Multidrug resistance among Gram-negative bacteria is a major global public health threat. Metallo-β-lactamases (MBLs) target the most widely used antibiotic class, the β-lactams, including the most recent generation of carbapenems. Interspecies spread renders these enzymes a serious clinical threat, and there are no clinically available inhibitors. We present the crystal structures of IMP-13, a structurally uncharacterized MBL from the Gram-negative bacterium Pseudomonas aeruginosa found in clinical outbreaks globally, and characterize the binding using solution nuclear magnetic resonance spectroscopy and molecular dynamics simulations.
KEYWORDS: IMP-13, metallo-β-lactamase, imipenemase, antibiotic resistance, solution NMR, X-ray crystallography, molecular dynamics, metalloenzyme, protein dynamics, β-lactam antibiotic, nuclear magnetic resonance
ABSTRACT
Multidrug resistance among Gram-negative bacteria is a major global public health threat. Metallo-β-lactamases (MBLs) target the most widely used antibiotic class, the β-lactams, including the most recent generation of carbapenems. Interspecies spread renders these enzymes a serious clinical threat, and there are no clinically available inhibitors. We present the crystal structures of IMP-13, a structurally uncharacterized MBL from the Gram-negative bacterium Pseudomonas aeruginosa found in clinical outbreaks globally, and characterize the binding using solution nuclear magnetic resonance spectroscopy and molecular dynamics simulations. The crystal structures of apo IMP-13 and IMP-13 bound to four clinically relevant carbapenem antibiotics (doripenem, ertapenem, imipenem, and meropenem) are presented. Active-site plasticity and the active-site loop, where a tryptophan residue stabilizes the antibiotic core scaffold, are essential to the substrate-binding mechanism. The conserved carbapenem scaffold plays the most significant role in IMP-13 binding, explaining the broad substrate specificity. The observed plasticity and substrate-locking mechanism provide opportunities for rational drug design of novel metallo-β-lactamase inhibitors, essential in the fight against antibiotic resistance.
INTRODUCTION
Multidrug-resistant bacteria pose a major challenge to human health, with mechanisms of resistance to all known classes of antibiotics now being identified. While much pharmaceutical research has focused on drugs to treat Gram-positive bacterial infections, multidrug resistance among Gram-negative pathogens remains a significant clinical challenge (1–3). β-Lactam antibiotics are used for the treatment of both Gram-negative and Gram-positive bacterial infections and are the most commonly prescribed antibiotics (4, 5). β-Lactam antibiotics act as inhibitors of cell wall biosynthesis, causing subsequent bacterial cell death (6). The success of the first β-lactam antibiotic, penicillin, discovered in 1928 by Alexander Fleming (7) and used clinically since 1943 (8), led to multiple developments of the β-lactam scaffold, providing new and more effective antibacterial compounds.
As a result of the widespread use of β-lactam antibiotics, resistance mechanisms against them have emerged (3). Resistance mechanisms can be divided into mutation of penicillin binding proteins (PBPs), which prevent the binding of β-lactams to their target protein; reduction of the antibiotic concentration in the cell due to increased efflux (through expression of efflux pumps) or decreased uptake (through altered expression of outer membrane proteins); and, most commonly and significantly, the production of β-lactamase enzymes (9). β-Lactamases hydrolyze the β-lactam ring, which distinguishes this class of antibiotics and which is key to its binding mechanism, thus preventing interaction of the antibiotic with its target (10). As a response to the emergence of enzyme-mediated resistances as early as the 1940s (11), cephalosporin and carbapenem-type β-lactam antibiotics were discovered, isolated, and developed (12–14). Carbapenems are formed of a core scaffold, consisting of a β-lactam ring fused to a pyrroline ring that is decorated with an exocyclic sulfur that links to the tail region of the molecule (see Fig. SI1 in the supplemental material).
However, β-lactamases that are capable of inactivating the most recent generation of carbapenems, often used as a last resort for the effective treatment of infections caused by multidrug-resistant bacteria, have now evolved (15) and spread rapidly (16). As such, carbapenem resistance is a hallmark of all three of the World Health Organization’s highest-priority pathogens (17).
On a structural basis, four main classes of β-lactamases can be defined: three classes of serine β-lactamases, distinguishable by their amino acid sequence and inhibitor susceptibility (Ambler classes A, C, and D), and one class of metallo-β-lactamases (MBLs; Ambler class B), requiring divalent zinc ions for their β-lactamase activity (18). MBLs have been shown to hydrolyze all bicyclic β-lactams, including the carbapenems (19). The existence of MBL genes on integron structures and plasmids, often coexpressed with other antibiotic resistance genes, renders MBLs a serious clinical challenge, due to the possibility of horizontal gene transfer (20, 21). Inhibitors of serine β-lactamases, e.g., clavulanic acid (22) and avibactam (23), are available and used clinically in combination with antibiotics (24). However, resistance to these treatments is already being seen in the clinic (25), and to date, there are no clinically available inhibitors of the class B MBLs, making MBLs a significant threat.
MBLs can be divided into three subclasses (subclasses B1, B2, and B3), based on sequence and structural similarities and the number of coordinated zinc ions, with the B1 class representing the most significant one clinically (26). Among the major B1 class enzymes are the imipenemases (IMP), Verona integron-encoded MBLs (VIM), and the New Delhi MBLs (NDMs), which can hydrolyze the most recent cephalosporins and carbapenems (20, 27, 28).
IMP-type MBLs were first identified in Japan, and the class now consists of at least 53 members (29, 30) identified in more than 26 species of Gram-negative bacteria from around the world (31). IMP-encoding genes have been shown to occur as resistance cassettes along with other resistance genes, including those for serine β-lactamases (32) and those for aminoglycoside (33) and streptomycin (34) resistance. IMPs can be divided into six subgroups based on phylogeny and sequence similarity. IMP-13, a member of subgroup 2, sharing 92.3% amino acid sequence similarity with IMP-2 and 82.5% with IMP-1 (Fig. SI2) (35), was first identified in the Gram-negative pathogen Pseudomonas aeruginosa from clinical samples in Italy (21) and is a common cause of carbapenem resistance, often involved in large outbreaks (36). IMP-13 has been detected in a number of other countries in Europe as well as South America (21, 36–38). While IMP-13 is most commonly associated with Pseudomonas aeruginosa infections, it has also been identified in other human pathogens, including Salmonella enterica; members of the Enterobacteriaceae, including Klebsiella and Enterobacter spp. (38, 39); as well as nonhuman pathogens in the environment e.g., Pseudomonas monteilii, related to the soil microbe P. putida (40). These studies indicate that IMP-13 is present in a variety of significant human pathogens, as well as in other nonhuman pathogens which can act as environmental reservoirs of antibiotic resistance.
The recombinant IMP-13 protein has been overexpressed, purified, and characterized biochemically (41), but no structural information concerning IMP-13 has yet been reported. Crystal structures of MBLs, such as NDM-1 and IMP-1, show a conserved αβ/βα fold, with an active site at the interface of the two αβ units involving one or two zinc ions (42, 43). Although the overall folds are expected to be very similar, divergence between the various structures makes these challenging targets for drug development. Currently, relatively few crystal structures are available for other members of the IMP class, with no structural information on the antibiotic binding mode. Thus, high-resolution structural information is essential to broaden overall knowledge of MBLs and their antibiotic binding modes and enable the design of novel β-lactamase inhibitors to fight antibiotic resistance. The plasticity of the active site is also seen to play a role in other MBL classes (44, 45), so analyzing a wide range of antibiotic binding modes will help to determine the key factors in this.
Here, we report two distinct apo IMP-13 structures and the structures of IMP-13 complexed with four clinically relevant carbapenem antibiotics bound in their hydrolyzed form (doripenem, ertapenem, imipenem, and meropenem). We also present backbone nuclear magnetic resonance (NMR) assignments and NMR relaxation measurements for IMP-13 in the apo and ertapenem-bound forms and molecular dynamics (MD) simulations for the apo and carbapenem-bound states. The structural information and dynamics presented here reveal important information about the mechanism of antibiotic binding, as well as a significant role for the active-site-covering loop (L1), indicating that the plasticity of the active-site region is important for the broad substrate recognition spectrum of these enzymes. The structural information presented here provides important information to further aid in the development of novel MBL inhibitors, essential to combat this significant bacterial threat.
RESULTS
Structure of the apo form of IMP-13.
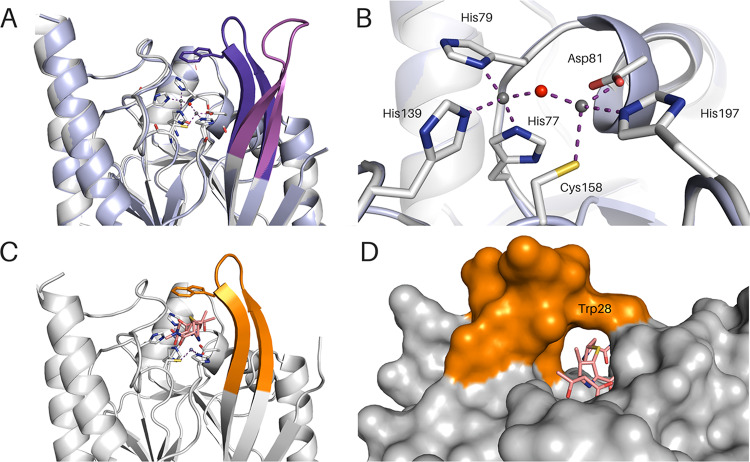
Two apo-form crystal structures of IMP-13, showing the L1 active-site loop (Fig. 1; see also Fig. SI3 and 4 in the supplemental material) in the open (apoopen) and closed (apoclosed) conformations, were solved to 1.9- and 2.2-Å resolutions, respectively (PDB accession numbers 6R79 and 6R78, respectively). We define the open conformation to be the loop pointing away from the protein toward the solvent and the closed conformation to be the loop positioned over the active site and pointing toward its rim. The distance between C-α atoms of Trp28 for the open and closed conformations is 8.8 Å. The overall protein architecture of the IMP-13 apo structure is consistent with that of the previously published metallo-β-lactamase fold (43), consisting of a global αβ/βα topology with a shallow active-site cleft at the border of the two β-sheets. In the apo structure presenting a closed L1 loop conformation (apoclosed conformation, PDB accession number 6R78), the two divalent zinc ions (Zn1 and Zn2) are found at a distance of 3.5 Å apart: one (Zn1) coordinates His77, His79, and His139 residues and a bridging water molecule in a tetrahedral geometry, while the other (Zn2) coordinates Asp81, Cys158, and His197 and the bridging water (Fig. 1B). The bridging water was previously proposed to be in the form of a hydroxide ion for activation of the β-lactam ring for hydrolysis (46, 47) and is seen to be about 3.3 Å from each of the oxygens of the Asp81 side chain, indicating that the hydroxide ion would be oriented by these residues, as seen in IMP-1 (48). In the apoopen structure (PDB accession number 6R79), the conformation is determined by interlocking with another loop, while in the closed conformation, these interactions are missing. The B-factor values of the loop residues are, in both cases, approximately 20 Å3 higher than the values for the rest of the protein molecule, indicating the high degree of flexibility of this region.
FIG 1.
IMP-13 apo and meropenem-bound structures. (A) Overlay of IMP-13 apo structures with open (magenta) and closed (violet) loops. The zinc-coordinating residues of the open state are shown as sticks. (B) Zoomed view of the image in panel A showing coordination of the two Zn(II) ions in the apo structures. (C) IMP-13 meropenem-bound structure. The loop is in orange, and the ligand is in salmon. (D) The closed loop forms a tunnel in the meropenem-bound structure. Zinc ions are shown as gray spheres, and water molecules are shown as red spheres.
Structure of carbapenem-bound IMP-13.
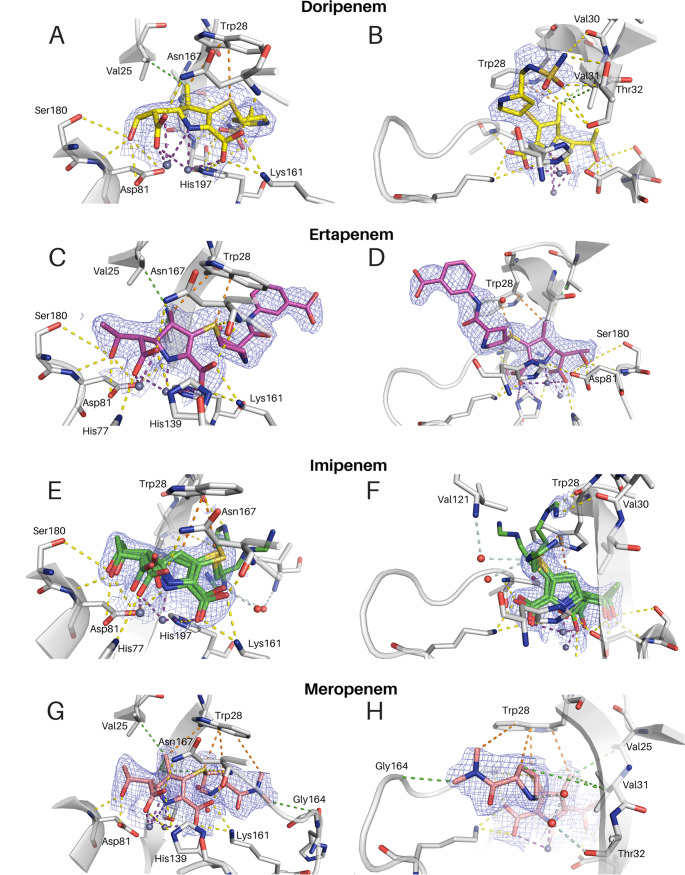
The crystal structures of IMP-13 bound to hydrolyzed doripenem (2.8 Å, PDB accession number 6S0H), ertapenem (2.2 Å, PDB accession number 6RZS), imipenem (1.9 Å, PDB accession number 6RZR), and meropenem (2.3 Å, PDB accession number 6R73) were solved by molecular replacement, with IMP-13–antibiotic complex crystals being prepared by cocrystallization. Both the tautomers with sp2 and sp3 carbons at the C-4 position of the carbapenems (46) (all without a hydrogen on N-6) were modeled into the ligand electron density and refined separately. As the resolution of the collected data is moderate and does not allow clear differentiation between the two tautomers and with an understanding that the crystal structure may be a weighted average of the two forms, the tautomer with the lowest B factor, the sp2 form, was deemed to be the most representative in all cases (Fig. 2), as C-4 attached to the S is not visibly tetrahedral. From this, with respect to the mechanisms shown by Lisa et al. (49) and Feng et al. (46), we believe that the primary state visible is that of the intermediate EI2. According to Lisa et al. (49), this would then become the Δ1 form after addition of the hydrogen via the sulfur-bound carbon atom and so could be a weighted average of these two states. In all the structures, the tail moiety of the carbapenem adopts different positions when bound to chain A or B of the crystal structure. Such an arrangement can be explained by the location of the tail, which experiences crystal packing contacts in one chain and solvent exposure in the other chain, resulting for the latter in high flexibility and rotational freedom, demonstrated by increased B-factor values compared to those for the core atoms of the carbapenem scaffold. Further description focuses on chain B of the doripenem-, ertapenem-, and imipenem-bound structures and chain A of the meropenem-bound structure, where crystal packing is not seen to affect ligand placement.
FIG 2.
2Fo − Fc maps at a contour level of 1σ for the presented complex crystal structures, showing interactions of the antibiotics’ carbapenem scaffold (A, C, E, G) and tails (B, D, F, H) with IMP-13. (E, F) Imipenem (green) shows three distinct tail conformations. All are depicted. (A to D, G, H) Doripenem (yellow) (A, B), ertapenem (magenta) (C, D), and meropenem (salmon) (G, H) are each seen in one conformation in each chain. Interactions are shown as dashed lines. Purple, zinc interactions; yellow, H bonds and charge-charge interactions; orange, aromatic interactions; green, hydrophobic interactions; pale blue, water network. Zinc ions are shown as gray spheres, and water molecules are shown as red spheres.
A comparison of the carbapenem-bound complex structures with apoclosed elicits very few distinct differences, with the L1 loop seen to be packed over the antibiotic binding pocket (Fig. 1A, C, and D). The root mean square deviation (RMSD) of the backbone C-α atoms (for 216 out of 217 residues), including the L1 loop, varied from 0.27 to 0.47 Å, showing a high level of structural similarity between the structures, with only a few differences being seen in the flexible loop regions. With the apoopen structure, on the other hand, the loop can be seen to point away from the active site, leaving the active site accessible to the substrate (Fig. 1A). The tunnel formation seen in the closed form is completely absent due to the different positioning of both backbone and side chain atoms. In the complex structures, the largest active-site-facing changes seen in the L1 loop occur between residues Val25 and Val31, with these two residues moving toward the carbapenems to form hydrophobic interactions (movements are in the range of 5 Å and 1 Å for Val25 and Val31, respectively). The residues located in the middle of the L1 loop, Val25 and Trp28, show more significant changes, moving approximately 9 to 10 Å in order to cover the substrate during catalysis, thereby closing the tunnel-like structure above the active site of the enzyme and acting as a gatekeeper between the ligand and the solvent. Further conformational differences between the apo and carbapenem-bound structures include the movement of Asn167 closer to the active site in order to facilitate the hydrogen bonding with the substrate (the rest of the L3 loop does not alter its conformation significantly). Strands B7 and B8 are also altered between the open and closed protein conformations, with Tyr123 and Trp124 showing the most pronounced changes.
Compared to the apo structures, the zinc ions in carbapenem-bound IMP-13 are located slightly farther apart, with the distance for the different antibiotics ranging from 3.8 Å (doripenem complex) to 4.2 Å (ertapenem complex), whereas the distance is 3.5 Å for the apo state, presumably to maximize interactions with the ligand. The zinc-protein coordination remains unchanged (Fig. SI5): Zn1 still coordinates the three histidines (His77, His79, and His139), while Zn2 coordinates Asp81, Cys158, and His197, but the bridging water is no longer observed due to the presence of the enzyme’s substrate.
Conserved binding mode of the carbapenem scaffold to IMP-13.
The binding of the carbapenem scaffold (Fig. 2; Fig. SI1) was similar for the four antibiotics investigated, with contacts to the surrounding residues being created via a network of hydrogen bonds and hydrophobic and electrostatic interactions. In the carbapenem complexes, two loops, L1 and L3 (Fig. 2; Fig. SI3 and 4), interact with the hydrolyzed substrates (Fig. SI6). Both zinc ions show an interaction with N-6 on the pyrroline ring, with Zn2 having a closer interaction (Table S1; Fig. SI5). In addition, there are interactions with carboxylic acid moieties O-9, O-26, and O-27. In all cases, Zn1 coordinates O-9, while Zn2 coordinates O-26 and O-27. Lys161 acts as a counterion to the carboxylate of the carbapenems (O-8, O-9). Hydrogen bonding is observed between the carbapenems’ hydroxyl groups (O-24) and the Asp81 backbone nitrogen. Interactions with the Asp81 side chain and between the side chain of Asn167 and the carbapenems’ hydroxyls (O-26, O-27) are also observed.
L1, the extended β loop that is conserved in β-lactamases, encompasses the active site, forming a tunnel-like structure of a hydrophobic nature (Fig. 1C and D); the amino acid composition of L1 results in a hydrophobicity index of 0.84 (50), whereas that for the overall protein is −0.32. This largely hydrophobic loop interacts with the β-lactam antibiotics, stabilizing their position during hydrolysis. The tryptophan (Trp28) at the tip of this loop is a key residue that bridges the gap between the loop backbone and the active-site residues, forming a closed tunnel. In IMP-1, the equivalent tryptophan is found to affect Km: for imipenem, a W64A mutation leads to a 5-fold increase in Km (51). The sulfur atom present in the linker region (S-10) of all carbapenems creates strong π-sulfur interactions with the aromatic ring of Trp28 (as well as interacting with the backbone of Asn167), thereby contributing to the position of the core scaffold of all presented carbapenem substrates. Due to the multiple tail conformations in imipenem, the orientation for the π-sulfur interaction in this case is not always optimal; however, the distance remains consistent. In addition, Trp28 shows interactions with the pyrroline methyl group (C-21), present in all the antibiotics studied other than imipenem. These interactions lead to restricted motion in the side chain of Trp28, which further rigidifies the L1 loop (Fig. 3). The lack of this interaction in imipenem could lead to a reduction in binding interactions to the loop and may contribute to the observed reduction in affinity (increase in Km) for imipenem (Table S2). It is likely that there is a hydrophobic interaction between the ring itself and the tryptophan in the absence of this methyl. Two other hydrophobic L1 residues, Val25 and Val31, also form alkyl interactions with C-21, where present.
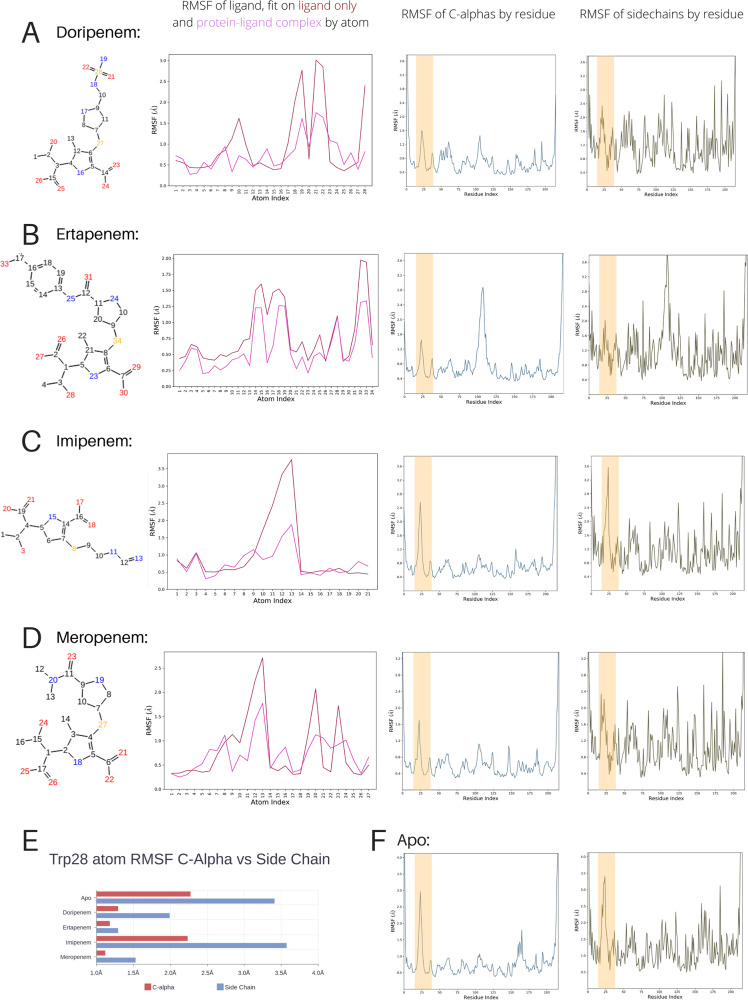
FIG 3.
Summary of molecular dynamics simulations for apo and carbapenem-bound IMP-13. The structures of the hydrolyzed antibiotics with the numbering used in the simulations are shown in column 1. Column 2 shows the root mean squared fluctuation (RMSF) of the ligand, fitted on the ligand only and on the protein-ligand complex, column 3 shows the RMSF of the protein C-α atoms, and column 4 shows the RMSF of the protein side chains. The L1 loop is marked by orange bars on all graphs. Results are shown for doripenem (A), ertapenem (B), imipenem (C), and meropenem (D). (F) Values for apo C-α and the side chain RMSF are shown for comparison. (E) C-α and side chain RMSF values for the key residue Trp28 are shown for all structures.
Binding modes of the antibiotic tails to IMP-13.
Analysis of the interactions between the carbapenem tail moieties and the surrounding residues and solvent molecules shows a complex network of position-dependent contacts. Due to the presence of more than one molecule in the unit cell, a representative molecule for which the crystal packing does not affect the antibiotic tail placement was chosen for discussion. Higher solvent accessibility of the tails leads to less restrained positions, characterized by increased B-factor values.
The most solvent-exposed parts of the four antibiotics are very distinct, while in addition, imipenem has no pyrrolidine ring in the antibiotic tail. The pyrrolidine ring of meropenem, doripenem, and ertapenem is suitably located to form aromatic, π-alkyl interactions with His197 and is further stabilized by the hydrophobic environment created by Val25 and Val31. The terminal nitromethyl group of meropenem is stabilized by direct as well as water-mediated interactions with the backbone of His163 and Gly164 and π-alkyl interactions with Trp28. The sulfonamide moiety of doripenem forms several hydrogen bonds: the nitrogen of the primary amine group interacts with Thr32 and the backbone of Val30, while the oxygen creates hydrogen bonds with backbone and side chain atoms of Thr32. The terminal part of the imipenem tail was modeled in three different conformations (50%:25%:25% occupancy), highlighting the extreme flexibility of this moiety. Due to the different conformers, the interaction network is different in every modeled position: conformer A creates a hydrogen bond with the backbone carbonyl of Val30 and water-mediated hydrogen bonds with the backbone of Thr32. The most solvent-exposed conformation, conformer B, lacks interactions with surrounding residues, most likely interacting with a water network, while conformer C interacts with the wider water network. In each case, a water molecule replaces the amine group from the other two conformer positions.
NMR measurements show altered dynamics in the presence and absence of antibiotic.
As discussed above, the L1 loop adopts very different conformations in the two apo structures (apoclosed and apoopen). In the apoclosed structure, the loop is folded over the active site, while in the apoopen structure, L1 is extended away from the protein, leaving the active site accessible. In complex with each of the hydrolyzed carbapenems, this loop forms a tightly locked, tunnel-like structure around the hydrolyzed antibiotic (Fig. 1D), with several hydrophobic interactions appearing to stabilize this state.
To further understand the role of the L1 loop in antibiotic binding, NMR spectra were acquired for IMP-13 in the apo state and for IMP-13 bound to ertapenem. An overlay of 1H,15N heteronuclear single quantum coherence (HSQC) spectra for the two forms (Fig. SI7) shows substantial chemical shift changes on addition of ertapenem, necessitating backbone assignment using triple resonance spectra in both forms. 1H,15N assignments from a total of 219 residues (excluding the 9 prolines) were achieved for 203 residues in the apo form (93%) and 195 residues in the ertapenem-bound form (89%). The assignments are shown in Fig. SI8 for the apo state and Fig. SI9 for the ertapenem-bound state.
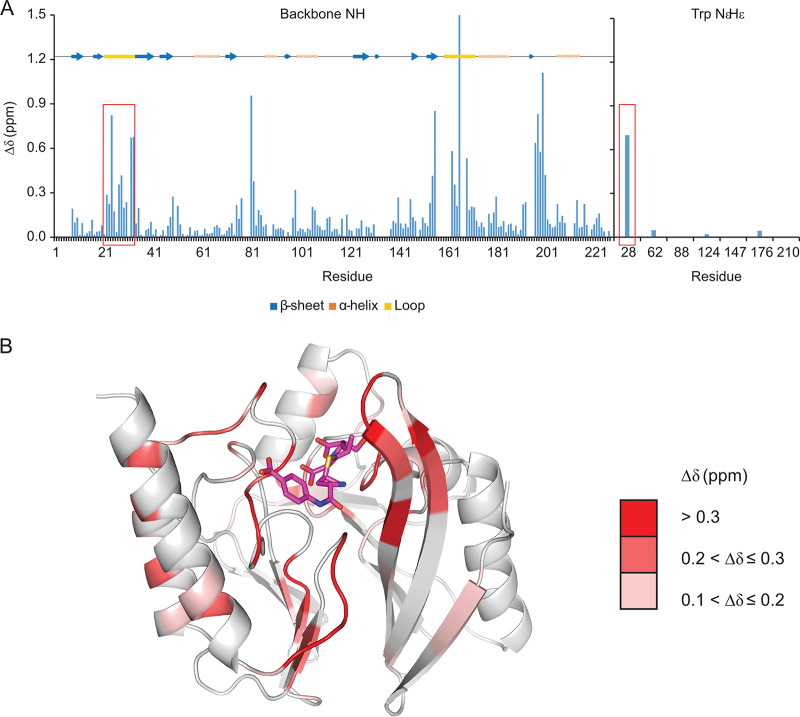
Chemical shift perturbations are shown in Fig. 4 and plotted on the ertapenem-bound structure (PDB accession numbers 6RZS). The largest changes are colored in red on the structure and predominantly localize to loops in the vicinity of the ertapenem-binding pocket. These changes are also marked with arrows on the spectra shown in Fig. SI7. As expected, significant shifts were seen for residues in the L1 loop (marked with red boxes in Fig. 4), in particular, residues Glu24, Gly27, Trp28, Thr32, and Lys33, as well as the side chain NεHε of Trp28; the L3 loop (residues 163 to 166, especially residue Gly166), which lies in the vicinity of the ertapenem tail; and residue Asp81, which coordinates Zn2, as well as in the linker between B11 and A5 (residues 197 to 201). Smaller changes were seen in the β-strands B9, B10, and B11. These changes are consistent with the observations in the crystal structure (see above).
FIG 4.
Chemical shift changes between the apo and ertapenem-bound forms of IMP-13. (A) 1H,15N backbone amide and tryptophan amide side chain chemical shift changes between the apo and ertapenem-bound forms of IMP-13 for the spectra shown in Fig. SI7 in the supplemental material (shown individually in Fig. SI8 and SI9) plotted against the residue number. A representation of the secondary structure of IMP-13 is shown above the plot. The L1 region is indicated by red boxes. (B) The shift changes (Δδ > 0.1) identified in panel A are shown on the ertapenem-bound crystal structure of IMP-13 (PDB accession number 6RZS).
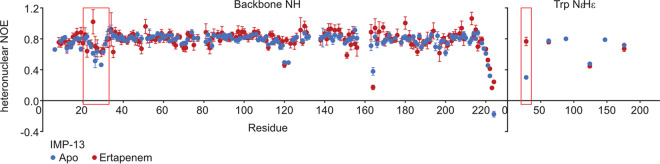
Heteronuclear 1H-15N nuclear Overhauser effect (hetNOE) experiments were acquired to detect fast (picosecond-nanosecond)-timescale motions (52, 53). Typically, structured regions of the protein show hetNOE values of >0.8, while flexible loops and the N and C termini show lower values (<0.8). Figure 5 shows an overlay of the hetNOE values for the apo and ertapenem-bound forms. Both states show similar values, with an average hetNOE value, taken across backbone residues Asp6 to Glu219, of 0.783 (standard deviation, 0.078) for the apo form and 0.799 (standard deviation, 0.096) for the ertapenem-bound form. However, notably lower values were recorded in the L1 loop in the apo state, with values of 0.51 and 0.46 for residues Asn26 and Gly29, respectively, whereas in the ertapenem-bound state, the values did not drop below 0.6 in this region. Most significantly, Trp28 NεHε had a hetNOE of 0.3 in the apo form, which rose to 0.77 in the ertapenem-bound state, comparable to the values for the backbone amides in structured regions of the protein, indicating a significant change in dynamic properties. This suggests that in the apo form the L1 loop is undergoing fast-timescale motions, while binding of antibiotic in the active site stabilizes the L1 loop. The restricted motion of Trp28 NεHε suggests that the antibiotic interacts with this residue, reducing the fast-timescale motions at this position. The hetNOE data show slightly more restriction in residues 165 to 168 in the presence of ertapenem, but residue Gly164 is considerably more flexible in both the apo and the ertapenem-bound states. It was not possible to assign residues at the beginning of the L3 loop, suggesting unfavorable dynamics in this region.
FIG 5.
Heteronuclear NOE data showing fast-timescale motions of apo and ertapenem-bound IMP-13. Heteronuclear NOE data for the backbone amides and tryptophan indole NεHε measured at 600 MHz (1H frequency) and 25°C for both the apo and ertapenem-bound forms. The loop 1 region is marked with a red rectangle. Residues above residue 224 were removed from the plot, as all showed negative heteronuclear NOEs in both the apo and ertapenem-bound forms.
Molecular dynamics simulations show significant variations in L1 loop dynamics between complex structures.
The distinct conformations observed in the two apo crystal structures and the NMR relaxation data indicate that the L1 loop is likely flexible in solution. We therefore performed molecular dynamics (MD) simulations to assess the movement and flexibility of this protein fragment on the nanosecond timescale for the apo and carbapenem-bound structures (Fig. 3). During a total simulation time of 50 ns for each system, none of the systems showed significant large-scale fluctuations, indicating that the solute systems were stable. In addition, the ligand RMSD values (Table S3) suggest that the conformation of the hydrolyzed ligands remains stable during both the 50-ns and 100-ns simulation times. Torsion angles were also generally maintained throughout both the simulation runs. Excluding the intrinsic flexibility of L1, the protein overall does not undergo significant conformational changes, aside from those in the active-site region.
However, the simulations reveal significant changes in L1 behavior between the different structures. The lower root mean squared fluctuations (RMSFs) for heavy atoms of L1 residues in the simulated doripenem-, ertapenem-, and meropenem-bound complex structures than for those of the apoclosed structure indicate that L1 is more rigid in the closed conformation when these ligands are bound, while in the apo protein, L1 can move with a higher degree of freedom. This is shown by the C-α RMSF being 1.4, 2.1, and 1.9 Å larger in the apo structure than in the doripenem-, ertapenem-, and meropenem-bound forms, respectively (1.1, 1.4, and 0.8 Å, respectively, for the replicas). However, while the behavior of L1 is comparable for the doripenem-, ertapenem-, and meropenem-bound structures and shows the greatest restriction for these structures, it differs significantly for the imipenem complex, which is comparable to that for the apo form (Fig. 3C and E). In the X-ray structures, the interaction of bound ertapenem and meropenem with the Trp28 side chain occurs via the sulfur adjacent to the β-lactam ring and, additionally, via the methyl group on the pyrroline ring. In contrast, imipenem lacks this additional methyl group, which reduces the strength of the lipophilic interaction with the Trp28 side chain, thus resulting in greater loop flexibility. Furthermore, as shown by both shorter and longer overall simulation times for all the complexes, the tail of imipenem shows high flexibility and occupies multiple rotamer positions; thus, it does not contribute further to the loop stability. Doripenem, however, does contain both a sulfur-π interaction and a methyl-π interaction, but the MD simulation shows a higher motility of the sulfonamide tail of the hydrolyzed antibiotic (atoms N-18, N-19, O-21, O-22, and S-28; Fig. 3A). This is in agreement with the chemical character of this moiety, which, due to a higher energy contribution to desolvation, is more prone to interact with nearby solvent molecules, thus leading to a markedly higher RMSF for the atoms in the antibiotic tail in comparison to the RMSFs for the ertapenem and meropenem structures.
DISCUSSION
We present the apo and complex structures of IMP-13 bound to hydrolyzed carbapenems. Imipenemases represent one of the major groups of class B1 metallo-β-lactamases found in Gram-negative pathogens, and these enzymes can hydrolyze all bicyclic β-lactam antibiotics. This includes carbapenems, which are often reserved for use as the treatment of last resort in cases of multidrug resistance. There are relatively few structures of imipenemase enzymes available, and no structures of imipenemase enzymes bound to carbapenems are available. Currently, no inhibitors of metallo-β-lactamases are available in the clinic. Consequently, understanding the structural features of carbapenem interactions with a member of the imipenemase class is essential to developing new inhibitors to treat infections caused by multidrug-resistant pathogens.
Our data demonstrate that the key interactions in the bound structures are found between the conserved carbapenem core (the β-lactam and pyrrole rings and the exocyclic sulfur) of the antibiotic and the divalent zinc ions, as well as the backbone and side chain residues of the IMP-13 active site, particularly the L1 and L3 loops. Notably fewer interactions are made with the antibiotic tail region, leading to high flexibility, which likely affects enzyme efficiency, rendering careful design of the tail section key in drug discovery efforts. That IMP-13 is not selective toward the antibiotic tail region likely contributes to its broad-spectrum activity, which makes the B1 class metallo-β-lactamases particularly challenging resistance determinants. A key feature of the binding mode is the interaction between the tryptophan of L1 and the carbapenem scaffold: the tryptophan forms a closed tunnel over the β-lactam ring, thus locking the loop and the antibiotic in place (Fig. 1D). In the apo-state crystal structures, two positions for the L1 loop, open and closed, were observed. Molecular dynamics simulations also showed different degrees of flexibility in the L1 loop region. These results are supported by the fast-timescale motion for loop L1 observed in the apo form in the hetNOE experiment, which was reduced in the presence of the antibiotic ertapenem (Fig. 5), and, in particular, the substantial reduction in the flexibility of Trp28HεNε between the apo and ertapenem-bound states. These observations are consistent with the results of previous NMR studies on a subclass B1 dizinc metallo β-lactamase from Bacteroides fragilis (54–56), where L1 loop residues show lower hetNOE values in the free form than in the presence of a tightly binding inhibitor, most notably, for the L1 tryptophan indole (Trp28 for IMP-13, Trp49 [56]), indicating a potentially important role of the L1 loop in substrate recruitment and stabilization during the hydrolysis reaction. Previous studies have discussed whether the tryptophan and other hydrophobic residues in the L1 loop may act as a recruiter, loosely binding the substrate in the open formation and then moving to the closed formation to aid substrate addition to the binding site (54, 56).
Nevertheless, it is clear from the MD simulations (Fig. 3) that differences in antibiotic structure affect the restriction of the L1 loop, with the doripenem, ertapenem, and meropenem complexes showing the greatest restriction in L1 loop mobility. This is consistent with the kinetic parameters reported for IMP-13 (see Table S2 in the supplemental material), showing tight binding for meropenem and ertapenem (Km values, in the low-micromolar and high-nanomolar ranges, respectively). In contrast, imipenem shows weaker binding (Km, ca. 50 μM), consistent with the higher L1 flexibility. Notably, the kcat for imipenem is 2 orders of magnitude higher than that for meropenem and ertapenem. Given that product release, preceded by the necessary L1 opening, likely determines the turnover rate, this indicates that tighter binding reduces the turnover rate of IMP-13. Consequently, an efficient, noncovalent inhibitor could interact with and stabilize the L1 loop in the closed conformation, forming a principle for inhibitor design.
IMP-13 shows 83% and 92% sequence identity with the IMP-1 and IMP-2 forms, respectively, and is quite divergent from other variants (41). Consequently, it is instructive to compare our structures to those of other available MBL structures (Fig. 6; Fig. SI10).
FIG 6.
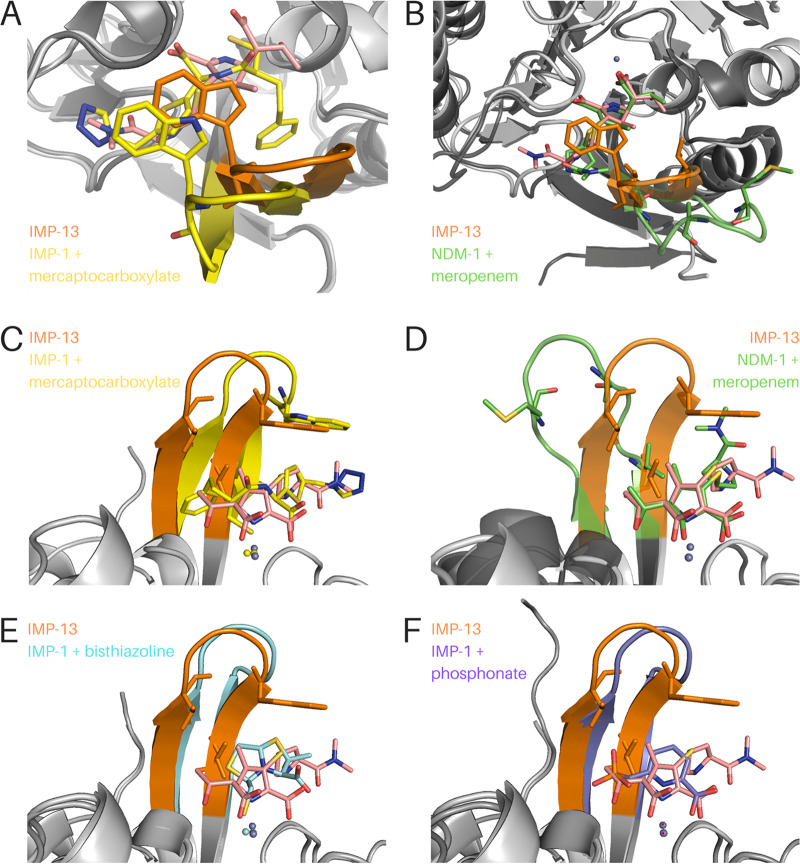
Comparison of the IMP-13 meropenem-bound structure with previously published metallo-β-lactamase structures. IMP-13 is always depicted with an orange loop, and the IMP-13-bound meropenem is depicted in salmon. (A and C) Two views of IMP-13 overlaid with IMP-1 bound to the mercaptocarboxylate inhibitor (yellow; PDB accession number 1DD6 [60]). (B and D) Overlay of IMP-13 with NDM-1 and hydrolyzed meropenem (green; PDB accession number 5N0H [67]). The phenylalanine residue at the tip of the loop is not resolved in the structure, but the backbone is shown as sticks. (E) Overlay of IMP-13 with IMP-1 bound to the bisthiazoline inhibitor L-VC26 (cyan; PDB accession number 5EWA [57]). (F) Overlay of IMP-13 with IMP-1 bound to a phosphonate-based inhibitor (purple; PDB accession number 5HH4 [61]).
Comparison of IMP-1 structures (PDB accession numbers 5Y5B and 5EV6 [57]) with both our apoclosed and carbapenem-bound IMP-13 structures yields differences in the L1 region. The amino acid sequences are highly conserved between the L1 regions of the two structures, with only one difference being seen, which is at the C-terminal end of the loop (Pro32 in IMP-1 is changed to Thr in IMP-13). In one of the IMP-1 structures, the β-strand of the loop at this point is seen to be straighter and farther out from the active site than in the case of the structures that we have presented, despite the tip of the loop being closer to the active site than in the open-conformation IMP-13 structure presented here. It is likely that the Pro32-to-Thr mutation leads to a more flexible loop in IMP-13 and a more restricted loop in IMP-1, as a result of the more constrained dihedral angles of proline. Mutagenesis analysis of IMP-18 (58) (a subclass B4 β-lactamase [31]) also indicates that this residue has a key effect, as the turnover rates of the enzyme are significantly altered (3- and 10-fold increases in the kcat for imipenem and meropenem, respectively) on mutation from threonine to proline. IMP-2 also lacks Pro32 at the end of L1, which is instead mutated to Ser. The IMP-2 structure (PDB accession number 4UBQ [59]) shows that the loop is found between the locations of the loop in the apoopen and apoclosed structures reported here. The varying extent of the β-strand structure (subclasses B2 and B3) on either side of the L1 loop may reflect the dynamic nature of this region, and thus, the loop can be captured in different conformations in different crystal structures.
The ligands crystallized previously with IMP-1 belong to different compound classes, and hence, we compared those structures with our carbapenem-bound structures to identify whether similar interactions are exploited. Comparison with the IMP-1 structure bound to the mercaptocarboxylate inhibitor (60) (PDB accession number 1DD6) shows that the benzyl ring is pointing toward the loop (residues 21 to 23). Were the loop in the same position as in IMP-13, this would clash with the loop position, particularly with Val31. The position of the loop in IMP-1 is shifted laterally by approximately 1 Å. This could indicate potential binding selectivity to IMP-1 or an alternative explanation of sterically induced loop movement. The free thiol of mercaptocarboxylate is coordinated by the two zinc atoms, displacing the nucleophilic water, equivalent to the carboxylate of β-lactams. In contrast, both the tertiary amine and the sulfur of the thiophene ring point in an orientation opposite to that seen in comparable antibiotic residues. This suggests that further structural optimization, based on a knowledge of antibiotic binding, could be used to optimize inhibitor interactions.
The structures of bisthiazolidine inhibitors bound to IMP-1 (PDB accession number 5EWA) (57) show a number of interactions mimicking those of antibiotic binding. The free thiol is coordinated by Zn1 and Zn2, displacing the nucleophilic water, while the thiazoline rings interact with the L1 tryptophan, creating stacking interactions. The carboxylate interacts with the lysine residue in L3, equivalent to the β-lactam carboxylate. In contrast, a phosphonate inhibitor (61) (PDB accession number 5HH4) does not displace the nucleophilic water, with the phosphonate group coordinating Ser119 (IMP-1 numbering) and the nucleophilic water. The pyridine nitrogen and carboxylate interact with Zn2 and the L3 lysine, again making interactions similar to those observed in our antibiotic-bound structures. The pyridine ring makes a T-shaped π-stacking interaction with the L1 tryptophan. These comparisons suggest that mimicking the key interactions found in the antibiotic complex structures presented in this paper is important in designing inhibitors.
Comparing our IMP-13 structures to the structure of the NDM-1 β-lactamase, another broad-spectrum MBL of clinical relevance, the key difference is the replacement of the tryptophan of L1 (Trp28) in IMP-13 with a phenylalanine in all 17 NDM variants (62). Consequently, this suggests an alternative mode of binding. In the published structure of NDM-1 in complex with hydrolyzed meropenem (PDB accession number 4EYL, which was rerefined with PDB accession number 5N0H) (63), the loop is shown in the open conformation and therefore does not form a closed tunnel covering the β-lactam ring. In contrast, Trp28 of IMP-13 shows direct interactions with the bound carbapenems, while the equivalent Phe70 of NDM-1 is more than 7 Å away and does not interact with the ligand. The conservation of this residue in all known NDM-1 variants indicates the importance of this amino acid for the proteins’ activity. The residue equivalent to Val25 (Met in NDM-1), which in IMP-13 flanks the flexible loop section and interacts with C-21 of meropenem, is seen in NDM-1 in a location where this interaction is removed altogether. The equivalent of Val31, the other flanking valine, is also seen in an altered position. This is farther away from C-21 of the antibiotic but is closer to the methylamine group, which could explain why this group of the meropenem molecule is in a different location in this structure, differing in position by about 5 Å.
We extended our comparison to structures of a variety of NDM proteins bound to various ligands that were deposited in the Protein Data Bank (PDB) in 2017 and 2018 (PDB accession numbers 4TYF, 4TZ9, 4TZB, 4TZE, 4TZF, and 5WIG [64], 5WIH [64], 5XP9 [65], 5A5Z [66], 5N0H [67], 5N0I and 5YPK [46], 5YPL [46], 5YPN [46], 6EX7 [67], 5YPM [46], and 5JQJ, 5K4M, and 5XP6 [65]). Notably, none of these structures show a fully closed-tunnel conformation like that seen in the IMP-13–carbapenem complexes presented here. The backbone is seen in a half-closed formation in many of these PDB accessions (e.g., PDB accession numbers 5JQJ, 5K4M, and 5XP6), but the chain never fully reaches over the substrate. This could lead to a reduced contact area between the ligand and the protein. The reduced hydrophobicity of this loop in NDM-1 could also explain the higher Km (lower affinity) of meropenem and imipenem (Table S2) relative to that of the IMP enzymes (28).
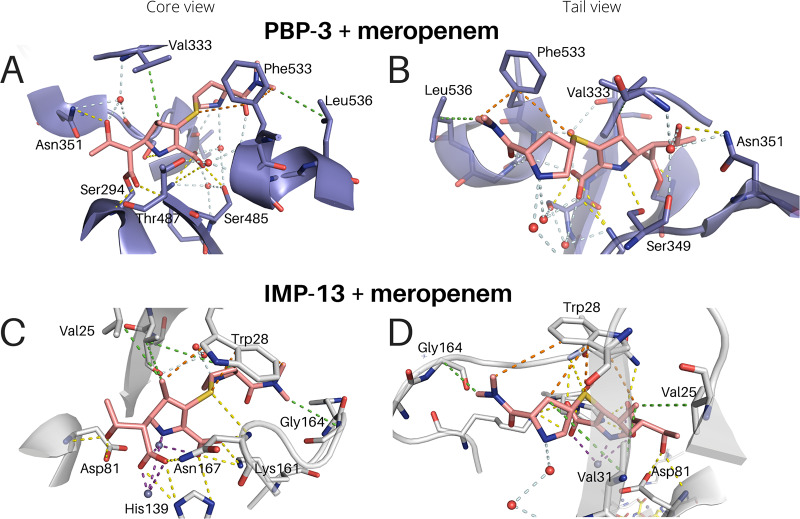
We also compared our results to the natural target of β-lactam antibiotics, the penicillin binding proteins (PBP). A structure of penicillin binding protein 3 (PBP-3 [3PBR]) (68) bound to meropenem is available, facilitating comparison between the interactions of meropenem with a β-lactam target protein (PBP) and the enzymes (β-lactamases) that degrade it (Fig. 7). From the point of view of drug development, comparative studies could highlight key similarities and differences, aiding with the development of new antibiotics in this class with lower susceptibility to the β-lactamase-driven degradation. The major interactions, primarily hydrophobic, between the protein and the antibiotic are maintained, but key differences are observed. First, the central Trp28 interaction of IMP-13, both to the sulfur and to C-21 of the core carbapenem scaffold, is replaced by a hydrophobic interaction with Phe533 of PBP-3. The interactions from His139 on L3 are partially emulated in PBP-3 by Gly486, while Asn167 and Cys158 provide interactions similar to those provided by Thr487 and Lys484 of PBP-3, respectively. However, while these interactions are different, they are very similar in character. It is also interesting to note the difference between the meropenem and imipenem interactions. In PBP, the imipenem scaffold (lacking C-21) interacts with Tyr532, while C-21 of meropenem displaces this and Phe533 rotates 180° around the chain to form an alternative hydrophobic environment in the vicinity. Despite the strong Tyr interaction and the fact that this is closer to the native position than it is in the meropenem-bound form, imipenem binds by a factor of 16 times weaker than meropenem (68), indicating that the tail plays a much stronger role in the binding to PBP than to IMP-13, where the carbapenem scaffold and alterations to it are more important. PBP-3 binding also utilizes an intricate water network to align for formation of the covalent bond with Ser294, which ultimately deactivates the protein. Neither covalent interactions nor a water network is seen in the IMP-13 or NDM-1 structure. The similarities at play highlight the challenge that drug discovery programs face in this area, but the differences may provide opportunities that can be exploited to deliver novel pharmaceutical solutions.
FIG 7.
Comparison of PBP-3 (3PBR) and IMP-13 binding to meropenem. View of the carbapenem scaffold (A and C) and of the tail moieties (B and D). Interactions are shown as dashed lines. Purple, zinc interactions; yellow, H bonds and charge-charge interactions; orange, aromatic interactions; green, hydrophobic interactions; pale blue, water network.
We report high-resolution structures of IMP-13 in diverse functional states and with different ligands bound. The structures explain the specificity of the enzymatic mechanism and the molecular recognition of substrates by the IMP-13 β-lactamase. The data presented and the comparisons presented above suggest that in IMP-13 the active-site loop plays a central role in antibiotic binding, with the primary interactions being to the core carbapenem scaffold. Consequently, such identified motifs that cause restriction in the L1 loop flexibility could form important parts of β-lactamase inhibitors engaging and stabilizing the loop in the closed conformation and blocking access of the natural substrates. The substantial chemical shift changes observed by NMR in the tryptophan indole region (Fig. 4; Fig. SI7) could be used in high-throughput screening to identify ligands with the potential to stabilize the active-site loop in the closed conformation, similar to previous two-dimensional screening approaches focusing on spectral properties specific to the system of interest (69).
On the other hand, the ability of the loop to adopt a fully open state, as observed in the apoopen structure of IMP-13, provides an alternative strategy for inhibitor development. Most drug development strategies based on these targets have so far been aimed at the active site itself. As the loop appears to play an active role in the binding of ligands to the protein, prevention of the loop closing by an allosteric inhibitor could have a similar inhibitory effect.
The presented crystal structures and experimental NMR data combined with our molecular dynamics simulations provide complementary information about changes in conformational dynamics linked to ligand binding that should be considered in the development of small-molecule inhibitors.
MATERIALS AND METHODS
Protein expression and purification.
The mature forms of the IMP-1 (residues 29 to 276) and IMP-13 (residues 21 to 246) proteins without a signal peptide were cloned into a pET-SUK vector (70). The constructs were transformed into Escherichia coli BL21(DE3) cells and plated on LB agar supplemented with kanamycin (50 μg/ml). The cells were grown in ZYM 5052 autoinduction medium (71) at 37°C until the optical density at 600 nm (OD600) was 2.0, and thereafter, the protein was expressed at 20°C overnight. The cells were collected by centrifugation, resuspended in lysis buffer [50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5 mM β-mercaptoethanol, 20 mM imidazole supplemented with 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF), DNase I, lysozyme], and lysed by sonication. The lysate was clarified by centrifuging for 45 min at 27,000 rpm, and the pH was adjusted to 8.0. The resulting supernatant was then passed twice over a HisTrap Excel column (GE Healthcare) that had been preequilibrated with lysis buffer. The column was washed with binding buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5 mM β-mercaptoethanol, 20 mM imidazole, 10 mM ZnCl2), and protein was eluted with elution buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5 mM β-mercaptoethanol, 300 mM imidazole). SUMO hydrolase (dtUD1) (72), provided by the Protein Expression and Purification Facility (PEPF; Helmholtz Zentrum München), was added to the eluted protein, and the components were gently mixed and incubated for 1 h at room temperature, followed by buffer exchange to binding buffer. A second step of affinity chromatography was performed to remove the SUMO tag and SUMO protease. The IMP-1/13-containing fractions were then concentrated and purified to homogeneity using a Superdex 75 size exclusion column preequilibrated with 5 mM Tris, pH 8.0, 50 mM NaCl, 5 mM β-mercaptoethanol, and 10 μM ZnCl2.
For isotopically labeled expression, cells were grown in M9 minimal medium supplemented with 15NH4Cl and [13C]glucose and induced as described above at an OD600 of 1, with overnight expression at 20°C. Cells were collected by centrifugation and resuspended in lysis buffer (100 mM Tris, 300 mM NaCl, 5 mM β-mercaptoethanol, pH 8.0) supplemented with DNase I and AEBSF. The supernatant was passed twice over Zn-nitrilotriacetic acid (NTA) beads. In our hands, NDM-1 was observed (as seen by paramagnetic effects in the NMR spectra) to bind Ni from the column in its active site; therefore, to prevent the possibility of the same occurring with IMP-13 and affecting the spectral quality, the NTA beads were loaded with Zn to ensure that the IMP-13 metal binding site was loaded with Zn. The column was preequilibrated with lysis buffer as described above, and SUMO hydrolase was added to the protein on the column and left overnight at room temperature. The cleaved IMP-13 was eluted with 5 ml lysis buffer and further purified using Superdex 75 size exclusion chromatography with a column that had been preequilibrated with NMR buffer (50 mM HEPES, 100 mM NaCl, pH 7.0). Samples were supplemented with 10% D2O for NMR spectroscopy.
Crystallization of IMP-13 in apo and carbapenem-bound forms.
Purified protein was concentrated to 12 mg/ml, and screening for crystallization conditions was performed using commercially available buffer sets in a sitting-drop vapor diffusion setup by mixing 0.2 μl of protein complex solution and 0.2 μl of buffer solution. For cocrystallization, meropenem or doripenem powder was added to the protein solution (final concentrations, 100 and 50 mM, respectively) and incubated for 30 min. For cocrystallization of IMP-13 with ertapenem and imipenem, antibiotic powder was dissolved in crystallization buffer and mixed with protein to a final concentration of 5 and 25 mM, respectively. All crystals were obtained at room temperature from solutions containing 0.1 M Tris, pH 8.5, 25% polyethylene glycol (PEG) 4000 (apoopen, in which the loop is open; PDB accession number 6R79), 0.1 M sucrose-phosphate-glutamic acid buffer, pH 8.0, 25% PEG 1500 (apoclosed, in which the loop is closed; PDB accession number 6R78), 0.1 M bis-Tris, pH 6.5, 25% PEG 3350 (meropenem complex; PDB accession number 6R73), 0.1 M bis-Tris, pH 5.5, 0.2 M ammonium sulfate, 25% PEG 3350 (imipenem complex; PDB accession number 6RZR), 0.1 M trisodium acetate, pH 5.6, 0.2 M ammonium acetate, 30% PEG 4000 (ertapenem complex; PDB accession number 6RZS), and 0.1 M sodium HEPES, pH 7.5, 25% PEG 2000 monomethyl ether (doripenem complex; PDB accession number 6S0H).
Structure determination and refinement.
Crystals were cryoprotected in 20% glycerol (apo), 2-methyl-2,4-pentanediol (meropenem complex), or 25% ethylene glycol (imipenem, ertapenem, and doripenem complexes) in the mother liquor and flash-frozen in liquid nitrogen. The diffraction data were collected at the id30b beamline at ESRF (Grenoble, France) and on the X06DA beamline at the Swiss Light Source (Paul Scherrer Institut, Villigen, Switzerland). The data were indexed and integrated using the XDS program package (73, 74) and scaled and merged using the Aimless program (75). The initial phases were obtained by molecular replacement, calculated using Phaser software (76) and the IMP-1 structure as a search model (PDB accession number 1DD6 [60]). The initial model was manually rebuilt according to the resulting electron density maps using the Coot program (77). The structures of IMP-13 in complex with hydrolyzed carbapenems were solved using the same approach and the IMP-13 apo structure as a search model. Carbapenem geometrical restraint files were created using the Grade web server (78). Restrained refinement was performed using the Phenix or Refmac program, with additional restraints being generated using the proSMART program (79–81). Five percent of the reflections were used for cross-validation analysis, and Rfree was employed to monitor the refinement strategy. Water molecules were added using the Coot program and afterwards were manually inspected. The final models were deposited in the Protein Data Bank under accession numbers 6R79 and 6R78 for the apo forms and 6R73, 6RZR, 6RZS, and 6S0H for the meropenem, imipenem, ertapenem, and doripenem bound forms, respectively. Interactions were visualized with Biovia Discovery Studio Visualizer software (82). All molecular graphics were prepared using the PyMOL (83) or Maestro (84) program. Crystallographic parameters are shown in Table S4 in the supplemental material.
NMR spectroscopy.
NMR experiments were recorded at 298 K on Bruker Avance III 600-MHz and 800-MHz spectrometers (1H frequency; 600 or 800 MHz, respectively) equipped with a 5-mm TCI or QCI cryoprobe. For assignments, 1H,15N HSQC, three-dimensional (3D) 15N-edited nuclear Overhauser effect spectroscopy (NOESY), HNCA, HN(CO)CA, HNCACB, HN(CO)CACB, and CBCACONH experiments were recorded on uniformly 15N,13C-labeled samples. Samples at 0.5 to 0.6 mM were prepared in NMR buffer (50 mM HEPES, 100 mM NaCl, pH 7.0) supplemented with 10% D2O. For the ertapenem assignment, the sample was supplemented with 5.7 mM ertapenem (a ca. 10-fold excess). Spectra were recorded with 2.9 mM and 5.7 mM ertapenem added, and with no further changes being observed between the two spectra, the protein was assumed to be saturated. Backbone assignment experiments (except for the 3D NOESY experiments) were recorded with 25% nonuniform sampling, using Poisson-gap sampling (85), and reconstructed using the Cambridge CS package and the CS-IHT algorithm (86). Heteronuclear NOE experiments were recorded at 600 MHz and 298 K using a sequence with interleaved saturated and unsaturated planes (53). Spectra were acquired with 2,048 by 300 complex points and a recycle delay of 1.2 s with 32 scans. NOEs were calculated as the ratio of the results of saturated to unsaturated experiments. Errors were calculated using the standard deviation of the noise. All spectra were processed with zero filling and Gaussian and/or sinebell window functions in the direct dimension and a sinebell window function in the indirect dimension. The water signal was removed by convolution with a sine function. The spectra were processed in the Azara program (W. Boucher, unpublished data) and analyzed using CcpNmr Analysis (87). Chemical shift perturbations (Δδ values) were calculated according to the following formula:
Antibiotic hydrolysis assay.
Enzymatic studies were carried out on the expressed proteins to confirm that the protein was in its active state. The enzymatic activity of the recombinantly produced metallo-β-lactamases was monitored as previously described (88) at 37°C in 75 mM HEPES buffer at pH 7.3, using 1 to 500 μM meropenem or imipenem as the substrate. β-Lactam hydrolysis was followed at 300 nm (Table S2).
Molecular dynamics simulations.
Molecular dynamics simulations were run using the Maestro Desmond molecular dynamics package (version 2017.3) (89, 90). The PDB accessions of the apoclosed and complex structures were prepared by adding missing side chains and hydrogens using the YASARA Structure’s built-in clean command (91). The structures were then imported into the Schrödinger Maestro (version 2017.3) program and further refined using the Maestro (version 11.1) protein preparation wizard (92). Protonation states were calculated using the PROPKA program (93, 94) at pH 7.0 ± 2.0, and minimization of hydrogen positions with a restrained backbone was performed using the OPLS3 force field (95) in order to optimize the hydrogen bonding network. Both the apoclosed and complex systems were then prepared for simulation using the Maestro (version 11.1) system builder graphical user interface (GUI) and the TIP4P (96) solvent model (crystallographic water molecules were deleted) in an automatically generated cubic cell with periodic boundary conditions. In addition to the solvated complex, Na+ and Cl− ions corresponding to those in a 150 mM buffer were placed in the cell in order to set the total net charge to zero. The coordination of the zinc metal centers was maintained by adding pseudo-bonds between the metals and the coordinating residues, using the default parameters for angles and charges of the OPLS-AA 2005 force field for the sake of the speed of the calculations. In the case of the apo protein, a tetrahedral coordination was chosen for both the zinc atoms in the active site. Furthermore, in the apo structure, no pseudo-bond was added to the bridging water molecule observed in the crystallographic structure, thus reducing the coordination of the metal centers to three residues to simulate the hydration sphere around the zinc atoms. For all the other complexes, the geometry of the zinc bound to histidines was considered tetrahedral (with a coordination number of 4) and that of the other zinc atom was considered to be octahedral (with a coordination number of 6).
Simulations of the systems were run using the Maestro Desmond molecular dynamics GUI for a total simulation time of 50 ns to ensure system convergence (this was checked on the RMSD plot of the simulations), recording at intervals of every 50 ps (1,000 snapshots in total) for xyz coordinates and 1.2 ps for potential energy calculations of the ensemble. Replicates of the simulations were performed using the Desmond molecular dynamics package (version 2019.3) in Maestro (version 11.8). One replica for each system was simulated for 100 ns, recording at intervals of every 100 ps (1,000 snapshots in total) for xyz coordinates and 2.5 ps for potential energy calculations. Both for the first run of production MD and for the replicates, the ensembles were set to a constant temperature (300 K) and pressure (1.01 × 105 Pa); the force cutoff radius was set to 9.0 Å, and each solvated model was relaxed with the Desmond default relaxation protocol before starting the simulation. Simulations were performed on a standard personal computer workstation (Intel Core i7 5960x, 32 GB random-access memory) using an Nvidia GeForce 1070 graphics processing unit.
Accession number(s).
Coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers 6R79 for the apo loop open form, 6R78 for the apo loop closed form, 6R73 for the meropenem complex, 6RZR for the imipenem complex, 6RZS for the ertapenem complex, and 6S0H for the doripenem complex. NMR assignments have been deposited in the BMRB under accession numbers 50012 for the apo form and 50013 for the ertapenem-bound form.
Supplementary Material
ACKNOWLEDGMENTS
This project was carried out with the support of BAG proposal MX-2088 at ESRF (Grenoble, France). We are grateful to the Protein Expression and Purification Facility and the X-ray Crystallography Platform at Helmholtz Zentrum München for support. We are thankful for all the discussion and help in structure refinement during the 2019 CCP4/APS School in Macromolecular Crystallography. We are also grateful to Rita Monson for useful discussions.
C.A.S., R.F., G.M.P., and M.S. acknowledge funding from the European Union’s Framework Program for Research and Innovation Horizon 2020 (2014 to 2020) under Marie Skłodowska-Curie grant agreement no. 675555, Accelerated Early staGe drug discovery (AEGIS). M.J.B. is supported by the Helmholtz Zentrum München. M.K. is supported by the Wrocław Centre of Biotechnology program, by the Leading National Research Centre (KNOW) for the years 2014 to 2018, and by a doctoral fellowship from the National Science Center (NCN) of Poland (grant no. 2018/28/T/NZ1/00337). R.M.-N. and K.M.Z. are supported by the German Federal Ministry of Research and Education (BMBF) program Targetvalidierung für die pharmazeutische Wirkstoffentwicklung (grant no. GFTARV38).
C.A.S., K.M.Z., M.J.B., R.X.Z., and M.K. carried out molecular biology and protein expression and purification. C.A.S., K.M.Z., M.K., and R.X.Z. carried out crystallization and data acquisition and analysis, and M.J.B. carried out NMR measurements and analysis. R.M.-N. carried out enzymatic assays, and R.F. performed molecular dynamics simulations. C.A.S., M.J.B., and K.M.Z. prepared the manuscript. H.M., M.S., and G.M.P. supervised the project.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
[This article was published on 21 May 2020 with a standard copyright line (“© 2020 American Society for Microbiology. All Rights Reserved.”). The authors elected to pay for open access for the article after publication, necessitating replacement of the original copyright line with the one above, and this change was made on 29 May 2020.]
REFERENCES
- 1.Exner M, Bhattacharya S, Christiansen B, Gebel J, Goroncy-Bermes P, Hartemann P, Heeg P, Ilschner C, Kramer A, Larson E, Merkens W, Mielke M, Oltmanns P, Ross B, Rotter M, Schmithausen RM, Sonntag H-G, Trautmann M. 2017. Antibiotic resistance: what is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg Infect Control 12:Doc05. doi: 10.3205/dgkh000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush K. 2010. Bench-to-bedside review: the role of β-lactamases in antibiotic-resistant Gram-negative infections. Crit Care 14:224. doi: 10.1186/cc8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tooke CL, Hinchliffe P, Bragginton EC, Colenso CK, Hirvonen VHA, Takebayashi Y, Spencer J. 2019. β-Lactamases and β-lactamase inhibitors in the 21st century. J Mol Biol 431:3472–3500. doi: 10.1016/j.jmb.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitout JD, Sanders CC, Sanders WE Jr. 1997. Antimicrobial resistance with focus on β-lactam resistance in Gram-negative bacilli. Am J Med 103:51–59. doi: 10.1016/S0002-9343(97)00044-2. [DOI] [PubMed] [Google Scholar]
- 5.Mölstad S, Lundborg CS, Karlsson A-K, Cars O. 2002. Antibiotic prescription rates vary markedly between 13 European countries. Scand J Infect Dis 34:366–371. doi: 10.1080/00365540110080034. [DOI] [PubMed] [Google Scholar]
- 6.Waxman D, Strominger JL. 1983. Penicillin-binding proteins and the mechanism of actions of the β-lactam antibiotics. Annu Rev Biochem 52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 7.Fleming A. 1929. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br J Exp Pathol 10:226–236. [Google Scholar]
- 8.Gaynes R. 2017. The discovery of penicillin—new insights after more than 75 years of clinical use. Emerg Infect Dis 23:849–853. doi: 10.3201/eid2305.161556. [DOI] [Google Scholar]
- 9.Drawz SM, Bonomo RA. 2010. Three decades of β-lactamase inhibitors. Clin Microbiol Rev 23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crowder MW, Spencer J, Vila AJ. 2006. Metallo-β-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc Chem Res 39:721–728. doi: 10.1021/ar0400241. [DOI] [PubMed] [Google Scholar]
- 11.Palumbi SR. 2001. Humans as the world’s greatest evolutionary force. Science 293:1786–1790. doi: 10.1126/science.293.5536.1786. [DOI] [PubMed] [Google Scholar]
- 12.Abraham EP, Newton GGF, Hale CW. 1954. Purification and some properties of cephalosporin N, a new penicillin. Biochem J 58:94–102. doi: 10.1042/bj0580094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton GG, Abraham EP. 1956. Isolation of cephalosporin C, a penicillin-like antibiotic containing d-α-aminoadipic acid. Biochem J 62:651–658. doi: 10.1042/bj0620651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birnbaum J, Kahan FM, Kropp H, MacDonald JS. 1985. Carbapenems, a new class of β-lactam antibiotics. Discovery and development of imipenem/cilastatin. Am J Med 78:3–21. doi: 10.1016/0002-9343(85)90097-X. [DOI] [PubMed] [Google Scholar]
- 15.Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, Shimokata K, Kato N, Ohta M. 1996. PCR detection of metallo-β-lactamase gene (blaIMP) in Gram-negative rods resistant to broad-spectrum β-lactams. J Clin Microbiol 34:2909–2913. doi: 10.1128/JCM.34.12.2909-2913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livermore D. 1995. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev 8:557–584. doi: 10.1128/CMR.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO. 2017. WHO priority pathogens list for R&D of new antibiotics. www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua=1.
- 18.Bush K, Jacoby GA, Medeiros AA. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother 39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meini M-R, Llarrull LI, Vila AJ. 2015. Overcoming differences: the catalytic mechanism of metallo-β-lactamases. FEBS Lett 589:3419–3432. doi: 10.1016/j.febslet.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito H, Arakawa Y, Ohsuka S, Wacharotayankun R, Kato N, Ohta M. 1995. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob Agents Chemother 39:824–829. doi: 10.1128/aac.39.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santella G, Pollini S, Docquier J-D, Mereuta AI, Gutkind G, Rossolini GM, Radice M. 2010. Intercontinental dissemination of IMP-13-producing Pseudomonas aeruginosa belonging in sequence type 621. J Clin Microbiol 48:4342–4343. doi: 10.1128/JCM.00614-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neu HC, Fu KP. 1978. Clavulanic acid, a novel inhibitor of β-lactamases. Antimicrob Agents Chemother 14:650–655. doi: 10.1128/aac.14.5.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehmann DE, Jahić H, Ross PL, Gu R-F, Hu J, Kern G, Walkup GK, Fisher SL. 2012. Avibactam is a covalent, reversible, non–β-lactam β-lactamase inhibitor. Proc Natl Acad Sci U S A 109:11663–11668. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Duin D, Bonomo RA. 2016. Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation β-lactam/β-lactamase inhibitor combinations. Clin Infect Dis 63:234–241. doi: 10.1093/cid/ciw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humphries RM, Yang S, Hemarajata P, Ward KW, Hindler JA, Miller SA, Gregson A. 2015. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 59:6605–6607. doi: 10.1128/AAC.01165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mojica MF, Bonomo RA, Fast W. 2016. B1-metallo-β-lactamases: where do we stand? Curr Drug Targets 17:1029–1050. doi: 10.2174/1389450116666151001105622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lauretti L, Riccio M, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini GM. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother 43:1584–1590. doi: 10.1128/AAC.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong DJ, Bae IK, Jang I-H, Jeong SH, Kang H-K, Lee K. 2015. Epidemiology and characteristics of metallo-β-lactamase-producing Pseudomonas aeruginosa. J Infect Chemother 47:81–97. doi: 10.3947/ic.2015.47.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bush K. 2018. Past and present perspectives on β-lactamases. Antimicrob Agents Chemother 62:e01076-18. doi: 10.1128/AAC.01076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao W-H, Hu Z-Q. 2011. IMP-type metallo-β-lactamases in Gram-negative bacilli: distribution, phylogeny, and association with integrons. Crit Rev Microbiol 37:214–226. doi: 10.3109/1040841X.2011.559944. [DOI] [PubMed] [Google Scholar]
- 32.Ho P-L, Lo W-U, Chan J, Cheung Y-Y, Chow K-H, Yam W-C, Lin C-H, Que T-L. 2014. pIMP-PH114 carrying bla IMP-4 in a Klebsiella pneumoniae strain is closely related to other multidrug-resistant IncA/C2 plasmids. Curr Microbiol 68:227–232. doi: 10.1007/s00284-013-0471-x. [DOI] [PubMed] [Google Scholar]
- 33.Mendes RE, Toleman MA, Ribeiro J, Sader HS, Jones RN, Walsh TR. 2004. Integron carrying a novel metallo-β-lactamase gene, blaIMP-16, and a fused form of aminoglycoside-resistant gene aac(6′)-30/aac(6′)-Ib′: report from the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother 48:4693–4702. doi: 10.1128/AAC.48.12.4693-4702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, Fang H, Wang L, Sun F, Wang Y, Yin Z, Yang H, Yang W, Wang J, Xia P, Zhou D, Liu C. 2014. IMP-1 encoded by a novel Tn402-like class 1 integron in clinical Achromobacter xylosoxidans, China. Sci Rep 4:7212. doi: 10.1038/srep07212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:(Web Server issue):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pagani L, Colinon C, Migliavacca R, Labonia M, Docquier J-D, Nucleo E, Spalla M, Bergoli ML, Rossolini GM. 2005. Nosocomial outbreak caused by multidrug-resistant Pseudomonas aeruginosa producing IMP-13 metallo-β-lactamase. J Clin Microbiol 43:3824–3828. doi: 10.1128/JCM.43.8.3824-3828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Queenan AM, Bush K. 2007. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chouchani C, Marrakchi R, Henriques I, Correia A. 2013. Occurrence of IMP-8, IMP-10, and IMP-13 metallo-β-lactamases located on class 1 integrons and other extended-spectrum β-lactamases in bacterial isolates from Tunisian rivers. Scand J Infect Dis 45:95–103. doi: 10.3109/00365548.2012.717712. [DOI] [PubMed] [Google Scholar]
- 39.Matsumura Y, Peirano G, Motyl MR, Adams MD, Chen L, Kresiwirth B, DeVinney R, Pitout J. 2017. Global molecular epidemiology of IMP-producing Enterobacteriaceae. Antimicrob Agents Chemother 61:e02729-16. doi: 10.1128/AAC.02729-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bogaerts P, Bouchahrouf W, Lissoir B, Denis O, Glupczynski Y. 2011. IMP-13-producing Pseudomonas monteilii recovered in a hospital environment. J Antimicrob Chemother 66:2434–2435. doi: 10.1093/jac/dkr294. [DOI] [PubMed] [Google Scholar]
- 41.Santella G, Docquier J-D, Gutkind G, Rossolini GM, Radice M. 2011. Purification and biochemical characterization of IMP-13 metallo-β-lactamase. Antimicrob Agents Chemother 55:399–401. doi: 10.1128/AAC.00421-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carfi A, Pares S, Duee E, Galleni M, Duez C, Frère J-M, Dideberg O. 1995. The 3-D structure of a zinc metallo-1-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J 14:4914–4921. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garau G, García-Sáez I, Bebrone C, Anne C, Mercuri P, Galleni M, Frère J-M, Dideberg O. 2004. Update of the standard numbering scheme for class B β-lactamases. Antimicrob Agents Chemother 48:2347–2349. doi: 10.1128/AAC.48.7.2347-2349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borra PS, Samuelsen O, Spencer J, Walsh TR, Lorentzen MS, Leiros H-K. 2013. Crystal structures of Pseudomonas aeruginosa GIM-1: active-site plasticity in metallo-β-lactamases. Antimicrob Agents Chemother 57:848–854. doi: 10.1128/AAC.02227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brem J, Struwe WB, Rydzik AM, Tarhonskaya H, Pfeffer I, Flashman E, van Berkel SS, Spencer J, Claridge TDW, McDonough MA, Benesch JLP, Schofield CJ. 2015. Studying the active-site loop movement of the São Paolo metallo-β-lactamase-1. Chem Sci 6:956–963. doi: 10.1039/c4sc01752h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng H, Liu X, Wang S, Fleming J, Wang D-C, Liu W. 2017. The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis. Nat Commun 8:2242. doi: 10.1038/s41467-017-02339-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page MI, Badarau A. 2008. The mechanisms of catalysis by metallo β-lactamases. Bioinorg Chem Appl 2008:576297. doi: 10.1155/2008/576297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamaguchi Y, Kuroki T, Yasuzawa H, Higashi T, Jin W, Kawanami A, Yamagata Y, Arakawa Y, Goto M, Kurosaki H. 2005. Probing the role of Asp-120(81) of metallo-β-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and X-ray crystallography. J Biol Chem 280:20824–20832. doi: 10.1074/jbc.M414314200. [DOI] [PubMed] [Google Scholar]
- 49.Lisa M-N, Palacios A, Aitha M, Gonzalez MM, Moreno DM, Crowder MW, Bonomo RA, Spencer J, Tierney DL, Llarrull LI, Vila AJ. 2017. A general reaction mechanism for carbapenem hydrolysis by mononuclear and binuclear metallo-β-lactamases. Nat Commun 8:538. doi: 10.1038/s41467-017-00601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peri S, Steen H, Pandey A. 2001. GPMAW—a software tool for analyzing proteins and peptides. Trends Biochem Sci 26:687–689. doi: 10.1016/s0968-0004(01)01954-5. [DOI] [PubMed] [Google Scholar]
- 51.Moali C, Anne C, Lamotte-Brasseur J, Groslambert S, Devreese B, Van Beeumen J, Galleni M, Frère JM. 2003. Analysis of the importance of the metallo-β-lactamase active site loop in substrate binding and catalysis. Chem Biol 10:319–329. doi: 10.1016/S1074-5521(03)00070-X. [DOI] [PubMed] [Google Scholar]
- 52.Jarymowycz VA, Stone MJ. 2006. Fast time scale dynamics of protein backbones: NMR relaxation methods, applications, and functional consequences. Chem Rev 106:1624–1671. doi: 10.1021/cr040421p. [DOI] [PubMed] [Google Scholar]
- 53.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 54.Scrofani SDB, Chung J, Huntley JJA, Benkovic SJ, Wright PE, Dyson HJ. 1999. NMR characterization of the metallo-β-lactamase from Bacteroides fragilis and its interaction with a tight-binding inhibitor: role of an active-site loop. Biochemistry 38:14507–14514. doi: 10.1021/bi990986t. [DOI] [PubMed] [Google Scholar]
- 55.Huntley JJA, Scrofani SDB, Osborne MJ, Wright PE, Dyson HJ. 2000. Dynamics of the metallo-β-lactamase from Bacteroides fragilis in the presence and absence of a tight-binding inhibitor. Biochemistry 39:13356–13364. doi: 10.1021/bi001210r. [DOI] [PubMed] [Google Scholar]
- 56.Huntley JJA, Fast W, Benkovic SJ, Wright PE, Dyson HJ. 2003. Role of a solvent-exposed tryptophan in the recognition and binding of antibiotic substrates for a metallo-β-lactamase. Protein Sci 12:1368–1375. doi: 10.1110/ps.0305303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hinchliffe P, Gonzalez MM, Mojica MF, Gonzalez JM, Castillo V, Saiz C, Kosmopoulou M, Tooke CL, Llarrull LI, Mahler G, Bonomo RA, Vila AJ, Spencer J. 2016. Cross-class metallo-β-lactamase inhibition by bisthiazolidines reveals multiple binding modes. Proc Natl Acad Sci U S A 113:E3745–E3754. doi: 10.1073/pnas.1601368113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furuyama T, Nonomura H, Ishii Y, Hanson ND, Shimizu-Ibuka A. 2016. Structural and mutagenic analysis of metallo-β-lactamase IMP-18. Antimicrob Agents Chemother 60:5521–5526. doi: 10.1128/AAC.00985-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamaguchi Y, Matsueda S, Matsunaga K, Takashio N, Toma-Fukai S, Yamagata Y, Shibata N, Wachino J-I, Shibayama K, Arakawa Y, Kurosaki H. 2015. Crystal structure of IMP-2 metallo-β-lactamase from Acinetobacter spp. Biol Pharm Bull 38:96–101. doi: 10.1248/bpb.b14-00594. [DOI] [PubMed] [Google Scholar]
- 60.Concha NO, Janson CA, Rowling P, Pearson S, Cheever CA, Clarke BP, Lewis C, Galleni M, Frere J-M, Payne DJ, Bateson JH, Abdel-Meguid SS. 2000. Crystal structure of the IMP-1 metallo-β-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor. Biochemistry 39:4288–4298. doi: 10.1021/bi992569m. [DOI] [PubMed] [Google Scholar]
- 61.Hinchliffe P, Tanner CA, Krismanich AP, Labbé G, Goodfellow VJ, Marrone L, Desoky AY, Calvopiña K, Whittle EE, Zeng F, Avison MB, Bols NC, Siemann S, Spencer J, Dmitrienko GI. 2018. Structural and kinetic studies of the potent inhibition of metallo-β-lactamases by 6-phosphonomethylpyridine-2-carboxylates. Biochemistry 57:1880–1892. doi: 10.1021/acs.biochem.7b01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khan AU, Maryam L, Zarrilli R. 2017. Structure, genetics and worldwide spread of new delhi metallo-β-lactamase (NDM): a threat to public health. BMC Microbiol 17:101. doi: 10.1186/s12866-017-1012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.King DT, Worrall LJ, Gruninger R, Strynadka NC. 2012. New Delhi metallo-β-lactamase: structural insights into β-lactam recognition and inhibition. J Am Chem Soc 134:11362–11365. doi: 10.1021/ja303579d. [DOI] [PubMed] [Google Scholar]
- 64.Stewart AC, Bethel CR, VanPelt J, Bergstrom A, Cheng Z, Miller CG, Williams C, Poth R, Morris M, Lahey O, Nix JC, Tierney DL, Page RC, Crowder MW, Bonomo RA, Fast W. 2017. Clinical variants of New Delhi metallo-β-lactamase are evolving to overcome zinc scarcity. ACS Infect Dis 3:927–940. doi: 10.1021/acsinfecdis.7b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang R, Lai T-P, Gao P, Zhang H, Ho P-L, Woo P-Y, Ma G, Kao R-T, Li H, Sun H. 2018. Bismuth antimicrobial drugs serve as broad-spectrum metallo-β-lactamase inhibitors. Nat Commun 9:439. doi: 10.1038/s41467-018-02828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klingler F-M, Wichelhaus TA, Frank D, Cuesta-Bernal J, El-Delik J, Müller HF, Sjuts H, Göttig S, Koenigs A, Pos KM, Pogoryelov D, Proschak E. 2015. Approved drugs containing thiols as inhibitors of metallo-β-lactamases: strategy to combat multi-drug resistant bacteria. J Med Chem 58:3626–3630. doi: 10.1021/jm501844d. [DOI] [PubMed] [Google Scholar]
- 67.Raczynska J, Shabalin I, Minor W, Wlodawer A, Jaskolski M. 2018. A close look onto structural models and primary ligands of metallo-β-lactamases. Drug Resist Updat 40:1–12. doi: 10.1016/j.drup.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han S, Zaniewski RP, Marr ES, Lacey BM, Tomaras AP, Evdokimov A, Miller JR, Shanmugasundaram V. 2010. Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 107:22002–22007. doi: 10.1073/pnas.1013092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jahnke W, Grotzfeld RM, Pellé X, Strauss A, Fendrich G, Cowan-Jacob SW, Cotesta S, Fabbro D, Furet P, Mestan J, Marzinzik AL. 2010. Binding or bending: distinction of allosteric abl kinase agonists from antagonists by an NMR-based conformational assay. J Am Chem Soc 132:7043–7048. doi: 10.1021/ja101837n. [DOI] [PubMed] [Google Scholar]
- 70.Helmholtz Zentrum Muenchen Protein Expression and Purification Facility. 2019. Bacterial expression vectors. Helmholtz Zentrum Muenchen Protein Expression and Purification Facility, Munich, Germany. [Google Scholar]
- 71.Studier FW. 2005. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif 41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 72.Weeks SD, Drinker M, Loll PJ. 2007. Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr Purif 53:40–50. doi: 10.1016/j.pep.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krug M, Weiss MS, Heinemann U, Mueller U. 2012. XDSAPP: a graphical user interface for the convenient processing of diffraction data using XDS. J Appl Crystallogr 45:568–572. doi: 10.1107/S0021889812011715. [DOI] [Google Scholar]
- 75.Evans PE, Murshudov GN. 2013. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smart OS, Womack TO, Sharff A, Flensburg C, Keller P, Paciorek W, Vonrhein C, Bricogne G. 2011. Grade web server. Global Phasing Ltd., Cambridge, United Kingdom. [Google Scholar]
- 79.Nicholls RA, Fischer M, McNicholas S, Murshudov GN. 2014. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr D Biol Crystallogr 70:2487–2499. doi: 10.1107/S1399004714016241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nicholls RA, Long F, Murshudov GN. 2012. Low-resolution refinement tools in REFMAC5. Acta Crystallogr D Biol Crystallogr 68:404–417. doi: 10.1107/S090744491105606X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Biovia DS. 2019. Discovery Studio Visualizer. Dassault Systemes, San Diego, CA. [Google Scholar]
- 83.Schrödinger. PyMOL molecular graphics system, version 2.0 LLC, v1.8. Schrödinger, New York, NY. [Google Scholar]
- 84.Schrödinger. 2017. L.L.C. Schrödinger Maestro release 2017.3. Schrödinger, New York, NY. [Google Scholar]
- 85.Hyberts SG, Takeuchi K, Wagner G. 2010. Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J Am Chem Soc 132:2145–2147. doi: 10.1021/ja908004w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bostock MJ, Holland DJ, Nietlispach D. 2012. Compressed sensing reconstruction of undersampled 3D NOESY spectra: application to large membrane proteins. J Biomol Nmr 54:15–32. doi: 10.1007/s10858-012-9643-4. [DOI] [PubMed] [Google Scholar]
- 87.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. 2005. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 88.King AM, Reid-Yu SA, Wang W, King DT, Pascale GD, Strynadka NC, Walsh TR, Coombes BK, Wright GD. 2014. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature 510:503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bowers KJ, Chow DE, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, Shaw DE. 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. SC'06. Proceedings of the ACM/IEEE SC 2006 Conference, Tampa, FL. doi: 10.1109/SC.2006.54. [DOI] [Google Scholar]
- 90.Bergdorf M, Baxter S, Rendlema CA, Shaw DE. 2016. Desmond/GPU performance as of November 2016. Technical report DESRES/TR D E Shaw Research, New York, NY. [Google Scholar]
- 91.Krieger E, Vriend G. 2014. YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics 30:2981–2982. doi: 10.1093/bioinformatics/btu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. 2013. Protein and ligand preparation: parameters, protocols and influence on virtual screening enrichments. J Comput Aided Mol Des 27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 93.Olsson MHM, Sondergaard CR, Rostkowski M, Jensen JH. 2011. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput 7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 94.Søndergaard CR, Olsson MHM, Rostkowski M, Jensen JH. 2011. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J Chem Theory Comput 7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 95.Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL, Kaus JW, Cerutti DS, Krilov G, Jorgensen WL, Abel R, Friesner RA. 2016. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 96.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. 1983. Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.