Coronaviruses employ a genetic economy by encoding multifunctional proteins that function in viral replication and also modify the host environment to disarm the innate immune response. The coronavirus papain-like protease 2 (PLP2) domain possesses protease activity, which cleaves the viral replicase polyprotein, and also DUB activity (deconjugating ubiquitin/ubiquitin-like molecules from modified substrates) using identical catalytic residues. To separate the DUB activity from the protease activity, we employed a structure-guided mutagenesis approach and identified residues that are important for ubiquitin binding. We found that mutating the ubiquitin-binding residues results in a PLP2 that has reduced DUB activity but retains protease activity. We engineered a recombinant murine coronavirus to express the DUB mutant and showed that the DUB mutant virus activated an earlier type I interferon response in macrophages and exhibited reduced replication in mice. The results of this study demonstrate that PLP2/DUB is an interferon antagonist and a virulence trait of coronaviruses.
KEYWORDS: DUB activity, IFN antagonist, PLP2, PLP2 structure, coronavirus, papain-like protease
ABSTRACT
Coronaviruses express a multifunctional papain-like protease, termed papain-like protease 2 (PLP2). PLP2 acts as a protease that cleaves the viral replicase polyprotein and as a deubiquitinating (DUB) enzyme which removes ubiquitin (Ub) moieties from ubiquitin-conjugated proteins. Previous in vitro studies implicated PLP2/DUB activity as a negative regulator of the host interferon (IFN) response, but the role of DUB activity during virus infection was unknown. Here, we used X-ray structure-guided mutagenesis and functional studies to identify amino acid substitutions within the ubiquitin-binding surface of PLP2 that reduced DUB activity without affecting polyprotein processing activity. We engineered a DUB mutation (Asp1772 to Ala) into a murine coronavirus and evaluated the replication and pathogenesis of the DUB mutant virus (DUBmut) in cultured macrophages and in mice. We found that the DUBmut virus replicates similarly to the wild-type (WT) virus in cultured cells, but the DUBmut virus activates an IFN response at earlier times compared to the wild-type virus infection in macrophages, consistent with DUB activity negatively regulating the IFN response. We compared the pathogenesis of the DUBmut virus to that of the wild-type virus and found that the DUBmut-infected mice had a statistically significant reduction (P < 0.05) in viral titer in liver and spleen at day 5 postinfection (d p.i.), although both wild-type and DUBmut virus infections resulted in similar liver pathology. Overall, this study demonstrates that structure-guided mutagenesis aids the identification of critical determinants of the PLP2-ubiquitin complex and that PLP2/DUB activity plays a role as an interferon antagonist in coronavirus pathogenesis.
IMPORTANCE Coronaviruses employ a genetic economy by encoding multifunctional proteins that function in viral replication and also modify the host environment to disarm the innate immune response. The coronavirus papain-like protease 2 (PLP2) domain possesses protease activity, which cleaves the viral replicase polyprotein, and also DUB activity (deconjugating ubiquitin/ubiquitin-like molecules from modified substrates) using identical catalytic residues. To separate the DUB activity from the protease activity, we employed a structure-guided mutagenesis approach and identified residues that are important for ubiquitin binding. We found that mutating the ubiquitin-binding residues results in a PLP2 that has reduced DUB activity but retains protease activity. We engineered a recombinant murine coronavirus to express the DUB mutant and showed that the DUB mutant virus activated an earlier type I interferon response in macrophages and exhibited reduced replication in mice. The results of this study demonstrate that PLP2/DUB is an interferon antagonist and a virulence trait of coronaviruses.
INTRODUCTION
Coronaviruses (CoVs) are members of the order Nidovirales, which includes enveloped viruses with large (∼30-kb), positive-sense single-stranded RNA genomes that yield a characteristic nested set of subgenomic mRNAs during replication in the cytoplasm of infected cells (1, 2). The genome organization for coronaviruses is highly conserved, with the 5′-most two-thirds of the genome encoding the replicase polyprotein, followed by sequences encoding the canonical structural proteins, i.e., spike, envelope, membrane, and nucleocapsid. Many CoVs contain accessory genes, which are interspersed among the genes for the structural proteins. Although these accessory genes are not necessarily required for virus replication and are, in general, not highly conserved within the virus family, many encode proteins that regulate the host response (3). Interestingly, coronavirus replicase proteins, which are highly conserved, can also act as antagonists to block or delay the host innate immune response to infection (1, 4–8). That a slew of coronavirus-encoded accessory and nonaccessory proteins have been shown to shape the host antiviral response suggests that virally mediated subversion of host defenses is an important element of infection. Here, we focus on the viral protease/deubiquitinase (DUB) with the goal of assessing the role of DUB activity in shaping the pathogenesis of mouse hepatitis virus (MHV), a model murine coronavirus.
Coronavirus proteases are essential for viral replication, as they are responsible for processing the replicase polyprotein (2, 9–11). The murine coronavirus MHV encodes three proteases: two papain-like proteases (PLP1 and PLP2) and one chymotrypsin-like protease (3CLpro, also termed Mpro). MHV PLP2 is similar to the single papain-like protease (termed PLpro) of severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) (9, 12–16). We and others have revealed that PLP2 or PLpro of multiple coronaviruses, including MHV, is multifunctional, not only capable of cleaving the viral polyprotein but also possessing deubiquitinase (DUB) and deISGylating (deconjugating interferon-stimulated gene 15 [ISG15] molecule from modified substrates) activities (9, 12, 13, 15–27). However, it has been challenging to study the effects of a mutated DUB on virus replication and pathogenesis because the protease and DUB activities share the same catalytic site, disruption of which is lethal for virus replication. Therefore, it was necessary for us to identify residue(s) within the MHV protease/DUB-ubiquitin (Ub) binding surface that can be mutated to result in reduced DUB activity without affecting polyprotein processing. In this report, we describe the X-ray structure-guided identification of such residues and evaluate the replication, interferon (IFN) antagonistic effect, and pathogenesis of a recombinant DUB mutant MHV.
RESULTS
Structure-guided identification of PLP2 residues interacting with ubiquitin.
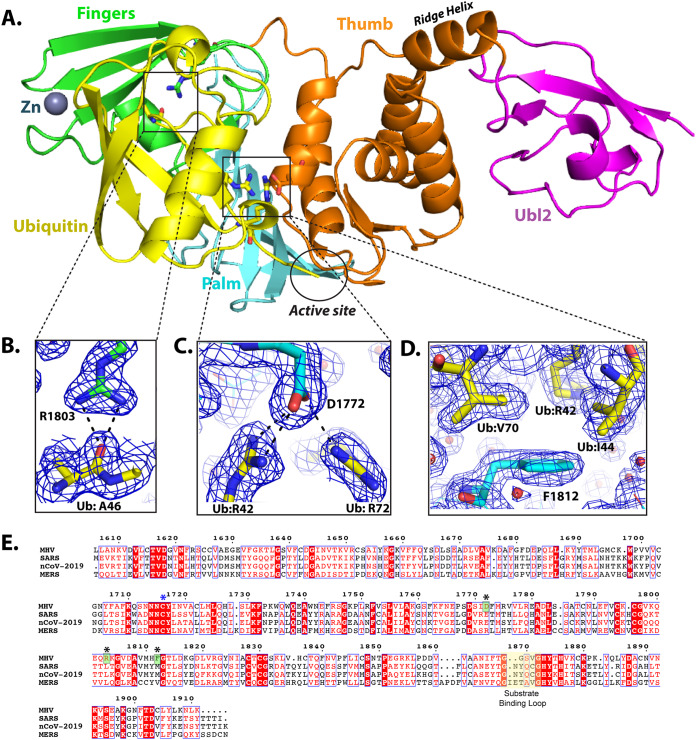
To investigate the role of viral deubiquitinating activity in coronavirus replication and pathogenesis, we first needed to identify amino acid residues that, when mutated, result in reduced DUB activity while preserving the enzyme’s deISGylating and protease activities, the latter being necessary for viral replication. X-ray structural studies of SARS-CoV and MERS-CoV papain-like protease/DUBs cocrystalized with ubiquitin modified at the C terminus with a covalent warhead (aldehyde, 3-bromo-propylamine, or propargylamine) allowed for identification of residues that are important for direct interaction with ubiquitin (17, 18, 20). Here, we took a slightly different approach and mutated the catalytic cysteine (C1716) of MHV PLP2 to a serine residue and cocrystalized it with free ubiquitin. The X-ray structure of MHV PLP2 (C1716S) in complex with ubiquitin (PDB code 5WFI) was determined to a resolution of 1.85 Å and an overall Rfree of 19.6% and Rwork of 15.8%. The overall structure of the MHV PLP2-ubiquitin complex is similar to other PLP2/PLpro ubiquitin-bound structures (Fig. 1A). Ubiquitin binds within the palm region and is gripped by the zinc finger motif, while the C terminus extends into the active site.
FIG 1.
X-ray structure of the MHV PLP2-ubiquitin complex and residues involved in ubiquitin binding. (A) Overall structure of MHV PLP2-C1716S-Ub complex. Domains are color coded as follows: yellow, Ub; purple, Ubl2 domain of PLP2; orange, thumb domain; cyan, palm domain; green, fingers domain. Residue D1772 of PLP2 is located outside the active site, which is circled in black. (B) Hydrogen bond interactions between MHV PLP2 R1803 and the backbone of A46 of ubiquitin. (C) Binding interactions between MHV PLP2 D1772 and two arginine residues (R42 and R72) of ubiquitin. (D) Hydrophobic interactions between F1812 of MHV PLP2 and ubiquitin residues I44 and V70. The 2Fo-Fc maps (blue) surrounding the residues are contoured at 1σ in each panel. The PDB coordinates for the MHV PLP2-C1716S-Ub complex are available under PDB code 5WFI. (E) Sequence alignment (MultAlign) of coronavirus papain-like protease/deubiquitinating domains from MHV (amino acids [aa] 1606 to 1911, GenBank accession number AAX23975), SARS (aa 1541 to 1854, accession number ACZ72209), SARS-CoV-2 (aa 1564 to 1878, accession number QHO60603), and MERS (aa 1480 to 1803, accession number AHY21467). Amino acids are colored according to similarity using the RISER coloring scheme. Numbering shown is based on MHV sequence. Amino acids mutated in this study are indicated with a black asterisk, the catalytic cysteine is indicated by a blue asterisk, and those amino acids that bind ubiquitin and were mutated in this study are boxed in green. The active site substrate binding loop also involved in binding inhibitors of SARS is shown highlighted in yellow. The sequence alignment was created using ESPript3.
Next, we aligned the primary amino acid sequence of the MHV PLP2 domain with the SARS-CoV and MERS-CoV papain-like protease domains (Fig. 1E). The sequence alignment and X-ray structure of the MHV PLP2–ubiquitin complex were then analyzed in conjunction with the previous structural and mutagenesis studies on SARS-CoVs and MERS-CoV to identify candidate residues that could be mutated to render a loss of DUB activity in vitro. From this analysis, we identified three residues (R1803, D1772, and F1812) in MHV PLP2 that form direct interactions with ubiquitin (Fig. 1B to D). Two of the side-chain guanidinium nitrogens of R1803 form direct hydrogen bonds with the backbone carbonyl oxygen of A46 in ubiquitin (Fig. 1B). The two side-chain carboxylate oxygens of D1772 in MHV PLP2 interact with ubiquitin by forming direct bonds with each of the guanidinium nitrogens of R42 and with one of the guanidinium nitrogens of R72 (Fig. 1C). Finally, F1812 forms Van der Waals contacts with the side chains of I44 and V70 and the delta-carbon of R42 of ubiquitin (Fig. 1D).
Biochemical analysis of PLP2 mutants’ activities.
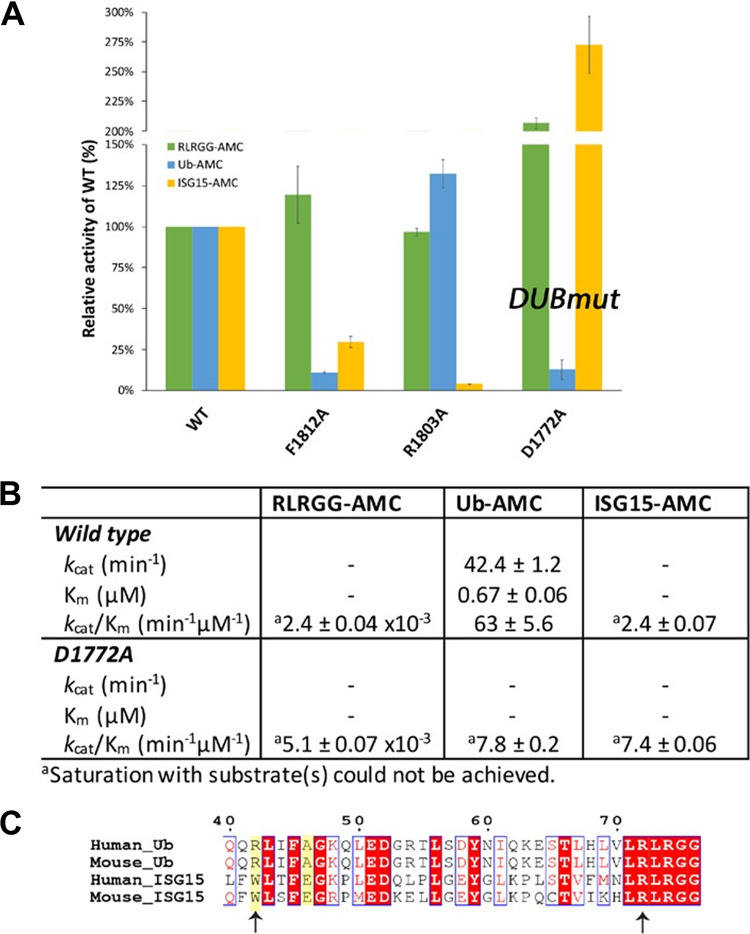
To identify a mutant MHV PLP2 enzyme that retains protease activity but exhibits reduced DUB and/or deISGylating activity, we performed site-directed mutagenesis on each of the residues (R1803, D1772, and F1812) by changing each to an alanine to disrupt interactions with ubiquitin. Each mutant enzyme was expressed, purified, and tested for its ability to hydrolyze three substrates: Z-RLRGG-AMC, Ub-AMC, and ISG15-AMC. The activity of each mutant enzyme toward each substrate relative to that of the wild-type (WT) enzyme is shown in Fig. 2A. All three mutants retained their ability to hydrolyze the peptide substrate, but each mutant had altered specificity toward Ub-AMC and ISG15-AMC substrates. Mutation of F1812 resulted in a substantial decrease in hydrolysis of both Ub-AMC and ISG15-AMC (class I), whereas mutation of R1803 resulted in loss of activity only toward ISG15-AMC (class II), and mutation of D1772 resulted in loss of activity only toward Ub-AMC (class III).
FIG 2.
Structure-guided mutagenesis of MHV PLP2 reveals that D1772A disrupts ubiquitin binding and reduces DUB activity. (A) Relative kinetic activities of three mutant MHV PLP2 enzymes toward three substrates: Z-RLRGG-AMC (green), Ub-AMC (blue), and ISG15-AMC (yellow) compared to the wild-type enzyme. (B) Steady-state kinetic parameters for wild-type and D1772A mutant enzymes. (C) Sequence alignment of Ub and ISG15 from human and mouse generated by Clustal Omega. The two arginine residues of Ub (R42 and R72) that interact with D1772 are indicated by arrows. R72 is conserved between Ub and ISG15, whereas R42 (shaded in yellow) is only present in Ub. Accession numbers are as follows: PDB code 1UBQ (Human_Ub); Uniprot accession number P62984 (Mouse_Ub); GenBank accession number AAH09507 (Human_ISG15); and GenBank accession number AAI09347 (Mouse_ISG15). The sequence alignment was created using ESPript.
Since one of the primary goals of this study is to understand the contribution of DUB activity to viral replication and pathogenesis, we next focused on further quantitating the effects of the D1772A mutant on the steady-state kinetic parameters of MHV PLP2 toward the three different substrates (Fig. 2B). The RLRGG-AMC peptide substrate is often used as a surrogate of the viral polyprotein substrate, and the kinetic data in Fig. 2B show that this substrate is still well recognized and cleaved by the D1772A mutant. In fact, we observed a small rate enhancement in the catalytic efficiency (i.e., kcat/Km) compared to that of the wild-type enzyme.
The Ub-AMC substrate, on the other hand, is more poorly recognized and cleaved by the D1772A mutant than by the wild-type enzyme. The wild-type enzyme normally interacts strongly with Ub-AMC, with a Km value of 0.67 μM. However, mutation of D1772 to an alanine significantly disrupts the interaction with ubiquitin, making it impossible to saturate MHV PLP2 under normal experimental conditions (Fig. 2B). The net result is a significant reduction in the catalytic efficiency (kcat/Km) compared to that of the wild-type enzyme, which was the goal of these trials.
The kinetic response of MHV PLP2 toward another substrate, ISG15-AMC, was also determined. ISG15 is an important ubiquitin-like modifier that is upregulated and used to ISGylate host proteins during viral infection. A number of viruses, including coronaviruses, engender ISGylation during infection, but the function(s) and importance of this activity are not clear (28–30). For MHV, neither the wild-type nor the D1772A mutant PLP2 enzyme can be saturated with ISG15-AMC, suggesting weak binding with this ubiquitin-like modifier (Fig. 2B). Moreover, the R1772A mutation does not disrupt the interaction with ISG15 but in fact enhances it to some degree. A potential explanation for the observed selective disruption of ubiquitin binding stems from our analysis of a primary sequence alignment of ubiquitin and ISG15 and the residues that interact with D1772 (Fig. 2C). The interaction between MHV PLP2 D1772 and the R42 residue in human and mouse ubiquitin is absent in human and mouse ISG15 since this residue is a tryptophan in human and mouse ISG15. Therefore, in line with our observations, D1772A mutation would not be expected to alter ISG15 binding. In contrast to R42, residue R72 is conserved in both ubiquitin and ISG15, and its interaction with MHV PLP2 for ubiquitin is likely weaker than that with ISG15.
PLP2 D1772A mutant exhibits reduced DUB activity and inhibition of IFN response in cell culture-based assays.
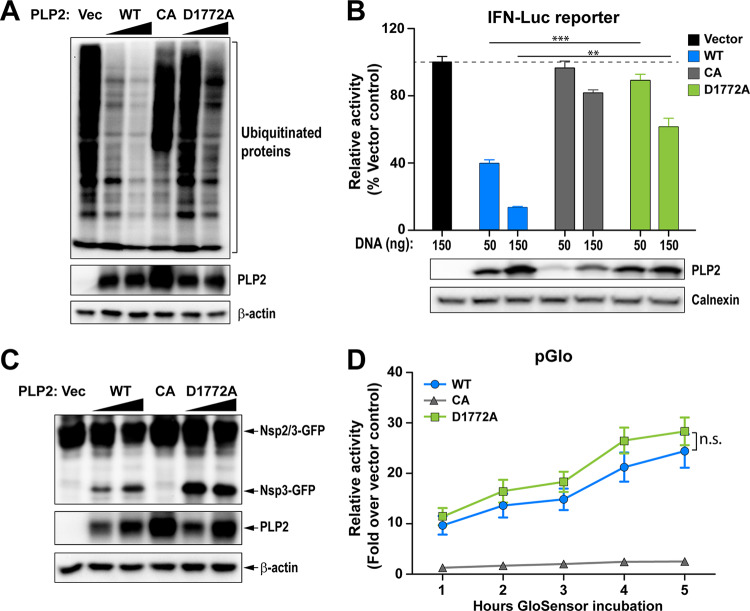
The in vitro biochemical studies presented here support the notion that we are able to use a structure-guided mutagenesis to uncouple the DUB enzymatic activity from MHV PLP2 while preserving the peptide hydrolysis and deISGylating activities of PLP2. Next, we focused on comparing the activity of the mutant enzyme to its wild-type counterpart for the ability to remove Flag-tagged-ubiquitin conjugated to host proteins in cultured cells (Fig. 3A). We found that in cells, wild-type PLP2 exhibits robust DUB activity and removes ubiquitin modifications from multiple cellular proteins. On the other hand, the PLP2-D1772A mutant exhibits reduced DUB activity, similar to that of the previously documented catalytic cysteine to alanine mutant, PLP2-CA (19). To determine if this impaired DUB activity altered the ability of PLP2 to act as an interferon antagonist, we transfected cells with a RIG-I expression plasmid, an interferon-luciferase (Luc) reporter construct, and either wild-type or mutant PLP2 plasmid and measured luciferase activity at 18 h posttransfection. In agreement with previous reports (13, 25, 31), we find that wild-type PLP2 acts as an interferon antagonist, reducing reporter activity by 50 to 80%. In contrast, PLP2-D1772A is unable to significantly reduce interferon activation in this assay despite similar expression levels of the wild-type and mutant versions of the protein (Fig. 3B). We also evaluated the protease activity of the enzymes in cells using two independent trans-cleavage assays and found that the wild-type and DUB-mutant enzymes produce similar levels of cleaved products. These results indicate that the D1772A substitution did not alter protease activity (Fig. 3C and D), in agreement with the in vitro kinetic results described above (Fig. 2). Together, these studies reveal that aspartic acid residue 1772 of MHV-PLP2 is important for DUB activity and interferon antagonism, but not for protease activity.
FIG 3.
D1772A substitution in the coronavirus papain-like protease Ub-binding site reduces DUB activity and interferon antagonism without reducing protease activity. (A) Western blot assessing the DUB activity of PLP2. (B) IFN antagonism of PLP2 was determined using an IFN-luciferase (Luc) reporter stimulated by N-RIG-I expression. The reporter activity of vector control was set to 100% (indicated by a dash line). Values are presented as means ± standard deviation (SD) and were statistically analyzed using an unpaired t test. **, P < 0.01; ***, P < 0.001. (C and D) Protease activity was evaluated using (C) a trans-cleavage assay that detects the cleaved products by Western blot and (D) a pGlo biosensor assay that is activated by PLP2-mediated cleavage of the substrate. Values are presented as means ± SD and were statistically analyzed using an unpaired t test at each time point. “n.s.” indicates that the values at the tested time points are not significantly different. Data are representative of at least two independent experiments.
Recombinant MHV harboring PLP2-D1772A activates an earlier IFN response in bone marrow-derived macrophages.
Since the D1772A substitution did not impact protease activity, we reasoned that we should be able to generate recombinant virus containing this substitution, thereby allowing us to determine if the mutation has any effect on viral replication kinetics and interferon antagonism in the context of the live virus. We engineered the mutant virus via reverse genetics (32), performed full-genome sequencing to verify the genotype (2 nucleotide changes at positions 5525 and 5526, resulting in D1772A substitution in the replicase polyprotein), and designated the virus DUBmut. Upon evaluating virus replication of the DUBmut virus by performing a growth kinetics experiment in parallel with wild-type virus, we found that the DUBmut virus replicates with essentially identical kinetics to those of the wild-type in a murine astrocytoma cell line (DBT cells) (Fig. 4A). These results are consistent with previous studies of coronavirus interferon antagonists, which showed in many cell lines that virally mediated interferon antagonism is not essential for virus replication (5, 6). Regarding the other ubiquitin-interacting residues identified in the structural analysis, we attempted to rescue virus with substitutions at the F1812 position, but we were unable to recover viable virus. These results indicate that F1812 may play a critical role within the polyprotein during virus replication. We were able to recover virus containing the R1803A substitution but found that it had no detectable phenotype, which we documented in our previous study (5). Here, we focus our efforts on evaluating replication and pathogenesis of the recovered DUBmut virus.
FIG 4.
Evaluating the replication kinetics of, and level of interferon activation by, WT MHV and DUBmut in cell culture. (A) Replication kinetics of WT and DUBmut virus in DBT cells. (B) IFN-α11 mRNA levels in WT- and DUBmut-infected BMDMs were assessed at indicated time points by reverse transcription-quantitative PCR (qRT-PCR). (C) IFN-α protein levels in the supernatants of infected BMDMs were evaluated at the times indicated. (D) Comparison of IFN-α11 mRNA levels in B6 versus MDA5−/− BMDMs at 12 h postinfection (h p.i.). (E) Assessment of levels of viral nucleocapsid (N) mRNA by qRT-PCR. (F) Replication kinetics of WT and DUBmut virus in BMDM cells. Data are representative of at least two independent experiments and are presented as means ± SD. Data in panels B and C were statistically analyzed using unpaired t tests. *, P < 0.05; **, P < 0.01.
To determine if the impaired DUB activity of the DUBmut virus had an effect on interferon antagonism, we infected primary bone marrow-derived macrophages (BMDMs) and evaluated viral replication kinetics and levels of interferon mRNA and protein. We observed significant activation of interferon alpha (IFN-α) mRNA expression (Fig. 4A) that is coupled with release of IFN-α protein into the supernatant, as detected by enzyme-limited immunosorbent assay (ELISA) (Fig. 4B). In addition, we performed an extensive transcriptional profiling of DUBmut-infected macrophages, which is described in our companion study (33), which revealed that multiple IFNs and ISGs, including IFN-β and ISG15 are activated during DUBmut infection. Here, we show that this activation of IFN-β is dependent on expression of the pattern recognition receptor MDA5 (Fig. 4D), in agreement with previous reports (5, 6, 34). To our surprise, we found that replication of DUBmut is not impaired relative to that of the wild-type virus in BMDMs, as measured by level of nucleocapsid RNA (Fig. 4E) and evaluation of infectious virus particle production over time in the kinetics assay (Fig. 4F). These results demonstrate that an elevated interferon response is generated during replication of the DUBmut virus but that this interferon profile is not associated with reduced production of infectious particles in either DBT cells or BMDMs.
Disrupting DUB activity mildly attenuates coronavirus pathogenesis in mice.
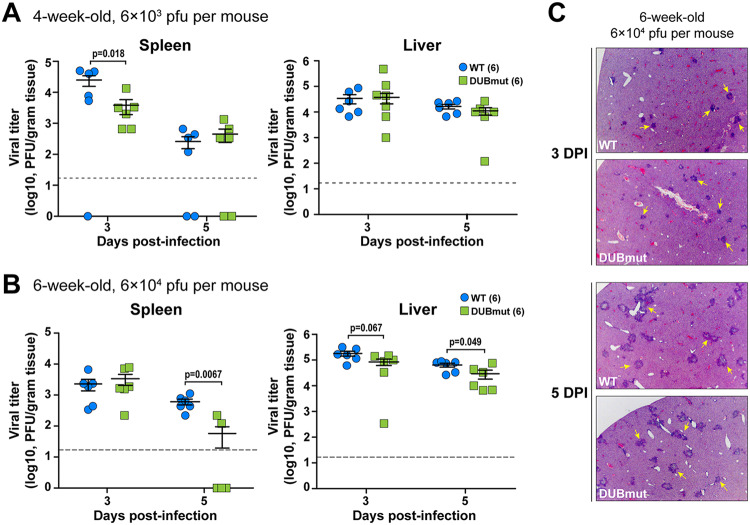
To complement our in vitro studies, we next sought to determine whether loss of DUB activity and the observed activation of interferon during the DUBmut virus infection in macrophages is associated with an attenuated pathogenesis in mice. To test this, we first used an intracranial infection model to evaluate pathogenesis in WT and DUBmut-infected mice. We found that intracranial infection of mice with either virus resulted in similar weight loss and 100% lethality (data not shown), suggesting that the loss of DUB activity does not affect the neuropathogenesis of MHV. To investigate the pathogenesis in the liver, we inoculated 4- (young) or 6-week-old (adult) mice intraperitoneally with a low dose (6 × 103 PFU) or a high dose (6 × 104 PFU) of the designated virus, respectively, and measured viral titer in the liver and spleen at the indicated days postinfection (d p.i.). We observed similar levels of infectious particles in young mice inoculated with a high dose of virus and adult mice with a low dose at 3 and 5 d p.i. (data not shown), but detected reduced viral titers in the spleens of young mice infected with a low dose of DUBmut virus at 3 d p.i. (Fig. 5A) and adult mice inoculated with a high dose at 5 d p.i. (Fig. 5B). Similar pathology in liver sections was observed at 3 and 5 d p.i. (Fig. 5C). These results indicate that the DUBmut virus is mildly attenuated compared to the wild-type virus, suggesting that the DUB activity does play a role in MHV pathogenesis and that PLP2 is a virulence factor.
FIG 5.
Evaluating replication and pathogenesis of MHV-DUBmut in mice. Four- (A) or 6-week-old (B) mice were infected with the indicated doses of MHV. Viral titer in livers and spleens isolated from WT- or DUBmut virus-infected mice was determined by plaque assay. The number of mice in each group is shown in parentheses. Data were statistically analyzed using unpaired t tests and are presented as means ± standard error of the mean (SEM). (C) H&E staining of liver sections from infected mice at 3 and 5 days postinfection (d p.i.). Representative MHV-associated liver lesions are indicated by arrows.
DISCUSSION
In the present study, we aimed to investigate the roles of PLP2 DUB activity during coronavirus infection. Through structure-guided mutagenesis, we identified residues of MHV PLP2 that mediate its interaction with ubiquitin. By mutating the residue Asp1772 to Ala (D1772A), we found that the DUB activity of PLP2 was greatly reduced, and a recombinant MHV carrying this mutation (DUBmut) activated an earlier IFN response in macrophages. Although we only observed a subtle attenuation of the DUBmut virus in the tested animal models, we demonstrated that PLP2 DUB activity does play a role in suppressing the host immune response and is a virulence trait, strengthening our earlier discoveries on SARS-CoV PLpro (4). We further investigated the differences in the interferon response generated in response to WT versus DUBmut virus in our companion study (33), which further supports a role for DUB activity in modulating the interferon response in macrophages.
The structure-guided approach used to generate the DUBmut virus allowed for characterization of the following three different classes of mutant enzymes: class I, deficient in both DUB and deISGylating activity; class II, deficient in deISGylating activity only; and class III, deficient in DUB activity but competent in protease and deISGylating activity. We utilized three unique biochemical substrates, each with a conjugated fluorescent 7-amino-4-methylcoumarin (AMC) reporter, to evoke the multifunctional activities of PLP2. Activity against the Z-RLRGG-AMC peptide substrate represents the polyprotein processing activity of MHV PLP2, while Ub-AMC and ISG15-AMC stimulate the deubiquitinating and deISGylating activities of the enzyme, respectively (17, 35). The kinetic data for D1772A DUBmut hydrolysis of the z-RLRGG-peptide provided in Fig. 2B indicate that the polyprotein processing ability of this mutant is likely not affected by the D1772A substitution. The deISGylating ability of the enzyme is also not affected. In contrast, the mutant enzyme’s deubiquitinating activity is significantly reduced relative to that of the wild type, which is most likely due to a lowered binding affinity for ubiquitin, as the enzyme could not be saturated with Ub-AMC as a substrate.
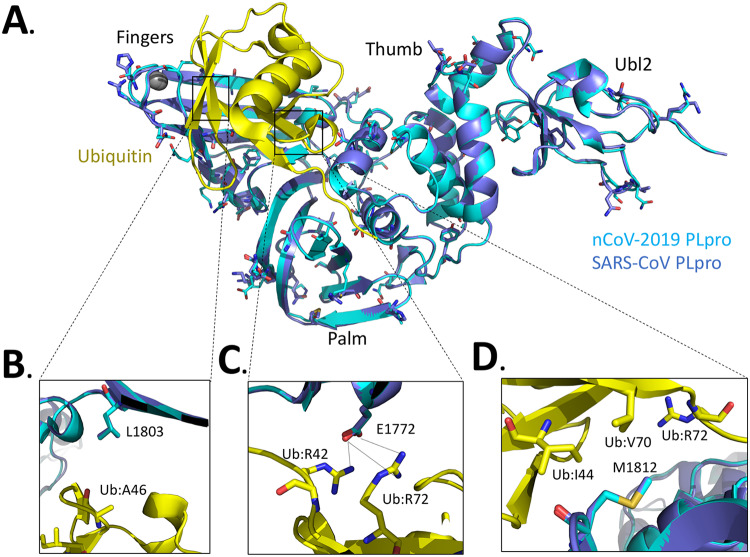
We can also use these structural, mutagenesis, and kinetic characterization studies on MHV PLP2 to guide future structure-based design studies of emerging coronaviruses, such as the newly emerged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (36–39). By aligning the X-ray structure of the PLpro domains of SARS-CoV bound to ubiquitin (17) with the sequence of SARS-CoV-2 PLpro (Fig. 6A) and comparing the differences in their residues with those of MHV PLP2 (Fig. 1E), we observed significant differences in the potential interactions between SARS-CoV-2 and ubiquitin. For example, we found that the hydrogen bonds between MHV PLP2 R1803 and the carbonyl oxygen of UbA46 (Fig. 1B) are lost in SARS-CoV-2 and SARS-CoV (Fig. 6B). In contrast, the hydrogen bonds between MHV PLP2 D1772 and Ub residues R42 and R72 (Fig. 1C) are likely preserved with the substitution of E1772 in SARS-CoV-2 and SARS-CoV PLpro (Fig. 6C). The hydrophobic interactions between Ub residues I44 and V70 and F1812 of MHV PLP2 (Fig. 1D) also appear to be preserved with M1812 of SARS-CoV-2 and SARS-CoV PLpro (Fig. 6D). The X-ray structures of MHV PLP2 and SARS-CoV PLpro, together with our structural model of SARS-CoV-2 PLpro, will provide testable hypotheses for the design and mutagenesis of the ubiquitin-binding domain (17) and diubiquitin-binding domains (20) of the recently emerged coronavirus SARS-CoV-2 PLpro.
FIG 6.
Alignment of the SARS-CoV-2 PLpro domain with the X-ray structure of the closely related SARS-CoV PLpro domain in complex with ubiquitin. (A) X-ray structure of SARS-CoV PLpro-ubiquitin-aldehyde complex (blue) (PDB code 4MM3) with each of its domains labeled as finger, palm, thumb, and Ubl2. Ubiquitin-aldehyde is colored yellow. The SARS-CoV-2 PLpro structure (cyan) was modeled by first mutating the residues of SARS PLpro in the X-ray structure to those of SARS-CoV-2 PLpro based upon the sequence alignment in Fig. 1E. The SARS-CoV-2 PLpro-ubiquitin-aldehyde complex was then refined using the structure-factor amplitudes and initial phases of the SARS PLP-ubiquitin-aldehyde complex (PDB code 4MM3). The residues that are different between SARS-CoV PLpro and SARS-CoV-2 PLpro are highlighted as sticks. (B) Potential interactions between L1803 of SARS-CoV PLpro and SARS-CoV-2 PLpro with residue A46 of ubiquitin. (C) Predicted interaction between E1772 of SARS-CoV PLpro and SARS-CoV-2 PLpro and residues R42 and R72 of ubiquitin. (D) Potential interactions between residues I44, V70, and R42 of ubiquitin with residues M1812 of SARS-CoV PLpro and SARS-CoV-2 PLpro.
We were able to reproduce the enzymatic profile of the purified PLP2-D1772A mutant protein when we expressed it in cell culture (Fig. 3). Therefore, our finding that the DUBmut virus containing the PLP2-D1772A substitution activates an elevated antiviral response in macrophages compared to that of the wild-type virus, but that this antiviral state results in only mild attenuation of disease in mice relative to WT infection, was unexpected. Previous studies demonstrated that ubiquitin has important roles in both the activation and the attenuation of innate antiviral pathways (40); therefore, we anticipated a more remarkable phenotype for a DUB mutant virus. We can imagine several possible explanations for our findings. First, it is possible that viral DUB activity has a relatively minor role in shaping pathogenesis in this system. In fact, our recent studies using SARS-CoV and SARS-related CoVs found that the papain-like protease domain/DUB is a virulence trait that varies among members of the SARS-coronavirus species (4). In that study, we found that replacing the SARS PLP2/DUB domain with a SARS-related PLP2/DUB domain reduced the ability of that virus to antagonize the innate immune response. This previous report, in conjunction with the current study, supports the concept that different PLP2/DUB domains may have distinct effects on antagonism of the innate immune response, depending on the virus and the host cell type. Another possibility is that the DUB mutant virus we generated may not have been sufficiently debilitated in its DUB activity to result in altered pathogenesis. We found that it was difficult to recover viable DUB mutant viruses; indeed, this D1772A mutant was the sole viable DUB mutant representative of our many attempts. Because an elevated interferon response was elicited from the DUBmut-infected cells, this mutant virus fulfilled our criteria for demonstrating the inactivation of an interferon antagonist. However, we speculate that if we are able to recover mutants that exhibit a range of DUB activities, we may be able to more fully assess the role of DUB activity as a contributor to coronaviral pathogenesis. In addition, the viral DUB activity may be important in specific cell types, such as epithelial cells that are infected by respiratory and enteric-tropic CoVs. Despite these caveats, the MHV DUB mutant generated in this study did exhibit a reproducible phenotype of eliciting an elevated interferon response in infected macrophages that was associated with mild attenuation of pathogenesis, with reduced titers in the livers and spleens of infected mice. Overall, we conclude that DUB activity is indeed a virulence trait and contributes to the ability of MHV to modulate the host innate immune response to infection. Further studies are needed to identify the targets of viral DUB activity and the detailed role of DUB activity in delaying the innate immune response to virus replication.
MATERIALS AND METHODS
Ethics statement.
The mouse experiment in this study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The experimental protocol was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at Loyola University Chicago (IACUC no. 2016-029). C57BL/6 female mice were purchased from The Jackson Laboratory and maintained in the Comparative Medicine Facility of Loyola University Chicago. Mice were consistently monitored for signs of distress over the course of the experiments in order to be removed from the experiment and euthanized using carbon dioxide inhalation to prevent unnecessary suffering.
Cells.
Human embryonic kidney (HEK) 293T cells were purchased the from the American Type Culture Collection (ATCC) (CRL-11268) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (product no. 10-017-CV; Corning) containing 10% fetal calf serum (FCS) and supplemented with 1% nonessential amino acids, 1% HEPES, 2% l-glutamine, 1% sodium pyruvate, and 1% penicillin/streptomycin. DBT cells were cultured in minimal essential medium (MEM) (catalog no. 61100-061; Thermo Fisher) supplemented with 5% FCS, 2% l-glutamine, and 10% tryptose phosphate broth (TPB). The 17Cl-1 cell line was maintained in DMEM containing 5% FCS. Baby hamster kidney cells expressing the MHV receptor (BHK-R) were kindly provided by Mark Denison (Vanderbilt University Medical Center) and maintained in DMEM supplemented with 10% FCS, 2% l-glutamine, and 0.8 μg/ml G418. Bone marrow-derived macrophages (BMDMs) were prepared and cultured as described previously (5).
Plasmids and mutagenesis.
The sequence of the PLP2 domain (amino acids 1525 to 1911 of MHV pp1ab) in frame with a V5 epitope tag was codon optimized, synthesized by GenScript (Piscataway, NJ) (sequence available upon request), and cloned into pCAGGS vector. For mutagenesis, an overlapping PCR strategy was used with synthetic primers (sequences available upon request). The introduced mutations were verified by sequencing. The RIG-I and nsp2/3-GFP expression plasmid was kindly provided Ralph Baric (University of North Carolina). The IFN-β-Luc reporter plasmid was a gift of John Hiscott (Jewish General Hospital, Montreal, Canada). The Flag-Ub plasmid was kindly provided by Adriano Marchese (University of Wisconsin-Milwaukee).
MHV PLP2 wild-type and mutant purification, kinetics, and X-ray structure.
The wild-type, C1716S, D1772A, R1803A, and F1812A mutant enzymes were expressed and purified similarly to in our previously published methods, except the MHV PLP2 construct used here (amino acids N1609 to N1909) did not include the DPUP domain (16). Crystallization and X-ray structure determination details will be published elsewhere. Steady-state kinetic studies on the wild-type, D1772A, R1803A, and F1812A mutant enzymes with substrates Z-RLRGG-AMC (where Z is a carboxybenzyl protecting group), ubiquitin-AMC, and ISG15-AMC were performed as described previously (16). Structure figures were generated with the software program UCSF Chimera (41).
Protease and deubiquitinating activity assays.
To determine the protease activity of PLP2, HEK293T cells grown to 70% confluence in 24-well plates (Corning) were transfected using TransIT-LT1 (catalog no. MIR2300; Mirus Bio) according to the manufacturer’s protocol. For the protease activity assay, HEK293T cells were transfected with 25 ng nsp2/3-green fluorescent protein (GFP) plasmid and 200 ng pCAGGS-PLP2-V5 expression plasmids (wild-type and mutant). To determine the deubiquitinating activity of the proteins, cells were transfected with 200 ng Flag-Ub plasmid and pCAGGS-PLP2-V5 expression plasmids (wild-type and mutant). Cells were lysed 24 h posttransfection with 100 μl of lysis buffer (comprising 20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride). Whole-cell lysates were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane in transfer buffer (0.025 M Tris, 0.192 M glycine, and 20% methanol) for 1 h at 60 V at 4°C. Following this, the membrane was blocked using 5% dried skim milk in TBST buffer (0.9% NaCl, 10 mM Tris-HCl [pH 7.5], and 0.1% Tween 20) overnight at 4°C. The membrane was incubated with either polyclonal rabbit anti-GFP antibody (catalog no. A11122; Life Technologies) for the protease assay, or mouse anti-Flag (catalog no. F3165; Sigma) for the DUB assay. The membrane was then washed 3 times for 15 min in TBST buffer, followed by incubation with either secondary donkey anti-rabbit-HRP antibody (catalog no. 711-035-152; Jackson ImmunoResearch) or goat anti-mouse-horseradish peroxidase (HRP) antibody (catalog no. 1010-05; SouthernBiotech). Then the membrane was washed 3 more times for 15 min in TBST buffer. Detection was performed using Western Lighting Chemiluminescence Reagent Plus (PerkinElmer) and visualized using a FluorChemE imager (Protein Simple). The expressions of PLP2, β-actin, and calnexin were probed with mouse anti-V5 antibody (catalog no. R960; Thermo Fisher), mouse anti–β-actin (catalog no. A00702; GenScript), or mouse anti-calnexin antibody (catalog no. 2433S; Cell Signaling), respectively.
Biosensor live cell assay.
The protease activity of PLP2 was also assessed using a biosensor live cell assay as described previously (14). Briefly, HEK293T cells in a 96-well black-wall plate were transfected with 37.5 ng pGlo-RLKGG construct and 50 ng PLP2 expression plasmids. GloSensor (catalog no. E1290; Promega) reagent diluted in DMEM plus 10% FCS was added at 18 hours postinfection (h p.i.). Plates were read using a luminometer (Veritas) every hour over a course of 5 h.
Generating DUB mutant MHV.
We used a previously described reverse genetics system of MHV-A59 (32) to generate the DUB mutant virus. Briefly, the nucleotides coding for the Asp1772 of the PLP2 domain were mutated via site-directed mutagenesis. Viral genomic RNA from in vitro transcription of ligated cDNA fragments using a mMESSAGE mMACHINE T7 transcription kit (catalog no. AM1344; Thermo Fisher) was electroporated into BHK-R cells. Cell supernatants were collected as viral stock following observation of cytopathic effects. Rescued virus was plaque purified, propagated on BHK-R cells, and titer was determined on 17Cl-1 cells. The stock virus was subjected to full-genome sequencing and the sequences were aligned to the parental strain, with the intended engineered mutation detected and no additional mutations detected (Kansas State University Diagnostic Laboratory).
Growth kinetics.
DBT cells or BMDMs were infected with wild-type icMHV-A59 or DUB mutant virus at a multiplicity of infection (MOI) of 1 in serum-free medium. After a 1-h incubation, the inoculum was replaced with fresh, complete medium. Cell culture supernatants were collected at the indicated time points and titer was determined by plaque assay on 17Cl-1 cells. Titers were obtained from three independent assays for each sample. Graphs of virus kinetics were generated using Prism software (GraphPad Software).
Quantification of IFN-α production by real-time quantitative PCR (RT-qPCR) and ELISA.
BMDMs in a 12-well plate were mock-infected or infected with MHV at an MOI of 1. At the indicated time points, monolayer cells were collected for RNA extraction and cell culture supernatants were harvested for ELISA analysis. To determine IFN-α11, β-actin, or MHV-A59 N gene mRNA levels, total RNA was extracted from collected cells using an RNeasy minikit (catalog no. 74104; Qiagen). The first-strand cDNA was synthesized from an equal amount of RNA using an RT2 HT First Strand kit (catalog no. 330401; Qiagen). Quantitative PCR (qPCR) was then performed with specific primers for mouse IFN-α11 (catalog no. PPM03050B-200; Qiagen), mouse β-actin (catalog no. PPM02945B-200; Qiagen), or the MHV-A59 N gene using RT2 SYBR green qPCR mastermix (catalog no. 330502; Qiagen) in the Bio-Rad CFX96 system. The thermocycler was set as follows: one step at 95°C (10 min), 40 cycles of 95°C (15 s), 60°C (1 min) and plate read, one step at 95°C (10 s), and a melt curve from 65°C to 95°C in increments of 0.5°C/0.05 s. Samples were evaluated in triplicate, and data are representative of three independent experiments. The levels of mRNA were reported relative to those of β-actin mRNA and expressed as 2−ΔCT [ΔCT = CT (gene of interest) − CT(β-actin)], where CT is the threshold cycle. The secreted amount of IFN-α in culture supernatants was assayed using a mouse IFN-α ELISA kit (catalog no. BMS6027; eBioscience) according to the manufacturer’s instructions.
Mouse experiments.
Evaluation of MHV pathogenesis in laboratory mouse was previously described (5, 42). Briefly, for intracranial infections, 6-week-old C57BL/6 female mice were inoculated with 600 PFU of virus in 20 μl phosphate-buffered saline (PBS). Infected mice were monitored for body weight daily and euthanized when weight loss surpassed 25%. Statistical analyses of survival rate were performed using the log rank test. For intraperitoneal infection, 6- or 4-week-old mice were injected with 6 × 103 or 6 × 104 PFU of virus in 100 μl PBS. Organs were collected at indicated time points and evaluated for viral burden. Liver pathology was evaluated using hematoxylin and eosin (H&E) staining by the Tissue Processing Core Facility at Loyola University Chicago.
Data availability.
Structural data are available in the Protein Data Bank under accession number 5WFI.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grant R01 AI085089 (to S.C.B. and A.D.M.). M.H. and R.C.M. were supported by an NIH T32 Training Grant for Experimental Immunology (no. AI007508), and R.C.M. was supported by an Arthur J. Schmitt Dissertation Fellowship in Leadership and Service (Arthur J. Schmitt Foundation). M.E.C. was supported by an NIH/NIGMS T32 Training Grant for Structural Biology and Biophysics (no. GM132024). Crystallization and DNA sequencing were partially supported by the Purdue Center for Cancer Research Macromolecular Crystallography and DNA Sequencing Shared Resources, which are supported by NIH grant P30 CA023168.
Conceptualization: X.D., A.D.M., and S.C.B. Investigation: X.D., Y.C., A.M.M., M.H., K.R.K., R.C.M., M.H., A.O., M.E.C. Formal Analysis: X.D., Y.C., A.M.M., M.H., K.R.K., R.C.M., A.O., A.D.M., and S.C.B. Writing – Original Draft Preparation: X.D., Y.C., A.M.M., A.D.M., and S.C.B. Writing – Review & Editing, with comments from X.D., Y.C., A.M.M., M.H., K.R.K., R.C.M., M.H., A.O., A.D.M. and S.C.B. Visualization: X.D., Y.C., K.R.K., A.O., M.E.C., A.D.M., and S.C.B. Funding Acquisition and Supervision: A.D.M. and S.C.B.
REFERENCES
- 1.Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enjuanes L, Almazán F, Sola I, Zuñiga S. 2006. Biochemical aspects of coronavirus replication and virus-host interaction. Annu Rev Microbiol 60:211–230. doi: 10.1146/annurev.micro.60.080805.142157. [DOI] [PubMed] [Google Scholar]
- 3.Totura AL, Baric RS. 2012. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol 2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niemeyer D, Mösbauer K, Klein EM, Sieberg A, Mettelman RC, Mielech AM, Dijkman R, Baker SC, Drosten C, Müller MA. 2018. The papain-like protease determines a virulence trait that varies among members of the SARS-coronavirus species. PLoS Pathog 14:e1007296. doi: 10.1371/journal.ppat.1007296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng X, Hackbart M, Mettelman RC, O’Brien A, Mielech AM, Yi G, Kao CC, Baker SC. 2017. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A 114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kindler E, Gil-Cruz C, Spanier J, Li Y, Wilhelm J, Rabouw HH, Züst R, Hwang M, V'kovski P, Stalder H, Marti S, Habjan M, Cervantes-Barragan L, Elliot R, Karl N, Gaughan C, van Kuppeveld FJM, Silverman RH, Keller M, Ludewig B, Bergmann CC, Ziebuhr J, Weiss SR, Kalinke U, Thiel V. 2017. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog 13:e1006195. doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menachery VD, Gralinski LE, Mitchell HD, Dinnon KH, Leist SR, Yount BL, McAnarney ET, Graham RL, Waters KM, Baric RS. 2018. Combination attenuation offers strategy for live-attenuated coronavirus vaccines. J Virol 92:e0071018. doi: 10.1128/JVI.00710-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kindler E, Thiel V, Weber F. 2016. Interaction of SARS and MERS coronaviruses with the antiviral interferon response. Adv Virus Res 96:219–243. doi: 10.1016/bs.aivir.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mielech AM, Chen Y, Mesecar AD, Baker SC. 2014. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res 194:184–190. doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanjanahaluethai A, Baker SC. 2000. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J Virol 74:7911–7921. doi: 10.1128/jvi.74.17.7911-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker SC, Shieh CK, Soe LH, Chang MF, Vannier DM, Lai MM. 1989. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J Virol 63:3693–3699. doi: 10.1128/JVI.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol 79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mielech AM, Kilianski A, Baez-Santos YM, Mesecar AD, Baker SC. 2014. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology 450–451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilianski A, Mielech A, Deng X, Baker SC. 2013. Assessing activity and inhibition of MERS-CoV papain-like and 3C-like proteases using luciferase-based biosensors. J Virol 87:11955–11962. doi: 10.1128/JVI.02105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng D, Chen G, Guo B, Cheng G, Tang H. 2008. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res 18:1105–1113. doi: 10.1038/cr.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Savinov SN, Mielech AM, Cao T, Baker SC, Mesecar AD. 2015. X-ray structural and functional studies of the three tandemly linked domains of non-structural protein 3 (nsp3) from murine hepatitis virus reveal conserved functions. J Biol Chem 290:25293–25306. doi: 10.1074/jbc.M115.662130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratia K, Kilianski A, Baez-Santos YM, Baker SC, Mesecar A. 2014. Structural basis for the ubiquitin-linkage specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog 10:e1004113. doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey-Elkin BA, Knaap RCM, Johnson GG, Dalebout TJ, Ninaber DK, van Kasteren PB, Bredenbeek PJ, Snijder EJ, Kikkert M, Mark BL. 2014. Crystal structure of the MERS coronavirus papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J Biol Chem 289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mielech AM, Deng X, Chen Y, Kindler E, Wheeler DL, Mesecar AD, Thiel V, Perlman S, Baker SC. 2015. Murine coronavirus ubiquitin-like domain is important for papain-like protease stability and viral pathogenesis. J Virol 89:4907–4917. doi: 10.1128/JVI.00338-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Békés M, van der Heden van Noort GJ, Ekkebus R, Ovaa H, Huang TT, Lima CD. 2016. Recognition of Lys48-linked di-ubiquitin and deubiquitinating activities of the SARS coronavirus papain-like protease. Mol Cell 62:572–585. doi: 10.1016/j.molcel.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey-Elkin BA, Knaap RCM, Kikkert M, Mark BL. 2017. Structure and function of viral deubiquitinating enzymes. J Mol Biol 429:3441–3470. doi: 10.1016/j.jmb.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clasman JR, Báez-Santos YM, Mettelman RC, O’Brien A, Baker SC, Mesecar AD. 2017. X-ray structure and enzymatic activity profile of a core papain-like protease of MERS coronavirus with utility for structure-based drug design. Sci Rep 7:40292. doi: 10.1038/srep40292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindner HA, Fotouhi-Ardakani N, Lytvyn V, Lachance P, Sulea T, Ménard R. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol 79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z, Wang Y, Ratia K, Mesecar AD, Wilkinson KD, Baker SC. 2007. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J Virol 81:6007–6018. doi: 10.1128/JVI.02747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clementz M, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, Li K, Mesecar AD, Baker SC. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol 84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xing Y, Chen J, Tu J, Zhang B, Chen X, Shi H, Baker SC, Feng L, Chen Z. 2013. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J Gen Virol 94:1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng X, Agnihothram S, Mielech AM, Nichols DB, Wilson MW, StJohn SE, Larsen SD, Mesecar AD, Lenschow DJ, Baric RS, Baker SC. 2014. A chimeric virus-mouse model system for evaluating the function and inhibition of papain-like proteases of emerging coronaviruses. J Virol 88:11825–11833. doi: 10.1128/JVI.01749-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt J. a, Rowland RR, Schmaljohn CS, Lenschow DJ, Snijder EJ, García-Sastre A, Virgin HW. 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, Osiak A, Levine B, Schmidt RE, Garcı A, Leib DA, Pekosz A, Knobeloch K, Horak I, Whiting H, Iv V. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci U S A 104:1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skaug B, Chen ZJ. 2010. Emerging role of ISG15 in antiviral immunity. Cell 143:187–190. doi: 10.1016/j.cell.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devaraj SG, Wang N, Chen Z, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng C-T, Baker SC, Li K. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem 282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yount B, Denison MR, Weiss SR, Ralph S, Baric RS. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J Virol 76:11065–11078. doi: 10.1128/jvi.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth-Cross JK, Bender SJ, Weiss SR. 2008. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J Virol 82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Báez-Santos YM, Mielech AM, Deng X, Baker S, Mesecar AD. 2014. Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the Middle East respiratory syndrome coronavirus. J Virol 88:12511–12527. doi: 10.1128/JVI.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heaton SM, Borg NA, Dixit VM. 2016. Ubiquitin in the activation and attenuation of innate antiviral immunity. J Exp Med 213:1–13. doi: 10.1084/jem.20151531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera? A visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.Deng X, StJohn SE, Osswald HL, O'Brien A, Banach BS, Sleeman K, Ghosh AK, Mesecar AD, Baker SC. 2014. Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J Virol 88:11886–11898. doi: 10.1128/JVI.01528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volk A, Hackbart M, Deng X, Cruz-Pulido Y, O’Brien A, Baker SC. 2020. Coronavirus endoribonuclease and deubiquitinating interferon antagonists differentially modulate the host response during replication in macrophages. J Virol 94:e00178-20. doi: 10.1128/JVI.00178-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Structural data are available in the Protein Data Bank under accession number 5WFI.