Type I interferons are important for controlling virus infection. We have found that the Epstein-Barr virus (EBV) BGLF2 tegument protein binds to a protein in the type I interferon signaling pathway Tyk2 and inhibits the expression of genes induced by type I interferons. Treatment of EBV-infected cells with type I interferon inhibits reactivation of the virus, while expression of EBV BGLF2 reduces the ability of type I interferon to inhibit virus reactivation. Thus, a tegument protein delivered to cells during virus infection inhibits the host’s antiviral response and promotes virus reactivation of latently infected cells. Therefore, EBV BGLF2 might protect virus-infected cells from the type I interferon response in cells undergoing lytic virus replication.
KEYWORDS: Epstein-Barr virus, tegument protein, BGLF2, interferon, Tyk2
ABSTRACT
Interferon alpha (IFN-α) and IFN-β are type I IFNs that are induced by virus infection and are important in the host’s innate antiviral response. EBV infection activates multiple cell signaling pathways, resulting in the production of type I IFN which inhibits EBV infection and virus-induced B-cell transformation. We reported previously that EBV tegument protein BGLF2 activates p38 and enhances EBV reactivation. To further understand the role of BGLF2 in EBV infection, we used mass spectrometry to identify cellular proteins that interact with BGLF2. We found that BGLF2 binds to Tyk2 and confirmed this interaction by coimmunoprecipitation. BGLF2 blocked type I IFN-induced Tyk2, STAT1, and STAT3 phosphorylation and the expression of IFN-stimulated genes (ISGs) IRF1, IRF7, and MxA. In contrast, BGLF2 did not inhibit STAT1 phosphorylation induced by IFN-γ. Deletion of the carboxyl-terminal 66 amino acids of BGLF2 reduced the ability of the protein to repress type I IFN signaling. Treatment of gastric carcinoma and Raji cells with IFN-α blocked BZLF1 expression and EBV reactivation; however, expression of BGLF2 reduced the ability of IFN-α to inhibit BZLF1 expression and enhanced EBV reactivation. In summary, EBV BGLF2 interacts with Tyk2, inhibiting Tyk2, STAT1, and STAT3 phosphorylation and impairs type I IFN signaling; BGLF2 also counteracts the ability of IFN-α to suppress EBV reactivation.
IMPORTANCE Type I interferons are important for controlling virus infection. We have found that the Epstein-Barr virus (EBV) BGLF2 tegument protein binds to a protein in the type I interferon signaling pathway Tyk2 and inhibits the expression of genes induced by type I interferons. Treatment of EBV-infected cells with type I interferon inhibits reactivation of the virus, while expression of EBV BGLF2 reduces the ability of type I interferon to inhibit virus reactivation. Thus, a tegument protein delivered to cells during virus infection inhibits the host’s antiviral response and promotes virus reactivation of latently infected cells. Therefore, EBV BGLF2 might protect virus-infected cells from the type I interferon response in cells undergoing lytic virus replication.
INTRODUCTION
Type I interferons (interferon alpha [IFN-α] and IFN-β) are important components of the host innate immune response that controls virus infection (1–4). The type I IFN signaling pathway is initiated when IFN-α and/or IFN-β bind to their cognate receptors IFN-α receptor 1 (IFNAR1) and IFNAR2, which trigger phosphorylation and activation of the receptor-associated Janus kinases JAK and tyrosine kinase 2 (Tyk2). The activated Janus kinases induce recruitment of the signal transducer and activator of transcription protein 1 (STAT1) and STAT2 to IFNAR1 and IFNAR2, respectively. The recruited STATs undergo phosphorylation, generating STAT1/2 heterodimers that associate with cytoplasmic IRF9, forming a heterotrimeric complex termed IFN-stimulating gene factor 3 (ISGF3) consisting of STAT1, STAT2, and IRF9 (5). ISGF3 translocates to the nucleus, binds to promoter sequences of ISGs, and induces their transcription, which result in antiviral activities at multiple stages of virus replication (2, 6–11).
Epstein-Barr virus (EBV) infects oropharyngeal epithelial cells where the virus is amplified, and B cells are subsequently infected. B cells, such as those in the tonsillar crypts may also be infected directly. The virus is shed from epithelial cells of infected persons and, subsequently, can infect EBV-naive persons. Thus, epithelial cells have an important role in the viral life cycle.
EBV is the primary cause of infectious mononucleosis and is associated with a number of malignancies, including Burkitt lymphoma and Hodgkin lymphoma. The virus is also associated with several epithelial cell tumors, including nasopharyngeal carcinoma and gastric carcinoma.
Infection of cells with EBV induces activation of multiple cell signaling pathways, such as Toll-like receptors (TLRs) (12–15), NF-κB (16), IFN regulatory factor 3 (IRF3), and IRF7 (17–20), that lead to the production of type I IFNs (21), phosphorylation of STAT1, and transcription of ISGs during EBV lytic replication and latency. Two EBV latency genes, EBV nuclear antigen-2 (EBNA-2) and latent membrane protein 1 (LMP1) induce the expression of IFNs (22–25). EBNA-2-mediated IFN production in lymphoblastoid cells lines (LCLs) results in the constitutive expression of type I IFN and activation of STAT1. LMP1 expression also induces the secretion of type I IFN (24), likely through activation of NF-κB and IRF7 (26). This induction of IFN by LMP1 results in phosphorylation of STAT1 (27) and IRF7 that may assist in establishment and/or maintenance of virus latency (28). EBV-encoded RNAs (EBERs), which are expressed abundantly in EBV latently infected cells, can be secreted from EBV-infected cells and recognized by Toll-like receptor 3 (TLR3), resulting in the production of type I IFN and inflammatory cytokines (29).
Type I IFN has complex effects on EBV early gene expression and transformation. IFN-α inhibits EBV early antigen when EBV replication is induced by chemicals or superinfection but does not inhibit early antigen in cells spontaneously undergoing lytic reactivation (30–33). IFN-α inhibits capping of CD21-EBV complexes (34), and EBV transformation of B cells (35–37). However, virus lytic replication and some EBV immortalized B cells show resistance to the antiproliferation effect of type I IFN due to virus proteins produced during replication and latency proteins, such as EBNA-2 (23, 25). IFN-β does not inhibit EBV early antigen in cells, unless EBV replication is induced by butyrate (32, 38). IFN-β also inhibits EBV transformation (37). Here, we show that EBV tegument protein BGLF2 binds to Tyk2, inhibits STAT1 and STAT3 phosphorylation, blocks type I IFN signaling, and counteracts the ability of IFN-α/β to inhibit reactivation of EBV.
RESULTS
BGLF2 associates with Tyk2.
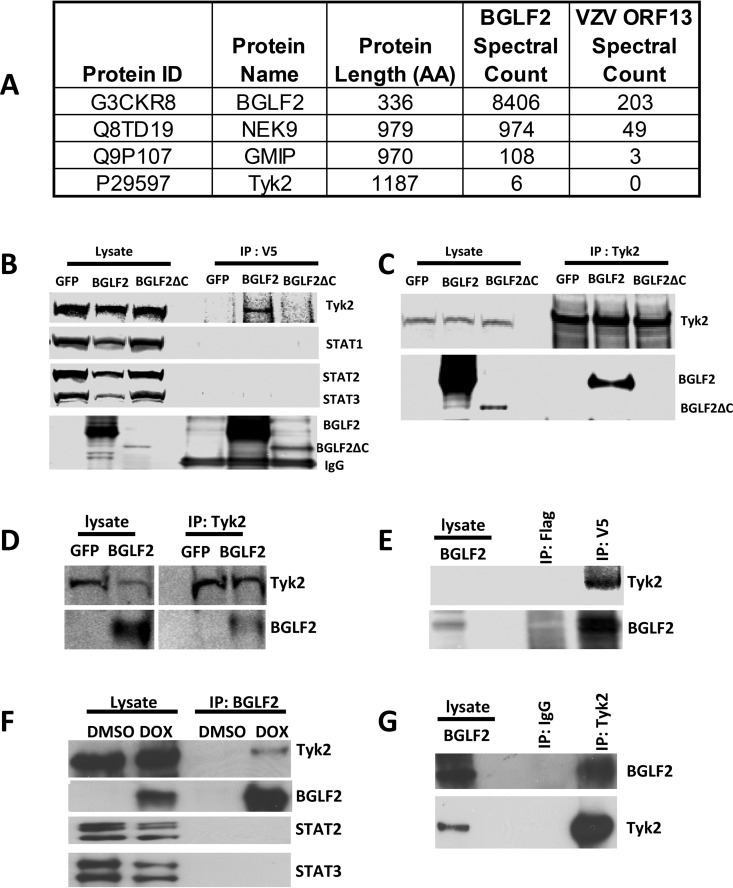
We reported previously that the EBV tegument protein BGLF2 activates p38 and JNK and enhances EBV reactivation (39), and BGLF2 was reported to activate AP-1 and increase EBV infectivity (40). To further understand the role of BGLF2 in EBV infection, we transfected 293T cells with plasmids expressing EBV BGLF2 or varicella-zoster virus (VZV) open reading frame 13 (ORF13) (as a control) with carboxyl-terminal V5 tags (BGLF2-V5 or ORF13-V5, respectively). The cell lysates were immunoprecipitated with V5-agarose beads, and immune complexes were analyzed by mass spectrometry. Tyk2 was found to associate with BGLF2, as well as two proteins, NEK9 and GMIP, previously shown to interact with BGLF2 (41) (Fig. 1A). Additional proteins interacting with EBV BGLF2, but not with the VZV ORF13 control, are shown in Table S1 in the supplemental material. To confirm the interaction of BGLF2 with Tyk2, 293T/17 SF cells were transfected with plasmids expressing GFP, V5-tagged BGLF2, or a V5-tagged carboxyl-terminal BGLF2 deletion mutant (BGLF2ΔC); cell lysates were immunoprecipitated with V5-magnetic beads; and immune complexes were immunoblotted with antibody to Tyk2 and other signaling molecules. BGLF2 associated with Tyk2, but not with STAT1, STAT2, or STAT3 (Fig. 1B), while BGLF2ΔC did not associate with Tyk2. Conversely, when cell lysates containing BGLF2 were immunoprecipitated with Tyk2 antibody, BGLF2 (but not BGLF2ΔC) was present in the immune complexes (Fig. 1C). BGLF2 and Tyk2 were also shown to associate in Raji cells (Fig. 1D and E) and 293T-BGLF2-V5 cells (Fig. 1F and G), in which BGLF2 expression is induced by doxycycline (39).
FIG 1.
BGLF2 associates with Tyk2. (A) 293T cells were transfected with plasmids expressing V5-tagged BGLF2 or V5-tagged VZV ORF13 as a control, and 24 h later the cells were washed with PBS and lysed with radioimmunoprecipitation assay (RIPA) buffer in the presence of protease and phosphatase inhibitors. After centrifugation, the supernatant was immunoprecipitated with V5-agarose beads and analyzed with mass spectrometry. Protein identification was based on UniProt, and spectral counts (total number of fragmentation spectra that map to peptides of the protein) for BGLF2, NEK9, GMIP, and Tyk2 from cells expressing BGLF2 or VZV ORF13 are shown. 293T/17 SF cells were transfected with GFP, BGLF2, or BGLF2ΔC for 48 h, and cell lysates were immunoprecipitated with mouse anti-V5 (B) or rabbit anti-Tyk2 antibody (C). The precipitated immune complexes were immunoblotted with antibody to Tyk2, STAT1, STAT2, STAT3, and V5 for BGLF2 and BGLF2ΔC. Raji cells were electroporated with plasmids expressing GFP or BGLF2-V5, cell lysates were immunoprecipitated with Tky2 antibody (D) or mouse anti-V5 or anti-flag (M2) antibody (E), and immunoblotting was performed with antibody to Tyk2 or V5 for BGLF2. 293T-BGLF2-V5 cells were induced to express BGLF2 with doxycycline; cell lysates were immunoprecipitated with antibody to V5 for BGLF2 (F), or Tyk2 or IgG (G); and immunoblotting was performed with antibody to Tyk2, V5 for BGLF2, STAT2, or STAT3.
BGLF2 inhibits phosphorylation of Tyk2, STAT1, and STAT3 induced by type I IFN.
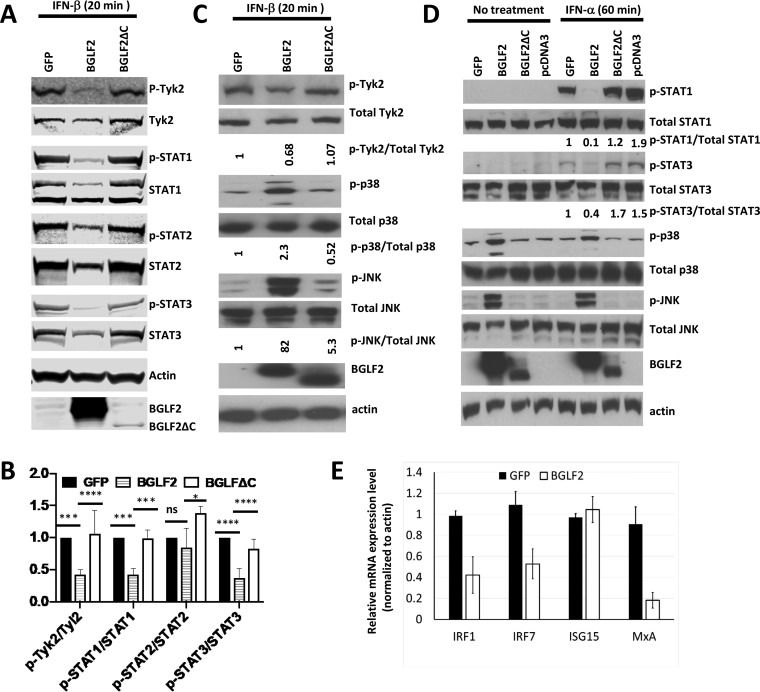
Since BGLF2 binds to Tyk2, we postulated that this interaction might modulate Tyk2 phosphorylation and subsequent JAK-STAT signaling in the type I IFN pathway. 293T/17 SF cells were transfected with plasmids expressing GFP, BGLF2, or BGLF2ΔC; 48 h after transfection, the cells were treated with IFN-β (1,000 U/ml) for 20 min and the levels of phosphorylated and total Tyk2, STAT1, STAT2, or STAT3 were assayed by immunoblotting. Expression of BGLF2 in IFN-β treated cells resulted in reduced levels of phosphorylated Tyk2 (p-Tyk2), phosphorylated STAT1 (p-STAT1), and phosphorylated STAT3 (p-STAT3), but not phosphorylated STAT2 (p-STAT2), compared with cells expressing GFP or BGLF2ΔC (Fig. 2A and B). In IFN-β-treated 293T cells, BGLF2 (but not BGLF2ΔC) increased levels of p-p38 and p-JNK relative to total p38 and JNK, as described previously (39), and reduced the phosphorylation of Tyk2 (Fig. 2C).
FIG 2.
BGLF2 suppresses type I interferon signaling. (A) 293T/17 SF cells were transfected with plasmids expressing GFP, BGLF2-V5, or BGLF2ΔC-V5 for 48 h and then treated with IFN-β (1000 U/ml) for 20 min. Cell lysates were prepared for detection of BGLF2 (using V5 antibody) or phosphorylated or total Tyk2, STAT1, STAT2, and STAT3. (B) One-way analysis of variance (ANOVA) statistics were used to quantitate the ratio of phosphoprotein to total protein in three separate experiments. (C) 293T cells were transfected with plasmids expressing BGLF2-V5, BGLF2ΔC-V5, or GFP for 24 h and then treated with IFN-β for 20 min. (D) 293T cells were transfected with plasmids expressing BGLF2-V5, BGLF2ΔC-V5, pcDNA3, or GFP for 48 h and treated with IFN-α (1000 U/ml) for 60 min; and cell lysates were immunoblotted with antibodies to various proteins or to the V5 epitope to detect BGLF2. (E) 293T cells were transfected with plasmids expressing GFP or BGLF2, and 24 h later the cells were treated with IFN-α (1,000 U/ml) for 3 h. ISG expression was measured by RT-PCR from cell lysates.
To verify that BGLF2 has similar effects on IFN-α-treated cells, 293T cells were transfected with plasmids expressing GFP, BGLF2, BGLF2ΔC, or empty vector; 48 h after transfection, cells were treated with IFN-α (1000 U/ml) for 60 min or left untreated and immunoblotting was performed. Levels of p-STAT1 and p-STAT3 in BGLF2, but not BGLF2ΔC-transfected cells, were reduced relative to the total levels of these proteins, while levels of p-p38 and p-JNK were increased in BGLF2, but not BGLF2ΔC-transfected cells, compared to total p38 and JNK (Fig. 2D). In cells not treated with IFN-α, p-STAT1 and pSTAT-3 were not detected, while p-p38 and p-JNK were also increased in cells expressing BGLF2.
Since BGLF2 inhibits type I IFN-induced STAT1 and STAT3 phosphorylation, we measured the expression of several IFN-stimulated genes (ISGs). IRF1, IRF7, ISG15, and MxA expression were assayed by real-time PCR (RT-PCR) in cells transfected with plasmids expressing BGLF2 or GFP and treated with IFN-α for 3 h. Transcription of IRF1, IRF7, and MxA, but not ISG15, was repressed by BGLF2 (Fig. 2E). Taken together, these results indicate that BGLF2 can block type I IFN signaling by inhibiting phosphorylation of Tyk2, STAT1, and STAT3; and BGLF2 reduces transcription of IRF1, IRF7, and MxA.
BGLF2 does not inhibit IFN-γ signaling.
IFN-γ signals through IFN-γ receptors α and β (IFNγ-Rα and IFNγ-Rβ, respectively) to trigger the phosphorylation of STAT1 via phosphorylation of JAK1 and JAK2. Since BGLF2 interferes with STAT1 phosphorylation induced by type I IFN signaling, we determined if BGLF2 is also able to reduce STAT1 phosphorylation induced by type II IFN-γ signaling. 293T cells were transfected with plasmids expressing BGLF2 or GFP and treated with IFN-α, IFN-β, or IFN-γ for 60 min; and levels of phosphorylated STAT1, total STAT1, BGLF2, and actin were measured. BGLF2 blocked IFN-α, and IFN-β induced STAT1 phosphorylation, but not IFN-γ induced STAT1 phosphorylation (Fig. 3).
FIG 3.
BGLF2 does not inhibit IFN-γ signaling. 293T cells were transduced with lentivirus expressing BGLF2-V5 or GFP, and after 4 days the cells were treated with 1,000 U/ml of IFN-α, IFN-β, or IFN-γ for 60 min. Lysates were immunoblotted with antibodies to p-STAT1, total STAT1, actin, and V5 to detect BGLF2.
BGLF2 inhibits type I IFN suppression of EBV lytic replication in gastric carcinoma cells.
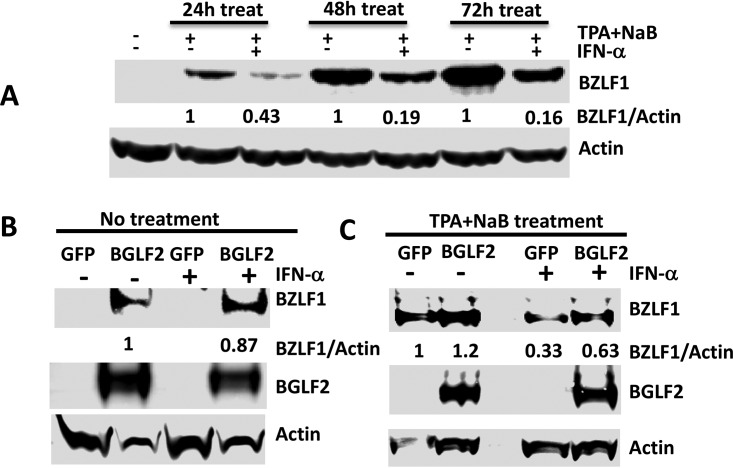
IFN is important for the innate immune response against virus infections. IFN-α represses EBV lytic replication and suppresses BZLF1 expression and EBV reactivation in latently infected Burkitt lymphoma cells when treated with a chemical inducer of lytic replication (38). Since BGLF2 represses type I IFN signaling and enhances lytic replication of EBV from latently infected cells (39), we postulated that BGLF2 inhibition of IFN signaling may contribute to the ability of BGLF2 to enhance lytic replication of EBV in the presence of type I IFN. We first determined if IFN-α inhibits EBV reactivation in AGS-EBV-GFP gastric carcinoma cells. AGS-EBV-GFP cells were pretreated with different doses of IFN-α, and then methotrexate was added for 3 days to induce EBV reactivation in the presence of IFN-α. EBV immediate early (IE) protein BZLF1 was induced in methotrexate-treated cells, indicating that lytic virus gene expression was occurring, while addition of IFN-α reduced methotrexate-induced BZLF1 expression (Fig. 4A). We next determined if type I IFN was also able to block BGLF2 activation of BZLF1. AGS-EBV-GFP and AGS-EBV-BGLF2 cells were treated with different doses of IFN-α for 2 days, and cells were collected for the detection of BZLF1 expression. In contrast to methotrexate treatment, IFN-α treatment did not suppress BGLF2-induced BZLF1 expression (Fig. 4B). To determine if expression of BGLF2 results in virus production in the presence of IFN-β, we treated AGS-EBV-GFP cells expressing luciferase, BGLF2, or BGLF2ΔC with IFN-β for 48 h and measured BZLF1 expression in the cell lysates and virus production in the supernatants. Supernatants were used to infect Raji cells, and expression of GFP in these cells indicated if infectious EBV had been released from the AGS-EBV-GFP cells. Expression of BGLF2, but not BGLF2ΔC, was associated with increased BZLF1 expression; BZLF1 was not suppressed by IFN-β in cells expressing BGLF2. Higher levels of infectious EBV were present in the supernatant of AGS-EBV-BGLF2 cells than in supernatants of AGS-EBV-GFP cells, as detected by GFP expression in Raji cells (Fig. 4D).
FIG 4.
BGLF2 counteracts the ability of IFN-α to inhibit EBV reactivation in AGS-EBV cells. (A) AGS-EBV-GFP cells were treated with different doses of IFN-α for 60 min, followed by methotrexate (MTX) for 2 days, and BZLF1 and β-actin was detected by immunoblot. (B) AGS-EBV-GFP and AGS-EBV-BGLF2-Flag cells were treated with different doses of IFN-α for 60 min, and BZLF1 and β-actin were detected by immunoblot. (C) AGS-EBV-GFP cells stably expressing luciferase, BGLF2-Flag, or BGLF2ΔC-Flag were treated with IFN-β for 48 h, and cell lysates were immunoblotted with antibody to BZLF1 and β-actin. (D) Supernatants from cells in C were filtered through a 0.45-μM filter and used to infect Raji cells. After 3 days, the cells were lysed and immunoblotted for GFP to determine if virus had been secreted from the cells. (E) AGS-EBV-GFP cells stably expressing luciferase or BGLF2-Flag were treated with MTX, MTX and IFN-α, or dimethyl sulfoxide (DMSO) (control) for 2 days, and cell lysates were immunoblotted with antibody to BZLF1, BGLF2, and actin.
BGLF2 counteracts the ability of IFN-α to inhibit methotrexate-induced EBV reactivation in gastric carcinoma cells.
Since type I IFN inhibits methotrexate-induced, but not BGLF2-induced EBV lytic replication in AGS-EBV-GFP cells, we further determined if BGLF2 can counteract the inhibitory effect of type I IFN on methotrexate-induced EBV reactivation. AGS-EBV-GFP cell lines stably expressing BGLF2 or luciferase (AGS-EBV-BGLF2 and AGS-EBV-Luc, respectively) were treated with IFN-α and then were incubated with methotrexate to reactivate EBV. Expression of BGLF2 induced higher levels of BZLF1 than cells expressing luciferase (Fig. 4E). Treatment with methotrexate induced BZLF1 expression in both luciferase and BGLF2-expressing cells, but BGLF2-expressing cells produced a greater amount of BZLF1 than luciferase-expressing cells. The presence of IFN-α antagonized the ability of methotrexate to induce expression of BZLF1 (Fig. 4E, lanes 3 and 5), while expression of BGLF2 counteracted the ability of IFN-α to inhibit methotrexate-induced BZLF1 expression (Fig. 4E, lanes 4 and 6).
BGLF2 counteracts the ability of IFN-α to inhibit EBV reactivation in Raji cells.
Since BGLF2 reduces the phosphorylation of Tyk2, STAT1, and STAT3 and counteracts the effects of type I IFN on EBV reactivation in AGS-EBV gastric carcinoma cells, we postulated that BGLF2 may also counteract type I IFN suppression of EBV reactivation in Raji cells, which are a Burkitt lymphoma cell line latently infected with EBV. Raji cells were treated with tumor promoter 12-O-tetradecanoylphorbol-13-acetate (TPA) and sodium butyrate for 24, 48, or 72 h in the presence of IFN-α TPA, and sodium butyrate treatment induced BZLF1 expression at 24 h; longer treatment induced higher expression of BZLF1 (Fig. 5A). Addition of IFN-α suppressed BZLF1 expression (EBV reactivation) induced by TPA and sodium butyrate (Fig. 5A) by 2.3-, 5.3-, or 6.3-fold at 24, 48, or 72 h of treatment, respectively. Expression of BGLF2 in Raji cells induced BZLF1 expression; treating these cells with IFN-α had little additional effect on BZLF1 expression (1.15-fold reduction) (Fig. 5B). Treating Raji cells expressing BGLF2 with TPA and sodium butyrate further induced BZLF1 expression (Fig. 5C, lanes 1 and 2); addition of IFN-α reduced BZLF1 expression in Raji cells expressing GFP by 3-fold, but addition of IFN-α resulted in only a 1.9-fold reduction of BZLF1 expression in Raji cells expressing BGLF2. In summary, IFN-α suppresses EBV reaction in gastric carcinoma cells as well as in Raji cells, and expression of BGLF2 counteracts the ability of IFN-α to suppress EBV reactivation.
FIG 5.
BGLF2 counteracts the ability of IFN-α to inhibit EBV reactivation in Raji cells. (A) Raji cells were treated with TPA (200 nM) and sodium butyrate (NaB; 3 mM) and IFN-α (1,000 U/ml) for 24 h, 48 h, or 72 h and immunoblotted for BZLF1 and β-actin. (B) Raji cells expressing GFP or BGLF2-V5 were treated with or without IFN-α. (C) Raji cells expressing GFP or BGLF2-V5 were treated with TPA and sodium butyrate with or without IFN-α for 48 h and immunoblotted for BZLF1, BGLF2, and β-actin.
BGLF2 inhibits STAT3 phosphorylation induced by IFN-β in Raji cells.
Since BGLF2 reduced STAT3 phosphorylation induced by IFN-β in 293T cells and inhibited EBV reactivation in Raji cells, we determined if BGLF2 had a similar effect on STAT3 phosphorylation in Raji cells. Raji cells were electroporated with plasmids expressing BGLF2 or GFP. EBV replication was induced with TPA and sodium butyrate for 48 h, and then the cells were treated with IFN-β (1 × 103 U/ml) for 20 min. BGLF2 reduced the level of p-STAT3 induced by IFN-β in Raji cells (Fig. 6A and B) and reduced the expression of p-STAT3 and STAT3 in Raji cells in the presence of TPA and sodium butyrate (Fig. 6C, D, and E).
FIG 6.
BGLF2 reduces p-STAT3 induced by IFN-β in Raji cells. (A) Raji cells were electroporated with a plasmid expressing BGLF2-V5 or GFP (control) and after 48 h were treated with 1,000 U/ml IFN-β for 20 min. (C) Raji cells were electroporated with plasmid expressing BGLF2-V5 or GFP, treated with TPA and sodium butyrate (NaB) for 48 h, and IFN-β (1,000 U/ml) was added for 20 min. (B, D, and E) t test statistics for the ratio of p-STAT3 to STAT3 from the experiment in panel A or the ratio of STAT3/actin and p-STAT3/actin from the experiment in panel C. The results shown in panels B, D, and E are based on three separate experiments.
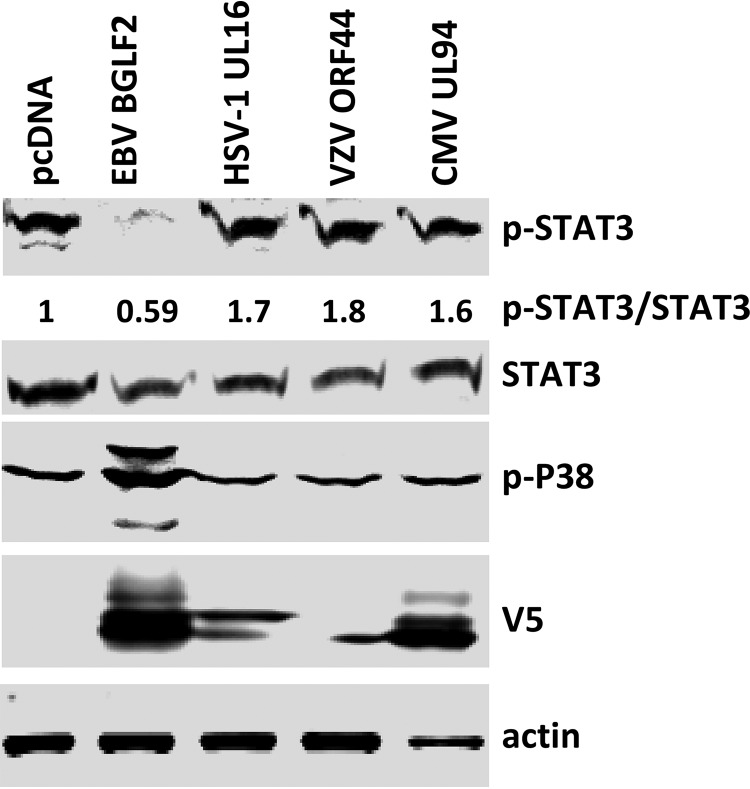
The ortholog of EBV BGLF2 in herpes simplex virus and human cytomegalovirus do not inhibit STAT3 phosphorylation or activate p38.
To determine if BGLF2 orthologs from other human herpesviruses might also inhibit type I interferon signaling, we constructed plasmids expressing EBV BGLF2 orthologs with V5 epitope tags in herpes simplex 1 (HSV1; UL16) and varicella-zoster virus (VZV; ORF44), both alphaherpesviruses, and in human cytomegalovirus (HCMV; UL94), a betaherpesvirus. These plasmids were individually transfected into 293T cells, and the cells were treated with IFN-β. Only HSV-1 UL16 and HCMV UL94 were expressed at levels similar to EBV BGLF2. While BGLF2 inhibited phosphorylation of STAT3 and activated p38, HSV-1 UL16 and HCMV UL94 did not inhibit STAT3 phosphorylation or activate p38 (Fig. 7).
FIG 7.
The effects of BGLF2 and its herpesvirus orthologs on p-STAT3 and p-p38. 293T cells were transfected with plasmids expressing EBV BGLF2, HSV-1 UL16, VZV ORF44, or CMV UL94 tagged with V5-tag at their C terminus or empty vector pcDNA3.1 (vector control). After 48 h, the cells were treated with IFN-β (1,000 U/ml) for 20 min, and cell lysates were immunoblotted with antibody to p-STAT3, STAT3, p-p38, V5, and actin.
DISCUSSION
We have found that EBV BGLF2 binds to Tyk2 and inhibits its phosphorylation, resulting in reduced phosphorylation of STAT1 and STAT3 and impaired type I IFN signaling. STAT1 is important for signaling through the IFN pathway and has a role both in immune surveillance of EBV-infected cells and in maintaining virus latency. STAT1 is critical for the control of EBV, and STAT1 gain of function has been associated with overwhelming and fatal EBV infection (42). Both EBNA1 (43) and LMP1 (44, 45) upregulate STAT1, and STAT1 is important to maintain latency (46). The ability of BGLF2 to inhibit phosphorylation of STAT1 may help to promote virus reactivation. BZLF1 inhibits phosphorylation and nuclear translocation of STAT1 (47).
Like STAT1, STAT3 is important for the control of EBV by the immune system and for maintaining virus latency. Patients with STAT3 dominant negative mutations have higher levels of EBV in their peripheral blood mononuclear cells and higher rates of lymphomas, some of which are EBV positive (48). LMP1 upregulates STAT3 (49) and EBNA-2 enhances the activity of STAT3 (50). STAT3 is required for EBV-induced B cell proliferation (51), and STAT3 inhibits lytic replication of EBV (52, 53). Thus, inhibition of STAT3 activation by BGLF2 may help to inhibit latency and promote virus reactivation of EBV.
BGLF2 inhibited several ISGs, including IRF1 and IRF7. Several other EBV proteins also inhibit IFN signaling and IRF7. EBV IE protein BZLF1 inhibits IFN-α/β production by its interaction with IRF7 (54). EBV IE protein BRLF1 inhibits transcription of IRF3 and IRF7 and suppresses induction of IFN-β (54, 55). EBV BGLF4, the virus-encoded protein kinase, interacts with IRF3 and reduces the amount of IRF3 recruited to ISREs, resulting in reduced induction of type I IFNs (56). EBV BCRF1, which encodes an interleukin-10 (IL-10) homolog (57) inhibits IFN-γ secretion from primary human B lymphocytes (58), while BARF1 inhibits IFN-α secretion from mononuclear cells (59). EBV tegument protein LF2 interacts with IRF7, inhibiting its ability to bind and activate the IFN-α promoter (60). EBV EBERs induce resistance to IFN-α-mediated apoptosis (61). EBNA-2 inhibits the ability of IFN signaling to produce ISGs (23, 25). LMP1 inhibits Tyk2 phosphorylation and induces cellular microRNA miR-146a to block IFN signaling in B cells (62, 63). LMP2A and LMP2B limit the activity of IFN by targeting IFN receptors for degradation (64). The observation that IFN-α did not inhibit BGLF2-induced EBV lytic replication in AGS-EBV cells may due to the fact that once EBV lytic replication is initiated, other virus proteins also antagonize the effect of IFN.
EBV BGLF2 has homologs in the other human herpesviruses, including herpes simplex virus (HSV) UL16, VZV ORF44, cytomegalovirus UL94, human herpesvirus 6 (HHV-6) and HHV-7 U65, and Kaposi’s sarcoma-associated herpesvirus ORF33. None of these proteins have been shown to definitely modulate IFN signaling pathways. A single study mapped a gene locus in the BamHI D fragment of HSV-2, which contains portions of UL14, UL15, and UL16, with resistance to mouse IFN-α/β (65). While we tested orthologs of BGLF2 in several other human herpesviruses, only HCMV UL94 and HSV-1 UL16 were expressed at levels comparable to BGLF2, and the HCMV UL94 and HSV-1 UL16 proteins did not inhibit STAT3 phosphorylation.
IFN-α inhibits the early stages of EBV reactivation (30–33) and suppresses shedding of virus from the oropharynx of immunosuppressed persons (66). As a tegument protein, BGLF2 is delivered to cells during infection. Therefore, EBV BGLF2 might protect virus-infected cells from the type I IFN response when the viral protein is present during initial EBV infection or during virus shedding from the oropharynx at a time when epithelial cells undergo lytic replication.
MATERIALS AND METHODS
Cells.
293T (human embryonic kidney) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS); HEK293T/17 SF cells (ATCC ACS-4500) were cultured in Gibco Expi293 expression medium (Gibco catalog no. A14350-01) as suspension cells at 8% CO2 at 37°C. EBV-infected Raji Burkitt lymphoma cells were grown in RPMI medium with 15% FBS, and EBV-infected AGS-EBV-GFP cells (67) were grown in Ham’s F-12 medium with 15% FBS. 293T-BGLF2-V5 cells, which express V5-tagged BGLF2 when incubated with doxycycline, were grown in DMEM with 10% FBS and described previously (39). Cell lines stably expressing BGLF2, termed 293T-BGLF2 and AGS-EBV-BGLF2, or expressing BGLF2ΔC, termed AGS-EBV-BGLF2ΔC, or expressing luciferase, termed AGS-EBV-Luc, were constructed by transducing cells with individual lentivirus expressing BGLF2-Flag, BGLF2ΔC-Flag, or luciferase and selecting with blasticidin (39).
Plasmids, lentiviral vectors, cell lines, and reagents.
Plasmids expressing V5-tagged BGLF2, V5-tagged BGLF2ΔC, BGLF2, or VZV ORF13 were reported previously (39). A lentiviral vector expressing GFP (pLJM1-EGFP) was from Addgene. IFN-α (Schering Corporation), IFN-β (Peprotech), and IFN-γ (Peprotech) were used to stimulate cells. Methotrexate (Teva Pharmaceuticals), or TPA (12-O-tetradecanoylphorbol-13-acetate; Cell Signaling Technology), and sodium butyrate were used to induce EBV lytic replication in Raji cells.
Immunoblotting and antibodies.
Cell lysates were fractionated in polyacrylamide gels, transferred to nitrocellulose membranes, and incubated with mouse anti-EBV BZLF1 (sc-53904; Santa Cruz), anti-Flag (M2; Sigma), anti-V5 (MAC1360; AbD Serotec), rabbit anti-p-p38, anti-p38, anti-p-JNK, anti-JNK, anti-p-Tyk2, anti-Tyk2, anti-p-STAT1, anti-STAT1, anti-p-STAT2, anti-STAT2, anti-p-STAT3, anti-STAT3, and anti-GFP (Cell Signaling Technology), or mouse or rabbit anti-actin antibody (Sigma).
RNA isolation, cDNA synthesis, and quantitative PCR.
Total RNA was isolated from 293T cells transfected with GFP or BGLF2-V5 plasmid for 24 h and treated with 1,000 U/ml IFN-β for an additional 3 h using an RNeasy minikit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RNA was treated with recombinant DNase I (5 units/100 μl) (Roche Diagnostics, Mannheim, Germany) on the column for 1 h at 37°C and eluted in 30 μl nuclease-free water. cDNA was synthesized from 10.4 μl RNA (30 μl of column eluent) in a 20-μl reaction using an oligo(dT)18 primer with a Transcriptor high-fidelity cDNA synthesis kit (Roche Diagnostics) according to the manufacturer’s instructions. Quantitative PCR (qPCR) was performed using 1 μl of cDNA in a 15-μl reaction in triplicate with FastStart universal SYBR green master mix (Rox) (Roche Diagnostics) in a 7500 real-time PCR system (Life Technologies). β-Actin was amplified from the cDNA as an internal control. The program used was the following: 1 cycle of 95°C hot start for 10 min and 45 cycles of 95°C for 15 sec, 55°C for 10 sec, and 68°C for 45 sec. A dissociation curve analysis from 60°C to 95°C was performed after every qPCR run to exclude primer-dimer formation and other nonspecific amplification. Primer sequences used to amplify IFN stimulated genes (ISGs) and actin were IRF1 (forward, 5′-GCT GGG AGA TCA ACA AGG AT-3′; reverse, 5′-CTT TCC TCT GGC TCT TGG TG-3′), IRF7 (forward, GCA GCG TGA GGG TGT GTC TT-3′; reverse, 5′-GCT CCA TAA GGA AGC ACT CGA T-3′), ISG15 (forward, 5′-TGG TGG ACA AAT GCG ACG AA-3′; reverse, 5′-CAG GCG CAG ATT CAT GAA C-3′), MxA (forward, 5′-TGA TCC ACC TGC TGC ATC CC-3′; reverse, 5′-GGC GCA CCT TCT CCT CAT AC-3′), and β-actinF961 (5′-GCA CCC AGC ACA ATG AAG A-3′), and β-actinR1024 (5′-CGA TCC ACA CGG AGT ACT TG-3′).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID). T.S. was supported by the Japan Herpesvirus Infections Forum.
We thank the Research Technologies Branch of NIAID for mass spectrometry analysis.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Katze MG, He Y, Gale M Jr. 2002. Viruses and interferon: a fight for supremacy. Nat Rev Immunol 2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 2.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird L. 2011. Viral immunity: shelter from interferons. Nat Rev Immunol 12:2. doi: 10.1038/nri3137. [DOI] [PubMed] [Google Scholar]
- 4.McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. 2015. Type I interferons in infectious disease. Nat Rev Immunol 15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivashkiv LB, Donlin LT. 2014. Regulation of type I interferon responses. Nat Rev Immunol 14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzgerald KA. 2011. The interferon inducible gene: viperin. J Interferon Cytokine Res 31:131–135. doi: 10.1089/jir.2010.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haller O, Kochs G. 2011. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J Interferon Cytokine Res 31:79–87. doi: 10.1089/jir.2010.0076. [DOI] [PubMed] [Google Scholar]
- 8.Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol 1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhury FZ, Farrar JD. 2013. STAT2: a shape-shifting anti-viral super STAT. JAKSTAT 2:e23633. doi: 10.4161/jkst.23633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morales DJ, Lenschow DJ. 2013. The antiviral activities of ISG15. J Mol Biol 425:4995–5008. doi: 10.1016/j.jmb.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perreira JM, Chin CR, Feeley EM, Brass AL. 2013. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol 425:4937–4955. doi: 10.1016/j.jmb.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Neill LAJ, Bowie AG. 2010. Sensing and signaling in antiviral innate immunity. Curr Biol 20:R328–R333. doi: 10.1016/j.cub.2010.01.044. [DOI] [PubMed] [Google Scholar]
- 13.Wilkins C, Gale M Jr. 2010. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol 22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arpaia N, Barton GM. 2011. Toll-like receptors: key players in antiviral immunity. Curr Opin Virol 1:447–454. doi: 10.1016/j.coviro.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barber GN. 2011. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr Opin Immunol 23:10–20. doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eliopoulos AG, Young LS. 2001. LMP1 structure and signal transduction. Semin Cancer Biol 11:435–444. doi: 10.1006/scbi.2001.0410. [DOI] [PubMed] [Google Scholar]
- 17.Ning S, Pagano JS, Barber GN. 2011. IRF7: activation, regulation, modification and function. Genes Immun 12:399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Pagano JS. 2001. Interferon regulatory factor 7: a key cellular mediator of LMP-1 in EBV latency and transformation. Semin Cancer Biol 11:445–453. doi: 10.1006/scbi.2001.0411. [DOI] [PubMed] [Google Scholar]
- 19.Ning S, Hahn AM, Huye LE, Pagano JS. 2003. Interferon regulatory factor 7 regulates expression of Epstein-Barr virus latent membrane protein 1: a regulatory circuit. J Virol 77:9359–9368. doi: 10.1128/jvi.77.17.9359-9368.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song YJ, Izumi KM, Shinners NP, Gewurz BE, Kieff E. 2008. IRF7 activation by Epstein-Barr virus latent membrane protein 1 requires localization at activation sites and TRAF6, but not TRAF2 or TRAF3. Proc Natl Acad Sci U S A 105:18448–18453. doi: 10.1073/pnas.0809933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikushima H, Negishi H, Taniguchi T. 2013. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harbor Symp Quant Biol 78:105–116. doi: 10.1101/sqb.2013.78.020321. [DOI] [PubMed] [Google Scholar]
- 22.Kanda K, Kempkes B, Bornkamm GW, von Gabain A, Decker T. 1999. The Epstein-Barr virus nuclear antigen 2 (EBNA2), a protein required for B lymphocyte immortalization, induces the synthesis of type I interferon in Burkitt’s lymphoma cell lines. Biol Chem 380:213–221. [DOI] [PubMed] [Google Scholar]
- 23.Kanda K, Decker T, Aman P, Wahlstrom M, von Gabain A, Kallin B. 1992. The EBNA2-related resistance towards alpha interferon (IFN-alpha) in Burkitt’s lymphoma cells effects induction of IFN-induced genes but not the activation of transcription factor ISGF-3. Mol Cell Biol 12:4930–4936. doi: 10.1128/mcb.12.11.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu D, Brumm K, Zhang L. 2006. The latent membrane protein 1 of Epstein-Barr virus (EBV) primes EBV latency cells for type I interferon production. J Biol Chem 281:9163–9169. doi: 10.1074/jbc.M511884200. [DOI] [PubMed] [Google Scholar]
- 25.Aman P, von Gabain A. 1990. An Epstein-Barr virus immortalization associated gene segment interferes specifically with the IFN-induced anti-proliferative response in human B-lymphoid cell lines. EMBO J 9:147–152. doi: 10.1002/j.1460-2075.1990.tb08090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ersing I, Bernhardt K, Gewurz BE. 2013. NF-kappaB and IRF7 pathway activation by Epstein-Barr virus Latent Membrane Protein 1. Viruses 5:1587–1606. doi: 10.3390/v5061587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Najjar I, Baran-Marszak F, Le Clorennec C, Laguillier C, Schischmanoff O, Youlyouz-Marfak I, Schlee M, Bornkamm GW, Raphaël M, Feuillard J, Fagard R. 2005. Latent membrane protein 1 regulates STAT1 through NF-kappaB-dependent interferon secretion in Epstein-Barr virus-immortalized B cells. J Virol 79:4936–4943. doi: 10.1128/JVI.79.8.4936-4943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Pagano JS. 2002. Structure and function of IRF-7. J Interferon Cytokine Res 22:95–101. doi: 10.1089/107999002753452700. [DOI] [PubMed] [Google Scholar]
- 29.Iwakiri D, Takada K. 2010. Role of EBERs in the pathogenesis of EBV infection. Adv Cancer Res 107:119–136. doi: 10.1016/S0065-230X(10)07004-1. [DOI] [PubMed] [Google Scholar]
- 30.Adams A, Strander H, Cantell K. 1975. Sensitivity of the Epstein-Barr virus transformed human lymphoid cell lines to interferon. J Gen Virol 28:207–217. doi: 10.1099/0022-1317-28-2-207. [DOI] [PubMed] [Google Scholar]
- 31.Lvovsky E, Levine PH, Fuccillo D, Ablashi DV, Bengali ZH, Armstrong GR, Levy HB. 1981. Epstein-Barr virus and Herpesvirus saimiri: sensitivity to interferons and interferon inducers. J Natl Cancer Inst 66:1013–1019. doi: 10.1093/jnci/66.6.1013. [DOI] [PubMed] [Google Scholar]
- 32.Tovey MG, Dron M, Gresser I. 1982. Interferon enhances the expression of epstein-Barr virus early antigen in Daudi cells. J Gen Virol 60:31–38. doi: 10.1099/0022-1317-60-1-31. [DOI] [PubMed] [Google Scholar]
- 33.Takimoto T, Ishikawa S, Umeda R, Ogura H, Hatano M. 1985. Effects of human interferon on cell proliferation and antigens induced by Epstein-Barr virus in epithelial hybrid cells derived from nasopharyngeal carcinoma. Otolaryngol Head Neck Surg 93:500–504. doi: 10.1177/019459988509300406. [DOI] [PubMed] [Google Scholar]
- 34.Delcayre AX, Lotz M, Lernhardt W. 1993. Inhibition of Epstein-Barr virus-mediated capping of CD21/CR2 by alpha interferon (IFN-alpha): immediate antiviral activity of IFN-alpha during the early phase of infection. J Virol 67:2918–2921. doi: 10.1128/JVI.67.5.2918-2921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin JC, Zhang ZX, Chou TC, Sim I, Pagano JS. 1989. Synergistic inhibition of Epstein-Barr virus: transformation of B lymphocytes by alpha and gamma interferon and by 3′-azido-3′-deoxythymidine. J Infect Dis 159:248–254. doi: 10.1093/infdis/159.2.248. [DOI] [PubMed] [Google Scholar]
- 36.Garner JG, Hirsch MS, Schooley RT. 1984. Prevention of Epstein-Barr virus-induced B-cell outgrowth by interferon alpha. Infect Immun 43:920–924. doi: 10.1128/IAI.43.3.920-924.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lotz M, Tsoukas CD, Fong S, Carson DA, Vaughan JH. 1985. Regulation of Epstein-Barr virus infection by recombinant interferons. Selected sensitivity to interferon-gamma. Eur J Immunol 15:520–525. doi: 10.1002/eji.1830150518. [DOI] [PubMed] [Google Scholar]
- 38.Sharp NA, Arrand JR, Clemens MJ. 1989. Epstein-Barr virus replication in interferon-treated cells. J Gen Virol 70:2521–2526. doi: 10.1099/0022-1317-70-9-2521. [DOI] [PubMed] [Google Scholar]
- 39.Liu X, Cohen JI. 2016. Epstein-Barr Virus (EBV) tegument protein BGLF2 promotes EBV reactivation through activation of the p38 mitogen-activated protein kinase. J Virol 90:1129–1138. doi: 10.1128/JVI.01410-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konishi N, Narita Y, Hijioka F, Masud H, Sato Y, Kimura H, Murata T. 2018. BGLF2 increases infectivity of Epstein-Barr virus by activating AP-1 upon de novo infection. mSphere 3:e00138-18. doi: 10.1128/mSphere.00138-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paladino P, Marcon E, Greenblatt J, Frappier L. 2014. Identification of herpesvirus proteins that contribute to G1/S arrest. J Virol 88:4480–4492. doi: 10.1128/JVI.00059-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharfe N, Nahum A, Newell A, Dadi H, Ngan B, Pereira SL, Herbrick JA, Roifman CM. 2014. Fatal combined immunodeficiency associated with heterozygous mutation in STAT1. J Allergy Clin Immunol 133:807–817. doi: 10.1016/j.jaci.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 43.Wood VHJ, O'Neil JD, Wei W, Stewart SE, Dawson CW, Young LS. 2007. Epstein-Barr virus-encoded EBNA1 regulates cellular gene transcription and modulates the STAT1 and TGFbeta signaling pathways. Oncogene 26:4135–4147. doi: 10.1038/sj.onc.1210496. [DOI] [PubMed] [Google Scholar]
- 44.Gires O, Kohlhuber F, Kilger E, Baumann M, Kieser A, Kaiser C, Zeidler R, Scheffer B, Ueffing M, Hammerschmidt W. 1999. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J 18:3064–3073. doi: 10.1093/emboj/18.11.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richardson C, Fielding C, Rowe M, Brennan P. 2003. Epstein-Barr virus regulates STAT1 through latent membrane protein 1. J Virol 77:4439–4443. doi: 10.1128/jvi.77.7.4439-4443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLaren JE, Zuo J, Grimstead J, Poghosyan Z, Bell AI, Rowe M, Brennan P. 2009. STAT1 contributes to the maintenance of the latency III viral programme observed in Epstein-Barr virus-transformed B cells and their recognition by CD8+ T cells. J Gen Virol 90:2239–2250. doi: 10.1099/vir.0.011627-0. [DOI] [PubMed] [Google Scholar]
- 47.Morrison TE, Mauser A, Wong A, Ting JP, Kenney SC. 2001. Inhibition of IFN-gamma signaling by an Epstein-Barr virus immediate-early protein. Immunity 15:787–799. doi: 10.1016/s1074-7613(01)00226-6. [DOI] [PubMed] [Google Scholar]
- 48.Siegel AM, Heimall J, Freeman AF, Hsu AP, Brittain E, Brenchley JM, Douek DC, Fahle GH, Cohen JI, Holland SM, Milner JD. 2011. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity 35:806–818. doi: 10.1016/j.immuni.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kung CP, Meckes DG Jr, Raab-Traub N. 2011. Epstein-Barr virus LMP1 activates EGFR, STAT3, and ERK through effects on PKCdelta. J Virol 85:4399–4408. doi: 10.1128/JVI.01703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muromoto R, Ikeda O, Okabe K, Togi S, Kamitani S, Fujimuro M, Harada S, Oritani K, Matsuda T. 2009. Epstein-Barr virus-derived EBNA2 regulates STAT3 activation. Biochem Biophys Res Commun 378:439–443. doi: 10.1016/j.bbrc.2008.11.053. [DOI] [PubMed] [Google Scholar]
- 51.Koganti S, de la Paz A, Freeman AF, Bhaduri-McIntosh S. 2014. B lymphocytes from patients with a hypomorphic mutation in STAT3 resist Epstein-Barr virus-driven cell proliferation. J Virol 88:516–524. doi: 10.1128/JVI.02601-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koganti S, Clark C, Zhi J, Li X, Chen EI, Chakrabortty S, Hill ER, Bhaduri-McIntosh S. 2015. Cellular STAT3 functions via PCBP2 to restrain Epstein-Barr Virus lytic activation in B lymphocytes. J Virol 89:5002–5011. doi: 10.1128/JVI.00121-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daigle D, Megyola C, El-Guindy A, Gradoville L, Tuck D, Miller G, Bhaduri-McIntosh S. 2010. Upregulation of STAT3 marks Burkitt lymphoma cells refractory to Epstein-Barr virus lytic cycle induction by HDAC inhibitors. J Virol 84:993–1004. doi: 10.1128/JVI.01745-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bentz GL, Liu R, Hahn AM, Shackelford J, Pagano JS. 2010. Epstein-Barr virus BRLF1 inhibits transcription of IRF3 and IRF7 and suppresses induction of interferon-beta. Virology 402:121–128. doi: 10.1016/j.virol.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu LH, Gao S, Jin R, Zhuang LL, Jiang L, Qiu LZ, Xu HG, Zhou GP. 2014. Repression of interferon regulatory factor 3 by the Epstein-Barr virus immediate-early protein Rta is mediated through E2F1 in HeLa cells. Mol Med Rep 9:1453–1459. doi: 10.3892/mmr.2014.1957. [DOI] [PubMed] [Google Scholar]
- 56.Wang JT, Doong SL, Teng SC, Lee CP, Tsai CH, Chen MR. 2009. Epstein-Barr virus BGLF4 kinase suppresses the interferon regulatory factor 3 signaling pathway. J Virol 83:1856–1869. doi: 10.1128/JVI.01099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsu DH, de Waal Malefyt R, Fiorentino DF, Dang MN, Vieira P, de Vries J, Spits H, Mosmann TR, Moore KW. 1990. Expression of interleukin-10 activity by Epstein-Barr virus protein BCRF1. Science 250:830–832. doi: 10.1126/science.2173142. [DOI] [PubMed] [Google Scholar]
- 58.Swaminathan S, Hesselton R, Sullivan J, Kieff E. 1993. Epstein-Barr virus recombinants with specifically mutated BCRF1 genes. J Virol 67:7406–7413. doi: 10.1128/JVI.67.12.7406-7413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cohen JI, Lekstrom K. 1999. Epstein-Barr virus BARF1 protein is dispensable for B-cell transformation and inhibits alpha interferon secretion from mononuclear cells. J Virol 73:7627–7632. doi: 10.1128/JVI.73.9.7627-7632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu L, Fossum E, Joo CH, Inn KS, Shin YC, Johannsen E, Hutt-Fletcher LM, Hass J, Jung JU. 2009. Epstein-Barr virus LF2: an antagonist to type I interferon. J Virol 83:1140–1146. doi: 10.1128/JVI.00602-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nanbo A, Inoue K, Adachi-Takasawa K, Takada K. 2002. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt’s lymphoma. EMBO J 21:954–965. doi: 10.1093/emboj/21.5.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geiger TR, Martin JM. 2006. The Epstein-Barr virus-encoded LMP-1 oncoprotein negatively affects Tyk2 phosphorylation and interferon signaling in human B cells. J Virol 80:11638–11650. doi: 10.1128/JVI.01570-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cameron JE, Yin Q, Fewell C, Lacey M, McBride J, Wang X, Lin Z, Schaefer BC, Flemington EK. 2008. Epstein-Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol 82:1946–1958. doi: 10.1128/JVI.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shah KM, Stewart SE, Wei W, Woodman CBJ, O'Neil JD, Dawson CW, Young LS. 2009. The EBV-encoded latent membrane proteins, LMP2A and LMP2B, limit the actions of interferon by targeting interferon receptors for degradation. Oncogene 28:3903–3914. doi: 10.1038/onc.2009.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Su YH, Oakes JE, Lausch RN. 1993. Mapping the genetic region coding for herpes simplex virus resistance to mouse interferon alpha/beta. J Gen Virol 74:2325–2332. doi: 10.1099/0022-1317-74-11-2325. [DOI] [PubMed] [Google Scholar]
- 66.Cheeseman SH, Henle W, Rubin RH, Tolkoff-Rubin NE, Cosimi B, Cantell K, Winkle S, Herrin JT, Black PH, Russell PS, Hirsch MS. 1980. Epstein-Barr virus infection in renal transplant recipients: effects of antithymocyte globulin and interferon. Ann Intern Med 93:39–42. doi: 10.7326/0003-4819-93-1-39. [DOI] [PubMed] [Google Scholar]
- 67.Feng WH, Cohen JI, Fischer S, Li L, Sneller M, Goldbach-Mansky R, Raab-Traub N, Delecluse HJ, Kenney SC. 2004. Reactivation of latent Epstein-Barr virus by methotrexate: a potential contributor to methotrexate-associated lymphomas. J Natl Cancer Inst 96:1691–1702. doi: 10.1093/jnci/djh313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.