Antibiotic resistance is a global concern; however, data on antibiotic-resistant Ureaplasma spp. and Mycoplasma hominis are limited in comparison to similar data on other microbes. A total of 492 Ureaplasma spp. and 13 M. hominis strains obtained in Hangzhou, China, in 2018 were subjected to antimicrobial susceptibility testing for levofloxacin, moxifloxacin, erythromycin, clindamycin, and doxycycline using the broth microdilution method.
KEYWORDS: antibiotic resistance, molecular dynamics simulations, Mycoplasma hominis, resistance mechanism, Ureaplasma spp.
ABSTRACT
Antibiotic resistance is a global concern; however, data on antibiotic-resistant Ureaplasma spp. and Mycoplasma hominis are limited in comparison to similar data on other microbes. A total of 492 Ureaplasma spp. and 13 M. hominis strains obtained in Hangzhou, China, in 2018 were subjected to antimicrobial susceptibility testing for levofloxacin, moxifloxacin, erythromycin, clindamycin, and doxycycline using the broth microdilution method. The mechanisms underlying quinolone and macrolide resistance were determined. Meanwhile, a model of the topoisomerase IV complex bound to levofloxacin in wild-type Ureaplasma spp. was built to study the quinolone resistance mutations. For Ureaplasma spp., the levofloxacin, moxifloxacin, and erythromycin resistance rates were 84.69%, 51.44%, and 3.59% in U. parvum and 82.43%, 62.16%, and 5.40% in U. urealyticum, respectively. Of the 13 M. hominis strains, 11 were resistant to both levofloxacin and moxifloxacin, and five strains showed clindamycin resistance. ParC S83L was the most prevalent mutation in levofloxacin-resistant Ureaplasma strains, followed by ParE R448K. The two mutations GyrA S153L and ParC S91I were commonly identified in quinolone-resistant M. hominis. A molecular dynamics-refined structure revealed that quinolone resistance-associated mutations inhibited the interaction and reduced affinity with gyrase or topoisomerase IV and quinolones. The novel mutations S21A in the L4 protein and G2654T and T2245C in 23S rRNA and the ermB gene were identified in erythromycin-resistant Ureaplasma spp. As fluoroquinolone resistance in Ureaplasma spp. and Mycoplasma hominis remains high in China, the rational use of antibiotics needs to be further enhanced.
INTRODUCTION
Ureaplasma spp. and Mycoplasma hominis are both genital mycoplasmas; 40% to 80% of sexually mature women have Ureaplasma spp. in their vagina/cervix and 20% to 50% of sexually mature women have M. hominis there (1). They are implicated in nongonococcal urethritis, infertility, chorioamnionitis, adverse pregnancy outcomes, and neonatal diseases (2–4). Antibiotics active against these mycoplasmas are limited to agents that interfere with protein synthesis and DNA replication via their unique physiological properties, such as fluoroquinolones, tetracyclines, and macrolide (5). The potential toxicities of quinolones and tetracyclines decrease the availability of therapeutic options for infants, children, and people allergic to tetracyclines and quinolones.
A rise of antibiotic resistance in Ureaplasma spp. and M. hominis has been reported in France, leading to global concerns (6). Studies of different regions or populations were conducted in succession. A high level of fluoroquinolone resistance in these organisms has been observed in China (7, 8), and high levels of tetracycline and erythromycin (ERY) resistance have been reported in Ureaplasma species isolated from South African women (9). Investigators agreed that ongoing monitoring to determine the susceptibility of these microbes to potentially active antibiotics was necessary (10, 11).
Bacterial resistance to fluoroquinolone is related to genetic alterations in DNA gyrase (GyrA and GyrB) and/or the topoisomerase IV complex (ParC and ParE) (12). The resistance of Ureaplasma spp. is mainly associated with the mutation of C248T (Ser83Leu) in the parC gene (13, 14). Acquired resistance to macrolides has rarely been reported in Ureaplasma spp., and this resistance can be linked to substitutions in domain II or V of 23S rRNA and/or in the ribosomal proteins L4 and L22 (15). However, M. hominis is intrinsically resistant to 14- and 15-membered macrolides (16).
Previous studies concerning antibiotic resistance in Ureaplasma spp. were mainly conducted before 2016. Furthermore, few studies have evaluated antibiotic resistance in China, and the commercial IST kit was the prevalent method for detecting resistance (17, 18). However, this method can lead to inaccurate reporting of antibiotic resistance, such as overestimations of antibiotic resistance (10, 11). In contrast, broth microdilution is an internationally accepted technique used to evaluate antibiotic resistance and is also recommended by the Clinical and Laboratory Standard Institute (CLSI) guidelines. In this study, we used the microdilution method to evaluate resistance to five antibiotics in 492 Ureaplasma spp. and 13 M. hominis strains isolated in China from 1 September to 30 December 2018. Meanwhile, a dynamic refined molecular structure of the topoisomerase IV (ParC-ParE) complex from wild-type Ureaplasma spp. bound to levofloxacin (LVX) was built to study the mechanisms underlying quinolone-resistant mutations.
RESULTS
Prevalence of resistance in Ureaplasma spp. and Mycoplasma hominis.
Of the 492 Ureaplasma spp. tested, 84.69% (354/418) of Ureaplasma parvum samples were LVX resistant, and 82.43% (61/74) of Ureaplasma urealyticum samples exhibited LVX resistance, with similar trends for MIC50 and MIC90 values in the two species. The resistance rate for moxifloxacin (MXF) ranged from 51.44% in U. parvum to 62.16% in U. urealyticum, but similar MIC50 values (4 μg/ml) were exhibited by the two species. MXF-resistant isolates were also resistant to LVX. The majority of Ureaplasma spp. had a MIC for ERY ranging from 1 μg/ml to 4 μg/ml. ERY resistance was observed in 15 (3.59%) U. parvum and 4 (5.40%) U. urealyticum strains. Since no CLSI breakpoint value for doxycycline (DOX) was available, DOX resistance in Ureaplasma spp. was unclear. The MIC90 values for DOX in U. parvum and U. urealyticum were both 1 μg/ml. Of the 13 M. hominis strains tested, 84.62% (11/13) of the strains showed resistance to both LVX and MXF, with MIC50 values of 16 μg/ml for LVX and of 4 μg/ml for MXF. Five strains (38.46%) were resistant to clindamycin (CLI), and MICs ranged from 1 to >32 μg/ml. Ten M. hominis strains showed MICs of 0.125 μg/ml for DOX (Table 1).
TABLE 1.
MIC distributions of Ureaplasma parvum, Ureaplasma urealyticum, and Mycoplasma hominis
| Antibiotic (MIC μg/ml)a | No. of isolates with an MIC (μg/ml) of: |
MIC (μg/ml) |
Resistance (n [%]) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | >32 | 50% | 90% | ||
| U. parvum (418 strains) | |||||||||||||
| LVX (≥4) | 0 | 4 | 4 | 22 | 34 | 49 | 141 | 82 | 48 | 34 | 8 | 32 | 354 (84.69) |
| MXF (≥4) | 3 | 7 | 28 | 57 | 108 | 105 | 64 | 30 | 11 | 5 | 4 | 16 | 215 (51.44) |
| ERY (≥6) | 7 | 3 | 30 | 114 | 122 | 85 | 42 | 9 | 4 | 2 | 2 | 8 | 15 (3.59) |
| DOX | 200 | 88 | 80 | 37 | 6 | 4 | 1 | 1 | 0 | 1 | 0.25 | 1 | NAb |
| U. urealyticum (74 strains) | |||||||||||||
| LVX (≥4) | 0 | 0 | 1 | 1 | 11 | 11 | 22 | 16 | 10 | 2 | 8 | 32 | 61 (82.43) |
| MXF (≥4) | 0 | 2 | 2 | 14 | 10 | 19 | 20 | 5 | 2 | 0 | 4 | 8 | 46 (62.16) |
| ERY (≥16) | 1 | 0 | 1 | 5 | 24 | 25 | 14 | 4 | 0 | 0 | 4 | 8 | 4 (5.40) |
| DOX | 16 | 18 | 19 | 15 | 5 | 1 | 0 | 0 | 0 | 0 | 0.5 | 1 | NA |
| M. hominis (13 strains) | |||||||||||||
| LVX (≥2) | 0 | 0 | 1 | 1 | 0 | 2 | 2 | 1 | 6 | 0 | 16 | 32 | 11 (84.62) |
| MXF (≥0.5) | 2 | 0 | 2 | 1 | 1 | 1 | 2 | 4 | 0 | 0 | 4 | 16 | 11 (84.62) |
| CLI (≥0.5) | 8 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0.125 | 16 | 5 (38.46) |
| DOX | 10 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0.125 | 0.5 | NA |
LVX, levofloxacin; MXF, moxifloxacin; ERY, erythromycin; DOX, doxycycline; CLI, clindamycin.
NA, not applicable (no CLSI breakpoint).
Molecular mechanism of quinolone resistance.
Of the 354 LVX-resistant U. parvum and 61 LVX-resistant U. urealyticum strains, 322 U. parvum and 46 U. urealyticum strains presented a single mutation within ParC, Ser83L. The majority of resistant strains had a MIC value of 8 μg/ml, which was twice the breakpoint of LVX. Nine U. parvum and three U. urealyticum strains showed double mutations, one was the prevalent mutation, and the other occurred at GyrA (Q104K or Q100L), GyrB (N481S, P462S, E482D, D443A, or N486Y), or ParE (A481T or A481V). The MICs for these strains ranged from 8 μg/ml to >32 μg/ml. There were two amino acid substitutions at the 83 position in ParC other than S83L: S83W and S83A. Two Ureaplasma strains harbored the S83W substitution, with MICs >4-fold increased against LVX. One U. parvum harbored the S83A mutation, which increased the MIC up to 2-fold. Moreover, some mutations occurred at the ParC codon for Glu87: E87K and E87Q. Two U. parvum strains had the E87K mutation, with a 4-fold increase in the MIC for LVX. One U. parvum strain carried E87Q, with a 1-fold increase in the MIC for LVX. In addition to the two loci in ParC, a mutation at ParC Ala136 (A136S) was detected in one less-resistant U. parvum strain with a MIC of 4 μg/ml. Although quinolone resistance mutations occurred most frequently in ParC, one substitution was responsible for LVX resistance in ParE (R448K), and six U. parvum strains contained this mutation, exhibiting MICs ranging from 4 μg/ml to >32 μg/ml.
For M. hominis, the double substitution ParC K144R/ParE V417I was identified in 2 LVX-sensitive and 11 LVX-resistant isolates. For the resistant mutations, one strain harbored the S91I mutation in ParC and had a MIC of 8 μg/ml for LVX. Two strains contained the double mutations ParC S91I/GyrA S153A, with MICs for LVX ranging from 8 to 16 μg/ml. Five strains had the double mutation ParC S91I/GyrA S153L and a similar MIC of 32 μg/ml for LVX. Moreover, three mutations were identified in one strain, ParC S91I, GyrA S153L, and GyrB A473V. This strain demonstrated a similar LVX-resistant profile as that for the five strains, while the MICs for MXF in this strain were lower (8 μg/ml) than those of the five strains (16 μg/ml). The combination of GyrA S153L and ParE R447K was observed in a strain with MICs of 4 μg/ml for LVX and MXF (Table 2).
TABLE 2.
MICs and genetic alternations of fluoroquinolone-resistant Ureaplasma parvum, Ureaplasma urealyticum, and Mycoplasma hominis
| No. of resistant strains | MIC (μg/ml)a |
QRDRb |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LVX |
MXF |
ERY/CLI |
DOX |
|||||||||
| 50% | Range | 50% | Range | 50% | Range | 50% | Range | GyrA | GyrB | ParC | ParE | |
| U. parvum | ||||||||||||
| 322 | 8 | 4 to >32 | 4 | 0.5 to >32 | 2 | 0.125 to >32 | 0.25 | 0.125 to >32 | C248T (S83L) | |||
| 3 | >32 | 32 to >32 | 32 | 8 to >32 | 1 | (1 to 8) | 0.125 | 0.125 to 0.5 | C310A (Q104K) | C248T (S83L) | ||
| 2 | 32 | 32 to >32 | 4 | (4 to 16) | 2 | (2 to 4) | 0.25 | (0.25 to 0.5) | A1442G (N481S) | C248T (S83L) | ||
| 1 | 16 | 8 | 4 | 0.5 | C1384T (P462S) | C248T (S83L) | ||||||
| 1 | 16 | 4 | 2 | 0.25 | A1446C (E482D) | C248T (S83L) | ||||||
| 1 | >32 | 16 | 0.5 | 0.125 | A1328C (D443A) | C248T (S83L) | ||||||
| 1 | 32 | 8 | 0.5 | 0.125 | C248T (S83L) | G1441A (A481T) | ||||||
| 1 | 16 | 8 | 2 | 0.25 | C248G (S83W) | |||||||
| 1 | 8 | 2 | 1 | 0.125 | T247G (S83A) | |||||||
| 2 | 16 | 16 to 32 | 4 | (4 to 8) | 2 | (2 to 2) | 0.125 | 0.125 to 4 | G259A (E87K) | |||
| 1 | 4 | 2 | 8 | 0.125 | G259C (E87Q) | |||||||
| 1 | 4 | 4 | 8 | 1 | G406T (A136S) | |||||||
| 5 | 4 | 4 to >32 | 4 | (1 to 16) | 4 | (0.25 to 32) | 0.25 | (0.125 to 1) | G1343A (R448K) | |||
| 1 | 8 | 4 | 2 | 0.125 | G406T (A136S) | G1343A (R448K) | ||||||
| 11 | 8 | 4 to >32 | 2 | (1 to 16) | 4 | (1 to 16) | 1 | (0.125 to 1) | N | N | N | N |
| U. urealyticum | ||||||||||||
| 46 | 8 | 4 to >32 | 4 | (1 to 32) | 4 | 0.125 to 16 | 0.5 | 0.125 to 8 | C248T (S83L) | |||
| 1 | >32 | 32 | 8 | 0.5 | C248G (S83W) | |||||||
| 1 | 32 | 8 | 2 | 0.125 | A299T (Q100L) | C248T (S83L) | ||||||
| 1 | 32 | 16 | 8 | 0.5 | A1456T (N486Y) | C248T (S83L) | ||||||
| 1 | 8 | 4 | 4 | 0.25 | C248T (S83L) | C1442T (A481V) | ||||||
| 11 | 4 | (4 to 32) | 2 | (1 to 8) | 4 | (1 to 16) | 0.5 | 0.125 to 1 | N | N | N | N |
| M. hominis | ||||||||||||
| 2c | 1 | (0.5 to 1) | 0.125 | 0.125 to 0.125 | 0.125 | 0.125 to 0.125 | 0.125 | 0.125 to 0.125 | A431G (K144R) | G1249A (V417I) | ||
| 1 | 8 | 0.5 | 0.125 | 0.125 | G272T (S91I) | |||||||
| 2 | 16 | (8 to 16) | 2 | 0.5 to 2 | 16 | (0.125 to 16) | 0.5 | 0.125 to 0.5 | T457G (S153A) | G272T (S91I) | G1249A (V417I) | |
| 3 | 32 | 32 to 32 | 16 | (8 to 16) | 0.125 | (0.125 to >32) | 0.125 | (0.125 to 0.5) | C458T (S153L) | G272T (S91I) & A431G (K144R) | G1249A (V417I) | |
| 2 | 32 | 32 to 32 | 16 | (16 to 16) | 1 | (0.125 to 1) | 0.125 | 0.125 to 0.125 | C458T (S153L) | G272T (S91I) | G1249A (V417I) | |
| 1 | 32 | 8 | 4 | 2 | C458T (S153L) | C1418T (A473V) | G272T (S91I) | |||||
| 1 | 4 | 4 | 0.125 | 0.125 | C458T (S153L) | A431G (K144R) | G1249A (V417I) and G1340A (R447K) | |||||
| 1 | 4 | 1 | 2 | 0.125 | A431G (K144R) | |||||||
LVX, levofloxacin; MXF, moxifloxacin; ERY, erythromycin; CLI, clindamycin; DOX, doxycycline.
N, no mutation detected. Numbers and letters in parentheses refer to amino acid changes based on Ureaplasma spp. or Mycoplasma hominis. Novel mutations are highlighted in boldface font. Mutations without association with fluoroquinolones resistance are underlined.
Two strains of M. hominis were sensitive.
Based on our analysis, there were still 22 Ureaplasma strains and 1 M. hominis strain without genetic alterations in their quinolone resistance-determining regions (QRDRs).
The MD-refined structure of topoisomerase IV bound to LVX in wild-type Ureaplasma spp.
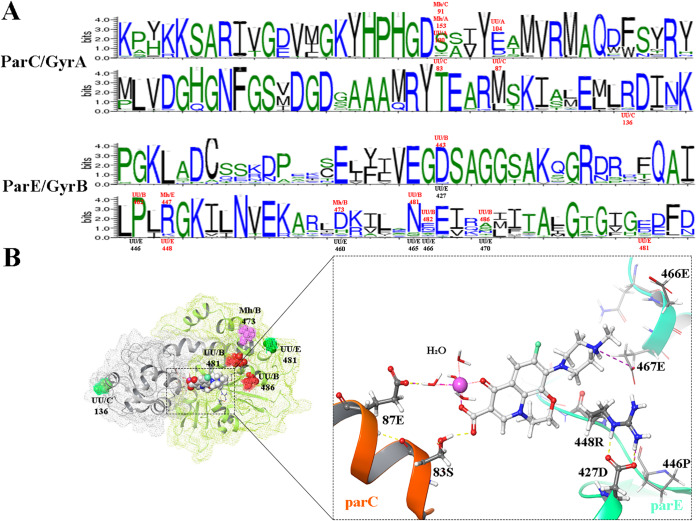
Figure 1A shows the alignment of amino acid sequences of ParC and GyrA and of ParE and GyrB. Conservation of amino acids at specific positions is associated with the height of the symbol within each stack. High sequence conservation was observed within the QRDR region of ParC and GyrA. Because the majority of the previously mentioned mutations occurred in a relatively conserved region, some hot spots were identified between different subunits (GyrA and ParC) or species (Ureaplasma spp. and M. hominis). For example, the mutation at ParC Ser83 in Ureaplasma spp. was equivalent to the alteration at GyrA Gln100 and corresponded to mutations in M. hominis, ParC Ser91 and GyrA Ser153. A similar corresponding relationship was observed for GyrA Gln104 and ParC Glu87 of Ureaplasma spp., for ParE Arg448 of Ureaplasma spp., and for ParE Arg447 of M. hominis.
FIG 1.
Sequence conservation of the quinolone resistance-determining region (QRDR) and Ureaplasma spp. ParC-ParE complex with levofloxacin. (A) The logos for ParC and GyrA and for ParE and GyrB were computed for 6,570 and 20,007 nonredundant sequences, respectively. The conservation of an amino acid is represented by the height of its symbol within the stack. The mutation sites that were based on the Ureaplasma spp. coordinates are indicated below the symbols by UU (Ureaplasma spp.) with a slash and with the position of the subunits (the subunit is before the position number; C represents ParC, and E represents ParE). The equivalent mutations in other subunits (A representing GyrA and B representing GyrB) or species (M. hominis [Mh]) were identified using the same naming. All the mutations mentioned in the body content are highlighted red. (B) The structure of Ureaplasma spp. topoisomerase IV bound to levofloxacin (LVX), and the interaction of ParC and ParE subunits with LVX. The mutations occurring outside the binding pocket were named as for panel A. The remaining mutations near the binding pocket were named based on the Ureaplasma spp. coordinates of ParC or of ParE. The yellow dashed lines represent hydrogen bonds, and the purple dashed lines represent an ionic bond. The magnesium ion is shown by the purple sphere, and it is coordinated by four water molecules.
A molecular dynamics (MD)-refined structure of the wild-type topoisomerase IV (ParC-ParE) complex bound to LVX in Ureaplasma spp. was made to identify the structural importance of the mutated amino acids. The analyses showed that ParC Ser83, ParC Glu87, ParE Asp427, ParE Arg448, and ParE Glu467 were the important residues involved in the binding pocket. The hydroxy residues of Ser83 (ParC) directly bound to the LVX C-3 carboxylic acid via hydrogen bonding. The ParC Glu87 formed a water-magnesium ion bridge with the C3/C4 keto acid of the quinolone by hydrogen bonds and charge interactions. The side chain of Glu467 (ParE) directly forms an ionic bond with R residues of LVX. Although no relationships were observed between ParE Asp427 or ParE Arg448 and LVX, the two amino acids form a strong ionic linkage and a hydrogen bond, avoiding the steric clashes between the bulky side chain of Arg448 and the LVX. These chemical bonds all worked together to stabilize the interaction between LVX and topoisomerase IV or gyrase.
In regard to the mutations that occurred at ParC Ser83 (S83L, S83W, and S83A) or its corresponding mutations in M. hominis (GyrA S153A, GyrA S153L, and ParC S91I), the alterations prevented the interaction with LVX. Meanwhile, the bulky side chains introduced by W, L, and I occupied the quinolone binding pocket of the LVX C-3 carboxylic acid. For the mutations ParC E87K (ParC), E87Q (ParC), Q104K (GyrA), and Q100L (GyrA) in Ureaplasma spp., which shared the same binding mode with LVX, variations in the electrical properties were observed among the first three mutations (E, “−”; K, “+”; Q, neutral [N]). Moreover, the K and L mutations prevented the formation of the hydrogen bond, consequently disrupting the water-magnesium ion bridge. For the mutations in ParE Arg448 (corresponding to ParE Arg447 in M. hominis) and ParE Asp427 (corresponding to GyrB Asp443 in Ureaplasma spp.), the former prevented the formation of hydrogen bonds and the latter weakened the ionic bond between the two amino acids (D443A: D, −; A, “N”). All of the above-mentioned mutations reduced the affinity between quinolone and gyrase or topoisomerase IV. Furthermore, mutations GyrB E482D and P462S and mutations occurring outside the quinolone binding pocket, which include ParC 136, GyrB 486, ParE 481, and GyrB 473 (in M. hominis), appeared to be similar to the differences in single nucleotide polymorphisms.
Molecular mechanism for the macrolide resistance of Ureaplasma spp.
Three ERY-resistant U. parvum strains contained two mutations in L22, A121S and T141I, with similar MICs of 16 μg/ml for ERY. One resistant strain harbored three substitutions, including two previously mentioned mutations and one S21A mutation in L4. A higher MIC for ERY was observed in this strain (>32 μg/ml). The D66N substitution in L22 was detected in one strain with a MIC of 16 μg/ml for ERY. Furthermore, two mutations within 23S rRNA, T2245C and G2654T, were detected in one strain with a MIC of 16 μg/ml (Table 3).
TABLE 3.
The MICs and genetic alternations of macrolide-resistant Ureaplasma parvum and Ureaplasma urealyticum
| No. of macrolide-resistant strains | MIC (μg/ml)a |
Macrolideb |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LVX |
MXFb |
ERYb |
DOXb |
||||||||
| 50% | Range | 50% | Range | 50% | Range | 50% | Range | L4 | L22 | 23S rRNA | |
| U. parvum | |||||||||||
| 3 | >32 | >32 to >32 | 32 | 16 to >32 | 16 | 16 to 16 | 0.5 | 1 to >32 | G361T (A121S) & C422T (T141I) | ||
| 1 | >32 | >32 | >32 | 4 | T61G (S21A) | G361T (A121S) & C422T (T141I) | |||||
| 1 | 32 | 16 | 16 | 1 | G196A (D66N) | ||||||
| 1 | 16 | 8 | 32 | 1 | T2245C ( E. coli numbering 2234T) | ||||||
| 9 | 32 | 4 to >32 | 16 | 2 to >32 | 16 | 16 to >32 | 1 | 0.5 to >32 | N | N | N |
| U. urealyticum | |||||||||||
| 1 | 16 | 4 | 16 | 2 | G2654T ( E. coli numbering 2643G) | ||||||
| 3 | 16 | (8 to 32) | 8 | (8 to 16) | 16 | (16 to 16) | 2 | (1 to 8) | N | N | N |
LVX, levofloxacin; MXF, moxifloxacin; ERY, erythromycin; DOX, doxycycline.
N, no mutation detected. For L4 and L22, numbers and letters in parentheses refer to amino acid changes based on Ureaplasma spp. For 23S rRNA, numbers and letters in parentheses are the corresponding numbers and letters in E. coli. Novel mutations are highlighted in boldface font.
Nine U. parvum and three U. urealyticum strains did not reveal any mutations in their 23S rRNA and/or ribosomal proteins L4 and L22. Among them, one U. parvum strain was detected with ermB, which is a subtype of the erm rRNA methyltransferase gene.
DISCUSSION
The rise of antibiotic resistance in Ureaplasma spp. and M. hominis is a global concern due to limited therapeutic options. Continued surveillance of the antibiotic susceptibility of these urogenital mycoplasmas has been recommended by many research studies (5, 10, 11). In previous studies, we evaluated the quinolone resistance of Ureaplasma spp. from 1999 to 2014, which indicated a high level of quinolone resistance in China (7, 19). In this study, we assessed the prevalence of antibiotic resistance among Ureaplasma spp. and M. hominis from September to December 2018. Moreover, the mechanisms of the quinolone resistance were also studied.
Fluoroquinolones have been extensively used to treat bacterial infections in respiratory, gastrointestinal, and urinary tracts over the past few decades in China, thereby resulting in serious antimicrobial resistance in many bacterial pathogens, i.e., Mycobacterium tuberculosis, Neisseria meningitidis, and Riemerella anatipestifer (20–22). High levels of fluoroquinolone resistance in Ureaplasma spp. were also observed with LVX resistance rates of 75.9% for U. parvum and 71.4% for U. urealyticum and with MXF resistance rates of 25.3% for U. parvum and 50% for U. urealyticum (7). Compared with that in the previous reports, an increasing trend of fluoroquinolone resistance has been observed in the present study, in which >80% of Ureaplasma spp. showed LVX resistance and >50% of Ureaplasma spp. were MXF resistant. The resistance rate in Ureaplasma spp. varied from country to country. In the United States, 6.4% of U. parvum and 5.2% of U. urealyticum strains were LVX resistant (23). In Switzerland, 19.4% and 9.7% of strains were resistant to ciprofloxacin (CIP) and ofloxacin (OFL), respectively (using a commercial kit) (24). In France, the LVX and MXF resistance rates of Ureaplasma spp. were 1.2% and 0.1%, respectively (11). Moreover, a high level of fluoroquinolone resistance has been observed in Japan (16/28 [57.14%] of Ureaplasma spp. for LVX resistance) and Korea (50.6% for OFL and 27.4% for CIP in Ureaplasma spp., using a commercial kit), which are both near China geographically (14, 17). In Italy, a CIP resistance rate of 77.1% was reported using a commercial kit (25). It is worth noting that commercial kits may lead to an overestimation of antibiotic resistance (11). For M. hominis, 84.62% of strains were LVX or MXF resistant. This finding is consistent with a previous report in Sichuan, China, where 85.7% of strains were LVX resistant and 73.8% of strains were MXF resistant (8). However, the resistance rates for M. hominis were 2.7% for LVX and 1.6% for MXF in France (11). Considering the levels of fluoroquinolone resistance in different reports, the resistance rates of Ureaplasma spp. and M. hominis in China is extremely high. The overuse and misuse of quinolones cause severe quinolone-resistance in China. Recent studies have shown that fluoroquinolones are still commonly used antibiotics in China (26–28), and even the concentrations of quinolones in livestock and environments are extremely high (29–32). Compared with the high levels of ERY resistance by Ureaplasma spp. (80%) in South Africa (9), the ERY resistance was not serious in China, observed in 3.59% of U. parvum and 5.4% of U. urealyticum strains.
Sequence analysis of the QRDRs in LVX-resistant Ureaplasma strains revealed that the ParC codon for Ser83 was a hot spot of mutation, with three amino acid substitutions (S83L, S83W, and S83A). Previous studies reported that the S83L substitution was the most frequent mutation (7, 13, 24); 93.50% of LVX-resistant U. parvum and 80.33% of LVX-resistant U. urealyticum carried this mutation in the present study. The S83W mutation has been reported in two Ureaplasma strains (7, 33), while the S83A substitution was first observed in Ureaplasma spp. The corresponding amino acid change has been reported in quinolone-resistant Escherichia coli (GyrA S83A) and Staphylococcus aureus (GyrA S84A) (34, 35). The second most frequent mutation was ParE R448K, which was first found in China (7). The previously reported mutations, ParC E87K and E87Q, were also observed in three U. parvum strains (23, 36). Moreover, a new substitution, ParC A136S, was observed in one U. parvum with low-level quinolone resistance. Nine mutations in GyrA, GyrB, or ParE were found along with ParC S83L. GyrA Q100R has been reported in a quinolone-resistant Ureaplasma strain (37); however, a new amino acid change (Q100L) was found in the present study. GyrA Q104K has been associated with MXF resistance (11); GyrB P462S and GyrB N481S appeared to act as neutral polymorphisms (7, 38). The remaining substitutions in GyrB (E482D, D443A, and N486Y) and ParE (A481T and A481V) were novel mutations, first identified in this study. For M. hominis, the ParC K144R mutation was observed in two susceptible strains, suggesting that this mutation did not contribute to fluoroquinolone resistance (8, 39). Six (54.55%) M. hominis strains carried the two mutations, ParC S91I and GyrA S153L, which were proved to be the most frequent genetic alterations in fluoroquinolone-resistant M. hominis (8, 11, 39). GyrA S153L substitution was associated with a high level of resistance (40). A novel mutation was identified in the same position, GyrA S153A. The previously reported R447K in ParE was detected in one strain (41). Moreover, a novel mutation, GyrB A473V, was observed in M. hominis.
DNA gyrase shares homology with topoisomerase IV (42). The alignment results revealed that the novel mutation GyrA S153A in M. hominis corresponds to the new alteration ParC S83A in Ureaplasma spp., and ParE Arg447 in M. hominis is consistent with ParE R448K in Ureaplasma spp. The MD-refined structure revealed that the fluoroquinolone-resistant mutations were aimed to disrupt the stable contacts within ParC (or GyrA) and fluoroquinolone. Disrupting the direct interaction was the most effective alteration to drug resistance. Consequently, ParC Ser83 in Ureaplasma spp. and ParC Ser91 and GyrA Ser153 in M. hominis, which shared a similar direct binding mode with LVX, were all mutation hot spots. The ParC Glu87, GyrA Gln100, and GyrA Gln104 in Ureaplasma spp. shared a similar binding mode to ParC Ser84 and Glu88 in Acinetobacter baumannii in a water-Mg2+-ligand fashion (43). An interaction between ParE Arg448 and ParE Asp427 was first found in Ureaplasma spp. The interactions between these codons and LVX are relatively weak, and mutations occurred at these codons have a low level of quinolone resistance. Moreover, even if the mutations occur in the same position, the level of resistance to quinolone varies with the different substituted amino acids.
The mechanism of macrolide resistance in clinical Ureaplasma spp. is less well characterized, since macrolide-resistant Ureaplasma spp. are uncommon at the international level (10, 24). A G2057A/U (E. coli numbering) substitution was detected in mutants of U. parvum selected in vitro with ERY, and this substitution was also associated with intrinsic resistance of M. hominis to 14- and 15-membered macrolides and with ERY-resistant Chlamydomonas reinhardtii, cutaneous propionibacteria, and E. coli (15, 44–47). The aforementioned mutation G2057A/U (E. coli numbering) was not detected in the 19 macrolide-resistant Ureaplasma strains in the present study; however, two novel alterations occurring in the 23S ribosome, T2245C and G2654T, were identified. Moreover, double alterations (A121S and T141I) or a single mutation (D66N) that occurred in L22 was observed within five strains, including one sample harboring an S21A substitution in L4 at the same time. These three mutations detected in the L22 protein were reported in macrolide-resistant U. parvum that carried five amino acid insertions in its L4 protein. However, the potential resistance function of these mutations has been ignored because of the polymorphisms at these positions within 14 serovars of Ureaplasma spp. (15). Notably, 13 serovars are both Asp at position 66 for the L22 protein; in contrast, serovar 1 (ATCC 27813) has an Asn at this position. The same phenomenon has been observed in serovar 14 (ATCC 33697), with Ser at L22 position 122 (others have Ala) and with Ile at L22 position 141 (others have Thr). Unfortunately, we do not have ATCC 27813 and ATCC 33697 strains in our laboratory, and their MICs to ERY and susceptibility/resistance to macrolide are unknown. S21A is a novel mutation in the L4 protein; there are nine U. parvum strains and three U. urealyticum isolates that did not reveal any mutations associated with macrolide resistance. Macrolide resistance is acquired in bacteria via three mechanisms, including drug inactivation, active drug efflux pumps, and modification of the target site by methylation or mutation (48, 49). Among the nine U. parvum strains, one strain was detected with a subtype of the erm rRNA methyltransferase gene, ermB. Lu et al. first observed the ermB gene in 21 Ureaplasma strains (50). The encoded protein of this gene can modify an analogous adenine residue in a highly conserved region of the 23S rRNA to yield macrolide-lincosamide-streptogramin B (MLS) resistance (51).
In conclusion, high levels of quinolone resistance in Ureaplasma spp. and M. hominis still exist in China, and the resistance showed an increasing trend in Ureaplasma spp. in 2018. The rational use of antibiotics needs to be further enhanced, and ongoing monitoring of antibiotic resistance is necessary.
MATERIALS AND METHODS
Clinical samples and strains.
Over a 4-month period (1 September to 30 December 2018), 492 clinical Ureaplasma species (418 Ureaplasma parvum and 74 Ureaplasma urealyticum) and 13 clinical M. hominis samples were obtained in the present study. Among these strains, 350 isolates were from female urogenital tracts, and 155 isolates were isolated from male urogenital tracts at Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University.
Determination of antibiotic resistance.
The antibiotics LVX, MXF, DOX, ERY, and CLI were selected and used at concentrations ranging from 0.125 μg/ml to 32 μg/ml. MICs were evaluated in duplicates by broth microdilution according to the CLSI guidelines. The breakpoints of antibiotics were interpreted, except for DOX, for which there is no current CLSI breakpoint. The antibiotics were purchased from Sigma-Aldrich (St. Louis, MO, USA), and Ureaplasma-selective culture broth was purchased from Hebei Zhongaisheng Biological Engineering Company (Hebei, China).
Identification of genetic mechanisms of resistant strains.
Amplifications of the QRDR of LVX-resistant Ureaplasma strains and all M. hominis strains were conducted using previously reported primers, and the sequences were then mapped to the corresponding sequences of U. parvum ATCC 700970, U. urealyticum ATCC 33699, or M. Hominis ATCC 23114 (40, 52, 53). PCR and analysis of the sequences of 23S rRNA, L4, and L22 in ERY-resistant Ureaplasma strains were performed as previously described (44). Moreover, genes involved in other potential resistance mechanisms associated with macrolide resistance, including erm rRNA methyltransferase genes, macrolide modification enzyme genes, and efflux pump genes, were screened in ERY-resistant Ureaplasma strains as previously described (15).
Multiple-sequence alignment analysis.
BLASTp was performed for protein sequences of ParC and ParE in Ureaplasma spp. using NCBI BLAST web services (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The nonredundant matched sequences were downloaded, with 6,570 sequences (containing GyrA) for ParC and 20,007 sequences (containing GyrB) for ParE. A subsequent multiple-sequence alignment analysis was performed using MAFFT software. Based on the Ureaplasma spp. coordinates, the amino acid sequences of ParC from residues 61 to 140 and of ParE from residues 405 to 484 were retrieved to generate the sequence logos in WebLogo (http://weblogo.berkeley.edu).
Refined molecular dynamics structure of topoisomerase IV in Ureaplasma spp.
A three-dimensional (3D) structure of topoisomerase IV in Ureaplasma spp. was built by homology modeling based on the quinolone-DNA cleavage complex of type IIA topoisomerases from Streptococcus pneumoniae (PDB code 3K9F) using Prime of Schrödinger Suite 2019-1 (54). The bound mode of the quinolone-DNA cleavage complex of Acinetobacter baumannii (PDB code 2XKK) was chosen for LVX. System equilibration was performed with the default parameters in Desmond. Fifty-nanosecond MD simulations were then carried out using the Desmond Molecular Dynamics module of Schrödinger 2019-1 (54).
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (grant no. 81572042), the Zhejiang Provincial Health Bureau Foundation (grant no. 2017KY406), and the Zhejiang Provincial Innovative Medical Discipline (grant no. 11-CX18). The funders had no role in the study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
We declare no conflict of interests.
T.Y., L.P., N.W., Z.L., and Y.K. performed the experiments. T.Y. analyzed the sequence data and drafted the manuscript. L.W. built the model of the topoisomerase IV complex bound to levofloxacin in wild-type Ureaplasma spp. Z.R., X.X., and J.Z. reviewed and edited the text. X.X. and J.Z. designed the study. All authors read and approved the final manuscript.
REFERENCES
- 1.Taylor-Robinson D. 2017. Mollicutes in vaginal microbiology: Mycoplasma hominis, Ureaplasma urealyticum, Ureaplasma parvum and Mycoplasma genitalium. Res Microbiol 168:875–881. doi: 10.1016/j.resmic.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Beeton ML, Payne MS, Jones L. 2019. The role of Ureaplasma spp. in the development of nongonococcal urethritis and infertility among men. Clin Microbiol Rev 32:e00137-18. doi: 10.1128/CMR.00137-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sweeney EL, Dando SJ, Kallapur SG, Knox CL. 2017. The human Ureaplasma species as causative agents of chorioamnionitis. Clin Microbiol Rev 30:349–379. doi: 10.1128/CMR.00091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waites KB, Katz B, Schelonka RL. 2005. Mycoplasmas and ureaplasmas as neonatal pathogens. Clin Microbiol Rev 18:757–789. doi: 10.1128/CMR.18.4.757-789.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beeton ML, Spiller OB. 2017. Antibiotic resistance among Ureaplasma spp. isolates: cause for concern? J Antimicrob Chemother 72:330–337. doi: 10.1093/jac/dkw425. [DOI] [PubMed] [Google Scholar]
- 6.Degrange S, Renaudin H, Charron A, Bebear C, Bebear CM. 2008. Tetracycline resistance in Ureaplasma spp. and Mycoplasma hominis: prevalence in Bordeaux, France, from 1999 to 2002 and description of two tet(M)-positive isolates of M. hominis susceptible to tetracyclines. Antimicrob Agents Chemother 52:742–744. doi: 10.1128/AAC.00960-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song J, Qiao Y, Kong Y, Ruan Z, Huang J, Song T, Zhang J, Xie X. 2015. Frequent topoisomerase IV mutations associated with fluoroquinolone resistance in Ureaplasma species. J Med Microbiol 64:1315–1320. doi: 10.1099/jmm.0.000153. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Zheng L, Zhao J, Ding S, Xia Y. 2019. Investigation of fluoroquinolone resistance mechanism in Mycoplasma hominis isolated from urogenital samples in a Chinese hospital. J Med Microbiol 68:206–210. doi: 10.1099/jmm.0.000913. [DOI] [PubMed] [Google Scholar]
- 9.Redelinghuys MJ, Ehlers MM, Dreyer AW, Lombaard HA, Kock MM. 2014. Antimicrobial susceptibility patterns of Ureaplasma species and Mycoplasma hominis in pregnant women. BMC Infect Dis 14:171. doi: 10.1186/1471-2334-14-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beeton ML, Chalker VJ, Jones LC, Maxwell NC, Spiller OB. 2016. Antibiotic resistance among clinical Ureaplasma isolates recovered from neonates in England and Wales between 2007 and 2013. Antimicrob Agents Chemother 60:52–56. doi: 10.1128/AAC.00889-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meygret A, Le Roy C, Renaudin H, Bebear C, Pereyre S. 2018. Tetracycline and fluoroquinolone resistance in clinical Ureaplasma spp. and Mycoplasma hominis isolates in France between 2010 and 2015. J Antimicrob Chemother 73:2696–2703. doi: 10.1093/jac/dky238. [DOI] [PubMed] [Google Scholar]
- 12.Drlica K, Zhao X. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev 61:377–392. doi: 10.1128/.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamiya Y, Shimada Y, Ito S, Kikuchi M, Yasuda M, Kawamura Y, Deguchi T. 2013. Analysis of the quinolone-resistance determining region of the gyrA gene and the analogous region of the parC gene in Ureaplasma parvum and Ureaplasma urealyticum detected in first-void urine of men with non-gonococcal urethritis. J Antimicrob Chemother 68:480–482. doi: 10.1093/jac/dks417. [DOI] [PubMed] [Google Scholar]
- 14.Kawai Y, Nakura Y, Wakimoto T, Nomiyama M, Tokuda T, Takayanagi T, Shiraishi J, Wasada K, Kitajima H, Fujita T, Nakayama M, Mitsuda N, Nakanishi I, Takeuchi M, Yanagihara I. 2015. In vitro activity of five quinolones and analysis of the quinolone resistance-determining regions of gyrA, gyrB, parC, and parE in Ureaplasma parvum and Ureaplasma urealyticum clinical isolates from perinatal patients in Japan. Antimicrob Agents Chemother 59:2358–2364. doi: 10.1128/AAC.04262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao L, Crabb DM, Duffy LB, Paralanov V, Glass JI, Hamilos DL, Waites KB. 2011. Mutations in ribosomal proteins and ribosomal RNA confer macrolide resistance in human Ureaplasma spp. Int J Antimicrob Agents 37:377–379. doi: 10.1016/j.ijantimicag.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 16.Pereyre S, Renaudin H, Charron A, Bebear C, Bebear CM. 2006. Emergence of a 23S rRNA mutation in Mycoplasma hominis associated with a loss of the intrinsic resistance to erythromycin and azithromycin. J Antimicrob Chemother 57:753–756. doi: 10.1093/jac/dkl026. [DOI] [PubMed] [Google Scholar]
- 17.Choi JB, Lee SJ, Lee MK, Lee SJ, Park DC, Kim HY, Lee DS, Choe HS. 2018. Prevalence and antimicrobial susceptibility of Ureaplasma spp. and Mycoplasma hominis in asymptomatic individuals in Korea. Microb Drug Resist 24:1391–1396. doi: 10.1089/mdr.2017.0431. [DOI] [PubMed] [Google Scholar]
- 18.Zhou YH, Ma HX, Yang Y, Gu WM. 2018. Prevalence and antimicrobial resistance of Ureaplasma spp. and Mycoplasma hominis isolated from semen samples of infertile men in Shanghai, China from 2011 to 2016. Eur J Clin Microbiol Infect Dis 37:729–734. doi: 10.1007/s10096-017-3167-5. [DOI] [PubMed] [Google Scholar]
- 19.Xie X, Zhang J. 2006. Trends in the rates of resistance of Ureaplasma urealyticum to antibiotics and identification of the mutation site in the quinolone resistance-determining region in Chinese patients. FEMS Microbiol Lett 259:181–186. doi: 10.1111/j.1574-6968.2006.00239.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z, Lu J, Wang Y, Pang Y, Zhao Y. 2014. Prevalence and molecular characterization of fluoroquinolone-resistant Mycobacterium tuberculosis isolates in China. Antimicrob Agents Chemother 58:364–369. doi: 10.1128/AAC.01228-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M, Zhang C, Zhang X, Chen M. 2019. Meningococcal quinolone resistance originated from several commensal Neisseria species. Antimicrob Agents Chemother 64:e01494-19. doi: 10.1128/AAC.01494-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu D, Zheng M, Xu J, Wang M, Jia R, Chen S, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Huang J, Liu Y, Zhang L, Yu Y, Pan L, Chen X, Cheng A. 2019. Prevalence of fluoroquinolone resistance and mutations in the gyrA, parC and parE genes of Riemerella anatipestifer isolated from ducks in China. BMC Microbiol 19:271. doi: 10.1186/s12866-019-1659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez J, Karau MJ, Cunningham SA, Greenwood-Quaintance KE, Patel R. 2016. Antimicrobial susceptibility and clonality of clinical Ureaplasma isolates in the United States. Antimicrob Agents Chemother 60:4793–4798. doi: 10.1128/AAC.00671-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider SC, Tinguely R, Droz S, Hilty M, Dona V, Bodmer T, Endimiani A. 2015. Antibiotic susceptibility and sequence type distribution of Ureaplasma species isolated from genital samples in Switzerland. Antimicrob Agents Chemother 59:6026–6031. doi: 10.1128/AAC.00895-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foschi C, Salvo M, Galli S, Moroni A, Cevenini R, Marangoni A. 2018. Prevalence and antimicrobial resistance of genital Mollicutes in Italy over a two-year period. New Microbiol 41:153–158. [PubMed] [Google Scholar]
- 26.Xiao M, Cheng JW, Kudinha T, Kong F, Xu YC. 2017. National antimicrobial stewardship and fluoroquinolone-resistant Clostridium difficile in China. Infect Drug Resist 10:329–331. doi: 10.2147/IDR.S149293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng S, Xu Z, Wang X, Liu W, Qian L, Chen X, Wei J, Zhu M, Gong Z, Yan Y. 2019. Time series analysis of antibacterial usage and bacterial resistance in China: observations from a tertiary hospital from 2014 to 2018. Infect Drug Resist 12:2683–2691. doi: 10.2147/IDR.S220183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Yan S, Li D, Gong Y, Lu Z, Yin X. 2019. Trends and patterns of outpatient and inpatient antibiotic use in China’s hospitals: data from the Center for Antibacterial Surveillance, 2012–16. J Antimicrob Chemother 74:1731–1740. doi: 10.1093/jac/dkz062. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Lu S, Guo W, Xi B, Wang W. 2018. Antibiotics in the aquatic environments: a review of lakes, China. Sci Total Environ 627:1195–1208. doi: 10.1016/j.scitotenv.2018.01.271. [DOI] [PubMed] [Google Scholar]
- 30.Chen L, Li H, Liu Y, Li Y, Yang Z. 2020. Occurrence and human health risks of twenty-eight common antibiotics in wild freshwater products from the Xiangjiang River and comparison with the farmed samples from local markets. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 2020:1–13. doi: 10.1080/19440049.2020.1730987. [DOI] [PubMed] [Google Scholar]
- 31.Zhao FK, Chen LD, Yang L, Fang L, Sun L, Li SJ. 2017. Composition and distribution of antibiotics in soils with different land use types in a typical peri-urban area of the Yangtze River Delta. Huan Jing Ke Xue 38:5237–5246. doi: 10.13227/j.hjkx.201705243 (In Chinese.) [DOI] [PubMed] [Google Scholar]
- 32.Jiang Z, Paudyal N, Xu Y, Deng T, Li F, Pan H, Peng X, He Q, Yue M. 2019. Antibiotic resistance profiles of Salmonella recovered from finishing pigs and slaughter facilities in Henan, China. Front Microbiol 10:1513. doi: 10.3389/fmicb.2019.01513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valentine-King MA, Brown MB. 2017. Antibacterial resistance in Ureaplasma species and Mycoplasma hominis Isolates from urine cultures in college-aged females. Antimicrob Agents Chemother 61:e01104-17. doi: 10.1128/AAC.01104-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hallett P, Maxwell A. 1991. Novel quinolone resistance mutations of the Escherichia coli DNA gyrase A protein: enzymatic analysis of the mutant proteins. Antimicrob Agents Chemother 35:335–340. doi: 10.1128/AAC.35.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goswitz JJ, Willard KE, Fasching CE, Peterson LR. 1992. Detection of gyrA gene mutations associated with ciprofloxacin resistance in methicillin-resistant Staphylococcus aureus: analysis by polymerase chain reaction and automated direct DNA sequencing. Antimicrob Agents Chemother 36:1166–1169. doi: 10.1128/AAC.36.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piccinelli G, Gargiulo F, Biscaro V, Caccuri F, Caruso A, De Francesco MA. 2017. Analysis of mutations in DNA gyrase and topoisomerase IV of Ureaplasma urealyticum and Ureaplasma parvum serovars resistant to fluoroquinolones. Infect Genet Evol 47:64–67. doi: 10.1016/j.meegid.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 37.Bebear CM, Renaudin H, Charron A, Clerc M, Pereyre S, Bebear C. 2003. DNA gyrase and topoisomerase IV mutations in clinical isolates of Ureaplasma spp. and Mycoplasma hominis resistant to fluoroquinolones. Antimicrob Agents Chemother 47:3323–3325. doi: 10.1128/AAC.47.10.3323-3325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao L, Crabb DM, Duffy LB, Paralanov V, Glass JI, Waites KB. 2012. Chromosomal mutations responsible for fluoroquinolone resistance in Ureaplasma species in the United States. Antimicrob Agents Chemother 56:2780–2783. doi: 10.1128/AAC.06342-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furnkranz U, Walochnik J, Henrich B. 2018. Mycoplasma hominis shows strain-dependent increase in resistance to selected antibiotics after symbiosis with Trichomonas vaginalis. J Glob Antimicrob Resist 14:169–175. doi: 10.1016/j.jgar.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Bebear CM, Bove JM, Bebear C, Renaudin J. 1997. Characterization of Mycoplasma hominis mutations involved in resistance to fluoroquinolones. Antimicrob Agents Chemother 41:269–273. doi: 10.1128/AAC.41.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng DY, Sun CJ, Yu JB, Ma J, Xue WC. 2014. Molecular mechanism of fluoroquinolones resistance in Mycoplasma hominis clinical isolates. Braz J Microbiol 45:239–242. doi: 10.1590/s1517-83822014000100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bush NG, Evans-Roberts K, Maxwell A. 2015. DNA topoisomerases. EcoSal Plus 6:ESP-0010-2014. doi: 10.1128/ecosalplus.ESP-0010-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wohlkonig A, Chan PF, Fosberry AP, Homes P, Huang J, Kranz M, Leydon VR, Miles TJ, Pearson ND, Perera RL, Shillings AJ, Gwynn MN, Bax BD. 2010. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat Struct Mol Biol 17:1152–1153. doi: 10.1038/nsmb.1892. [DOI] [PubMed] [Google Scholar]
- 44.Pereyre S, Metifiot M, Cazanave C, Renaudin H, Charron A, Bebear C, Bebear CM. 2007. Characterisation of in vitro-selected mutants of Ureaplasma parvum resistant to macrolides and related antibiotics. Int J Antimicrob Agents 29:207–211. doi: 10.1016/j.ijantimicag.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 45.Harris EH, Burkhart BD, Gillham NW, Boynton JE. 1989. Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross JI, Eady EA, Cove JH, Jones CE, Ratyal AH, Miller YW, Vyakrnam S, Cunliffe WJ. 1997. Clinical resistance to erythromycin and clindamycin in cutaneous propionibacteria isolated from acne patients is associated with mutations in 23S rRNA. Antimicrob Agents Chemother 41:1162–1165. doi: 10.1128/AAC.41.5.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ettayebi M, Prasad SM, Morgan EA. 1985. Chloramphenicol-erythromycin resistance mutations in a 23S rRNA gene of Escherichia coli. J Bacteriol 162:551–557. doi: 10.1128/JB.162.2.551-557.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vester B, Douthwaite S. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother 45:1–12. doi: 10.1128/AAC.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weisblum B. 1995. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother 39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu C, Ye T, Zhu G, Feng P, Ma H, Lu R, Lai W. 2010. Phenotypic and genetic characteristics of macrolide and lincosamide resistant Ureaplasma urealyticum isolated in Guangzhou, China. Curr Microbiol 61:44–49. doi: 10.1007/s00284-009-9574-9. [DOI] [PubMed] [Google Scholar]
- 51.Arthur M, Brisson-Noēl A, Courvalin P. 1987. Origin and evolution of genes specifying resistance to macrolide, lincosamide and streptogramin antibiotics: data and hypotheses. J Antimicrob Chemother 20:783–802. doi: 10.1093/jac/20.6.783. [DOI] [PubMed] [Google Scholar]
- 52.Bebear CM, Renaudin H, Charron A, Gruson D, Lefrancois M, Bebear C. 2000. In vitro activity of trovafloxacin compared to those of five antimicrobials against mycoplasmas including Mycoplasma hominis and Ureaplasma urealyticum fluoroquinolone-resistant isolates that have been genetically characterized. Antimicrob Agents Chemother 44:2557–2560. doi: 10.1128/AAC.44.9.2557-2560.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bebear CM, Renaudin H, Charron A, Bove JM, Bebear C, Renaudin J. 1998. Alterations in topoisomerase IV and DNA gyrase in quinolone-resistant mutants of Mycoplasma hominis obtained in vitro. Antimicrob Agents Chemother 42:2304–2311. doi: 10.1128/AAC.42.9.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jacobson MP, Friesner RA, Xiang Z, Honig B. 2002. On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol 320:597–608. doi: 10.1016/S0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]