How a nonenveloped virus escapes from a host membrane to promote infection remains enigmatic. In the case of the nonenveloped polyomavirus SV40, penetration of the ER membrane to reach the cytosol is a decisive virus infection step. In this study, we found a new host factor called Ubqln4 that facilitates escape of SV40 from the ER into the cytosol, thereby providing a path for the virus to enter the nucleus to cause infection.
KEYWORDS: endoplasmic reticulum, membrane transport, protein chaperone, simian virus 40, virus entry
ABSTRACT
The nonenveloped polyomavirus simian virus 40 (SV40) must penetrate the host endoplasmic reticulum (ER) membrane to enter the cytosol in order to promote infection. How this is accomplished is not entirely clear. Here, we demonstrate that the cytosolic chaperone Ubiquilin4 (Ubqln4) binds directly to the ER membrane J proteins B12 and B14. Strategically localized at the ER-cytosol interface, Ubqln4 captures SV40 emerging from the ER, thereby facilitating escape of the virus from the ER into the cytosol, which leads to infection. Strikingly, Ubqln4 engages the J proteins in a J-domain-independent manner, in contrast to the previously reported Hsc70-Hsp105-SGTA-Bag2 cytosolic complex that also mediates SV40 ER-to-cytosol transport. Our results also reveal that the H domain and STI1 motif (1-2) of Ubqln4 support J protein binding, essential for SV40 infection. Together, these data further clarify the molecular basis by which a nonenveloped virus escapes a host membrane during infectious entry.
IMPORTANCE How a nonenveloped virus escapes from a host membrane to promote infection remains enigmatic. In the case of the nonenveloped polyomavirus SV40, penetration of the ER membrane to reach the cytosol is a decisive virus infection step. In this study, we found a new host factor called Ubqln4 that facilitates escape of SV40 from the ER into the cytosol, thereby providing a path for the virus to enter the nucleus to cause infection.
INTRODUCTION
During entry of nonenveloped viruses, the viral particles must penetrate a host membrane to reach the cytosol, and often the nucleus, to cause infection. How they breach a cellular membrane in order to escape into the cytosol remains an enigma (1–3). Despite this, recent studies have provided greater clarity to the molecular basis of membrane penetration by nonenveloped viruses, with the polyomavirus (PyV) simian virus 40 (SV40) being a salient example (4, 5).
Structurally, SV40 is composed of 72 pentamers of the structural coat protein VP1 (6, 7), with each pentamer harboring an internal hydrophobic protein, VP2 or VP3, that is normally hidden beneath the VP1 pentamers (8). The 5-kbp circular DNA genome is in turn buried inside the viral particle. To enter host cells, SV40 undergoes receptor-mediated endocytosis (9, 10), reaching the endoplasmic reticulum (ER) (11), where it penetrates the ER membrane to enter the cytosol and then the nucleus in order to cause infection (12, 13). Our group and others have started to elucidate the molecular basis on which SV40 penetrates the ER membrane in order to escape into the cytosol.
Specifically, upon reaching the lumen of the ER from the cell surface, ER-resident protein disulfide isomerase (PDI) family proteins impart conformational changes to the virus by reducing and isomerizing the disulfide bonds formed between the VP1 pentamers (14–16). This structural change exposes the hydrophobic proteins VP2 and VP3 (14, 17, 18). Hydrophobic SV40 is rendered soluble by interacting with the ER luminal chaperones BiP and Grp170 (19, 20). When BiP and Grp170 are released from SV40, the hydrophobic virus binds to and inserts into the ER membrane (20, 21). Within the ER membrane, the EMC1 transmembrane chaperone stabilizes the viral particle to prevent premature disassembly of the virus (22). For the membrane-integrated SV40 to enter the cytosol, a cytosolic protein complex composed of Hsc70, Hsp105, SGTA, and Bag2 is recruited to the ER membrane via binding to the J domain of the ER transmembrane J proteins B12, B14, and C18 (19, 23–28). Strategically localized at the ER-cytosol interface, the Hsc70-Hsp105-SGTA-Bag2 complex extracts the virus into the cytosol (23–27). However, the precise mechanism by which SV40 is ejected into the cytosol and whether other host factors are involved in this process remain unknown.
In this report, we identify the cytosolic chaperone Ubiquilin4 (Ubqln4) (29) as a new binding-partner of B12 and B14. At the ER-cytosol junction, Ubqln4 engages SV40 when the virus exits the ER to promote ER escape of the virus into the cytosol, which is essential for infection. Surprisingly, Ubqln4 interacts with the J proteins in a J-domain-independent fashion, in stark contrast to the Hsc70-Hsp105-SGTA-Bag2 complex. Additionally, our results pinpoint the H domain and STI1 motif (1-2) within Ubqln4 as crucial for supporting J protein binding and SV40 infection. Together, these findings further illuminate the molecular basis by which a nonenveloped virus escapes a host membrane during infectious entry.
RESULTS
Ubqln4 binds directly to the ER membrane B12 J protein in a J-domain-independent manner.
An RNA interference (RNAi) screen led to the identification of the ER membrane J proteins B12, B14, and C18 as crucial host factors that promote ER escape of SV40 into the cytosol (19). Based on this finding, we hypothesized that cytosolic binding partners of these J proteins function to extract SV40 from the ER into the cytosol. To test this, we used a proteomics approach to identify the J protein binding partners. In this approach, FLAG-tagged J proteins stably expressed in Flp-in 293T-REx cells were immunoprecipitated, and the precipitated material was subjected to mass spectrometry analysis. This analysis revealed the cytosolic Hsc70-Hsp105-SGTA-Bag2 protein complex to be a J-protein-binding partner that extracts ER membrane-embedded SV40 into the cytosol, leading to infection (23–28).
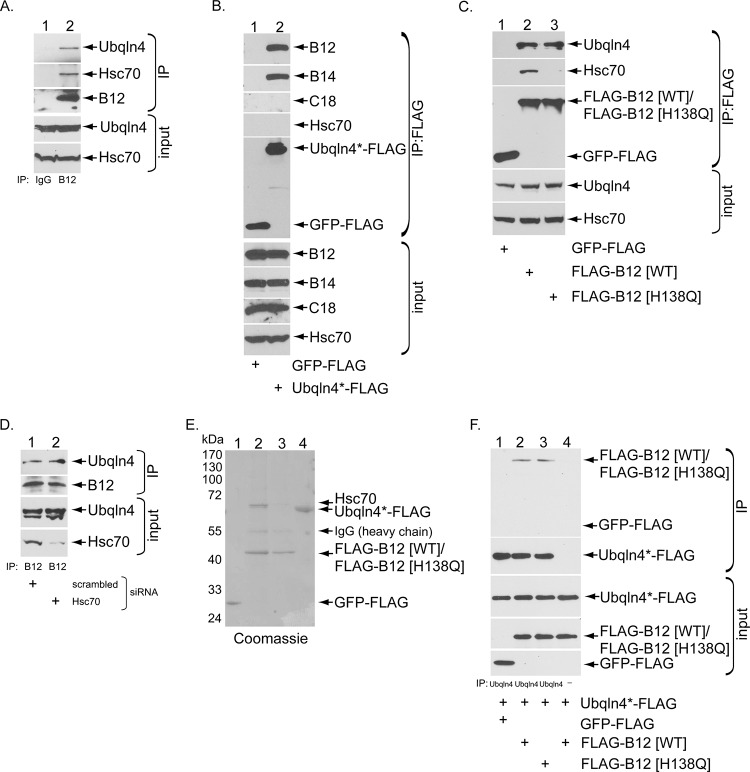
In the same immunoprecipitation (IP)-mass spectrometry data, a unique peptide corresponding to Ubqln4 was found in the B12 precipitated material (28). Importantly, when endogenous B12 in simian CV-1 cells was immunoprecipitated and the precipitated sample was subjected to SDS-PAGE, followed by immunoblotting, Ubqln4 was found in the precipitated material (Fig. 1A, top); Hsc70 was also found in the precipitated sample (Fig. 1A, second from top), as expected. We then evaluated the B12-Ubqln4 interaction in reverse. Specifically, when transiently expressed FLAG-tagged Ubqln4 (Ubqln4*-FLAG) or the control green fluorescent protein (GFP)-FLAG was pulled down, endogenous B12 and B14, but not C18 or Hsc70, were found in the Ubqln4*-FLAG precipitated sample (Fig. 1B, top four gels). Together, these data identify Ubqln4 as a new binding partner of B12 and B14.
FIG 1.
Ubqln4 binds directly to the ER membrane B12 J protein in a J-domain-independent manner. (A) CV-1 cell extracts were used for immunoprecipitation with control IgG or antibodies against endogenous B12. The precipitated material was subjected to SDS-PAGE, followed by immunoblotting with the indicated antibodies. (B) COS-7 cells were transfected with Ubqln4-FLAG designed to be resistant to Ubqln4 no. 1 siRNA, Ubqln4*-FLAG, or control GFP-FLAG. Immunoprecipitation from the cell extracts was performed using anti-FLAG agarose beads, with subsequent immunoblot analysis with the indicated antibodies. (C) As in panel B, except the cells were transfected with GFP-FLAG, FLAG-B12 (WT), and FLAG-B12 (H138Q). (D) COS-7 cells were transfected with control siRNA or Hsc70 siRNA (50 nM) for 3 days. The cells were lysed, and the resulting extract was subjected to immunoprecipitation with endogenous B12 antibody, followed by SDS-PAGE and immunoblotting with the indicated antibodies. (E) Coomassie staining of the purified FLAG-tagged proteins. (F) Direct binding of Ubqln4 and B12 in vitro. Purified GFP-FLAG, FLAG-B12 (WT), or FLAG-B12 (H138Q) was incubated with Ubqln4*-FLAG. Ubqln4*-FLAG in the reaction mixture was immunoprecipitated by the addition of endogenous Ubqln4 antibody where indicated. The precipitated material was subjected to SDS-PAGE and immunoblotting.
We then asked if the B12-Ubqln4 interaction is mediated by the J domain of B12, a domain characteristic of J proteins that is typically used to recruit Hsc70 chaperones (30). To test this, we generated FLAG-tagged wild-type B12 [FLAG-B12 (WT)], as well as a FLAG-tagged B12 mutant defective in Hsc70 binding due to a mutation in the highly conserved H138 residue within the B12 J domain [FLAG-B12 (H138Q)]. As expected, precipitation of FLAG-B12 (WT) pulled down endogenous Ubqln4 and Hsc70 (Fig. 1C, top two gels). In contrast, although FLAG-B12 (H138Q) could not bind to Hsc70 as anticipated, it nonetheless interacted with Ubqln4 (Fig. 1C, top two gels). These findings indicate that binding of Ubqln4 to B12 does not require an intact B12 J domain and suggest that Hsc70 recruitment to B12 is not essential in supporting the B12-Ubqln4 interaction. To further support this idea, we knocked down Hsc70 using a small interfering RNA (siRNA) approach and found that the B12-Ubqln4 interaction was unaffected in cells depleted of Hsc70 (Fig. 1D, top gel). Together, these data demonstrate that Ubqln4 associates with B12 in a J-domain- and Hsc70-independent manner.
We used a purified reconstituted system to assess whether the B12-Ubqln4 interaction is direct. To this end, we purified FLAG-B12 (WT), FLAG-B12 (H138Q), and Ubqln4*-FLAG, as well the control protein GFP-FLAG, from mammalian cells (Fig. 1E). While Hsc70 copurified with FLAG-B12 (WT), Hsc70 did not copurify with FLAG-B12 (H138Q), as expected (Fig. 1E, compare lane 2 to lane 3). Ubqln4*-FLAG was incubated with either FLAG-B12 (WT)/Hsc70, FLAG-B12 (H138Q), or GFP-FLAG, and the samples were then incubated with (or without) an antibody against Ubqln4 to precipitate Ubqln4*-FLAG. The precipitated material was subjected to SDS-PAGE and immunoblotted with the indicated antibodies. Importantly, we found that FLAG-B12 (WT) and FLAG-B12 (H138Q), but not GFP-FLAG, were pulled down to the same level (Fig. 1F, top gel); this binding occurred only if Ubqln4*-FLAG was precipitated using an anti-Ubqln4 antibody. The findings indicate that B12 binds directly to Ubqln4 independently of Hsc70.
Ubqln4 facilitates SV40 infection.
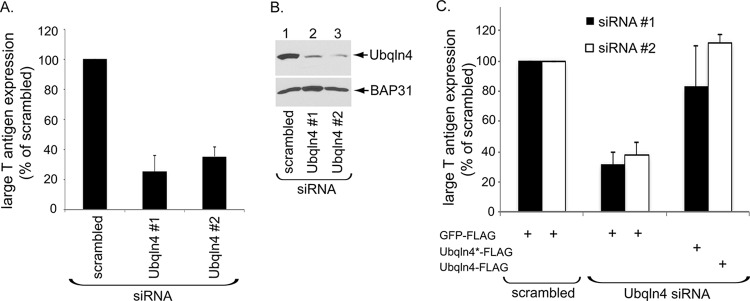
We next evaluated the role of Ubqln4 during SV40 infection in CV-1 cells, which are used classically to study SV40 entry. To monitor infection, we assessed the expression of the virus-encoded large T antigen (TAg), which occurs only when SV40 reaches the host nucleus. In cells transfected with either of two different siRNAs against Ubqln4 (Ubqln4 no. 1 and no. 2 siRNAs), SV40 infection was robustly decreased compared to cells transfected with the control scrambled siRNA (Fig. 2A); the extent of Ubqln4 knockdown is shown in Fig. 2B (top gel). These results suggest that Ubqln4 plays an important role during SV40 infection.
FIG 2.
Ubqln4 facilitates SV40 infection. (A) CV-1 cells transfected with the indicated siRNA for 72 h were infected with SV40 (MOI, 0.5) for 20 to 24 h, fixed, and immunostained for large TAg. Infection was scored using immunofluorescence microscopy (counting >1,000 cells for each condition). The data were normalized to the scrambled siRNA. The values represent means and standard deviations (SD) (n ≥ 3). (B) CV-1 cells were transfected with the indicated siRNA for 72 h and lysed, and the resulting extract was subjected to SDS-PAGE and immunoblotting with the indicated antibodies. (C) CV-1 cells were transfected with the indicated siRNA for 48 h prior to transfection with the indicated plasmids for 24 h. The cells were then infected with SV40, fixed, and immunostained for anti-FLAG and anti-TAg antibodies. The percentage of TAg-positive cells was determined only in cells (≥500) expressing the indicated FLAG-tagged protein by immunofluorescence microscopy. The data were normalized to the scrambled siRNA.
To verify that this phenotype is not due to off-target effects of the siRNAs, we performed knockdown-rescue experiments. Accordingly, cells transfected with scrambled, Ubqln4 no. 1, or Ubqln4 no. 2 siRNA were cotransfected with the control GFP-FLAG, Ubqln4*-FLAG, or Ubqln4-FLAG. The cells were then infected with SV40, and large T antigen expression was scored only in FLAG-expressing cells. (Ubqln4*-FLAG was used to restore Ubqln4 expression in Ubqln4 no. 1 siRNA-transfected cells because the construct is resistant to the siRNA. Since Ubqln4 no. 2 siRNA targeted the noncoding region of Ubqln4, Ubqln4-FLAG was used to restore Ubqln4 expression in Ubqln4 no. 2 siRNA-transfected cells.) Importantly, using this approach, we found that the block in SV40 infection due to either of the Ubqln4 siRNAs could be largely restored by expression of Ubqln4 (Fig. 2C). These findings demonstrate that the phenotype caused by the Ubqln4 siRNAs was not due to off-target effects, unambiguously establishing a critical role for Ubqln4 during SV40 infection.
Ubqln4 promotes arrival of SV40 in the cytosol from the ER.
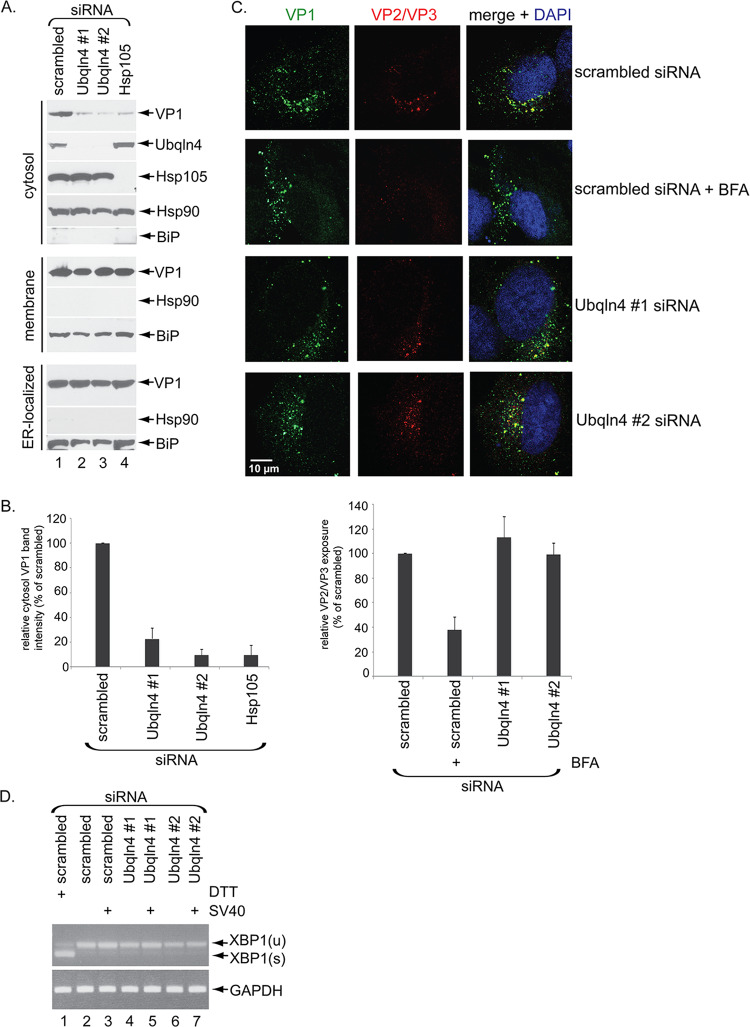
Because Ubqln4 is positioned at the ER-cytosol interface via binding to the ER membrane J proteins (Fig. 1), we hypothesized that it facilitates entry of SV40 from the ER into the cytosol. To test this hypothesis, we used a well-established cell-based, semipermeabilized assay (12) in which SV40-infected CV-1 cells transfected with the indicated siRNA were harvested and incubated with a low concentration of digitonin to permeabilize the plasma membrane without compromising internal membranes. The cells were then centrifuged to generate two fractions: a supernatant fraction containing cytosolic proteins and virus that reaches the cytosol (the cytosol fraction) and a pellet fraction that harbors membranes, including the ER and membrane-associated virus (the membrane fraction). The membrane fraction can be further processed (see Materials and Methods) to isolate ER-localized virus. The integrity of this protocol can be verified by the presence of the cytosolic marker Hsp90 in the cytosol, but not the membrane fraction (Fig. 3A, compare the fourth gel from the top to the seventh gel), and the appearance of the ER luminal BiP protein in the membrane, but not the cytosol fraction (Fig. 3A, compare the eighth gel to the fifth gel).
FIG 3.
Ubqln4 promotes arrival of SV40 in the cytosol from the ER. (A) CV-1 cells transfected with the indicated siRNAs for 72 h were infected with SV40 (MOI, 5), harvested 16 h postinfection (hpi), and subjected to the ER-to-cytosol transport assay (see Materials and Methods). The ER luminal protein BiP served as the membrane marker, while cytosolic Hsp90 served as the cytosol marker. (B) Relative VP1 band intensities in the cytosol fraction from panel A were determined by using FIJI/ImageJ (NIH) software. The data were normalized to scrambled siRNA. The values represent means and SD of the results of three independent experiments. (C) SV40-infected cells transfected with the indicated siRNAs (and incubated with or without BFA) were fixed, stained with VP2/VP3 and VP1 antibodies, and analyzed by confocal microscopy. Relative VP2/VP3 exposure was determined by quantifying VP2/VP3 signals normalized to the VP1 signals by FIJI/ImageJ and is displayed as the percentage of scrambled siRNA-treated sample. (D) CV-1 cells were transfected with the indicated siRNAs for 72 h and infected or not with SV40 (MOI, 5) for 16 h. RNA was isolated from the cells, and RT-PCR was performed to identify splicing of the XBP1 mRNA. Cells treated with DTT served as a positive control. The top gel shows the level of unspliced XBP1 [XBP1(u)] and spliced XBP1 [XBP1(s)], while the bottom gel shows the level of the internal control GAPDH mRNA.
Using this approach, we found that knockdown of Ubqln4 or, as a positive control, Hsp105 (a member of the Hsc70 extraction complex) decreased the SV40 VP1 level in the cytosol (Fig. 3A, top gel; the VP1 intensity level in the cytosol fraction in Fig. 3A is quantified in Fig. 3B). This decrease is not due to a block in arrival of the virus in the ER from the cell surface because the level of ER-localized SV40 VP1 was unaffected by depletion of Ubqln4 (Fig. 3A, ninth gel from the top). Additionally, we found that exposure of VP2/VP3 (which occurs only when SV40 reaches the ER from the plasma membrane) was also unaffected by Ubqln4 depletion, in contrast to cells treated with brefeldin A (BFA), which disrupts retrograde transport to the ER (Fig. 3C; see quantification below). These findings thus demonstrate that depletion of Ubqln4 selectively decreases ER-to-cytosol escape of SV40 without perturbing virus arrival at the ER from the cell surface.
We next tested the possibility that loss of Ubqln4 might cause a general induction of ER stress, which in turn indirectly impaired arrival of SV40 in the cytosol from the ER. However, this is unlikely to be the case, because splicing of the XBP1 transcript, a marker of ER stress, was not observed in Ubqln4 knockdown cells regardless of the presence of SV40 (Fig. 3D, top gel, compare lanes 4 to 7 to lanes 2 and 3), in contrast to cells treated with the chemical reductant dithiothreitol (DTT), which triggers ER stress (Fig. 3D, top gel, lane 1); XBP1 splicing was also not induced due to loss of ubiquilin in Drosophila cells (31). We conclude that Ubqln4 selectively promotes arrival of SV40 in the cytosol from the ER, consistent with its role in supporting virus infection (Fig. 2).
The H domain and STI1 motif (1-2) of Ubqln4 are required to support B12 binding and SV40 infection.
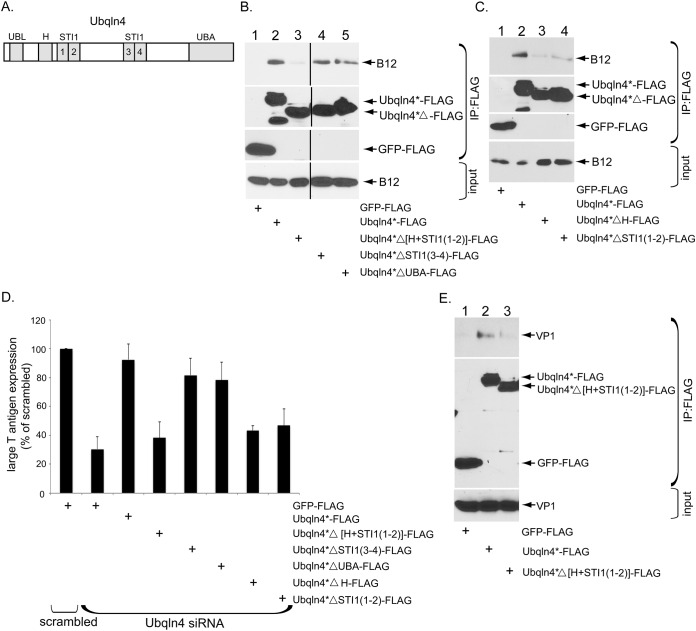
We next sought to identify structural features in Ubqln4 that support B12 interaction and SV40 infection. Structurally, Ubqln4 contains an N-terminal ubiquitin-like (UBL) domain, followed by a hydrophobic (H) domain, four repeats of the stress-inducible heat shock chaperonin-binding (STI1) motif (1-4), and a C-terminal ubiquitin-associated (UBA) domain (29) (Fig. 4A). Accordingly, we generated Ubqln4 deletion constructs lacking either the UBL domain (Ubqln4*ΔUBL-FLAG), the H domain and STI1 motif (1-2) {Ubqln4*Δ[H+STI1(1-2)]-FLAG}, the STI1 motif (3-4) [Ubqln4*ΔSTI1(3-4)-FLAG], or the UBA domain (Ubqln4*ΔUBA-FLAG). Because Ubqln4*ΔUBL-FLAG was expressed poorly in cells (not shown), we did not characterize the construct further. Importantly, whereas Ubqln4*Δ STI1(3-4)-FLAG and Ubqln4*ΔUBA-FLAG bound to endogenous B12 to an extent similar to that of Ubqln4*-FLAG, Ubqln4*Δ[H+ STI1(1-2)]-FLAG did not (Fig. 4B, top gel). Consistent with this, using the same knockdown-rescue strategy shown in Fig. 2, we found that while expression of either Ubqln4*Δ STI1(3-4)-FLAG or Ubqln4*ΔUBA-FLAG could largely restore the block in SV40 infection due to depletion of Ubqln4 to a level similar to that of Ubqln4*-FLAG, expressing Ubqln4*Δ[H+ STI1(1-2)]-FLAG did not (Fig. 4D). These findings strongly suggest that the H domain and STI1 motif (1-2) of Ubqln4 are necessary for the B12 interaction that is essential to support SV40 infection.
FIG 4.
The H domain and STI1 motif (1-2) of Ubqln4 are required to support B12 binding and SV40 infection. (A) Schematic representation of Ubqln4, which contains the UBL domain, H domain, 4 repeated STI1 domains (1 to 4), and the UBA domain. (B) COS-7 cells were transfected with control GFP-FLAG, Ubqln4*-FLAG, Ubqln4*Δ[H+STI1(1-2)]-FLAG, Ubqln4*ΔSTI1(3-4)-FLAG, or Ubqln4*ΔUBA-FLAG for 48 h. Immunoprecipitation from the cell extracts was performed using anti-FLAG agarose beads. The precipitated material was subjected to SDS-PAGE and immunoblotting with the indicated antibodies. (C) As in panel B, except the cells were transfected with the indicated plasmids. (D) CV-1 cells were transfected with the indicated siRNA for 48 h prior to transfection with the indicated plasmids. The cells were then infected with SV40, fixed, and immunostained for anti-FLAG and anti-TAg antibodies. The percentages of TAg-positive cells were determined only in cells (≥500) expressing the indicated FLAG-tagged protein by immunofluorescence microscopy. The data were normalized to the scrambled siRNA. The values represent means and SD (n ≥ 3). (E) COS-7 cells in 10-cm dishes were transfected with no. 1 siRNA for 48 h to deplete endogenous Ubqln4; transfected with control GFP-FLAG, Ubqln4*FLAG, or Ubqln4*Δ[H+STI1(1-2)]-FLAG for 24 h; and infected with SV40 (MOI, 15) for 16 h. Cells were harvested and lysed, and the resulting extract was subjected to immunoprecipitation using anti-FLAG agarose beads. The precipitated material was subjected to SDS-PAGE and immunoblotting with the indicated antibodies.
To further pinpoint whether the H domain, STI1 motif (1-2), or both regions of Ubqln4 are responsible for binding to B12, we generated Ubqln4*ΔH-FLAG and Ubqln4*ΔSTI1(1-2)-FLAG. Our analysis revealed that neither Ubqln4*ΔH-FLAG nor Ubqln4*ΔSTI1(1-2)-FLAG bound to B12 efficiently (Fig. 4C, top gel). Not surprisingly, neither construct restored SV40 infection in Ubqln4 knockdown cells (Fig. 4D). We conclude that both the H domain and the STI1 motif (1-2) of Ubqln4 support B12 binding that is crucial for promoting virus infection.
Ubqln4 binds to SV40 during entry.
Our results thus far depict a scenario in which Ubqln4, via its H domain and STI1 motif (1-2), is recruited to the ER membrane protein B12 (and B14). We posit that, strategically positioned at the ER-cytosol interface, Ubqln4 binds to SV40 as the viral particle emerges from the ER, thereby facilitating escape into the cytosol (Fig. 3). To test this, (CV-1-derived) COS-7 cells were first depleted of endogenous Ubqln4 by siRNA knockdown. The cells were then transfected with either GFP-FLAG, Ubqln4*-FLAG, or Ubqln4*Δ[H+STI1(1-2)]-FLAG and infected with SV40. COS-7 cells were used for this experiment due to the high transfection efficiency required for the study. The resulting cell extract was subjected to immunoprecipitation by FLAG antibodies, and the precipitated sample was subjected to immunoblotting. We found that Ubqln4*-FLAG, but not GFP-FLAG or Ubqln4*Δ[H+STI1(1-2)]-FLAG), pulled down SV40 VP1 (Fig. 4E, top gel). These results demonstrate that Ubqln4 binds to SV40 escaping the ER and that proper positioning of Ubqln4 next to the J proteins is critical for this interaction.
DISCUSSION
The molecular mechanism of membrane penetration by nonenveloped viruses remains enigmatic. This study further clarifies how the nonenveloped SV40 penetrates the ER membrane to reach the cytosol, a decisive infection step. Specifically, we found that the cytosolic chaperone Ubqln4 is a host factor that facilitates escape of SV40 from the ER into the cytosol (Fig. 5). Ubqln4 executes this role by interacting with the ER membrane J proteins B12 (and B14), which effectively positions Ubqln4 at the ER-cytosol interface. Strategically located at this juncture, Ubqln4 captures SV40 as the viral particle emerges from the ER into the cytosol—this engagement enables SV40 to successfully enter the cytosol.
FIG 5.
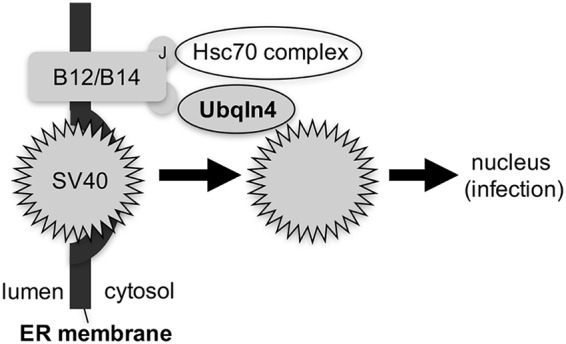
Ubqln4 promotes escape of SV40 into the cytosol from the ER. Ubqln4 is recruited to the cytosolic surface of the ER membrane via binding to the B12 (and B14) ER membrane J proteins. Strategically positioned at the ER-cytosol interface, Ubqln4 captures SV40 emerging from the ER to facilitate viral dislocation into the cytosol. Because Ubqln4 interacts with the J protein via a J-domain-independent mechanism, an individual J protein can recruit both Ubqln4 and a Hsc70 complex (via the J domain). We propose that Ubqln4 and the Hsc70 complex cooperate to extract SV40 into the cytosol.
One critical finding presented here is that Ubqln4 binds directly to the B12 J protein in a J-domain-independent manner. All J proteins are defined by the presence of a signature J domain, which is used to bind to Hsc/Hsp70 chaperones that stimulate their ATPase activities (30). However, we found that a B12 J domain mutant incapable of binding to Hsc70 nonetheless interacts with Ubqln4, suggesting that Ubqln4 binds to a region in B12 outside its J domain. This observation raises an intriguing scenario in which B12 can recruit two distinct cytosolic protein machineries to drive ER-to-cytosol escape of SV40. First, via its J domain, B12 recruits an Hsc70 protein complex (composed of Hsc70, Hsp105, SGTA, and Bag2) that we previously found to be essential in promoting SV40 ER membrane penetration (23–27). Second, via a J-domain-independent mechanism, B12 engages Ubqln4, which also facilitates viral escape into the cytosol.
Why two distinct cytosolic protein machineries are required to eject SV40 from the ER into the cytosol is an outstanding question that remains to be investigated. One possibility is that the Hsc70 protein complex actively extracts the ER membrane-embedded hydrophobic virus into the cytosol; simultaneously, Ubqln4 binds to and shields the exposed hydrophobic region of the same viral particle emerging from the ER, thereby protecting it against aggregation. The cooperative effects of extraction and shielding successfully dislocate SV40 into the cytosol. This idea is not without precedent, as the ubiquilins were previously shown to bind to cellular transmembrane proteins in the cytosol, shielding their hydrophobic domains to preclude aggregation (32).
Interestingly, our analysis also revealed that the H domain and ST1 motif (1-2) of Ubqln4 mediate B12 engagement. This observation is not entirely surprising, as the domain and motif are thought to support protein-protein interactions (33). Additionally, because the H domain can interact with hydrophobic proteins, such as the small hydrophobic protein of the mumps virus (34), the domain might also have a critical function in directly capturing the hydrophobic SV40 particle when it emerges from the ER.
ER-to-cytosol transport of SV40 likely exploits an endogenous ER protein quality control process called ER-associated degradation (ERAD). During ERAD, misfolded ER proteins are targeted to the cytosol, ubiquitinated, and degraded by the cytosolic proteasome (35). Because ubiquilins have been shown to function during ERAD (36, 37), how SV40 can coopt the action of Ubqln4 during ER escape into the cytosol yet avoid a degradative fate upon reaching the cytosol remains enigmatic and is clearly a fruitful area for future studies.
MATERIALS AND METHODS
Reagents.
Endogenous Ubiqln4, monoclonal large T antigen, Hsp90, and Hsp105 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Polyclonal DnaJ B12 and B14 antibodies were purchased from the Proteintech Group (Chicago, IL). Polyclonal DnaJ C18 antibody was generated by GenScript (Piscataway, NJ). Monoclonal VP1 antibody was kindly provided by Walter Scott (University of Miami). BiP and VP2/3 antibodies were purchased from Abcam (Cambridge, MA). Hsc70 was purchased from Invitrogen (Carlsbad, CA). Anti-FLAG antibody, FLAG M2 antibody-conjugated beads, Triton X-100, 2-mercaptoethanol, and BFA were purchased from Sigma-Aldrich (St. Louis, MO). Opti-MEM and 0.25% trypsin were purchased from Invitrogen (Carlsbad, CA). Prolong diamond antifade mount with DAPI (4′,6-diamidino-2-phenylindole) mounting reagent was purchased from Thermo Fisher (Carlsbad, CA). Phosphate-buffered saline (PBS) (1×) was purchased from Gibco (Carlsbad, CA). Digitonin, Deoxy Big CHAP (DBC), and phenylmethylsulfonyl fluoride (PMSF) were purchased from EMD Millipore Chemicals (San Diego, CA). Fugene HD was purchased from Promega (Madison, WI). CV-1 and COS-7 (ATCC) cells were cultured in complete Dulbecco’s modified Eagle medium (cDMEM) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Atlanta, GA), 10 U/ml penicillin, and 10 μg/ml streptomycin (Thermo Fisher Scientific).
siRNA transfection and DNA plasmid.
Allstar negative-control siRNA (Qiagen, Hilden, Germany) was used as a scrambled control siRNA. Two siRNAs specific to human Ubqln4 were synthesized by Sigma-Aldrich, and the target sequences were as follows: siRNA no. 1, CAAUGGAGCUUGCUCGGAA; siRNA no. 2, CAAACAGCAGGGUGACUUU. CV-1 and COS-7 cells were reverse transfected with each siRNA at 20 nM using Lipofectamine RNAi Max (Thermo Fisher Scientific) and incubated for 72 h prior to SV40 infection. A construct expressing human Ubqln4 was a gift from Henry L. Paulson (University of Michigan) and was cloned with a C-terminal FLAG tag into pcDNA3.1 minus (Invitrogen). To generate an siRNA-resistant construct, the following silent mutations (underlined) of siRNA no. 1 were introduced by PCR: CAATGGAACTAGCACGGAA. All truncated Ubqln4 mutants were constructed using the Ubqln4-FLAG-resistant construct as a template and the following primers: ΔUBLF262–284, GCTCAAGATCCAGCTGCTGCCAC; ΔUBLR35–57, GGTCTTGACGGTGACCCGAATGG; ΔHSTI1(1-2)F778–799, GAGATGATGCGGAACCAGGACG; ΔHSTI1(1-2)R383–408, CCGGCTTCCACTGCCAGCATCTGAAG; ΔSTI1(1-2)R552–576, ATTGGACATCAGCTGCCGCTGCATC; ΔHF574–597, AATCCTGAGATGCTGTCACAGATC; ΔSTI1(3-4)F1426–1446, CTACAGACCTTGCAGACCGAG; STI1(3-4)R1157–1179, GTTCTCAGAGATCTGCTGGAGGA; ΔUBAF1786–1806, CTGGGCTCCCAGCTCTCCTAA; and ΔUBAR1634–1656, CACCTGTGAGTTTCCACTTCCAG.
Preparation of SV40.
Preparation of purified SV40 using the OptiPrep gradient system (Sigma) has been described previously (12).
Immunoprecipitation and purification of FLAG-tagged protein.
For immunoprecipitation of endogenous B12, CV-1 cells in 15-cm dishes were lysed in lysis buffer containing 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM PMSF, and 1% DBC for 20 min and then centrifuged at 13,200 rpm at 4°C for 10 min. The cleared extracts were incubated with anti-B12 antibody overnight at 4°C with rotation. Protein A/G agarose beads were used to capture anti-B12 antibody, washed 4 times with lysis buffer containing 0.1% DBC, and boiled in 1× SDS buffer before being subjected to SDS-PAGE. For immunoprecipitation of FLAG-tagged proteins, COS-7 cells were seeded in 10-cm plates and transfected with FLAG-tagged constructs in polyethyleneimine (PEI) and Opti-MEM for 48 h. The cells were lysed as described above, and the supernatant was collected and rotated at 4°C for 2 h with M2 FLAG-conjugated beads. The beads were washed 4 times with lysis buffer containing 0.1% DBC, and the bound FLAG-tagged proteins were eluted with 1× FLAG peptide at 0.1 mg/ml. For purification of the FLAG-tagged proteins, 1% and 0.1% Triton X-100 were used (instead of 1% and 0.1% DBC) in the lysis buffer and washing buffer described above, respectively.
SV40 infection.
CV-1 cells were seeded and transfected with 20 nM Ubqln4 siRNA, 50 nM Hsp105 siRNA, or 50 nM scrambled siRNA (along with Opti-MEM and RNAiMax) onto glass coverslips in 12-well plates at 37°C for 72 h. For knockdown-rescue experiments, after 48 h of siRNA transfection as described above, the CV-1 cells were washed and transfected with the indicated FLAG-tagged constructs using Fugene and Opti-MEM for 24 h. Seventy-two hours posttransfection, the cells were infected with SV40 (multiplicity of infection [MOI], 0.5) for 20 to 24 h, fixed, and stained using antibodies against SV40 TAg, as described previously (12). For evaluating SV40 infection in cells without DNA transfection, at least 1,000 cells were counted per condition. To assess SV40 infection in rescue experiments, at least 150 FLAG-expressing cells were counted per condition. For each infection experiment, at least 500 FLAG-expressing cells were counted.
VP2/VP3 exposure assay.
CV-1 cells were seeded and transfected with 20 nM Ubqln4 siRNA or 20 nM scrambled siRNA onto glass coverslips in 12-well plates at 37°C for 72 h. The cells were then infected with SV40 (MOI, 1) for 6 h. For the BFA-treated cells, BFA (5 μg/ml) was added to the cells before virus infection for 2 h, and then the cells were incubated with virus for 6 h. The cells were fixed and subjected to immunofluorescence microscopy using VP1 and VP2/3 antibodies. Microscopy was performed using a Zeiss LSM 800 confocal laser scanning microscope with a Plan-Apochromat 40×/1.4 oil differential interference contrast M27 objective. For each coverslip, at least three images were taken, and three independent experiments were performed. The VP1 and VP2/3 signals were measured by FIJI distribution of ImageJ (FIJI/ImageJ) (National Institutes of Health).
XBP1 splicing assay.
Reverse transcription (RT)-PCR was performed to detect XBP1 splicing as described previously (38) using the following primers: 5′-CGCGGATCCGAATGTGAGGCCAGTGG-3′ and 5′-GGGGCTTGGTATATATGTGG-3′.
ER-to-cytosol transport assay.
The ER-to-cytosol transport assay was performed as described previously (12). Briefly, 5 × 105 CV-1 cells were seeded in 6-cm plates and transfected with 20 nM Ubqln4 siRNA, 50 nM scrambled siRNA, or 50 nM Hsp105 siRNA for 72 h. The cells were then infected with purified SV40 (MOI, 5) for 16 h, semipermeabilized using 0.1% digitonin for 10 min, and then centrifuged. The resulting supernatant fraction represented the cytosol fraction, while the pellet fraction represented the membrane fraction. The membrane fraction was further treated with 1% Triton X-100 and subjected to centrifugation—the resulting supernatant contained ER-localized SV40 (12). The VP1 band intensity was quantified using FIJI/ImageJ software and normalized to the Hsp90 loading control band.
ACKNOWLEDGMENTS
We thank Chelsey Spriggs for her critical comments on the manuscript and Parikshit Bagchi for technical assistance.
This work was funded by grants from the National Institutes of Health (RO1 AI064296 and R21 AI140449) to B.T.
REFERENCES
- 1.Spriggs CC, Harwood MC, Tsai B. 2019. How non-enveloped viruses hijack host machineries to cause infection. Adv Virus Res 104:97–122. doi: 10.1016/bs.aivir.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar CS, Dey D, Ghosh S, Banerjee M. 2018. Breach: host membrane penetration and entry by nonenveloped viruses. Trends Microbiol 26:525–537. doi: 10.1016/j.tim.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Moyer CL, Nemerow GR. 2011. Viral weapons of membrane destruction: variable modes of membrane penetration by non-enveloped viruses. Curr Opin Virol 1:44–49. doi: 10.1016/j.coviro.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toscano MG, de Haan P. 2018. How simian virus 40 hijacks the intracellular protein trafficking pathway to its own benefit … and ours. Front Immunol 9:1160. doi: 10.3389/fimmu.2018.01160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen YJ, Liu X, Tsai B. 2019. SV40 hijacks cellular transport, membrane penetration, and disassembly machineries to promote infection. Viruses 11:E917. doi: 10.3390/v11100917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC. 1991. Structure of simian virus 40 at 3.8-A resolution. Nature 354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 7.Stehle T, Gamblin SJ, Yan Y, Harrison SC. 1996. The structure of simian virus 40 refined at 3.1 A resolution. Structure 4:165–182. doi: 10.1016/s0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 8.Chen XS, Stehle T, Harrison SC. 1998. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J 17:3233–3240. doi: 10.1093/emboj/17.12.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai B, Gilbert JM, Stehle T, Lencer W, Benjamin TL, Rapoport TA. 2003. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J 22:4346–4355. doi: 10.1093/emboj/cdg439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert J, Benjamin T. 2004. Uptake pathway of polyomavirus via ganglioside GD1a. J Virol 78:12259–12267. doi: 10.1128/JVI.78.22.12259-12267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kartenbeck J, Stukenbrok H, Helenius A. 1989. Endocytosis of simian virus 40 into the endoplasmic reticulum. J Cell Biol 109:2721–2729. doi: 10.1083/jcb.109.6.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue T, Tsai B. 2011. A large and intact viral particle penetrates the endoplasmic reticulum membrane to reach the cytosol. PLoS Pathog 7:e1002037. doi: 10.1371/journal.ppat.1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geiger R, Andritschke D, Friebe S, Herzog F, Luisoni S, Heger T, Helenius A. 2011. BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat Cell Biol 13:1305–1314. doi: 10.1038/ncb2339. [DOI] [PubMed] [Google Scholar]
- 14.Schelhaas M, Malmstrom J, Pelkmans L, Haugstetter J, Ellgaard L, Grunewald K, Helenius A. 2007. Simian virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131:516–529. doi: 10.1016/j.cell.2007.09.038. [DOI] [PubMed] [Google Scholar]
- 15.Walczak CP, Tsai B. 2011. A PDI family network acts distinctly and coordinately with ERp29 to facilitate polyomavirus infection. J Virol 85:2386–2396. doi: 10.1128/JVI.01855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue T, Dosey A, Herbstman JF, Ravindran MS, Skiniotis G, Tsai B. 2015. ERdj5 reductase cooperates with protein disulfide isomerase to promote simian virus 40 endoplasmic reticulum membrane translocation. J Virol 89:8897–8908. doi: 10.1128/JVI.00941-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniels R, Rusan NM, Wadsworth P, Hebert DN. 2006. SV40 VP2 and VP3 insertion into ER membranes is controlled by the capsid protein VP1: implications for DNA translocation out of the ER. Mol Cell 24:955–966. doi: 10.1016/j.molcel.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Kuksin D, Norkin LC. 2012. Disassembly of simian virus 40 during passage through the endoplasmic reticulum and in the cytoplasm. J Virol 86:1555–1562. doi: 10.1128/JVI.05753-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodwin EC, Lipovsky A, Inoue T, Magaldi TG, Edwards AP, Van Goor KE, Paton AW, Paton JC, Atwood WJ, Tsai B, DiMaio D. 2011. BiP and multiple DNAJ molecular chaperones in the endoplasmic reticulum are required for efficient simian virus 40 infection. mBio 2:e00101-11. doi: 10.1128/mBio.00101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue T, Tsai B. 2015. A nucleotide exchange factor promotes endoplasmic reticulum-to-cytosol membrane penetration of the nonenveloped virus simian virus 40. J Virol 89:4069–4079. doi: 10.1128/JVI.03552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rainey-Barger EK, Magnuson B, Tsai B. 2007. A chaperone-activated nonenveloped virus perforates the physiologically relevant endoplasmic reticulum membrane. J Virol 81:12996–13004. doi: 10.1128/JVI.01037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bagchi P, Inoue T, Tsai B. 2016. EMC1-dependent stabilization drives membrane penetration of a partially destabilized non-enveloped virus. Elife 5:e21470. doi: 10.7554/eLife.21470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walczak CP, Ravindran MS, Inoue T, Tsai B. 2014. A cytosolic chaperone complexes with dynamic membrane J-proteins and mobilizes a nonenveloped virus out of the endoplasmic reticulum. PLoS Pathog 10:e1004007. doi: 10.1371/journal.ppat.1004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagchi P, Walczak CP, Tsai B. 2015. The endoplasmic reticulum membrane J protein C18 executes a distinct role in promoting simian virus 40 membrane penetration. J Virol 89:4058–4068. doi: 10.1128/JVI.03574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravindran MS, Bagchi P, Inoue T, Tsai B. 2015. A non-enveloped virus hijacks host disaggregation machinery to translocate across the endoplasmic reticulum membrane. PLoS Pathog 11:e1005086. doi: 10.1371/journal.ppat.1005086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dupzyk A, Williams JM, Bagchi P, Inoue T, Tsai B. 2017. SGTA-dependent regulation of Hsc70 promotes cytosol entry of simian virus 40 from the endoplasmic reticulum. J Virol 91:e00232-17. doi: 10.1128/JVI.00232-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dupzyk A, Tsai B. 2018. Bag2 is a component of a cytosolic extraction machinery that promotes membrane penetration of a nonenveloped virus. J Virol 92:e00607-18. doi: 10.1128/JVI.00607-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue T, Tsai B. 2017. Regulated Erlin-dependent release of the B12 transmembrane J-protein promotes ER membrane penetration of a non-enveloped virus. PLoS Pathog 13:e1006439. doi: 10.1371/journal.ppat.1006439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee DY, Brown EJ. 2012. Ubiquilins in the crosstalk among proteolytic pathways. Biol Chem 393:441–447. doi: 10.1515/hsz-2012-0120. [DOI] [PubMed] [Google Scholar]
- 30.Kampinga HH, Andreasson C, Barducci A, Cheetham ME, Cyr D, Emanuelsson C, Genevaux P, Gestwicki JE, Goloubinoff P, Huerta-Cepas J, Kirstein J, Liberek K, Mayer MP, Nagata K, Nillegoda NB, Pulido P, Ramos C, De Los Rios P, Rospert S, Rosenzweig R, Sahi C, Taipale M, Tomiczek B, Ushioda R, Young JC, Zimmermann R, Zylicz A, Zylicz M, Craig EA, Marszalek J. 2019. Function, evolution, and structure of J-domain proteins. Cell Stress Chaperones 24:7–15. doi: 10.1007/s12192-018-0948-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Şentürk M, Lin G, Zuo Z, Mao D, Watson E, Mikos AG, Bellen HJ. 2019. Ubiquilins regulate autophagic flux through mTOR signalling and lysosomal acidification. Nat Cell Biol 21:384–396. doi: 10.1038/s41556-019-0281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itakura E, Zavodszky E, Shao S, Wohlever ML, Keenan RJ, Hegde RS. 2016. Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol Cell 63:21–33. doi: 10.1016/j.molcel.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lassle M, Blatch GL, Kundra V, Takatori T, Zetter BR. 1997. Stress-inducible, murine protein mSTI1. Characterization of binding domains for heat shock proteins and in vitro phosphorylation by different kinases. J Biol Chem 272:1876–1884. doi: 10.1074/jbc.272.3.1876. [DOI] [PubMed] [Google Scholar]
- 34.Woznik M, Rodner C, Lemon K, Rima B, Mankertz A, Finsterbusch T. 2010. Mumps virus small hydrophobic protein targets ataxin-1 ubiquitin-like interacting protein (ubiquilin 4). J Gen Virol 91:2773–2781. doi: 10.1099/vir.0.024638-0. [DOI] [PubMed] [Google Scholar]
- 35.Sun Z, Brodsky JL. 2019. Protein quality control in the secretory pathway. J Cell Biol 218:3171–3187. doi: 10.1083/jcb.201906047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim PJ, Danner R, Liang J, Doong H, Harman C, Srinivasan D, Rothenberg C, Wang H, Ye Y, Fang S, Monteiro MJ. 2009. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J Cell Biol 187:201–217. doi: 10.1083/jcb.200903024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim TY, Kim E, Yoon SK, Yoon JB. 2008. Herp enhances ER-associated protein degradation by recruiting ubiquilins. Biochem Biophys Res Commun 369:741–746. doi: 10.1016/j.bbrc.2008.02.086. [DOI] [PubMed] [Google Scholar]
- 38.Uemura A, Oku M, Mori K, Yoshida H. 2009. Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J Cell Sci 122:2877–2886. doi: 10.1242/jcs.040584. [DOI] [PubMed] [Google Scholar]