscopy to visualize basal GFP-INF2 localization. Asterisks indicate cells with perinuclear actin ring. (D) Quantification of perinuclear-to-cortical actin intensity ratios. Cells expressing INF2 wt and INF2 ΔDID were used as positive and negative controls, respectively (black boxes). Benign INF2 variants, FSGS-only, and FSGS/CMT-linked mutations are labeled in green, orange, and red, respectively. The dashed line indicates the mean+2 SD of the wt value. Classification of cells exhibiting intracellular actin rings (brown boxes) and/or filopodia-like structures (red boxes) is shown. Arrow and light brown box indicate INF2 E184Q variant with intermediate actin reorganization. Scale bars, 10 µm.
In addition to the expected ER localization of the INF2-CAAX isoform (Supplemental Figure 1), active INF2 ΔDID localized to the tips of cellular protrusions that were filled with actin (Figure 2C, Supplemental Figure 3, A and B, Supplemental Video 3). These structures were distinct from INF2-free filopodia in control cells (Supplemental Figure 3D), were also seen in primary podocytes (Supplemental Figure 3, C and D) and have been reported for HepG2 cells.42 We found filopodial recruitment of INF2 for many disease variants (Figure 2C, Supplemental Figure 3, A–C), including all FSGS/CMT mutants. Interestingly, some FSGS-only mutants did not induce filopodia (Figure 2C). We quantified the actin signal within transformed cells compared with cortical actin levels at the basal cell surface. Expression of INF2 ΔDID and disease-associated INF2 variants led to significant increases in intracellular actin filaments (Figure 2D, dashed line corresponds to the 75% quantile). The A13T variant and the two FH2-located mutants resembled wt distribution. In summary, all INF2 mutations that are associated with disease phenotypes lead to reorganization of actin consistent with deregulated INF2 activation. This can be monitored by accumulation of actin around the nucleus and by formation of filopodia. The extent of these effects varies, with E184Q in particular exhibiting intermediate activation (Figure 2D, arrow).
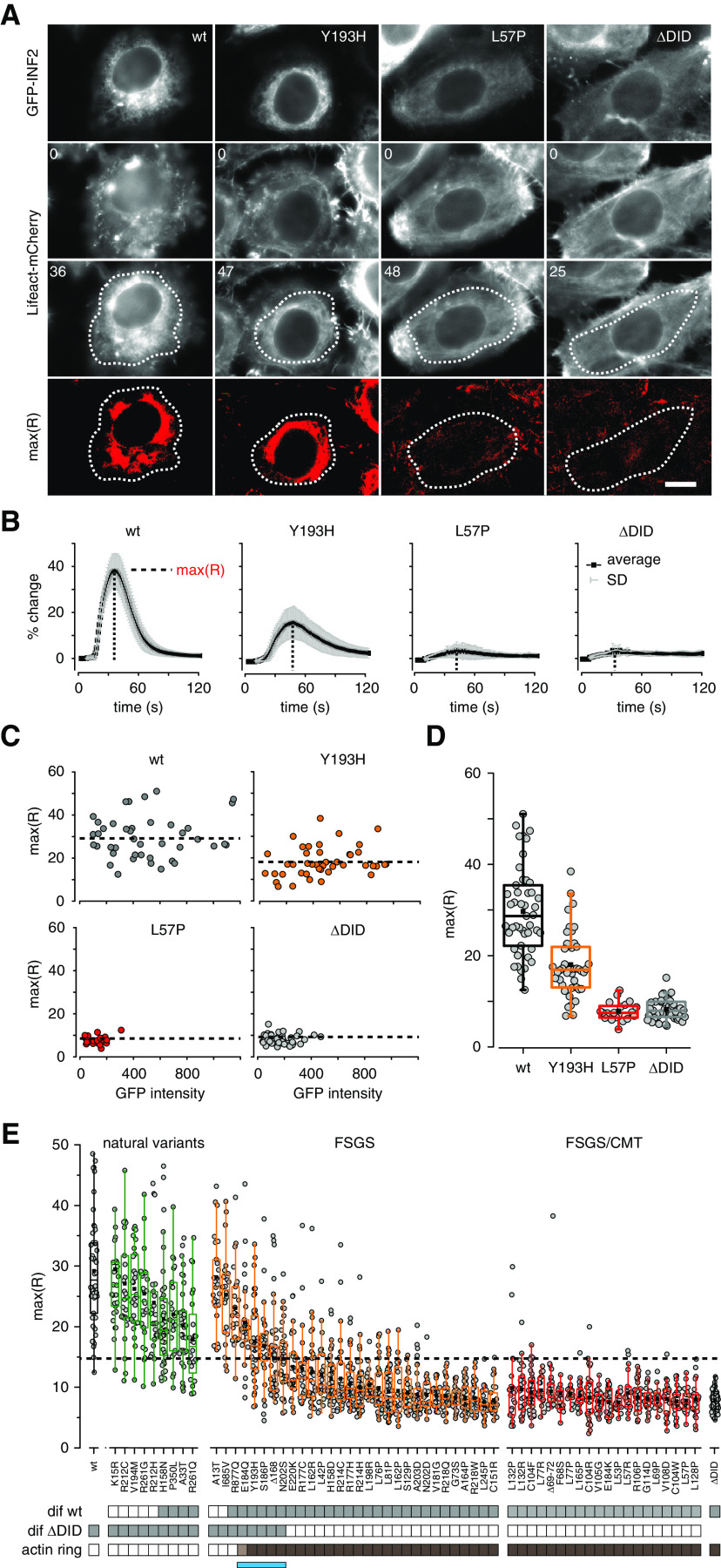
To better discriminate between the effects of different INF2 mutants, we proceeded to establish CaAR as a live cell quantitative readout for analysis of INF2 function.3 To this end we transfected HeLa INF2 KO cells that stably express Lifeact-mCherry with INF2 mutants and triggered CaAR via laser ablation.4 We established an automated image analysis routine using the maximal actin change during CaAR [max(R)] as characteristic parameter (Figure 3, A and B, Supplemental Video 4). Although GFP-INF2-CAAX fully rescued the CaAR reaction3 (Figure 3, A and B), cells expressing the ΔDID mutant showed no response to calcium (Figure 3, A and B). When we expressed FSGS/CMT-associated variants, we found two types of responses. A subset of mutants, exemplified by Y193H, exhibited reduced actin reorganization upon calcium stimulation (Figure 3, A and B). In others, such as the FSGS/CMT-associated L57P, no change in actin distribution could be detected at all (Figure 3, A and B). We found no correlation between GFP-INF2 expression and the corresponding max(R) of the CaAR reaction (Figure 3C). Our results, together with previous reports,31 indicate that INF2 variants exhibit slightly reduced expression compared with INF2 wt (Figure 3C, Supplemental Figure 4). Importantly, the effects of FSGS/CMT-associated INF2 variants could already be discerned at low expression levels of the mutant protein (Figure 3C). In summary, the magnitude of the CaAR reaction was independent of INF2 expression and could therefore be used for robust quantification of INF2 function (Figure 3D).
Figure 3.
Quantitative analysis of the functionality of INF2 mutant proteins on the basis of the CaAR assay. (A) HeLa INF2 KO cells stably expressing Lifeact-mCherry were transfected with the indicated GFP-INF2 constructs. We then monitored actin organization in cells after laser-induced calcium influx at t=8 seconds. Images depict actin organization in cells at t=0 second and at the time point of maximal actin reorganization. Regions used for intensity measurements in (B) are delineated by dashed lines. Scale bar, 10 µm; times are given in seconds. (B) Kinetics of actin reorganization observed in (A), given in terms of percent change (mean±SD). (C) Graphs of maximal actin reorganization values max(R) for cells expressing the indicated GFP-INF2 constructs, plotted against average fluorescence intensities of the respective GFP-INF2 variant. Dashed line represents mean of all max(R) for given variant. (D) Box plots of max(R) values in (C). (E) Box plots of max(R) values for all analyzed INF2 mutations (n≥21). INF2 wt and ΔDID were used as positive and negative controls (black boxes), respectively. Benign INF2 variants, FSGS-only, and FSGS/CMT-linked mutations are labeled in green, orange, and red, respectively. Values are ordered first by the type of INF2 mutation and then by how much they differ from the ΔDID value (largest to smallest). Gray boxes below the graph indicate that max(R) values were significantly different relative to either wt or ΔDID (one-way ANOVA test with Holm–Bonferroni post hoc correction). Brown boxes indicate GFP-INF2 expression leading to actin reorganization at t=0 second (see also Figure 2D). The blue bar indicates the group of INF2 variants with intermediate actin reorganization phenotypes.
We therefore extended our study and comprehensively analyzed all reported INF2 mutations. On the basis of clinical reports and database information (Figure 2A, Table 1), we separated INF2 variants into benign variants (green), FSGS-only mutations (orange), and FSGS/CMT-linked mutations (red). All tested benign variants reacted to calcium and displayed CaAR reactions with averaged max(R) values that were significantly above those of ΔDID-expressing cells (Figure 3E, dotted line). In contrast, all tested FSGS/CMT variants hardly showed any response, with max(R) values similar to ΔDID-expressing cells (Figure 3E). FSGS-only variants displayed a broad distribution of reorganization phenotypes, resulting in max(R) values ranging from wt-like (A13T) to nonresponding mutants (C151R). Results for all INF2 mutations examined are summarized in Table 1. Calcium-induced actin reorganization in the HeLa wt background was similar to that observed in KO cells (Supplemental Figure 5) or cultured INF2 KO podocytes (Supplemental Figure 6). This confirms the robust utility of CaAR for evaluation of INF2 mutations.
Statistical analysis confirmed that benign variants differed significantly from INF2 ΔDID, whereas disease-linked INF2 variants (except A13T and I685V) were significantly different from wt INF2 (Figure 3E, gray boxes). A subset of intermediate variants resulted in max(R) values that were significantly different from both wt and ΔDID controls (Figure 3E). Of these, the four benign variants and R877Q did not exhibit any intracellular actin accumulation before stimulation, indicative of reduced INF2 functionality, rather than deregulated activation. The remaining five members were all FSGS-only mutants and included E184Q, Y193H, S186P, Δ168, and N202S (Figure 3E, blue bar). They exhibited intracellular actin accumulation (Figure 3E, dark brown boxes) independently of calcium, with E184Q showing a weaker phenotype than the others (Figures 2D and 3E, light brown box). Our results indicate that disease-linked dominant INF2 mutations can be divided into distinct subsets using cell-based profiling.
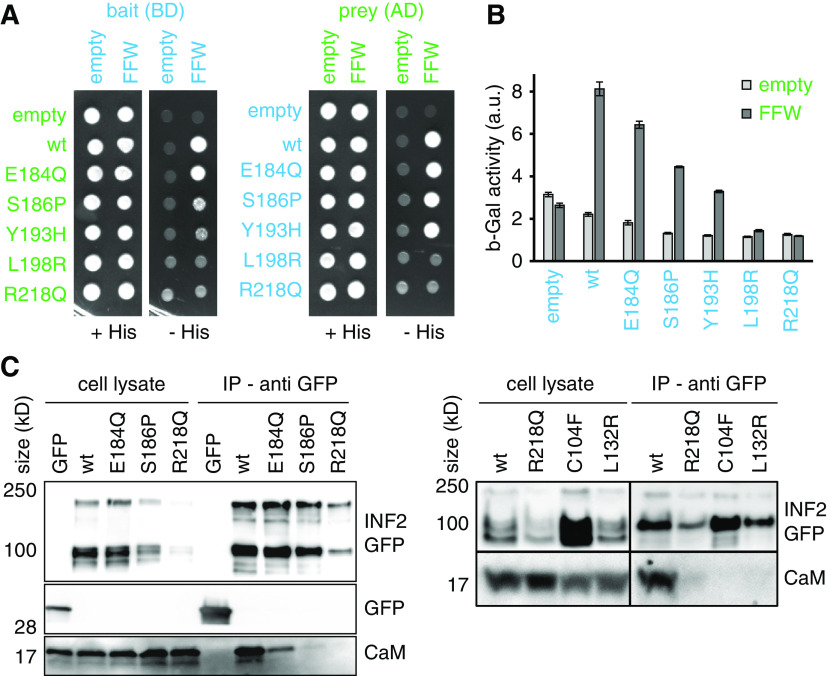
To further explore the differences between intermediate and fully activated INF2 variants, we initially focused on biochemical interactions and subcellular localization of the formin. Using yeast two-hybrid assays, we detected interaction between the wt DID (aa 1–420) domain and the C-terminal segment of INF2 (FFW, aa 421–1008, Figure 4, A and B). Active INF2 mutants, such as L198R and R218Q, showed no interaction with the FFW domain, either as bait or prey (Figure 4, A and B). In contrast, intermediate INF2 variants (E184Q, S186P, and Y193H) retained weak interactions leading to reduced growth on interaction indicator plates (Figure 4A) and reduced production of β-Gal (Figure 4B). We have previously shown that INF2 binds to recombinant calmodulin.4 To test whether activation of INF2 affected this interaction we performed coimmunoprecipitation experiments in HEK293 cells. GFP-tagged INF2 wt bound to calmodulin in the presence of calcium (CaM, Figure 4C). This interaction was completely abolished in the case of the strong FSGS and FSGS/CMT mutants (Figure 4C). In contrast, the intermediate mutant E184Q was able to bind to CaM with reduced affinity (Figure 4C).
Figure 4.
Biochemical interactions of INF2 variants. (A and B) Yeast two-hybrid analysis of auto-inhibitory INF2 DID/diaphanous autoregulatory domain interaction. Fusions of the INF2 DID (aa 1–420) and INF2 FFW (aa 421–1008) to the GAL4 activation domain (AD; prey) and the GAL4 binding domain (BD; bait) were expressed in the yeast strain PJ69–4A. The transformed cells were tested for growth on SC-Leu-Trp-His plates (−His) (A) and for β-galactosidase activity (B), which indicate positive two-hybrid interactions. (C) Western blot showing coimmunoprecipitation of calmodulin with GFP-INF2 variants expressed in HEK293 cells. Soluble GFP was used as negative control in left blot. Immunoprecipitation was performed using anti-GFP trap. Signal was detected using anti-GFP antibody (INF2-GFP, GFP) and anti-calmodulin antibody (CaM).
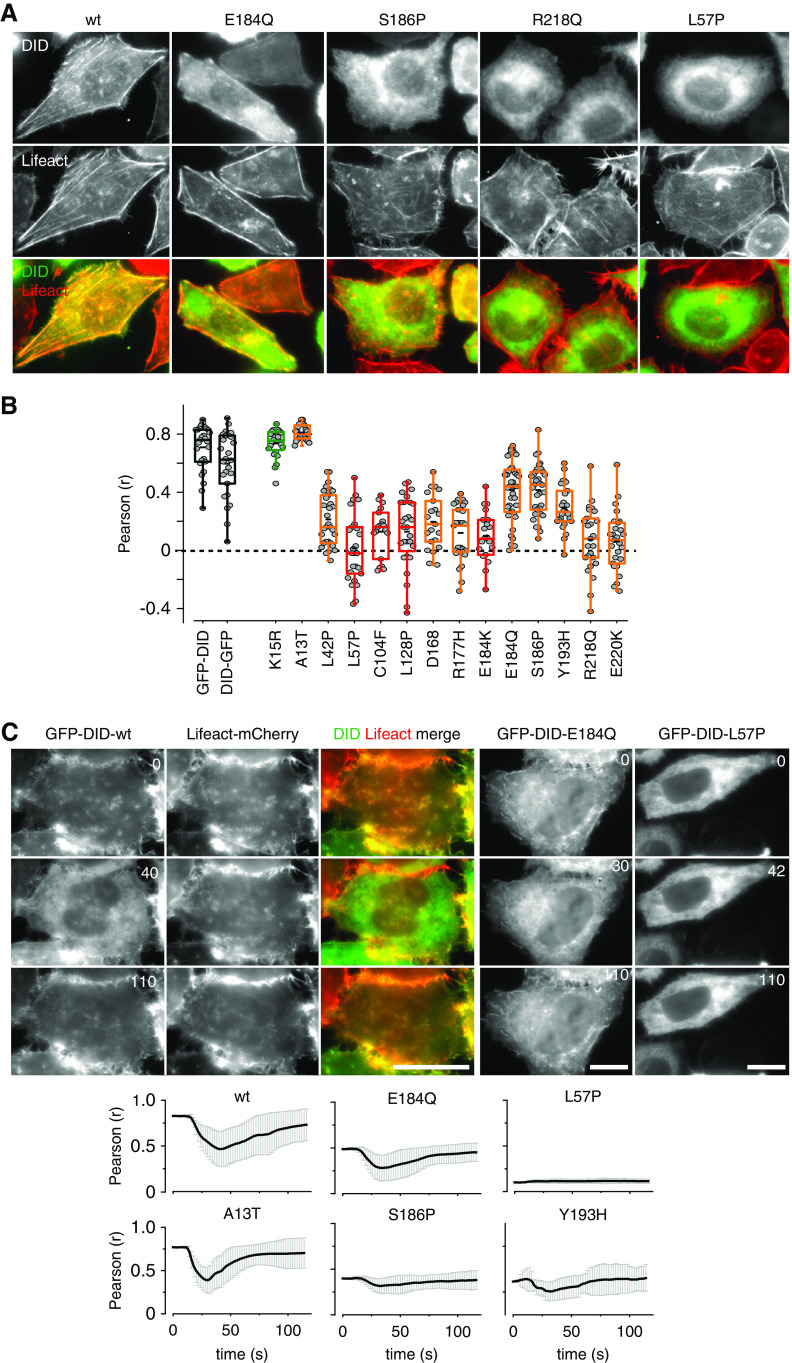
To further evaluate the cellular consequences of DID domain mutations, we expressed GFP fusions to the DID domain in Hela INF2 KO cells and analyzed their localization. Surprisingly, we found very strong colocalization of GFP-DID with Lifeact-mCherry (Figure 5, A and B). This colocalization was lost for strong DID mutants (R218Q, L57P), whereas intermediate DID mutants retained partial colocalization with actin (S186P, E184Q, Figure 5, A and B). The localization patterns of wt and mutant constructs suggested that the actin-binding of the INF2 DID domain might be regulated by calcium. We therefore stimulated HeLa INF2 KO cells by laser ablation and monitored GFP-DID and Lifeact-mCherry signals over time. We observed relocalization of the GFP-DID domain from actin structures to the cytosol, which was reversed within 2 minutes (Figure 5C, Supplemental Video 5). We used the Pearson correlation coefficient to describe the kinetics of relocalization (Figure 5C). Mutants of the intermediate group were partially able to relocalize upon calcium stimulation, whereas strong DID mutants did not (Figure 5C, Supplemental Video 6). The putative benign variant A13T behaved identically to the wt DID (Figure 5C). In summary, our results suggest that intermediate INF2 mutants retain reduced capacity for regulation by calcium as indicated by modulations in intramolecular interactions and subcellular localization.
Figure 5.
Subcellular localization of the INF2 DID domain. (A) HeLa INF2 KO cells stably expressing Lifeact-mCherry were transfected with the indicated GFP-INF2 DID domain constructs (aa 1–420) and analyzed for GFP-INF2 DID localization. (B) Quantification of colocalization of wt (black boxes), benign (green), FSGS-linked only (orange), and FSGS/CMT-linked GFP-INF2 DID variants (red) with Lifeact-mCherry was quantified by Pearson correlation coefficient (r) for n≥20 cells. (C) Kinetics of GFP-INF2 DID relocalization from actin structures to the cytosol after calcium stimulation, as indicated by the Pearson correlation coefficient (r) of time lapse images with respect to image at t=0 second. Scale bars, 10 µm.
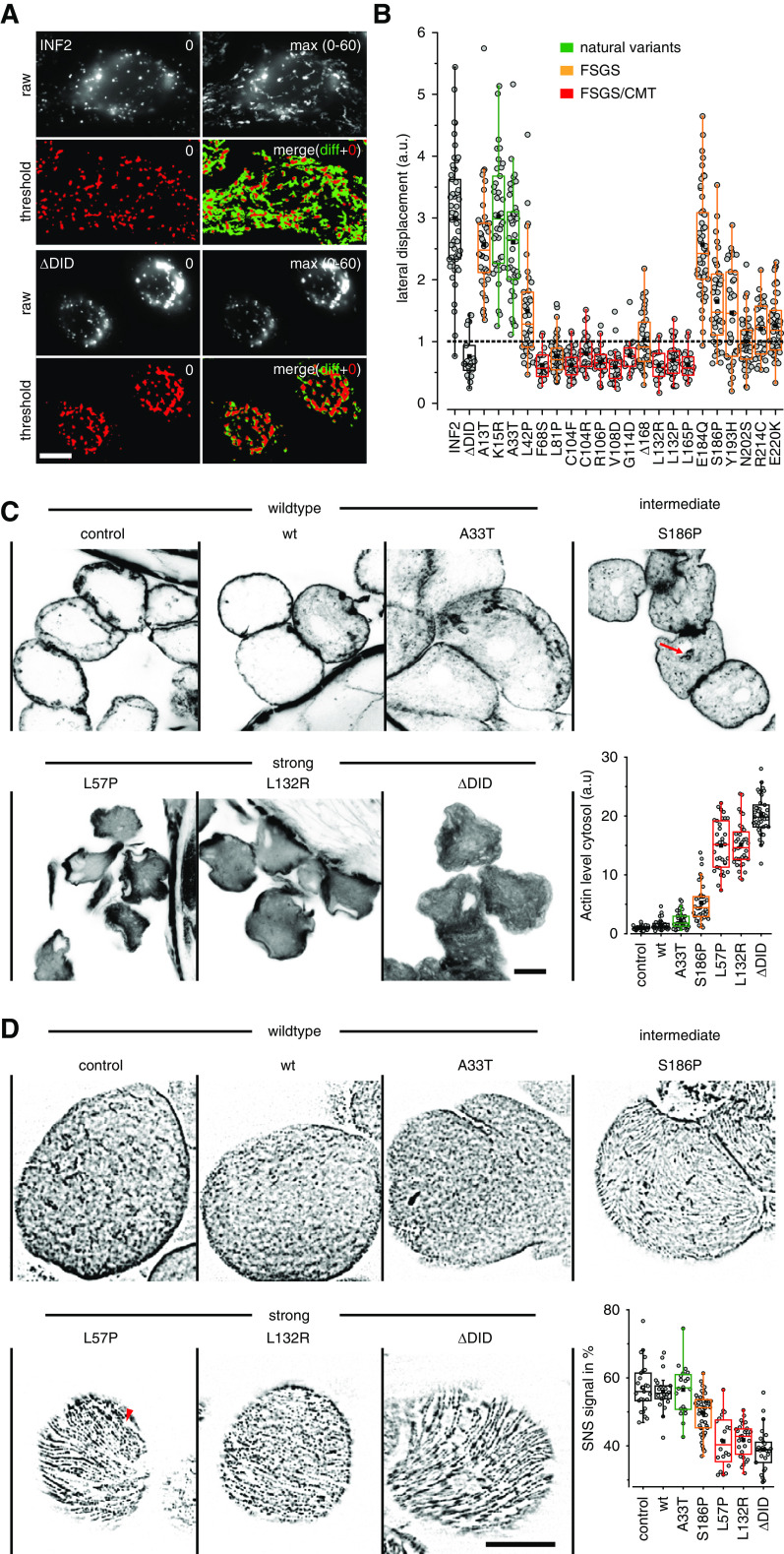
Actin polymerization during CaAR leads to transient inhibition of organelle mobility.4 To test whether this type of stress response is also induced in cells expressing deregulated disease variants of INF2, we followed displacement of lysosomes over time. In HeLa INF2 KO cells expressing wt INF2, lysosomes were highly mobile (Figure 6A). In cells expressing the INF2 ΔDID lysosomes were concentrated around the nuclear periphery, and exhibited very little motion (Figure 6, A and B). All strong INF2 variants behaved like INF2 ΔDID, whereas intermediate variants exhibited moderately reduced lysosome mobility (Figure 6B).
Figure 6.
Physiologic consequences of disease-linked INF2 mutations. (A and B) HeLa INF2 KO cells were transfected with the indicated GFP-INF2 constructs, stained with LysoTracker red and monitored for the indicated periods. To analyze the distribution of lysosomes in cells, images were thresholded at t=0 second (red signal) and the area of lysosomal signal at t=0 second (red in merged image) was compared with the additional area occupied over the period covered by the image series (maximum intensity projection of t=0 to t=60 seconds minus t=0 seconds, green in merged image). Box plots in (B) indicate the relative change in the area covered by lysosomes in the course of 60 seconds, with one representing 100% area change (dashed line). wt GFP-INF2 and INF2 ΔDID were used as positive and negative controls (black boxes), respectively. Benign INF2 variants, FSGS, and FSGS/CMT-linked mutations are labeled in green, orange, and red, respectively. (C) Fly nephrocytes stably expressing the indicated Myc-tagged INF2 variants were formaldehyde fixed and stained with Alexa Fluor 647 phalloidin to analyze actin organization. Graph shows quantification of intracellular versus cortical actin (n≥31 cells). Images taken at midsection of nephrocytes. (D) Fly nephrocytes stably expressing the indicated Myc-INF2 mutants were heat fixed and analyzed for nephrin (Sns) localization using anti-Sns antibody. Images represent surface planes of shown nephrocytes. See Supplemental Figure 7 for medial sections. Graph shows quantification of nephrin density (n≥22 cells). Scale bars, 10 µm.
For validation of our results in vivo, we used the UAS/Gal4 system to express INF2 variants specifically in fly nephrocytes, which are functionally homologous to the podocytes of vertebrates.43 Neither development nor nephrocyte morphology were affected by INF2 expression (Figure 6C). Alexa Fluor 647 phalloidin staining of untransfected nephrocytes or nephrocytes expressing wt INF2 showed the expected cortical concentration of actin filaments (Figure 6C). In contrast, expression of the activated INF2 variants ΔDID, L57P, and L132R led to intracellular accumulation of polymerized actin and reduced cortical staining (Figure 6C). Nephrocytes expressing the intermediate INF2 mutant S186P exhibited punctate intracellular actin staining (Figure 6C, red arrow), but retained continuous cortical actin (Figure 6C). The ratio of cytosolic to cortical actin correlated with the suggested degree of INF2 activation (Figure 6C). To test whether altered actin distribution in nephrocytes had consequences on the structural organization of slit diaphragms, we performed immunofluorescence analysis of the fly nephrin homolog Sns (Sticks and stones). We found the typical dense network organization of Sns on the plasma membrane of control and INF2 wt-expressing nephrocytes (Figure 6D). Intermediate and strong INF2 variants were associated with reduced Sns staining (Figure 6D, Supplemental Figure 7), with particularly large areas that were devoid of nephrin seen for the strong variants (Figure 6D, red arrowhead, Supplemental Figure 7). The discontinuous labeling of membranes with nephrin is comparable to the pattern observed in glomeruli of human patients with FSGS.44 Our results indicate that redistribution of cytosolic actin driven by disease-linked INF2 variants has consequences for podocyte/nephrocyte organization.
Discussion
We have performed a systematic analysis of actin reorganization in cells expressing INF2 variants. Our study firmly establishes that INF2 mutations that are linked to familial FSGS and CMT result in aberrant—partial or constitutive—activation of the formin. This activation leads to perinuclear and ER-associated actin accumulation, presumably because of weakening of the autoinhibitory interaction between the DID domain and the diaphanous autoregulatory domain. This constitutive activity can be observed in primary podocytes and in cells obtained from patients with FSGS, and is consistent with the observed autosomal dominant nature of these hereditary conditions. In particular, we have shown that several variants linked to FSGS/CMT reduce cortical actin staining (R177H, L165P and R106P,6 E184K and R218Q5) when transiently expressed in cultured cells. These variants were also shown to have increased affinity for constitutively active Cdc426 and decreased affinity for mDia2.45
Our evaluation of INF2 mutations using the CaAR response as a quantitative readout revealed that the degree of INF2 activation varies between different mutant forms of the protein. Mutations associated with FSGS/CMT always resulted in full INF2 activation. In contrast, mutations exclusively associated with FSGS exhibited a broader range of actin responses. Several intermediate INF2 variants, in particular E184Q, Y193H, and S186P, retained partial autoinhibition and were susceptible to further activation by calcium. Interestingly, the S186P variant was reported to retain the ability to bind to mDia245 and not to induce intracellular actin accumulation.5 We were also able to identify potential false-positive associations of INF2 variants with disease phenotypes. In all our assays, the A13T variant behaved like the wt INF2, confirming its recent classification as a benign polymorphism.10 The two FH2 domain variants I685V and R877Q26 also behaved like wt INF2 in our assays. Our results suggest that CaAR can be used as sensitive cellular assay for INF2 function and for robust evaluation of disease-linked variants of the formin. A more precise correlation of cellular readouts with disease parameters is currently difficult because of the low case numbers (Table 1) and inconsistent or often nonquantitative patient data available.
During our characterization of molecular interactions of INF2, we surprisingly found that the DID domain closely associated with cortical F-actin in a calcium-sensitive manner. Interestingly, a proteolytic N-terminal fragment of INF2 has recently been shown to associate with the cell cortex.44 It is at present unclear how calcium-regulated actin-binding of INF2-DID contributes to the function of the full-length protein.
We performed our systematic analyses of INF2 variants in the established HeLa cell line, which is very convenient for genetic manipulations, but it may not reflect the responses of differentiated cells. However, CaAR and intracellular actin accumulation upon expression of active INF2 variants, were also seen in immortalized podocytes, primary podocytes, and cells isolated from the urine of human patients. Importantly, intracellular actin accumulation quantitatively correlated with perturbed distribution of nephrin in the plasma membrane of fly nephrocytes, which is reminiscent of other nephropathy models in fly.46 This defect could be linked to a function of cortical actin in the organization of the slit diaphragm47 or owing to defects in trafficking. The latter notion is supported by our findings on the cessation of organelle mobility during the CaAR reaction4 and on expression of active INF2 variants. A role for INF2 in vesicular trafficking of MAL2 has been previously proposed.42
Many aspects of INF2 regulation have been linked to FSGS, including INF2 expression levels,31 INF2 interaction with acetylated actin30 or MAL,48 proteolytic processing,44 or effects on microtubule organization.49,50 On the basis of our systematic evaluation, we favor the idea that INF2-linked FSGS reflects global reorganization of the actin cytoskeleton and thereby a deregulated cellular stress response, rather than specific molecular interactions. This hypothesis would be consistent with aberrant functions of other dominant genetic factors that lead to FSGS. ACTN4 variants have been shown to exhibit calcium-independent bundling of actin, whereas TRPC6 variants associated with FSGS mediate excessive influx of calcium into cells.3 Accumulation of intracellular actin could therefore globally affect intracellular trafficking or the ability of cells to react to acute stress. This interpretation is consistent with the defects seen in transgenic mice that express the strong INF2 FSGS variant R218Q. Although kidney development and morphology are not affected in these mice, the ability of podocytes to recover from acute damage induced by protamine sulfate is severely impaired.28
Disclosures
All authors have nothing to disclose.
Funding
This work was supported by DFG grant SFB1009-B20 to RWS and HPP and by the IZKF Münster Wed2/022/18 to RWS. Dr. Pavenstädt reports grants from Deutsche Forschungsgemeinschaft, during the conduct of the study. Dr. Wedlich-Söldner reports grants from German Research Foundation and grants from IZKF Münster, during the conduct of the study.
Supplementary Material
Acknowledgments
We thank Hartmut Schmitt for help with isolation of urine cells and Paul Hardy for critical reading of the manuscript. We are indebted to Thomas Zobel and the Münster Imaging Network for help with microscopy.
R. Wedlich-Söldner, H. Pavenstädt, and C. Schuberth designed the study and supervised the project. S. Bayraktar, J. Nehrig, E. Menis, K. Karli, A. Janning, T. Struk, and C. Schuberth performed experiments and analyzed data. M. Krahn and U. Michgehl helped with experimental design. J. Halbritter provided FSGS patient cells. S. Bayraktar, C. Schuberth, and R. Wedlich-Söldner wrote the paper, with help from all authors.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2019111174/-/DCSupplemental.
Supplemental Figure 1. INF2 variant localization.
Supplemental Figure 2. Systematic evaluation of INF2 mutations on actin organization in HeLa wt cells.
Supplemental Figure 3. Filopodia-like structures induced by INF2 variants.
Supplemental Figure 4. Expression analysis of INF2 variants.
Supplemental Figure 5. Quantitative analysis of CaAR upon expression of INF2 variants in HeLa wt cells.
Supplemental Figure 6. Quantitative analysis of CaAR upon expression of INF2 variants in AB8 INF2 KO podocytes.
Supplemental Figure 7. Sns distribution in fly nephrocytes.
Supplemental Video 1. CaAR in primary podocytes.
Supplemental Video 2. INF2 variants in primary podocytes.
Supplemental Video 3. Filopodia formation by INF2 variants.
Supplemental Video 4. CaAR with INF2 variants.
Supplemental Video 5. Actin association of INF2 DID.
Supplemental Video 6. Actin association of INF2 DID variants.
References
- 1.Rood IM, Deegens JK, Wetzels JF: Genetic causes of focal segmental glomerulosclerosis: Implications for clinical practice. Nephrol Dial Transplant 27: 882–890, 2012. [DOI] [PubMed] [Google Scholar]
- 2.Wiggins RC: The spectrum of podocytopathies: A unifying view of glomerular diseases. Kidney Int 71: 1205–1214, 2007. [DOI] [PubMed] [Google Scholar]
- 3.Pollak M: Genetics of familial FSGS. Semin Nephrol 36: 467–472, 2016. [DOI] [PubMed] [Google Scholar]
- 4.Wales P, Schuberth CE, Aufschnaiter R, Fels J, García-Aguilar I, Janning A, et al.: Calcium-mediated actin reset (CaAR) mediates acute cell adaptations. eLife 5: e19850, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown EJ, Schlöndorff JS, Becker DJ, Tsukaguchi H, Tonna SJ, Uscinski AL, et al.: Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis [published correction appears in Nat Genet 42: 361, 2010]. Nat Genet 42: 72–76, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer O, Benoit G, Gribouval O, Nevo F, Tête MJ, Dantal J, et al.: Mutations in INF2 are a major cause of autosomal dominant focal segmental glomerulosclerosis. J Am Soc Nephrol 22: 239–245, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roos A, Weis J, Korinthenberg R, Fehrenbach H, Häusler M, Züchner S, et al.: Inverted formin 2-related Charcot-Marie-Tooth disease: Extension of the mutational spectrum and pathological findings in Schwann cells and axons. J Peripher Nerv Syst 20: 52–59, 2015. [DOI] [PubMed] [Google Scholar]
- 8.Toyota K, Ogino D, Hayashi M, Taki M, Saito K, Abe A, et al.: INF2 mutations in Charcot-Marie-Tooth disease complicated with focal segmental glomerulosclerosis. J Peripher Nerv Syst 18: 97–98, 2013. [DOI] [PubMed] [Google Scholar]
- 9.Mademan I, Deconinck T, Dinopoulos A, Voit T, Schara U, Devriendt K, et al.: De novo INF2 mutations expand the genetic spectrum of hereditary neuropathy with glomerulopathy. Neurology 81: 1953–1958, 2013. [DOI] [PubMed] [Google Scholar]
- 10.Barua M, Brown EJ, Charoonratana VT, Genovese G, Sun H, Pollak MR: Mutations in the INF2 gene account for a significant proportion of familial but not sporadic focal and segmental glomerulosclerosis. Kidney Int 83: 316–322, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez PQ, Lohkamp B, Celsi G, Mache CJ, Auer-Grumbach M, Wernerson A, et al.: Novel INF2 mutation p. L77P in a family with glomerulopathy and Charcot-Marie-Tooth neuropathy. Pediatr Nephrol 28: 339–343, 2013. [DOI] [PubMed] [Google Scholar]
- 12.Boyer O, Nevo F, Plaisier E, Funalot B, Gribouval O, Benoit G, et al.: INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N Engl J Med 365: 2377–2388, 2011. [DOI] [PubMed] [Google Scholar]
- 13.Caridi G, Lugani F, Dagnino M, Gigante M, Iolascon A, Falco M, et al.: Novel INF2 mutations in an Italian cohort of patients with focal segmental glomerulosclerosis, renal failure and Charcot-Marie-Tooth neuropathy. Nephrol Dial Transplant 29[Suppl 4]: iv80-iv86, 2014. [DOI] [PubMed] [Google Scholar]
- 14.Jin S, Wang W, Wang R, Lv H, Zhang W, Wang Z, et al.: INF2 mutations associated with dominant inherited intermediate Charcot-Marie-Tooth neuropathy with focal segmental glomerulosclerosis in two Chinese patients. Clin Neuropathol 34: 275–281, 2015. [DOI] [PubMed] [Google Scholar]
- 15.Park HJ, Kim HJ, Hong YB, Nam SH, Chung KW, Choi BO: A novel INF2 mutation in a Korean family with autosomal dominant intermediate Charcot-Marie-Tooth disease and focal segmental glomerulosclerosis. J Peripher Nerv Syst 19: 175–179, 2014. [DOI] [PubMed] [Google Scholar]
- 16.Münch J, Grohmann M, Lindner TH, Bergmann C, Halbritter J: Diagnosing FSGS without kidney biopsy - a novel INF2-mutation in a family with ESRD of unknown origin. BMC Med Genet 17: 73, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Büscher AK, Celebi N, Hoyer PF, Klein HG, Weber S, Hoefele J: Mutations in INF2 may be associated with renal histology other than focal segmental glomerulosclerosis. Pediatr Nephrol 33: 433–437, 2018. [DOI] [PubMed] [Google Scholar]
- 18.Rood IM, Bongers EM, Lugtenberg D, Klein IH, Steenbergen EJ, Wetzels JF, et al.: Familial focal segmental glomerulosclerosis: Mutation in inverted formin 2 mimicking alport syndrome. Neth J Med 74: 82–85, 2016. [PubMed] [Google Scholar]
- 19.Challis RC, Ring T, Xu Y, Wong EK, Flossmann O, Roberts IS, et al.: Thrombotic microangiopathy in inverted formin 2-mediated renal disease. J Am Soc Nephrol 28: 1084–1091, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gbadegesin RA, Lavin PJ, Hall G, Bartkowiak B, Homstad A, Jiang R, et al.: Inverted formin 2 mutations with variable expression in patients with sporadic and hereditary focal and segmental glomerulosclerosis. Kidney Int 81: 94–99, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan W, Lovric S, Ashraf S, Rao J, Schapiro D, Airik M, et al.: Analysis of 24 genes reveals a monogenic cause in 11.1% of cases with steroid-resistant nephrotic syndrome at a single center. Pediatr Nephrol 33: 305–314, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santín S, Bullich G, Tazón-Vega B, García-Maset R, Giménez I, Silva I, et al.: Clinical utility of genetic testing in children and adults with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 6: 1139–1148, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Safarikova M, Stekrova J, Honsova E, Horinova V, Tesar V, Reiterova J: Mutational screening of inverted formin 2 in adult-onset focal segmental glomerulosclerosis or minimal change patients from the Czech Republic. BMC Med Genet 19: 147, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HK, Han KH, Jung YH, Kang HG, Moon KC, Ha IS, et al.: Variable renal phenotype in a family with an INF2 mutation. Pediatr Nephrol 26: 73–76, 2011. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez-Ares M, Garcia-Vidal M, Antucho EE, Julio P, Eduardo VM, Lens XM, et al.: A novel mutation, outside of the candidate region for diagnosis, in the inverted formin 2 gene can cause focal segmental glomerulosclerosis. Kidney Int 83: 153–159, 2013. [DOI] [PubMed] [Google Scholar]
- 26.Lipska BS, Iatropoulos P, Maranta R, Caridi G, Ozaltin F, Anarat A, et al.; PodoNet Consortium: Genetic screening in adolescents with steroid-resistant nephrotic syndrome. Kidney Int 84: 206–213, 2013. [DOI] [PubMed] [Google Scholar]
- 27.Chhabra ES, Higgs HN: INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem 281: 26754–26767, 2006. [DOI] [PubMed] [Google Scholar]
- 28.Subramanian B, Sun H, Yan P, Charoonratana VT, Higgs HN, Wang F, et al.: Mice with mutant Inf2 show impaired podocyte and slit diaphragm integrity in response to protamine-induced kidney injury. Kidney Int 90: 363–372, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun H, Al-Romaih KI, MacRae CA, Pollak MR: Human kidney disease-causing INF2 mutations perturb rho/dia signaling in the glomerulus. EBioMedicine 1: 107–115, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mu A, Fung TS, Kettenbach AN, Chakrabarti R, Higgs HN: A complex containing lysine-acetylated actin inhibits the formin INF2. Nat Cell Biol 21: 592–602, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rollason R, Wherlock M, Heath JA, Heesom KJ, Saleem MA, Welsh GI: Disease causing mutations in inverted formin 2 regulate its binding to G-actin, F-actin capping protein (CapZ α-1) and profilin 2. Biosci Rep 36: e00302, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moeller MJ, Sanden SK, Soofi A, Wiggins RC, Holzman LB: Podocyte-specific expression of cre recombinase in transgenic mice. Genesis 35: 39–42, 2003. [DOI] [PubMed] [Google Scholar]
- 33.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L: A global double-fluorescent Cre reporter mouse. Genesis 45: 593–605, 2007. [DOI] [PubMed] [Google Scholar]
- 34.Möckli N, Auerbach D: Quantitative beta-galactosidase assay suitable for high-throughput applications in the yeast two-hybrid system. Biotechniques 36: 872–876, 2004. [DOI] [PubMed] [Google Scholar]
- 35.Groth AC, Fish M, Nusse R, Calos MP: Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhuang S, Shao H, Guo F, Trimble R, Pearce E, Abmayr SM: Sns and Kirre, the Drosophila orthologs of Nephrin and Neph1, direct adhesion, fusion and formation of a slit diaphragm-like structure in insect nephrocytes. Development 136: 2335–2344, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hochapfel F, Denk L, Mendl G, Schulze U, Maaßen C, Zaytseva Y, et al.: Distinct functions of Crumbs regulating slit diaphragms and endocytosis in Drosophila nephrocytes. Cell Mol Life Sci 74: 4573–4586, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao X, Li Q, Mogilner A, Bershadsky AD, Shivashankar GV: Mechanical stimulation induces formin-dependent assembly of a perinuclear actin rim. Proc Natl Acad Sci U S A 112: E2595–E2601, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramabhadran V, Korobova F, Rahme GJ, Higgs HN: Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol Biol Cell 22: 4822–4833, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chhabra ES, Ramabhadran V, Gerber SA, Higgs HN: INF2 is an endoplasmic reticulum-associated formin protein. J Cell Sci 122: 1430–1440, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramabhadran V, Gurel PS, Higgs HN: Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing. J Biol Chem 287: 34234–34245, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madrid R, Aranda JF, Rodríguez-Fraticelli AE, Ventimiglia L, Andrés-Delgado L, Shehata M, et al.: The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev Cell 18: 814–827, 2010. [DOI] [PubMed] [Google Scholar]
- 43.Weavers H, Prieto-Sánchez S, Grawe F, Garcia-López A, Artero R, Wilsch-Bräuninger M, et al.: The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature 457: 322–326, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian B, Chun J, Perez-Gill C, Yan P, Stillman IE, Higgs HN, et al.: FSGS-causing INF2 mutation impairs cleaved INF2 N-fragment functions in podocytes. J Am Soc Nephrol 31: 374–391, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun H, Schlondorff JS, Brown EJ, Higgs HN, Pollak MR: Rho activation of mDia formins is modulated by an interaction with inverted formin 2 (INF2). Proc Natl Acad Sci U S A 108: 2933–2938, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hermle T, Braun DA, Helmstädter M, Huber TB, Hildebrandt F: Modeling monogenic human nephrotic syndrome in the Drosophila garland cell nephrocyte. J Am Soc Nephrol 28: 1521–1533, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dlugos CP, Picciotto C, Lepa C, Krakow M, Stöber A, Eddy ML, et al.: Nephrin signaling results in integrin β1 activation. J Am Soc Nephrol 30: 1006–1019, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrés-Delgado L, Antón OM, Madrid R, Byrne JA, Alonso MA: Formin INF2 regulates MAL-mediated transport of Lck to the plasma membrane of human T lymphocytes. Blood 116: 5919–5929, 2010. [DOI] [PubMed] [Google Scholar]
- 49.Bartolini F, Andres-Delgado L, Qu X, Nik S, Ramalingam N, Kremer L, et al.: An mDia1-INF2 formin activation cascade facilitated by IQGAP1 regulates stable microtubules in migrating cells [published correction appears in Mol Biol Cell 28: 356, 2017]. Mol Biol Cell 27: 1797–1808, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrés-Delgado L, Antón OM, Bartolini F, Ruiz-Sáenz A, Correas I, Gundersen GG, et al.: INF2 promotes the formation of detyrosinated microtubules necessary for centrosome reorientation in T cells. J Cell Biol 198: 1025–1037, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.