Four influenza pandemics in humans (the Spanish flu of 1918 [H1N1], the Asian flu of 1957 [H2N2], the Hong Kong flu of 1968 [H3N2], and the swine origin flu of 2009 [H1N1]) are all proposed to have been caused by avian or swine influenza viruses that acquired virulence factors through adaptive mutation or reassortment with circulating human viruses. Currently, influenza viruses circulating in animals are repeatedly transmitted to humans, posing a significant threat to public health. However, the molecular properties accounting for interspecies transmission of influenza viruses remain unclear. In the present study, we demonstrated that PA-X plays an important role in cross-species transmission of influenza viruses. At least three human-specific amino acid substitutions in PA-X dramatically enhanced the adaptation of animal influenza viruses in mammals. In particular, PA-X 195K might have contributed to cross-species transmission of H7N9, H5N6, and H1N1/2009 viruses from animal reservoirs to humans.
KEYWORDS: influenza virus, PA-X, cross-species transmission, virulence
ABSTRACT
In the 21st century, the emergence of H7N9 and H1N1/2009 influenza viruses, originating from animals and causing severe human infections, has prompted investigations into the genetic alterations required for cross-species transmission. We previously found that replacement of the human-origin PA gene segment in avian influenza virus (AIV) could overcome barriers to cross-species transmission. Recently, it was reported that the PA gene segment encodes both the PA protein and a second protein, PA-X. Here, we investigated the role of PA-X. We found that an H9N2 avian influenza reassortant virus bearing a human-origin H1N1/2009 PA gene was attenuated in mice after the loss of PA-X. Reverse genetics analyses of PA-X substitutions conserved in human influenza viruses indicated that R195K, K206R, and P210L substitutions conferred significantly increased replication and pathogenicity on H9N2 virus in mice and ferrets. PA-X R195K was present in all human H7N9 and H1N1/2009 viruses and predominated in human H5N6 viruses. Compared with PA-X 195R, H7N9 influenza viruses bearing PA-X 195K showed increased replication and transmission in ferrets. We further showed that PA-X 195K enhanced lung inflammatory responses, potentially due to decreased host shutoff function. A competitive transmission study in ferrets indicated that 195K provides a replicative advantage over 195R in H1N1/2009 viruses. In contrast, PA-X 195K did not influence the virulence of H9N2 AIV in chickens, suggesting that the effects of the substitution were mammal specific. Therefore, future surveillance efforts should scrutinize this region of PA-X because of its potential impact on cross-species transmission of influenza viruses.
IMPORTANCE Four influenza pandemics in humans (the Spanish flu of 1918 [H1N1], the Asian flu of 1957 [H2N2], the Hong Kong flu of 1968 [H3N2], and the swine origin flu of 2009 [H1N1]) are all proposed to have been caused by avian or swine influenza viruses that acquired virulence factors through adaptive mutation or reassortment with circulating human viruses. Currently, influenza viruses circulating in animals are repeatedly transmitted to humans, posing a significant threat to public health. However, the molecular properties accounting for interspecies transmission of influenza viruses remain unclear. In the present study, we demonstrated that PA-X plays an important role in cross-species transmission of influenza viruses. At least three human-specific amino acid substitutions in PA-X dramatically enhanced the adaptation of animal influenza viruses in mammals. In particular, PA-X 195K might have contributed to cross-species transmission of H7N9, H5N6, and H1N1/2009 viruses from animal reservoirs to humans.
INTRODUCTION
Influenza viruses circulating in animal reservoirs represent a significant public health threat as a potential source of pandemic viruses. The influenza pandemics in humans that occurred in 1918, 1957, 1968, and 2009 are all proposed to have been caused by avian influenza virus (AIV) or swine influenza virus (SIV) that acquired virulence factors through adaptive mutation or reassortment with human influenza viruses to overcome the species barrier (1–3). For an influenza virus to cause a pandemic, it must be (i) antigenically novel to the human immune system, (ii) virulent in human hosts, and (iii) transmissible from person to person (4). As all animal influenza viruses possess novel antigenicity in humans, identification of molecular determinants of virulence and transmission would allow rational steps toward early detection and control of novel influenza viruses emerging at the animal-human interface. The most recent severe outbreak of animal influenza viruses in humans was the H7N9 outbreak that emerged in 2013 in China, which caused 1,568 human infections, with a mortality rate of approximately 39% reported by July 2019 (http://www.fao.org/ag/againfo/programmes/en/empres/H7N9/situation_update.html). Although efficient human-to-human transmission of H7N9 influenza viruses was not observed, some human isolates were highly transmissible in ferrets via respiratory droplet transmission (5). The novel reassortant H7N9 virus comprised the waterfowl H7 and N9 genes, and all of its six internal genes originated from an H9N2 virus (6). It was proposed that the adaptability of H9N2 viruses in mammals might confer on H7N9 viruses the ability to easily infect humans (7, 8). However, the specific molecular properties that account for the infection and resulting severe illness caused by H7N9 viruses in humans remain poorly defined.
Influenza A virus is an enveloped negative-strand RNA virus whose genome comprises eight viral RNA (vRNA) segments. The polymerase acidic (PA) gene is a significant determinant of viral tropism, and introducing a human-origin PA gene dramatically enhanced the adaptation of AIV to mammalian cells and animals (9–11). Recently, it was revealed that the PA gene encodes both the PA protein and a novel protein, PA-X (12, 13). PA-X is a fusion protein containing the endonuclease domain of the viral PA protein and a unique C-terminal domain derived from the +1 frameshift open reading frame (X-ORF). PA-X plays an important role in host shutoff that inhibits cellular gene expression and hinders the induction of antiviral responses (14–19). Previous studies using global gene expression profiling reported that PA-X-deficient influenza viruses markedly upregulated the expression of apoptosis-, inflammation-, and immune response-related genes in the lungs of mice and chickens (20–23). Comprehensive genetic analysis showed that the PA-X genes of diverse influenza A viruses can be divided into two groups according to ORF length (24). Avian, equine, and human seasonal influenza viruses express a full-length PA-X protein, whereas H1N1/2009 and canine and most simian immunodeficiency viruses (SIVs) have a stop codon at X-ORF position 42, resulting in a truncated PA-X protein. It was further demonstrated that H1N1/2009, avian H5N1, and avian H9N2 viruses bearing full-length PA-X were more virulent and caused more severe inflammatory responses in mice and birds (25–28). However, SIVs bearing truncated PA-X showed increased replication and pathogenicity in swine (25), indicating that associations between PA-X length and virulence may be host dependent.
Here, we evaluated two hypotheses: (i) PA-X contributes to cross-species transmission of influenza virus, and (ii) genetic changes in the PA gene that do not alter the coding sequence of the PA protein but result in PA-X amino acid substitutions play a crucial role in cross-species transmission of influenza viruses. Previously, we demonstrated that a reassortant virus bearing the H1N1/2009 PA gene segment in a H9N2 background showed significantly increased replication ability and pathogenicity in mice compared with the corresponding wild-type H9N2 (wtH9N2) virus (9). Here, we found that the virulence of this reassortant virus was attenuated to a level similar to that of wtH9N2 virus after the loss of PA-X expression, indicating that PA-X was crucial for the adaptation of AIVs to mammalian hosts. We further revealed that individually introducing several human-specific PA-X substitutions into the X-ORF of an H9N2 virus significantly increased its replication and pathogenicity in mice and ferrets. Among these substitutions, PA-X 195K, which is present in all human H7N9 and H1N1/2009 viruses, enhanced the immunopathology and respiratory droplet transmission of H7N9 viruses and conferred selective advantages on H1N1/2009 viruses in mammals. Therefore, both of our hypotheses were confirmed by these data. Together, our results show that specific adaptations of the PA-X protein may represent a strategy used by influenza viruses to enhance replication, pathogenicity, and transmissibility in human hosts.
RESULTS
PA-X contributes to the increased virulence of H9N2 AIV in mammals.
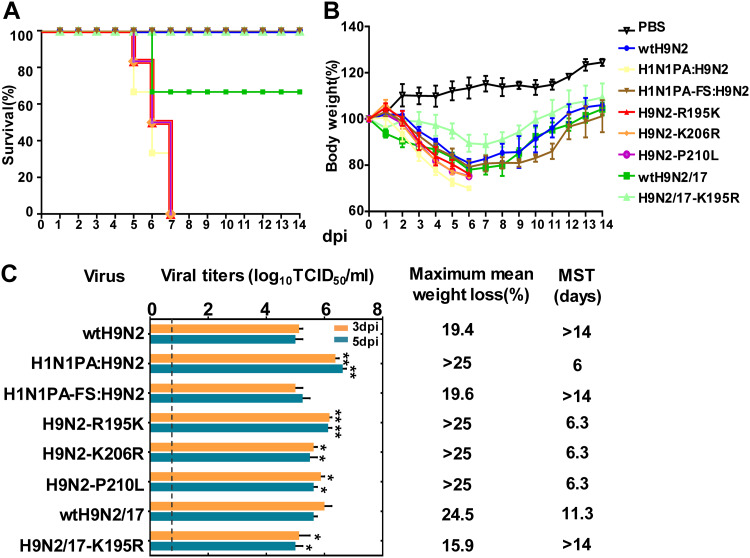
In our previous study, we demonstrated that introduction of the H1N1/2009 PA gene into an H9N2 AIV (A/chicken/Hebei/LC/2008 [wtH9N2]) enhanced its virulence in mice: the reassortant virus (H1N1PA:H9N2) had a 50% lethal dose (LD50) 1.5-fold lower than that of the wtH9N2 virus (9). To investigate whether the PA-X protein encoded by the PA gene contributed to this effect, we generated an H1N1PA-FS:H9N2 virus in which PA-X expression was abolished without altering the PA ORF. As shown in Fig. 1, 100% of mice inoculated with 106 times the 50% tissue culture infective dose (TCID50) of the H1N1PA:H9N2 virus died by 7 days postinfection (dpi). In contrast, the H1N1PA-FS:H9N2 virus had significantly lower pathogenicity, with infection resulting in only 19.6% of the maximum body weight loss of mice infected with H1N1PA:H9N2, similar to wtH9N2 virus (Fig. 1). These results indicated that PA-X played an important role in the adaptation of AIVs to mammalian hosts.
FIG 1.
Virulence of H9N2 viruses in mice. Mice were intranasally inoculated with 106 TCID50 of each test virus or with PBS. (A and B) The survival rates and body weights of six inoculated mice were measured and are represented as percentages of the weight on the day of inoculation (day 0). (C) Mean viral titers in the lungs of three mice. The error bars indicate standard deviations of the virus titers in the lungs. The dashed line indicates the lower limit of detection. Maximum mean weight loss (percent weight loss relative to 0 dpi) and the mean survival time (MST) are shown. Statistical significance relative to the corresponding wild-type H9N2 virus was assessed by two-way ANOVA (*, P < 0.05; **, P < 0.01).
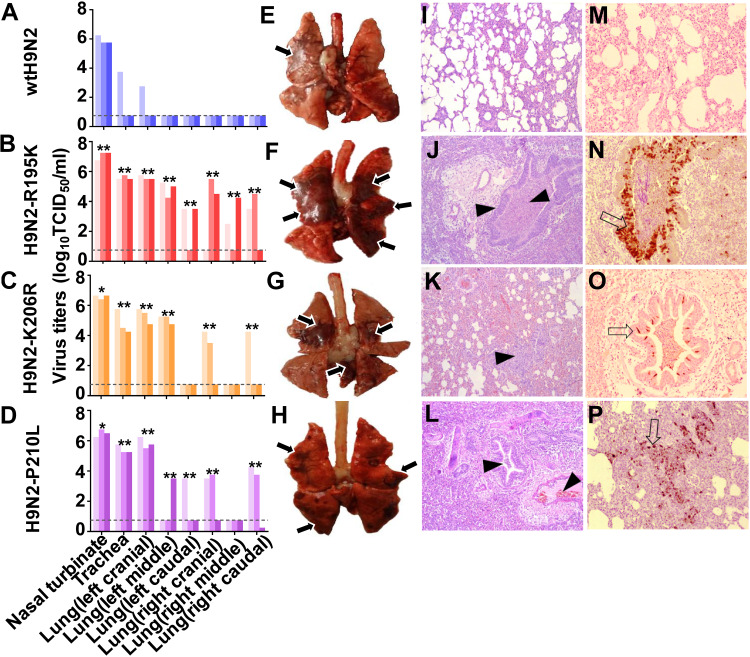
To further identify the amino acid residues responsible for the increased virulence of avian-human reassortant viruses bearing human-origin PA-X proteins, we compared the sequences of the unique X domain of PA-X proteins among avian and human influenza viruses available in the National Center for Biotechnology Information database (https://www.ncbi.nlm.nih.gov). Three amino acid substitutions were identified by the comparison of avian and human influenza viruses: R195K, K206R, and P210L (avian versus human) (Table 1). We introduced these three substitutions individually into A/chicken/Hebei/LC/2008 using reverse genetics techniques. Although none of them (R195K, K206R, or P210L) altered the PA ORF, they significantly increased viral replication and pathogenicity (P < 0.05) (Fig. 1). We further assessed the pathogenicity of H9N2-R195K, H9N2-K206R, and H9N2-P210L viruses compared with wtH9N2 in ferrets (Fig. 2). Each group of three ferrets was infected intranasally with 106 TCID50 of the viruses. The nasal turbinates, trachea, and various lung lobes were collected from each ferret at 5 dpi for virus enumeration. The wtH9N2 virus was detected in the nasal turbinates of all three ferrets, with a mean titer of 5.9 log10 TCID50/ml, and was also detected in the trachea and lung (left cranial) of one of three ferrets. In contrast, virus could be detected in the nasal turbinates, tracheas, and some lung lobes of the ferrets inoculated with H9N2-R195K, H9N2-K206R, and H9N2-P210L, with titers ranging from 2.3 to 7.3 log10 TCID50/ml (Fig. 2B, C, and D). Severe bronchopneumonia was observed via gross and histopathological lesions in the lung tissues of ferrets inoculated with H9N2-R195K, H9N2-K206R, and H9N2-P210L viruses, which were characterized by consolidation, hemorrhages, and edema (Fig. 2F to H and J to L). In contrast, little or no lung consolidation was observed in ferrets infected with wtH9N2 virus (Fig. 2E and I). The viral nucleoproteins (NPs) of H9N2-R195K, H9N2-K206R, and H9N2-P210L viruses, but not wtH9N2 virus, could be readily detected in the bronchioles, terminal bronchioles, and alveoli of infected ferrets (Fig. 2M to P). These results indicated that a single amino acid substitution in PA-X could significantly enhance the virulence of AIVs in mammals.
TABLE 1.
PA-X polymorphism frequencies at selected positions of the X domain
| Site in PA-X | Polymorphism (frequency [%]) |
||
|---|---|---|---|
| Avian influenza virus | Human influenza virus |
||
| H1N1/2009 | Seasonal | ||
| 195 | R (55.6) | K (100.0) | K (99.0) |
| K (44.3) | R (1.0) | ||
| E, N (≤0.1) | |||
| 206 | K (98.2) | R (95.4) | K (98.7) |
| R (1.5) | K (4.4) | R (1.3) | |
| E, I, M, S, L, T (≤0.3) | L, Q (≤0.2) | ||
| 210 | P (95.2) | L (96.7) | L (84.9) |
| Q (3.1) | Q (2.7) | P (10.9) | |
| L (1.6) | A, C, P (≤0.6) | Q (4.1) | |
| A, C, R, S, T (≤0.1) | R, I (≤0.1) | ||
FIG 2.
Viral replication and pathological findings in ferrets at 5 dpi. (A to D) Virus replication in the indicated organ. Three ferrets from each group were infected intranasally with 106 TCID50 of each virus and euthanized at 5 dpi for virus enumeration. Viral titers in the nasal turbinates, tracheas, and lungs were determined by the infection of MDCK cells. The dashed lines indicate the lower limit of detection. Statistical significance relative to H7N9-K195R virus was assessed using two-way ANOVA (*, P < 0.05; **, P < 0.01). (E to H) Representative bronchopneumonia changes in the gross lesions in lung tissues. Severe bronchopneumonia with hemorrhages, edema, and diffuse consolidation were evident in ferrets inoculated with H9N2-R195K, H9N2-K206R, or H9N2-P210L virus (arrows). (I to L) Representative histopathological changes in H&E-stained lung tissues. The wtH9N2 virus induced moderate bronchopneumonia, whereas H9N2-R195K, H9N2-K206R, and H9N2-P210L viruses induced severe bronchopneumonia with hemorrhages, edema, and diffuse consolidation (arrowheads). (M to P) Immunohistochemical staining of lung sections. Extensive viral NP localization in bronchioles and alveoli was observed for H9N2-R195K, H9N2-K206R, and H9N2-P210L viruses (arrows). (I to P) The images were taken by microscopy at ×200 magnification.
Increased frequency of PA-X 195K in different influenza viruses.
Because the R195K, K206R, and P210L substitutions increased the virulence of H9N2 AIVs bearing the internal genes of H7N9 influenza viruses in mammals, we further analyzed the frequencies of these substitutions among H9N2 and H7N9 influenza viruses. Our sequence searches revealed that PA-X 206R and 210L were infrequent in H9N2 and H7N9 influenza viruses; however, an obvious increase of PA-X 195K was observed in both H9N2 and H7N9 influenza viruses. Prior to 2000, nearly all H9N2 viruses possessed the avian-like PA-X 195R residue. From 2001 to 2008, 34.7% of H9N2 influenza viruses bore the PA-X 195K residue. The percentage of H9N2 viruses carrying PA-X 195K increased dramatically to 80.5% over the period from 2013 to 2015, eventually coming to predominate (Fig. 3A). Since PA-X 195K was increased and became predominant in recent H9N2 viruses, we further evaluated the effect of PA-X K195R in a later H9N2 virus, A/chicken/Guangdong/28/2017 (wtH9N2/17). It was observed that PA-X K195R also considerably reduced the replication ability and pathogenicity of this strain in mice (Fig. 1). During the first wave of the emergence of H7N9 in China, 64.2% of avian H7N9 isolates bore the human-specific PA-X residue 195K (Fig. 3B). The percentage of isolates bearing PA-X 195K reached 100% in the second wave of the H7N9 outbreak, during which more than twice the number of human cases and deaths occurred compared with the first wave. The percentage of H7N9 isolates bearing PA-X 195K has remained at 100% since that time. Importantly, all of the human H7N9 influenza viruses observed during the five waves of the outbreak bore PA-X 195K. Additionally, novel H5N6 reassortant viruses have caused a total of 22 human infections in China from 2014 to 2018 (https://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en). Interestingly, 78.9% of these human H5N6 viruses bore PA-X 195K; in contrast, 18.5% of avian H5N6 isolates bore 195K over the same period in China (Fig. 3C). Swine origin H1N1/2009 viruses bore PA genes of North American AIV origin (29). Interestingly, we noticed that 57.0% of SIVs with PA genes of North American AIV origin bore 195K in PA-X compared with 100% of H1N1/2009 viruses (Fig. 3D). Given these findings, we hypothesized that PA-X 195K may be important for the adaptation of influenza viruses to mammalian hosts and for cross-species transmission from avian reservoirs to human populations.
FIG 3.
Frequencies of the PA-X mutations 195K and 195R in different influenza viruses. (A) The actual numbers of PA-X sequences of avian influenza viruses are as follows: 1994 to 2000, n = 83; 2001 to 2008, n = 303; 2009 to 2012, n = 422; 2013 to 2015, n = 421; 2016 to 2018, n = 189. (B) The numbers of PA-X sequences in avian H7N9 influenza viruses were as follows: wave 1, n = 54; wave 2, n = 346; wave 3, n = 264; wave 4, n = 244; wave 5, n = 105. The actual numbers of PA-X sequences in human H7N9 influenza viruses were as follows: wave 1, n = 94; wave 2, n = 207; wave 3, n = 127; wave 4, n = 118; wave 5, n = 356. (C) The numbers of PA-X sequences in H5N6 influenza viruses were as follows: avian H5N6, n = 292; human H5N6, n = 31. (D) The numbers of PA-X sequences in SIVs with the North American avian PA gene and H1N1/2009 were 24 and 183, respectively.
PA-X R195K enhances the replication of H7N9 influenza viruses in human cells.
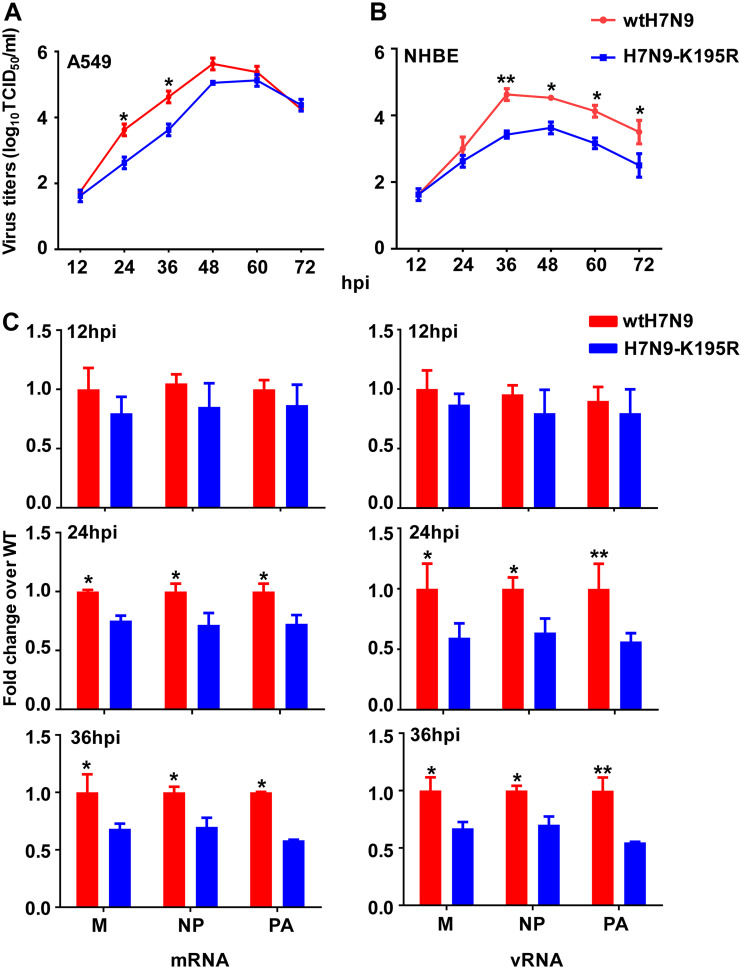
To determine the role of PA-X 195K in the virulence of H7N9 viruses, we compared a human H7N9 virus (A/Anhui/1/2013; wtH7N9 with PA-X 195K) (30) with a virus containing a back-mutation (K195R) in PA-X but an unaltered PA sequence (H7N9-K195R). We assessed the replication kinetics of these viruses in human lung epithelial (A549) cells and normal human bronchial epithelial (NHBE) cells by infecting them at multiplicities of infection (MOI) of 0.01 and 0.001, respectively. At early time points following the infection of A549 cells, wtH7N9 and H7N9-K195R viruses showed similar growth kinetics, whereas at 24 h postinfection (hpi) and 36 hpi, the titers of H7N9-K195R in the culture medium were significantly lower than those of wtH7N9 (P < 0.05) (Fig. 4A). Similarly, the titers of wtH7N9 viruses in NHBE cells were significantly (up to 10-fold) higher than those of H7N9-K195R viruses at 36 hpi (P < 0.01) (Fig. 4B).
FIG 4.
Viral growth kinetics and abundances of mRNAs and vRNAs. (A) Multistep growth curves of H7N9 viruses in A549 cells inoculated at an MOI of 0.01. (B) Multistep growth curves of H7N9 viruses in NHBE cells inoculated at an MOI of 0.001. (C) Relative abundances of viral M, NP, and PA mRNAs and vRNAs. A549 cells were infected with wtH7N9 and H7N9-K195R virus at an MOI of 0.01. The abundances of mRNAs and vRNAs encoding the M, NP, and PA proteins of the indicated viruses are expressed as a fold change in copy number compared with the wild-type group at the corresponding time point. Data are presented as the means ± standard deviations of three independent experiments. Statistical significance was assessed using two-way ANOVA (*, P < 0.05; **, P < 0.01).
To further understand whether the decreased viral growth associated with PA-X K195R was due to differences in the efficiencies of viral transcription and replication, we examined the levels of mRNA and vRNA in A549 cells infected with wtH7N9 or H7N9-K195R virus. Quantitative real-time PCR (qRT-PCR) analyses using mRNA- or vRNA-specific primers showed that levels of mRNAs encoding the matrix (M), NP, and PA proteins in wtH7N9-infected cells were dramatically increased (1.25- to 1.72-fold) from 24 to 36 hpi compared with H7N9-K195R-infected cells (P < 0.05) (Fig. 4C). A549 cells infected with wtH7N9 virus produced at least 1.5-fold higher levels of M, NP, and PA vRNAs from 24 to 36 hpi compared with H7N9-K195R-infected cells (P < 0.05) (Fig. 4C).
Taken together, these results indicated that H7N9 viruses carrying PA-X 195K showed an increased ability to replicate in human cells, as well as increased genome transcription and replication, compared with viruses carrying PA-X 195R.
PA-X 195K reduces the shutdown of host protein synthesis and promotes the induction of inflammatory responses.
PA-X protein plays a major role in host shutoff (31–35), which suppresses host protein synthesis in infected cells. To determine the impact of PA-X 195K in virus-induced shutoff of exogenous protein expression, we compared the activities of PA-X 195K and PA-X 195R from H9N2, H7N9, or H1N1/2009 virus in suppressing nonviral protein synthesis. Following cotransfection of green fluorescent protein (GFP)-encoding cDNA or a pRL-SV40 vector encoding Renilla luciferase, together with expression vectors encoding PA-X (195K) or PA-X (195R), we found that GFP and Renilla luciferase expression levels were significantly increased in the presence of PA-X 195K. This result indicated that PA-X 195K conferred a decreased ability for host shutoff compared with PA-X 195R (Fig. 5A and B).
FIG 5.
Effects of PA-X substitutions in suppressing nonviral protein expression and cytokine responses. (A) GFP expression measured at 24 h posttransfection. 293T cells were cotransfected with expression vectors encoding GFP, together with expression vectors carrying either the PA-X (195K) or PA-X (195R) gene of the indicated virus. Untransfected 293T cells or 293T cells cotransfected with GFP expression plasmids and the empty pCAGGS vector served as controls. (B) Luciferase expression from a pRL-SV40 vector encoding Renilla luciferase in 293T cells cotransfected with either empty pCAGGS plasmid (Vector) or pCAGGS encoding the indicated genes after 24 h. The results are shown as means and standard deviations (n = 3 biological replicates; n = 3 technical replicates). (C) Effect of H7N9 PA-X (195K) or H7N9 PA-X (195R) on cytokine mRNA abundance in A549 cells. Total RNA was extracted from transfected cells at 24 h posttransfection, and the abundances of IL-1β, IL-6, IL-12, TNF-α, and NF-κB mRNAs were quantified by qRT-PCR. (D) Effect of PA-X on cytokine mRNA abundance in A549 cells. A549 cells were infected with wtH7N9 virus or H7N9-K195R virus at an MOI of 1. Total RNA was extracted from infected cells at 12 hpi, and the abundances of IL-1β, IL-6, IL-12, TNF-α, and NF-κB mRNAs were quantitated by qRT-PCR. The data are presented as the means and standard deviations of the results of three independent experiments. Statistical significance was assessed using two-way ANOVA (*, P < 0.05; **, P < 0.01).
Previous studies indicated that the function of PA-X in host shutoff was associated with its impact on the innate immune response (20). Since PA-X 195K led to decreased inhibition of host protein synthesis, we explored the direct effects of PA-X 195K on the inflammatory response by expressing H7N9 PA-X (195K) or H7N9 PA-X (195R) protein ectopically in A549 cells and stimulating an innate immune response using lipopolysaccharide (LPS). The abundances of mRNAs encoding interleukin 1β (IL-1β), IL-6, IL-12, tumor necrosis factor alpha (TNF-α), and nuclear factor κB (NF-κB) in A549 cells were analyzed by qRT-PCR. We found that IL-1β, IL-6, IL-12, TNF-α, and NF-κB transcript levels in cells expressing H7N9 PA-X (195K) were about 2- to 4-fold higher than those in cells expressing H7N9 PA-X (195R) (P < 0.01) (Fig. 5C). We also analyzed the abundances of IL-1β, IL-6, IL-12, TNF-α, and NF-κB mRNAs in A549 cells infected with wtH7N9 virus or H7N9-K195R virus at an MOI of 1. The abundance of mRNAs encoding IL-6 in wtH7N9-infected cells was 31.4-fold higher than that in H7N9-K195R-infected cells at 12 hpi (P < 0.01). Moreover, the abundances of IL-1β, IL-12, TNF-α, and NF-κB transcripts were all significantly increased in wtH7N9-infected cells (P < 0.05) (Fig. 5D).
In general, these findings illustrated that PA-X 195K confers decreased efficiency of host shutoff and induces significantly higher proinflammatory cytokine expression in vitro.
PA-X R195K increases the replication and pathogenicity of H7N9 human influenza virus in mice and ferrets.
To examine the effects of PA-X 195K on the replication and pathogenicity of H7N9 in mice, BALB/c mice were inoculated with 105 TCID50 or 106 TCID50 of wtH7N9 or H7N9-K195R. When 105 TCID50 was used for inoculation, all of the mice infected with wtH7N9 died by 9 dpi, whereas 2 of 5 mice infected with H7N9-K195R died over the same period (a 40% mortality rate) (Fig. 6A and B). When 106 TCID50 was used for inoculation, all of the mice infected with wtH7N9 died by 5 dpi, whereas it took until 8 dpi for 100% of the mice infected with H7N9-K195R to succumb to their infections (Fig. 6C and D). Viral titers in the lungs were at least 10-fold higher in mice infected with 105 TCID50 of the wtH7N9 virus than in mice infected with the H7N9-K195R virus at 3 and 5 dpi (P < 0.05) (Fig. 6E). Furthermore, as shown in Fig. 6F, infection with the H7N9-K195R virus caused mild pathology, whereas severe bronchopneumonia was observed in hematoxylin-eosin (H&E)-stained lung tissues of mice infected with the wtH7N9 virus. We assessed the protein levels of IL-1β, IL-6, TNF-α, gamma interferon (IFN-γ), monocyte chemoattractant protein 1 (MCP-1), macrophage inflammatory protein 1α (MIP-1α), and chemokines (KC) in the lungs of infected mice at 1, 3, and 5 dpi. At 1 dpi, mice infected with wtH7N9 viruses showed levels of all cytokines dramatically higher than those of mice infected with H7N9-K195R (Fig. 6G), consistent with the increased pathogenicity and replication of the former virus. Quantitation of immune cells in bronchoalveolar lavage fluid (BALF) at 3 dpi showed that mice infected with H7N9-K195R had significantly reduced numbers of white blood cells compared with wtH7N9-infected mice (Fig. 7A). This difference was also apparent when specifically assessing the influx of either neutrophils or macrophages in BALF (Fig. 7B and C). However, no significant differences in the numbers of BALF dendritic cells were observed between wtH7N9- and H7N9-K195R-infected mice (Fig. 7D), suggesting that this cell type was not a major contributor to the increase in white blood cells.
FIG 6.
Mortality and body weight changes, viral titers, histopathological changes, and cytokine levels in the lungs of mice infected with H7N9 viruses. (A and C) Survival of mice (n = 5/group) infected with 105 TCID50 or 106 TCID50 of wtH7N9 or H7N9-K195R virus. (B and D) Body weight changes in mice inoculated with 105 TCID50 or 106 TCID50 of wtH7N9 or H7N9-K195R virus. (E) Viral titers in the lungs at 3 and 5 dpi from three mice in each group infected with 105 TCID50 wtH7N9 or H7N9-K195R virus. (F) H&E-stained lung sections from mice infected with 105 TCID50 of the indicated viruses at 5 dpi. The H7N9-K195R virus caused mild pathology, whereas edema of the bronchial and vessel walls and infiltration of inflammatory cells, including lymphocytes and monocytes, were observed in wtH7N9-infected mice (arrows). The image was obtained by microscopy at ×200 magnification. (G) Detection of cytokine/chemokine proteins in the lungs of mice infected with 105 TCID50 wtH7N9 or H7N9-K195R virus at 1, 3, and 5 dpi (n = 3/group). The data are presented as the means ± standard deviations of the results for three mice. Statistical significance was assessed using two-way ANOVA (*, P < 0.05; **, P < 0.01).
FIG 7.
Lung cell characterization following infection with wtH7N9 and H7N9-K 195R viruses. Shown are the numbers of white blood cells (A), neutrophils (B), macrophages (C), and dendritic cells (D). Mice were infected with 103 TCID50 of influenza viruses and euthanized at 3 dpi. BALF from the animals was assayed by flow cytometry to determine the mean numbers of white blood cells, neutrophils, macrophages, and dendritic cells in mice infected with wtH7N9 or H7N9-K195R virus compared with BALB/c mice (n = 10) exposed to PBS (as a control). Statistical significance was assessed using two-way ANOVA (*, P < 0.05; **, P < 0.01). The error bars indicate standard deviations.
Next, we tested the replication abilities of wtH7N9 and H7N9-K195R in ferrets. Each group of three ferrets was infected intranasally with 106 TCID50 of wtH7N9 or H7N9-K195R virus, and the nasal turbinates, trachea, and various lung lobes were collected from each ferret at 5 dpi for virus enumeration. Viral titers in the turbinates and tracheas of wtH7N9-infected ferrets were 7-fold and 263-fold lower, respectively, than those of H7N9-K195R-infected ferrets (P < 0.05). Moreover, viruses were detected in all of the left-lung lobes of ferrets infected with wtH7N9 virus, but only in the left cranial lungs of H7N9-K195R-infected ferrets (Fig. 8A and B). The lungs of wtH7N9-infected ferrets showed severe bronchopneumonia, interstitial pneumonia, interstitial edema, and thickening of the alveolar walls, and the alveolar lumen was flooded with detached alveolar cells, erythrocytes, and inflammatory cells (Fig. 8C and E). In contrast, little or no lung consolidation was observed in H7N9-K195R-infected ferrets (Fig. 8D and F). Viral-antigen-positive cells were distributed among the inflammatory lesions, and many of these cells were elongated, with thin cytoplasms and hemming observed around the alveolar wall, indicating the presence of type I pneumocytes in wtH7N9-infected ferrets (Fig. 8G). In contrast, only a few NP-positive cells were observed in the lung inflammatory lesions of H7N9-K195R-infected ferrets (Fig. 8H). To confirm whether any back mutations occurred in H7N9 viruses during their replication in ferrets, viruses harvested from the lungs of ferrets infected with wtH7N9 and H7N9-K195R at 5 dpi were plaque purified and sequenced. All of the viruses harvested from the wtH7N9-infected ferrets carried PA-X 195K, and all of the viruses harvested from the H7N9-K195R-infected ferrets carried PA-X 195R.
FIG 8.
Viral replication in and gross histopathology of the lungs of ferrets infected with wtH7N9 and H7N9-K195R. (A and B) Virus replication in the indicated organs. Three ferrets from each group were infected intranasally with 106 TCID50 of each virus and euthanized at 5 dpi for virus enumeration. Viral titers in the nasal turbinates, tracheas, and lungs were determined by infection of MDCK cells. Statistical significance relative to H7N9-K195R virus was assessed using two-way ANOVA (*, P < 0.05; **, P < 0.01). (C and D) Representative gross lung pathology from ferrets infected with wtH7N9 and H7N9-K195R influenza viruses. Severe bronchopneumonia with hemorrhages, edema, and diffuse consolidation was observed in ferrets inoculated with wtH7N9 virus (arrows). (E and F) Corresponding histological (H&E) staining of lung sections. The wtH7N9 virus induced severe bronchopneumonia (E, arrowheads), whereas the H7N9-K195R virus induced only moderate bronchopneumonia (F, arrowhead). (G and H) Immunohistochemical staining of lung sections of ferrets infected with wtH7N9 and H7N9-K195R influenza viruses. Extensive viral NP localization to bronchioles and alveoli was observed for wtH7N9 viruses (arrows). (E to H) Images were obtained by microscopy at ×200 magnification.
Collectively, these data showed that PA-X 195K increased the replication and pathogenicity of H7N9 influenza viruses in mice and ferrets, indicating the important role of the residue in the virulence of H7N9 viruses in mammals.
PA-X 195K improves the transmissibility of H7N9 human influenza virus in ferrets.
To investigate the transmissibility of wtH7N9 and H7N9-K195R viruses in ferrets, a group of three ferrets were infected intranasally with 106 TCID50 of the viruses. Twenty-four hours later, the three inoculated animals were individually paired and cohoused with a direct-contact ferret; an aerosol-spread animal was also housed in a wire frame cage adjacent to the infected ferret. Nasal washes from the directly infected and transmission animals were collected every 2 days beginning at 2 dpi for virus enumeration. The viral titers in the H7N9-K195R inoculation and contact groups were significantly decreased compared with the equivalent wtH7N9 animals (P < 0.05) (Fig. 9). Furthermore, virus was detected in all three aerosol-spread ferrets in the wtH7N9 group, whereas it could be detected in only one of three aerosol-spread ferrets in the H7N9-K195R group. These findings demonstrated that PA-X 195R decreased the transmissibility of H7N9 viruses in ferrets.
FIG 9.
Horizontal transmission and respiratory droplet transmission of wtH7N9 and H7N9-K195R viruses in ferrets. Shown are viral titers in nasal washes from ferrets infected with wtH7N9 (A) or H7N9-K195R (B) virus. At 1 dpi, one inoculated ferret was moved to a clean isolator containing a naive ferret in direct contact. Additionally, another naive ferret was placed in the same isolator so that no direct contact was possible, with transmission occurring only via respiratory droplets. To monitor viral shedding, nasal washes were collected from all the animals every other day for 14 days. No data are displayed when the virus was not isolated from all three ferrets. The dashed lines indicate the lower limit of virus detection. Statistical significance relative to H7N9-K195R virus was assessed using two-way ANOVA (*, P < 0.05; **, P < 0.01).
PA-X 195K confers selective advantages on H1N1/2009 influenza viruses.
We next evaluated the effects of PA-X 195K on H1N1/2009 viruses by competitive transmission studies in ferrets. Three ferrets (one per cage) were infected intranasally with a 1:1 virus mixture consisting of wtH1N1 carrying PA-X 195K and an H1N1-K195R PA-X mutant. One day later, individual ferrets were placed in a cage adjacent to an infected ferret (“contact” ferret) to monitor viral transmission via aerosols. In the infected animals, the relative amounts of wtH1N1 virus increased over time. At 2, 4, and 6 dpi, the proportions of wtH1N1 virus gradually increased in nasal washes from the three ferrets (means, 64.3%, 74.0%, and 87.3%, respectively) concomitant with decreased proportions of the H1N1-K195R virus (35.7%, 26.0%, and 12.7%, respectively) (Fig. 10). The selective advantage of PA-X 195K was more obvious in the contact group, in which the relative proportions of virus carrying PA-X 195K at 4, 6, and 8 dpi were 83.6%, 88.0%, and 90.6%, respectively. The percentage of 195K reached 100% in one ferret at 6 dpi. These findings indicated that PA-X 195K conferred selective advantages during H1N1/2009 infection in mammals.
FIG 10.
Competitive transmission studies of wtH1N1 and H1N1-K195R viruses in ferrets. The wtH1N1 and H1N1-K195R viruses were mixed at a 1:1 ratio, and three ferrets were infected with 106 TCID50 of the mixture. One day later, each ferret was placed in a cage adjacent to an infected ferret. Virus populations in the nasal washes of infected and contact ferrets were assessed at the indicated time points. ND, not determined (nasal wash samples with viral titers of less than 101.75 TCID50 were not processed for sequence analysis).
PA-X 195K does not alter the replication ability or transmission of AIVs in chickens.
Since PA-X 195K exists in H9N2 and H5N6 AIVs and the frequency of the residue gradually increased in emerging H9N2 viruses, we evaluated the effect of this substitution on the shutoff activity of PA-X from A/chicken/Hebei/LC/2008 H9N2 virus in DF-1 cells. As shown in Fig. 11A and B, there were no statistically significant differences in the suppression ability of PA-X with 195R or 195K (P > 0.05).
FIG 11.
Effect of PA-X 195K on the suppression of nonviral protein expression of H9N2 PA-X in avian cells and replication of H9N2 virus in chickens. (A) GFP expression in DF-1 cells cotransfected with expression vectors encoding GFP, together with expression vectors encoding either the PA-X (195K) or PA-X (195R) gene of H9N2 virus at 24 h posttransfection. 293T cells, untransfected or cotransfected with GFP expression plasmids and empty pCAGGS vector, served as controls. (B) Luciferase expression from pCAGGS-Luc in DF-1 cells cotransfected with either empty pCAGGS plasmid (Vector) or pCAGGS containing the indicated genes for 24 h. The results are shown as means and standard deviations (n = 3 biological replicates; n = 3 technical replicates). (C) Three 6-week-old SPF chickens per group were inoculated intranasally with 106 EID50 of the indicated viruses. The next day, the inoculated animals were individually paired by cohousing them with direct-contact chickens. To monitor viral shedding, tracheal swabs were collected from all the animals every other day for 8 days. The dashed line indicates the lower limit of detection.
We then verified whether this substitution also enhanced the replication of AIVs. Six-week-old specific-pathogen-free (SPF) chickens were intranasally inoculated with 106 times the 50% egg infectious dose (EID50) of A/chicken/Hebei/LC/2008 (H9N2) virus and then cohoused with naive chickens at 24 h postinoculation. No significant differences in viral titers in the tracheal swabs were observed in either the inoculation or contact groups (P > 0.05) (Fig. 11C), suggesting that PA-X 195K had no effect on the virulence of AIVs in chickens.
DISCUSSION
Understanding the molecular mechanisms that shape the biological characteristics of viruses is essential to unravel fundamental aspects of influenza virus evolution, including successful adaptation to new host species. In the present study, we identified a series of virulence markers in the PA-X protein, which dramatically increase the virulence of influenza viruses in mammalian hosts. Of note, the results of sequence analyses and biological characterization studies indicated that PA-X 195K might have contributed to the adaptation of novel H7N9, H5N6, and H1N1/2009 influenza viruses to humans.
Successful replication and transmission in a novel host are key requirements for cross-species transmission of influenza viruses, which might be tied to multiple genetic variations (36–38). Various single amino acid substitutions have enormous effects in potentiating cross-species transmission. Position 627 in polymerase basic protein 2 (PB2) is a particularly remarkable host-associated genetic signature. In different highly pathogenic AIVs, the presence of PB2-627K confers high virulence in experimentally infected mice, guinea pigs, and ferrets (39–42) and has led to fatal outcomes in several zoonotic human infections (43, 44). The PA gene is also important for cross-species infection by influenza viruses. The introduction of a human influenza virus PA gene was able to enhance the replication, pathogenicity, and transmission of different AIVs in mice and ferrets (9, 10, 45). It was proposed that increased polymerase activity in mammalian cells resulting from introduction of the human PA protein was responsible for this effect. In 2012, Jagger et al. reported that the PA gene encodes both the PA protein and a novel protein, PA-X (12). When we inhibited the expression of PA-X, but not the PA protein, the replication and pathogenicity of a reassortant virus bearing an H1N1/2009 PA gene in an H9N2 AIV background were reduced dramatically to levels comparable to those of wtH9N2 virus. This suggested that PA-X plays a significant role in interspecies transmission of AIVs in mammals.
PA-X is a fusion protein consisting of the N-terminal domain of the PA protein (191 amino acids) and a unique C-terminal domain resulting from the X-ORF (12). To identify the amino acids responsible for the effects of PA-X on cross-species transmission of influenza viruses, we compared the X-ORFs of avian and human influenza viruses. Three amino acid substitutions (R195K, K206R, and P210L) were observed whose frequencies differed between these viruses (avian versus human influenza viruses). Each human-specific substitution at these three positions individually enhanced the virulence of H9N2 AIVs in mice and ferrets, but none of them altered the coding sequence of the PA protein. Because viruses carrying these substitutions might represent a threat to public health, we further analyzed whether the substitutions occurred in AIV isolates. PA-X 195K was present in the H5N6 and H9N2 AIVs, and its frequency gradually increased in H9N2 viruses. The six internal genes of novel H7N9 influenza viruses were all derived from H9N2 viruses (5, 6). Interestingly, 64.2% of H7N9 AIVs in poultry carried 195K in the first wave of an outbreak, whereas 100% of human H7N9 isolates carried 195K over the same period. In the second wave of the outbreak, PA-X 195K was present in all of the H7N9 viruses observed. The PA gene of an H1N1/2009 virus was derived from a Eurasian avian-like lineage of SIV (29). Of note, 57.0% of these SIVs carried PA-X 195K, whereas all of the H1N1/2009 viruses observed carried PA-X 195K. Thus, the results of our genetic and epidemiological analyses provided strong evidence of the relationship between PA-X 195K and influenza virus adaptation in humans.
The clear advantage of PA-X 195K in both of the human outbreaks caused by animal influenza viruses (H7N9 and H1N1/2009) in the 21st century prompted us to evaluate the effect of this substitution on the biological characteristics of the viruses. We found that PA-X 195K enhanced the virulence of H7N9 viruses in human cells and mammalian animals, as well as viral transmissibility between ferrets. The increased replication of viruses carrying PA-X 195K might have resulted from improvements in viral transcription and replication. Competitive transmission studies of H1N1/2009 viruses carrying either PA-X 195K or 195R in ferrets showed that after inoculation with the same dose of each virus, the proportion of viruses carrying 195K gradually increased; this selective advantage was even more obvious among contact animals. Our study also suggested that PA-X sequence hallmarks were associated with immunopathology. Enhanced immunopathology, as shown by extensive infiltration of inflammatory cells and increased cytokine expression in the lungs, which might have been induced by a macrophage- and neutrophil-dominated inflammatory response, was observed for some viruses, depending on their PA-X sequences. H1N1/2009 viruses and novel H7N9 viruses usually cause more severe pneumonia than seasonal influenza viruses in human clinical infections (46–48). Previous studies showed that human H7N9 viruses had higher replication and transmission capacity than an avian H7N9 virus in animal models and also caused more severe inflammation (46). H5 viruses are typical examples of viruses that can be highly pathogenic by causing inflammatory storms (43, 49). H5N1 and 1918 pandemic influenza virus infections led to excessive infiltration of macrophages and neutrophils in the lungs of mice in the early stages of infection (50). These results suggested that the R195K substitution might exacerbate pneumonia in infected humans, thereby representing an increased threat to public health.
Although gradual increases in the frequency of PA-X 195K were observed in H9N2, H5N6, and H7N9 AIVs, we found that PA-X did not affect the replication of an H9N2 virus in chickens. Thus, the emergence of PA-X 195K may be stochastic. Additionally, the internal genes of H7N9 AIVs were derived from H9N2 (8), and frequent reassortment occurred among H5, H7, and H9 AIVs that cocirculated in poultry in China (51–53). Thus, the possibility that PA-X 195K emerged in one subtype and was transmitted to other subtypes by gene reassortment cannot be excluded.
We found that PA-X 195K did not influence the polymerase activity in 293T cells (data not shown), potentially because the residue is not located in the region of PA that interacts with PB1 (54–56). An important function of the PA-X protein is in host cell shutoff (12). It has been demonstrated that the endonuclease active site in the N-terminal domain of PA-X is responsible for cellular mRNA degradation (19, 34), and the unique C-terminal X domain is required for high shutoff activity (33, 57). Because host shutoff can inhibit cellular gene expression and hinder the induction of antiviral responses, diverting ribosomes toward translation of vRNAs (16), high shutoff activity might facilitate viral replication. Our previous studies demonstrated that truncation of 20 amino acids at the PA-X C terminus in H1N1/2009, H5N1, and H9N2 influenza viruses reduced their host shutoff activity concomitant with decreased replication capability (27). However, PA-X R195K clearly reduced host shutoff activity, which might lead to uncontrolled host immune responses. Exaggerated inflammation can lead to dysregulated immune responses that do not efficiently inhibit and clear virus but instead cause increased levels of tissue damage. Similar effects were also observed in 1918 H1N1, H1N1/2009, and H5N1 viruses that lost their PA-X genes (12, 26). Thus, an excessively strong or weak host shutoff function mediated by PA-X could exacerbate the virulence of influenza viruses.
Although we demonstrated that PA-X R195K was able to enhance replication and pathogenicity in two H9N2 AIVs in mice, not all H9N2 strains with 195K possessed higher virulence than those with 195R in a study performed by Li et al. (7). It should be noted that the genotypes of the tested strains in this study were varied, indicating that there are multiple other amino acid substitutions. It is known that the virulence of influenza viruses is determined by various genes or substitutions. Thus, the amino acid in position 195 in the PA-X protein combined with other substitutions contributed together to the phenotype of the virus, which may explain why PA-X 195K has been dominant in H9N2 AIVs in China, while recent H9N2 isolates did not generally show obvious increased virulence in mammals. Nevertheless, our results suggest that we should pay close attention to the contributions of newly discovered proteins to the interspecies transmission of AIVs. There are likely other currently known viral proteins that may play similar important roles. If changes to the biological characteristics of viruses induced by such proteins are not assessed during epidemiological monitoring, it may hinder our ability to recognize potential public health threats in a timely manner.
MATERIALS AND METHODS
Ethics statement.
This study was carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People’s Republic of China. The protocols for animal studies were approved by the Committee on the Ethics of Laboratory Animals of China Agricultural University (approval SKLAB-B-2010-003).
Viruses and cells.
The influenza viruses A/Beijing/16/2009 (H1N1/2009), A/Anhui/1/2013 (H7N9), and A/chicken/Hebei/LC/2008 (H9N2) were described previously (9, 30). A/chicken/Guangdong/28/2017 (H9N2) was isolated from a diseased chicken in Guangdong, China, in June 2017 and was propagated in 10-day-old SPF embryonated chicken eggs. Human embryonic kidney 293 (HEK293T) cells, Madin-Darby canine kidney (MDCK) cells, and human lung epithelial A549 cells were obtained from the National Infrastructure of Cell Line Resource. DF-1 cells were kindly provided by Guozhong Zhang at the China Agricultural University. The cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C under a humidified atmosphere containing 5% CO2. NHBE cells (Lonza, Allendale, NJ, USA) were cultured in bronchial epithelial cell growth medium (Lonza) at the air-liquid interface, as previously described (58, 59). All experiments with live viruses were performed in a biosafety level 3 laboratory.
Generation of recombinant viruses by reverse genetics.
All eight gene segments were amplified by reverse transcription-PCR from A/Beijing/16/2009 (H1N1/2009), A/Anhui/1/2013 (H7N9), A/chicken/Hebei/LC/2008 (H9N2), and A/chicken/Guangdong/28/2017 (H9N2) viruses and then cloned into the dual-promoter plasmid pHW2000. Mutations were introduced into the PA gene using a QuikChange site-directed mutagenesis kit (Agilent) in accordance with the manufacturer’s instructions. PCR primer sequences are available upon request. The PA-X-deficient virus H1N1PA-FS:H9N2 was created by site-directed mutagenesis (QuikChange mutagenesis kit; Agilent) of the corresponding PA gene of H1N1PA:H9N2, which converted the frameshifting motif from UCC UUU CGU to AGC UUC AGA (U592A, C593G, U597C, C598A, and U600A) to prevent the formation of PA-X (12). Rescued viruses were detected using hemagglutination assays. Viruses were purified by sucrose density gradient centrifugation. Viral RNA was extracted and analyzed by reverse transcription-PCR, and each viral segment was sequenced.
Viral titration and growth kinetics in cells.
The TCID50 was determined in MDCK cells by inoculation of 10-fold serially diluted viruses at 37°C for 72 h. The TCID50 value was calculated by the Reed-Muench method (60). Multistep replication kinetics were determined using A549 and NHBE cells. A549 cells were infected with viruses at an MOI of 0.01, overlaid with serum-free DMEM containing 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Sigma-Aldrich), and incubated at 37°C. NHBE cells were infected with viruses at an MOI of 0.001 and cultured in B-ALI growth medium (Lonza) at 37°C. The supernatants were sampled at 12, 24, 36, 48, 60, and 72 hpi and titrated by inoculating MDCK cells in 96-well plates. Three independent experiments were performed.
Host shutoff assay.
293T cells or DF-1 cells were cotransfected with 125 ng of pCAGGS containing PA-X, together with enhanced GFP (eGFP) expression plasmid. Fluorescence images indicative of GFP expression at 24 h posttransfection were captured under identical exposure conditions.
The 293T or DF-1 cells were transfected with 200 ng pCAGGS containing PA-X, together with the pRL-SV40 vector (Promega) encoding Renilla luciferase, using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, luciferase production was measured using a dual-luciferase reporter assay system (Promega) as previously described (19).
Relative expression of mRNA and vRNA.
The levels of mRNA and vRNA were assessed in A549 cells infected with H7N9 viruses at an MOI of 0.01. Total RNA was extracted from infected A549 cells using TRIzol reagent (Invitrogen) in accordance with the manufacturer’s instructions. To detect mRNA and vRNA, oligo(dT) and uni-12 (5ʹ-AGCAAACGACC-3ʹ) primers, respectively, were used to generate cDNAs by reverse transcription of 1 μg of total RNA per sample with Superscript III first-strand synthesis SuperMix (Invitrogen). The qRT-PCR mixture for each reaction consisted of 10 μl of 2× SYBR green PCR master mix (Applied Biosystems), 7 μl of nuclease-free water, 0.5 μl of each primer, and 2 μl of cDNA template (diluted 1:100). The levels of the mRNAs and vRNAs encoding the M, NP, and PA proteins, as well as β-actin, were quantified using a 7500 real-time PCR system (Applied Biosystems) and the following cycling program: 1 cycle at 95°C for 2 min followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. Expression values for each transcript relative to the expression of β-actin were calculated using the 2−ΔΔCT method. In all qRT-PCR experiments, samples (assayed in duplicate), no-template control, RNA standards, and positive and negative controls were included. Each experiment comprised three technical replicates for each sample, and three experimental replicates were performed.
Mouse studies.
Groups of 6- to 8-week-old female BALB/c mice (Charles River Laboratories, Beijing, China) were anesthetized with Zoletil (tiletamine-zolazepam; Virbac; 20 μg/g) and intranasally inoculated with 50 μl of diluted infectious virus in phosphate-buffered saline (PBS). For H9N2 viruses, 106 TCID50 of each test virus was used for inoculation. Three mice from each group were euthanized at 3 and 5 dpi, and viral titers in the lung were determined by infection of MDCK cells. The remaining six mice were monitored daily for 14 days for weight loss and mortality. Mice that lost >25% of their body weight were euthanized. For H7N9 viruses, 105 TCID50 or 106 TCID50 of test virus was used for inoculation. Three mice from the 105-TCID50 group were euthanized at 1, 3, and 5 dpi for quantitation of cytokine/chemokine expression levels. Three mice each were euthanized at 3 and 5 dpi, and their lungs were collected for virus enumeration. Additionally, a portion of each lung tissue from mice infected with the indicated viruses at 5 dpi was immersed in 10% neutral buffered formalin solution, processed, and embedded in paraffin. Sections (3 μm) were stained with H&E.
Enumeration of BALF cells.
BALB/c mice (n = 10) were infected with 103 TCID50 of influenza viruses and euthanized by CO2 inhalation at 3 dpi. The trachea was exposed and cannulated with a 24-gauge plastic catheter (BDInsyte; Becton, Dickinson, Sandy, UT, USA). The lungs were lavaged three times with 1 ml of cold, sterile PBS. Flow cytometry (FACSCalibur; Becton, Dickinson, San Jose, CA, USA) was performed, and the number of white blood cells per milliliter in the BALF suspension was determined (Hemavet 3700; Drew Scientific, Dallas, TX, USA) after red blood cell depletion using red cell lysis solution (Sigma). Briefly, cells were stained with 1 μl/106 cells of anti-Gr1 (fluorescein isothiocyanate [FITC]) and anti-F480 (peridinin chlorophyll protein [Per-CP]) antibodies for 20 min, washed, and then suspended in 100 μl of fluorescence-activated cell sorter (FACS) wash buffer. The proportions of neutrophils (CD11b+, CD11c−, Ly6G/c+, CD4−, and CD8−), macrophages (CD11b+, CD11c−, Ly6G/c−, CD4−, and CD8−), and dendritic cells (CD11b−, CD11c+, Ly6G/c−, CD4−, and CD8−) were assessed as a proportion of total cellular events.
Quantitation of cytokine/chemokine expression levels.
The A549 cells were transfected with 125 ng of pCAGGS expression vectors encoding H7N9 PA-X (195K) or H7N9 PA-X (195R). Mock-infected A549 cells were transfected with empty pCAGGS vector. Twenty-four hours later, 100 ng of LPS (Beyotime, Shanghai, China) was added to the A549 cells to stimulate cytokine production. Twenty-four hours poststimulation, cell lysates were prepared for RNA extraction, and the levels of IL-1β, IL-6, IL-12, TNF-α, and NF-κB mRNAs in A549 cells were analyzed by qRT-PCR. All results are shown as the averages and standard deviations of three independent experiments.
A549 cells were either uninfected or infected with H7N9 virus at an MOI of 1. Total RNA was extracted from A549 cells using TRIzol reagent (Invitrogen) at specified time points according to the manufacturer’s instructions. The abundances of IL-1β, IL-6, IL-12, TNF-α, and NF-κB mRNAs in A549 cells were analyzed by qRT-PCR. The sequences of primers for the amplification of cytokine transcripts and for qRT-PCR analysis are available upon request.
The levels of cytokines/chemokines, including IFN-γ, IL-1β, IL-6, KC, TNF-α, MIP-1α, and MCP-1, in the lungs of mice were determined by cytometric bead array assays using the BD cytometric bead array (CBA) mouse inflammation kit (BD Bioscience). Briefly, 50 μl of mouse inflammation capture bead suspension and 50 μl of detection reagent were added to an equal volume of sample and incubated in the dark for 2 h at room temperature. Subsequently, each sample was washed with 1 ml of wash buffer and centrifuged at 200 × g at room temperature for 5 min. The supernatants were discarded, and a further 300 μl of wash buffer was added. Samples were analyzed on a FACS Array bioanalyzer (BD Bioscience). Data were analyzed using BD CBA software (BD Bioscience). The quantity of each chemokine or cytokine was determined in picograms per milliliter of homogenate.
Ferret studies.
Six-month-old female ferrets (Wuxi Cay Ferret Farm, Jiangsu, China) that were serologically negative for influenza viruses were used in these studies. The animals were anesthetized via intramuscular injection of ketamine (20 mg/kg of body weight) and xylazine (1 mg/kg) and infected intranasally with 106 TCID50 of test viruses in a 500-μl volume (250 μl per nostril). The ferrets were euthanized at 5 dpi, and the nasal turbinates, tracheas, and lungs were collected for virus enumeration in MDCK cells. The lung tissue was also collected for pathological examination.
For transmission studies, each experiment was conducted using three ferrets for each virus. One ferret was inoculated intranasally with 106 TCID50 of virus. Twenty-four hours later, two naive ferrets were added to the cage. One ferret was placed in direct contact with a naive ferret, and the second naive ferret was added to the other half of the cage separated by two layers of thin wire mesh, allowing only respiratory contact. Nasal washes were collected at 2-day intervals beginning at 2 dpi and were titrated in MDCK cells. The ambient conditions for these studies were 20 to 22°C and 30% to 40% relative humidity. The airflow in the isolator was horizontal at a speed of 0.1 m/s; the airflow direction was from the inoculated animals toward the exposed animals.
For the ferret competition transmission study, three ferrets were intranasally inoculated with 106 TCID50 of a 1:1 mixture of wtH1N1 and H1N1-K195R viruses. One day postinfection, each of three naive ferrets was placed in a cage adjacent to an inoculated ferret. Nasal washes from inoculated and contact ferrets were collected for titration in MDCK cells. A total of 50 purified virus clones from each nasal wash sample were sequenced and analyzed to determine the residue at PA-X position 195.
Histopathology.
Excised tissues from animal organs preserved in 10% phosphate-buffered formalin were processed for paraffin embedding and cut into 3-μm-thick sections. One section from each tissue sample was stained using a standard H&E procedure; another was processed for immunohistological staining with a rabbit polyclonal antibody specific for type A influenza nucleoprotein antigen (AA5H; Abcam, Hong Kong). Specific antigen-antibody reactions were visualized using 3,3ʹ-diaminobenzidine tetrahydrochloride staining and the Dako Envision system (Dako Cytomation).
Chicken study.
Six-week-old SPF chickens were obtained from Boehringer Ingelheim Animal Health Care Corporation in Beijing, China. Three chickens per group were infected intranasally with 106 EID50 of each stock virus. At 1 dpi, the inoculated chickens were housed together with three contact chickens. Tracheal swabs from each chicken were taken at 2, 4, 6, and 8 dpi for virus enumeration. The virus titer detection limit was 0.75 log10 EID50/ml.
Statistical analyses.
Differences between experimental groups were assessed using analysis of variance (ANOVA). P values of <0.05 were considered statistically significant.
Accession number(s).
The nucleotide sequences of the virus are available in the GenBank database under accession numbers MT032405 to MT032412.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31761133003 and 31961130381), the National Natural Science Fund for Outstanding Young Scholars (31522058), Newton Advanced Fellowships from the Royal Society (NAF\R1\191166), the National Key Research and Development Program of China (2016YFD0501601), and grants from the Chang Jiang Scholars Program.
REFERENCES
- 1.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 2.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RAM, Pappas C, Alpuche-Aranda CM, López-Gatell H, Olivera H, López I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilbourne ED. 2006. Influenza pandemics of the 20th century. Emerg Infect Dis 12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowen AC, Mubareka S, Tumpey TM, Garcia-Sastre A, Palese P. 2006. The guinea pig as a transmission model for human influenza viruses. Proc Natl Acad Sci U S A 103:9988–9992. doi: 10.1073/pnas.0604157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, Wang G, Chen Y, Liu L, Liang L, Li Y, Fan J, Wang J, Li W, Guan L, Li Q, Yang H, Chen P, Jiang L, Guan Y, Xin X, Jiang Y, Tian G, Wang X, Qiao C, Li C, Bu Z, Chen H. 2013. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 341:410–414. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- 6.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Shi J, Guo J, Deng G, Zhang Q, Wang J, He X, Wang K, Chen J, Li Y, Fan J, Kong H, Gu C, Guan Y, Suzuki Y, Kawaoka Y, Liu L, Jiang Y, Tian G, Li Y, Bu Z, Chen H. 2014. Genetics, receptor binding property, and transmissibility in mammals of naturally isolated H9N2 avian influenza viruses. PLoS Pathog 10:e1004508. doi: 10.1371/journal.ppat.1004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pu J, Wang S, Yin Y, Zhang G, Carter RA, Wang J, Xu G, Sun H, Wang M, Wen C, Wei Y, Wang D, Zhu B, Lemmon G, Jiao Y, Duan S, Wang Q, Du Q, Sun M, Bao J, Sun Y, Zhao J, Zhang H, Wu G, Liu J, Webster RG. 2015. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci U S A 112:548–553. doi: 10.1073/pnas.1422456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y, Qin K, Wang J, Pu J, Tang Q, Hu Y, Bi Y, Zhao X, Yang H, Shu Y, Liu J. 2011. High genetic compatibility and increased pathogenicity of reassortants derived from avian H9N2 and pandemic H1N1/2009 influenza viruses. Proc Natl Acad Sci U S A 108:4164–4169. doi: 10.1073/pnas.1019109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Zhang Q, Kong H, Jiang Y, Gao Y, Deng G, Shi J, Tian G, Liu L, Liu J, Guan Y, Bu Z, Chen H. 2013. H5N1 hybrid viruses bearing 2009/H1N1 virus genes transmit in guinea pigs by respiratory droplet. Science 340:1459–1463. doi: 10.1126/science.1229455. [DOI] [PubMed] [Google Scholar]
- 11.Mehle A, Doudna JA. 2009. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci U S A 106:21312–21316. doi: 10.1073/pnas.0911915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yewdell JW, Ince WL. 2012. Frameshifting to PA-X influenza. Science 337:164–165. doi: 10.1126/science.1225539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng KH, Sun M, Iketani S, Holmes EC, Parrish CR. 2016. Comparing the functions of equine and canine influenza H3N8 virus PA-X proteins: suppression of reporter gene expression and modulation of global host gene expression. Virology 496:138–146. doi: 10.1016/j.virol.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaucherand L, Porter BK, Levene RE, Price EL, Schmaling SK, Rycroft CH, Kevorkian Y, McCormick C, Khaperskyy DA, Gaglia MM. 2019. The influenza A virus endoribonuclease PA-X usurps host mRNA processing machinery to limit host gene expression. Cell Rep 27:776–792.e777. doi: 10.1016/j.celrep.2019.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber F, Haller O. 2007. Viral suppression of the interferon system. Biochimie 89:836–842. doi: 10.1016/j.biochi.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oishi K, Yamayoshi S, Kawaoka Y. 2019. Identification of amino acid residues in influenza A virus PA-X that contribute to enhanced shutoff activity. Front Microbiol 10:432. doi: 10.3389/fmicb.2019.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohei O, Seiya Y, Hiroko K-H, Masaaki O, Yoshihiro K. 2018. N-Terminal acetylation by NatB is required for the shutoff activity of influenza A virus PA-X. Cell Rep 24:851–860. doi: 10.1016/j.celrep.2018.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desmet EA, Bussey KA, Stone R, Takimoto T. 2013. Identification of the N-terminal domain of the influenza virus PA responsible for the suppression of host protein synthesis. J Virol 87:3108–3118. doi: 10.1128/JVI.02826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi T, MacDonald LA, Takimoto T. 2015. Influenza A virus protein PA-X contributes to viral growth and suppression of the host antiviral and immune responses. J Virol 89:6442–6452. doi: 10.1128/JVI.00319-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu J, Mo Y, Wang X, Gu M, Hu Z, Zhong L, Wu Q, Hao X, Hu S, Liu W, Liu H, Liu X, Liu X. 2015. PA-X decreases the pathogenicity of highly pathogenic H5N1 influenza A virus in avian species by inhibiting virus replication and host response. J Virol 89:4126–4142. doi: 10.1128/JVI.02132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rigby RE, Wise HM, Smith N, Digard P, Rehwinkel J. 2019. PA-X antagonises MAVS-dependent accumulation of early type I interferon messenger RNAs during influenza A virus infection. Sci Rep 9:7216. doi: 10.1038/s41598-019-43632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiao H, Chunxi M, Xiufan L. 2018. PA-X: a key regulator of influenza A virus pathogenicity and host immune responses. Med Microbiol Immunol 207:1–15. doi: 10.1007/s00430-018-0548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi M, Jagger BW, Wise HM, Digard P, Holmes EC, Taubenberger JK. 2012. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J Virol 86:12411–12413. doi: 10.1128/JVI.01677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu G, Zhang X, Sun Y, Liu Q, Sun H, Xiong X, Jiang M, He Q, Wang Y, Pu J, Guo X, Yang H, Liu J. 2016. Truncation of C-terminal 20 amino acids in PA-X contributes to adaptation of swine influenza virus in pigs. Sci Rep 6:21845. doi: 10.1038/srep21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao H, Sun Y, Hu J, Qi L, Wang J, Xiong X, Wang Y, He Q, Lin Y, Kong W, Seng LG, Sun H, Pu J, Chang KC, Liu X, Liu J. 2015. The contribution of PA-X to the virulence of pandemic 2009 H1N1 and highly pathogenic H5N1 avian influenza viruses. Sci Rep 5:8262. doi: 10.1038/srep08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao H, Sun H, Hu J, Qi L, Wang J, Xiong X, Wang Y, He Q, Lin Y, Kong W, Seng LG, Pu J, Chang KC, Liu X, Liu J, Sun Y. 2015. Twenty amino acids at the C-terminus of PA-X are associated with increased influenza A virus replication and pathogenicity. J Gen Virol 96:2036–2049. doi: 10.1099/vir.0.000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J, Yu H, Li Y, Ma J, Lang Y, Duff M, Henningson J, Liu Q, Li Y, Nagy A, Bawa B, Li Z, Tong G, Richt JA, Ma W. 2017. Impacts of different expressions of PA-X protein on 2009 pandemic H1N1 virus replication, pathogenicity and host immune responses. Virology 504:25–35. doi: 10.1016/j.virol.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann G, Noda T, Kawaoka Y. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bi Y, Xiao H, Chen Q, Wu Y, Fu L, Quan C, Wong G, Liu J, Haywood J, Liu Y, Zhou B, Yan J, Liu W, Gao GF. 2016. Changes in the length of the neuraminidase stalk region impact H7N9 Virulence in mice. J Virol 90:2142–2149. doi: 10.1128/JVI.02553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khaperskyy DA, Emara MM, Johnston BP, Anderson P, Hatchette TF, McCormick C. 2014. Influenza a virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog 10:e1004217. doi: 10.1371/journal.ppat.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi T, Chaimayo C, Takimoto T. 2015. Impact of influenza PA-X on host response. Oncotarget 6:19364–19365. doi: 10.18632/oncotarget.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oishi K, Yamayoshi S, Kawaoka Y. 2015. Mapping of a region of the PA-X protein of influenza A virus that is important for its shutoff activity. J Virol 89:8661–8665. doi: 10.1128/JVI.01132-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khaperskyy DA, Schmaling S, Larkins-Ford J, McCormick C, Gaglia MM. 2016. Selective degradation of host RNA polymerase II transcripts by influenza A virus PA-X host shutoff protein. PLoS Pathog 12:e1005427. doi: 10.1371/journal.ppat.1005427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaimayo C, Dunagan M, Hayashi T, Santoso N, Takimoto T. 2018. Specificity and functional interplay between influenza virus PA-X and NS1 shutoff activity. PLoS Pathog 14:e1007465. doi: 10.1371/journal.ppat.1007465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Wit E, Munster VJ, van Riel D, Beyer WE, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. 2010. Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. J Virol 84:1597–1606. doi: 10.1128/JVI.01783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lloren KKS, Lee T, Kwon JJ, Song MS. 2017. Molecular markers for interspecies transmission of avian influenza viruses in mammalian hosts. Int J Mol Sci 18:E2706. doi: 10.3390/ijms18122706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zanin M, Wong SS, Barman S, Kaewborisuth C, Vogel P, Rubrum A, Darnell D, Marinovapetkova A, Krauss S, Webby RJ. 2017. Molecular basis of mammalian transmissibility of avian H1N1 influenza viruses and their pandemic potential. Proc Natl Acad Sci U S A 114:11217–11222. doi: 10.1073/pnas.1713974114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao P, Watanabe S, Ito T, Goto H, Wells K, McGregor M, Cooley AJ, Kawaoka Y. 1999. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J Virol 73:3184–3189. doi: 10.1128/JVI.73.4.3184-3189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Govorkova EA, Rehg JE, Krauss S, Yen HL, Guan Y, Peiris M, Nguyen TD, Hanh TH, Puthavathana P, Long HT, Buranathai C, Lim W, Webster RG, Hoffmann E. 2005. Lethality to ferrets of H5N1 influenza viruses isolated from humans and poultry in 2004. J Virol 79:2191–2198. doi: 10.1128/JVI.79.4.2191-2198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatta M, Gao P, Halfmann P, Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 42.Steel J, Lowen AC, Mubareka S, Palese P, Baric RS. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog 5:e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJD, Chau TNB, Hoang DM, Chau NVV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha DQ, Guan Y, Peiris JSM, Chinh NT, Hien TT, Farrar J. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A 101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimble JB, Sorrell E, Shao H, Martin PL, Perez DR. 2011. Compatibility of H9N2 avian influenza surface genes and 2009 pandemic H1N1 internal genes for transmission in the ferret model. Proc Natl Acad Sci U S A 108:12084–12088. doi: 10.1073/pnas.1108058108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, Wang D, Gao R, Zhao B, Song J, Qi X, Zhang Y, Shi Y, Yang L, Zhu W, Bai T, Qin K, Lan Y, Zou S, Guo J, Dong J, Dong L, Zhang Y, Wei H, Li X, Lu J, Liu L, Zhao X, Li X, Huang W, Wen L, Bo H, Xin L, Chen Y, Xu C, Pei Y, Yang Y, Zhang X, Wang S, Feng Z, Han J, Yang W, Gao GF, Wu G, Li D, Wang Y, Shu Y. 2013. Biological features of novel avian influenza A (H7N9) virus. Nature 499:500–503. doi: 10.1038/nature12379. [DOI] [PubMed] [Google Scholar]
- 47.Yu H, Cowling BJ, Feng L, Lau EH, Liao Q, Tsang TK, Peng Z, Wu P, Liu F, Fang VJ, Zhang H, Li M, Zeng L, Xu Z, Li Z, Luo H, Li Q, Feng Z, Cao B, Yang W, Wu JT, Wang Y, Leung GM. 2013. Human infection with avian influenza A H7N9 virus: an assessment of clinical severity. Lancet 382:138–145. doi: 10.1016/S0140-6736(13)61207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujimoto Y, Hasegawa S, Matsushige T, Wakiguchi H, Nakamura T, Hasegawa H, Nakajima N, Ainai A, Oga A, Itoh H, Shirabe K, Toda S, Atsuta R, Morishima T, Ohga S. 2017. Pulmonary inflammation and cytokine dynamics of bronchoalveolar lavage fluid from a mouse model of bronchial asthma during A(H1N1)pdm09 influenza infection. Sci Rep 7:9128. doi: 10.1038/s41598-017-08030-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szretter KJ, Gangappa S, Lu X, Smith C, Shieh WJ, Zaki SR, Sambhara S, Tumpey TM, Katz JM. 2007. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J Virol 81:2736–2744. doi: 10.1128/JVI.02336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrone LA, Plowden JK, Garcia-Sastre A, Katz JM, Tumpey TM. 2008. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog 4:e1000115. doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bi Y, Chen Q, Wang Q, Chen J, Jin T, Wong G, Quan C, Liu J, Wu J, Yin R, Zhao L, Li M, Ding Z, Zou R, Xu W, Li H, Wang H, Tian K, Fu G, Huang Y, Shestopalov A, Li S, Xu B, Yu H, Luo T, Lu L, Xu X, Luo Y, Liu Y, Shi W, Liu D, Gao GF. 2016. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe 20:810–821. doi: 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 52.Gu M, Xu L, Wang X, Liu X. 2017. Current situation of H9N2 subtype avian influenza in China. Vet Res 48:49. doi: 10.1186/s13567-017-0453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cui L, Liu D, Shi W, Pan J, Qi X, Li X, Guo X, Zhou M, Li W, Li J, Haywood J, Xiao H, Yu X, Pu X, Wu Y, Yu H, Zhao K, Zhu Y, Wu B, Jin T, Shi Z, Tang F, Zhu F, Sun Q, Wu L, Yang R, Yan J, Lei F, Zhu B, Liu W, Ma J, Wang H, Gao GF. 2014. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun 5:3142. doi: 10.1038/ncomms4142. [DOI] [PubMed] [Google Scholar]
- 54.He X, Zhou J, Bartlam M, Zhang R, Ma J, Lou Z, Li X, Li J, Joachimiak A, Zeng Z, Ge R, Rao Z, Liu Y. 2008. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454:1123–1126. doi: 10.1038/nature07120. [DOI] [PubMed] [Google Scholar]
- 55.Obayashi E, Yoshida H, Kawai F, Shibayama N, Kawaguchi A, Nagata K, Tame JR, Park SY. 2008. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454:1127–1131. doi: 10.1038/nature07225. [DOI] [PubMed] [Google Scholar]
- 56.Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 57.Hayashi T, Chaimayo C, McGuinness J, Takimoto T. 2016. Critical role of the PA-X C-terminal domain of influenza A virus in its subcellular localization and shutoff activity. J Virol 90:7131–7141. doi: 10.1128/JVI.00954-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hudy MH, Traves SL, Wiehler S, Proud D. 2010. Cigarette smoke modulates rhinovirus-induced airway epithelial cell chemokine production. Eur Respir J 35:1256–1263. doi: 10.1183/09031936.00128809. [DOI] [PubMed] [Google Scholar]
- 59.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci U S A 101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]