Wild aquatic birds are the primary natural reservoir of influenza A viruses (IAVs) and are therefore responsible for the dispersal and maintenance of IAVs representing a broad range of antigenic and genetic diversity. The aims of IAV surveillance in waterfowl not only relate to understanding the risk of spillover risk to humans, but also to improving our understanding of basic questions related to IAV evolution and ecology. By evaluating several decades of surveillance data from wild aquatic birds sampled along North American migratory flyways, we discovered an annual trend of increasing subtype diversity during southbound migration, peaking on southern wintering grounds. Winter sampling revealed the highest proportion of mixed and rare infections that suggest higher opportunity for spillover. These findings allow improvements to surveillance efforts to robustly capture IAV diversity that will be used for vaccine development and cultivate a more thorough understanding of IAV evolution and persistence mechanisms.
KEYWORDS: ecology, hemagglutinin, influenza A virus, neuraminidase, season, subtype diversity, wild birds, waterfowl
ABSTRACT
The discovery in 1976 of waterfowl as the primary reservoir of influenza A viruses (IAVs) has since spurred decades of waterfowl surveillance efforts by researchers dedicated to understanding the ecology of IAV and its subsequent threat to human and animal health. Here, we employed a multidecade, continental-scale approach of surveillance data to understand trends of seasonal IAV subtype diversity. Between 1976 and 2015, IAVs were detected in 8,427 (10.8%) of 77,969 samples from migratory waterfowl throughout the Central and Mississippi Migratory Flyways in the United States and Canada. A total of 96 hemagglutinin (HA)/neuraminidase (NA) subtype combinations were isolated, which included most HA (H1 to H14) and all 9 NA subtypes. We observed an annual trend of high influenza prevalence, involving a few dominant subtypes, on northern breeding grounds during summer with progressively lowered influenza prevalence, comprised of a highly diverse profile of subtypes, as waterfowl migrate toward southern wintering grounds. Isolates recovered during winter had the highest proportion of mixed and rare HA/NA combinations, indicating increased opportunity for reassortment of IAVs. In addition, 70% of H5 and 49% of H7 IAV isolates were recovered from samples collected during fall and spring, respectively; these are subtypes that can have significant implications for public health and agriculture sectors. Annual cyclical dominance of subtypes on northern breeding grounds is revealed through the longitudinal nature of this study. Our novel findings exhibit the unrealized potential for discovery using existing IAV surveillance data.
IMPORTANCE Wild aquatic birds are the primary natural reservoir of influenza A viruses (IAVs) and are therefore responsible for the dispersal and maintenance of IAVs representing a broad range of antigenic and genetic diversity. The aims of IAV surveillance in waterfowl not only relate to understanding the risk of spillover risk to humans, but also to improving our understanding of basic questions related to IAV evolution and ecology. By evaluating several decades of surveillance data from wild aquatic birds sampled along North American migratory flyways, we discovered an annual trend of increasing subtype diversity during southbound migration, peaking on southern wintering grounds. Winter sampling revealed the highest proportion of mixed and rare infections that suggest higher opportunity for spillover. These findings allow improvements to surveillance efforts to robustly capture IAV diversity that will be used for vaccine development and cultivate a more thorough understanding of IAV evolution and persistence mechanisms.
INTRODUCTION
The significance of influenza A virus (IAV; family Orthomyxoviridae) to public health, wildlife health, and the agriculture industry, exemplifies the need of a One Health approach to understanding the epidemiology of such multihost pathogens, (1, 2). The IAV genome is composed of 8 unique RNA segments, which allows for frequent genomic reassortment and increased viral diversity (3, 4). In waterfowl, the primary reservoir for IAVs, these viruses exhibit subtype diversity that results from reassortment of two segments that encode surface glycoproteins hemagglutinin (HA) and neuraminidase (NA). IAVs are classified by various subtype combinations on the basis of antigenic variations within HA and NA. Currently, 16 HA and 9 NA subtypes have been isolated from wild aquatic birds, predominantly from the orders Anseriformes (ducks, geese, and swans) and Charadriiformes (gulls, terns, and waders) (5–7). In waterfowl, IAV replicates in cells lining the intestinal tract and is excreted in high concentrations in the feces, typically causing no apparent signs of infection (3). These viral characteristics, combined with the propensity of the host species for long-distance migration, create abundant opportunities for viral transmission (within and between species), dispersal, and reassortment.
In 1968, the detection of IAV antibodies in sera of wild birds provided initial evidence that migratory birds were involved in the epidemiology of these viruses (8). This observation was confirmed by isolations of IAVs from wild ducks in North America in 1973 (9). Subsequent isolations of IAVs from seabirds in Australia provided the first evidence that wild birds were infected with IAV globally (10). Surveillance activities in wild bird populations have been ongoing since these findings. Numerous collaborative research groups have since focused on understanding the ecology of IAVs and factors that increase the risk of pandemic influenza emergence at the human-animal interface.
The zoonotic and pandemic potentials of IAVs, coupled with the economic and production threats they pose to the poultry and livestock industries, have kept IAVs at the forefront of global attention (11). All past human pandemic influenza viruses originated through reassortment of avian gene segments with contemporary human strains (12–14). Further, the pathogenicity and destructive capacity of IAVs in commercial poultry often results in enormous global economic burden and losses. In 2014 to 2015, several outbreaks of highly pathogenic avian influenza H5Nx viruses were reported in the United States, resulting in the depopulation of 50.5 million commercial birds across 21 states and an estimated cost of $3.3 billion to the economy (15). Given these threats, IAV surveillance in wild birds is centered on understanding the dynamics of viral ecology in the natural reservoir and capturing the viral diversity in these reservoirs, both of which can improve our understanding of IAV risk in other species.
In this study, we aggregated 4 decades of IAV-related data to investigate ecological and epidemiological trends over time. Our exploratory analysis used IAV surveillance data from migratory waterfowl in the Mississippi Migratory Flyway (MMF) and Central Migratory Flyway (CMF) between 1976 and 2015. We examined patterns of seasonal subtype diversity as they correspond to waterfowl migration. Our finding of a progressive increase in subtype diversity during southbound migration indicates that subtype diversity can be effectively captured through year-round IAV surveillance in waterfowl, with an emphasis on winter sampling.
RESULTS
The aggregate data set, resulting from the merging of data from three research groups, included a total of 77,969 samples from wild birds collected over 40 years from 14 U.S. states and 3 Canadian provinces at 295 sampling sites concentrated along the MMF and CMF (Fig. 1A). Fifty species from orders Anseriformes (n = 42 species, waterfowl), Gruiformes (n = 6, coots), and Podicipediformes (n = 2, grebes) were represented (Table 1). A total of 8,427 wild bird-origin IAV isolates were recovered from the 77,969 wild birds sampled, yielding an overall virus isolation frequency of 11%.
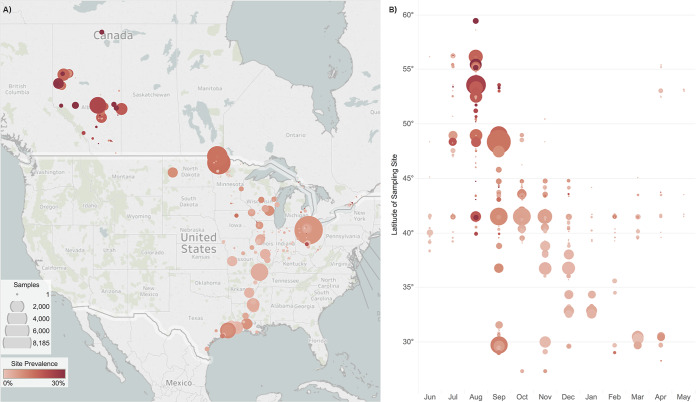
FIG 1.
Spatial and temporal distribution of influenza A virus (IAV) sampling sites of waterfowl in North America between 1976 and 2015. Latitude of map on the left (A) approximately aligns with the latitude shown on the y axis on the right (B) for a clear context. The red color gradation of circles gives IAV prevalence, with the darkest shade indicating ≥30% prevalence. The size of circles represents the relative number of samples collected at a specific study site or latitude. The average latitude of sampling sites was 51.2°N during summer (June to August), 41.0°N during fall (September to November), 35.5°N during winter (December to February), and 28.3°N during spring (March to May). Migratory activity in fall and spring relates to the wider range of sampling latitudes: summer range, 21.2°; fall, 26.2°; winter, 15.7°; and spring, 27.1°.
TABLE 1.
Samples collected and influenza A virus prevalence in individual waterfowl species in North America between 1976 and 2015
| Group | Species common name (scientific name) | No. samples | Percent IAV prevalence (no. isolates) |
|---|---|---|---|
| Dabbler | Mallard (Anas platyrhynchos) | 30,823 | 17 (5,287) |
| Blue-winged teal (Anas discors) | 18,423 | 8 (1,474) | |
| American green-winged teal (Anas crecca carolinensis) | 7,113 | 5 (377) | |
| Northern pintail (Anas acuta) | 4,409 | 19 (822) | |
| Wood duck (Aix sponsa) | 2,974 | 0.6 (18) | |
| Gadwall (Anas strepera) | 2,947 | 2 (60) | |
| Northern shoveler (Anas clypeata) | 2,379 | 6 (151) | |
| American wigeon (Anas americana) | 984 | 4 (44) | |
| American black duck (Anas rubripes) | 489 | 9 (46) | |
| American black duck X mallard hybrid (Anas rubripes X platy) | 92 | 8 (7) | |
| Mottled duck (Anas fulvigula) | 35 | ||
| Unidentified teal | 25 | 12 (3) | |
| Cinnamon teal (Anas cyanoptera) | 7 | 29 (2) | |
| Black-bellied whistling duck (Anas Dendrocygna autumnalis) | 3 | ||
| Mallard X gadwall hybrid (Anas platy X strepera) | 1 | ||
| Divers | Ring-necked duck (Aythya collaris) | 984 | 2 (17) |
| Redhead (Aythya americana) | 792 | 5 (36) | |
| Lesser scaup (Aythya affinis) | 491 | 3 (13) | |
| Canvasback (Aythya valisineria) | 312 | 2 (6) | |
| Greater scaup (Aythya marila) | 147 | 3 (4) | |
| Hooded merganser (Lophodytes cucullatus) | 137 | 0.7 (1) | |
| Ruddy duck (Oxyura jamaicensis) | 73 | 4 (3) | |
| Common merganser (Mergus merganser) | 30 | ||
| Red-breasted merganser (Mergus serrator) | 16 | ||
| Unidentified scaup | 12 | ||
| Sea ducks | Long-tailed duck (Clangula hyemalis) | 349 | 3 (10) |
| Bufflehead (Bucephala albeola) | 212 | 8 (17) | |
| Common goldeneye (Bucephala clangula) | 140 | 4 (6) | |
| White-winged scoter (Melanitta deglandi) | 29 | 3 (1) | |
| Black scoter (Melanitta americana) | 12 | 8 (1) | |
| Surf scoter (Melanitta perspicillata) | 3 | ||
| Other | Canada goose (Branta canadensis) | 2,172 | 0.2 (5) |
| Snow goose (Chen caerulescens) | 648 | 0.2 (1) | |
| American coot (Fulica americana) | 349 | 1 (5) | |
| Mute swan (Cygnus olor) | 85 | ||
| Sandhill crane (Grus canadensis) | 67 | ||
| Greater white-fronted goose (Anser albifrons) | 58 | 2 (1) | |
| Ross's goose (Chen rossii) | 39 | ||
| Virginia rail (Rallus limicola) | 22 | ||
| Red-necked grebe (Podiceps grisegena) | 19 | 32 (6) | |
| Unidentified duck | 17 | 18 (3) | |
| Pied-billed grebe (Podilymbus podiceps) | 14 | ||
| Domestic duck | 7 | ||
| Domestic goose | 6 | ||
| Trumpeter swan (Cygnus buccinator) | 6 | ||
| Sora rail (Porzana carolina) | 4 | ||
| Lesser White-Fronted Goose (Anser erythropus) | 3 | ||
| Unidentified grebe | 3 | ||
| Common gallinule (Gallinula galeata) | 2 | ||
| Brant (Branta bernicla) | 1 | ||
| Cackling goose (Branta hutchinsii) | 1 | ||
| King rail (Rallus elegans) | 1 | ||
| Red-breasted goose (Branta ruficollis) | 1 | ||
| Tundra swan (Cygnus columbianus) | 1 |
Surveillance distribution.
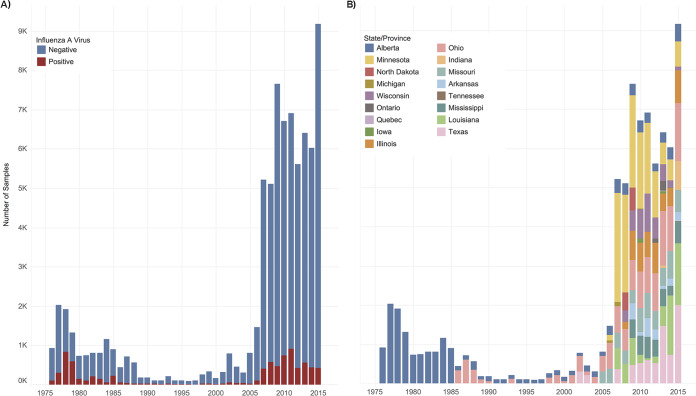
IAV surveillance efforts and success understandably varied over the 4 decades of data collection, due to inherent year-to-year variations in waterfowl migration and external factors, such as variations in funding allocations, research priorities, and site access. In the first decade of surveillance (1976 to 1985), an average of 1,136 samples were collected per year, exclusively from Alberta (Fig. 1B; Table 2). Ninety-five percent (n = 10,799 samples) of samples collected between 1976 and 1985 were obtained during the summer months (June 1 to August 31), with the remaining 2% (n = 214) and 3% (n = 348) collected during fall (September 1 to November 30) and spring (March 1 to May 31), respectively. An average of 272 and 362 samples per year were collected in Alberta, Ohio, Missouri, and Texas during the second (1986 to 1995) and third (1996 to 2005) decades, respectively. In 1986 to 1995, 48% and 50% of samples were collected during summer and fall, respectively, and only 3% during spring sampling months. In 1996 to 2005, 50% of samples were collected during summer, 39% during fall, 10% during winter (December 1 to February 28), and 1% during spring months. The average number of samples collected per year was the highest (n = 6,026) in the fourth decade (2006 to 2015) and was most inclusive of water or wetland habitats along the extent of the MMF and CMF. This decade was also most inclusive of sampling throughout the year, with 16% of samples collected during summer, 59% during fall, 15% during winter, and 9% during spring.
TABLE 2.
Samples collected and influenza A virus prevalence from migratory waterfowl in North America between 1976 and 2015, stratified by various predictors
| Predictor | No. samples | Percent IAV prevalence (no. isolates) |
|---|---|---|
| Decade | ||
| 1976–1985 | 11,361 | 24 (2,749) |
| 1986–1995 | 2,721 | 11 (304) |
| 1996–2005 | 3,624 | 8 (303) |
| 2006–2015 | 60,263 | 8 (5,071) |
| Location | ||
| Alberta | 16,523 | 22 (3,626) |
| Arkansas | 1,772 | 2 (37) |
| Illinois | 4,857 | 3 (150) |
| Indiana | 804 | 0.3 (2) |
| Iowa | 183 | 14 (26) |
| Louisiana | 5,725 | 4 (247) |
| Michigan | 190 | 6 (11) |
| Minnesota | 14,396 | 15 (2,219) |
| Mississippi | 3,176 | 3 (94) |
| Missouri | 5,181 | 3 (156) |
| North Dakota | 1,050 | 9 (96) |
| Ohio | 12,731 | 8 (975) |
| Ontario | 357 | 5 (18) |
| Quebec | 7 | |
| Tennessee | 3 | |
| Texas | 7,226 | 7 (520) |
| Wisconsin | 3,788 | 7 (250) |
| Gender | ||
| Female | 29,242 | 12 (3,520) |
| Male | 42,981 | 11 (4,620) |
| Unknown | 5,746 | 5 (287) |
| Age | ||
| Juvenile | 41,296 | 15 (6,387) |
| Adult | 31,439 | 6 (1,779) |
| Unknown | 5,234 | 5 (261) |
| Season | ||
| Summer | 23,829 | 19 (4,457) |
| Fall | 38,792 | 9 (3,501) |
| Winter | 9,507 | 3 (253) |
| Spring | 5,841 | 4 (216) |
Of the 77,969 samples, 31% (n = 23,829 samples) were collected in summer, 50% (38,792) in fall, and 12% (9,507) and 7% (5,841) were collected in winter and spring, respectively (Table 2). August and September were the overall peak sampling months, representing 25% (n = 19,632) and 29% (22,737) of all samples collected, respectively, due to increased waterfowl banding and the first early waterfowl hunting seasons, both of which accounted for increased access to wild birds. As birds inhabit northern breeding grounds during summer months for breeding, marshaling, and initiation of migration, the average summer collection site latitude was 51.2°N, which approximately passes through the city of Calgary in Alberta (summer sampling range: 59.7° to 38.3°N). As migrating birds retreat toward southern wintering grounds, the average collection latitude during winter months was 35.5°N, which defines the southern border of Tennessee (winter sampling range: 49.7° to 29.0°N) (Fig. 1B). Due to the expanse of migratory activity that occurs during fall and spring months, sample collection spanned a wider range of latitudes during these seasons (fall sampling range: 59.74° to 27.3°N; spring sampling range: 55.4° to 28.3°N). The association between time and sampling location demonstrates that both these variables are associated with bird ecology within this aggregate data set.
Of all samples, 81% (n = 63,039 samples) were collected by cloacal swabs, 14% (n = 11,129) by paired cloacal and oropharyngeal swabs from the same bird, and 5% (n = 3,801) by various combinations of sampling methods or tissue collections. Of the 8,427 IAV isolates, 56% (4,736 isolates) were subtyped by genomic methods, 42% (3,568) by antigenic methods, and 2% (123) by both methods. There was an apparent bias by subtype characterization method relative to sampling season, and all pairwise two-sample tests of proportions by season were significant (P ≤ 0.01; Table 3).
TABLE 3.
Organizations that provided influenza A virus surveillance data from migratory waterfowl in North America between 1976 and 2015 displayed by subtyping method and season of sample collection
| Organization | No. summer | No. fall | No. winter | No. spring | No. grand total (% of total samples) |
|---|---|---|---|---|---|
| The Ohio State University (OSU) | |||||
| Negative IAV | 3,416 | 18,201 | 8,159 | 917 | 30,693 (39.37) |
| Positive IAV | 368 | 1,098 | 221 | 12 | 1,699 (2.18) |
| Antigenic characterization | 162 | 433 | 56 | 11 | 662 |
| Genomic characterization | 206 | 665 | 165 | 1 | 1,037 |
| OSU Total | 3,784 | 19,299 | 8,380 | 929 | 32,392 (41.54) |
| St. Jude Children's Research Hospital (SJ) | |||||
| Negative IAV | 12,294 | 256 | 347 | 12,897 (16.54) | |
| Positive IAV | 3,547 | 78 | 1 | 3,626 (4.65) | |
| Antigenic characterization | 2,736 | 53 | 1 | 2,790 | |
| Genomic characterization | 709 | 23 | 732 | ||
| Both required | 102 | 2 | 104 | ||
| SJ Total | 15,841 | 334 | 348 | 16,523 (21.19) | |
| The University of Georgia (UGA) | |||||
| Negative IAV | 3,662 | 16,834 | 1,095 | 4,361 | 25,952 (33.29) |
| Positive IAV | 542 | 2,325 | 32 | 203 | 3,102 (3.98) |
| Antigenic characterization | 14 | 94 | 6 | 2 | 116 |
| Genomic characterization | 528 | 2,212 | 26 | 201 | 2,967 |
| Both required | 19 | 19 | |||
| UGA Total | 4,204 | 19,159 | 1,127 | 4,564 | 29054 (37.26) |
| Grand Totals | 23,829 | 38,792 | 9,507 | 5,841 | 77,969 (100) |
Spatiotemporal frequency of IAV recovery.
By season, frequency of IAV recovery peaked in late summer, declined progressively in fall and winter, and slightly increased in spring (Table 2). The frequency of IAV recovery was related to sampling location, with Alberta (summer) and Minnesota (early fall) having the highest virus recovery (22% and 15%, respectively) compared with the relatively lower frequency of virus recovery in southern U.S. states such as Louisiana and Mississippi (4% and 3% in winter, respectively). Figure 1B shows a trend of IAV prevalence decreasing in a north-to-south gradient as migration progresses across migratory seasons.
The frequency of IAV recovery was significantly higher in the first 3 decades of surveillance than in the most recent decade (χ2 = 1,603, P < 0.001). Between 1976 and 2005, there were 17,706 samples collected and the total IAV recovery was 18.9%. These 3 decades consisted of primarily summer and fall sampling in Northern latitudes. In contrast, between 2006 and 2015, there were 60,263 samples collected and the IAV recovery was 8.4% (Fig. 2A). This decade consisted of sampling schema more evenly dispersed throughout the flyways and all 4 seasons.
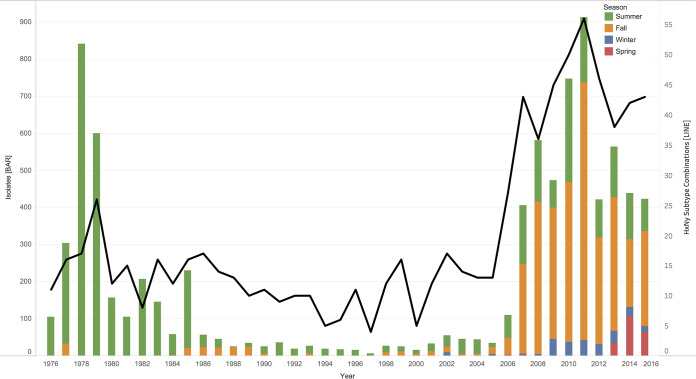
FIG 2.
Stacked bar graphs showing total number of IAV samples collected from waterfowl per year between 1976 and 2015 in North America. (A) The number of IAV isolates recovered each year is indicated in red, and IAV-negative samples are indicated in blue. IAV isolates recovered by decade are as follows: 1976 to 1985, 2,749 (24%) IAV isolates from 11,361 waterfowl; 1986 to 1995, 304 (11%) IAV isolates from 2,721 waterfowl; 1996 to 2005, 303 (8%) IAV isolates from 3,624 waterfowl; and 2006 to 2015, 5,071 (8%) IAV isolates recovered from 60,263 waterfowl. (B) The number of samples collected each year by state or province (see key for color coding). Locations are given in descending order from north to south by latitude of the sampling site. Sampling expanded to locations throughout the extent of the Mississippi Migratory Flyway in 2006.
Frequency of virus recovery by species, age, and gender.
The frequency of IAV recovery was significantly different (χ2 = 28.9, P < 0.001) by gender, with a slightly higher prevalence in female than male birds (Table 2). Prevalence of viral recovery also significantly differed by age, being higher in juvenile than adult birds (χ2 = 1,703, P ≤ 0.001; Table 2).
Dabbling ducks accounted for 91% of all samples. Specifically, samples from mallards (Anas platyrhynchos), blue-winged teals (Anas discors), American green-winged teals (A. crecca), northern pintails (A. acuta), and wood ducks (Aix sponsa) represent 82% of all samples. Table 1 gives the number of samples and IAV prevalence by bird species. By functional group, 12% of dabblers, 3% of divers, 5% of sea ducks, and 1% of other birds were IAV positive. IAV prevalence was significantly higher in dabblers than in all other functional groups (χ2 = 1,103; P ≤ 0.001). Of note, the Canada goose (Branta canadensis) was relatively well represented (2,172 [3%] of all samples), but yielded only 5 (0.2%) IAV isolates over the 4 decades. Similarly, the wood duck was well represented (2,974 [4%] of all samples), but yielded only 18 (0.6%) IAV isolates over the 4 decades. The only IAV isolates recovered from hosts outside the Anatidae family were from 5 American coots (Fulica americana) and 6 red-necked grebes (Podiceps grisegena).
Subtype distribution.
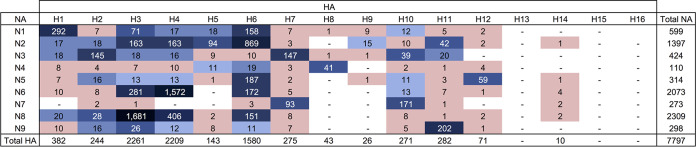
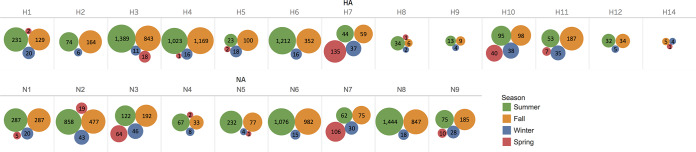
Subtypes of IAV isolates included H1 to H12 and H14 and N1 to N9. Of the 8,427 isolates, 7,797 (93%) were classified as a single HA subtype and a single NA subtype, representing 96 unique combinations (Fig. 3). Mixed infections, indicated by the presence of multiple HA and/or NA subtypes, were detected in 282 (3%) isolates, whereas 348 (4%) isolates could not be fully subtyped for various reasons.
FIG 3.
Distribution of 96 IAV virus subtype combinations isolated from migratory waterfowl in North America between 1976 and 2015. All subtype combinations shown in salmon color are classified as rare (recovered ≤10 times in the data set). The remaining subtype combinations in blue darken relative to the frequency of recovery.
Of the 7,797 single-subtype combinations, H3N8, H4N6, and H6N2 were the most frequently identified, representing 22%, 20%, and 11% among the isolates, respectively (Fig. 3). H13 was recovered once in the form of a mixed infection with H4 in a blue-winged teal sampled in 1989. H15 and H16 were not identified in these projects during the surveillance period.
Of the 282 isolates with apparent mixed subtypes, H3/H4 (64 isolates, 23% of all mixed isolates), H3/H6 (19 isolates, 7% of mixed isolates), and H4/H6 (9 isolates, 3% of mixed isolates) were most often isolated in combination, and N6/N8 (36 isolates, 13% of mixed isolates), N1/N4 (29 isolates, 10% of mixed isolates), and N2/N8 (29 isolates, 10% of mixed isolates) were most often isolated together. By season, the proportion of isolates with mixed HA and/or NA subtypes in winter (32/253 [13%]) was significantly higher than in all other seasons (summer, 117/4,457 [3%]; fall, 130/3,501 [4%]; spring, 3/216 [1%], where all proportions were significant [P ≤ 0.001] compared to winter).
Of the 96 HA-NA subtype combinations in this study, 50 subtype combinations from 220 total isolates were classified as being sporadically recovered (recovered ≤10 times) within this sampling period (light red cells in Fig. 3). The proportion of sporadically recovered isolates in winter (30/253 [12%]) was significantly higher than in all other seasons (summer, 78/4,457 [2%]; fall, 101/3,501 [3%]; spring, 11/216 [5%], where all proportions were significant [P ≤ 0.01] compared to winter).
Distribution of HA subtypes and apparent cyclical patterns.
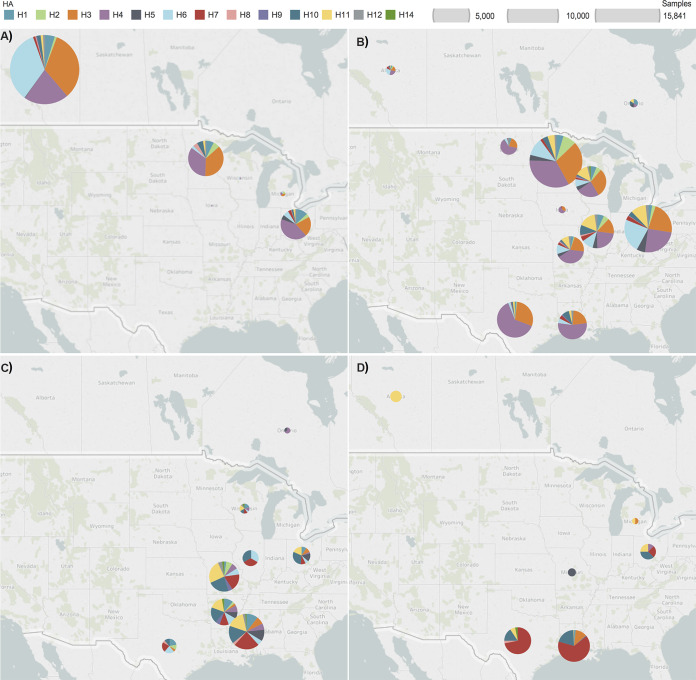
As observed in Fig. 1B, sampling during summer occurred mainly on northern premigratory breeding and staging grounds in Alberta, Minnesota, and Ohio. During this time, H3, H4, and H6 (82% of all summer isolates) were the dominant HA subtype infecting waterfowl, with the other 18% of isolates representing H1, H2, H5, H7, H8, H9, and various mixed HA subtypes. (Fig. 4A). As the southern fall migration progressed, the proportion of non-H3, -H4, and -H6 isolates began to increase. However, H3, H4, and H6 still constituted 75% of all HA subtypes recovered in fall (Fig. 4B). IAVs recovered in winter showed a higher proportion of non-H3, -H4, and -H6 subtypes with no dominant HA, as shown in pie charts in Fig. 4C. Dominant HA subtypes during winter differed from those circulating in previous months. Of the winter isolates, 19% were H10, 18% were H7, and 17% were H11. Total sampling numbers and frequency of IAV isolates recovered were both lowest in spring. Of the few spring isolates, 63% were H7 and 20% were H10 (Fig. 4D). Notably, 92% (198/216) of all spring isolates were from blue-winged teals. Blue-winged teals were also heavily sampled during spring, accounting for 69% (4,033/5,841) of all sampled waterfowl.
FIG 4.
Influenza A virus hemagglutinin (HA) subtype diversity from migratory waterfowl sampled during (A) summer (June to August), (B) fall (September to November), (C) winter (December to February), and (D) spring (March to May) between 1976 and 2015. Each pie segment represents the proportion of the respective HA subtype (color-coded) isolated in the respective location and season. Size of circles are relative to the number of samples collected at a location. An annual trend is evident of progressively increased HA subtype diversity during southbound migration, peaking during winter.
Subtypes H1, H3, H6, H8, and H9 and N2, N4, N5, N6, and N8 were most frequently detected during summer (Fig. 5). Subtypes H2, H4, H5, H10, H11, H12, and H14 and N3 and N9 were most frequently detected during fall. Subtypes H7 and N7 were most frequently detected during spring. Several subtypes (H2, H6, H9, and H12 and N6 and N8) were never detected during spring. All subtypes were detected in fall and winter. All subtypes except H14 were detected during summer.
FIG 5.
Temporal distribution of IAV subtypes of HA and NA recovered from migratory waterfowl in North America between 1976 and 2015. The 7,797 single-subtype combination isolates are color-coded by season of recovery (3-month intervals; summer: June to August).
The frequency of IAV recovery increased little over the years, despite a 5-fold increase in annual sample collections (Fig. 2A). Annual viral diversity, as measured by the number of subtype combinations recovered, in recovered isolates increased over the years (Fig. 6). In the first decade (1976 to 1985) in which there was primarily summer sampling in Alberta, an average of 275 isolates and 15 subtype combinations were obtained per year (95% confidence interval [CI] 11.9 to 18). In comparison, in the last decade (2006 to 2015) in which sampling was more evenly distributed throughout the year and throughout the flyways, an average of 507 isolates and 43 subtype combinations were recovered per year (95% CI 37.7 to 47.5).
FIG 6.
Stacked bars representing IAV isolates per year from waterfowl sampled in North America between 1976 and 2015, stratified by season sampled (3-month intervals; summer: June to August). The black line indicates subtype diversity, calculated by the number of distinct subtype combinations detected each year. Expansion of IAV surveillance efforts to locations associated with fall migration, winter, and spring migration led to higher diversity of IAV subtypes recovered than in years with predominant summer sampling.
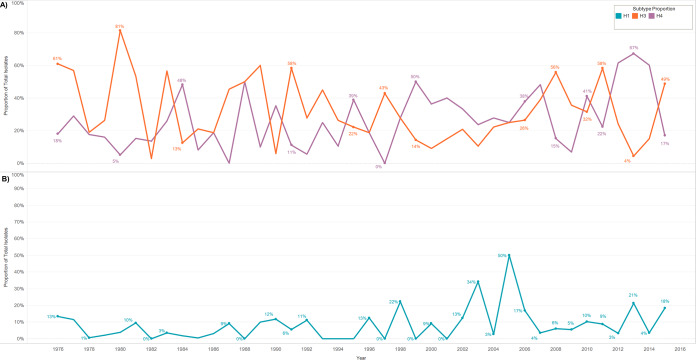
The longitudinal nature of this study also provided evidence that certain HA subtypes display annual cyclical patterns of subtype dominance on breeding grounds in summer. Of note, H3 and H4 appeared to demonstrate an inverse cycle wherein one subtype accounts for a higher proportion of isolates 1 year and the other subtype dominates the following year (Fig. 7A). Similarly, the proportion of H1 isolates also appeared to cycle and peak every other year (Fig. 7B). No other HA subtypes appeared to cycle during summer, and data were not efficiently consistent to identify cyclical patterns at a similar multidecade level during other seasons or in other regions.
FIG 7.
Cyclical trends in the annual proportion of respective IAV HA subtypes isolated during summer (June to August) from migratory waterfowl in North America. (A) H3 and H4 demonstrated an apparent inverse cyclical trend and (B) H1 demonstrated an apparent cyclical trend. No other HA subtypes appeared to cycle during summer, and data for other seasons were limited. Restriction by season assumes that waterfowl were sampled in regionally similar areas each year.
DISCUSSION
On the basis of our findings and those from similar studies, we infer the following annual IAV trends of increasing HA subtype diversity contrasted by decreasing IAV prevalence across a migration season (16, 17). Segregated waterfowl populations on northern breeding grounds, largely consisting of immunologically naive juveniles, would be particularly susceptible to H3, H4, and H6, the most prevalent circulating IAV subtypes. Summer has been characterized by predominately homosubtypic intraspecies (between the same avian species) transmission (18). However, phylogenetic evidence suggests even antigenically related, or homosubtypic, IAVs may display transient and divergent genomic constellations (19). During early southbound migration, the immune systems of juvenile waterfowl likely matures through interactions with new populations, resulting in decreased IAV prevalence and increased HA subtype diversity. The bottlenecking of previously unassociated populations congregating on common stopover sites and wintering grounds could explain, and drive, the mixing of IAVs, which accounts for the diverse profile of circulating subtypes recovered during winter (18, 20).
The proportion of mixed infections (13%) and uncommonly isolated subtype combinations (12%) recovered in winter isolates was higher than that for other seasons. Further, peak reassortment and interspecies (between different avian species) transmission of IAVs have been previously associated with winter isolates, which suggests an increased risk of spillover into domestic species and humans (18, 21, 22). Finally, H7 and H10 dominated HA subtype proportion in the few spring isolates recovered. The highest number of spring isolates were recovered from blue-winged teals, although they were oversampled during this season. As blue-winged teals travel exceedingly far to wintering grounds in South America, researchers have suggested they can serve as a link for the dispersal of IAVs from North America to the neotropics and may differ from other migratory waterfowl in their temporal population immunity (23, 24). Studies strongly support that low-pathogenic H7 IAVs are particularly abundant in blue-winged teals during spring, which supports our findings and has important implications for public health and agriculture sectors (25, 26). The shift to less common subtypes dominating the landscape during spring migration may be a critical component in the maintenance of this annual IAV diversity cycle (27). However, overall spring sampling is relatively minimal and might not be adequately robust to represent encompassing seasonal trends.
Substantial evidence suggests that winter sampling will yield the highest IAV HA subtype diversity (4, 17, 20, 25, 28, 29). However, due to decreased viral prevalence in waterfowl during the winter months, larger sample sizes are required for comparable yields of isolates. Currently, fall sampling accounts for most of the IAV surveillance because of accessibility of waterfowl during the hunting season (7, 30). Formation of the collaborative and organized research network NIAID CIERS in 2007 helped expand surveillance efforts to locations associated with fall migration, wintering, and spring waterfowl migration. Clearly, the expansion of IAV surveillance in fall and winter led to recovering a higher diversity of IAV subtypes than during years when predominant surveillance occurred in summer (Fig. 6). Thus, fall could be considered a favorable sampling season to balance the recovery of isolates and maximize subtype diversity from waterfowl.
Various host population immunity and reassortment events throughout a migratory season can influence the annually maintained trends of IAV diversity (4). The probability of infection with a heterosubtypic infection increases over time, while developed immune responses provide protection against homosubtypic infection (16). As evidenced in European mallards, the H3 clade (H3, H4, and H14) dominates fall infections and the H1 clade (H6, H1, H5, H2) predominates later in the season (16). Studies comparing viral diversity at opposite ends of flyways support that reassortment contributes to different profiles of subtype diversity in breeding versus wintering grounds (28). Hill et al. recovered 42 IAV subtype combinations from dabbling ducks wintering in California and only 17 IAV subtype combinations from dabbling ducks on Alaskan breeding grounds over a 2-year cross-seasonal study (17). In surveillance at Louisiana in 1990, 9 of the 13 currently known HA subtypes were recovered from only 28 total isolates from the variety of species wintering on coastal marshes (20).
The uneven distribution of sample collections inherently skewed the broad generalizations shown in Fig. 5. However, viewing results in this manner allows us to gain a unique perspective on seasonal subtype prevalence. Surprisingly, 70% of H5 and 49% of H7 isolates were recovered during fall and spring, respectively, which has important implications because these subtypes may become highly pathogenic and cause severe outbreak in poultry (6). Also, our finding that H14 was isolated 10 times (5 in fall, 4 in winter, 1 in spring, and 0 in summer) since 2010 is remarkable, as H14 had been undocumented since its original recovery in 1982 (31–33).
Several studies have suggested cyclical subtype dominance, but no study has yet sampled consistently in the same region over enough time to adequately support these observations (7). Periodicity in annual proportions of subtype isolations, particularly H3, H4, and H6, has been shown in overlapping samples from 1976 to 2001 (7). In a 6- to 8-year study conducted in Alberta and New York, subtypes rarely sustained dominance over consecutive years or between flyways (34). We observed that often in summers (June to August) when H3 was the dominant subtype, H4 was rarely isolated and vice versa. Distinction by season ensured that these viruses circulated in the same area at the same time of year. This inverse cycle of subtype dominance could provide insights into functions of herd immunity and our capability to predict the annual dominant subtype. H1 dominance was also cyclical, accounting for 3%, 21%, 4%, and 18% of subtypes captured in 2012, 2013, 2014, and 2015, respectively. Again, this apparent cycle points to a pertinent mechanism underlying immunity, population structure, or an unknown variable in viral ecology. However, subtype dominance returns to being unpredictable as we return to the level of individual sampling locations, indicating the complexity of IAV dynamics and the extensive yet-unknown mechanisms driving this design.
Higher IAV prevalence in juvenile birds than in adult birds is likely driven by naive immunological responses due to lack of previous exposure and an immature immune system (6, 18, 35). The influx of hatchlings on breeding grounds corresponds to high IAV prevalence in northern latitudes in late summer before birds begin southbound migration. Although the 1% difference in IAV prevalence between males and females was statistically significant in our study, other studies have reported no difference by gender (17, 36). One possible explanation for this observation is the bias introduced by sampling hunter-harvested birds (e.g., targeting drakes for colorful plumage and hunting restrictions on hens).
In our study, 12% of dabbling ducks were IAV positive, and this functional group accounted for 98% of all isolates in the data set. Transmission of IAV in duck populations has been linked to fecal-oral transmission via contaminated water, and IAVs can remain infective in this water for extended periods (37). The use of shallow water habitats during surface “dabbling” feeding behavior could provide increased opportunity for exposure in heavily populated water bodies (6, 38). However, dabbling ducks were overrepresented in our data set (91% of all samples), indicating an obvious sampling bias. Mallards were both the primary host of IAV infection as well as the most-sampled species, which is consistent with results from several studies (17, 35, 36). Also, IAV prevalence in the Canada goose and wood duck was lower than that in other waterfowl species. These results are consistent with previous findings and further support hypotheses that behavior and life history strategies can influence IAV prevalence (6, 38–41).
Although the benefits of aggregating years of surveillance are noteworthy, they inherently introduce limitations and challenges in comparability among data sets and standardization of diagnostic assays. Primarily, lab and sampling techniques change over time and differ among organizations. The subtyping method significantly differed between seasons, which could affect the identification of mixed infections and might result in their underestimation or uneven distribution (17). The advent of genomic subtyping characterization techniques might have also confounded the higher recovery of diverse subtypes and mixed infections in more recent years. Embryonated chicken eggs (ECEs) were used here as the exclusive method for virus isolation. The earlier years of research occurred before the advent of PCR technology and ECEs were preferred over cell culture for IAVs from avian species due to their reliable ability to propagate influenza viruses and high specificity for influenza A viruses (42, 43). Lastly, priorities and project objectives also changed over the years, leading to potential species, temporal, or location sampling biases. Therefore, it is important to appreciate the insights a multidecade and robust data set may provide, but also to refrain from overinterpretation due to the variable consistency inherent in a retrospective study design.
In conclusion, we plan in future studies to look at this diversity trend at the phylogenetic level to strengthen and validate our findings. Similar IAV surveillance data should be collated globally to reevaluate strategies that can effectively advance the study of IAV ecology. Identifying the possible mechanisms underlying the persistence of particular subtypes to regions, seasons, or species year after year is of utmost importance, and could involve expanding research in under-surveilled seabirds or uncovering an elusive environmental reservoir key to the mechanism of IAV persistence.
Our data collected over multiple decades provided unique insights and a glimpse of the big picture with regard to epidemiological trends of IAVs in waterfowl over time. We found a recurring pattern in IAV subtype diversity in waterfowl in North America, with a high IAV prevalence of a few dominant subtypes in the northern breeding grounds that then shifts to progressively decreased IAV prevalence and increased subtype diversity profiles as waterfowl migrate toward southern wintering grounds. Evidence of cyclical subtype dominance over consecutive years on a regional scale can prompt future investigations on whether cycles are similarly retained at a local level. We therefore recommend continued year-round sampling at multiple sites after migration to accomplish the surveillance goals of capturing IAV diversity and understanding the evolution and movement of IAVs.
MATERIALS AND METHODS
Surveillance and sample collection.
Three research groups within the Centers of Excellence for Influenza Research and Surveillance at St. Jude Children’s Research Hospital (Memphis, TN) contributed data for waterfowl samples collected in the MMF and CMF between August 1976 and December 2015 (44) (Table 3). Samples were collected from study sites in 14 U.S. states (Minnesota, North Dakota, Michigan, Wisconsin, Iowa, Illinois, Ohio, Indiana, Missouri, Tennessee, Arkansas, Mississippi, Louisiana, and Texas) and 3 Canadian provinces (Alberta, Ontario, and Quebec) over 4 decades: 1976 to 1985, 1986 to 1995, 1996 to 2005, and 2006 to 2015. In 1976 to 1985, surveillance was performed in Alberta, and sampling at Ohio was added in 1986. In 1986 to 2005, surveillance locations included Alberta and Ohio, as well as Missouri and Texas. In 2006 to 2015, surveillance efforts expanded to sites throughout the MMF (Fig. 2B), which correlated with the 2007 initiation of the National Institute of Allergy and Infectious Diseases Centers of Excellence for Influenza Research and Surveillance (NIAID CEIRS).
Cloacal swabs, oropharyngeal swabs, tracheal swabs, and tissue samples were collected from birds at 295 study sites. Birds were trapped by standard live-capture methods or hunter harvested (45, 46). Surveillance efforts were linked with bird-banding efforts on breeding and staging grounds in summer and in wintering areas during spring migration. Hunter-harvested birds were sampled in waterfowl hunting seasons during southbound migration and in wintering areas. All sample collections, when required, were done in compliance with the US Fish and Wildlife Service, Environment and Climate Change Canada, state or provincial permits, and after being approved by the Institutional Animal Care and Use Committee. Species, gender, age (estimated as juvenile or adult using wing plumage techniques), sample collection date, and location were recorded for each bird sampled (47).
Laboratory testing.
IAVs were detected by means of previously described laboratory-specific methods (22, 48). Virus isolation in 9- to 11-day-old serum pathogen-free (SPF) ECEs was considered the determinate test for IAV-positive samples (43). Viral isolates were subtyped by antigenic methods using hemagglutination and neuraminidase inhibition assays or genomic methods via PCR and sequencing methods, depending on the lab and year of recovery (49–51). Subtype of a given isolate was preferentially assigned using the genomic subtype over the antigenic subtype, if both were available.
Data analyses.
Each sample was classified by host taxonomy, sampling season, and functional group of host species (dabbler, diver, sea duck, or other) (52). Seasons were structured in 3-month intervals to approximately reflect the general temporal behavior of migratory birds (e.g., summer as June 1 to August 31). Year was designated by calendar year beginning January 1. Subtype diversity was defined as the number of distinct HA subtypes or HA-NA subtype combinations.
All descriptive epidemiological analyses were performed using Microsoft Excel (version 15.32, Microsoft, CA). Pearson chi-square tests of independence were used to determine statistically significant associations between predictor variables (decade, gender, age, and functional group) and IAV recovery. Proportions of rare isolates (isolated ≤10 times) and isolates with mixed infections were compared in a pairwise fashion between seasons with a two-sample test of proportions, using a significance level of = 0.05. Statistical tests were performed using Stata (version 15.0, StataCorp, College Station, TX). Data visualizations and charts were curated in Tableau (Desktop version 10.4, Tableau Software, Inc., Seattle, WA).
ACKNOWLEDGMENTS
We thank Bruce Turner, Paul Pryor, Frank Baldwin, Garnet Raven, David Walker, and Angela Danner.
This work was supported by Centers of Excellence for Influenza Research and Surveillance, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Department of Health and Human Services (DHHS), contract number HHSN272201400006C, and the American Lebanese Syrian Associated Charities.
REFERENCES
- 1.Olson SH, Parmley J, Soos C, Gilbert M, Latorre-Margalef N, Hall JS, Hansbro PM, Leighton F, Munster V, Joly D. 2014. Sampling strategies and biodiversity of influenza A subtypes in wild birds. PLoS One 9:e90826. doi: 10.1371/journal.pone.0090826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dwyer DE, Kirkland PD. 2011. Influenza: One Health in action. N S W Public Health Bull 22:123–126. doi: 10.1071/NB11005. [DOI] [PubMed] [Google Scholar]
- 3.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fries AC, Nolting JM, Bowman AS, Lin X, Halpin RA, Wester E, Fedorova N, Stockwell TB, Das SR, Dugan VG, Wentworth DE, Gibbs HL, Slemons RD. 2015. Spread and persistence of influenza A viruses in waterfowl hosts in the North American Mississippi migratory flyway. J Virol 89:5371–5381. doi: 10.1128/JVI.03249-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fouchier RAM, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus A. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol 79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus ADME, Fouchier RAM. 2006. Global patterns of influenza A virus in wild birds. Science 312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 7.Krauss S, Walker D, Pryor SP, Niles L, Chenghong L, Hinshaw VS, Webster RG. 2004. Influenza A viruses of migrating wild aquatic birds in North America. Vector Borne Zoonotic Dis 4:177–189. doi: 10.1089/vbz.2004.4.177. [DOI] [PubMed] [Google Scholar]
- 8.Easterday BC, Trainer DO, Tůmová B, Pereira HG. 1968. Evidence of infection with influenza viruses in migratory waterfowl. Nature 219:523–524. doi: 10.1038/219523a0. [DOI] [PubMed] [Google Scholar]
- 9.Slemons RD, Johnson DC, Osborn JS, Hayes F. 1974. Type-A influenza viruses isolated from wild free-flying ducks in california. Avian Dis 18:119–124. doi: 10.2307/1589250. [DOI] [PubMed] [Google Scholar]
- 10.Downie JC, Laver WG. 1973. Isolation of a type A influenza virus from an Australian pelagic bird. Virology 51:259–269. doi: 10.1016/0042-6822(73)90426-1. [DOI] [PubMed] [Google Scholar]
- 11.Machalaba CC, Elwood SE, Forcella S, Smith KM, Hamilton K, Jebara KB, Swayne DE, Webby RJ, Mumford E, Mazet JAK, Gaidet N, Daszak P, Karesh WB. 2015. Global avian influenza surveillance in wild birds: a strategy to capture viral diversity. Emerg Infect Dis 21:e1–e7. doi: 10.3201/eid2104.141415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 13.Runstadler J, Hill N, Hussein ITM, Puryear W, Keogh M. 2013. Connecting the study of wild influenza with the potential for pandemic disease. Infect Genet Evol 17:162–187. doi: 10.1016/j.meegid.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taubenberger JK, Kash JC. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7:440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.USDA APHIS. 2016. Final report for the 2014–2015 outbreak of highly pathogenic avian influenza (HPAI) in the United States. United States Department of Agriculture, Washington, DC: https://www.aphis.usda.gov/animal_health/emergency_management/downloads/hpai/2015-hpai-final-report.pdf. [Google Scholar]
- 16.Latorre-Margalef N, Grosbois V, Wahlgren J, Munster VJ, Tolf C, Fouchier RAM, Osterhaus A, Olsen B, Waldenström J. 2013. Heterosubtypic immunity to influenza A virus infections in mallards may explain existence of multiple virus subtypes. PLoS Pathog 9:e1003443. doi: 10.1371/journal.ppat.1003443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill NJ, Takekawa JY, Cardona CJ, Meixell BW, Ackerman JT, Runstadler JA, Boyce WM. 2012. Cross-seasonal patterns of avian influenza virus in breeding and wintering migratory birds: a flyway perspective. Vector Borne Zoonotic Dis 12:243–253. doi: 10.1089/vbz.2010.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill NJ, Ma EJ, Meixell BW, Lindberg MS, Boyce WM, Runstadler JA. 2016. Transmission of influenza reflects seasonality of wild birds across the annual cycle. Ecol Lett 19:915–925. doi: 10.1111/ele.12629. [DOI] [PubMed] [Google Scholar]
- 19.Dugan VG, Chen R, Spiro DJ, Sengamalay N, Zaborsky J, Ghedin E, Nolting J, Swayne DE, Runstadler JA, Happ GM, Senne DA, Wang R, Slemons RD, Holmes EC, Taubenberger JK. 2008. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog 4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stallknecht DE, Shane SM, Zwank PJ, Senne DA, Kearney MT. 1990. Avian influenza viruses from migratory and resident ducks of coastal Louisiana. Avian Dis 34:398–405. doi: 10.2307/1591427. [DOI] [PubMed] [Google Scholar]
- 21.Steel J, Lowen AC. 2014. Influenza A virus reassortment, p 377–401. In Current Topics in Microbiology and Immunology. Springer International Publishing, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- 22.Hinshaw VS, Webster RG, Turner B. 1978. Novel influenza A viruses isolated from Canadian feral ducks: including strains antigenically related to Swine influenza (Hsw1N1) viruses. J Gen Virol 41:115–127. doi: 10.1099/0022-1317-41-1-115. [DOI] [PubMed] [Google Scholar]
- 23.Afanador-Villamizar A, Gomez-Romero C, Diaz A, Ruiz-Saenz J. 2017. Avian influenza in Latin America: a systematic review of serological and molecular studies from 2000–2015. PLoS One 12:e0179573. doi: 10.1371/journal.pone.0179573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramey AM, Walther P, Link P, Poulson RL, Wilcox BR, Newsome G, Spackman E, Brown JD, Stallknecht DE. 2016. Optimizing surveillance for South American origin influenza A viruses along the United States gulf coast through genomic characterization of isolates from blue-winged teal (Anas discors). Transbound Emerg Dis 63:194–202. doi: 10.1111/tbed.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramey AM, Poulson RL, González-Reiche AS, Wilcox BR, Walther P, Link P, Carter DL, Newsome GM, Müller ML, Berghaus RD, Perez DR, Hall JS, Stallknecht DE. 2014. Evidence for seasonal patterns in the relative abundance of avian influenza virus subtypes in blue-winged teal (Anas discors). J Wildl Dis 50:916–922. doi: 10.7589/2013-09-232. [DOI] [PubMed] [Google Scholar]
- 26.Ferro PJ, Budke CM, Peterson MJ, Cox D, Roltsch E, Merendino T, Nelson M, Lupiani B. 2010. Multiyear surveillance for avian influenza virus in waterfowl from wintering grounds, Texas Coast, USA. Emerg Infect Dis 16:1224–1230. doi: 10.3201/eid1608.091864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nolting JM, Fries AC, Gates RJ, Bowman AS, Slemons RD. 2016. Influenza A viruses from overwintering and spring-migrating waterfowl in the Lake Erie Basin, United States. Avian Dis 60:241–244. doi: 10.1637/11138-050815-ResNoteR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearce JM, Ramey AM, Flint PL, Koehler AV, Fleskes JP, Franson JC, Hall JS, Derksen DV, Ip HS. 2009. Avian influenza at both ends of a migratory flyway: characterizing viral genomic diversity to optimize surveillance plans for North America. Evol Appl 2:457–468. doi: 10.1111/j.1752-4571.2009.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latorre-Margalef N, Tolf C, Grosbois V, Avril A, Bengtsson D, Wille M, Osterhaus ADME, Fouchier RAM, Olsen B, Waldenström J. 2014. Long-term variation in influenza A virus prevalence and subtype diversity in migratory mallards in northern Europe. Proc Biol Sci 281:20140098. doi: 10.1098/rspb.2014.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoye BJ, Munster VJ, Nishiura H, Klaassen M, Fouchier R. 2010. Surveillance of wild birds for avian influenza virus. Emerg Infect Dis 16:1827–1834. doi: 10.3201/eid1612.100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nolting J, Fries AC, Slemons RD, Courtney C, Hines N, Pedersen J. 2012. Recovery of H14 influenza A virus isolates from sea ducks in the Western Hemisphere. PLoS Curr 4:RRN1290. doi: 10.1371/currents.RRN1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fries AC, Nolting JM, Danner A, Webster RG, Bowman AS, Krauss S, Slemons RD. 2013. Evidence for the circulation and inter-hemispheric movement of the H14 subtype influenza A virus. PLoS One 8:e59216. doi: 10.1371/journal.pone.0059216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawaoka Y, Yamnikova S, Chambers TM, Lvov DK, Webster RG. 1990. Molecular characterization of a new hemagglutinin, subtype H14, of influenza a virus. Virology 179:759–767. doi: 10.1016/0042-6822(90)90143-f. [DOI] [PubMed] [Google Scholar]
- 34.Hinshaw VS, Wood JM, Webster RG, Deibel R, Turner B. 1985. Circulation of influenza viruses and paramyxoviruses in waterfowl originating from two different areas of North America. Bull World Health Organ 63:711–719. [PMC free article] [PubMed] [Google Scholar]
- 35.Bevins SN, Pedersen K, Lutman MW, Baroch JA, Schmit BS, Kohler D, Gidlewski T, Nolte DL, Swafford SR, DeLiberto TJ. 2014. Large-scale avian influenza surveillance in wild birds throughout the United States. PLoS One 9:e104360. doi: 10.1371/journal.pone.0104360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munster VJ, Baas C, Lexmond P, Waldenström J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WEP, Schutten M, Olsen B, Osterhaus A, Fouchier R. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog 3:e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lebarbenchon C, Yang M, Keeler SP, Ramakrishnan MA, Brown JD, Stallknecht DE, Sreevatsan S. 2011. Viral replication, persistence in water and genetic characterization of two influenza A viruses isolated from surface lake water. PLoS One 6:e26566. doi: 10.1371/journal.pone.0026566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ely CR, Hall JS, Schmutz JA, Pearce JM, Terenzi J, Sedinger JS, Ip HS. 2013. Evidence that life history characteristics of wild birds influence infection and exposure to influenza A viruses. PLoS One 8:e57614. doi: 10.1371/journal.pone.0057614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris MT, Brown JD, Goekjian VH, Page Luttrell M, Poulson RL, Wilcox BR, Swayne DE, Stallknecht DE. 2010. Canada geese and the epidemiology of avian influenza viruses. J Wildl Dis 46:981–987. doi: 10.7589/0090-3558-46.3.981. [DOI] [PubMed] [Google Scholar]
- 40.van Dijk JGB, Verhagen JH, Wille M, Waldenström J. 2018. Host and virus ecology as determinants of influenza A virus transmission in wild birds. Curr Opin Virol 28:26–36. doi: 10.1016/j.coviro.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 41.Kistler WM, Gibbs SEJ, Stallknecht DE, Yabsley MJ. 2015. Wood ducks (Aix sponsa) as potential reservoirs for avian influenza and avian paramyxoviruses. Avian Pathol 44:169–174. doi: 10.1080/03079457.2015.1020757. [DOI] [PubMed] [Google Scholar]
- 42.Moresco KA, Stallknecht DE, Swayne DE. 2012. Evaluation of different embryonating bird eggs and cell cultures for isolation efficiency of avian influenza A virus and Avian paramyxovirus serotype 1 from real-time reverse transcription polymerase chain reaction–positive wild bird surveillance samples. J Vet Diagn Invest 3:563–567. doi: 10.1177/1040638712440991. [DOI] [PubMed] [Google Scholar]
- 43.Brauer R, Chen P. 2015. Influenza virus propagation in embryonated chicken eggs. J Vis Exp 97:1–6. doi: 10.3791/52421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.U.S. Fish and Wildlife Service. 2008. Flyways. U.S. Fish and Wildlife Service, Department of the Interior, Washington, DC: https://www.fws.gov/birds/management/flyways.php. [Google Scholar]
- 45.Hunt GS, Dahlka KJ. 1953. Live trapping of diving ducks. J Wildl Manage 17:92–95. doi: 10.2307/3796814. [DOI] [Google Scholar]
- 46.Bowman AS, Nolting JM, Massengill R, Baker J, Workman JD, Slemons RD. 2015. Influenza A virus surveillance in waterfowl in Missouri, USA, 2005–2013. Avian Dis 59:303–308. doi: 10.1637/11002-121014-Reg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carney SM. 1964. Preliminary keys to waterfowl age and sex identification by means of wing plumage. Special Scientific Report-Wildlife No. 82, p 7–16. Washington, DC. [Google Scholar]
- 48.Slemons RD, Easterday BC. 1978. Virus replication in the digestive tract of ducks exposed by aerosol to type-A influenza. Avian Dis 22:367–377. doi: 10.2307/1589291. [DOI] [PubMed] [Google Scholar]
- 49.Lee C, Senne D. a, Suarez DL. 2006. Development and application of reference antisera against 15 hemagglutinin subtypes of influenza virus by DNA vaccination of chickens. Clin Vaccine Immunol 13:395–402. doi: 10.1128/CVI.13.3.395-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chander Y, Jindal N, Stallknecht DE, Sreevatsan S, Goyal SM. 2010. Full length sequencing of all nine subtypes of the neuraminidase gene of influenza A viruses using subtype specific primer sets. J Virol Methods 165:116–120. doi: 10.1016/j.jviromet.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 52.De Graaf RM, Tilghman NG, Anderson SH. 1985. Foraging guilds of North American birds. Environ Manage 9:493–536. doi: 10.1007/BF01867324. [DOI] [Google Scholar]