Ebola virus (EBOV) remains a high-priority pathogen since it continues to cause outbreaks with high case fatality rates. Although it is well established that EBOV results in severe organ damage, our understanding of tissue injury in the liver, adrenal glands, and lymphoid tissues remains limited. We begin to address this knowledge gap by conducting longitudinal gene expression studies in these tissues, which were collected from EBOV-infected cynomolgus macaques. We report robust and early gene expression changes within these tissues, indicating they are primary sites of EBOV infection. Furthermore, genes involved in metabolism, coagulation, and adaptive immunity were downregulated, while inflammation-related genes were upregulated. These results indicate significant tissue damage consistent with the development of hemorrhagic fever and lymphopenia. Our study provides novel insight into EBOV-host interactions and elucidates how host responses within the liver, adrenal glands, and lymphoid tissues contribute to EBOV pathogenesis.
KEYWORDS: Ebola virus, NHP, adrenal, hemorrhagic fever, liver, lymph nodes, pathogenesis, spleen, tissues, transcriptomics
ABSTRACT
Ebola virus (EBOV) continues to pose a significant threat to human health, as evidenced by the 2013–2016 epidemic in West Africa and the ongoing outbreak in the Democratic Republic of the Congo. EBOV causes hemorrhagic fever, organ damage, and shock culminating in death, with case fatality rates as high as 90%. This high lethality combined with the paucity of licensed medical countermeasures makes EBOV a critical human pathogen. Although EBOV infection results in significant damage to the liver and the adrenal glands, little is known about the molecular signatures of injury in these organs. Moreover, while changes in peripheral blood cells are becoming increasingly understood, the host responses within organs and lymphoid tissues remain poorly characterized. To address this knowledge gap, we tracked longitudinal transcriptional changes in tissues collected from EBOV-Makona-infected cynomolgus macaques. Following infection, both liver and adrenal glands exhibited significant and early downregulation of genes involved in metabolism, coagulation, hormone synthesis, and angiogenesis; upregulated genes were associated with inflammation. Analysis of lymphoid tissues showed early upregulation of genes that play a role in innate immunity and inflammation and downregulation of genes associated with cell cycle and adaptive immunity. Moreover, transient activation of innate immune responses and downregulation of humoral immune responses in lymphoid tissues were confirmed with flow cytometry. Together, these data suggest that the liver, adrenal gland, and lymphatic organs are important sites of EBOV infection and that dysregulating the function of these vital organs contributes to the development of Ebola virus disease.
IMPORTANCE Ebola virus (EBOV) remains a high-priority pathogen since it continues to cause outbreaks with high case fatality rates. Although it is well established that EBOV results in severe organ damage, our understanding of tissue injury in the liver, adrenal glands, and lymphoid tissues remains limited. We begin to address this knowledge gap by conducting longitudinal gene expression studies in these tissues, which were collected from EBOV-infected cynomolgus macaques. We report robust and early gene expression changes within these tissues, indicating they are primary sites of EBOV infection. Furthermore, genes involved in metabolism, coagulation, and adaptive immunity were downregulated, while inflammation-related genes were upregulated. These results indicate significant tissue damage consistent with the development of hemorrhagic fever and lymphopenia. Our study provides novel insight into EBOV-host interactions and elucidates how host responses within the liver, adrenal glands, and lymphoid tissues contribute to EBOV pathogenesis.
INTRODUCTION
Ebola virus disease (EVD) is marked by extensive virus replication and systemic spread. Lymphopenia, hemorrhage, organ damage and dysfunction, and a dysregulated inflammatory response are hallmarks of infection (1–4). EVD remains a global health threat, with the largest documented outbreaks occurring within the past decade. The 2013–2016 Ebola virus (EBOV) epidemic in West Africa was the largest on record, with >28,000 confirmed/suspected cases and >11,000 deaths (5). The current EBOV outbreak in the northeastern provinces of the Democratic Republic of the Congo (which was declared July 2018) is the second largest outbreak, with over 3,428 cases and 2,250 reported deaths as of 2 February 2020 (6). Given the paucity of licensed countermeasures for prevention or treatment of EVD, it is critical to expand our knowledge of EVD pathogenesis.
Liver, adrenal gland, and lymphoid tissues are important early targets for EBOV. Longitudinal studies in experimentally infected nonhuman primates (NHPs), the gold standard model for filovirus studies, indicate that monocytes, macrophages, and dendritic cells are the primary targets of EBOV infection and play a critical role in viral dissemination (7, 8). Virus particles migrate from the initial infection site into the bloodstream and are quickly taken up by highly vascularized organs, including liver, spleen, and adrenal glands. Immune recruitment from surveilling cell populations from the lymphatic system and bloodstream help to establish infections in the lymph nodes. In situ hybridization and electron microscopic analyses of tissues from fatal human cases and experimentally infected NHPs show that endothelial cells, fibroblasts, hepatocytes, and adrenal cortical cells all support EBOV replication (7, 9–12). Copious viral antigen and high infectious viral titers have been observed in the lymph nodes, liver, spleen, and adrenal glands of infected NHPs and humanized mice (1, 9, 10, 13–16), with detection as early as 2 to 3 days postinfection (DPI) in the macaque model (7). Histological changes in human cases and macaques include hepatocellular degeneration, necrosis, and viral inclusions within hepatocytes (7, 9, 11, 17); congestion and necrosis in adrenal glands (9, 10); and lymphoid depletion and numerous apoptotic bodies within the spleen and lymph nodes (9, 10).
These pathological alterations interfere with the normal function of these organs as evidenced by aberrations in liver enzyme levels and physiological derangements reflective of hormonal deregulation. The liver functions in the excretion, detoxification and metabolism of lipids, carbohydrates and proteins, and is responsible for the synthesis of coagulation factors. Adrenal glands are the main producers of steroid hormones that regulate blood pressure and metabolism. Furthermore, secondary lymphoid tissues (lymph nodes and the spleen) are major sites of antigen presentation and T cell and B cell activation. Given the importance of these organs in regulating coagulation, blood pressure, and host immunity, EBOV-induced damage within these tissues presumably contributes significantly to disease. However, the molecular basis of tissue injury within these target tissues is largely unknown.
EVD gene expression studies have mostly been limited to analysis of peripheral blood samples. These studies demonstrated large transcriptional changes associated with metabolic and hemostatic dysregulation following infection, as well as severe inflammatory signaling, including the sustained expression of interferon-stimulated genes (ISG) (8, 18–20). In contrast, transcripts involved in adaptive immunity were significantly repressed. In terms of tissue expression, one study reported upregulated expression of innate immune genes in the liver, spleen, adrenal gland, and axillary lymph nodes of macaques infected via the aerosol route (19). However, this study analyzed the expression of only 125 genes previously shown to be strongly upregulated in peripheral blood mononuclear cell (PBMCs) of cynomolgus macaques intramuscularly infected with Ebola. The authors did not specify whether these changes were unique to tissue or contributed by blood circulating throughout the tissue.
In this study, we utilized RNA sequencing (RNA-Seq) to determine the longitudinal transcriptional profiles of liver, adrenal gland, spleen, and lymph node samples obtained from cynomolgus macaques challenged with EBOV-Makona, the variant responsible for the West African epidemic. Analysis of tissue-specific genes revealed increased expression of immune-related genes in addition to dysregulation of metabolic, coagulation, hormone synthesis and vascular function pathways in both the liver and the adrenal glands. In lymphoid tissue, we observed increased expression of genes involved in innate immunity and inflammation. Conversely, genes related to transcription, cell cycle, antigen presentation, and T and B cell activation were downregulated. Flow cytometric analysis of lymphoid tissue corroborated the lack of adaptive immune activation at the protein level. These findings provide novel insights into the detailed molecular mechanisms contributing to the pathophysiology of EBOV in key tissues and provide a foundation to identify potential targets for intervention strategies.
RESULTS
EBOV-Makona infection results in liver injury.
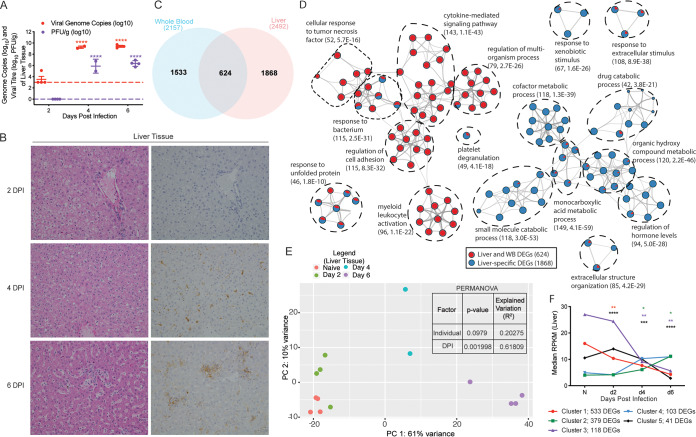
Consistent with viremia in whole blood (8), viral loads (genome copies and infectious virus titers) significantly increased 4 and 6 DPI in the liver relative to the limit of detection (1,000 viral copies/g, and 1 PFU/g), represented as a dashed line in Fig. 1A. Examination of gross pathology and histopathology of the liver revealed no significant lesions 2 DPI in any of the animals (Table 1; Fig. 1B). In line with viral loads, histologic lesions were noted in animals euthanized 4 and 6 DPI (Table 1; Fig. 1B). Specifically, mild hepatic pallor, reticulation, and mild hepatitis were observed 4 DPI in 2 of 2 of the animals (Fig. 1B). Additionally, specific antifilovirus immunoreactivity was apparent in scattered mononuclear cells 4 DPI (2 of 2 animals; Fig. 1B). By 6 DPI, hepatic pallor (2 of 4 animals) and reticulation (3 of 4 animals) increased in severity and was accompanied by mild necrotizing hepatitis (4 of 4 animals). Numerous isolated and clustered mononuclear cells and hepatocytes stained positive for EBOV antigen in the liver at this time point (4 of 4 animals; Fig. 1B).
FIG 1.
Makona infection results in significant histological and transcriptional changes in the liver. (A) Mean ± standard deviation (SD) of EBOV infectious virus titer and genome copy numbers in liver were quantified using plaque assay and RT-qPCR for each animal at each time point. Significance was determined using one-way ANOVA with Dunnett’s multiple-comparison test relative to the limit of detection, which is represented as a dashed line. ****, P < 0.0001. (B) Hematoxylin and eosin staining (left) and immunohistochemistry staining (right) using EBOV-specific anti-VP40 antibody of liver at 2, 4, and 6 DPI (brown indicates reactivity); magnification ×20. (C) Venn diagram comparing differentially expressed genes (DEGs) detected in whole blood (WB) and liver pooled from all the time points analyzed. (D) Network image showing functional enrichment of DEGs detected in both liver and WB, and liver only generated using Metascape and visualized using Cytoscape. Each circle represents a theme of highly related gene ontology (GO) terms. The edges (lines) link similar GO terms, and the thickness of the line indicates the relatedness of the GO terms. Each GO term is shown as a pie chart that depicts the relative contribution of DEGs detected in both liver and WB (red) and liver only (blue). (E) Principal component analysis (PCA) of liver-specific DEGs over time. (F) STEM analysis identified five distinct and significant temporal expression clusters of genes. The median RPKM of each cluster over time is plotted. The Bonferroni-corrected P values for each cluster in order are 2.83e-24, 1.05e-6, 1.10e-20, 9.70e-17, and 1.30e-4. Statistical significance between time points was determined using one-way ANOVA with Dunnett’s multiple-comparison test relative to N (naive). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
TABLE 1.
Histopathology and immunohistochemistry findingsa
| Tissue type | Day 2 |
Day 4 |
Day 6 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Animal no. 1 | Animal no. 2 | Animal no. 3 | Animal no. 4 | Animal no. 5 | Animal no. 6 | Animal no. 7 | Animal no. 8 | Animal no. 9 | Animal no. 10 | |
| Liver | 0, 0 | 0, 0 | 0, 0 | 0, 0 | 1, 1 | 1, 1 | 2, 2 | 2, 2 | 2, 2 | 2, 2 |
| Adrenal gland | 0, 0 | 0, 0 | 0, 0 | 0, 0 | 1, 1 | 0, 1 | 1, 2 | 1, 2 | 1, 2 | 1, 2 |
| Axillary lymph node | 0, 0 | 0, 0 | 0, 0 | 0, 0 | 1, 2 | 1, 2 | 1, 1 | 1, 1 | 1, 3 | 1, 3 |
| Inguinal lymph node | 0, 0 | 0, 0 | 0, 0 | 0, 0 | 1, 2 | 1, 2 | 1, 1 | 1, 1 | 1, 3 | 1, 3 |
| Spleen | 0, 0 | 0, 0 | 0, 0 | 0, 0 | 0, 2 | 0, 2 | 2, 2 | 2, 1 | 1, 3 | 1, 3 |
Severity scoring: the first score is for H&E, and the second score is for IHC.
EBOV-Makona infection induces transcriptional changes in the liver involved in metabolism, coagulation, hormone synthesis, and inflammation.
Longitudinal transcriptional changes in the liver were assessed using RNA-Seq. Since the tissues were not perfused at the time of necropsy, we contrasted differentially expressed genes (DEGs) detected in the liver to those identified in whole blood (WB) in our prior study (8) (Fig. 1C). This comparison identified 1,868 DEGs that were designated liver-specific DEGs (Fig. 1C). Using Enrichr, a Web-based tool for analyzing gene sets to annotate biological features and tissue protein expression (21, 22), we confirmed that DEGs detected in both liver and WB originate mainly from WB, CD14+ monocytes, and CD33+ myeloid cells (adjusted P value, 3.92e-40, 5.47e-22, and 4.03e-21, respectively), while the 1,868 DEGs designated liver-specific overwhelmingly enriched to prototypical bulk liver tissue (adjusted P value, 5.14e-142). Functional enrichment of the 624 DEGs detected in both liver and WB using Metascape (23) showed overrepresentation of gene ontology (GO) terms associated with innate immunity (“cytokine-mediated signaling pathway,” “myeloid leukocyte activation,” and “cellular response to tumor necrosis factor”) (Fig. 1D). In contrast, functional enrichment of the 1,868 liver-specific DEGs revealed predominant mapping to metabolic processes (e.g., “drug catabolic process,” “small molecule catabolic process”, and “regulation of hormone levels”) (Fig. 1D).
Next, we performed principal-component analysis (PCA) to represent the multivariate data as a 2-dimensional plot. PCA of liver-specific DEGs showed distinct clustering at each time point, with 61% of the variation explained by loading along PC1 (DPI), while PC2 (intragroup variability) only explained 10% of the variance (Table 2; Fig. 1E). A permutational multivariate analysis of variance (PERMANOVA) test also showed the variation between animals at each time point was nonsignificant (P = 0.0979; R2 = 0.20275), whereas the DPI significantly explained the variance in the data (P = 0.001998; R2 = 0.61809). To better understand the temporal patterns of gene expression changes in liver-specific DEGs, we used Short Time-series Expression Minor (STEM) software (24), which identified five distinct and significant clusters (Fig. 1F). Functional enrichment of DEGs in each cluster was carried out using MetaCore and Ingenuity Pathway Analysis (IPA) (Qiagen).
TABLE 2.
DEG comparisons relative to naive (N)a
| Comparison | No. of DEGs | No. of liver-, adrenal gland-, or lymphoid tissue-specific DEGs |
|---|---|---|
| Liver | ||
| Day 2 vs N | 162 | 114 |
| Day 4 vs N | 800 | 545 |
| Day 6 vs N | 2,251 | 1,653 |
| Adrenal glands | ||
| Day 2 vs N | 713 | 590 |
| Day 4 vs N | 735 | 490 |
| Day 6 vs N | 1,641 | 1,065 |
| Lymphoid tissues | ||
| Infection vs N | 3,282 | 2,737 |
DEGs are defined as those with at least 2-fold change in expression relative to naive tissue with an FDR-corrected P value of <0.05 and an average RPKM of >1. The DEGs listed are protein-coding and human homolog genes.
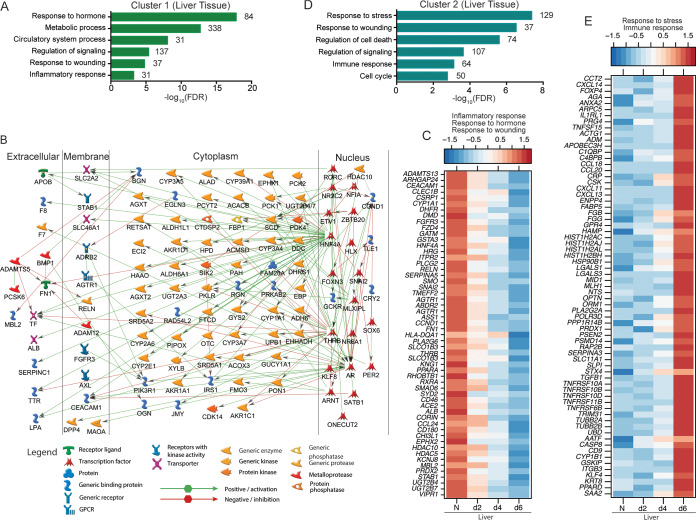
Expression of DEGs in cluster 1 decreased 2 to 6 DPI (Fig. 1F). Functional enrichment using MetaCore indicated these DEGs mapped to GO terms associated with hormonal responses, metabolic processes, blood circulation, signaling, and inflammation (Fig. 2A). Of the 338 genes that enriched to the GO term “metabolic process,” 111 were predicted to directly interact with each other (Fig. 2B). A notable downregulated transcription factor within this network was HNF4A, which is involved in liver development and regulates many genes and transcription factors involved in glucose metabolism (PCK1, PCK2, PDK4, and PKLR); fatty acid and cholesterol synthesis (APOB, CYP39A1, and CYP2E1); xenobiotic, intermediary and drug metabolism (e.g., CYP1A1, CYP3A7, and CYP2A6) (25–28); and coagulation (F7, F8, TF, F3, LPA, TTR, and SERPINC1) (29, 30) (Fig. 2B). Moreover, decreased expression of HNF4A is correlated with hepatic fibrosis, coagulopathy, and progression of liver failure in humans (31). Other downregulated liver-specific DEGs mapping to the GO term “inflammatory response” play a role in pathogen sensing (CD180), antigen presentation (HLA-DQA1), chemotaxis (CCL24 and STAB1), and complement regulation (CD46). Genes enriching to “response to wounding” included various adhesion molecules (e.g., CLEC1B) and vascularization factors (FZD4, DHFR). Finally, “response to hormone” DEGs encoded potent vasopressors and blood pressure regulators, e.g., albumin (ALB), angiotensin II receptor type 1 (AGTR1), angiotensin-converting enzyme 2 (ACE2), and corin enzyme (CORIN) (Fig. 2C).
FIG 2.
DEGs in the liver play a role in inflammation and metabolism. (A) Functional enrichment of DEGs within cluster 1 generated using MetaCore. The bar graph indicates the –log10 FDR corrected P value. Numbers next to the bars indicate the number of DEGs that mapped to each GO term. (B) Network image showing DEGs that directly interact with one another from cluster 1 that mapped to the GO term “metabolic process” generated using MetaCore. (C) Heatmap of DEGs within cluster 1 that mapped to the GO terms “response to wounding,” “response to hormone,” and “inflammatory response.” Each column represents the median normalized transcript counts (RPKM) for each gene at each time point as follows: N (naive; n = 4), d2 (2 DPI; n = 4), d4 (4 DPI; n = 2), d6 (6 DPI; n = 4). The range of colors is based on scaled and centered RPKM values of the entire set of genes, with red indicating highly expressed and blue indicating lowly expressed genes. (D) Functional enrichment of DEGs within cluster 2 generated using MetaCore. (E) Heatmap of DEGs in cluster 2 that enriched to “response to stress” and “immune response.” Each column represents the median normalized transcript counts (RPKM) for each gene at each time point: N (naive; n = 4), d2 (2 DPI; n = 4), d4 (4 DPI; n = 2), d6 (6 DPI; n = 4). The range of colors is based on scaled and centered RPKM values of the entire set of genes, with red indicating highly expressed genes and blue indicating lowly expressed genes.
Cluster 2 DEGs were more abundantly expressed at 4 to 6 DPI (Fig. 1F) and were associated with cellular stress, host defense, cell death, and cell cycle signaling (Fig. 2D). Some noteworthy upregulated genes function in chemotaxis (CCL18, CCL20, CXCL11, CXCL13, and CXCL14), cell death (CASP8, TNFSF15, and TNFRSF10A), and vasodilation (ADM) (Fig. 2E). Several genes encoding positive acute-phase proteins were elevated, including CRP, TNFRSF, IL1RL1, HAMP, C4BP, SERPINA3, ORM1, C1QBP, and SAA2 (Fig. 2E). While numerous cluster 1 and 3 DEGs encoding clotting factors and negative acute-phase proteins were downregulated, we observed upregulation of fibrinogen genes (FGG, FGB) in cluster 2. These results indicate substantial dysregulation of the coagulation cascade and likely unbalanced consumption of hemostatic components that contribute to disseminated intravascular coagulation (DIC).
Similar to cluster 1, expression of cluster 3 DEGs declined during the course of infection (Fig. 1F) and enriched to metabolic processes and hormone regulation (Fig. 3A). This cluster of genes participates in detoxification and drug metabolism (ALDH7A1, CES2, and SULT2A1), maintaining the integrity of extracellular matrix or basement membrane (COL4A5 and HYAL3), fatty acid metabolism (CERS4, ELOVL2, LIPC, and LCAT), hepatocyte growth (HGFAC), and coagulation (HPN) (Fig. 3B). Enrichment using IPA indicated that DEGs in clusters 1 and 3 were associated with a decrease in the disease and in the biological functions “fatty acid metabolism,” “transport of molecule,” “synthesis of lipid,” “steroid metabolism” and the toxicological function “increased levels of hematocrit.” Conversely, the biological functions “morbidity and mortality” and “glucose metabolism disorder,” as well as the toxicological function “inflammation of liver,” were predicted to increase (see Table S1 in the supplemental material).
FIG 3.
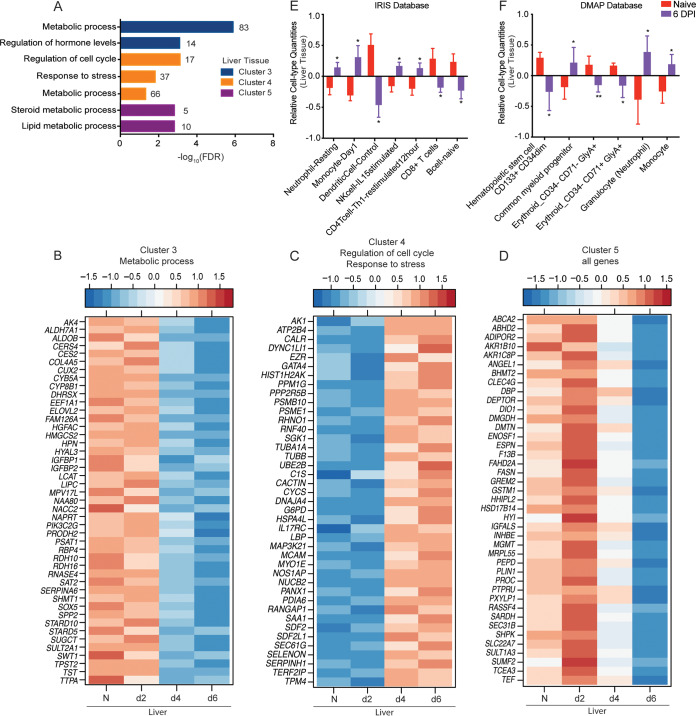
Liver-specific DEGs play a role in metabolism and cell cycle. (A) Functional enrichment of liver DEGs within clusters 3, 4, and 5 determined using MetaCore. The bar graph indicates the –log10 FDR-corrected P value. The numbers next to the bars indicate the number of genes that mapped to each of the GO terms. (B to D) Heatmaps of genes in cluster 3 that enriched to “metabolic process” (B), genes in cluster 4 that enriched to “regulation of cell cycle” and “response to stress” (C), and all DEGs within cluster 5 (D). Each column represents the median normalized transcript counts (RPKM) for each gene at each time point as follows: N (naive; n = 4), d2 (2 DPI; n = 4), d4 (4 DPI n = 2); d6 (6 DPI; n = 4). The range of colors is based on scaled and centered RPKM values of the entire set of genes, with red indicating highly expressed genes and blue indicating lowly expressed genes. (D and E) Bar graph depicting the mean ± SD of immune-cell type frequencies predicted using ImmQuant software using the IRIS (D) and DMAP (E) databases in naive samples (n = 4) and those obtained 6 DPI (n = 4). Significance between naive and 6 DPI samples was determined using unpaired two-tailed t test with Welch’s correction. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Cluster 4 DEGs also increased in expression 4 to 6 DPI (Fig. 1F) and mapped to “cell cycle,” “response to stress,” and “metabolic process” (Fig. 3A). DEGs associated with the cell cycle participate in mitosis (DYNC1LI1), negative control of cell division (PPP2R5B), and regulation of telomere function (TERF2IP) (Fig. 3C). “Response to stress” genes are implicated in DNA damage (RHNO1), protection against oxidative stress (SELENON and NOS1AP), and the unfolded protein response (UBE2B, PDIA6, and HSPA4L) (Fig. 3C). Lastly, upregulated metabolic-specific DEGs are important for cholesterol homeostasis (SAA1), nucleotide metabolism (AK1), and glycolysis (G6PD) (Fig. 3C). IPA enrichment of DEGs in clusters 2 and 4 revealed upregulation of the canonical pathways “IL-8 signaling,” “IL-6 signaling,” and “acute phase signaling,” indicating induction of inflammatory processes. Disease and biological functions included “infection by RNA viruses” and the toxicological function “apoptosis of liver cells.” The canonical pathways “PPAR signaling” and “PPARa/RXRa activation,” as well as the toxicological functions “hepatic injury” and “liver damage,” were downregulated in line with the significant pathology that EBOV infection inflicts in the liver (Table S1).
Expression of DEGs in cluster 5 transiently surged 2 DPI before returning to baseline levels (Fig. 1F). These genes enriched to metabolic, steroid hormone, and lipid processes (Fig. 3A). Notable genes within this cluster are involved in glucose and lipid metabolism (ADIPOR2, FASN, PLIN1, and SHPK), mitochondrial function (ENOSF1, MRPL55), and cellular growth and differentiation (IGFALS, INHBE, and PTPRU) (Fig. 3D). Other noteworthy genes within this cluster include F13B and PROC, which encode coagulation factor XIII B subunit and anticoagulant protein C, respectively. IPA enrichment also showed that transcriptional changes in cluster 5 are associated with upregulation of the canonical pathways “extrinsic prothrombin activation pathway” and “coagulation system,” as well as the toxicological function “inflammation of liver.” In contrast, the canonical pathway “PPARa/RXRa activation” and the toxicological function “primary hepatocellular carcinoma” were predicted to be downregulated (Table S1).
Next, to infer whether liver-specific DEGs at 6 DPI were associated with changes in immune cell frequencies, we performed digital cell quantification (DCQ) analyses using ImmQuant software and the DMAP and IRIS databases (32–34). These algorithms predict human cell subset quantities based on transcriptional signatures. We observed a significant increase in frequencies of neutrophils, monocytes (including early-stimulated monocytes), common myeloid progenitors, interleukin-15 (IL-15)-stimulated NK cells, and early-stimulated T helper cells (Fig. 3E and F). Frequencies of hematopoietic stem cells, erythroid subsets, resting dendritic cells, CD8+ cells, and naive B cells were instead predicted to decrease (Fig. 3E and F).
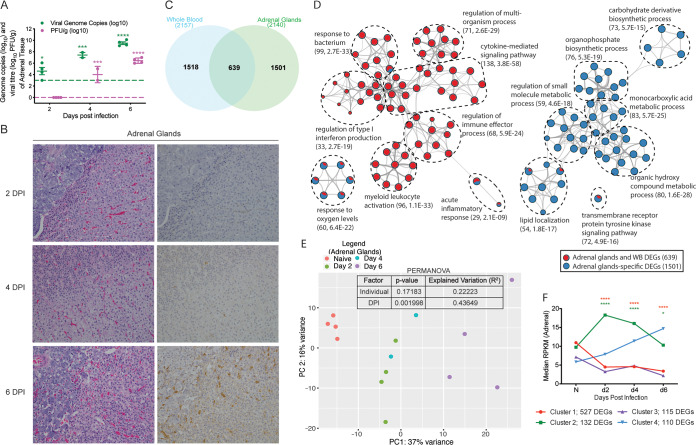
EBOV infection induces significant damage in adrenal glands.
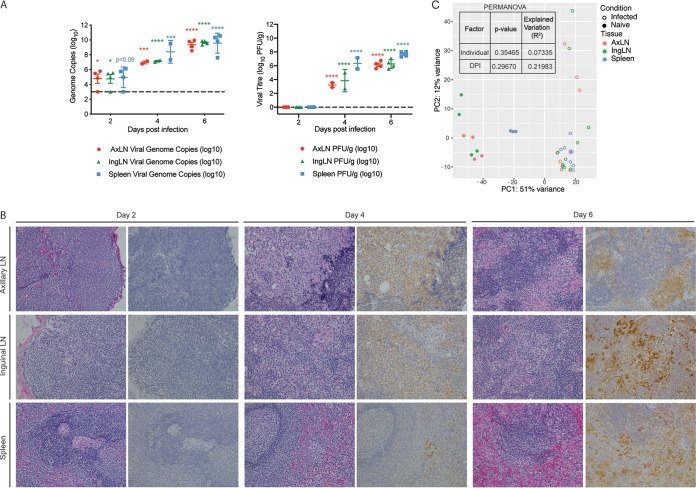
As described for the liver, we detected a significant increase in viral genome copies 2 to 6 DPI and infectious virus titers at 4 to 6 DPI in adrenal gland tissue relative to the assays’ limits of detection (Fig. 4A). No substantial gross or histologic lesions were noted 2 DPI (Fig. 4B; Table 1). At 4 DPI, minimal adrenalitis was observed in 1 of 2 animals. Scattered mononuclear cells in the adrenal gland exhibited positive EBOV-specific immunoreactivity in both animals (Fig. 4B). On gross examination, reticulation (3 of 4 animals) and friable parenchyma (2 of 4 animals) with mild adrenalitis (4 of 4 animals) were evident 6 DPI. Virus antigen was observed in individualized and scattered mononuclear cells in all 4 macaques at this time point (4 of 4) (Fig. 4B; Table 1).
FIG 4.
Makona infection results in significant organ damage and transcriptional changes in the adrenal gland. (A) Mean ± SD of EBOV infectious virus titer and genome copy numbers in adrenal gland were quantified using a plaque assay and RT-qPCR for each animal and each time point. Significance was determined using one-way ANOVA with Dunnett’s multiple-comparison test relative to the limit of detection, which is represented as a dashed line. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (B) Hematoxylin and eosin staining (left) and immunohistochemistry staining (right) (brown indicates reactivity) using anti-VP40-specific antibody of adrenal glands at 2, 4, and 6 DPI; ×20 magnification. (C) Venn diagram comparing differentially expressed genes (DEGs) detected in whole blood (WB) and adrenal gland pooled DEGs from all time points. (D) Network image showing functional enrichment of DEGs detected in both adrenal glands and WB, and adrenal-specific DEGs using Metascape and visualized using Cytoscape. Each circle represents a theme of highly related gene ontology (GO) terms. The edges (lines) link similar GO terms, and the thickness of the lines indicates the relatedness of the GO terms. Each GO term is shown as a pie chart that depicts the relative transcriptional contribution of DEGs detected in both adrenal gland and WB (red) and adrenal gland only (blue). (E) Principal component analysis (PCA) of adrenal gland-specific DEGs over time. (F) STEM analysis identified four distinct and significant temporal expression clusters of genes. The median RPKM of each cluster over time is plotted. The Bonferroni-corrected P values for each cluster in order are 4.67e-14, 7.37e-5, 3.00e-4, and 1.66e-4. Statistical significance between time points was determined using one-way ANOVA with Dunnett’s multiple-comparison test relative to N (naive). *, P < 0.05; ****, P < 0.0001.
Gene expression changes in the adrenal gland are associated with disruptions in metabolism and regulation of blood circulation.
DEGs were detected as early as 2 DPI in adrenal gland and in greater numbers than the liver (Table 2). We identified 1,501 adrenal gland-specific DEGs after exclusion of those originating from WB (Fig. 4C). Using Enrichr, we confirmed that DEGs detected in both adrenal gland and WB originate mainly from WB, CD14+ monocytes, and CD33+ myeloid cells (adjusted P value, 2.82e-36, 1.57e-28, and 4.23e-22, respectively), while adrenal tissue specificity was confirmed using Enrichr (adjusted P value, 3.35e-20). Enrichment analysis of the 639 DEGs detected in both the adrenal gland and WB using Metascape revealed overrepresentation of processes largely associated with innate immunity (e.g., “cytokine-mediated signaling pathway,” “myeloid leukocyte activation,” and “regulation of type I interferon production”). Functional enrichment of the adrenal gland-specific DEGs showed overrepresentation of GO terms involved in lipid, carbohydrate, and monocarboxylic acid metabolic processes (Fig. 4D). PCA revealed that adrenal gland-specific DEGs clustered based on DPI, with the lowest dimensional separation at 2 and 4 DPI and the highest variance, corresponding to the highest transcriptional changes, at 6 DPI (Fig. 4E; Table 2). As noted for the liver, PC1 (DPI) explained more variance (37%) than PC2 (intragroup differences) (Fig. 4E). Similarly, a PERMANOVA test indicated no significant variation between animals at each time point (P = 0.17183; R2 = 0.22223), while DPI significantly impacted variance (P = 0.001998; R2 = 0.43649). STEM analysis identified four distinct and significant clusters (Fig. 4F).
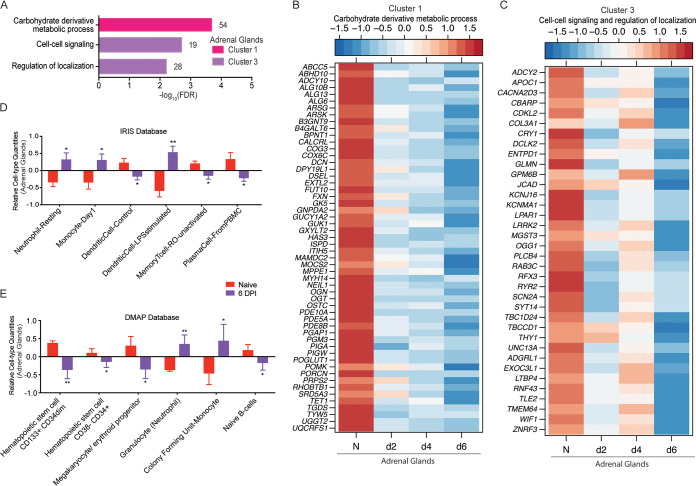
Cluster 1 DEGs decreased in expression throughout infection (Fig. 4F) and enriched to the GO term “carbohydrate derivative metabolic processes” (Fig. 5A), which consists of genes that play a role in glycosylation (ALG10B, GXYLT2, and OGT), mitochondrial respiration (FXN and UQCRFS1), and extracellular matrix support (HAS3, DCN, ECM, and POMK) (Fig. 5B). Additionally, DEGs that regulate cyclic nucleotide functions in signaling (PDE10A, ADCY10, and GUK1) were also suppressed (Fig. 5B). DEGs in cluster 3 had a comparable pattern (Fig. 4F) and mapped to the GO terms “cell-cell signaling” and “regulation of localization” (Fig. 5A). Some of the cluster 3 genes participate in tissue and blood vessel support (COL3A1 and GLMN), Wnt signaling (TLE2, WIF1, ZNRF3), Ca2+ mediated signaling (DCLK2, RYR2, TMEM64), and extracellular matrix development (LTBP4) (35) (Fig. 5C). IPA-based functional enrichment of these DEGs indicated upregulation of the disease and biological functions “incidence of tumor” and “liver lesion,” as well as the toxicological function “destruction of glomerular capillary.” Downregulated toxicological functions included “histopathological changes of liver,” “alcoholic cirrhosis,” and “ventricular fibrillation” (Table S1).
FIG 5.
Adrenal-specific DEGs play a role in metabolic processes and signaling. (A) Functional enrichment of adrenal-specific DEGs within clusters 1 and 3 carried out using MetaCore. The bar graph indicates the –log10 FDR-corrected P value. Numbers next to the bars indicate the number of genes that mapped to each of the GO terms. (B and C) Heatmap of genes in cluster 1 that enriched to “carbohydrate derivative metabolic process” (B) and genes in cluster 3 that enriched to “cell to cell signaling” and “regulation of localization” (C). Each column represents the median normalized transcript counts (RPKM) for each gene at each time point: N (naive; n = 4), d2 (2 DPI; n = 4); d4 (4 DPI; n = 2); d6 (6 DPI; n = 4). The range of colors is based on scaled and centered RPKM values of the entire set of genes, with red indicating highly expressed genes and blue indicating lowly expressed genes. (D to E) Bar graph depicting the mean ± SD of immune-cell-type frequencies predicted using ImmQuant software and the IRIS (D) and DMAP (E) databases in naive samples (n = 4) and those obtained 6 DPI (n = 4). Significance between naive and 6 DPI samples was determined using unpaired two-tailed t test with Welch’s correction. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
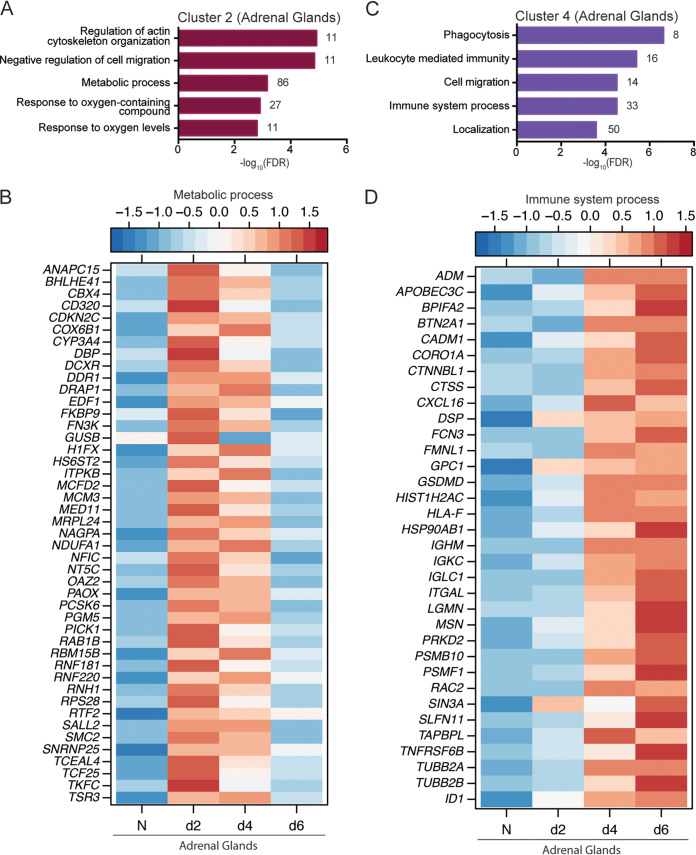
Cluster 2 DEGs, the expression of which peaked 2 DPI (Fig. 4F), enriched to GO terms associated with cell migration, metabolism, and response to oxygen (Fig. 6A). Metabolic-affiliated DEGs aid in cell division (ANAPC15, CDKN2C, and SMC2), mitochondrial respiration (COX6B1 and NDUFA1), and coagulation (MCFD2) (Fig. 6B). Additionally, DEGs involved in ubiquitination and proteasomal degradation (RNF181 and RNF220), as well as transcriptional regulation (DRAP1, TCEAL4, and MED11), were also transiently upregulated (Fig. 6B). Additional enrichment of these DEGs using IPA pointed to increased “oxidative phosphorylation” canonical signaling as well as the biological functions “transactivation of RNA” and “viral infection” (Table S1).
FIG 6.
Adrenal-gland specific DEGs play a role in metabolism and immunity. (A and C) Functional enrichment of adrenal gland-specific DEGs within cluster 2 (A) and cluster 4 (C) generated using MetaCore. The bar graph indicates the –log10 FDR-corrected P value. Numbers next to the bars indicate the number of genes that mapped to each of the GO terms. (B and D) Heatmap of DEGs within cluster 2 that enriched to the GO term “metabolic process” (B) and DEGs in cluster 4 that enriched to the GO term “immune system process” (D). Each column represents the median normalized transcript counts (RPKM) for each gene at each time point as follows: N (naive; n = 4), d2 (2 DPI; n = 4); d4 (4 DPI; n = 2), d6 (6 DPI; n = 4). The range of colors is based on scaled and centered RPKM values of the entire set of genes, with red indicating highly expressed genes and blue indicating lowly expressed genes.
Expression of cluster 4 DEGs increased steadily following infection (Fig. 4F). These genes were associated with host defense, cell migration, and localization (Fig. 6C). DEGs that enriched to “immune system process” function in antigen presentation (e.g., CTSS, HLA-F, TAPBPL), chemotaxis and migration (e.g., CORO1A, FMNL1, CXCL16, and MSN), cell death (SLFN11 and TNFRSF6B), and localization (e.g., TUBB2A and TUBB2B) (Fig. 6D). Additionally, immunoglobulin-related genes (IGHM, IGKC, and IGLC1) and adrenomedullin (ADM), a potent hypotensive and vasodilator agent, were also upregulated (Fig. 6D). Enrichment using IPA showed upregulation of the disease and biological functions “migration of vascular endothelial cells,” “adhesion of phagocytes,” “cell movement of leukocytes,” and “cell movement of monocytes” in addition to toxicological functions related to cardiac injury. Disease and biological functions associated with apoptosis of immune cells were downregulated, e.g., “apoptosis of lymphocytes” and “apoptosis of leukocytes.” Upstream regulators of DEGs in this cluster include INFG, NF-κB, and STAT1 (Table S1).
ImmQuant DCQ analysis suggested that infection led to increased frequencies of neutrophils, monocytes (resting and activated phenotypes), and activated dendritic cells at 6 DPI in adrenal gland tissue (Fig. 5D and E). In contrast, lower frequencies of various hematopoietic stem cell subsets, megakaryocyte and erythroid progenitor, resting dendritic cells, memory T cells, naive B cells, and plasma cells were predicted (Fig. 5D and E).
EBOV-Makona infection dysregulates host responses in spleen and lymph nodes characterized by inflammation and lymphopenia.
Virus titers and histological changes were next examined within the spleen, axillary lymph nodes, and inguinal lymph nodes. Viral genome copies were significantly increased as early as 2 DPI in the axillary and inguinal lymph nodes, while infectious EBOV titers were significantly elevated at both 4 and 6 DPI in these tissues (Fig. 7A). No significant gross or histologic lesions were observed in any animals 2 DPI (Fig. 7B; Table 1). On gross examination, mild enlargement of the axillary and inguinal lymph nodes was evident in 1 of 2 animals at 4 DPI. Additionally, minimal subscapular and medullary histiocytosis of both the axillary and inguinal lymph nodes were noted in both animals 4 DPI. At this time point, EBOV antigen was detected in scattered individual to clustered mononuclear cells in axillary lymph node, inguinal lymph node, and spleen sections (Fig. 7B). By 6 DPI, enlargement of the axillary lymph node (3 of 4 animals), inguinal lymph node (2 of 4 animals), and spleen (3 of 4 animals) was apparent in a majority of subjects. These changes were accompanied by subscapular and medullary histiocytosis of both the axillary and inguinal lymph nodes (4 of 4 animals), mild distortion of normal splenic white pulp architecture (3 of 4 animals), and minimal to mild expansion of splenic red pulp (4 of 4 animals). This coincided with the detection of EBOV antigen in individualized to sheets of mononuclear cells in the axillary lymph node (4 of 4 animals), inguinal lymph node (4 of 4 animals), and splenic red pulp (4 of 4 animals) and white pulp (2 of 4 animals) (Fig. 7B).
FIG 7.
EBOV-Makona infection results in viremia, histopathological changes, and large a transcriptional response in lymphoid tissue. (A) The mean ± SD of EBOV infectious virus titer and genome copy numbers were quantified in lymphoid tissues using a plaque assay and RT-qPCR for each animal. Significance was determined using one-way ANOVA with Dunnett’s multiple-comparison test relative to the limit of detection, which is represented as a dashed line. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (B) Hematoxylin and eosin staining (left) and immunohistochemistry staining (right) (brown indicates reactivity) using anti-VP40-specific antibody of lymphoid tissues at 2, 4, and 6 DPI; magnification ×20. (C) Principal component analysis of lymphoid-specific DEGs over time.
Transcriptional profiles of spleen and axillary and inguinal lymph nodes from EBOV-infected animals revealed a clear distinction between samples obtained from naive and infected animals when examined using PCA but no appreciable time-dependent or tissue-dependent gene expression changes in any of the lymphoid tissues (Fig. 7C). Moreover, a PERMANOVA test revealed no significant variation between any of the three lymphoid tissues at any given DPI (P = 0.29670; R2 = 0.21983). Similarly, a PERMANOVA test indicated no significant variation between animals at each time point in lymphoid tissues (P = 0.35465; R2 = 0.07335). Therefore, all three infected lymphoid tissue data sets were merged and compared to naive tissues.
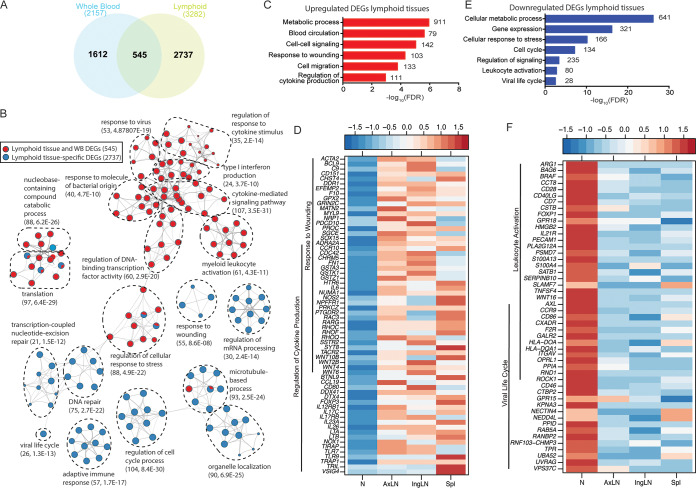
We identified 545 DEGs common between lymphoid tissues and WB and 2,737 DEGs designated lymphoid tissue-specific DEGs (Fig. 8A). DEGs detected in both lymphoid tissues and WB were predominantly involved in antiviral immunity, cytokine signaling, and myeloid leukocyte activation, while lymphoid tissue-specific DEGs were implicated in cell cycle (e.g., “regulation of cell cycle process” and “DNA repair”) in addition to host defense (e.g., “response to wounding” and “adaptive immune response”) (Fig. 8B). A more in-depth functional enrichment analysis of showed that upregulated lymphoid-specific DEGs enriched to metabolic processes, blood circulation, signaling, and host defense (Fig. 8C). Specifically, DEGs that mapped to “response to wounding” and “regulation of cytokine production” were involved in inflammation (C9, PRKCZ, and LTA), chemotaxis and migration (CD151, CCR10, PTGDR2, and RHOC), antigen recognition or presentation (TIRAP, TLR7, TLR9, and CD80), and lymphocyte homing (CCL19). Interestingly, we also observed increased expression of genes associated with the differentiation and maintenance of T helper 17 (Th17) cells (IL17RB, IL17C, IL6, IL23A, and IL-26) as well as genes that negatively regulate adaptive immunity (FOXP3 and VSIG4) (Fig. 8D). Additional DEGs upregulated in lymphoid tissues include genes that control blood pressure (ACTA2 and MYL9), extracellular matrix assembly (DDR1, MATN2, and SGCE), coagulation (EFEMP2, F10, PROC, and FN1), cell death (PDCD10), and cell cycle (CDC42 and NUMA1) (Fig. 8D). Furthermore, we detected DEG signatures consistent with generation of, or response to, reactive oxygen species (GPX2, NOS2, and NOX1) (Fig. 8D).
FIG 8.
Transcriptional changes in lymphoid tissues are associated with metabolism, regulation of gene expression, inflammation, and lymphopenia. (A) Venn diagram comparing pooled differentially expressed genes (DEGs) detected in whole blood (WB) and lymphoid tissues from all time points. (B) Network image showing functional enrichment of DEGs detected in both lymphoid tissues and WB and in lymphoid tissue only obtained using Metascape and visualized using Cytoscape. Each circle represents a theme of highly related gene ontology (GO) terms. The edges (lines) link similar GO terms, and the thickness of the line indicates the relatedness of the GO terms. Each GO term is shown as a pie chart that depicts the relative transcriptional contribution of DEGs detected in both lymphoid tissue and WB (red) and lymphoid tissue only (blue). (C and E) Functional enrichment of the upregulated (C) and downregulated (E) DEGs detected in lymphoid tissues was carried out using MetaCore. The bar graph indicates the –log10 FDR-corrected P value. Numbers next to the bars indicate the number of genes that mapped to each of the GO terms. (D and F) Heatmaps of upregulated (D) and downregulated (F) genes mapping to the indicated GO terms. Each column represents the median normalized transcript counts (RPKM) for each gene within each lymphoid organ: N (naive; n = 12), AxLN (axillary lymph node; n = 10), IngLN (inguinal lymph node; n = 10), Spl (spleen; n = 10). The range of colors is based on scaled and centered RPKM values of the entire set of genes, with red indicating highly expressed genes and blue indicating lowly expressed genes.
Lymphoid tissue-specific DEGs that were downregulated also enriched to GO terms associated with metabolism and cell cycle progression and, to a lesser extent, cellular stress and leukocyte activation (Fig. 8E). DEGs that mapped to “viral life cycle” and “leukocyte activation” included genes important for antigen presentation (HLA-DOA and HLA-DQA1) and lymphocyte activation and proliferation (CD40LG, CD7, CD28, CD46, TNFSF4, and FOXOP1). Other downregulated genes play a role in cell adhesion (ITGAV and NECTIN4), ubiquitin-mediated proteasomal degradation (NEDD4L, RANBP2, and UBA52), and vesicle transport (RAB5A and VPS37C) (Fig. 8F). Additionally, IPA revealed upregulation of diseases and functions associated with viral infection, such as “infection by RNA virus,” and the canonical pathways “lymphotoxin b receptor signaling,” “production of nitric oxide and reactive oxygen species in macrophages,” and “sphingosine-1-phosphate signaling.” The disease and function term “formation of lymphoid tissue,” as well as canonical pathways “PTEN signaling” and “Aryl hydrocarbon receptor signaling,” were predicted to be downregulated (Table S1).
EBOV-Makona infection alters frequency and phenotype of immune cells in spleen and lymph nodes.
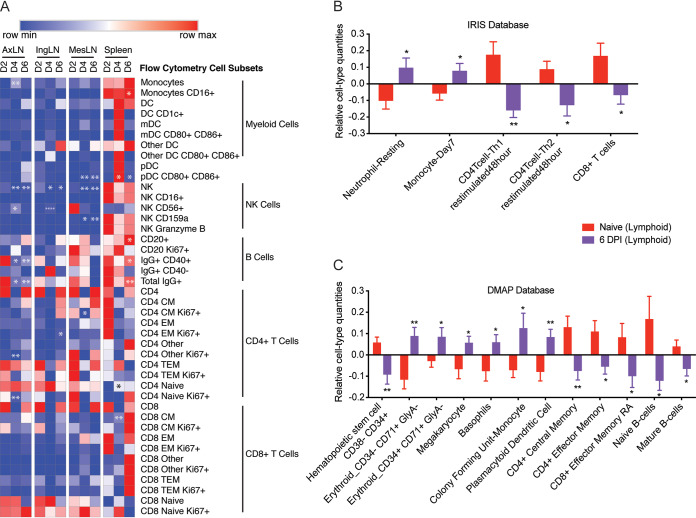
To characterize the immune response to EBOV-Makona, we used flow cytometry to measure changes in frequency of key innate and adaptive immune cells in the spleen and axillary, inguinal, and mesenteric lymph nodes. The frequency of monocytes increased 4 DPI in axillary lymph nodes relative to 2 DPI (P = 0.01) while remaining relatively stable in inguinal and mesenteric lymph nodes and the spleen (Fig. 9A). Although the overall frequency of monocytes did not change in the spleen, the relative abundance of nonclassical CD16+ monocytes diminished, while that of classical CD16– monocytes increased 6 DPI (P = 0.02) (Fig. 9A). Frequencies of total myeloid dendritic cells (DCs) (mDC) and plasmacytoid DCs (pDC) remained unchanged throughout infection in all four lymphoid tissues. Interestingly, expression of the costimulatory markers CD80 and CD86 increased on mesenteric and splenic pDCs at 4 and 6 DPI (P = 0.02 and 0.04, respectively) (Fig. 9A). The frequency of total natural killer (NK) cells was lower in axillary, inguinal, and mesenteric lymph nodes at 4 (P = 0.001, 0.037, and 0.005, respectively) and 6 DPI (P = 0.006, 0.043, and 0.008, respectively) relative to 2 DPI (Fig. 9A). This decrease in overall frequency was accompanied by an increase in the relative abundance of CD56+ NK cells 4 DPI in axillary and inguinal lymph nodes (P = 0.0122 and 0.0001, respectively). No significant change in relative abundance of CD16+, CD159+, or granzyme B+ NK cells was observed at any time point for lymphoid tissues (Fig. 9A).
FIG 9.
Immune cell frequencies within lymphoid tissues. (A) Heatmap of immune cell subset frequencies measured using flow cytometry. Each column indicates the median percentage frequency of the various subsets within each tissue. The range of colors is based on scaled row values of each cell subset, with red representing high cell frequency and blue representing low cell frequency. Significance was determined using one-way ANOVA relative to 2 DPI. *, P < 0.05; **, P < 0.01; ***, P < 0.01; ****, P < 0.0001. (B and C) Bar graph depicting mean ± SD immune-cell type frequencies predicted using ImmQuant software using the IRIS (B) and DMAP (C) databases in naive and 6 DPI samples. Significance was determined using unpaired two-tailed t test with Welch’s correction. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, we analyzed changes in the frequency of T and B lymphocytes within secondary lymphoid organs. The frequency of spleen B cells increased 6 DPI relative to 2 DPI (P = 0.0217). Conversely, the relative abundance of total IgG+ and IgG+CD40+ B cells decreased relative to that at 2 DPI in axillary lymph nodes 4 and 6 DPI (P = 0.0152 and 0.009; 0.014 and 0.01) and in the spleen 6 DPI (P = 0.0062 and 0.035) (Fig. 9A). Overall, frequencies of CD4 and CD8 T cells or their naive/memory subsets did not change significantly with DPI in any of the lymphoid tissues, with the exception of an increase in frequency of splenic naive CD4 T cells (P = 0.036) and an increase in splenic CD8 CM T cells 4 DPI (P = 0.005) (Fig. 9A). However, the frequency of proliferating (Ki67+) axillary naive CD4 T cells (P = 0.002) and mesenteric central memory CD4 T cells (P = 0.03) was reduced 4 DPI. The CD4 T cell inguinal effector memory subset increased DPI 6 (P = 0.041).
To complement our limited flow analysis and capture shifts in erythrocyte and granulocyte populations, we performed ImmQuant DCQ. Our analysis predicted that gene expression changes at 6 DPI were associated with increased frequencies of erythroid cell subsets, megakaryocytes, basophils, resting neutrophils, monocytes (immature and activated), and plasmacytoid dendritic cells (Fig. 9B). Frequencies of hematopoietic stem cells, memory CD4+ and CD8+ T cell subsets, as well as naive and mature B cells were predicted to decrease (Fig. 9C). In line with our flow cytometry data, both the DMAP and IRIS databases predicted decreases in CD4 and CD8 T-cell subsets.
DISCUSSION
The liver, adrenal gland, and lymphoid tissue are important early targets for EBOV as evidenced by the detection of viral antigen and high viral titers in these organs (7, 9, 14, 15). However, studies investigating the impact of EBOV infection within these tissues have been limited to histopathological examination. In this study, we sought to define for the first time longitudinal transcriptional changes within these key organs following EBOV-Makona infection using RNA-Seq.
The magnitude of transcriptional changes in the liver increased as infection progressed and correlated with increasing viral replication within mononuclear cells and hepatocytes. Functional enrichment of early transcriptional changes revealed significant downregulation of transcripts involved in glucose and lipid metabolism, drug metabolism, coagulation, and blood pressure regulation indicative of an early loss of liver function. These gene expression signatures are likely due to substantial virus-induced tissue damage and corresponded with onset of thrombocytopenia and increased plasma levels of the liver enzymes AST, ALT, ALP, and GGT in these animals as we reported previously (20). It is possible that early dysregulation in the expression of clotting factor genes in the liver may initiate the events that result in the development of systemic coagulopathy, a hallmark of EVD (4). At later time points, transcriptional changes were indicative of a cellular response to stress characterized by heightened inflammation and increased cell death. These transcriptional changes correlated with an increasing severity of necrotizing hepatitis. This contrasts with previous studies of EBOV-infected Huh7 cells or HepG2 cells, which exhibited small changes in host gene expression and poor induction of innate immune responses (36, 37). This observation emphasizes the importance of infiltrating immune cells that can only be studied using in vivo studies and suggests caution in overinterpretation of in vitro studies of cell lines used to model filovirus-impacted tissues.
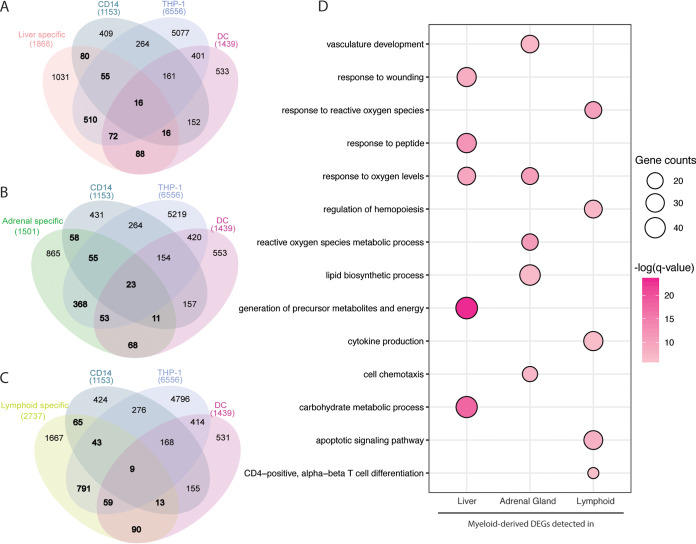
Our in silico analysis using ImmQuant predicted that the transcriptional changes observed at 6 DPI were associated with increased frequency of monocytes in the liver, which is consistent with our immunohistochemistry (IHC) report of immunoreactive mononuclear cells within this tissue. This observation suggests that monocytes infected in the blood or the draining lymph nodes may be infiltrating the liver. Additional comparison to publicly available data sets obtained from either in vivo-infected CD14+ monocytes or in vitro-infected THP1 cells or human monocyte-derived dendritic cells (8, 38, 39) indicates that a large number of liver-specific DEGs (45%) overlapped with DEGs detected as a result of EBOV infection in these myeloid cells (Fig. 10A). These DEGs enriched to metabolic pathways and inflammation (Fig. 10D). These data could reflect both an infection of liver-resident Kupfer cells or infiltration of EBOV-infected monocytes.
FIG 10.
A significant number of liver, adrenal, and lymphoid tissue DEGs originate from myeloid cells. (A to C) Venn diagrams depicting the comparison of tissue-specific DEGs and those detected in CD14+ monocytes obtained from infected macaques as well as in vitro-infected THP-1 and monocyte-derived dendritic cells in (A) liver, (B) adrenal gland, (C) and lymphoid tissues. (D) Bubble plot representing functional enrichment of myeloid-derived DEGs detected in liver, adrenal gland, and lymphoid tissues. The size of the bubbles represents gene counts for each GO process. The colors represent the levels of significance relative to each GO process.
Similar to the liver, early gene expression changes were evident in the adrenal gland following EBOV infection. Expression of a large number of genes important for carbohydrate metabolism, extracellular matrix support, and signal transduction was reduced. As expected, these changes became more pronounced later during infection and correlated with increased viremia and histopathological changes. Genes that increased in expression throughout infection were involved in inflammatory pathways, innate immunity, and cell death. Thus, these gene expression signatures reflect extensive cell death and decline of adrenal gland function as a direct and/or indirect effect of viral replication, which could explain the adrenalitis observed 4 to 6 DPI. As described for liver, ImmQuant analysis predicted that transcriptional changes in the adrenal gland were associated with an increase of neutrophils, activated dendritic cells, and monocytes, which supports these findings. A comparison against publicly available data sets obtained from either in vivo-infected CD14+ monocytes or in vitro-EBOV-infected THP1 cells or human monocyte-derived dendritic cells (8, 38, 39) showed a 42% overlap common DEGs enriching to oxidative damage, lipid biosynthesis, and vasculature development processes (Fig. 10B and D). In addition to gene expression changes associated with inflammation, we also observed significant upregulation of genes associated with cardiac injury and vasodilation in this tissue. Given the role of the adrenal gland in regulating blood pressure and cardiovascular function, these gene expression changes are in line with the circulatory failure that is a hallmark of EVD and ensuing hypotension, shock, and multiorgan damage.
Concordant with both liver and adrenal gland, the lymphoid tissue (axillary lymph node, inguinal lymph node, and spleen) exhibited differential gene expression as early as 2 DPI. However, in contrast to the liver and adrenal gland, no time-dependent changes in gene expression were detected. DEGs upregulated in lymphoid tissue in response to infection play a role in inflammation, chemotaxis, and antigen presentation, which is consistent with the induction of inflammatory genes reported in the spleen of BALB/c mice infected with mouse-adapted EBOV (40). Furthermore, these transcriptional changes correlate with histiocytosis 4 to 6 DPI and a predicted increased frequency of neutrophils, monocytes, and plasmacytoid DCs. Consistent with lymphoid depletion and identification of apoptotic bodies within lymph nodes (7, 41), we observed decreased expression of several lymphocyte-associated transcripts and genes involved in antigen presentation. This observation is in line with the lack of induction of T and B cell responses in EBOV fatal cases in both humans and NHP (8, 42, 43), including the peripheral blood of the animals from which tissues were collected for the current study (20, 44). Similarly, in silico ImmQuant analysis predicted that cells important for adaptive immunity, such as lymphocytes and dendritic cells, were predicted to decrease in both liver and adrenal gland. Importantly, the decreased expression of T and B cell-associated genes in all tissues analyzed indicates a systemic loss of lymphocytes rather than recruitment of lymphocytes into tissues. Finally, nitric oxide levels have been detected in humans and animal models in response to EBOV infection (45–47). We detected increased expression of transcripts associated with generating reactive oxygen species, notably NOS2, which is important in synthesizing nitric oxide (48) and has been shown to induce apoptosis (49), suggesting that generation of reactive oxygen species may contribute to lymphocyte death.
Interestingly, both viral loads and transcriptional changes were detected earlier in tissues than in WB of these same animals (20), suggesting these tissues are initial sites of infection from which the virus can disseminate systemically following intramuscular challenge. Indeed, by 6 DPI, viral titers in the blood were 2 log10 higher than in liver, adrenal gland, and lymphoid tissues (20). Similar to what we reported recently in the blood (8), the source of inflammation is likely myeloid cells, notably monocytes/macrophages, as recently shown in human cases (50). Macrophages may amplify the pathogenic cascade by secreting proinflammatory cytokines (IL-1, IL-6, tumor necrosis facto [TNF]), chemokines, growth factors, and reactive oxygen species, thereby increasing vascular permeability and contributing to endothelial dysfunction. ImmQuant analysis for all tissues predicted increased frequencies of monocytes, which supports our flow cytometry data and previous complete blood cell counts from these animals. Moreover, 39% of the DEGs detected in lymphoid tissues overlapped with those obtained from CD14+ monocytes obtained from EBOV-infected macaques or THP1 and human monocyte-derived macrophages infected with EBOV in vitro (Fig. 10C). These common DEGs were involved in the response to reactive oxygen species, regulation of hemopoiesis, cytokine production, apoptotic signaling, and CD4 cell differentiation (Fig. 10D).
As described for EBOV-Kikwit-challenged macaques (9, 10), we detected viral replication, immunoreactive mononuclear cells, and histopathological lesions in liver, adrenal gland, and lymphoid tissues starting 4 DPI. However, compared to animals intramuscularly challenged with the EBOV-Kikwit variant (10), these changes are less severe following EBOV-Makona challenge. At 6 DPI, viral titers in these animals were 1 to 2 log10 lower than viral loads seen in the respective tissues of EBOV-Kikwit-infected macaques (10). Moreover, histological changes were more mild, with reduced severity of liver necrosis and inflammation compared to the significant degeneration, necrosis and congestion observed in Kikwit-infected macaques (10). These results are consistent with previous reports that indicate that EBOV-Makona infection results in delayed disease progression (13, 20).
In summary, EBOV-Makona infection results in early and robust gene expression changes within the liver, adrenal gland, and lymphoid tissues. These tissues appear to serve as initial reservoirs for the virus. Infection results in extensive dysregulation of metabolism and coagulation in addition to the impairment of antigen presentation and adaptive immunity. This coupled with increased inflammation contributes to cell death and injury within these organs. Consequently, these organs are prevented from carrying out their normal function while supporting the dissemination of the virus, which ultimately contributes to the development of EVD.
MATERIALS AND METHODS
Study design, virus, and challenge.
A detailed description of the study design, clinical findings, and virus challenge were provided in our previous study (8, 20). Briefly, 10 healthy, filovirus-negative male cynomolgus macaques, 3 to 5 years of age and weighing 4 to 8 kg, were challenged intramuscularly (i.m.) with 1,000 PFU of EBOV-Makona (isolate C07). The dose was equally divided into the left and right quadriceps. Animals were monitored postchallenge for clinical signs of disease. Animals were euthanized at 2 DPI (n = 4), 4 DPI (n = 2), and 6 DPI (n = 4). At these time points, liver, adrenal glands, and lymphoid tissues (axillary lymph nodes, inguinal lymph nodes, and spleen) were collected for histopathology, immunohistochemistry, virology, and RNA-Seq. Historical tissues obtained earlier from naive cynomolgus macaques (n = 4) used in a previous study were used as the reference for transcriptomic analysis. Fresh lymphoid tissue was additionally analyzed using flow cytometry to substantiate predicted immune-related gene expression changes. All work was conducted at the biosafety level 4 (BSL-4) containment level at the Galveston National Laboratory (Galveston, TX) at the University of Texas Medical Branch (UTMB). All animal experiments and protocols were approved by the Institutional Animal Care and Use Committee (IACUC) and the Institutional Biosafety Committee at UTMB.
Virus detection and quantification.
Infectious virus titers were measured using plaque assay on Vero E6 cells. Cells were seeded in 6-well plates and grown to confluence. Tissue samples from infected macaques were titrated from 10−1 to 10−6 in duplicate wells. Plaques were counted using neutral red stain.
RNA was isolated from tissues using a viral RNA minikit (Qiagen, Valencia, CA). As previously described, primers/probe targeting the VP30 gene of ZEBOV-Makona were used for reverse transcriptase quantitative PCR (RT-qPCR) (51). EBOV RNA was detected using the CFX96 detection system (Bio-Rad Laboratories, Hercules, CA) using One-Step Probe qRT-PCR kits (Qiagen, Valencia, CA) with the following cycle conditions: 50°C for 10 minutes, 95°C for 10 seconds, and 40 cycles of 95°C for 10 seconds followed by 59°C for 30 seconds. Threshold cycle (CT) values corresponding to the ZEBOV genome were determined with CFX Manager software. Data were transformed into genome equivalents (GEq) per milliliter. The GEq standard was calculated using Avogadro’s number, the molecular weight of the viral genome, and known concentrations of EBOV RNA.
Histology and immunohistochemistry.
Tissues were fixed in 10% neutral buffered formalin with 2 changes for a minimum of 21 days. Following fixation and inactivation, tissues were placed in cassettes and processed with a Shandon Excelsior instrument on a 12-hour automated schedule using a graded series of ethanol, xylene, and ParaPlast Extra. Embedded tissues were sectioned at 5 μm and dried overnight at 60°C prior to staining. Immunohistochemistry staining was accomplished using a Dako autostainer. Specific antifilovirus immunoreactivity was detected using an anti-EBOV VP40 primary antibody at a 1:4,000 dilution for 60 minutes, a biotinylated goat anti-rabbit IgG (BA-1000; Vector Labs) at 1:200 for 30 minutes, and Dako LSAB2 streptavidin-HRP (K1016) for 15 minutes. Slides were developed with Dako DAB chromogen (K3468) for 5 minutes and counterstained with Harris hematoxylin for 1 minute.
Severity scoring was awarded for each hematoxylin and eosin (H&E)-stained tissue based on a 0 to 4 scale (Table 1). A score of 0 indicates no lesions; 1 indicates minimal lesions (less than 25% of the tissue affected); 2 indicates mild lesions (26 to 50% of the tissue affected); 3 indicates moderate lesions (51 to 75% of the tissue affected); and 4 indicates marked lesions (76 to 100% of the tissue affected). A similar scoring system was instituted for immunohistochemistry positivity in each tissue section as follows: 0 indicates no immunoreactivity; 1 indicates 1 to 25% immunoreactivity; 2 indicates 26 to 50% immunoreactivity; 3 indicates 51 to 75% immunoreactivity; and 4 indicates 76 to 100% immunoreactivity.
Tissue processing and flow cytometry.
To obtain single-cell suspensions, each lymphoid tissue was mechanically forced through a stainless-steel mesh screen using a sterile syringe plunger and collected into a 50-ml conical tube containing RPMI 1640 medium (Gibco). The suspension was then strained through a 70- to 100-μm filter and centrifuged at 800 × g for 5 minutes. Cells were washed in RPMI, treated briefly with ACK lysis buffer (Sigma) to eliminate erythrocytes, and washed twice in RPMI prior to staining.
To determine cell subset frequencies, fresh samples were stained with fluorescent antibodies against the following markers: monocytes (CD3, CD20, CD14, HLA-DR, CD16, CD11c, and CD123), dendritic cells (CD3, CD20, CD14, HLA-DR, CD16, CD11c, CD123, and CD80), NK cells (CD3, CD20, CD8a, CD159a, granzyme B, and CD16), T cells (CD4, CD8b, CD28, CD95, CCR7, and Ki67), and B cells (CD20, IgD, CD27, and Ki67). Surface and intracellular staining was accomplished using the manufacturer recommendations as described previously (20). For panels containing only surface markers (monocytes and DCs), cells were fixed in a 4% paraformaldehyde solution (fixation buffer; BioLegend, San Diego, CA) following staining and resuspended in bovine serum albumin (BSA) staining buffer (Becton, Dickinson Biosciences, San Jose, CA). For panels requiring intracellular stains, cells were fixed and permeabilized using a CytoFix/CytoPerm kit (BD Biosciences, San Jose, CA) before addition of granzyme B (NK cell panel) or Ki67 (T and B cell panels) antibodies. Samples were analyzed with a BD FACS Canto-II flow cytometer and BD FACS Diva software (Becton, Dickinson Biosciences, San Jose, CA). Live cells were identified by forward scatter and side scatter properties. A minimum of 10,000 and an average of 300,000 events were collected for each sample. Data were analyzed using FlowJo version 10 (FlowJo LLC, Ashland, OR) and Prism version 8 software (GraphPad, San Diego, CA). Gating strategies and antibody information were previously described (20) (Table 3).
TABLE 3.
Antibodies used for flow cytometric analysis of immune cell subsetsa
| Flow panels | |||
|---|---|---|---|
| Antibody | Cloneb | Companyb | Catalog no.b |
| Monocyte/DC basic | |||
| Mouse anti-human CD3 FITC | SP34-2 | BD | 556611 |
| Mouse anti-human CD20 FITC | 2H7 | BD | 556632 |
| Mouse anti-human CD16 PE | 3G8 | BioLegend | 302008 |
| Mouse anti-human CD11c PE-Cy7 | 3.9 | BioLegend | 301608 |
| Mouse anti-human CD14 APC | M5E2 | BioLegend | 301808 |
| Mouse anti-human CD123 PerCP-Cy5.5 | 6H6 | BioLegend | 306016 |
| Mouse anti-human HLA-DR APC-Cy7 | L243 | BioLegend | 307618 |
| NK | |||
| Mouse anti-human CD3 FITC | – | – | – |
| Mouse anti-human CD20 FITC | – | – | – |
| Mouse anti-human CD14 FITC | M5E2 | BioLegend | 301804 |
| Mouse anti-human CD56 PE-Cy7 | B159 | BD | 557747 |
| Mouse anti-human granzyme B Alex647* | GB11 | BioLegend | 515405 |
| Mouse anti-human CD159a PE | Z199 | Beckman Coulter | IM3291U |
| Mouse anti-human CD8a APC-Cy7 | RPA-T8 | BioLegend | 301016 |
| Mouse anti-human CD16 PerCP-Cy5.5 | 3G8 | BioLegend | 302028 |
| DC Activation | |||
| Mouse anti-human CD3 FITC | – | – | – |
| Mouse anti-human CD20 FITC | – | – | – |
| Mouse anti-human CD14 FITC | M5E2 | BioLegend | 301804 |
| Mouse anti-human HLA-DR APC-Cy7 | – | – | – |
| Mouse anti-human CD11c PE-Cy7 | – | – | – |
| Mouse anti-human CD123 PerCpCy5.5 | – | – | – |
| Mouse anti-human CD1c APC | AD5-8E7 | Miltenyi | 130-090-903 |
| Mouse anti-human CD86 PE | IT2.2 | BioLegend | 305406 |
| T cells | |||
| Mouse anti-human CD4 PerCp-Cy5.5 | OKT4 | Tonbo | 65-0048 |
| Mouse anti-human CD8b PE | 2ST8.5H7 | BD | 641057 |
| Mouse anti-human CD28 PE-Cy7 | CD28.2 | Tonbo | 60-0289 |
| Mouse anti-human CD95 APC | DX2 | BD | 558814 |
| Mouse anti-human CCR7 APC-Cy7 | G043H7 | BioLegend | 353212 |
| Mouse anti-human Ki67 FITC* | B56 | BD | 556026 |
| B cells | |||
| Mouse anti-human CD20 PE-Cy7 | 2H7 | eBioscience | 25-0209-73 |
| Mouse anti-human CD27 APC | O323 | Tonbo | 20-0279 |
| Mouse anti-human IgD PE | IA6-2 | Biolegend | 348204 |
| Mouse anti-human CD40 APC-Cy7 | 5C3 | BioLegend | 334324 |
| Mouse anti-human Ki67 FITC* | – | – | – |
| Mouse anti-human IgG PerCP-Cy5.5 | HP6017 | BioLegend | 409312 |
*Denotes intracellular antibody.
–, Same as monocyte/DC basic panel.
RNA extraction and library preparation.
Two to five whole lymph nodes and multiple pieces of each of spleen, liver, and adrenal gland from each animal were collected and homogenized with mesh filters. RNA was isolated using TRIzol reagent (spleen and lymph nodes) or RLT buffer (liver and adrenal glands) and Direct-zol RNA mini-prep kits (Zymo) and Qiagen RNeasy minikits (Qiagen), respectively. RNA quality and concentration were assessed using an Agilent 2100 bioanalyzer. A total of 100 ng of each sample was used for library preparation. Total RNA samples were treated with 1 U of DNase I (New England BioLabs) at 37°C for 10 min and cleaned with RNAClean XP beads (Beckman Coulter). Next, ribosomal RNAs were depleted using rRNA removal beads (Illumina). The remaining RNA was fragmented at 94°C for 8 min to yield a median fragment size of 155 nucleotides (nt) and a final library of 309 nt. Libraries were prepared using the TruSeq stranded total RNA library prep kit according to the instructions provided by the manufacturer. Final library quality was confirmed with an Agilent high-sensitivity DNA kit. Libraries with unique barcoded adaptors were pooled and sequenced in a single-end read format with adaptors trimmed by default. The adrenal and liver samples were sequenced on the Illumina HiSeq 2500 platform (100-bp reads), whereas the lymphoid tissues were sequenced on the NextSeq 500 platform (86-bp reads).
RNA sequencing bioinformatics analysis.
Data analysis was performed using the RNA-Seq workflow module of the systemPipeR package available on Bioconductor (52) as previously described (20). RNA-Seq reads were demultiplexed, quality filtered, and trimmed using Trim Galore with an average phred score cutoff of 30 and minimum length of 65 bp. Quality reports were generated with the FastQC function. Trimmed reads were mapped to the Macaca fascicularis reference genome (Macaca_fascicularis.Macaca_fascicularis_5.0.dna.toplevel.fa) using HISAT2 and the corresponding gene annotation (Macaca_fascicularis.Macaca_fascicularis_5.0.94.gtf) from Ensembl. Uniquely mapped reads that overlap exonic regions of genes were counted using summarizeOverlaps in strand-specific mode. Normalization and statistical validation of differentially expressed genes (DEGs) was performed using the EdgeR package (53). Disease-free historical naive tissues were used as the reference. Only DEGs with at least a 2-fold change in expression over naive tissue (n = 4 cynomolgus macaques) and a multiple hypothesis Benjamini-Hochberg false discovery rate (FDR) corrected P value less than 0.05 were included. Since the tissues were not perfused at the time of necropsy, we combined the DEGs detected at 2 DPI (n = 4), 4 DPI (n = 2), and 6 DPI (n = 4) and compared them to the concatenated DEGs detected at DPI 2, 4, and 6 in whole blood (WB) from the same animals as previously reported (8). This comparison allowed us to identify tissue-specific DEGs.
Short Time-series Expression Minor (STEM) software (24) was used in tissues that exhibited temporal changes in gene expression. DEGs were clustered into distinct and significant temporal expression clusters using default settings (minimum correlation of gene clustering of 0.7 with the significance level reported as a Bonferroni-corrected P value less than 0.05).
Publicly available RNA-Seq data obtained from CD14+ monocytes isolated from Makona-infected cynomolgus macaques (8) and human monocyte-derived dendritic cells infected in vitro with EBOV (39) were imported and quality filtered and trimmed using Trim Galore with an average phred score cutoff of 30. CD14+ monocyte sequences were mapped to the Macaca fascicularis genome using HISAT2, and DEGs were determined using EdgeR as described above. Human monocyte-derived dendritic cells were aligned to the human genome (Homo_sapiens.GRCh38.p10.dna.primary_assembly.fa) using HISAT2 and the corresponding gene annotation (Homo_sapiens.GRCh38.91.gtf) from Ensembl. DEGs were determined using EdgeR’s pairwise function. A list of DEGs obtained from THP1 cells infected in vitro with EBOV (38) was filtered to include only protein-coding DEGs with at least a 2-fold change in expression, a multiple hypothesis Benjamini-Hochberg false discovery rate (FDR) corrected P value less than 0.05, and an average of at least 1 read per kilobase of transcript per million mapped reads (RPKM).
Functional enrichment and data visualization.
DEGs were first mapped to human homologs using BioMart (Ensemble Gene 94). Only protein-coding genes with human homologs and an average of at least 1 read per kilobase of transcript per million mapped reads (RPKM) were included for further analysis. The functional enrichment of tissue-specific DEGs was assessed using Metascape (23) and visualized using Cytoscape. Functional enrichment of gene clusters identified using STEM were enriched using MetaCore (Thomson Reuters, New York, NY) and Ingenuity Pathway Analysis. Significant functional enrichment gene ontology (GO) terms were defined as those with an FDR-corrected P value of less than 0.05.
Principal Component Analysis (PCA), heatmaps, and Venn diagrams were generated using the following R packages: DESeq2, VennDiagram, and ggplot2. Heatmaps are scaled and represent normalized expression values (RPKM) of the gene set.
To infer immune-cell-type frequencies in tissues based on RNA-Seq-derived transcriptional signatures, we used the ImmQuant and DMAP/IRIS databases (33, 34).
Statistical analysis.
Statistical analysis of the viral loads and ImmQuant data was carried out using Prism version 8 (GraphPad, San Diego, CA). For the viral load and flow cytometry results, significance was determined using a one-way ANOVA with a Dunnett’s multiple-comparison test. Non-normally distributed data were analyzed using a Kruskal Wallis and a Dunn’s multiple-comparison test. ImmQuant data significance was determined using a two-tailed unpaired t test with Welch’s correction.
Data availability.
The RNA sequencing data presented in this article are available in the National Center for Biotechnology Information Sequence Read Archive (accession no. PRJNA606902).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, and NIH grant U19A109945. Preparation of the Ebola virus seed stock was supported by NIAID/NIH grant U19AI109711. Operational support of the Galveston National Laboratory was provided by NIAID/NIH grant UC7AI094660.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Larsen T, Stevens EL, Davis KJ, Geisbert JB, Daddario-DiCaprio KM, Jahrling PB, Hensley LE, Geisbert TW. 2007. Pathologic findings associated with delayed death in nonhuman primates experimentally infected with Zaire Ebola virus. J Infect Dis 196:S323–S328. doi: 10.1086/520589. [DOI] [PubMed] [Google Scholar]
- 2.Wauquier N, Becquart P, Padilla C, Baize S, Leroy EM. 2010. Human fatal Zaire Ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl Trop Dis 4:e837. doi: 10.1371/journal.pntd.0000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Messaoudi I, Amarasinghe GK, Basler CF. 2015. Filovirus pathogenesis and immune evasion: insights from Ebola virus and Marburg virus. Nat Rev Microbiol 13:663–676. doi: 10.1038/nrmicro3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldmann H, Geisbert TW. 2011. Ebola haemorrhagic fever. Lancet 377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. 2016. Ebola situation report - 30 March 2016. http://apps.who.int/ebola/current-situation/ebola-situation-report-30-march-2016.
- 6.WHO. 2020. Ebola virus disease, Democratic Republic of the Congo, external situation report 78. https://www.who.int/publications-detail/ebola-virus-disease-democratic-republic-of-congo-external-situation-report-78-2019.
- 7.Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, Davis KJ. 2003. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol 163:2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menicucci AR, Versteeg K, Woolsey C, Mire CE, Geisbert JB, Cross RW, Agans KN, Jankeel A, Geisbert TW, Messaoudi I. 2017. Transcriptome analysis of circulating immune cell subsets highlight the role of monocytes in Zaire Ebola virus Makona pathogenesis. Front Immunol 8:1372–1372. doi: 10.3389/fimmu.2017.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Twenhafel NA, Mattix ME, Johnson JC, Robinson CG, Pratt WD, Cashman KA, Wahl-Jensen V, Terry C, Olinger GG, Hensley LE, Honko AN. 2013. Pathology of experimental aerosol Zaire ebolavirus infection in rhesus macaques. Vet Pathol 50:514–529. doi: 10.1177/0300985812469636. [DOI] [PubMed] [Google Scholar]
- 10.Geisbert TW, Young HA, Jahrling PB, Davis KJ, Larsen T, Kagan E, Hensley LE. 2003. Pathogenesis of Ebola hemorrhagic fever in primate models: evidence that hemorrhage is not a direct effect of virus-induced cytolysis of endothelial cells. Am J Pathol 163:2371–2382. doi: 10.1016/S0002-9440(10)63592-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaax NK, Davis KJ, Geisbert TJ, Vogel P, Jaax GP, Topper M, Jahrling PB. 1996. Lethal experimental infection of rhesus monkeys with Ebola-Zaire (Mayinga) virus by the oral and conjunctival route of exposure. Arch Pathol Lab Med 120:140–155. [PubMed] [Google Scholar]
- 12.Zaki SR, Shieh W-J, Greer PW, Goldsmith CS, Ferebee T, Katshitshi J, Tshioko FK, Bwaka MA, Swanepoel R, Calain P, Khan AS, Lloyd E, Rollin PE, Ksiazek TG, Peters CJ, for the Commission de Lutte contre les Epidémies à K. 1999. A novel immunohistochemical assay for the detection of Ebola virus in skin: implications for diagnosis, spread, and surveillance of Ebola hemorrhagic fever. J Infect Dis 179:S36–S47. doi: 10.1086/514319. [DOI] [PubMed] [Google Scholar]
- 13.Marzi A, Feldmann F, Hanley PW, Scott DP, Günther S, Feldmann H. 2015. Delayed disease progression in cynomolgus macaques infected with Ebola virus Makona strain. Emerg Infect Dis 21:1777–1783. doi: 10.3201/eid2110.150259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spengler JR, Saturday G, Lavender KJ, Martellaro C, Keck JG, Nichol ST, Spiropoulou CF, Feldmann H, Prescott J. 2017. Severity of disease in humanized mice infected with Ebola virus or Reston virus is associated with magnitude of early viral replication in liver. J Infect Dis 217:58–63. doi: 10.1093/infdis/jix562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bird BH, Spengler JR, Chakrabarti AK, Khristova ML, Sealy TK, Coleman-McCray JD, Martin BE, Dodd KA, Goldsmith CS, Sanders J, Zaki SR, Nichol ST, Spiropoulou CF. 2016. Humanized mouse model of Ebola virus disease mimics the immune responses in human disease. J Infect Dis 213:703–711. doi: 10.1093/infdis/jiv538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reed C, Alves DA, Fritz EA, Geisbert JB, Hensley LE, Geisbert TW, Larsen T. 2011. Pathogenesis of Marburg hemorrhagic fever in cynomolgus macaques. J Infect Dis 204:S1021–S1031. doi: 10.1093/infdis/jir339. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez AG, Feldmann H. 2006. Filoviridae: Marburg and Ebola viruses, p 1409–1448. In Knipe DM, Howley PM (ed), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 18.Liu X, Speranza E, Muñoz-Fontela C, Haldenby S, Rickett NY, Garcia-Dorival I, Fang Y, Hall Y, Zekeng E-G, Lüdtke A, Xia D, Kerber R, Krumkamp R, Duraffour S, Sissoko D, Kenny J, Rockliffe N, Williamson ED, Laws TR, N’Faly M, Matthews DA, Günther S, Cossins AR, Sprecher A, Connor JH, Carroll MW, Hiscox JA. 2017. Transcriptomic signatures differentiate survival from fatal outcomes in humans infected with Ebola virus. Genome Biol 18:4. doi: 10.1186/s13059-016-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caballero IS, Honko AN, Gire SK, Winnicki SM, Mele M, Gerhardinger C, Lin AE, Rinn JL, Sabeti PC, Hensley LE, Connor JH. 2016. In vivo Ebola virus infection leads to a strong innate response in circulating immune cells. BMC Genomics 17:707. doi: 10.1186/s12864-016-3060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Versteeg K, Menicucci AR, Woolsey C, Mire CE, Geisbert JB, Cross RW, Agans KN, Jeske D, Messaoudi I, Geisbert TW. 2017. Infection with the Makona variant results in a delayed and distinct host immune response compared to previous Ebola virus variants. Sci Rep 7:9730. doi: 10.1038/s41598-017-09963-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14:128–128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A. 2016. Enrichr: a comprehensive gene set enrichment analysis Web server 2016 update. Nucleic Acids Res 44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LCF, Yángüez E, Andenmatten D, Pache L, Manicassamy B, Albrecht RA, Gonzalez MG, Nguyen Q, Brass A, Elledge S, White M, Shapira S, Hacohen N, Karlas A, Meyer TF, Shales M, Gatorano A, Johnson JR, Jang G, Johnson T, Verschueren E, Sanders D, Krogan N, Shaw M, König R, Stertz S, García-Sastre A, Chanda SK. 2015. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ernst J, Bar-Joseph Z. 2006. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA. 2004. Control of pancreas and liver gene expression by HNF transcription factors. Science 303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waxman DJ, Holloway MG. 2009. Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol 76:215–228. doi: 10.1124/mol.109.056705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohkawa H, Miura H, Suzuki M, Nakanishi M, Tomaru Y, Kimura Y, Hayashizaki Y, Ohta Y. 2009. Identification of an inter-transcription factor regulatory network in human hepatoma cells by matrix RNAi. Nucleic Acids Res 37:1049–1060. doi: 10.1093/nar/gkn1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez FJ. 2008. Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab Pharmacokinet 23:2–7. doi: 10.2133/dmpk.23.2. [DOI] [PubMed] [Google Scholar]
- 29.Burdin DV, Kolobov AA, Brocker C, Soshnev AA, Samusik N, Demyanov AV, Brilloff S, Jarzebska N, Martens-Lobenhoffer J, Mieth M, Maas R, Bornstein SR, Bode-Böger SM, Gonzalez F, Weiss N, Rodionov RN. 2016. Diabetes-linked transcription factor HNF4α regulates metabolism of endogenous methylarginines and β-aminoisobutyric acid by controlling expression of alanine-glyoxylate aminotransferase 2. Sci Rep 6:35503. doi: 10.1038/srep35503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reijnen MJ, Sladek FM, Bertina RM, Reitsma PH. 1992. Disruption of a binding site for hepatocyte nuclear factor 4 results in hemophilia B Leyden. Proc Natl Acad Sci U S A 89:6300–6303. doi: 10.1073/pnas.89.14.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guzman-Lepe J, Cervantes-Alvarez E, Collin de l’Hortet A, Wang Y, Mars WM, Oda Y, Bekki Y, Shimokawa M, Wang H, Yoshizumi T, Maehara Y, Bell A, Fox IJ, Takeishi K, Soto-Gutierrez A. 2018. Liver-enriched transcription factor expression relates to chronic hepatic failure in humans. Hepatol Commun 2:582–594. doi: 10.1002/hep4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frishberg A, Brodt A, Steuerman Y, Gat-Viks I. 2016. ImmQuant: a user-friendly tool for inferring immune cell-type composition from gene-expression data. Bioinformatics 32:3842–3843. doi: 10.1093/bioinformatics/btw535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, Frampton GM, Drake ACB, Leskov I, Nilsson B, Preffer F, Dombkowski D, Evans JW, Liefeld T, Smutko JS, Chen J, Friedman N, Young RA, Golub TR, Regev A, Ebert BL. 2011. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, Fong S, van Lookeren Campagne M, Godowski P, Williams PM, Chan AC, Clark HF. 2005. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun 6:319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- 35.Fallon JR, McNally EM. 2018. Non-glycanated biglycan and LTBP4: leveraging the extracellular matrix for Duchenne muscular dystrophy therapeutics. Matrix Biol 68–69:616–627. doi: 10.1016/j.matbio.2018.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kash JC, Muhlberger E, Carter V, Grosch M, Perwitasari O, Proll SC, Thomas MJ, Weber F, Klenk HD, Katze MG. 2006. Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J Virol 80:3009–3020. doi: 10.1128/JVI.80.6.3009-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuzmin IV, Schwarz TM, Ilinykh PA, Jordan I, Ksiazek TG, Sachidanandam R, Basler CF, Bukreyev A. 2017. Innate immune responses of bat and human cells to filoviruses: commonalities and distinctions. J Virol 91:e02471-16. doi: 10.1128/JVI.02471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosworth A, Dowall DS, Armstrong S, Liu X, Dong X, Bruce BC, Ng LFP, Carroll WM, Hewson R, Hiscox AJ. 2019. Investigating the cellular transcriptomic response induced by the Makona variant of Ebola virus in differentiated THP-1 cells. Viruses 11:1023. doi: 10.3390/v11111023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ilinykh PA, Lubaki NM, Widen SG, Renn LA, Theisen TC, Rabin RL, Wood TG, Bukreyev A. 2015. Different temporal effects of Ebola virus VP35 and VP24 proteins on global gene expression in human dendritic cells. J Virol 89:7567–7583. doi: 10.1128/JVI.00924-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasmussen AL, Okumura A, Ferris MT, Green R, Feldmann F, Kelly SM, Scott DP, Safronetz D, Haddock E, LaCasse R, Thomas MJ, Sova P, Carter VS, Weiss JM, Miller DR, Shaw GD, Korth MJ, Heise MT, Baric RS, Pardo-Manuel de Villena F, Feldmann H, Katze MG. 2014. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science 346:987–991. doi: 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baize S, Leroy EM, Mavoungou E, Fisher-Hoch SP. 2000. Apoptosis in fatal Ebola infection. Does the virus toll the bell for immune system? Apoptosis 5:5–7. doi: 10.1023/a:1009657006550. [DOI] [PubMed] [Google Scholar]
- 42.Wong G, Richardson JS, Pillet S, Patel A, Qiu X, Alimonti J, Hogan J, Zhang Y, Takada A, Feldmann H, Kobinger GP. 2012. Immune parameters correlate with protection against Ebola virus infection in rodents and nonhuman primates. Sci Transl Med 4:158ra146. doi: 10.1126/scitranslmed.3004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saphire EO, Schendel SL, Fusco ML, Gangavarapu K, Gunn BM, Wec AZ, Halfmann PJ, Brannan JM, Herbert AS, Qiu X, Wagh K, He S, Giorgi EE, Theiler J, Pommert KBJ, Krause TB, Turner HL, Murin CD, Pallesen J, Davidson E, Ahmed R, Aman MJ, Bukreyev A, Burton DR, Crowe JE Jr, Davis CW, Georgiou G, Krammer F, Kyratsous CA, Lai JR, Nykiforuk C, Pauly MH, Rijal P, Takada A, Townsend AR, Volchkov V, Walker LM, Wang C-I, Zeitlin L, Doranz BJ, Ward AB, Korber B, Kobinger GP, Andersen KG, Kawaoka Y, Alter G, Chandran K, Dye JM, Viral Hemorrhagic Fever Immunotherapeutic C. 2018. Systematic analysis of monoclonal antibodies against Ebola virus GP defines features that contribute to protection. Cell 174:938–952.e13. doi: 10.1016/j.cell.2018.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubins KH, Hensley LE, Wahl-Jensen V, Daddario DiCaprio KM, Young HA, Reed DS, Jahrling PB, Brown PO, Relman DA, Geisbert TW. 2007. The temporal program of peripheral blood gene expression in the response of nonhuman primates to Ebola hemorrhagic fever. Genome Biol 8:R174. doi: 10.1186/gb-2007-8-8-r174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cross RW, Fenton KA, Geisbert JB, Mire CE, Geisbert TW. 2015. Modeling the disease course of Zaire ebolavirus infection in the outbred guinea pig. J Infect Dis 212:S305–S315. doi: 10.1093/infdis/jiv237. [DOI] [PubMed] [Google Scholar]
- 46.Cross RW, Mire CE, Borisevich V, Geisbert JB, Fenton KA, Geisbert TW. 2016. The domestic ferret (Mustela putorius furo) as a lethal infection model for 3 species of Ebolavirus. J Infect Dis 214:565–569. doi: 10.1093/infdis/jiw209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez A, Lukwiya M, Bausch D, Mahanty S, Sanchez AJ, Wagoner KD, Rollin PE. 2004. Analysis of human peripheral blood samples from fatal and nonfatal cases of Ebola (Sudan) hemorrhagic fever: cellular responses, virus load, and nitric oxide levels. J Virol 78:10370–10377. doi: 10.1128/JVI.78.19.10370-10377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo FH, De Raeve HR, Rice TW, Stuehr DJ, Thunnissen FB, Erzurum SC. 1995. Continuous nitric oxide synthesis by inducible nitric oxide synthase in normal human airway epithelium in vivo. Proc Natl Acad Sci U S A 92:7809–7813. doi: 10.1073/pnas.92.17.7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snyder CM, Shroff EH, Liu J, Chandel NS. 2009. Nitric oxide induces cell death by regulating anti-apoptotic BCL-2 family members. PLoS One 4:e7059. doi: 10.1371/journal.pone.0007059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McElroy AK, Shrivastava-Ranjan P, Harmon JR, Martines RB, Silva-Flannery L, Flietstra TD, Kraft CS, Mehta AK, Lyon GM, Varkey JB, Ribner BS, Nichol ST, Zaki SR, Spiropoulou CF. 2019. Macrophage activation marker soluble CD163 associated with fatal and severe Ebola virus disease in humans. Emerg Infect Dis 25:290–298. doi: 10.3201/eid2502.181326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thi EP, Mire CE, Lee ACH, Geisbert JB, Zhou JZ, Agans KN, Snead NM, Deer DJ, Barnard TR, Fenton KA, MacLachlan I, Geisbert TW. 2015. Lipid nanoparticle siRNA treatment of Ebola-virus-Makona-infected nonhuman primates. Nature 521:362–365. doi: 10.1038/nature14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Backman TWH, Girke T. 2016. systemPipeR: NGS workflow and report generation environment. BMC Bioinformatics 17:388. doi: 10.1186/s12859-016-1241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data presented in this article are available in the National Center for Biotechnology Information Sequence Read Archive (accession no. PRJNA606902).