Epstein-Barr virus (EBV) has a well-described tropism for B cells and epithelial cells. Recently, we described the ability of a second strain of EBV, EBV type 2, to infect mature peripheral T cells. Using a neutralizing antibody assay, we determined that EBV uses the viral glycoprotein gp350 and the cellular protein CD21 to gain entry into mature peripheral T cells. CRISPR-Cas9 deletion of CD21 on the Jurkat T-cell line confirmed that CD21 is required for EBV infection. This study has broad implications, as we have defined a function for CD21 on mature peripheral T cells, i.e., as a receptor for EBV. In addition, the requirement for gp350 for T-cell entry has implications for EBV vaccine studies currently targeting the gp350 glycoprotein to prevent EBV-associated diseases.
KEYWORDS: CD21, EBV, T cells
ABSTRACT
Epstein-Barr virus (EBV) is associated with a number of T-cell diseases, including some peripheral T-cell lymphomas, hemophagocytic lymphohistiocytosis, and chronic active EBV disease. The tropism of EBV for B cells and epithelial cell infection has been well characterized, but infection of T cells has been minimally explored. We have recently shown that the EBV type 2 (EBV-2) strain has the unique ability to infect mature T cells. Utilizing an ex vivo infection model, we sought to understand the viral glycoprotein and cellular receptor required for EBV-2 infection of T cells. Here, using a neutralizing-antibody assay, we found that viral gp350 and complement receptor 2 (CD21) are required for CD3+ T-cell infection. Using the HB5 anti-CD21 antibody clone but not the Bly-4 anti-CD21 antibody clone, we detected expression of CD21 on both CD4+ and CD8+ T cells, with the highest expression on naive CD4 and CD8+ T-cell subsets. Using CRISPR to knock out CD21, we demonstrated that CD21 is necessary for EBV entry into the Jurkat T-cell line. Together, these results indicate that EBV uses the same viral glycoprotein and cellular receptor for both T- and B-cell infection.
IMPORTANCE Epstein-Barr virus (EBV) has a well-described tropism for B cells and epithelial cells. Recently, we described the ability of a second strain of EBV, EBV type 2, to infect mature peripheral T cells. Using a neutralizing antibody assay, we determined that EBV uses the viral glycoprotein gp350 and the cellular protein CD21 to gain entry into mature peripheral T cells. CRISPR-Cas9 deletion of CD21 on the Jurkat T-cell line confirmed that CD21 is required for EBV infection. This study has broad implications, as we have defined a function for CD21 on mature peripheral T cells, i.e., as a receptor for EBV. In addition, the requirement for gp350 for T-cell entry has implications for EBV vaccine studies currently targeting the gp350 glycoprotein to prevent EBV-associated diseases.
INTRODUCTION
Epstein-Barr virus (EBV) is a human gammaherpesvirus with a well-characterized tropism for B cells and epithelial cells. The predilection of EBV to infect B cells and epithelial cells is reflected in the EBV-associated B-cell lymphoproliferative diseases and lymphomas (1–4) and epithelial cell malignancies (5, 6). Less well characterized is the tropism of EBV for T cells. In 1988, EBV was detected in tumor cells in three cases of T-cell lymphoma (7), sparking early interest in a potential role for EBV in T-cell diseases. Subsequent studies have shown that EBV-infected T cells are found in both peripheral T-cell lymphomas and T-cell lymphoproliferative diseases, including hemophagocytic lymphohistiocytosis, hydroa vacciniforme, and chronic active EBV disease (8–12). The presence of EBV in a subset of peripheral T-cell lymphomas has been linked to an unfavorable prognosis, suggesting that the virus contributes to the poor outcome of these malignancies (13). Detection of EBV in T cells of healthy children (14) and in tonsils of infectious mononucleosis patients (15, 16) suggests that infection of T cells is part of EBV’s life cycle.
Understanding T-cell susceptibility to EBV infection and the role of EBV in associated T-cell diseases has been limited due to the lack of an in vitro model to study infection of primary T cells. EBV type 1 (EBV-1), the predominant strain of EBV, was found to infect human thymocytes, with the viral genome being detected through 6 weeks postinfection (17). CD8+ T cells could not be infected with EBV-1 even though viral binding occurred (18). T-cell lines have also been reported to be resistant to EBV infection (19) or susceptible (20, 21), but follow-up studies were not done. Recently, we reported that in contrast to the EBV-1 strain, the less common EBV type 2 strain (EBV-2) can latently infect primary mature CD3+ T cells in vitro with a higher frequency of CD8+ T cell infection than with CD4+ T cells (22, 23). Infection is characterized by proliferation, upregulation of activation markers and inflammatory cytokines, and expression of EBV latent but not lytic genes. EBV-2 can also infect CD3+ T cells in a humanized mouse model, confirming both in vitro and in vivo susceptibility with this EBV strain (24).
The mechanism that allows virus attachment and entry into B cells and epithelial cells has been well characterized. Initial attachment to B cells occurs through the most abundant viral glycoprotein on the surface of the virion, gp350, and its receptors, either CD21 (complement receptor 2 [CR2]) or CD35 (complement receptor 1 [CR1]) on the cell surface (25–30). This initial attachment event induces endocytosis of the virion (31). The next step involves the viral glycoprotein gp42, in a trimeric complex with gH and gL, binding to HLA class II (32–34). This allows fusion with the endocytic membrane by the EBV glycoprotein gB (35, 36). In contrast, neither gp350 or gp42 is required for epithelial cell infection. The initial attachment to epithelial cells is with the dimeric complex of gH and gL, with gH binding to αvβ5, αvβ6, or αvβ8 integrin (37, 38) or (as was more recently reported) ephrin receptor A2 (39, 40). This induces fusion directly at the plasma membrane with gB, which has been shown to bind neuropilin-1 (41). In this study, we asked what viral glycoproteins and cellular receptors are required for T-cell infection.
RESULTS
EBV infection of CD3+ T cells is neutralized by antibodies against viral gp350 and cellular CD21.
Two viral neutralization assays have been used to identify the viral glycoproteins and cellular receptors used for EBV entry into B cells. The first is a cord blood transformation assay based on EBV’s ability to immortalize B cells (42). An alternative assay was developed that relies on the insertion of the gene for green fluorescent protein (GFP) into the EBV-1 genome and infection of the Raji B-cell line assessed by flow cytometry (43). Because EBV-2 infection of T cells does not result in cell immortalization and there is no recombinant EBV-2 expressing GFP, a quantitative PCR (qPCR)-based neutralization assay developed for B-cell infection (44) was adapted for evaluation of T-cell infection.
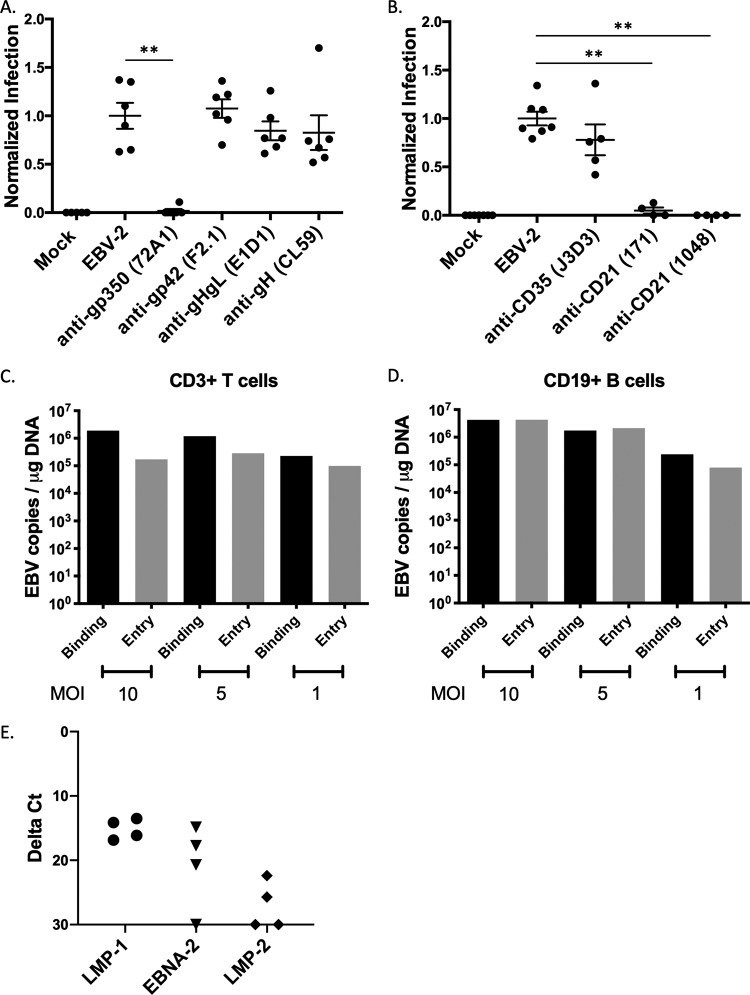
EBV-2 was incubated with monoclonal antibodies against gp350 (clone 72A1), gp42 (clone F2.1), gHgL (clone E1D1), or gH (clone CL59). These antibodies were previously shown to block infection of B cells (clones 72A1 and F2.1) and epithelial cells (clones E1D1 and CL59) at the concentrations used in our study (45–48). Following a 2-h infection, the cells were washed and cultured for 3 days. DNA was extracted, and the viral load was analyzed by a qPCR assay. At 3 days postinfection (dpi), only the anti-gp350 antibody (P = 0.0022) neutralized T-cell infection (Fig. 1A), suggesting that the viral gp350, not gp42, gHgL, or gH, is utilized for entry into CD3+ T cells.
FIG 1.
Neutralization assay to assess viral glycoproteins and cellular receptors for EBV-2 infection of T cells. CD3+ T cells were infected with EBV incubated with antibodies to either EBV glycoproteins (A) or complement receptors (B). (A) Neutralization assay after incubation of virus with 10 μg monoclonal antibodies against EBV glycoproteins. Mock, uninfected cells; EBV-2, no antibody; anti-gp350, clone 72A1; anti-gp42, clone F2.1; anti-gHgL, clone E1D1; anti-gH, clone CL59. Viral loads were normalized to the uninhibited EBV-2 infection for each experiment and are presented as normalized infection. 72A1, P = 0.0022; F2.1, P = 0.6991; E1D1, P = 0.4610; CL59, P = 0.3939). (B) Neutralization assay with incubation of purified T cells with 20 μg monoclonal antibodies against CD35 (clone J3D3) or CD21 (clones 171 and 1048). Viral loads were normalized to infection with uninhibited EBV-2 infection (J3D3, P = 0.1174; 171, P = 0.0061; 1048, P = 0.0061). Infections were performed with 5 EBV genome copies/cell (n = 4 to 6). (C) CD3+ T cells were incubated with EBV-2 at different MOI for 1 h at 4°C (binding) or for 1 h at 4°C with a subsequent shift to 37°C (entry). Cells were analyzed for viral load by qPCR at 24 hpi. Data are from replicate wells. (D) CD19+ B cells were incubated with EBV-2 at different MOI for 1 h at 4°C (binding) or for 1 h at 4°C with a subsequent shift to 37°C (entry). Cells were analyzed for viral load by qPCR at 24 hpi. Data are from replicate wells. (E) CD3+ T cells were infected with EBV-2, and viral gene expression was analyzed by RT-PCR at 24 hpi. Data shown are from four replicates.
The EBV gp350 has two known cellular receptors, the well-characterized CD21 (also known as complement receptor 2 [CR2]) and the more recently described CD35 (CR1) (25, 26, 28–30). Therefore, we used the 3-day neutralization assay to determine the requirement for these receptors in EBV-2 entry into T cells. We incubated the purified CD3+ T cells with blocking antibodies against CD35 (clone J3D3) or CD21 (clone 171 or clone 1048) (49, 50) and observed that both antibodies against CD21 significantly neutralized EBV-2 infection of T cells (clone 171, P = 0.0061; clone 1048, P = 0.0061) (Fig. 1B). Incubation with anti-CD35 antibody prior to T-cell infection reduced the EBV load, but this reduction was not significant. Data from these assays suggest that, like B cells, the viral gp350 interaction with CD21 was important for T-cell infection by EBV-2.
In the qPCR-based neutralization assay, DNA from EBV-infected CD3+ T cells was evaluated for viral load at 3 dpi. However, it is possible that at 3 dpi, we could have been measuring only bound virus and not internalized viral genomes. To test this possibility, CD3+ T cells were incubated with EBV-2 at three different multiplicities of infection (MOI; 10, 5, or 1) either at 4°C for 3 h to measure viral binding or at 4°C incubation for 3 h followed by a temperature shift to 37°C for 1 h to measure viral entry. DNA was extracted and analyzed by qPCR for EBV DNA load to quantify viral genome copy number. At all MOI tested, EBV genomes were detected in the entry assay (Fig. 1C), even when cells were infected at the low MOI of 1 genome copy per cell. Consistent with a model where more virions bind to the cell than gain entry into the cell, the number of EBV copies per microgram of total DNA was higher in the binding assay than in the entry assay at all MOI tested.
We next compared the ability of EBV-2 to bind to and enter CD19+ cells using the same assay. As shown in Fig. 1D, EBV-2 was more efficient at entry into B cells at the higher MOI (10 and 5 genome copies/cell) than at entry into T cells. However, at lower MOI (1 genome copy/cell), numbers of EBV-2 genome copies were comparable when entry into T and B cells was measured. As a further demonstration of viral entry into CD3+ T cells, we next measured the EBV latent gene transcripts, LMP-1, LMP-2, and EBNA-2, at 24 hpi by a probe-based reverse transcription-PCR (RT-PCR). To determine the relative levels of transcripts, the cycle threshold (CT) of each transcript was normalized to the CT of the housekeeping gene β2 microglobulin. Consistent with our previously published data (22), we were able to detect the EBV latent gene expression in EBV-2 infected T cells. These data suggest that the 3-day neutralization assay detected internalized genomes by 3 days postinfection.
CD21 is expressed on mature peripheral T-cell subsets.
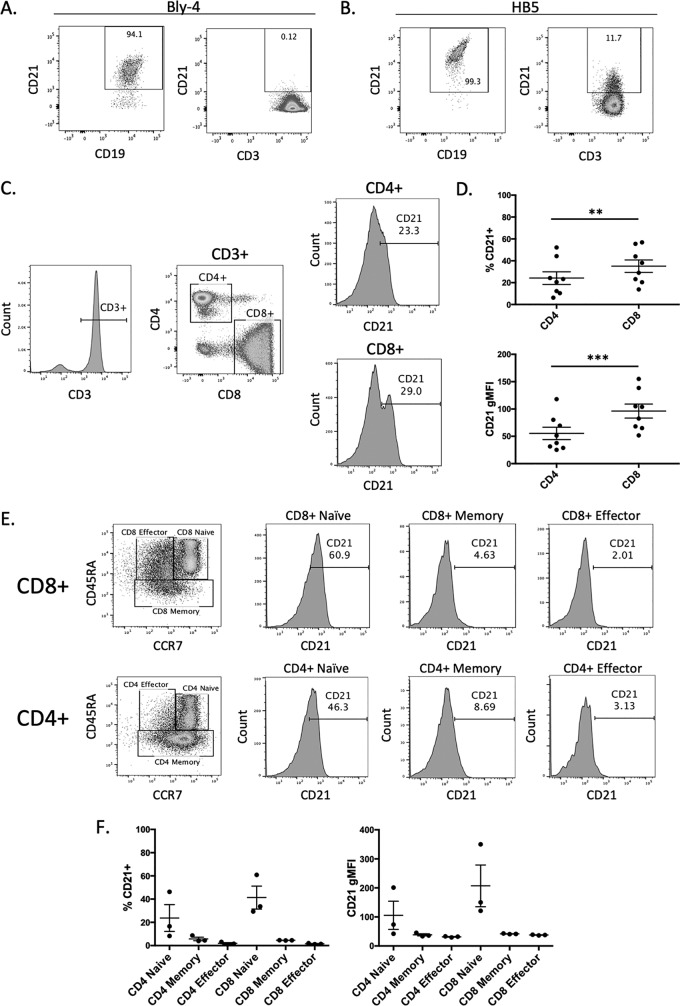
The observation that blocking CD21 could prevent EBV-2 infection of T cells was a paradox, as the expression of CD21 on mature T cells is controversial: some studies detected CD21 expression (51–53), while others did not detect any CD21 (54). Consistent with the latter observation, we tested whether an anti-CD21 monoclonal antibody (clone Bly4) could detect CD21 on CD19+ B cells and CD3+ T cells. When this antibody was used, the majority of peripheral CD19+ B cells expressed CD21; however, no expression of CD21 was detected on peripheral CD3+ T cells (Fig. 2A). Interestingly, studies that described CD21 expression on mature, peripheral T cells (51) all used the anti-CD21 monoclonal antibody clone HB5 (52, 53). When this antibody was used to evaluate CD21 expression by flow cytometry, we were able to detect expression of CD21 on a subset of peripheral CD3+ T cells, as well as CD19+ B cells (Fig. 2B), although the percentage of cells expressing CD21 was low in T cells (mean, 34.4% CD21+) compared to the nearly ubiquitous expression on B cells.
FIG 2.
Expression of CD21 on peripheral T cell subsets. Representative flow cytometry plot of PBMCs labeled with FITC-conjugated anti-CD19 monoclonal antibody (MAb) or FITC-conjugated anti-CD3 MAb and either anti-CD21 antibody clone Bly4 (A) or clone HB5 (B) conjugated with APC or PE, respectively. CD21 expression of CD19+ B cells (left) and CD3+ T cells (right) was determined. Gates were set based on an unstained control. (C) Representative gating strategy for CD21 analysis of CD8+ and CD4+ T cells. (D) Comparison of the percent CD21-positive cells in the total CD4+ and CD8+ T-cell populations (P = 0.0012) and gMFI of CD21 expression in CD4+ and CD8+ T cells (P = 0.0005) (n = 8). (E) Representative gating strategy for CD21 expression analysis of CD4+ and CD8+ T-cell subsets. (F) Comparison of the percent CD21-positive cells and gMFI in CD4+ and CD8+ T-cell subsets. The data are from 3 donors.
Given the detection of CD21 on mature peripheral CD3+ T cells, we next evaluated CD21 expression on peripheral T-cell subsets using the HB5 anti-CD21 antibody and flow-cytofluorimetric analysis. CD21 was detected on both CD4+ and CD8+ T cells (Fig. 2C), with a higher level of CD21 detected on CD8+ T cells than CD4+ T cells, in terms of both the percentage of cells that were CD21+ (P = 0.0012) and the higher geometric mean fluorescent intensity (gMFI) of the CD21+ cells (P = 0.0005) (Fig. 3D and E). This is consistent with a higher level of EBV-2 infection in the CD8+ T cells than CD4+ T cells, which we previously reported (22). To further evaluate CD21 on T-cell subsets, we stained peripheral blood mononuclear cells (PBMCs) with fluorescently conjugated anti-CCR7 and anti-CD45RA antibodies to distinguish the naive (CD45RA+ CCR7+), memory (CD45RA− CCR7+/−), and effector (CD45RA+ CCR7−) T-cell subsets. CD21 was expressed on naive, memory, and effector CD4+ and CD8+ T cells (Fig. 2E), with a higher percentage of CD21+ cells and a higher gMFI of CD21 expression in both the CD4+ and CD8+ naive T-cell populations than in the effector or memory T-cell subsets (Fig. 2F).
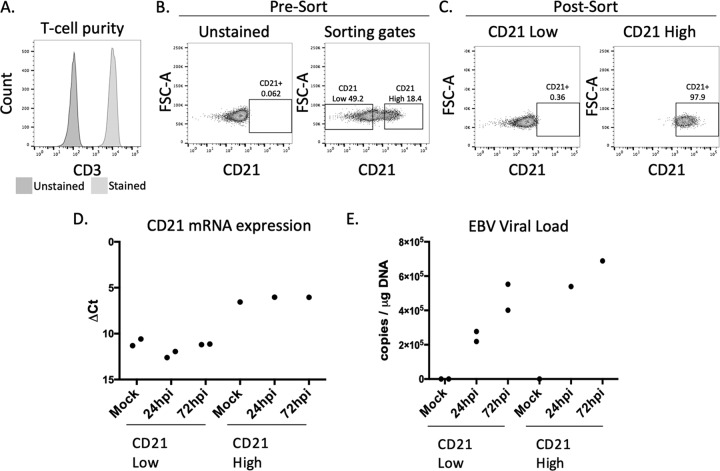
FIG 3.
EBV-2 infection of CD21hi and CD21lo T cells. (A) T-cell purity analysis of T cells for CD21 sorting. (B). Sorting gates used for CD21 T-cell sorting. Left, unstained control; right, gating strategy. (C) Postsorting analysis of cell populations analyzing CD21 expression. (D) CD21 gene expression analysis postinfection. Mock, uninfected cells. (E) EBV viral load at 24 and 72 hpi in sorted CD21lo and CD21hi populations. Infections were done at 5 EBV genome copies/cell.
CD21 is required for EBV infection of T cells.
To assess the requirement of CD21 for EBV infection of mature T cells, CD3+ T cells were negatively selected, confirmed for high purity, and then sorted based on CD21 expression (Fig. 3A and B). Following cell sorting, the CD21lo population showed no detectable expression of CD21 and the CD21hi population was 97.9% CD21 positive as measured by flow cytometry (Fig. 3C). However, there was still detectable CD21 mRNA in the CD21lo population (Fig. 3D), although CD21 mRNA was 21-fold higher in the CD21hi population. To determine if there was a differential susceptibility in the CD21lo and CD21hi cells to EBV-2 infection, sorted cell subsets were infected at 5 EBV genome copies/cell. DNA was extracted at 1 and 3 dpi, and qPCR was performed to measure EBV DNA (Fig. 3E). While we still detected the EBV-2 genome in both the CD21lo and CD21hi populations, the amount of viral genome detected at both 1 and 3 dpi was higher in the CD21hi cells, suggesting that the level of CD21 on the surface of the mature CD3+ T cells was critical to a robust infection.
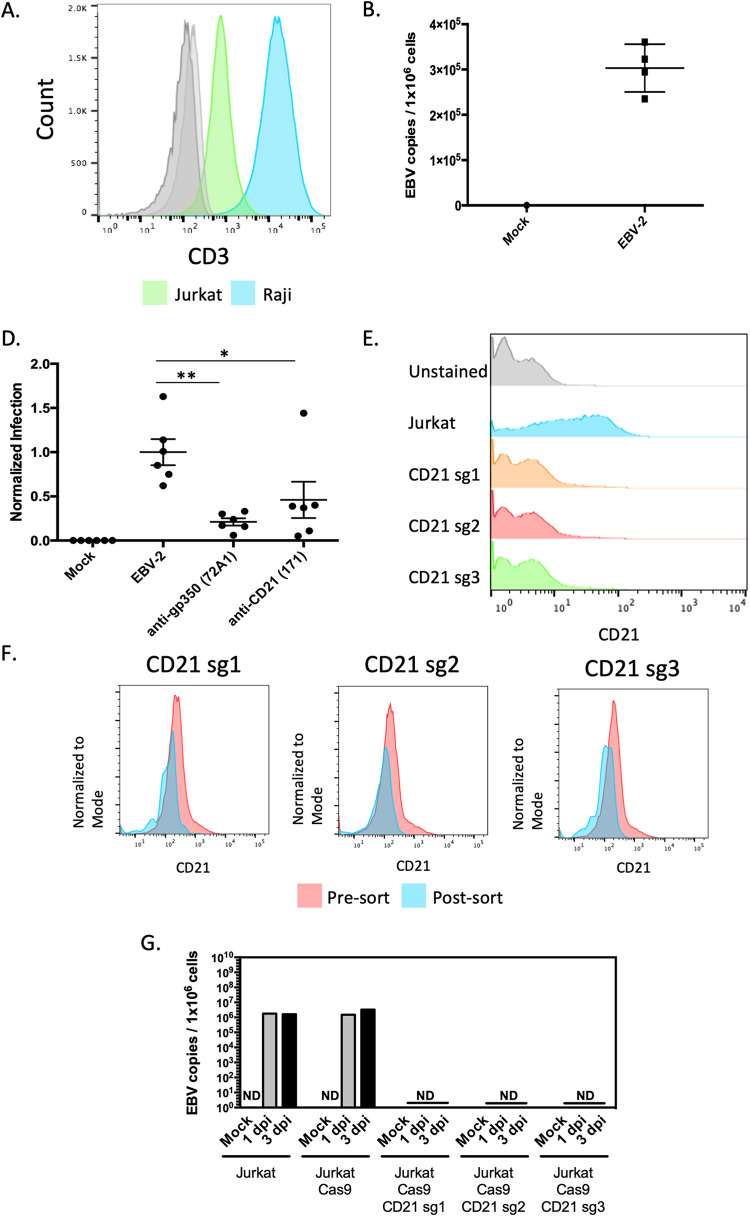
To test our hypothesis that CD21 was required for EBV-2 infection of T cells, we needed a more manipulatable model system. Consistent with earlier reports, we confirmed the expression of CD21 on the Jurkat T cell line (53, 55) using the anti-CD21 antibody (clone HB5) (Fig. 4A). Similar to what was observed in primary cells, CD21 expression was lower on the Jurkat T cell line than on a representative B-cell line, Raji (Fig. 4A). To evaluate the susceptibility of Jurkat cell line to EBV infection, Jurkat cells were infected with EBV-2 at an MOI of 5 genome copies/cell. The EBV-2 genome was detected in cell extracts at 24 hpi (Fig. 4B). To test the requirement for gp350 and CD21 for viral entry, we utilized the in vitro neutralization assay and quantified viral genome copies following infection of Jurkat cells with virus incubated with the anti-gp350 neutralizing antibody or with cells incubated with a CD21-blocking antibody (Fig. 4C). Both incubation with anti-gp350 antibodies or anti-CD21 antibodies significantly reduced the amount of EBV genomes detected in the Jurkat cells, suggesting that, similar to infection of peripheral CD3+ T cells, infection of the Jurkat T-cell line was mediated by binding of the viral gp350 to the cellular receptor CD21.
FIG 4.
Requirement for CD21 for EBV-2 infection of Jurkat T cells. (A) CD21 (clone HB5) expression on unstained Jurkat (light gray), unstained Raji (dark gray), Jurkat (green), and Raji (blue) cells. (B) Viral load 24 h following infection of Jurkat cells with EBV-2 virus (5 genome copies/cell). Uninfected cells (mock) were a negative control. Data are presented as log(copies/microgram). (C) Neutralization assay using Jurkat cells and infecting with EBV-2. Viral loads were normalized to the uninhibited EBV-2 infection of Jurkat cells. Virus was incubated with 10 μg of anti-gp350 antibody (clone 72A1) (P = 0.0022), or the cells were incubated with 10 μg of anti-CD21 antibody (clone 171) (P = 0.0411) (5 genome copies/cell). (D) CD21 expression of Jurkat CAS9 cells and CD21 knockout cells. (E) Pre- and postsorting analysis of CD21 knockout cells. (F) EBV viral load at 1 dpi and 3 dpi following infection of Jurkat, Jurkat Cas9, and the three CD21 knockout cell lines with EBV-2 virus (10 genome copies/cell). ND, not detected.
To conclusively determine if CD21 is required for EBV infection of T cells, we used CRISPR-Cas9 to knock out CD21 expression in Jurkat cells. To ensure that the effects were not due to off-target activity, we created CD21 knockout cell pools with three distinct single guide RNAs (sgRNAs) targeting the CD21 gene. Furthermore, to ensure the purity of the CD21 knockout cells, the CD21-negative cells were purified by fluorescence-activated cell sorting (Fig. 4E). Jurkat CD21− and Jurkat Cas9 cells were then infected with EBV-2 (10 genome copies/cell) for 1 or 3 days. DNA was extracted, and viral genomes were quantified by qPCR. Viral genomes were detected in Jurkat and Jurkat Cas9 cells but not in cells in which CD21 was knocked out (Fig. 4F).
EBV-2 gp350 differs in the CD21 binding domain.
Our data indicated that EBV-2, unlike EBV-1, has the unique ability to infect primary T cells in vitro and in vivo (14, 22). An unanswered question is why EBV-2 readily infects mature peripheral T cells while EBV-1 cannot. Our initial analysis and published phylogenetic analysis of 225 EBV strains (56) showed that EBV-1 and EBV-2 were distinct in the gp350 gene. Upon analysis of the amino acid differences between two common EBV isolates, the EBV-1 Akata strain and the EBV-2 Jijoye strain, we observed 12 amino acid changes within the first 504 N-terminal amino acids of the gp350 gene (Fig. 5). Three of these changes fell within one of the known CD21 binding regions for gp350 (57). Notably, the second binding region, also the epitope for the anti-gp350 neutralizing antibody 72A1 used in this and other studies (58), was not altered.
FIG 5.
Comparison of gp350 amino acid sequences. The amino acid sequence (amino acids 1 to 504) of the gp350 protein of the EBV-1 strain Akata is shown. The known CD21 binding regions are underlined. The amino acid differences in gp350 of the EBV-2 strain Jijoye are in bold above the EBV-1 sequence.
DISCUSSION
These data identified CD21 as a cellular receptor for EBV-2 entry into mature CD3+ T cells. The demonstration that T-cell infection is blocked following incubation of the virus with a gp350-neutralizing antibody is also consistent with this result. Together, these data suggest that EBV uses the same glycoprotein and cellular receptor for entry into B cells and T cells.
While several older studies reported expression of CD21 on peripheral T cells (52, 53), others have reported that CD21 is not expressed on peripheral T cells (54). Our data suggest that the discrepancies in these findings could be due to the variability in commercial antibody binding to CD21 on CD19+ B cells compared to CD3+ T cells. The anti-CD21 HB5 antibody has been mapped to short consensus repeats 3 and 4 of CD21, but the Bly4 antibody has yet to be epitope mapped (59), so it is unknown why Bly4 detects CD21 only on B cells. It is possible that the HB5 antibody was generated with pure CD21 antigen while the Bly4 clone was selected for CD21 from B cells. On B cells, CD21 is complexed with CD19, CD81, and Leu-13 or with CD35 (60, 61). But, as CD19 is a B-cell-specific protein, CD21 either is complexed with different cell surface proteins on T cells or is a stand-alone molecule, as suggested in one study (62).
Early studies on the pattern of CD21 expression reported that thymocytes were CD21+ (63) and that anti-CD21 antibody blocked EBV infection of thymocytes (17). More recently, recent thymic immigrants were found to express CD21 (64), which would be consistent with our observations that CD21 was expressed on both naive CD4+ and CD8+ mature peripheral T cells. This suggests a model whereby EBV uses CD21 to infect both immature and mature T cells.
Both strains of EBV encode gp350 and express it on the viral envelope, facilitating attachment to CD21 on B cells. Why only EBV-2 and not EBV-1 is able to readily infect T cells is unknown. We hypothesize that differences in the gp350 protein, specifically in the CD21 binding region, may create a higher-affinity or -avidity interaction between gp350 and CD21, allowing the lower level of CD21 expression on T cells to be sufficient for viral entry of the EBV-2 strain. Consistent with this hypothesis, we identified sequence differences in the CD21 binding regions of the EBV-1 and EBV-2 gp350 genes. It is also possible that differences in the level of CD21 expression on T cells, in combination with sequence differences in the CD21 binding region of gp350, could allow EBV-2 infection of T cells. For example, Levy et al. (53) found that in patients with systemic lupus erythematosus, more than 80% of T cells express CD21. It could also be that binding of the EBV-2 gp350 to CD21 could be greater due to potential structural differences of CD21 on T cells compared to B cells (51).
In sum, our study shows that CD21 is expressed on mature peripheral CD3+ T cells, with a higher level of expression on the naive CD4+ and CD8+ T-cell subsets. These studies point the way to future mechanistic studies of the function of CD21 on T cells and raise the question of whether CD21 expression differs among high-risk populations or in people with EBV-associated T-cell diseases. This could provide insight into the susceptibility of T cells to EBV infection in unique populations and their potential for increased risk of EBV-associated T-cell malignancies. In addition, we show that EBV gains access to T cells and B cells through a common pathway utilizing the viral glycoprotein gp350 and the cellular complement receptor CD21. This suggests that EBV vaccines targeting gp350 will prevent both B- and T-cell infection (65). Further studies are needed to determine if other steps in the viral entry pathway are similar between B cells and T cells. Finally, the availability of a model system to evaluate EBV infection of T cells will be of importance for understanding how EBV drives oncogenesis and aid in the identification of potential targets for therapeutic intervention of EBV-associated T-cell diseases.
MATERIALS AND METHODS
Blood processing and T-cell purification.
After consent had been obtained, peripheral blood was taken from healthy U.S. adults as per a protocol approved by the Institutional Review Board of the University of Colorado Anschutz Medical Campus and according to the Declaration of Helsinki. Ficoll-Paque Plus (GE Healthcare, Little Chalfont, United Kingdom) was layered under peripheral blood to isolate peripheral blood mononuclear cells (PBMCs). CD3+ T cells and CD19+ B cells were isolated by negative enrichment using the human pan-T-cell isolation kit and the pan-B-cell isolation kit, respectively (Miltenyi Biotec, Bergisch Gladbach, Germany). Purity analysis of isolated cell populations was performed following cell isolation via flow cytometry to stain for CD3 and CD19 using anti-CD3+ (BD Biosciences, Franklin Lanes, NJ) or anti-CD19+ (BD Biosciences) monoclonal antibodies and analyzed on a BD LSRFortessa cell analyzer (BD Biosciences). Flow cytometry data were analyzed by FlowJo software (Tree Star, Ashland, OR). The purity of isolated CD3+ T cells and that of CD19+ B cells were greater than 95% and 90%, respectively. Primary cell cultures were grown at 37°C and 5% CO2 in complete RPMI (Fisher Scientific, Hampton, NH) supplemented with 10% fetal bovine serum (Equafetal; Atlas Biological, Fort Collins, CO), 1% l-glutamine (Fisher Scientific), and 1% penicillin and streptomycin (Fisher Scientific).
Cell lines and virus production.
The EBV-2-positive Burkitt’s lymphoma cell line Jijoye (P-2003, P-3J; ATCC, Manassas, VA) and the EBV-negative cell line BL41 (gift of J. Sixbey, St. Jude Children's Research Hospital) were maintained in complete RPMI at 37°C and 5% CO2. The Jurkat T cell line (American Type Culture Collection, Baltimore, MD) was maintained in complete RPMI with 10 mM HEPES. Jijoye cells were used for production of virus, as virus produced from this cell line has been shown to infect T cells in vitro (22). EBV stocks were generated as described before (66). In short, Jijoye cells were treated with sodium butyrate (Sigma-Aldrich, St. Louis, MO) and tetradecanoyl phorbol acetate (Sigma-Aldrich) (4 mM and 25 ng/ml, respectively). Cell debris was removed by centrifugation at 4,000 × g for 10 min at 4°C, and supernatant was passed over a 0.45-μm sterile filter (Fisher Scientific). Viral particles were then pelleted by ultracentrifugation at 16,000 × g for 90 min at 4°C. Virus pellets were resuspended in 1/200 of the original volume in complete RPMI with 100 μg/ml bacitracin (Fisher Scientific). Virus was DNase treated (100 μg/ml) (Sigma-Aldrich), and encapsulated genomes were quantitated by quantitative PCR (qPCR) using primers and probes designed to amplify a 70-bp region of the EBV BALF5 gene and β-actin gene as a control for DNA input, as previously described (67). Infections were done with 5 to 10 DNase-resistant EBV-2 genome copies per cell.
EBV neutralization assay.
A qPCR-based neutralization assay was adapted for T-cell infection from the assay described by Weiss and colleagues for B-cell infection (44). For analysis of neutralization of EBV entry using anti-EBV monoclonal antibodies, 5 × 106 copies of EBV-2 virus were incubated with 10 μg of monoclonal antibody against EBV glycoproteins for 2 h at 37°C in a total of 100 μl complete RPMI. Antibody clones against EBV glycoproteins were targeting gp350 (72A1), gp42 (F2.1), gHgL (E1D1), and gH (CL59) (45–48). These antibodies were received as a gift from Lindsey Hutt-Fletcher (Louisiana State University Health Sciences Center, Shreveport, LA). The antibody-virus mixture was added to 1 × 106 CD3+ cells in a total volume of 200 μl in a 96-well tissue culture plate (Fisher Scientific) and incubated for 2 h at 37°C. For analysis of neutralization using anti-cell surface receptor antibodies, 1 × 106 purified CD3+ T cells were incubated with 20 μg of blocking antibody clones against CD35 (clone J3D3) (Beckman-Coulter) and CD21 (clones 171 and 1048) in 100 μl complete RPMI for 2 h at 37°C (49). CD21 antibodies were received as a gift from Michael Holers (University of Colorado Anschutz Medical Campus, Aurora, CO). Following incubation with antibodies, the virus with or without antibodies was added to CD3+ T cells for 2 h at 37°C. Following infection, the T cells were washed 3 times with phosphate-buffered saline (PBS) (Fisher Scientific) to remove any free virus, plated in 1 ml complete RPMI in a 24-well tissue culture plate (Fisher Scientific), and incubated for 3 days at 37°C with 5% CO2. After 3 days, cells were washed again three times with PBS, pelleted by centrifugation at 200 × g at room temperature, and frozen at −80°C. DNA was extracted from the cells using the Qiagen DNeasy kit according to the manufacturer’s protocol (Qiagen, Hilden, Germany). EBV viral load was determined as previously described by qPCR for the BALF5 gene and β-actin (67). The viral load following infection with virus or cells incubated with each monoclonal antibody was normalized to the viral load following infection with no antibodies. Experimental conditions were performed in triplicate for each donor and repeated using 2 or 3 donors. Normalized infection was calculated by dividing the viral load of an antibody-inhibited sample by the mean viral load of the uninhibited samples for each donor.
Measurement of viral gene expression.
CD3+ T cells were infected with EBV-2 at a multiplicity of infection (MOI) of 5 to 10 genome copies/cell. At 24 h postinfection (hpi), RNA was extracted using the RNeasy kit (Qiagen). Epstein-Barr nuclear antigen 2 (EBNA-2), latent membrane protein 1 (LMP-1), and LMP-2 gene expression was measured using specific primer and probes and normalized to β2 microglobulin expression, as previously described (68). RNA was extracted from infected T cells at 24 hpi. This PCR was performed using the ITaq Universal Probes Supermix and run on the IQ5 real-time PCR platform (Bio-Rad). The PCR protocol was as follows: 95°C for 10 min and 45 cycles of 95°C for 15 s and 60°C for 1 min (68). An EBV-2 lymphoblastoid cell line, LCL-10, was used as a positive control (69). Delta cycle threshold (CT) analysis was used to evaluate EBV transcript expression relative to β2 microglobulin expression by subtracting the CT value of the EBV target gene from the CT value of β2 microglobulin.
Binding and entry assay.
CD3+ T cells or CD19+ B cells isolated by negative enrichment were generated as described above. The virus binding and entry assay was done as described (70, 71). Essentially, for the virus binding assay, purified T or B cells were incubated with various EBV genome copies for 3 h at 4°C. Cells were washed 3 times with cold PBS, and then DNA was extracted using the Qiagen DNeasy kit as per the manufacturer’s protocol for the binding assay (Qiagen). For the entry assay, after the 3-h incubation on ice, cells were washed 3 times with cold PBS and moved to 37°C for 1 h to allow for virus entry. After 1 h at 37°C, cells were washed again and incubated with 1 mg/ml proteinase K for 1 h on ice. Cells were then washed 3 times with PBS, and DNA was extracted with the Qiagen DNeasy kit. EBV viral load was determined as previously described by qPCR for the BALF5 gene and β-actin (67).
Flow cytometry and cell sorting.
Cells were analyzed for CD21 expression by flow-cytometric analysis. PBMC or Jurkat cells were washed in flow cytometry buffer (PBS, 0.5% bovine serum albumin, and 2 mM EDTA). Cells were then incubated with Fc Block (BD Biosciences) for 20 min. After washing, cells were stained for 20 min at room temperature with fluorophore-conjugated antibodies against CD3 (BD Biosciences), CD19 (BD Biosciences), CD4 (eBioscience), CD8 (eBioscience), and CD21 (clone Bly4 or HB5). For T-cell subset analysis, the following antibody panel was used: fluorescein isothiocyanate (FITC)-conjugated anti-CD3, anti-CD4–AF700, allophycocyanin (APC)-conjugated anti-CD8, anti-CCR7–PeCy7 (BD Biosciences), anti-CD45RA–BV421 (BD Biosciences), and phycoerythrin (PE)-conjugated anti-CD21 (HB5). All experiments were run on the BD-LSR Fortessa X-20 (BD Biosciences). Data analysis was done using FlowJo 10.
For sorting, negatively isolated T cells or Jurkat cells were blocked with Fc Block (BD Biosciences) for 20 min. Cells were then stained for 20 min with a CD21-PE antibody (clone HB5). Cells were sorted on the BD Aria Fusion sorter (BD Biosciences). Gates were determined based on the unstained control.
CRISPR-Cas9.
Jurkat cells with stable expression of Cas9 were generated by lentivirus transduction and selection with blasticidin as previously described (72). Cas9 activity was confirmed using pXPR-011 (gift from John Doench; Addgene plasmid number 59702), which encodes GFP and a short guide RNA (sgRNA) that targets GFP, as previously described (73). CD21-targeting sgRNA oligonucleotides from the Broad Avana sgRNA library were generated by Integrated DNA Technologies and cloned into the pLentiGuide-Puro vector (gift from Feng Zhang, Massachusetts Institute of Technology; Addgene plasmid number 52963), as previously described (72). The sequences of the sgRNAs were as follows: sg1, GCACTTCCTATGATCCACAA; sg2, TTGCAAAGCTGATAACACCT; and sg3, TCTGACTATCAACTGTACAA. Puromycin selection (3 μg/ml) was added 48 h after lentivirus transduction, and loss of CD21 protein expression was measured by flow-cytometric analysis for cell surface CD21.
Phylogenetic analysis.
Amino acid sequences for gp350 were downloaded from GenBank using previously published, annotated amino acid sequences (74). Sequences were uploaded into Molecular Evolutionary Genetics Analysis (MEGA) 7 and aligned using the MUSCLE alignment algorithm (75). Phylogenetic trees were generated using the maximum-likelihood method. Genetic differences in gp350 between EBV-1 and EBV-2 were identified manually from aligned sequences.
Statistical analysis.
Statistical analysis was performed using Prism version 6.0 (GraphPad, San Diego, CA). Comparison of EBV viral loads in the neutralization assays was carried out using the Mann-Whitney U test. Comparison of CD21 expression on CD4+ and CD8+ T cells, as determined by percent positivity and gMFI, was performed using a paired t test. Differences in EBV viral load following infection of Jurkat clones was determined by a t test.
ACKNOWLEDGMENTS
We thank Lindsey Hutt-Fletcher and Michael V. Holers for antibodies used in this study against the EBV glycoproteins and CD21 and Gary Chan for the EBV entry and binding protocols.
This work was supported by NIH R01 CA102667 (R.R.), R21 A1122670 (R.R.), 5T32AI007405-27 (N.A.S.), NIH R01 AI137337 (B.E.G.), a Burroughs Wellcome Medical Scientist Career Award (B.E.G.), and an American Cancer Society Research Scholar Award (B.E.G.).
We declare no conflicts of interest.
REFERENCES
- 1.Bibas M, Antinori A. 2009. EBV and HIV-related lymphoma. Mediterr J Hematol Infect Dis 1:e2009032. doi: 10.4084/MJHID.2009.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Epstein MA, Achong BG, Barr YM. 1964. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet 283:702–703. doi: 10.1016/S0140-6736(64)91524-7. [DOI] [PubMed] [Google Scholar]
- 3.Flavell KJ, Murray PG. 2000. Hodgkin’s disease and the Epstein-Barr virus. Mol Pathol 53:262–269. doi: 10.1136/mp.53.5.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green M, Michaels MG. 2013. Epstein-Barr virus infection and posttransplant lymphoproliferative disorder. Am J Transplant 13(Suppl 3):41–54; quiz, 54. doi: 10.1111/ajt.12004. [DOI] [PubMed] [Google Scholar]
- 5.Iizasa H, Nanbo A, Nishikawa J, Jinushi M, Yoshiyama H. 2012. Epstein-Barr Virus (EBV)-associated gastric carcinoma. Viruses 4:3420–3439. doi: 10.3390/v4123420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young LS, Dawson CW. 2014. Epstein-Barr virus and nasopharyngeal carcinoma. Chin J Cancer 33:581–590. doi: 10.5732/cjc.014.10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones JF, Shurin S, Abramowsky C, Tubbs RR, Sciotto CG, Wahl R, Sands J, Gottman D, Katz BZ, Sklar J. 1988. T-cell lymphomas containing Epstein-Barr viral DNA in patients with chronic Epstein-Barr virus infections. N Engl J Med 318:733–741. doi: 10.1056/NEJM198803243181203. [DOI] [PubMed] [Google Scholar]
- 8.Gru AA, Haverkos BH, Freud AG, Hastings J, Nowacki NB, Barrionuevo C, Vigil CE, Rochford R, Natkunam Y, Baiocchi RA, Porcu P. 2015. The Epstein-Barr virus (EBV) in T cell and NK cell lymphomas: time for a reassessment. Curr Hematol Malig Rep 10:456–467. doi: 10.1007/s11899-015-0292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujiwara S, Kimura H, Imadome K, Arai A, Kodama E, Morio T, Shimizu N, Wakiguchi H. 2014. Current research on chronic active Epstein-Barr virus infection in Japan. Pediatr Int 56:159–166. doi: 10.1111/ped.12314. [DOI] [PubMed] [Google Scholar]
- 10.Kasahara Y, Yachie A. 2002. Cell type specific infection of Epstein-Barr virus (EBV) in EBV-associated hemophagocytic lymphohistiocytosis and chronic active EBV infection. Crit Rev Oncol Hematol 44:283–294. doi: 10.1016/S1040-8428(02)00119-1. [DOI] [PubMed] [Google Scholar]
- 11.Park S, Ko YH. 2014. Epstein-Barr virus-associated T/natural killer-cell lymphoproliferative disorders. J Dermatol 41:29–39. doi: 10.1111/1346-8138.12322. [DOI] [PubMed] [Google Scholar]
- 12.Okuno Y, Murata T, Sato Y, Muramatsu H, Ito Y, Watanabe T, Okuno T, Murakami N, Yoshida K, Sawada A, Inoue M, Kawa K, Seto M, Ohshima K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Narita Y, Yoshida M, Goshima F, Kawada JI, Nishida T, Kiyoi H, Kato S, Nakamura S, Morishima S, Yoshikawa T, Fujiwara S, Shimizu N, Isobe Y, Noguchi M, Kikuta A, Iwatsuki K, Takahashi Y, Kojima S, Ogawa S, Kimura H. 2019. Defective Epstein-Barr virus in chronic active infection and haematological malignancy. Nat Microbiol 4:404–413. doi: 10.1038/s41564-018-0334-0. [DOI] [PubMed] [Google Scholar]
- 13.Haverkos BM, Coleman C, Gru AA, Pan Z, Brammer J, Rochford R, Mishra A, Oakes CC, Baiocchi RA, Freud AG, Porcu P. 2017. Emerging insights on the pathogenesis and treatment of extranodal NK/T cell lymphomas (ENKTL). Discov Med 23:189–199. [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman CB, Daud II, Ogolla SO, Ritchie JA, Smith NA, Sumba PO, Dent AE, Rochford R. 2017. Epstein-Barr virus type 2 infects T cells in healthy Kenyan children. J Infect Dis 216:670–677. doi: 10.1093/infdis/jix363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anagnostopoulos I, Hummel M, Kreschel C, Stein H. 1995. Morphology, immunophenotype, and distribution of latently and/or productively Epstein-Barr virus-infected cells in acute infectious mononucleosis: implications for the interindividual infection route of Epstein-Barr virus. Blood 85:744–750. doi: 10.1182/blood.V85.3.744.bloodjournal853744. [DOI] [PubMed] [Google Scholar]
- 16.Barros MHM, Vera-Lozada G, Segges P, Hassan R, Niedobitek G. 2019. Revisiting the tissue microenvironment of infectious mononucleosis: identification of EBV infection in T cells and deep characterization of immune profiles. Front Immunol 10:146. doi: 10.3389/fimmu.2019.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watry D, Hedrick JA, Siervo S, Rhodes G, Lamberti JJ, Lambris JD, Tsoukas CD. 1991. Infection of human thymocytes by Epstein-Barr virus. J Exp Med 173:971–980. doi: 10.1084/jem.173.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauvageau G, Stocco R, Kasparian S, Menezes J. 1990. Epstein-Barr virus receptor expression on human CD8+ (cytotoxic/suppressor) T lymphocytes. J Gen Virol 71:379–386. doi: 10.1099/0022-1317-71-2-379. [DOI] [PubMed] [Google Scholar]
- 19.Menezes J, Seigneurin JM, Patel P, Bourkas A, Lenoir G. 1977. Presence of Epstein-Barr virus receptors, but absence of virus penetration, in cells of an Epstein-Barr virus genome-negative human lymphoblastoid T line (Molt 4). J Virol 22:816–821. doi: 10.1128/JVI.22.3.816-821.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shapiro IM, Volsky DJ, Saemundsen AK, Anisimova E, Klein G. 1982. Infection of the human T-cell-derived leukemia line Molt-4 by Epstein-Barr virus (EBV): induction of EBV-determined antigens and virus reproduction. Virology 120:171–181. doi: 10.1016/0042-6822(82)90015-0. [DOI] [PubMed] [Google Scholar]
- 21.Koizumi S, Zhang XK, Imai S, Sugiura M, Usui N, Osato T. 1992. Infection of the HTLV-I-harbouring T-lymphoblastoid line MT-2 by Epstein-Barr virus. Virology 188:859–863. doi: 10.1016/0042-6822(92)90542-W. [DOI] [PubMed] [Google Scholar]
- 22.Coleman CB, Wohlford EM, Smith NA, King CA, Ritchie JA, Baresel PC, Kimura H, Rochford R. 2015. Epstein-Barr virus type 2 latently infects T cells, inducing an atypical activation characterized by expression of lymphotactic cytokines. J Virol 89:2301–2312. doi: 10.1128/JVI.03001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabson M, Gradoville L, Heston L, Miller G. 1982. Non-immortalizing P3J-HR-1 Epstein-Barr virus: a deletion mutant of its transforming parent, Jijoye. J Virol 44:834–844. doi: 10.1128/JVI.44.3.834-844.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coleman CB, Lang J, Sweet LA, Smith NA, Freed BM, Pan Z, Haverkos B, Pelanda R, Rochford R. 2018. Epstein-Barr virus type 2 infects T cells and induces B-cell lymphomagenesis in humanized mice. J Virol 92:e00813-18. doi: 10.1128/JVI.00813-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fingeroth JD, Weis JJ, Tedder TF, Strominger JL, Biro PA, Fearon DT. 1984. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc Natl Acad Sci U S A 81:4510–4514. doi: 10.1073/pnas.81.14.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frade R, Barel M, Ehlin-Henriksson B, Klein G. 1985. gp140, the C3d receptor of human B lymphocytes, is also the Epstein-Barr virus receptor. Proc Natl Acad Sci U S A 82:1490–1493. doi: 10.1073/pnas.82.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D, Kieff E. 2004. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci U S A 101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nemerow GR, Mold C, Schwend VK, Tollefson V, Cooper NR. 1987. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J Virol 61:1416–1420. doi: 10.1128/JVI.61.5.1416-1420.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nemerow GR, Wolfert R, McNaughton ME, Cooper NR. 1985. Identification and characterization of the Epstein-Barr virus receptor on human B lymphocytes and its relationship to the C3d complement receptor (CR2). J Virol 55:347–351. doi: 10.1128/JVI.55.2.347-351.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogembo JG, Kannan L, Ghiran I, Nicholson-Weller A, Finberg RW, Tsokos GC, Fingeroth JD. 2013. Human complement receptor type 1/CD35 is an Epstein-Barr Virus receptor. Cell Rep 3:371–385. doi: 10.1016/j.celrep.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanner J, Weis J, Fearon D, Whang Y, Kieff E. 1987. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 50:203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- 32.Haan KM, Kwok WW, Longnecker R, Speck P. 2000. Epstein-Barr virus entry utilizing HLA-DP or HLA-DQ as a coreceptor. J Virol 74:2451–2454. doi: 10.1128/JVI.74.5.2451-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haan KM, Longnecker R. 2000. Coreceptor restriction within the HLA-DQ locus for Epstein-Barr virus infection. Proc Natl Acad Sci U S A 97:9252–9257. doi: 10.1073/pnas.160171697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, Spriggs MK, Kovats S, Turk SM, Comeau MR, Nepom B, Hutt-Fletcher LM. 1997. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J Virol 71:4657–4662. doi: 10.1128/JVI.71.6.4657-4662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haan KM, Lee SK, Longnecker R. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106–114. doi: 10.1006/viro.2001.1141. [DOI] [PubMed] [Google Scholar]
- 36.Neuhierl B, Feederle R, Adhikary D, Hub B, Geletneky K, Mautner J, Delecluse HJ. 2009. Primary B-cell infection with a deltaBALF4 Epstein-Barr virus comes to a halt in the endosomal compartment yet still elicits a potent CD4-positive cytotoxic T-cell response. J Virol 83:4616–4623. doi: 10.1128/JVI.01613-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chesnokova LS, Nishimura SL, Hutt-Fletcher LM. 2009. Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins alphavbeta6 or alphavbeta8. Proc Natl Acad Sci U S A 106:20464–20469. doi: 10.1073/pnas.0907508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chesnokova LS, Hutt-Fletcher LM. 2011. Fusion of Epstein-Barr virus with epithelial cells can be triggered by alphavbeta5 in addition to alphavbeta6 and alphavbeta8, and integrin binding triggers a conformational change in glycoproteins gHgL. J Virol 85:13214–13223. doi: 10.1128/JVI.05580-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J, Sathiyamoorthy K, Zhang X, Schaller S, Perez White BE, Jardetzky TS, Longnecker R. 2018. Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nat Microbiol 3:172–180. doi: 10.1038/s41564-017-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Li Y, Wang HB, Zhang A, Chen ML, Fang ZX, Dong XD, Li SB, Du Y, Xiong D, He JY, Li MZ, Liu YM, Zhou AJ, Zhong Q, Zeng YX, Kieff E, Zhang Z, Gewurz BE, Zhao B, Zeng MS. 2018. Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat Microbiol 3:164–168. doi: 10.1038/s41564-017-0080-8. [DOI] [PubMed] [Google Scholar]
- 41.Wang HB, Zhang H, Zhang JP, Li Y, Zhao B, Feng GK, Du Y, Xiong D, Zhong Q, Liu WL, Du H, Li MZ, Huang WL, Tsao SW, Hutt-Fletcher L, Zeng YX, Kieff E, Zeng MS. 2015. Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat Commun 6:6240. doi: 10.1038/ncomms7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller G, Niederman JC, Stitt DA. 1972. Infectious mononucleosis: appearance of neutralizing antibody to Epstein-Barr virus measured by inhibition of formation of lymphoblastoid cell lines. J Infect Dis 125:403–406. doi: 10.1093/infdis/125.4.403. [DOI] [PubMed] [Google Scholar]
- 43.Sashihara J, Burbelo PD, Savoldo B, Pierson TC, Cohen JI. 2009. Human antibody titers to Epstein-Barr Virus (EBV) gp350 correlate with neutralization of infectivity better than antibody titers to EBV gp42 using a rapid flow cytometry-based EBV neutralization assay. Virology 391:249–256. doi: 10.1016/j.virol.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss ER, Alter G, Ogembo JG, Henderson JL, Tabak B, Bakis Y, Somasundaran M, Garber M, Selin L, Luzuriaga K. 2017. High Epstein-Barr virus load and genomic diversity are associated with generation of gp350-specific neutralizing antibodies following acute infectious mononucleosis. J Virol 91:e01562-16. doi: 10.1128/JVI.01562-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffman GJ, Lazarowitz SG, Hayward SD. 1980. Monoclonal antibody against a 250,000-dalton glycoprotein of Epstein-Barr virus identifies a membrane antigen and a neutralizing antigen. Proc Natl Acad Sci U S A 77:2979–2983. doi: 10.1073/pnas.77.5.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller N, Hutt-Fletcher LM. 1988. A monoclonal antibody to glycoprotein gp85 inhibits fusion but not attachment of Epstein-Barr virus. J Virol 62:2366–2372. doi: 10.1128/JVI.62.7.2366-2372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molesworth SJ, Lake CM, Borza CM, Turk SM, Hutt-Fletcher LM. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J Virol 74:6324–6332. doi: 10.1128/JVI.74.14.6324-6332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oba DE, Hutt-Fletcher LM. 1988. Induction of antibodies to the Epstein-Barr virus glycoprotein gp85 with a synthetic peptide corresponding to a sequence in the BXLF2 open reading frame. J Virol 62:1108–1114. doi: 10.1128/JVI.62.4.1108-1114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guthridge JM, Young K, Gipson MG, Sarrias MR, Szakonyi G, Chen XS, Malaspina A, Donoghue E, James JA, Lambris JD, Moir SA, Perkins SJ, Holers VM. 2001. Epitope mapping using the X-ray crystallographic structure of complement receptor type 2 (CR2)/CD21: identification of a highly inhibitory monoclonal antibody that directly recognizes the CR2-C3d interface. J Immunol 167:5758–5766. doi: 10.4049/jimmunol.167.10.5758. [DOI] [PubMed] [Google Scholar]
- 50.Schwartz JT, Barker JH, Long ME, Kaufman J, McCracken J, Allen LA. 2012. Natural IgM mediates complement-dependent uptake of Francisella tularensis by human neutrophils via complement receptors 1 and 3 in nonimmune serum. J Immunol 189:3064–3077. doi: 10.4049/jimmunol.1200816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsoukas CD, Lambris JD. 1993. Expression of EBV/C3d receptors on T cells: biological significance. Immunol Today 14:56–59. doi: 10.1016/0167-5699(93)90059-T. [DOI] [PubMed] [Google Scholar]
- 52.Fischer E, Delibrias C, Kazatchkine MD. 1991. Expression of CR2 (the C3dg/EBV receptor, CD21) on normal human peripheral blood T lymphocytes. J Immunol 146:865–869. [PubMed] [Google Scholar]
- 53.Levy E, Ambrus J, Kahl L, Molina H, Tung K, Holers VM. 2008. T lymphocyte expression of complement receptor 2 (CR2/CD21): a role in adhesive cell-cell interactions and dysregulation in a patient with systemic lupus erythematosus (SLE). Clin Exp Immunol 90:235–244. doi: 10.1111/j.1365-2249.1992.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Braun M, Melchers I, Peter HH, Illges H. 1998. Human B and T lymphocytes have similar amounts of CD21 mRNA, but differ in surface expression of the CD21 glycoprotein. Int Immunol 10:1197–1202. doi: 10.1093/intimm/10.8.1197. [DOI] [PubMed] [Google Scholar]
- 55.Sinha SK, Todd SC, Hedrick JA, Speiser CL, Lambris JD, Tsoukas CD. 1993. Characterization of the EBV/C3d receptor on the human Jurkat T cell line: evidence for a novel transcript. J Immunol 150:5311–5320. [PubMed] [Google Scholar]
- 56.Correia S, Palser A, Elgueta Karstegl C, Middeldorp JM, Ramayanti O, Cohen JI, Hildesheim A, Fellner MD, Wiels J, White RE, Kellam P, Farrell PJ. 2017. Natural variation of Epstein-Barr virus genes, proteins, and primary microRNA. J Virol 91:e00375-17. doi: 10.1128/JVI.00375-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szakonyi G, Klein MG, Hannan JP, Young KA, Ma RZ, Asokan R, Holers VM, Chen XS. 2006. Structure of the Epstein-Barr virus major envelope glycoprotein. Nat Struct Mol Biol 13:996–1001. doi: 10.1038/nsmb1161. [DOI] [PubMed] [Google Scholar]
- 58.Tanner JE, Coincon M, Leblond V, Hu J, Fang JM, Sygusch J, Alfieri C. 2015. Peptides designed to spatially depict the Epstein-Barr virus major virion glycoprotein gp350 neutralization epitope elicit antibodies that block virus-neutralizing antibody 72A1 interaction with the native gp350 molecule. J Virol 89:4932–4941. doi: 10.1128/JVI.03269-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lowell CA, Klickstein LB, Carter RH, Mitchell JA, Fearon DT, Ahearn JM. 1989. Mapping of the Epstein-Barr virus and C3dg binding sites to a common domain on complement receptor type 2. J Exp Med 170:1931–1946. doi: 10.1084/jem.170.6.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fearon DT, Carter RH. 1995. The CD19/CR2/TAPA–1 complex of B lymphocytes: linking natural to acquired immunity. Annu Rev Immunol 13:127–149. doi: 10.1146/annurev.iy.13.040195.001015. [DOI] [PubMed] [Google Scholar]
- 61.Matsumoto AK, Martin DR, Carter RH, Klickstein LB, Ahearn JM, Fearon DT. 1993. Functional dissection of the CD21/CD19/TAPA-1/Leu-13 complex of B lymphocytes. J Exp Med 178:1407–1417. doi: 10.1084/jem.178.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prodinger WM, Larcher C, Schwendinger M, Dierich MP. 1996. Ligation of the functional domain of complement receptor type 2 (CR2, CD21) is relevant for complex formation in T cell lines. J Immunol 156:2580–2584. [PubMed] [Google Scholar]
- 63.Tsoukas CD, Lambris JD. 1988. Expression of CR2/EBV receptors on human thymocytes detected by monoclonal antibodies. Eur J Immunol 18:1299–1302. doi: 10.1002/eji.1830180823. [DOI] [PubMed] [Google Scholar]
- 64.Pekalski ML, Garcia AR, Ferreira RC, Rainbow DB, Smyth DJ, Mashar M, Brady J, Savinykh N, Dopico XC, Mahmood S, Duley S, Stevens HE, Walker NM, Cutler AJ, Waldron-Lynch F, Dunger DB, Shannon-Lowe C, Coles AJ, Jones JL, Wallace C, Todd JA, Wicker LS. 2017. Neonatal and adult recent thymic emigrants produce IL-8 and express complement receptors CR1 and CR2. JCI Insight 2:e93739. doi: 10.1172/jci.insight.93739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cohen JI. 2015. Epstein-barr virus vaccines. Clin Trans Immunol 4:e32. doi: 10.1038/cti.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hutt-Fletcher LM, Turk SM. 2001. Virus isolation. Methods Mol Biol 174:119–123. doi: 10.1385/1-59259-227-9.119. [DOI] [PubMed] [Google Scholar]
- 67.Piriou E, Asito AS, Sumba PO, Fiore N, Middeldorp JM, Moormann AM, Ploutz-Snyder R, Rochford R. 2012. Early age at time of primary Epstein-Barr virus infection results in poorly controlled viral infection in infants from Western Kenya: clues to the etiology of endemic Burkitt lymphoma. J Infect Dis 205:906–913. doi: 10.1093/infdis/jir872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kubota N, Wada K, Ito Y, Shimoyama Y, Nakamura S, Nishiyama Y, Kimura H. 2008. One-step multiplex real-time PCR assay to analyse the latency patterns of Epstein-Barr virus infection. J Virol Methods 147:26–36. doi: 10.1016/j.jviromet.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 69.Simbiri KO, Smith NA, Otieno R, Wohlford EE, Daud II, Odada SP, Middleton F, Rochford R. 2015. Epstein-Barr virus genetic variation in lymphoblastoid cell lines derived from Kenyan pediatric population. PLoS One 10:e0125420. doi: 10.1371/journal.pone.0125420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chan G, Nogalski MT, Yurochko AD. 2009. Activation of EGFR on monocytes is required for human cytomegalovirus entry and mediates cellular motility. Proc Natl Acad Sci U S A 106:22369–22374. doi: 10.1073/pnas.0908787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chan GC, Yurochko AD. 2014. Analysis of cytomegalovirus binding/entry-mediated events. Methods Mol Biol 1119:113–121. doi: 10.1007/978-1-62703-788-4_8. [DOI] [PubMed] [Google Scholar]
- 72.Ma Y, Walsh MJ, Bernhardt K, Ashbaugh CW, Trudeau SJ, Ashbaugh IY, Jiang S, Jiang C, Zhao B, Root DE, Doench JG, Gewurz BE. 2017. CRISPR/Cas9 screens reveal Epstein-Barr virus-transformed B cell host dependency factors. Cell Host Microbe 21:580–591 e7. doi: 10.1016/j.chom.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. 2014. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palser AL, Grayson NE, White RE, Corton C, Correia S, Ba Abdullah MM, Watson SJ, Cotten M, Arrand JR, Murray PG, Allday MJ, Rickinson AB, Young LS, Farrell PJ, Kellam P. 2015. Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J Virol 89:5222–5237. doi: 10.1128/JVI.03614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]