Boid inclusion body disease (BIBD), caused by reptarenavirus infection, affects captive snake populations worldwide, but the reservoir hosts of reptarenaviruses remain unknown. Here, we report the identification of novel reptarenaviruses, hartmaniviruses, and a chuvirus in captive Brazilian boas with BIBD. Three of the four snakes studied showed coinfection with all three viruses, and one of the snakes harbored three novel reptarenavirus L segments and one novel S segment. The samples originated from collections with Brazilian indigenous snakes only, which could indicate that these viruses circulate in wild snakes. The findings could further indicate that boid snakes are the natural reservoir of reptarena- and hartmaniviruses commonly found in captive snakes. The snakes infected with the novel chuvirus all suffered from BIBD; it is therefore not possible to comment on its potential pathogenicity and contribution to the observed changes in the present case material.
KEYWORDS: arenavirus, boa constrictor, boid inclusion body disease, chuvirus, hartmanivirus, reptarenavirus
ABSTRACT
Boid inclusion body disease (BIBD) is a transmissible viral disease of captive snakes that causes severe losses in snake collections worldwide. It is caused by reptarenavirus infection, which can persist over several years without overt signs but is generally associated with the eventual death of the affected snakes. Thus far, reports have confirmed the existence of reptarenaviruses in captive snakes in North America, Europe, Asia, and Australia, but there is no evidence that it also occurs in wild snakes. BIBD affects boa species within the subfamily Boinae and pythons in the family Pythonidae, the habitats of which do not naturally overlap. Here, we studied Brazilian captive snakes with BIBD using a metatranscriptomic approach, and we report the identification of novel reptarenaviruses, hartmaniviruses, and a new species in the family Chuviridae. The reptarenavirus L segments identified are divergent enough to represent six novel species, while we found only a single novel reptarenavirus S segment. Until now, hartmaniviruses had been identified only in European captive boas with BIBD, and the present results increase the number of known hartmaniviruses from four to six. The newly identified chuvirus showed 38.4%, 40.9%, and 48.1% amino acid identity to the nucleoprotein, glycoprotein, and RNA-dependent RNA polymerase, respectively, of its closest relative, Guangdong red-banded snake chuvirus-like virus. Although we cannot rule out the possibility that the found viruses originated from imported snakes, the results suggest that the viruses could circulate in indigenous snake populations.
IMPORTANCE Boid inclusion body disease (BIBD), caused by reptarenavirus infection, affects captive snake populations worldwide, but the reservoir hosts of reptarenaviruses remain unknown. Here, we report the identification of novel reptarenaviruses, hartmaniviruses, and a chuvirus in captive Brazilian boas with BIBD. Three of the four snakes studied showed coinfection with all three viruses, and one of the snakes harbored three novel reptarenavirus L segments and one novel S segment. The samples originated from collections with Brazilian indigenous snakes only, which could indicate that these viruses circulate in wild snakes. The findings could further indicate that boid snakes are the natural reservoir of reptarena- and hartmaniviruses commonly found in captive snakes. The snakes infected with the novel chuvirus all suffered from BIBD; it is therefore not possible to comment on its potential pathogenicity and contribution to the observed changes in the present case material.
INTRODUCTION
The global decline in biodiversity is a topic of concern also for members of the class Reptilia. The worldwide transportation of wild-caught, farm- and captive-bred reptiles also facilitates the transmission of pathogens. Thus, further information on reptilian pathogens is required to enable efficient screening of transported animals in order to secure, e.g., zoological collections and to avoid spread of infectious agents into private and commercial breeding collections. Boid inclusion body disease (BIBD), known to affect captive constrictor snakes, was recognized in the 1970s (1, 2), and arenaviruses were identified as the causative agents in the early 2010s (3–10). BIBD affects nonvenomous constrictor snakes inhabiting biotopes in the neotropics and tropics. The natural habitats of boid species within the subfamily Boinae include Central and South America and Madagascar, while pythons, i.e., species within the family Pythonidae, are inherent in Africa, Asia, and Australia. Although the habitats of boas and pythons do not naturally overlap geographically, snake species from several continents are housed together or in close proximity in zoological and private collections all around the world. As the name implies, BIBD manifests by the formation of eosinophilic and electron-dense inclusion bodies (IBs) within almost all cell types (2, 3, 5, 11). In fact, antemortem BIBD diagnosis relies on the detection of IBs in cytological specimens, e.g., blood smears (12, 13), or liver biopsy specimens (1, 14). The identification of reptarenaviruses as the causative agents for BIBD has enabled reverse transcriptase PCR (RT-PCR)-based diagnostic procedures and screening of collections (12, 13, 15). For unknown reasons, BIBD is diagnosed more often in boas than in pythons (1, 10, 14). The disease can manifest itself with central nervous system (CNS) signs, which include opisthotonus (“stargazing”), head tremors, disorientation, regurgitation, and “corkscrewing” (1, 2). However, during the past decades, boas with BIBD and clinical CNS signs have become rare, and even snakes with extensive IB formation often appear clinically healthy (10, 12, 14). Instead, snakes with BIBD seem to emaciate progressively and become terminally ill due to secondary, usually bacterial, infections, presumably due to BIBD-associated immunosuppression (13).
In 2015, the BIBD-associated arenaviruses were grouped to form the genus Reptarenavirus in the family Arenaviridae, and the formerly known arenaviruses of rodents and bats formed the genus Mammarenavirus (16). The mamm- and reptarenavirus genomes are a bisegmented negative-sense RNAs with an ambisense coding strategy (17). The S segment encodes the glycoprotein precursor (GPC) and nucleoprotein (NP), and the L segment encodes the zinc finger matrix protein (ZP) and the RNA-dependent RNA polymerase (RdRp) (17). Coincidentally, we identified Haartman Institute snake virus 1 (HISV-1) in a snake with BIBD (9) and later demonstrated that the genome of HISV-1 is similar to those of mamm- and reptarenaviruses except that it lacks the ZP gene (18). The identification of HISV-1 led to the formation of a third arenavirus genus, Hartmanivirus (19). The most recent addition to the family Arenaviridae is the genus Antennavirus, the representatives of which were identified in fish and carry three instead of two genome segments (20). We and others have demonstrated that snakes with BIBD often show coinfection with several reptarenaviruses (8, 9). We also identified further hartmaniviruses and showed that hartmaniviruses can coinfect snakes with BIBD (18). However, so far it is not clear whether hartmaniviruses contribute to BIBD pathogenesis.
The origin of reptarenaviruses and hartmaniviruses is still unknown, as reports have described reptarenaviruses only in captive snakes in North America, Europe, Asia, and Australia (3–7, 21, 22). However, in order to gather information about whether boid snakes themselves can be the viral reservoirs, it is of particular interest to see whether BIBD occurs within boid snake populations in the natural habitats. Boa constrictors are indigenous in Brazil, and the knowledge on reptarenavirus occurrence is limited to a single case report of a suspected BIBD case in Corallus annulatus kept in a zoological garden (23). In 2017, we diagnosed the first cases of BIBD in captive Brazilian Boa constrictor, and we undertook the present study to investigate the nature and phylogeny of the causative viruses involved.
RESULTS
Case descriptions.
(i) Clinical histories. Animals 1 and 4 died after unsuccessful therapeutic attempts (antibiotic and fluid therapy and catheter feeding), chronic inflammatory processes in the oral cavity and sinuses, and a period of apathy, animal 3 died after a prolonged period of apathy and neurological signs, and animal 2 was found dead without prior clinical signs (Table 1).
TABLE 1.
Animals and pathological findingsa
| Animal no. | Age (yr) | Sex | Origin | Captivity | Clinical history | Diagnoses |
|---|---|---|---|---|---|---|
| 1 | 12 | F | Novo Arão (Amazonas, Brazil) | Private owner | Regurgitation and ulcerative lesion in oral cavity for 3 mo; antibiotic therapy, fluid therapy, catheter feeding for 2 mo; no improvement and then apathy and death | Chronic ulcerative stomatitis and osteomyelitis (maxilla); chronic ulcerative dermatitis and myositis; BIBD |
| 2 | 10 | F | Novo Arão (Amazonas, Brazil) | Private owner | Found dead without prior clinical signs | Fibrinonecrotizing cloacitis, embolic-metastatic nephritis; hepatic lipidosis; BIBD |
| 3 | 12 | M | Novo Arão (Amazonas, Brazil) | Private owner | Apathy and neurological signs (e.g., disorientation, stargazing) for 5 mo; euthanasia | Focally ulcerated granulomatous stomatitis and osteomyelitis (maxilla); BIBD |
| 4 | 13 | M | Unknown | Botanic Zoo Foundation of Rio Grande | Oral mucosal bleeding, nasal discharge, lethargy, anorexia for approx 5 mo; hospitalization and antibiotic therapy, fluid therapy, catheter feeding; no improvement; after 45 days, apathy and death | Emaciation; chronic suppurative sinusitis; BIBD |
All animals were Boa constrictor constrictor snakes held in captivity in Porto Alegre, Brazil.
(ii) Post mortem findings. At necropsy, animals 1 to 3 exhibited good body condition, whereas animal 4 was emaciated. All four snakes exhibited overt inflammatory processes: a chronic ulcerative stomatitis and osteomyelitis of the maxilla (animals 1 and 3) (Fig. 1), a multifocal ulcerative deep dermatitis and myositis extending to the vertebral bones (animal 1), a fibrino-necrotizing cloacitis (animal 2), and a chronic suppurative sinusitis (animal 4). Histological examination confirmed the findings. In animal 1, the stomatitis was predominantly heterophilic (i.e., suppurative); bacteriology and mycology isolated Enterobacter gergoviae, Providencia spp., Proteus spp., and Candida albicans from the lesions. The inflammatory infiltrate of the cloacitis in animal 2 was heterophil dominated, with abundant aggregates of coccoid bacilli within the superficial layer of fibrin and necrotic debris. This animal exhibited multifocal areas of necrosis with embedded aggregates of coccoid bacilli in the kidneys, consistent with embolic-metastatic nephritis as a consequence of bacteremia. A bacteriological examination was not performed. In animal 3, the stomatitis was granulomatous, with multinucleated giant cells; only nonspecific flora (Klebsiella spp.) was isolated from the lesion. In the sinusitis of animal 4, heterophils were the predominant inflammatory cells; the bacteriological examination of a sample from this lesion yielded exuberant mixed growth (Enterobacter gergoviae and Pseudomonas spp.). In addition, this animal exhibited a mild multifocal heterophil-dominated pneumonia.
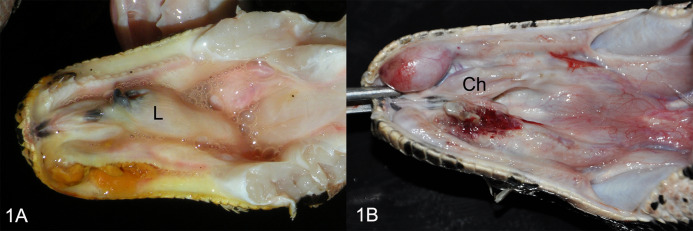
FIG 1.
Oral cavities of snakes with confirmed BIBD. (A) Animal 1, showing chronic ulcerative stomatitis. L, larynx. (B) Animal 3, showing focally ulcerated chronic stomatitis. Ch, choana.
In all snakes, histology served to confirm BIBD. The characteristic eosinophilic intracytoplasmic IBs were found in parenchymal cells in a range of organs (brain [Fig. 2A] and spinal cord, liver [Fig. 2E], pancreas, lungs, and kidneys) in all animals. The IBs varied in size distribution, indicating a chronic stage of the disease (Fig. 2A and E).
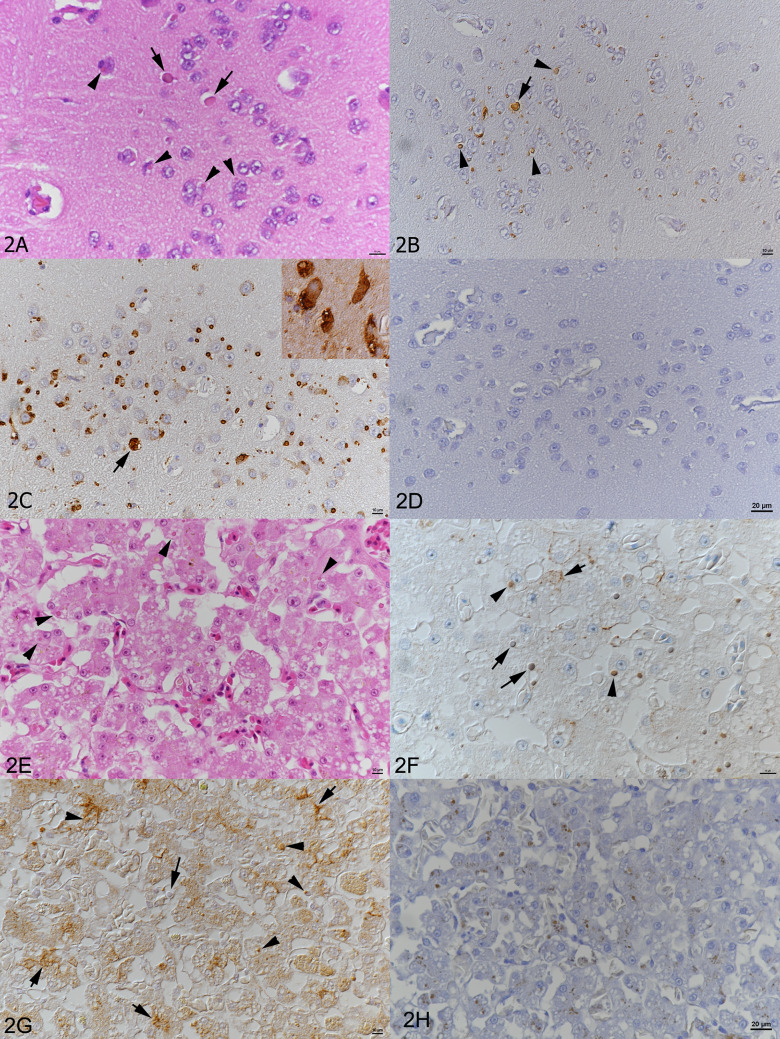
FIG 2.
Histological and immunohistological findings in the brain of animal 3 (A to C) and in the liver of animal 4 (E to G) and staining of tissues from animals without reptarena- or hartmanivirus infection (D and H). (A) Neurons exhibit the typical cytoplasmic eosinophilic IBs (arrowheads), which vary in size and can reach the size of and obscure the nucleus (arrows). HE stain. (B) Staining with the anti-pan-reptarenavirus antibody highlights the IBs depicted in the HE stain. Immunohistology, hemalaun counterstain. (C) Staining with the anti-pan-hartmani antibody highlights the IBs but also shows the presence of NP within the entire cytoplasm of infected cells (arrow and inset). Immunohistology, hemalaun counterstain. (D) Staining of reptrena- and hartmanivirus-negative tissue with anti-pan-reptarenavirus antibody. (E) Numerous hepatocytes exhibit a cytoplasmic eosinophilic IB of variable size (arrowheads). HE stain. (F) Staining with the anti-UHV NP antibody highlights individual IBs (arrowheads) and shows that some cells contain several small IBs (short arrow). Some larger IBs appear negative (large arrows). Immunohistology, hemalaun counterstain. (G) Staining with the anti-pan-reptarenavirus antibody shows the presence of abundant individual (arrowheads) and multiple (short arrows) IBs within hepatocytes. Again, a few larger IBs appear negative (large arrow). Immunohistology, hemalaun counterstain. (H) Staining of reptrena- and hartmanivirus-negative tissue with anti-pan-reptarenavirus antibody.
Identification of reptarenaviruses, hartmaniviruses, and a chuvirus.
To identify the infecting viruses, we isolated RNA from liver samples and performed a metatranscriptomic analysis, an approach we have successfully applied in earlier studies (9, 18, 24–26). We used the Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify the viral sequences. The sequencing confirmed all snakes to be reptarenavirus infected, and similarly to earlier observations (8, 9, 24), all snakes harbored several L segments; however, we identified only a single S segment in each snake (Table 2). In addition to reptarenaviruses, the analysis revealed the presence of one hartmanivirus S-L segment pair in three of the four snakes (animals 1 to 3) studied (Table 2). We recovered complete coding sequences (CDSs) for one L segment and two S segments and a nearly complete CDS (covering >95% of the segment) of an additional hartmanivirus L segment. Genome de novo assembly using the sequence data obtained from these three snakes also produced close-to-identical contigs, varying in length from 10,549 to 10,718 nucleotides (nt) (Table 2), that showed highest matches in the BLAST analysis to chuvirus-like viruses. Table 2 contains the virus names, contig lengths, GenBank accession numbers, and average coverages for the viruses identified.
TABLE 2.
Sequences recovered from NGS data by de novo genome assemblya
| Animal no. | Virus | Segment | Contig length (nt) | GenBank accession no. | Avg coverage (no. of reads) | Closest match by PASC and BLAST (% identity, GenBank accession no.) |
|---|---|---|---|---|---|---|
| 1 | ArBV-1 | S | 3,372 | MN567042 | 14,173.9 | PASC, 74.0, KP071473.1; BLAST, 76.3, KP071671.1 |
| L | 6,883 | MN567044 | 5,232.6 | PASC, 72.7, KR870030.1; BLAST, 74.5, KR870030.1 | ||
| KaBV-1 | L | 7,010 | MN567043 | 7,105.8 | PASC, 72.4, KP071677.1; BLAST, 97.2, KX527584.1 | |
| ArSV-1 | L | 6,780 | MN567045 | 1,413.6 | PASC, 63.3, KR870020.1; BLAST, 67.1, KP071562.1 | |
| UHV-2 | L | 6,813 | MN567046 | 2,765.8 | PASC, 86.4, KR870030.1; BLAST, 87.1, KR870030.1 | |
| FStV-1 | L | 6,831 | MN567047 | 2,233.2 | PASC, 74.1, KR870030.1; BLAST, 75.8, MN567046.1 | |
| SPVV-1 | S | 3,560 | MN567048 | 268.3 | PASC, 21.8, FJ607031.1 BLAST, 69.7, MH483027.1 | |
| L | 3,578 and 2,223 | MN567049 and MN567050 | 35.1 | PASC, NA; BLAST, 65.7, MH778629.1 and 71.8, MH483040.1 | ||
| HFrV-1 | N.A. | 10,718 | MN567051 | 1,698.9 | PASC, NA; BLAST, 66.4, MG600009.1 (query coverage, 28%) | |
| 2 | ArBV-1 | S | 3,372 | MN567052 | 26,110.1 | PASC, 73.7, KP071473.1; BLAST, 76.5, KP071506.1 |
| L | 6,948 | MN567054 | 9,586.7 | PASC, 72.8, KR870030.1; BLAST, 74.4, KR870030.1 | ||
| KaBV-1 | L | 7,009 | MN567053 | 6,688.2 | PASC, 72.3, KP071567.1; BLAST, 97.1, KX527584.1 | |
| AHeV-1 | S | 3,534 | MN567055 | 99.3 | PASC, 22.0, FJ607031.1; BLAST, 64.7, MH483026.1 | |
| L | 5,953 | MN567056 | 28.5 | PASC, 18.7, MG812678.1; BLAST, 67.7, MH483044.1 | ||
| HFrV-1 | NA | 10,549 | MN567057 | 1,073.9 | PASC, NA; BLAST, 67.7, MG600009.1 (query coverage, 28%) | |
| 3 | ArBV-1 | S | 3,351 | MN567058 | 11,236.5 | PASC, 74.0, KP071671.1; BLAST, 75.1, MH503957.1 |
| L | 6,926 | MN567060 | 3,443.9 | PASC, 72.7, KR870030.1; BLAST, 73.9, MH503952.1 | ||
| KaBV-1 | L | 7007 | MN567059 | 2,913.8 | PASC, 72.2, KR870030.1 BLAST, 97.2, KX527584.1 | |
| AHeV-1 | S | 3,598 | MN567061 | 1,433.3 | PASC, 21.8, NC_004294.1; BLAST, 64.7, MH483026.1 | |
| L | 5,961 | MN567062 | 548.3 | PASC, 18.9, MG812678.1; BLAST, 67.5, MH483044.1 | ||
| HFrV-1 | N.A. | 10,574 | MN567063 | 126.8 | PASC, NA; BLAST, 66.4, MG600009.1 (query coverage, 29%) | |
| 4 | PAV-1 | S | 3,464 | MN567064 | 2,071.6 | PASC, 73.3, KP071559.1; BLAST, 72.2, KP071473.1 |
| L | 6,911 | MN567065 | 1,158.3 | PASC, 79.6, KP071549.1; BLAST, 80.0, MH503952.1 | ||
| SauV-1 | L | 6,835 | MN567066 | 523.1 | PASC, 73.2, KP071479.1; BLAST, 74.6, MH483084.1 | |
| GauV-1 | L | 7,203 | MN567067 | 438.3 | PASC, 72.8, KR870030.1; BLAST, 75.5, KP071564.1 |
Abbreviations: ArBV-1, Aramboia boa virus 1; KaBV-1, Kaltenbach virus 1; ArSV-1, Arabuta snake virus 1; UHV-2, University of Helsinki virus 2; FStV-1, Frankfurter Strasse virus 1; SPVV-1, SetPatVet virus 1; HFrV-1, Herr Frank virus 1; AHeV-1, Andere Heimat virus 1; PAV-1, Porto Alegre virus 1; SauV-1, Saudades virus 1; GauV-1, Gaucho virus 1; NA, not available.
Immunohistology served to detect reptarenavirus and hartmanivirus NP in cells with IBs. Reptarenavirus NP expression was mainly restricted to the IBs (Fig. 2B, D, and G), whereas hartmanivirus NP was also detected throughout the cytoplasm (Fig. 2C).
Analysis of the identified reptarenavirus sequences.
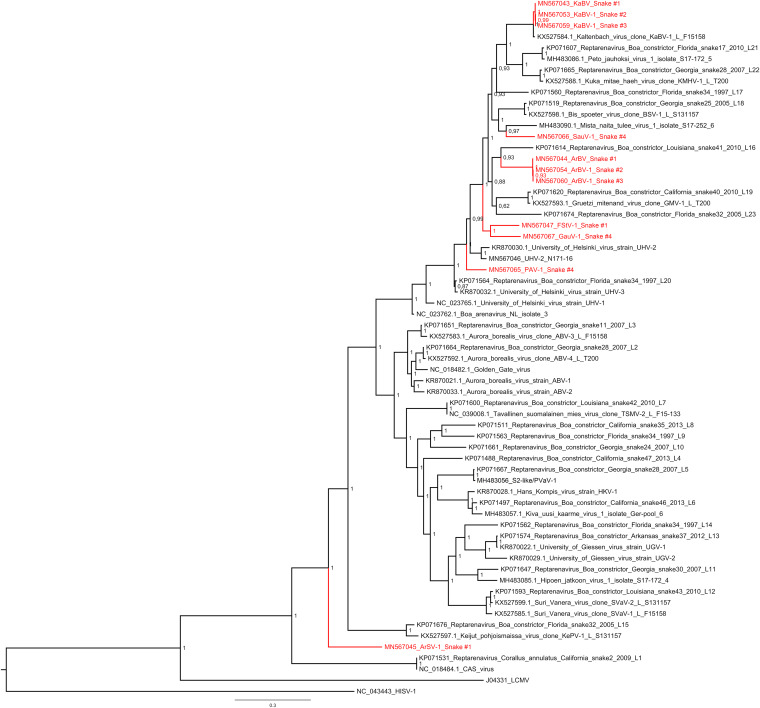
We used the PAirwise Sequence Comparison (PASC) web tool (available at https://www.ncbi.nlm.nih.gov/sutils/pasc/viridty.cgi?textpage=overview), recommended by the Arenaviridae study group of the International Committee on Taxonomy of Viruses (ICTV) for arenavirus classification (16), to analyze the identified reptarenavirus segments. The PASC results showed that we had recovered CDSs for seven novel L segments and two novel S segments (Table 2). The PASC analysis identified one of the L segments in snake 1 as University of Helsinki virus 2 (UHV-2) (86.4% nucleotide identity), whereas in BLAST analysis three L segments in snakes 1 to 3 were >97% identical to Kaltenbach virus 1 (KaBV-1) (24), which is apparently not included in the PASC reference data set. Six L segments had less than 76% nucleotide identity to any currently known reptarenavirus sequences; Tables 3 and 4 show the nucleotide identity matrixes of the reptarenavirus segments. The analyses confirmed that we had recovered L segment CDSs for six novel reptarenaviruses (Tables 2 to 4): Aramboia boa virus 1 (ArBV-1) (in animals 1 to 3), Arabuta Snake virus 1 (ArSV-1) (in animal 1), Frankfurter Strasse virus 1 (FStV-1) (in animal 1), Porto Alegre virus 1 (PAV-1) (in animal 4), Saudades virus 1 (SauV-1) (in animal 4), and Gaucho virus 1 (GauV-1) (in animal 4). We found only a single S segment CDS for each of the studied snakes, and we chose to name the S segments according to the L segment with highest coverage found in the same snake (Table 2): ArBV-1 (in animals 1 to 3) and PAV-1 (in animal 4). Phylogenetic analysis of the reptarenavirus L (Fig. 3) and S (Fig. 4A and B) segments showed the viruses to be distant from those present in GenBank, suggesting that they would represent segments of novel reptarenavirus species. Notably, the reptarenavirus sequences recovered from the Brazilian snakes did not show geographical clustering but were interspersed among the viruses detected from captive snakes in Europe and the United States.
TABLE 3.
Nucleotide identities between reptarenavirus L segments identified in this studya
| Animal no., virus segment | Identity (%) with: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1, ArBV-1 L | 2, ArBV-1 L | 3, ArBV-1 L | 1, KaBV-1 L | 2, KaBV-1 L | 3, KaBV-1 L | 1, ArSV-1 L | 1, UHV-2 L | 1, FStV-1 L | 4, PAV L | 4, SauV-1 L | 4, GauV-1 L | |
| 1, ArBV-1 L | ||||||||||||
| 2, ArBV-1 L | 99 | |||||||||||
| 3, ArBV-1 L | 99 | 100 | ||||||||||
| 1, KaBV-1 L | 71 | 71 | 71 | |||||||||
| 2, KaBV-1 L | 71 | 71 | 71 | 100 | ||||||||
| 3, KaBV-1 L | 71 | 72 | 71 | 100 | 100 | |||||||
| 1, ArSV-1 L | 63 | 64 | 64 | 63 | 63 | 63 | ||||||
| 1, UHV-2 L | 73 | 74 | 74 | 73 | 73 | 73 | 63 | |||||
| 1, FStV-1 L | 73 | 73 | 73 | 73 | 73 | 73 | 63 | 74 | ||||
| 4, PAV L | 72 | 73 | 73 | 72 | 72 | 72 | 64 | 77 | 74 | |||
| 4, SauV-1 L | 73 | 73 | 73 | 72 | 72 | 72 | 63 | 73 | 72 | 73 | ||
| 4, GauV-1 L | 69 | 69 | 69 | 70 | 70 | 70 | 60 | 70 | 70 | 71 | 68 | |
Dark gray, >76% identity between sequences; light gray, <76% identity between sequences.
TABLE 4.
Nucleotide identities between reptarenavirus S segments identified in this studya
| Animal no., virus segment | Identity (%) with: |
|||
|---|---|---|---|---|
| 1, ArBV-1 S | 2, ArBV-1 S | 3, ArBV-1 S | 4, PAV S | |
| 1, ArBV-1 S | ||||
| 2, ArBV-1 S | 97 | |||
| 3, ArBV-1 S | 96 | 99 | ||
| 4, PAV S | 72 | 73 | 73 | |
Dark gray, >76% identity between sequences; light gray, <76% identity between sequences.
FIG 3.
Maximum clade credibility tree of reptarenavirus L segments. The tree was constructed from amino acid sequences of the reptarenavirus representatives available from GenBank and those identified in this study, using the Bayesian MCMC method with the Jones model of amino acid substitution. Posterior probabilities are shown in each node.
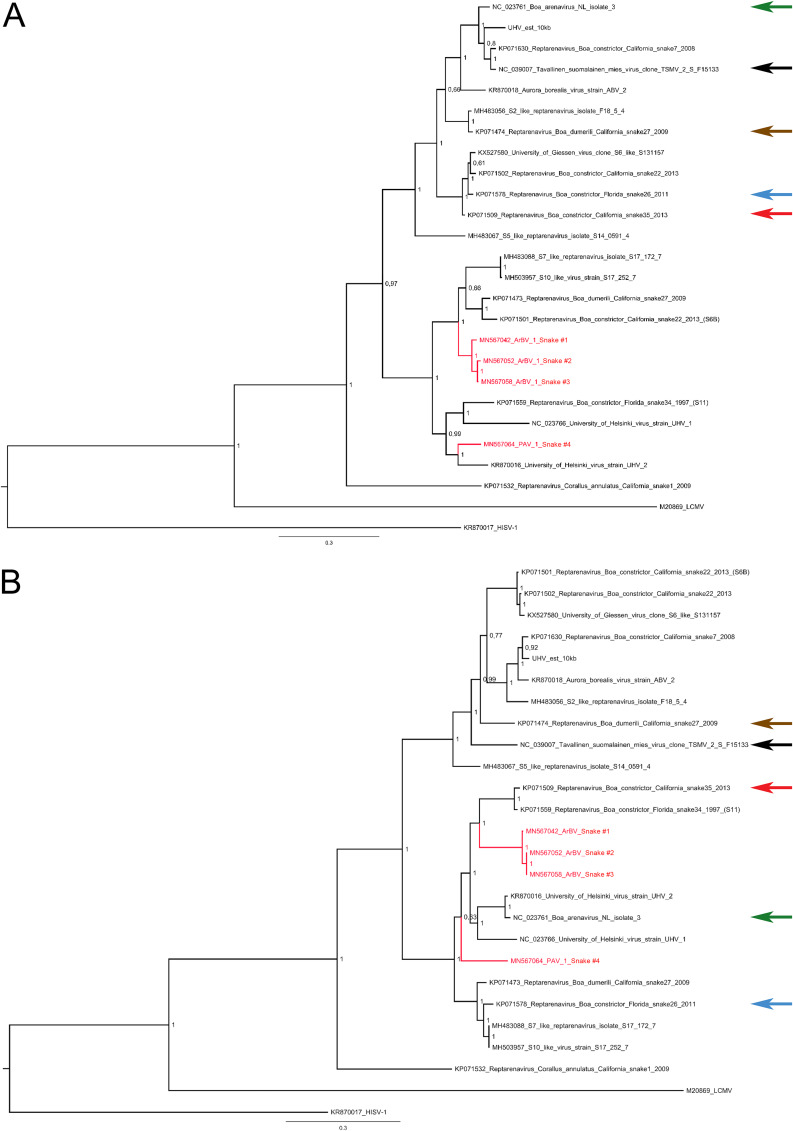
FIG 4.
Maximum clade credibility trees of reptarenavirus GPCs and NPs. (A) The phylogenetic tree based on the GPC amino acid sequences of the viruses identified in this study and those available in GenBank was constructed using the Bayesian MCMC method with the Blosum model of amino acid substitution. (B) The phylogenetic tree based on the NP amino acid sequences of the viruses identified in this study and those available in GenBank was constructed using the Bayesian MCMC method with the Jones model of amino acid substitution. The arrows indicate the S segments with the software-predicted recombination events listed in Table 5.
In line with a previous study (8), recombination analysis suggested five highly supported recombination events in the S segment, detected by at least five out of seven methods (Table 5). All of the estimated recombination break points were located in the intergenic region between the NP- and GPC-coding regions. Consistently, the phylogenetic trees constructed based on the NP- and GPC-coding regions of reptarenavirus had incongruent topologies (Fig. 4A and B).
TABLE 5.
Recombination analyses of reptarenavirus S segments
| Breakpoint positions |
Recombinant sequence | Minor parental sequence(s) | Major parental sequence(s) |
P value by detection method: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| In alignment |
In recombinant sequence |
||||||||||||
| Begin | End | Begin | End | RDP | GENECONV | Bootscan | Maxchi | Chimaera | SiSscan | 3Seq | |||
| 1618 | 4014 | 1565 | 3677 | KP071578 | MH48088, MH503957 | KP071502, KP071501, KX527580 | NSc | 3.28 × 10−58 | 4.69 × 10-55 | 8.43 × 10−40 | 2.36 × 10−10 | 1.16 × 10−61 | 5.62 × 10−190 |
| 3998 | 1827 | 3351 | 1443 | NC_023761 | KP071630 | KR870016 | NS | 2.38 × 10−88 | 3.24 × 10−74 | 1.61 × 10−44 | 6.54 × 10−48 | 1.19 × 10−55 | 2.67 × 10−183 |
| 4002 | 1910 | 3483 | 1528 | KP071474b | MH483056 | KX527580, KP071501, KP071502 | NS | 2.16 × 10−69 | 6.59 × 10−49 | 1.71 × 10−35 | 5.99 × 10−21 | 4.73 × 10−51 | 3.12 × 10−7 |
| 1828 | 3940 | 1407 | 3326 | NC_039007b | Unknown | KP071630 | NS | NS | 4.59 × 10−11 | 4.10 × 10−18 | 1.19 × 10−7 | 4.62 × 10−36 | 8.88 × 10−50 |
| 1744a | 1916 | 1402a | 1545 | KP071559b | MN567064 | KP071473 | NS | 3.55 × 10−9 | 6.75 × 10−5 | 8.359 × 10−7 | 1.06 × 10−4 | 7.70 × 10−7 | NS |
The actual breakpoint position is undetermined.
The recombinant sequence may have been misidentified (one of the identified parents might be the recombinant).
NS, nonsignificant.
Analysis of the identified hartmanivirus sequences.
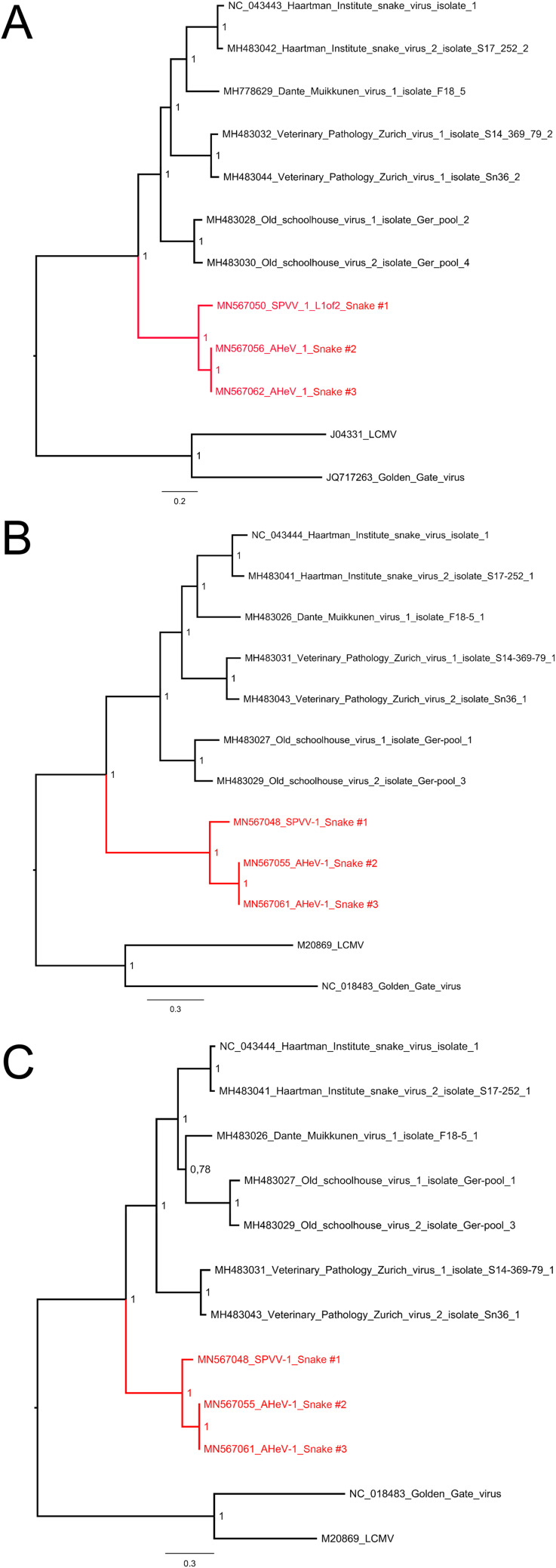
We also used the PASC tool for analyzing the identified hartmanivirus S and L segment CDSs; however, the analyses returned matches with very low sequence identities (22% and below) (Table 2). To compare the sequences to those of known hartmaniviruses, we aligned the identified sequences with those found in GenBank and generated nucleotide identity matrixes (Tables 6 and 7). The analysis showed that the sequences are distant enough from each other and those of the known hartmaniviruses to represent new species: SetVetPat virus 1 (SPVV-1) (in animal 1) and Andere Heimat virus 1 (AHeV-1) (in animals 2 and 3). The phylogenetic analysis of the hartmanivirus L and S segments suggested that these two viruses form a sister clade to the previously known hartmaniviruses (Fig. 5A to C).
TABLE 6.
Nucleotide identities between hartmanivirus L segments identified in this and earlier studiesa
| Animal no., virus segment or virus, accession no. | Identity (%) with: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1, SPVV-1 L piece1/2 | 1, SPVV-1 L piece2/2 | 2, AHeV-1 L | 3, AHeV-1 L | HISV-1, NC_043443.1 | HISV-2, MH483042.1 | OScV-1, MH483028.1 | OScV-2, MH483025.1 | VPZV-1, MH483040.1 | VPZV-2, MH483044.1 | DaMV-1, MH778629.1 | |
| 1, SPVV-1 L piece1/2 | |||||||||||
| 1, SPVV-1 L piece2/2 | 2 | ||||||||||
| 2, AHeV-1 L | 83 | 70 | |||||||||
| 3, AHeV-1 L | 83 | 82 | 100 | ||||||||
| HISV-1, NC_043443.1 | 60 | 56 | 59 | 59 | |||||||
| HISV-2, MH483042.1 | 61 | 56 | 59 | 60 | 89 | ||||||
| OScV-1, MH483028.1 | 62 | 55 | 59 | 59 | 65 | 65 | |||||
| OScV-2, MH483025.1 | 61 | 56 | 59 | 59 | 65 | 65 | 85 | ||||
| VPZV-1, MH483040.1 | 61 | 58 | 60 | 60 | 65 | 65 | 65 | 65 | |||
| VPZV-2, MH483044.1 | 61 | 56 | 59 | 59 | 65 | 65 | 64 | 64 | 86 | ||
| DaMV-1, MH778629.1 | 60 | 56 | 58 | 59 | 69 | 69 | 65 | 65 | 65 | 65 | |
Dark gray, >76% identity between sequences; light gray, <76% identity between sequences.
TABLE 7.
Nucleotide identities between hartmanivirus S segments identified in this and earlier studies a
| Animal no., virus segment or virus, accession no. | Identity (%) with: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1, SPVV-1 S | 2, AHeV-1 S | 3, AHeV-1 S | HISV-1, NC_043444.1 | HISV-2, MH483041.1 | OScV-1, MH483027.1 | OScV-2, MH483029.1 | VPZV-1, MH483039.1 | VPZV-2, MH483043.1 | DaMV-1, MH483026.1 | |
| 1, SPVV-1 S | ||||||||||
| 2, AHeV-1 S | 77 | |||||||||
| 3, AHeV-1 S | 77 | 100 | ||||||||
| HISV-1, NC_043444.1 | 52 | 53 | 54 | |||||||
| HISV-2, MH483041.1 | 53 | 53 | 54 | 86 | ||||||
| OScV-1, MH483027.1 | 52 | 53 | 53 | 61 | 61 | |||||
| OScV-2, MH483029.1 | 53 | 52 | 52 | 61 | 61 | 79 | ||||
| VPZV-1, MH483039.1 | 51 | 51 | 52 | 62 | 62 | 60 | 60 | |||
| VPZV-2, MH483043.1 | 51 | 52 | 52 | 62 | 62 | 60 | 59 | 84 | ||
| DaMV-1, MH483026.1 | 52 | 52 | 53 | 65 | 65 | 60 | 63 | 61 | 61 | |
Dark gray, >76% identity between sequences; light gray, <76% identity between sequences.
FIG 5.
Maximum clade credibility trees for hartmanivirus RdRp, GPC, and NP. (A) The phylogenetic tree based on the RdRp amino acid sequences of the viruses identified in this study and those available in GenBank was constructed using the Bayesian MCMC method with the Blosum model of amino acid substitution. (B) The phylogenetic tree based on the GPC amino acid sequences of the viruses identified in this study and those available in GenBank was constructed using the Bayesian MCMC method with the Blosum model of amino acid substitution. (C) The phylogenetic tree based on the NP amino acid sequences of the viruses identified in this study and those available in GenBank was constructed using the Bayesian MCMC method with the Wag model of amino acid substitution.
Analysis of the novel member of the family Chuviridae.
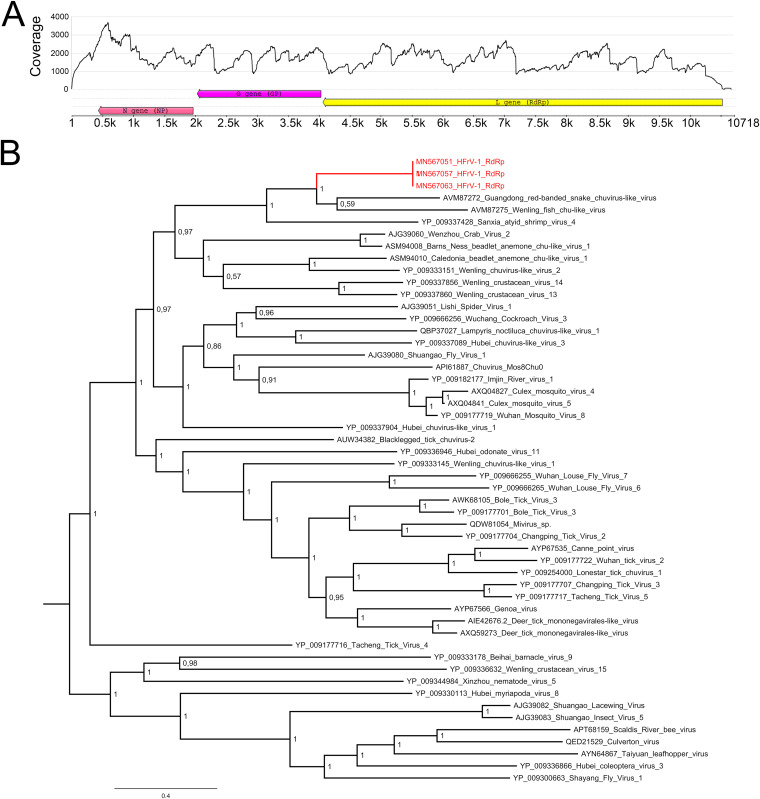
BLAST analysis identified three contigs that showed similarities to chuvirus-like viruses (family Chuviridae, genus Mivirus). These sequences had three open reading frames (ORFs) in antigenomic orientation, representing the L, G, and N genes, with RNA-dependent RNA polymerase (RdRp), glycoprotein (GP), and nucleoprotein (NP) as the respective protein products (Fig. 6A). We named the novel virus Herr Frank virus-1 (HFrV-1). BLAST analysis identified the Guangdong red-banded snake chuvirus-like virus L protein (GenBank accession no. AVM87272.1) as the closest match (48.56% amino acid identity) for the HFrV-1 L gene product (Table 8) and the putative GP (AVM87273.1) and NP (AVM87274.1) of Guangdong red-banded snake chuvirus-like virus as the closest matches for the HFrV-1 GP and N gene products (respective amino acid identities of 43.06% and 41.97%) (Table 8). In addition to BLAST analysis, we employed HMMSCAN (available at https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan) to study the ORFs of HFrV-1. The analyses (Table 8) further confirmed the annotation of the ORFs. The phylogenetic analysis (Fig. 6B) of HFrV and the representatives of other chuvirus-like virus RdRp sequences suggested that HFrV clusters together with Guangdong red-banded snake chuvirus-like virus and Wenling fish chuvirus-like virus (27). Sanxia atyid shrip virus 4 formed an outgroup for these three vertebrate-associated chuvirus-like viruses (Fig. 6B).
FIG 6.
Genome organization, similarity analyses, and phylogenetic tree of HFrV-1. (A) Genome organization and coverage (sequence from snake 1) of HFrV-1. The arrows represent the orientation of the open reading frames (ORFs). The L gene encodes RNA-dependent RNA polymerase (RdRp), the G gene encodes glycoprotein (GP), and the N gene encodes nucleoprotein (NP). The coverage (y axis) show the sequencing depth at each nucleotide position (x axis). (B) A maximum clade credibility tree based on the RdRp amino acid sequences of chuvirus-like viruses and chuviruses. The tree was constructed using the Bayesian MCMC method with the Blosum model of amino acid substitution. Posterior probabilities are shown in each node.
TABLE 8.
Similarity analyses of HFrV-1 open reading frames by BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and HMMSCAN (https://www.ebi.ac.uk/Tools/hmmer/)
| Parameter | ORF1 (RdRp, L gene) | ORF2 (glycoprotein, G gene) | ORF3 (nucleoprotein, N gene) |
|---|---|---|---|
| Protein size (amino acids) | 2,144 | 660 | 503 |
| Signal peptide | No | 1–17 | No |
| Transmembrane domain | 2125–2142 | 542–563 | No |
| Closest match by protein BLAST (identity at amino acid level) | L protein of Guangdong red-banded snake chuvirus-like virus, AVM87272.1 (48.56) | Putative glycoprotein of Guangdong red-banded snake chuvirus-like virus, AVM87273.1 (43.06) | Putative nucleoprotein of Guangdong red-banded snake chuvirus-like virus, AVM87274.1 (41.97) |
| Domains identified by protein BLAST (domain, amino acid region, E value) | Mononegavirales RNA-dependent RNA polymerase (72–1002, 4.25E−60), Mononegavirales mRNA-capping region V (1038–1282, 2.03E−12), mRNA-capping enzyme, Paramyxovirus family (1615–1855, 8.69E−3) | None | None |
| Protein Information Resource Superfamily (PIRSF) matches (accession no.; amino acid region; E value) | RNA-directed RNA polymerase, Paramyxoviridae type (PIRSF000830; 21–584, 594–1066, 1703–1985; 1.7E−24) | None | None |
| Pfam matches (accession no; amino acid region; E value) | Mononeg_RNA_pol (PF00946.19; 55–1013, 1708–1734; 2.1E−73, 0.22) | None | Hypoxia-inducible factor 1 (PF11413.8; 154–167; 3.8E−06) |
| Mononeg_mRNAcap (PF14318.6; 1038–1284; 4.1E−19) | Arginine repressor, DNA binding domain (PF01316.21; 39–65, 283–310; 0.79, 6.4E−05) |
DISCUSSION
This study aimed to confirm the presence of BIBD and reptarenaviruses in boa constrictors, which are indigenous to the Brazilian wildlife. We initially used immunohistology to confirm the presence of reptarenavirus-induced IBs in all four studied snakes, thus confirming the BIBD diagnosis. The subsequent metatranscriptomic analysis of the livers confirmed the presence of reptarenaviruses and helped to identify two novel hartmanivirus species and a novel chuvirus in the snakes. The affected boas had lived in captivity, with contact with other snakes. However, three of the snakes originated from the Amazon region in Brazil, which could indicate that both reptarenaviruses and hartmaniviruses exist in wild snakes in this region.
At the time of sampling/euthanasia, all snakes suffered from bacterial (and fungal) infections caused by opportunistic agents that are part of the mucosal flora in various species, including snakes. These agents occur in the oral cavities of captive snakes with stomatitis or in snakes with septicemia (28, 29); some, such as Enterobacter gergoviae, exist in water or soil (30). All can cause disease, particularly in immunocompromised patients. Secondary infections with related inflammatory processes are common in snakes with BIBD, indicating an immunosuppressive effect at least in later, chronic stages of BIBD. Much less is known about the effect of acute reptarenavirus infections. Based on our experience with in vitro studies, we assume that larger IBs, often more amphophilic than eosinophilic, represent late stages of IB formation. Such IBs often exhibit only a peripheral positive immunohistological reaction for viral NP, similar to the reaction seen in the tissues of the snakes in this study. In vitro, small irregular IBs appear at early stages (approximately 3 days postinfection [dpi]) of reptarenavirus infection, and from 12 dpi onwards the IBs become larger and more electron dense (unpublished data). Thus, we assume that the snakes included in this study were chronically reptarenavirus infected and therefore immunocompromised (the hartmanivirus and chuvirus coinfections may also have contributed), which in turn led to the observed secondary infections.
Metatranscriptomic analysis of the animals revealed the presence of multiple reptarenavirus L segments but only a single S segment per snake. The finding is well in line with earlier observations of reptarenavirus coinfections being common in snakes with BIBD (8, 9). One of the identified segments, the KaBV-1 L segment (animals 1 to 3), showed a striking 97% identity to a previously identified (24) reptarenavirus genome segment. In addition, one L segment showed approximately 86% identity to the UHV-2 L segment, but the other segments differed enough from the previously identified reptarenavirus segments to warrant classification as novel reptarenavirus species. Interestingly, we did not find University of Giessen virus or the “S6-like” S segment in any of the studied snakes, even though the segment is most often reported in captive snakes with BIBD (8, 9, 13, 24). In the phylogenetic trees, most of the reptarenaviruses identified from the Brazilian snakes were interspersed among the viruses previously detected from captive snakes in Europe and the United States. Given the high genetic diversity between the virus species within genus Reptarenavirus, it is likely that the evolutionary history of reptarenaviruses exceeds the time that boid snakes have been housed in captivity. Therefore, the most likely origin of these viruses lies in wild snakes. However, due to the limited sequence data available from wild snakes, our current analysis is not sufficient to assess the origin of reptarenaviruses. Most of the identified viruses were divergent enough to represent new species, and therefore, it is not possible to pinpoint the potential viral transmission events from wild host population to captive snakes. Notably, however, one of the reptarenaviruses, KaBV-1, showed high identity to a strain identified from captive snakes in Switzerland (24). This may indicate that reptarenaviruses have been introduced into captive snake populations by wild boas and then have been exported with them, due to the lack of clinical signs of infection/disease (13). A great number of snakes are traded annually; according to the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES), 20,000 snakes (6,600 pythonids and 3,100 boids) were transported in 2018 alone (https://trade.cites.org/en/cites_trade/#). While the trading and transport may technically follow CITES regulations, a great number of transported animals are likely not captive bred but wild caught. In addition to the transport of animals following CITES regulations, smuggling of wild-caught snakes occurs frequently. The spread would further be aided by the fact that reptarenavirus infection does not induce clinical signs rapidly, especially not in boas (10), and is vertically transmitted (24). The studies on snakes with BIBD strongly suggest that reptarenavirus L and S segments are able to pair with each other rather freely, since most often the individuals harbor more L segments than S segments (8, 9, 24). Assuming that snakes, or more accurately boas and pythons, are the reservoir hosts of reptarenaviruses and that reptarenaviruses have coevolved with their reservoir hosts, then multiple cross-species transmission events could explain the status quo in captive snakes. However, with the current set of data we cannot rule out the possibility that the wild-caught boas included in the study had not been infected during cohousing.
The identification of novel hartmaniviruses in Brazilian B. constrictor snakes is interesting, since until now hartmanivirus infection has been reported only in European captive snakes (9, 18). The hartmanivirus-infected snakes included in the present study had developed BIBD as confirmed by the presence of IBs in both blood smear and tissues, which is in accordance with our earlier findings (9, 18). They showed strong expression of hartmanivirus NP in parenchymal cells in various organs. We have thus far detected hartmaniviruses mainly in snakes with BIBD; however, the fact that we look for viruses mainly in diagnostic cases might introduce a bias and could explain the seeming correlation between hartmanivirus infection and BIBD. In fact, when studying samples collected from a single breeding colony for the presence of IBs, reptarenaviruses, and hartmaniviruses, we did not find a significant correlation between hartmanivirus infection and BIBD (13). Although hartmanivirus infection appears to most often occur simultaneously with reptarenavirus infection, hartmaniviruses can infect and replicate without a coinfecting reptarenavirus (18), and further studies need to address their pathogenicity. Like that of reptarenaviruses, the origin of hartmaniviruses remains unknown. In addition to snakes being the reservoir hosts of the viruses, one could speculate that blood-feeding parasites, e.g., mites, ticks, mosquitoes, etc., would serve as reservoirs and/or vectors in virus transmission.
The novel chuvirus, HFrV-1, found in three BIBD-positive snakes originating from a snake sanctuary in the Amazonas region, but housed in a smaller colony for several years afterwards, was an unexpected finding. By amino acid identity, the closest relative to the newly found mononegavirus is the Guandong red-banded snake chuvirus, which was identified from a liver sample of a Chinese snake (27). In general, the identification of chuvirus-like viruses in fish and snakes from different continents (Asia and the Americas) suggests that chuviruses might be common and geographically widespread. Due to bacterial and viral coinfections in the snakes with HFrV-1 infection, we cannot draw conclusions on the potential morbidity of HFrV-1. The fact that the identified viruses showed nearly identical sequences suggests that the chuvirus infection may have occurred during captivity.
According to Gibbon et al., the major threats causing declining reptilian populations are habitat loss and degradation, introduction of invasive species, environmental pollution, disease, unsustainable use, and global climate change (31). Our results raise an obvious question: from where did the reptarenaviruses that infected the diseased snakes come? Both cohoused imported snakes and local wild snakes are a potential source of infection. Animals 1 to 3 originated from the Amazonas region, where they resided in a snake sanctuary before moving to a private collection in Porto Alegre. It was not possible to obtain information on other snake species housed in the sanctuary, since it was closed several years before. We also lack more specific information on the origin of the snakes. Similarly, the origin of animal 4 remained unknown. However, all animals studied were B. constrictor, indigenous to Brazil, and it is therefore possible that they originated from the wild. If snakes are not the reservoir hosts of reptarenaviruses, then the occurrence of BIBD-positive B. constrictor in Brazil is an alarming signal posing a potential threat to Brazilian wild B. constrictor populations.
MATERIALS AND METHODS
Animals.
The study was undertaken with four captive adult B. constrictor constrictor snakes. Three were from a single private owner (animals 1 to 3), and the fourth was from a zoological garden (Table 1). All animals were submitted for diagnostic postmortem examination to the Department of Veterinary Pathology in Porto Alegre. Tissue specimens were fixed in 10% buffered formalin for histological and immunohistological examination. Additional sets of samples were stored frozen in RNAlater stabilization solution (Thermo Fisher Scientific) for RNA extraction. For animals 1, 3, and 4, samples from the oral and nasal lesions were subjected to a routine bacteriological examination; for animal 1, a routine mycological examination was also performed.
Histology and immunohistology.
Formalin-fixed tissue specimens were trimmed and routinely paraffin wax embedded. Consecutive sections (3 to 5 μm) were prepared and stained with hematoxylin-eosin (HE) and special stains (periodic acid-Schiff [PAS] reaction and Grocott methenamine silver stains) when appropriate. Further sections were subjected to immunohistological staining for reptarenavirus and hartmanivirus NP as described previously (5).
Antibodies, protein expression, and immunization.
The anti-UHV NP and anti-UHV NP C-terminal antibodies were described earlier (5, 32). To generate broadly cross-reactive antiserum against reptarenavirus NPs, we performed amino acid alignment for the reptarenavirus NPs available in GenBank. Based on homology between the sequences, we selected the following regions: amino acids 47 to 140 from UGV-1 (GenBank accession no. YP_009508464.1), 173 to 224 from UHV-1 (YP_009019205.1), 233 to 270 from UHV-1 (YP_009019205.1), 286 to 359 from UGV-1 (YP_009508464.1), 208 to 280 from UGV-1 (YP_009508464.1), and 494–567 from UHV-1 (YP_009019205.1). To generate a cross-reactive antiserum against hartmanivirus NPs, we used the same approach and selected the following regions: amino acids 199 to 256 from Veterinary Pathology Zürich virus 1 (VPZV-1) (AZI72586.1), 132 to 180 from Haartman Institute Snake virus 2 (HISV-2) (AZI72594.1), 257 to 299 from VPZV-1 (AZI72586.1), and 312 to 364 from VPZV-2 (AZI72596.1). We included five glycine residues between the selected epitopes and ordered the engineered proteins as synthetic genes optimized for Escherichia coli expression in plasmid pET-20b(+) from GenScript. We transformed One Shot BL21(DE3) Chemically Competent (Thermo Scientific) E. coli with the plasmids following the manufacturer’s protocol and performed protein expression and purification via His tag as described previously (25, 32). Antisera against the purified proteins were raised by BioGenes, as described in earlier studies (25, 26, 32). We designated the novel antisera anti-pan-reptarenavirus and anti-pan-hartmani.
NGS and genome assembly.
We extracted RNAs for next-generation sequencing (NGS) from liver samples stored frozen in RNAlater (the sample from animal 1 had been kept at ambient temperature for a few weeks prior to extraction) as described previously (24), prepared NGS libraries, and performed sequencing and subsequent genome assembly as described previously (24, 25).
RT-PCR and Sanger sequencing.
For the sample that had been stored in RNAlater at ambient temperature (animal 1), we obtained open reading frames (ORFs) for several NP, ZP, GPC, and RdRp genes instead of complete L and S segments by NGS and de novo assembly. To complete the L and S segment sequences, we designed the following primers to amplify the missing intergenic regions: Br_GPC1 (5′-ACACTTGGATTCTATGGGAGT-3′), Br_NP1 (5′-ACTGCATGGTGTTCTCAAG-3′), Br_ZP1 (5′-GAGTCTAACCAATCCCAGAA-3′), Br_ZP2 (5′-CATGCCTAATGGCAAAAC-3′), Br_ZP3 (5′-CAGAATGTAGGGCAACAC-3′), Br_ZP4 (5′-AGGGTCTAAATCAACATCCC-3′), Br_UHV_RdRp (5′-GTCAGAATATCACTCCTGGAG-3′), Br_RdRp2 (5′-TAGGGTGACACTTTTGAAGG-3′), Br_RdRp3 (5′-GAACATTAGGGTATCACTCCTC-3′), and Br_RdRp4 (5′-AGAGTCTAAGGGTCCTGGA-3′). We performed reverse transcriptase PCR (RT-PCR) with all primer combinations (ZPs with RdRps, and GPC with NP) as described previously (24) and used GeneJET gel extraction kit (Thermo Fisher Scientific) to purify the RT-PCR products, which were further cleaned with AMPure XP beads (Beckman Coulter) before ligation to plasmid using the Zero Blunt TOPO PCR cloning kit (Thermo Fisher Scientific); all steps were according to the manufacturer’s instructions. Chemically competent E. coli (TOP10; Thermo Fisher Scientific) transformed with the ligated plasmids were grown on LB plates with 100 μg/ml of ampicillin overnight at 37°C, and colonies were picked and grown in 5 ml of 2× YT medium with 100 μg/ml of ampicillin overnight at 37°C. The plasmids were purified from 2 ml of the overnight culture using the GeneJET plasmid miniprep kit (Thermo Fisher Scientific) and sent for Sanger sequencing to DNA Sequencing and Genomics, Institute of Biotechnology, University of Helsinki.
Phylogenetic analysis.
The amino acid sequences of the representatives of all reptarena- and hartmaniviruses were downloaded from GenBank, and aligned with the amino acid sequences of viruses identified in this study using the MAFFT E-INS-i algorithm (33). For chuvirus-like sequences, the 100 closest BLASTx matches for the putative RdRp amino acid sequence were downloaded from GenBank and aligned as indicated above. Redundant sequences (fragmental and identical sequences) were removed from the data set.
The best-fit amino acid substitution models and phylogenetic trees were inferred using the Bayesian Monte Carlo Markov chain (MCMC) method implemented in MrBayes v3.2.6. (34). MrBayes was run for 500,000 generations and sampled every 5,000 steps, with final standard deviations between two runs of <0.02 for all analyses. The analyses were carried out at the CSC server (IT Center for Science Ltd., Espoo, Finland).
Recombination analysis.
Recombination events were sought using the RDP (35), bootscan (36), maxchi (37), chimera (38), 3seq (39), geneconv (40), and siscan (41) methods implemented in RDP4 software (42).
Data availability.
The names for newly sequenced viruses with corresponding abbreviations and GenBank accession numbers are provided in Table 2.
ACKNOWLEDGMENTS
We are grateful to the laboratory staff of the Histology Laboratory, Institute of Veterinary Pathology, Vetsuisse Faculty, University of Zürich, and to colleagues and professors of Universidade Federal do Rio Grande do Sul for excellent technical support. We also thank L. Sonne and S. P. Pavarini, who assisted in the snake necropsies during their undergraduate studies.
This study received financial support from the Leading House for the Latin American Region, University of St. Gallen, Switzerland, and the Academy of Finland (grant numbers 1308613 and 1314119).
This work is dedicated to the memory of Herbert Frank and to his colleagues at the Institute of Veterinary Pathology, Faculty of Veterinary Medicine, Justus-Liebig-University Giessen, Germany, where the three senior authors of this publication were trained in veterinary pathology.
REFERENCES
- 1.Chang L, Jacobson ER. 2010. Inclusion body disease, a worldwide infectious disease of boid snakes: a review. J Exotic Pet Med 19:216–225. doi: 10.1053/j.jepm.2010.07.014. [DOI] [Google Scholar]
- 2.Schumacher J, Jacobson ER, Homer BL, Gaskin JM. 1994. Inclusion body disease in boid snakes. J Zoo Wildl Med 25:511–524. [Google Scholar]
- 3.Stenglein MD, Sanders C, Kistler AL, Ruby JG, Franco JY, Reavill DR, Dunker F, Derisi JL. 2012. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: candidate etiological agents for snake inclusion body disease. mBio 3:e00180-12. doi: 10.1128/mBio.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodewes R, Kik MJ, Raj VS, Schapendonk CM, Haagmans BL, Smits SL, Osterhaus AD. 2013. Detection of novel divergent arenaviruses in boid snakes with inclusion body disease in The Netherlands. J Gen Virol 94:1206–1210. doi: 10.1099/vir.0.051995-0. [DOI] [PubMed] [Google Scholar]
- 5.Hetzel U, Sironen T, Laurinmaki P, Liljeroos L, Patjas A, Henttonen H, Vaheri A, Artelt A, Kipar A, Butcher SJ, Vapalahti O, Hepojoki J. 2013. Isolation, identification, and characterization of novel arenaviruses, the etiological agents of boid inclusion body disease. J Virol 87:10918–10935. doi: 10.1128/JVI.01123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodewes R, Raj VS, Kik MJ, Schapendonk CM, Haagmans BL, Smits SL, Osterhaus AD. 2014. Updated phylogenetic analysis of arenaviruses detected in boid snakes. J Virol 88:1399–1400. doi: 10.1128/JVI.02753-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hetzel U, Sironen T, Laurinmaki P, Liljeroos L, Patjas A, Henttonen H, Vaheri A, Artelt A, Kipar A, Butcher SJ, Vapalahti O, Hepojoki J. 2014. Reply to “Updated phylogenetic analysis of arenaviruses detected in boid snakes.” J Virol 88:1401–1413. doi: 10.1128/JVI.03044-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stenglein MD, Jacobson ER, Chang LW, Sanders C, Hawkins MG, Guzman DS, Drazenovich T, Dunker F, Kamaka EK, Fisher D, Reavill DR, Meola LF, Levens G, DeRisi JL. 2015. Widespread recombination, reassortment, and transmission of unbalanced compound viral genotypes in natural arenavirus infections. PLoS Pathog 11:e1004900. doi: 10.1371/journal.ppat.1004900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hepojoki J, Salmenpera P, Sironen T, Hetzel U, Korzyukov Y, Kipar A, Vapalahti O. 2015. Arenavirus coinfections are common in snakes with boid inclusion body disease. J Virol 89:8657–8660. doi: 10.1128/JVI.01112-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stenglein MD, Sanchez-Migallon Guzman D, Garcia VE, Layton ML, Hoon-Hanks LL, Boback SM, Keel MK, Drazenovich T, Hawkins MG, DeRisi JL. 2017. Differential disease susceptibilities in experimentally reptarenavirus-infected boa constrictors and ball pythons. J Virol 91:e00451-17. doi: 10.1128/JVI.00451-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang LW, Fu A, Wozniak E, Chow M, Duke DG, Green L, Kelley K, Hernandez JA, Jacobson ER. 2013. Immunohistochemical detection of a unique protein within cells of snakes having inclusion body disease, a world-wide disease seen in members of the families Boidae and Pythonidae. PLoS One 8:e82916. doi: 10.1371/journal.pone.0082916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang L, Fu D, Stenglein MD, Hernandez JA, DeRisi JL, Jacobson ER. 2016. Detection and prevalence of boid inclusion body disease in collections of boas and pythons using immunological assays. Vet J 218:13–18. doi: 10.1016/j.tvjl.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Windbichler K, Michalopoulou E, Palamides P, Pesch T, Jelinek C, Vapalahti O, Kipar A, Hetzel U, Hepojoki J. 2019. Antibody response in snakes with boid inclusion body disease. PLoS One 14:e0221863. doi: 10.1371/journal.pone.0221863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keilwerth M, Buhler I, Hoffmann R, Soliman H, El-Matbouli M. 2012. Inclusion body disease (IBD of boids)—a haematological, histological and electron microscopical study. Berl Munch Tierarztl Wochenschr 125:411–417. [PubMed] [Google Scholar]
- 15.Aqrawi T, Stohr AC, Knauf-Witzens T, Krengel A, Heckers KO, Marschang RE. 2015. Identification of snake arenaviruses in live boas and pythons in a zoo in Germany. Tierarztl Prax Ausg K 43:239–247. doi: 10.15654/TPK-140743. [DOI] [PubMed] [Google Scholar]
- 16.Radoshitzky SR, Bao Y, Buchmeier MJ, Charrel RN, Clawson AN, Clegg CS, DeRisi JL, Emonet S, Gonzalez JP, Kuhn JH, Lukashevich IS, Peters CJ, Romanowski V, Salvato MS, Stenglein MD, de la Torre JC. 2015. Past, present, and future of arenavirus taxonomy. Arch Virol 160:1851–1874. doi: 10.1007/s00705-015-2418-y. [DOI] [PubMed] [Google Scholar]
- 17.Radoshitzky SR, Buchmeier MJ, Charrel RN, Clegg JCS, Gonzalez JJ, Gunther S, Hepojoki J, Kuhn JH, Lukashevich IS, Romanowski V, Salvato MS, Sironi M, Stenglein MD, de la Torre JC, ICTV Report Consortium. 2019. ICTV virus taxonomy profile: Arenaviridae. J Gen Virol 100:1200–1201. doi: 10.1099/jgv.0.001280. [DOI] [PubMed] [Google Scholar]
- 18.Hepojoki J, Hepojoki S, Smura T, Szirovicza L, Dervas E, Prahauser B, Nufer L, Schraner EM, Vapalahti O, Kipar A, Hetzel U. 2018. Characterization of Haartman Institute snake virus-1 (HISV-1) and HISV-like viruses—the representatives of genus Hartmanivirus, family Arenaviridae. PLoS Pathog 14:e1007415. doi: 10.1371/journal.ppat.1007415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maes P, Alkhovsky SV, Bao Y, Beer M, Birkhead M, Briese T, Buchmeier MJ, Calisher CH, Charrel RN, Choi IR, Clegg CS, de la Torre JC, Delwart E, DeRisi JL, Bello PLD, Serio FD, Digiaro M, Dolja VV, Drosten C, Druciarek TZ, Du J, Ebihara H, Elbeaino T, Gergerich RC, Gillis AN, Gonzalez JJ, Haenni AL, Hepojoki J, Hetzel U, Ho T, Hong N, Jain RK, Jansen van Vuren P, Jin Q, Jonson MG, Junglen S, Keller KE, Kemp A, Kipar A, Kondov NO, Koonin EV, Kormelink R, Korzyukov Y, Krupovic M, Lambert AJ, Laney AG, LeBreton M, Lukashevich IS, Marklewitz M, Markotter W, Martelli GP, Martin RR, Mielke-Ehret N, Muhlbach HP, Navarro B, Ng TFF, Nunes MRT, Palacios G, Paweska JT, Peters CJ, Plyusnin A, Radoshitzky SR, Romanowski V, Salmenpera P, Salvato MS, Sanfacon H, Sasaya T, Schmaljohn C, Schneider BS, Shirako Y, Siddell S, Sironen TA, Stenglein MD, Storm N, Sudini H, Tesh RB, Tzanetakis IE, Uppala M, Vapalahti O, Vasilakis N, Walker PJ, Wang G, Wang L, Wang Y, Wei T, Wiley MR, Wolf YI, Wolfe ND, Wu Z, Xu W, Yang L, Yang Z, Yeh SD, Zhang YZ, Zheng Y, Zhou X, Zhu C, Zirkel F, Kuhn JH. 2018. Taxonomy of the family Arenaviridae and the order Bunyavirales: update 2018. Arch Virol 163:2295–2310. doi: 10.1007/s00705-018-3843-5. [DOI] [PubMed] [Google Scholar]
- 20.Abudurexiti A, Adkins S, Alioto D, Alkhovsky SV, Avšič-Županc T, Ballinger MJ, Bente DA, Beer M, Bergeron É, Blair CD, Briese T, Buchmeier MJ, Burt FJ, Calisher CH, Cháng C, Charrel RN, Choi IR, Clegg JCS, de la Torre JC, de Lamballerie X, Dèng F, Di Serio F, Digiaro M, Drebot MA, Duàn X, Ebihara H, Elbeaino T, Ergünay K, Fulhorst CF, Garrison AR, Gāo GF, Gonzalez J-PJ, Groschup MH, Günther S, Haenni A-L, Hall RA, Hepojoki J, Hewson R, Hú Z, Hughes HR, Jonson MG, Junglen S, Klempa B, Klingström J, Kòu C, Laenen L, Lambert AJ, Langevin SA, Liu D, Lukashevich IS, Luò T, Lü C, Maes P, de Souza WM, Marklewitz M, Martelli GP, Matsuno K, Mielke-Ehret N, Minutolo M, Mirazimi A, Moming A, Mühlbach H-P, Naidu R, Navarro B, Nunes MRT, Palacios G, Papa A, Pauvolid-Corrêa A, Pawęska JT, Qiáo J, Radoshitzky SR, Resende RO, Romanowski V, Sall AA, Salvato MS, Sasaya T, Shěn S, Shí X, Shirako Y, Simmonds P, Sironi M, Song J-W, Spengler JR, Stenglein MD, Sū Z, Sūn S, Táng S, Turina M, Wáng B, Wáng C, Wáng H, Wáng J, Wèi T, Whitfield AE, Zerbini FM, Zhāng J, Zhāng L, Zhāng Y, Zhang Y-Z, Zhāng Y, Zhou X, Zhū L, Kuhn JH. 2019. Taxonomy of the order Bunyavirales: update 2019. Arch Virol 164:1949–1965. doi: 10.1007/s00705-019-04253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abba Y, Hassim H, Hamzah H, Ibrahim OE, Ilyasu Y, Bande F, Mohd Lila MA, Noordin MM. 2016. In vitro isolation and molecular identification of reptarenavirus in Malaysia. Virus Genes 52:640–650. doi: 10.1007/s11262-016-1345-7. [DOI] [PubMed] [Google Scholar]
- 22.Hyndman TH, Marschang RE, Bruce M, Clark P, Vitali SD. 2019. Reptarenaviruses in apparently healthy snakes in an Australian zoological collection. Aust Vet J 97:93–102. doi: 10.1111/avj.12792. [DOI] [PubMed] [Google Scholar]
- 23.Turchetti A, Tinoco H, de Campos Cordeiro Malta M, Loyola Teixeira da Costa E, Pessanha A, Soave S, Paixao A, Santos R. 2013. Inclusion body disease in a Corallus hortulanus. Braz J Vet Pathol 6:15–18. [Google Scholar]
- 24.Keller S, Hetzel U, Sironen T, Korzyukov Y, Vapalahti O, Kipar A, Hepojoki J. 2017. Co-infecting reptarenaviruses can be vertically transmitted in boa constrictor. PLoS Pathog 13:e1006179. doi: 10.1371/journal.ppat.1006179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dervas E, Hepojoki J, Laimbacher A, Romero-Palomo F, Jelinek C, Keller S, Smura T, Hepojoki S, Kipar A, Hetzel U. 2017. Nidovirus-associated proliferative pneumonia in the green tree python (Morelia viridis). J Virol 91:e00718-17. doi: 10.1128/JVI.00718-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hetzel U, Szirovicza L, Smura T, Prahauser B, Vapalahti O, Kipar A, Hepojoki J. 2019. Identification of a novel deltavirus in boa constrictors. mBio 10:e00014-19. doi: 10.1128/mBio.00014-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi M, Lin XD, Chen X, Tian JH, Chen LJ, Li K, Wang W, Eden JS, Shen JJ, Liu L, Holmes EC, Zhang YZ. 2018. The evolutionary history of vertebrate RNA viruses. Nature 556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 28.Pees M, Schmidt V, Marschang RE, Heckers O, Thielebein J, Seybold J, Krautwald ME. 2009. Untersuchungen auf infektiöse Erkrankungen in unauffälligen Boiden-Beständen, p 33–38. 1 DVG-Tagung Über Vogel- Und Reptilienkrankeiten. [Google Scholar]
- 29.Grego KF, Carvalho MPNd, Cunha MPV, Knöbl T, Pogliani FC, Catão-Dias JL, Sant'Anna SS, Ribeiro MS, Sellera FP. 2017. Antimicrobial photodynamic therapy for infectious stomatitis in snakes: clinical views and microbiological findings. Photodiagn Photodyn Ther 20:196–200. doi: 10.1016/j.pdpdt.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 30.dos Santos GS, Solidônio EG, Costa MCVV, Melo ROA, de Souza IFAC, Silva GR, Sena KXFR. 2015. Study of the Enterobacteriaceae group CESP (Citrobacter, Enterobacter, Serratia, Providencia, Morganella and Hafnia): a review, p 794–805. In Méndez-Vilas A. (ed), The battle against microbial pathogens: basic science, technological advances and educational programs. Formatex Research Center S.L, Badajoz, Spain. [Google Scholar]
- 31.Gibbons JW, Scott DE, Ryan TJ, Buhlmann KA, Tuberville TD, Metts BS, Greene JL, Mills T, Leiden Y, Poppy S, Winne CT. 2000. The global decline of reptiles, déjà vu amphibians: reptile species are declining on a global scale. Six significant threats to reptile populations are habitat loss and degradation, introduced invasive species, environmental pollution, disease, unsustainable use, and global climate change. Bioscience 50:653–666. doi: 10.1641/0006-3568(2000)050[0653:TGDORD]2.0.CO;2. [DOI] [Google Scholar]
- 32.Hepojoki J, Kipar A, Korzyukov Y, Bell-Sakyi L, Vapalahti O, Hetzel U. 2015. Replication of boid inclusion body disease-associated arenaviruses is temperature sensitive in both boid and mammalian cells. J Virol 89:1119–1128. doi: 10.1128/JVI.03119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin D, Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 36.Salminen MO, Carr JK, Burke DS, McCutchan FE. 1995. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses 11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 37.Smith JM. 1992. Analyzing the mosaic structure of genes. J Mol Evol 34:126–129. doi: 10.1007/bf00182389. [DOI] [PubMed] [Google Scholar]
- 38.Posada D, Crandall KA. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A 98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boni MF, Posada D, Feldman MW. 2007. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047. doi: 10.1534/genetics.106.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Padidam M, Sawyer S, Fauquet CM. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- 41.Gibbs MJ, Armstrong JS, Gibbs AJ. 2000. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582. doi: 10.1093/bioinformatics/16.7.573. [DOI] [PubMed] [Google Scholar]
- 42.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The names for newly sequenced viruses with corresponding abbreviations and GenBank accession numbers are provided in Table 2.