Significance Statement
Although antibiotics have been associated with an increased risk of kidney stones, particularly early in life, perturbations of the gut microbiome and metabolome in early-onset nephrolithiasis have not been investigated. Using shotgun metagenomic sequencing and untargeted metabolomics of stool samples in a study of 44 children with kidney stones and 44 controls matched for age, sex, and race, the authors found that 31 bacterial taxa—including seven butyrate-producing taxa and three that degrade oxalate—were less abundant among children with calcium oxalate stones. Levels of 18 metabolites differed between cases and controls and correlated with the fecal bacteria that were less abundant among children with nephrolithiasis. Such disruptions in the gut microbiome and metabolome may thus be determinants of early-onset disease and may explain the association between antibiotics and nephrolithiasis.
Keywords: kidney stones, metabolism, intestine, pediatric nephrology
Abstract
Background
The relationship between the composition and function of gut microbial communities and early-onset calcium oxalate kidney stone disease is unknown.
Methods
We conducted a case-control study of 88 individuals aged 4–18 years, which included 44 individuals with kidney stones containing ≥50% calcium oxalate and 44 controls matched for age, sex, and race. Shotgun metagenomic sequencing and untargeted metabolomics were performed on stool samples.
Results
Participants who were kidney stone formers had a significantly less diverse gut microbiome compared with controls. Among bacterial taxa with a prevalence >0.1%, 31 taxa were less abundant among individuals with nephrolithiasis. These included seven taxa that produce butyrate and three taxa that degrade oxalate. The lower abundance of these bacteria was reflected in decreased abundance of the gene encoding butyryl-coA dehydrogenase (P=0.02). The relative abundance of these bacteria was correlated with the levels of 18 fecal metabolites, and levels of these metabolites differed in individuals with kidney stones compared with controls. The oxalate-degrading bacterial taxa identified as decreased in those who were kidney stone formers were components of a larger abundance correlation network that included Eggerthella lenta and several Lactobacillus species. The microbial (α) diversity was associated with age of stone onset, first decreasing and then increasing with age. For the individuals who were stone formers, we found the lowest α diversity among individuals who first formed stones at age 9–14 years, whereas controls displayed no age-related differences in diversity.
Conclusions
Loss of gut bacteria, particularly loss of those that produce butyrate and degrade oxalate, associates with perturbations of the metabolome that may be upstream determinants of early-onset calcium oxalate kidney stone disease.
Kidney stone disease (nephrolithiasis) is highly prevalent, increasingly common, and is characterized by painful stone events that cause considerable morbidity. In addition, as a disorder of mineral metabolism, nephrolithiasis is associated with increased risks of kidney function loss,1,2 decreased bone2pt mineral density and fracture,3−5 and cardiovascular disease.6−8 Kidney stones affect one in 11 people in the United States9 and result in annual healthcare costs >$10 billion.10 The prevalence of nephrolithiasis has increased by 70% over 20 years.9,11 Our group and others have discovered disproportionate increases in the incidence of nephrolithiasis among children, adolescents, and women.12−14 The shift in nephrolithiasis to a younger age of onset has caused increasing hospitalizations, surgeries, and healthcare expenditures.15 In addition, the morbidity associated with nephrolithiasis appears to be more pronounced in younger individuals, and stone recurrence rates may be higher in children than adults.4,16 The reasons for this shift in the epidemiology of nephrolithiasis are unclear. However, the rapidity of the change suggests that the driving forces are external exposures such as diet and antibiotics. Many of these exposures may disrupt the gut-kidney axis, which is the complex interplay between the intestinal and urinary tracts in human health and disease.17
Prior investigations have demonstrated perturbations of the gut microbiome among adults with nephrolithiasis. These studies found that the gut microbiome of those who formed kidney stones is less diverse than controls18,19 and that bacteria that degrade oxalate were less abundant in the stool of adults with kidney stones.20 Currently, the composition and function of the gut microbiome and metabolome among individuals with early-onset calcium oxalate kidney stone disease, the most common form of nephrolithiasis, is unknown.21 Discovery of the identity and function of microbial communities and downstream metabolites perturbed in early-onset calcium oxalate kidney stone disease could reveal targets for novel therapeutics for kidney stone prevention across the life span and help determine causes of the rapid shift in the epidemiology of nephrolithiasis.
Methods
Study Design and Population
We conducted a matched case-control study of 88 individuals aged 4–18 years who received care in The Children’s Hospital of Philadelphia (CHOP) healthcare system. We excluded individuals who took antibiotics in the last 3 months and those with inflammatory bowel disease, prior bariatric surgery, monogenic causes of kidney stones, cancer, immobility, cystic fibrosis, celiac disease, diabetes, congenital anomalies of the kidney and urinary tract, urinary tract obstruction, and renal tubular acidosis. The Institutional Review Board at CHOP approved this study.
Cases included individuals with incident and recurrent kidney stones consisting of 100% calcium (of which at least 50% was calcium oxalate) that spontaneously passed or were removed surgically within the prior 3 years (stone analysis done by mass spectroscopy at various clinical laboratories). We used a 3-year time window since the last stone event to capture individuals most likely to have active stone disease.16 Cases were recruited during outpatient visits to the CHOP Kidney Stone Center.
Controls were healthy volunteers matched to cases on age (±2 years), sex, race, and ethnicity. The study was nested within the CHOP healthcare system so that cases and controls arose from the same source population, and equal application of eligibility criteria for both cases and controls ensured that both groups were healthy, with the exception that cases were individuals with kidney stones. To increase study efficiency, participants with kidney stones were first matched to healthy participants (n=17) who had enrolled as controls in an independent study that used the same stool collection and exposure questionnaire as our study. These control participants were recruited from the CHOP emergency and dermatology departments, oral surgery clinic, and urgent care centers. The remaining 27 control participants were recruited from seven suburban and urban practices in the Pediatric Research Consortium, which includes the network of CHOP primary care practices in Pennsylvania and New Jersey.
Study Procedures
Participants completed a baseline questionnaire, including past medical history, early lifetime exposures (e.g., vaccinations), tobacco use, recent hospitalizations, and probiotic use. In addition, research nutritionists administered 24-hour dietary recalls over the telephone on 3 days (2 weekdays, 1 weekend day) to estimate participants’ daily nutrient and mineral intake. The 24-hour dietary recalls were collected using the Nutrition Data System for Research developed at the University of Minnesota.22 Dietary intake data were gathered by a multiple-pass interview approach,23 and values for 165 nutrients, nutrient ratios, and other food components were generated from a database that includes >18,000 foods.24,25 Antibiotics, including route and class, and medications taken over the last 1 year were ascertained by (1) self-report or interview of caregivers, (2) prescriptions from CHOP, and (3) antibiotics reconciled in the electronic health record. Age at nephrolithiasis diagnosis was determined through chart review. Stool specimens were collected at the participants’ homes, kept chilled, and shipped in an insulated container on ice packs. Stool stabilizer was not used. All specimens were shipped by same-day delivery and arrived at the CHOP Microbiome Center within 12–24 hours. At the time of receipt, specimen temperature was checked. Any specimen >20°C was rejected. Of the 88 original specimens received, only one had to be recollected due to a temperature upon arrival of >20°C. All study procedures were completed within 3 months of enrollment.
Shotgun Metagenomics
Upon arrival, stool samples were maintained at −80°C until being processed. Genomic DNA was extracted from stool and prepared for shotgun metagenomic sequencing using the Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA). DNA extraction blanks and DNA-free water were included as negative control samples to assess environmental and reagent contamination. Laboratory-generated mock communities consisting of DNA from Vibrio campbellii, Cryptococcus diffluens, and λ phage were included as positive controls. DNA sequencing was carried out on an Illumina HiSeq 2500 instrument, generating 125 bp paired-end sequence reads.
Reads were quality filtered and trimmed to remove adapter sequences using Trimmomatic version 0.36.26 Reads aligning to the human host genome (version hg38) were removed using BWA version 0.7.17-r1188.27 Taxonomic annotations were generated by Kraken version 1.0 using the standard database with all complete bacterial, archaeal, and viral genomes in the National Center for Biotechnology Information’s RefSeq.28 To assess gene function, reads were aligned to the KEGG database of gene ortholog sequences29 using Diamond version 0.9.24.30 Oxalate-degrading organisms were identified using the lists compiled in Ticinesi et al.20 and Miller and Dearing.31 Butyrate-producing organisms were identified by searching bacterial species data in Bergey’s Manual of Systematics of Archaea and Bacteria.32
The Shannon index and richness for α diversity analysis was calculated using the vegan package version 2.5-6 (https://CRAN.R-project.org/package=vegan) in R (version 3.6.0). The association between α diversity and study group was evaluated using a paired t test. To test for the association of taxon abundance with study group, we log-transformed the taxon proportions and applied a paired t test. Taxa were tested if the abundance in any sample exceeded 0.1%. Oxalobacter formigenes and Oxalobacteraceae were also tested, despite lower abundance, due to their role in oxalate degradation. To correct for multiple comparisons, we adjusted P values using the method of Benjamini–Hochberg to control the false discovery rate (FDR).33 To identify bacteria increasing or decreasing in abundance with age of first stone among participants with kidney stones, a linear model was used to test the association between the log-transformed relative abundance of taxa with linear and quadratic terms of the age at stone disease diagnosis for cases and their corresponding matched controls. Taxa were included in the latter analysis only if their relative abundance was at least 0.1% in any individual who formed kidney stones.
Untargeted Metabolomics
Metabolomic profiling (Metabolon, Inc., Durham, NC) was performed using a Waters Acquity Ultra-Performance Liquid Chromatography and a Thermo Scientific Q-Exactive high-resolution/accurate mass spectrometer interfaced with a heated electrospray ionization source and Orbitrap mass analyzer operated at 35,000 mass resolution. Metabolites were identified by comparison to library entries of purified standards of >3300 commercially available purified standard compounds, and concentrations were quantified using the area under the curve. Missing values were imputed with the minimum value, and metabolite values were normalized by registering the run-day medians to equal one to correct for variation resulting from instruments.34
Principal components analysis (PCA) and linear discriminant analysis (LDA) were carried out to identify fecal metabolomic characteristics of the study groups. The PCA and LDA included 841 out of 867 measured, named biochemicals after excluding noninformative (largely xenobiotic drug) metabolites. For PCA and LDA, metabolite values were normalized such that the mean equaled zero and the SD equaled one. The paired-sample t test was also performed to test the association between metabolite level and study group. We correlated metabolites and species abundances that differed between groups and validated the correlations by permutation testing to evaluate the possibility that spurious correlations may have been detected by coincidence, particularly when a metabolite and taxon were both identified as higher in cases and lower in controls. Specifically, we generated the null distribution of correlation values by randomly shuffling the taxon abundances within cases and controls. Accordingly, we assessed the significance of the correlation between metabolite level and abundance of bacterial species using P values obtained from permutation tests and identified as statistically significant only metabolite-taxon pairs that were more correlated than expected while controlling for the study group.
Results
Characteristics of Study Population
The study consisted of 44 individuals with incident or recurrent calcium oxalate kidney stones that occurred at ≤18 years of age and 44 healthy controls (Table 1). Those who were stone formers were representative of the pediatric kidney stone population in the United States,35 with a median age of 15.6 years at time of study, 13 years at stone diagnosis, 52% female and 98% white race. Of the participants with kidney stones, 59% had recurrent kidney stones and 68% had a family history of nephrolithiasis. Antibiotic exposures are summarized in Table 2, and dietary intakes and current medications of the study population are presented in Supplemental Tables 1 and 2. Dietary oxalate (P=0.05), fructose (P=0.03), and protein (P=0.04) intake was higher among control participants, and antibiotic exposure within 3–12 months of enrollment was higher among those with kidney stones (P<0.001). For the 36 participants with kidney stones and 31 controls for whom data were available on antibiotic exposure within the first 3 years of life, this exposure was also higher among those who formed kidney stones (P=0.03).
Table 1.
Population characteristics
| Characteristics | Participants with Kidney Stones (n=44) | Controls (n=44) |
|---|---|---|
| Median age at enrollment, yr (IQR) | 15.6 (11.8, 17.1) | 15.6 (12.4, 16.5) |
| Median age at first stone, yr (IQR) | 13 (8.5, 15) | N/A |
| Sex, no. (%) | ||
| Male | 21 (47.7) | 21 (47.7) |
| Female | 23 (52.3) | 23 (52.3) |
| Ethnicity, no. (%) | ||
| Hispanic or Latino | 3 (6.8) | 2 (4.5) |
| Not Hispanic or Latino | 40 (90.9) | 42 (95.5) |
| Missing | 1 (2.3) | 0 |
| Race, no. (%) | ||
| Black | 1 (2.3) | 2 (4.5) |
| White | 43 (97.7) | 40 (90.9) |
| Multiple | 0 | 2 (4.5) |
| Median body mass index, percentile (IQR) | 67.6 (28.5, 91.8) | 61.4 (40.3, 80.1)a |
| Stone activity, no. (%)b | ||
| Quiescent | 21 (47.7) | N/A |
| Active | 23 (52.3) | N/A |
| Recurrent, no. (%)c | ||
| No | 18 (40.9) | N/A |
| Yes | 26 (59.1) | N/A |
| Family history, no. (%) | ||
| No | 14 (31.8) | N/A |
| Yes | 30 (68.2) | N/A |
IQR, interquartile range; N/A, not applicable.
Three missing, n=41.
Quiescent is the absence of growth of an existing stone and no formation of a new stone in the year before specimen collection. Active is the growth of an existing stone or formation of a new stone in the year before specimen collection.
Recurrent stone formers are participants with a new symptomatic stone or new stone formation on imaging between their first stone and the date of specimen collection.
Table 2.
Antibiotic exposure within 3–12 months
| Exposure | Count (%) | P Valuea | |
|---|---|---|---|
| Participants with Kidney Stones (n=39) | Controls (n=37) | ||
| Any antibiotic | 28 (72) | 9 (24) | <0.001 |
| 1 | 7 (18) | 5 (14) | |
| 2 | 11 (28) | 4 (11) | |
| 3 | 6 (15) | 0 (0) | |
| 4 | 1 (3) | 0 (0) | |
| 6 | 1 (3) | 0 (0) | |
| 7 | 2 (5) | 0 (0) | |
| Sulfa | 7 (18) | 0 (0) | 0.06 |
| 1 | 6 (15) | 0 (0) | |
| 2 | 1 (3) | 0 (0) | |
| Cephalosporin | 18 (46) | 1 (3) | <0.001 |
| 1 | 8 (21) | 1 (3) | |
| 2 | 8 (21) | 0 (0) | |
| 3 | 1 (3) | 0 (0) | |
| 4 | 1 (3) | 0 (0) | |
| Fluoroquinolone | 8 (21) | 0 (0) | 0.02 |
| 1 | 4 (10) | 0 (0) | |
| 2 | 3 (8) | 0 (0) | |
| 3 | 1 (3) | 0 (0) | |
| Nitrofurantoin | 0 (0) | 0 (0) | — |
| Penicillin | 7 (18) | 3 (8) | 0.29 |
| 1 | 6 (15) | 3 (8) | |
| 2 | 1 (3) | 0 (0) | |
| Broad spectrum penicillin | 0 (0) | 0 (0) | — |
| Metronidazole | 0 (0) | 0 (0) | — |
| Macrolide | 3 (8) | 5 (14) | 0.73 |
| 1 | 2 (5) | 3 (8) | |
| 2 | 1 (3) | 2 (5) | |
| Helicobacter pylori treatment | 0 (0) | 0 (0) | — |
| Tetracycline | 2 (5) | 0 (0) | 0.50 |
| 1 | 1 (3) | 0 (0) | |
| 2 | 1 (3) | 0 (0) | |
| Antimycobacterial | 0 (0) | 0 (0) | — |
| Lincosamide | 1 (3) | 0 (0) | >0.99 |
| 2 | 1 (3) | 0 (0) | |
| Aminoglycoside | 2 (5) | 0 (0) | 0.50 |
| Unknown class | 0 (0) | 1 (3) | >0.99 |
| 2 | 0 (0) | 1 (3) | |
Exact McNemar significance probability.
Shotgun Metagenomic Sequencing Reveals Taxonomic and Gene Alterations in Children with Kidney Stones
The overall taxonomic profile of the gut microbiome among participants with kidney stones and controls was similar to that observed in previous studies,36 where the Bacteroidetes and Clostridia accounted for most of the bacterial population (Supplemental Figure 1). Among the Bacteroidetes, the species Bacteroides vulgatus was especially prominent in a subset of samples, with abundance values >40%.
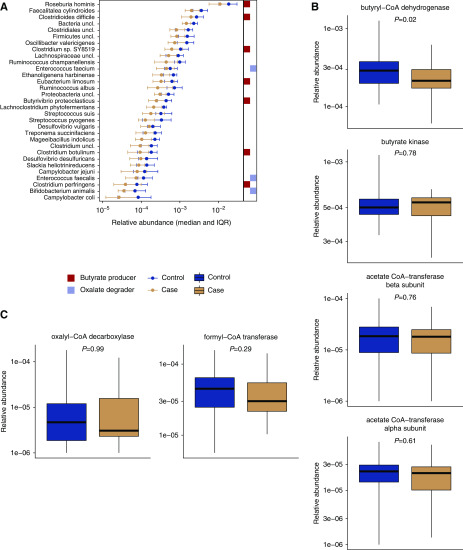
To compare the taxonomic composition of all participants, we tested 91 bacterial taxa that had >0.1% abundance in at least one sample. We found that 31 bacterial taxa differed in abundance between participants with kidney stones and controls at a pre-defined threshold of FDR adjusted P<0.05 (Figure 1A). Of the taxa identified, all were less abundant among participants who were stone formers than controls. These included seven taxa that produce the short-chain fatty acid butyrate, including several Roseburia and Clostridium species. Correspondingly, the gene abundance of butyryl-coA dehydrogenase, a key bacterial enzyme in the butyrate production pathway, was lower among those who were stone formers (P=0.02, Figure 1B).
Figure 1.
The relative abundance of 32 bacterial taxa and the abundance of the bacterial gene butyryl-CoA dehydrogenase were lower among participants with kidney stone disease. (A) Taxa identified as differentially abundant in those with kidney stones compared with controls. Butyrate-producing and oxalate-degrading species are marked to the right of the plot. Relative abundance of bacterial genes for (B) butyrate production and (C) oxalate degradation. IQR, interquartile range.
Sequencing also revealed a lower abundance of three oxalate-degrading bacterial taxa in participants with kidney stones: Enterococcus faecalis, Enterococcus faecium, and Bifidobacterium animalis. We detected the oxalate-degrading species O. formigenes at low relative abundance in two subjects from the control group (Supplemental Figure 1). Neither the presence/absence nor relative abundance of O. formigenes were different between participants with kidney stones and controls due to the small number of samples where it was detected (P=0.4 for relative abundance, P=0.5 for presence/absence). We detected a small proportion of reads assigned to the Oxalobacteraceae family in all controls and in all but one participant with kidney stones. There was no difference in Oxalobacteraceae abundance (P=0.4). We tested for an association between oxalate intake and bacterial species abundance and found no correlations after correction for multiple comparisons. None of the oxalate-degrading species were associated with oxalate intake, even before accounting for multiple comparisons. We aligned reads to a database of bacterial gene sequences and tested for a difference in the abundance of oxalyl-CoA decarboxylase or formyl-CoA transferase, bacterial genes involved in oxalate degradation (Figure 1C). We found that the abundance of these genes was not different between groups (P≥0.29).
Fecal Metabolome of Those with Kidney Stones Is Correlated with Bacterial Abundance
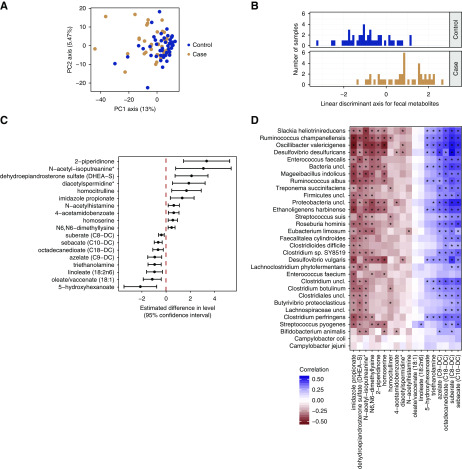
We carried out untargeted metabolomics of fecal samples to identify metabolite products that may be associated with differences in the microbiome. As with the taxonomic analysis, the overall profile of metabolites was similar between participants with kidney stones and controls (Figure 2A). We carried out an LDA to determine if a subset of the metabolites could be used to distinguish the two groups, and found that a linear discriminant separated those with kidney stones from controls (Figure 2B) with 77% accuracy. Thus, we found a subset of fecal metabolites that differed in participants with kidney stones.
Figure 2.
The fecal metabolome is distinct in participants with kidney stone disease. (A) PCA of untargeted fecal metabolite survey. (B) LDA distinguishes those with kidney stones from controls with 77% accuracy. (C) Metabolites increased (right) and decreased (left) in those who form kidney stones. (D) Correlation of metabolite concentration and bacterial abundance. Stars indicate significant correlations, after controlling for study group. PC, principal component.
We compared metabolite abundances and identified 18 metabolites that were significantly different between participants with kidney stones and controlsat a pre-specified nominal P<0.01 (Figure 2C). Ten metabolites were more abundant in those who were stone formers versus controls, whereas eight metabolites were less abundant in those with kidney stones than controls. Most of the metabolites that were more abundant among those with kidney stones were classified under the superpathway of amino acids and derivatives, whereas metabolites less abundant in those with kidney stones were classified as lipids and lipid-like molecules.
We hypothesized that the levels of these metabolites could be associated with the relative abundance of bacterial taxa identified in our study. We computed a correlation matrix between metabolites and species abundances (Figure 2D), which revealed strong correlations for many of the taxon-metabolite pairs. E. faecalis was positively correlated with three amino acid derivatives as well as other metabolites. Other bacterial species were correlated with several metabolites, in both a positive and negative direction. Thus, we found a set of metabolites that distinguished those with kidney stones and controls, and which were highly correlated with a select group of bacterial taxa.
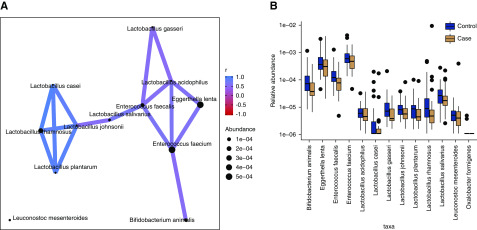
Oxalate-Degrading Bacteria in Those with Kidney Stones Form a Modular Network
Following our untargeted analysis, we focused on oxalate-degrading bacteria, which have received considerable attention in the literature on nephrolithiasis given the high prevalence of calcium oxalate kidney stones.37−39 We constructed an abundance correlation network for oxalate-degrading bacteria detected in this study (Figure 3A), and found that it consisted of positive correlations among oxalate-degrading taxa and lacked any strong negative correlations. The network consisted of two modules: one populated by relatively high-abundance, oxalate-degrading taxa and the other by low-abundance Lactobacilli (Figure 3B). In the high-abundance module, we observed that E. feacium was correlated with both Eggerthella lenta and E. faecalis. B. animalis was also connected to E. faecium, but was not otherwise attached to the module.
Figure 3.
Oxalate-degrading bacterial taxa identified as decreased among kidney stone formers were components of a larger correlation network. (A) Correlation network of oxalate-degrading bacteria observed in the study. Taxa are connected if the absolute correlation was >0.5. Node size corresponds to mean taxon abundance. (B) Abundance of oxalate-degrading species.
Three of the species attached to the high-abundance module were identified as lower in those who were stone formers versus controls: E. faecalis, E. faecium, and B. animalis. Moreover, these species were associated with metabolites that distinguished the groups. Thus, we observed that a module of oxalate-degrading bacteria varied in a coordinated manner that tracked with metabolite differences.
Children with Kidney Stones Exhibit a Unique Age-Dependent Microbiota Profile
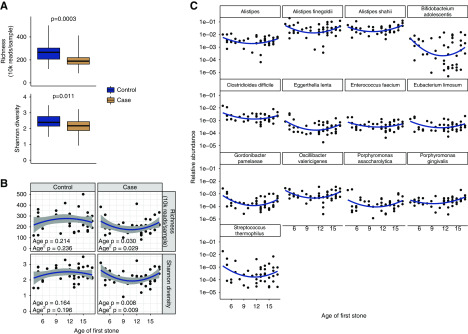
As expected, α diversity of the gut microbiome was lower in participants with kidney stones compared with controls, assessed by richness (P<0.001) and Shannon index (P=0.01; Figure 4A). In addition to an overall lower value in cases, the α diversity exhibited an age-dependent association in those with kidney stones but not in controls (Figure 4B). The association was not described by a simple linear relationship: bacterial diversity first decreased and then increased with age among those who were kidney stone formers (linear term P=0.030, quadratic term P=0.029 for richness, linear term P=0.008, quadratic term P=0.009 for Shannon index). The lowest α diversity was found among individuals who first formed kidney stones between 9 and 14 years of age. In contrast, the α diversity of the microbiome of control participants was similar across the age spectrum, and there were no significant associations found with age.
Figure 4.
The lowest diversity of the gut microbiome was found among individuals who first formed kidney stones between 9 and 14 years of age. (A) Species richness and Shannon diversity of bacterial communities in participants with kidney stones and controls. (B) Quadratic fits of diversity versus age of stone formation. (C) Candidate taxa associated with age-dependent diversity profile in those who form kidney stones.
In an exploratory analysis, we performed a regression analysis of bacterial taxon abundance to determine which bacteria might be associated with the age-dependent pattern of α diversity in those with kidney stones. We tested 77 taxa with a relative abundance of at least 0.1% in any participant who was a kidney stone former, and identified 13 taxa that exhibited a similar pattern as the overall α diversity (Figure 4C). For these taxa, the uncorrected P values for their associations with linear or quadratic terms for age at first stone were <0.05, but none were significantly associated after FDR correction. Two of the species, E. lenta and E. faecium, are oxalate degraders that were identified in our correlation network analysis. We observed partial overlap between age-dependent species and species identified as less abundant in participants with kidney stones.
Discussion
We found that the gut microbiome of children and adolescents with calcium oxalate kidney stone disease is less diverse than that of controls, consistent with prior studies in adults.18,19 In particular, butyrate-producing bacteria and a network of oxalate-degrading species were less abundant in children with calcium oxalate kidney stones compared with controls. In addition, levels of 18 metabolites, largely in the amino acid and lipid superfamilies, were different in the stool of youth with early-onset kidney stone disease compared with healthy children. These metabolites were correlated with the fecal bacteria that were less abundant in those with kidney stones versus controls. We also identified an age dependence of microbial diversity among those who were kidney stone formers, and we identified candidate bacteria that may underlie the age-dependent pattern observed. Collectively, our results highlight the function of the gut-kidney axis in early-onset calcium oxalate kidney stone disease and suggest that loss of bacteria, particularly those that produce butyrate and degrade oxalate, may act synergistically to influence gut metabolism and favor kidney stone formation.
Calcium oxalate kidney stone disease, which is the most common form of nephrolithiasis,21 occurs when calcium and oxalate become supersaturated in the urine and crystallization occurs. To date, evaluation and treatment of nephrolithiasis has focused on the kidney rather than upstream perturbations that may affect urine chemistries and other mediators of stone formation. The gut-kidney axis represents a potential causal pathway between the gut microbiome, intestinal metabolites, urine metabolites, and urine chemistries.17 In the context of kidney stone disease, this axis provides a framework for understanding how exposures that perturb the composition of the gut microbiome might have downstream effects on the intestinal and urinary tracts. Our group recently demonstrated that exposure to certain oral antibiotics was associated with a 1.3- to 2.3-fold increased odds of developing kidney stones.40 The risk was greatest for those exposed at younger ages, which is consistent with reports that exposure to antibiotics early in life produce greater alterations of host metabolism than those later in life.41 This study, in which prior antibiotic exposure was also higher among individuals with nephrolithiasis, provides a potential link between antibiotic exposure, gut dysbiosis,18,19 and nephrolithiasis.
In addition to an overall lower diversity of the gut microbiome, shotgun metagenomics revealed two themes of altered bacterial metabolism (decreased butyrate production and decreased oxalate degradation) among individuals with early-onset calcium oxalate kidney stone disease. Butyrate-producing organisms that were lower in abundance among those who formed kidney stones included Roseburia, which accounts for 1% of all bacteria present in the gut microbiome and is particularly sensitive to dietary intake.42 Butyrate is a short-chain fatty acid that is a mediator of inflammation43,44 and, importantly, helps maintain the gut mucosal barrier and regulates expression of SLC26 oxalate transporters in the intestine.45−47 These functions of butyrate suggest that loss of butyrate production would increase absorption of oxalate in the gut and lead to increased urinary oxalate excretion.
We also found lower abundance of oxalate-degrading bacteria, namely E. faecalis, E. faecium, and B. animalis. These three taxa were part of an oxalate-degrading network, which included lower abundance taxa in the Lactobacillus genus. However, we found no differences in the abundance of O. formigenes, which is an anaerobic gram-negative bacteria that has been the focus of much attention37,38,48 due to its use of oxalate as its carbon and energy source. Miller et al.49 have also reported a microbial network associated with oxalate degradation among adults, which included Ruminococcus, Bifidobacterium, and Oscillospira. In contrast to the untargeted approach we took, they focused their analysis on microbial networks involved in oxalate homeostasis (taxa associated with Oxalobacter species or those stimulated by oxalate in a rodent model). The absence of Oxalobacter in our samples could explain the differences in the composition of the oxalate-degrading network between studies, as could differences in the gut microbiome between children and adults. A recent study found that bacterial genes involved in oxalate degradation were less abundant in the stool of adults with kidney stones and showed that these genes were represented in several bacterial species (but not Oxalobacter), whose cumulative abundance inversely correlated with urine oxalate excretion.20 Although our study did not identify overall differences in gene abundance, we identified several oxalate-degrading species that were reduced in abundance, indicating a species-dependent difference in oxalate degradation function for the microbiome of children with kidney stones. Cumulatively, these results suggest that the community of oxalate-degrading bacteria might influence the development of calcium oxalate stone disease across the life span and is one that includes far more bacteria than O. formigenes.
We found strong evidence of perturbations in the gut metabolome among children with kidney stone disease. In particular, 18 metabolites differed between cases and controls, with ten metabolites more abundant and eight metabolites less abundant among individuals with nephrolithiasis. Most of the metabolites that were more abundant among individuals with nephrolithiasis were in the amino acid or derivatives superfamily, whereas most of the metabolites that were less abundant were in the lipid superfamily. These differences in fecal metabolites were largely explained by the lower abundance of the 31 specific bacterial taxa among individuals with early-onset nephrolithiasis, with oleate/vaccinate being the only metabolite not associated with any bacterial taxa. These metabolites could help focus future investigations that seek to identify the causal pathway between external exposures that perturb the gut microbiome and nephrolithiasis.
Our results provide the first evidence of an altered gut microbiome in early-onset kidney stone disease and the first evidence of an associated altered gut metabolome in individuals of any age with calcium oxalate nephrolithiasis. This study is also the first to use shotgun metagenomics to investigate the gut microbiome in nephrolithiasis, which improves resolution of species-level identification of bacteria and their biologic functions compared with 16S ribosomal RNA sequencing. This resolution allowed us to investigate overall and species-level differences in gut microbiome diversity by the age of disease onset. In particular, the lowest microbial diversity was found among children who first formed kidney stones between 9 and 14 years of age, with 13 taxa nominally associated with the age of nephrolithiasis onset. We found no association between age and α diversity in matched control samples. These results are consistent with our prior study, which demonstrated that the incidence rates of nephrolithiasis begin to increase at 10 years of age.14 We hypothesize that this nadir of microbial diversity may increase susceptibility to other well described exposures that increase the risk of incident nephrolithiasis, such as increased dietary sodium or low fluid intake, which were similar between participants with kidney stones and controls in this study. Although more work is needed to follow up on age-dependent associations in children who form kidney stones, we have identified a set of bacterial candidates worthy of further investigation.
Our findings provide insight into understanding the increase in the prevalence of nephrolithiasis and the shift toward an earlier age of onset.14 The rapidity of this change suggests that changes in external, potentially modifiable, factors are driving this change. It seems unlikely that traditional risk factors for nephrolithiasis, such as low fluid intake, have changed that dramatically to cause both a 70% increase in the prevalence of nephrolithiasis over the past two decades and rising incidence during childhood. However, antibiotic use has increased over this time period. Over 250 million antibiotic courses were prescribed in the United States in 2011, with the highest rates of administration for children <10 years old and women.50 Children receive more antibiotics than any other age group, and 30% of antibiotics prescribed during ambulatory care visits are inappropriate.51−53 Future studies should determine if the differences found in the gut microbiome and metabolome in early-onset kidney stone disease are due to antibiotic exposure. If confirmed in other studies, the unique microbial communities and their associated functions we identified could lead to new therapies for kidney stone prevention. For example, recent murine studies have demonstrated that downregulating the NLRP1-mediated inflammasome pathway can expand the community of organisms that produce butyrate, which may help prevent inflammatory bowel disease.54
We acknowledge there are several limitations to this study. As in all observational studies, unmeasured confounding and bias are possible. We reduced selection bias by matching on age, sex, and race, and nesting controls in the same health system from which participants with kidney stones were derived. We also extensively phenotyped participants and collected information on antibiotics and other exposures that affect the microbiome (e.g., diet and other medications). However, the reliance on self-report/interview for some of the medication exposures is a limitation, particularly for more remote prescriptions for which missingness was greater. Although the current sample size is too small to determine how diet and antibiotics perturb the gut microbiome and metabolome and affect kidney stone disease, we are currently expanding the population to perform these mediation analyses. It is also possible that those who form kidney stones, particularly children with a strong family history of kidney stones, could have genetic differences in intestinal physiology that affect the microbiome and metabolome as an alternate explanation for the differences observed. Second, this study could not assess the structure and function of the microbiome before stone formation. Third, microbiome data are high dimensional and thus require special considerations to reduce spurious results. In addition, many species and biochemicals appear rarely in microbiome and metabolomic data, respectively. Therefore, when conducting multiple tests, we controlled for a prespecified FDR, did not test rare species, and confirmed the significance of the taxon-metabolite associations with permutation testing. Fourth, butyrate, which is volatile, could not be directly measured in the untargeted metabolomics assay. We also did not analyze the urine microbiome or measure urine metabolites and chemistries. Future studies should extend analyses to urine to examine the effect of perturbations of the gut microbiome on the kidney. Finally, imaging was not obtained for controls. Thus, some may have had asymptomatic stones, reported to be found among 4% of adults, but would likely be markedly less common among children.55 The presence of stones in controls would bias results to the null, and would cause us to potentially underestimate associations with kidney stone formation.
Loss of gut bacteria, particularly those that produce butyrate and degrade oxalate, is associated with perturbations of the metabolome that may be upstream determinants of early-onset kidney stone disease.
Disclosures
Dr. Denburg reports grants from CHOP, National Center for Complementary and Integrative Health, NIDDK, Mallinckrodt Pharmaceuticals, Patient-Centered Outcomes Research Institute (PCORI), outside the submitted work. Dr. Tasian reports grants from CHOP (Foerderer grant), National Institutes of Health (NIH)/NIDDK, PCORI, Pennsylvania Department of Health, during the conduct of the study; and personal fees from Allena Pharmaceuticals and Lumenis Inc., outside the submitted work. All remaining authors have nothing to disclose.
Funding
This study was supported by funding from the CHOP Foerderer grant, the Commonwealth Universal Research Enhancement (CURE) program’s Tobacco Formula grant SAP #4100068710, and NIH/NIDDK grant K23DK106428 (to Dr. Tasian).
Supplementary Material
Acknowledgments
The views expressed in this article are those of the authors and do necessarily represent the official view of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2019101131/-/DCSupplemental.
Supplemental Table 1. Daily dietary intake in kidney stone formers versus controls.
Supplemental Table 2. Current/recent medication use in kidney stone formers versus controls.
Supplemental Figure 1. Heatmap of bacterial taxa in children with kidney stone disease and matched healthy controls.
References
- 1.Alexander RT, Hemmelgarn BR, Wiebe N, Bello A, Morgan C, Samuel S, et al.: Alberta Kidney Disease Network: Kidney stones and kidney function loss: A cohort study. BMJ 345: e5287, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denburg MR, Jemielita TO, Tasian GE, Haynes K, Mucksavage P, Shults J, et al.: Assessing the risk of incident hypertension and chronic kidney disease after exposure to shock wave lithotripsy and ureteroscopy. Kidney Int 89: 185–192, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carbone LD, Hovey KM, Andrews CA, Thomas F, Sorensen MD, Crandall CJ, et al. : Urinary tract stones and osteoporosis: Findings from the women’s health initiative. J Bone Miner Res 30: 2096–2102, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denburg MR, Leonard MB, Haynes K, Tuchman S, Tasian G, Shults J, et al.: Risk of fracture in urolithiasis: A population-based cohort study using the health improvement network. Clin J Am Soc Nephrol 9: 2133–2140, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor EN, Feskanich D, Paik JM, Curhan GC: Nephrolithiasis and risk of incident bone fracture. J Urol 195: 1482–1486, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexander RT, Hemmelgarn BR, Wiebe N, Bello A, Samuel S, Klarenbach SW, et al.; Alberta Kidney Disease Network: Kidney stones and cardiovascular events: A cohort study. Clin J Am Soc Nephrol 9: 506–512, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferraro PM, Taylor EN, Eisner BH, Gambaro G, Rimm EB, Mukamal KJ, et al.: History of kidney stones and the risk of coronary heart disease. JAMA 310: 408–415, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rule AD, Roger VL, Melton LJ 3rd, Bergstralh EJ, Li X, Peyser PA, et al.: Kidney stones associate with increased risk for myocardial infarction. J Am Soc Nephrol 21: 1641–1644, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scales CD Jr, Smith AC, Hanley JM, Saigal CS; Urologic Diseases in America Project: Prevalence of kidney stones in the United States. Eur Urol 62: 160–165, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearle MS, Calhoun EA, Curhan GC; Urologic Diseases of America Project: Urologic diseases in America project: Urolithiasis. J Urol 173: 848–857, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Stamatelou KK, Francis ME, Jones CA, Nyberg LM, Curhan GC: Time trends in reported prevalence of kidney stones in the United States: 1976-1994. Kidney Int 63: 1817–1823, 2003 [DOI] [PubMed] [Google Scholar]
- 12.Dwyer ME, Krambeck AE, Bergstralh EJ, Milliner DS, Lieske JC, Rule AD: Temporal trends in incidence of kidney stones among children: A 25-year population based study. J Urol 188: 247–252, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scales CD Jr, Curtis LH, Norris RD, Springhart WP, Sur RL, Schulman KA, et al.: Changing gender prevalence of stone disease. J Urol 177: 979–982, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Tasian GE, Ross ME, Song L, Sas DJ, Keren R, Denburg MR, et al.: Annual incidence of nephrolithiasis among children and adults in South Carolina from 1997 to 2012. Clin J Am Soc Nephrol 11: 488–496, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang HH, Wiener JS, Lipkin ME, Scales CD Jr, Ross SS, Routh JC: Estimating the nationwide, hospital based economic impact of pediatric urolithiasis. J Urol 193[Suppl 5]: 1855–1859, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tasian GE, Kabarriti AE, Kalmus A, Furth SL: Kidney stone recurrence among children and adolescents. J Urol 197: 246–252, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robijn S, Hoppe B, Vervaet BA, D’Haese PC, Verhulst A: Hyperoxaluria: A gut-kidney axis? Kidney Int 80: 1146–1158, 2011 [DOI] [PubMed] [Google Scholar]
- 18.Stern JM, Moazami S, Qiu Y, Kurland I, Chen Z, Agalliu I, et al.: Evidence for a distinct gut microbiome in kidney stone formers compared to non-stone formers. Urolithiasis 44: 399–407, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang R, Jiang Y, Tan A, Ye J, Xian X, Xie Y, et al.: 16S rRNA gene sequencing reveals altered composition of gut microbiota in individuals with kidney stones. Urolithiasis 46: 503–514, 2018 [DOI] [PubMed] [Google Scholar]
- 20.Ticinesi A, Milani C, Guerra A, Allegri F, Lauretani F, Nouvenne A, et al.: Understanding the gut-kidney axis in nephrolithiasis: An analysis of the gut microbiota composition and functionality of stone formers. Gut 67: 2097–2106, 2018 [DOI] [PubMed] [Google Scholar]
- 21.Lieske JC, Rule AD, Krambeck AE, Williams JC, Bergstralh EJ, Mehta RA, et al.: Stone composition as a function of age and sex. Clin J Am Soc Nephrol 9: 2141–2146, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feskanich D, Sielaff BH, Chong K, Buzzard IM: Computerized collection and analysis of dietary intake information. Comput Methods Programs Biomed 30: 47–57, 1989 [DOI] [PubMed] [Google Scholar]
- 23.Johnson RK, Driscoll P, Goran MI: Comparison of multiple-pass 24-hour recall estimates of energy intake with total energy expenditure determined by the doubly labeled water method in young children. J Am Diet Assoc 96: 1140–1144, 1996 [DOI] [PubMed] [Google Scholar]
- 24.Sievert YA, Schakel SF, Buzzard IM: Maintenance of a nutrient database for clinical trials. Control Clin Trials 10: 416–425, 1989 [DOI] [PubMed] [Google Scholar]
- 25.Schakel SF, Sievert YA, Buzzard IM: Sources of data for developing and maintaining a nutrient database. J Am Diet Assoc 88: 1268–1271, 1988 [PubMed] [Google Scholar]
- 26.Bolger AM, Lohse M, Usadel B: Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouyang Y, Ye F, Liang Y: A modified electronegativity equalization method for fast and accurate calculation of atomic charges in large biological molecules. Phys Chem Chem Phys 11: 6082–6089, 2009 [DOI] [PubMed] [Google Scholar]
- 28.Wood DE, Salzberg SL: Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15: R46, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S: KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchfink B, Xie C, Huson DH: Fast and sensitive protein alignment using DIAMOND. Nat Methods 12: 59–60, 2015 [DOI] [PubMed] [Google Scholar]
- 31.Miller AW, Dearing D: The metabolic and ecological interactions of oxalate-degrading bacteria in the mammalian gut. Pathogens 2: 636–652, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitman WB, editor: Bergey’s Manual of Systematics of Archaea and Bacteria, Hoboken, NJ, Wiley, 2015 [Google Scholar]
- 33.Benjamini Y, Hochberg Y: Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300, 1995 [Google Scholar]
- 34.Evans AM, Bridgewater BR, Liu Q, Mitchell MW, Robinson RJ, Dai H, et al. : High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics 4: 132, 2014 [Google Scholar]
- 35.Ward JB, Feinstein L, Pierce C, Lim J, Abbott KC, Bavendam T, et al.: NIDDK Urologic Diseases in America Project: Pediatric urinary stone disease in the United States: The urologic diseases in America project. Urology 129: 180–187, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Human Microbiome Project Consortium: Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufman DW, Kelly JP, Curhan GC, Anderson TE, Dretler SP, Preminger GM, et al.: Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J Am Soc Nephrol 19: 1197–1203, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwak C, Kim HK, Kim EC, Choi MS, Kim HH: Urinary oxalate levels and the enteric bacterium Oxalobacter formigenes in patients with calcium oxalate urolithiasis. Eur Urol 44: 475–481, 2003 [DOI] [PubMed] [Google Scholar]
- 39.Siener R, Bangen U, Sidhu H, Hönow R, von Unruh G, Hesse A: The role of Oxalobacter formigenes colonization in calcium oxalate stone disease. Kidney Int 83: 1144–1149, 2013 [DOI] [PubMed] [Google Scholar]
- 40.Tasian GE, Jemielita T, Goldfarb DS, Copelovitch L, Gerber JS, Wu Q, et al.: Oral antibiotic exposure and kidney stone disease. J Am Soc Nephrol 29: 1731–1740, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al.: Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158: 705–721, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Louis P, Flint HJ: Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 294: 1–8, 2009 [DOI] [PubMed] [Google Scholar]
- 43.Puddu A, Sanguineti R, Montecucco F, Viviani GL: Evidence for the gut microbiota short-chain fatty acids as key pathophysiological molecules improving diabetes. Mediators Inflamm 2014: 162021, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vieira EL, Leonel AJ, Sad AP, Beltrão NR, Costa TF, Ferreira TM, et al.: Oral administration of sodium butyrate attenuates inflammation and mucosal lesion in experimental acute ulcerative colitis. J Nutr Biochem 23: 430–436, 2012 [DOI] [PubMed] [Google Scholar]
- 45.Canani RB, Terrin G, Elce A, Pezzella V, Heinz-Erian P, Pedrolli A, et al.: Genotype-dependency of butyrate efficacy in children with congenital chloride diarrhea. Orphanet J Rare Dis 8: 194, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang Z, Asplin JR, Evan AP, Rajendran VM, Velazquez H, Nottoli TP, et al.: Calcium oxalate urolithiasis in mice lacking anion transporter Slc26a6. Nat Genet 38: 474–478, 2006 [DOI] [PubMed] [Google Scholar]
- 47.Landry GM, Hirata T, Anderson JB, Cabrero P, Gallo CJ, Dow JA, et al.: Sulfate and thiosulfate inhibit oxalate transport via a dPrestin (Slc26a6)-dependent mechanism in an insect model of calcium oxalate nephrolithiasis. Am J Physiol Renal Physiol 310: F152–F159, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azcarate-Peril MA, Bruno-Bárcena JM, Hassan HM, Klaenhammer TR: Transcriptional and functional analysis of oxalyl-coenzyme A (CoA) decarboxylase and formyl-CoA transferase genes from Lactobacillus acidophilus. Appl Environ Microbiol 72: 1891–1899, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller AW, Choy D, Penniston KL, Lange D: Inhibition of urinary stone disease by a multi-species bacterial network ensures healthy oxalate homeostasis. Kidney Int 96: 180–188, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hicks LA, Bartoces MG, Roberts RM, Suda KJ, Hunkler RJ, Taylor TH Jr, et al.: US outpatient antibiotic prescribing variation according to geography, patient population, and provider specialty in 2011. Clin Infect Dis 60: 1308–1316, 2015 [DOI] [PubMed] [Google Scholar]
- 51.Pichichero ME: Dynamics of antibiotic prescribing for children. JAMA 287: 3133–3135, 2002 [DOI] [PubMed] [Google Scholar]
- 52.Hicks LA, Taylor TH Jr, Hunkler RJ: U.S. outpatient antibiotic prescribing, 2010. N Engl J Med 368: 1461–1462, 2013 [DOI] [PubMed] [Google Scholar]
- 53.Fleming-Dutra KE, Hersh AL, Shapiro DJ, Bartoces M, Enns EA, File TM Jr, et al.: Prevalence of inappropriate antibiotic prescriptions among US ambulatory care visits, 2010-2011. JAMA 315: 1864–1873, 2016 [DOI] [PubMed] [Google Scholar]
- 54.Tye H, Yu CH, Simms LA, de Zoete MR, Kim ML, Zakrzewski M, et al.: NLRP1 restricts butyrate producing commensals to exacerbate inflammatory bowel disease. Nat Commun 9: 3728, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bansal AD, Hui J, Goldfarb DS: Asymptomatic nephrolithiasis detected by ultrasound. Clin J Am Soc Nephrol 4: 680–684, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.