ALV-J can cause severe immunosuppression and myeloid leukemia in infected chickens. However, no vaccine or antiviral drug is available against ALV-J, and the mechanism for ALV-J pathogenesis needs to be elucidated. It is generally believed that gp85 and LTR of ALV contribute to its pathogenesis. Here, we found that the C terminus and the tyrosine motifs (YxxM, ITIM, and ITAM-like) in the CTD of Gp37 of ALV-J could affect the pathogenicity of ALV-J in vitro and in vivo. The pathogenicity of ALV-J with Gp37 containing ITIM only was significantly less than ALV-J with Gp37 containing both YxxM and ITIM and ALV-J with Gp37 containing both YxxM and ITAM-like. This study highlights the vital role of the C terminus of Gp37 in the pathogenesis of ALV-J and thus provides a new perspective to elucidate the interaction between ALV-J and its host and a molecular basis to develop efficient strategies against ALV-J.
KEYWORDS: avian leukosis virus, gp37, tyrosine motif, C terminus, pathogenesis, Gp37, tyrosine motifs
ABSTRACT
Different from other subgroups of avian leukosis viruses (ALVs), ALV-J is highly pathogenic. It is the main culprit causing myeloid leukemia and hemangioma in chickens. The distinctiveness of the env gene of ALV-J, with low homology to those of other ALVs, is linked to its unique pathogenesis, but the underlying mechanism remains unclear. Previous studies show that env of ALV-J can be grouped into three species based on the tyrosine motifs in the cytoplasmic domain (CTD) of Gp37, i.e., the inhibitory, bifunctional, and active groups. To explore whether the C terminus or the tyrosine motifs in the CTD of Gp37 affect the pathogenicity of ALV-J, a set of ALV-J infectious clones containing different C termini of Gp37 or the mutants at the tyrosine sites were tested in vitro and in vivo. Viral growth kinetics indicated not only that ALV-J with active env is the fastest in replication and ALV-J with inhibitory env is the lowest but also that the tyrosine sites essentially affected the replication of ALV-J. Moreover, in vivo studies demonstrated that chickens infected by ALV-J with active or bifunctional env showed higher viremia, cloacal viral shedding, and viral tissue load than those infected by ALV-J with inhibitory env. Notably, the chickens infected by ALV-J with active or bifunctional env showed significant loss of body weight compared with the control chickens. Taken together, these findings reveal that the C terminus of Gp37 plays a vital role in ALV-J pathogenesis, and change from inhibitory env to bifunctional or active env increases the pathogenesis of ALV-J.
IMPORTANCE ALV-J can cause severe immunosuppression and myeloid leukemia in infected chickens. However, no vaccine or antiviral drug is available against ALV-J, and the mechanism for ALV-J pathogenesis needs to be elucidated. It is generally believed that gp85 and LTR of ALV contribute to its pathogenesis. Here, we found that the C terminus and the tyrosine motifs (YxxM, ITIM, and ITAM-like) in the CTD of Gp37 of ALV-J could affect the pathogenicity of ALV-J in vitro and in vivo. The pathogenicity of ALV-J with Gp37 containing ITIM only was significantly less than ALV-J with Gp37 containing both YxxM and ITIM and ALV-J with Gp37 containing both YxxM and ITAM-like. This study highlights the vital role of the C terminus of Gp37 in the pathogenesis of ALV-J and thus provides a new perspective to elucidate the interaction between ALV-J and its host and a molecular basis to develop efficient strategies against ALV-J.
INTRODUCTION
Avian leukosis virus (ALV) belongs to the genus Alpharetrovirus, family Retrovirus. Based on the characteristics of its viral envelope protein, ALV was recently divided into 7 subgroups (A, B, C, D, E, J, and K) in chickens (1–3). Among them, ALV-E is endogenous ALV, which exhibits low pathogenicity and oncogenicity, while the other subgroups are exogenous viruses with different pathogenicity to chickens (4). Due to its characteristics of tumorigenesis and immunosuppression, ALV has caused enormous economic losses to the poultry industry (5, 6). Differently from other subgroups of ALVs, ALV-J is the main culprit causing hematopoietic malignancy with myeloid leukemia and hemangioma in chickens (5, 7). Moreover, ALV-J possesses a higher pathogenicity and is more prevalent than the other exogenous subgroups of ALV. The unique pathogenicity of ALV-J is considered to be closely related to its unusual env gene, which is distinct from the other ALV subgroups (8, 9). ALV-J env has approximately 97% homology to the endogenous EAV-HP sequence, the endogenous retrovirus element ubiquitous in the genomes of animals of the Gallus genus (10). Thus, it is believed that the ALV-J env gene originates from the env of EAV-HP.
Similarly to other retroviruses, the glycosylated envelope protein (Env) of ALV-J is encoded by its env gene and can be divided into Gp85 and Gp37 (1). The Gp85 protein, with high variability, is responsible for binding to host cell receptors, whereas the Gp37 protein is responsible for mediating the fusion of viral protein with host cell membrane and is more conserved than Gp85 (11–16). Although the distinctive symptoms and pathogenesis of ALV-J are thought to be closely related to its unique Env protein, the exact molecular mechanism for the pathogenesis of ALV-J Env is still not clear. Previously, ALV-J Env proteins were divided into three types (i.e., inhibitory, bifunctional, and active Env) according to their different tyrosine-based motifs including immune tyrosine-based inhibitory motif (ITIM), YxxM, and immune tyrosine-based active motif-like (ITAM-like) that are within the C terminus of Gp37 (16). To evaluate the effect of the C terminus of Gp37 and those tyrosine motifs on the pathogenicity of ALV-J, nine novel ALV-J infectious clones containing three types of Env with different tyrosine-based motifs and their mutants at the tyrosine sites were tested in vitro and in vivo in this study. Viral kinetic curves and animal infection study revealed that the C terminus of ALV-J Gp37 plays a vital role in the pathogenesis of ALV-J.
RESULTS
Rescue of ALV-J viruses with different C termini and tyrosine motifs in the C-terminal domain (CTD) of Gp37.
To investigate the roles of the C terminus of Gp37 and those tyrosine motifs in the pathogenicity of ALV-J, ALV-J infectious clones containing different C termini of Gp37, including EAV-HP (J1 backbone with C terminus of EAV-HP gp37), 4817 (J1 backbone with C terminus of 4817 gp37), and J1 (one kindly gifted by Yixin Wang, Shangdong Agriculture University, China), and their variants of tyrosine motif were first obtained through homologous recombination as shown in Fig. 1. EAV-HP, 4817, and J1 represented ALV-J inhibitory env, active env, and bifunctional env, respectively. The nine novel infectious clones were designated EAV-HP, 4817, NY-EAV-HP, YF-EAV-HP, YF-J1, YN-J1, YN-4817, 2YF-4817, and NFF-4817 (Fig. 1). The amino acid (aa) sequence 74 to 197 or 209 of Gp37 in the green frame in Fig. 1B represented the ALV-J C terminus of Gp37, and the tyrosine motifs YxxM, ITIM, and ITAM-like within the C terminus of Gp37 were indicated using purple, red, and blue, respectively (Fig. 1B). For viral rescuing, the infectious clones was separately transfected into DF-1 cells, followed by immunofluorescence assay (IFA) to identify the rescued viruses. As shown in Fig. 2, the DF-1 cells infected with EAV-HP, 4817, and J1 viruses and their tyrosine site mutants NY-EAV-HP, YF-EAV-HP, YF-J1, YN-J1, YN-4817, 2YF-4817, and NFF-4817 showed specific green fluorescence, but the uninfected cells did not. All the rescued viruses were further confirmed by sequencing analysis (data not shown). These data demonstrate that a total of 10 ALV-J viruses were successfully rescued with those infectious constructs.
FIG 1.
Strategy for construction of the recombinant ALV-J with different gp37 genes. (A) The original backbone infectious clone of ALV-J J1 is shown. The other infectious clones, EAV-HP, 4817, NY-EAV-HP, YF-EAV-HP, YF-J1, YN-J1, YN-4817, 2YF-4817, and NFF-4817, were cloned by recombination through replacing the C terminus of Gp37 (aa 74 to 197, J1) or mutating the tyrosine sites. Sequences which are identical are shown in the same color. (B) Sequence alignment for the Gp37 sequences of the ALV-J infectious clones used in this study. The sequences in green, purple, and light blue frames are the C terminus of Gp37 in this study and the YxxM motif and ITAM-like motif in the CTD of Gp37, respectively. The sequence with red line below it is ITIM.
FIG 2.
Rescue of the recombinant ALV-J expressing different gp37 genes. DF-1 cells were transfected with the indicated ALV-J infectious clones for 7 days, and the virus-containing supernatants were blindly passaged in DF-1 cells. DF-1 cells were infected with the rescued EAV-HP (A), NY-EAV-HP (B), YF-EAV-HP (C), 4817 (D), YN-4817 (E), 2YF-4817 (F), NFF-4817 (G), J1 (H), YF-J1 (I), and YN-J1 (J) viruses, and the uninfected DF-1 cells were set as control (K). These infected cells were identified by IFA using JE9 MAb against ALV-J Gp85.
ALV-J with active env replicated faster than ALV-J with bifunctional or inhibitory env in vitro.
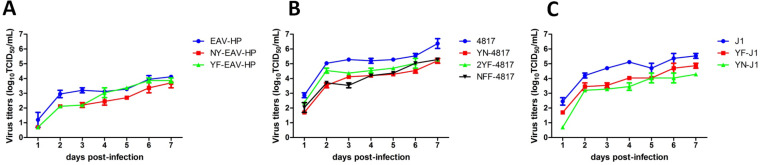
To explore the growth characteristics of the rescued viruses in vitro, DF-1 cells were inoculated with the rescued viruses at a multiplicity of infection (MOI) of 0.01, and the replication kinetics of these viruses in the DF-1 cells were determined by titration. As shown in Fig. 3, 4817 virus replicated more efficiently and yielded higher viral titers than EAV-HP and J1 viruses did. The viral titer of 4817 could reach to 5.336 and 6.669 log10 50% tissue culture infectious doses (TCID50)/ml at days 3 and 7 postinfection, respectively, whereas those of EAV-HP and J1 were less than 4.199 and 5.699 log10 TCID50/ml at those days, respectively. Notably, the peak titer of 4817 or J1 in DF-1 cells was more than 100 or 10 times that of EAV-HP, respectively. Thus, the viral growth curves demonstrated that the C terminus of Gp37 was critical to the replication of ALV-J in DF-1 cells. Notably, as shown in Fig. 3, the viral titers of 4817 at 1 to 7 days postinfection (dpi) were higher than those of the 4817 tyrosine mutants (i.e., YN-4817, 2YF-4817, and NFF-4817). The peak titer of 4817 was at least 25 times higher than those of the 4817 mutants. In addition, the viral titer at 4 dpi and the peak titer of J1 virus could reach 5.199 and 5.699 log10 TCID50/ml, respectively. In contrast, the peak titers of EAV-HP and J1 mutants were less than 5.032 log10 TCID50/ml. Moreover, mutation from Y to F within ITIM obviously decreased the replication of EAV-HP virus, but mutation from NxxM to YxxM only slightly attenuated the replication of EAV-HP virus. Taken together, the replication ability of 4817 and J1 viruses was decreased due to the mutation from the tyrosine amino acid to N or F, but this situation was not the same as in EAV-HP virus.
FIG 3.
Growth curves of the recombinant ALV-J in DF-1 cells. The rescued EAV-HP, 4817, J1, NY-EAV-HP, YF-EAV-HP, YF-J1, YN-J1, YN-4817, 2YF-4817, and NFF-4817 viruses were inoculated into DF-1 cells at an MOI of 0.01, and the virus-containing supernatants were collected at the indicated time points for virus titration. (A) Growth curves for EAV-HP, NY-EAV-HP, and YF-EAV-HP viruses in DF-1 cells. (B) Growth curves for 4817, YN-4817, 2YF-4817, and NFF-4817 viruses in DF-1 cells. (C) Growth curves for J1, YF-J1, and YN-J1 viruses in DF-1 cells.
ALV-J 4817 and J1 caused higher cloacal viral shedding and tissue viral load than EAV-HP in vivo.
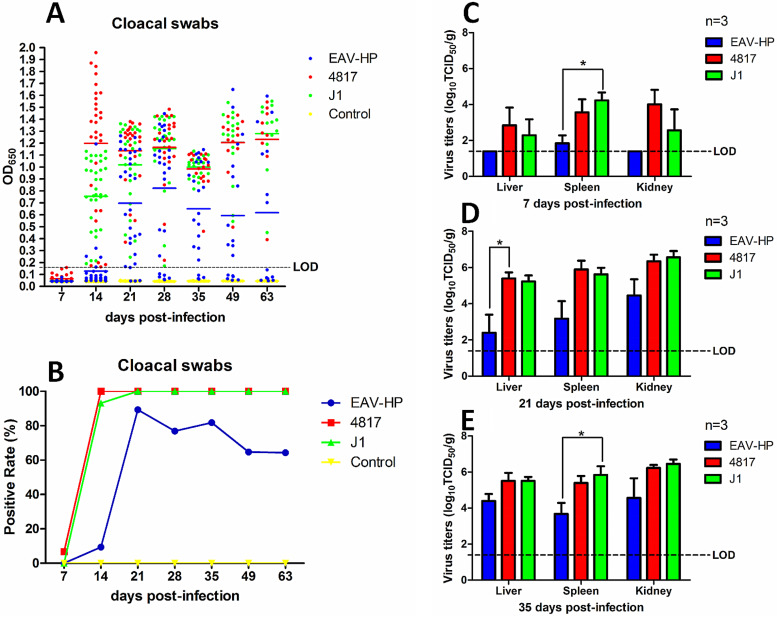
To further evaluate the pathogenesis of ALV-J with a different C terminus of Gp37 in vivo, specific-pathogen-free (SPF) chickens were inoculated with EAV-HP, 4817, and J1 viruses. The chickens inoculated with phosphate-buffered saline (PBS) were used as a control. The cloacal swabs and organs from the different groups were collected and analyzed at the indicated time points. According to the viral shedding in the cloaca, the positive rate for the 4817 group was 6.7% at 7 dpi and reached 100% with average values of optical density at 650 nm (OD650) values of 1.20, 1.14, 1.16, 0.98, 1.21, and 1.23 from 14 to 63 dpi (Fig. 4A and B). Similar to the high positive rate of 4817, the cloacal positive rate for THE J1 group was 0% at 7 dpi but rapidly increased to 93.1% with an average OD650 of 0.75 at 14 dpi and reached 100% with average OD650 values of 1.02, 1.15, 1.00, 1.21, and 1.28 for the later time points. In contrast, the positive rate for the EAV-HP group was only 9.4% with an average OD650 value of 0.13 at 14 dpi but reached peak with an average OD650 value of 0.82 at 28 dpi, followed by a gradual decrease to 81.8%, 64.7%, and 64.3% with average OD650 values of 0.65, 0.59, and 0.62 at 35, 49, and 63 dpi, respectively. In the control group, no virus was detected from the cloacal swabs. According to tissue viral load, the chickens infected with 4817 virus had the highest viral titers in the liver and kidney at 7 dpi, and their titers were slightly higher than those of chickens infected with J1 virus (Fig. 4C). Moreover, the virus could be detected only in the spleens of chickens infected with EAV-HP at 7 dpi. Furthermore, the viral titers in the liver, spleen, and kidney from chickens infected with 4817 were similar to those in the J1 group at 21 dpi and 35 dpi. However, the viral titers in the chickens infected with EAV-HP virus were about 10 to 100 times lower than those in the chickens infected with 4817 or J1 virus at 21 dpi and 35 dpi (Fig. 4D and E). All the results demonstrated that the chickens infected with J1 and 4817 viruses had more virus shed in the cloaca and higher viral loads in tissues than those infected with EAV-HP.
FIG 4.
Viral cloacal shedding and viral tissue load in infected chickens. The blue, red, green, and yellow spots or columns represent the EAV-HP, 4817, J1, and control groups, respectively. (A and B) Comparison of viral P27 antigen levels (A) or viral positive rate (B) of the cloacal swabs assayed by ELISA from chickens infected with EAV-HP, 4817, and J1 viruses. (C to E) Comparison of viral loads in organs from chickens infected with EAV-HP, 4817, and J1 at 7, 21, and 35 dpi, respectively. The data were analyzed with a Student t test. A P value of <0.05 was considered significant. LOD, limit of detection.
ALV-J 4817 and J1 caused significant loss of body weight and severe viremia compared with EAV-HP.
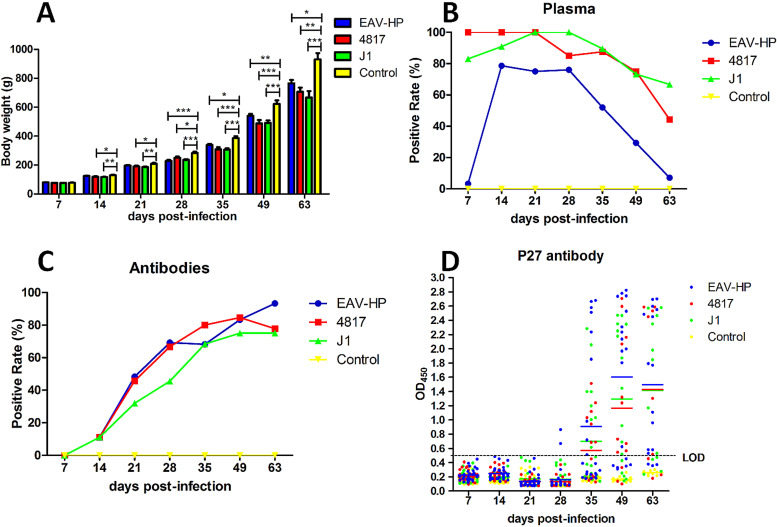
To evaluate the effect of ALV-J with different env genes on the body weights of the infected chickens, the body weights of the chickens were monitored at the indicated time points. As shown in Fig. 5A, the average body weights of the chickens in the EAV-HP, 4817, J1, and control groups were 765.2 g, 706.3 g, 666.7 g, and 930.2 g at 63 dpi, respectively, which indicated that different viruses caused loss of body weight in the infected chickens. Notably, 4817 and J1 viruses caused more loss of body weight than EAV-HP virus. To determine the viremia and antibody titers, blood samples from chickens inoculated with different ALVs were collected and analyzed at indicated time points. As shown in Fig. 5B, the viremia-positive rates in the chickens infected with 4817 were 100%, 100%, 100%, 85%, 87.5%, 75%, and 44.4% at 7, 14, 21, 28, 35, 49, and 63 dpi, respectively. Accordingly, the rates were 83%, 90.9%, 100%, 100%, 89.5%, 73.3%, and 66.7% for J1 virus, respectively. Interestingly, for the EAV-HP group, the viremia-positive rate at 7 dpi was only 3.3%, but the rates rapidly rose to 78.6%, 75%, and 76% at 14, 21, and 28 dpi, respectively, followed by dramatic decreases to 52.4%, 29.4%, and 7.1% at 35, 49, and 63 dpi, respectively. In regard to antibody against ALV, the overall positive rate for EAV-HP was higher than that for 4817 and J1 viruses (Fig. 5C). At 63 dpi, the positive rate for EAV-HP reached 93.3% whereas the rates for 4817 and J1 viruses were 77.8% and 75%, respectively. To confirm this, the antibody against ALV P27 was also detected in the infected chickens. At 35, 49, and 63 dpi, the OD450 values for P27 antibody titers to EAV-HP, 4817, and J1 were 0.91, 1.6, and 1.50; 0.57, 1.17, and 1.43; and 0.70, 1.30, and 1.41, respectively. It is noteworthy that two serum samples from chickens infected with EAV-HP were positive at 28 dpi, whereas the chickens infected with 4817 and J1 were all negative. Therefore, EAV-HP induced antibody against ALV P27 earlier and to higher titers than both 4817 and J1 (Fig. 5D). All these data clearly demonstrate that the ALV-J with active or bifunctional env caused significant growth inhibition and severe viremia in the infected chickens compared with inhibitory env-carrying ALV-J.
FIG 5.
Body weight loss, viremia, and antibody against ALV in infected chickens. The blue, red, green, and yellow columns or curves represent the EAV-HP, 4817, J1, and control groups, respectively. (A) Comparison of body weights measured at different time points. (B) Comparison of the viral positive rates in plasma from chickens infected with EAV-HP, 4817, and J1 viruses. (C) Comparison of positive rates for antibody against ALV from chickens infected with EAV-HP, 4817, and J1 viruses, determined by IFA. (D) Comparison of levels of antibody against ALV P27 from chickens infected with EAV-HP, 4817, and J1 viruses, determined by ELISA. Statistical analysis was performed with one-way ANOVA using GraphPad 5 software. A P value of <0.05 was considered significant. *, **, and *** indicate P values of less than 0.05, 0.01, and 0.001, respectively.
DISCUSSION
ALV-J was first identified in 1988 from meat-type chickens in the United Kingdom, but it became more virulent and could infect both layers and indigenous chickens subsequently in China (5, 6, 17–21). ALV-J mainly induces myeloid leukemia, hemangioma, and immunosuppression in chickens, which have caused significant economic losses to the chicken industry worldwide (5). It is noteworthy that the env gene of ALV-J has less than 40% homology to those of other ALV subgroups; however, it has extremely high homology to that of EAV-HP. The env gene is considered to play a vital role in the pathogenesis of ALV-J (10). EAV-HP is a type of ancient avian retrovirus element that has existed in the genomes of animals of the genus Gallus for millions of years (22–24). Previously, our group found that the Env protein of ALV-J could be divided into three types (i.e., inhibitory, bifunctional, and active Env) according to the tyrosine-based motifs located in the CTD of Gp37 (16). The inhibitory Env contains ITIM only, the bifunctional Env contains both ITIM and YxxM motifs, and the active Env carries YxxM and ITAM-like motifs. Based on this, about 37%, 62%, and 1% of ALV-J Env sequences from GenBank are grouped as inhibitory, bifunctional, and active Env, respectively. Notably, the Env proteins from the ALV-J prototype strain (HPRS-103) and from most EAV-HP sequences belong to the inhibitory Env type. We also hypothesize that the motifs in ALV-J Gp37 are possibly functional and play vital roles in ALV-J pathogenesis and oncogenesis (16). However, the mechanism underlying the role of Gp37 in ALV-J pathogenesis is unclear. To explore the role of Gp37 C-terminal sequence, nine ALV-J infectious clones were in vitro and in vivo tested in this study (Fig. 1). All the infectious ALV clones were successfully rescued in DF-1 cells (Fig. 2). Viral growth curves for ALV-J containing the C terminus of Gp37 from three types of env genes revealed that 4817 virus replicated faster than J1 virus in DF-1 cells, and the EAV-HP virus was the slowest in replication (Fig. 3A to C). These results clearly demonstrated that the C terminus of Gp37 could affect the replication ability of ALV-J in vitro. Notably, the main target cells for ALV-J are hemocytes; however, the DF-1 cell line is still the most widely used cell line in studies on ALV replication kinetics, since testing for viral growth curves using DF-1 cells is more reliable. Moreover, ALV-J viruses with point mutations at the tyrosine sites also revealed that the tyrosine sites are important for viral replication of 4817 and J1 viruses (Fig. 3). Surprisingly, although the recombination of J1 with the C terminus of Gp37 derived from EAV-HP could significantly decrease viral replication efficiency, mutation from NxxM to YxxM or from Y to F in the ITIM of EAV-HP Gp37 did not cause significant change in viral replication efficiency. The reason behind this is possibly due to some other amino acid sites in the C terminus of Gp37 masking the effect of the tyrosine motifs in EAV-HP. It is noteworthy that the proteins with tyrosine motifs like YxxM, ITIM, or ITAM play significant roles in regulating the activity of immune cells such as T, B, and NK cells (25–28), and these immune cells can be target cells of ALV-J. However, whether ALV-J could affect the regulation of these cells through the tyrosine motifs of Gp37 needs to be further studied. In addition, although several ALV strains from other subgroups, such as ALV-A (SDAU09E2), ALV-B (SDAU09C2), and ALV-C (RSV-C), have the YxxM motif in the CTD of their Gp37, no ITIM or ITAM-like was found in the Gp37 from the other subgroups of ALV. The roles of the YxxM motif in other ALV subgroups also need to be tested.
To further study the effect of the three types of env gene on viral virulence in chickens, J1, EAV-HP, and 4817 viruses were studied in vivo. The SPF chickens infected with 4817 or J1 virus showed higher viremia (Fig. 5B) and higher cloacal viral P27 and cloacal viral positive rate (Fig. 4A and B) than those infected with EAV-HP. The data on body weight loss (Fig. 5A) and viral load also support the notion that 4817 and J1 viruses possess higher pathogenicity than EAV-HP virus (Fig. 4C to E). It is noteworthy that although the pathogenicity of EAV-HP virus was weaker than both 4817 and J1 viruses, the positive rate of antibodies against ALV-J at 63 dpi and the titers of antibody against ALV P27 for J1 and 4817 viruses were lower than those for EAV-HP virus (Fig. 5C and D). This result suggested that Gp37 of ALV-J may affect not only virus shedding in chickens but also antibody production. Notably, the results of detecting antibodies against ALV and ALV P27 by IFA and enzyme-linked immunosorbent assay (ELISA), respectively, did not completely match each other at each time point. This discrepancy is probably due to the differences in the methods and their sensitivity in detecting the antibodies. Moreover, it should be mentioned that no tumors were observed in all of the infected chickens during this study. In line with this, hematoxylin-eosin (HE) staining analysis on the livers and spleens of the chickens at 63 dpi showed no typical histopathological symptoms of ALV-J infection in the tissues (data not shown). It is possible that a 63-day infection with ALV-J was not long enough to develop tumors in the infected chickens. On the other hand, the inoculation method could also affect the possibility of tumor formation, as some studies suggest that the inoculation of chicken embryos with ALV is more likely to cause tumors (29, 30).
Previous studies on ALV-J mainly revealed the critical effect of the long terminal repeat (LTR) and gp85 on the pathogenesis of ALV-J (11–13, 31, 32); the effect of gp37 on the pathogenicity of ALV-J has not been investigated. In this study, we not only find some pathogenic determinants in Gp37 of ALV-J but also systematically show the comprehensive data from an animal study on ALV-J. To our knowledge, this is the first report that clearly demonstrates that not only the C terminus of Gp37 protein but also the tyrosine motifs in its CTD could affect the replication ability and pathogenesis of ALV-J. The data from this research have clearly revealed that ALV-J with inhibitory env showed lower pathogenicity than ALV-J with bifunctional or active env. Notably, EAV-HPs in the genomes of animals of the Gallus genus are ancient endogenous elements responsible for the emergence of highly pathogenic ALV-J via recombination (10, 22–24). Thus, it is possible that the evolution from endogenous EAV-HP env to exogenous ALV-J env is a switch point from immune inhibitory receptor to immune active receptor. This study may also provide an insight into the variation and functional transformation of the env gene in the evolution from EAV-HP env to exogenous ALV-J env. In addition, the findings from this study could facilitate development of a new approach to controlling the diseases caused by ALV-J. Further studies should focus on determining what roles tyrosine motifs play in signaling pathways in the cells infected with ALV-J carrying different tyrosine motifs in Gp37 and whether ALV-J Gp37 can be phosphorylated at the tyrosine sites.
MATERIALS AND METHODS
Cells and plasmid.
DF-1 cells (from ATCC, kept in our lab) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 5% fetal bovine serum (FBS), 100 U/ml penicillin, and 0.1 mg/ml streptomycin in a cell incubator with 5% CO2 at 37°C. pcDNA3.1-EAV-HP-env and pcDNA3.1-4817-env, which contained the env gene from endogenous avian retrovirus EAV-HP (GenBank accession number AF125528) and the ALV-J 4817 strain (GenBank accession number AF247385), respectively, were preserved in our lab. 4817 is an ALV-J field strain isolated from meat-type chickens from an American chicken farm (33). The ALV-J infectious clone with a bifunctional env gene was kindly provided by Yixin Wang, Shangdong Agriculture University.
Construction of ALV-J infectious clone mutants.
The construction strategy for all the infectious clones is shown in Fig. 1. Different ALV-J infectious clones with mutations in their CTDs were constructed as previously described (34) using the primer pairs listed in Table 1. Briefly, the target gene fragment and the linearized vector were amplified by PCR using the indicated primers and template. Then, the amplified gene fragment and the linearized vector were ligated and recombined by the ClonExpress II one-step cloning kit as previously described (34). Lowercase bases in the primers in Table 1 were those designed to generate the target mutation site. The underlined sequences in the primers were the homologous complementary sequence, and the normal part of the primers were the sequence of the viral genome itself or its reverse complement.
TABLE 1.
Primers for generating ALV-J infectious clones through homologous recombination technique
| Target infectious clone | Template/primer name and sequence (position of primer) (fragment size) for amplifying: |
|
|---|---|---|
| Target gene fragments | Linearized vectors | |
| EAV-HP | pc-EAV-HP-env/J-gp37-IC-F: CGGCACGCGGTGTTGCAGAATCGAGCAGCCATCGATTTC (bp 220–243 of EAV-HP-gp37); EAV-gp37-IC-R: CGCGTTCGGAACCTACAGCTGCTCCCTAATTCTAT (bp 572–591 of EAV-HP-gp37) (402 bp) | J1/S2-IC-F: TAGGTTCCGAACGCGATGTAACGGGGCAAG (bp 7009–7038 of J1); S2-IC-R: CAACACCGCGTGCCGGATGCTGTTCATGTC (bp 6607–6636 of J1) (9,921 bp) |
| 4817 | pc-4817-env/J-gp37-IC-F: CGGCACGCGGTGTTGCAGAATCGAGCAGCCATCGATTTC (bp 220–243 of 4817-gp37); 4817-gp37-IC-R: CGCGTTCGGAACCTATACTGCCCCATTTTCGGGCTG (bp 220–243 of 4817-gp37) (438 bp) | J1/S2-IC-F: TAGGTTCCGAACGCGATGTAACGGGGCAAG (bp 7009–7038 of J1); S2-IC-R: CAACACCGCGTGCCGGATGCTGTTCATGTC (bp 6607–6636 of J1) (9,921 bp) |
| YF-J1 | J1/J-gp37-IC-F: CGGCACGCGGTGTTGCAGAATCGAGCAGCCATCGATTTC (bp 6622–660 bp of J1); YF-J1R: TCCCTAATTCTATGAaATCTTATGCGTTCATCCATGAACTG (bp 6961–7001 of J1) (380 bp) | J1/YF-S2-IC-F: TCATAGAATTAGGGAGCAACTGTAGGTTCC (bp 6987–7016 of J1); S2-IC-R: CAACACCGCGTGCCGGATGCTGTTCATGTC (bp 6607–6636 of J1) (9,943 bp) |
| YN-J1 | J1/J-gp37-IC-F: CGGCACGCGGTGTTGCAGAATCGAGCAGCCATCGATTTC (bp 6622–6660 of J1); YN-J1R: CATCCATGAACTGATtCATTGTTCTTGATAGACAGTCCTGG (bp 6633–6973 of J1) (352 bp) | J1/YN-S2-IC-F: ATCAGTTCATGGATGAACGCATAAGATATC (bp 6959–6988 of J1); S2-IC-R: CAACACCGCGTGCCGGATGCTGTTCATGTC (bp 6607–6636 of J1) (9,971 bp) |
| NY-EAV-HP | EAV-HP/NY-EAV-HP-F: CTATTGAGAACAATGtATCAGTTTATGGATGAACGC (bp 6943–6978 of EAV-HP); NY-EAV-HP-R: GTACCACCTTACTTCCACCAATCAACGGGTACAGTGCC (bp 7341–7378 of EAV-HP) (436 bp) | EAV-HP/NY-EAV-HP-IC-F: GAAGTAAGGTGGTACGGTCATGGTATGATC (bp 7364–7393 of EAV-HP); NY-EAV-HP-IC-R: CATTGTTCTCAATAGACAATCCTGAAAGCAC (bp 6927–6957 of EAV-HP) (9,887 bp) |
| YF-EAV-HP | EAV-HP/J-gp37-IC-F: CGGCACGCGGTGTTGCAGAATCGAGCAGCCATCGATTTC (bp 6622–6660 of EAV-HP); YF-EAV-HP-R: TCCCTAATTCTATGAaATTTTATGCGTTCATCCATAAACTG (bp 6967–7001 of EAV-HP) 380 bp | EAV-HP/YF-EAV-HP-IC-F: TCATAGAATTAGGGAGCAGCTGTAGGTTC (bp 6987–7015 of EAV-HP); S2-IC-R: CAACACCGCGTGCCGGATGCTGTTCATGTC (bp 6607–6636 of EAV-HP) (9,943 bp) |
| YN-4817 | 4817/J-gp37-IC-F: CGGCACGCGGTGTTGCAGAATCGAGCAGCCATCGATTTC (bp 6622–6660 of 4817); YN-4817-R: CATCCATAAACTGATtCATTGTTCTCGATAGACAATCCTGG (bp 6933–6957 of 4817) (336 bp) | 4817/YN-4817-IC-F: ATCAGTTTATGGATGAACGCATCAGCTATCAC (bp 6959–6990 of 4817); S2-IC-R: CAACACCGCGTGCCGGATGCTGTTCATGTC (bp 6607–6636 bp of 4817) (10,007 bp) |
| 2YF-4817 | 4817/J-gp37-IC-F: CGGCACGCGGTGTTGCAGAATCGAGCAGCCATCGATTTC (bp 6622–6660 of 4817); 2YF-4817-R: ACAAGCCTTTTGCAACTTCTTGaATTCCGCGTGAaAGCT (bp 6982–7020 of 4817) | 4817/2YF-4817-IC-F: TTGCAAAAGGCTTGTAGGCAGCCCGAAAATG (bp 7006–7036 of 4817); S2-IC-R: CAACACCGCGTGCCGGATGCTGTTCATGTC (bp 6607–6636 of 4817) (9,960 bp) |
| NFF-4817 | 2YF-4817/J-gp37-IC-F: CGGCACGCGGTGTTGCAGAATCGAGCAGCCATCGATTTC (bp 6622–6660 of 2YF-4817); YN-4817-R: CATCCATAAACTGATTCATTGTTCTCGATAGACAATCCTGG (bp 6933–6957 of 2YF-4817) (336 bp) | 2YF-4817/NFF-4817-IC-F: ATCAGTTTATGGATGAACGCATCAGCTtTCAC (bp 6959–6990 of 2YF-4817); S2-IC-R: CAACACCGCGTGCCGGATGCTGTTCATGTC (bp 6607–6636 of 2YF-4817) (10,007 bp) |
Rescue of ALV-J.
Four micrograms of 10 different ALV-J infectious clones was respectively transfected into fresh DF-1 cells with 70% confluence using Mirus as previously described (35). At 18 h posttransfection (hpt), the cell supernatant with transfection mixtures was replaced by fresh DMEM with 1% FBS. Seven days posttransfection (dpt), the supernatants containing viruses were blindly inoculated into fresh DF-1 cells followed by serial passages. The rescued viruses were all identified by PCR, IFA, and sequencing.
Indirect IFA.
DF-1 cells were first infected with different ALV variants for 6 days. Then, the infected or uninfected cells were fixed with an ice-cold acetone-ethanol (3:2) mixture for 5 min and washed using PBS. Then, the cells were incubated with mouse monoclonal antibody (MAb) JE9 specific to Gp85 of ALV-J for 45 min. After three washes with PBS, the cells were then incubated using goat anti-mouse antibody labeled with fluorescein isothiocyanate (FITC) for another 45 min. After three washes with PBS, the cell plates were observed under an inverted fluorescence microscope.
Growth curves for the rescued viruses.
The viral replication kinetics were measured as previously described (35). Briefly, DF-1 cells in 6-well plates were inoculated with different rescued ALV-J viruses at an MOI of 0.01. At 2 h postinfection (hpi), the cells were washed with PBS and fresh DMEM with 1% FBS was added to maintain the cells. Two hundred microliters of the infected cell cultures was collected at various time points, and an equal amount of fresh medium was added to each well. The viral titers were determined in DF-1 cells using IFA, and TCID50s of the supernatants were calculated using the Reed-Muench method. The final viral growth curves were constructed with GraphPad Prism 5 software.
Chicken infection study.
A total of 140 1-day-old SPF chickens were randomly divided into four groups (35 chickens per group; group I, chickens infected with EAV-HP; group II, chickens infected with 4817; group III, chickens infected with J1; group IV, chickens inoculated with PBS). All the chickens in the infection groups were inoculated with 104 TCID50s of the indicated virus in 0.2 ml PBS by leg muscle injection. At 7, 14, 21, 28, 35, 49, and 63 dpi, the body weights of chickens were monitored, and blood and cloacal swab samples were collected from the chickens for virus detection. At 7, 21, and 35 dpi, three chickens in each group were euthanized, and the liver, spleen, and kidney were collected for determination of viral titers.
Determination of viral titer in organs and blood.
The homogenates of liver, spleen, and kidney from the chickens were treated with 10× penicillin and streptomycin for 1 h and centrifuged to obtain supernatant. The virus-containing supernatants were inoculated into DF-1 cells, and the TCID50s of these supernatants were determined by the Reed-Muench method. For detection of viremia, the collected blood was inoculated into DF-1 cells and cultured for 6 days. Then, the infected DF-1 cells were fixed and detected by IFA using JE9 MAb as described above.
Sandwich ELISA for detection of P27 antigen.
Five hundred microliters of PBS was added to the collected cloacal swab sample. After three cycles of repeated freeze-thaw and vortexing, the samples were directly subjected to a sandwich ELISA against P27 that could efficiently detect the P27 of ALV as previously described (35). The OD650 value of the cutoff for the ELISA was 0.15.
IFA for detection of antibodies against ALV.
DF-1 cells in a 96-well plate were transfected with the J1 infectious clone for 48 h. Cells were fixed as described above. Chicken serum (1:400 diluted in PBS) samples were added to the plates, and rabbit anti-chicken antibody labeled with fluorescein isothiocyanate (FITC) was used as the secondary antibody. The result was observed under an inverted fluorescence microscope.
Indirect ELISA for detection of antibody against ALV P27.
The chicken serum samples were first diluted 1:500 in dilution buffer (phosphate-buffered saline with Tween 20 [PBST] with a 1/200 volume of 3 mg/ml of Escherichia coli BL21 total lysate) and incubated at 37°C for 30 min. One hundred microliters of treated samples was then directly subjected to the indirect ELISA detecting antibody against ALV P27 as previously described (36). The OD450 value of the cutoff for the ELISA was 0.5.
Statistical analysis.
All the result are presented as means ± standard deviations. The statistical analysis in this study was performed with a Student t test or one-way analysis of variance (ANOVA) using GraphPad 5 software. A P value of <0.05 was considered significant. *, **, and *** indicate P values of less than 0.05, 0.01, and 0.001, respectively.
Ethics approval and consent to participate.
All animal experiments complied with institutional animal care guidelines and were approved by the Animal Care Committee of Yangzhou University. At the end of the experiment, all the chickens were euthanized with CO2.
Availability of data and materials.
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
ACKNOWLEDGMENTS
We thank Yixin Wang (Shangdong Agriculture University, China) for kindly providing us with ALV-J infectious clone p-ALV-J1, and we thank Tuoyu Geng (College of Animal Science and Technology, Yangzhou University) for proofreading the manuscript.
We declare that we have no competing interests.
This study was supported by the National Natural Science Foundation of China (31972661 and 31472171), NCFC-RCUK-BBSRC (31761133002 and BB/R012865/1), the National Key Research & Development (R&D) Plan (2018YFD0500106 and 2016YFD0501605), the Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agrifood Safety and Quality (26116120), the Research Foundation for Talented Scholars in Yangzhou University, and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
J.Y. and T.L. designed the project. T.L., X.Y., C.L., J.Z., W.W., H.L., H.F., Q.X., W.G., and L.L. carried out the experiments. T.L., X.Y., J.X., and A.Q. analyzed the data. T.L., J.Y., and H.S. drafted the manuscript. J.Y. supervised all the experiments and participated in the data analysis. T.L., W.G., and A.Q. discussed and prepared the final report. All of the authors have read and approved the final manuscript.
REFERENCES
- 1.Payne LN, Brown SR, Bumstead N, Howes K, Frazier JA, Thouless ME. 1991. A novel subgroup of exogenous avian leukosis virus in chickens. J Gen Virol 72:801–807. doi: 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- 2.Payne LN, Howes K, Gillespie AM, Smith LM. 1992. Host range of Rous sarcoma virus pseudotype RSV(HPRS-103) in 12 avian species: support for a new avian retrovirus envelope subgroup, designated J. J Gen Virol 73:2995–2997. doi: 10.1099/0022-1317-73-11-2995. [DOI] [PubMed] [Google Scholar]
- 3.Dong X, Zhao P, Xu B, Fan J, Meng F, Sun P, Ju S, Li Y, Chang S, Shi W, Cui Z. 2015. Avian leukosis virus in indigenous chicken breeds, China. Emerg Microbes Infect 4:e76. doi: 10.1038/emi.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adkins HB, Blacklow SC, Young JA. 2001. Two functionally distinct forms of a retroviral receptor explain the nonreciprocal receptor interference among subgroups B, D, and E avian leukosis viruses. J Virol 75:3520–3526. doi: 10.1128/JVI.75.8.3520-3526.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Payne LN, Nair V. 2012. The long view: 40 years of avian leukosis research. Avian Pathol 41:11–19. doi: 10.1080/03079457.2011.646237. [DOI] [PubMed] [Google Scholar]
- 6.Gao YL, Qin LT, Pan W, Wang YQ, Le Qi X, Gao HL, Wang XM. 2010. Avian leukosis virus subgroup J in layer chickens, China. Emerg Infect Dis 16:1637–1638. doi: 10.3201/eid1610.100780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng Z, Liu J, Cui Z, Zhang L. 2010. Tumors associated with avian leukosis virus subgroup J in layer hens during 2007 to 2009 in China. J Vet Med Sci 72:1027–1033. doi: 10.1292/jvms.09-0564. [DOI] [PubMed] [Google Scholar]
- 8.Weiss RA, Vogt PK. 2011. 100 years of Rous sarcoma virus. J Exp Med 208:2351–2355. doi: 10.1084/jem.20112160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chesters PM, Howes K, Petherbridge L, Evans S, Payne LN, Venugopal K. 2002. The viral envelope is a major determinant for the induction of lymphoid and myeloid tumours by avian leukosis virus subgroups A and J, respectively. J Gen Virol 83:2553–2561. doi: 10.1099/0022-1317-83-10-2553. [DOI] [PubMed] [Google Scholar]
- 10.Bai J, Payne LN, Skinner MA. 1995. HPRS-103 (exogenous avian leukosis virus, subgroup J) has an env gene related to those of endogenous elements EAV-0 and E51 and an E element found previously only in sarcoma viruses. J Virol 69:779–784. doi: 10.1128/JVI.69.2.779-784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan W, Gao Y, Qin L, Ni W, Liu Z, Yun B, Wang Y, Qi X, Gao H, Wang X. 2012. Genetic diversity and phylogenetic analysis of glycoprotein GP85 of ALV-J isolates from Mainland China between 1999 and 2010: coexistence of two extremely different subgroups in layers. Vet Microbiol 156:205–212. doi: 10.1016/j.vetmic.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 12.Jiang L, Zeng X, Hua Y, Gao Q, Fan Z, Chai H, Wang Q, Qi X, Wang Y, Gao H, Gao Y, Wang X. 2014. Genetic diversity and phylogenetic analysis of glycoprotein gp85 of avian leukosis virus subgroup J wild-bird isolates from Northeast China. Arch Virol 159:1821–1826. doi: 10.1007/s00705-014-2004-8. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Cui Z. 2006. Evolution of gp85 gene of subgroup J avian leukosis virus under the selective pressure of antibodies. Sci China C Life Sci 49:227–234. doi: 10.1007/s11427-006-0227-y. [DOI] [PubMed] [Google Scholar]
- 14.Cui Z, Du Y, Zhang Z, Silva RF. 2003. Comparison of Chinese field strains of avian leukosis subgroup J viruses with prototype strain HPRS-103 and United States strains. Avian Dis 47:1321–1330. doi: 10.1637/6085. [DOI] [PubMed] [Google Scholar]
- 15.Thu WL, Wang CH. 2003. Phylogenetic analysis of subgroup J avian leucosis virus from broiler and native chickens in Taiwan during 2000–2002. J Vet Med Sci 65:325–328. doi: 10.1292/jvms.65.325. [DOI] [PubMed] [Google Scholar]
- 16.Ye J, Fan Z, Shang J, Tian X, Yang J, Chen H, Shao H, Qin A. 2015. ALV-J GP37 molecular analysis reveals novel virus-adapted sites and three tyrosine-based Env species. PLoS One 10:e0122887. doi: 10.1371/journal.pone.0122887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Y, Cui Z, Qin A. 1999. Subgroup J avian leukosis viruses in China. China Poult Sci 3:1. [Google Scholar]
- 18.Wang P, Lin L, Li H, Yang Y, Huang T, Wei P. 2018. Diversity and evolution analysis of glycoprotein GP85 from avian leukosis virus subgroup J isolates from chickens of different genetic backgrounds during 1989–2016: coexistence of five extremely different clusters. Arch Virol 163:377–389. doi: 10.1007/s00705-017-3601-0. [DOI] [PubMed] [Google Scholar]
- 19.Meng F, Li Q, Zhang Y, Zhang Z, Tian S, Cui Z, Chang S, Zhao P. 2018. Characterization of subgroup J avian leukosis virus isolated from Chinese indigenous chickens. Virol J 15:33. doi: 10.1186/s12985-018-0947-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su Q, Li Y, Li W, Cui S, Tian S, Cui Z, Zhao P, Chang S. 2018. Molecular characteristics of avian leukosis viruses isolated from indigenous chicken breeds in China. Poult Sci 97:2917–2925. doi: 10.3382/ps/pex367. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Meng F, Li W, Wang Y, Chang S, Zhao P, Cui Z. 2018. Characterization of avian leukosis virus subgroup J isolated between 1999 and 2013 in China. Poult Sci 97:3532–3539. doi: 10.3382/ps/pey241. [DOI] [PubMed] [Google Scholar]
- 22.Borisenko L. 2003. Avian endogenous retroviruses. Folia Biol 49:177–182. [PubMed] [Google Scholar]
- 23.Weiss RA. 2006. The discovery of endogenous retroviruses. Retrovirology 3:67. doi: 10.1186/1742-4690-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borisenko L, Rynditch AV. 2004. Complete nucleotide sequences of ALV-related endogenous retroviruses available from the draft chicken genome sequence. Folia Biol 50:136–141. [PubMed] [Google Scholar]
- 25.Vanhaesebroeck B, Stephens L, Hawkins P. 2012. PI3K signalling: the path to discovery and understanding. Nat Rev Mol Cell Biol 13:195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- 26.Okkenhaug K, Vanhaesebroeck B. 2003. PI3K in lymphocyte development, differentiation and activation. Nat Rev Immunol 3:317–330. doi: 10.1038/nri1056. [DOI] [PubMed] [Google Scholar]
- 27.Humphrey MB, Lanier LL, Nakamura MC. 2005. Role of ITAM-containing adapter proteins and their receptors in the immune system and bone. Immunol Rev 208:50–65. doi: 10.1111/j.0105-2896.2005.00325.x. [DOI] [PubMed] [Google Scholar]
- 28.Ravetch JV, Lanier LL. 2000. Immune inhibitory receptors. Science 290:84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 29.Justice JF IV, Morgan RW, Beemon KL. 2015. Common viral integration sites identified in avian leukosis virus-induced B-cell lymphomas. mBio 6:e01863-15. doi: 10.1128/mBio.01863-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Gao Y, Wang Y, Qin L, Qi X, Qu Y, Gao H, Wang X. 2012. A 205-nucleotide deletion in the 3′ untranslated region of avian leukosis virus subgroup J, currently emergent in China, contributes to its pathogenicity. J Virol 86:12849–12860. doi: 10.1128/JVI.01113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zavala G, Cheng S, Jackwood MW. 2007. Molecular epidemiology of avian leukosis virus subgroup J and evolutionary history of its 3′ untranslated region. Avian Dis 51:942–953. doi: 10.1637/0005-2086(2007)51[942:MEOALV]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 32.Gao Y, Guan X, Liu Y, Li X, Yun B, Qi X, Wang Y, Gao H, Cui H, Liu C, Zhang Y, Wang X, Gao Y. 2015. An avian leukosis virus subgroup J isolate with a Rous sarcoma virus-like 5’-LTR shows enhanced replication capability. J Gen Virol 96:150–158. doi: 10.1099/vir.0.071290-0. [DOI] [PubMed] [Google Scholar]
- 33.Silva RF, Fadly AM, Hunt HD. 2000. Hypervariability in the envelope genes of subgroup J avian leukosis viruses obtained from different farms in the United States. Virology 272:106–111. doi: 10.1006/viro.2000.0352. [DOI] [PubMed] [Google Scholar]
- 34.Shao H, Fan Z, Wan Z, Tian X, Chen H, Perez DR, Qin A, Ye J. 2015. An efficient and rapid influenza gene cloning strategy for reverse genetics system. J Virol Methods 222:91–94. doi: 10.1016/j.jviromet.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Li T, Xie J, Lv L, Sun S, Dong X, Xie Q, Liang G, Xia C, Shao H, Qin A, Ye J. 2018. A chicken liver cell line efficiently supports the replication of ALV-J possibly through its high level viral receptor and efficient protein expression system. Vet Res 49:41. doi: 10.1186/s13567-018-0537-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu Y, Qian K, Shen H, Jin W, Qin A. 2011. Development and validation of an indirect enzyme-linked immunosorbent assay for the detection of avian leukosis virus antibodies based on a recombinant capsid protein. J Vet Diagn Invest 23:991–993. doi: 10.1177/1040638711416966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.