Plants employ multiple defense mechanisms to restrict viral infections, among which RNA interference is the best understood. The activation of innate immunity often leads to both local and systemic necrotic responses, which confine the virus to the infected cells and can also provide resistance to distal, noninfected parts of the organism. Systemic necrosis, which is regarded as a special form of the local hypersensitive response, results in necrosis of the apical stem region, usually causing the death of the plant. Here, we provide evidence that the dsRNA-binding protein 2 of Nicotiana benthamiana plays an important role in virus-induced systemic necrosis. Our findings are not only compatible with the recent hypothesis that DRB proteins act as viral invasion sensors but also extends it by proposing that DRBs play a critical role in establishing the dominant antiviral measures that are triggered during virus infection.
KEYWORDS: DRB2, PAMP-triggered immunity, RDR6, antiviral RNA interference, double-stranded RNA
ABSTRACT
Double-stranded RNA (dsRNA) is a common pattern formed during the replication of both RNA and DNA viruses. Perception of virus-derived dsRNAs by specialized receptor molecules leads to the activation of various antiviral measures. In plants, these defensive processes include the adaptive RNA interference (RNAi) pathway and innate pattern-triggered immune (PTI) responses. While details of the former process have been well established in recent years, the latter are still only partially understood at the molecular level. Nonetheless, emerging data suggest extensive cross talk between the different antiviral mechanisms. Here, we demonstrate that dsRNA-binding protein 2 (DRB2) of Nicotiana benthamiana plays a direct role in potato virus X (PVX)-elicited systemic necrosis. These results establish that DRB2, a known component of RNAi, is also involved in a virus-induced PTI response. In addition, our findings suggest that RNA-dependent polymerase 6 (RDR6)-dependent dsRNAs play an important role in the triggering of PVX-induced systemic necrosis. Based on our data, a model is formulated whereby competition between different DRB proteins for virus-derived dsRNAs helps establish the dominant antiviral pathways that are activated in response to virus infection.
IMPORTANCE Plants employ multiple defense mechanisms to restrict viral infections, among which RNA interference is the best understood. The activation of innate immunity often leads to both local and systemic necrotic responses, which confine the virus to the infected cells and can also provide resistance to distal, noninfected parts of the organism. Systemic necrosis, which is regarded as a special form of the local hypersensitive response, results in necrosis of the apical stem region, usually causing the death of the plant. Here, we provide evidence that the dsRNA-binding protein 2 of Nicotiana benthamiana plays an important role in virus-induced systemic necrosis. Our findings are not only compatible with the recent hypothesis that DRB proteins act as viral invasion sensors but also extends it by proposing that DRBs play a critical role in establishing the dominant antiviral measures that are triggered during virus infection.
INTRODUCTION
Plants possess a sophisticated multilayered immune system which protects them against a variety of invading pathogens, including viruses (1, 2). This immune system includes processes which can broadly be classified as innate and adaptive immune responses. Double-stranded RNAs (dsRNAs) are generally associated with the replication of both RNA and DNA viruses and can activate both types of immunity. Pattern-triggered immunity (PTI) is a form of innate immune response, which is activated following the recognition of evolutionarily conserved pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs). Virus-derived dsRNAs are recognized as PAMPs by specific PRRs, resulting in the activation of downstream signaling cascades. These events eventually trigger various defense measures, including local or systemic necrosis. In animal cells, viral dsRNAs are sensed by membrane-bound Toll-like receptors (TLRs) and intracellular retinoic acid-inducible I-like proteins (RIG-I) (3, 4). The dsRNA-binding PRRs of plants have not been identified yet (5, 6).
In addition to PTI, virus-derived dsRNAs can also activate an adaptive immune response in plants, known as antiviral RNA interference (RNAi). Though antiviral RNAi was discovered much more recently, our understanding of it at the molecular level is more complete than that of most of the processes of innate immunity (7–9). Briefly, viral dsRNAs of various origins (replicative intermediates or genomic intramolecular fold-back structures) are processed into primary viral small interfering RNAs (vsiRNAs) by RNase III-like enzymes (Dicer-like 2 [DCL2], DCL3, and DCL4) with the assistance of dsRNA-binding domain (dsRBD)-possessing cofactors (DRBs). Endogenous RNA-dependent RNA polymerases (RdRps) also synthesize dsRNAs using virus-derived single-stranded RNAs (ssRNAs) as templates, thereby providing substrates for the production of secondary vsiRNAs. Both classes of vsiRNAs are eventually incorporated into argonaute (AGO)-protein containing RNA-induced silencing complexes (RISCs), which can limit virus replication via a variety of mechanisms. A portion of the vsiRNAs leaves the infected cells and enters into the vascular system of the plant, thereby limiting the systemic spread of viruses. In turn, viruses have developed a number of countermeasures to avoid neutralization by the above-mentioned mechanisms, including the deployment of viral proteins that possess the ability to interfere with RNAi at various steps (viral suppressors of RNA silencing [VSRs]) (10).
Increasing amounts of data suggest extensive cross talk between the dsRNA-activated innate and adaptive immune responses, and dsRBD-containing proteins may represent the potential points of contact (6, 11). Of the four principal protein families involved in antiviral RNAi Dicer-like proteins (DCLs), DRBs and RdRps possess dsRBDs (12). Due to their extensive similarity to vertebrate RIG-I proteins, DCLs seemed to be the obvious candidates as the universal plant dsRNA-binding PRRs. A recent report, however, has clearly demonstrated that, unlike antiviral RNAi, dsRNA-induced PTI is not impaired in dcl mutant plants (5). Several members of the DRB protein family are obligate binding partners of DCLs, indicating their intimate involvement in RNA silencing (13, 14). For example, DRB1 (HYL1) and DRB4 are required by DCL1 and DCL4 to efficiently process their structurally distinct dsRNA substrates in the microRNA (miRNA) and siRNA biogenesis pathways, respectively. Functions of the remaining members of the DRB family (DRB2, DRB3, and DRB5) are more obscure although sporadic data have also implicated them in miRNA biogenesis and action (15–18). A recent live-cell imaging study revealed that DRB2, DRB3, and DRB5 relocated from their uniform cytoplasmic distribution to nascent viral replication complexes (VRCs) that developed in the cell following virus infection (19). Although no mechanistic details were provided, the authors hypothesized that DRB proteins might act as viral invasion sensors and contribute to the triggering of various defense measures. Prompted by this result, we reexamined the involvement of members of the DRB protein family in antiviral responses. Our findings indicate that DRB2 plays a direct role in potato virus X (PVX)-induced systemic necrosis and thus may be an integral component of virus elicited PTI.
RESULTS
Cloning of N. benthamiana DRBs.
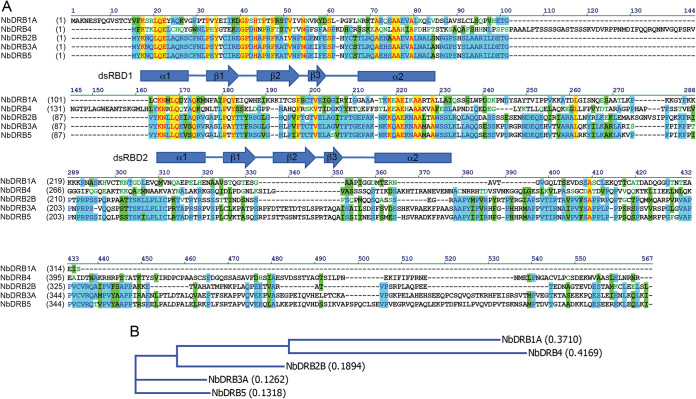
Available transcriptome data sets indicate that the DRB protein family has five members in Nicotiana benthamiana, similar to findings in Arabidopsis thaliana (20, 21). The role of DRB1 (HYL1) in miRNA biogenesis has been firmly established, but its direct involvement in antiviral RNAi has not been documented. Therefore, DRB1 was not included in our subsequent analyses; instead, we were mostly focusing on the remaining members of the DRB protein family. The full-length cDNAs of DRB2, DRB3, DRB4, and DRB5 were cloned and inserted into binary expression vectors along with N-terminal hemagglutinin (HA) epitope tags. N. benthamiana is an allotetraploid species; consequently, it expresses highly related homeologs of many genes, including DRB2 and DRB3. Of these, only one of the homeologs was studied in detail (DRB2B for DRB2 and DRB3A for DRB3). The N. benthamiana DRB proteins exhibit the same overall domain architecture as reported previously for the A. thaliana DRB proteins; viz. the two highly conserved N-terminal dsRBD domains are followed by a more divergent C-terminal tail region (Fig. 1) (13, 14). The latter segments of DRB1 and DRB4 are unique, exhibiting no recognizable similarity to the remaining members of the DRB family or to each other. In contrast, the C-terminal tails of DRB3 and DRB5 are highly homologous, suggesting that their functions are also closely related. DRB2 in its C-terminal region shares strings of identical amino acids with DRB3 and DRB5; nonetheless, the presence of several large deletions may be an indication of DRB2’s distinct function.
FIG 1.
(A) Multiple alignment of the cloned N. benthamiana (Nb) DRB proteins. Positions of the two conserved N-terminal dsRBD domains are indicated. (B) Phylogenetic tree of the N. benthamiana DRB proteins. Calculated distance values according to the neighbor joining method are given in parentheses.
DRB2 inhibits antiviral RNAi.
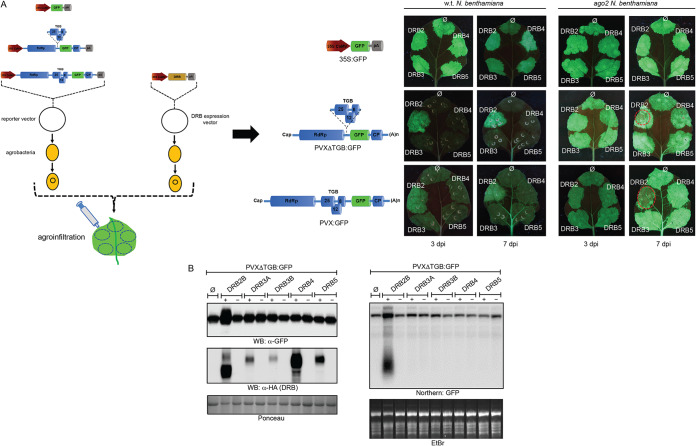
Effects of DRB proteins on virus replication were tested in transient agroinfiltration-based assays (Fig. 2A). Previously, using a similar approach, antiviral activities of different AGO proteins were successfully demonstrated, and functionally important regions of the main antiviral argonaute protein, AGO2, were also identified (22, 23). Virus infections were initiated in N. benthamiana leaves from agroinfiltrated binary vectors which contained either the intact or the VSR-deficient PVX genome in green fluorescent protein (GFP)-tagged forms (PVX-GFP or PVXΔTGB-GFP, where TGB is triple gene block). DRB proteins were also coexpressed with PVX using coinfiltrated binary expression vectors. To visualize GFP expression, at 3 and 7 days postinfiltration (dpi) the infiltrated leaves were examined under UV light. In most of the infiltrated leaf patches, GFP fluorescence was low regardless of whether or not a DRB protein was coexpressed with the virus. This observation is consistent with the strong silencing response generally elicited by viruses encoding weak or mutant VSR proteins (10). Unlike expression of the other DRBs, however, coexpression of DRB2 with PVXΔTGB-GFP reproducibly resulted in a higher GFP level than that of the control not expressing DRB. The increased GFP signal intensity was evident with PVX-GFP as well, especially at 7 dpi. None of the DRBs affected the GFP levels significantly produced by a Cauliflower mosaic virus (CaMV) promoter-driven GFP control vector, underscoring the specific effect of DRB2 on PVX (Fig. 2A). The elevated virus-encoded GFP levels were followed closely by those of the viral genomic RNA (gRNA) (Fig. 2B).
FIG 2.
Analysis of the effects of DRB proteins on PVX replication. (A) An agroinfiltration-based transient virus replication assay was used to evaluate the effects of ectopic expression of DRB proteins on PVX replication. Suspensions of agrobacteria carrying either PVX-GFP- or PVXΔTGB-GFP-encoding binary vectors were infiltrated into leaves of wild-type (wt) or ago2 N. benthamiana. As a control a CaMV 35S promoter-driven GFP expression vector was used. Along with the reporter constructs, agrobacteria carrying binary expression vectors for DRB proteins were also codelivered. As a negative control, a coinfiltrated suspension of empty agrobacteria (∅) was employed. The bacterial suspensions (optical density at 600 nm of 1) were mixed at a 1:1 ratio. GFP expression in the infiltrated leaf patches was monitored using an appropriate UV light source at 3 and 7 dpi. Pictures of representative leaves are shown. At 7 dpi, PVX- and DRB2-coinfiltrated leaf patches frequently exhibited necrosis, which resulted in the apparent fading of the GFP signal. These necrotic areas are circled in red. Experiments were repeated three times. (B) Expression of PVXΔTGB-GFP-encoded GFP was monitored by Western blotting (WB). Leaves of wild-type N. benthamiana were agroinfiltrated with a PVXΔTGB-GFP reporter either alone (right side of leaves) or combined with an expression vector for one of the indicated DRB proteins (left side of leaves). At 5 dpi samples were collected and pooled from three identically infiltrated leaves. Protein lysates were prepared from the pooled samples and analyzed by GFP antibody. To verify the expression of the DRB proteins, the same filter was probed with HA antibody. As a loading control, the Ponceau-stained filter is shown. From the above-described infiltrated leaves, total RNA samples were also prepared, and viral RNA levels were monitored by Northern blotting (right panel). The Northern blot was probed with a radioactively labeled GFP DNA fragment. The ethidium bromide (EtBr)-stained gel is shown as a loading control. Protein lysate or total RNA prepared from PVXΔTGB-GFP and empty agrobacteria (∅)-infiltrated leaves were used as negative controls in the Western and Northern blots.
The increased PVX accumulation observed in the presence of overexpressed DRB2 relative to that of the other DRBs could be a consequence of either accelerated virus replication or diminished antiviral silencing. The main antiviral silencing effector targeting PVX is AGO2 (24, 25). If DRB2 acts as a factor promoting virus replication, its positive effect on PVX levels should be seen not only in wild-type but also in ago2 plants as well. Unlike that observed in wild-type plants, infiltration of PVXΔTGB-GFP into leaves of ago2 plants resulted in high GFP fluorescence signal intensity, consistent with the strongly diminished antiviral silencing. Importantly, coexpression of none of the DRBs affected the GFP levels in the ago2 plants (Fig. 2B). This finding indicates that DRB2 interferes with antiviral silencing and is unlikely to act as a host replication factor for PVX.
The dsRBD1 domain of DRB2 is necessary and sufficient for the inhibition of antiviral RNAi.
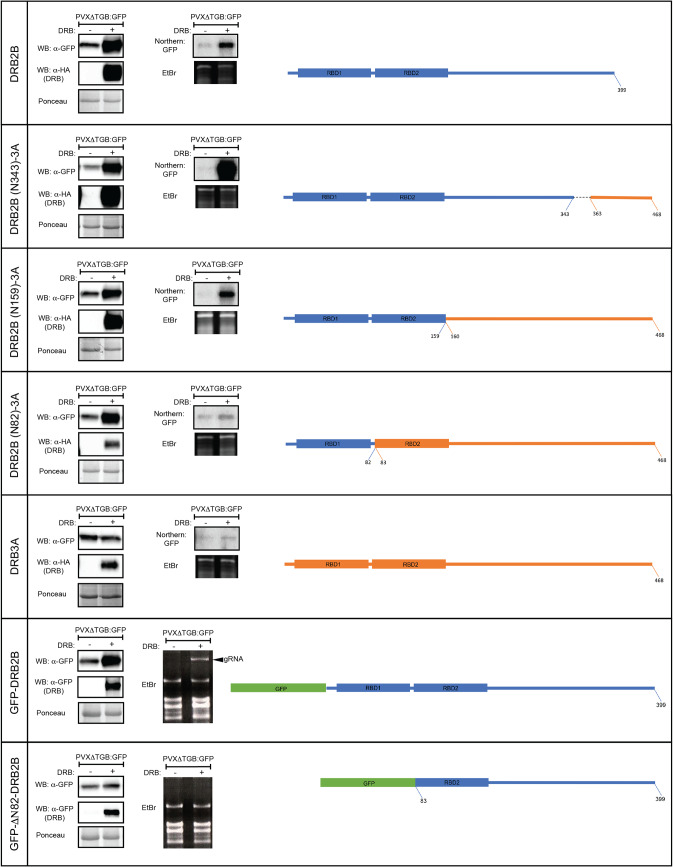
To better understand the mechanism DRB2 employs to inhibit RNA silencing, a series of domain-swapping and deletion mutants were created. Starting from the C termini, equivalent segments of DRB2B and DRB3A were exchanged. Binary vectors expressing the chimeric DRB molecules were coinfiltrated with PVXΔTGB-GFP into N. benthamiana leaves, and GFP levels were monitored by Western blotting at 5 dpi (Fig. 3). We found that the dsRBD1 domain of DRB2B (amino acids [aa] 1 to 82) was necessary and sufficient to abolish the silencing of GFP expression. Rescue of the viral gRNA level was also dependent on the same domain. Analysis of N-terminal deletion mutants of DRB2 corroborated the above results.
FIG 3.
Mapping of the domain of DRB2 necessary for the inhibition of antiviral RNAi. A PVXΔTGB-GFP binary vector was agroinfiltrated either alone or along with binary constructs expressing the depicted mutant DRB proteins into leaves of wild-type N. benthamiana (left or right sides of the same leaves, respectively). At 5 dpi samples were collected and pooled from three identically infiltrated leaves. From the collected samples, protein lysates and total RNA were prepared. GFP and the DRB mutant protein expression levels were monitored by Western blotting (WB) using GFP and HA antibodies, respectively (left panels). PVX gRNA levels were detected by Northern blotting using radioactively labeled GFP probe (right panels). In the case of the bottom two constructs, a GFP tag was used instead of an HA tag (the GFP tag was employed because the HA tag could not stabilize the N-terminally deleted mutant of DRB2). Accordingly, GFP antibody was used to detect DRB expression as well. In these cases, no Northern blots were performed; instead, images of ethidium bromide (EtBr)-stained gels are shown in which the position of the PVX gRNA is indicated by an arrowhead. DRB2B- and DRB3A-derived segments of the chimeric DRB molecules are indicated by blue and orange colors, respectively. The green box represents GFP. The corresponding Ponceau-stained protein filters and EtBr-stained RNA gels are shown as loading controls for the Western and Northern blots, respectively.
Inhibition of antiviral RNAi by DRB2 does not depend on RDR6.
The most distinguishing feature of DRB proteins is the presence of two highly conserved dsRBD domains at their N termini. Although the two dsRBDs exhibit high sequence similarity and adopt the same canonical α-β-β-α fold, functionally they do not seem to be equivalent. Available structural and functional data on DRB1 and DRB4 indicate that while dsRBD1 is predominantly responsible for high-affinity dsRNA binding, dsRBD2 is mostly mediating protein-protein interactions (26–31). DRB2 binds dsRNA that conceivably also depends on its dsRBD1, the same domain that is necessary for silencing inhibition (see above). Therefore, one might hypothesize that DRB2 inhibits RNAi as a consequence of its ability to compete with components of the silencing pathway that exhibit similar substrate specificity (such as dsRNA binding). The most likely candidates are RdRps and DCLs. To test this assumption, first we examined whether DRB2 overexpression can interfere with silencing triggered by a hairpin RNA, which can directly be converted into siRNAs by DCLs in an RdRp-independent fashion (Fig. 4A). A PVXΔTGB-GFP-expressing binary vector was agroinfiltrated into N. benthamiana leaves either alone or in combination with a GFP hairpin-expressing binary construct (GFP-inverted repeat [GFP-IR]). Coinfiltration of GFP-IR strongly reduced the amount of virus-derived GFP. Coexpression of DRB2 was able to restore the GFP level and, partially, the viral gRNA level as well. This finding strongly indicates that DRB2 blocks the RNA silencing pathway downstream of the actions of RdRps. To confirm this result, we examined the ability of DRB2 to inhibit RNAi in an rdr6 N. benthamiana plant line, which we have created recently using genome editing (32). An efficient defense response of N. benthamiana against PVX infection was shown to be dependent on RDR6 (32–35). This RdRp produces the long dsRNAs from PVX-derived ssRNAs (subgenomic RNAs [sgRNAs] and aberrant RNA fragments), which are subsequently diced by DCLs into secondary vsiRNAs. Importantly, DRB2 overexpression was able to diminish antiviral silencing even in this plant, as evidenced by the restored PVX coat protein (PVX-CP) and viral gRNA levels (Fig. 4B). Combined, these results provide conclusive proof that DRB2 inhibits antiviral RNAi in an RDR6-independent fashion.
FIG 4.
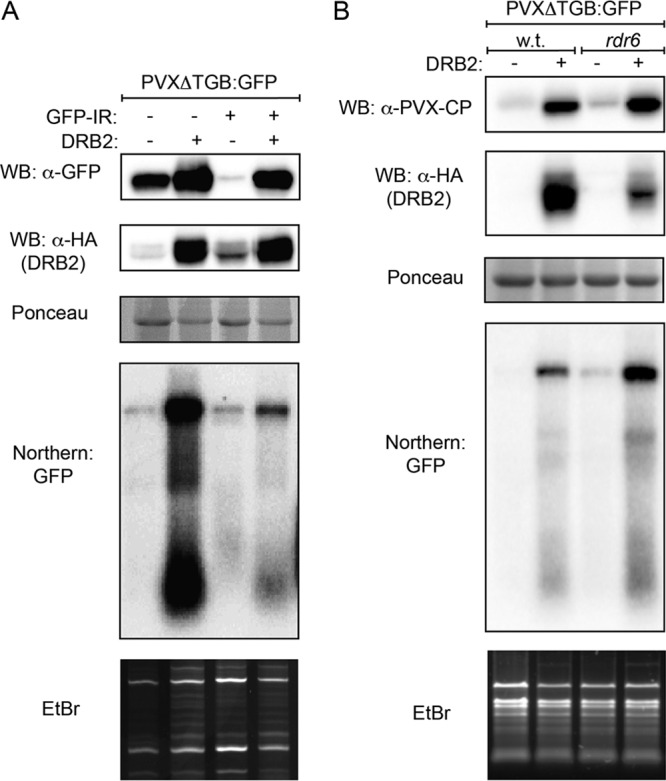
Inhibition of RNAi by DRB2 does not depend on RDR6. (A) Inverted-repeat-initiated posttranscriptional gene silencing is inhibited by DRB2. The indicated binary expression constructs were agroinfiltrated into leaves of wild-type N. benthamiana. At 5 dpi, samples were collected and pooled from three identically infiltrated leaves. From the collected samples protein lysates and total RNA were prepared. Viral protein synthesis and DRB2 protein levels were monitored by Western blotting using GFP and HA antibodies, respectively. Note that the HA antibody detects nonspecific bands in the first and third lanes at positions comparable to those of DRB2. The PVXΔTGB-GFP virus level was followed in Northern blotting using a radioactively labeled GFP probe. (B) DRB2 inhibits RNAi in rdr6 mutant N. benthamiana. Leaves of wild-type (wt) and rdr6 N. benthamiana were agroinfiltrated with a PVXΔTGB-GFP reporter either alone (left side of leaves) or combined with a DRB2 expression vector (right side of leaves). At 5 dpi, samples were collected and pooled from three identically infiltrated leaves. From the collected samples protein lysates and total RNA were prepared. Viral protein synthesis and DRB2 protein levels were monitored by Western blotting using PVX-CP and HA antibodies, respectively. PVXΔTGB-GFP virus level was followed by Northern blotting using a radioactively labeled GFP probe. The corresponding Ponceau-stained protein filters and ethidium bromide (EtBr)-stained RNA gels are shown as loading controls for the Western and Northern blots, respectively.
DRB2 inhibits DCL activity.
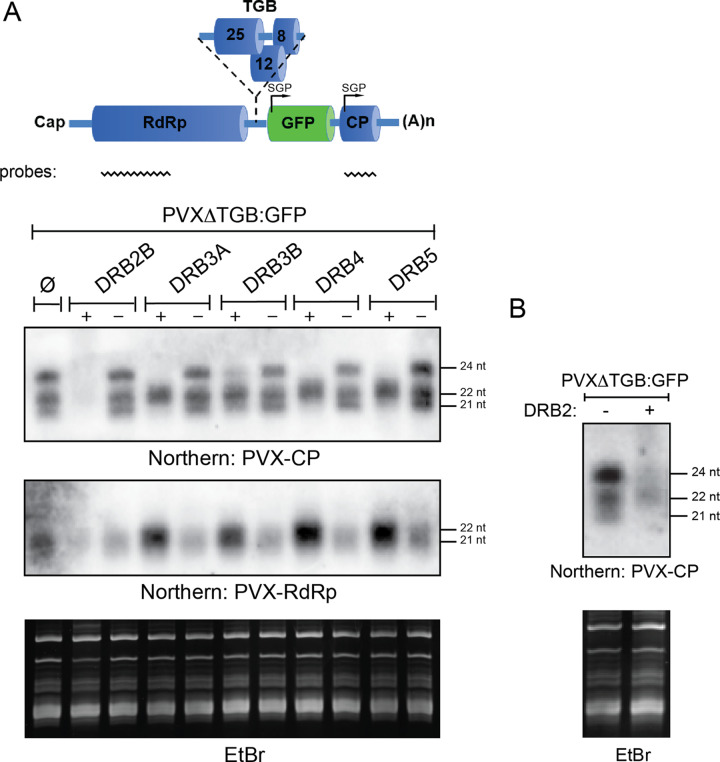
Since DRB2 does not target RDR6, we examined the possibility that it may influence the activity of DCLs. To this end, the effect of DRB2 overexpression on the production of vsiRNAs was examined. A PVXΔTGB-GFP-expressing binary vector was infiltrated into wild-type N. benthamiana leaves either alone or in combination with a DRB2 expression vector. As controls, the rest of the cloned DRBs were also included in the experiment. Production of vsiRNAs that originated from two different regions of the virus was monitored in small-RNA Northern blotting (Fig. 5A). A PVX-CP-specific probe detected vsiRNAs of all three size classes (21, 22, and 24 nucleotides [nt]) implicated in antiviral silencing. Interestingly, the PVX-RdRp region spawned predominantly 21-nt vsiRNAs. This difference is likely due to the differential accessibility of the CP encoding viral subgenomic RNAs (sgRNAs) and the RdRp encoding gRNAs to the various DCLs involved in vsiRNA production. Regardless, overexpression of DRB2 almost completely abrogated the production of vsiRNAs of all size classes, consistent with its strong inhibitory effect on antiviral silencing. The same was observed in rdr6 N. benthamiana as well, confirming that DRB2 predominantly targets DCLs (Fig. 5B). Interestingly, overexpression of DRB3, DRB4, and DRB5 also reduced the accumulation of the 21-nt and the 24-nt vsiRNAs. However, their effect on the buildup of the 22-nt vsiRNAs was the opposite, with the most substantial increase observable in the level of vsiRNAs originating from the RdRp region. The inverse correlation between the amounts of the 21-nt and 22-nt vsiRNAs is consistent with the known substrate competition between DCL2 and DCL4 (36). Importantly, this phenomenon can also explain how efficient silencing is maintained upon the overexpression of DRB3, DRB4, and DRB5. Combined, the above results indicate that, unlike the other DRBs, DRB2 can inhibit antiviral silencing efficiently due to its ability to block the dicing activity of all three DCLs involved in vsiRNA production. However, as a supplementary mechanism, direct inhibition of vsiRNA-loaded RISCs by DRB2 cannot be discounted either, particularly since dicing and RISC loading are biochemically closely coupled.
FIG 5.
DRB2 inhibits the activity of all DCLs involved in antiviral RNAi. (A) The RNA samples analyzed in the experiment shown in Fig. 1 were used for small RNA Northern blots to examine the vsiRNA production from different regions of PVXΔTGB-GFP in the presence or absence of DRB proteins. Two different probes were used to monitor vsiRNA levels. The positions of the probes are indicated at the top of the cartoon. The filter was first probed with a radioactively labeled PVX-RdRp fragment and subsequently with a PVX-CP fragment (after stripping). The ethidium bromide (EtBr)-stained gel is shown as a loading control. SGP, subgenomic promoter; (A)n, poly(A) tail. (B) DRB2 overexpression inhibits the activity of all DCLs involved in antiviral RNAi in rdr6 N. benthamiana. Leaves of rdr6 N. benthamiana were agroinfiltrated with PVX:ΔTGB-GFP either alone (left side of leaves) or together with a DRB2 binary expression construct (right side of leaves). At 5 dpi samples were collected and pooled from three infiltrated leaves, and total RNAs were prepared. RNAs were analyzed on a small RNA blot as described above. The filter was probed with radioactively labeled PVX-CP probe. The EtBr-stained gel is shown as a loading control.
DRB2 induces necrosis in the presence of replicating PVX.
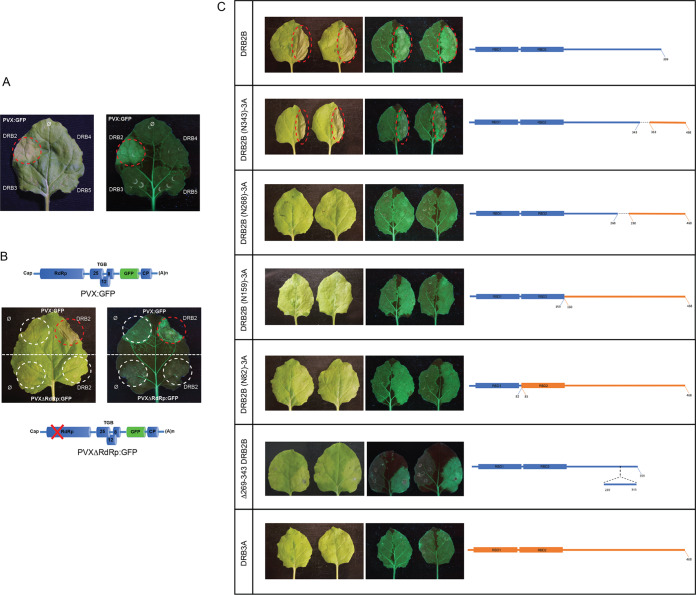
Leaf patches coinfiltrated with DRB2 and PVX-GFP (or PVXΔTGB-GFP) became necrotic at 7 dpi. The other DRBs had no effect under the same conditions (Fig. 6A). Importantly, DRB2 on its own or combined with a replication-incompetent mutant PVX (carrying a frameshift mutation in the RdRp gene) was also unable to induce necrosis (Fig. 6B). We hypothesized that these observations might bear relevance concerning the role of DRB2 in virus infections. Therefore, we decided to examine this phenomenon more closely. First, we wanted to narrow down the protein region responsible for the induction of necrosis (Fig. 6C). Using domain-swapping mutants, the C-terminal DRB2B segment consisting of aa 269 to 343 was shown to be necessary for PVX-dependent necrosis. This result was confirmed using a DRB2B mutant with deletion of aa 269 to 343 (Δ269–343-DRB2B) which was unable to induce necrosis but could still suppress RNAi. The above finding was complemented by the observation that Δ1–82-DRB2A (a deletion mutant lacking only dsRBD1) was inactive with respect to both silencing inhibition and necrosis induction (data not shown). In summary, these results clearly establish that the ability of DRB2 to suppress RNAi is a necessary but insufficient prerequisite to elicit PVX-dependent necrosis. A protein segment clearly separable from the N-terminal dsRBDs is also essential for the induction of necrosis.
FIG 6.
DRB2 induces HR-like local necrosis in N. benthamiana leaves. (A) PVX-GFP was agroinfiltrated into leaves of wild-type N. benthamiana either alone or in combination with the indicated DRB binary expression vectors. At 7 dpi, infiltrated leaves were photographed under visible (left panel) and UV (right panel) light. The DRB2-PVX-GFP-coinfiltrated necrotic patch is circled. (B) Replication of PVX is necessary for DRB2-dependent necrosis. Wild-type N. benthamiana leaves were agroinfiltrated with PVX-GFP and PVXΔRdRp-GFP (a PVX mutant carrying a frameshift mutation leading to a premature stop codon in the RdRp gene). The viral reporters were infiltrated either alone (left side of leaves) or along with a DRB2 binary expression vector (right side of leaves). Leaves were photographed as described above at 7 dpi. Infiltrated leaf patches are circled (red circle indicates the DRB2-PVX-GFP-coinfiltrated necrotic area). (C) Mapping of domains of DRB2 required for PVX-dependent necrosis. Leaves of wild-type N. benthamiana were agroinfiltrated with PVX-GFP either alone (left side of leaves) or combined with the indicated DRB binary expression vectors (right side of leaves). Photographs of leaves were taken under visible and UV light at 7 dpi. Every experiment was repeated at least twice. In each experiment, a total of six leaves were infiltrated in the same manner (two plants, with three leaves each). Pictures of representative leaves are shown. The necrotic areas of leaves are circled in red. DRB2B- and DRB3A-derived segments of the chimeric DRB molecules are indicated by blue and orange colors, respectively.
Next, we wanted to assess the effect of DRB2 on virus infection in a more physiological setting. To this end, PVX binary vectors were engineered that express the full-length DRB2B or DRB3A open reading frame (ORF) from the viral CP promoter (PVX-DRB2 and PVX-DRB3). The use of these recombinant PVX constructs offers the opportunity to study the roles of DRBs in bona fide systemic virus infections. Agroinfiltration of the above plasmids into N. benthamiana leaves was used to initiate virus infections. The empty PVX vector and recombinant PVX constructs expressing strong viral suppressor proteins (VSRs) (PVX-TEV-HcPro and PVX-CIRV-p19, where TEV is Tobacco etch virus, CIRV is Carnation Italian ringspot virus, and HcPro is helper component proteinase) were also employed as controls. Signs of systemic infection were noted with each virus construct at 5 dpi. By 14 dpi significant differences had been developed between the plants infected with the different viruses (Fig. 7A). The empty-PVX-infected and PVX-DRB3-infected plants were phenotypically comparable, displaying moderate stunting relative to growth of the mock-infected plants. Their apical leaves exhibited gradually reduced crinkling and mottling as a sign of recovery. In these plants no necrosis (local or systemic) was evident. The PVX-DRB2-infected plants were significantly more stunted than the empty- PVX (or PVX-DRB3)-infected ones. The infiltrated leaves were completely necrotized, and the adjacent ones also showed extensive necrosis. The apical leaves were not necrotic; however, they remained strongly symptomatic. In general, the PVX-DRB2-infected plants resembled the PVX-TEV-HcPro- and PVX-CIRV-p19-infected plants more than the empty-PVX (or PVX-DRB3)-infected ones. The disparate phenotypes of plants infected with the different viruses were not due to variable levels of virus replication, as evidenced by the comparable viral gRNA and CP accumulations in their systemically infected leaves (Fig. 7B). Expression of DRB2 from PVX itself strongly inhibited vsiRNA production similarly to CIRV-p19, while DRB3 and TEV-HcPro had only moderate effects on the vsiRNA levels.
FIG 7.
DRB2 overexpressing PVX induces necrosis. (A) Infections of wild-type N. benthamiana with the indicated recombinant PVX viruses were initiated by agroinfiltration. Pictures of infected plants were taken at 14 dpi. (B) At 7 dpi protein lysates and total RNA were prepared from systemically infected leaves of N. benthamiana. Presence of PVX RNA was monitored by Northern blotting using radioactively labeled PVX-CP probe. The ethidium bromide (EtBr)-stained gel is shown as a loading control. Viral CP and DRB protein expression levels were analyzed by Western blotting using the indicated antibodies. The Ponceau-stained filter is shown as a loading control. PVX vsiRNA levels were analyzed by small-RNA Northern blotting. The filter was probed with radioactively labeled PVX-CP probe. The ethidium bromide-stained gel is shown as a loading control.
Knockdown of DRB2 rescues PVX-induced systemic necrosis.
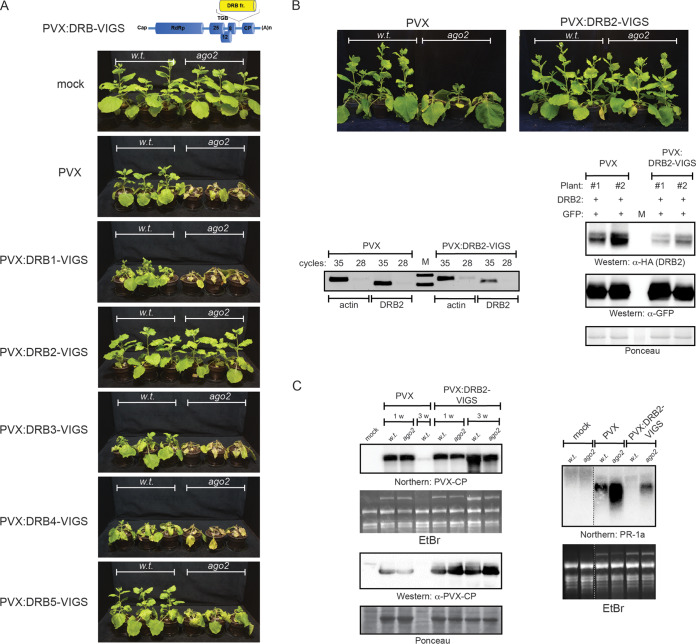
Recently, we reported the generation of ago2 mutant N. benthamiana that exhibited increased sensitivity toward a variety of viruses (32). Challenge of this plant line with PVX elicited apical necrosis that frequently resulted in the death of the infected plants. Based on the temporal and spatial separation of the infection event and the necrotic response, one may regard this phenomenon as virus-induced systemic necrosis (SN). The accompanying transcriptome changes corroborated this assumption (unpublished data). The PVX and DRB2-dependent local necrotic reaction described in the previous section is reminiscent of the hypersensitive response (HR) seen during incompatible host-virus interactions. Growing amounts of data indicate that despite their distinctly different manifestations and disparate roles, SN and HR share many underlying molecular attributes (1, 37, 38). Prompted by these findings, we decided to explore whether DRB2 was also involved in the PVX-induced SN of ago2 N. benthamiana. The endogenous DRB2 level was knocked down by virus-induced gene silencing (VIGS) for which PVX itself was employed as a vector. An ∼400-bp segment of the DRB2B ORF was cloned into a PVX binary vector downstream of the CP promoter in antisense orientation. For comparison, similar VIGS constructs were created for the remaining DRB family members (Fig. 8A). Leaves of wild-type and ago2 N. benthamiana plants were infiltrated with the above-mentioned PVX-DRB-VIGS constructs to initiate virus infections. As a control, the empty PVX vector was also included in the experiment. All of the viruses replicated efficiently, as evidenced by the accumulation of comparable amounts of viral gRNA in systemically infected leaves (data not shown). As reported before, the empty PVX elicited apical necrosis in ago2 plants by 21 dpi, whereas wild-type plants exhibited ongoing recovery by that time (25). PVX-DRB5-VIGS-infected wild-type and ago2 plants were phenotypically indistinguishable from the empty-PVX-infected ones. PVX-DRB3-VIGS resulted in slightly exacerbated symptoms compared to those of infection with the empty PVX, consistent with the recently proposed antiviral role for DRB3 (19). PVX-DRB4-VIGS produced more severe symptoms in both wild-type and ago2 N. benthamiana plants than the control virus, which agrees well with the fundamental role of DRB4 in siRNA biogenesis. PVX-DRB1-VIGS-infected ago2 plants were indistinguishable from the equivalent empty-PVX-infected ones. In wild-type plants the same virus produced strong leaf deformities, which were likely the consequence of the essential role of DRB1 in miRNA biogenesis. In stark contrast to knockdown of the other DRBs, knocking down DRB2 expression significantly diminished signs of PVX infection in general, and, remarkably, it was also able to prevent the apical necrosis of ago2 plants (Fig. 8B). The reduced symptoms were not due to decreased virus replication as both viral gRNA and CP were present in comparable quantities in the systemically infected leaves of all plants at 7 dpi (Fig. 8C). Furthermore, PVX-DRB2-VIGS persisted at high levels in both wild-type and ago2 plants at least up until 21 dpi. By that time the empty-PVX-infected ago2 plants usually necrotized while the wild-type infected plants contained barely detectable amounts of virus, consistent with their advanced recovery. Virus-induced SN is accompanied by the robust induction of numerous pathogenesis-related genes, of which PR-1a is a frequently used marker for necrosis. The control PVX resulted in significantly stronger induction of PR-1a in ago2 plants than in the wild-type plants, consistent with the more severe symptoms they exhibit. No PR-1a induction was observed in the PVX-DRB2-VIGS-infected wild-type plants. In the ago2 plants only a weak signal was detectable, which did not even reach the level seen in the empty-PVX-infected wild-type plants. In summary, downregulation of DRB2 significantly reduced the symptoms of PVX infection and was also able to hinder the development of SN in ago2 N. benthamiana.
FIG 8.
Effects of knockdown of DRB proteins on PVX infection. (A) Wild-type and ago2 N. benthamiana plants were agroinfiltrated with the indicated PVX-DRB-VIGS recombinant constructs. Pictures of mock- and virus-infected plants were taken at 21 dpi. The cartoon at the top depicts the structure of the VIGS constructs. DRB fr., DRB VIGS fragment. (B) Downregulation of DRB2 rescues PVX-induced systemic necrosis of ago2 N. benthamiana. Pictures of PVX-DRB2-VIGS-infected wild-type and ago2 N. benthamiana plants were taken at 21 dpi (top panels). Downregulation of endogenous DRB2 mRNA level was verified by semiquantitative reverse transcription-PCR (bottom left panels). As a control, a fragment of the actin mRNA was amplified. Cleavage-independent translational repression can also contribute to the effects of VIGS. To take this into account, the ability of PVX-DRB2-VIGS to knock down the DRB2 protein level was also assessed (bottom right panels). N. benthamiana plants were infected with PVX-DRB2-VIGS and with empty PVX (as control). At 14 dpi, apical systemically infected leaves of the plants were coinfiltrated with suspensions of agrobacteria expressing epitope-tagged DRB2 and GFP (as negative control). Three days later, the expression levels of HA-DRB2 and GFP were monitored in the infiltrated leaves using Western blotting. The Ponceau-stained filter is shown as a loading control. M, marker lane. (C) Replication of PVX-DRB2-VIGS was verified by Northern blotting (using radioactive PVX-CP probe) and Western blotting (using PVX-CP specific antibody) in systemically infected leaves (left panels). The PR-1a necrotic marker gene expression level was analyzed by Northern blotting (right panel). As loading controls for Northern and Western blots, the corresponding ethidium bromide (EtBr)-stained gel and Ponceau-stained filter are shown, respectively. Each experiment was repeated at least three times with similar results. w, week.
Knockdown of DRB2 blocks PVX-PPV synergism induced SN.
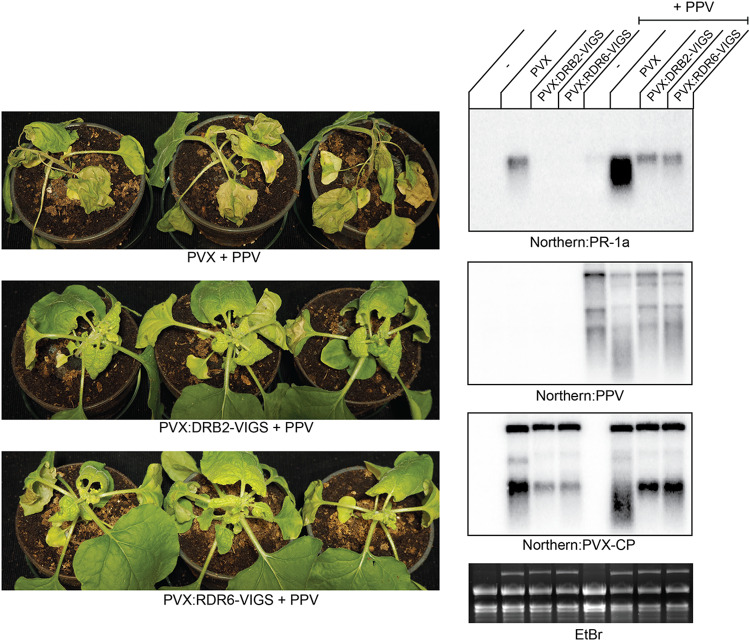
A likely interpretation of the results described in the previous section is that DRB2 deficiency compromises the plants’ ability to sense the replicating virus as a pathogen, which may in turn halt the appropriate defense measures (e.g., HR or SN) to be taken. We hypothesize that the above DRB2-dependent mechanism would come into play under circumstances in which the symptoms of virus infection become unusually severe and thereby seriously threaten the organism’s or the population’s integrity. Situations like this can be produced experimentally by inactivating components of the antiviral RNAi pathway (e.g., mutagenesis of ago2) or may also occur under natural circumstances, for example, during viral synergisms. Simultaneous infection of plants by multiple unrelated viruses is a frequent event in nature, and many diseases are attributable to synergistic interactions (39, 40). Synergy is often manifested as a remarkable increase in both virus accumulation and symptom expression compared to levels with single infections. Mechanistically, synergisms are suggested to be the result of the suppression of host defense processes (e.g., antiviral RNAi) by strong VSRs (41). One of the best-described synergisms is the interaction of PVX with various potyviruses (Plum pox virus [PPV], Tobacco etch virus [TEV], etc.), in which case the exacerbation of symptoms may even lead to SN (42–44). In the next set of experiments, we aimed to answer the question of whether downregulation of DRB2 could alleviate the consequences of PVX-potyvirus synergism as well. To initiate virus infections, wild-type N. benthamiana plants were infiltrated with binary constructs expressing PPV, PVX, and PVX-DRB2-VIGS either individually or in combinations (PPV plus PVX and PPV plus PVX-DRB2-VIGS). PVX-infected plants produced the usual PVX-specific symptoms compared to which PVX-DRB2-VIGS-infected plants were almost symptomless (see above). PPV also produced relatively mild symptoms in wild-type N. benthamiana, which included slight growth retardation, leaf crinkling, and mottling (data not shown). Coinfection of plants with PVX and PPV caused a remarkable enhancement of symptoms, which culminated in the death of the infected plants by 14 dpi (Fig. 9). Using PVX-DRB2-VIGS instead of the empty PVX alleviated the infection symptoms. Though the plants were still strongly stunted, no necrosis was noticeable in either the infiltrated leaves or shoot apices. Induction of PR-1a reflected the visual symptoms of the infected plants. Viral gRNAs were accumulated to comparable levels, as verified by Northern blotting. In summary, these results indicate that a DRB2-mediated mechanism may contribute to the activation of SN under physiologically relevant circumstances, such as mixed virus infections.
FIG 9.
Knockdown of DRB2 and RDR6 inhibits PPV-PVX synergism-induced SN. Wild-type N. benthamiana plants were infected by agroinfiltration using binary plasmid constructs expressing the indicated viruses. Pictures of the infected plants were taken at 14 dpi. Total RNAs were prepared from systemically infected plants at 7 dpi. Samples collected from three identically infected plants were pooled. Viral RNA levels and necrotic marker gene expression levels were analyzed by Northern blotting using the indicated radioactively labeled virus-specific and PR-1a-specific probes. The ethidium bromide (EtBr)-stained gel is shown as a loading control. The experiment was repeated three times with the same outcomes.
RDR6-dependent viral dsRNAs may be able to trigger SN.
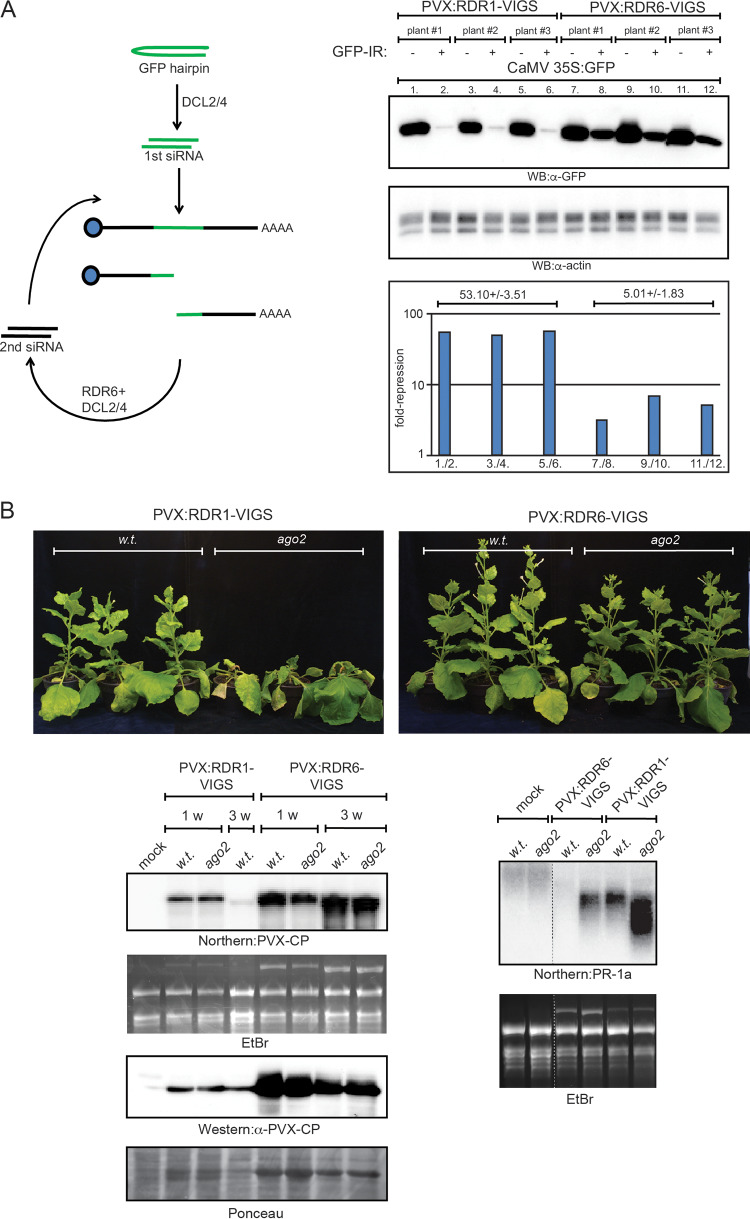
The dsRBD1 domain of DRB2 is necessary for the induction of necrosis. Since the dsRBD1 domain possesses the ability to bind dsRNAs, one may assume that binding of DRB2 to viral dsRNAs may serve as a cue for the triggering of SN. During PVX infection, the RDR6-dependent amplification cycle is the most abundant source of virus-derived dsRNAs. Thus, downregulation of RDR6 activity might be able to interfere with virus-induced necrosis. The aim of the subsequent experiment was to test this hypothesis. RDR6 expression was downregulated by VIGS employing the same approach as described above (i.e., using PVX itself as a VIGS vector). Using an inverted repeat RNA (IR-RNA)-triggered functional silencing assay, we verified that PVX-RDR6-VIGS was able to decrease RDR6 activity considerably compared to the level with PVX-RDR1-VIGS (this virus was used as a negative control because the RDR1 gene of N. benthamiana is a dysfunctional pseudogene due to a 72-bp insertion in its first exon) (Fig. 10A). Wild-type and ago2 N. benthamiana plants were infected with PVX-RDR6-VIGS and the control PVX-RDR1-VIGS expressing binary vectors using agroinfiltration. Both viruses replicated and spread in the plants efficiently, as evidenced by the accumulation of viral gRNA and CP in the systemically infected leaves (Fig. 10B). As expected, by 21 dpi PVX-RDR1-VIGS infection resulted in SN in the ago2 plants, while recovery was observable in the wild-type cohort. In contrast, PVX-RDR6-VIGS elicited much milder symptoms in the infected plants and resulted in no SN even in the ago2 plants. Similar to PVX-DRB2-VIGS, this virus also persisted at high levels in the systemically infected leaves, at least up until 21 dpi. Consistent with the mild symptoms, PR-1a induction was also much lower in the PVX-RDR6-VIGS-infected plants than in PVX-RDR1-VIGS-infected ones. Finally, we tested whether downregulation of RDR6 could alleviate symptoms of PVX-PPV synergism (Fig. 9). Coinfection of wild-type N. benthamiana with PVX-RDR6-VIGS and PPV resulted in no necrosis, and PR-1a induction was also much reduced compared to that in plants coinfected with the PVX and PPV. To sum up, the above results suggest that RDR6-dependent virus-derived dsRNAs contribute to the triggering of SN.
FIG 10.
Downregulation of RDR6 rescues PVX-induced SN of ago2 N. benthamiana. (A) VIGS-mediated downregulation of RDR6 activity was verified by a functional gene silencing assay. The assay is based on the observation that the inverted repeat (IR)-initiated posttranscriptional gene silencing is significantly enhanced by RDR6-dependent secondary siRNAs. Wild-type N. benthamiana plants were infected with either PVX-RDR6-VIGS or PVX-RDR1-VIGS recombinant PVX viruses (three plants each). Infections were initiated by agroinfiltration of a binary vector expressing the appropriate virus. At 14 dpi, systemically infected leaves of the plants were infiltrated with a 35S CaMV-GFP binary vector either alone (left side of leaves) or together with a GFP-IR binary construct (right side of leaves). Three days later, samples were collected from the leaves, and total protein lysates were prepared. GFP levels were analyzed by quantitative Western blotting using GFP antibody. GFP-IR-induced repression (plotted in the chart at the bottom) was obtained by dividing the GFP band intensities measured in the absence of GFP-IR by the intensities detected in its presence (lane pairs are indicated on the x axis). GFP band intensities were normalized for actin signals. Errors are given as standard deviations. Note that in PVX-RDR6-VIGS-infected plants, efficiency of IR posttranscriptional gene silencing decreased by approximately 1 order of magnitude compared to that of the control PVX-RDR1-VIGS-infected plants, indicating efficient downregulation of RDR6 activity. (B) PVX-RDR6-VIGS and PVX-RDR1-VIGS (negative control) virus infections were initiated in wild-type and ago2 N. benthamiana plants using agroinfiltration. Pictures of infected plants were taken at 21 dpi. Virus levels were monitored by Northern and Western blotting in systemically infected leaves (bottom left panels). The Northern blot was hybridized with radioactively labeled PVX-CP probe, while the Western blot was probed with PVX-CP antibody. As loading controls for Northern and Western blots, the corresponding ethidium bromide-stained gel and Ponceau-stained filter are shown, respectively. Necrotic marker gene expression was analyzed by Northern blotting using radioactively labeled PR-1a-specific probe (bottom right panels). The ethidium bromide-stained gel is shown as a loading control. The experiment was repeated three times with the same outcomes. w, week.
DISCUSSION
As obligate intracellular parasites, viruses entirely rely on their host’s synthetic apparatus for their reproduction. Most of the molecular patterns generated during virus replication are hardly distinguishable from the cell’s own biochemical features; therefore, identifying them as pathogen derived poses a difficult challenge for the organism. One of the best-characterized foreign features generally associated with the replication of both RNA and DNA viruses is the double-stranded structure of RNA. It has long been recognized that dsRNA acts as PAMP in animals (45). The PRRs and the downstream innate immune responses they can activate have also been identified and extensively characterized. In contrast, in plants dsRNA has only very recently been demonstrated to function as a PAMP (5, 6). SERK (somatic embryogenesis receptor-like kinase) kinases are involved in the dsRNA-induced signaling process; however, the dsRNA-binding receptor has not been identified yet. In plants, viral dsRNAs can also activate an adaptive defense process known as antiviral RNAi (7–9). Even though evidence for the cross talk between the two antiviral pathways is rapidly emerging, there are still numerous outstanding questions. For example, there are uncertainties regarding the factors that influence the choice of the dominant pathway that is activated during a viral infection. From the standpoint of the host, this is often a life-or-death verdict; therefore, it must be controlled strictly. Since RNAi and PTI deviate from each other in their initial steps, the above decision may happen as early as the recognition of their common inducer, the virus-derived dsRNA. Understanding the details of this process, therefore, is of considerable importance.
Three of the four protein families involved in antiviral RNAi possess dsRNA-binding domains: DCLs, RdRps, and DRBs (8). Of these, DCLs are the first that engage viral dsRNA and consequently set the RNAi machinery in motion. Interestingly, DCLs exhibit extensive sequence homology to the vertebrate dsRNA helicases of the RIG‐I family, which also act as dsRNA-binding PRRs in animals. Thus, DCLs are also the obvious candidates for being the plant dsRNA receptors that are responsible for triggering PTI. A recent report, however, refutes this assumption by demonstrating that virus-induced PTI is not compromised in dcl mutant plants (5). Members of the DRB protein family also contain dsRBDs, and many of them are indispensable for the proper functions of DCLs. A novel paper from Barton et al. reports that DRB2, DRB3, and DRB5 can relocate from the cytoplasm to nascent VRCs in virus-infected cells (19). These observations raise the intriguing possibility that DRBs may play a novel, unanticipated function in antiviral responses by acting as viral invasion sensors. Encouraged by these data, we explored the potential roles of DRBs in virus infections. The principal findings we report here are the following:
-
1.
Ectopic expression of DRB2 increases the levels of PVX gRNA and virus-encoded GFP in N. benthamiana. This increase is the result of inhibition of RNAi.
-
2.
DRB2 interferes with antiviral RNAi by hindering DCL-mediated dicing of virus-derived dsRNAs. This feature of DRB2 is dependent on its dsRBD1 domain.
-
3.
DRB2 can elicit HR-like local necrosis in the presence of replicating PVX in N. benthamiana. In addition to the dsRBD1 domain, a C-terminal segment of the protein is also indispensable for the induction of necrosis.
-
4.
Downregulation of DRB2 by VIGS is able to rescue PVX-triggered SN in ago2 mutant N. benthamiana.
-
5.
DRB2 deficiency alleviates the symptoms of viral synergism induced by PVX-PPV mixed infection.
-
6.
RDR6-dependent viral dsRNAs may be the trigger for virus-induced SN in PVX-infected N. benthamiana.
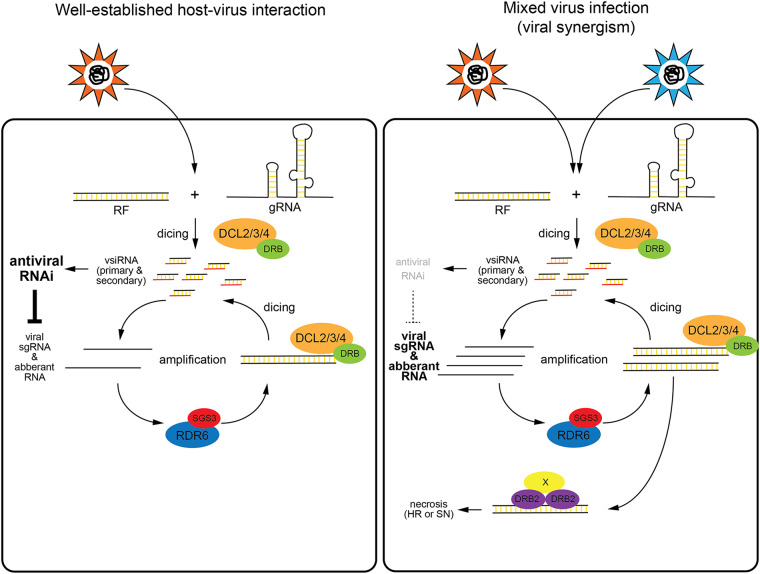
Based on the above findings, we propose a model for the physiological role of DRB2 in the context of virus infection (Fig. 11). According to the model, virus-derived dsRNA can activate two competing, but at the same time complementary, antiviral mechanisms: RNAi and DRB2-dependent PTI. In a well-established host-virus relationship, antiviral RNAi can keep virus replication under a certain threshold, which allows the survival of both partners. This type of stable interaction is mutually beneficial for both the plant and the virus for a number of reasons (2). What happens if this equilibrium is lost? This may occur when a virus infects a new host which is not well adapted to it or during mixed virus infections when synergism results in severely enhanced symptoms (e.g., PVX-potyvirus synergism). Experimentally, these situations can be modeled by inactivating critical components of the antiviral RNAi pathway (e.g., ago2). In such cases antiviral RNAi is no longer able to keep virus replication under check, which, due to the limited DCL activity, may lead to the buildup of a large pool of virus-derived long dsRNAs. The excess dsRNA can interact with DRB2, resulting in the formation of necrosis-inducing complexes, which may trigger either HR or SN. HR can block the spread of the virus from the site of infection to distal parts of the plant while SN is employed as a last measure to prevent the transmission of infection from diseased to healthy individuals within the population.
FIG 11.
Model for the role of DRB2 in virus infection. In a well-established host-virus interaction, the robust antiviral RNAi response can efficiently limit virus amplification and prevent the buildup of significant amounts of viral dsRNA (left panel). During a mixed virus infection, due to the hampered antiviral RNAi response, virus infection gets out of control, resulting in the accumulation of virus-derived dsRNAs. The excess viral dsRNA interacts with DRB2, triggering necrotic responses (right panel). See further details in the text. RF, replicative form RNA.
While the DRB2-dsRNA complex can activate a PTI response, a similar complex of DRB4 is responsible for triggering the RNAi pathway. The adequate activation of the two pathways may be ensured by the different affinities of DRB2 and DRB4 toward viral dsRNA. In this respect, it is of interest that colocalization of the two DRBs with viral dsRNA (inside nascent cytoplasmic VRCs) has recently been described (46). Additionally, consistent with their proposed antagonistic role, transgenic plants overexpressing DRB2 mimicked drb4 mutants at both the morphological and molecular levels (47). Plausibly, under default conditions viral dsRNAs preferentially interact with DRB4 (or DRB4-DCL4), ensuring efficient activation of antiviral RNAi. At physiological protein concentrations, the DRB2-dsRNA complexes are likely to form only when RNAi can no longer restrain virus amplification, and virus-derived dsRNAs accumulate to high levels. Whether virus infection also affects the DRB2 protein level is unknown. Our transcriptome analysis indicates that the DRB2 mRNA level does not change during PVX infection; however, upregulation of the DRB2 protein level by a posttranscriptional mechanism cannot be excluded (unpublished data). This issue can be unequivocally resolved only once appropriate antibodies become available. Interestingly, while overexpression of DRB2 inhibited the activity of all the DCLs involved in vsiRNA production, ectopic overproduction of other DRBs (DRB3, DRB4, and DRB5) was also able to hinder the activity of DCL3 and DCL4. In these cases, however, due to the elevated levels of 22-nt vsiRNAs, efficient antiviral RNAi was still maintained. Whether this observation has any physiological relevance is not yet clear. Nonetheless, of the tested DRBs, only the downregulation of DRB2 was able to rescue virus-induced SN, underscoring the specific regulatory role of DRB2 in virus-induced PTI.
In the A-form duplex, which is the predominant structure adopted by dsRNA, contacts with proteins are mainly mediated by the sugar-phosphate backbone and are thus largely sequence independent (48). Consequently, the dsRNA-DRB2 interaction is likely influenced by factors such as stoichiometry, cellular localization, etc., rather than by the primary sequence of the dsRNA per se. The importance of stoichiometry as the principal driving force is supported by the finding that VIGS-mediated downregulation of RDR6 (the main supplier of virus-derived dsRNA during PVX infection) can substantially alleviate PVX infection-elicited disease symptoms and even rescue SN. Importantly, the less severe symptoms elicited by PVX infection in rdr6 N. benthamiana compared to those of ago2 plants are also consistent with the above assumption (rdr6 plants exhibit stunting but rarely SN while in ago2 plants SN is the predominant symptom [unpublished results]).
In summary, the work presented here suggests that DRB2 plays an important role in PVX-induced necrotic responses in N. benthamiana. Our findings are not only compatible with the recent hypothesis that DRB proteins act as viral invasion sensors (19) but also extend it by proposing that DRBs play a critical role in establishing the dominant antiviral pathways that are activated during virus infection. Finally, it should be noted that DRB2 is mainly expressed in the apical meristem (16). Given that viruses are generally excluded from this territory of the plant, one may theorize that DRB2 plays a role in the protection of meristemic tissues from viral invasion. Testing of the above hypotheses in addition to the generalization of the presented model (extending it to additional viruses and hosts) is an important issue to be tackled in the future.
MATERIALS AND METHODS
Plasmid construction.
Plasmids were generated using standard techniques (49). Full-length N. benthamiana DRB ORFs were amplified from cDNAs prepared from leaves using PCR. The full-length DRB ORFs were inserted into pGEM-T easy plasmid vector, and the resulting plasmids were sequenced. The correct DRB cDNAs were subsequently cloned into a pBIN61 binary expression vector. HA epitope tags were added to the N termini of the proteins using oligonucleotides. The DRB2B-DRB3A domain-swapping and DRB2B deletion mutants were created by overlapping PCR using appropriate oligonucleotide primers. The PVX-VIGS constructs were generated using a pgR107 binary vector. Approximately 0.4-kb segments of the DRB ORFs and the RDR6 ORF were PCR amplified and subsequently cloned in reverse orientation between ClaI and SalI sites of pgR107. To obtain DRB2B- and DRB3A-expressing PVX constructs, the appropriate HA epitope-tagged full-length ORFs were inserted into the polylinker region of pgR107. The structures of all constructs were verified by sequencing. Sequences of oligonucleotides used in the study are available upon request.
Agroinfiltration and protein analysis.
Agroinfiltration of N. benthamiana leaves and Western blot analysis of protein lysates were carried out as described earlier (23, 25, 32). For Western analyses, the following primary antibodies were used: rat monoclonal HA 3F10 (Roche), mouse monoclonal plant actin (Sigma-Aldrich), rabbit polyclonal GFP (Sigma-Aldrich), and rabbit polyclonal PVX-CP (kind gift from Helena Plchova).
RNA analyses.
RNA preparations and conventional and small-RNA analyses were performed as described before (23, 25, 32).
ACKNOWLEDGMENTS
We thank Peter Moffett, Helena Plchova, and Fabio Pasin for reagents.
This work was supported by the National Research Development and Innovation Office, Hungary (K124705).
REFERENCES
- 1.Mandadi KK, Scholthof KB. 2013. Plant immune responses against viruses: how does a virus cause disease? Plant Cell 25:1489–1505. doi: 10.1105/tpc.113.111658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paudel DB, Sanfaçon H. 2018. Exploring the diversity of mechanisms associated with plant tolerance to virus infection. Front Plant Sci 9:1575. doi: 10.3389/fpls.2018.01575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hornung V. 2014. SnapShot: nucleic acid immune sensors, part 1. Immunity 41:868–868.e. doi: 10.1016/j.immuni.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Hornung V. 2014. SnapShot: nucleic acid immune sensors, part 2. Immunity 41:1066–1066.e1. doi: 10.1016/j.immuni.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Niehl A, Wyrsch I, Boller T, Heinlein M. 2016. Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol 211:1008–1019. doi: 10.1111/nph.13944. [DOI] [PubMed] [Google Scholar]
- 6.Niehl A, Heinlein M. 2019. Perception of double-stranded RNA in plant antiviral immunity. Mol Plant Pathol 20:1203–1210. doi: 10.1111/mpp.12798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding SW. 2010. RNA-based antiviral immunity. Nat Rev Immunol 10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 8.Pumplin N, Voinnet O. 2013. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol 11:745–760. doi: 10.1038/nrmicro3120. [DOI] [PubMed] [Google Scholar]
- 9.Guo Z, Li Y, Ding SW. 2019. Small RNA-based antimicrobial immunity. Nat Rev Immunol 19:31–44. doi: 10.1038/s41577-018-0071-x. [DOI] [PubMed] [Google Scholar]
- 10.Csorba T, Kontra L, Burgyán J. 2015. Viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology 479–480:85–103. doi: 10.1016/j.virol.2015.02.028. [DOI] [PubMed] [Google Scholar]
- 11.Moon JY, Park JM. 2016. Cross-talk in viral defense signaling in plants. Front Microbiol 7:2068. doi: 10.3389/fmicb.2016.02068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bologna NG, Voinnet O. 2014. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol 65:473–503. doi: 10.1146/annurev-arplant-050213-035728. [DOI] [PubMed] [Google Scholar]
- 13.Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T. 2005. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol 57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- 14.Curtin SJ, Watson JM, Smith NA, Eamens AL, Blanchard CL, Waterhouse PM. 2008. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett 582:2753–2760. doi: 10.1016/j.febslet.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Eamens AL, Wook Kim K, Waterhouse PM. 2012. DRB2, DRB3 and DRB5 function in a non-canonical microRNA pathway in Arabidopsis thaliana. Plant Signal Behav 7:1224–1229. doi: 10.4161/psb.21518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eamens AL, Kim KW, Curtin SJ, Waterhouse PM. 2012. DRB2 is required for microRNA biogenesis in Arabidopsis thaliana. PLoS One 7:e35933. doi: 10.1371/journal.pone.0035933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reis RS, Hart-Smith G, Eamens AL, Wilkins MR, Waterhouse PM. 2015. Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nat Plants 1:14027. doi: 10.1038/nplants.2014.27. [DOI] [PubMed] [Google Scholar]
- 18.Reis RS, Eamens AL, Roberts TH, Waterhouse PM. 2015. Chimeric DCL1-partnering proteins provide insights into the microRNA pathway. Front Plant Sci 6:1201. doi: 10.3389/fpls.2015.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barton DA, Roovers EF, Gouil Q, da Fonseca GC, Reis RS, Jackson C, Overall RL, Fusaro AF, Waterhouse PM. 2017. Live cell imaging reveals the relocation of dsRNA binding proteins upon viral infection. Mol Plant Microbe Interact 30:435–443. doi: 10.1094/MPMI-02-17-0035-R. [DOI] [PubMed] [Google Scholar]
- 20.Nakasugi K, Crowhurst RN, Bally J, Wood CC, Hellens RP, Waterhouse PM. 2013. De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS One 8:e59534. doi: 10.1371/journal.pone.0059534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakasugi K, Crowhurst R, Bally J, Waterhouse P. 2014. Combining transcriptome assemblies from multiple de novo assemblers in the allo-tetraploid plant Nicotiana benthamiana. PLoS One 9:e91776. doi: 10.1371/journal.pone.0091776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brosseau C, Moffett P. 2015. Functional and genetic analysis identify a role for arabidopsis Argonaute5 in antiviral RNA silencing. Plant Cell 27:1742–1754. doi: 10.1105/tpc.15.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fátyol K, Ludman M, Burgyán J. 2016. Functional dissection of a plant Argonaute. Nucleic Acids Res 44:1384–1397. doi: 10.1093/nar/gkv1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaubert M, Bhattacharjee S, Mello AF, Perry KL, Moffett P. 2011. Argonaute2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol 156:1556–1564. doi: 10.1104/pp.111.178012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludman M, Burgyán J, Fátyol K. 2017. Crispr/Cas9 Mediated inactivation of Argonaute 2 reveals its differential involvement in antiviral responses. Sci Rep 7:1010. doi: 10.1038/s41598-017-01050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang SW, Chen HY, Yang J, Machida S, Chua NH, Yuan YA. 2010. Structure of Arabidopsis Hyponastic Leaves1 and its molecular implications for miRNA processing. Structure 18:594–605. doi: 10.1016/j.str.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukudome A, Kanaya A, Egami M, Nakazawa Y, Hiraguri A, Moriyama H, Fukuhara T. 2011. Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4. RNA 17:750–760. doi: 10.1261/rna.2455411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machida S, Chen HY, Adam Yuan Y. 2011. Molecular insights into miRNA processing by Arabidopsis thaliana Serrate. Nucleic Acids Res 39:7828–7836. doi: 10.1093/nar/gkr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Ren W, Zhao Q, Zhang P, Wu F, He Y. 2014. Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA. Nucleic Acids Res 42:12224–12236. doi: 10.1093/nar/gku907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baranauskė S, Mickutė M, Plotnikova A, Finke A, Venclovas Č, Klimašauskas S, Vilkaitis G. 2015. Functional mapping of the plant small RNA methyltransferase: HEN1 physically interacts with HYL1 and Dicer-like 1 proteins. Nucleic Acids Res 43:2802–2812. doi: 10.1093/nar/gkv102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiliveri SC, Aute R, Rai U, Deshmukh MV. 2017. DRB4 dsRBD1 drives dsRNA recognition in Arabidopsis thaliana tasi/siRNA pathway. Nucleic Acids Res 45:8551–8563. doi: 10.1093/nar/gkx481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludman M, Fátyol K. 2019. The virological model plant, Nicotiana benthamiana expresses a single functional RDR6 homeolog. Virology 537:143–148. doi: 10.1016/j.virol.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 33.Qu F, Ye X, Hou G, Sato S, Clemente TE, Morris TJ. 2005. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J Virol 79:15209–15217. doi: 10.1128/JVI.79.24.15209-15217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwach F, Vaistij FE, Jones L, Baulcombe DC. 2005. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol 138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaistij FE, Jones L. 2009. Compromised virus-induced gene silencing in RDR6-deficient plants. Plant Physiol 149:1399–1407. doi: 10.1104/pp.108.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouché N, Lauressergues D, Gasciolli V, Vaucheret H. 2006. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.García-Marcos A, Pacheco R, Martiáñez J, González-Jara P, Díaz-Ruíz JR, Tenllado F. 2009. Transcriptional changes and oxidative stress associated with the synergistic interaction between Potato virus X and Potato virus Y and their relationship with symptom expression. Mol Plant Microbe Interact 22:1431–1444. doi: 10.1094/MPMI-22-11-1431. [DOI] [PubMed] [Google Scholar]
- 38.Komatsu K, Hashimoto M, Ozeki J, Yamaji Y, Maejima K, Senshu H, Himeno M, Okano Y, Kagiwada S, Namba S. 2010. Viral-induced systemic necrosis in plants involves both programmed cell death and the inhibition of viral multiplication, which are regulated by independent pathways. Mol Plant Microbe Interact 23:283–293. doi: 10.1094/MPMI-23-3-0283. [DOI] [PubMed] [Google Scholar]
- 39.Latham JR, Wilson AK. 2008. Transcomplementation and synergism in plants: implications for viral transgenes? Mol Plant Pathol 9:85–103. doi: 10.1111/j.1364-3703.2007.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Syller J. 2012. Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol Plant Pathol 13:204–216. doi: 10.1111/j.1364-3703.2011.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruss G, Ge X, Shi XM, Carrington JC, Bowman Vance V. 1997. Plant viral synergism: the potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9:859–868. doi: 10.1105/tpc.9.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vance VB. 1991. Replication of potato virus X RNA is altered in coinfections with potato virus Y. Virology 182:486–494. doi: 10.1016/0042-6822(91)90589-4. [DOI] [PubMed] [Google Scholar]
- 43.Vance VB, Berger PH, Carrington JC, Hunt AG, Shi XM. 1995. 5’ proximal potyviral sequences mediate potato virus X/potyviral synergistic disease in transgenic tobacco. Virology 206:583–590. doi: 10.1016/s0042-6822(95)80075-1. [DOI] [PubMed] [Google Scholar]
- 44.González-Jara P, Tenllado F, Martínez-García B, Atencio FA, Barajas D, Vargas M, Díaz-Ruiz J, Díaz-Ruíz JR. 2004. Host-dependent differences during synergistic infection by potyviruses with potato virus X. Mol Plant Pathol 5:29–35. doi: 10.1111/j.1364-3703.2004.00202.x. [DOI] [PubMed] [Google Scholar]
- 45.Hur S. 2019. Double-stranded RNA sensors and modulators in innate immunity. Annu Rev Immunol 37:349–375. doi: 10.1146/annurev-immunol-042718-041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Incarbone M, Monsion B, Kuhn L, Scheer H, Poignavent V, Dunoyer P, Ritzenthaler C. Immunocapture of dsRNA-bound proteins provides insight into Tobacco rattle virus replication complexes. bioRxiv https://www.biorxiv.org/content/10.1101/842666v1. [DOI] [PMC free article] [PubMed]
- 47.Pélissier T, Clavel M, Chaparro C, Pouch-Pélissier MN, Vaucheret H, Deragon JM. 2011. Double-stranded RNA binding proteins DRB2 and DRB4 have an antagonistic impact on polymerase IV-dependent siRNA levels in Arabidopsis. RNA 17:1502–1510. doi: 10.1261/rna.2680711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peisley A, Hur S. 2013. Multi-level regulation of cellular recognition of viral dsRNA. Cell Mol Life Sci 70:1949–1963. doi: 10.1007/s00018-012-1149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]